Summary
The green alga Chlamydomonas proliferates by ‘multiple fission’: a long G1 with >10-fold cell growth followed by multiple rapid divisions. Cells above a critical size threshold are ‘committed’ and will divide independent of light and further cell growth. The number of divisions carried out depends on the initial size of the committed mother cell. Here, I show that CDKA1, the ortholog of the yeast/animal mitotic inducer CDK1, regulates the critical size for commitment. The Rb/E2F/Dp1 pathway regulates division number as well as commitment size. Epistasis analysis indicated that CDKA1 and Rb/E2F/Dp1 regulate multiple fission by distinct mechanisms. Rb-E2F/Dp1 regulates G1/S gene expression in animals and land plants. Transcriptome analysis showed that mat3 or dp1 disruption altered regulation of a large group of cell division-associated genes with respect to cell size, but not with respect to synchronization timing. In contrast, cdka1 inactivation disturbed both temporal and cell size regulation of expression. These defects were enhanced by double inactivation of cdka1 and dp1, suggesting interaction between CDKA1 and the Rb-E2F/Dp1 pathways in regulating cell-cycle-specific gene expression and cell cycle initiation. In the context of a theoretical model for regulation of Chlamydomonas multiple fission, these results suggest that CDKA1 may promote a switch into a division-competent state, and E2F/Dp1 may promote maintenance of this state.
eTOC Blurb
Cross reports that in Chlamydomonas, the CDK1 ortholog CDKA1 controls commitment to entry into multiple fission, the earliest regulatory step in the cell cycle. CDKA1 also regulates induction of the mitotic transcriptional regulon. E2F/Dp1 also may control commitment and mitotic transcription, but via a distinct pathway from CDKA1.
Graphical Abstract

Introduction
In Chlamydomonas, cell growth is dramatically separated from cell division. After a long G1 growth period, Chlamydomonas undergoes multiple fission; rapid and synchronous divisions, with all progeny remaining within the mother cell wall. After divisions are complete, cells regrow flagellae and hatch.
Newborn cells cannot divide without a brief period of photoautotrophic growth [1]. After this period, cells are ‘committed’ to divide even if transferred to dark, although division follows commitment by many hours. Longer light exposure supports more divisions upon dark transfer. Two size controls, a minimal mother size to carry out any divisions, and a ‘target’ daughter size that controls the number of divisions carried out, may keep final daughter cell size within a two-fold range independent of initial mother cell size. This process can be coupled to the diurnal cycle, with photosynthetic growth during daylight, and divisions restricted to dark when cell growth ceases [1].
Chlamydomonas MAT3, a homolog of animal retinoblastoma protein (Rb), negatively regulates commitment and division number [2] by inhibiting the E2F1/DP1 transcription factor complex [3][4]. E2F1/DP1 are orthologs of the primary animal Rb target [6]. The cyclin-dependent kinase CDKG1, activated by D-type cyclins, may inhibit MAT3 [5]. In Chlamydomonas, unlike in other systems, the Rb/E2F/Dp circuit may function independently of transcriptional regulation [3]. MAT3 did not dissociate from E2F1/DP1 [4], unlike animal Rb [6]). Animal Rb and related proteins regulate transcription by multiple pathways [6] in addition to direct regulation of E2F-Dp. However, epistasis of e2f1/dp1 to mat3 [3] for the mat3 small cell-size phenotype argues against additional MAT3 pathways in Chlamydomonas.
In yeast and animals, CDK1 is required for mitosis. However, the plant kingdom ortholog CDKA1 is dispensable for mitosis, and instead regulates timing of cell cycle initiation, in both Chlamydomonas and the land plant Arabidopsis [7,8][9]. Arabidopsis CDKA may inactivate Rb, promoting transcription of many genes including CDKB [9], a plant- kingdom-specific CDK required for mitosis [7,8,10].
Chlamydomonas CDKA1 is required for timely and efficient induction of the mitotic regulon, a large group of genes including most genes required for cell cycle progression 15 [11]. How CDKA1-dependent promotion of cell cycle entry and mitotic transcription relates to commitment is unknown, as is the relationship between CDKA1 and the MAT3-E2F/DP1 regulatory circuit. Here I constructed single and multiple mutants in cdka1, mat3 and dp1 and quantified both commitment and induction of the mitotic transcriptional regulon, to determine dependencies of CDKA1 and MAT3-E2F/DP1 in regulating these key features of cell division control in Chlamydomonas.
Results
Growth and division in WT and cdka1 mutants.
cdka1 mutant cells exhibit a long delay before division [7,8]; however, overall culture doubling time is near-WT. To understand how the cdka1 division cycle compensates for the long G1, I carried out DNA flow cytometry analysis in cultures of WT and cdka1 synchronized by nitrogen deprivation/refeeding, and examined correlated cell size and DNA content by flow cytometry (Figure 1; compare WT and cdka1).
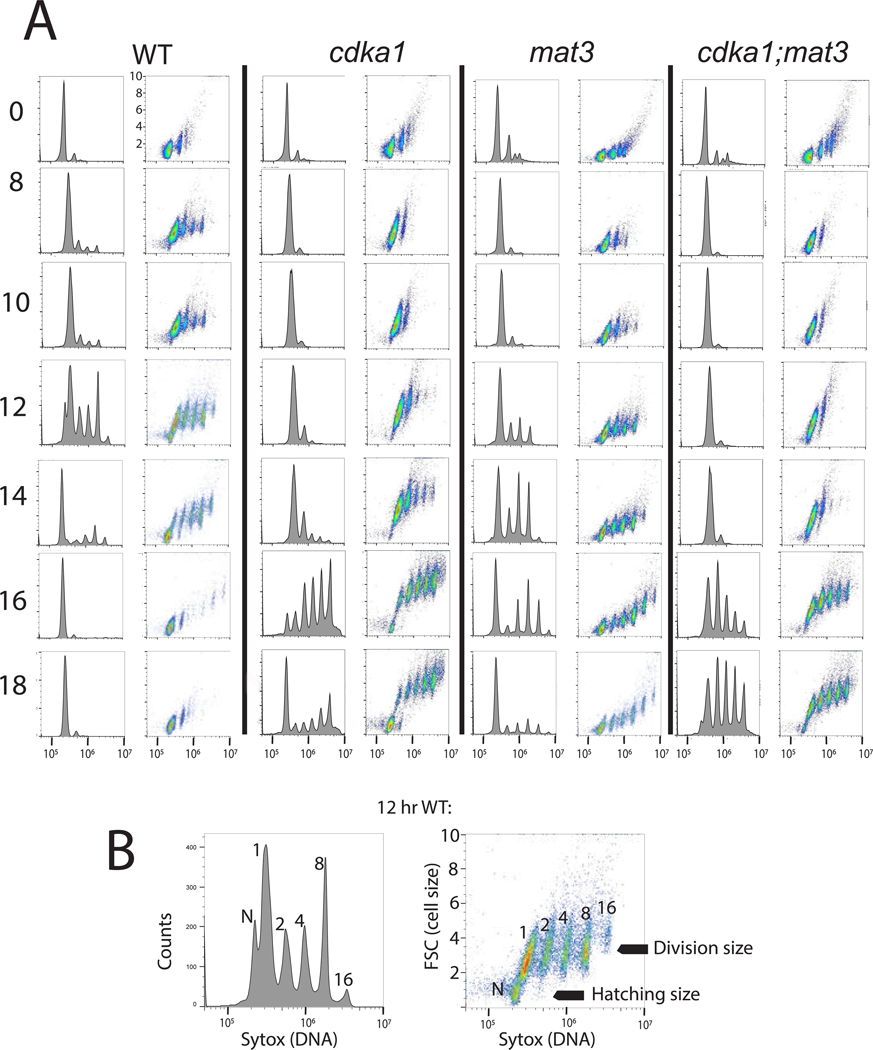
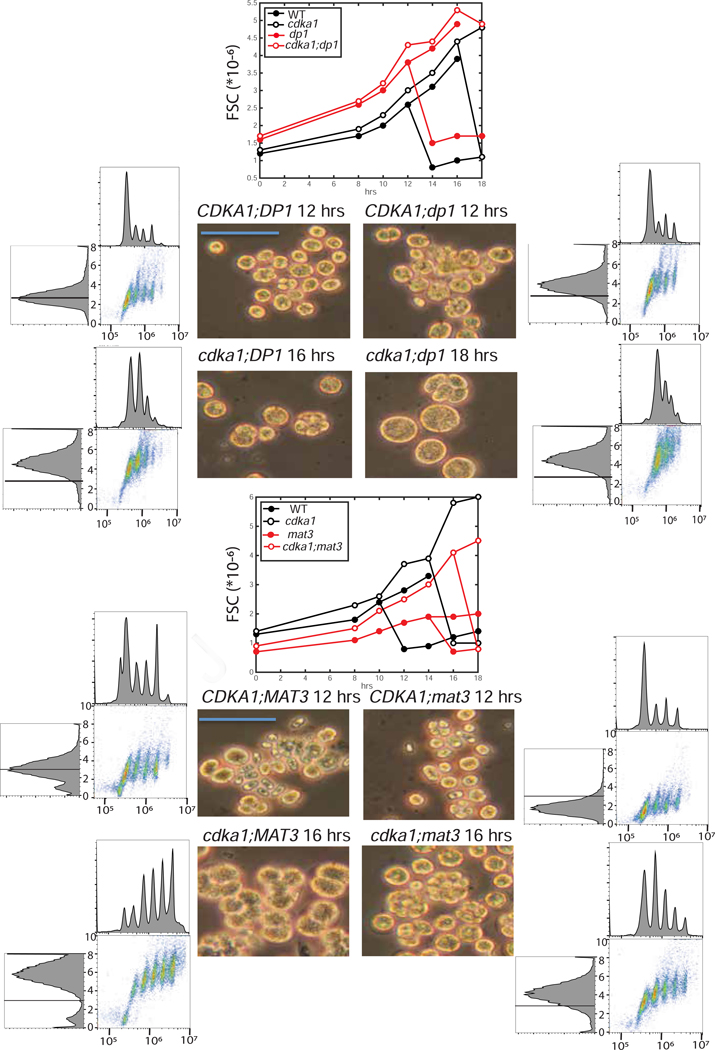
Figure 1. Flow cytometric analysis of WT and cdka1 cell cycles, and interaction of cdka1 and mat3.
