Abstract
Purpose
To construct a prognostic model of breast cancer using ferroptosis-related lncRNAs and explore novel therapeutic targets.
Materials and Methods
A prognostic characteristic model based on differential expression of ferroptosis-related lncRNAs in breast cancer was established based on TCGA data.
Results
Eleven ferroptosis-related lncRNAs associated with breast cancer prognosis were identified. Kaplan–Meier analysis suggested that high-risk lncRNA signatures correspond to a poor prognosis. The AUC of the signature lncRNAs was 0.682, demonstrating that it is accurate in predicting BC prognosis. GSEA showed that ferroptosis-related lncRNAs in high-risk individuals are mainly enriched in cell cycle, cell adhesion and tumor pathways. Immunity and gene expression analysis revealed that APC co-inhibition, check-point, HLA, inflammation-promoting and T cell co-stimulation among others were significantly different between the high-and low-risk group. Three immune checkpoints were highly expressed in the high-risk group.
Conclusion
Ferroptosis-related lncRNAs can be used as a prognostic feature to construct a prognostic model of breast cancer, based on which early detection markers, therapeutic targets and anti-tumor immune microenvironment can be studied, and clinical treatment can also be instructive.
Keywords: breast cancer, ferroptosis, immune microenvironment, lncRNA
Introduction
Breast cancer is the most common type of cancer in women and the second-leading cause of cancer death among women.1 About 1.2 million women are diagnosed with breast cancer each year, and morbidity is rising by 5% to 20% a year.1 The pathogenesis of breast cancer is complex, involving gene mutation, signal molecule change and pathway abnormality. Risk factors for the disease include postmenopausal obesity, the use of hormone replacement therapy, increased mammary gland tissue and a family history of the disease.2 Treatment of breast cancer depends on histological type, stage, and hormone sensitivity. At present, the main treatments are surgery combined with chemotherapy or radiotherapy, hormone therapy and targeted therapy. However, the effectiveness of clinical treatment is limited by the high recurrence and metastasis rates. Therefore, it is urgent to find new early diagnostic markers and therapeutic targets.
Ferroptosis is a novel cell death mode, which is iron-dependent cell death dependent on intracellular reactive oxygen species (ROS), different from apoptosis.3 The proliferation of cancer cells showed an iron dependence compared to normal cells.4 Some studies have found that activating the ferroptosis process in cancer cells could be a new strategy for treating tumors, especially for cancers that lack therapeutic targets.5,6 Ma et al. Found that Siramesine and Lapatinib might activate the ferroptosis process in breast cancer cells, which may be a new treatment option for refractory breast cancer.7
Long non-coding RNAs (lncRNAs) are defined as a class of RNAs with more than 200 nucleotides in length and without protein-coding ability.8 More and more studies have been conducted on LncRNA in recent years. Although lncRNA cannot encode proteins, it is involved in a variety of biological processes, such as cell proliferation, migration, invasion and resistance to apoptosis. These processes are usually realized by interacting with DNA,9 RNA10,11 and/or proteins to regulate transcription and translation. A number of studies have shown that lncRNA is abnormally expressed in a variety of tumors, which may promote tumor progression by promoting tumor cell proliferation, migration and invasion.12–14 Recent research has shown that some lncRNAs can activate the ferroptosis process of cancer cells and thus play an anticancer role, meaning that a new tumor treatment scheme may be created. However, studies on ferroptosis-related lncRNAs are still lacking. Therefore, it is urgent to explore the expression and regulation of ferroptosis-related lncRNAs in cancer cells. In addition, immune microenvironment is also closely associated with ferroptosis in tumor cells. In this study, we constructed a prognostic characteristic model of differential expression for ferroptosis-related lncRNAs in breast cancer based on TCGA data, and performed enrichment analyses of the differential genes for their function and the pathways involved. Subsequently, differences in immune cells, immune function and immune checkpoints in prognostic models were analyzed.
Methods
Data Collection
RNA-sequence data of 1222 patients were downloaded from the TCGA-GDC database, including 1109 breast cancer patients and 113 normal ones. The clinical characteristics of the patients are revealed in Table 1. We obtained the corresponding ferroptosis-related genes from an online database called FerrDb that supplied ferroptosis markers, their regulatory molecules and associated diseases. In total, 382 ferroptosis-related genes containing 150 Drivers; 109 suppressors; 123 markers were confirmed. The co-expression genes between ferroptosis-related lncRNAs and breast cancer and their relationship were analyzed using Pearson coefficient. The correlation coefficient |R 2|>0.3 at P<0.001 was considered statistically significant. The clinical-pathological data of breast cancer patients containing gender, age, stage, TMN, survival status and survival time were downloaded from TCGA-GDC database. Differentially expressed ferroptosis-related lncRNAs were screened based on FDR<0.05 and |log2FC|≥1. Function of ferroptosis-related differentially expressed genes (DEGs) including biological processes (BP), molecular functions (MF) and cellular components (CC) were analyzed by Gene ontology (GO). Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis (KEGG) was used to analyze the signaling pathways involved in DEGs. In this study, both GO and KEGG were carried out based on the Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool (https://david.ncifcrf.gov/).
Table 1.
Clinical Characteristics of Patients in TCGA Dataset
| Variable | Number of Samples |
|---|---|
| Gender | |
| Male/Female | 12/1085 |
| Age at diagnosis | |
| ≤65/>65 | 776/321 |
| Stage | |
| I/II/III/IV/NA | 183/621/249/20/24 |
| T | |
| T0/T1/T2/T3/T4/NA | 0/281/635/138/40/3 |
| M | |
| M0/M1/NA | 912/22/163/ |
| N | |
| N0/N1/N2/N3/NA | 516/364/120/77/20 |
Survival Analysis and Prognostic Model Construction of the Ferroptosis-Related lncRNAs
Lasso‐penalized Cox regression and univariate Cox regression analyses were used to explore the ferroptosis-related lncRNA signature, and classification of these lncRNAs was performed based on risk score (Coefficient lncRNA1 × expression of lncRNA1) + (Coefficient lncRNA2 × expression of lncRNA2) + ⋯ + (Coefficient lncRNAn × expression lncRNAn). These lncRNAs were classified in low-risk (<median number) or high-risk (≥ median number) group according to the median score. Then, we constructed the prognostic model of the ferroptosis-related lncRNAs. Univariate and multivariate COX analysis were used to analyze independent prognostic factors of breast cancer.
