Abstract
Purpose
With the increasing incidence of thyroid cancer (TC), associations between genetic polymorphisms and TC risk have attracted a lot of attention. Considering that the results of associations of genetic variants with TC were usually inconsistent based on publications until now, we attempted to comprehensively evaluate the real evidence of associations between single nucleotide polymorphisms (SNPs) and TC risk.
Method
We performed meta-analyses on 36 SNPs in 23 genes associated with TC susceptibility based on the data from 99 articles and comprehensively valued the epidemiological evidence of significant associations through the Venice criteria and false-positive report probability (FPRP) test. OR and P value were also calculated for 19 SNPs in 13 genes based on the insufficient data from 22 articles.
Results
19 SNPs were found significantly associated with TC susceptibility. Of these, strong epidemiological evidence of associations was identified for the following seven SNPs: POU5F1B rs6983267, FOXE1 rs966423, TERT rs2736100, NKX2-1 rs944289, FOXE1 rs1867277, FOXE1 rs2439302, and RET rs1799939, in which moderate associations were found in four SNPs and weak associations were found in eight SNPs. In addition, probable significant associations with TC were found in nine SNPs.
Conclusion
Our study systematically evaluated associations between SNPs and TC risk and offered reference information for further understanding of polymorphisms and TC susceptibility.
1. Introduction
Thyroid cancer (TC) is the most common endocrine malignant tumor with the increasing incidence worldwide. Besides radiation exposure, TC is also closely related to family inheritance and genetic variant risk [1]. As early as 2009, Gudmundsson et al. firstly pointed out that variants on 9q22.33 (FOXE1) and 14q13.3 (NK2 homeobox 1 (NKX2-1)) might increase the risk of papillary thyroid cancer and follicular thyroid cancer [2]. BRAF V600E mutation is comparatively common and widely used in the detection of papillary thyroid cancer [3]. However, still most of the genetic variation remains uncharacterized with TC susceptibility.
So far, the research on associations between genetic variation and cancer risk received a lot of attention. Quite a few pooled studies and reviews have expounded the relationship between TC and genetic variation [4–6], but it is difficult to interpret the inconsistent results between the same variants and TC risk. A small sample size may not have sufficient ability to detect the true associations. Meta-analyses can comprehensively conduct secondary research by collecting the effective data from single study, which can increase the statistical power and reliability of the causality [7, 8]. However, there are still inconsistent results in the meta-analyses updated until now, which indicates the existence of false-positive report caused by unnecessary overlap. Moreover, the Venice rating standard, firstly proposed by Ioannidis et al. [9], has been used to systematically grade the cumulative evidence of genetic associations, so as to help understand associations between genetic variants and disease [10, 11]. Herein, we collected data updated until now and performed meta-analyses to comprehensively evaluate the evidence for further understanding of associations between genetic variation and TC risk.
2. Materials and Methods
Our study was performed based on the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement and the Human Genome Epidemiology Network for the systematic review of genetic association studies [12, 13].
We searched publications about genetic variation and TC risk on PubMed, MEDLINE, Web of Science, and CNKI before December 31, 2020, using the keywords as follows: (“thyroid”), (“cancer” or “carcinoma”), (“genetic” or “single nucleotide polymorphism (SNP)” or “SNP” or “polymorphism” or “genotype” or “variation” or “variant” or “mutation” or “susceptibility”), (“association” or “associate”), using “and” collect each keyword as well. A total of 3887 records were searched, as well as 157 records from relevant reference publications. As a result, 99 relevant publications with available data were included in our study. The articles included in our study must meet the following inclusion criteria: (i) the object of study must be thyroid cancer, (ii) studying associations between genetic variants and etiology of TC using human-related case-control or cohort or cross-sectional study, and (iii) offering sufficient data to perform meta-analyses. Repetitive and unrelated articles were excluded by browsing titles and abstracts. The articles were excluded (i) if the interests were not concentrated on variants with TC risk, (ii) if there is lack of necessary data, and (iii) if the articles were just letters to editors.
2.1. Data Extraction
Data extraction was carried out by two people independently and exchanged to check with each other after extraction. Any inconsistent was duplicately checked and discussed to reach an agreement with the corresponding author. For the variants reported in articles, we extracted data as follows: PMID of articles, first author, year of publication, country or region, ethnicity, name of variants, polymorphisms, study design, genotyping method, case and control, Hardy–Weinberg equilibrium (HWE) status, genotype counts, and minor allelic frequency (MAF). According to the results of previous meta-analyses, we divided the major ethnic groups into 3 group categories: Caucasians, Asians, and African Americans. The overall population was defined as two or more populations as above. As for the name of SNP, which often has many different naming methods, we selected the most common and well-known name of the SNP as the representative by querying on NCBI.
2.2. Statistical Analysis
For each SNP, we sorted out allelic, dominant, and recessive models according to the included ethnicities. Then, meta-analyses based on models and ethnicities were performed using STATA, version 12 (Stata, College Station, TX, USA) only if two or more studies were included. Crude ORs with the corresponding 95% CIs were used to assess the strength of the association between SNPs and TC risk. The I2 test was performed to quantitatively assess possible heterogeneity in the combined studies as follows: I2 ≤ 25 indicated no or mild heterogeneity, 25% < I2 < 50% indicated moderate heterogeneity, and I2 ≥ 50% indicated large heterogeneity [14]. Sensitivity analysis was performed by removing the first published study from the total or studies deviated from the HWE in the controls and reanalyzing the remainder. In addition, publication bias was assessed by Egger's test and P > 0.05 indicated no publication bias existed [15, 16].
2.3. Evaluation of Epidemiological Evidence
We evaluated the evidence of significant associations between SNPs and TC by the Venice guideline first based on three criteria as follows: amount of evidence, replication of association, and protection from bias [17, 18]. The amount of evidence was related to the number of alleles or genotypes and graded as A (N > 1000), B (100 ≤ N ≤ 1000), or C (N < 100). The replication of association was graded as A (I2 ≤ 25%), B (25% < I2 < 50%), or C (I2 ≥ 50%) based on heterogeneity statistics. The protection from bias was determined by various potential sources of bias, including sensitivity analysis, publication bias, and small study bias, as well as an excess of significant findings. A was graded when there was no demonstrable bias or the bias was unlikely to invalidate the association. B was graded when there was no obvious bias without sufficient information on identifying evidence. C was graded when there was obvious bias or the bias was likely to explain the presence of association. Furthermore, C was graded in any one of the following situations: (1) association lost with exclusion of first study or studies deviated from HWE in sensitive analysis; (2) a low magnitude of the association (0.87 < OR < 1.15) only if the association had been identified by GWAS or several studies with no evidence of publication bias; and (3) evidence of obvious publication bias (P value in Egger's test<0.05). In summary, the cumulative epidemiological evidence of significant associations was graded as follows: strong associations (all above three grades were A), weak associations (any grade was C), and moderate associations (all other conditions).