A. Strains were synchronized by nitrogen deprivation/refeeding, and analyzed by flow cytometry. Hours after release on left. Histograms: DNA (Sytox; log scale). Scatterplots: DNA (log scale, x) vs. forward scatter (FSC, linear scale; units * 10^−6, y). All Sytox and FSC plots have the same axis values indicated by hatch marks. B: subpopulations from the 12 hr WT sample: 1,2,4,8: large cells after 0,1,2,3 rounds of synchronous DNA replication; N: newly hatched, with low FSC (small size) and slightly lower DNA signal than the large 1C cells due to large-cell background. Divisions are highly synchronous and multiple-fission daughters stay in the mother cell wall until hatching, so DNA peaks at 1,2,…2^n are observed (Chlamydomonas FACS analysis discussed in STAR Methods). Figure S1: similar experiment showing interaction of cdka1 and dp1; Tables S1,S3: quantified cell sizes.
Since all descendants of a mother cell undergoing multiple fission stay within the mother cell wall before hatching, multiple cycles of replication can be scored by the total ploidy of the mother cell (Figure 1, bottom; see STAR Methods). During their long G1, cdka1 cells grew to large size. However, they ultimately underwent as many or more cycles of DNA replication and division as WT (Figure 1, Table S1). Newly hatched cdka1 cells are only slightly larger than WT, but due to their long G1, cdka1 2C cells are much larger than WT 2C (Table S1). Thus cdka1 cells may carry out an additional division to attain the final newborn size, compensating for the longer multiple-fission cycle time, since more daughters are likely produced per cycle.
Thus, disrupting CDKA1 strongly delays transition from G1, raising the possibility of CDKA1 involvement in the ‘commitment’ regulatory step (Introduction). Once dividing, cdka1 cells have only minor delays [8], and may undergo even more divisions that WT.
Single-cell imaging of commitment.
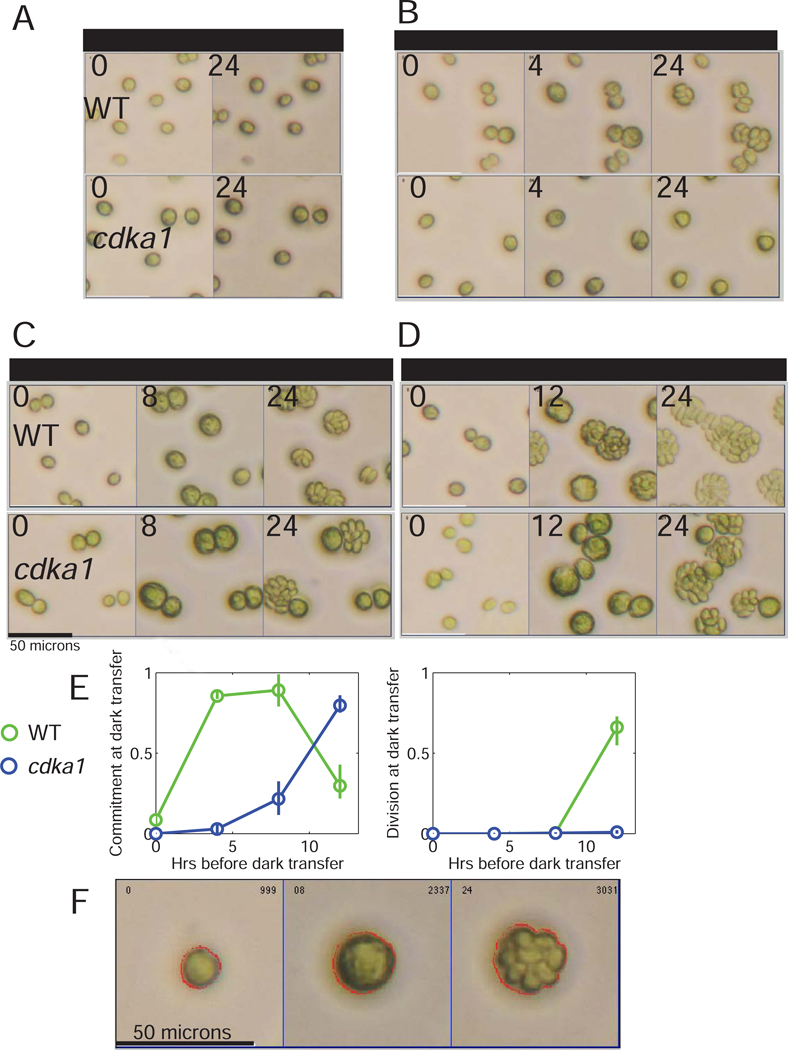
Timelapse microscopy allows correlation of commitment to division and cell size in individual cells [12]. Light-grown cultures are incubated on photoautotrophic medium in the dark, to allow completion of divisions. Daughter cells are then plated on solid medium and allowed varying periods of light incubation before transfer to dark. Fields of cells are imaged at plating, dark transfer, and after 24–30 hrs when divisions are complete (Figure 2A–D). From the micrographs, cells are sized by automated segmentation, and number of divisions estimated manually (Figure 2F).
Figure 2. CDKA1 regulates commitment timing.
A-D. Light-grown cells were incubated in dark for 18 hrs, then after varying times of light incubation transferred back to dark. Brightfield images were taken at plating, at dark transfer, and at 24 hrs. (E) Proportion of cells committed (undivided at dark transfer, divided at 24 hr; left) or divided at dark transfer (right). WT and cdka1 strains (2 each) were tested in parallel. Error bars: spread between genotype replicates. F: Example of segmentation of cell boundaries (red lines) for one cell incubated in light for 8 hrs, which divided 3 times after dark transfer. Table S2: Segmented cell sizes after first dark incubation.
Most dark-starved WT cells transferred directly to dark failed to divide, but if first incubated in light for ~4 hrs, most cells divided once or twice after dark transfer (2A,B). With longer light incubation, both the proportion of dividing cells and the number of divisions increased (Figure 2A–D) as expected (Introduction).
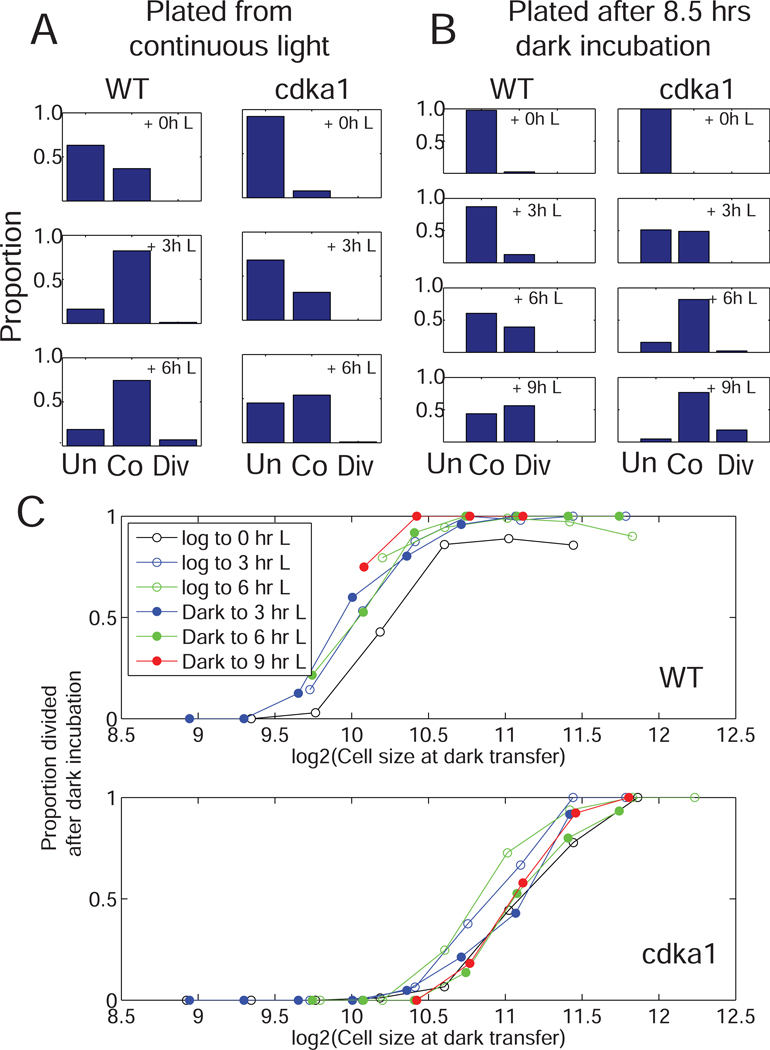
Cultures grown in continuous light had more committed cells than cultures incubated in dark before plating; for both, increasing light incubations after plating increased the proportion of committed cells (Figure 3A,B). The proportion of committed cells was sharply dependent on cell size at the time of dark transfer, with similar curves for all conditions of light pre-incubation (Figure 3C, top). This suggests that the major effect of time in light is to allow sufficient cell growth to pass the commitment size threshold. Consistently, primacy of cell size or growth rate, rather than time, in regulating commitment, was concluded from measuring commitment under widely varying growth conditions [13,14].
