Abstract
Much has been learned about the protein corona and their biological implications within the context of nanomedicine and nanotoxicology. Yet no data is available about the protein corona associated with nanoparticles undergoing spontaneous surface-energy minimization, a common phenomenon during the synthesis and shelf life of nanomaterials. Accordingly, here we employed gold nanoparticles (AuNPs) possessing the three initial states of spiky, mid-spiky and spherical shapes and determined their acquisition of human plasma protein coronae with label-free mass spectroscopy. The AuNPs collected coronal proteins that were different in abundance, physicochemical parameters, and interactive biological network. Furthermore, the AuNPs induced endothelial leakiness to different degrees, which was partially negated by their protein coronae as revealed by confocal fluorescence microscopy, in vitro and ex vivo transwell assays, as well as signalling pathway assays. This study filled a knowledge void concerning the dynamic protein corona of nanoparticles possessing an evolving morphology and shed light on their implication for future nanomedicine harnessing the paracellular pathway.
Keywords: protein corona, morphology, proteomics, endothelial leakiness, nanomedicine
Table of Content Graphic
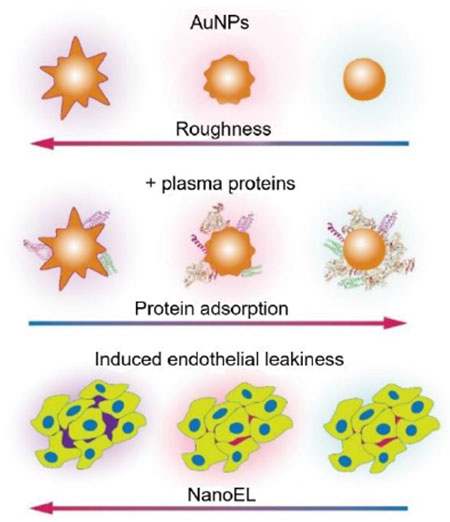
This study examined the dynamic protein corona of gold nanoparticles assuming an evolving morphology and further evaluated their effect on the paracellular transport of nanoparticles.
Introduction
Engineered nanoparticles (NPs) interface with biological systems to acquire a protein “corona”, which subsequently dictates the biological fate of the NPs downstream.1–4 This spontaneous protein association is driven by free-energy minimization,5 mediated by Columbic and van der Waals forces, hydrogen bonding and hydrophobic interactions. Temporally, the adsorption and desorption of proteins are dictated by the Vroman effect,6 where proteins of high abundance (i.e., soft corona) are replaced over time by proteins of high binding affinity (i.e., hard corona) for the NP substrate.7 Spatially, the protein corona evolves as NPs navigate through different biological compartments in vivo to accumulate their protein fingerprints,8 and the NP-protein corona is scrutinized by the immune system through opsonization to impact the biodistribution, delivery, and clearance of the NPs.
The formation of the protein corona is largely determined by both the NPs and their biological hosts.3, 9 Specifically, intrinsic NP properties pertinent to protein adsorption include their size, surface charge, chemical composition, morphology, as well as surface functionalization.10–17 Serum, plasma, and cell culture media, for example, are representative in vitro environments, while in vivo environments afford more abundant biomolecular species and complex anatomy.18–22 In addition, exposure time and temperature also contribute to evolution of the protein corona.23, 24
While the protein corona has been extensively studied for different nanomaterials and biological environments,10, 11, 18, 25–29 NPs in aqueous suspensions often undergo agglomeration to change their size and shape mediated by van der Waals forces and hydrophobic interactions. Furthermore, NPs themselves may also undergo energy minimization to evolve in surface roughness and shape during their synthesis and shelf life,30–32 yet their associated dynamic protein corona according to such transformation has not been reported to date. Accordingly, in the current study we characterized the dynamic plasma protein corona of gold nanoparticles (AuNPs) evolving from spiky to mid-spiky and to spherical shapes along the free-energy landscape. AuNPs are commonly employed in nanomedicine and nanobiotechnology with their applications ranging from tumor sensing and imaging33, 34 to photothermal,35 radiation36 and antiangiogenic therapies.37 This experimental design allowed us to single out NP surface roughness/protrusion as a focused physical parameter in the examination of the protein corona while maintaining other physicochemical factors of the NPs (i.e., chemical composition and surface functionalization) constant. As an application of the dynamic protein corona, we evaluated their effect on modulating nanomaterial-induced endothelial leakiness (NanoEL) in a human umbilical vein endothelial cell (HUVEC) monolayer in vitro and in swine vessels ex vivo. This study filled a knowledge void in the literature of the protein corona from a unique NP perspective and should benefit the design of future nanomedicine exploiting the NP surface morphology and their entailed suspendibility, biodistribution, paracellular transport, and opsonization.
