Abstract
Background/Aim:
Diallyl trisulfide (DATS) has been shown to prevent and inhibit breast carcinogenesis. CCL2/MCP-1 has been shown to play a significant role in breast cancer. This study explored DATS efficacy on triplenegative breast cancer (TNBC) cells.
Materials and Methods:
DATS efficacy on TNF-α induced TNBC cells were examined via trypan blue exclusion test, wound-healing assay, human cytokine arrays, ELISA, and RT-PCR.
Results:
DATS significantly induced cell death and inhibited cell migration. Expression of CCL2/MCP-1, IL-6, PDGF-BB, NT-3, and GM-CSF in TNF-α-treated cells increased. However, DATS significantly decreased the expression of CCL2/MCP-1 in TNF-α-treated MDA-MB-231 but not in MDA-MB-468 cells. DATS significantly down-regulated mRNA expression of IKBKE and MAPK8 in both cell lines, indicating a possible effect in genes involved in the NF-κB and MAPK signaling.
Conclusion:
DATS may have a role in TNBC therapy and prevention by targeting CCL2.
Keywords: Breast cancer, CCL2, diallyl trisulfide, IKBKE, MAPK8, MCP-1, tumor necrosis factor alpha
Globally, breast cancer in women is ranked the second leading cause of cancer-related deaths and has claimed the lives of 685,000, with 2.3 million new cases in 2020 (1). Breast cancers are classified into distinct groups according to differential gene expression patterns, each with a unique set of phenotypes. Estrogen/progesterone receptor-positive (ER+/PR+), HER2+, and triple-negative breast cancer (TNBC) (estrogen, progesterone, and HER2 receptor negative) are the three major subtypes of breast cancer (2-4). Compared to their Caucasian, African American (AA) women are more likely to develop TNBC. This aggressive and invasive subtype accounts for 15 to 20% of all breast cancer cases (4, 5) and is correlated with a poor prognosis, a mutation in the BRCA1 gene, and a higher mortality rate amongst AA women. There is no clinical therapy specifically for TNBC patients (6, 7).
Also, an estimated 15 to 20% of all cancer-related deaths have been linked to inflammation, where proinflammatory cytokines have been associated with the tumorigenesis process (4). Cancer-linked inflammation consists of an intricate network of immune system processes, which result in the growth and metastasis of many cancers (8). In humans, almost every type of cancer, including renal, prostate, and breast, produce and release high levels of monocyte chemotactic protein-1 (MCP-1) also known as C-C motif chemokine ligand-2 (CCL2). CCL2/MCP-1 is a tumor-promoting chemokine, which recruits monocytes to the tumor site through its receptors such as monocyte G-coupled CCL2 receptors (CCR2A/2B) (8-15). In TNBC, inflammation and immune response plays a major role in the progression and metastasis of the disease. TNF-α is released by macrophages to destroy the tumor due to its toxic effect on cancer cells. Unfortunately, if TNF-α is unable to destroy the cancer cells, macrophages begin to mimic the characteristics of cancer, eventually turning into tumor-associated macrophages (TAM). Therapeutic targeting of the release of CCL2 or its receptor (monocyte CCR2 receptor) may be used to inhibit the mobilization of infiltrating monocytes to the tumor site (8, 16, 17).
Despite some promising advances against breast cancer, TNBC responds differently to various treatments, unlike the other subtypes, creating new challenges in developing effective targeted therapy and chemotherapy that would be safe and effective (2, 4, 18). The more invasive forms of breast cancer treatments include radical mastectomy, chemotherapy, and irradiation. These treatments can be administered individually or in various combinations. Less invasive treatments include endocrine and targeted therapy. Although endocrine therapy has had positive results in hormone receptor (HR) positive breast cancer, it is ineffective in TNBC.
Garlic has been used medicinally in Asia and the middle east for many centuries. It is reported to have many biological activities, including antioxidant, antihypertensive, antidiabetic, anti-atherosclerotic, antibacterial, antifungal, and anticarcinogenic. Diallyl trisulfide (DATS), the most effective active ingredient found in garlic, has been shown to have antioxidant properties, the ability to reduce DNA double-strand breaks and induce cell cycle arrest and apoptosis (19-26). DATS is a secondary metabolite of allicin; an organosulfur compound produced from the crushing of Allium sativum (garlic) (27). Alliin is converted by the alliinase enzyme to allicin, producing its three main products diallyl sulfide, disulfide, and diallyl trisulfide (27). DATS was reported to suppress metastasis of breast cancer cell lines (MDA-MB-231 and HS 578T) by down-regulating ERK/NFκB/MMP-2/MMP-9 activities and expression (28). In MCF-7 cells, DATS stimulated apoptosis, which was associated with elevated levels of cyclin B1. Real-time PCR assays indicated that DATS-enhanced FAS and cyclin D1 expression levels and down-regulated the expression levels of Akt and Bcl-2. Induction of apoptosis by DATS was linked to induction of pro-apoptotic Bax protein and increase in p53 protein expression, suggesting that DATS might offer a novel strategy for the treatment of human breast cancer (29).
Therefore, the current study was developed to investigate the effects of the organosulfur compound DATS on cell survival and its modulatory effects on releasing proinflammatory cytokines in triple-negative human breast tumor (MDA-MB-231 and MDA-MB-468) cells. The data from this research contribute to the literature concerning the potential role of DATS in TNBC therapy and prevention.
Materials and Methods
Cell line, chemicals, and reagents.
MDA-MB-231 (Caucasian American TNBC cell line) and MDA-MB-468 (African American TNBC cell line) were acquired from the American Type Culture Collection (ATCC) (Rockville, MD, USA). Dimethyl sulfoxide (DMSO), Fetal Bovine Serum (FBS), Dulbecco’s Modified Eagles Medium (DMEM) high glucose, diallyl trisulfide (DATS), and Alamar blue were acquired from Sigma (St. Louis, MO, USA). Penicillin/Streptomycin, trypsin-EDTA, and Hanks Balanced Salt Solution (HBSS) were obtained from Invitrogen (Carlsbad, CA, USA). Ethanol was acquired from Cruzan International (Deerfield, IL, USA). Human cytokine arrays, human recombinant TNF-α, and ELISA assays for MCP-1 (Cat# ELH-MCP1–1) were acquired from RayBiotech (Norcross, GA, USA). The PCR primers and iScript advanced reverse transcriptase kit were obtained from Bio-Rad (Hercules, CA, USA).
Maintenance of cells.
MDA-MB-231 and MDA-MB-468 TNBC cells were cultivated in DMEM media with fetal bovine serum (10%) and 1% penicillin (100 U/ml)/streptomycin (0.1 mg/ml) and incubated in a humidified atmosphere of 5% CO2 at 37°C. The complete media was replaced every 3-4 days, and the cells were sub-cultured every 4-7 days. Cells were sub-cultured in T-175 flasks until 90% confluency before plating the cells to start each experiment.
Cell viability assay.
Cytotoxicity in both MDA-MB-231 and MDA-MB-468 cells was determined using the trypan blue exclusion test. Cells were seeded in 5 ml of media at a density of 1×105 cells in a T-25 flask. The cells were stabilized overnight in the incubator at 37°C in 5% CO2. Cells were treated with or without DATS (25, 50, 75, 100, 150, and 200 μM) for different periods (24, 48, and 72 h). DATS was dissolved in dimethyl sulfoxide (DMSO) and diluted in 10% FBS complete media; the final concentration of DMSO did not exceed 0.1%. An equal volume of DMSO was added to the control. All treatments were performed in triplicate. At the end of the incubation period, the cells were pelleted and resuspended in 200 μl fresh media, 500 μl of 0.4% trypan blue solution, and 300 μl of phosphate-buffered saline (PBS) solution. The solution was then incubated for 15 min at room temperature. At least 300 cells were counted after the incubation period to determine cell survival. The data were expressed as a percentage of live untreated controls.
Wound healing assay.