Figure 3. CDKA1 regulates cell size dependence of commitment.
Photoautotrophic cultures of WT and cdka1 were plated directly from light (A) or after 8.5 hrs dark incubation (B). Plates received the indicated periods of light before transfer to dark. Images were taken at plating, dark transfer and 30 hrs after plating. ‘Unco’ (uncommitted): no division at 30 hrs; ‘Co’ (committed): no division at time of dark transfer, but division at 30 hrs; ‘Div’ (divided): divided at time of dark transfer. C. Cell sizes at dark transfer were determined by automated segmentation, and cells sorted into 0.25-log2 size bins. Proportions of binned cells that divided after dark incubation was determined (pooled data for two strains of each genotype).
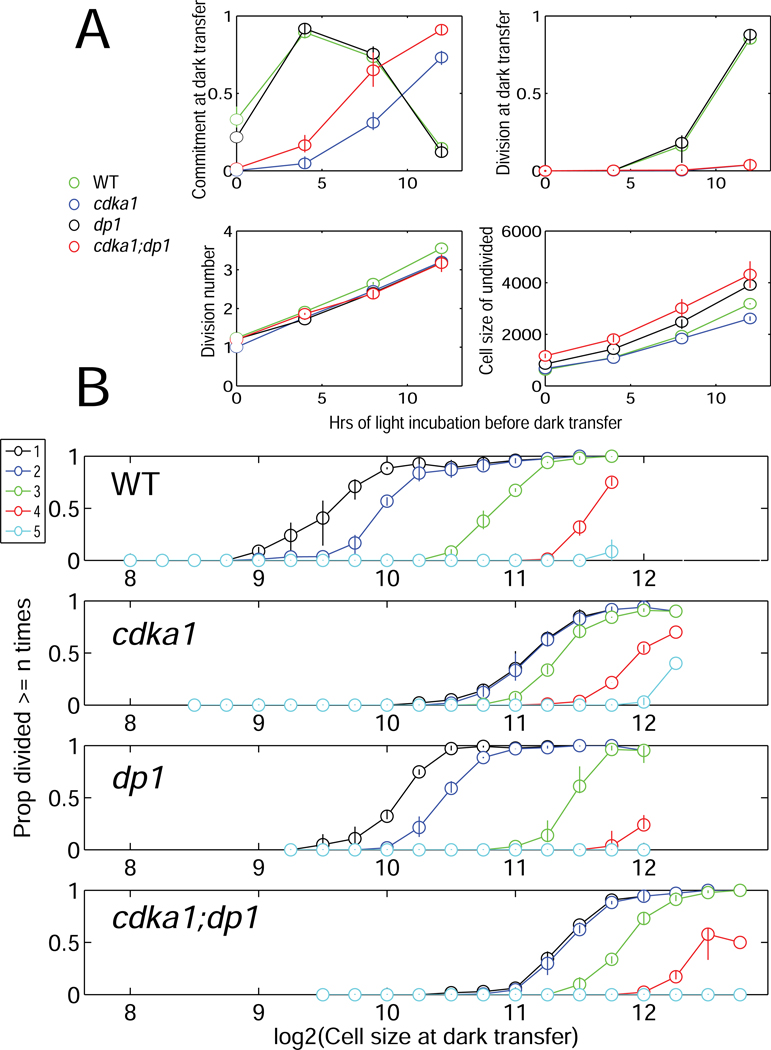
Plotting the proportion of cells dividing at least 1,2,... times as a function of size at dark transfer showed a strong effect of cell size at dark transfer on probability of increasing numbers of divisions, independent of time of light incubation (Figure 4).
Figure 4. Synergism of dp1 and cdka1 inactivation.
A. Kinetics of commitment, division and cell growth in strains of the indicated genotype. B. Cells were binned by size as in Figure 3. Proportions of cells in each bin dividing ≥1, ≥2,..≥5 times (color key, upper left) were determined, pooling all timepoints. Error bars: spread between two genotype replicates. Statistical significance of genotype differences is demonstrated by an independent repeat with a different set of strains, with very similar results (data not shown) and by Monte Carlo simulation (Figure S2A). Figure S2B–E shows corresponding analysis with mat3 and with cyca;d2;d3 rather than dp1. Table S2: Measured cell sizes after first dark incubation.
CDKA1 sets the critical size for commitment.
cdka1 null cells exhibited a long delay in commitment (Figures 2,3,4). The delay (~4–6 hrs) is similar to the delay in division itself [8]. Consistently, continuous-light cdka1 cultures had a much lower proportion of committed cells than WT, implying a longer pre-commitment interval (Figure 3A, top left).
Dark-incubated cdka1 cells were moderately larger than WT (Table S2), and grew similarly to WT in the light (Figures 1,2,4A). As with WT, cell size at dark transfer was a strong predictor of commitment and ultimate cdka1 division. However, the threshold size was about two-fold larger (Figure 3). Since cdka1 cells started at near-WT size, and grew at a similar rate, this shift in critical size accounts for their delayed commitment.
Size-by-division number plots (Figures 4B, S2A,B) show that cdka1 mutant cells had a much higher size requirement than WT for dividing at least once or twice, but are similar to WT in the relationship between size and probability of dividing ≥3 times. Thus cdka1 cells have a similar relationship of cell size to multiple division probability as WT once committed: if small-to-medium, they will not divide at all; but if large they will divide multiple times (see Figure 2).
A dynamical systems framework for Chlamydomonas [12] suggests a hysteretic bistable system switching between states permissive or non-permissive for cell division. A high critical cell size leads to divisions; with sufficient sequential divisions cells reach a lower critical size and switch back to the non-dividing state. In this framework, CDKA1 may be required to set the higher cell size threshold controlling transition between non-dividing and dividing states.
Test of potential cyclin activators of CDKA1 for commitment.
Disruption of CYCA1, encoding the sole Chlamydomonas cyclin A, eliminated in vitro histone H1 protein kinase activity co-immunoprecipitated with CDKA1 [8]. However, deletion of cyclin A had little effect on kinetics or cell size control of commitment (Figure S2C–E). Either CDKA1 has other cyclin activators driving commitment (that do not score in the in vitro kinase assay), or CDKA1 has a cyclin-independent role. CYCB1, the single B-type cyclin, is an unlikely candidate since in Chlamydomonas it appears specialized for activation of CDKB1 [8]. Chlamydomonas has other cyclins: 4 D-type, an SDS-type and a hybrid A/B [1]. I tested a triple cyca1;cycd2;cycd3 strain using candidate cycd2 and cycd3 null alleles [10]. This strain exhibited increased cell size at commitment (Figure S2C–E), but kinetics were distinct from cdka1, resembling results of inactivation of DP1; see below). Thus cyclin activator(s) for CDKA1 controlling commitment cannot yet be identified.
CDKA1 and Rb/E2F/Dp1 act in distinct but overlapping pathways to regulate multiple fission.
MAT3 and its target E2F1/DP1 regulate both commitment size and number of divisions of committed cells [2,3]. A dp1 null mutant was similar to WT in kinetics of commitment (Figure 4A). (dp1 was used because available E2F1 alleles are thought to be dominant-negative, complicating interpretation [3]). However, at a given cell size, dp1 mutants carried out fewer divisions than WT (Figure 4B).
mat3 disruption also yielded roughly similar kinetics of commitment to WT, but exhibited more divisions from a given size. mat3 cells grew more slowly than WT, possibly slowing commitment at later times (Figure S2). Consistently, mat3 disruption reduced daughter cell size (especially following gametogenesis) and dp1 disruption increased it (Tables S1–S3). These findings confirm previous results [2][3].