The Predictive Nomogram of Clinical Features Combined with Prognostic Model
We combined the ferroptosis-related lncRNA signatures with clinicopathological features obtained from the TCGA database and draw a predictive nomogram using R software, survival and regplot packages.
The Relationship Between Ferroptosis-Related lncRNAs and mRNA
The interaction network between ferroptosis-related lncRNAs and mRNA was shown by Cytoscape software.
Gene Set Enrichment Analysis (GSEA)
Gene set enrichment analysis (GSEA) was carried out to pick out the pathway that ferroptosis-related lncRNAs enriched based on TCGA. We considered P<0.05 as statistical significance.
Immunity and Gene Expression Analysis
We use TIMER, CIBERSORT, CIBERSORT−ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms to estimate cellular components and cell immune responses between high- and low-risk group based on ferroptosis-related lncRNA signature. A heatmap was obtained to show the differences in immune response under different algorithms. In addition, the differences of immune cell subsets infiltrating the tumor and related functions between high-risk and low-risk group were analyzed by single-sample gene set enrichment analysis (ssGSEA), and the immune checkpoint-related genes retrieved from previous literature were analyzed for differential expression between high-risk and low-risk group.
Statistical Analysis
R software, version 4.1.1 was used to perform data analysis. We use the unpaired Student’s t-test and the Wilcoxon test to analyze the normally and non-normally distributed variables, respectively. The differently expressed lncRNAs were confirmed using Benjamini–Hochberg method based on FDR. We also plotted operating characteristic curve (ROC) to evaluate the accuracy of the model in predicting patient prognosis.15 Logistic regression analysis was used to evaluate the relationship between ferroptosis-related lncRNAs and clinicopathological features. In addition, we performed the Kaplan–Meier survival analysis to plot the survival curves of BC patients based on the ferroptosis-related lncRNA signature. For each analysis, P<0.05 was considered statistically significant.
Results
GO and KEGG Analysis of Ferroptosis-Related DEGs
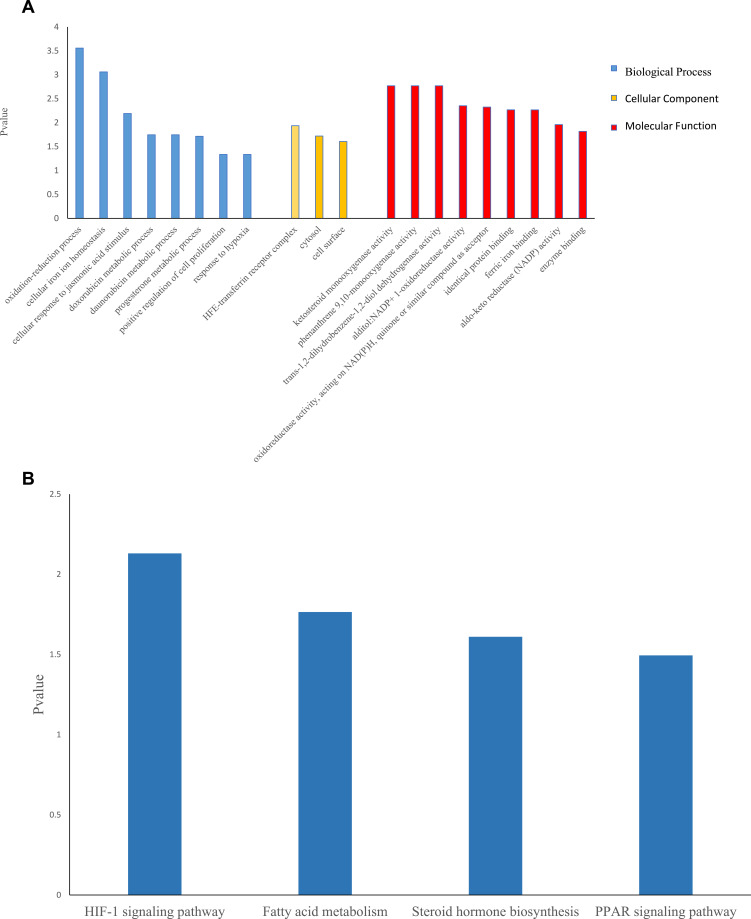
Thirty-eight ferroptosis-related DEGs were identified including 15 upregulated genes and 23 downregulated ones. GO analysis revealed that Biological Process enriched in oxidation-reduction process, cellular iron ion homeostasis, progesterone metabolic process and positive regulation of cell proliferation among others. Cellular Component mainly contained HFE-transferrin receptor complex, cytosol and cell surface. Molecular Function participated in ketosteroid monooxygenase activity, alditol: NADP+1-oxidoreductase activity, identical protein binding, ferric iron binding and so on. KEGG analysis suggested that ferroptosis-related DEGs mainly involved in HIF-1 signaling pathway, fatty acid metabolism, steroid hormone biosynthesis and PPAR signaling pathway (Figure 1A and B).
Figure 1.
GO and KEGG analyses for ferroptosis-related differentially expressed genes.
Notes: (A) Barplot graph for GO (the longer bar means the more genes enriched. Blue: biological process; yellow: cellular component; red: molecular function). (B) Barplot graph for KEGG (the longer bar means the more genes enriched).
Abbreviations: GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis.
The Ferroptosis-Related lncRNAs Prognostic Model
In total, 1089 ferroptosis-related lncRNAs were identified. Twenty-five significant ferroptosis-related lncRNAs were confirmed using univariate COX analysis, which were then involved in the multivariate COX analysis. We considered 11 differentially expressed lncRNAs (AC137630.3, KLHDC7B-DT, AC012213.3, LIPE-AS1, SIDT1-AS1, AC009171.2, HSD11B1-AS1, LINC02446, TFAP2A-AS1, YTHDF3-AS1, AC079298.3) as independent prognosis predictors of BC. Afterwards, we performed risk scores and constructed a prognostic model based on these lncRNAs, which divided patients into high-risk and low-risk group.
