Abstract
We report here a novel fluorescent protein-based screen to identify small, synthetic internal ribosome entry site (IRES) elements in vivo. A library of bicistronic plasmids encoding the enhanced blue and green fluorescent proteins (EBFP and EGFP) separated by randomized 50-nucleotide-long sequences was amplified in bacteria and delivered into mammalian cells via protoplast fusion. Cells that received functional IRES elements were isolated using the EBFP and EGFP reporters and fluorescence-activated cell sorting, and several small IRES elements were identified. Two of these elements were subsequently shown to possess IRES activity comparable to that of a variant of the encephalomyocarditis virus IRES element in a context-independent manner both in vitro and in vivo, and these elements functioned in multiple cell types. Although no sequence or structural homology was apparent between the synthetic IRES elements and known viral and cellular IRES elements, the two synthetic IRES elements specifically blocked poliovirus (PV) IRES-mediated translation in vitro. Competitive protein-binding experiments suggested that these IRES elements compete with PV IRES-mediated translation by utilizing some of the same factors as the PV IRES to direct translation. The utility of this fluorescent protein-based screen in identifying IRES elements with improved activity as well as in probing the mechanism of IRES-mediated translation is discussed.
Internal ribosome entry site (IRES)-mediated translation was first recognized in picornaviruses as a mechanism of translation that is distinct from cap-dependent translation of cellular RNAs (19, 27). Picornaviruses such as poliovirus (PV), encephalomyocarditis virus (EMCV), and rhinovirus were shown to possess a cis-acting region, the IRES, within the viral 5′ untranslated region (5′ UTR) that could mediate translation in the absence of a cap structure at the 5′ end of the RNA. Subsequently, it was shown that a variety of other viruses, including hepatitis C virus (HCV), as well as certain cellular mRNAs, such as those encoding BiP, c-myc, and the G subunit of eukaryotic initiation factor 4 (eIF4G), also directed translation via IRES elements (21, 38, 42). All of these IRES elements span hundreds of nucleotides in length and possess complex secondary and tertiary structures.
An understanding of the mechanisms of IRES-mediated translation has come about in recent years due mainly to in vitro studies of viral IRES elements. In particular, an in vitro assay for the formation of 48S preinitiation complexes has demonstrated that viral IRES elements do not require eIF4E, the cap-binding protein, for complex formation. In addition, various viruses require different combinations of other canonical translation factors; the EMCV IRES, for example, requires eIF4A and a portion of eIF4G for 48S complex formation, while the HCV IRES does not require either of these factors (28, 29). Although these studies have shed light on the minimum required factors for viral IRES-mediated translation, it is apparent that trans-acting factors play an important role in modulating IRES activity. Cellular proteins such as La, PTB, PCBP2, and unr have been shown to interact with viral IRES elements and stimulate IRES-mediated translation (2, 6, 7, 16). Since these proteins are multimeric or contain multiple RNA-binding domains, it is hypothesized that they may act as “RNA chaperones,” stabilizing IRES secondary and tertiary structure to allow efficient translation to take place (5).
Although progress has been made in understanding the role of trans-acting factors in IRES-mediated translation, the sequence and structural requirements of IRES elements remain unclear. Little, if any, sequence homology has been found between the various IRES elements (18). Picornaviral IRES elements, which span approximately 450 nucleotides (nt) in length, terminate at the 3′ end with an element consisting of a conserved UUUC stretch followed by a polypyrimidine-rich tract and a G-poor space and ending in an AUG triplet. Maintenance of proper (approximately 25 nt) spacing between the UUUC region and the AUG appears to be essential for picornaviral IRES activity; PVs constructed with insertions that increase the spacing in this region tend to grow slowly, if at all, and revertants that exhibit wild-type growth properties have eliminated these insertions or have introduced an additional AUG to maintain proper spacing between cis elements (1). RNA structure also appears to play an important role in viral IRES-mediated translation. For example, phenotypic revertants of point mutations in picornaviral IRES elements often include second-site suppressor mutations that restore the wild-type base pairing, suggesting that maintenance of RNA structure is crucial for IRES activity (12, 13). Also, maintenance of a phylogenetically conserved stem-loop structure was found to be important for the ability of the HCV IRES to mediate translation (14). Taken together, these observations point to the importance of both sequence and structure in viral IRES-mediated translation. Less is known regarding the cis-acting requirements of cellular IRES elements. It has been proposed that a number of cellular IRES elements possess a Y-type stem-loop structure (22), but the ability of these RNAs to mediate translation has not been correlated with maintenance of the Y-shaped structure (15, 36, 43). Recently, the 196-nt 5′ UTR of the mouse Gtx mRNA was found to contain a 9-nt segment that can function as an IRES element. Because this stretch of 9 nt is complementary to 18S rRNA, it was suggested that IRES activity was due to the ability of the 9-nt segment to base-pair with rRNA, thereby recruiting the ribosome to the RNA (8). However, comparison of other IRES elements with rRNA sequences has not, as yet, revealed regions of obvious complementarity.
Many of the studies investigating cis-acting requirements for IRES elements have focused on mutational analysis of viral 5′ UTRs (12–14). However, the 5′ regions of viral RNAs are involved in other functions crucial for the viral life cycle, including replication and packaging (30). Often, cis-acting signals for different viral functions overlap in the 5′ UTR, making it difficult to identify a viral sequence that is solely responsible for IRES-mediated translation (30). In this study, we wished to screen for small synthetic IRES elements for a number of reasons. First, these synthetic elements would be selected only for their ability to promote IRES-mediated translation and not for other roles commonly played by the 5′ UTRs of viral RNAs. As a result, sequence and structural analyses of synthetic IRES elements should reveal motifs specific to IRES-mediated translation. Second, small RNAs are easier to study in terms of structure and protein-binding properties than longer RNAs; the identification of small IRES elements may shed light on structural requirements and trans-acting factors for IRES-mediated translation. Also, in addition to addressing mechanisms of IRES-mediated translation, small synthetic IRES elements may be of practical use in biotechnological applications. Finally, the growing identification of numerous cellular IRES elements leads to the question of how common it is for a particular stretch of RNA to possess IRES activity. A screen for IRES activity among a randomized population of RNA may provide insight into this question.
Here, we have developed a novel fluorescent protein-based screen to identify small (approximately 50 nt in length) synthetic IRES elements in vivo. By adapting methods recently developed by Tan and Frankel (37), we delivered a library of bicistronic plasmids which had been amplified in bacteria into mammalian cells via protoplast fusion. Positive cells were isolated using enhanced blue and green fluorescent protein (EBFP and EGFP) reporters and fluorescence-activated cell sorter (FACS) analysis, and several small functional IRES elements were identified. Two of these elements were subsequently shown to possess IRES activity in a context-independent manner in vivo and in vitro and functioned in multiple cell types. Although no sequence or structural homology is apparent between these synthetic IRES elements and known IRESs, these elements may promote IRES-mediated translation in a manner similar to PV.
MATERIALS AND METHODS
Construction of plasmids and randomized library.
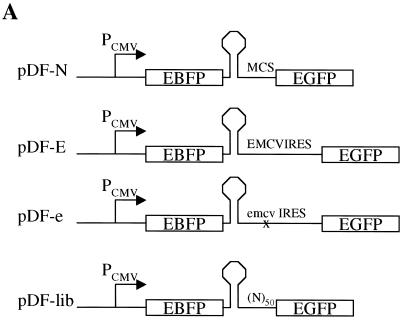
pDF-N contains EBFP (Clontech) and a highly thermodynamically stable stem-loop structure (less than −40 kcal/mol) upstream of the EGFP gene in the pEGFP-N1 plasmid (Clontech). EBFP was cloned between the HindIII and NotI sites, while the stem-loop was placed between the NotI and EcoRI sites. pDF-E is identical to pDF-N, except that the EMCV IRES (from pIREShyg; Clontech) was inserted between the EcoRI and BamHI sites. pDF-e was made by inserting an EMCV IRES that was mutated by site-directed mutagenesis at positions 299 to 302, from 5′-GCGA-3′ to 5′-AAAG-3′, into pDF-N.
