Abstract
Characterization of COVID-19 antibodies has largely focused on memory B cells; however, it is the antibody-secreting plasma cells that are directly responsible for the production of serum antibodies, which play a critical role in resolving SARS-CoV-2 infection. Little is known about the specificity of plasma cells, largely because plasma cells lack surface antibody expression, thereby complicating their screening. Here, we describe a technology pipeline that integrates single-cell antibody repertoire sequencing and mammalian display to interrogate the specificity of plasma cells from 16 convalescent patients. Single-cell sequencing allows us to profile antibody repertoire features and identify expanded clonal lineages. Mammalian display screening is used to reveal that 43 antibodies (of 132 candidates) derived from expanded plasma cell lineages are specific to SARS-CoV-2 antigens, including antibodies with high affinity to the SARS-CoV-2 receptor-binding domain (RBD) that exhibit potent neutralization and broad binding to the RBD of SARS-CoV-2 variants (of concern/interest).
Keywords: antibody, plasma cell, B cell, single-cell sequencing, antibody repertoire, COVID-19, SARS-CoV-2, VoC, CRISPR, mammalian display
Graphical abstract
Ehling et al. identify SARS-CoV-2 specific and neutralizing antibodies from plasma cells of patients who recovered from COVID-19 using an integrated pipeline of single-cell sequencing and mammalian display. The method described herein enables the functional profiling of expanded plasma cells.
Introduction
Serum antibody responses against severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) play a critical role in resolving viral infection (Hung et al., 2020). SARS-CoV-2 consists of four major structural components, with the multi-domain Spike (S) representing a relevant target for antibodies. The S protein contains the S1 subunit, including the receptor-binding domain (RBD) and the S2 subunit. S1 is highly immunogenic and a target for immunoglobulin (Ig) seroconversion and neutralization, as the RBD directly interacts with human surface receptor angiotensin-converting enzyme 2 (ACE2), thereby initiating attachment and viral entry (Hoffmann et al., 2020; Kawase et al., 2012). The infusion of SARS-CoV-2-specific antibodies through convalescent plasma from recovered individuals has been investigated and received Emergency Use Authorization (EUA) by the US Food and Drug Administration (FDA) (Agarwal et al., 2020; Simonovich et al., 2020) (NCT04383535 and CTRI/2020/04/024775). Several therapeutic monoclonal antibodies (mAbs), most notably bamlanivimab and etesevimab (Eli Lilly), originally derived from the B cells of infected or convalescent patients and targeting the RBD have successfully progressed through clinical development (Hoffmann et al., 2020). Earlier this year, the emergence of variants such as Beta and Gamma that showed resistance to several mAbs, including bamlanivimab, led to EUA revocation by the FDA (Food and Drug Administration, 2021). An original strain of SARS-CoV-2 (Pango lineage B/B.1) relinquished its global dominance to the Alpha (B.1.1.1, first described in the United Kingdom) around April 2021. Alpha was subsequently replaced by Delta (B.1.612.2, which came to prominence in India in February 2021), and as of September 2021, it accounts for >80% of all new infections. Thus, it is important to assess the binding antibodies derived from a B.1 convalescent response (date of infection March–April 2020) to currently circulating variants.
Additional antibodies discovered from patients with coronavirus disease 2019 (COVID-19) are being developed as therapeutics (Chen et al., 2020; Hansen et al., 2020; Pinto et al., 2020; Rogers et al., 2020; Wec et al., 2020), as highlighted by their rapid clinical progression (e.g., NCT04452318, NCT04519437). The isolation of SARS-CoV-2-specific antibodies has focused overwhelmingly on memory B cells (Bmem), which due to their surface bound B cell receptor (BCR) can be labeled with soluble antigen (e.g., S1, RBD) and screened by fluorescence-activated cell sorting (FACS) (Boonyaratanakornkit and Taylor, 2019). While integrated pipelines incorporating single-cell sequencing are emerging to expedite antibody discovery (Cao et al., 2020; Gilchuk et al., 2020), the isolation of antibodies from Bmem is still technically challenging due to a variety of factors such as the rarity of antigen-specific clones (e.g., 0.07% RBD-binding Bmem in patients with COVID-19; Robbiani et al., 2020]) and drop out during single-cell sorting (Maeshima et al., 2017). Crucially, antibodies present in serum are not directly produced by Bmem but instead by antibody-secreting plasma cells (PCs).
Antigen-specific PCs can arise as early as the first week of infection (Carter et al., 2017) and thus can be profiled at an early time point (Nutt et al., 2015). Recent studies have shown that many SARS-CoV-2-specific antibodies are germline-like, often possessing stereotypical sequence patterns (Kim et al., 2021; Kreer et al., 2020); with this in mind, it is possible that early or transitional PCs present in blood may encode specific and neutralizing antibodies. This transitional phenotype ranges from plasmablasts to short-lived PCs, before migration into bone marrow and differentiation into long-lived PCs (Ribatti, 2017). The frequency of transitional PCs in peripheral blood mononuclear cells (PBMCs) of healthy donors is estimated at <5% of all B cells, but has been shown to increase to >20% during COVID-19 (Wen et al., 2020). This is a more dynamic response than in Bmem, in which the fraction appears unchanged at 40%–50% of total B cells during COVID-19 (Wen et al., 2020). Previous research has shown that clonally expanded PCs following infection or immunization are often highly enriched for specificity to target antigens (Chen et al., 2018; Henry Dunand et al., 2016; Reddy et al., 2010; Wrammert et al., 2008); however, it has yet to be determined whether a similar phenomenon occurs in patients infected with SARS-CoV-2. A major challenge, however, is that in contrast to Bmem, PCs cannot be pre-enriched for antigen binding due to low (IgA, IgM, IgE) or non-existent (IgG) surface expression of BCRs (Pinto et al., 2013).
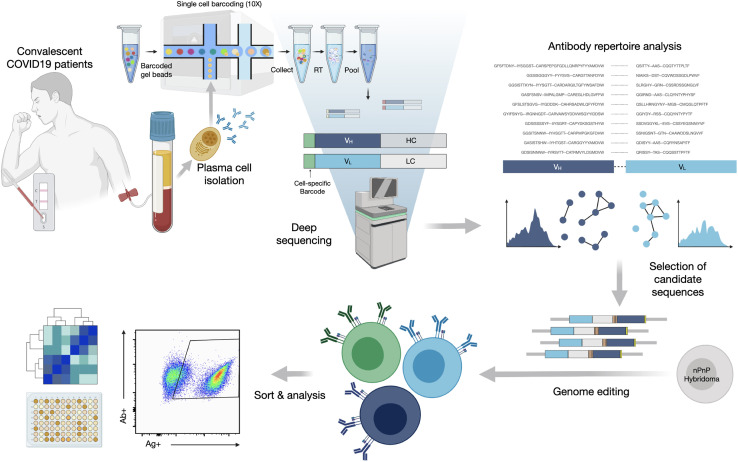
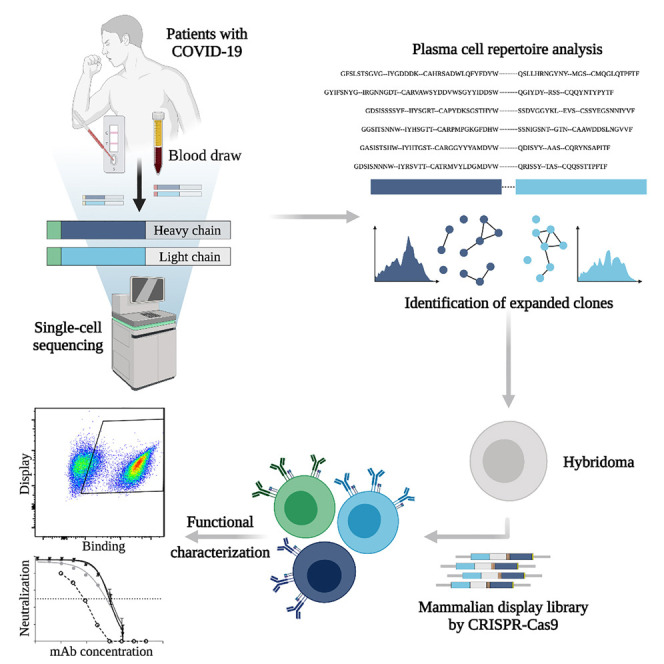
To interrogate the specificity of PCs in convalescent patients recovered from COVID-19, we describe a pipeline that integrates single-cell antibody repertoire sequencing, genome editing, and high-throughput mammalian display screening (Figure 1 ). We performed single-cell antibody sequencing of PCs isolated from the blood of 16 convalescent patients, enabling the identification of endogenously paired variable heavy (VH) and light (VL) chains and bioinformatic analysis of clonally expanded PCs. Both repertoires and expanded PC lineages show broad germline usage and diversity. We next selected 132 clonally expanded PC lineages across all patients and designed a genome-editing strategy to rapidly integrate antibody genes into a mammalian display platform (Mason et al., 2018; Parola et al., 2019; Pogson et al., 2016). Selection based on FACS and deep sequencing led to the discovery of 43 unique antibodies with specificity to SARS-CoV-2 (S1, S2, and RBD). Antibodies are derived from clonal lineages across 11 of the patients; notably 3 antibodies with specificity to the RBD exhibit potent neutralization in viral-pseudotype assays, and maintain binding to a number of SARS-CoV-2 variants (e.g., Alpha, Beta, Gamma, Delta). Thus, our integrated workflow is able to demonstrate that convalescent patients produce highly expanded PCs with specific and neutralizing antibodies for SARS-CoV-2.
Figure 1.
An integrated workflow for interrogating the antibody specificity of PCs from convalescent patients with COVID-19
Serum and PBMCs are collected from patients (with confirmed SARS-CoV-2 PCR positive test). The serum is assayed with IgA and IgG ELISAs as well as POCTs. From a subset of 16 patients, PCs are isolated from PBMCs by magnetic cell sorting and to then undergo gel encapsulation and barcoding for single-cell sequencing of their antibody heavy (HC) and light chain (LC) transcripts. Antibody repertoire analysis is performed to identify expanded PC clonal lineages, which are then reformatted into single open reading frame (ORF) full-length synthetic antibody genes, including homology arms, to allow for single-step cloning-free genome editing. The resulting mammalian display library then undergoes high-throughput screening for SARS-CoV-2 binding by flow cytometry and deep sequencing to recover the identity of the corresponding clonal lineages. Supernatant is used to determine the cross-reactivity of antibodies within the library with coronavirus antigens.