Survival Analysis and Multifactorial Testing
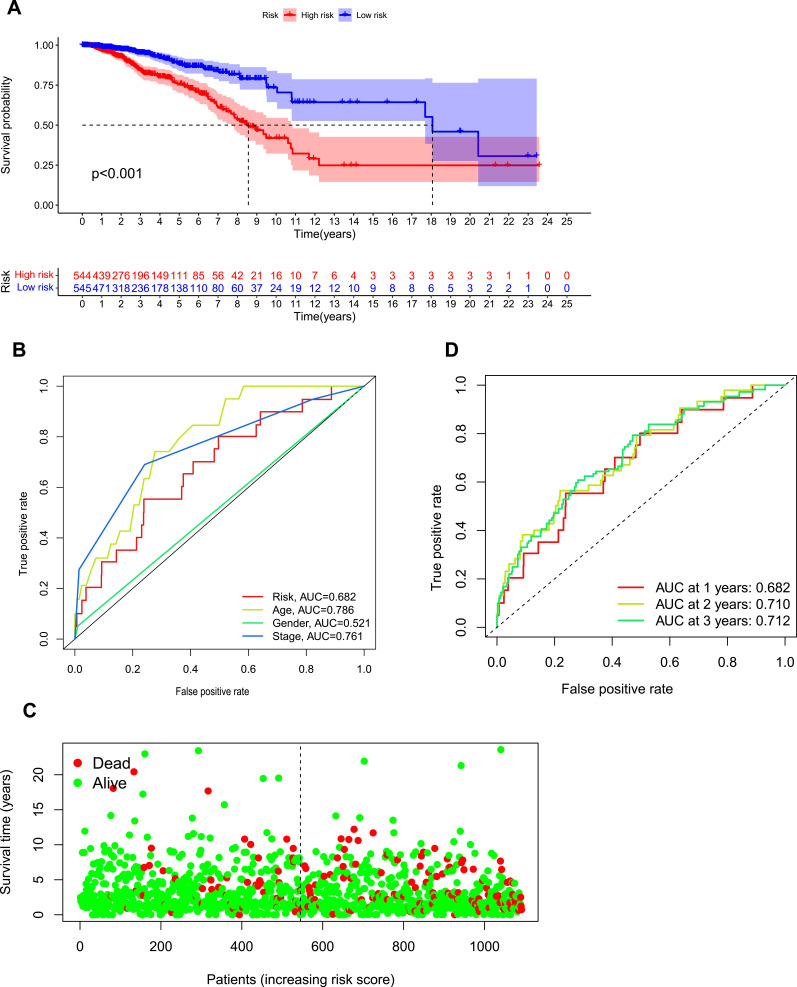
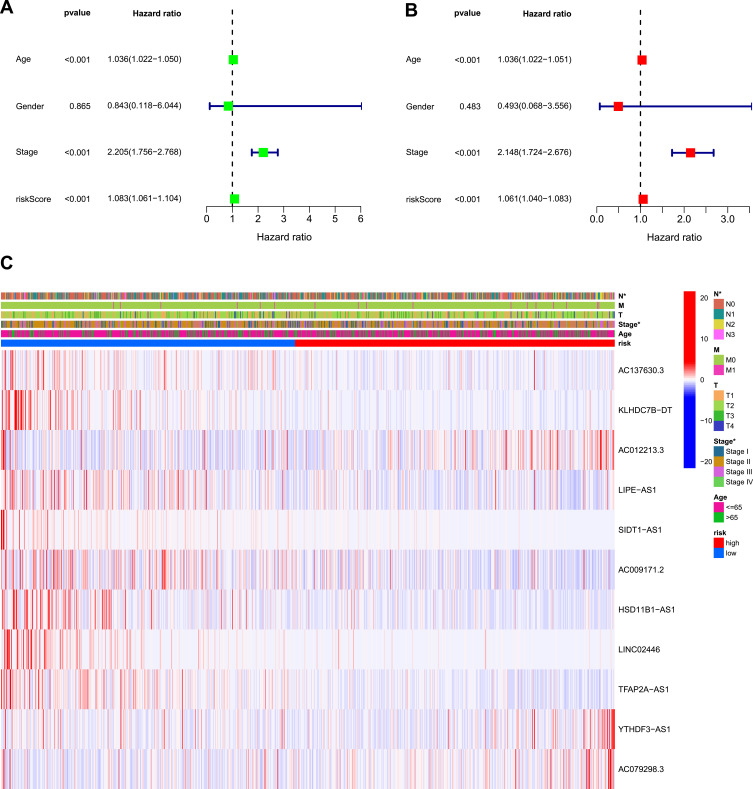
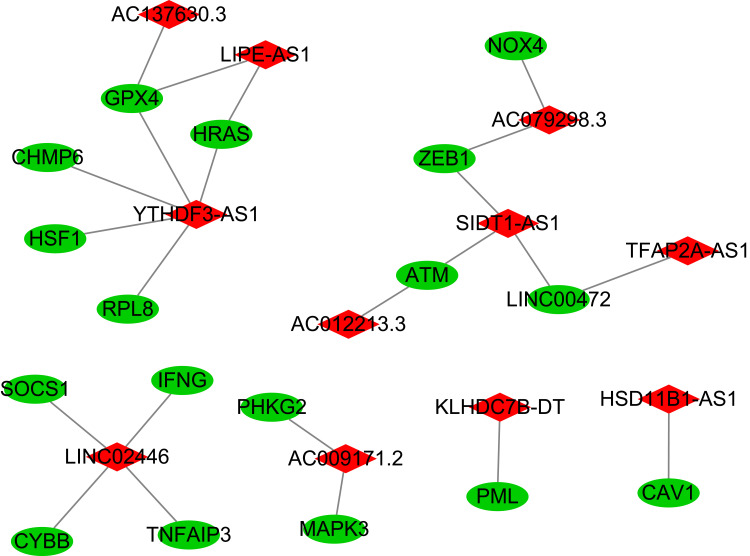
Kaplan–Meier analysis suggested that the expression of high-risk lncRNAs signatures is associated with low survival rate (P<0.001, Figure 2A). In addition, the AUC of the signature lncRNAs was 0.682, suggesting that it along with age and stage is accurate in predicting BC prognosis because their Area Under the Curve (AUC) is greater than 0.6 (Figure 2B). Patients’ risk score–survival status diagram revealed that the survival rate of BC patients was inversely proportional to the risk score in 10 years (Figure 2C). The AUC predictive value for 1, 2 and 3-year survival rate of signature lncRNAs was 0.682, 0.71 and 0.712, respectively (Figure 2D), suggesting their accuracy in predictive prognosis. Univariate and multivariate COX analysis demonstrated that lncRNA signature (HR: 1.08,95CI: 1.06–1.10), age (HR: 1.04,95CI: 1.02–1.05) and tumor stage (HR: 2.21,95CI: 1.76–2.77) were independent prognosis factors of overall survival for BC patients (Figure 3A and B). The hazard ratio (HR) >1, the more the expression level, the higher the risk of patients. The association between ferroptosis-related lncRNAs prognostic signature and clinicopathological features was shown in a heatmap, which showed that three lncRNAs (AC012213.3, YTHDF3−AS1, AC079298.3) were highly expressed in the high-risk group, with a more red color, indicating that they were high-risk genes for BC. On the contrary, the expression of AC137630.3, KLHDC7B−DT, LIPE−AS1, SIDT1−AS1, AC009171.2, HSD11B1−AS1, LINC02446, TFAP2A−AS1 were reduced in high-risk group, with a less red color, suggesting that they were low-risk genes for BC. Additionally, there were significant differences in tumor stage between the high- and low-risk groups (Figure 3C). We also exhibit the interaction between signature lncRNA and ferroptosis-related mRNA in Figure 4. The