Abstract
R-loops are non-B DNA structures that form during transcription when the nascent RNA anneals to the template DNA strand forming a RNA:DNA hybrid. Understanding the genomic distribution and function of R-loops is an important goal, since R-loops have been implicated in a number of adaptive and maladaptive processes under physiological and pathological conditions. Based on R-loop mapping datasets, we propose the existence of two main classes of R-loops, each associated with unique characteristics. Promoter-paused R-loops (Class I) are short R-loops that form at high frequency during promoter-proximal pausing by RNA polymerase II. Elongation-associated R-loops (Class II) are long structures that occur throughout gene bodies at modest frequencies. We further discuss the relationships between each R-loop class with instances of genome instability and suggest that increased class I R-loops, resulting from enhanced promoter-proximal pausing, represent the main culprits for R-loop mediated genome instability under pathological conditions.
INTRODUCTION
R-loops are three-stranded non-B DNA structures that form during transcription upon reannealing of the nascent RNA to the template DNA strand, forming an RNA:DNA hybrid and causing the non-template DNA strand to loop out in a single-stranded state. R-loops were first recognized to form at the replication origins of bacterial ColE1-type plasmids, where they serve to open the DNA double helix and the RNA strand can be processed upon Ribonuclease H digestion into a primer for leading strand replication (Itoh and Tomizawa, 1980; Masukata and Tomizawa, 1990). A similar mechanism was shown to mediate DNA replication initiation in bacteriophage T4 (Belanger and Kreuzer, 1998; Carles-Kinch and Kreuzer, 1997; Kreuzer and Brister, 2010) and in the mitochondrial genome (Lee and Clayton, 1996, 1998; Xu and Clayton, 1995). R-loops were then recognized to form in the chromosomes of mammalian B cells upon induction of transcription at specialized class switch regions (Yu et al., 2003). In this case, R-loop formation is associated with the formation of programmed double-stranded DNA breaks (DSBs) that are required to initiate immunoglobulin class switch recombination. Thus, from early on, it became apparent that transcription-mediated R-loop formation could play important physiological roles from E. coli to mammals, and that R-loops represent a novel type of cis-acting biological signal.
The study of E. coli RNase H mutant strains, however, provided evidence that R-loops, if left to accumulate, could cause significant problems. rnhA mutants have the unique ability to replicate their genome independently of the chromosomal replication origin, oriC, and of the DnaA replication initiation protein (Kogoma and von Meyenburg, 1983; Ogawa et al., 1984). A similar ability was also observed for knockout mutants of the recG gene, which encodes a helicase capable of resolving R-loops (Hong et al., 1995). This mode of replication termed constitute stable DNA replication (cSDR) is strictly dependent on the recombinase activity of the RecA protein (Kogoma et al., 1985). It arises due to the formation of RecA-catalyzed R-loops that persist due to their reduced resolution in the absence of RNase H or RecG activity, and initiate DNA replication at alternative replication origins termed oriKs, distributed along the chromosome (Drolet and Brochu, 2019; Kogoma, 1997). rnhA− and recG− E. coli mutants show sluggish growth, and increased genome instability, consistent with the induction of replication forks from oriKs causing global alterations of replication for migration patterns (Maduike et al., 2014; Wimberly et al., 2013). A similar induction of alternative replication origins was observed at highly transcribed rDNA regions in yeast cells that accumulate R-loops due to deficiency in RNase H activity and DNA topoisomerase I (Stuckey et al., 2015). Therefore, it became clear early on that R-loop levels must be tightly controlled to avoid deleterious consequences on genome stability and that cells have evolved enzymes such as RNase H and helicases to promote R-loop resolution.
Over the last decade, our understanding of R-loops, including the mechanisms that control their formation and resolution, their genomic distribution, and their functional consequences has dramatically increased. R-loops represent a prevalent class of non-B DNA structures in all genomes including from yeasts, plants, flies, and worms (Alecki et al., 2020; El Hage et al., 2014; Hartono et al., 2018; Wahba et al., 2016; Xu et al., 2020; Xu et al., 2017; Zeller et al., 2016). In mammalian genomes, R-loops collectively occur over tens of thousands of conserved genic loci (Chen et al., 2017; Crossley et al., 2019b; Ginno et al., 2012; Sanz et al., 2016; Wang et al., 2021b; Yan et al., 2019), highlighting the fact that R-loops are well-tolerated by cells under normal conditions. In addition, a variety of functional roles such as that described above for prokaryotic replication origins, have been assigned to R-loops, further suggesting that they play adaptive roles under physiological situations (see below).
At the same time, many studies have suggested that under pathological conditions, harmful R-loops arise from defective cellular processes and trigger DNA damage and genomic instability. Defects in co-transcriptional processes such as RNA export, cleavage, and splicing have been particularly associated with harmful R-loops (Aguilera and Garcia-Muse, 2012; Chan et al., 2014; Kaneko et al., 2007; Li and Manley, 2005, 2006; Paulsen et al., 2009; Stirling et al., 2012). One of the key pieces of evidence supporting the idea of harmful R-loops is that cellular over-expression of Ribonuclease H1 (RNase H1), an enzyme with a clear biochemical ability to resolve RNA:DNA hybrids and R-loops (Cerritelli and Crouch, 2009), can at least partially suppress a variety of genome instability phenotypes (Huertas and Aguilera, 2003; Paulsen et al., 2009). Harmful R-loops, in turn, were proposed to affect genome stability by causing or exacerbating transcription-replication collisions (Hamperl et al., 2017; Hamperl and Cimprich, 2014, 2016; Lang et al., 2017), triggering replicative stress (Barroso et al., 2019; Crossley et al., 2019a; Herold et al., 2019; Landsverk et al., 2019; Morales et al., 2016), or inducing nuclease-mediated DNA breakage (Sollier and Cimprich, 2015; Sollier et al., 2014). We note, however, that the association between harmful R-loops and genome instability relied in many instances on observations of excessive R-loop levels by S9.6 immunofluorescence microscopy. Recent evidence, however, suggests that these observations may need to be revisited given the likelihood of significant confounding artefacts in S9.6 imaging studies (Smolka et al., 2021). At the genomic level, harmful R-loops remain poorly characterized. Similarly, the spatiotemporal relationship between harmful R-loop formation, DNA damage initiation and their suppression by RNase H1 expression has for the most part never been directly assessed. Thus, significant questions remain surrounding the identities of harmful R-loops and their mechanism of action. Here, we focus on reviewing recent R-loop mapping efforts in mammalian cells. We suggest that these studies can be most easily reconciled in light of the existence of distinct R-loop classes, each with unique characteristics. We further propose that events of genome instability may be connected to specific R-loop sub-types.
R-loop mapping efforts suggest the existence of two classes of R-loops.