Furthermore, we used a false-positive report probability (FPRP) assay suggested by Wacholder et al. [17] with a prior probability of 0.05 and an FPRP cutoff value of 0.2 to detect the potential false-positive results among significant associations, in order to confirm whether there was a real association between SNPs and TC risk. The evidence of FPRP was graded as strong (FPRP < 0.05), moderate (0.05 ≤ FPRP ≤ 0.2), and weak (FPRP > 0.2), which indicated upgrading of cumulative evidence one level (from moderate to strong or from weak to moderate), maintaining of the original level, and downgrading of cumulative evidence one level (from strong to moderate or from moderate to weak), respectively.
3. Results
As presented in Figure 1, a total of 3887 records were searched, as well as 157 records from relevant reference publications. Of these, 1821 duplicate records were removed, and 1931 irrelevant records were excluded via scanning the title or abstract. Of the 292 publications assessed for eligibility, 121 publications were excluded due to no etiology of TC, 34 publications were excluded due to no genetic polymorphism, 13 publications were excluded due to no case-control or cohort or cross-sectional study, 16 publications were excluded due to lack of necessary data, and 9 publications were excluded for letter to editors. At last, a total of 99 eligible publications were included in our study. The data from those publications involving 36 SNPs in 23 genes were used to perform meta-analyses and value the cumulative epidemiological evidence with the Venice criteria and FPRP test. Additionally, 22 publications including 19 SNPs in 13 genes with insufficient data were also used to calculate OR and P value.
Figure 1.
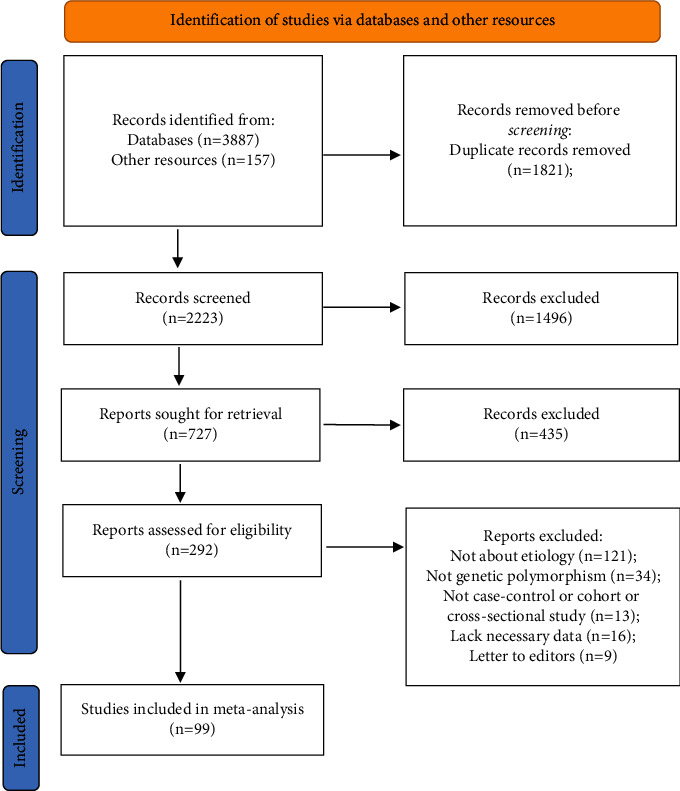
Flow diagram of search strategy and study selection.
In the result of meta-analyses, 19 SNPs were significantly associated with TC risk as follows: POU5F1B rs6983267, miR-146a rs2910164, FOXE1 rs71369530, FOXE1 rs907580, NKX2-1 rs944289, FOXE1 rs965513, FOXE1 rs966423, FOXE1 rs1443434, FOXE1 rs1867277, FOXE1 rs2439302, FOXE1 rs30215269, MTHFR rs1801133, RET rs1800858, RET rs1799939, RET rs1800862, RET rs1800863, TERT rs2736100, XRCC3 rs1799794, and XRCC3 rs861539 (Table 1). 17 SNPs had no obvious association with TC as follows: ATM rs189037, ATM rs664677, ATM rs1801516, CYP1A1m1 rs4646903, CYP1A1m2, GSTM1 null/present, GSTP1, GSTT1 null/present, NAT rs10419839, P53 rs1042522, RET rs2565206, RET rs1800861, XRCC1 rs25487, XRCC1 rs25489, XRCC1 rs1799782, XRCC2 rs3218536, and XRCC3 rs1799796 (Supplementary Table 1). Of the significantly associated SNPs, RET rs1800858 was found inversely associated with TC risk (OR = 0.898 under allelic model and 0.867 under dominant model). All other significant associations of SNPs could increase the risk of TC.
Table 1.
Statistically significant variants from meta-analysis, false-positive report probabilities (FPRPs), and cumulative epidemiological evidence.
| Gene | Variant | Alleles | Ethnicity | MAF† | Studies | Number evaluation | Risk of meta-analysis | PQ | Amount of evidence | Replication | Protection from bias | Reason for bias exemption | Venice criteria grade§ | FPRP values at prior probability of 0.05 and OR of 1.5 | Cumulative epidemiological evidence¶ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample size (case/control) | Genetic models | Effect model | OR (95% CI) | Pvalue | I (%) | N‡minor | Grade | Grade | Pegger | ||||||||||||
| POU5F1B | rs6983267 | G > T | Overall | 0.47 | 10 | 30673 (7504/23169) | Allelic | F | 1.129 (1.086 − 1.174) | ≤0.01 | 28.1 | 0.19 | 29188 | A | B | Ca | 0.25 | No | ABC | ≤0.01 | Moderate |
| 9 | 27428 (7018/20410) | Dominant | F | 1.175 (1.107 − 1.247) | ≤0.01 | 0.0 | 0.48 | 20223 | A | A | A | 0.33 | No | AAA | ≤0.01 | Strong | |||||
| 9 | 27428 (7018/20410) | Recessive | F | 1.158 (1.091 − 1.228) | ≤0.01 | 27.6 | 0.19 | 6725 | A | B | A | 0.28 | No | ABA | ≤0.01 | Strong | |||||
| miR-146a | rs2910164 | G > C | Overall | 0.32 | 12 | 16737 (4857/11880) | Dominant | R | 1.144 (1.003 − 1.304) | ≤0.01 | 57.0 | ≤0.01 | 8639 | A | C | Ca | 0.89 | No | ACC | 0.46 | Weak |
| FOXE1 | rs71369530 | >14-Ala vs. ≤14-Ala | Caucasian | 0.24 | 4 | 1271 (576/695) | Allelic | R | 1.836 (1.353 − 2.492) | ≤0.01 | 64.5 | 0.04 | 757 | B | C | A | 0.27 | No | BCA | 0.02 | Moderate |
| FOXE1 | rs907580 | C > T | Caucasian | 0.26 | 3 | 6884 (497/6387) | Allelic | R | 1.593 (1.184 − 2.145) | ≤0.01 | 67.6 | 0.05 | 3733 | A | C | A | 0.80 | No | ACA | 0.11 | Weak |
|
| |||||||||||||||||||||
| NKX2-1 | rs944289 | C > T | Overall | 0.55 | 17 | 65182 (9467/55715) | Allelic | F | 1.304 (1.255 − 1.355) | ≤0.01 | 45.5 | 0.02 | 72278 | A | B | A | 0.40 | No | ABA | ≤0.01 | Strong |