The effect of DATS on cell migration was determined via wound-healing assay. IBIDI inserts were placed in a 12-well plate and seeded at 3×104-5×104 cells per insert in 100 μl of complete media. Each experiment was performed in triplicate. Once cells were confluent, the complete media was discarded and replaced with serum-free media for another 24 h. The inserts were then removed, creating a wound in the bottom of the well. Treatments included control (cells only), DATS (75 μM), TNF-α (40 ng/ml), and the combination of DATS (75 μM)+TNF-α (40 ng/ml). Finally, the cells were washed with media and observed under a light microscope with an attached camera using the ToupeView software to measure the closure of the wound over 0, 12, and 24 h.
Human cytokine arrays.
The effect of DATS on 60 cytokine proteins released by TNF-α-activated TNBC cells was assessed using RayBiotech human cytokine arrays. The experiments were conducted according to the manufacturer’s instructions in triplicate. Briefly, antibody-coated array membranes were first added to a 1 ml blocking buffer and incubated for 30 min. The blocking buffer was then aspirated and substituted with treated cell supernatant (1 ml). These cells were subjected to different treatments for 24 h. Treatments included control (cells only), DATS (75 μM), TNF-α (40 ng/ml), and the combination of DATS (75 μM)+TNF-α (40 ng/ml). The membranes were placed on a shaker and incubated overnight at 4°C. The following day, the media was aspirated; membranes were washed and incubated for 2 h with 1 ml biotin-conjugated antibodies. Finally, biotin-conjugated antibodies were aspirated, and membranes were washed and incubated for another 2 h with HRP-conjugated streptavidin. The chemiluminescent reagent was used, and Flour-S Max Multi-imager (Bio-Rad Laboratories) was used to capture the images of spots. Quantity One Software established the density of each spot (Bio-Rad Laboratories). Human Cytokine Array software C1000 (CODE: S02-AAH-CYT-1000) from RayBiotech was used to develop an Excel-based data analysis.
Human CCL2/MCP-1 ELISA and quantification.
The supernatants of cells exposed to the various treatments for a 24 h period were collected. Treatments included control (cells only), DATS (75 μM), TNF-α (40 ng/ml), and the combination of DATS (75 μM)+TNF-α (40 ng/ml). Each experiment was conducted according to the Raybiotech ELISA MCP-1 kit instructions in triplicate. TNBC cells were acquired and centrifuged at 4°C for 4 min at 1,000 rpm (67×g). A specific ELISA for human CCL2/MCP-1 detection was applied in compliance with the manufacturer’s instructions. Briefly, the supernatants (100 μl) from each standard and samples were added to 96 well plates pre-coated with capture antibody and incubated for 2.5 h at room temperature under shaking. After washing, a biotinylated antibody mixture (100 μl) was prepared, added to each well, and incubated for 1 h. Next, the mixture was aspirated, and into each well, streptavidin solution (100 μl) was added and incubated for 45 min. One hundred μl of substrate reagent was then pipetted into each well and incubated for 30 min, followed by the addition of 50 μl of stop solution. Synergy HTX Multi-Reader (BioTek, Winooski, VT, USA) was used to measure the optical density of the samples at 450 nm.
Real-time polymerase chain reaction (RT-PCR)
RNA extraction.
Cells were exposed to the various treatments for 24 h, then harvested, and cell pellets were collected. Each experiment was conducted in triplicate. Treatments included control (cells only), DATS (75 μM), TNF-α (40 ng/ml), and the combination of DATS (75 μM) + TNF-α (40 ng/ml). One ml of TRIzol reagent was used to lyse the cell pellets. Then, 0.2 ml of chloroform was combined with the lysed samples; the tubes were shaken, incubated for 2-3 min at 15-30°C, and centrifuged at 10,000 rpm (6,720×g) at 2-8°C for 15 min. The newly lysed samples (aqueous phase) were transferred into a new tube and combined with 0.5 ml isopropyl alcohol to precipitate the RNA. After 15 min of incubation, samples were centrifuged, the supernatant was aspirated, 75% ethanol was used to wash the RNA pellets, and then centrifuged at 7,500 rpm (3,780×g) at 2-8°C for 5 min. The RNA pellet was left to dry at room temperature, resuspended in RNAase-free water, and placed on ice for 30 min. RNA purity and quantity were evaluated using Nanodrop (Thermo Fischer Scientific, Wilmington, DE, USA).
cDNA synthesis and RT-PCR.
iScript advanced reverse transcriptase from Bio-Rad was used to synthesize cDNA strands from mRNA. Briefly, 4 μl of 5X iScript advanced reaction mix, 1 μl reverse transcriptase, 7.5 μl of the sample (1.5 μg/reaction), and 7.5 μl water were mixed in a tube to a final volume of 20 μl. According to the manufacturer’s protocol for the reverse transcription, the thermal cycling program included two steps: 46°C for 20 min and then 95°C for 1 min. RT-PCR amplification was conducted following the manufacturer’s protocol (BioRad). In each well, 1 μl of the sample (200 ng cDNA/reaction), 10 μl of the master mix, 1 μl of primers, and 8 μl of water were mixed. According to the manufacturer, the thermal cycling process required an initial hold step at 95°C for 2 min and denaturation at 95°C for 10 s, followed by 39 cycles of 60°C for 30 s (annealing/extension), and 65°C-95°C for 5 s/step (melting curve) using the Bio-Rad CFX96 Real-Time System. Specific primers were determined for the genes of interest. The Unique Assay ID for CCL2/MCP1 primer was qHsa-CID0011608, for MAPK8 primer was qHsaCIP002749 and for IKBKE primer was qHsaCID0014831 0.
Statistical analysis.
The results of at least three biological replicates, tested in triplicates and averaged, are presented as mean±SEM and analyzed using GraphPad Prism (San Diego, CA, USA). LC50 was determined via non-linear regression using GraphPad Prism. The significance of differences was determined by one-way ANOVA and compared by Dunnett’s test or using a Student’s t-test. Values of p<0.05 were considered statistically significant. Gene expression was analyzed using the CFX 3.1 Manager software (Bio-Rad).
Results
The cytotoxic effect of DATS on MDA-MB-231 and MDA-MB-468 TNBC cells.
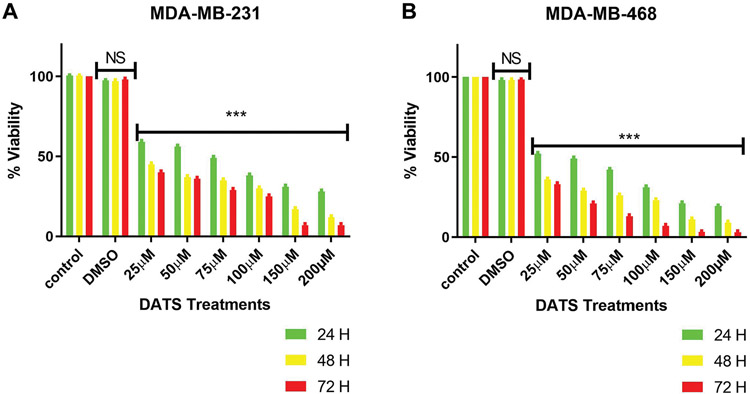
DATS effect at various concentrations was investigated in MDA-MB-231 and MDA-MB-468 cell lines after 24, 48, and 72 h treatment. The viability results showed time-dependent toxicity (Figure 1A and B) in both cell lines. Treatment with DATS concentrations equal to or higher than 25 μM significantly decreased cell viability after 24 h in both cell lines compared to the control. Cells treated with DMSO showed no cytotoxicity in comparison to control. After 24 h of treatment, the LC50s were 24.84±0.10 μM in MDA-MB-231 and 22.47±0.54 μM in MDA-MB-468 cells. After 48 h- and 72 h-treatment, the LC50s decreased to 17.31±0.19 and 18.22±0.90 μM in MDA-MB-231, and 11.68±0.07 and 15.04±0.26 μM in MDA-MB-468 cells, respectively. Statistical analysis comparing the LC50s for DATS cytotoxic effect on both cell lines showed that MDA-MB-468 cells were more sensitive to the compound and had a lower percentage of viable cells after 24 h (p<0.05) and 48 h (p<0.01), showing no significant effect after 72 h when compared to MDA-MB-231 cells. The data from cell viability assays were used to establish DATS cytotoxicity and concentration in further studies.
Figure 1.