To create a randomized library, a polyacrylamide gel electrophoresis (PAGE)-purified 50-nucleotide-long randomized oligonucleotide library (Integrated DNA Technologies) with the sequence 5′-GCGCACTGATGAATTC-N50-GGATCCTCAGACTCCG-3′ was obtained. The phosphoramidite ratio for random-sequence DNA synthesis was normalized to account for differing coupling rates (40). The oligonucleotide pool was amplified by 10 cycles of PCR as described by Tuerk (39), and the amplified DNA was cut with EcoRI and BamHI and ligated to EcoRI-BamHI-digested pDF-N in 10 separate reactions. Each ligation reaction mixture was divided into two parts and electroporated into DH-5α cells. Each transformation yielded approximately 50,000 colonies; colonies were combined to yield a total pool of approximately 106 transformants. The library of plasmids represented by the 106 trasformants, termed pDF-lib, was analyzed for sequence diversity. Twenty-five individual clones were sequenced, and the nucleotide content of the approximately 1,225 bases was found to be evenly distributed (25% G, 26% A, 24% T, 25% C). None of these clones had the same sequence. The lengths of the randomized inserts ranged from 48 to 50 nt, possibly due to incomplete separation of the oligonucleotide library during PAGE purification. To facilitate discussion, the library will be referred to as containing 50-nt insertions. One of the clones sequenced possessed, in its 50-nt insert, a palindromic sequence recognized by the restriction enzyme KpnI, while another clone possessed a SmaI site. As an additional check for insert diversity in our library, 100 other randomly selected clones were tested for the presence of KpnI or SmaI sites within their inserts and were found to be lacking both of these sites. It should be noted that because EcoRI and BamHI sites were used to construct the plasmid library, we expected that the sequences 5′-GAATTC-3′ and 5′-GGATCC-3′ would be underrepresented.
Protoplast fusion.
Protoplast fusion was performed essentially as described by Tan and Frankel (37). Protoplasts from plasmid-containing DH-5α cultures were prepared as described previously (33); conversion of rod-shaped bacteria to round protoplasts was monitored by phase-contrast microscopy. Protoplasts were slowly diluted with room temperature serum-free Dulbecco's modified Eagle's medium (DMEM) containing 10% sucrose and 10 mM MgCl2 and were held at room temperature for 15 min. Protoplasts (2 to 4 ml of suspension at approximately 1.5 × 109 protoplasts/ml) were added to HEK293 cells that had been grown to approximately 75% confluence in 6-well plates, plates were centrifuged at 1,650 × g for 10 min at room temperature, and supernatants were removed. Two milliliters of room temperature 50% (wt/vol) polyethylene glycol 1500 was added to each well. After 2 min, the polyethylene glycol 1500 was removed, cells were gently washed twice with 2 ml of serum-free DMEM, and 4 ml of DMEM containing 10% fetal bovine serum, penicillin, and ampicillin was added. Cells were examined for fluorescence 24 h later.
FACS analysis.
Transfected or protoplast-fused cells were resuspended 24 h later at a concentration of 1 × 106 to 2 × 106 cells/ml in phosphate-buffered saline. Samples were analyzed or sorted by FACS using a dual-laser flow cytometer. An argon laser was used to excite cells at 488 nm, and a 530 ± 15-nm band-pass filter was used to detect EGFP expression, while a second laser tuned to 355 nm was used to excite EBFP, which was detected by a 424 ± 22-nm collection filter. It should be noted that EGFP fluoresced at a much higher intensity than EBFP (approximately 30-fold-higher intensity), consistent with other reports (44). Both FACS analysis (5,000 to 10,000 cells per sample) and FACS sorting (2,500 cells/s) were performed by using the UCLA Core Flow Cytometry Laboratory FACStarplus cell sorter (Becton Dickinson). FACS data were analyzed using CELLQUEST software.
Screening of randomized library.
Protoplasts containing pDF-e, pDF-E, and pDF-lib were fused to HEK293 cells. After 24 h, cells fused with pDF-E and pDF-e protoplasts were analyzed by FACS to establish a sorting window that would exclude most cells fused with pDF-e while including many of the cells fused with pDF-E. Subsequently, approximately 3 × 107 cells fused with pDF-lib were sorted by FACS, and positive cells were collected and mixed with 50,000 unfused HEK293 cells. Plasmids were recovered by alkaline lysis essentially as described elsewhere (37) and electroporated into DH-5a cells; resulting colonies were used to make protoplasts for the next round of screening. The cycle was repeated for three rounds, after which a significant proportion (>50%) of cells exhibited a fluorescence profile similar to that of cells fused with pDF-E.
Transfection, fluorescence microscopy, and luciferase assays.
HEK293 and HuH7 cells were seeded into 6-well or 96-well plates and allowed to reach 70% confluency. Transfection was performed by addition of a mixture containing 0.2 to 2.0 μg of DNA and either 0.5 to 3 μl of Cytofectene (Bio-Rad) or 1 to 4 μl of Lipofectin and 1 to 8 μl of Plus Reagent (Gibco-BRL) to cells. For fluorescence microscopy, 96-well plates were visualized directly on a Nikon Diaphot 200 inverted microscope using the UV-2A filter cube to detect EBFP and the GFP filter cube to detect EGFP expression. For dual luciferase assays, 6-well plates were washed in phosphate-buffered saline and resuspended in Cell Culture Lysis Buffer (Promega) before analysis of Luc and Rluc expression by the Promega Dual Luciferase System on a Monolight 2010 Luminometer (Analytical Luminescence Laboratory).
In vitro translation, UV cross-linking, and RNA structural analysis.
HeLa cell extracts were prepared as described previously (9), and in vitro translation was performed under standard conditions (9). Briefly, 40 μg of HeLa translation extract was incubated at 37°C for 1 h with 0.5 to 2.0 μg of template RNA prepared by standard in vitro transcription before analysis by sodium dodecyl sulfate-PAGE. In competitive translation experiments, a two- to fourfold molar excess of competitor RNA was preincubated with HeLa extract and all other components necessary for translation at 37°C for 10 min before the template RNA was added. UV-cross-linking experiments were also performed as previously described (9); 32P-labeled RNA was incubated with HeLa translation extract for 10 min before being subjected to short-wave UV irradiation. After RNase treatment, samples were analyzed by sodium dodecyl sulfate-PAGE. In competitive UV-cross-linking experiments, a 100- to 250-fold molar excess of competitor RNA was preincubated with HeLa extract before addition of 32P-labeled RNA.
Structural analysis of RNA, which involved free energy minimization modeling and nuclease probing, was performed essentially as described previously (41). Secondary structure predictions were obtained using the RNA folding program MFOLD; for each RNA, all structures within 10% of the minimum predicted free energy were retained as possible candidates. Nuclease probing was performed by equilibrating RNAs with heating to 65°C for 5 min and slow cooling to 37°C. Digestions with nuclease S1 (Promega) and RNase T1 (Boehringer Mannheim) were performed in an RNA buffer (50 mM sodium acetate [pH 4.5], 280 mM NaCl, and 4.5 mM ZnSO4) at 37°C for 10 min, and primer extension with radiolabeled oligonucleotide primers was performed essentially as described elsewhere (35). For position markers, sequencing ladders were generated from plasmid DNA using the same primers and a Sequenase kit (version 2.0; U.S. Biochemicals), and all samples were analyzed via denaturing PAGE.