| 10 | 16277 (5187/11090) | Dominant | F | 1.609 (1.465 − 1.767) | ≤0.01 | 26.3 | 0.20 | 12706 | A | B | A | 0.24 | No | ABA | ≤0.01 | Strong | |||||
| 10 | 16277 (5187/11090) | Recessive | F | 1.414 (1.301 − 1.536) | ≤0.01 | 40.5 | 0.09 | 5073 | A | B | A | 0.14 | No | ABA | ≤0.01 | Strong | |||||
|
| |||||||||||||||||||||
| FOXE1 | rs965513 | G > A | Overall | 0.34 | 18 | 61943 (8167/53776) | Allelic | R | 1.703 (1.575 − 1.842) | ≤0.01 | 66.7 | ≤0.01 | 43701 | A | C | A | 0.13 | No | ACA | ≤0.01 | Moderate |
| 9 | 14348 (3680/10668) | Dominant | R | 1.694 (1.429 − 2.010) | ≤0.01 | 68.4 | ≤0.01 | 7542 | A | C | A | 0.17 | No | ACA | ≤0.01 | Moderate | |||||
| 9 | 14348 (3680/10668) | Recessive | F | 1.954 (1.729 − 2.208) | ≤0.01 | 46.4 | 0.06 | 1599 | A | B | Cd | 0.02 | No | ABC | ≤0.01 | Moderate | |||||
| DIRC3 | rs966423 | T > C | Overall | 0.50 | 5 | 9604 (4953/4651) | Allelic | F | 1.227 (1.153 − 1.306) | ≤0.01 | 0.0 | 0.44 | 9931 | A | A | A | 0.13 | No | AAA | ≤0.01 | Strong |
| FOXE1 | rs1443434 | T > G | Caucasian | 0.39 | 4 | 9627 (2453/7174) | Allelic | R | 1.392 (1.084 − 1.787) | ≤0.01 | 78.7 | ≤0.01 | 7800 | A | C | A | 0.54 | No | ACA | 0.20 | Weak |
|
| |||||||||||||||||||||
| FOXE1 | rs1867277 | G > A | Caucasian | 0.39 | 12 | 21820 (5654/16166) | Allelic | F | 1.503 (1.426 − 1.583) | ≤0.01 | 43.7 | 0.05 | 18169 | A | B | A | 0.47 | No | ABA | ≤0.01 | Strong |
| 6 | 10702 (2958/7744) | Dominant | R | 1.702 (1.352 − 2.143) | ≤0.01 | 65.4 | ≤0.01 | 6892 | A | C | A | 0.21 | No | ACA | ≤0.01 | Moderate | |||||
| 6 | 10702 (2958/7744) | Recessive | F | 1.703 (1.498 − 1.937) | ≤0.01 | 44.4 | 0.11 | 1809 | A | B | A | 0.20 | No | ABA | ≤0.01 | Strong | |||||
| FOXE1 | rs2439302 | C > G | Overall | 0.35 | 4 | 9265 (3146/6119) | Allelic | F | 1.325 (1.240 − 1.415) | ≤0.01 | 25.6 | 0.26 | 7409 | A | B | A | 0.42 | No | ABA | ≤0.01 | Strong |
| FOXE1 | rs30215269 | T > C | Caucasian | 0.39 | 3 | 6997 (684/6313) | Allelic | R | 1.634 (1.254 − 2.127) | ≤0.01 | 62.0 | 0.07 | 5555 | A | C | A | 0.99 | No | ACA | 0.97 | Weak |
|
| |||||||||||||||||||||
| MTHFR C677T | rs1801133 | C > T | Overall | 0.30 | 8 | 6267 (2902/3365) | Allelic | R | 1.418 (1.114 − 1.806) | ≤0.01 | 71.4 | ≤0.01 | 4276 | A | C | A | 0.39 | No | ACA | 0.12 | Weak |
| 9 | 7454 (3447/4007) | Dominant | R | 1.383 (1.081 − 1.769) | ≤0.01 | 69.0 | ≤0.01 | 4337 | A | C | A | 0.31 | No | ACA | 0.20 | Weak | |||||
| 8 | 6267 (2902/3365) | Recessive | F | 1.258 (1.081 − 1.464) | ≤0.01 | 43.1 | 0.09 | 843 | B | B | A | 0.95 | No | BBA | 0.06 | Moderate | |||||
|
| |||||||||||||||||||||
| RET A45A | rs1800858 | G > A | Overall | 0.31 | 8 | 4620 (1867/2753) | Allelic | F | 0.898 (0.818 − 0.987) | 0.03 | 18.9 | 0.28 | 2885 | A | A | Cab | 0.24 | No | AAC | 0.33 | Weak |
| 7 | 4462 (1809/2653) | Dominant | F | 0.867 (0.764 − 0.984) | 0.03 | 11.0 | 0.35 | 2358 | A | A | Cb | 0.55 | No | AAC | 0.34 | Weak | |||||
|
| |||||||||||||||||||||
| RET G691S | rs1799939 | G > A | Overall | 0.21 | 12 | 6643 (2853/3790) | Allelic | R | 1.352 (1.171 − 1.561) | ≤0.01 | 53.8 | ≤0.01 | 2934 | A | C | A | 0.12 | No | ACA | ≤0.01 | Moderate |
| 12 | 6643 (2853/3790) | Dominant | R | 1.386 (1.155 − 1.664) | ≤0.01 | 55.6 | ≤0.01 | 2475 | A | C | A | 0.63 | No | ACA | ≤0.01 | Moderate | |||||
| 12 | 6643 (2853/3790) | Recessive | R | 1.535 (1.224 − 1.924) | ≤0.01 | 0.0 | 0.90 | 459 | B | A | A | 0.29 | No | BAA | ≤0.01 | Strong | |||||
|
| |||||||||||||||||||||
| RET S836S | rs1800862 | C > T | Caucasian | 0.04 | 14 | 6654 (2701/3953) | Allelic | F | 1.129 (1.008 − 1.409) | 0.04 | 21.8 | 0.22 | 637 | B | B | Cab | 0.74 | No | BBC | 0.88 | Weak |
| 9 | 5791 (2250/3541) | Dominant | F | 1.283 (1.058 − 1.557) | ≤0.01 | 36.6 | 0.13 | 500 | B | B | Cd | ≤0.01 | No | BBC | 0.19 | Weak | |||||
| RET S904S | rs1800863 | C > G | Overall | 0.19 | 6 | 3073 (1178/1895) | Recessive | F | 1.578 (1.090 − 2.286) | 0.02 | 0.0 | 0.75 | 124 | B | A | A | 0.48 | No | BAA | 0.43 | Weak |
|
| |||||||||||||||||||||
| TERT | rs2736100 | T > G | Asian | 0.39 | 5 | 10104 (5052/5052) | Allelic | F | 1.430 (1.352 − 1.512) | ≤0.01 | 22.6 | 0.27 | 8802 | A | B | A | 0.49 | No | ABA | ≤0.01 | Strong |
| 5 | 10104 (5052/5052) | Dominant | F | 1.535 (1.411 − 1.668) | ≤0.01 | 15.5 | 0.32 | 6768 | A | A | A | 0.95 | No | AAA | ≤0.01 | Strong | |||||
| 5 | 10104 (5052/5052) | Recessive | F | 1.666 (1.509 − 1.839) | ≤0.01 | 0.0 | 0.72 | 2034 | A | A | A | 0.64 | No | AAA | ≤0.01 | Strong | |||||
|
| |||||||||||||||||||||
| XRCC3 A17893G | rs1799794 | A > G | Overall | 0.29 | 4 | 2477 (1106/1371) | Allelic | R | 1.275 (1.008 − 1.613) | 0.04 | 71.2 | 0.02 | 1519 | A | C | Cc | 0.33 | No | ACC | 0.47 | Weak |
| 4 | 2477 (1106/1371) | Dominant | R | 1.321 (1.023 − 1.705) | 0.03 | 55.4 | 0.08 | 1184 | A | C | Cc | 0.73 | No | ACC | 0.43 | Weak | |||||
