Abstract
Retinoblastoma (RB) tumor suppressor family pocket proteins induce cell cycle arrest by repressing transcription of E2F-regulated genes through both histone deacetylase (HDAC)-dependent and -independent mechanisms. In this study we have identified a stable complex that accounts for the recruitment of both repression activities to the pocket. One component of this complex is RBP1, a known pocket-binding protein that exhibits both HDAC-dependent and -independent repression functions. RB family proteins were shown to associate via the pocket with previously identified mSIN3-SAP30-HDAC complexes containing exclusively class I HDACs. Such enzymes do not interact directly with RB family proteins but rather utilize RBP1 to target the pocket. This mechanism was shown to account for the majority of RB-associated HDAC activity. We also show that in quiescent normal human cells this entire RBP1-mSIN3-SAP30-HDAC complex colocalizes with both RB family members and E2F4 in a limited number of discrete regions of the nucleus that in other studies have been shown to represent the initial origins of DNA replication following growth stimulation. These results suggest that RB family members, at least in part, drive exit from the cell cycle by recruitment of this HDAC complex via RBP1 to repress transcription from E2F-dependent promoters and possibly to alter chromatin structure at DNA origins.
The retinoblastoma (RB) tumor suppressor gene product, pRB, regulates transcriptional events important for cell proliferation (for reviews, see references 17 and 45). A major target of pRB is the E2F family of transcription factors that control expression of many genes required for DNA synthesis and cell cycle progression. Binding of pRB to E2F species inhibits expression of E2F-regulated genes, resulting in withdrawal from the cell cycle (for reviews, see references 3, 26, and 45). pRB and related pocket proteins p107 and p130 utilize multiple mechanisms to elicit this effect. With certain promoters, binding via the pocket to the activation domain of E2F inhibits E2F-mediated transactivation (23, 27, 28). But this mechanism does not explain how, with other promoters, E2F binding sites function as negative regulatory elements (13, 31, 47, 61). In these cases, RB family members function as transcriptional repressors, which utilize E2F proteins as DNA-docking factors. Studies using pRB fused to heterologous DNA binding domains indicated that the pRB pocket functions as an active repressor (1, 7, 52, 61, 62). This repression function, and not pRB-mediated inhibition of the E2F transactivation domain, was shown recently to be required for G1 arrest triggered by transforming growth factor β, p16INK4a, and contact inhibition (67) and for blockage of the cell cycle by pRB (25, 52). Thus, active repression by RB family members is important for exit from the cell cycle.
Several mechanisms have been proposed to account for repression by pRB. Dean and coworkers suggested that the pocket might prevent transactivators from interacting with components of the TFIID complex (41, 61). Rose et al. proposed that E2F itself may be affected in this way by the pocket (50). The pocket is now known to interact simultaneously with E2F and other cellular factors, and our work and that of others suggested that RB-binding proteins might utilize conserved LXCXE motifs to recruit repression activities to the pocket (6, 16, 20, 21, 31, 39, 40, 41, 42, 57, 62). Some of the models proposed in these studies (6, 16, 41, 42, 57) stressed the importance of chromatin structure in regulating transcriptional activity (for reviews, see references 4, 36, and 53). Active repression by pRB therefore could involve a mechanism by which condensed chromatin structure is enhanced through the creation of hypoacetylated histones or other target proteins.
Understanding of the enzyme complexes involved in histone acetylation and deacetylation has increased greatly in recent years (for reviews see references 2 and 37). The catalytic subunits (Rpd3, HDA, and HOS) of histone deacetylase (HDAC) complexes from yeast were cloned (8, 51), and several mammalian versions have now been identified within two classes (14, 18, 44, 55, 60, 65, 66). HDAC1, HDAC2, and HDAC3 are class I enzymes that share homology with yeast Rpd3, whereas HDAC4, HDAC5, HDAC6, and HDAC7 are class II enzymes that share homology with yeast HDA1. At least two distinct HDAC complexes (mSIN3-HDAC and NURD) were isolated in mammalian cells (58, 59, 64, 68, 69, 70, 71). Both complexes contain the class I HDACs, HDAC1 and HDAC2. Most of the subunits in the NURD and mSIN3-HDAC complexes have now been identified. Some subunits, like RBAP46, RBAP48, HDAC1, and HDAC2, are present in both complexes; however, the majority of subunits in these complexes are distinct. The NURD complex contains an ATPase subunit, Mi2, MTA2, and MBD3, whereas the mSIN3 contains the mSIN3A/B, SAP30, and SAP18 components (2, 64, 68, 69, 70, 71). The mSIN3-HDAC complexes are recruited by many transcriptional repressors in a pattern evolutionarily conserved from yeasts to humans (32, 54, 71). In contrast to the mSIN3-HDAC complex, the NURD complex is able to deacetylate nucleosomal templates, and it is recruited to methylated DNA, suggesting a role in gene silencing by DNA methylation (46, 59, 69).
The pocket domains of RB family members were shown not long ago to repress E2F-dependent transcription by recruiting HDAC1, HDAC2, and HDAC3 (5, 6, 39, 41, 42). These results suggested that HDACs are important for the active repression function of RB family members. However, drug inhibition studies by our group and others indicated that both HDAC-dependent and -independent activities function at the pocket simultaneously, suggesting that other components are also involved with active repression by RB family proteins (39, 41).
We have previously demonstrated that an LXCXE-containing RB-binding protein, RBP1, functions as an adapter protein by recruiting class I HDACs to the pocket. In addition, RBP1 possesses an HDAC-independent repression activity. Therefore, the recruitment of RBP1 to the pocket provides a model that explains both HDAC-dependent and -independent repression by RB family members (39, 40). Furthermore, it has recently been suggested that RBP1 is present as a component of the mSIN3-HDAC complex (D. Reinberg, unpublished data). In this report, we present evidence that HDAC activity present in the mSIN3-HDAC complex is recruited to RB family members via RBP1 and not via a direct interaction between the pocket and an IXCXE motif found in HDAC1 and HDAC2, as suggested previously (6, 21, 41, 42). We further demonstrate the physiological importance of the recruitment of the mSIN3-HDAC complex by RB family members in quiescent cells.
MATERIALS AND METHODS
Plasmids and antibodies.
Bacterial expression plasmids expressing glutathione S-transferase (GST)–pRB (379-928), GST-pRB(379-928-C706F), GST-p107(252-936), and GST-p107(252-936-C713F) were gifts from Bill Kaelin and Mark Ewen (19, 33). Plasmid constructs expressing GST-SAP30 and pET28-SAP30 for in vitro translation have been described before (71). pVZmSIN3B and pVZmSIN3A constructs for in vitro translation were gifts from Bob Eisenman. pGEM-RbAp48 and pcDNA3-Flag-HDAC1 plasmids used for in vitro translation were provided by Eva Lee (49) and Ed Seto, respectively. Plasmids expressing GST-HDAC1 and GST-HDAC3 have been described elsewhere (56). All GST fusion proteins with various truncations of RBP1, including GST-R1, GST-R2, GST-ARID, GST-dl1208C, and GST-dl747C, were generated by subcloning cDNAs from the Gal4 fusion mammalian expression plasmids expressing the corresponding mutants (39) into plasmid pGEX-2TK. Plasmid expressing GST-E1A in bacteria was described before (39).
Monoclonal antibodies against RBP1 used in coimmunoprecipitation and immunofluorescence studies (LY11, LY32, and LY48) were gifts from Bill Kaelin and Jim DeCaprio. LY11 and LY32 have been described previously (39, 40). Amino-terminal RBP1 polyclonal antiserum was produced commercially by injecting an amino-terminal RBP1 peptide (CLKQDNTTQLVQDDQVKGPLRV) into rabbits (Genemed Synthesis Inc.). Monoclonal antibody NM11 against p300 was kindly provided by Betty Moran (12). Antibodies against pRB purchased commercially included XZ91 and G3-245 (Pharmingen), IF8 (Santa Cruz Biotechnology), and polyclonal C-15 (Santa Cruz); monoclonal antibody C36 was obtained from Ed Harlow. Rabbit polyclonal antibodies against HDAC1, HDAC2, and HDAC3 were described previously, and goat polyclonal antibodies against HDAC1 (C-19) and HDAC2 (C-19) were purchased from Santa Cruz. Monoclonal antibody 15G12 against RBAP46/48 (GeneTex) and polyclonal antibodies AK11 and AK12 against mSIN3A and mSIN3B, respectively, were purchased from Santa Cruz. Rabbit polyclonal antibody against SAP30 has been described elsewhere (71). Finally, monoclonal antibody against the His6 epitope was purchased commercially (Amersham Pharmacia).