combined nomogram of ferroptosis-related lncRNAs prognostic signature and clinicopathologic characteristics (Figure 5) was able to accurately predict the survival of each patient by calculating the contribution of risk models and clinical features to the prognosis of BC, which might be applied in clinical treatment of BC patients.
Figure 2.
Ferroptosis-related lncRNAs signature based on TCGA.
Notes: (A) Kaplan–Meier curves for BC patients in high- and low-risk group. (B) The AUC values showed the predictive efficiency of the risk factors (the AUC>0.6 means high accuracy of prediction). (C) The survival status for each patient (low-risk population: on the left side of the dotted line; high-risk population: on the right side of the dotted line). (D) ROC curve indicated the predictive efficiency of risk model for BC in 1, 2, 3-year.
Abbreviations: AUC, area under the curve; ROC, receiver operating characteristic; BC, breast cancer.
Figure 3.
Univariate and multivariate COX analysis for risk model and clinical features.
Notes: (A) Univariate analysis for the risk model and clinical features. (B) Multivariate analysis for the risk model and clinical features (P value less than 0.01 were considered statistically significant). (C) Heatmap showed the differences between the high- and low-risk groups for ferroptosis-related lncRNAs prognostic signature and clinicopathological manifestations (*means significant difference).
Figure 4.
The interaction between signature lncRNA and ferroptosis-related mRNA expression.
Notes: Red represents signature lncRNA, green represents ferroptosis-related mRNA.
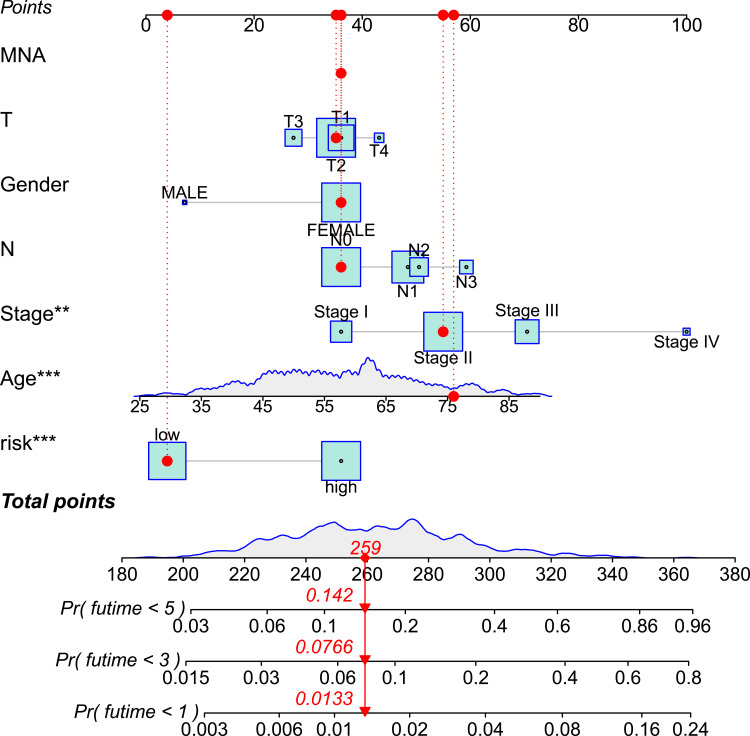
Figure 5.
A combined nomogram of ferroptosis-related lncRNAs prognostic signature and clinicopathologic characteristics.
Note: The nomogram predicts patient survival based on the clinical characteristics and risk model score of each patient. “*” represents the contribution to survival prediction, the more*, the greater the contribution.