The effect of DATS on viability oϕ MDA-MB-231 (A) and MDA-MB-468 (B) TNBC cells. MDA-MB-231 and MDA-MB-468 were treated with various concentrations (0-200 μM) of DATS for 24, 48, and 72 h. The effect of DATS was both dose- and -time-dependent. Treatment with a low dose of 25 μM of DATS significantly decreased cell viability compared to control. DMSO treatment showed no cytotoxicity at any time point. The lethal concentration 50 (LC50) value was acquired from GraphPad Prism using mean values of data points at the previously mentioned concentrations. All experiments were performed at least three times with n=3. The data are presented as the mean±SEM. Statistically significant differences between control vs. treatments were evaluated by a one-way ANOVA, followed by’ ‘Dunnett’s multiple comparison test or Student’s t-test to compare the results between the two cell lines with ***p<0.001.
The anti-migration effect of DATS on MDA-MB-231 and MDA-MB-468 TNBC cells.
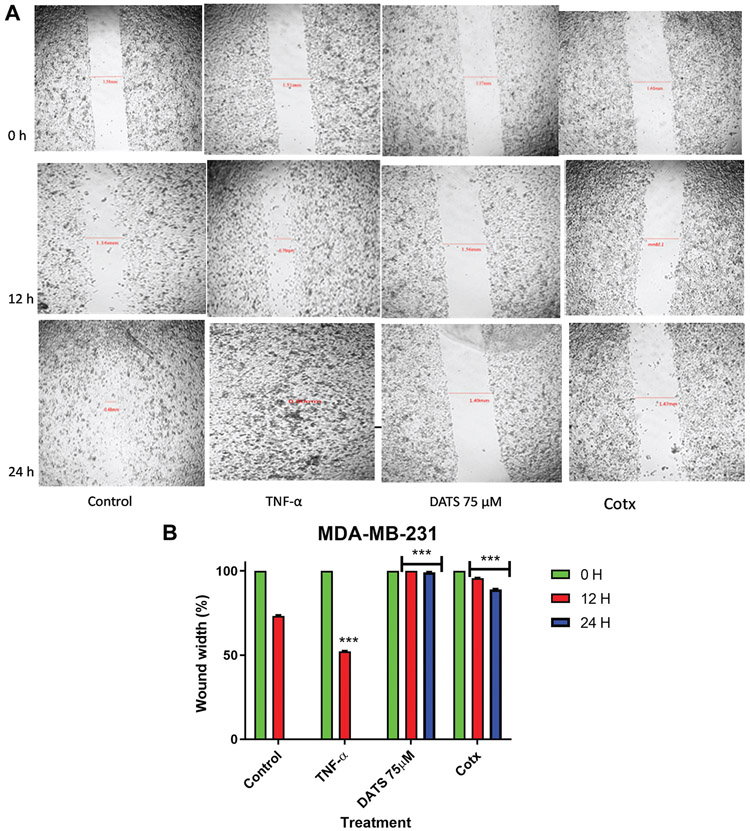
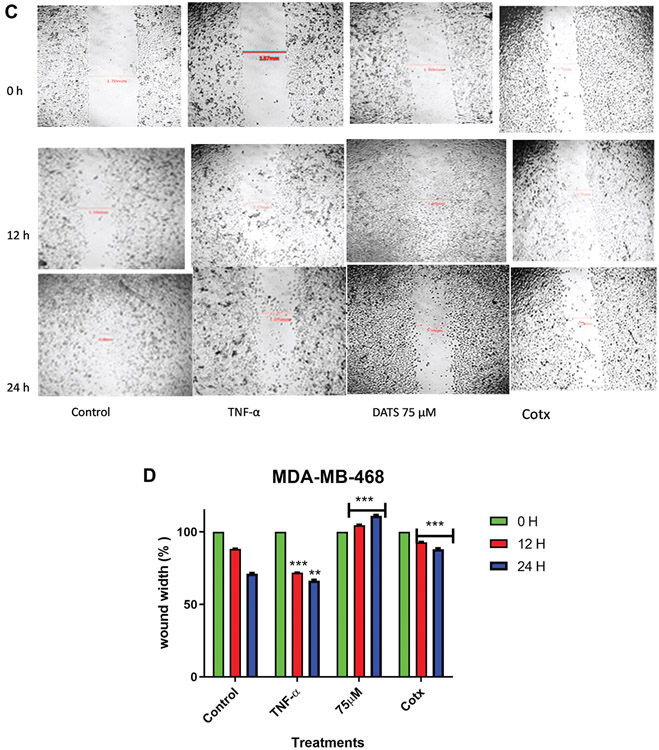
In the present study, a wound-healing assay was used to study cell migration in vitro. The assay allowed us to examine DATS’s ability to inhibit migration of MDA-MB-231 and MDA-MB-468 cells. TNF-α treatment (40 ng) significantly increased the percent rate of recovery in MDA-MB-231 and MDA-MB-468 cells (47.8% and 28.17% respectively) (Figure 2) in comparison to the untreated wound control (26.7% and 11.79% respectively) after 12 h. DATS significantly inhibited the migration of cells after a 12 h exposure to 75 μM DATS in both cell lines (0% - MDA-MB-231 cells; and −4.47 - MDA-MB-468 cells). In MDA-MB-468 cells, the wound size significantly increased after 24 h (−11.04%), showing higher inhibition in migration (p<0.001). This could be because MDA-MB-468 cells are more sensitive to DATS cytotoxicity. The combination of DATS and TNF-α treatment (75 μM DATS and 40 ng TNF-α) showed that DATS significantly inhibits the wound size after 24 h in both cell lines decreasing the TNF-α migration effect. Compared to control (100%), the combination treatment wound size was significantly larger. These results showed that TNF-α and the combination of DATS and TNF-α had time-dependent effects.
Figure 2.
The effect of DATS on TNF-α induced MDA-MB-231 and MDA-MB-468 breast cancer cells. A wound-healing assay was used to examine the effect of diallyl trisulfide (DATS) on cell migration in TNF-α induced MDA-MB-231 and MDA-MB-468 breast cancer cells (A and C) at various time points (0, 12, and 24 h). Wound healing assay representative image and quantification of MDA-MB-231 and MDA-MB-468 cell migration with a magnification of 40X. (B and D) Wound width % was statistically analyzed. At least 3 independent experiments were performed with n=3. The data are presented as the mean±S.E.M. Statistically significant differences between control vs. treatments were evaluated by a one-way ANOVA, followed by Dunnett’s multiple comparison test with **p<0.01 and ***p<0.001.
The effect of DATS on cytokine expression in TNF-α-activated MDA-MB-231 and MDA-MB-468 TNBC cells.