Results
A patient cohort of convalescent patients with COVID-19
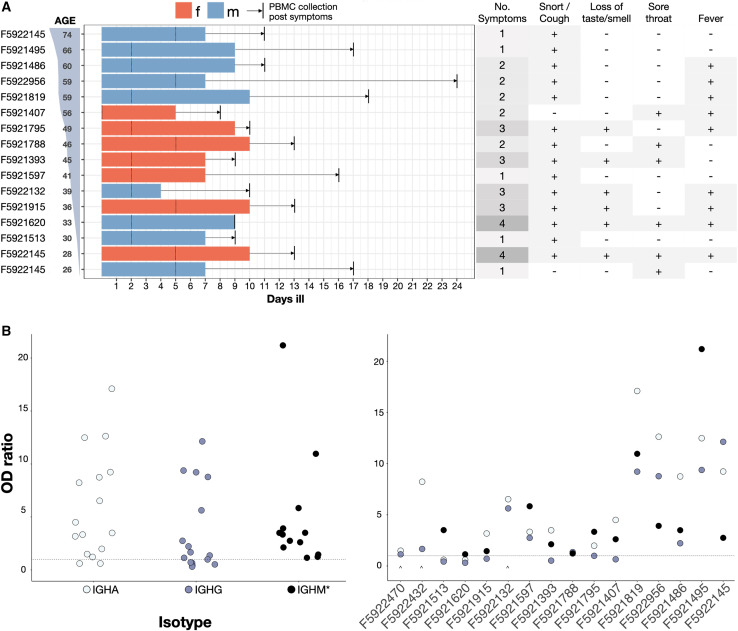
PBMCs were isolated from a set of 16 convalescent patients, which was part of a larger cohort biobank (Kaltenbach et al., 2020). Among the selected group of 16, all of the patients were confirmed to be SARS-CoV-2+ by RT-PCR testing; point-of-care lateral flow tests (POCT) confirmed serum IgM and IgG antibody responses to SARS-CoV-2 S1 and nucleocapsid protein (NCP) antigens (data not shown). Clinical characteristics of the patients are summarized in Figure 2 ; all of the patients displayed mild symptoms (no hospitalization required) and with the exception of patients 2, 14 and 15, none of the patients required any assistance (Figure 2A, left). PBMCs were collected within 9–24 days after the onset of symptoms (average: 13 days) or 0–17 days after resolution of the disease (average ∼5.1 days). A follow-up ELISA on patient serum (Figure 2B) failed to detect S1-specific IgA + IgG titers in 2 patients (F5921513, F5931620), and IgG titers in 4 patients (F5921407, F5921795, F5921393, F5921915), thus highlighting a discordance to POCTs. Sensitivity and specificity of the particular POCT used were >92% and 99%, respectively (Rudolf et al., 2020). The 2 patients who lacked both S1-specific IgA and IgG titers (F5921513, F5931620) showed IgM specificity to SARS-CoV-2 NCP (Figure 2B).
Figure 2.
Demographics and symptoms of patients in this study
(A) Patients sorted by age. Bars, colored by sex, show number of days ill total, before and after RT-PCR confirmed diagnosis (marked with a dotted line). Time between resolution of the disease and blood draw (vertical black line) is highlighted with an arrow. Summary of standardized symptoms per patient (right table).
(B) IgA-IgG and IgM (∗) ELISA results overall patients (left) and per-patient sorted by age (right) showing optical density (OD) values over background for IGHA (light blue), IGHG (dark blue), and IGHM (black) titers. The dotted line represents classification cutoff. The caret denotes missing datasets for IGHM titers.
Single-cell antibody repertoire sequencing and analysis of PCs from convalescent patients
To interrogate the antibody repertoire of PCs, we performed single-cell sequencing. First, PCs were isolated from PBMCs based on CD138 expression using magnetic beads. Second, single-cell sequencing libraries were prepared using the 10X Genomics Chromium system and the V(D)J protocol, which combines gel encapsulation and DNA barcoding to tag antibody transcripts of both heavy chain (HC) and light chain (LC) originating from the same cell. Processing the raw reads resulted in ∼8.3 million reads per patient PC sample, with a mean depth of 176,000 reads per cell (Table S1). Contigs were then assembled, filtered, and re-aligned. Only cells with a productive HC and LC were considered for the final analysis. A PC clone was defined by 100% amino acid identity of paired HC and LC complementarity determining region 3 (CDRH3-CDRL3) sequences. Clones with an identical HC germline and at least 80% similarity between their CDRH3 amino acid sequences were defined to be clonally related variants, forming a clonal lineage.
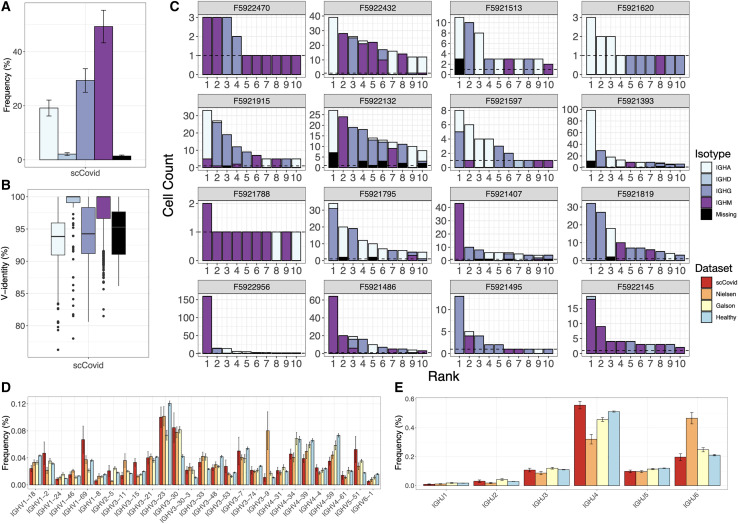
The number of cells in which both HC and LC antibody sequences could be recovered varied across the 16 single-cell sequencing patient datasets, ranging from 30 to 1,482 cells per sample. Analysis of the isotype usage across repertoires showed that the majority of clones were of IGHM (49.7% ± 6.0%, mean ± SE), although a substantial fraction of class-switched IGHG (29.6% ± 4.4%) and IGHA (19.4% ± 3.0%) was also observed (Figures 3A and S2). Next, we measured the percentage identity to the germline V-gene, which served as a proxy for somatic hypermutation (SHM); this showed as expected that IGHG (94.4% ± 0.1%) and IGHA (93.4% ± 0.1%) had lower V-gene identity than IGHM (98.1% ± 0.05%); however, on a repertoire level, this indicated that class-switched antibodies had only a minor degree of SHM relative to repertoires from other viral infections (e.g., lymphocytic choriomeningitis virus) (Neumeier et al., 2021a) (Figure 3B). We then examined the clonal expansion profiles per patient, identifying various degrees of clonal expansion, with several patients showing highly expanded IGHG and IGHA clones (Figure 3C). The observed isotype distribution among expanded clones is not necessarily correlated to the serum titer as it can also be a result of a secondary infection. Clonal expansion of PCs is also not necessarily predictive of antigen specificity, as recently shown in a study performed on immunized mice (Neumeier et al., 2021b). Finally, differences in quality and cell recovery per sample inevitably lead to unevenness in the distributions of major repertoire features and are thus descriptive in nature.
Figure 3.
Single-cell antibody repertoire features and clonal expansion analysis
(A) SHM rates per isotype across all datasets. Error bars represent means ± SEs.
(B) Isotype usage in single-cell repertoires sequencing data of our patient cohort (error bars represent means ± SEs).
(C) Clonal expansion profiles across all patients showing the 10 most expanded clones, colored by isotype. Error bars represent means ± SEs.
(D and E) V (D) and (E) J gene usage of HC compared to previously reported repertoire sequencing datasets from patients with COVID-19 and a healthy control. Error bars represent means ± SEs.
Recent reports describe antibody repertoire sequencing on B cells from individuals infected with SARS-CoV-2 (Galson et al., 2020; Nielsen et al., 2020). While these studies performed bulk sequencing (no VH and VL pairing) on peripheral B cells (mixture of B cell subsets) and did not interrogate antigen specificity, they are a valuable reference for repertoire features such as germline gene usage during SARS-CoV-2 infection. We therefore compared the germline profiles from our single-cell repertoire data to the bulk repertoire data from these two studies and observed very similar trends in IGHV gene usage, including an increased usage of IGHV3-30, IGHV3-30-3, IGHV3-33, and IGHV5-51 relative to a healthy control repertoire dataset (Ghraichy et al., 2020) (Figure 3D). While increased IGHJ6 usage was observed for the dataset from Nielsen et al. (2020), the overall usage distribution is similar for all datasets (Figure 3E). Analysis of the CDRH3 length distribution also showed no major differences between COVID-19 repertoire studies and the healthy control nor across isotype (Figures S2–S6).
We then compared the pairing ratios of IGHV with IGKV or IGLV from our patient cohort with that of pairing data from healthy individuals reported by DeKosky et al. (2016). The COVID-19 and healthy pairing ratios were generally very similar—notably IGKV3-20 and IGKV1-39 were both shown to pair with a wide range of IGHV, while others such as IGLV1-47 were observed only in combination with a limited number of IGHV (i.e., IGHV1-2) (Figure S1). However, this can at least partly be explained by the increased relative frequencies at which LCs such as IGKV3-20 are observed in overall repertoires. Additional analysis of paired CDRL3 and CDRH3 length distributions did not reveal any trends or preferences in COVID-19 antibody repertoires (Figures S2A–S2G).
Selection of expanded PC clonal lineages for mammalian display antibody screening
One major advantage of performing single-cell sequencing was that it provided us with an opportunity to reconstruct full-length antibodies with natural VH and VL pairing, which allowed us to then interrogate the specificity of PCs against SARS-CoV-2 antigens. This approach requires the construction of synthetic antibody genes for recombinant expression, and thus it is not cost-effective to screen thousands of candidates. Although we implemented a streamlined design and workflow here, the gene synthesis of thousands of antibody sequences became prohibitively expensive, emphasizing the importance of appropriate in silico analysis and selection. Despite the limitations in throughput, one of the critical questions that we could nevertheless answer is whether highly expanded PC clonal lineages produced antibodies specific to SARS-CoV-2 antigens, as well as antibodies with neutralizing function. We started by selecting the IGHG clone with the highest abundance (cell count) in each of the 16 patients (Figure S2H). We then selected an additional 20 antibody sequences associated with the most abundant clonal lineages from the 4 patients who had the highest single-cell sequencing depth (based on the number of cells with both VH and VL sequences recovered). This resulted in 36 unique antibody sequences and PC clonal lineages, which were pooled together for subsequent antibody expression and screening experiments (pool A). We selected and pooled an additional 96 antibody sequences associated with the most expanded clonal lineages across all 16 patients (pool B). The resulting, highly diverse set of 132 candidate antibody sequences (Table S2A) covered 104 unique HC-LC combinations (34 HCs and 30 LCs) (Figure S2I), with CDRH3 and CDRL3 lengths ranging from 6 to 27 and from 8 to 15 amino acids, respectively. The average germline identity to IGHV and IGKV/IGLV was 94.2% and 96.4%, respectively, including 26 variants that did not have any SHM (100% germline identity). To screen these pools efficiently, we adapted our previously developed mammalian cell antibody surface display screening system: the plug-and-(dis)play (PnP) hybridoma platform, which facilitates the rapid generation of stable mammalian cell lines that display and secrete mAbs (Pogson et al., 2016). PnPs take advantage of CRISPR-Cas9 and homology-directed repair (HDR) to integrate a fluorescence reporter gene (mRuby) and subsequently synthetic antibody (sAb) genes into the endogenous IGHV genomic locus. In addition, the stable integration of Cas9 into the genome and modification of HDR templates further improves integration efficiencies and enables the generation and screening of large mutagenesis libraries in specific regions such as the CDRH3 (Mason et al., 2018), as well as entire variable gene libraries (Parola et al., 2019) and therapeutic antibody optimization by machine learning (Mason et al., 2021).