In Arabidopsis, it was proposed that CDKA1 might primarily inhibit Rb [9], thereby activating E2F/DP. If this is so in Chlamydomonas, then cdka1;mat3 and cdka1;dp1 double mutants should resemble mat3 and dp1 respectively. Alternatively, E2F/DP1 might function primarily to activate CDKA1. (For example, in animals, E2F/DP1 promotes transcription of cyclin E, which activates Cdk2 [6].) If so, cdka1;mat3 and cdka1;dp1 should both resemble cdka1.
cdka1;dp1 and cdka;mat3 cells delayed commitment similarly to cdka1 (Figure 4A,S2). However, cdka1;dp1 cells committed to divisions at significantly larger cell sizes than either single mutant (Figures 4A, S2A,C). Conversely, cdka1;mat3 cells were moderately shifted to smaller cell size for a given division number compared to cdka1 (Figure S2B,C), and are also intermediate between the single mutants in other assays (Tables S1–3; Figure 1 [discussed below]).
These results argue against linear pathways with CDKA1 upstream or downstream of MAT3-E2F/DP1. Mutations in the MAT3-E2F/DP1 pathway affected number of divisions as a function of cell size [2,3,5] (see Introduction), and this was confirmed in the single-cell commitment assays above. MAT3 and E2F/DP1 also affect commitment size [2,3,5], though this effect is quantitatively smaller. In contrast, cdka1 has a much stronger effect on commitment size, while large committed cdka1 cells undergo similar numbers of divisions to WT. Double mutant phenotypes are nearly additive. For example, the suppression of low division number characteristic of cdka1 is observed in cdka1;dp1 (overlap of cumulative ≥1 and ≥2 division graphs); and the shift of size for a given number of divisions characteristic of dp1 is seen in cdka1;dp1 compared to cdka1 (Figure 4B, S2A,C).
Thus, CDKA1 and the MAT3-E2F/DP1 pathway influence cell size and commitment in distinct ways. Condensing the size relationships for given final division numbers (Figure S2E) shows that cdka1 disruption increases the size requirement only for sizes at which WT will undergo 1–2 divisions; at higher division numbers cdka1 and WT largely overlap. In contrast, dp1 and mat3 disruptions shift all thresholds, to larger and smaller sizes respectively.
cyca1;cycd2;cycd3 resembles dp1 rather than cdka1 in these kinetics (Figure S2C–E). This is consistent with activation of CDKG1 by D-type cyclins [5], since CDKG1-CYCD might be required to inactivate MAT3 and thereby activate DP1.
CDKA1 may overlap with the MAT3-E2F/Dp1 pathway in activating transcription of the mitotic regulon
Rb-E2F/Dp1 regulates cell-cycle-periodic gene expression in animals [6], but this has been questioned in Chlamydomonas [3]. I carried out three RNAseq experiments with synchronized cultures. The experiments compared: (1) WT, cdka1, dp1 and cdka1;dp1; (2) WT, cdka1, mat3 and cdka1;mat3; (3) WT, cdka1, dp1, cdka1;dp1, cdka1;mat3 and cdka1;mat3;dp1. Samples were every 2 hrs from 8–18 hrs (1, 2), or every 2.5 hrs from 10–17.5 hrs (3). Each experiment employed different congenic isolates from mating and tetrad analysis.
Synchronization was by nitrogen deprivation/refeeding. Flow cytometry analysis of cdka1 vs. WT in this procedure (Figures 1 and S1, left 2 columns; discussed above) showed delay of replication to larger cell sizes, followed by multiple rounds of efficient replication. Perturbations of cell size and division number control in mat3 and dp1 (Figures 1,S1,5, Table S1) were consistent with commitment assays and previous results [2,3,5]: dp1 and mat3 mutants initiated division cycles at times similar to WT, but at greater and lesser cell sizes, yielding larger and smaller daughter cells, respectively. Additional disruption of cdka1 superimposed a large delay in DNA replication and division, with attendant increase in the size of cells at division (Figures 1,S1,5; Table S1). As with the commitment assays (Figures 4, S2), cdka1;mat3 and cdka1;dp1 double mutants had phenotypes distinct from the single mutants, suggesting distinct CDKA1 and MAT3-E2F/DP1 pathways.
Figure 5. Synchronization of cdka1, mat3, dp1 mutants.
Single, double and triple mutants between cdka1, mat3 and dp1 were synchronized (FACS analysis in Figures 1,S1). Approximate modal FSC values were estimated and plotted for two experiments (center line graphs). Graph bifurcation is from appearance of a newborn daughter cluster. Brightfield images and flow cytometry profiles at an early timepoint with significant DNA replication (Figures 1,S1) are shown. Blue bars: 50 microns. Top: cdka1 x dp1 (experiment 1); bottom: cdka1 x mat3 (experiment 2). WT peak is marked on all FSC histograms (left). Key size values for the three experiments are reported in Table S1. Table S3 contains daughter cell sizes prepared by a different protocol for comparison.
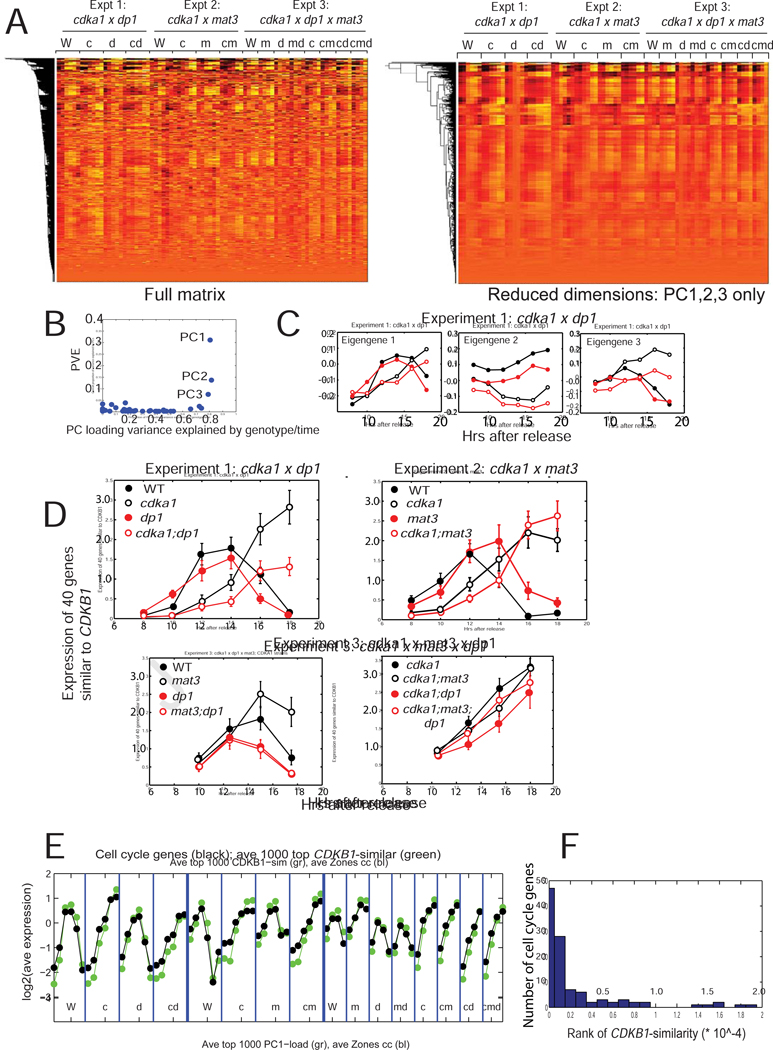
After RNA extraction, sequencing and alignment, hierarchical clustering of the log2-transformed readcounts (Figure 6A, left) showed similar variation for many genes (common pattern extending down columns). Principal components (PC) were determined. The first three PCs, accounting for 54% of variation, gave similar results for replicates of genotype/time in different experiments (Figure 6B). Higher PCs, each accounting for minor variation, were more variable between replicates (Figures 6A,B). A dimension-reduced dataset with only contributions from PC1,2,3 has a pattern similar to the complete dataset (Figure 6A, right). PCs 1–3 were similar when calculated from random subsets of 10% of genes (data not shown). Therefore these PCs are likely due to large numbers of genes following very similar patterns of expression (Figures 6A, S3).
Figure 6. Transcriptomics.
RNA was extracted from synchronous cultures (Figures 1, S1, 5), sequenced and aligned. A. Left: Clustergram of the log2-transformed read-count matrix. Genotype abbreviations: W: WT; c: cdka1; m: mat3; d: dp1; multiple mutants indicated by multiple letters. Right: clustergram of the dimension-reduced matrix (only PC1–3). B. Proportion of overall variance explained against proportion of variation in loadings due to reproducible differences between genotypes/timepoints. C. Loadings for PCs1–3 were separated out for experiment 1. D. Pattern of expression of the mitotic regulon. The 40 genes closest to CDKB1 (canonical regulon member [16][11]) in the log2-transformed matrix were extracted; mean and standard deviation of their expression in the three experiments is shown. Note similarity of eigengene 1 (Figure 6B) to this pattern. Figure S3 presents additional statistical analysis. E. From ref. [16],108 curated cell-cycle-specific genes, most of which show strong periodic expression, were extracted, averaged and plotted together with the 1000 genes most similar to CDKB1 across the experiments. F: Histogram of the rank order of the 108 cell cycle genes for CDKB1 similarity. The random expectation is for flat representation across the range, so the clustering to low ranks demonstrates very strong over-representation of cell-cycle-annotated genes. Similar results were obtained indexing by loading of eigengene 1 (data not shown). Figure S3 contains supporting statistical analysis. Raw readcount data in supplemental Data S1.