Two main types of R-loop mapping methodologies have been developed to provide population-average views of genomic R-loop distributions. These strategies rely either on the S9.6 anti RNA:DNA hybrid monoclonal antibody (Boguslawski et al., 1986), or on catalytically inactive variants of Ribonuclease H1 (dRNase H1) that are still binding-competent due to the RNase H1 hybrid-binding domain (Chen et al., 2017). Despite significant concerns about the use of S9.6 in imaging applications, it permits accurate R-loop mapping in genomics applications after DNA:RNA ImmunoPrecipitation (DRIP) (Smolka et al., 2021). Several variations of the initial DRIP-seq method (Ginno et al., 2012) with various degrees of resolution and strand-specificity have been published (Crossley et al., 2019b; Sanz et al., 2016; Smolka et al., 2021; Xu et al., 2017) and generally produce highly congruent maps in human cells (Chedin et al., 2021). Importantly, S9.6-based methods require initial steps of DNA extraction and fragmentation which allow the pre-treatment of extracted nucleic acids by exogenous RNase H. As expected, DRIP-seq maps are highly sensitive to RNase H pre-treatment, providing an essential specificity control. In addition, DRIP-based maps have been independently validated using approaches based on non-denaturing sodium bisulfite in an S9.6-independent manner (Malig et al., 2020). Nonetheless, S9.6-based mapping methodologies can be considered as mapping R-loops ex vivo since they require initial nucleic acid extraction from cells. By contrast, dRNase H1-based approaches rely on mapping R-loops either through mapping the binding sites of dRNase H1 expressed in vivo (such as in RNase H1 ChIP, or R-ChIP (Chen et al., 2017)) or by liberating R-loops from native chromatin via methodologies derived from CUT&RUN and CUT&TAG (Wang et al., 2021b; Yan et al., 2019). One key advantage of such methods is that R-loops are profiled under more native conditions without the need to extract nucleic acids or chromatin from cells prior to mapping.
Distribution of R-loops from native mapping methodologies.
Major differences have emerged between dRNase H1- and S9.6-based R-loop maps. dRNase H1-based maps consistently identify R-loops over GC-rich and GC-skewed promoter-proximal pause regions of numerous transcribed genes. Of the twelve thousand or so R-ChIP-seq peaks recovered, nearly 60% mapped to promoter-proximal regions, significantly higher than observed in gene bodies (17%), or over gene terminal regions (6%) (Chen et al., 2017). In addition to genic R-loops, dRNase H1-based methods have consistently detected the presence of several thousand intergenic R-loops mapping to active enhancer regions (Chen et al., 2017; Wang et al., 2021b; Wulfridge and Sarma, 2021; Yan et al., 2019). tRNA genes also represent among the strongest hotspots for dRNase H1 binding by R-ChIP (Chen et al., 2017). This observation is also true in yeast (El Hage et al., 2014; Hartono et al., 2018; Legros et al., 2014), which suggests that high R-loop loads may associate with these short genes. Median R-loop peak sizes reported by R-ChIP were relatively short, around 200-300 bp (Chen et al., 2017; Wang et al., 2021b). Importantly, dRNase H1 recruitment to transcription start sites (TSSs) was dynamically correlated with transcriptional pausing, suggesting a mechanistic connection between pausing and R-loop formation (Chen et al., 2017).
Distribution of R-loops ex vivo.
S9.6-based maps, on the other hand, show that R-loops are predominantly distributed along transcribed genic regions and correlate with both gene expression levels and gene length (Sanz et al., 2016). Tens of thousands of conserved peaks of R-loop formation have consistently been recovered from a variety of human cell lines and from multiple studies (Chedin et al., 2021; Sanz et al., 2016). About half of these peaks map to transcribed gene bodies, with hotspots observed downstream of GC-skewed CpG island promoters (13%) and terminal genic regions (19%)(Figure 1) (Sanz et al., 2016). By contrast with native R-loops that are confined to a short region immediately downstream of the transcription start site (TSS), R-loops revealed by DRIP approaches only reach a maximum representation about 1-1.5 kb downstream of the TSS. While ex vivo promoter R-loops clearly associate with GC skew, we note that this sequence property progressively decreases past the exon1 / intron 1 junction (Hartono et al., 2015), suggesting that other properties in addition to the thermodynamic stability of RNA:DNA hybrids may be facilitating R-loop formation downstream of the TSS. It is possible that as the RNA polymerase II (RNAPII) enters productive elongation, it introduces negative supercoiling on the DNA template, driving the formation of R-loops to relieve the associated topological stress (Stolz et al., 2019). Interestingly, gene body and terminal ex vivo R-loops associate with variable levels of GC skew, suggesting that such an interplay between DNA sequence features and topological considerations may be at play for many loci (Chedin and Benham, 2020; Sanz et al., 2016). In contrast to native R-loops, ex vivo R-loops show little signal over tRNA genes and are not readily detected over intergenic enhancers. Similarly, ex vivo R-loops often define much larger peaks, with median lengths of 1.5 kilobases (Sanz et al., 2016). Single-molecule R-loop footprinting analysis revealed that such large peaks are caused by the clustering of smaller individual R-loops over larger R-loop zones (Malig et al., 2020).
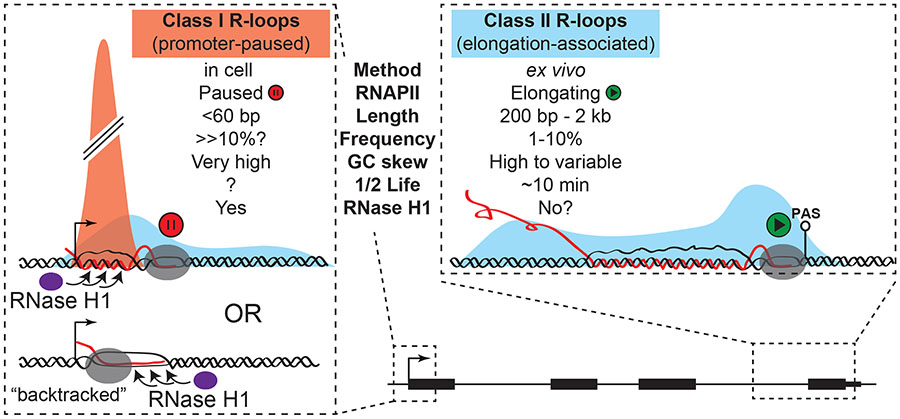
Figure 1. Schematic of proposed classes of R-loops: “promoter-paused” R-loops and “elongation-associated” R-loops.
Shading represents population distribution of R-loops for the two classes. Promoter-paused R-loops are best captured under native conditions and occur in the immediate surrounding of the promoter region and transcription start site. Elongation-associated R-loops form throughout the gene body and transcription termination site.
Reconciling R-loop classes: paused- versus elongation-associated R-loops.
The variation between S9.6- and dRNase H1-based methods could be explained by differences in specificities between RNase H1 and S9.6 and/or by the possibility that RNase H1 was targeted to R-loops found at paused promoters. To clarify the differences between dRNaseH1- and S9.6-mapping methods, a recent study profiled R-loops using the CUT&TAG technology, taking advantage of the N-terminal hybrid binding domain (HBD) of RNase H1 and of the S9.6 antibody as R-loop sensors that were fused to GST- and His6-tagged moieties (Wang et al., 2021b). Each sensor protein was then used for both native and ex vivo R-loop profiling. The HBD sensor protein, when used ex vivo, generated maps similar to those obtained using the S9.6 sensor, recapitulating previous high-resolution strand-specific profiling results using the DRIPc-seq methodology (Sanz and Chedin, 2019; Sanz et al., 2016). This suggests that the RNase H1 HBD and S9.6 can recognize the same subset of R-loops. Strikingly, when used in CUT&TAG approaches for native and fragmentation-free R-loop mapping, both sensor proteins generated results consistent with other dRNase H1-based R-loop profiles. This establishes that the primary difference between S9.6-based and dRNase H1-based R-loop mapping derives from the application of these reagents to mapping R-loops in a native context versus ex vivo. Methods that capture native R-loops like MapR, R-ChIP, and R-loop CUT&TAG better reflect R-loops formed near paused promoter regions, while methods that capture R-loops ex vivo like DRIP-seq and its derivatives identify R-loops that form through gene body regions and therefore associate with transcription elongation. The mechanistic connections between these two R-loop types and transcriptional pausing versus transcription elongation are well-reflected in their response to drugs such as DRB that enforce heightened promoter pausing by blocking the release of RNAPII into elongation. DRB treatment caused increased dRNase H1 recruitment to promoter regions, reflecting increased pausing-associated R-loops. Conversely, washes following DRB treatment, caused the reduction of dRNase H1 binding and promoter-associated R-loops, as expected from the release of previously paused RNAPII complexes into elongation (Chen et al., 2017). In sharp contrast, DRB treatment caused a rapid reduction of R-loops 1-2 kilobases downstream of promoters as profiled by DRIP-qPCR (Sanz et al., 2016), reflecting a rapid decrease in elongation complexes. Prolonged DRB treatment progressively suppressed all instances of R-loop formation along gene bodies (Crossley et al., 2019a; Sanz et al., 2016). As expected, washes following DRB treatment caused a rapid return of R-loops as measured by DRIP-qPCR, consistent with the resumption of elongation (Sanz et al., 2016). Thus, emerging data suggest that there are two distinct classes of R-loops that: (i) associate with two distinct states of the transcription cycle; and (ii) are best profiled through different approaches.