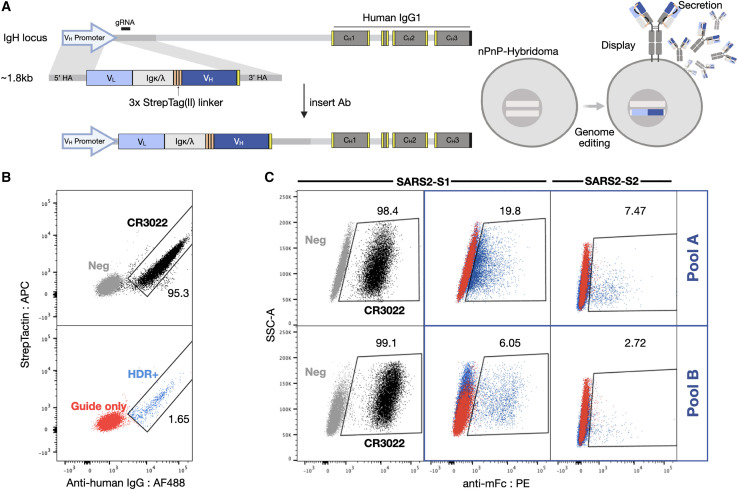
We started by adapting the PnP cell line by removing the fluorescent reporter mRuby from the VH locus, thereby creating a negative PnP cell line (nPnP) (data not shown). We hypothesized that this would improve HDR rates for templates with short homology arms, thus providing a cloning-free approach for antibody library generation and screening. Next, to design sAb templates for Cas9-induced HDR integration into nPnP cells, the following 5 gene modules were assembled: (1) the 5′ homology arm containing a partial VH1-82 promoter, followed by (2) each VL chain region (V-J) matched with its respective constant human light chain, either constant kappa (IGKC) or constant lambda (IGLC1); (iii) a flexible 57-amino acid linker, containing 3 tandem Strep-Tag(II) motifs providing both a connection to the HC as well as a peptide tag for phenotypic detection of HDR integration and antibody expression (Moffett et al., 2019); (4) the VH chain region (V-D-J), followed by (5) a 3′ homology arm (consisting of the downstream intron). The full assembly of the modules resulted in a gene fragment ∼1.8 kb in size.
Commercial gene synthesis was used to generate sAb HDR templates of pool A (mAbs 1–36) and pool B (mAbs 40–135) and the control CR3022, which is a previously identified antibody that binds SARS-CoV-1 (ter Meulen et al., 2006) and SARS-CoV-2 and has high affinity (<0.1–6.3 nM) to a cryptic, non-neutralizing epitope on the S protein RBD (Tian et al., 2020; Yuan et al., 2020a). The sAb HDR templates and guide RNA (gRNA) were co-transfected into nPnP cells and subsequently screened. Following successful HDR, mRNA from the sAb splices with the downstream IgH constant HC (CH1) to assemble into a full-length antibody; thus, only targeted integration results in antibody expression (Figure 4A, left). Due to alternative splicing inherent to the endogenous IgH locus (Pogson et al., 2016), sAbs are both displayed and secreted into the supernatant (Figure 4A, right). Cells displaying antibodies are thus derived from HDR events and can be selected by FACS when staining for anti-human IgG. The Strep-Tag (II) is visualized on the surface by staining with Strep-Tactin (Figure 4B). To screen a large number of antibody sequences at the same time, pools A and B were integrated separately into the genome and sorted. Ab+ (HDR+) pools were then selected for binding to SARS-CoV-2 antigens, either to the S1 or S2 subunit, resulting in ∼5%–19% and ∼2%–7% binding populations, respectively (Figure 4C). These populations were subsequently resorted to increase purity.
Figure 4.
Mammalian display enables antibody screening of selected clonal lineages
(A) HDR templates are synthesized and transfected into PnP-hybridoma cell lines through CRISPR-Cas9 into the endogenous VH locus.
(B and C) After sorting by flow cytometry for successful integration with Strep-Tactin and anti-human IgG (B), (C) enriched hybridoma pools (pools A and B) are sorted for binding to SARS-CoV-2 S1 or S2.
Validation of antibodies with specificity to SARS-CoV-2 antigens by deep sequencing and flow cytometry
To rapidly assess and validate the specificity of candidate antibodies, we performed deep sequencing on pool A and B libraries following FACS. Genomic DNA was extracted from PnP cells of both pools after sorting for antibody expression (Ab+), as well as antigen binding (Ag+). Libraries were then prepared based on PCR amplification of the VH gene. Samples were subjected to deep sequencing (Illumina) followed by alignment and post-processing using MiXCR (Bolotin et al., 2015). Sequencing only the VH region was still sufficient to determine the PC clonal lineage, as all of the candidates that were chosen possessed unique combinations of CDRH1, H2, and H3. Hence, the sequence of the VH region provides information to match it with the cognate VL and thus recover the full candidate antibody sequence.
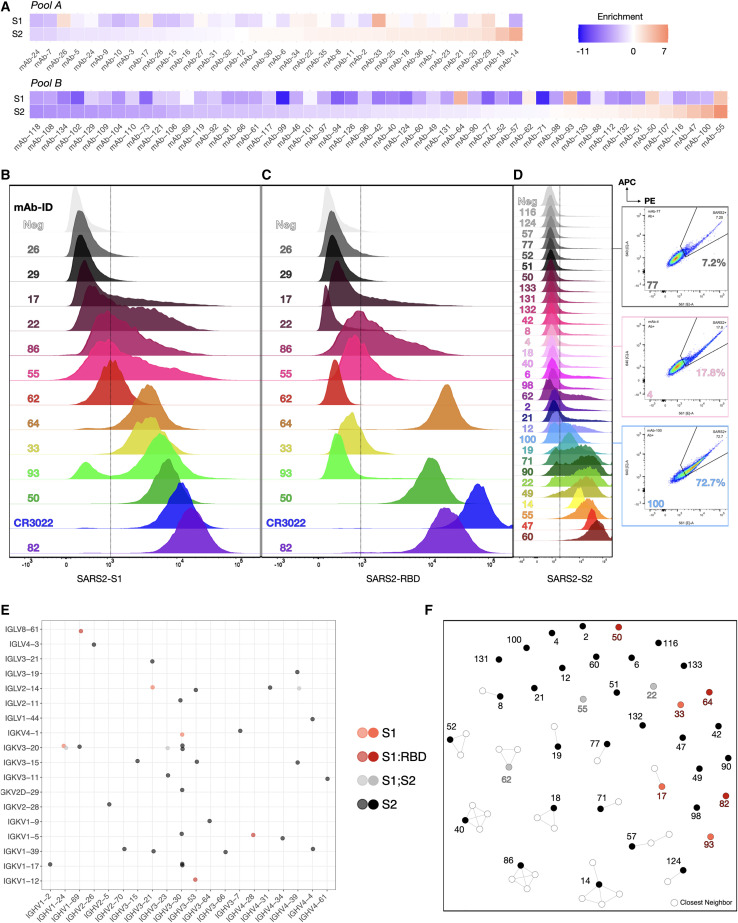
Of 132 total antibody candidates present in both pools, 118 sequences (100% for pool A, 85% for pool B) were found in the deep sequencing data of the Ab+ population. The loss of some candidate sequences was initially believed to stem from re-formatting (linker), but it is more likely a result of an uneven distribution of sequences within the pools due to subsampling inaccuracies, further confounded by the bottleneck of limited HDR efficiencies (Figure S3A). From the deep sequencing data, enrichment ratios were calculated by taking the log2 of the fraction of sequence counts in the Ag+ population and Ab+ population (Figure 5A, Table S2B). This statistical method aids in determining true binders from background (false negatives of binders) and accounts for an uneven distribution of sequences in the parental Ab+ populations. Highly enriched antibody sequences, as well as sequences differentially enriched for one of the S1 or S2 subunits were considered binders to the respective SARS-CoV-2 antigen and selected for downstream characterization. In addition, neutral or weakly depleted sequences were chosen to help determine the enrichment cutoffs for binding versus non-binding antibodies.
Figure 5.
Validation of SARS-CoV-2 reactive antibodies by deep sequencing and flow cytometry
(A) Sorted cells were deep sequenced (see Figure S4B). Enrichment ratios are calculated to determine SARS-CoV-2 binders. Red tiles indicate highly enriched sequences; blue tiles correspond to strongly depleted sequences post-FACS.
(B) RBD staining of S1 enriched candidates.
(C) S1 staining of enriched candidates.
(D) S2 staining of enriched candidates. Three example sequences shown on the side stained for both SARS-CoV-2 S2-PE and APC.
(E) Dot plot showing the VH/VL gene usage across all verified reactive (S1, S1-RBD, and S2) clonal lineages.
(F) Sequence similarity network of SARS-CoV-2-reactive sequences and their closest neighbors found in the patients’ repertoires (based on CDRH3 sequences; edges are drawn between sequences with Levenshtein distance ≤3).
Based on enrichment, a subset of 43 sequences was selected and Cas9 HDR was performed on nPnP cells to generate mAb cell lines. Flow cytometry on the 43 monoclonal PnP cell lines was performed to verify specificity to either SARS-CoV-2 S1, RBD, or S2 antigen (Figures 5B–5D). Of 43 tested candidates, 37 displayed strong or sufficient Stokes shifts in the fluorescence channel (561 nm) to confirm their specificity. For some candidates, strongly trailing peaks indicated an incomplete staining, possibly due to differential antibody expression and stability. Nevertheless, these antibodies can still be considered specific to SARS-CoV-2 antigen due to a fluorescence signal in both 561- and 633-nm channels (Figure S4A). In addition, an unrelated mFc-tagged antigen (here: anti-hCD69-mFc) was used to verify that the observed signal was not a result of mFc interactions (selected samples remained negative for the unrelated mFc-tagged antigen, while positive for S1 and S2 antigens) (Figure S4B). S1 binders (Figure 5B) were also labeled with RBD antigen, revealing that at least three clones appeared to bind to the RBD (Figure 5C) and could be neutralizing antibodies. Flow cytometry confirmed that most of the highly enriched clones, but surprisingly also some differentially depleted clones, showed specificity to SARS-CoV-2 antigens, with varying degrees of intensity (mean fluorescence intensity [MFI] = 383–17,165 for S1 [background = 388], MFI = 376–65,396 for RBD [background = 354], and MFI = 923–50,408 for S2 [background = 826]) (Figure 5D). Both S1- and S2-specific clones displayed a breadth of signal, most likely derived from the dynamic range of antibody surface expression on PnP cells and a large range of affinities.
To investigate any potential cross-reactivity to other common human coronaviruses (HCoVs), supernatant from pools A and B was collected and ELISAs were performed with antigens derived from 4 other HCoVs: 229E, HKU1, OC43, and NL63, in addition to SARS-CoV-2 antigens S1, S2, and NCP (Figure S5). As a reference, the CR3022 supernatant showed strong binding to SARS-CoV-1 and SARS-CoV-2 Spike, while showing no cross-reactivity with any other CoV antigen. The antibodies from pool A showed more binding to all CoV antigens, while the pool B antibodies appeared to be more focused on S1 and NCP antigens, which was correlated with what was observed by FACS.
From the previously missing sequences from pool B, sAb HDR templates were subsequently repeated for Cas9 HDR integration into nPnP cells (in bulk), followed by FACS to determine their antigen specificity. While no candidates were checked individually, Ab+ sorting revealed that all but 1 of the 13 mAbs were found to express productive antibody on their surface; antigen sorting and deep sequencing analysis on RBD, S1, and S2 positive events led to the identification of 2 additional RBD binders (mAb-56, mAb-76) and 4 additional S2 binders (mAb-78, -80, -111, -115) (Figures S3B and S3C).