RESULTS
Development of fluorescent reporter-based screen to identify IRES activity in vivo.
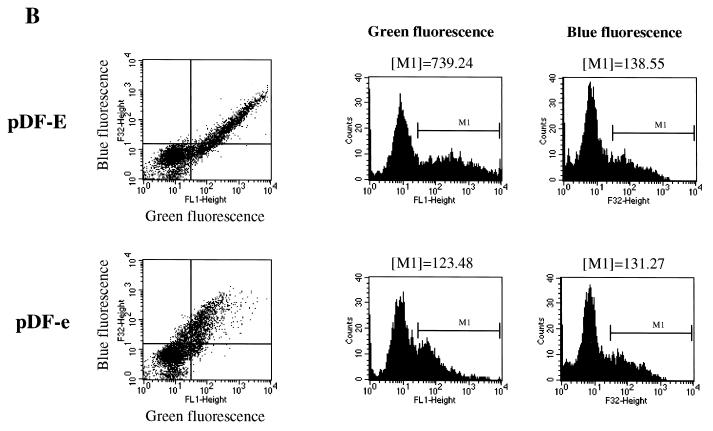
As a first step in developing a screening procedure to identify IRES elements in vivo, we constructed bicistronic plasmids encoding the two reporter proteins EBFP and EGFP (Clontech). These reporters were separated either by a multiple cloning site that has been demonstrated to not possess significant IRES activity (pDF-N) (A. Venkatesan and A. Dasgupta, submitted for publication), a variant of the EMCV IRES (pDF-E) (Clontech), or a mutant form of this EMCV IRES in which four nucleotides have been altered to greatly diminish IRES activity (pDF-e) (Fig. 1A) (31). Upon introduction of these plasmids into mammalian cells, the cytomegalovins promoter is expected to drive expression of a single transcriptional unit encompassing both cistrons. The first cistron, EBFP, will typically be translated via cap-dependent translation. In contrast, the second cistron, EGFP, should only be efficiently translated if a functional IRES element is present upstream. Twenty-four hours after transfection of the pDF plasmids into HEK293 cells, the cells were analyzed for fluorescent protein expression via flow cytometry. As shown in Fig. 1B, the expression profiles of pDF-E-transfected cells differed significantly from those of cells transfected with pDF-e. Although the mean blue fluorescence, representative of cap-dependent translation, of cells transfected with pDF-E or pDF-e was almost identical, the mean green fluorescence of cells transfected with pDF-E was approximately six times greater than that of cells transfected with pDF-e, indicating that pDF-E is a much stronger IRES than pDF-e (Fig. 1B). The fluorescent protein expression of pDF-e is very similar to that of pDF-N (data not shown for transfection; see Fig. 3), which also contains a poor IRES element. Thus, flow cytometry allowed us to distinguish between plasmids containing poor IRES elements (pDF-e and pDF-N) and those containing strong IRES elements (pDF-E).
FIG. 1.
Establishment of a fluorescent protein-based reporter system to study IRES-mediated translation in vivo. (A) All plasmids contain the EBFP gene at the 5′ end, followed by a highly stable stem-loop structure that has been previously used in bicistronic constructs (24) and the EGFP gene at the 3′ end. pDF-N contains a multiple cloning site (MCS) derived from the pEGFP-N1 plasmid. pDF-E contains a variant of the EMCV IRES obtained from pIREShyg (Clontech). In pDF-e, a 4-base mutation has been introduced in a proposed GNRA tetraloop of the EMCV IRES (31) (see Materials and Methods). pDF-lib is a library of plasmids that contain a 48- to 50-base randomized insertion just upstream of the EGFP gene. (B) Flow cytometric analysis of HEK293 cells transfected with pDF-E and pDF-e. Dot plots represent blue fluorescence (y axis) versus green fluorescence (x axis). Approximately 30% of the cells were transfected in this experiment. Region M1 in the histogram plots represents transfected cells, and the mean fluorescence value of these cells is shown above each histogram.
FIG. 3.
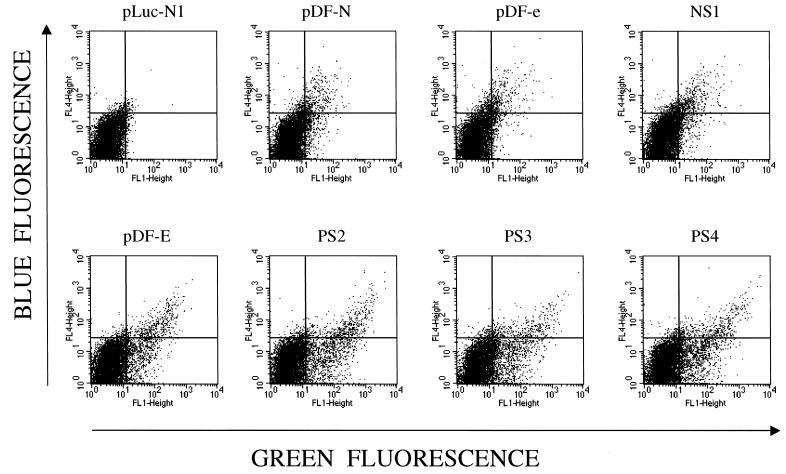
Flow cytometric analysis of clones recovered from screen. Protoplasts were made from selected plasmids and fused to HEK293 cells. After 24 h, cells were analyzed via flow cytometry for blue (y axis) and green (x axis) fluorescence. PS2, PS3, and PS4 are positively selected clones, while NS1 is a randomized 50-nt element that does not possess significant IRES activity. As a negative control, protoplasts from a control plasmid (pLuc-N1) that does not express fluorescent proteins were fused to HEK293 cells. In this experiment, approximately 10% of HEK293 cells were productively fused with protoplasts.
Several methods to deliver libraries into mammalian cells have been described, including transfection, retroviral transfer, electroporation, and protoplast fusion (25, 33). In order to find a suitable method to conduct a fluorescence-based screen for IRES elements, we tested whether transfection of plasmids into HEK293 cells, followed by FACS analysis to isolate desired cells, resulted in an enrichment of strong IRES elements. To mimic the situation encountered in a library screen, we attempted to recover plasmids containing a strong IRES element after cotransfecting a large amount of plasmid DNA containing a poor IRES element (pDF-e) with a small amount of plasmid DNA encoding the strong IRES element (pDF-E). Plasmid DNA mixed in a ratio of 104:1 (pDF-e to pDF-E) was transfected into HEK293 cells, and 24 h later cells were subjected to FACS analysis to recover cells that expressed significant levels of both EBFP and EGFP, indicative of cells containing the pDF-E plasmid. Plasmid DNA was recovered from sorted cells, amplified in Escherichia coli, and transfected into 293 cells for another round of screening. After three rounds of screening, 54 individual clones were sequenced; all possessed the pDF-e sequence, indicating that significant enrichment of strong IRES activity had not occurred. To further confirm that transfection was not a suitable method to screen a library for IRES elements, we pooled the approximately 1,000 recovered colonies following the third round of screening and sequenced the pooled DNA. Again, no significant enrichment of the pDF-E sequence was noted (data not shown). Since it is well established that transfection of a combination of plasmids often leads to the delivery of all of them into the same cell, we reasoned that in our case cells containing the pDF-E plasmid also contained large numbers of the pDF-e plasmid (due to the overabundance of pDF-e in our experiment) and that this background prevented us from successfully enriching our population for pDF-E.