Eigengene 1 (corresponding to PC1) showed strong induction on entering division cycles, and a drop upon completion of divisions (Figure 6C). This is the pattern of the ‘mitotic regulon’, which includes most genes involved in cell cycle control, DNA replication and mitosis [11,15,16]. I collected the 40 genes most similar in expression to CDKB1, a canonical member of the mitotic regulon. Expression of this cluster in the three experiments is highly similar to eigengene 1 (compare Figure 6D, experiment 1, to Figure 6C). Although the pattern of eigengenes 2 and 3 differed from the mitotic regulon (Figure 6C), nevertheless expression of genes with high loading of each of eigengenes 1–3 was strongly correlated with the mitotic regulon (Figures 6, S3A). Eigengene 1 loading and CDKB1 similarity identified a common expression pattern ramifying through >5000 transcripts (of 19526 total), though the pattern had decreasing amplitude as eigengene 1 loading or similarity to CDKB1 decreased (Figure S3B,C). These genes are strongly enriched for cell cycle-related annotations (Fig 6E and data not shown).
Time of expression of this large battery of genes (and eigengene 1) was highly responsive to CDKA1 genotype; almost independent of mat3 and dp1, expression was delayed in cdka1 strains for ~4 hours, comparable to the delay in time to commitment, DNA replication and cell division (Figures 1, S1, 6D, S3).
Eigengenes 2 and 3 showed more complex responsiveness to CDKA1, MAT3 and DP1 genotype (Figure 6C; data not shown). MAT3 and DP1 inactivation modestly affected expression of the mitotic regulon (Figure 6D), increasing and lowering expression respectively.
Thus disruption of the MAT3-E2F/DP1 system has little effect on timing of gene expression, confirming previous results [3]. However, the mutants exhibit a strong gene expression phenotype with respect to cell size: the mitotic regulon is induced at a large cell size compared to WT in dp1, and at a small size in mat3 (Figure 6, S3 [3]).
The cdka1 mutant exhibits misregulation with respect to both time and cell size (Figures 1,2,3,4). Deletion of dp1 in the cdka1 background further decreased the rate and/or level of induction of the S/M regulon, and cdka1;dp1 cells grew larger than cdka1 cells without dividing (Figures 4,S1,Table S1). As with commitment assays, the lack of epistasis in these transcriptional effects suggests that CDKA1 and E2F/Dp1 collaborate to regulate these genes by distinct pathways.
Discussion
The mitotic regulon, cell size, and G1 timing
The mitotic regulon is under tight control: very low during the long G1, and rising sharply as cells begin division cycles [16][11]. Mutations in the E2F/DP1 pathway specifically affect cell size at which the regulon is induced with little effect on timing or expression level in synchronization experiments [3]; Figures 6, S3). Cells mutated in this pathway have smaller (mat3) or bigger (dp1) daughter sizes than WT, and the size differential persists through G1 ([2,3]; Figures 1,S1,Table S1; data not shown). Therefore, threshold cell size for transcriptional activation may be altered by disruption of MAT3 and DP1, and coincident timing with WT follows from the cells starting at different initial sizes. In contrast, regulon induction is delayed in cdka1 until cells have grown to about double the cell size at WT induction (Figure S4, 6).
In animals and plants, ablation of Rb results in excess proliferation and small cell sizes [9,17], and this is likely associated with misregulation of target genes with respect to cell size, as in Chlamydomonas. Like animal Rb, MAT3 binds to E2F/DP1, but MAT3 does not seem to dissociate at any point in the cell cycle [4], suggesting a divergent mechanism. In Drosophila, inactivation of dE2F slows cell division and dE2F over-expression accelerates division [18]; neither manipulation alters rates of cell growth, resulting in greatly increased or reduced cell sizes respectively. Drosophila Rb/E2F may couple cell division to cell size, as we observe in Chlamydomonas.
The large mitotic regulon is coordinately regulated across the multiple mutant timecourses tested here. A very substantial fraction of the transcriptome may be regulated in this way (Figures 6, S3B), though the amplitude of regulation is low for many of these genes. We do not know how many of these genes are directly regulated by upstream E2F/Dp1 binding, as opposed to indirect or global mechanisms such as chromatin modification, or regulation of other transcription factors.
Commitment, transcription and bistability
The commitment assay and transcriptional induction of the mitotic regulon show similar responses to MAT3, DP1 or CDKA1 disruption: altered commitment/gene induction with respect to cell size but not time in mat3 or dp1 mutants, in contrast to alteration of both timing and cell size control of commitment/gene induction in the absence of CDKA1. This suggests that commitment and induction of the mitotic regulon are mechanistically coupled. A problem with this view, though, is that commitment occurs many hours earlier than induction of the regulon [16].
A resolution could come from the consideration that ‘commitment’ is operational: a cell is ‘committed’, by definition, if it will ultimately divide without further photosynthesis-dependent cell growth. Recently, we proposed that passing the CP corresponded to attaining a critical cell size that, upon transfer to darkness and given considerable additional time, allows an irreversible change in the cell corresponding to flipping a bistable switch [12]. Thus, ‘biochemical commitment’ can occur many hours after operationally defined passage of the CP. This viewpoint allows speculative alignment of biochemical commitment with induction of the mitotic regulon. This regulon includes almost every gene required for cell division. Thus its expression is surely necessary for division, so expression may create a cell-division-permissive environment, which will persist until sufficient divisions have occurred to reduce cell size (or more likely the ratio of cell size to DNA content, which replicates in every cycle) to turn the regulon off again.
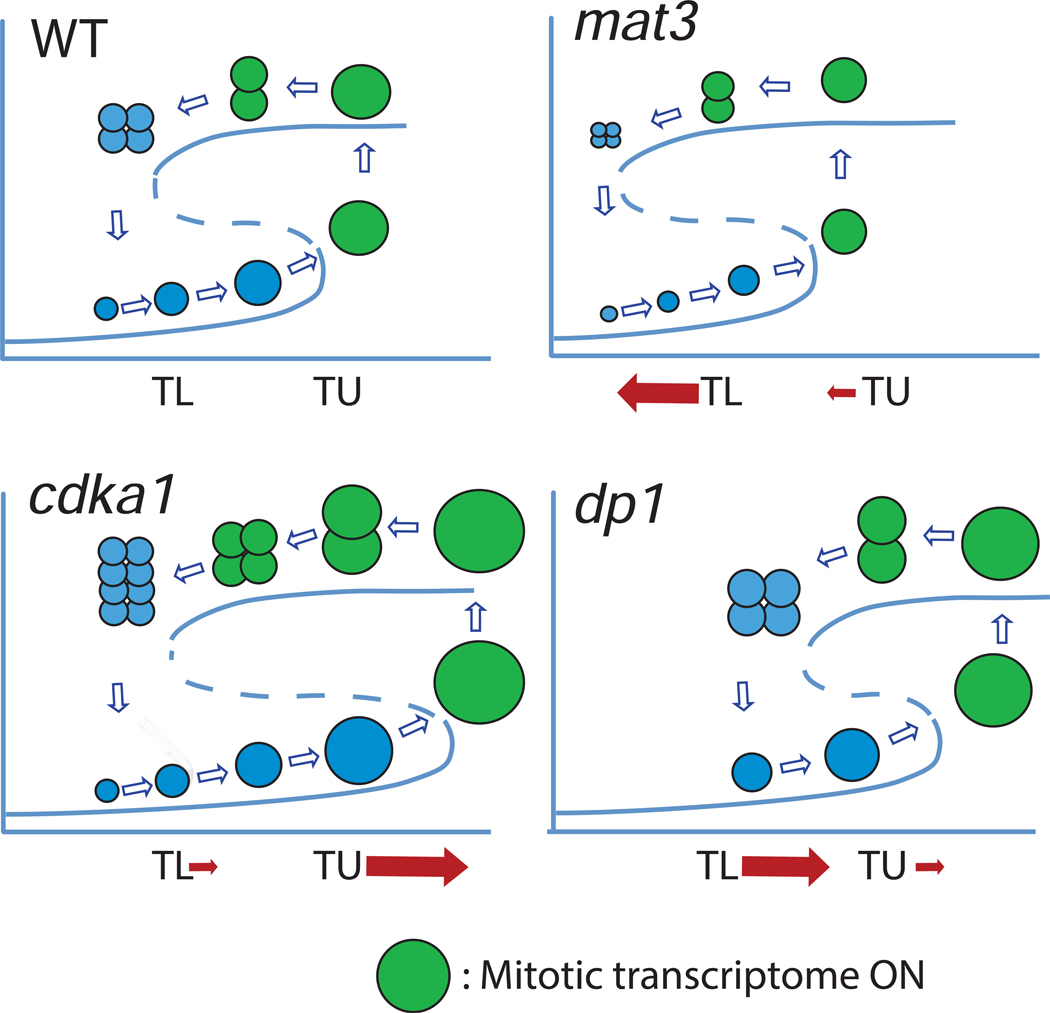
This idea can be abstractly described by a hysteretic bistability diagram, where the controlling variable is coupled to cell size [12]. Transition to the high branch is permissive for cell division; cell division (by necessity) reduces cell size, ultimately sending the system back to the lower, division-incompetent branch. The model has two critical sizes (Figure 7): TU, beyond which a jump to the upper branch is inevitable; and TL, below which residence on the upper branch cannot be maintained. Such a scheme compactly accounts for the Chlamydomonas multiple division cycle, provided the the hysteretic diagram has a very broad range of bistability: multiple cell divisions are required to transition back to the monostable non-dividing regime [12].