Materials and methods
Synthesis of gold nanoparticles
Citrate-coated AuNP seeds were synthesized by step-wise seeded growth Turkevich-Frens protocol38 with slight modifications. Specifically, aqueous chloroauric acid (HAuCl4, 150 mL, 0.25 mM, Sigma Aldrich) was brought to boil at 100 °C for 15 min under continuous stirring, then sodium citrate solution (Na3C6H5O7, 38.75 mM, Sigma Aldrich) was added to initiate the reduction reaction. Approximately 2.9 mL and 4.8 mL of Na3C6H5O7 solutions were added to render AuNP seeds of 13 nm and 18 nm, respectively. The solutions were kept in boiling condition with vigorous stirring until a wine-red color emerged. The resultant Au seed solutions were kept at room temperature and used for AuNP formation. The spherical AuNPs with a size of 13 nm were used as seeds to produce AuNPs of 30 nm. Surface roughness was introduced to the AuNPs seeds using 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Sigma Aldrich) aided branch growth. Spiky 30 nm AuNPs (termed as AuNP1 hereafter) were synthesized by adding 1.6 mL of as-synthesized 13 nm spherical AuNP seeds into 23.1 mL of HEPES aqueous solution (50 mM, pH 7). Thereafter, 0.3 mL of freshly prepared hydroxylammonium chloride (NH2OH · HCl, 40 mM, Sigma Aldrich) was added to the mixture and surface roughness of the AuNPs was achieved through the addition of 5 mL of aqueous solution HAuCl4 (1 mM) in a dropwise manner. The reaction was maintained in stirring condition for another 2 h at room temperature to allow the growth of sufficient surface roughness.
Different surface roughness was achieved by controlling the working concentrations of the HEPES solution. Spherical AuNPs (termed as AuNP3 hereafter) were produced in the absence of HEPES solution, whereas the mid-spiky AuNPs (AuNP2) were produced in the presence of 5 mM HEPES. The different-sized AuNPs were dialyzed with Cellu·Sep T1/nominal MWCO: 3.5~5 kDa (Adelab Scientific) against 10 mM HEPES solution. The dialyzed AuNPs were kept at 4 °C and used as the stock for subsequent experiments.
Protein corona formation and isolation
Plasma was prepared by collecting the top layer following centrifugation of fresh blood at 900 g, for 15 min, without brake. The blood was collected from a healthy human volunteer into Greiner Bio-One sodium heparin vacuettes, following the University of Melbourne Human ethics approval 1443420 and the Australian National Health and Medical Research Council Statement on Ethical Conduct in Human Research.
AuNPs were mixed with excess human plasma at a mass ratio of 1:10 and incubated at 37 °C for 24 h under shaking conditions. The sample suspensions were centrifuged at 16 300 g for 15 min at room temperature to isolate the hard corona complexes and were washed thrice with 1× phosphate-buffered saline (PBS, pH 7, Sigma Aldrich) to remove unbound proteins. The total amount of proteins bound to different AuNPs was determined by the Pierce™ BCA Protein Assay Kit (Thermo Scientific) according to the manufacturer’s instructions and various concentrations of bovine serum albumin (BSA) as the standard. For uniform measurements, AuNPs (1 mg) in excess of the plasma concentration were used. The resulting pellets were resuspended in 100 μL of 1× PBS buffer and 25 μL of sample volume was used for a 96 well plate assay. The adsorption was measured at 562 nm using a fluorescence microplate reader (PerkinElmer EnSight HH33400).