Patient and SARS-CoV-2-specific antibody sequence analysis
Of the 132 antibody sequences selected from the highly expanded PC clonal lineages, 43 (32.5%) could be confirmed to be specific for SARS-CoV-2 S1, RBD, or S2 antigen (Table S2C). By examining these SARS-CoV-2-specific sequences and which patients they originated from, we observed patient-related trends (Table S3). For instance, ∼67% (6/9) of the antibody sequences selected from patient F5921407 were SARS-CoV-2 antigen specific, while for another patient, only 10% (1/10) of the tested antibody sequences demonstrated antigen binding. As the number of selected sequences varied per patient, the mean hit rate per patient was slightly higher, at 37%. From our cohort, serum from 2 of the patients (F5921513 and F5921620) showed a complete lack of S protein-reactive antibodies by ELISA (Figure 2B), which is in stark contrast with previous studies showing that all patients develop some S protein-specific antibody response (Shrock et al., 2020; Zhu et al., 2020). To further corroborate this, none of the 4 antibody sequences (mAb-1, mAb-9, mAb-104, and mAb-135) from highly expanded PCs of these 2 patients were shown to bind to S1 or S2 antigens. The 37 initially observed SARS-CoV-2-specific antibody sequences were distinct from one another and did not strongly correlate with trends observed in the overall repertoire. For example, there were 34 unique VH-VL germline combinations and a broad distribution of V- and J-gene usage (Figures 5E and S6A). The most frequently observed VH gene (IGHV3-30, seen in 8 binders) was also present at an increased frequency in both the overall repertoires as well as the pool of 132 selected sequences. The CDRH3 lengths ranged from 6 to 26 (CDRL3: 8 and 15) (Figure S6B), and the set of binders exhibited a broad range of SHM with V-gene identities between 85% and 100% (Figure S6C). Notably, 11 of the antibodies possessed no SHM, with a germline identity of 100% in both their VH and VL sequences. Comparing our results to previously published SARS-CoV-2-specific antibody sequences, we found that 2 clones, mAb-64 and mAb-93, were present in a similar form in other available datasets (Galson et al., 2020; Raybould et al., 2020). Specifically, mAb-64 has 6 clonal neighbors with ∼85% similarity in the CoV-AbDab (a database compiling known CoV-binding antibody sequences) (Raybould et al., 2020), while the CDRH3 sequence of mAb-93 was also found in the previously published COVID-19 repertoire dataset from Galson et al. (2020). Notably, these SARS-CoV-2-specific antibody sequences have only a limited number of close neighbors (Levenshtein distance ≤3) in our patient repertoires (Figure 5F). The sparsity of the similarity network, however, is likely in large part due to the limited size of the single-cell sequencing datasets.
Clonally expanded PCs produce high-affinity antibodies to the RBD that are capable of neutralizing SARS-CoV-2
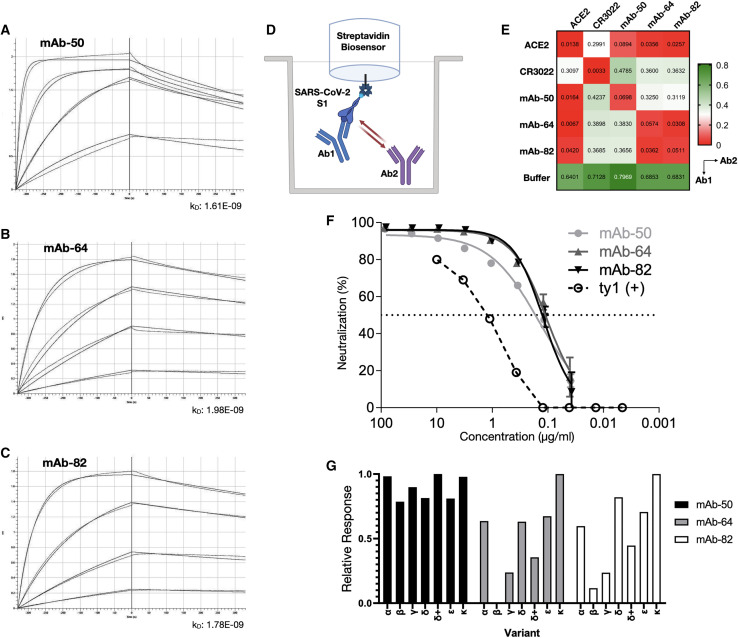
Of special interest are the PCs and their corresponding antibodies with specificity to the RBD, as binding to the RBD is often associated with neutralization of SARS-CoV-2. The 3 antibodies in this group used different combinations of VH and VL germline genes, namely IGHV4-28-IGKV1-5 (mAb-50), IGHV3-53-IGKV1-12 (mAb-64), and IGHV1-69-IGLV8-61 (mAb-82). Interestingly, IGHV3-53 and IGHV1-69 have previously been reported in association with several other neutralizing antibodies to SARS-CoV-2 (Brouwer et al., 2020; Collins et al., 2020; Kim et al., 2021; Wec et al., 2020; Yuan et al., 2020b). Previous work has established that Bmem can produce antibodies that are germline or have very few somatic mutations and be specific for SARS-CoV-2 antigens, including the RBD (Kreer et al., 2020). Thus, similar to Bmem, we found this similar trend in PCs, as 2 of the 3 antibodies (mAb-64 and mAb-82) were completely germline sequences in both VH and VL chains, while the third (mAb-50) had a V-gene identity of 94%, which was the same as the average for the whole repertoire. Next, we measured the binding affinities of the RBD-specific antibodies by using a biotinylated prefusion stabilized SARS-CoV-2 S1 antigen and biolayer interferometry (BLI). All 3 antibodies exhibited similar binding kinetics, with apparent equilibrium dissociation constants (K D) values of 1.61 nM (mAb-50), 1.96 nM (mAb-64), and 1.78 nM (mAb-82) (R2 = 0.99) (Figures 6A–6C). Antibodies derived from Bmem have thus far been reported to routinely display affinities in the nanomolar (KD ≈ 1 × 10−9) to subnanomolar (KD < 1 × 10−9) range, and even in some cases the low picomolar range (KD ≈ 1 × 10−12) (Cao et al., 2020; Jones et al., 2021; Kreer et al., 2020; Pinto et al., 2013; Robbiani et al., 2020; Rogers et al., 2020). While the RBD constitutes only a small subsection of the S1 antigen, it contains at least 4 distinct epitopes (Liu et al., 2020) for antibody binding, which can affect the capacity for neutralization in different ways. Targeting multiple different (non-overlapping) epitopes has been shown to potentiate neutralization and prevent viral escape in SARS-CoV-1 and, recently, SARS-CoV-2 (Koenig et al., 2021; ter Meulen et al., 2006). Therefore, we performed epitope mapping of our PC-derived antibodies by using an in-tandem competition assay: SARS-CoV-2 S1 was immobilized to Streptavidin biosensors, followed by incubation with an antibody 1 and a secondary, competing antibody 2 at a lower concentration. As controls, we used the CR3022 antibody or the extracellular domain of the ACE2 receptor (Figure 6D). As expected, competition and self-blocking occurred between pairs of the same antibody. None of the PC antibodies competed with CR3022, but all of the antibodies were able to block the binding of a secondary ACE2. For CR3022, the binding intensity of the secondary antibody was found to be approximately half of the competition-free response, indicating a weak interaction or overlap. Given that CR3022 binds a cryptic epitope that requires a conformational change of the RBD (Yuan et al., 2020a), binding of another antibody may reduce the stability of such a change. The germline-like antibodies mAb-64 and mAb-82 displayed similar behavior and were unable to bind in the presence of one another, thus indicating an overlapping epitope. The epitope targeted by mAb-50 did not overlap with either mAb-64 or mAb-82 (Figure 6E). Finally, to determine whether RBD-specific antibodies were indeed able to neutralize SARS-CoV-2, we performed neutralization assays using HEK293T-ACE2 cells and SARS-CoV-2 pseudotyped lentivirus (Hanke et al., 2020). An alpaca-derived single-domain antibody (ty1) was used as a positive control for neutralization (Hanke et al., 2020) (Figure 6F). Half-maximal inhibitory concentration (IC50) values were determined to be 145.0 ng/mL (mAb-50), 104.7 ng/mL (mAb-64), and 120.2 ng/mL (mAb-82), while neutralization saturates below 100% at 93.4%, 95.9%, and 96.1%, respectively. These results verify that transitional PCs in blood are antigen specific and demonstrate that single-cell sequencing on PCs can be used to select and identify high-affinity and potently neutralizing antibodies against SARS-CoV-2. Due to the ongoing evolution of SARS-CoV-2, viral variants have emerged, including ones possessing RBDs with mutations that enable escape from several mAbs, including clinically approved therapeutics (e.g., bamlanivimab). Therefore, we tested 3 of our mAbs (mAb-50, -64, and -82) for binding to SARS-CoV-2 RBD proteins derived from several variants: Alpha, Beta, Gamma, Delta, Delta+, Epsilon and Kappa (see key resources table). Remarkably, mAb-50 maintained high-level binding to all RBD variants, while mAb-64 and -82, consistent with sharing at least a partial epitope, either failed (mAb-64) or had only low levels of binding to Beta, or strongly reduced binding to Gamma. While mAb-64 and -82 maintain binding to Delta, the addition of K417N in Delta+ strongly reduces their binding, suggesting that K417 is part of the recognized epitope (Figure 6G).
Figure 6.
Characterization of RBD-specific antibodies for affinity, epitope binning, and neutralization
(A–C) Purified antibodies were assayed at different concentrations (shown here: 125, 37.5, 7.8, and 1.9 nM). Software-calculated fits are shown in red. Binding kinetics (apparent affinity constant kD) to SARS-CoV-2 S1 for FACS-confirmed RBD binders, determined by biolayer interferometry.
(D) Epitope binning setup of binders with ACE2.
(E) Additional binding by the secondary molecule indicates an unoccupied epitope (non-competitor, green), while no binding indicates epitope blocking (competitor, red). Self-blocking confirmation can be found on the diagonal.
(F) Viral neutralization, reduction in luciferase signal indicates successful viral neutralization.
(G) Relative response of RBD binders to variants Alpha, Beta, Gamma, Delta, Delta+, Epsilon, and Kappa.
Discussion
Here, we elucidated the antibody response of PCs in convalescent patients by performing single-cell antibody repertoire sequencing combined with mammalian display antibody screening. Single-cell sequencing revealed a feature landscape that was highly consistent with previously reported repertoires of patients with COVID-19 with respect to germline gene usage and CDR3 length distribution (Galson et al., 2020; Nielsen et al., 2020). We also observed a comparatively low rate of SHM for Ig class-switched antibodies on a repertoire level, a finding in line with previous reports on antibodies associated with SARS-CoV-2 infection (Nielsen et al., 2020; Rogers et al., 2020). Comparing sequences of our patient repertoires with sequences from the CoV-AbDab, we were able to identify 45 antibodies with an amino acid sequence similarity >80% in CDRH3 (Table S4). In addition, we found that mAb-64 shared high sequence similarity (up to 92%) to recently described stereotypical sequences of RBD-binding antibodies found to be commonly present in patients infected with SARS-CoV-2 (Kim et al., 2021). The recovered HC and LC pairing information was in line with previously reported data from healthy donors, with an increased frequency of common heavy and light pairs. IgG isotype-resolved lineage expansion analysis enabled the identification of 132 highly expanded PC clonal lineages, representing a diverse sequence space, to be tested for specificity to SARS-CoV-2 antigens.