Gene Set Enrichment Analyses
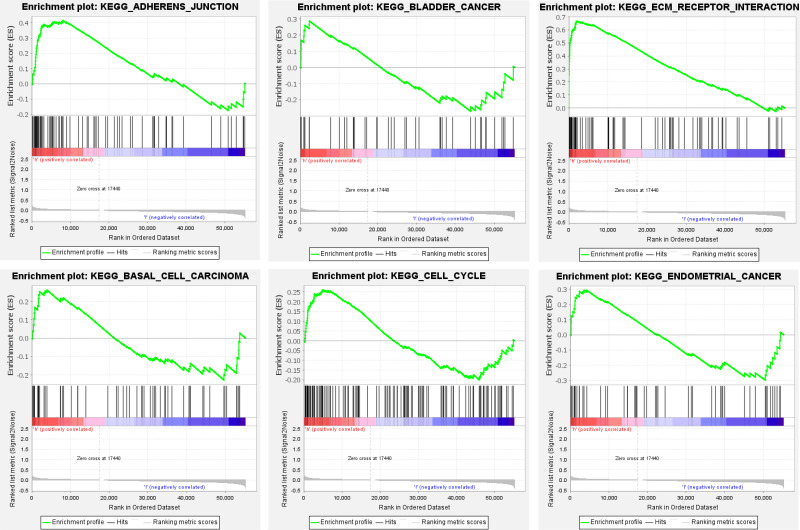
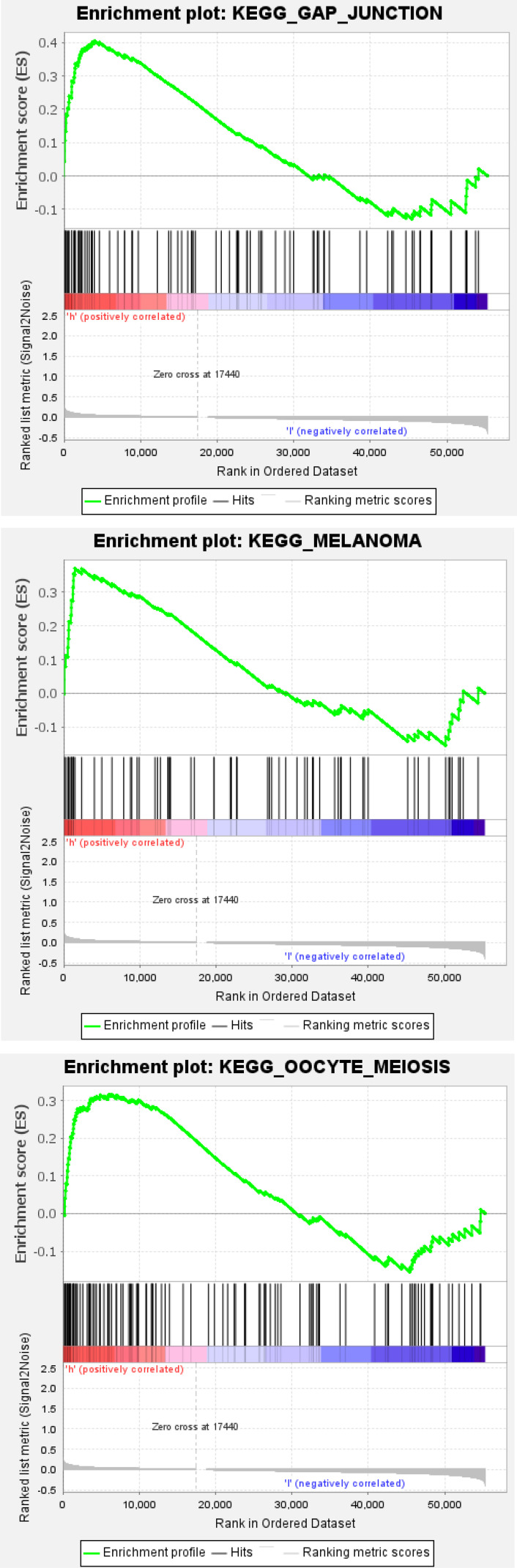
Gene set enrichment analysis (GSEA) demonstrated that a mass of the ferroptosis-related lncRNAs-enriched pathways was active in the high-risk group, which mainly involved in cell cycle, cell adhesion and cancer, such as adherens junction, basal cell carcinoma, bladder cancer, cell cycle, ECM receptor interaction, endometrial cancer, gap junction melanoma and oocyte meiosis (Figure 6), suggesting that these ferroptosis-related lncRNAs are highly likely to be directly related to the occurrence and development of BC.
Figure 6.
Continue.
Figure 6.
Gene enrichment analysis for ferroptosis-related lncRNAs based on TCGA.
Notes: Red: high risk, the darker the red, the higher the risk; blue: low risk, the darker the blue, the lower the risk.
Immunity and Gene Expression Analysis
A heatmap obtained by TIMER, CIBERSORT, CIBERSORT−ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms revealed the differential expression of immune cells between high-risk and low-risk group. Among them, the expression of macrophage M1 and cancer-associated fibroblast significantly increased in the high-risk group, while NK cell, T cell and B cell mainly infiltrated in the low-risk group, which can determine the correlation between immune cells and breast cancer (Figure 7). Correlation analysis of immune cell subgroups and relevant function showed that APC co-inhibition, check-point, cytolytic activity, HLA, inflammation-promoting, MHC class 1, parainflammation, T cell co-inhibition, T cell co-stimulation and type 1 IFN response were significantly reduced in high-risk group (Figure 8), indicating that the deletion of these immune functions increased the risk of BC. To make better use of immune checkpoint suppression therapy, differential expression of immune checkpoints between the two groups was studied. The results showed that most checkpoints were significantly reduced in high-risk group, while only four checkpoints (TNFSF4, TNFSF15, NRP1 and CD276) increased in the high-risk group (Figure 9), which suggests that immune checkpoint inhibition can reduce the risk of breast cancer, and checkpoints expressed in high-risk groups can help guide ICIs research and optimize immunotherapy.
Figure 7.
Heatmap for immune cells infiltration based on TIMER, CIBERSORT, CIBERSORT−ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms between high and low risk group.
Notes: Red: increased infiltration; blue: reduced infiltration.
Figure 8.
ssGSEA for the association between immune cell subgroups and related functions.
Figure 9.
Expression of immune checkpoints among high and low BC risk groups.
Note: The blue bar represents checkpoint gene expression in the low-risk group, and the red bar represents checkpoint gene expression in the high-risk group. *Stands for significant difference (P<0.05), **stands for more significant difference (P<0.01), ***stands for much more significant difference (P<0.001).