Purification of His6-tagged and GST fusion proteins.
His6-tagged HDAC1, HDAC2, and HDAC3 proteins were purified from SF9 insect cells infected with baculovirus expressing these proteins under a constitutive promoter (a gift from Ed Seto and Yi Zhang). All GST fusion proteins and His-SAP30 were purified from Escherichia coli BL21-DE3. Competent bacterial cells were transformed with pGEX-2TK plasmids containing cDNAs encoding appropriate proteins. Transformed cells were grown in 2YT medium at 30°C with agitation until the optical density at 595 nm reached 1.2. Isopropyl-β-d-thiogalactopyranoside (50 mg/liter, final concentration) was used to induce expression of GST fusion proteins for another 1.5 h. Cells were harvested and lysed by sonication in buffer B (50 mM Tris-HCl [pH 7.5] containing 200 mM NaCl, 5 mM EDTA, 10 mM mercaptoethanol, 1% [vol/vol] Triton X-100, and 1 mM protease inhibitor cocktail). GST fusion proteins were isolated from extracts by incubation with 1 ml of glutathione-Sepharose 4B (Pharmacia) per liter for 2 to 4 h. Proteins bound to beads were washed six times with buffer B and eluted by incubation with 20 mM reduced glutathione (Sigma). Eluted proteins were spin dialyzed and concentrated by Centricon spin columns (Millipore). Concentrations of purified proteins were determined by standard Bradford assays.
In vitro binding assays.
mSIN3A, mSIN3B, RBAP48, HDAC1, SAP30, and luciferase proteins were labeled and synthesized in the presence of [35S]methionine protein labeling mix (NEN) using the TnT coupled reticulocyte lysate system (Promega). Three-microgram aliquots of GST fusion proteins purified from bacteria were incubated with 5 μl of in vitro-translated proteins with 10 μl of glutathione-Sepharose 4B beads (Pharmacia) in 1 ml of buffer A (1× phosphate-buffered saline, 0.1% NP-40, 1 mM aprotinin, 1 mM leupeptin, 1 mM pepstatin) for 2 h at 4°C. GST pull-down assays were performed using six washes, and the resulting proteins associated with the beads were eluted with 2X sample buffer. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using gels containing 10% polyacrylamide. Gels were fixed by dimethyl sulfoxide–2,5-diphenyloxazole treatment and then dried and analyzed by autoradiography using Kodak X-Omat film. In vitro binding assays using purified His6-tagged proteins were performed similarly, except that instead of in vitro-translated proteins, 3-μg aliquots of His6-tagged proteins were incubated with 3 μg of GST fusion proteins. GST pull-down assays were performed, and the samples were resolved by SDS-PAGE using 10% polyacrylamide gels. Proteins were then transferred to polyvinylidene difluoride membranes (Millipore), which were probed with anti-His6 monoclonal antibody (Amersham Pharmacia) and then by horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Jackson Laboratories). Binding was detected by enhanced Luminol reagent (NEN Life Science).
HDAC assays.
Three-microgram aliquots of GST proteins purified from bacteria were incubated with H1299 cell nuclear extracts in 10 μl of glutathione-Sepharose 4B beads (Pharmacia). GST pull-down assays were performed, and the resulting beads were incubated with 10,000 cpm of 3H-labeled histones extracted from HeLa cells, as described previously (60), in 0.1 ml of buffer H (50 mM Tris-HCl [pH 7.5] containing 100 mM NaCl, 0.1 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride). The reaction mixtures were incubated at 37°C for 2 h, and reactions were stopped by the addition of 50 μl of 0.1 M HCl–0.16 M acetic acid solution. Then 0.5 ml of ethyl acetate was used to extract [3H]acetate. The resulting mixtures were separated into aqueous and organic phases by centrifugation, and 0.4 ml of the upper organic phase was quantified by scintillation counting.
Immunodepletion.
Lysates from one half of a 150-mm-diameter plate of H1299 cells were incubated with 5 to 10 μl of antibody in 10 μl of protein G- or protein A-Sepharose in buffer A containing 1 μM protease inhibitor cocktail for 4 h at 4°C. Immunoprecipitated proteins were washed six times with buffer A and subjected to HDAC assays to detect HDAC activity. The remaining lysates were subjected to another round of immunoprecipitation using the same quantity of antibodies and fresh protease inhibitor cocktails. Between 7 to 10 rounds of immunodepletion were performed until background levels of HDAC activity were detected. Such lysates were incubated with 100 μl of Pansorbin cells (Calbiochem) for another 4 h at 4°C to remove excess antibodies. These extracts were used to detect associated HDAC activity.
In vivo binding assays.
H1299 cells were cultured, harvested, and lysed as described before (39). Five to 10 μl of antibody against different endogenous proteins was used to perform immunoprecipitations using lysates from 150-mm-diameter plates of cells. Immunoprecipitation was performed at 4°C for 12 h, and samples were washed and eluted as described previously (39). Associating proteins were detected by Western blot analysis using appropriate antibodies.
Immunofluorescence.
WI38 cells were seeded on plates containing coverslips and serum-starved in Dulbecco modified Eagle medium containing 0.1% fetal bovine serum for 72 h. Cells were incubated with cell proliferation labeling reagents containing bromodeoxyuridine (BrdU) and fluorodeoxyuridine (Amersham), fixed in 4% paraformaldehyde, and permeabilized in 1× phosphate-buffered saline containing 0.5% Triton X-100. Indirect immunofluorescence was performed using a monoclonal antibody against BrdU (Amersham). Fluorescein- or Texas red-conjugated secondary antibodies (Vector Laboratories) were used to detect BrdU incorporation. Cells were visualized under a light microscope and counted to evaluate BrdU incorporation. No BrdU staining was detected in serum-starved cells, whereas significant amounts of staining were detected in asynchronously growing cells. To detect the staining patterns of endogenous proteins, appropriate mouse monoclonal and rabbit polyclonal antibodies were used, and the same fluorescein- or Texas red-conjugated secondary antibodies (Vector Laboratories) were employed to detect staining. Imaging was performed using a charge-coupled device (CCD) digital camera and deconvolution microscopy using a 100× objective lens (Scanalytics).
RESULTS
RB family members do not interact directly with HDAC.
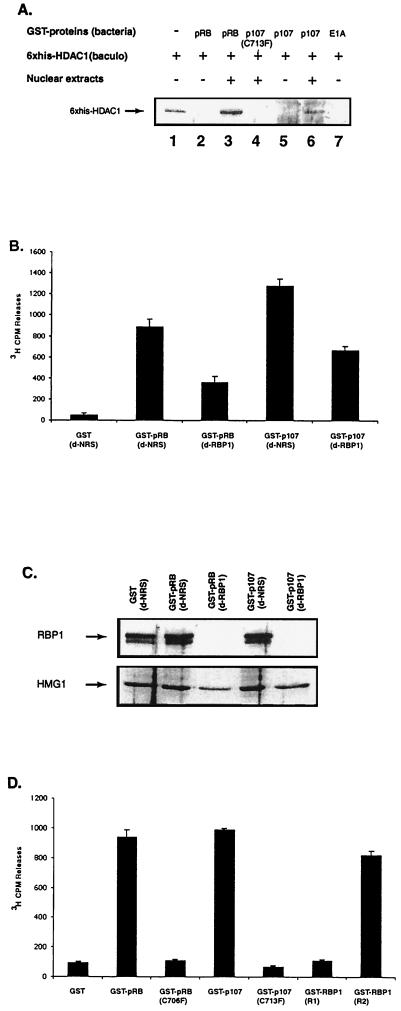
All members of the RB family of proteins appear to associate with class I HDACs, including HDAC1, HDAC2, and HDAC3 (6, 21, 39, 41, 42). A direct association between HDAC1 and RB family proteins has been suggested, based on interactions observed between purified GST-RB and in vitro-translated HDAC1 (21, 42). To examine the interaction between RB family proteins and HDAC1 further, we synthesized GST-pRB(pocket) and GST-p107(pocket) proteins in bacteria and combined each purified product with purified histidine-tagged HDAC1 isolated from SF9 insect cells infected with a baculovirus that overexpressess His6-HDAC1. Although we believe that results similar to those described below would be obtained using the pocket of p130, we have not been successful in obtaining suitable p130(pocket) material to conduct this type of experiment. Following incubation of GST fusion proteins in vitro with purified HDAC1, we performed GST pull-down experiments and detected associations by Western blotting using anti-His antibody. Figure 1A shows that no interaction between purified HDAC1 and the pocket of either pRB or p107 was detectable; however, addition of nuclear extract along with the purified proteins resulted in a significant amount of association. These results differed from previous assessments in that they suggested that another nuclear factor(s) could act as a bridge in this in vitro interaction. Similar results were also obtained using purified HDAC3 (data not shown). The association observed previously (and demonstrated below in Fig. 6B) between pRB and in vitro-translated HDAC1 had been proposed to occur via interactions between the pocket and a degenerate IXCXE motif in HDAC1 (21, 42). Because HDAC3 lacks such a motif, HDAC3 must require other factors to associate with the pRB pocket, and the data in Fig. 1 suggest that such is also the case with HDAC1. An explanation for the disparity between our results and those reported previously might relate to the fact that in vitro-translated HDAC1 is defective in HDAC activity, whereas HDAC1 purified from SF9 cells has been shown to be enzymatically active (43). But the simplest interpretation is that both HDAC1 and HDAC3 require an additional linker protein present both in nuclear extracts and the in vitro translation mixture used in the earlier studies. While we cannot exclude the possibility that the pocket can bind directly to HDACs via the IXCXE motif, this interaction may not represent the major mechanism of binding between RB family members and HDACs.