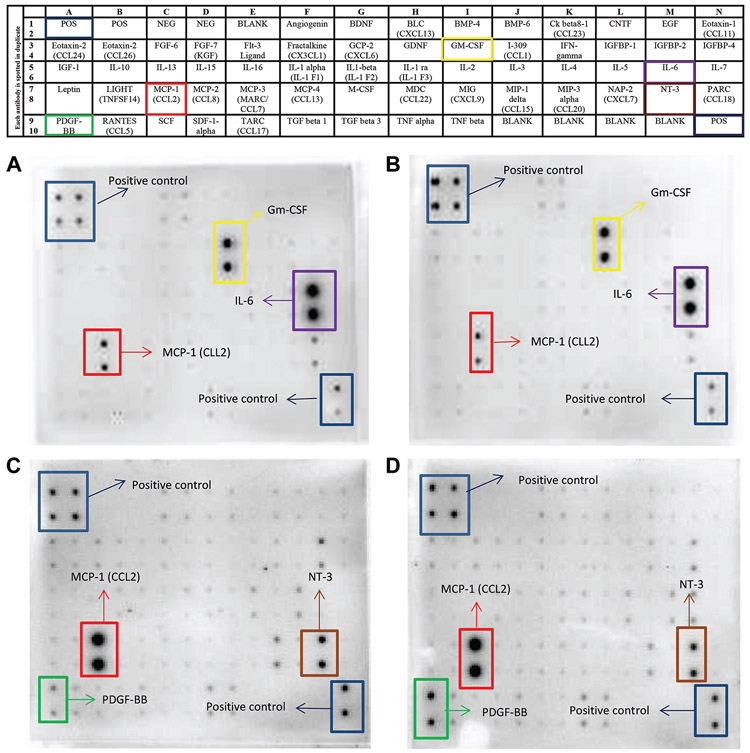
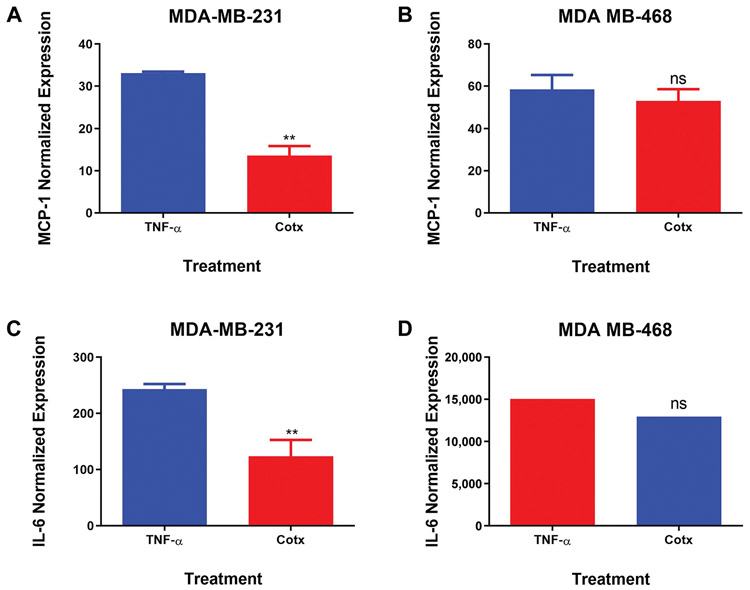
Cytokine array assay was used to compare DATS anticancer effects on TNF-α-induced release of proinflammatory cytokines by MDA-MB-231 and MDA-MB-468 TNBC cells (Figure 3). The results showed that TNF-α induced the up-regulation of five specific cytokines: chemokine (C-C motif) ligand 2 (CCL2/MCP-1), GM-CSF, and interleukin-6 (IL-6) by MDA-MB-231 cells (Figure 3A and B). In contrast, NT-3 and PDGF-BB were up-regulated in MDA-MB-468 cells (Figure 3C and D). MCP-1 was the only common up-regulated protein observed in both cell lines. Although TNF-α induced MCP-1 expression in both cell lines, the expression was two times higher in the MDA-MB-468 compared to the MDA-MB-231 cells. However, DATS treatment resulted in differential cytokine expression in the two cell lines, inhibiting MCP-1 expression in MDA-MB-231 but not in MDA-MB-468 cells. Normalized results obtained from TNF-α-stimulated cells and cells co-treated with DATS and TNF-α confirmed that DATS attenuated TNF-α-induced CCL2/MCP-1 release significantly in MDA-MB-231 cells (3-fold inhibition), but not in MDA-MB-468 cells (Figure 4A and B), showing that the distinct cell lines respond differently to DATS treatment.
Figure 3.
Comparative effect of diallyl trisulfide (DATS) on cytokine expression in TNF-alpha induced MDA-MB-231 and MDA-MB-468 breast cancer cells (n=3). The array layout used to assess chemokines/cytokines expression in supernatants of treated cells, showing the cytokines map, and highlighting CCL2/MCP-1, IL-6, PDGF-BB, NT-3, and positive controls. (A) (C) supernatant of cells treated with TNF-α and (B) (D) supernatant of cells co-treated with TNF-α and DATS Chemiluminescent spot intensity derived indicates changes in cytokine expression after 24 h.
Figure 4.
Normalized protein expression of CCL2/MCP-1 and IL-6 in MDA-MB-231 (A and C) and MDA-MB-468 (B and D) triple negative breast cancer cells. Data represent normalized dot spot intensities from the cytokine arrays based on the positive controls found in each of the membranes’ corners using RAYBIO®ANALYSIS software (RayBiotech). Data are expressed as % of control (mean±S.E.M. n=3), representing treatment with TNF-α (40 ng/ml) and co-treatment with DATS (75 μM)+TNF-α (40 ng/ml). Student’s t-test evaluated the statistically significant differences between TNF-α vs. co-treatment, with **p<0.01. NS: Non-significant.
The inhibitory effect of DATS on Human MCP-1 (CCL2) and IL-6 protein expression.
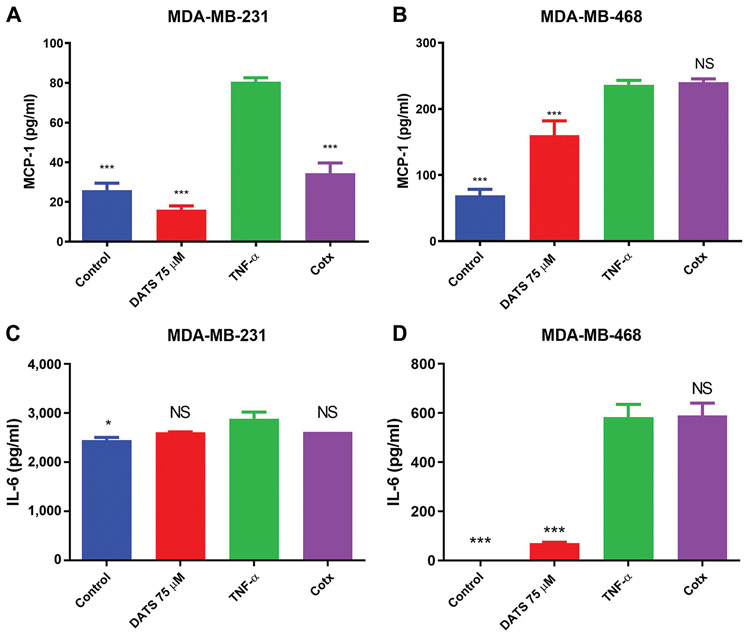
CCL2/MCP-1 and IL-6 specific ELISAs were used to validate the cytokine array findings. The results confirmed that TNF-α induces up-regulation of CCL2/MCP-1 and IL-6 expression in MDA-MB-231 and MDA-MB-468 breast cancer cell lines. DATS treatment could down-regulate CCL2/MCP-1 protein expression only in MDA-MB-231 cells, with no significant effect on MDA-MB-468 cells, corroborating with the cytokine arrays finding (Figure 5A and B). However, the ELISA for IL-6 showed that DATS had no significant effect on TNF-α-induced IL-6 expression in either the cell lines (Figure 5C and D).
Figure 5.
ELISA protein expression quantification in MDA-MB-231 and MDA-MB-468 triple negative breast cancer cells (TNBC). MDA-MB-231 and MDA-MB-468 TNBC cells were treated with diallyl trisulfide (DATS) (75 μM), TNF-α (40 ng/ml), the combination of DATS (75 μM)+TNF-α (40 ng/ml) or left untreated. CCL2/MCP-1 (A, B) and IL-6 (C, D) protein expression were assayed by ELISA. Each data point represents the mean±S.E.M. of three independent experiments (n=3). Statistically significant differences between TNF-α vs. treatments were evaluated by a one-way ANOVA, followed by Dunnett’s multiple comparison test with *p<0.05 and ***p<0.001. NS: Non-significant.
The inhibitory effect of DATS on MCP-1 (CCL2), IKBKE, and MAKP8 mRNA expression in MDA-MB-231 and MDA-MB-468 TNBC cells.