Characterization of gold nanoparticles
The synthesized AuNPs were imaged using a transmission electron microscope (FEI Tecnai G2 F20, Eindhoven) operated at a voltage of 200 kV. Briefly, the AuNPs were dropped onto a carbon-coated TEM grid (Formvar) and viewed under an accelerating voltage of 200 kV. The primary sizes of the AuNPs were determined by measuring for at least 50 randomly selected AuNPs with ImageJ software.39 The absorbance of the AuNPs was obtained through spectral scanning from wavelength of 300 to 800 nm with a UV-Vis spectrophotometer (UV-3600, Shimadzu). The zeta potential of the AuNPs was determined through dynamic light scattering (DLS) analysis with a Zetasizer (Malvern). The samples were dispersed in ultrapure water (pH = 7.2) under probe sonication for 1 min before DLS analysis.
In-gel proteolytic digestion, LC-MS/MS label-free quantitation, analysis, and protein informatics
The isolated hard corona was resuspended in 2× reducing loading dye (Bio-Rad), incubated at 95 °C for 5 min and spun down. The supernatant (100 μL) was resolved on an SDS-PAGE gel (Mini-PROTEAN TGX, Bio-Rad), stained using Instant Blue Stain (Expedeon) and destained using Milli-Q water. The resolved region of the gel was cut and subjected to an in-gel trypsin digestion procedure, as described previously.40 The extracted peptides were dried and resuspended in 20 μL of 2% acetonitrile and 0.1% formic acid and stored at −20 °C until analysis. Liquid chromatography with tandem mass spectrometry (LC-MS/MS) was carried out as described previously,41 with minor modifications. Briefly, LC-MS/MS was performed using a Q Exactive™ HF Hybrid Quadrupole-Orbitrap mass spectrometer. Samples were loaded at a flow rate of 15 μL/min onto a reverse-phase trap column (75 μm × 2 cm) Acclaim PepMap media (Dionex) in 2% acetonitrile (Sigma Aldrich) and 0.1% trifluoroacetic acid (Sigma Aldrich). Peptides were then eluted from the trap column at a flow rate of 0.25 μL/min through a reverse-phase capillary column (75 μm × 50 cm) (LC Packings, Dionex). The HPLC gradient was set to 128 min using a gradient that reached 30% of ACN after 93 min, then 34% of ACN after 96 min, 79.2% of ACN after 101 min, and 2% after 108 min for a further 20 min. The mass spectrometer was operated in data-dependent mode with 2 microscan Fourier transform mass spectrometry scan events at 60,000 resolution (MS) over the m/z range of 375~1575 Da in the positive-ion mode, and up to 30 data-dependent higher energy collision dissociated MS/MS scans. Peptide sequences (and protein identity) were determined using MaxQuant software (version 1.6.0.1) by matching the human protein database (Homo sapiens, UniProt-proteome_UP000005640.fasta) and label-free quantification of identified proteins was then performed as previously described.40
Experiments were done in triplicate and proteins detected in at least two replicates (intensity >0) were used for further analyses. The total intensity was used to approximate the relative protein abundance between the different types of AuNPs and protein abundance in each type individually. A student’s t-test was used to evaluate the significance of differences observed across the three independent replicates of spiky, mid-spiky and spherical AuNPs and P-values < 0.05 were considered. MetaboAnalyst was used to normalize the coronal protein intensity data into the log transformed auto scaled intensities and further used for univariate and multivariate analyses, including hierarchical clustering using heatmaps, PCA 2D plots, partial least squares discriminant analysis (PLS-DA) plots, and volcano plots to derive the fold change variations between the AuNP series.