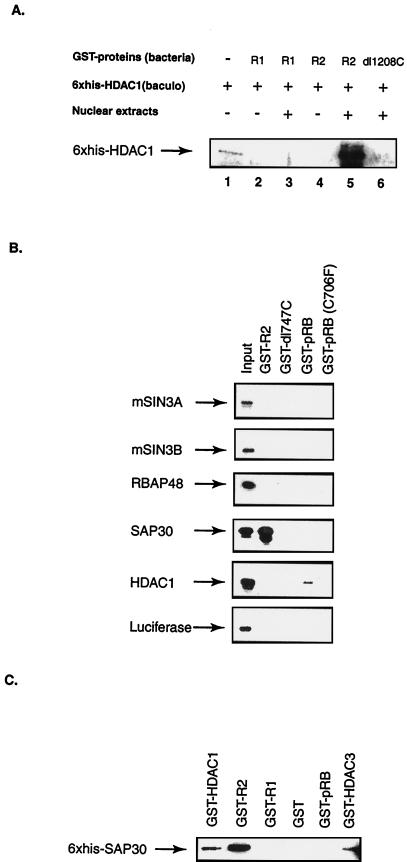
FIG. 1.
RBP1 recruits HDAC activity to RB family members via the R2 repression domain. (A) RB family members do not interact directly with HDAC1. GST fusion proteins were purified from bacteria, whereas His6-HDAC1 protein was purified from baculovirus (baculo)-infected SF9 insect cells. Mixtures were incubated in vitro, and binding was assessed as described below following separation by SDS-PAGE and western blotting using anti-His antibody. Lanes 3, 4, and 6 are samples that were incubated following addition of nuclear extracts from H1299 cells. pRB(pocket) and p107(pocket) contain only the pocket regions of these proteins. The E1A protein used contains only residues encoded by the first exon, including conserved region 1 and 2. (B) RBP1 constitutes half of the HDAC activity recruited by the pocket of RB family members. d-NRS refers to extracts depleted with NRS, and d-RBP1 denotes extracts depleted with monoclonal antibody LY11 against RBP1. Extracts were subjected to GST pull-down with or without the indicated immunodepletion and then assessed for HDAC activity by measuring the amount of [3H]acetate extractable by ethyl acetate, as described in the text. (C) RBP1 is absent in extracts depleted with monoclonal antibody LY11 against RBP1. Immunodepletion was performed 7 to 10 times until LY11 immunoprecipitates contained only background levels of HDAC activity, 40-μg aliquots of immunodepleted extracts quantified by Bradford assay were combined with various GST fusion proteins (listed at the top), as described in Materials and Methods, and loaded into each lane. RBP1 was detected by Western blotting using antibody LY32. HMG1 (bottom) was detected in these extracts using a polyclonal antibody (Pharmingen). (D) pRB and p107-associated HDAC activities are pocket dependent, and RBP1-associated HDAC activity requires the R2 repression domain. HDAC activity was determined as in Fig. 1B. Values in panels B and D represent the averages of duplicate samples from three independent experiments.
FIG. 6.
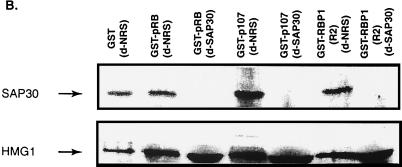
SAP30 is required for the RBP1 R2 repression domain to recruit HDAC activity. (A) SAP30 constitutes at least half of the HDAC activity recruited by the pocket of RB family members. Immunodepletion was performed as in Fig. 1 and 2, using anti-SAP30 antibodies (d-SAP30) or control NRS (d-NRS). HDAC activity was measured as in Fig. 1 and 2. (B) SAP30 is absent in extracts depleted using antibodies against SAP30. Immunodepletion was performed 7 to 10 times until anti-SAP30 immunoprecipitates associated with background levels of HDAC activity; 40 μg of immunodepleted extract quantified by Bradford assays was loaded into each lane. SAP30 was detected by the same polyclonal antibody used for immunodepletion by Western blotting following SDS-PAGE. HMG1 was detected using a polyclonal antibody (Pharmingen).
RBP1 is a factor that recruits HDAC activity to RB family members.
We have demonstrated previously that the RB-binding protein RBP1 may function as a bridging factor to recruit class I HDACs (39). Studies were therefore conducted to estimate the importance of RBP1 in this association. GST-pRB fusion proteins were shown previously to be able to recruit HDAC activity from nuclear extracts (6, 42). We have modified this assay by immunodepleting RBP1 from nuclear extracts using an anti-RBP1 antibody, LY11, and then incubating such extracts with GST-pRB protein. As shown in Fig. 1C, after seven rounds of immunodepletion with LY11 virtually no detectable RBP1 remained, as determined by Western blotting of samples containing equal amounts of protein using another anti-RBP1 antibody, LY32. With extracts depleted using nonspecific rabbit serum (NRS), significant amounts of RBP1 remained. Figure 1C also shows that such immunodepletion caused little change in the levels of another nuclear protein, HMG1. Samples were then subjected to GST pull-down analysis and assessed directly for HDAC activity to quantify the level of HDAC following LY11 or NRS immunodepletion. Figure 1B shows that significant amounts of HDAC activity were associated with the pocket of pRB and p107 after immunodepletion with NRS; however, with both pRB and p107, HDAC activity decreased to less than half that in samples immunodepleted with LY11 anti-RBP1. These results suggested that RBP1-associated HDAC activity constitutes a significant proportion of the HDAC activity associated with RB family members. It was possible that this effect might have resulted from a depletion of the overall pool of HDAC activity in the extract; however, no similar loss of RB-associated HDAC activity was observed with extracts immunodepleted using antibodies against individual class I HDACs that removed an even greater fraction of the overall HDAC pool (Fig. 2 and data not shown). The inability to abolish RB-associated HDAC activity completely may suggest that RBP1 may not be the only bridging factor recruiting HDACs to RB family members. This contention is consistent with other findings that showed that RBAP48, another RB-binding protein, and c-Ski, an LXCXE-containing RB-binding protein, are also able to recruit HDAC1 to RB family members (56; B. K. Kennedy, personal communication). In addition, the failure to abolish all activity could result from the fact that RBP1 exists in multiple isoforms (48), not all of which may be recognized by antibody LY11.
FIG. 2.