Abbreviation: BC, breast cancer.
Discussion
With the development of medical technology, the five-year survival rate of breast cancer has improved in the past few decades.16 However, the treatment of advanced breast cancer is still difficult. Therefore, it is imperative to explore biomarkers for early risk prediction and targeted therapy.
In this study, we identified 11 ferroptosis-related lncRNAs associated with breast cancer based on TCGA database, and a prognostic model was constructed according to the characteristics of these lncRNAs. Patients were then divided into high-risk group and low-risk group on the basis of this model. We compared the gene expression differences between the two groups and analyzed the cell function and signaling pathway enrichment of the DEGs. DEGs were mainly enriched in cellular iron ion homeostasis, progesterone metabolic process and positive regulation of cell proliferation, indicating that ferroptosis, progesterone metabolism and cell proliferation disorders are likely to affect the occurrence and development of BC. Survival analysis of both groups showed that the high-risk group was associated with lower patient survival. We also demonstrated that the model was accurate in predicting patient outcomes through ROC curve. Although its AUC value is lower than some clinical features, this may be related to sample size or other factors, which need to be further studied. Interestingly, we uncovered some differences in immune cells, immune function and checkpoints between high- and low-risk group.
Several new studies have found that lncRNAs play an important role in the occurrence and development of tumors, which can be a tumor carcinogen or tumor suppressor. RNA2 (HOXA- as2) is a novel tumor-associated lncRNA being found abnormally expressed in a variety of malignancies containing breast cancer.17 Abnormally expressed lncRNA usually affects disease progression by regulating transcription and translation, but recent studies have found that it can also affect cancer by regulating ferroptosis. For instance, Wang et al discovered that lncRNA LINC00336 could inhibit ferroptosis in lung cancer.18 Meanwhile, another researcher found that LINC00618 could promote ferroptosis in human leukemia.19 In addition, data suggested that lncRNA could be a potential target for breast cancer treatment.20 lncRNA LINC00908 could encode ASRPS, a small regulatory peptide of STAT3, whose downregulation blocks angiogenesis in triple-negative breast cancer.21 Due to the high efficiency, high tissue specificity and high stability of lncRNA, it might be a potential early diagnostic marker and therapeutic target. Ferroptosis is a new way of cell death, and cancer cells depend on iron to proliferate, meaning that activating the ferroptosis process in cancer cells may be a novel treatment for cancer.5,6 Lu et al reported that reduced overall survival in colorectal cancer patients is partly attributable to inhibition of ferroptosis in colorectal cancer.22 Recent study indicated that prominin2 (PROM2) could reduce the ferroptosis of breast cancer cells by stimulating iron export, thus promoting tumor progression.23 Some breast cancer drugs cause cell death by inducing cancer cells to produce too much ROS,24,25 while ROS is essential for ferroptosis. However, most refractory patients have drug resistance, which requires us to open up novel therapeutic paths, such as promoting ferroptosis of cancer cells in these patients. Ma et al found that Siramesine and Lapatinib could be a potential treatment because of their induction of ferroptosis in breast cancer cells.7 Recent studies have found some novel regulatory factors for ferroptosis, such as P53 and ACSL4, which coincidentally are regulated by lncRNA.26 Therefore, it is reasonable for us to construct a prognostic model of BC based on the difference of ferroptosis-related lncRNAs expression between BC patients and normal samples, although this model is calculated. Of course, the model has limitations because it was not applied to actual clinical treatment and evaluation, and we need more clinical data and experiments to prove its credibility. In this study, we screened out three lncRNAs (AC012213.3, YTHDF3−AS1 and AC079298.3) expressed highly in high-risk group, suggesting that they might be high-risk oncogene for breast cancer. Hongmei Zhong et al proved that lncRNA AC012213.3 was highly expressed in breast cancer tissues and cell lines.27 In addition, they also found that RAD54B was a downstream molecule of AC012213.3, overexpression of the lncRNA AC012213.3 promoted proliferation, migration and invasion of breast cancer via RAD54B/PI3K/AKT axis and was associated with worse patient prognosis.27 However, reports on the other two high-risk lncRNAs (YTHDF3−AS1 and AC079298.3) are scarce, which needs further study. So far, there are few studies on ferroptosis-related lncRNAs, especially relationship between it and breast cancer needs further research, so as to find more novel therapeutic targets for breast cancer, particularly for those refractory cancer patients.
GSEA revealed that ferroptosis-related lncRNAs associated with breast cancer high-risk patients mainly enriched in some pathways like cell cycle, cell adhesion and various cancers, further proving that ferroptosis-related lncRNA is connected to the occurrence of cancer. Meanwhile, cell cycle confusion is the main cause of infinite cell proliferation, which is consistent with tumor growth. On the other hand, the destruction of cell adhesion is the driving force of tumor metastasis and invasion. These results suggest that ferroptosis-related lncRNA is also closely related to tumor progression.