| 4 | 2477 (1106/1371) | Recessive | F | 1.383 (1.092 − 1.750) | ≤0.01 | 29.8 | 0.23 | 335 | B | B | Cc | 0.74 | No | BBC | 0.15 | Weak | |||||
|
| |||||||||||||||||||||
| XRCC3 | rs861539 | C > T | Overall | 0.24 | 11 | 5978 (2413/3565) | Allelic | R | 1.363 (1.193 − 1.559) | ≤0.01 | 53.9 | 0.02 | 3142 | A | C | Cd | 0.03 | No | ACC | ≤0.01 | Moderate |
| 11 | 5978 (2413/3565) | Dominant | R | 1.357 (1.135 − 1.622) | ≤0.01 | 56.9 | ≤0.01 | 2570 | A | C | A | 0.35 | No | ACA | 0.62 | Weak | |||||
| 11 | 5978 (2413/3565) | Recessive | F | 1.709 (1.428 − 2.046) | ≤0.01 | 19.6 | 0.26 | 572 | B | A | Cd | ≤0.01 | No | BAC | ≤0.01 | Moderate | |||||
F: meta-analysis was performed under the fixed-effects model. R: meta-analysis was performed under the random-effects model. Overall: two or more ethnicities were reported in the study. †Frequency of minor allele in controls. ‡Number of test allele or genotype. §Venice criteria grades are amount of evidence, replication of the association, and protection from bias. ¶Cumulative epidemiological evidence as graded by the combination of results from the Venice criteria and FPRP. aThe grade of C is given because the OR value is between 0.87 and 1.15, and the association is not replicated by GWAS or GWAS meta-analysis. bThe grade of C is given for no significant association existed by excluding the first published study. cThe grade of C is given for no significant association existed by excluding studies deviated from the HWE in the controls. dThe grade of C is given for significant publication bias (Pegger ≤0.05)
Furthermore, in the result of subgroup analysis for 19 SNPs based on ethnicity, 11 SNPs were significantly associated with TC risk as follows: 3POU5F1B rs6983267, NKX2-1 rs944289, FOXE1 rs965513, DIRC3 rs966423, FOXE1 rs966423, FOXE1 rs2439302, MTHFR rs1801133, RET rs1800858, RET rs1799939, XRCC1 rs1799782, and XRCC3 rs861539 (Table 2), and 8 SNPs were not (miR-146a rs2910164, P53 rs1042522, XRCC1 rs25487, XRCC1 rs25489, XRCC3 rs1799794, XRCC3 rs1799796, RET rs1800861, and RET rs1800863) (Supplementary Table 2). In the result of merely calculating OR and P value, probable significant associations with TC were found in 9 SNPs (CYP1A2F rs762551, FTO rs1477196, FTO rs8047395, FTO rs11642841, FTO rs17817288, IL-18 rs360717, miR-608 rs4919510, TSHR rs1991517, and XRCC3 rs56377012) (Table 3).
Table 2.
Significant variants in subtype analysis from meta-analysis, false-positive report probabilities (FPRPs), and cumulative epidemiological evidence.
| Gene | Variant | Alleles | Ethnicity | MAF† | Studies | Number evaluation | Risk of meta-analysis | PQ | Amount of evidence | Replication | Protection from bias | Reason for bias exemption | Venice criteria grade§ | FPRP values at prior probability of 0.05 and OR of 1.5 | Cumulative epidemiological evidence¶ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample size (case/control) | Genetic models | Effect model | OR (95% CI) | Pvalue | I (%) | N‡minor | Grade | Grade | Pegger | ||||||||||||
| POU5F1B | rs6983267 | G > T | Caucasian | 0.50 | 7 | 47420 (11450/35970) | Allelic | F | 1.139 (1.090 − 1.190) | ≤0.01 | 31.2 | 0.19 | 24106 | A | B | Ca | 0.32 | No | ABC | ≤0.01 | Moderate |
| Caucasian | 7 | 23710 (5725/17985) | Dominant | F | 1.202 (1.117 − 1.294) | ≤0.01 | 0.0 | 0.65 | 17952 | A | A | A | 0.29 | No | AAA | ≤0.01 | Strong | ||||
| Caucasian | 7 | 23710 (5725/17985) | Recessive | F | 1.177 (1.097 − 1.261) | ≤0.01 | 45.3 | 0.09 | 6154 | A | B | A | 0.31 | No | ABA | ≤0.01 | Strong | ||||
| Asian | 0.35 | 3 | 13926 (3558/10368) | Allelic | F | 1.095 (1.009 − 1.189) | 0.03 | 36.3 | 0.21 | 5082 | A | B | Ca | 0.23 | No | ABC | 0.37 | Weak | |||
| Asian | 2 | 3718 (1293/2425) | Dominant | F | 1.123 (1.015 − 1.242) | 0.02 | 38.5 | 0.20 | 2271 | A | B | Ca | 0.20 | No | ABC | 0.31 | Weak | ||||
|
| |||||||||||||||||||||
| NKX2-1 | rs944289 | C > T | Caucasian | 0.56 | 13 | 59074 (7732/51342) | Allelic | F | 1.273 (1.220 − 1.329) | ≤0.01 | 31.1 | 0.13 | 66909 | A | B | A | 0.96 | No | ABA | ≤0.01 | Strong |
| Caucasian | 7 | 13356 (3957/9399) | Dominant | F | 1.468 (1.311 − 1.644) | ≤0.01 | 0.0 | 0.72 | 10675 | A | A | A | 0.45 | No | AAA | ≤0.01 | Strong | ||||
| Caucasian | 7 | 13356 (3957/9399) | Recessive | F | 1.378 (1.255 − 1.514) | ≤0.01 | 47.2 | 0.08 | 4399 | A | B | A | 0.34 | No | ABA | ≤0.01 | Strong | ||||
| Asian | 0.41 | 4 | 6108 (1735/4373) | Allelic | F | 1.429 (1.315 − 1.553) | ≤0.01 | 50.2 | 0.11 | 5369 | A | C | A | 0.97 | No | ACA | ≤0.01 | Moderate | |||
| Asian | 3 | 2921 (1230/1691) | Dominant | F | 1.968 (1.663 − 2.328) | ≤0.01 | 0.0 | 0.80 | 2031 | A | A | A | 0.82 | No | AAA | ≤0.01 | Strong | ||||
| Asian | 3 | 2921 (1230/1691) | Recessive | F | 1.544 (1.296 − 1.840) | ≤0.01 | 19.0 | 0.29 | 674 | B | A | A | 0.25 | No | BAA | ≤0.01 | Strong | ||||
|