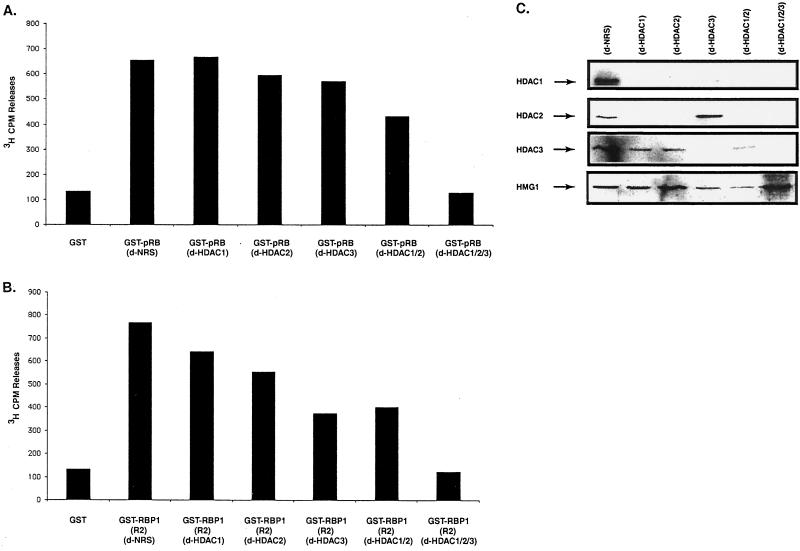
Both pRB and RBP1 recruit class I HDAC enzymes. Immunodepletion was performed using NRS (d-NRS), goat polyclonal antibody against HDAC1 (d-HDAC-1) or HDAC-2 (d-HDAC2), or rabbit polyclonal antibody against HDAC3 (d-HDAC-3). In some experiments, these antibodies were combined and used together for immunodepletion (d-HDAC1/2 and d-HDAC1/2/3). Following GST pull-down, HDAC activity was measured. Values in panels A and B represent the averages of two independent experiments. Immunodepletion was performed 7 to 10 times until anti-HDAC immunoprecipitates associated only with background levels of HDAC activity detected in HDAC assays. (A) pRB-associated HDAC activities from extracts were completely abolished with extracts depleted with HDAC1, HDAC2, and HDAC3. (B) RBP1 R2-associated HDAC activities were completely abolished in extracts depleted for HDAC1, HDAC2, and HDAC3. (C) Detection of the presence of different class I HDACs in extracts depleted using different HDAC-specific antibodies. Forty-microgram aliquots of immunodepleted extracts, as quantified by Bradford assays, were loaded into each lane, and HDACs (or control HMG1) were detected by Western blotting following SDS-PAGE.
R2 but not the R1 region of RBP1 recruits HDAC to the pocket of RB.
To determine if the HDAC activity associated with RB family members is dependent on the pocket, point mutants of pRB (C706F) and p107 (C713F), which abolish binding of RBP1 (Fig. 4C and data not shown), were used to detect pRB- and p107-associated HDAC activities. Figure 1D shows that both mutants were unable to recruit significant levels of HDAC activity, indicating that a functional pocket domain is of great importance for the association. We have shown previously that association of class I HDACs with RBP1 requires the R2 but not the R1 repression domain of RBP1. Consistent with our previous results, Figure 1D shows that R2 recruited high levels of HDAC activity, whereas R1 associated only with background levels. Thus, repression by R1 does not involve HDACs that are measurable by the present assay or that are inhibited by the HDAC inhibitor trichostatin A (TSA). All HDAC activity associated with RB family members or the R2 repression domain of RBP1 was inhibited by TSA (data not shown), in keeping with our previous in vivo repression studies (39). In addition, transcriptional repression activities associated with pRB and full-length RBP1 are known to be only partially sensitive to TSA (39, 41), suggesting that repression by RB family members may also utilize the HDAC-independent activity associated with the R1 repression domain of RBP1.
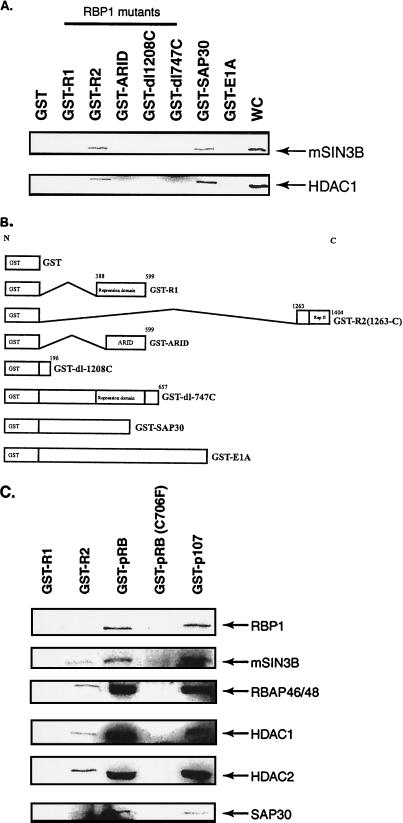
FIG. 4.
RBP1 recruits the mSIN3-HDAC complex via the R2 repression domain. GST pull-down experiments were performed using extracts from H1299 cells and the indicated GST fusion proteins bound to glutathione-Sepharose beads. (A) Only the R2 repression domain of RBP1 binds mSIN3B and HDAC1 in vitro. GST pull-downs assays were performed, and mSIN3B and HDAC1 were detected by Western blotting using appropriate antibodies. WC represents whole-cell extracts; 20 μg of GST fusion protein was used in each pull-down experiment presented in each lane. (B) Schematic diagram of RBP1 truncation mutants of GST-RBP1 fusion proteins. (C) The R2 repression domain, pRB, and p107 bind other components of mSIN3-HDAC complex, including mSIN3A, mSIN3B, HDAC1, HDAC2, RBAP46/48, and SAP30. GST pull-down experiments were performed, and binding of the indicated proteins was assessed by Western blotting using appropriate antibodies. Mutant C706F is defective in the pRB pocket.
pRB and RBP1 recruit only class I HDAC activities.
Both RB family members and RBP1 have been shown to associate with the class I HDACs, HDAC1, HDAC2, and HDAC3. To determine if either can associate with other HDACs (class II or unidentified HDACs), experiments similar to those in Fig. 1B were performed except that antibodies against specific class I HDAC species were used in seven rounds of immunodepletion. Western blotting analysis shown in Fig. 2C indicated that antibodies against either HDAC1 or HDAC2 depleted both of these species but not much HDAC3. Similar results were obtained with a mixture of anti-HDAC1 and anti-HDAC2 sera. This observation was consistent with the idea that HDAC1 and HDAC2 are present together as a complex. Anti-HDAC3 antibodies depleted not only HDAC3 but also some amounts of HDAC1, but not HDAC2. This observation suggested that human H1299 cells may contain complexes involving both HDAC1 and HDAC3, unlike Jurkat and HeLa cells studied previously (24). Addition of a mixture of antibodies against all three class I enzymes eliminated all three but had little effect on the control nuclear protein HMG1. Figure 2A shows that immunodepletion using any single anti-class I HDAC antibody had only a small effect on the levels of HDAC activity associated with pRB, whereas treatment with a mixture of antibodies recognizing all three eliminated essentially all of the pRB-associated HDAC activity. Almost identical results were obtained using the R2 domain of RBP1 (Fig. 2B), indicating that both pRB and RBP1 associate with HDAC1, HDAC2, and HDAC3 but not with other members of the HDAC family, including class II enzymes, which we have found are abundant in the extracts used in these experiments (data not shown). Furthermore, immunoprecipitation experiments using antibodies against HDAC4 and HDAC6 did not coimmunoprecipitate any RBP1 or RB family members (data not shown). These data strengthen the idea that RBP1 plays an important role as a bridge between RB family members and class I HDACs.
RBP1 recruits the mSIN3-HDAC complex.
Mammalian cells contain two known class I HDAC complexes, the mSIN3-related and NURD HDAC complexes (58, 59, 64, 68, 69, 70, 71). Antibodies against mSIN3 and the SAP30 component of the mSIN3A-HDAC complex were used to determine the association between RBP1 and this complex. Figure 3A shows that RBP1 was coimmunoprecipitated from nuclear extracts with AK11 (anti-mSIN3A) and AK12 (anti-mSIN3B) antibodies. Interestingly, large amounts of RBP1 were also detected using anti-SAP30 antibodies. An antibody that recognizes the p300 histone acetyltransferase (NM11) did not coimmunoprecipitate any RBP1 from these extracts. Figure 3B shows that immunoprecipitation of RBP1 using antibody LY11 results in the coprecipitation of mSIN3A, mSIN3B, low levels of pRB, and HDAC1. In addition, Figure 3C shows that another component of all class I HDAC complexes, RBAP46/48, is also present in immunoprecipitates prepared using anti-RBP1 or anti-pRB antibodies but not with those against p300. HDAC activity was also measured directly using these immunoprecipitates, and significant levels were detected using antibody LY11 but not with anti-p300 antibody NM11 (data not shown). These results indicated that RBP1 associates with the mSIN3-HDAC complex in vivo.