Using various algorithms, we calculated that immune cells, including multiple B cells, T cell subsets and natural killer (NK) cells, were less infiltrated in the high-risk group for breast cancer. In contrast, M2 macrophage infiltration increased in the high-risk group. These findings uncovered that there was a close relationship between anti-tumor and immune microenvironment. Zhang et al reported that immune-related pathways, such as NK cell-mediated cytotoxicity and T cell receptor signaling, were significantly down-regulated in high-risk patients with breast cancer.28 M2 macrophage, a tumor microenvironment immune suppressor cell, could down-regulate or suppress the systemic immune response and thus inhibit anti-tumor immunity.29–31 Recent study reported that signals released by ferroptotic cells could recruit antigen presenting cells to the site of ferroptosis occurring.32 Wang et al found that CD8 + T cells could act against tumors by activating ferroptosis in tumor cells.33 These are all consistent with our findings. Many new mechanisms of ferroptosis induction of CD8 + T cells in tumor cells have also been found, which may be a new way to explore anti-tumor immunity.34–36
One hallmark of cancer is its ability to evade the immune system through tumor-mediated immune evasion.37 Immune evasion can occur through a number of mechanisms, including the regulation of immune checkpoints regulating the adaptive immune system, containing CTLA-4, PD-1, and PD-L1. Immune checkpoint inhibitors (ICIs) have been shown to respond to various cancers and have anticancer benefits.38,39 Although the percentage of patients who benefited from single-agent ICIs was demonstrated to be low, these monotherapy studies also suggest that immunotherapy can induce long-lasting antitumor responses. Therefore, exploring the relationship between the immune system and cancer is conducive to further optimizing immunotherapy. Clinical classification of breast cancer is mainly based on the existence of endocrine receptors (ERs) and HER2.40 Different clinical subtypes of breast cancer respond differently to immunotherapy. Earlier studies have shown that breast cancers lacking these major receptors, known as triple negative breast cancer (TNBC), may be the most sensitive of all subtypes to immunotherapy. A clinical trial of 26 patients with estrogen receptor (ER)–positive metastatic breast cancer treated with the CTLA-4 inhibitors tremelimumab and exemestane showed that CD4+ and CD8+ T cells relative to FOXP3-positive regulatory T cells increased in peripheral, which might suggest a strong anti-tumor immune response.41 KEYNOTE-012 phase Ib trial explored the use of pembrolizumab in patients with PD-L1 positive chemotherapy-resistant metastatic TNBC, which showed that the overall response rate (ORR) of the 27 evaluable patients was 18.5%.42 Results of ICI monotherapy in patients with HER2 positive breast cancer are rarely reported. ICIs rarely work against tumors that are relatively deficient in CD8+ T cells, which are also known as “cold” tumors.43 In theory, increasing the number of CD8+T cells in breast cancer, thus making the tumor “hot” would make ICIs work better. Some combinations of measures that may make the tumor hot are as follows: dual-checkpoint blockade,44 induction chemotherapy,45 PARP inhibitors,46 radiation therapy47 and oncolytic virotherapy.48 Some therapies that combine chemotherapy with ICIs have yielded encouraging results. For instance, ENHANCE 1 (KEYNOTE-150) trial investigating the combination of eribulin mesylate with pembrolizumab in metastatic TNBC patients demonstrated that the ORR of the entire TNBC population regardless of PD-L1 status was 26%. Given the critical role of immune checkpoints in anti-tumor therapy, we identified checkpoint expression levels for patients in the model and found that multiple immune checkpoints were significantly increased in the low-risk group based on ferroptosis-related-lncRNAs signature, including TNFRSE25, CD48, TNFSF14, TNFSF4, and so on. On the contrary, only four checkpoints (TNFSF4, TNFSF15, NRP1 and CD276) expressed highly in the high-risk group. Targeting these four immune checkpoints in high-risk groups with ICIs may provide a novel immunotherapy strategy for breast cancer patients. However, more clinical studies are needed to determine the specific use of ICIs and whether they should be combined with other therapies.
Although we have identified some novel ferroptosis-related lncRNAs associated with breast cancer, the specific mechanism of action remains unclear. Therefore, further studies are needed on the relationship between ferroptosis-related lncRNAs and anti-tumor immunity. There are two limitations to this study: Firstly, our study is a retrospective study based on previous data, so the results are limited. On the other hand, due to the lack of data for each clinical subtype of breast cancer, it is not possible to analyze the characteristics of immune microenvironment and anti-tumor immunity for each subtype. Further clinical studies and basic trials are needed to verify the practicality and accuracy of our model. However, our research is also promising in that the ferroptosis-related lncRNAs we have identified can help assess the risk of breast cancer patients and explore strategies to optimize immunotherapy using selected immune checkpoints.
Conclusion
Ferroptosis-related lncRNAs can be used as a prognostic feature to construct a prognostic model of breast cancer, based on which early detection markers, therapeutic targets and anti-tumor immune microenvironment can be studied, and clinical treatment can also be instructive.
Statement
The data in this paper came from TCGA public database, and the original data had passed the ethical review and obtained the written informed consent of the patients when provided. Our use of the data has passed the review of the Ethical Review Committee of Weifang Medical College again.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sun YS, Zhao Z, Yang ZN, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387–1397. doi: 10.7150/ijbs.21635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0 [DOI] [PubMed] [Google Scholar]
- 3.Cao JY, Dixon SJ. Mechanisms of Ferroptosis. Cell Mol Life Sci. 2016;73(11–12):2195–2209. doi: 10.1007/s00018-016-2194-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manz DH, Blanchette NL, Paul BT, Torti FM, Torti SV. Iron and cancer: recent insights. Ann N Y Acad Sci. 2016;1368(1):149–161. doi: 10.1111/nyas.13008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassannia B, Vandenabeele P, Vanden BT. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35(6):830–849. doi: 10.1016/j.ccell.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 6.Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12(1):34. doi: 10.1186/s13045-019-0720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7(7):e2307. doi: 10.1038/cddis.2016.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postepska-Igielska AA, Giwojna L, Gasri-Plotnitsky L, Schmitt N, Dold A. lncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol Cell. 2015;60:626–636. doi: 10.1016/j.molcel.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 10.Kleaveland B, Shi CY, Stefano J, Bartel DP. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zealy RW, Fomin M, Davila S, et al. Long noncoding RNA complementarity and target transcripts abundance. Biochim Biophys Acta GeneRegul Mech. 2018;1861:224–234. doi: 10.1016/j.bbagrm.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Pajares V. Long non-coding RNA regulation of gene expression during differentiation. Pflugers Arch. 2016;468:971–981. doi: 10.1007/s00424-016-1809-6 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Tian H, Yang J, et al. Long noncoding RNAs regulate cell growth, proliferation and apoptosis. DNA Cell Biol. 2016;35:459–470. doi: 10.1089/dna.2015.3187 [DOI] [PubMed] [Google Scholar]