| |||||||||||||||||||||
| FOXE1 | rs965513 | G > A | Caucasian | 0.35 | 16 | 59202 (7025/52177) | Allelic | R | 1.747 (1.613 − 1.891) | ≤0.01 | 64.1 | ≤0.01 | 42714 | A | C | A | 0.54 | No | ACA | ≤0.01 | Moderate |
| Caucasian | 7 | 11607 (2538/9069) | Dominant | R | 1.765 (1.437 − 2.168) | ≤0.01 | 69.6 | ≤0.01 | 6683 | A | C | A | 0.16 | No | ACA | ≤0.01 | Moderate | ||||
| Caucasian | 7 | 11607 (2538/9069) | Recessive | F | 2.027 (1.780 − 2.307) | ≤0.01 | 49.8 | 0.06 | 1471 | A | B | A | 0.05 | No | ABA | ≤0.01 | Strong | ||||
|
| |||||||||||||||||||||
| DIRC3 | rs966423 | T > C | Caucasian | 0.42 | 4 | 7794 (4108/3686) | Allelic | F | 1.214 (1.135 − 1.298) | ≤0.01 | 0.8 | 0.39 | 6979 | A | A | A | 0.12 | No | AAA | ≤0.01 | Strong |
|
| |||||||||||||||||||||
| FOXE1 | rs966423 | T > C | Caucasian | 0.42 | 4 | 7794 (4108/3686) | Allelic | F | 1.214 (1.135 − 1.298) | ≤0.01 | 0.8 | 0.39 | 6979 | A | A | A | 0.12 | No | AAA | ≤0.01 | Strong |
|
| |||||||||||||||||||||
| FOXE1 | rs2439302 | C > G | Caucasian | 0.48 | 3 | 6006 (2611/3395) | Allelic | F | 1.326 (1.233 − 1.426) | ≤0.01 | 50.4 | 0.13 | 6088 | A | C | A | 0.05 | No | ACA | ≤0.01 | Moderate |
|
| |||||||||||||||||||||
| MTHFR C677T | rs1801133 | C > T | Caucasian | 0.23 | 5 | 1728 (514/1214) | Allelic | R | 1.434 (1.148 − 1.791) | ≤0.01 | 36.5 | 0.18 | 897 | B | B | A | 0.24 | No | BBA | 0.04 | Strong |
| Caucasian | 5 | 1728 (514/1214) | Dominant | R | 1.423 (1.024 − 1.977) | 0.04 | 50.3 | 0.09 | 771 | B | C | A | 0.43 | No | BCA | 0.52 | Weak | ||||
| Caucasian | 5 | 1728 (514/1214) | Recessive | F | 2.279 (1.545 − 3.363) | ≤0.01 | 0.0 | 0.89 | 126 | B | A | A | 0.70 | No | BAA | 0.04 | Strong | ||||
|
| |||||||||||||||||||||
| RET A45A | rs1800858 | G > A | Caucasian | 0.28 | 6 | 3808 (1440/2368) | Allelic | F | 0.895 (0.804 − 0.997) | 0.04 | 0.0 | 0.45 | 2134 | A | A | Cab | 0.69 | No | AAC | 0.46 | Weak |
| Caucasian | 5 | 3650 (1382/2268) | Dominant | F | 0.857 (0.746 − 0.985) | 0.03 | 19.2 | 0.29 | 1782 | A | A | Cb | 0.49 | No | AAC | 0.36 | Weak | ||||
|
| |||||||||||||||||||||
| RET G691S | rs1799939 | G > A | Caucasian | 0.22 | 8 | 4971 (1992/2979) | Allelic | R | 1.322 (1.103 − 1.584) | ≤0.01 | 60.0 | ≤0.01 | 2371 | A | C | A | 0.24 | No | ACA | 0.05 | Moderate |
| Caucasian | 8 | 4971 (1992/2979) | Dominant | R | 1.356 (1.060 − 1.736) | 0.02 | 64.3 | ≤0.01 | 1979 | A | C | A | 0.55 | No | ACA | 0.27 | Weak | ||||
| Caucasian | 8 | 4971 (1992/2979) | Recessive | R | 1.437 (1.117 − 1.849) | ≤0.01 | 0.0 | 0.91 | 392 | B | A | A | 0.65 | No | BAA | 0.13 | Moderate | ||||
| Asian | 0.15 | 4 | 1672 (861/811) | Allelic | R | 1.426 (1.116 − 1.822) | ≤0.01 | 40.5 | 0.17 | 563 | B | B | A | 0.12 | No | BBA | 0.12 | Moderate | |||
| Asian | 4 | 1672 (861/811) | Dominant | R | 1.444 (1.098 − 1.899) | ≤0.01 | 34.5 | 0.21 | 496 | B | B | A | 0.25 | No | BBA | 0.21 | Weak | ||||
| Asian | 4 | 1672 (861/811) | Recessive | R | 2.002 (1.192 − 3.362) | ≤0.01 | 0.0 | 0.61 | 67 | C | A | Cd | ≤0.01 | No | CAC | 0.55 | Weak | ||||
|
| |||||||||||||||||||||
| XRCC1 | rs1799782 | C > T | Asian | 0.24 | 5 | 3388 (1404/1984) | Recessive | R | 1.777 (1.245 − 2.537) | ≤0.01 | 53.7 | 0.07 | 321 | B | C | Cd | 0.93 | No | BCC | 0.14 | Weak |
|
| |||||||||||||||||||||
| XRCC3 | rs861539 | C > T | Caucasian | 0.31 | 6 | 2611 (1029/1582) | Allelic | R | 1.252 (1.060 − 1.478) | ≤0.01 | 37.8 | 0.15 | 1641 | A | B | A | 0.12 | No | ABA | 0.13 | Moderate |
| Caucasian | 6 | 2611 (1029/1582) | Recessive | F | 1.427 (1.114 − 1.826) | ≤0.01 | 0.0 | 0.42 | 321 | B | A | Cd | 0.02 | No | BAC | 0.12 | Weak | ||||
| Asian | 0.19 | 5 | 3367 (1384/1983) | Allelic | R | 1.507 (1.256 − 1.808) | ≤0.01 | 51.8 | 0.08 | 1501 | A | C | A | 0.78 | No | ACA | ≤0.01 | Moderate | |||
| Asian | 5 | 3367 (1384/1983) | Dominant | R | 1.485 (1.178 − 1.871) | ≤0.01 | 54.3 | 0.68 | 1250 | A | C | A | 0.31 | No | ACA | 0.03 | Moderate | ||||
| Asian | 5 | 3367 (1384/1983) | Recessive | F | 2.103 (1.614 − 2.739) | ≤0.01 | 0.0 | 0.54 | 251 | B | A | A | 0.28 | No | BAA | ≤0.01 | Strong | ||||
F: meta-analysis was performed under the fixed-effects model. R: meta-analysis was performed under the random-effects model. †Frequency of minor allele in controls. ‡Number of test allele or genotype. §Venice criteria grades are amount of evidence, replication of the association, and protection from bias. ¶Cumulative epidemiological evidence as graded by the combination of results from the Venice criteria and FPRP. aThe grade of C is given because the OR value is between 0.87 and 1.15, and the association is not replicated by GWAS or GWAS meta-analysis. bThe grade of C is given for no significant association existed by excluding the first published study. dThe grade of C is given for significant publication bias (Pegger ≤0.05)
Table 3.