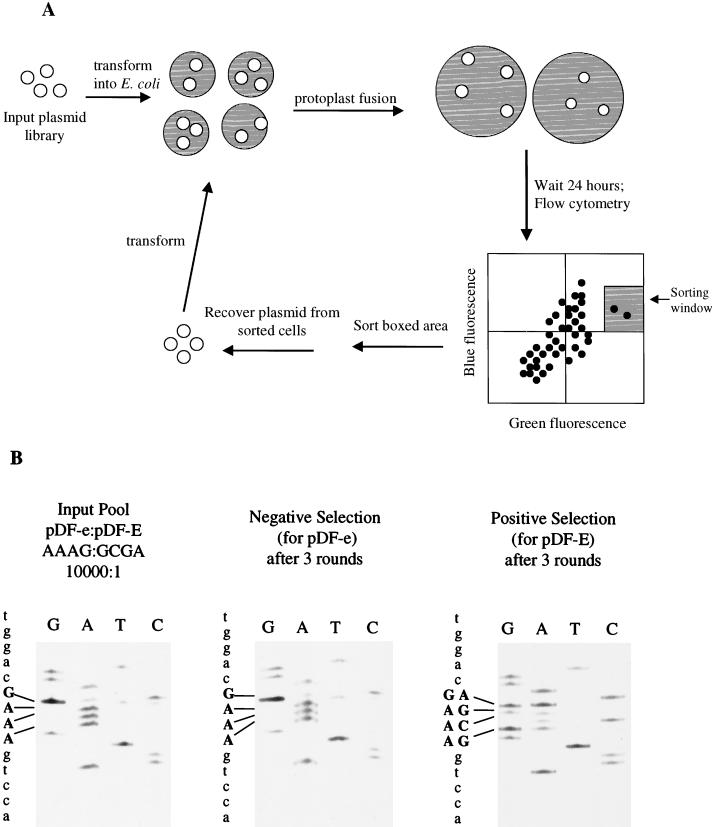
We next decided to employ protoplast fusion to deliver plasmids into HEK293 cells, as it has been observed that this technique results in near-clonal delivery of plasmids into cells (37). To confirm this, we transformed E. coli cells with plasmids encoding EBFP and EGFP in a 1:1 ratio, made protoplasts, and fused them with 293 cells. We found, like Tan and Frankel (37), that protoplast fusion is indeed a near-clonal delivery process; fused cells expressed, for the most part, either EBFP or EGFP but not both. In contrast, cells cotransfected with plasmids expressing EBFP and EGFP in a 1:1 ratio expressed, for the most part, both EBFP and EGFP (data not shown). These observations suggested that protoplast fusion may be a suitable method to deliver a library of plasmids in a near-clonal fashion into cells to ensure a low background during a screen. As a pilot experiment, we mixed DNA encoding pDF-e and pDF-E in a 104:1 ratio, made protoplasts, fused them with HEK293 cells, and sorted positive cells 24 h later via FACS. Plasmid DNA from positive cells (those expressing significant levels of both EBFP and EGFP) was isolated and transformed into E. coli, and protoplasts were again made and fused with 293 cells (Fig. 2A). After three rounds of selection, significant enrichment of pDF-E had occurred; 14 of the 20 clones (70%) sequenced had the pDF-E sequence (data not shown). In addition, the approximately 700 colonies recovered after three rounds of selection were pooled, and plasmid DNA was isolated and sequenced from this pool. A large percentage (>60%, as judged by quantitation of sequencing band intensities) of the clones recovered after three rounds of screening possessed the pDF-E sequence, 5′-GCGA-3′, rather than the pDF-e sequence, 5′-AAAG-3′, at EMCV IRES positions 299 to 302 (Fig. 2B, positive selection sequence). In contrast, sequencing of the input pool consisting of a pDF-e/pDF-E ratio of 104:1, as well as of a population of clones selected for poor IRES activity (Fig. 2B, input pool and negative selection sequences), both displayed the population sequence 5′-AAAG-3′ at positions 299 to 302. Thus, our screening method allows us to identify rare, strong IRES elements under conditions that mimic a library screen.
FIG. 2.
(A) Strategy for screening for IRES elements. The input plasmid library containing two fluorescent reporters is introduced into eukaryotic cells by protoplast fusion. Small shaded circles represent E. coli, and large shaded circles represent HEK293 cells. A suitable sorting window is used to isolate double-positive cells via FACS analysis, and plasmids are extracted by alkaline lysis and electroporated into bacteria. Protoplasts are made from the enriched population, and the cycle is repeated (adapted from Tan and Frankel [37]). (B) Pilot experiment to establish screening conditions. The input pool, whose collective sequence is shown in the left panel, contained a mixture of plasmids with a pDF-e/pDF-E ratio of 10,000:1. After three rounds of screening for strong (pDF-E) IRES elements, 700 positively selected clones were obtained; their collective sequence is shown in the right panel. In the middle panel is the collective sequence obtained when the sorting window was positioned so as to select poor (pDF-e) IRES elements. IRES positions 299 to 302, which have been mutated to create pDF-e (see Materials and Methods), are shown in boldface; the sequence of pDF-e at these positions is 5′-AAAG-3′, while the sequence of pDF-E is 5′-GCGA-3′.
Screening of randomized library for IRES activity.
Having successfully established a method for selecting IRES elements in vivo, we constructed a library of plasmids, called pDF-lib, that contained a randomized sequence of 48 to 50 bases (see Materials and Methods) between the two fluorescent reporters (Fig. 1A). Protoplasts were made from pDF-lib and were screened as diagrammed in Fig. 2A. Three rounds of screening were performed, and increasing numbers of EGFP-expressing cells were observed in each succeeding round (data not shown). Plasmids recovered after the third round of selection were individually transfected into HEK293 cells and scored via fluorescence microscopy for EBFP and EGFP expression. As seen in Table 1, over 60% of the clones (97 out of 158 clones) recovered after three rounds of selection expressed significant levels of EGFP, indicating that successful enrichment of IRES activity had occurred. In comparison, none of the 142 clones assayed from the original randomized library, pDF-lib, expressed significant amounts of EGFP, although all clones expressed similar amounts of EBFP. Of the clones recovered after three rounds of selection that expressed significant levels of EGFP, five different sequences were found (Table 1); these sequences were named PS1 through PS5 and were subsequently analyzed by a variety of methods to determine whether they indeed encoded IRES activity.
TABLE 1.
Results after three rounds of screening of pDF-lib for IRES elementsa
| Sequence name | No. of times identified | Sequence |
|---|---|---|
| PS1 | 5 | 5′ CACAgTACgTAAgCTTAAgCTAAgCgTAgATAAgggTATATTTTTgCg 3′ |
| PS2 | 21 | 5′ gAAATAgCTATCCTCCATCACTgCACCgAgACTACggTTgCgCgTgTCgT 3′ |
| PS3 | 52 | 5′ TgACAAACTgTACATgCCgTTAACTgTAATTTTgCgTgATTTTTTTgTAg 3′ |
| PS4 | 7 | 5′ AggTggTAgCCgCAAACATAgTTCAATACAAACTTgCTgTCTCggCgg 3′ |
| PS5 | 12 | 5′ AggCAgTATAATCAgTTCCCACATAgAAAACCAggACTgTATCAAAgTgT 3′ |
After three rounds of selection, 158 individual clones were assayed by transfection. Of these, 97 were identified as a strong IRES element (i.e., expressed a significant level of EGFP). Five different sequences were found.