Contrasting properties of R-loop classes.
Based on the R-loop mapping data, the proposed two R-loop classes possess distinct properties that may account for their differential ability to be detected. We note that RNAPI-driven and RNAPIII-driven R-loops, both of which likely correspond to important R-loop classes, are not being discussed here. Similarly, the formation of RNA:DNA hybrids or R-loops at sites of DNA double-stranded breaks (Cohen et al., 2018; D'Alessandro et al., 2018; Ohle et al., 2016) is not considered here.
Length and stability.
Promoter-associated R-loops (referred to here as Class I) are expected to be small, reaching 60 bp at most given the lengths of RNA transcripts at promoter-proximal pause sites (Adelman and Lis, 2012). As suggested (Chedin et al., 2021), the short lengths of such R-loops may result in lower stability during genome fragmentation in DRIP-based approaches. It is also possible that such small R-loops owe their stability in situ to the presence of large protein complexes nearby, including the paused RNAPII machinery and associated pausing and pause-regulating factors. If so, deproteinization during ex vivo DNA extraction may further destabilize them. This, together with minimal size thresholds (>100 bp) enforced during DRIP library construction steps, may account for significant recovery losses over these regions in ex vivo approaches. Non-denaturing bisulfite-based approaches are similarly challenged in identifying Class I R-loops due to their short size and paucity of cytosines on the displaced strand (Chedin et al., 2021; Malig et al., 2020). It is possible that Class I R-loops can only be captured under native conditions. The difficulties associated with recovering and detecting Class I R-loops in ex vivo approaches may in fact have allowed the detection of more stable, but less abundant, elongation-associated (Class II) R-loops (see below). We note that paused RNA polymerases are often backtracked (Noe Gonzalez et al., 2021; Sheridan et al., 2019), and it was recently proposed that small “anterior R-loops” may form ahead of backtracked RNA polymerases (Zatreanu et al., 2019) (Figure 1). The exact molecular features of promoter-associated Class I R-loops therefore remain to be clarified. In contrast to Class I R-loops, single-molecule R-loop footprinting approaches revealed that elongation-associated R-loops show median lengths of about 300 base-pairs and can extend to kilobase-length structures (Malig et al., 2020). Thus, the two R-loop classes show nearly an order of magnitude difference in length. The large sizes of Class II R-loops may account for their relative stability to DNA extraction and fragmentation, allowing ex vivo profiling. As noted previously, however, it is likely that some Class II R-loops are unstable in the face of DNA fragmentation, especially when negative DNA supercoiling played a prominent role in driving their formation (Chedin and Benham, 2020; Stolz et al., 2019).
Frequency of formation.
While Class I and Class II R-loops show clear differences in length and in their association with paused versus elongating RNAPII, much less is known regarding the frequency at which they form. The average yields for elongation-associated R-loops, measured by DRIP-qPCR as a percentage of input, range from 1-10% at positive loci (Sanz et al., 2016). Yields from RChIP-qPCR are notably lower (Chen et al., 2017). We suspect, however, that this may not reflect the true frequency distribution of Class I and Class II R-loops. ChIP experiments, which involve crosslinking and harsh sonication prior to immunoprecipitation may be limited in their ability to efficiently recover Class I R-loops. This may be further compounded if only a portion of Class I R-loops are RNase H1-bound at any given time. Issues of epitope accessibility may further complicate recovery given the presence of large macromolecular complexes over paused promoter regions (Core and Adelman, 2019). While future experiments will be necessary to accurately quantify the relative amounts of Class I and Class II R-loops, we suggest that Class I R-loops formed over paused promoters are much more abundant than Class II R-loops are at any given position. If correct, this proposal suggests that “native” approaches are limited in their ability to recover Class II R-loops simply because the bulk of R-loops in a cell correspond to Class I R-loops formed at promoters. This proposal follows the well-accepted notion that the highest RNAPII density measured by ChIP-seq approaches, and the highest transcriptional activity measured by profiling nascent transcription, are primarily found over paused promoters compared to transcribed gene bodies (Henriques et al., 2013; Rahl et al., 2010; Wissink et al., 2019). Thus, the proposed high frequency of Class I R-loop formation may simply reflect the prevalence of promoter-proximal paused RNAPII complexes. Importantly, pause sites, particularly over CpG island promoters associate with very high, R-loop-favorable, GC skew levels (Chen et al., 2017; Hartono et al., 2015). In addition, the presence of a nearby free 5’-end may further facilitate R-loop initiation during promoter pausing (Chen et al., 2017; Roy et al., 2010). Finally, the fact that RNAPII machinery itself is paused may provide a long kinetic window for an R-loop to arise. Overall, we propose that Class I R-loops dominate the R-loop landscape by virtue of their association with abundant paused RNAPII complex, the presence of favorable sequence characteristics, and the availability of a free 5’-end.
Half-lives.
Class II R-loops have an estimated half-life of about 10 minutes (Crossley et al., 2019b; Sanz et al., 2016); by contrast, the half-life of Class I R-loops is not known. It is reasonable to propose, however, that Class I R-loops may show a half-life similar to that of paused RNAPII complexes. Measurements of RNAPII pausing indicate that pause duration has a median value of 7 minutes (Jonkers et al., 2014), but that there exists considerable variation from 2 to 30 minutes depending on the promoter being considered (Core and Adelman, 2019). Enhancers may display an even shorter pause duration (Henriques et al., 2018). Thus, in all cases R-loop formation is a dynamic process but the half-lives, and potentially the types of enzymatic activities associated with R-loop resolution may vary between Class I and Class II R-loops.
Functional consequences of Class I and Class II R-loops.