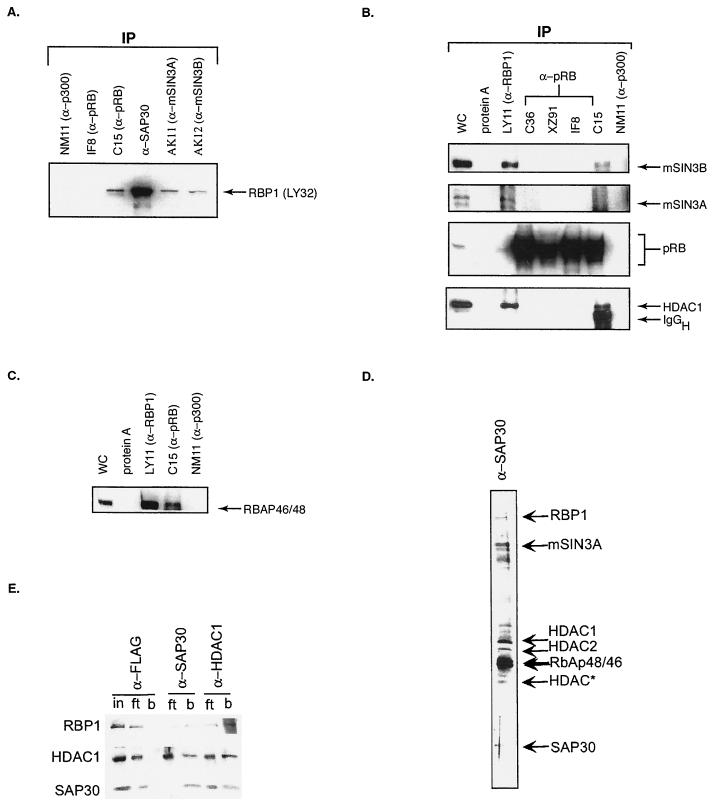
FIG. 3.
pRB associates with mSIN3-HDAC complex via RBP1. Coimmunoprecipitations (IP) were performed with H1299 extracts using the indicated antibodies against components of the mSIN3-HDAC complex, or against pRB, RBP1, or p300, under low-stringency conditions. WC represents whole-cell extracts; 20 μg of cell protein were loaded in each lane, and apart from panel E, proteins were identified by Western blotting using the antibodies indicated. (A) RBP1 coimmunoprecipitates with components of the mSIN3-HDAC complex. (B) Immunoprecipitation of RBP1 or pRB coprecipitates components of the mSIN3-HDAC complex. Antibody C-15 recognizes the carboxy terminus of pRB, whereas C36, XZ91, and IF8 recognize regions of the pRB pocket. IgGH, immunoglobulin G heavy chain. (C) Immunoprecipitation of RBP1 coprecipitates RBAP46/48. Polyclonal antibody C-15 or monoclonal antibody LY11 was used to immunoprecipitate RBP1. Anti-p300 antibody NM11 was used as a control. (D) Immunoaffinity-purified SAP30 complexes. HeLa cell extracts were immunoaffinity purified using anti-SAP30 antibody and separated by SDS-PAGE, and the purified proteins were visualized by silver staining. The input for immunoaffinity purifications was the 0.35 M KCl eluate of DE-52 columns prepared from the 0.5 M KCl phosphocellulose fraction of HeLa cell nuclear extracts. Proteins were washed with a buffer containing 0.5 M KCl and 0.05% NP-40 and eluted with Tris-glycine (pH 2.6). (E) Western blotting of input (in), flowthrough (ft), and bound (b) fractions of HeLa cell nuclear extracts subjected to immunoaffinity purification using anti-SAP30 and anti-HDAC1 antibodies. Anti-FLAG immunoaffinity columns were used as a negative control. (F) Immunoprecipitation of RBP1 in RB−/− H596 lung cancer cells. Anti-pRB antibody C-15, anti-p107 antibody SD9, and anti-p130 antibody C-20 were used to immunoprecipitate RB family members in asynchronously growing H596 cell extracts. Coimmunoprecipitation of RBP1 was detected by Western blotting analysis using LY32 anti-RBP1 antibody.
It is interesting that the only anti-pRB antibody capable of coimmunoprecipitating RBP1, RBAP46/48, HDAC1, mSIN3A, and mSIN3B was C-15, despite the fact that all RB antibodies are capable of immunoprecipitating large amounts of the various forms of pRB (Fig. 3A and B). More interestingly, only antibody C-15 was able to coimmunoprecipitate HDAC1, suggesting that the majority of HDAC1 associated with pRB may be via RBP1. Antibody C-15 targets the carboxy terminus of pRB and thus may not disrupt the pocket region, which is crucial for recruitment of HDAC activity both in vitro and in vivo and for binding RBP1 (6, 39, 41, 42). The other anti-pRB antibodies tested were either raised against regions of the pocket or capable of disrupting the integrity of the pocket upon binding. These antibodies might therefore either disrupt RBP1-HDAC binding to the pocket or fail to recognize pRB molecules in which the pocket is occupied by the RBP1 complexes. To demonstrate that coprecipitation of RBP1 by antibody C-15 results from its specificity for pRB and not from any cross-reactivity with RBP1, immunoprecipitation experiments similar to those shown in Fig. 3A and B were performed with H596 lung cancer cells lacking pRB. Figure 3F demonstrates that no RBP1 was present in immunoprecipitates from these cells prepared using antibody C-15, whereas with anti-p107 antibody SD9, RBP1 was coprecipitated. Figure 3F also shows that anti-p130 antibody C-20 failed to coprecipitate RBP1, presumably because p130 levels are extremely low in asynchronously growing cells (reference 10 and data not shown). Thus, these results further support the idea that the mSIN3-HDAC complex is recruited to pRB via a pocket-dependent association with RBP1.
Further evidence for the role of RBP1 in recruiting the SIN3-HDAC complex was obtained in an analysis of material that had been immunoaffinity purified from HeLa cells using antibodies against SAP30, a component of this complex. Figure 3D shows an SDS-PAGE profile and silver staining of members of the complex. A separate study using ion trap mass spectroscopy confirmed the presence of a series of HDACs, mSIN3A, RBAP48/46, and a 180-kDa species found to represent RBP1 (68, 71). Figure 3E shows similar affinity purifications of HeLa cell extracts using anti-FLAG (control), anti-SAP30, and anti-HDAC1 antibodies, which were analyzed following SDS-PAGE by Western blotting using anti-RBP1, anti-HDAC1, and anti-SAP30 antibodies. None of these species were retained (Fig. 3E, lanes b) using anti-FLAG antibodies, but all were bound in the anti-SAP30 and anti-HDAC1 samples. These data confirmed RBP1 as a stable member of this complex. The 180-kDa RBP1 species was not detected in a parallel study using anti-Mi-2 antibodies, thus confirming that RBP1 is not present in the NURD-HDAC complex (references 68 and 71 and data not shown). Interestingly, RB family members and E2F were not detected in this stable complex (data not shown). The absence of these species may be explained in three ways. First, the complex was isolated from HeLa cells, which express human papillomavirus (HPV) E7 protein. HPV E7 has been shown to prevent the pocket of pRB from recruiting HDAC activity. These observations were consistent with our previous results showing that RBP1 and HDACs fail to associate at high levels with RB family members in 293 and 293T cells, which express adenovirus E1A protein and simian virus 40 large T antigen (39). Second, both class I HDACs and RBP1 associate only with the hypophosphorylated form of pRB, and pRB is known to be highly phosphorylated in HeLa cells. Finally, pRB-RBP1-mSIN3-HDAC complex formation appears to be cell cycle dependent (10) and may represent only a small fraction of the pool of HDAC complexes in this rapidly growing population of cells. Nevertheless, these results suggested that RBP1 recruits the mSIN3-HDAC complex to RB family members in a pocket -dependent manner.
R2 repression domain recruits mSIN3-HDAC complex.
We generated a series of RBP1 deletion mutants and expressed them as GST fusion proteins (Fig. 4B). Using these mutant GST-RBP1 proteins as well as GST fusion products containing either SAP30 or adenovirus E1A protein, the binding of components of the mSIN3-HDAC complex was examined further. Figure 4A shows that the only RBP1 product that associated with mSIN3B and HDAC1 was GST-R2, thus confirming that the R2 region is uniquely responsible for binding of the mSIN3-HDAC complex to RBP1. This finding was further strengthened by results shown in Fig. 4C, which indicated that GST-R2 but not GST-R1 was able to bind to mSIN3B, RBAP46/48, HDAC1/2, and Sap30 present in nuclear extracts. In addition, Fig. 4C shows that both GST-pRB and GST-p107 recognized these same components of the mSIN3-HDAC complex. Failure of such binding by the pRB pocket mutant C706F confirmed the requirement of the pocket for this association with RBP1 and the mSIN3-HDAC complex. These experiments demonstrated that the R2 repression domain of RBP1 is responsible for recruiting the entire mSIN3-HDAC complex.
RBP1 binds to HDAC via SAP30.