OR and P value for probable significant variants with insufficient data.
| Gene | Variant | Alleles | Ethnicity | MAF† | Studies | Number evaluation | Risk of meta-analysis | PQ | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample size (case/control) | Genetic models | Effect model | OR (95%CI) | P value | I (%) | |||||||
| CYP1A2F | rs762551 | A > C | Caucasian | 0.44 | 2 | 688 (170/518) | Recessive | F | 1.921 (1.163 − 3.174) | ≤0.01 | 0.0 | 0.55 |
| FTO | rs1477196 | G > A | Overall | 0.32 | 2 | 2188 (1044/1144) | Allelic | F | 1.176 (1.037 − 1.334) | ≤0.01 | 43.7 | 0.18 |
| FTO | rs8047395 | A > G | Overall | 0.42 | 2 | 2191 (1046/1145) | Allelic | R | 1.235 (1.009 − 1.511) | 0.04 | 61.8 | 0.11 |
| FTO | rs11642841 | C > A | Overall | 0.18 | 2 | 2195 (1046/1149) | Allelic | F | 0.784 (0.653 − 0.941) | ≤0.01 | 38.3 | 0.20 |
| FTO | rs17817288 | G > A | Overall | 0.44 | 2 | 2194 (1045/1149) | Recessive | F | 1.410 (1.148 − 1.732) | ≤0.01 | 0.0 | 0.60 |
| IL-18-127C-T | rs360717 | C > T | Overall | 0.34 | 2 | 721 (130/591) | Allelic | F | 1.652 (1.192 − 2.288) | ≤0.01 | 25.4 | 0.27 |
| miR-608 | rs4919510 | G > C | Asian | 0.42 | 2 | 2975 (1193/1782) | Recessive | F | 0.813 (0.664 − 0.997) | 0.05 | 0.0 | 0.40 |
| TSHR | rs1991517 | A > C | Overall | 0.27 | 2 | 1239 (566/673) | Allelic | F | 1.250 (1.046 − 1.494) | ≤0.01 | 0.0 | 0.83 |
| XRCC3 | rs56377012 | A > G | Asian | 0.07 | 2 | 1229 (459/770) | Recessive | F | 9.421 (4.581 − 19.378) | ≤0.01 | 0.0 | 0.86 |
F: meta-analysis was performed under the fixed-effects model. R: meta-analysis was performed under the random-effects model. Overall: two or more ethnicities were reported in the study. †Frequency of minor allele in controls.
Sensitivity analysis was performed for all significantly associated SNPs and significant SNPs in subgroup analysis by removing the first published study from the total publications or studies deviated from the HWE in the controls. As a result of removing the first published study, RET rs1800858 was no longer significantly associated with TC under all models, neither was RET 1800862 under the allelic model. In addition, only XRCC3 rs1799794 lost the significant association with TC when removing studies deviating from the HWE. Meanwhile, publication bias was assessed by Egger's test. Obvious publication bias was shown in FOXE1 rs965513 under recessive model for the overall population, RET rs1799939 under recessive model for the Asian population, RET rs1800862 under dominant model for the Caucasian population, XRCC1 rs1799782 under recessive model for the Asian population, and XRCC3 rs861539 under recessive model for the overall population and the Caucasian population.
Next, we assessed the cumulative epidemiological evidence of significant associations through the Venice criteria. Of all the 19 SNPs significantly associated with TC, 3 SNPs were found strongly associated with TC risk (POU5F1B rs6983267, FOXE1 rs966423, and TERT rs2736100), 6 SNPs were found moderately associated with TC risk (NKX2-1 rs944289, FOXE1 rs1867277, FOXE1 rs2439302, MTHFR rs1801133, RET rs1799939, and RET rs1800863), and 10 SNPs were found weakly associated with TC (miR-146a rs2910164, FOXE1 rs71369530, FOXE1 rs907580, FOXE1 rs965513, FOXE1 rs1443434, FOXE1 rs30215269, RET rs1800858, RET rs1800862, XRCC3 rs1799794, and XRCC3 rs861539). As for the subgroup analysis of ethnicity, 4 SNPs were assessed as a strong association with TC (POU5F1B rs6983267, NKX2-1 rs944289, DIRC3 rs966423, and FOXE1 rs966423), 4 SNPs were assessed as a moderate association with TC (MTHFR rs1801133, RET rs1799939, XRCC3 rs861539, and FOXE1 rs965513), and 3 SNPs were assessed as a weak association with TC (FOXE1 rs2439302, RET rs1800858, and XRCC1 rs1799782).
Furtherly, we valued the cumulative epidemiological evidence of associations according to the FPRP value calculated at a prior probability of 0.05 and used the statistical power to detect an odds ratio of 1.5. As a result, 7 SNPs (POU5F1B rs6983267, FOXE1 rs966423, TERT rs2736100, NKX2-1 rs944289, FOXE1 rs1867277, FOXE1 rs2439302, and RET rs1799939) were graded as strong cumulative epidemiological evidence of association with TC risk and 4 SNPs (NKX2-1 rs944289, FOXE1 rs1867277, FOXE1 rs2439302, and RET rs1799939) therein were upgraded from moderate to strong as the FPRP value. 4 SNPs were graded as a moderate association with TC (FOXE1 rs71369530, FOXE1 rs965513, MTHFR rs1801133, and XRCC3 rs861539). Of these, the cumulative epidemiological evidence of 3 SNPs (FOXE1 rs71369530, FOXE1 rs965513, and XRCC3 rs861539) was upgraded from weak to moderate and 1 SNP (MTHFR rs1801133) was maintained as a moderate association. 8 SNPs were graded as weak associations with TC risk (miR-146a rs2910164, FOXE1 rs907580, FOXE1 rs1443434, FOXE1 rs30215269, RET rs1800858, RET rs1800862, XRCC3 rs1799794, and RET rs1800863). Only 1 SNP (RET rs1800863) was downgraded from moderate to weak, and all others were still maintained as weak associations with TC risk.
In addition, in the subgroup analysis, 7 SNPs were graded as strong associations with TC after calculating FPRP value (POU5F1B rs6983267, NKX2-1 rs944289, DIRC3 rs966423, FOXE1 rs966423, MTHFR rs1801133, XRCC3 rs861539, and FOXE1 rs965513), in which the cumulative epidemiological evidence of MTHFR rs1801133, XRCC3 rs861539, and FOXE1 rs965513 was upgraded from moderate to strong. 2 SNPs were graded as a moderate association with TC (FOXE1 rs2439302 and RET rs1799939), and the association of FOXE1 rs2439302 was upgraded from weak to moderate. 2 SNPs were still maintained as weak association with TC based on the FPRP value (RET rs1800858 and XRCC1 rs1799782).
4. Discussion
In this study, we collected data about associations between polymorphisms and TC from publications, performed meta-analyses, and valued the cumulative epidemiological evidence of associations by the Venice criteria and FPRP test, which extended our understanding of true associations between SNPs and TC etiology.