Class II R-loops have been associated with several important functions under normal conditions in mammalian cells (Chedin, 2016). Whether they occur in promoter-distal regions, gene bodies, or terminal genic regions, Class II R-loops correspond to regions of increased RNAPII density (Sanz et al., 2016). This suggests that they help to slow or stall the transcription machinery, as observed in vitro (Belotserkovskii et al., 2017; Belotserkovskii et al., 2018). Towards the beginning of genes, where Class II R-loops are prominent, slower elongation is expected to favor the recruitment of chromatin modifying enzymes to the C-terminal domain of RNAPII. This, in turn, may account for the increased deposition of several transcription-coupled histone modifications such as histone H3 lysine 36 trimethylation observed for R-loop-positive genes compared to expression-matched, but R-loop-negative, genes (Sanz et al., 2016). At the end of genes, which also correspond to Class II R-loop hotspots, R-loop-positive regions show dramatically elevated RNAPII stalling compared to expression-matched R-loop-negative terminal regions. Stalling, in turn, associates with efficient transcription termination, which is a property preferentially observed for genes with close neighbors (Sanz et al., 2016). Mechanistically, slower transcription elongation downstream of the polyadenylation site may tip the kinetic competition between the XRN2 ribonuclease and RNAPII in favor of XRN2-mediated transcript degradation (Saldi et al., 2018). Class II R-loop formation also generally correlates with regions of increased chromatin accessibility, consistent with the notion that rigid A-like form RNA:DNA hybrids do not wrap around nucleosomes (Dunn and Griffith, 1980). Finally, R-loops were proposed to absorb large amounts of negative superhelicity, contributing to the transient relaxation of topological stresses in the genome (Chedin and Benham, 2020; Stolz et al., 2019). Thus, under normal conditions, Class II R-loops have been assigned roles in chromatin patterning, transcription regulation, and topological management. Class I R-loops have been associated with chromatin features typical of highly active promoters over the promoter-proximal pause sites, such as high GC skew and G quadruplex motifs, high levels of H3K4 trimethylation and histone acetylation, and high RNA polymerase II occupancy (Chen et al., 2017). Interestingly, similar enrichments were observed around the TSSs of Class II R-loop-forming genes (Sanz et al., 2016), consistent with the notion that genes undergoing R-loop formation during transcription elongation also correspond to genes that accumulate Class I R-loops over their promoter-proximal pause regions. Indeed, Class II R-loop formation downstream of promoters was shown to be significantly associated with RNA polymerase II pausing (Zhang et al., 2017).
Harmful R-loops and genome instability.
Harmful R-loops, whether they correspond to Class I or Class II R-loops, should in principle be revealed using appropriate genomic mapping techniques. It is therefore worth reviewing studies that credibly examined global R-loop patterns in cellular models of genome instability for any evidence that might clarify the nature of such structures and their spatiotemporal relationship to phenomena associated with DNA damage.
Class II R-loops and genome instability, an elusive connection.
DRIP-type approaches have been used in a variety of cellular models. DNA topoisomerase I (Top I) is widely thought to suppress R-loop formation by relaxing the R-loop-favorable negative superhelicity that propagates upstream of the active transcription machinery (Kouzine et al., 2004; Pommier et al., 2016). Long-term Top I depletion in human HeLa cells leads to elevated DNA damage and globally slower replication fork progression due to transcription-replication conflicts (Tuduri et al., 2009). Following up on this work, Promonet et al., (2020) mapped R-loop distributions, DNA breaks, and the location of DNA damage markers including phosphorylated RPA, a replicative stress marker, and γH2AX. In control cells, R-loops were observed broadly over promoter distal regions, gene bodies, and terminal regions, consistent with the distribution of elongation-associated R-loops. By contrast, phosphorylated RPA accumulation was only observed over the terminal regions of expressed genes that are replicated in a head-on (HO) orientation relative to transcription. This indicates that stalled forks marked by phosphorylated RPA occurred as a result of HO replication-transcription interactions, which coincide with naturally R-loop-rich terminal genic regions. The vast majority of Class II R-loops therefore do not interfere with DNA replication under normal conditions (Promonet et al., 2020). In Top I-depleted cells, R-loops showed a slight increase over terminal regions which was accompanied by increased γH2AX and DSBs formation. Importantly, replication fork speeds were uniformly reduced by 30-40% in Top I-depleted cells, even though R-loop increases were minor and localized. Thus, it is unlikely that “excessive” R-loop formation can account for the global replication slowdown. Instead, it was proposed that the stalled forks that naturally occur at a subset of HO genes are further challenged in the absence of Top I, leading to fork collapse, DSBs, and the activation of the ATR kinase to slow S phase progression globally (Promonet et al., 2020). Overall, this study suggests that Class II R-loops may not be directly involved in events of genome instability even upon Top I depletion.
Interestingly, RNase H1 over-expression was able to suppress the slow replication fork phenotype observed upon Top I depletion (Promonet et al., 2020; Tuduri et al., 2009). To account for this, Promonet et al., (2020) proposed that RNase H1 may degrade RNA:DNA hybrid structures that form over stalled forks and prevent fork rescue or remodeling. Through this activity, RNase H1 was suggested to reduce ATR activation and thereby counteract a global replication slowdown over undamaged forks, consistent with prior observations (Mutreja et al., 2018; Seiler et al., 2007). This is an exciting possibility that needs to be further tested. It also suggests that the sensitivity of a phenotype to RNase H1 over-expression may not necessarily implicate co-transcriptional R-loops and could instead reflect a novel effect of RNase H1 on replication fork rescue.
Excessive R-loop formation was hypothesized early on to be responsible for the increased γH2AX deposition observed upon splicing factor depletion or mutation (Li and Manley, 2006; Li et al., 2005; Paulsen et al., 2009). We recently used DRIP-seq to profile Class II R-loops upon inhibition of the U2 spliceosome SF3B1 subunit using the splicing inhibitor Pladienolide B (PladB) (Kotake et al., 2007; Yokoi et al., 2011). This treatment causes widespread intron retention as well as γH2AX accumulation and sensitivity to ATR inhibitors (Nguyen et al., 2018). Surprisingly, PladB treatment resulted in a dramatic genome-wide R-loop loss (Castillo-Guzman et al., 2020). This loss was caused by a profound negative feedback on transcription elongation caused by increased promoter-proximal pausing and increased premature transcription termination (Caizzi et al., 2021; Castillo-Guzman et al., 2020; Sousa-Luis et al., 2021). For a small subset of ~400 genes, PladB triggered a transcription termination defect, leading to readthrough transcription far downstream of genes. Such readthrough transcription events were associated with new instances of R-loop formation, defining a class of de novo excessive R-loops (Castillo-Guzman et al., 2020). Importantly, the γH2AX accumulation caused by PladB was significantly delayed compared to de novo R-loop accumulation and was not spatially enriched over regions of increased R-loops (Castillo-Guzman et al., 2020). It therefore appears that excessive Class II R-loops generated during splicing inhibition did not associate with DNA damage events. More broadly, DNA damage events induced by U2 spliceosome inhibition occurred against the backdrop of a dramatic loss of Class II R-loop.