To enable the rapid screening and discovery of SARS-CoV-2 reactive candidates within highly expanded PC lineages, we adapted a mammalian display platform based on the previously reported PnP-hybridoma system (Pogson et al., 2016). In a cloning-free, CRISPR-Cas9 HDR-mediated process, we created 2 antibody libraries (pool A and pool B) and screened for binding to SARS-CoV-2 antigens by flow cytometry. Through the use of deep sequencing and enrichment analysis, we could effectively determine the identity of antibodies binding to SARS-CoV-2. A subset of these antibody candidates was then integrated individually into the PnP system to generate monoclonal cell lines to verify and further characterize specificity to SARS-CoV-2 antigens. Thereby, we could show that in most of the patients in our cohort (11/16), there were highly expanded PCs producing antibodies with specificity to SARS-CoV-2. While we were unable to discover antibodies from all of the patients with specificity to S1 or S2, this could be explained by the fact that some patients had low or no detectable serum antibody titers against S protein (Figure 2B), or in some patients there was low single-cell sequencing depth that may have compromised clonal expansion analysis (Figure 3C). In both pools A and B, antibodies showed cross-reactivity to other human CoV antigens, including HCoV-229E, -OC43, -HKU1, and -NL63, but they did not show any specificity to Middle Eastern respiratory syndrome (MERS)-CoV-Spike or SARS-CoV-1 Spike, which is in contrast to the cross-specific antibodies discovered by others (Wec et al., 2020; Pinto et al., 2020), in which a large fraction of candidates potently bound both SARS-CoV-1 and SARS-CoV-2. Importantly, Pinto and colleagues successfully screened B cells from recovered SARS-CoV-1 patients for SARS-CoV-2 binding, proving that certain epitopes are preserved; Wec et al. followed a slightly different approach, in which a convalescent patient was chosen who had also survived a SARS-CoV-1 infection in the 2003 SARS epidemic. While SARS-CoV-1 patients thus seem to contain SARS-CoV-2 reactive antibodies, other discovery efforts that did not include SARS-CoV-1 experienced donors have been unable to discover antibodies with cross-reactivity (Ju et al., 2020). However, a recent study by Walker and colleagues that applied protein mutagenesis and directed evolution suggests that it is possible to engineer SARS-CoV-2-specific antibodies to have cross-reactivity and neutralization across a large panel of human CoV variants (Rappazzo et al., 2021).
Similar to the study by Ju and colleagues, there were no generalizable repertoire or sequence features within the discovered SARS-CoV-2-specific antibodies, further supporting the diversity of antibody repertoires that develop in patients with COVID-19. Despite this heterogeneity, we did find sequences that appear to be similar to those already reported (Galson et al., 2020; Robbiani et al., 2020), hinting at the importance of a germline-driven response in reaction to an infection with SARS-CoV-2 (Collins et al., 2020; Kreer et al., 2020). The overall number of antibodies that were identified to be specific for SARS-CoV-2 in this study is likely an underestimation, as not all of the pooled candidates enriched by flow cytometry and deep sequencing were tested separately. Based on the 81% success rate (35/43) of individually expressed antibodies, we believe that a number of as yet untested candidates are likely to be positive for SARS-CoV-2 binding. In addition, antibodies were only screened for binding to the S protein subunits, while there are possibly a number of antibodies that also bind to the NCP antigen based on ELISA reactivity of the pool A supernatants (Figure S5). Candidate sequences that were missing from the library based on deep sequencing data were believed to have had stability and expression issues in the PnP system. While the linker-based design of the HDR construct improved and facilitated the generation of stable cell lines, reformatting typically does come with its limitations, similar to known issues of converting antibody fragments (e.g., single-chain variable fragments and fragment antigen binding) to full-length IgG (Koerber et al., 2015; Xiao et al., 2017), as well as affinity, stability, and expression issues. Evaluating the RBD binders in the non-linker format increased the affinity and neutralizing potential, emphasizing the negative influence of a linker on antigen binding. However, after performing HDR integration into nPnP cells of previously missing mAbs, the reason behind the loss of sequences was found to be primarily due to error propagation of subsampling, mixing, and HDR, leading to an uneven distribution of sequences within the pool and, in some cases, the complete loss of sequences. This bottleneck can be overcome by using evenly resuspended DNA (resuspend by the supplier) or increasing the incubation post-resuspension, increasing the number of transfections or creating smaller subsets (i.e., here, instead of 96 sequences per transfection, working in 36 sequence increments).
Recently, Cao et al. (2020) have demonstrated how clonal expansion of Bmem can be used as a proxy for antigen specificity. However, despite finding two RBD-specific antibodies among the 132 candidates that were expressed, only 1 of them appeared to be (weakly) neutralizing, leading to the conclusion that antigen pre-selection was required. In this study, we found 3 potent neutralizing antibodies targeting the RBD from 132 selected clonal lineages, and while this is only a slight improvement in the success rate compared to that of Cao et al., it did demonstrate the potential to discover antibodies from PCs by single-cell sequencing. Given the unique physiology of PCs, this is particularly relevant at early stages of an infection as well as in the first weeks and months of a pandemic, when expediency is required and antigen-based selection may not be possible due to the delayed availability of antigen or reduced Bmem formation. It is important to note that serum antibodies represent an exceptional source for clinical antibody candidates (Williams et al., 2017) to which plasmablasts and short- and long-lived PCs are the sole contributors (Nutt et al., 2015). In the future, different strategies of selecting PC clonal lineages could allow for improvement in the successful identification of antigen-specific antibodies from single-cell sequencing data. In addition to clonal expansion, other factors could be incorporated into the selection criteria such as Ig isotype, SHM, and convergence in sequence space (Friedensohn et al., 2020).
Limitations of the study
As highlighted above, there are numerous caveats when creating libraries by HDR, including library coverage and costs associated with synthesizing genes for hundreds of candidate antibodies. In addition, using deep sequencing-based enrichment does not fully recover the relationship between binding and non-binding selections by FACS, as this is dependent on the underlying affinities of the interrogated antibody subset and antigen-labeling conditions. For future analysis, antibody pools could be screened at various antigen concentrations, which may improve deep sequencing enrichment analysis. The usage of a linker sequence instead of a dual-promoter or 2A system may also have affected antibody stability and expression, so that it may be prudent to further improve the sAb design strategy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human IgG-AF488 | Jackson ImmunoResearch | 109-545-003; RRID: AB_2337831 |
| mAbs 1-36, 40-135 | This paper. | N/A |
| CR3022 | ter Meulen et al., 2006 | PDB : 6W41 |
| Anti-mouse IgG1-PE [RMG1-1] | BioLegend | 406608; RRID:AB_10551618 |
| Anti-mouse IgG1-APC [RMG1-1] | BioLegend | 406610; RRID:AB_10696420 |
| Anti-human CD69 (mouse-IgG1) [FN50] | BioLegend | 310902; RRID:AB_314837 |
| Bacterial and virus strains | ||
| Pseudotyped SARS-CoV-2 | Hanke et al., 2020 | N/A |
| Biological samples | ||
| PBMCs and Serum | Patient specific. Part of SERO-BL-COVID19 biobank. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Strep-Tactin-APC | Iba-lifesciences | 6-5010-001 |
| Strep-Tactin-HRP | Iba-lifesciences | 2-1502-001 |
| Human ACE2 (His tagged) | SinoBiological | 10108-H08H |
| SARS-CoV-2 S1 (Avi-tag, biotinylated) | AcroBiosystems | S1N-C82E8 |
| SARS-CoV-2 S1-mFc | SinoBiological | 40591-V05H1 |
| SARS-CoV-2 S2-mFc | SinoBiological | 40590-V05B |
| SARS-CoV-2 S (Ectodomain) | SinoBiological | 40589-V08B1 |
| SARS-CoV-2 RBD-mFc | SinoBiological | 40592-V05H |
| SARS-CoV-2 Nucleocapsid | SinoBiological | 40588-V08B |
| SARS-CoV-2 RBD (Alpha) biotinylated (N501Y) | AcroBiosystems | SPD-C82E6 |
| SARS-CoV-2 RBD (Beta) biotinylated (K417N, E484K, N501Y) | AcroBiosystems | SPD-C82E5 |
| SARS-CoV-2 RBD (Gamma) biotinylated (K417T, E484K, N501Y) | AcroBiosystems | SPD-C82E7 |
| SARS-CoV-2 RBD (Delta) biotinylated (L452R, T478K) | AcroBiosystems | SPD-C82ED |
| SARS-CoV-2 RBD (Delta+) biotinylated (K417N, L452R, T478K) | AcroBiosystems | SPD-C82EG |
| SARS-CoV-2 RBD (Epsilon) biotinylated (L452R) | AcroBiosystems | SPD-C82E3 |
| SARS-CoV-2 RBD (Kappa) biotinylated (L452R, E484Q) | AcroBiosystems | SPD-C82EC |
| SARS-CoV-1 S1 | SinoBiological | 40150-V08B1 |
| MERS-CoV Spike | SinoBiological | 40069-V08B |
| HCoV-229E Spike | SinoBiological | 40605-V08B |
| HCoV-OC43 Spike | SinoBiological | 40607-V08B |
| HCoV-NL63 Spike | SinoBiological | 40604-V08B |
| HCoV-HKU1 | SinoBiological | 40606-V08B |
| DMEM GlutaMax | Gibco | 61965-026 |
| RPMI 1640 | Gibco | A10491-01 |
| Ultra-low IgG Fetal bovine serum | Gibco | 16250-078 |
| HEPES 1M | Gibco | 15630-080 |
| Penicillin/Streptamycin | Gibco | 15140-122 |
| 2-beta-mercaptoethanol | Gibco | 31350-010 |
| Critical commercial assays | ||
| Strep-Tactin® Buffer W | Iba-lifesciences | 2-1003-100 |
| Strep-Tactin® Buffer E (desthiobiotin) | Iba-lifesciences | 2-1000-025 |
| Strep-Tactin® Buffer R (HABA) | Iba-lifesciences | 2-1002-100 |
| Strep-Tactin® Sepharose | Iba-lifesciences | 2-1201-002 |
| Protein G Agarose | Pierce | 20399 |
| Protein G Elution Buffer | Pierce | 21004 |
| SF Nucleofector® Kit S | Lonza | V4XC-2032 |
| SF Nucleofector® Kit L | Lonza | V4XC-2024 |
| Octet Streptavidin Biosensors | ForteBio | 18-5019 |
| 10X Kinetics Buffer | ForteBio | 18-1105 |
| Chromium Single Cell 5’ Library & Gel Bead Kit | 10x Genomics | PN-1000006 |
| Chromium Single Cell 5’ Library Construction Kit | 10x Genomics | PN-1000020 |
| Chromium Single Cell V(D)J Enrichment Kit, Human B cell | 10x Genomics | PN-1000016 |
| Chromium Single Cell A Chip Kit | 10x Genomics | PN-1000009 |
| Chromium i7 Multiplex Kit | 10x Genomics | PN-120262 |
| EasySep Human CD138 Pos Selection Kit II | STEMCELL | 17877 |
| Illumina MiSeq v3 kit (600 cycle) | Illumina | MS-102-3003 |
| Deposited data | ||
| Fully annotated extracted antibody sequences and single cell results | This paper, Mendeley data | Table S1. CellRanger metrics—resulting sequencing reads and cell numbers determined from CellRanger version 3.1.1.0, for all patient samples, related to STAR, Table S4. Sequence similar hits in databases, https://dx.doi.org/10.17632/rvd7999gk4.1 |
| Patient VDJ data, raw FASTQ | Patients from SERO-BL-COVID19 study. This paper. | http://www.ncbi.nlm.nih.gov/bioproject/782883 |
| Enriched hybridoma NGS, raw FASTQ | This paper. | http://www.ncbi.nlm.nih.gov/bioproject/782992 |
| Experimental models: Cell lines | ||
| HEK293T-ACE2 | Hanke et al., 2020 | N/A |
| mRuby+ Cas9+ PnP Hybridoma | Pogson et al., 2016 | N/A |
| nPnP Hybridoma | This paper. | N/A |
| Expi293F | ThermoFisher | Cat#A14635 |
| Oligonucleotides | ||
| crRNA-3, ATATGACTCCTTCGACTCGA | This paper. IDT | N/A |
| Recombinant DNA | ||
| Patient-derived mAb HDR constructs | This paper | N/A |
| pTWIST transient expression constructs | This paper. TWIST bioscience | N/A |
| Software and algorithms | ||
| FlowJo X | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| GraphPad Prism 9.2.0 | GraphPad | https://www.graphpad.com/ |
| Original code | This paper | https://github.com/LSSI-ETH/Ehling_Covid_2021, zenodo: https://dx.doi.org/10.5281/zenodo.5781348 |
| R 4.0.1 (+ ggplot2, ComplexHeatmap, RColorBrewer, stringdist, igraph packages) | R Core Team, Gu et al., 2016, Neuwirth, 2014, van der Loo, 2014, Csardi and Nepusz, 2006) | N/A |
| CellRanger v3.1.1.0 | 10x Genomics | https://github.com/10XGenomics/cellranger |
| Immcantation v3.0.0 (pRESTO/IgBlast) | Vander Heiden et al., 2014 | https://hub.docker.com/r/kleinstein/immcantation |
| MiXCR | Bolotin et al., 2015 | https://mixcr.readthedocs.io/en/master/assemble.html |
Resource availability
Lead contact
Requests for resources and reagents should be directed to the lead contact, Sai Reddy (sai.reddy@ethz.ch).