First, protoplasts made from E. coli containing PS1 through PS5 were fused to HEK293 cells and analyzed by flow cytometry for expression of EBFP and EGFP. Shown in Fig. 3 are the results of such an experiment, in which the fluorescence profiles of three positively selected clones are displayed along with the profiles of pDF-e and pDF-E. The mean green fluorescence of all five of the positively selected clones was equal to or higher than that of pDF-E and was 6 to 10 times higher than that of pDF-N or pDF-e, suggesting that the PS clones possess functional IRES elements that promote translation of the downstream cistron, EGFP. Some of the cells receiving plasmids containing strong IRES elements appeared to express EGFP but not EBFP (Fig. 3); this is most probably due to the fact that the intensity of EGFP fluorescence is approximately 30 times higher than that of EBFP (see Materials and Methods). Therefore, cells receiving low numbers of plasmids containing strong IRES elements would express enough EGFP to be detected by flow cytometry, while the intensity of EBFP fluorescence would not be detectable. Upon transfection of the pDF-ps plasmids into both HEK293 cells and Huh7 cells followed by flow cytometric analysis, the mean green fluorescence of the PS clones was also 6 to 10 times higher than that of pDF-N or pDF-e, indicating that the activity of these IRES elements is not restricted to a single cell type or to a particular method of plasmid delivery into mammalian cells.
Context dependence of selected IRES elements.
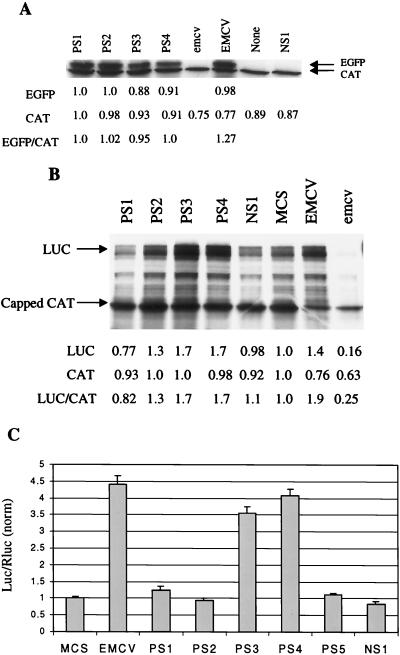
We next sought to determine whether the PS elements could function as IRES elements in different contexts, since it has been reported that the context of an IRES element can play a role in its activity (4, 17, 26). First, we eliminated the upstream cistron, EBFP, leaving the stem-loop structure upstream of the IRES element followed by EGFP. Uncapped mRNA was transcribed and translated in vitro, and the results are depicted in Fig. 4A. The four PS elements pictured, as well as PS5, all promoted levels of EGFP translation that were significantly higher than the virtually undetectable translation levels directed either by a randomized element that does not possess significant IRES activity (NS1) or by a construct in which there are no intervening nucleotides between the stem-loop and EGFP (Fig. 4A, None). In vitro-transcribed and 5′-capped monocistronic chloramphenicol acetyltransferase (CAT) RNA was added to each reaction mixture in order to compare translation directed by the various PS elements with cap-dependent translation. Quantitation of bands in Fig. 4A indicates that levels of EGFP translation directed by the PS elements are similar to that directed by the EMCV IRES variant. As has been previously noted (10, 11, 34), addition of the EMCV IRES to in vitro translation reactions inhibits capped mRNA translation, presumably by the ability of the J-K region to sequester cellular factors needed for translation; such an effect was also seen here with the mutant form of the EMCV IRES, which also contains the J-K region. Similar results to those seen in Fig. 4A were obtained in in vitro translation reactions where both the upstream cistron and the stem-loop were removed, leaving a monocistronic construct consisting of the IRES followed by EGFP (data not shown). Thus, it appears that the PS elements found in our bicistronic screen also function as IRES elements in a monocistronic context in the absence of upstream sequences.
FIG. 4.
(A) Effects of removal of upstream cistron on IRES activity of recovered elements. Two micrograms of uncapped monocistronic RNAs consisting of the stem-loop structure followed by either the 50-nt sequences (PS1-4, NS1), no intervening sequence (None), the EMCV IRES variant (EMCV), or its mutant form (emcv) was used in in vitro HeLa translation reactions. Each reaction mixture also contained 500 ng of capped CAT RNA for comparative purposes, and the results of densitometric analysis of bands representing translated proteins, normalized to levels of translation in the PS1 reaction, are shown below. Translation of EGFP directed by NS1, None, and emcv was undetectable by densitometric quantitation. (B) Demonstration of context independence of elements PS3 and PS4. Two micrograms of uncapped monocistronic RNAs, consisting of either the 50-nt sequences (PS1-4, NS1), the multiple cloning site from pDF-N (MCS), the EMCV IRES from pDF-E (EMCV), or the mutant EMCV IRES from pDF-e (emcv) upstream of firefly luciferase (Luc), was used in in vitro HeLa translation reactions. Each reaction mixture also contained 500 ng of capped CAT RNA, and densitometric analysis of translated proteins is shown below. (C) PS3 and PS4 possess context-independent IRES activity in vivo. The 50-nt elements, MCS, or EMCV IRES were placed in a bicistronic plasmid between Renilla luciferase (Rluc) and firefly luciferase (Luc). Twenty-four hours after transfection into HEK293 cells, cells were analyzed for Luc and Rluc activity. The Luc/Rluc ratios, normalized to that found in cells transfected with the MCS-containing plasmid, are shown. The experiment was performed two separate times in triplicate.
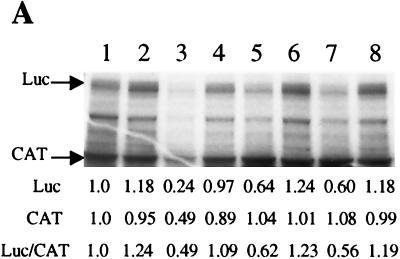
Next, we determined whether the nature of the downstream cistron affected the ability of the PS elements to direct IRES-mediated translation. Monocistronic RNA consisting of the IRES element followed by the firefly luciferase gene (Luc) rather than EGFP was prepared in vitro and translated in HeLa cell extracts. As shown in Fig. 4B, both PS1 and PS2 lost IRES activity when placed before Luc, as did PS5 (data not shown). On the other hand, elements PS3 and PS4 were still able to significantly direct IRES-mediated translation when placed before Luc, suggesting that these two elements function in vitro as IRES elements independent of the context in which they are placed. Because the bands corresponding to Luc were somewhat diffuse (Fig. 4B), we also quantitated Luc activity via a luminometer; similar results were obtained as with the densitometric quantification in Fig. 4B, and PS3 and PS4 were the only two selected elements to show significant (>1.5-fold, as compared to either NS1 or MCS) IRES activity (data not shown). Again, addition of the EMCV IRES repressed cap-dependent translation of CAT (compare Fig. 4B with A). We also wished to determine whether elements PS3 and PS4 can function in vivo as IRES elements in a context different from that in which they were isolated. Bicistronic plasmids consisting of the reporter genes Renilla luciferase (Rluc) and Luc separated by the PS elements were constructed and called pDL-ps. Upon transfection of these pDL-ps plasmids into HEK293 cells, PS3 and PS4 again showed levels of IRES activity comparable to that of the EMCV IRES, as assayed by the Luc/Rluc ratio (Fig. 4C). Thus, the selected elements PS3 and PS4 can promote IRES-mediated translation from bicistronic constructs in vivo and from monocistronic constructs in vitro in a context-independent manner.
Translation directed by PS3 and PS4 is similar to PV IRES-mediated translation.