STRING (v10.5, string-DB.org/) was employed to analyze, classify, and visualize the network of biological processes related to the identified coronal proteins.42 The string annotations and interactions were imported to open-source software Cytoscape to get biomolecular interaction networks integrated with the fold change data obtained from MetaboAnalyst volcano plots.43
HUVEC cell culture, confocal microscopy, transwell, ROS and viability assays
Human umbilical vein endothelial cells (HUVEC, Sigma Aldrich) were cultured in EGM™ Endothelial Cell Growth Medium (Lonza), supplemented with EGM™ SingleQuots™ Supplement Pack (Lonza). Glass cover slides were placed in 24 well plates (Corning Costar), 50×104 cells were seeded to each well and let grow for 5-6 days to reach ~100 % confluency to form an intact monolayer. The spent media were removed and the cells were treated for 30 min with AuNPs dispersed in 200 μL of media with a final concentration of 100 μM. After the treatment, the media were removed and the cells were washed twice with Hanks’ Balanced Salt Solution (HBSS, Sigma Aldrich). The cells were fixed in 4% paraformaldehyde (PFA, Sigma Aldrich) for 15 min and rinsed off with 500 μL of wash buffer (0.1% azide, Sigma Aldrich) in 1× PBS. The cells were permeabilized and blocked using 200 μL blocking buffer containing 0.1% saponin (Sigma Aldrich) and 5% horse serum (Sigma Aldrich) for 60 min at room temperature and washed twice with wash buffer. The cells were incubated with primary antibody Rabbit anti-VE-cadherin (Abcam; 1:400 in wash buffer containing 5% horse serum) overnight at 4 °C. The primary antibody solution was washed off using wash buffer and the cells were incubated with a secondary antibody solution (Donkey anti-rabbit Alexa Fluor 647/594 (Abcam) in wash buffer containing 5% phalloidin (Sigma Aldrich) for 2 h at room temperature. The cells were then washed twice with wash buffer to remove secondary and Hoechst 33342/DAPI (Thermo Fisher Scientific) in wash buffer was added to cells. The cells were then washed with wash buffer and excess liquid was dried. A cover-glass was placed on top of the glass slide and mounted with ProLong Gold (Thermo Fisher Scientific). All the slides were imaged using a confocal fluorescence microscope (SP8 LIGHTNING, Leica Microsystems). Semiquantitative image analysis was done using ImageJ39 where intercellular gaps in immunofluorescence images were counted and gap areas were measured and normalized to total image areas.
In vitro transwell assay:
HUVEC cells were seeded (50,000 cells) on transwell inserts (polycarbonate membrane, 0.4 μm pore diameter; Corning Costar, USA) in a 24-well plate until the formation of a monolayer (4 days). The cells were treated with different AuNPs and their respective protein coronae for 1 h. After the treatment, all groups were incubated with FITC—dextran (0.1 mg/mL, 40 kDa; Sigma Aldrich) for 20 min. Subsequently, culture media from the bottom compartment of each well were collected and the fluorescence reading for FITC-dextran was measured by a microplate reader (PerkinElmer EnSight HH33400 plate reader) at wavelengths of 490/520 nm (excitation/emission).
Viability assay:
A 96 well plate (Costar black/clear bottom) was preincubated with 70 μL Poly-L-lysine (Sigma, 0.01%), at 37 °C for 30 min. After removing Poly-L-lysine, the wells were washed thrice using PBS and HUVEC were cultured at a density of ~ 50,000 cells per well and incubated at 37 °C and with 5% CO2 for 48 h to reach ~ 80% of confluency. The cell culture medium was then changed to fresh media containing 1 × 10 −6 M propidium iodide (PI) dye and incubated for 30 min. Subsequently the cells were treated with different AuNPs with and without their coronae and their PI fluorescence was measured using Operetta (PerkinElmer, 20× PlanApo microscope objective, numerical aperture NA = 0.7) in a live cell chamber (37 °C, 5% CO2) for 15 h of treatment. The percentage of dead cells labelled by PI-positive to total cell count was determined by a built-in bright-field mapping function of Harmony High-Content Imaging and Analysis software (PerkinElmer).