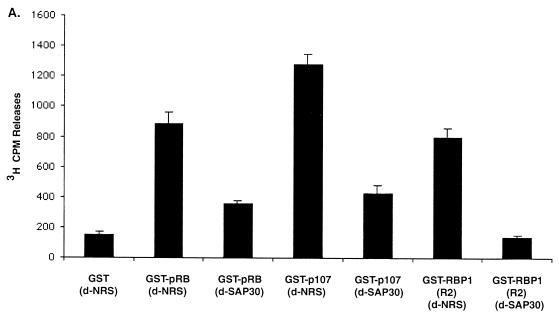
To identify the protein that is directly responsible for binding of the mSIN3-HDAC complex to the R2 domain of RBP1, purified GST-R2 protein was incubated in vitro with various components of the complex, and binding was assessed in GST pull-down experiments as in Fig. 4. Figure 5A shows results obtained by Western blotting using anti-His antibody and indicated that His6 HDAC1, which had been synthesized in and purified from SF9 insect cells, failed to associate with RBP1-R1 or with a deletion mutant of RBP1 (dl 1208C) that lacks the carboxy-terminal R2 domain, even in the presence of nuclear extracts. Binding was not apparent with the R2 domain either, unless nuclear extract was added. These results suggested that HDAC1 does not interact directly with R2 but rather does so via another factor found in nuclear extracts. Figure 5B shows results of similar experiments carried out with other components of the mSIN3-HDAC complex which had been translated in vitro using appropriate cDNAs in rabbit reticulocyte lysates in the presence of [35S]methionine and detected by autoradiography following SDS-PAGE. Interestingly, only in vitro-translated SAP30 was able to associate with the R2 domain of RBP1. Some level of association of HDAC1 with GST-pRB in a pocket -dependent manner was also observed, as also seen by others. It should be noted that SAP30 did not interact with GST-dl747C, a truncation mutant of RBP1 that contains R1 but not R2, nor with GST-pRB, suggesting that utilizing R2, RBP1 truly functions as a bridge to recruit mSIN3-HDAC complex to the pocket of pRB. To strengthen this contention further, various GST fusion proteins and His-SAP30 were synthesized in and purified from bacteria and incubated together in vitro. GST pull-down assays were performed, and binding of His-SAP30 was assessed by Western blotting using anti-His antibody. GST-HDAC1 was demonstrated previously by others to bind directly with SAP30 (71) and was used as a positive control in this experiment. Figure 5C shows that GST-R2 interacted directly with purified SAP30 protein as did GST-HDAC3; however, GST-R1, GST-pRB, and GST alone did not. These results indicated that the association of the mSIN3-HDAC complex with the R2 domain of RBP1 likely occurs via SAP30. In addition, the association seen between HDAC3 and SAP30 in vitro may suggest that there exists a pool of mSIN3-SAP30-HDAC complex that contains HDAC3 as a subunit. This observation further supported results in Fig. 2C, which suggested that there may be a pool of complex containing both HDAC1 and HDAC3 as subunits that constitute part of the HDAC activity recruited by pRB and RBP1.
FIG. 5.
The R2 repression domain associates with mSIN3-HDAC complex via a direct interaction with SAP30. (A) R2 repression domain does not interact directly with HDAC1. Lane 1 is loaded with 0.2 μg of His6-HDAC1 protein. Lanes 3, 5, and 6 contain samples in which purified proteins were also incubated in the presence of H1299 nuclear extracts. Binding of HDAC1 was assessed in GST pull-down experiments followed by Western blotting using anti-His antibody. (B) [35S]methionine-labeled in vitro-translated SAP30 associates with the R2 repression domain specifically. Labeled in vitro-translated SAP30 (lane Input, containing 1/10 of the total) was incubated with the indicated GST fusion proteins. Binding was assessed by autogradiography. (C) Purified SAP30 binds directly with purified RBP1 R2 domain, HDAC1, and HDAC3. Binding was assessed by Western blotting using anti-His antibody.
RBP1 and RB family members require SAP30 to recruit HDAC activity.
To determine if SAP30 is essential for pRB and p107 to recruit HDAC activity, an immunodepletion study similar to that described in Fig. 1B was carried out using a polyclonal antibody that recognizes SAP30. Figure 6B shows that anti-SAP30 antibody removed SAP30 from the extracts while not affecting levels of the control nuclear protein HMG1. As shown in Fig. 6A, immunodepletion of SAP30 completely abolished the association of HDAC activity with the R2 domain of RBP1. Consistent with our earlier results with RBP1 immunodepletion, more than half of the HDAC activity recruited by the pockets of both p107 and pRB was abolished in SAP30-depleted extracts. This experiment therefore demonstrated that the SAP30-containing mSIN3-HDAC complex recruited by the R2 region of RBP1 is responsible for at least 50% of the HDAC activity present at the pocket of RB family members.
pRB colocalizes with RBP1 and mSIN3-HDAC complex in quiescent cells.
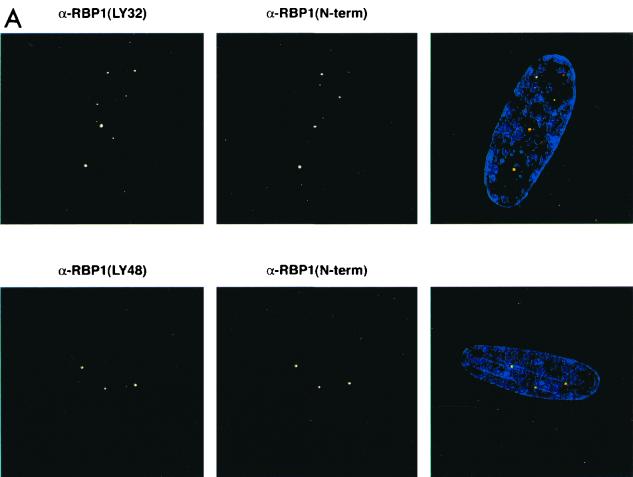
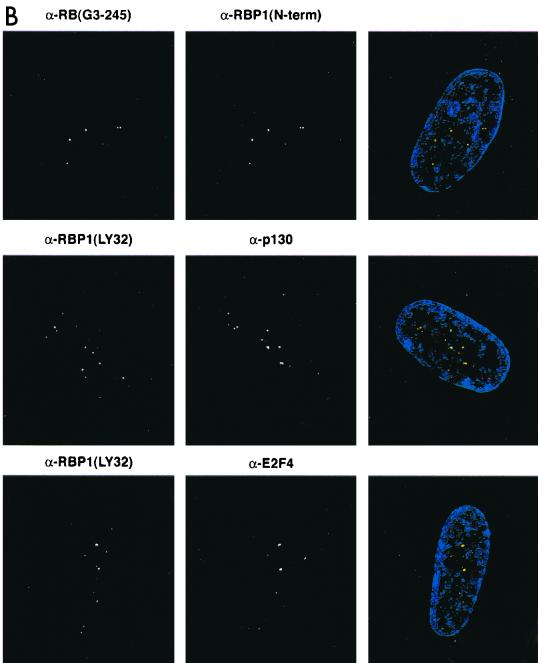
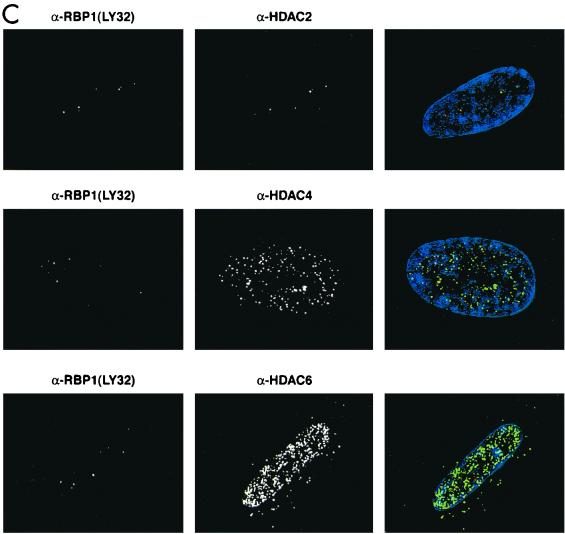
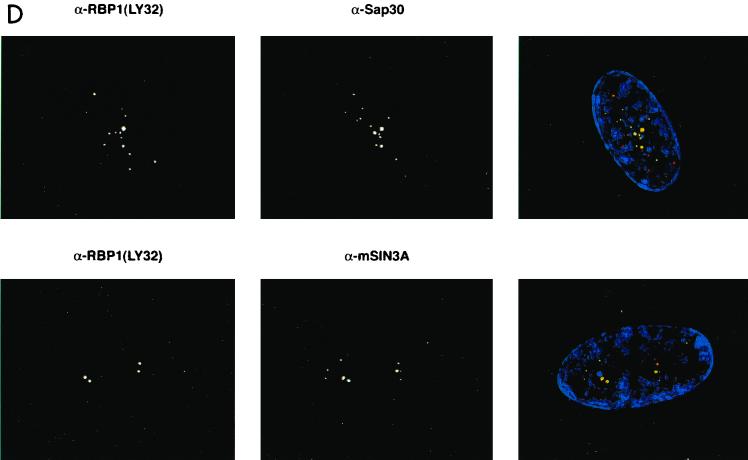
The biochemical evidence presented in this study suggested that pRB-E2F complexes can recruit mSIN3-HDAC complex via RBP1. If such complexes exist in vivo, individual components should colocalize in cells within the nucleus. To determine the relative cellular localization of RBP1 and various binding partners, quiescent primary human diploid fibroblasts were studied because we previously detected pRB-RBP1-E2F complexes in growth-arrested cells (40). WI38 human diploid fibroblasts were serum starved for 3 days, and BrdU incorporation studies were performed to determine if the cells had exited the cell cycle. Immunofluoresence studies were also performed using various monoclonal and polyclonal antibodies against BrdU, RBP1, pRB, E2F4, HDAC1, HDAC2, HDAC3, HDAC4, HDAC6, mSIN3A, and SAP30. High-resolution deconvolution microscopy utilizing a CCD digital camera was used to image these stained cells. Figure 7A shows that using two different monoclonal antibodies (LY32 and LY48) and one polyclonal (amino-terminal) antibody against RBP1, similar staining patterns showing very discrete regions within nuclei were observed in these cells. When the green and red channels were merged, the yellow spots represented the highest concentrations of RBP1, and thus the most representative of the true location of RBP1. BrdU staining was not detected in these cells, indicating that they were in full growth arrest (data not shown). Figure 7B shows that pRB, p130, and E2F4 also colocalized in these RBP1-containing discrete nuclear regions. Similar studies were not carried out with p107, as we and others have found p107 levels to be undetectable in quiescent cells (10, 40). As shown in separate colocalization studies (35; B. K. Kennedy, D. A. Barbie, A. Lai, M. Classon, N. Dyson, P. E. Branton, and E. Harlow, unpublished results). these regions appear to occupy perinucleolar sites and do not appear to be those containing promyelocytic leukemia (PML) bodies. Rather, these same regions were found to contain the initial origins of DNA replication following serum stimulation (35; Kennedy et al., unpublished) (see Discussion). As biochemical studies implied a specific association of RBP1 with class I HDACs, similar colocalization studies were performed comparing RBP1 to HDAC1 to -3. Figure 7C shows that HDAC2 also largely colocalizes to these discrete RBP1-containing regions, as did HDAC1 and HDAC3 (data not shown). Interestingly, Figure 7C also shows that class II HDAC4 and HDAC6 exhibit quite different staining patterns and did not colocalize with RBP1 at significant levels, as was consistent with our immunodepletion studies. Finally, and consistent with the previous biochemical data, Figure 7D shows that a significant number of stained foci of both mSIN3A and SAP30 also colocalized with RBP1. These results confirmed that RBP1 exists in resting cells in association with pRB, p130, and components of the mSIN3-HDAC complex.