To gain a preliminary understanding of the mechanism by which these 50-nt stretches of RNA direct IRES-mediated translation, we first sought to determine whether PS3 or PS4 utilize similar mechanisms as other viral IRES elements. By introducing these small RNAs into in vitro translation reactions programmed by different viral IRES elements, we found that PS3 and PS4 can compete significantly with PV but not HCV or EMCV IRES-mediated translation. Shown in Fig. 5A are the results of a competitive in vitro translation experiment in which Luc translation was directed by the PV IRES while CAT was made by cap-dependent mechanisms. Preincubation of elements PS3 and PS4 in these translation reaction mixtures specifically led to a decrease in PV IRES-mediated translation of Luc, while cap-dependent translation of CAT was not significantly affected (Fig. 5A, lanes 4 to 7). While a twofold molar excess of PS3 and PS4 did not reduce Luc translation significantly, addition of a fourfold molar excess of competitor RNAs reduced Luc translation significantly and reproducibly (Fig. 5A and data not shown). The reason for this nonlinear behavior of competitor RNAs in translation is not known at present. Addition of the poor IRES NS1 at fourfold molar excess (Fig. 5A, lane 8) did not affect either PV IRES-mediated or cap-dependent translation. It should be noted that preincubation with PV IRES as the competitor (Fig. 5A, lanes 2 and 3) led to a decrease in both PV IRES-directed Luc translation as well as cap-dependent translation. The precise reason for this is not known but may be due to the binding by PV IRES of general translation factors that are also required for cap-dependent translation.
FIG. 5.
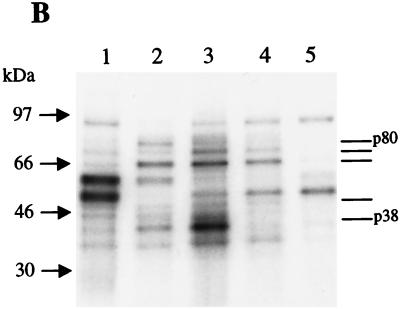
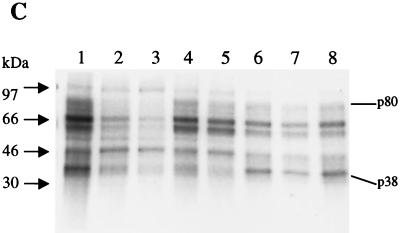
(A) IRES elements PS3 and PS4 compete with PV IRES-mediated translation in vitro. One microgram of a monocistronic construct containing the PV IRES upstream of Luc was translated in vitro in the absence (lane 1) or presence (lanes 2 to 8) of competitor RNAs. Competitor RNAs are PV IRES (lanes 2 and 3), PS3 (lanes 4 and 5), PS4 (lanes 6 and 7) and NS1 (lane 8). Either a twofold (lanes 2, 4, and 6) or fourfold (lanes 3, 5, 7, and 8) molar excess of competitor RNAs was used in each reaction mixture. Also, 250 ng of capped CAT RNA was added to each reaction mixture. (B) PS3 and PS4 exhibit a binding profile similar to that of PV IRES. 32P-labeled RNA (lane 1, EMCV IRES; lane 2, PV IRES; lane 3, PS3; lane 4, PS4; lane 5, NS1) was cross-linked to polypeptides in HeLa translation extract. Numbers to the left correspond to migration of marker proteins (in kilodaltons). Bars on right correspond to proteins of same apparent molecular mass that are bound by PV, PS3, and PS4. (C) Competitive UV cross-linking. 32P-labeled PV IRES was cross-linked to HeLa translation extract polypeptides in the absence (lane 1) or presence (lanes 2 to 8) of competitor RNAs. Competitor RNAs are PV IRES (lanes 2 and 3), PS3 (lanes 4 and 5), PS4 (lanes 6 and 7), and NS1 (lane 8) and were added at either a 100-fold (lanes 2, 4, and 6) or 250-fold (lanes 3, 5, 7, and 8) molar excess.
These observations raised the possibility that PS3 and PS4 may utilize some of the same trans-acting factors as PV for ribosome recruitment. Therefore, we analyzed the protein binding profiles of PS3 and PS4 by UV cross-linking these 32P-labeled RNAs to proteins in HeLa translation extract. As shown in Fig. 5B, PS3 and PS4 bind many proteins of the same apparent molecular mass as does the PV IRES. In particular, polypeptides of apparent molecular masses of 80, 75, 66, 50, and 35 kDa are bound by PS3, PS4, and PV IRES (Fig. 5B, lanes 2 to 4). Also, a protein of an apparent molecular mass of 38 kDa was bound very strongly by PS3; a protein of a similar apparent mass was bound by the PV IRES, too (Fig. 5B, lanes 2 and 3). The binding profile of a 50-nt RNA that does not promote significant IRES-mediated translation, NS1, was included for comparison. As can be seen in Fig. 5B (lane 5), there is no significant similarity between the protein binding profile of NS1 and those of PV IRES, PS3, and PS4. To determine whether the identities of some of the proteins bound by PS3 and PS4 are the same as those bound by the PV IRES, we performed a competitive UV-cross-linking assay in which unlabeled PS3 and PS4 were preincubated with HeLa extract before UV-cross-linking with 32P-labeled PV IRES RNA. As seen in Fig. 5C, PS3 and PS4 did indeed compete significantly for certain proteins that are bound by the PV IRES. In particular, PS3, which bound a 38-kDa protein very strongly (Fig. 5B), efficiently competed out p38 binding by PV IRES (Fig. 5C, lanes 4 and 5). PS4 appears to specifically compete for the binding of a wider range of PV IRES-bound proteins, including p80, p50, and to a lesser extent, p38 (Fig. 5C, lane 7). The nonspecific RNA (NS1) did compete with PV IRES binding to some extent (Fig. 5C, lane 8) at the highest concentration tested. Such competition of IRES binding by high concentrations of nonspecific RNAs has been noted previously by our laboratory and others (9). Overall, these data suggest that PS3 and PS4 may employ some of the same trans-acting factors as PV to efficiently direct IRES-mediated translation.
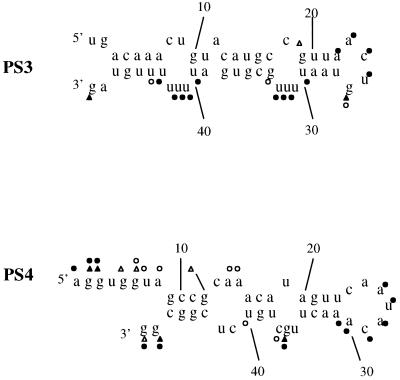
Determination of secondary structures of PS3 and PS4.
Close inspection of the sequences of PS3 and PS4 (see Table 1) revealed no significant homology to each other or to known viral or cellular IRES elements, except for the presence of a polypyrimidine tract at the 3′ end of PS3 (see Discussion). Also, no regions of potential complementarity between these IRES elements and 18S rRNA were noted (see Discussion), suggesting no obvious mechanism by which these elements can directly bind 18S rRNA through base-pairing interactions. Since it is hypothesized that RNA structure plays an important role in IRES activity, we decided to further characterize PS3 and PS4 by determining their secondary structures by using a combination of enzymatic cleavage analysis and free energy minimization modeling. Depicted in Fig. 6 are the proposed structures of PS3 and PS4 based on fitting the results of nuclease cleavage of the PS3 and PS4 RNAs with MFOLD-predicted secondary structures (45). The proposed structure of PS3 is the one predicted by the MFOLD algorithm to have the lowest free energy (−6.9 kcal/mol), while that of PS4 is the one predicted to have the second lowest free energy (−7.7 kcal/mol). The MFOLD-predicted structure for PS4 with the lowest free energy (−8.4 kcal/mol) was excluded, based on its nuclease cleavage pattern. For example, we observed strong RNase T1 cleavage of nt 36 and almost complete protection from cleavage of nt 44; these results are compatible with the structure of PS4 displayed in Fig. 6 but not with the lowest free energy structure (data not shown). No obvious secondary structural homology was noted between these two small IRES elements and other known viral IRES elements. It has been proposed that a number of cellular IRES elements contain a Y-type stem-loop motif that may be involved in their function (22). However, this motif is also absent in the secondary structures of PS3 and PS4. Therefore, it appears that we have selected for two independent synthetic IRES elements that promote translation in a PV-like fashion but which do not possess obvious sequence or structural homologies with known viral or cellular IRES elements.
FIG. 6.