Reactive oxygen species (ROS) assay:
HUVEC cells (50,000 cells/well) were seeded into a 96-well black plate and cultured overnight to reach 80% confluency. ROS detection was performed using an OxiSelectTM intracellular ROS detection kit. The cells were stained with H2DCFDA (20 μg/mL) for 30 min and subsequently treated by the AuNPs samples. ROS levels were then measured indirectly by the oxidation of non-fluorescent DCFDA to fluorescent DCF on a fluorescence microplate reader (PerkinElmer EnSight HH33400), at wavelengths of 488/535 nm (excitation/emission). Untreated cells were used as negative control and H2O2 (100 μM) as positive control.
Endothelial signalling pathway assay
HUVECs were cultured in 6-cm cell culture dishes and exposed to solutions containing the three types of AuNPs and their respective protein coronae. After incubation for 1 h, the cells were washed thrice with cold PBS and collected after cell lysis in SDS-PAGE sample buffer which was supplemented with a mixture of 1% protease and phosphatase inhibitors. The collected samples were subjected to gel electrophoresis using 10% resolving polyacrylamide gels (Mini Protean, Bio-Rad, USA) and were transferred onto nitrocellulose membranes. The membranes were then blocked with 5% BSA solution for 1 h, before incubation in a solution of a relevant primary antibody at 4 °C overnight. The membranes were washed and incubated in corresponding horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h. Protein bands on the membranes were visualized and captured in a chemiluminescence imaging setup. The images of protein bands were analyzed in a semi-quantitative manner through ImageJ software.
Ex vivo transwell assay
An ex vivo vascular leakiness assay was performed using swine vessels as a transwell insert. Swine vessels were obtained from a local slaughterhouse in Chongqing, China. Specifically, the original membranes from a commercial transwell chamber were replaced with the blood vessels from the coronary artery which were cut into individual transverse membranes. AuNPs and their corona counterparts were added to the swine vessel transwell device and incubated for 6 h at 37 °C. After the treatment, the AuNPs-containing media were discarded and replaced with media containing Evans blue dye (100 mM) to each well, and further incubated for 1 h. The media from the lower chamber was collected and fluorescence signals were quantified at 624/680nm (excitation/emission) with a microplate reader. Negative controls were used for normalization of the fluorescence readings.
Statistical analysis.
Data are represented as means (n=3) ± standard errors of the mean (SEM). Statistical analysis was performed via one-way ANOVA. A p-value < 0.05 was considered statistically significant.
Results and Discussion
Characterization of AuNPs and their protein coronae
Spiky AuNPs (AuNP1) synthesized from 13 nm Au seeds were imaged for their primary size and roughness with transmission electron microscopy (TEM) (Figure 1a). AuNP1 underwent dynamic changes in its primary shape and surface roughness over time and evolved into spherical morphologies in ~24 h (Figure 1a). Mid-spiky AuNPs (AuNP2) were observed as an intermediate state during the transition of spiky AuNP1 to spherical AuNPs (AuNP3), and a primary size of 30 nm was confirmed for all three morphological states (Figure 1b). UV-Vis absorption spectra displayed distinct plasmon resonance peaks for the AuNPs of three morphologies (Figure 1c). The largest peak wavelength (i.e., 658 nm) for AuNPs at time zero, as compared to the AuNPs at 6 h (573 nm) and especially at 24 h (522 nm), could be due to the most irregular and least efficient morphology of the AuNPs at time zero for exciting surface plasmon resonance.
Figure 1.