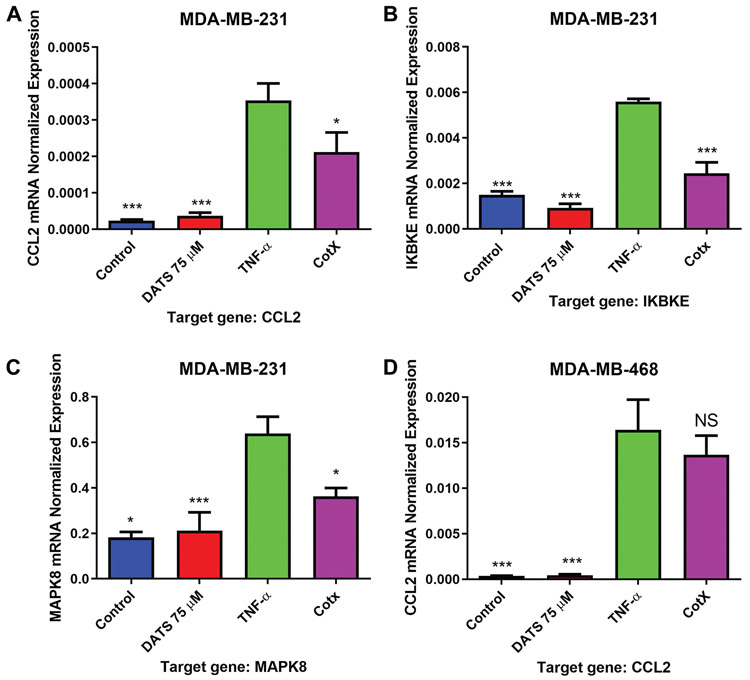
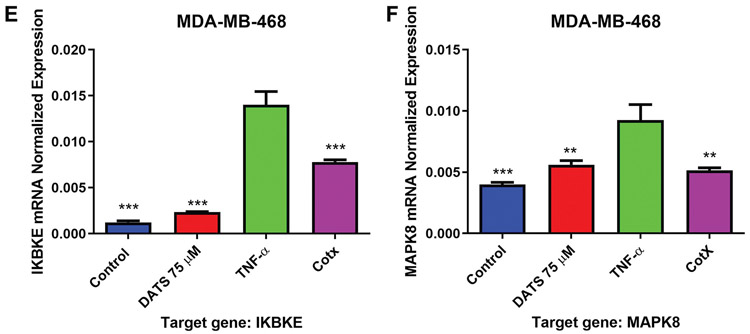
Quantitative real-time PCR was used to investigate the DATS effect on CCL2, IKBKE, and MAKP8 mRNA expression in MDA-MB-231 and MDA-MB-468 breast cancer cell lines. CCL2 pattern of expression showed similar to that of the cytokine arrays and ELISA experiments. TNF-α-induced a significant increase in CCL2 expression in both cell lines compared to control. DATS significantly (p<0.05) reduced CCL2 expression in MDA-MB-231 cells, reducing mRNA levels by more than 40% (Figure 6A and D). The inhibitory effect of DATS over CCL2 expression at the transcription level followed a pattern similar to that observed at the translational level. TNF-α also up-regulated IKBKE and MAPK8 mRNA expression in MDA-MB-231 and MDA-MB-468 cells, compared to the control (Figure 6B, C, E, and F). DATS inhibited the expression of IKBKE and MAPK8 in both cell lines (Figure 6B, C, E, and F).
Figure 6.
Quantification of mRNA expression in MDA-MB-231 and MDA-MB-468 triple negative breast cancer cells using RT-PCR. MDA-MB-231 and MDA-MB-468 TNBC cells were treated with diallyl trisulfide (DATS) (75 μM), TNF-α (40 ng/ml), the combination of DATS (75 μM)+TNF-α (40 ng/ml) or left untreated. The normalized levels of CCL2, IKBKE, and MAPK8 mRNA were assayed by using RT-PCR. Each data point represents the mean±S.E.M. of three independent experiments (n=3). Statistically significant differences between TNF-α vs. treatments were evaluated by a one-way ANOVA, followed by Dunnett’s multiple comparison test with *p<0.05, **p<0.01, and ***p<0.001.
Discussion
TNBC, which accounts for about 20% of breast cancers, is characterized by the lack of ER, PR, and the HER2/neu receptor expression (28, 30-32). Currently, there are limited therapeutic options available for TNBC patients; the poor prognosis, aggressive tumor behavior, and lack of targeted therapy make TNBC treatment challenging (28, 30-34). African American women experience a 40% higher TNBC mortality rate than Caucasian American women (35). For TNBC patients, chemotherapy is currently the only available fundamental therapeutic strategy (28, 30-34). Therefore, the standard treatment for TNBC currently includes non-targeted drugs, such as anthracyclines, taxanes, or a combination of broad-spectrum anticancer drugs. Unfortunately, these treatments are susceptible to tumor resistance and induce cellular toxicity (28, 30-32). Furthermore, TNBC is exceedingly more metastatic and invasive than receptor-specific types of breast cancer. Thus, it is essential to identify innovative targeted antitumor agents with minimal resistance and toxicity. Researchers have postulated that the utilization of natural compounds or drugs can suppress oncogenic cytokines (e.g., CXCL1, CCL18, CCL8, CCL2, IL-8, IL-6, etc.) by providing therapeutic advantages against aggressive inflammatory breast cancers (36-41). The organic sulfur compounds such as DAS/DADS/DATS have demonstrated antitumor activity in various types of tumor cells, including breast, lung, colorectal, and prostate cancer cells (20, 23-25).
In the present study, we investigated DATS effects on the viability of MDA-MB-231 and MDA-MB-468 breast cancer cell lines after 24, 48, and 72 h treatments. DATS in concentrations of 25 μM and higher significantly decreased cell viability in both cell lines time and dose-dependent manner. The statistical analysis comparing the LC50s for both cell lines, calculated after 24 and 48 h treatment with DATS, showed that MDA-MB-468 cells are more sensitive to DATS treatment. These findings are in agreement with those of Byeong-Chel et al. (42) and Chandra-Kuntal et al. (43), confirming that DATS treatment resulted in a dose and time-dependent cytotoxicity in MDA-MB-231 cells. Unlike DATS, the vehicle control DMSO caused no cytotoxicity, as has also been reported by Byeong-Chel et al. (42).
Cell motility is a crucial factor in cancer metastasis. In this study, the motility of MDA-MB-231 and MDA-MB-468 human breast cancer cells was determined by the wound-healing assay. Combination treatment with DATS and TNF-α, as well as DATS treatment alone, showed an inhibitory effect on vertical cell migration of tumor cells, compared to the control in both cell lines (Figure 2A-D). Interestingly, based on the significant decrease in vertical migration (indicative of cellular toxicity), MDA-MB-468 cells were more sensitive to the effect of DATS (Figure 2C and D). Similarly, Lui et al. (28) reported that DATS inhibited horizontal and vertical migration of MDA-MB-231 and HS 578T cells, as well as invasion. In addition, DATS could alter the morphology of MDA-MB-231 and HS 578T cells, therefore, possibly having antimetastatic potential. Targeting migration and invasion of cancer cells can potentially treat metastatic breast cancer (28, 44).
This investigation also showed that DATS could inhibit TNF-α-induced CCL2 release. Several studies have shown the important role of CCL2 signaling in breast cancer cells (4, 45, 46), indicating that targeting the CCL2 signaling pathway may modulate various mechanisms involved in cancer progression, hence making CCL2 a promising therapeutic target (4, 47). CCL2 is a cancer-promoting chemokine capable of enhancing malignant properties (12). Once enabled to differentiate into tumor-associated macrophages (TAMs), these TAMs can promote metastasis largely by matrix remodeling. TAMs have crucial roles in tumor progression and breast cancer metastasis. They enroll tumor cell factors, including matrix-degrading enzymes, and therefore promote invasion and metastasis of breast tumors. TAMs are known to produce CCL2, a key determinant of macrophage infiltration and angiogenesis that has been demonstrated to have a role in breast cancer, contributing to the recruitment of macrophages and inducing proliferation and migration (48-50). TAMs and elevated expression of CCL2 have been linked to various types of human cancer (14, 48, 51). The data from this study showed that DATS inhibited, at the transcriptional and translational level, TNF-α-induced expression of CCL2 in MDA-MB-231 but not in MDA-MB-468 cells suggesting the cytotoxic effect on MDA-MB-468 cells is not associated with CCL2 regulation. It is well-understood that malignant tissues can release chemokines such as CCL2 amongst chemoattractants and growth factors, which is a key component in the progression of cancer cell proliferation, migration, and invasion (12). CCL2 controls the signaling for the recruitment and mobilization of monocytes, macrophages, and other inflammatory responses (16). CCL2 recruits CD14+ and CD16+ monocytes, which are able to produce the proinflammatory cytokine TNF-α (52). TNFα-induced CCL2 expression is mediated via MAPK and NFκB signaling pathways. NFκB has been described as a factor promoting tumor development, changing cell homeostasis, and inducing inflammatory processes (53). In addition, it is well known that cell proliferation alone does not cause cancer; however, it is the combination of an unrestrained proliferation in an environment rich in inflammatory cells, growth factors, and DNA damage inducers that increases the risk of tumor development and can lead to cancer (54).