Proposed secondary structures of PS3 and PS4, based on an enzymatic digestion map. Triangles represent nucleotides reactive to RNAse T1, and circles mark nucleotides reactive to nuclease S1. Solid symbols represent strong reactivities, while open symbols represent weak reactivities. RNAse V1, which we have previously used to identify base-paired regions of other RNAs (41), is not currently available to the scientific community and therefore was not used in this study.
DISCUSSION
In this study, we have developed a novel fluorescence-based screen to select IRES elements in vivo. By screening a library of plasmids containing randomized sequences placed between two fluorescent reporters, we have successfully identified 50-nt-long stretches of RNA that promote IRES activity. Two of these 50-nt RNAs promote IRES-mediated translation both in vitro and in vivo and appear to do so in a context-independent manner in multiple cell types. To our knowledge, this is the first screen conducted that has successfully identified synthetic IRES elements.
In our approach to finding synthetic IRES elements, we chose to randomize a 50-nt-long region for a variety of reasons. First, we wanted to ensure that our synthetic RNAs would be able to assume stable secondary structures, as it has been established that structure plays an important role in IRES activity (10, 12, 14). A stretch of 50 nt of RNA would be long enough to assume a variety of secondary structures that would not be accessible by smaller RNAs. In addition, we hoped that structures formed by 50 nt are stable enough to be maintained when the RNA is placed in different contexts; indeed, preliminary experiments have suggested that the structures of PS3 and PS4 remain unaltered when placed in different environments (data not shown), perhaps explaining their context independence. It is possible that PS1, PS2, and PS5 did not function as context-independent IRES elements because their secondary structures were altered when the downstream reporter was changed from EGFP to Luc; however, at this time we cannot distinguish this possibility from that of sequences within EGFP contributing to the IRES activity of these three elements. Also entering into our decision of the length of randomized nucleotides to use was that we did not want to randomize too large a stretch of RNA, as our ability to sample the possible combinations of RNA sequence, known as “sequence space” (3), would drop considerably. Here, we have screened approximately 106 different sequences, which is a very small percentage of the sequence space encompassed by a 50-nt randomized region. Although we had no expectations a priori as to the number of different IRES sequences we would identify, it is possible that the reason we only identified five positive IRES elements is that our coverage of sequence space was poor. Had we used a longer randomized region, our coverage would have been significantly poorer. However, it should be noted that the 106 different sequences screened, though covering a small percentage of sequence space, may have been able to assume quite a number of different structures, thereby allowing us to identify the five positive IRES elements in our screen.
Regardless of our ability to cover the sequence space encompassed by a 50-nt randomized region, the results of our study indicate that RNAs of this length do not commonly possess IRES activity. For instance, none of the 142 clones picked at random from our initial library displayed significant IRES activity. Using a Bayesian statistical approach with the unlikely initial assumption of a uniform prior distribution (i.e., an assumption that the chances of obtaining a strong IRES element are equal to the chances of obtaining a poor IRES element), our inability to find any strong IRES elements among the 142 clones sampled would indicate that the probability of a strong IRES element occurring in our randomized library is 1 in 143 (0.7%). The use of more realistic prior distributions, in which the chances of finding a poor IRES element are greater than the chances of finding a strong IRES element, would result in even lower predicted probabilities of a strong IRES element occurring in our library. Indeed, other evidence suggests that the frequency of a strong IRES element occurring in our randomized library is much lower than 0.7%. In the first round of our screen, where we analyzed approximately 2 × 106 protoplast-fused cells, we estimated that we would include in our sort about 10,000 false-positive cells (i.e., those cells that receive poor IRES elements) based upon our sorting window (data not shown). The fact that we obtained approximately the same number of sorted cells in the first round of our screen (8,741 cells) as the number of estimated false positives again indicates that the number of strong IRES elements in our library is very low and almost definitely comprises less than 0.7% of all the randomized elements. Thus, we have found that significant IRES activity is not a common property of RNAs that are on the order of 50 bases in length. Although it is tempting to speculate that one reason that most naturally occuring viral and cellular IRES elements are hundreds of nucleotides in length is because 50-base-long RNA stretches rarely tend to possess IRES activity, there is currently no evidence regarding the frequency of IRES activity in longer stretches of RNA.
Sequence and structural requirements of IRES elements are poorly understood. The IRESs of picornaviruses consist of approximately 450 nt that are highly structured and possess, at the 3′ end, a conserved UUUC motif followed by a polypyrimidine tract, a G-poor spacer, and an AUG triplet (1). None of the five IRES elements that we identified possess the UUUC motif, two have a polypyrimidine tract at the 3′ end, and none are notably lacking in G residues. Our synthetic IRES elements, besides lacking homology with viral IRES elements, also appear to have different cis requirements than cellular IRES elements. Recently, a 9-nt stretch of RNA from the mouse Gtx gene was found to possess IRES activity (8). Since this sequence was found to be exactly complementary to a segment of 18S rRNA, it was suggested that base-pairing interactions between the 9-nt RNA and the 18S rRNA were involved in recruitment of ribosomes to the IRES. Comparison of the sequences of our selected IRES elements to rRNA database sequences did not reveal any significant homology of stretches of 9 nt or longer. However, we cannot rule out the possibility that smaller stretches of RNA in our synthetic IRES elements mediate base-pairing interactions with rRNA. The identification of more synthetic IRES elements may allow us to determine whether small stretches of RNA complementary to rRNA occur in a statistically significant manner. Another proposed mechanism by which cellular IRES elements direct translation is through a Y-type stem-loop structure. Our structural analyses of IRES elements PS3 and PS4 indicate that no such structural motif is present; rather, the structures consist of alternating stems and bulges. In addition, alignment of the PS elements with GenBank sequences revealed no significant homologies.
Despite the absence of notable similarities between the PS elements and known sequences, our UV-cross-linking and in vitro translation studies suggest that PS3 and PS4 interact with cellular proteins that are also bound by the PV IRES. Certainly, these results are preliminary and additional careful experimentation with purified proteins must be conducted before conclusions regarding the nature of proteins involved in ribosome recruitment by the small IRESs can be made. An interesting question, however, is how a 50-nt RNA can be capable of interacting with a multitude of proteins normally bound by much longer (450 to 600 nt) viral IRES elements. The precise answer to this question is not known, but this phenomenon is not surprising to us, as we have previously isolated other small RNAs that bind to a multitude of IRES-binding proteins (9, 41). One possibility is that binding of functionally redundant proteins to different molecules of RNA (or small IRESs) may lead to the formation of distinct RNA-protein complexes capable of ribosome recruitment. Alternatively, protein-protein interaction in addition to RNA-protein interaction may ultimately lead to the binding of a multitude of proteins to a relatively small RNA. Thus, although PS3 and PS4 appear to mediate translation in a PV-like manner, elucidation of the precise mechanism by which they promote IRES-mediated translation awaits further experimentation.