A number of studies nonetheless provide qualified support for the notion that increased formation of Class II R-loops triggers or enhances genome instability phenomena. Stork et al., (2016) showed that addition of the hormone estrogen (E2) to breast cancer cells triggers rapid expression of E2-responsive genes and an increase in elongation-associated R-loops over these targets (Stork et al., 2016). E2 treatment also causes rapid cellular proliferation and increased deposition of the γH2AX DNA damage marker during S phase, indicating that R-loops may be driving this response. Indeed, rearrangements observed in breast tumors were enriched over E2-responsive genes. However, proximity ligation assays using S9.6 and γH2AX antibodies suggested that the majority of DNA damage events induced by E2 were located at a distance from R-loops (Stork et al., 2016). An alternative source of endogenous damage could come from the introduction of DSBs by Topoisomerase 2 beta at promoters, where it functions to facilitate transcription initiation in response to estrogens (Ju et al., 2006; Morimoto et al., 2019). Gorthi et al., (2018) showed that Ewing sarcoma cells driven by the EWS-FLI1 oncoprotein display globally elevated transcription levels and R-loop loads. These tumors were additionally characterized by increased replicative stress and sensitivity to ATR inhibitors and genotoxic agents (Gorthi et al., 2018). However, the direct involvement of R-loops in these responses remains unclear. Evidence suggests that the hyper-transcription caused by EWS-FLI1 results in the sequestration of BRCA1 with elongating RNA polymerase complexes, phenocopying a BRCA1 deficiency, and leading to a DNA damage response (DDR) deficiency (Gorthi et al., 2018). Two recent studies analyzed the impact of knocking out or depleting subunits of mammalian SWI/SNF chromatin remodeling complexes. Knockout of PBRM1, encoding the BAF180 subunit of the polybromo-associated BAF complex (PBAF), led to increased γH2AX foci formation, replication stress, and DNA breaks (Chabanon et al., 2021). This was correlated with an increased genic R-loop burden measured by DRIP-seq and the involvement of R-loops was further suggested by the ability of RNase H1 to rescue many of the above phenotypes. However, the spatial overlap of such excessive R-loops with DNA damage events was not assessed. In addition, authors noted that PRBM1 deficiency led to significant reductions in the protein levels of multiple genome stability factors, such as members of the Fanconi Anemia complex and of the BLM helicase (Chabanon et al., 2021). Thus, the genome instability observed in the absence of PBRM1 may reflect an intrinsically reduced DDR capacity in addition to elevated R-loop levels. Depletion of the BRG1 subunit common to all SWI/SNF complexes led to an array of RNase H1-sensitive genome instability phenotypes, including global decrease in replication fork velocity (Bayona-Feliu et al., 2021). DRIP-based profiling of Class II R-loops confirmed an increased burden of genic R-loops over 3,200 loci enriched for BRG1 binding sites. Analysis of transcription-replication conflicts showed that HO conflicts were marked by elevated DNA damage markers γH2AX and FANCD2, when co-directional (CD) conflicts were not, even though the R-loop levels observed at HO and CD regions were not significantly different (Bayona-Feliu et al., 2021). Thus, consistent with earlier statements, many R-loops outside of HO conflicts do not associate with DNA damage markers. HO interactions between transcription and replication, however, are particularly responsible for stalled forks and DNA damage. Whether the R-loops observed over these regions are causally involved in further enhancing the fragility of HO replication-transcription interactions currently rests on the interpretation of the sensitivity of instability phenotypes to RNase H1 over-expression. As discussed above, RNase H1 binding sites mapped by R-ChIP mostly map to promoter proximal regions while Class II R-loops mapped by DRIP approaches primarily map to transcribed gene bodies. The deployment of R-ChIP approaches in models of genome instability such as those described above may clarify whether RNase H1 can gain access to new R-loop subsets under these conditions or if it can be recruited directly to stalled forks, as suggested (Promonet et al., 2020).
Overall, the association of Class II R-loops and genome instability phenomena remains elusive. Part of the issue is that only few studies have both mapped R-loops and analyzed the presence of DNA breaks at sufficient resolution to reach definitive conclusions regarding the roles of R-loops as causes of genome instability. In addition, the observations that instability phenomena have been linked to global R-loop increases and losses in various models suggests that instability may arise through a variety of mechanisms. Indirect effects linked to reduced DDR responses under pathological conditions further complicate matters. To date, no specific R-loops subset has been associated directly with events of DNA breakage at high resolution. Furthermore, the distinguishing molecular features of harmful R-loops remain to be defined, an important task given the consensus finding that most Class II R-loops do not cause instability even under altered conditions.
Class I R-loops as harmful R-loops candidates.
Overexpression of nuclear RNase H1 suppresses a diversity of genome instability phenotypes. Common sense dictates that for RNase H1 to mediate these effects, it needs to gain access to the loci causing these altered phenotypes in the first place. Based on the few available datasets where RNase H1 genomic binding sites were mapped in human cell lines, these loci predominantly correspond to Class I R-loops that form at the promoter-proximal regions of paused promoters (Chen et al., 2018; Chen et al., 2017). By contrast, most Class II R-loops are not bound by RNase H1 in vivo. It is therefore difficult to conceive how RNase H1 might relieve instability phenotypes if these were driven by elongation-associated R-loops.
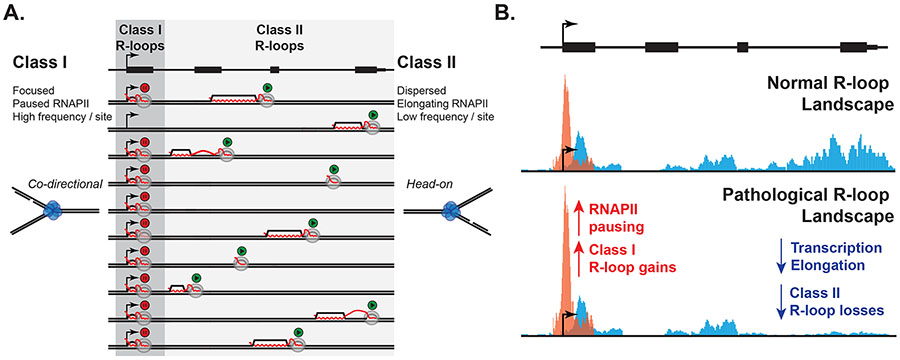
One interpretation of the data is therefore that harmful R-loops predominantly correspond to Class I R-loops. This model is attractive for several reasons. It has been well documented that the deleterious effects of harmful R-loops are linked to conflicts with the replication machinery (Garcia-Muse and Aguilera, 2016; Gomez-Gonzalez and Aguilera, 2019; Hamperl and Cimprich, 2016; Zeman and Cimprich, 2014). Class I R-loops possess multiple properties that might make them much more formidable replication obstacles than Class II R-loops. First, they are proposed to arise at high frequencies compared to Class II R-loops, consistent with the much greater RNAPII density at paused promoters compared to gene bodies. Second, Class I R-loops consistently occur in a narrow near-TSS genomic window, while Class II R-loops can occur almost anywhere along R-loop hotspots distributed throughout gene bodies (Figure 2A). Thus, encounters between replication forks and R-loops will be more likely to involve Class I R-loops, and these encounters are expected to be focused over paused promoters. Third, one major difference between Class I and Class II R-loops is that the former is associated with a paused, and possibly backtracked, RNAPII transcription machinery (Sheridan et al., 2019). By extension, Class I R-loops will also be proximal to the large general transcription factor (GTF) complexes that recruit RNAPII to promoter sequences (Verger et al., 2021). By contrast, Class II R-loops occur during elongation behind an actively translocating RNAPII, far away from GTF complexes. One can even envision that RNAPII has moved away from an R-loop once R-loop extension is terminated (Figure 2A).
Figure 2. Pause-associated R-loops as candidate harmful R-loops.
A. Schematic of a gene and its hypothetical R-loop formation state in 10 independent chromosomes. Class I R-loops are suggested to frequently arise at promoters as a result of promoter-proximal pausing. By contrast, elongation-associated Class II R-loops are spread throughout the gene body and therefore occur at much lower frequencies at any given site. RNA is only depicted when in a R-loop bound state or exiting RNAP (full transcript not shown). B. Under pathological conditions caused for instance by defective mRNA processing, RNAPII pausing is proposed to increase, causing a focal increase in Class I R-loops. Class II R-loops are by contrast proposed to be reduced as a result of lower transcription initiation due to increased pausing. Under such conditions, paused RNAPII complexes anchored to Class I R-loops may represent significant obstacles to replication fork progression, causing DNA damage.