NFκB activation induces cell growth and programmed cell death in multiple cell lines and has been associated with proliferative pathways and cell death signals (55-57). TNF-α induces activation of NFκB, which translocates from the cytoplasm into the nucleus. NFκB localization is dictated by IκBS, which bind to NFκB and prevent nuclear localization. IκBs are modulated by phosphorylation, which can stimulate ubiquitin-dependent degradation, allowing free NFκB to translocate into the nucleus and initiate gene transcription. IKKi/IKKε plays a vital role in implementing TNFα signaling by acting as a serine-threonine kinase (58). IKKε/IKBKE has been classified as an oncogene because of its association with cell proliferation and transformation (59). Additionally, MAPK families, including JNK, ERK, and P38 kinases, also have been shown to play a key role in cell invasion and cancer metastasis (60).
The data presented in this study showed DATS’s ability to down-regulate both IKBKE and MAPK8 mRNA expression in both MDA-MB-231 and MDA-MB-468 cells. DATS’s ability to inhibit CCL2 expression in MDA-MB-231 cells could be due to its ability to down-regulate IKBKE and MAPK8 mRNA expression. Bauer et al. showed that IKBKE down-regulation attenuated CCL2 expression in MDA-MB-231 TNBC cells (61). The IKBKE gene is linked to cancer and inflammation; and is over-expressed in approximately 30% of human breast tumors (56, 57). In addition, the IKBKE gene stimulates cytokine release and pro-survival signaling via the activation of NFκB and JAK-STAT signaling pathways (56). In summary, the present investigation demonstrated DATS anticancer potential in two different TNBC cell lines: MDA-MB-231 and MDA-MB-468. DATS showed anti-migration effects on both cell lines and higher cytotoxicity in MDA-MB-468 compared to MDA-MB-231 cells. In addition, DATS down-regulated the mRNA expression of TNF-α-stimulated MCP-1 in MDA-MB-231 (Caucasian) cells but not in MDA-MB-468 (African Americans) cells. Our data indicate that the inhibitory effects of DATS on IKBKE and MAPK8 mRNA expression could be a possible molecular mechanism involved in the down-regulation of MCP-1.
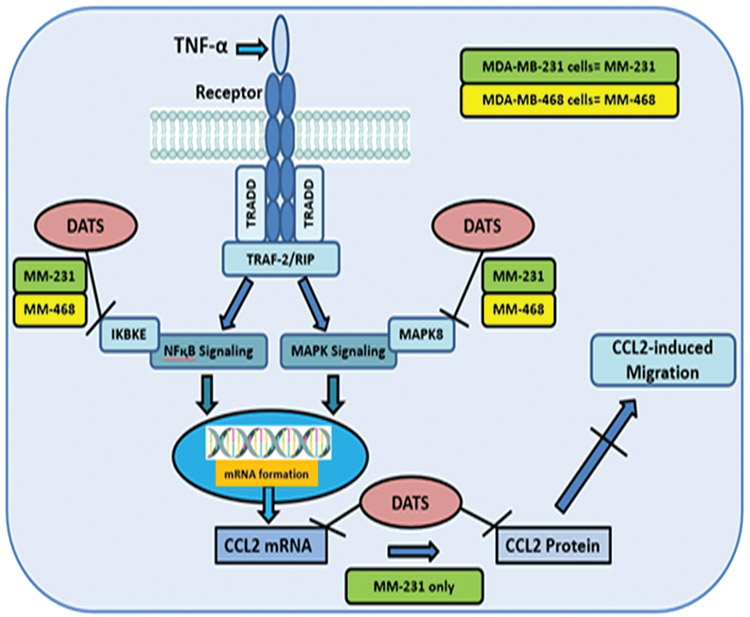
In conclusion, this study indicates that DATS targets CCL2 in MDA-MB-231 but not in MDA-MB-468 cells, suggesting that genetically different cell lines may respond in diverse ways to DATS. The findings show that DATS may have a potential role in TNBC therapy and prevention by targeting CCL2 (Figure 7). In addition, these results provide more evidence regarding the role of garlic in cancer prevention and in reducing the incidence rate of several cancers.
Figure 7.
Schematic diagram of our proposed mechanism of action for diallyl trisulfide (DATS) in TNF-α induced TNBC illustrating proinflammatory genes involved in different signaling pathways that mediate the release of CCL2 in MDA-MB-231 (green color) and MDA-MB-468 (yellow color) TNBC cells induced by TNF-α. DATS can inhibit CCL2, IKBKE, and MAPK8.
Acknowledgements
The present study was supported by the National Institute of Minority Health and Health Disparity through the U54 MD007582 and P20 MD006738 grants.
Footnotes
Conflicts of Interest
The Authors declare no competing interests concerning this study.
References
- 1.American Cancer Society. Cancer Statistics Center. Available at: http://cancerstatisticscenter.cancer.org. 2020. [Last accessed on September 20, 2021] [Google Scholar]
- 2.Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar AF, Minna JD and Pollack JR: Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One 4(7): e6146, 2009. PMID: 19582160. DOI: 10.1371/journal.pone.0006146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121(7): 2750–2767, 2011. PMID: 21633166. DOI: 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendonca P, Horton A, Bauer D, Messeha S and Soliman KFA: The inhibitory effects of butein on cell proliferation and TNF-α-induced CCL2 release in racially different triple negative breast cancer cells. PLoS One 14(10): e0215269, 2019. PMID: 31665136. DOI: 10.1371/journal.pone.0215269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF and Ellis IO: Prognostic markers in triple-negative breast cancer. Cancer 109(1): 25–32, 2007. PMID: 17146782. DOI: 10.1002/cncr.22381 [DOI] [PubMed] [Google Scholar]
- 6.Elias AD: Triple-negative breast cancer: a short review. Am J Clin Oncol 33(6): 637–645, 2010. PMID: 20023571. DOI: 10.1097/COC.0b013e3181b8afcf [DOI] [PubMed] [Google Scholar]
- 7.Foulkes WD, Smith IE and Reis-Filho JS: Triple-negative breast cancer. N Engl J Med 363(20): 1938–1948, 2010. PMID: 21067385. DOI: 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 8.Bauer D, Mazzio E, Soliman KF, Taka E, Oriaku E, Womble T and Darling-Reed S: Diallyl disulfide inhibits TNFα-induced CCL2 release by MDA-MB-231 cells. Anticancer Res 34(6): 2763–2770, 2014. PMID: 24922637. [PMC free article] [PubMed] [Google Scholar]
- 9.Wong MP, Cheung KN, Yuen ST, Fu KH, Chan AS, Leung SY and Chung LP: Monocyte chemoattractant protein-1 (MCP-1) expression in primary lymphoepithelioma-like carcinomas (LELCs) of the lung. J Pathol 186(4): 372–377, 1998. PMID: 10209485. DOI: [DOI] [PubMed] [Google Scholar]
- 10.Varney ML, Johansson SL and Singh RK: Tumour-associated macrophage infiltration, neovascularization and aggressiveness in malignant melanoma: role of monocyte chemotactic protein-1 and vascular endothelial growth factor-A. Melanoma Res 15(5): 417–425, 2005. PMID: 16179869. DOI: 10.1097/00008390-200510000-00010 [DOI] [PubMed] [Google Scholar]
- 11.Soria G and Ben-Baruch A: The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett 267(2): 271–285, 2008. PMID: 18439751. DOI: 10.1016/j.canlet.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 12.Craddock JA, Lu A, Bear A, Pule M, Brenner MK, Rooney CM and Foster AE: Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b. J Immunother 33(8): 780–788, 2010. PMID: 20842059. DOI: 10.1097/CJI.0b013e3181ee6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ksiazkiewicz M, Gottfried E, Kreutz M, Mack M, Hofstaedter F and Kunz-Schughart LA: Importance of CCL2-CCR2A/2B signaling for monocyte migration into spheroids of breast cancer-derived fibroblasts. Immunobiology 215(9-10): 737–747, 2010. PMID: 20605053. DOI: 10.1016/j.imbio.2010.05.019 [DOI] [PubMed] [Google Scholar]
- 14.Daurkin I, Eruslanov E, Stoffs T, Perrin GQ, Algood C, Gilbert SM, Rosser CJ, Su LM, Vieweg J and Kusmartsev S: Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res 71(20): 6400–6409, 2011. PMID: 21900394. DOI: 10.1158/0008-5472.CAN-11-1261 [DOI] [PubMed] [Google Scholar]