Figure 7. Bistability in control of multiple fission.
A sketch of levels of a division-inducer as a function of cell size. Inducer is low in very small cells; growth above a lower threshold TL makes low inducer metastable, but the system stays on the lower branch until growth past a higher threshold TU, where loss of the lower curve enforces a jump to the higher branch. Inducer remains high until division has reduced size to below TL. See [12]. Mutations in mat3, dp1 or cdka1 can be conceptualized as moving the thresholds TL and TU as indicated. cdka1 mutation predominantly increases TU; a minor effect on TL is reflected in moderately increased cdka1 daughter size. The predominant effects of mat3 or dp1 mutation is to shift TL down or up respectively, reflecting the known modulation of daughter cell size; a minor effect on TU in the same directions is also indicated, reflecting effects on commitment size [5][3][2]. Green: expression of the mitotic regulon. Figure S4 graphs cell size vs. synchronization timing for expression of the regulon for all genotypes tested.
In this framework, cdka1 mutation may primarily increase TU, the higher critical size (Figure 7), such that cells must be bigger to jump to the upper branch. This yields the key cdka1 phenotype of an abnormally long G1, followed by excess divisions so that final daughter sizes are close to WT. This is consistent with the observation that most of the G1 increase is due to expansion of the pre-CP period. CDKA1 mainly affects size at which commitment occurs, though a small effect on number of divisions for a given cell size is observed in most experiments, leading to a moderate increase in final daughter size compared to WT. This is a possible prediction of the model [12], since TU and entry into the dividing state is controlled by a ‘Starter’ activity, while TL and maintenance of the dividing state is controlled by a sum of ‘Starter’ and ‘Maintenance’ activities. Thus reduction in a ‘Starter’ module might cause a large shift in TU and a small shift in TL. In contrast, dp1 disruption can be interpreted as increasing TL, and mat3 disruption as decreasing it, thus altering the number of divisions carried out by cells of a given size ([2][3]; Figures 3, S1). dp1 and mat3 mutation have a moderate effect on size at commitment ([2][3]), implying an effect of these mutations on TU as well (Figures 4, 5, S2).
Figure 7 illustrates a possible position of expression of the mitotic regulon on this diagram (green), which corresponds to the jump to the higher branch. A plot of size vs. timing for regulon expression (Figure S4) corresponds to this scheme, at least qualitatively.
Heldt et al. [12] proposed a specific biochemical model, with transcription factor TF and inhibitor IN; reduction of these activities altered the lower critical size TL for maintenance of the dividing state, yielding decreases or increases in number of divisions for reductions in TF or IN, respectively. TF and IN could align to E2F/DP1 and MAT3 [12]. The model has an activity called ‘Starter kinase’ SK; reductions in SK should increase the upper critical size for initiation of the dividing state. CDKA1 gives an excellent fit to the properties of SK. (However, viability of single and multiple mutants indicates that there must be ‘backup’ pathways).
CDKA1 may promote entry into ‘division phase’ in cells above a critical size, perhaps by activating transcription of the mitotic regulon; E2F/DP1 may then maintain cells in division phase until cell division reduces cell size below a lower threshold. This idea maps CDKA1 function to commitment, the earliest known cell cycle step in Chlamydomonas, with no strong CDKA1 requirement once cells begin divisions. In Arabidopsis, CDKA1 is also not required for mitosis, and controls an early cell cycle event inferred to be upstream of mitotic transcription [9]. In Arabidopsis and in Chlamydomonas, the regulon of cell-division-associated genes is specifically expressed at dusk [19]. CDKA1 and RB/E2F may control this regulon, throughout the plant kingdom.
CDK1 and its close relative CDK2 are involved in early cell cycle decisions in yeast and animals, perhaps similarly to plant kingdom CDKA, so this role of CDK1 may be very ancient. However, in Chlamydomonas, and probably in land plants as well, mitosis is promoted by CDKB rather than by CDK1 [7–9], most likely reflecting a plant lineage-specific innovation.
Transcriptional control by Rb, E2F and Dp family members in animals is complex, since there are multiple members of each family and they associate differentially with other transcription-regulatory complexes [6,21]. Rb/E2F/Dp1 in Chlamydomonas may represent a minimal version of this system, which was likely present in the last eukaryotic common ancestor [20][22]. Since the system was lost in most fungi [22], Chlamydomonas may be the best available microbial model to understand this ancient regulatory network.
In natural environments, division may be suppressed during the day, and accumulated growth is rapidly partitioned into multiple daughter cells by the multiple fission cycle after dark [1]. Flagellar motility (important for photosynthesis) is incompatible with the basal bodies serving as microtubule organizing centers for coordination of spindle and cytokinesis geometries [27,28], so restricting division to night could be advantageous. CDKA and RB/E2F may be key to achieving this control.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Fred Cross (fcross@mail.rockefeller.edu). All unique/stable reagents, databases and code generated in this study are available from the Lead Contact without restriction.
Materials availability statement:
All unique/stable reagents, databases and code generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Strains and culture conditions.
All strains were congenic with the congenic pair CC-4403 and CC-4402 (Chlamydomonas Resource Center). dp1::PARO and mat3::PARO strains were provided by Jim Umen, and backcrossed multiple times into the CC-4402 background before use. cdka1::HYGRO and cycA1::PARO are disruption (null) alleles described previously [8]. Crosses and tetrad dissections were by standard methods. Strains were grown routinely on TAP, and for experiments involving light deprivation/restoration, on photoautotrophic medium HS (medium recipes at (https://www.chlamycollection.org/methods/media-recipes/). For gamete induction, M-N medium was used with 1/5 the level of Huntner’s trace elements. For growth on HS, light was provided with blue/red LED panels yielding approximately 100 micro-einsteins/m^2/sec in photsynthetically available radiation, determined with an Apogee quantum meter. Slight variations in schedules of light, dark and medium were tested throughout these experiments (e.g., pre-incubation in liquid TAP medium or TAP-agar; different times of dark incubation). Such variations were not found to have strong effects on results; in any individual experiment all strains were treated identically. Strains for each experiment are indicated in Table S4. Some variability was noted across experiments in the proportion of dark-starved WT cells that could divide on transfer to fresh medium in the dark (time of dark transfer zero in quantitative plots throughout); this was not obviously traceable to a specific experimental variation, but does not affect the conclusions.
All commitment experiments followed the same basic template. Cultures pregrown on TAP (acetate-containing, so not photoautotrophic) in the light were grown for a period in HS (photo-autotrophic medium), then transferred to dark in HS for a period intended to be sufficient for completion of all divisions ‘committed’ or in progress (in most experiments, 18–24 hrs). Cells were then plated on solid HS medium, incubated in light for various intervals and transferred to dark, for a total interval before plating of 24–30 hrs. In all experiments, light was provided by red and blue LED growlights, at ~100–150 micro-einsteins per sec per m^2 (some variability in illumination, mostly dependent on position on the incubator shelf, is inevitable in every setup I’ve tried). In different experiments, the HS light pre-growth interval, whether the HS pregrowth was in liquid or solid medium, and the total incubation time after plating were varied (all strains in a given experiment were treated identically). Preculture conditions and time of final image acquisition were: Figure 2, agar 18 hrs in dark at 29°C; 24 hrs; Figure 3 (left), liquid medium continuous light, shaking 120 rpm at 29°C, 30 hrs; Figure 3 (right), liquid medium continuous light, then 8.5 hrs dark, shaking 120 rpm at 29°C, 30 hrs; Figure 4, agar 12 hrs light then 20 hr dark at 29°C, 30 hrs; Figure S2B, agar 18 hrs light then 12 hr dark at 29°C, 30 hrs; Figures S2C,D, agar 18 hrs light then 18 hr dark at 29°C.
For nitrogen starve-refeed synchrony, TAP-grown cells were plated on TAP-N (nitrogen-free) supplemented with 10% the normal ammonium sulfate level (a cell suspension equivalent to 1 ml at OD750 2.0 was spread on a 25 ml agar plate). These were grown for two days at 29°C in continuous light, undergoing several divisions, thereby depleting the limited nitrogen and arresting as small daughters. They were then resuspended and plated on TAP with nitrogen at 29oC in light (~150 micro-einsteins/m^2/sec in photsynthetically available radiation) to release the block.
METHOD DETAILS
Imaging and scoring for the commitment assays.