DIRC3, first identified as a fusion transcript in familial renal carcinoma as early as 2003, was identified to affect thyroid-stimulating hormone levels and promote TC development through decreasing thyroid epithelium differentiation [18, 19]. The SNP rs966423 located in 2q35 of the DIRC3 gene, within a lncRNA, was valued as strong evidence for association with TC risk in our study. The allele C mutation increased TC risk in the overall population and the Caucasian population compared with the wild-type allele T (OR = 1.227 and OR = 1.214, respectively). However, lack of data resulted in ambiguous associations for the Asian population. As susceptibility genetic loci of DIRC3 were also commonly found in GWAS in the Korean population [20], further investigation for SNPs on DIRC3 in the Asian population is necessary.
The TERT gene is a catalytic subunit of telomerase and plays an essential part in cellular immortality by maintaining telomere length at the end of chromosomes, which exhibited low or no expression in normal cells but highly expressed in 85%–90% of tumor cells and stem cells [21–23]. The SNP rs2736100 is located in intron No. 2 of TERT gene and has a genotype-specific impact on TERT expression [24]. In our meta-analyses, 5 studies with a sample size of over 10000 subjects demonstrated its true evidence of strongly increasing TC risk in the Asian population, especially for the Chinese population. GWAS conducted by Julius Gudmundsson et al. confirmed the similar result in populations of European ancestry (rs2736100(C): OR = 1.11; P = 7.3 × 10^4) [25].
The SNP rs6983267 is located in chromosome 8q24 and has been identified to be associated with several cancers, such as prostate, ovary, colon, and several other carcinomas [26, 27]. POU5F1B (also known as POU5F1P1) is the nearest gene of rs6983267, which can probably encode a functional protein contributing to carcinogenesis by acting as a weak transcriptional activator [28]. We found strong epidemiological evidence of increasing TC risk among the overall population, especially in the Caucasian population. A higher TC risk among Caucasians than Asians was demonstrated in our study, and it was consistent with the result of the meta-analyses performed by Zhu et al. [26], which may be related to the lower mutation of risk allele G among Asians than Caucasians.
The SNP rs944289 is located in a 249 kb LD region near the gene of NK2 homeobox 1 neighborhood (NKX2-1), which plays a vital role in thyroid morphogenesis regulating via encoding thyroid transcription factor 1 (TTF1) [27]. Previous studies found that it is significantly associated with TC risk in the Japanese and Icelandic populations, but not associated with that in the Belarusian population [27, 29]. Strong evidence of increasing TC risk among three populations was confirmed in our meta-analyses with over 10000 subjects. Previous publication is referred to a probable relationship between rs944289 and female TC susceptibility for a higher prevalence of allele T in female patients of TC [30].
In our study, RET rs1799939 was found significantly increasing with the TC risk by 1.535-fold and had strong epidemiological evidence in the overall population. A change from allele G to allele A of rs1799939 may activate RET via leading to an amino acid change from glycine to serine, which played a vital role in thyroid carcinogenesis [31, 32].
Forkhead factor E1 (FOXE1), also called TTF2 for thyroid transcription factor (2), was firstly isolated from cDNA of mouse and modified the development of the thyroid gland and their expression in thyroid tumors through encoding thyroid-specific transcription factors [33, 34]. For both rs1867277 and rs2439302, strong accumulative epidemiological evidence of increasing TC risk was demonstrated among Caucasians in our study. Previous publications have referred that allele A of rs1867277 was significantly related to TC risk in Poles [35]. As one of the most specific thyroid transcription factors, FOXE1 could identify thyroperoxidase and thyroglobulin, which contributed a lot in tumor transformation [36], but lack of sufficient data resulted in the ambiguous association among the Asian population, which need further accumulation and investigation about other ethnicities.
4 SNPs in our study were demonstrated as a moderate association with TC risk and 8 SNPs as weak associations. For SNPs such as FOXE1 rs965513, MTHFR rs1801133, XRCC3 rs861539, and XRCC3 rs1799794, a different epidemiological evidence for associations was observed in different ethnicities or genetic models. In addition to ethnic heterogeneity, the influence of diverse genetic behaviors and multiple environments should also be considered in further well-designed studies. Due to insufficient data, only OR and P value were calculated for 19 SNPs in which 7 SNPs revealed probably increased TC risk, while 2 SNPs might decrease the risk of TC. Further large size studies were expected to identify the actual association for these SNPs.
A total of 17 SNPs showed no association with TC risk in meta-analyses. A similar result was also found in the meta-analysis of Kang et al. that ATM variants might not be important dominants of TC susceptibility [37]. Besides, 5 SNPs had a sample size of more than 6000 subjects with the MAFs ranging from 10% to 30%. Based on the detection level or value setting at 1.15 in the additive model, the meta-analyses can provide about 86% power with a MAF of 10% and improve 97% power with a MAF of 20%. Therefore, no significant results may be presented for these five SNPs in the future TC susceptibility investigation with a similar sample size.
Certain inevitable limitations existed in this study: (i) despite the full trade-off between inclusion and exclusion criteria, some articles may have been missed; (ii) owing to the insufficient of some data, meta-analyses could not be performed for SNPs included in each ethnicity and genetic model; (iii) study was designed only for associations among SNPs and TC susceptibility, but not involved in tumor progression, metastasis, and prognosis of TC; and (iv) factors included in this study were only ethnicity and genetic models, and other factors such as pathological types of TC and radiation exposure should be considered to further assess the association. Despite these limitations, our study provides an updated and comprehensive evaluation of the TC susceptibility and provides a reference for further genetic research.
In conclusion, our study comprehensively assesses the cumulative epidemiological evidence of significant associations among SNPs and TC susceptibility based on the Venice criteria and FPRP test. Seven SNPs were identified as strong evidence of associations with TC risk, as well as four SNPs with moderate evidence. We provided an updated understanding of TC susceptibility and inspired further investigation into gene polymorphism and clinic strategy of TC.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Disclosure
Ran Ran and Gang Tu are co-first authors.
Conflicts of Interest
The authors confirm that there are no conflicts of interest.
Authors' Contributions
Ran Ran and Gang Tu contributed equally to this work.
Supplementary Materials
Supplementary Table 1: insignificant variants from meta-analysis. Supplementary Table 2: insignificant variants in subtype analysis from meta-analysis.