- 15.Zollo M, Di Dato V, Spano D, De Martino D, Liguori L, Marino N, Vastolo V, Navas L, Garrone B, Mangano G, Biondi G and Guglielmotti A: Targeting monocyte chemotactic protein-1 synthesis with bindarit induces tumor regression in prostate and breast cancer animal models. Clin Exp Metastasis 29(6): 585–601, 2012. PMID: 22484917. DOI: 10.1007/s10585-012-9473-5 [DOI] [PubMed] [Google Scholar]
- 16.Perry JA, Thamm DH, Eickhoff J, Avery AC and Dow SW: Increased monocyte chemotactic protein-1 concentration and monocyte count independently associate with a poor prognosis in dogs with lymphoma. Vet Comp Oncol 9(1): 55–64, 2011. PMID: 21303454. DOI: 10.1111/j.1476-5829.2010.00235.x [DOI] [PubMed] [Google Scholar]
- 17.Sanford DE, Belt BA, Panni RZ, Mayer A, Deshpande AD, Carpenter D, Mitchem JB, Plambeck-Suess SM, Worley LA, Goetz BD, Wang-Gillam A, Eberlein TJ, Denardo DG, Goedegebuure SP and Linehan DC: Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin Cancer Res 19(13): 3404–3415, 2013. PMID: 23653148. DOI: 10.1158/1078-0432.CCR-13-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitra S: MicroRNA therapeutics in triple negative breast cancer. Archives of Pathology and Clinical Research 1(1): 009–017, 2021. DOI: 10.29328/journal.hjpcr.1001003 [DOI] [Google Scholar]
- 19.Xiao D, Herman-Antosiewicz A, Antosiewicz J, Xiao H, Brisson M, Lazo JS and Singh SV: Diallyl trisulfide-induced G(2)-M phase cell cycle arrest in human prostate cancer cells is caused by reactive oxygen species-dependent destruction and hyperphosphorylation of Cdc 25 C. Oncogene 24(41): 6256–6268, 2005. PMID: 15940258. DOI: 10.1038/sj.onc.1208759 [DOI] [PubMed] [Google Scholar]
- 20.Antosiewicz J, Herman-Antosiewicz A, Marynowski SW and Singh SV: c-Jun NH(2)-terminal kinase signaling axis regulates diallyl trisulfide-induced generation of reactive oxygen species and cell cycle arrest in human prostate cancer cells. Cancer Res 66(10): 5379–5386, 2006. PMID: 16707465. DOI: 10.1158/0008-5472.CAN-06-0356 [DOI] [PubMed] [Google Scholar]
- 21.Das A, Banik NL and Ray SK: Garlic compounds generate reactive oxygen species leading to activation of stress kinases and cysteine proteases for apoptosis in human glioblastoma T98G and U87MG cells. Cancer 110(5): 1083–1095, 2007. PMID: 17647244. DOI: 10.1002/cncr.22888 [DOI] [PubMed] [Google Scholar]
- 22.Kim JM, Noh EM, Song HK, Lee M, Lee SH, Park SH, Ahn CK, Lee GS, Byun EB, Jang BS, Kwon KB and Lee YR: Salvia miltiorrhiza extract inhibits TPA-induced MMP-9 expression and invasion through the MAPK/AP-1 signaling pathway in human breast cancer MCF-7 cells. Oncol Lett 14(3): 3594–3600, 2017. PMID: 28927117. DOI: 10.3892/ol.2017.6638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu XJ, Hu Y, Lamy E and Mersch-Sundermann V: Apoptosis induction in human lung adenocarcinoma cells by oil-soluble allyl sulfides: triggers, pathways, and modulators. Environ Mol Mutagen 50(3): 266–275, 2009. PMID: 19197990. DOI: 10.1002/em.20467 [DOI] [PubMed] [Google Scholar]
- 24.Chandra-Kuntal K and Singh SV: Diallyl trisulfide inhibits activation of signal transducer and activator of transcription 3 in prostate cancer cells in culture and in vivo. Cancer Prev Res (Phila) 3(11): 1473–1483, 2010. PMID: 20959517. DOI: 10.1158/1940-6207.CAPR-10-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsubura A, Lai YC, Kuwata M, Uehara N and Yoshizawa K: Anticancer effects of garlic and garlic-derived compounds for breast cancer control. Anticancer Agents Med Chem 11(3): 249–253, 2011. PMID: 21269259. DOI: 10.2174/187152011795347441 [DOI] [PubMed] [Google Scholar]
- 26.Nkrumah-Elie YM, Reuben JS, Hudson AM, Taka E, Badisa R, Ardley T, Israel B, Sadrud-Din SY, Oriaku ET and Darling-Reed SF: The attenuation of early benzo(a)pyrene-induced carcinogenic insults by diallyl disulfide (DADS) in MCF-10A cells. Nutr Cancer 64(7): 1112–1121, 2012. PMID: 23006051. DOI: 10.1080/01635581.2012.712738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosono T, Sato A, Nakaguchi N, Ozaki-Masuzawa Y and Seki T: Diallyl Trisulfide Inhibits Platelet Aggregation through the Modification of Sulfhydryl Groups. J Agric Food Chem 68(6): 1571–1578, 2020. PMID: 31927886. DOI: 10.1021/acs.jafc.9b05557 [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Zhu P, Wang Y, Wei Z, Tao L, Zhu Z, Sheng X, Wang S, Ruan J, Liu Z, Cao Y, Shan Y, Sun L, Wang A, Chen W and Lu Y: Antimetastatic therapies of the polysulfide diallyl trisulfide against triple-negative breast cancer (TNBC) via suppressing MMP2/9 by blocking NF-κB and ERK/MAPK signaling pathways. PLoS One 10(4): e0123781,2015. PMID: 25927362. DOI: 10.1371/journal.pone.0123781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malki A, El-Saadani M and Sultan AS: Garlic constituent diallyl trisulfide induced apoptosis in MCF7 human breast cancer cells. Cancer Biol Ther 8(22): 2175–2185, 2009. PMID: 19823037. DOI: 10.4161/cbt.8.22.9882 [DOI] [PubMed] [Google Scholar]
- 30.Irvin WJ Jr and Carey LA: What is triple-negative breast cancer? Eur J Cancer 44(18): 2799–2805, 2008. PMID: 19008097. DOI: 10.1016/j.ejca.2008.09.034 [DOI] [PubMed] [Google Scholar]
- 31.Tan AR and Swain SM: Therapeutic strategies for triple-negative breast cancer. Cancer J 14(6): 343–351, 2008. PMID: 19060597. DOI: 10.1097/PPO.0b013e31818d839b [DOI] [PubMed] [Google Scholar]
- 32.Arslan C, Dizdar O and Altundag K: Pharmacotherapy of triple-negative breast cancer. Expert Opin Pharmacother 10(13): 2081–2093, 2009. PMID: 19640211. DOI: 10.1517/14656560903117309 [DOI] [PubMed] [Google Scholar]
- 33.Garlapati C, Joshi S, Sahoo B, Kapoor S and Aneja R: The persisting puzzle of racial disparity in triple negative breast cancer: looking through a new lens. Front Biosci (Schol Ed) 11: 75–88, 2019. PMID: 30844737. DOI: 10.2741/S527 [DOI] [PubMed] [Google Scholar]
- 34.Newman LA, Jenkins B, Chen Y, Oppong JK, Adjei E, Jibril AS, Hoda S, Cheng E, Chitale D, Bensenhaver JM, Awuah B, Bekele M, Abebe E, Kyei I, Aitpillah F, Adinku M, Nathanson SD, Jackson L, Jiagge E, Merajver S, Petersen LF, Proctor E, Gyan KK, Martini R, Kittles R and Davis MB: Hereditary susceptibility for triple negative breast cancer associated with Western sub-Saharan African ancestry: Results from an international surgical breast cancer collaborative. Ann Surg 270(3): 484–492, 2019. PMID: 31356281. DOI: 10.1097/SLA.0000000000003459 [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Susick L, Davis M, Bensenhaver J, Nathanson SD, Burns J and Newman LA: Evaluation of triple-negative breast cancer early detection via mammography screening and outcomes in African American and white American patients. JAMA Surg 155(5): 440–442, 2020. PMID: 32074266. DOI: 10.1001/jamasurg.2019.6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta SC, Sundaram C, Reuter S and Aggarwal BB: Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta 1799(10-12): 775–787, 2010. PMID: 20493977. DOI: 10.1016/j.bbagrm.