Materials availability
Reagent generated in this study will be made available on request, but we may require a completed Materials Transfer Agreement.
Experimental model and subject details
Convalescent patients post-COVID19
Non-hospitalized patients older than 18 with a normal immune system and positive RT-PCR test were derived from the Swiss project “COVID-19 in Baselland Investigation and Validation of Serological Diagnostic Assays and Epidemiological Study of SARS-CoV-2 specific Antibody Responses (SERO-BL-COVID-19)”, as reported earlier (Kaltenbach et al., 2020). Participants consisted of patients from the SERO-BL-COVID-19 study sponsored by the Department of Health, Canton Basel-Land, Switzerland. SARS-CoV-2 infection was confirmed by RT-PCR of nasopharyngeal swab samples. Importantly, participants had mild to moderate symptoms without hospitalization. Over 300 patients were characterized and recruited for PBMC and serum collection. Patients included in this subset were selected based on POCTs assessing the presence of IgG and IgM SARS-CoV-2-specific antibodies (Qingdao Hightop Biotech, #H100), performed at the time of blood collection, after resolution of symptoms as seen from Figure 1. PMBCs were isolated as described previously (Kaltenbach et al., 2020). Sex, age distribution and displayed symptoms can be found in Figure 2 (upper). Ethics permission was granted by the ethics board “Ethikkommission Nordwest-und. Zentralschweiz (EKNZ)”. Every participant has signed a written informed consent as described previously (Kaltenbach et al., 2020)
Cells and virus
Cas9+ VH (−/−)VL (−/−) Hybridoma (nPnP) were grown in Dulbecco's modified Eagle's medium (DMEM-GlutaMAX, 61,965-026) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Thermo Cat#16000-044), 10 mM HEPES (Gibco Cat#15630-080), 50 μM 2-mercaptoethanol (Sigma-Aldrich Cat#M3148), 10 mM HEPES buffer (Thermo Cat#15630-056) and 100 U/mL penicillin-streptomycin (Thermo Cat#15140-122) at 37°C 5% CO2. Cells were subcultured every 48-72 h.
ACE2+ Human Embryonic Kidney cells (HEK293T-ACE2, derived from ATCC CRL-3216) were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco Cat#11960-085) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 1% penicillin-streptomycin at 37°C 5% CO2. Pseudotyped virus was produced as reported previously (Hanke et al., 2020). In brief, pseudotyped viruses were generated by the co-transfection of HEK293T cells with plasmids encoding the SARS-CoV-2 spike protein, a plasmid encoding firefly luciferase, and a lentiviral packaging plasmid (Addgene cat#8455) using Lipofectamine 3000 (Invitrogen). Media was changed 12–16 h after transfection, and pseudotyped viruses were harvested at 48 and 72 h post transfection, filtered through a 0.45 μm filter, and stored at −80°C until use.
Expi293F (Thermo Cat#A14635) were maintained in Expi293 serum free expression medium in a humidified incubator at 37°C 8% CO2 in vented 125 mL PETG Erlenmeyer, shaking at 125 rpm (25 mm orbital shaker), and passaged according to the manufacturer's instructions.
Method details
PBMC isolation and serum ELISA
In brief, PBMCs were isolated from EDTA blood using density gradient separation in Ficoll Paque Plus reagent (GE Healthcare, #17-1440-02). After separation, plasma was collected for ELISA detection of IgG and IgA SARS-CoV-2-S1 specific antibodies (Euroimmun Medizinische Labordiagnostika, #EI2668-9601G, #EI2606-9601A), as well as detection of IgM SARS-CoV-2-N specific antibodies (EDI™ Bencard Diagnostics KT-1033 231,002). PMBCs were then resuspended in freezing medium (RPMI 1640, 10%(v/v) FBS, 10%(v/v) dimethyl sulfoxide) and cryopreserved in liquid nitrogen.
Immunomagnetic isolation of PCs
PBMC samples were thawed, washed in complete media (RPMI 1640, 10%(v/v) FBS), followed by centrifugation. The cell pellets were resuspended in 0.5 mL complete media, counted and treated with 10 U mL−1 DNase I (STEMCELL Technologies, #07900) for 15 min at RT to prevent cell clumping. Cells were then washed again and pelleted by centrifugation. After resuspension in 0.5 mL flow cytometry buffer (PBS, 2%(v/v) FBS, 2 mM EDTA), the cells were filtered through a 40 μm cell strainer prior to immunomagnetic isolation, using an EasySep Human CD138 Pos. Selection kit II (STEMCELL Technologies, #17877) according to the manufacturer's instructions. Whenever possible, cells were adjusted to a concentration of 1 × 106 live cells/mL in PBS, 0.04%(v/v) BSA before proceeding with droplet generation.
Single-cell antibody repertoire library preparation
PCs were encapsulated with DNA-barcoded gel beads using a 10x Chromium controller (PN-110203). Samples were counted using a BioRad TC-10 automated cell counter and normalized to 1.7 × 104 cells in reverse transcription (RT) mix to ensure the recovery of 1 × 104 cells post-loading. After recovery, target enrichment RT and library preparation were performed according to the manufacturer's instructions (CG000086 manual, RevM, 10x Genomics) using the following kits: Chromium Single Cell 5’ Library & Gel Bead Kit (PN-1000006), Chromium Single Cell 5′ Library Construction Kit (PN-1000020), Chromium Single Cell V(D)J Enrichment Kit, Human B Cell (PN-1000016), Chromium Single Cell A Chip Kit (PN-1000009), Chromium i7 Multiplex Kit (PN-120262). Sequencing subsequently revealed that only approximately 0.3–0.4 × 104 cells were recovered after loading, probably due to inaccuracies in cell counting.
Deep sequencing
Single-cell sequencing
Single-cell antibody repertoire libraries were sequenced on an Illumina NovaSeq 6000 system (Illumina RTA Version: V3.4.4) using a 26 × 91 bp read configuration.
Mammalian display library sequencing
Samples of VH libraries for deep sequencing were prepared as described previously (Mason et al., 2018). Briefly, genomic DNA was extracted from 1 × 106 cells using a PureLink™ Genomic DNA mini Kit (Thermo, Cat: K182001). Extracted DNA was PCR amplified with a forward primer binding to the end of the Strep-Tag-Linker and a reverse primer annealing to the intronic region directly 3′ of the J segment. PCRs were performed with Q5® High-fidelity DNA polymerase (NEB, M0491L) plus GC buffer with the following reaction conditions: 1 cycle of 98°C for 30 s; 20 cycles of 98°C, 60°C for 20 s, 72°C for 30 s; and a final extension at 72°C for 1 min; followed by 10°C for temporary storage. Products were checked on an agarose gel and cleaned-up through a PCR-clean up kit (DNA Clean and Concentrator-5, Zymo, D4013), followed by a secondary PCR to add Illumina indices (Figure S4C). PCR products were then gel purified (2% TAE).
Design of homology-directed repair (HDR) donors
Single-ORF antibody sequences were designed as reported previously (Koerber et al., 2015; Moffett et al., 2019). Briefly, selected paired VJ-IgK/IgL and VDJ were linked through 57–amino acid glycine-serine linker containing three tandem Strep-Tag II motifs. A universal leader sequence was added as well as 250-350 bp homology arms to the 5′ and -3′ end.
Mammalian cell culture, transfections
An iteration of the established Cas9+ PnP-mRuby cell line was generated by flanking and deleting the mRuby-pA gene with a pair of guides (crRNA1 and 2), as well as providing a bridging ssODN at 500 pmols to facilitate the integration of a new guide sequencing as well as an XhoI restriction site for detection of HDR on single-cell clones.
ssODN Sequence (homology arms, insert, XhoI restriction site): GAGCCTATGGTAGTAAATACAGGCATGCCCACACTGTGAAAACAACATATGACTCCTTC GACTCGAGGGT CAGGTCTTTATTTTTAACCTTTGTTATGGAGTTTTCTGAGCATTGCAGACTAATCTTGGA
crRNA1: TCTGGGACTGTGGAGAAGAC
crRNA2: TTAAAAATAAAGACCTGGAG
crRNA3: ATATGACTCCTTCGACTCGA.
Electroporation was performed with the 4D-Nucleofector™ System (Lonza) using SF Cell line 4D-Nucleofector® X Kit L or X Kit S (Lonza, V4XC-2024, V4XC-2032) with the program CQ-104. Previous to nucleofection, cells were centrifuged at 150xg for 5 min and washed once with pre-warmed (37°C) Opti-MEM® I Reduced Serum Medium (Thermo, 31,984-062) at 1 mL per 1 × 106 cells. The resulting cell pellet was then resuspended in SF buffer according to the manufacturer's description, after which Alt-R gRNA and (3.5-4.5 pmoles per 1 × 106 cells) of HDR donors were added. Cells were analyzed for HDR events approx. 3-4 days post transfection.