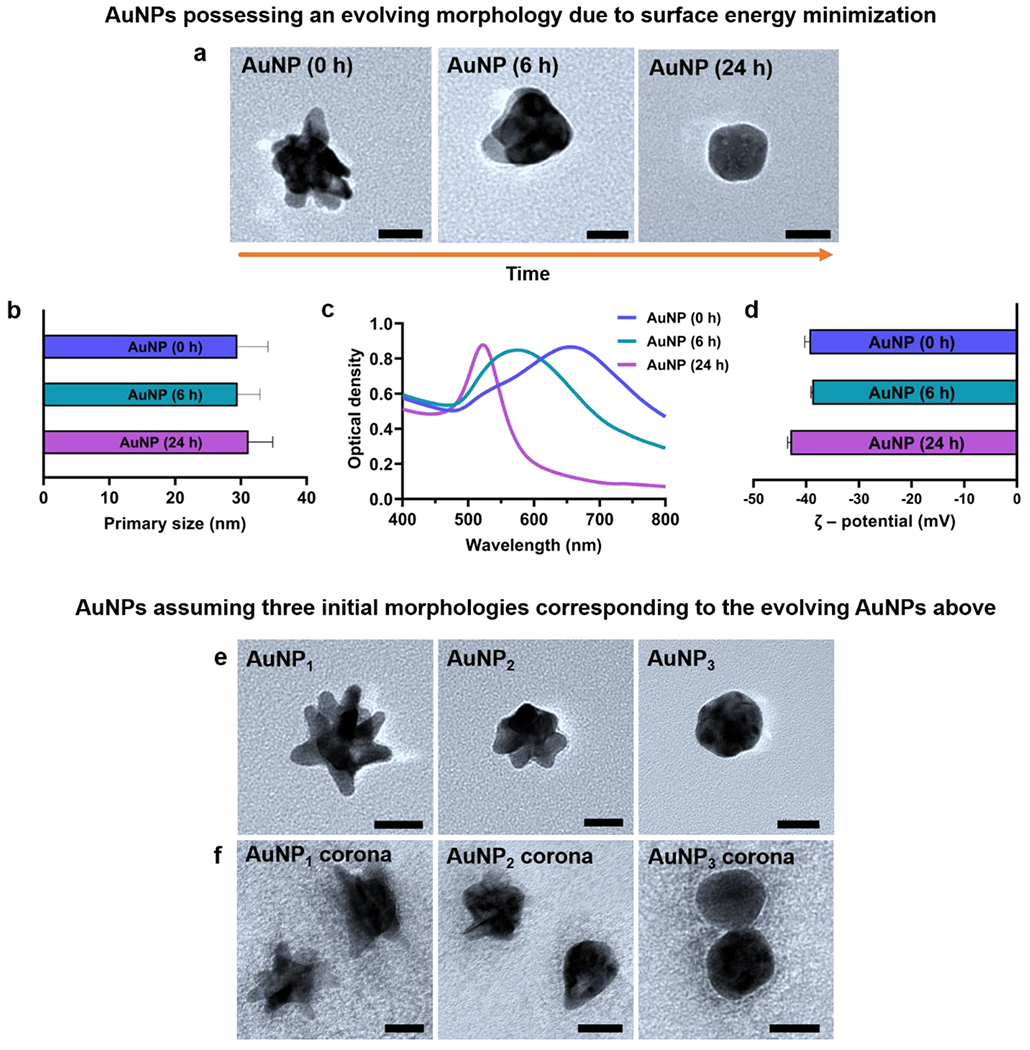
Characterizations of AuNPs possessing a changing morphology (a-d) and AuNPs of three initial given morphologies with and without their human plasma coronae (e, f). a) TEM imaging revealed the AuNPs evolving from spiky AuNPs (AuNP1) to mid-spiky AuNPs (AuNP2) and spherical AuNPs (AuNP3) over 24 h. b) Primary size distributions of the AuNPs. c) UV-Vis absorption spectra for the three types of AuNPs. d) Zeta potentials of the three types of AuNPs in 10 mM HEPES buffer. TEM imaging confirmed the AuNPs assuming the morphologies comparable to e) AuNP1, AuNP2, AuNP3 and their respective protein coronae f) AuNP1 corona, AuNP2 corona, and AuNP3 corona (All scale bars: 20 nm).