Recent in vitro work suggests that R-loops by themselves do not represent a strong impediment for the E. coli replication machinery, while the presence of transcription complexes led to potent blockages, particularly in the head-on orientation (Bruning and Marians, 2020). In addition, R-loop-anchored transcription complexes arrested at UV lesions were proposed to represent the main cause of head-on replication blocks in RNase H-deficient E. coli mutants (Kouzminova and Kuzminov, 2021). Class I R-loops, associated with paused RNAPII complexes, therefore represent attractive “harmful” R-loop candidates. We note that human genes are generally thought to be replicated co-directionally from origins located upstream of promoter regions (Petryk et al., 2016). Thus, one would expect encounters between Class I R-loops and replication forks to be mostly co-directional. These interactions, while potentially harmful (Hamperl et al., 2017), are thought to play a lesser role compared to head-on conflicts in driving genome instability (Gomez-Gonzalez and Aguilera, 2019; Hamperl and Cimprich, 2016). It remains possible, however, that Class I R-loops might also arise from antisense transcripts that frequently originate from promoters, setting up head-on clashes with incoming forks. In addition, recent evidence suggests that antisense-associated promoter regions display delayed replication characteristics that may factor in possible fragility events (Wang et al., 2021a). Studies aimed at dissecting transcription-replication encounters, and their intersection with Class I R-loop formation, will be important to further delineate the role of R-loops in genome fragility.
A growing number of studies provide support to the notion that Class I R-loops increase in frequency under pathological instances. Using R-ChIP, Chen and colleagues profiled Class I R-loops in HEK293T cells harboring mutations in the splicing factors SRSF2, U2AF1, and U2AF2 (Chen et al., 2018). Mutations in SRSF2 and U2AF1 caused cellular growth defects, γH2AX induction, and replicative stress as evidenced by slower replication forks and ATR activation. Such defects could be at least partially alleviated by RNase H1over-expression. R-ChIP revealed increased Class I R-loops over promoter regions. SRSF2 and U2AF1 mutations also caused increased RNAPII promoter-pausing (Chen et al., 2018), further linking promoter-proximal pausing and Class I R-loop formation. Pharmacological splicing inhibition also triggers increased promoter pausing (Caizzi et al., 2021; Castillo-Guzman et al., 2020; Sousa-Luis et al., 2021) suggesting that Class I R-loops may generally increase under these conditions. Release from the promoter-proximal pause is regulated by the pTEFb complex which is recruited in part via the general BRD4 co-activator protein (Kwak and Lis, 2013). Loss of BRD4 function triggers global transcriptional pausing (Muhar et al., 2018). Pharmacological inhibition of BRD4 and BRD4 depletion cause S phase-dependent γH2AX deposition, DSB formation, and slow replication fork progression in a manner that was compensated by RNase H1 expression (Edwards et al., 2020; Lam et al., 2020). Performing R-ChIP, Edwards et al., (2020) showed that BRD4 loss of function caused global increase in Class I R-loops over promoters, as expected, and also in gene bodies, which is counter-intuitive given the reduction of the elongating form of RNAPII (Edwards et al., 2020). While none of studies cited above mapped DSBs and therefore did not directly address the connection between R-loops and DNA breakage, this recent work supports the notion that Class I R-loops may represent harmful obstacles to replication progression and a source of DNA damage.
More broadly, we propose that defects in mRNA processing, particularly splicing dysfunction, may feedback on transcription by increasing promoter pausing and therefore, increasing Class I R-loops (Figure 2B). The increased focal burden of paused transcriptional complexes anchored to Class I R-loops is expected to enhance replication-transcription conflicts over promoter regions and may lead to replication stress and R-loop-induced DNA damage. Given that increased promoter pausing associates with reduced transcription initiation (Gressel et al., 2019; Gressel et al., 2017; Shao and Zeitlinger, 2017), the model further predicts that Class II R-loops may undergo progressive losses as a result of reduced elongation (Figure 2B), as observed upon U2 spliceosome inhibition (Castillo-Guzman et al., 2020).
Concluding remarks and future directions.
The notion that RNAPII-driven R-loops can be broken into pausing-associated and elongation-associated structures hopefully serves to reconcile and rationalize the seemingly discordant results obtained through ex vivo and native R-loop mapping approaches. With the right methodologies now at hand, new investigations will test the proposal that Class I R-loops significantly contribute to genome instability phenotypes associated with RNA processing defects. We suggest that future work should include integrative strategies that reveal the distributions of both Class I and Class II R-loops under relevant cellular models of R-loop dysfunction in combination with DNA double-strand break profiling and nascent transcription analysis. Such integrative studies are the most likely to reveal the mechanisms that lead to genome destabilization when gene expression programs are deregulated. Given the importance of RNase H1 over-expression as a tool and its broad ability to suppress genomic stresses, it will also be essential to ascertain how suppression is mechanistically achieved. If Class I R-loops truly emerge as source of DNA damage under pathological conditions, it will become important to understand how RNase H1 activity at paused promoters can lower transcription-replication conflicts. One possibility is that RNase H1, when over-expressed, facilitates the release of paused RNAPII into elongation (Sridhara et al., 2017). Alternatively, RNase H1 may mediate its effect by facilitating the processing and restart of stalled replication forks (Promonet et al., 2020) in the vicinity of paused promoters. Interestingly, recent evidence suggests that the persistence of RNAPII at TSSs characterized by strong antisense transcription prevents the timely replication of these loci until G2/M (Wang et al., 2021a). In agreement, mapping of transcription-replication interactions in murine B cells revealed that such interactions primarily occurred over regions characterized by bidirectional promoters and focal accumulation of markers of replicative stress and DNA breakage (St Germain et al., 2021). Future work will be required to tease out these interesting possibilities.
Acknowledgments
Work in the Chedin lab is supported by the National Institutes of Health (R35 GM139549). D.C.G. was supported in part by the NIH T32 predoctoctoral training program in Molecular and Cellular Biology (GM007377) and by an NIH F31 individual fellowship (GM136143). We thank members of the Chedin lab for useful discussion and suggestions.