Expi293 culture and transfections were carried out according to the manufacturer's specifications (ThermoFisher, Cat#A14525 and as described previously (Vazquez-Lombardi et al., 2018)
Flow cytometry
Transfected cells were sorted for HDR + by staining for StrepTactin-APC 1:100 (IBA lifesciences, Cat: 6-5010-001) and anti-hIgG : AF488 1:100 (Jackson ImmunoResearch, Cat: 109-545-003). Enriched cells were then selected for antigen binding by incubating with SARS-CoV-2 S1-mFc 1:75 (Sinobiological, Cat: 40591-V05H1), S2-mFc 1:75 (Sinobiological, Cat: 40,590-V05B) on ice for 20 min, followed by a single wash and a secondary staining with anti-mFc IgG1: PE and/or APC [clone RMG1-1] at 1:100 (Biolegend, Cat: 406,608/406,610). After enrichment for S1, cells were also tested for binding to SARS-CoV-2 RBD-mFc 1:375 (Sinobiological, Cat: 40,592-V05H), and the same secondary staining. FACS was analyzed using FlowJo X software. At least 20′000 live events were acquired per sample to ensure sufficient statistical sampling.
Antibody purification
Dense Hybridoma or Expi293F cultures were centrifuged at 300xg for 5 min to pellet the cells. Supernatant was filtered using Steriflip® 0.22 μm (Merck, SCGP00525) filter units.
Strep-Tag purification was carried out according to the manufacturer's instructions (Strep-Tactin®-Sepharose®, iba-lifesciences). First supernatant was adjusted to pH > 7 using Buffer W. Sepharose columns were equilibrated twice using 1 column volume (CV) of Buffer W. Supernatant was then added to the column, the flow through was collected and placed on ice. After washing the column with 5x 1 CV of Buffer W, mAbs were eluted off the column and collected in 6 fractions of 0.5 CV Buffer E each.
For Protein G purification, Expi293F supernatant was directly loaded onto Protein G Agarose (Pierce, Cat# 20,399) gravity columns, washed twice with PBS and eluted using Protein G Elution Buffer (Pierce, Cat# 21,004). The eluted fractions were immediately neutralized with 1M TRIS-Buffer (pH = 8) to physiological pH.
For both Strep-Tag and Protein G purifications, A280 nm absorption was quantified by Nanodrop™ 2000c. Protein containing fractions were pooled and buffer exchanged using SnakeSkin dialysis tubing (10 MWCO, Pierce Cat#68100) followed by further dialysis and concentration using Amicon Ultra-4 10kDa centrifugal units (Merck, Cat# UFC801096), as described previously (Vazquez-Lombardi et al., 2018)
Cross-specificity ELISA
Cell supernatant containing secreted antibodies was analyzed for the presence of coronavirus specific responses. First, supernatant was collected and filtered with 0.2 μm filters (Sartorius, 16534-K). 96-well microtiter plates were coated with recombinant coronavirus antigens purchased from SinoBiological: SARS2-Spike S1+S2 (40,589-V08B1), SARS1-S1 (40,150-V08B1), MERS-CoV Spike (40,069-V08B), HCoV-229E Spike (40,605-V08B), HCoV-OC43 Spike (40,607-V08B), HCoV-HKU1 Spike (40,606-V08B), HCoV-NL63 Spike (40,604-V08B), SARS-CoV-2 Nucleocapsid (40,588-V08B) at 8 μg/mL in PBS at 4°C overnight. After blocking with 2% milk, 0.05% tween PBS (PBSTM) for 1 h at RT, cell supernatants were diluted 1:2 in 2% milk PBS (PBSM) and added to the microtiter plate for 1 h at RT. Antigen-specific responses were detected using Strep-Tactin®-HRP (iba lifesciences, #2-1502-001) diluted 1:10′000 in PBSM, followed by repeated washing and using TMB (3,3′,5,5′-tetramethylbenzidine) (1-Step™ Ultra TMB, #34028) as a substrate. The absorbance of each sample was measured at 450 nm as well as 630 nm. All samples reported here were interrogated in the same plate.
Octet biolayer interferometry
Kinetics: Biolayer interferometry assays were performed on the Octet Red96 (ForteBio) at 25°C, shaking at 1000 rpm. Kinetics assays were performed with Streptavidin (SA) Biosensors (ForteBio, Cat-No. 18-5019) with the following steps: (0) Hydration of SA Biosensors in 1X kinetics buffer for 30 min (ForteBio, Cat-No. 18-1105). (1) Baseline equilibration in 1X kinetics buffer for 60s. (2) Regeneration of sensors (3x) in 10 mM Glycine (3) Baseline for 300s (4) Loading of antigen (SARS-CoV-2 S1 Avi-tag, Cat-No. S1N-C82E8) at 100 nM for 300 s (5) Quenching/Blocking of sensors in 50 μg/mL hIgG in 1X KB for 30s (6) Antibody binding: sensors immersed into purified antibody at 1.9–500 nM in 1X KB for 350 s (7) Dissociation in 1X KB for 700 s (8) Regeneration of sensors. Curve fitting was performed using the ForteBio Octet HTX data analysis software using a 1:1 model, and a baseline correction using a reference sensor.
Epitope Binning: Kinetic analysis of antibody pairs was performed using Streptavidin (SA) Biosensors (ForteBio, Cat-No. 18-5019) using the “binning assay” definition with the following steps: (0) Hydration of SA Biosensors in 1X kinetics buffer for 30 min (ForteBio, Cat-No. 18-1105). (1) Baseline equilibration in 1X kinetics buffer for 60s. (2) Regeneration of sensors (3x) in 10 mM Glycine (3) Baseline for 300s. (4) Loading of antigen (SARS-CoV-2 S1 Avi-tag, Cat-No. S1N-C82E8) at 41.6 nM for 45 s (5) Quenching/Blocking of sensors in 50 μg/mL hIgG in 1X KB for 30s. (6) Association of Ab1, ACE2 (SinoBiological, Cat-No. 10108-H08H) or Buffer (1X KB) at 166 nM for 800 s, followed by (7) short dissociation step for 10 s and then (8) the association of Ab2 at 78 nM for 400 s (9) Regeneration of sensors. Curve fitting was performed using the ForteBio Octet HTX data analysis software using a 1:1 model, and a baseline correction with a reference sensor.
Variant Response: RBD-variants were acquired from AcroBiosystems (alpha: SPD-C82E6; beta: SPD-C82-E5; gamma: SPD-C82E7; delta: SPD-C82ED; delta+: SPD-C82EG; epsilon: SPD-C82E3; kappa: SPD-C82EC) and loaded onto Streptavidin (SA) biosensors (ForteBio, Cat-No. 18-5019) using a standard kinetics layout with the following steps: (0) Hydration of SA Biosensors in 1X kinetics buffer for 30 min (ForteBio, Cat-No. 18-1105). (1) Baseline equilibration in 1X kinetics buffer for 60s. (2) Loading of biotinylated variant RBDs onto the sensor at 2 μg/mL for 60s with a threshold for all but the baseline channel at 0.5 nm. (3) Another baseline acquisition in 1X kinetics buffer for 120s. Association of mAbs diluted in 1X kinetics buffer at 20 nM (mAb-50) and 60 nM (mAb-64, -82) for 60s, followed by (4) a dissociation step in 1X kinetics buffer for 300s. Fitting was performed using the ForteBio Octet HTX software package as described above.
Viral neutralization assays
Neutralization assays were performed as described previously (Hanke et al., 2020). Briefly, SARS-CoV-2 spike-pseudotyped lentiviruses incorporating a transfer plasmid encoding firefly luciferase were produced from HEK293T cells. Pseudotyped virions standardized to an input producing ∼100,000 RLUs were incubated with serial dilutions of recombinant antibodies for 60 min at 37°C prior to the addition of ∼15,000 HEK293T-ACE2 cells and incubation for 48h. Luminescence was measured using Bright-Glo (Promega) on a GM-2000 luminometer.
Quantification and statistical analysis
Data preprocessing
Raw sequencing results were processed with CellRanger (v3.1.1.0, 10x Genomics). Assembled and filtered contigs were subsequently re-aligned using Immcantation's pRESTO/IgBlast pipeline (https://hub.docker.com/r/kleinstein/immcantation, v3.0.0 (Vander Heiden et al., 2014). Sequences were aligned to the IMGT database. Only cells with a productive heavy and light chain were considered for the final analysis.
Statistical analysis and plots
Statistical analysis was performed using R 4.0.1 (R Core Team). Graphics were generated using the ggplot2 (Wickham et al., 2020), ComplexHeatmap (Gu et al., 2016), and RColorBrewer (Neuwirth, 2014) R package.
Enrichment
Repeated sorting to achieve pure populations (Ab + or Ag+) was performed to determine the enrichment of specific sequences in the Ag-selected fraction and, hence, their specificity. This statistical approach aids in determining true binders from false background. Enrichment (En) of a sequence (s) was calculated by taking the log2 of the ratio between the clonal frequency (f) in the post selection sample (sel) and the frequency observed in the background (Ab+) sample (bkg). Only sequences observed in all three (Background, S1 and S2 selected) samples are reported.
Overlap calculation
Overlap was determined as previously described (Greiff et al., 2017a; 2017b). Briefly the number of shared sequences was divided by the size of the smaller repertoire.
Sequence similarity networks
Network plots were generated using the igraph package [v.1.2.4.2 (Csardi and Nepusz, 2006),] as previously described (Miho et al., 2019), using the layout option ‘layout-nicely’ with edges drawn between amino acid sequences closer than edit distance 3. Edit distances were calculated using the R package stringdist [v.0.9.6.3(van der Loo, 2014)] using the method “hamming”.
Viral neutralization data processing
Luciferase signals for viral neutralization were measured on GM-2000 luminometerand analyzed using GraphPad Prism. ELISA signals were measured on a Tecan infinite-pro 2000 and processed using GraphPad Prism.
Acknowledgments
We acknowledge the ETH Zurich D-BSSE Single Cell Unit and the Genomics Facility Basel for excellent support and assistance. This work was supported by the European Research Grant CoroNAb (to S.T.R. and B.M.), Personalized Health and Related Technologies (to S.T.R. and R.V-L.), and the Botnar Research Centre for Child Health (to S.T.R.).
Author contributions
R.A.E., F.B., R.V-L., and S.T.R. designed the experiments; D.M.M., B.W., F.B., R.V-L., R.B.D.R., A.P.C., F.R., and M.S. assisted in the patient sample collection and analysis and the cell isolation experiments; R.A.E., K.-L.H, M.D.O., M.P., and C.R.W. performed genome editing and the antibody screening experiments; D.J.S. and B.M. designed and performed the viral neutralization experiments; R.A.E., C.R.W., and S.F. analyzed the sequencing data; R.A.E., C.R.W,. and S.T.R. wrote the paper, with feedback and input from all of the authors.
Declaration of interests
The authors declare no competing interests.
Published: December 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110242
Supplemental information
Methods
(A) Fully annotated sequences of the selected sequences, related to Figures 2, 3, 4, 5, and 6. (B) The 132 selected variants were examined for enrichment. (C) Summary of confirmed binders per patient, related to Figures 4 and 5.
Three categories of patients: No reactive sequences identified. Only S2 reactive and both S1 and S2 reactive (37/132 selected, ∼28% were reactive) (mean hits/selected per repertoire 35%), related to Figures 2 and 5.
Binders in comparison with sequences found in the CovAbDab, related to Figure 3
Data and code availability
-
•
VDJ Raw FASTQ files have been uploaded to SRA (NCBI) and are publicly available as of the date of publication, see key resources table.
-
•
Original code for the analysis of repertoire data has been deposited at https://github.com/LSSI-ETH
-
•
Any additional data that support the findings of this study are available from the lead contract upon reasonable request.