The imaging setup was described previously [8][12]: a Zeiss Axioskop 40 Tetrad dissection microscope was modified in-house to allow accurate manual alignment to positions at spacing of a standard 384 microplate array. The microscope has a connected Canon Rebel digital camera which was used for image acquisition. Images were aligned precisely by custom MATLAB software, and a custom GUI allowed examination of individual cells for scoring divisions. The software included a segmentation routine to determine boundaries of cells in brightfield. To accommodate irregularities in the morphology of the cells in brightfield, I found an empirical solution to be iterative finding of a conservative boundary, followed by erosion of the dimmest pixels in the periphery, iterated until the area was reduced by 10% from starting values. This gave a satisfactory approximation of cell area judging by eye. Cases where the segmentation went significantly awry (e.g., overly large borders around cells, segmentation including neighboring cells) were excluded during manual scoring. 0,1, 2 and sometimes 3 divisions could be unambiguously scored (if 8 individual cells could be counted; Figure 2, bottom, shows an ideal case). Assignments of additional divisions were based on a visual approximation, assuming that all divisions were symmetrical (as is known to be the case in general) and carried out by all cells in a cluster (so that all clusters were assumed to contain 2^n cells). Uncertain cases were excluded during scoring. The GUI output was a data matrix indicating number of divisions detected at each time, and estimated cell size (pixel area) at each time, for every cell scored. Cell sizes were considered unreliable once cells divided, since the hatching process frequently results in an immediate large increase in area [8].
Please note that the scoring procedure has subjective elements at several points, but good reproducibility of scoring between genotype replicates within an experiment, and between experiments for the same genotypes, was attained. Software, original images and data matrices available on request.
Cell numbers scored per genotype and timepoint were variable, due mainly to plating variability (too-sparse or too-dense areas yielded few cells to score). All genotype/timepoint combinations are represented by at least ~100 up to >500 cells scored. The Monte Carlo simulation method for evaluating statistical significance embeds variable sample size within the calculation.
Chlamydomonas flow cytometry
FACS analysis was performed to determine DNA content and cell size (Figure 1). Cells were fixed in acetic acid-ethanol, RNase A-treated and stained with 500 nM Sytox Green nucleic acid stain (Thermo Fisher Scientific). Analysis was in an Accuri C6 (BD Biosciences) as described [7], following the manufacturer’s instructions.
This assay in Chlamydomonas has some special features requiring explanation. In Chlamydomonas sequential rounds of DNA replication, nuclear division and cytokinesis occur within the mother cell wall; flagellar growth and hatching occur only after all divisions are complete. Therefore, the synchronous division cycles within a given mother cell yield strong peaks at 1, 2, 4, … 2^n times the haploid DNA content, evenly spaced on a log plot. Newly hatched cells have Sytox (DNA) signal lower than the 1C cells just prior to the first round of nuclear DNA replication, because the latter cells have grown very large, and have significant background. Forward scatter (FSC) is responsive to cell size; therefore, newly hatched cells have a characteristic position on a 2-D Sytox X FSC scatterplot (‘N’ in Figure 1, bottom). Division cycles lead to an almost horizontal series of clusters with 1, 2, 4… 2^n DNA content, because cells have little time to grow during the rapid divisions. The FSC value of 2C cells thus approximates the cell size at which division cycles are initiated. FSC of the starting population of nitrogen-starved cells is similar to that of newly hatched cells, as is expected since the starting population represents the product of the last division before arrest.
For Figure 5, modal FSC values were estimated by visual inspection of the graphs (Figures 1, S1; the FACS graphs for experiment 3 are not shown due to space limitations). For Table S1, the complete raw datafile from the Accuri C6 flow cytometer was downloaded and analyzed with custom MATLAB code that allowed selection of a region in the 2D plot (using log scales for FSC and for DNA), then calculated a data-smoothed modal value and an average value for all cells in the region. This allowed determination of cell size for exact subpopulations (Figure 1, bottom). Code is available on request. Note that slight differences are expected between Figure 5 and Table S1; Table S1 reflects an accurate quantification of modal value for cells of an exact DNA content, with a clustered cell size (Figure 1, bottom).
RNA extraction and sequencing
RNA was isolated as follows: cells on agar plates were scraped off the plate into warm TAP medium using a sharp razor blade, transferred to a 15 ml conical centrifuge tube containing crushed ice (enough to chill the sample when melted), and spun down in a tabletop centrifuge. Pellets were resuspended in TE (10 mM Tris pH 7.5 1 mM EDTA), transferred to screw-cap freezer tubes and pelleted. Pellets were frozen at −70°C until RNA extraction. For extraction, the tubes with pellets were transferred to ice, and to the pellet was added, in this order, 400 microliters of acid-washed glass beads, 400 microliters of phenol-chloroform, and 400 microliters of NETS buffer (TE+0.2M NaCl + 0.2% SDS). Tubes were processed in a FastPrep bead-beater device, setting 5, 2 X 20sec, then spun in a microfuge 15 min. The aqueous phase was ethanol-precipitated and dissolved in ETS buffer (TE+0.2% SDS). RNA was immediately purified over a Qiagen RNA purification column (RNEasy) with on-column DNase digestion following the manufacturer’s instructions. RNA was then processed using the Illumina TruSeq RNA Sample Prep kit v.2 following the manufacturer’s instructions. Paired-end sequencing was by Genewiz.
QUANTIFICATION AND STATISTICAL ANALYSIS
Monte Carlo test of statistical significance.
The plots of cell size vs. division probability do not naturally fit standard statistical tests, since the assumptions of these tests are violated: numbers of cells for each data point are irregular, since total number of cells scored is variable, as is population size of each size bin across samples, and the different division-number curves for a given genotype are only partially independent of each other. I devised a Monte Carlo procedure to test for the significance of effects of genotype. The basis for the test is to use actual data independent of genotype to populate random datasets matched for sample size with the real ones, and then to examine how frequently the actual genotype-dependent distributions depart from a collection of such random datasets. For example, in the cdka1 x dp1 test (Figure 4), if cdka1 genotype is irrelevant for these plots, then the WT and cdka1 cells (CDKA1;DP1, cdka1;DP1) could have been drawn from a random mix of CDKA1;DP1 and cdka1;DP1 or CDKA1;dp1 and cdka1;dp1, respectively. So I made synthetic pools of DP1;CDKA1 and DP1;cdka1 by replacement at random from the combined set of DP1;CDKA1 and DP1;cdka1, matching the actual number of cells tested for each genotype. (Similarly for dp1;CDKA1 and dp1;cdka1). I selected 10,000 such random data sets and calculated the curves for each, then plotted a ‘band’ at each size bin containing 99% of the curves at that point. In principle, if CDKA1 is irrelevant to the shape of these curves, then all of the actual data points should be inside of the bands 99% of the time. This procedure should subsume the effects of variable number of data points at each size and genotype, potentially different variability of the data at different cell sizes, etc. A similar procedure was employed to test the effects of DP1 and MAT3 genotype in CDKA1 and cdka1 backgrounds. Note that by the logic of the test, if any point on a curve is outside of the 99% band, this is in principle a result supporting P<0.01. The conclusions drawn here for genotype significance are all supported by multiple points on multiple curves that are on or entirely outside of the 99% borders. cdka1 disruption clearly shifts the curves to higher cell sizes, especially (perhaps exclusively) for lower division numbers, in MAT3;DP1, mat3;DP1 and MAT3;dp1 backgrounds (P<0.01). dp1 disruption shifts the curves to higher cell sizes in CDKA1 and cdka1 backgrounds (P<0.01). mat3 disruption shifts the curves to lower cell sizes in the CDKA1 background (P<0.01); in a cdka1 background effects of mat3 disruption are marginal, though still detectable. These procedures were carried out on two independent cdka1 x dp1 experiments, with very similar results; this deals with any multiple hypothesis testing issues, and yields P<< 0.01 for the combined set of conclusions. Only one cdka1 x mat3 experiment is presented; a second was performed with similar results overall, but it had technical limitations due to high inviability in one of the WT strains, for unknown causes, therefore is not presented (data not shown). MATLAB code for these calculations is available on request.
Quantitative analysis of transcriptome data.
Reads were aligned to the reference annotated transcript models found at https://phytozome.jgi.doe.gov, [S4,S5] using the bowtie2 aligner [S6]. The .bam format output from bowtie2 was processed with custom awk software to filter reads and obtain a total readcount for each transcript model. Filtering: only reads aligned with a CIGAR string of 150M (end-to-end alignment), aligning to the same gene model in the mate pair, and with a mapping quality score >9 were included. The mapping quality requirement was eliminated for the 9% of genes annotated with alternative transcript models from the same gene model, since the aligner assigns ambiguous reads from such genes randomly to one of the alternative transcripts with 0 mapping quality. Raw readcount data is provided in supplementary file Data S1.
The raw data matrix (rows: transcripts; columns: genotype/timepoint) was processed by converting zero cells to 1, dividing each column (sample) by its total readcount, and dividing each row (transcript) by its mean, and log2-transforming. Hierarchical clustering and PCA analysis were carried out in MATLAB (clustergram and svd commands).MATLAB scripts used for analysis and figure generation are available upon request.
Principal components (PCs) were determined by: MATLAB [U,S,V]=svd(data,’econ’). The columns of V are ‘eigengenes’, linear combinations of which go to make up data for each actual gene. Columns of V are perpendicular to each other. The columns are ordered by how much of the total variability in the data set each accounts for.