FIG. 7.
RBP1 colocalizes with pRB-E2F and components of the mSIN3-HDAC complex in quiescent WI38 human diploid fibroblasts. All photographs were taken using a CCD camera attached to a deconvolution microscope with a 100× objective lens. (Left panels) Mouse monoclonal antibody was detected by Texas red-conjugated secondary antibody against mouse immunoglobulin G, which appears fluorescent in the red channel. (Middle panels) Rabbit polyclonal antibody was detected by fluorescein-conjugated secondary antibody against rabbit immunoglobulin G, which appears fluorescent in the green channel. 4′,6-Diamidino-2-phenylindole staining was used to stain the nucleus and appears fluorescent at the blue channel. (Right panels) Merged pictures represent superimposed blue, red, and green channels, and yellow fluorescence indicates positions of colocalization. (A) Localization pattern of RBP1 within the nucleus, using multiple antibodies against RBP1. (B) RBP1 colocalizes with pRB, p130, and E2F4 in discrete regions of the nucleus. (C) RBP1 colocalizes with class I HDACs but not class II HDACs within the nucleus. (D) RBP1 colocalizes with both mSIN3A and SAP30 within the nucleus.
DISCUSSION
Active repression function of pRB requires RBP1.
This study demonstrates the role of RBP1 as a bridging factor that recruits mSIN3-HDAC complexes to the pocket of RB family members. One of the important biological functions of the RB family is to regulate expression of genes required for cell cycle progression and DNA synthesis. RB family members utilize the pocket domain to recruit HDAC activities to repress transcription of these genes in early G1 or when cells exit the cell cycle. We have provided evidence here that RB family members are able to recruit the mSIN3-HDAC complex via a pocket -dependent association with RBP1 to actively repress transcription. Although not all of the HDAC activity associated with pRB can be accounted for by the recruitment of RBP1 as a corepressor, this mechanism may account for over half of such activity. The remaining activity may associate through interactions with other bridging factors, possibly including RBAP46/48 and c-Ski, or some other, unidentified pocket-binding protein that associate with HDAC complexes. Nonetheless, RBP1 is the first molecule described that appears to contribute two separate classes of transcriptional repression activities important for transcriptional repression by RB family members, both by recruiting the mSIN3-HDAC complex via R2 and via an as yet unidentified repression mechanism utilizing the R1 domain (Fig. 8). These dual repression activities of RBP1 may account for the ability of pRB to repress transcription in a variety of promoters, including those that have been reported to be insensitive or only partially sensitive to the HDAC inhibitor TSA.
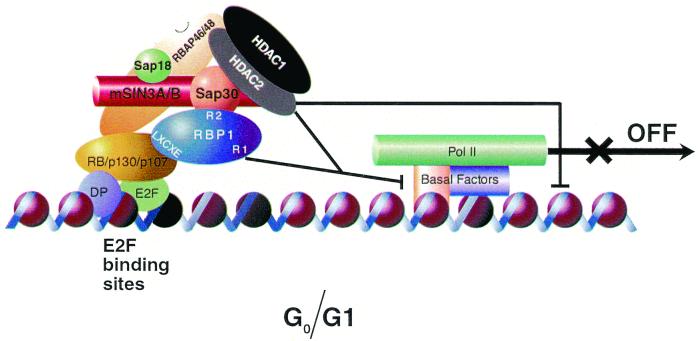
FIG. 8.
Model of transcriptional repression by pRB-E2F complexes during quiescence and the early G1 phase of the cell cycle involving recruitment of the mSIN3-HDAC complex through the pocket-dependent association of RBP1.
The structure of the pRB pocket in association with a peptide containing the HPV E7 LXCXE sequence has allowed three independent groups to introduce point mutations that appear to abolish the ability of the pRB pocket to interact with the LXCXE motif without affecting binding to E2F (9, 11, 15). Interestingly, two groups observed that LXCXE-mediated binding is important for the ability of pRB to induce growth arrest (9, 11), whereas the other group found that such did not appear to be the case (15). This difference could stem from the different assay systems used or perhaps from subtle effects of the different pocket mutations used. The same two groups (9, 11) also showed that HDAC1 interactions with the pocket were abolished with their mutants. Thus, our result that RBP1 bridges the interaction between HDAC1/2 and pRB supports this finding, as RBP1 utilizes an LXCXE motif to associate with the pocket. Evidence suggesting that the HDAC3-pRB interaction remains intact with the pocket mutants has also been obtained (11). This effect is somewhat unexpected, as we found that HDAC3 also interacts with RBP1, and we have proposed that HDAC3 is also recruited in a pocket -dependent manner despite the absence of an LXCXE motif (39). HDAC3 has recently been shown to be part of the NCoR complex, which is distinct from the SIN3-SAP30-HDAC1/2 complex. It is possible that HDAC3 associated with this complex could be recruited to pRB via contacts other than the LXCXE-interacting regions. Thus, we continue to believe that HDAC3 found in the SIN3 complex associates with the pocket via RBP1 contact, but that HDAC3 may able to associate with RB via different vehicles. In general, our results support results from the two groups that present evidence that LXCXE pocket-binding proteins are important for the ability for RB to induce growth arrest and active repression.
pRB and p130 recruit the RBP1-mSIN3-HDAC complex in quiescent cells.
In previous studies we had detected stable p130-E2F-RBP1 and pRB-E2F-RBP1 complexes capable of binding consensus E2F-specific DNA sequences in quiescent cell extracts (40). Consistent with these previous findings, we have now found that in quiescent cells, RBP1 and mSIN3-HDAC complexes colocalize to regions of the nucleus containing RB family members and E2F4. Other studies (35; Kennedy et al., unpublished) indicated that pRB redistributes to other regions of the nucleus later in the cell cycle and that it no longer colocalizes with either class I HDAC or RBP1 in cells in late S phase. Such observations suggest that induction of gene expression required for S-phase progression may involve a mechanism that relocates the RBP1-mSIN3-HDAC complexes. One interesting finding was that class II HDACs had very different staining patterns and did not colocalize with RBP1 or RB family members. This observation distinguishes the pRB- or RBP1-mediated mechanism of transcriptional repression from that of nuclear hormone receptors, which appear to utilize both classes of HDACs (29, 34).