References
- Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P. Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020;371:m3939. doi: 10.1136/bmj.m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit J., Taylor J.J. Techniques to study antigen-specific B cell responses. Front. Immunol. 2019;10:1694. doi: 10.3389/fimmu.2019.01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., Straten K.van der, Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.J., Mitchell R.M., Meyer Sauteur P.M., Kelly D.F., Trück J. The antibody-secreting cell response to infection: kinetics and clinical applications. Front. Immunol. 2017;8:630. doi: 10.3389/fimmu.2017.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N. Engl. J. Med. 2020;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-Q., Wohlbold T.J., Zheng N.-Y., Huang M., Huang Y., Neu K.E., Lee J., Wan H., Rojas K.T., Kirkpatrick E., et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. 2018;173:417–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A.M., Yaari G., Shepherd A.J., Lees W., Watson C.T. Germline immunoglobulin genes: disease susceptibility genes hidden in plain sight? Curr. Opin. Syst. Biol. 2020;24:100–108. doi: 10.1016/j.coisb.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G., Nepusz T. The igraph software package for complex network research. Interjournal. 2006;Complex Systems(1695) [Google Scholar]
- DeKosky B.J., Lungu O.I., Park D., Johnson E.L., Charab W., Chrysostomou C., Kuroda D., Ellington A.D., Ippolito G.C., Gray J.J., et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. U S A. 2016;113:E2636–E2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Drug Administration Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab. 2021. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab
- Friedensohn S., Neumeier D., Khan T.A., Csepregi L., Parola C., Vries A.R.G.de, Erlach L., Mason D.M., Reddy S.T. Convergent selection in antibody repertoires is revealed by deep learning. BioRxiv. 2020 doi: 10.1101/2020.02.25.965673v1. [DOI] [Google Scholar]
- Galson J.D., Schaetzle S., Bashford-Rogers R.J.M., Raybould M.I.J., Kovaltsuk A., Kilpatrick G.J., Minter R., Finch D.K., Dias J., James L.K., et al. Deep sequencing of B cell receptor repertoires from COVID-19 patients reveals strong convergent immune signatures. Front. Immunol. 2020;11:3283. doi: 10.3389/fimmu.2020.605170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghraichy M., Galson J.D., Kovaltsuk A., von Niederhäusern V., Pachlopnik Schmid J., Recher M., Jauch A.J., Miho E., Kelly D.F., Deane C.M., et al. Maturation of the human immunoglobulin heavy chain repertoire with age. Front. Immunol. 2020;11:1734. doi: 10.3389/fimmu.2020.01734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk P., Bombardi R.G., Erasmus J.H., Tan Q., Nargi R., Soto C., Abbink P., Suscovich T.J., Durnell L.A., Khandhar A., et al. Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nat. Biomed. Eng. 2020;4:1030–1043. doi: 10.1038/s41551-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiff V., Weber C.R., Palme J., Bodenhofer U., Miho E., Menzel U., Reddy S.T. Learning the high-dimensional immunogenomic features that predict public and private antibody repertoires. J. Immunol. 2017;199:2985–2997. doi: 10.4049/jimmunol.1700594. [DOI] [PubMed] [Google Scholar]
- Greiff V., Menzel U., Miho E., Weber C., Riedel R., Cook S., Valai A., Lopes T., Radbruch A., Winkler T.H., et al. Systems analysis reveals high genetic and antigen-driven predetermination of antibody repertoires throughout B cell development. Cell Rep. 2017;19:1467–1478. doi: 10.1016/j.celrep.2017.04.054. [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinforma. Oxf. Engl. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Dunand C.J., Leon P.E., Huang M., Choi A., Chromikova V., Ho I.Y., Tan G.S., Cruz J., Hirsh A., Zheng N.-Y., et al. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.-N., Cheng V.C.-C., Li X., Tam A.R., Hung D.L.-L., Chiu K.H.-Y., Yip C.C.-Y., Cai J.-P., Ho D.T.-Y., Wong S.-C., et al. SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: a case series. Lancet Infect. Dis. 2020;20(9):1051–1060. doi: 10.1016/S1473-3099(20)30364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:593. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kaltenbach H.-M., Rudolf F., Linnik J., Deichmann J., Ruf T., Altamura R., Kapetanovic E., Mason D., Wagner B., Goetz T., et al. Initial characterisation of ELISA assays and the immune response of the clinically correlated SARS-CoV-2 biobank SERO-BL-COVID-19 collected during the pandemic onset in Switzerland. MedRxiv. 2020 doi: 10.1101/2020.07.05.20145888. [DOI] [Google Scholar]
- Kawase M., Shirato K., Hoek L.van der, Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., Lee H., Jung J., Kang C.K., Song K.-H., et al. Stereotypic neutralizing VH antibodies against SARS-CoV-2 spike protein receptor binding domain in COVID-19 patients and healthy individuals. Sci. Transl. Med. 2021;13:eabd6990. doi: 10.1126/scitranslmed.abd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P.-A., Das H., Liu H., Kümmerer B.M., Gohr F.N., Jenster L.-M., Schiffelers L.D.J., Tesfamariam Y.M., Uchima M., Wuerth J.D., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:6530. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber J.T., Hornsby M.J., Wells J.A. An improved single-chain Fab platform for efficient display and recombinant expression. J. Mol. Biol. 2015;427:576–586. doi: 10.1016/j.jmb.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- van der Loo M.P.J. The stringdist package for approximate string matching. R. J. 2014;6:111–122. [Google Scholar]
- Maeshima N.M., Jovanovic V.K., Ewen C.L., Clarke S.J., Kokaji A.I., Woodside S.M., Thomas T.E., Eaves A.C. Fast and easy isolation of CD27-positive human memory B cells using EasySep Releasable RapidSpheres. J. Immunol. 2017;198:144–152. [Google Scholar]
- Mason D.M., Weber C.R., Parola C., Meng S.M., Greiff V., Kelton W.J., Reddy S.T. High-throughput antibody engineering in mammalian cells by CRISPR/Cas9-mediated homology-directed mutagenesis. Nucleic Acids Res. 2018;46:7436–7449. doi: 10.1093/nar/gky550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D.M., Friedensohn S., Weber C.R., Jordi C., Wagner B., Meng S., Ehling R., Bonati L., Dahinden J., Gainza P., et al. Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nat. Biomed. Eng. 2021;5:600–612. doi: 10.1038/s41551-021-00699-9. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. Plos Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miho E., Roškar R., Greiff V., Reddy S.T. Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat. Commun. 2019;10:1321. doi: 10.1038/s41467-019-09278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett H.F., Harms C.K., Fitzpatrick K.S., Tooley M.R., Boonyaratanakornkit J., Taylor J.J. B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 2019;4:35. doi: 10.1126/sciimmunol.aax0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumeier D., Pedrioli A., Genovese A., Sandu I., Ehling R., Hong K.-L., Papadopoulou C., Agrafiotis A., Kuhn R., Robbiani D., et al. Single-cell sequencing reveals clonally expanded plasma cells during chronic viral infection produce virus-specific and cross-reactive antibodies. BioRxiv. 2021 doi: 10.1101/2021.01.29.428852. [DOI] [Google Scholar]
- Neumeier D., Yermanos A., Agrafiotis A., Csepgregi L., Chowdhury T., Ehling R.A., Kuhn R., Brisset-Di Roberto R., Di Tacchio M., Antonialli R., et al. Phenotypic determinism and stochasticity in antibody repertoires of clonally expanded plasma cells. BioRxiv. 2021 doi: 10.1101/2021.07.16.452687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwirth E. 2014. RColorBrewer: ColorBrewer Palettes. [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.-Y., Rustagi A., Rogers A.J., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- Parola C., Neumeier D., Friedensohn S., Csepregi L., Tacchio M.D., Mason D.M., Reddy S.T. Antibody discovery and engineering by enhanced CRISPR-Cas9 integration of variable gene cassette libraries in mammalian cells. MAbs. 2019;11:1367–1380. doi: 10.1080/19420862.2019.1662691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Montani E., Bolli M., Garavaglia G., Sallusto F., Lanzavecchia A., Jarrossay D. A functional BCR in human IgA and IgM plasma cells. Blood. 2013;121:4110–4114. doi: 10.1182/blood-2012-09-459289. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Pogson M., Parola C., Kelton W.J., Heuberger P., Reddy S.T. Immunogenomic engineering of a plug-and-(dis)play hybridoma platform. Nat. Commun. 2016;7:12535. doi: 10.1038/ncomms12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2020;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S.T., Ge X., Miklos A.E., Hughes R.A., Kang S.H., Hoi K.H., Chrysostomou C., Hunicke-Smith S.P., Iverson B.L., Tucker P.W., et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotechnol. 2010;28:965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- Ribatti D. The discovery of plasma cells: an historical note. Immunol. Lett. 2017;188:64–67. doi: 10.1016/j.imlet.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf F., Kaltenbach H.-M., Linnik J., Ruf M.-T., Niederhauser C., Nickel B., Gygax D., Savic M. Clinical Characterisation of Eleven Lateral Flow Assays for Detection of COVID-19 Antibodies in a population. MedRxiv. 2020 doi: 10.1101/2020.08.18.20177204. [DOI] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:6520. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovich V.A., Burgos Pratx L.D., Scibona P., Beruto M.V., Vallone M.G., Vázquez C., Savoy N., Giunta D.H., Pérez L.G., Sánchez M. del L., et al. A randomized trial of convalescent plasma in covid-19 severe pneumonia. N. Engl. J. Med. 2020;384:619–629. doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden J.A., Yaari G., Uduman M., Stern J.N.H., O’Connor K.C., Hafler D.A., Vigneault F., Kleinstein S.H. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Lombardi R., Nevoltris D., Luthra A., Schofield P., Zimmermann C., Christ D. Transient expression of human antibodies in mammalian cells. Nat. Protoc. 2018;13:99–117. doi: 10.1038/nprot.2017.126. [DOI] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., Chang W., Henry L., Thomas L.P., Kohske T., Wilke C., Woo K., Yutani H., Dunnington D. 2020. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. [Google Scholar]
- Williams L.D., Ofek G., Schätzle S., McDaniel J.R., Lu X., Nicely N.I., Wu L., Lougheed C.S., Bradley T., Louder M.K., et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol. 2017;2:eaal2200. doi: 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.-Y., Mays I., Garman L., Helms C., et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Douthwaite J.A., Chen Y., Kemp B., Kidd S., Percival-Alwyn J., Smith A., Goode K., Swerdlow B., Lowe D., et al. A high-throughput platform for population reformatting and mammalian expression of phage display libraries to enable functional screening as full-length IgG. MAbs. 2017;9:996–1006. doi: 10.1080/19420862.2017.1337617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 2020;6:eabc9999. doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods
(A) Fully annotated sequences of the selected sequences, related to Figures 2, 3, 4, 5, and 6. (B) The 132 selected variants were examined for enrichment. (C) Summary of confirmed binders per patient, related to Figures 4 and 5.
Three categories of patients: No reactive sequences identified. Only S2 reactive and both S1 and S2 reactive (37/132 selected, ∼28% were reactive) (mean hits/selected per repertoire 35%), related to Figures 2 and 5.
Binders in comparison with sequences found in the CovAbDab, related to Figure 3
Data Availability Statement
-
•
VDJ Raw FASTQ files have been uploaded to SRA (NCBI) and are publicly available as of the date of publication, see key resources table.
-
•
Original code for the analysis of repertoire data has been deposited at https://github.com/LSSI-ETH
-
•
Any additional data that support the findings of this study are available from the lead contract upon reasonable request.