The entries in the columns of V state a weight of each sample (genotype/timepoint) to make up the eigengene. If the eigengene reflects reproducible variation across experiments, then replicate entries within that column of V will have similar values; if on the other hand the eigengene reflects an unreproduced batch effect between experiments, then there can be high variation between replicate entries within that eigengene. Thus genotype/timepoint reproducibility for each eigengene was estimated by determining the proportion of variability in the entries of the eigengene that was accounted for by mean values for identical genotype/timepoint from different experiments. (For this purpose, the slightly different timepoints from experiment 3 were grouped with experiments 1 and 2 by aligning 12.5 hrs with 12 hrs, 15 hrs with 14 hrs, and 17.5 hrs with 18 hrs). The relevant formula is Var=mean(Var_within) + Var(means), where Var refers to total variance of the eigengene’s entries, Var(means) refers to variance of the mean values of the entries for genotype/timepoint replicates, and Var_within refers to variance of eigengene entries for each set of genotype/timepoint replicates.
For functional categories in transcriptome analysis, two datasets were used: a curated set of 108 cell-cycle-specific genes, most of which show strong periodic expression [16], and a set of broad functional categories covering large numbers of genes (cell cycle, cell wall, photosynthesis, etc.) assembled previously [11].
DATA AND CODE AVAILABILITY
All data and code are freely available on request.
KEY RESOURCES TABLE
Uploaded separately.
Supplementary Material
Data S1: Read count file from RNAseq experiments. Related to STAR Methods and Figures 6, S4. File contains number of reads assigned to each of the 19,526 transcripts following Bowtie_2 alignment [S6] to a .fa file containing spliced transcript sequences downloaded from Phytozome https://phytozome.jgi.doe.gov [S4,S5] as described in STAR Methods. The .fa file was modified to correct the sequence of CYCB1 [8]; file is available on request. File columns are: standard transcript names (e.g. Cre08.g372550.t1.1); readcounts assigned to this transcript in the 80 samples, in the order shown in the figures (indicated in the header line; w=WT, c=cdka1, d=dp1, m=mat3; followed by number of hours of incubation; e.g. w_8: WT 8 hrs; cd_12: cdka1;dp1 double mutant 12 hrs); all of experiment 1, followed by experiment 2, followed by experiment 3. Read count columns are followed by rank order of loading of PC1, PC2, PC3 and CYCB1 similarity (Euclidean distance); followed by annotations downloaded from Phytozome, as indicated in the header line.
Highlights.
Chlamydomonas CDKA1/CDK1 controls cell-size-dependent commitment to division.
CDKA1 and the E2F/DP1 transcription factor control division in distinct pathways.
The mitotic transcriptional regulon may define commitment to division
Acknowledgments
Thanks to Kresti Pecani for excellent technical assistance, and to Kristi Lieberman, Jim Roberts and Jim Umen for discussions. Thanks to Jim Umen for providing strains. Funding was from PHS GM-78153-10 from the National Institutes of Health.
Footnotes
Declaration of interests: The author declares no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cross FR, and Umen JG (2015). The Chlamydomonas cell cycle. Plant J. Cell Mol. Biol. 82, 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umen JG, and Goodenough UW (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15, 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang S-C, de los Reyes C, and Umen JG (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2, e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olson BJSC, Oberholzer M, Li Y, Zones JM, Kohli HS, Bisova K, Fang SC, Meisenhelder J, Hunter T, and Umen JG (2010). Regulation of the Chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. Plant Cell 22, 3331–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Liu D, López-Paz C, Olson BJ, and Umen JG (2016). A new class of cyclin dependent kinase in Chlamydomonas is required for coupling cell size to cell division. eLife 5, e10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer M, and Müller GA (2017). Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 52, 638–662. [DOI] [PubMed] [Google Scholar]
- 7.Tuli F., and Cros FR. (2014). A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26, 4019–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkins KC, and Cross FR (2018). Interregulation of CDKA/CDK1 and the Plant- Specific Cyclin-Dependent Kinase CDKB in Control of the Chlamydomonas Cell Cycle. Plant Cell 30, 429–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, and Schnittger A (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22, 1030–1040. [DOI] [PubMed] [Google Scholar]
- 10.Breker M, Lieberman K, and Cross FR (2018). Comprehensive Discovery of Cell-Cycle-Essential Pathways in Chlamydomonas reinhardtii. Plant Cell 30, 1178–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tulin F, and Cross FR (2015). Cyclin-Dependent Kinase Regulation of Diurnal Transcription in Chlamydomonas. Plant Cell 27, 2727–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heldt FS, Tyson JJ, Cross FR, and Novak B (2020). A single light- responsive sizer can control multiple-fission cycles in Chlamydomonas. Curr. Biol. CB 30, 1–11. [DOI] [PubMed] [Google Scholar]
- 13.ítová M, Bišová K, Umysová D, Hlavová M, Kawano S, Zachleder V, and Cížková M (2011). Chlamydomonas reinhardtii: duration of its cell cycle and phases at growth rates affected by light intensity. Planta 233, 75–86. [DOI] [PubMed] [Google Scholar]
- 14.Vítová M, Bišová K, Hlavová M, Kawano S, Zachleder V, and Cížková M (2011). Chlamydomonas reinhardtii: duration of its cell cycle and phases at growth rates affected by temperature. Planta 234, 599–608. [DOI] [PubMed] [Google Scholar]
- 15.Panchy N, Wu G, Newton L, Tsai C-H, Chen J, Benning C, Farré EM, and Shiu S-H (2014). Prevalence, evolution, and cis-regulation of diel transcription in Chlamydomonas reinhardtii. G3 Bethesda Md 4, 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zones JM, Blaby IK, Merchant SS, and Umen JG (2015). High-Resolution Profiling of a Synchronized Diurnal Transcriptome from Chlamydomonas reinhardtii Reveals Continuous Cell and Metabolic Differentiation. Plant Cell 27, 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, and Jacks T (2000). Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14, 3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neufeld TP, de la Cruz AF, Johnston LA, and Edgar BA (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193. [DOI] [PubMed] [Google Scholar]
- 19.de Los Reyes P, Romero-Campero FJ, Ruiz MT, Romero JM, and Valverde F (2017). Evolution of Daily Gene Co-expression Patterns from Algae to Plants. Front. Plant Sci. 8, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogozi IB., Bas MK., Csürö M., and Kooni EV. (2009). Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol. Evol. 1, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Pei C, Wang X, Xiang J, Sun B-F, Cheng Y, Qi X, Marchetti M, Xu J-W, Sun Y-P, et al. (2017). A Balance of Yki/Sd Activator and E2F1/Sd Repressor Complexes Controls Cell Survival and Affects Organ Size. Dev. Cell 43, 603–617.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina EM, Turner JJ, Gordân R, Skotheim JM, and Buchler NE (2016). Punctuated evolution and transitional hybrid network in an ancestral cell cycle of fungi. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao G, Lee TJ, Mori S, Nevins JR, and You L (2008). A bistable Rb-E2F switch underlies the restriction point. Nat. Cell Biol. 10, 476–482. [DOI] [PubMed] [Google Scholar]
- 24.Skotheim JM, Di Talia S, Siggia ED, and Cross FR (2008). Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charvin G, Oikonomou C, Siggia ED, and Cross FR (2010). Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 8, e1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross FR, Buchler NE, and Skotheim JM (2011). Evolution of networks and sequences in eukaryotic cell cycle control. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 366, 3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehler LL, and Dutcher SK (1998). Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 40, 193–207. [DOI] [PubMed] [Google Scholar]
- 28.Ehler LL, Holmes JA, and Dutcher SK (1995). Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 141, 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heldt FS, Tyson JJ, Cross FR, and Novák B (2019). A single light-responsive sizer can control multiple-fission cycles in Chlamydomonas. bioRxiv, 648436. [DOI] [PubMed] [Google Scholar]
- 30.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaby IK, Blaby-Haas CE, Tourasse N, Hom EFY, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, et al. (2014). The Chlamydomonas genome project: a decade on. Trends Plant Sci. 19, 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan S-C., and Ume JG. (2008). A suppressor screen in chlamydomonas identifies novel components of the retinoblastoma tumor suppressor pathway . Genetics 178, 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Read count file from RNAseq experiments. Related to STAR Methods and Figures 6, S4. File contains number of reads assigned to each of the 19,526 transcripts following Bowtie_2 alignment [S6] to a .fa file containing spliced transcript sequences downloaded from Phytozome https://phytozome.jgi.doe.gov [S4,S5] as described in STAR Methods. The .fa file was modified to correct the sequence of CYCB1 [8]; file is available on request. File columns are: standard transcript names (e.g. Cre08.g372550.t1.1); readcounts assigned to this transcript in the 80 samples, in the order shown in the figures (indicated in the header line; w=WT, c=cdka1, d=dp1, m=mat3; followed by number of hours of incubation; e.g. w_8: WT 8 hrs; cd_12: cdka1;dp1 double mutant 12 hrs); all of experiment 1, followed by experiment 2, followed by experiment 3. Read count columns are followed by rank order of loading of PC1, PC2, PC3 and CYCB1 similarity (Euclidean distance); followed by annotations downloaded from Phytozome, as indicated in the header line.
Data Availability Statement
All data and code are freely available on request.