While this work was in progress, the development of another selection system for IRES elements was reported (32). In that system, the reporter for IRES activity is a cell surface-expressed epitope whose expression is selected for by antibody-coated magnetic beads. Belsham and coworkers randomized a 4-base region of the EMCV IRES that is proposed to form a GNRA tetraloop and transfected a library of these 256 IRES elements into COS-7 cells. They were able to identify the sequence RNRA in that portion of the IRES as being optimal for translation activity. They found that, after three rounds of selection, a maximum of 10 to 20% of their selected clones contained strong IRES elements. Since, at a minimum, their initial library contained 16 positive elements (representing all combinations of the sequence RNRA) out of 256 total elements, they achieved, at most, a threefold increase (from 6 to 20%) in desirable IRES elements after three rounds of screening. We found that even with a far more complex library (over 3 orders of magnitude larger) and, as previously discussed, much rarer instances of positive IRES elements, 60 to 70% of our selected clones contained the desired IRES activity. A large degree of enrichment was also observed in our pilot experiment, where the presence of pDF-E was increased by more than 1,000-fold (Fig. 2B) after three rounds of screening. Our use of protoplast fusion, rather than transfection, to deliver the plasmid library into cells is at least in part responsible for the greater efficiency of our screen. Belsham and coworkers acknowledge that the high levels of background they observed may be due to the entry of many different plasmids into the same cell and that plasmids may adhere to the outside of cells during transfection. We found that significant enrichment of rare IRES elements occurred only when we introduced plasmids via protoplast fusion and not by transfection (see Results). Besides our use of protoplast fusion to deliver plasmids for screening, our use of fluorescent reporters also offers advantages during selection. For example, since we can simultaneously monitor both cap-dependent translation of EBFP and IRES-mediated translation of EGFP in individual cells by flow cytometry, we can ensure that selected cells do not contain plasmids with gene rearrangements or deletions that result in strong EGFP expression in the absence of EBFP expression. Additionally, we can readily choose and adjust the levels of IRES activity that we wish to select for by our choice of sorting windows. Although in this study we attempted to select for IRES elements whose activities were comparable to that of the EMCV IRES variant, it is conceivable that future studies using our system could yield a wide range of activities for IRES elements if based on different sorting gates.
Besides serving as tools for exploring the cis requirements of IRES elements and the trans-acting factors that they interact with, small IRESs may be of practical use. Currently, viral IRES elements are often used in gene therapy applications. Commonly, a retrovirus is engineered to contain a bicistronic construct encoding both a gene of interest that is translated via cap-dependent mechanisms and a reporter gene whose translation is directed by the viral IRES (23), ensuring that both proteins are expressed in infected cells. One limitation of this approach is that viruses used to deliver genes are constrained by the amount of RNA that they can efficiently package into a virion (25). We envision that our small synthetic IRES elements, which are at least 10 times smaller than the commonly used EMCV IRES element, may allow for the incorporation of larger genes or multiple (three or more) genes into a single retroviral vector. Our screening approach may have other applications as well. For instance, randomly mutagenized viral IRES elements may be screened for attenuated or strengthened IRES activity, providing insight into RNA determinants of viral translation. Additionally, libraries of 5′ UTRs of cellular mRNAs can be screened for IRES activity. Such an approach may complement that of Johannes, Sarnow, and coworkers (20) in rapidly identifying many cellular IRES elements.
ACKNOWLEDGMENTS
We thank Weimin Tsai for his superb technical help, Steve Landt for discussions regarding protoplast fusion, Stephen Smale for comments on the manuscript, and Thomas Belin for helpful discussions on Bayesian statistics. We also thank Iris Williams and Ingrid Schmid of the UCLA Core Flow Cytometry Laboratory.
This work was supported by NIH grant AI 45733 to A.D. A.V. was supported by NIH Medical Scientist Training Program grant NRSA/GM 08042-15 and the UCLA Microbial Pathogenesis Training Grant.
REFERENCES
- 1.Agol V. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D P, Szostak J W. Study of RNA-protein recognition by in vitro selection. In: Nagai K, Mattaj I W, editors. RNA-protein interactions. Oxford, United Kingdom: IRL Press; 1994. pp. 248–268. [Google Scholar]
- 4.Belsham G. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1106–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belsham G J, Sonenberg N. Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol. 2000;8:330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 6.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chappell S A, Edelman G M, Mauro V P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das S, Coward P, Dasgupta A. A small yeast RNA selectively inhibits internal initiation of translation programmed by poliovirus RNA: specific interaction with cellular proteins that bind to the viral 5′-untranslated region. J Virol. 1994;68:7200–7211. doi: 10.1128/jvi.68.11.7200-7211.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duke G M, Hoffman M A, Palmenberg A C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evstafieva A G, Ugarova T Y, Chernov B K, Shatsky I N. A complex RNA sequence determines the internal initiation of encephalomyocarditis virus RNA translation. Nucleic Acids Res. 1991;19:665–671. doi: 10.1093/nar/19.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haller A A, Semler B L. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J Virol. 1992;66:5313–5319. doi: 10.1128/jvi.66.8.5075-5086.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman M A, Palmenberg A C. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honda M, Beard M R, Ping L-H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huez I, Creancier L, Audigier S, Gensac M-C, Prats A-C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt S L, Hsuan J J, Totty N, Jackson R J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt S L, Kaminski A, Jackson R J. The influence of viral coding sequences on the efficiency of internal initiation of translation of cardiovirus RNAs. Virology. 1993;197:801–807. doi: 10.1006/viro.1993.1661. [DOI] [PubMed] [Google Scholar]
- 18.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 19.Jang S K, Krausslich H G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le S-Y, Maizel J V., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Salas E. Internal ribosome entry site biology and its use in expression vectors. Curr Opin Biotechnol. 1999;10:458–464. doi: 10.1016/s0958-1669(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 24.Negulescu D, Leong L E-C, Chandy K G, Semler B L, Gutman G A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 25.Nolan G P, Shatzman A R. Expression vectors and delivery systems. Curr Opin Biotechnol. 1999;9:447–450. doi: 10.1016/s0958-1669(98)80027-x. [DOI] [PubMed] [Google Scholar]
- 26.Ohlmann T, Jackson R J. The properties of chimeric picornavirus IRESes show that discrimination between internal translation initation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA. 1999;5:764–778. doi: 10.1017/s1355838299982158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 28.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of hepatitis C virus and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 477–522. [Google Scholar]
- 31.Roberts L O, Belsham G J. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the coexpression of fragments of the IRES. Virology. 1997;227:53–62. doi: 10.1006/viro.1996.8312. [DOI] [PubMed] [Google Scholar]
- 32.Robertson M E M, Seamons R A, Belsham G J. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandri-Goldin R M, Goldin A L, Levine M, Glorioso J C. High-frequency transfer of cloned herpes simplex virus type 1 sequences to mammalian cells by protoplast fusion. Mol Cell Biol. 1981;1:743–752. doi: 10.1128/mcb.1.8.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheper G C, Thomas A A, Voorma H O. The 5′ untranslated region of encephalomyocarditis virus contains a sequence for very efficient binding of eukaryotic initiation factor eIF-2/2B. Biochim Biophys Acta. 1991;1089:220–226. doi: 10.1016/0167-4781(91)90011-a. [DOI] [PubMed] [Google Scholar]
- 35.Stern S, Moazed D, Noller H. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 36.Stoneley M, Paulin F E M, Quesne J P C L, Chappell S A, Willis A E. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 37.Tan R, Frankel A D. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Proc Natl Acad Sci USA. 1998;95:4247–4252. doi: 10.1073/pnas.95.8.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuerk C. Using the SELEX combinatorial chemistry process to find high affinity nucleic acid ligands to target molecules. Methods Mol Biol. 1997;67:219–230. doi: 10.1385/0-89603-483-6:219. [DOI] [PubMed] [Google Scholar]
- 40.Unrau P J, Bartel D P. RNA-catalysed nucleotide synthesis. Nature. 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- 41.Venkatesan A, Das S, Dasgupta A. Structure and function of a small RNA that selectively inhibits internal ribosome entry site-mediated translation. Nucleic Acids Res. 1999;27:562–572. doi: 10.1093/nar/27.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang T T, Sinai P, Green G, Kitts P A, Chen Y T, Lybarger L, Chervenak R, Patterson G H, Piston D W, Kain S R. Improved fluorescence and dual color detection with enhanced blue and green variants of the green fluorescent protein. J Biol Chem. 1998;273:8212–8216. doi: 10.1074/jbc.273.14.8212. [DOI] [PubMed] [Google Scholar]
- 45.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]