REFERENCES
- Adelman K, and Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics 13, 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, and Garcia-Muse T (2012). R loops: from transcription byproducts to threats to genome stability. Molecular cell 46, 115–124. [DOI] [PubMed] [Google Scholar]
- Alecki C, Chiwara V, Sanz LA, Grau D, Arias Perez O, Boulier EL, Armache KJ, Chedin F, and Francis NJ (2020). RNA-DNA strand exchange by the Drosophila Polycomb complex PRC2. Nature communications 11, 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso S, Herrera-Moyano E, Munoz S, Garcia-Rubio M, Gomez-Gonzalez B, and Aguilera A (2019). The DNA damage response acts as a safeguard against harmful DNA-RNA hybrids of different origins. EMBO reports 20, e47250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Feliu A, Barroso S, Munoz S, and Aguilera A (2021). The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nature genetics. [DOI] [PubMed] [Google Scholar]
- Belanger KG, and Kreuzer KN (1998). Bacteriophage T4 initiates bidirectional DNA replication through a two-step process. Molecular cell 2, 693–701. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii BP, Soo Shin JH, and Hanawalt PC (2017). Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic acids research 45, 6589–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskii BP, Tornaletti S, D'Souza AD, and Hanawalt PC (2018). R-loop generation during transcription: Formation, processing and cellular outcomes. DNA repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, and Carrico RJ (1986). Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods 89, 123–130. [DOI] [PubMed] [Google Scholar]
- Bruning JG, and Marians KJ (2020). Replisome bypass of transcription complexes and R-loops. Nucleic acids research 48, 10353–10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caizzi L, Monteiro-Martins S, Schwalb B, Lysakovskaia K, Schmitzova J, Sawicka A, Chen Y, Lidschreiber M, and Cramer P (2021). Efficient RNA polymerase II pause release requires U2 snRNP function. Molecular cell 81, 1920–1934 e1929. [DOI] [PubMed] [Google Scholar]
- Carles-Kinch K, and Kreuzer KN (1997). RNA-DNA hybrid formation at a bacteriophage T4 replication origin. J Mol Biol 266, 915–926. [DOI] [PubMed] [Google Scholar]
- Castillo-Guzman D, Hartono SR, Sanz LA, and Chédin F (2020). SF3B1-targeted Splicing Inhibition Triggers Global Alterations in Transcriptional Dynamics and R-Loop Metabolism. bioRxiv, 2020.2006.2008.130583. [Google Scholar]
- Cerritelli SM, and Crouch RJ (2009). Ribonuclease H: the enzymes in eukaryotes. Febs J 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabanon RM, Morel D, Eychenne T, Colmet-Daage L, Bajrami I, Dorvault N, Garrido M, Meisenberg C, Lamb A, Ngo C, et al. (2021). PBRM1 Deficiency Confers Synthetic Lethality to DNA Repair Inhibitors in Cancer. Cancer research. [DOI] [PubMed] [Google Scholar]
- Chan YA, Hieter P, and Stirling PC (2014). Mechanisms of genome instability induced by RNA-processing defects. Trends in genetics : TIG 30, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F (2016). Nascent Connections: R-Loops and Chromatin Patterning. Trends in genetics : TIG 32, 828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, and Benham CJ (2020). Emerging roles for R-loop structures in the management of topological stress. J Biol Chem 295, 4684–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, Hartono SR, Sanz LA, and Vanoosthuyse V (2021). Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J 40, e106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen JY, Huang YJ, Gu Y, Qiu J, Qian H, Shao C, Zhang X, Hu J, Li H, et al. (2018). The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Molecular cell 69, 412–425 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen JY, Zhang X, Gu Y, Xiao R, Shao C, Tang P, Qian H, Luo D, Li H, et al. (2017). R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Molecular cell 68, 745–757 e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Puget N, Lin YL, Clouaire T, Aguirrebengoa M, Rocher V, Pasero P, Canitrot Y, and Legube G (2018). Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nature communications 9, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L, and Adelman K (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33, 960–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley MP, Bocek M, and Cimprich KA (2019a). R-Loops as Cellular Regulators and Genomic Threats. Molecular cell 73, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley MP, Bocek M, Hamperl S, Swigut T, and Cimprich KA (2019b). qDRIP: Quantitative differential RNA:DNA hybrid immunoprecipitation sequencing. bioRxiv. [Google Scholar]
- D'Alessandro G, Whelan DR, Howard SM, Vitelli V, Renaudin X, Adamowicz M, Iannelli F, Jones-Weinert CW, Lee M, Matti V, et al. (2018). BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nature communications 9, 5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet M, and Brochu J (2019). R-loop-dependent replication and genomic instability in bacteria. DNA repair 84, 102693. [DOI] [PubMed] [Google Scholar]
- Dunn K, and Griffith JD (1980). The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic acids research 8, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DS, Maganti R, Tanksley JP, Luo J, Park JJH, Balkanska-Sinclair E, Ling J, and Floyd SR (2020). BRD4 Prevents R-Loop Formation and Transcription-Replication Conflicts by Ensuring Efficient Transcription Elongation. Cell Rep 32, 108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Webb S, Kerr A, and Tollervey D (2014). Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS genetics 10, e1004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T, and Aguilera A (2016). Transcription-replication conflicts: how they occur and how they are resolved. Nature reviews Molecular cell biology 17, 553–563. [DOI] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, and Chedin F (2012). R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Molecular cell 45, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, and Aguilera A (2019). Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev 33, 1008–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, Iniguez AB, Bernard X, Masamsetti VP, Roston S, et al. (2018). EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 555, 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel S, Schwalb B, and Cramer P (2019). The pause-initiation limit restricts transcription activation in human cells. Nature communications 10, 3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel S, Schwalb B, Decker TM, Qin W, Leonhardt H, Eick D, and Cramer P (2017). CDK9-dependent RNA polymerase II pausing controls transcription initiation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Bocek MJ, Saldivar JC, Swigut T, and Cimprich KA (2017). Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170, 774–786 e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, and Cimprich KA (2014). The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA repair 19, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, and Cimprich KA (2016). Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell 167, 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartono SR, Korf IF, and Chedin F (2015). GC skew is a conserved property of unmethylated CpG island promoters across vertebrates. Nucleic acids research 43, 9729–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartono SR, Malapert A, Legros P, Bernard P, Chedin F, and Vanoosthuyse V (2018). The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J Mol Biol 430, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, and Adelman K (2013). Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Molecular cell 52, 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, and Adelman K (2018). Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 32, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Kalb J, Buchel G, Ade CP, Baluapuri A, Xu J, Koster J, Solvie D, Carstensen A, Klotz C, et al. (2019). Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 567, 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X, Cadwell GW, and Kogoma T (1995). Escherichia coli RecG and RecA proteins in R-loop formation. Embo J 14, 2385–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, and Aguilera A (2003). Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular cell 12, 711–721. [DOI] [PubMed] [Google Scholar]
- Itoh T, and Tomizawa J (1980). Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77, 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, and Lis JT (2014). Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 3, e02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, and Rosenfeld MG (2006). A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Chu C, Shatkin AJ, and Manley JL (2007). Human capping enzyme promotes formation of transcriptional R loops in vitro. Proc Natl Acad Sci U S A 104, 17620–17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T (1997). Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, Skarstad K, Boye E, von Meyenburg K, and Steen HB (1985). RecA protein acts at the initiation of stable DNA replication in rnh mutants of Escherichia coli K-12. J Bacteriol 163, 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T, and von Meyenburg K (1983). The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J 2, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, and Mizui Y (2007). Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3, 570–575. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Liu J, Sanford S, Chung HJ, and Levens D (2004). The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol 11, 1092–1100. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA, and Kuzminov A (2021). Ultraviolet-induced RNA:DNA hybrids interfere with chromosomal DNA synthesis. Nucleic acids research 49, 3888–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN, and Brister JR (2010). Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virology journal 7, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, and Lis JT (2013). Control of transcriptional elongation. Annual review of genetics 47, 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FC, Kong YW, Huang Q, Vu Han TL, Maffa AD, Kasper EM, and Yaffe MB (2020). BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nature communications 11, 4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk HB, Sandquist LE, Sridhara SC, Rodland GE, Sabino JC, de Almeida SF, Grallert B, Trinkle-Mulcahy L, and Syljuasen RG (2019). Regulation of ATR activity via the RNA polymerase II associated factors CDC73 and PNUTS-PP1. Nucleic acids research 47, 1797–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, Woodward JJ, Dreifus JE, and Merrikh H (2017). Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 170, 787–799 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, and Clayton DA (1996). Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J Biol Chem 271, 24262–24269. [DOI] [PubMed] [Google Scholar]