It was also observed that SAP30 does not colocalize perfectly with RBP1, despite the fact that RBP1 recruitment of HDAC activity absolutely requires the presence of SAP30. These results suggested that not all SAP30-containing complexes contain RBP1; consistent with this notion, SAP30 was also found in HDAC complexes involved in the action of nuclear hormone receptors through association with NCoR/SMRT and in other complexes (38). SAP30-containing complexes may be involved in multiple cellular processes, whereas RBP1-containing SAP30-HDAC complexes may specifically function with RB family members.
Model of cell cycle exit and progression control by RB family members.
Zhang et al. had previously demonstrated the importance of the transcriptional repression function of pRB in cells induced into growth arrest by p16, contact inhibition, or transforming growth factor β (67). Their work suggested that blocking of E2F transactivation by pRB is not sufficient to induce such growth arrest. Here we have presented evidence that a complex containing HDAC activity associates with RB family members in quiescent cells. This complex appeared to be largely the previously identified mSIN3-SAP30-containing HDAC complex that also contains a newly identified subunit, RBP1, which appears to bridge it to RB family members. Figure 8 illustrates this new model in which RB family members induce growth arrest by recruiting the mSIN3-HDAC complex to the pocket via a direct association with the RBP1 subunit of the complex. Biochemical evidence presented in this study suggested that RBP1 contacts SAP30 directly via the R2 repression domain, whereas SAP30 interacts directly with HDAC1 and HDAC2 within the complex. RBAP46 and RBAP48, which are also found in the complex, may also have contacts with regions of pRB; however, experiments in Fig. 4 and 5B and those by others (Kennedy, personal communication) failed to detect any interaction in vitro between pRB and RBAP48. mSIN3A and mSIN3B do not interact directly with RBP1 and may be associated with RBP1 through direct interactions with SAP30 and SAP18. The precise functions of individual components of this complex in transcriptional repression function are still unclear, but RBP1 may serve to target this complex to pRB and also provide a second HDAC-independent repression activity that may be of critical importance in some cases. Further studies to address this issue are under way.
An additional role for the E2F-pRB-RBP1-SAP30-mSIN3-HDAC complexes might be proposed based on recent studies on the localization of such complexes in normal human cells (35; Kennedy et al., unpublished). These studies indicated that the discrete regions occupied by such complexes are those that first incorporate BrdU following serum stimulation of growth-arrested cells and thus represent the initial origins of DNA replication. It is possible therefore that the HDAC complexes targeted to these regions by RBP1 function in cell cycle exit not only by repressing transcription but also by remodeling chromatin at these critical sites, thus blocking the origins and possibly serving as targets for initiation of DNA synthesis. Again, further studies will be required to test this model.
ACKNOWLEDGMENTS
We thank Bill Kaelin and Jim DeCaprio for providing monoclonal antibodies against RBP1. We also thank Eva Lee, Betty Moran, and Bob Eisenman for providing additional reagents. Wei-Ming Yang and Ed Seto provided class I HDAC-related reagents; those for class II HDAC were from Xiang Jiao Yang and Nick Bertos. Deconvolution microscopy was performed at the Whitehead Institute Microscopy Facility.
This work was supported through grants to P.E.B. from the National Cancer Institute of Canada and the Canadian Institutes for Health Research and to D.R. from NIH (GM485180) and the HHMI. A. L. is the recipient of Terry Fox Biomedical Studentship supported by National Cancer Institute of Canada. M.-C.T is supported by a scholarship from the Fonds pour la Formation de Chercheurs et l' Aide à la Recherche (FRSQ-FCAR-Santé). B.K.K. is supported by a Leukemia Society of America Fellowship, D.A.B. is supported by a Karen Grunebaum Cancer Research Fellowship, and Y.Z. is supported by an NIH Fellowship.
REFERENCES
- 1.Adnane J, Shao Z, Robbins P D. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. doi: 10.1074/jbc.270.15.8837. [DOI] [PubMed] [Google Scholar]
- 2.Ayer D E. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 3.Bernards R. E2F: a nodal point in cell cycle regulation. Biochim Biophys Acta. 1997;1333:M33–M40. doi: 10.1016/s0304-419x(97)00027-9. [DOI] [PubMed] [Google Scholar]
- 4.Björklund S, Almouzni G, Davidson I, Nightingale K P, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. doi: 10.1016/s0092-8674(00)80586-3. [DOI] [PubMed] [Google Scholar]
- 5.Brehm A, Kouzarides T. Retinoblastoma protein meets chromatin. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 9.Chen T T, Wang J Y. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol Cell Biol. 2000;20:5571–5580. doi: 10.1128/mcb.20.15.5571-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbeil H B, Branton P E. Characterization of an E2F-p130 complex formed during growth arrest. Oncogene. 1997;15:657–668. doi: 10.1038/sj.onc.1201224. [DOI] [PubMed] [Google Scholar]
- 11.Dahiya A, Gavin M R, Luo R X, Dean D C. Role of the LXCXE binding site in Rb function. Mol Cell Biol. 2000;20:6799–6850. doi: 10.1128/mcb.20.18.6799-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas P B, Yaciuk P, Moran E. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J Virol. 1997;71:1726–1731. doi: 10.1128/jvi.71.2.1726-1731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 15.Dick F A, Sailhamer E, Dyson N J. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol Cell Biol. 2000;20:3715–3727. doi: 10.1128/mcb.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 17.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 18.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ewen E M, Xing Y G, Lawrence J B, Livingston D M. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 20.Fattaey A R, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7802–7812. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemington E K, Speck S H, Kaelin W G J. E2F-1 mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamel P A, Gill R M, Phillips R A, Gallie B L. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He S, Cook B L, Deverman B E, Weihe U, Zhang F, Prachand V, Zheng J, Weintraub S J. E2F is required to prevent inappropriate S-phase entry of mammalian cells. Mol Cell Biol. 2000;20:363–371. doi: 10.1128/mcb.20.1.363-371.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 27.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiebert S W. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRb-mediated growth suppression. Mol Cell Biol. 1993;13:3384–3391. doi: 10.1128/mcb.13.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda M A, Nevins J R. Identification of distinct roles for separate E1A domains in disruption of E2F complexes. Mol Cell Biol. 1993;13:7029–7035. doi: 10.1128/mcb.13.11.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 32.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 33.Kaelin W J, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 34.Kao H-Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy B K, Barbie D A, Dyson N, Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 37.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 38.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J M, Mullen T M, Davie J R, Rose D W, Glass C K, Rosenfield M G, Ayer D E, Eisenman R N. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 39.Lai A, Lee J M, Yang W M, DeCaprio J A, Kaelin W G, Jr, Seto E, Branton P E. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999;19:6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai A, Marcellus R C, Corbeil H B, Branton P E. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999;18:2091–2100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- 41.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 42.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 43.Martínez-Balbás M A, Bauer U-M, Nielsen J S, Brehm A, Kouzarides K. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 46.Ng H H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 47.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–2150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otterson G A, Kratzke R A, Lin A Y, Johnston P G, Kaye F J. Alternative splicing of the RBP1 gene clusters in an internal exon that encodes potential phosphorylation sites. Oncogene. 1993;8:949–957. [PubMed] [Google Scholar]
- 49.Qian Y W, Wang Y C, Hollingsworth R J, Jones D, Ling N, Lee E Y. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 50.Ross J F, Liu X, Dynlacht B D. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- 51.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sellers W R, Rodgers J W, Kaelin W G J. A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 54.Sun Z W, Hampsey M. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999;152:921–932. doi: 10.1093/genetics/152.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 56.Tokitou F, Nomura T, Khan M M, Kaul S C, Wadhwa R, Yasukawa T, Kohno I, Ishii S. Viral ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion. J Biol Chem. 1999;274:4485–4458. doi: 10.1074/jbc.274.8.4485. [DOI] [PubMed] [Google Scholar]
- 57.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 59.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 60.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng J, Han J, Yang X J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 62.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 63.Welch P J, Wang J Y. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Indian J Pediatr. 1993;60:193–201. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 64.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 65.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 67.Zhang S H, Postigo A A, Dean D C. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4A, TGF-β, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, Sun Z W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]