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, Song J, Zhang F, Zhang X, Wang Q and Wang Z: CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis 9(9): 880, 2018. PMID: 30158589. DOI: 10.1038/s41419-018-0876-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, Soong DYH, Cotechini T, Anur P, Lin EY, Fidanza A, Lopez-Yrigoyen M, Millar MR, Urman A, Ai Z, Spellman PT, Hwang ES, Dixon JM, Wiechmann L, Coussens LM, Smith HO and Pollard JW: Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell 35(4): 588–602.e10, 2019. PMID: 30930117. DOI: 10.1016/j.ccell.2019.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Little AC, Pathanjeli P, Wu Z, Bao L, Goo LE, Yates JA, Oliver CR, Soellner MB and Merajver SD: IL-4/IL-13 stimulated macrophages enhance breast cancer invasion via rho-GTPase regulation of synergistic VEGF/CCL-18 signaling. Front Oncol 9: 456, 2019. PMID: 31214501. DOI: 10.3389/fonc.2019.00456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tandon I and Sharma NK: Macrophage flipping from foe to friend: A matter of interest in breast carcinoma heterogeneity driving drug resistance. Curr Cancer Drug Targets 19(3): 189–198, 2019. PMID: 29952260. DOI: 10.2174/1568009618666180628102247 [DOI] [PubMed] [Google Scholar]
- 41.Sudhakaran M, Sardesai S and Doseff AI: Flavonoids: New frontier for immuno-regulation and breast cancer control. Antioxidants (Basel) 8(4): 103, 2019. PMID: 30995775. DOI: 10.3390/antiox8040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee BC, Park BH, Kim SY and Lee YJ: Role of Bim in diallyl trisulfide-induced cytotoxicity in human cancer cells. J Cell Biochem 112(1): 118–127, 2011. PMID: 21053278. DOI: 10.1002/jcb.22896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandra-Kuntal K, Lee J and Singh SV: Critical role for reactive oxygen species in apoptosis induction and cell migration inhibition by diallyl trisulfide, a cancer chemopreventive component of garlic. Breast Cancer Res Treat 138(1): 69–79, 2013. PMID: 23412769. DOI: 10.1007/s10549-013-2440-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckhardt BL, Francis PA, Parker BS and Anderson RL: Strategies for the discovery and development of therapies for metastatic breast cancer. Nat Rev Drug Discov 11(6): 479–497, 2012. PMID: 22653217. DOI: 10.1038/nrd2372 [DOI] [PubMed] [Google Scholar]
- 45.Kopfstein L and Christofori G: Metastasis: cell-autonomous mechanisms versus contributions by the tumor microenvironment. Cell Mol Life Sci 63(4): 449–468, 2006. PMID: 16416030. DOI: 10.1007/s00018-005-5296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joyce JA and Pollard JW: Microenvironmental regulation of metastasis. Nat Rev Cancer 9(4): 239–252, 2009. PMID: 19279573. DOI: 10.1038/nrc2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P and Cheng N: CCL2/CCR2 chemokine signaling coordinates survival and motility of breast cancer cells through Smad3 protein- and p42/44 mitogen-activated protein kinase (MAPK)-dependent mechanisms. J Biol Chem 287(43): 36593–36608, 2012. PMID: 22927430. DOI: 10.1074/jbc.M112.365999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra P, Banerjee D and Ben-Baruch A: Chemokines at the crossroads of tumor-fibroblast interactions that promote malignancy. J Leukoc Biol 89(1): 31–39, 2011. PMID: 20628066. DOI: 10.1189/jlb.0310182 [DOI] [PubMed] [Google Scholar]
- 49.Leung SY, Wong MP, Chung LP, Chan AS and Yuen ST: Monocyte chemoattractant protein-1 expression and macrophage infiltration in gliomas. Acta Neuropathol 93(5): 518–527, 1997. PMID: 9144591. DOI: 10.1007/s004010050647 [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Patel L and Pienta KJ: CC chemokine ligand 2 (CCL2) promotes prostate cancer tumorigenesis and metastasis. Cytokine Growth Factor Rev 21(1): 41–48, 2010. PMID: 20005149. DOI: 10.1016/j.cytogfr.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazur G, Jaskuła E, Kryczek I, Dłubek D, Butrym A, Wróbel T, Lange A and Kuliczkowski K: Proinflammatory chemokine gene expression influences survival of patients with non-Hodgkin’s lymphoma. Folia Histochem Cytobiol 49(2): 240–247, 2011. PMID: 21744323. DOI: 10.5603/fhc.2011.0033 [DOI] [PubMed] [Google Scholar]
- 52.Feng AL, Zhu JK, Sun JT, Yang MX, Neckenig MR, Wang XW, Shao QQ, Song BF, Yang QF, Kong BH and Qu X: CD16+ monocytes in breast cancer patients: expanded by monocyte chemoattractant protein-1 and may be useful for early diagnosis. Clin Exp Immunol 164(1): 57–65, 2011. PMID: 21361908. DOI: 10.1111/j.1365-2249.2011.04321.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanahan D and Weinberg RA: The hallmarks of cancer. Cell 100(1): 57–70, 2000. PMID: 10647931. DOI: 10.1016/s0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 54.Dvorak HF: Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med 315(26): 1650–1659, 1986. PMID: 3537791. DOI: 10.1056/NEJM198612253152606 [DOI] [PubMed] [Google Scholar]
- 55.Barkett M and Gilmore TD: Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene 18(49): 6910–6924, 1999. PMID: 10602466. DOI: 10.1038/sj.onc.1203238 [DOI] [PubMed] [Google Scholar]
- 56.Karin M and Lin A: NF-kappaB at the crossroads of life and death. Nat Immunol 3(3): 221–227, 2002. PMID: 11875461. DOI: 10.1038/ni0302-221 [DOI] [PubMed] [Google Scholar]
- 57.Biswas DK, Martin KJ, McAlister C, Cruz AP, Graner E, Dai SC and Pardee AB: Apoptosis caused by chemotherapeutic inhibition of nuclear factor-kappaB activation. Cancer Res 63(2): 290–295, 2003. PMID: . [PubMed] [Google Scholar]
- 58.Häcker H and Karin M: Regulation and function of IKK and IKK-related kinases. Sci STKE 2006(357): re13, 2006. PMID: 17047224. DOI: 10.1126/stke.3572006re13 [DOI] [PubMed] [Google Scholar]
- 59.Verhelst K, Verstrepen L, Carpentier I and Beyaert R: IκB kinase ε (IKKκ): a therapeutic target in inflammation and cancer. Biochem Pharmacol 85(7): 873–880, 2013. PMID: 23333767. DOI: 10.1016/j.bcp.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong S, Park KK, Magae J, Ando K, Lee TS, Kwon TK, Kwak JY, Kim CH and Chang YC: Ascochlorin inhibits matrix metalloproteinase-9 expression by suppressing activator protein-1-mediated gene expression through the ERK1/2 signaling pathway: inhibitory effects of ascochlorin on the invasion of renal carcinoma cells. J Biol Chem 280(26): 25202–25209, 2005. PMID: 15863510. DOI: 10.1074/jbc.M413985200 [DOI] [PubMed] [Google Scholar]
- 61.Bauer D, Redmon N, Mazzio E and Soliman KF: Apigenin inhibits TNFα/IL-1α-induced CCL2 release through IKBK-epsilon signaling in MDA-MB-231 human breast cancer cells. PLoS One 12(4): e0175558, 2017. PMID: 28441391. DOI: 10.1371/journal.pone.0175558 [DOI] [PMC free article] [PubMed] [Google Scholar]