- 14.Cai B, Ma W, Bi C, et al. Long noncoding RNA H19 mediates melatonin inhibition of premature senescence of c-kit (+) cardiac progenitor cells by promoting miR-675. J Pineal Res. 2016;61:82–95. doi: 10.1111/jpi.12331 [DOI] [PubMed] [Google Scholar]
- 15.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi: 10.1177/0272989X06295361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–448. doi: 10.3322/caac.21412 [DOI] [PubMed] [Google Scholar]
- 17.Fang Y, Wang J, Wu F, et al. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget. 2017;8:46090–46103. doi: 10.18632/oncotarget.17552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Mao C, Ouyang L, et al. Long Noncoding RNA LINC00336 Inhibits Ferroptosis in Lung Cancer by Functioning as a Competing Endogenous RNA. Cell Death Differ. 2019a;26(11):2329–2343. doi: 10.1038/s41418-019-0304-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Liu C, Li Z. Regulation of GSK3β/Nrf2 Signaling Pathway Modulated Erastin-Induced Ferroptosis in Breast Cancer. Mol Cell Biochem. 2020;473(1–2):217–228. doi: 10.1007/s11010-020-03821-8 [DOI] [PubMed] [Google Scholar]
- 20.Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234(6):9105–9117. doi: 10.1002/jcp.27587 [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Wu S, Zhu X, et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med. 2020;217(3):e20190950. doi: 10.1084/jem.20190950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu D, Yang Z, Xia Q, et al. ACADSB Regulates Ferroptosis and Affects the Migration, Invasion, and Proliferation of Colorectal Cancer Cells. Cell Biol Int. 2020;44(11):2334–2343. doi: 10.1002/cbin.11443 [DOI] [PubMed] [Google Scholar]
- 23.Brown CW, Amante JJ, Chhoy P, et al. Prominin2 drives ferroptosis resistance by stimulating iron export. Dev Cell. 2019;51(5):575–586. doi: 10.1016/j.devcel.2019.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Zhang XW, Liao QL, Wu WT. Electrochemical Monitoring of Paclitaxel-Induced ROS Release from Mitochondria inside Single Cells. Small. 2019;15(48):e1901787. doi: 10.1002/smll.201901787 [DOI] [PubMed] [Google Scholar]
- 25.Vernier M, Dufour CR, McGuirk S, et al. Estrogen-related Receptors Are Targetable ROS Sensors. Genes Dev. 2020;34(7–8):544–559. doi: 10.1101/gad.330746.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X, Long D. Nrf2 and ferroptosis: a new research direction for 42. neurodegenerative diseases. Front Neurosci. 2020;14:267. doi: 10.3389/fnins.2020.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong H, Zeng G, Lang H. Overexpression of the lncRNA AC012213.3 Promotes Proliferation, Migration and Invasion of Breast Cancer via RAD54B/PI3K/AKT Axis and is Associated with Worse Patient Prognosis. Cancer Manag Res. 2021;13:7213–7223. doi: 10.2147/CMAR.S322195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiming Z, Liqin P, Tian D, et al. A Ferroptosis-Related lncRNAs Signature Predicts Prognosis and Immune Microenvironment for Breast Cancer. Front Mol Biosci. 2021;8:678877. doi: 10.3389/fmolb.2021.678877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suarez-Lopez L, Sriram G, Kong YW, et al. MK2 contributes to tumor progression by promoting M2 macrophage polarization and tumor angiogenesis. Pro Natl Acad Sci USA. 2018;115:4236–4244. doi: 10.1073/pnas.1722020115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wing JB, Tanaka A, Sakaguchi S. Human FOXP 3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 2019;50:302–316. doi: 10.1016/j.immuni.2019.01.020 [DOI] [PubMed] [Google Scholar]
- 31.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the Crossroads of Cancer-Acquired Drug Resistance and Immune Evasion. Nat Rev Cancer. 2019;19(7):405–414. doi: 10.1038/s41568-019-0149-1 [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Green M, Choi JE, et al. CD8+ T Cells Regulate Tumour Ferroptosis during Cancer Immunotherapy. Nature. 2019b;569(7755):270–274. doi: 10.1038/s41586-019-1170-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Zhang Y, Xia S, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Wang Y, Ding J, et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579:421–426. doi: 10.1038/s41586-020-2079-1 [DOI] [PubMed] [Google Scholar]
- 36.Lang X, Green MD. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019;9:1673–1685. doi: 10.1158/2159-8290.CD-19-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn GP, Bruce AT, Ikeda H, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991 [DOI] [PubMed] [Google Scholar]
- 38.Robert C, Schachter J, Long GV, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- 39.Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- 40.Tobin NP, Harrell JC, Lovrot J, et al. The molecular subtype and tumor characteristics of breast cancer metastases significantly influence patient post-relapse survival. Ann Oncol. 2015;26:81–88. doi: 10.1093/annonc/mdu498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dirix LY, Takacs I, Jerusalem G, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat. 2018;167:671–686. doi: 10.1007/s10549-017-4537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanda R, Chow LQ, Dees EC, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34(21):2460. doi: 10.1200/JCO.2015.64.8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 44.Nolan E, Savas P, Policheni AN, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. 2017;9(393):eaal4922. doi: 10.1126/scitranslmed.aal4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kok M, Voorwerk L, Horlings H, et al. Adaptive Phase II randomized trial of nivolumab after induction treatment in triple negative breast cancer (TONIC trial): final response data stage I and first translational data. JCO;2018. 1012. doi: 10.1200/JCO.2018.36.15_suppl.1012 [DOI] [Google Scholar]
- 46.McCann KE, Hurvitz SA. Advances in the use of PARP inhibitor therapy for breast cancer. Drugs Context. 2018;7:212540. doi: 10.7573/dic.212540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu ZI, Ho AY, McArthur HL. Combined radiation therapy and immune checkpoint blockade therapy for breast cancer. Int J Radiat Oncol Biol Phys. 2017;99(1):153–164. doi: 10.1016/j.ijrobp.2017.05.029 [DOI] [PubMed] [Google Scholar]
- 48.Force J, Holl E, Brown M, et al. Recombinant oncolytic poliovirus combined with checkpoint blockade for breast cancer therapy. JCO. 2018;36:e12641. doi: 10.1200/JCO.2018.36.15_suppl.e12641 [DOI] [Google Scholar]