References
- 1.Jones A. M., Howarth K. M., Martin L., et al. Thyroid cancer susceptibility polymorphisms: confirmation of loci on chromosomes 9q22 and 14q13, validation of a recessive 8q24 locus and failure to replicate a locus on 5q24. Journal of Medical Genetics . 2012;49(3):158–163. doi: 10.1136/jmedgenet-2011-100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gudmundsson J., Sulem P., Gudbjartsson D. F., et al. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nature Genetics . 2009;41(4):460–464. doi: 10.1038/ng.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park K. S., Saindane M., Yang E. Y., et al. Selective inhibition of V600E-mutant BRAF gene induces apoptosis in thyroid carcinoma cell lines. Annals of Surgical Treatment and Research . 2021;100(3):127–136. doi: 10.4174/astr.2021.100.3.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J., Li C., Wang C., Ai Z. Common genetic variant on 14q13.3 contributes to thyroid cancer susceptibility: evidence based on 12 studies. Molecular Genetics and Genomics . 2015;290(3):1125–1133. doi: 10.1007/s00438-014-0981-7. [DOI] [PubMed] [Google Scholar]
- 5.Ai L., Liu X., Yao Y., Yu Y., Sun H., Yu Q. Associations between rs965513/rs944289 and papillary thyroid carcinoma risk: a meta-analysis. Endocrine . 2014;47(2):428–434. doi: 10.1007/s12020-014-0256-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y.-H., Zhang Y.-Q. Exploration of the association between FOXE1 gene polymorphism and differentiated thyroid cancer: a meta-analysis. BMC Medical Genetics . 2018;19(1):p. 83. doi: 10.1186/s12881-018-0604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau J., Ioannidis J. P., Schmid C. H. Summing up evidence: one answer is not always enough. The Lancet . 1998;351(9096):123–127. doi: 10.1016/s0140-6736(97)08468-7. [DOI] [PubMed] [Google Scholar]
- 8.Ma X., Zhang B., Zheng W. Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut . 2014;63(2):326–336. doi: 10.1136/gutjnl-2012-304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ioannidis J. P., Boffetta P., Little J., et al. Assessment of cumulative evidence on genetic associations: interim guidelines. International Journal of Epidemiology . 2008;37(1):120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 10.Langevin S. M., Ioannidis J. P. A., Vineis P., Taioli E. Assessment of cumulative evidence for the association between glutathione S-transferase polymorphisms and lung cancer: application of the Venice interim guidelines. Pharmacogenetics and Genomics . 2010;20(10):586–597. doi: 10.1097/fpc.0b013e32833c3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian J., Liu C., Liu G., Zuo C., Chen H. Cumulative evidence for association between genetic polymorphisms and esophageal cancer susceptibility: a review with evidence from meta‐analysis and genome‐wide association studies. Cancer Medicine . 2019;8(3):1289–1305. doi: 10.1002/cam4.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moher D., Liberati A., Tetzlaff J., Altman D. G. Preferred reporting Items for systematic reviews and meta-analyses: the PRISMA statement. Journal of Clinical Epidemiology . 2009;62(10):1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Sagoo G. S., Little J., Higgins J. P. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Medicine . 2009;6:p. e28. doi: 10.1371/journal.pmed.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins J. P. T., Thompson S. G., Deeks J. J. Measuring inconsistency in meta-analyses. BMJ . 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begg C. B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics . 1994;50(4):1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 16.Egger M., Smith G. D., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ . 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacholder S., Chanock S., Garcia-Closas M., El ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. Journal of the National Cancer Institute: Journal of the National Cancer Institute . 2004;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khoury M. J., Bertram L., Boffetta P., et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. American Journal of Epidemiology . 2009;170(3):269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudmundsson J., Sulem P., Gudbjartsson D. F., et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nature Genetics . 2012;44(3):319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwangbo Y., Park Y. J. Genome-Wide association studies of autoimmune thyroid diseases, thyroid function, and thyroid cancer. Endocrinology and Metabolism . 2018;33(2):175–184. doi: 10.3803/enm.2018.33.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z., Li Q., Li K., et al. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene . 2013;32(36):4203–4213. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- 22.Blasco M. A. Telomeres and human disease: ageing, cancer and beyond. Nature Reviews Genetics . 2005;6(8):611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 23.González-Suárez. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. The EMBO Journal . 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge M., Shi M., An C., et al. Functional evaluation of TERT-CLPTM1L genetic variants associated with susceptibility of papillary thyroid carcinoma. Scientific Reports . 2016;6(1) doi: 10.1038/srep26037.26037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudmundsson J., Thorleifsson G., Sigurdsson J. K., et al. A genome-wide association study yields five novel thyroid cancer risk loci. Nature Communications . 2017;8(1):p. 14517. doi: 10.1038/ncomms14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu M., Wen X., Liu X., Wang Y., Liang C., Tu J. Association between 8q24 rs6983267 polymorphism and cancer susceptibility: a meta-analysis involving 170,737 subjects. Oncotarget . 2017;8(34):57421–57439. doi: 10.18632/oncotarget.18960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogounovitch T. I., Bychkov A., Takahashi M., et al. The common genetic variant rs944289 on chromosome 14q13.3 associates with risk of both malignant and benign thyroid tumors in the Japanese population. Thyroid . 2015;25(3):333–340. doi: 10.1089/thy.2014.0431. [DOI] [PubMed] [Google Scholar]
- 28.Panagopoulos I., Möller E., Collin A., Mertens F. The POU5F1P1 pseudogene encodes a putative protein similar to POU5F1 isoform 1. Oncology Reports . 2008;20:1029–1033. [PubMed] [Google Scholar]
- 29.Matsuse M., Takahashi M., Mitsutake N., et al. The FOXE1 and NKX2-1 loci are associated with susceptibility to papillary thyroid carcinoma in the Japanese population. Journal of Medical Genetics . 2011;48(9):645–648. doi: 10.1136/jmedgenet-2011-100063. [DOI] [PubMed] [Google Scholar]
- 30.Chen L., Jiang Y., Chen C. Correlation of FOXE1 gene polymorphism with papilliary thyroid carcinoma in Yunnan China. Modern Oncology . 2013;21(9):1958–1963. [Google Scholar]
- 31.Arighi E., Borrello M. G., Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine & Growth Factor Reviews . 2005;16(4–5):441–467. doi: 10.1016/j.cytogfr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Durbec P., Marcos-Gutierrez C. V., Kilkenny C., et al. GDNF signalling through the Ret receptor tyrosine kinase. Nature . 1996;381(6585):789–793. doi: 10.1038/381789a0. [DOI] [PubMed] [Google Scholar]
- 33.Zannini M., Avantaggiato V., Biffali E. TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. The EMBO Journal . 1997;16(11):3185–3197. doi: 10.1093/emboj/16.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maillard S., Damiola F., Clero E., et al. Common variants at 9q22.33, 14q13.3, and ATM loci, and risk of differentiated thyroid cancer in the French Polynesian population. PLoS One . 2015;10(4) doi: 10.1371/journal.pone.0123700.e0123700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikitski A. V., Rogounovitch T. I., Bychkov A., et al. Genotype analyses in the Japanese and Belarusian populations reveal independent effects of rs965513 and rs1867277 but do not support the role of FOXE1 polyalanine tract length in conferring risk for papillary thyroid carcinoma. Thyroid . 2017;27(2):224–235. doi: 10.1089/thy.2015.0541. [DOI] [PubMed] [Google Scholar]
- 36.Fernández L. P., López-Márquez A., Martínez Á. M., Gómez-López G., Santisteban P. New insights into FOXE1 functions: identification of direct FOXE1 targets in thyroid cells. PLoS One . 2015;8 doi: 10.1371/journal.pone.0062849.e62849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang J., Deng X.-Z., Fan Y.-B., Wu B. Relationships of FOXE1 and ATM genetic polymorphisms with papillary thyroid carcinoma risk: a meta-analysis. Tumor Biology . 2014;35(7):7085–7096. doi: 10.1007/s13277-014-1865-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: insignificant variants from meta-analysis. Supplementary Table 2: insignificant variants in subtype analysis from meta-analysis.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