- Lee DY, and Clayton DA (1998). Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem 273, 30614–30621. [DOI] [PubMed] [Google Scholar]
- Legros P, Malapert A, Niinuma S, Bernard P, and Vanoosthuyse V (2014). RNA processing factors Swd2.2 and Sen1 antagonize RNA Pol III-dependent transcription and the localization of condensin at Pol III genes. PLoS genetics 10, e1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, and Manley JL (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122, 365–378. [DOI] [PubMed] [Google Scholar]
- Li X, and Manley JL (2006). Cotranscriptional processes and their influence on genome stability. Genes Dev 20, 1838–1847. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, and Manley JL (2005). Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev 19, 2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduike NZ, Tehranchi AK, Wang JD, and Kreuzer KN (2014). Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol 91, 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malig M, Hartono S, Giafaglione J, Sanz LA, and Chedin F (2020). Ultra-Deep Coverage Single-Molecule R-loop Footprinting Reveals Principles of R-loop Formation J Mol Biol in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata H, and Tomizawa J (1990). A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62, 331–338. [DOI] [PubMed] [Google Scholar]
- Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, and Boothman DA (2016). XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS genetics 12, e1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto S, Tsuda M, Bunch H, Sasanuma H, Austin C, and Takeda S (2019). Type II DNA Topoisomerases Cause Spontaneous Double-Strand Breaks in Genomic DNA. Genes (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, Rescheneder P, Lipp JJ, Herzog VA, Reichholf B et al. (2018). SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 360, 800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja K, Krietsch J, Hess J, Ursich S, Berti M, Roessler FK, Zellweger R, Patra M, Gasser G, and Lopes M (2018). ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Rep 24, 2629–2642 e2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HD, Leong WY, Li W, Reddy PNG, Sullivan JD, Walter MJ, Zou L, and Graubert TA (2018). Spliceosome Mutations Induce R Loop-Associated Sensitivity to ATR Inhibition in Myelodysplastic Syndromes. Cancer research 78, 5363–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe Gonzalez M, Blears D, and Svejstrup JQ (2021). Causes and consequences of RNA polymerase II stalling during transcript elongation. Nature reviews Molecular cell biology 22, 3–21. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Pickett GG, Kogoma T, and Kornberg A (1984). RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A 81, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, and Fischer T (2016). Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 167, 1001–1013 e1007. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. (2009). A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Molecular cell 35, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d'Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, and Hyrien O (2016). Replication landscape of the human genome. Nature communications 7, 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nature reviews Molecular cell biology 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promonet A, Padioleau I, Liu Y, Sanz L, Biernacka A, Schmitz AL, Skrzypczak M, Sarrazin A, Mettling C, Rowicka M et al. (2020). Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nature communications 11, 3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, and Young RA (2010). c-Myc regulates transcriptional pause release. Cell 141, 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Zhang Z, Lu Z, Hsieh CL, and Lieber MR (2010). Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Molecular and cellular biology 30, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T, Fong N, and Bentley DL (2018). Transcription elongation rate affects nascent histone pre-mRNA folding and 3' end processing. Genes Dev 32, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, and Chedin F (2019). High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat Protoc 14, 1734–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz LA, Hartono SR, Lim YW, Steyaert S, Rajpurkar A, Ginno PA, Xu X, and Chedin F (2016). Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Molecular cell 63, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler JA, Conti C, Syed A, Aladjem MI, and Pommier Y (2007). The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Molecular and cellular biology 27, 5806–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, and Zeitlinger J (2017). Paused RNA polymerase II inhibits new transcriptional initiation. Nature genetics 49, 1045–1051. [DOI] [PubMed] [Google Scholar]
- Sheridan RM, Fong N, D'Alessandro A, and Bentley DL (2019). Widespread Backtracking by RNA Pol II Is a Major Effector of Gene Activation, 5' Pause Release, Termination, and Transcription Elongation Rate. Molecular cell 73, 107–118 e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka JA, Sanz LA, Hartono SR, and Chedin F (2021). Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. The Journal of cell biology 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, and Cimprich KA (2015). Breaking bad: R-loops and genome integrity. Trends in cell biology 25, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Stork CT, Garcia-Rubio ML, Paulsen RD, Aguilera A, and Cimprich KA (2014). Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Molecular cell 56, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Luis R, Dujardin G, Zukher I, Kimura H, Weldon C, Carmo-Fonseca M, Proudfoot NJ, and Nojima T (2021). POINT technology illuminates the processing of polymerase-associated intact nascent transcripts. Molecular cell 81, 1935–1950 e1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara SC, Carvalho S, Grosso AR, Gallego-Paez LM, Carmo-Fonseca M, and de Almeida SF (2017). Transcription Dynamics Prevent RNA-Mediated Genomic Instability through SRPK2-Dependent DDX23 Phosphorylation. Cell Rep 18, 334–343. [DOI] [PubMed] [Google Scholar]
- St Germain CP, Zhao H, Sinha V, Sanz L, Chedin F, and Barlow JH (2021). Genomic patterns of transcription-replication interactions in mouse primary B cells. biorXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, and Hieter P (2012). R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz R, Sulthana S, Hartono SR, Malig M, Benham CJ, and Chedin F (2019). Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc Natl Acad Sci U S A 116, 6260–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork CT, Bocek M, Crossley MP, Sollier J, Sanz LA, Chedin F, Swigut T, and Cimprich KA (2016). Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey R, Garcia-Rodriguez N, Aguilera A, and Wellinger RE (2015). Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proc Natl Acad Sci U S A 112, 5779–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11, 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A, Monte D, and Villeret V (2021). Take Your PIC. Trends Biochem Sci. [DOI] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, and Koshland D (2016). S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30, 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rojas P, Mao J, Muste Sadurni M, Garnier O, Xiao S, Higgs MR, Garcia P, and Saponaro M (2021a). Persistence of RNA transcription during DNA replication delays duplication of transcription start sites until G2/M. Cell Rep 34, 108759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang H, Li C, Yin Z, Xiao R, Li Q, Xiang Y, Wang W, Huang J, Chen L, et al. (2021b). Genomic profiling of native R loops with a DNA-RNA hybrid recognition sensor. Sci Adv 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, and Hastings PJ (2013). R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nature communications 4, 2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink EM, Vihervaara A, Tippens ND, and Lis JT (2019). Nascent RNA analyses: tracking transcription and its regulation. Nature reviews Genetics 20, 705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfridge P, and Sarma K (2021). A nuclease- and bisulfite-based strategy captures strand-specific R-loops genome-wide. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, and Clayton DA (1995). A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Molecular and cellular biology 15, 580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Li K, Li S, Hou Q, Zhang Y, Liu K, and Sun Q (2020). The R-Loop Atlas of Arabidopsis Development and Responses to Environmental Stimuli. Plant Cell 32, 888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Xu H, Li K, Fan Y, Liu Y, Yang X, and Sun Q (2017). The R-loop is a common chromatin feature of the Arabidopsis genome. Nat Plants 3, 704–714. [DOI] [PubMed] [Google Scholar]
- Yan Q, Shields EJ, Bonasio R, and Sarma K (2019). Mapping Native R-Loops Genome-wide Using a Targeted Nuclease Approach. Cell Rep 29, 1369–1380 e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, Sugi NH, Sagane K, Hamaguchi M, Iwata M et al. (2011). Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J 278, 4870–4880. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, and Lieber MR (2003). R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4, 442–451. [DOI] [PubMed] [Google Scholar]
- Zatreanu D, Han Z, Mitter R, Tumini E, Williams H, Gregersen L, Dirac-Svejstrup AB, Roma S, Stewart A, Aguilera A, et al. (2019). Elongation Factor TFIIS Prevents Transcription Stress and R-Loop Accumulation to Maintain Genome Stability. Molecular cell 76, 57–69 e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P, Padeken J, van Schendel R, Kalck V, Tijsterman M, and Gasser SM (2016). Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nature genetics 48, 1385–1395. [DOI] [PubMed] [Google Scholar]
- Zeman MK, and Cimprich KA (2014). Causes and consequences of replication stress. Nat Cell Biol 16, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chiang HC, Wang Y, Zhang C, Smith S, Zhao X, Nair SJ, Michalek J, Jatoi I, Lautner M, et al. (2017). Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nature communications 8, 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]