To ensure repeatability and practicality (due to the extended time required for the LC-MS/MS procedures), we employed AuNPs of three given initial morphologies (Figure 1e) to infer the corona formation for AuNPs of a changing morphology (Figure 1a). AuNPs with morphologies comparable to AuNP2 and AuNP3 were synthesized with slight modifications to buffer concentrations, and their primary size and shape were again confirmed by TEM (Figure 1e). The three types of AuNPs were then incubated with human plasma at physiological conditions (pH 7.5, 37 °C) with constant shaking for 24 h and their hard NP-protein coronae were collected by centrifugal separation followed by multiple washes. For AuNP3, we observed a small increase in aggregation after 24 h of incubation (Figure 1f). TEM analysis using ImageJ indicated increases in primary size and DLS measurements showed decreases in zeta potential for the AuNPs, most evidently for AuNP3, understandably resulting from their protein corona acquisition (Table 1). The total amounts of proteins associated with the hard coronae of the three types of AuNPs were quantified by the bicinchoninic acid (BCA) assay (Figure S1). Consistently with Table 1, the total amounts of hard coronal proteins associated with AuNP1 and AuNP2 were not markedly affected by their morphology changes, but a significant increase occurred in the total amount of coronal proteins on the surface of spherical AuNP3 (Figure S1). This may be due to the larger available surface area and less steric hindrance of AuNP3 for protein binding in comparison with that of AuNP1 or AuNP2.
Table 1.
Physiochemical characteristics of AuNPs-protein coronae.
| AuNPs | Peak absorbance (nm) | Primary size (nm) | ζ – potential (mV) | ||
|---|---|---|---|---|---|
| Bare NPs | NP-protein corona | Bare NPs | NP-protein corona | ||
| AuNP1 | 658 | 31.4±4.5 | 31.6±5.6 | −39.40±0.88 | −12.47±0.28 |
| AuNP2 | 573 | 30.7±2.8 | 33.6±2.8 | −38.86±0.20 | −11.70±0.70 |
| AuNP3 | 522 | 30.2±1.5 | 39.3±3.6 | −43.00±0.52 | −9.78±0.27 |
Qualitative and quantitative evolution of the AuNP-protein coronae
The compositions and abundance levels of the hard protein coronae around different AuNPs isolated from centrifugal spin were characterized qualitatively and quantitatively using label-free shotgun LC-MS/MS with triplicate. A student’s t-test was done to test the statistical significance of differences across independent triplicate. For the abundance ratio calculation and analysis, only those proteins with a P-value of <0.05 were considered significant and included in further enrichment analysis. To identify the differences in coronal abundance among the different types of AuNPs, the mean fold difference for the MS/MS intensities was calculated by taking the average of the three independent quantitative experiments and by keeping the fold change difference thresholds as 0.5 and 1.5 for comparing relative abundance between the coronal proteins formed on different AuNPs.
For the coronae of the AuNPs, 338 distinct serum proteins were identified by LC-MS/MS, of which 275 were present in all groups, 13 unique proteins each for AuNP1 and AuNP2 and 5 unique proteins for AuNP3 were identified (Figure 2a&Supplementary Excel file). Overall, the types of coronal proteins were not largely different across the three types of NPs, but the amounts of the coronal proteins did significantly differ. The top-10 proteins for each type of AuNPs are enlisted in Table 2. The most abundant proteins for different AuNPs appeared similar in name, while the order and relative abundance of top-10 proteins across the AuNP species largely varied (Figure 2c). Specifically, serum albumin was the most abundant in the protein coronae of AuNP2 and AuNP3, whereas apolipoprotein B-100 was the top coronal protein for AuNP1 and was 2.4 times more abundant for AuNP1 than for AuNP3. Immunoglobulin heavy constant mu, immunoglobulin heavy constant gamma 1, complement C4-B, complement C3, and C4b-binding protein alpha chain were the other top proteins found across all samples.
Figure 2.
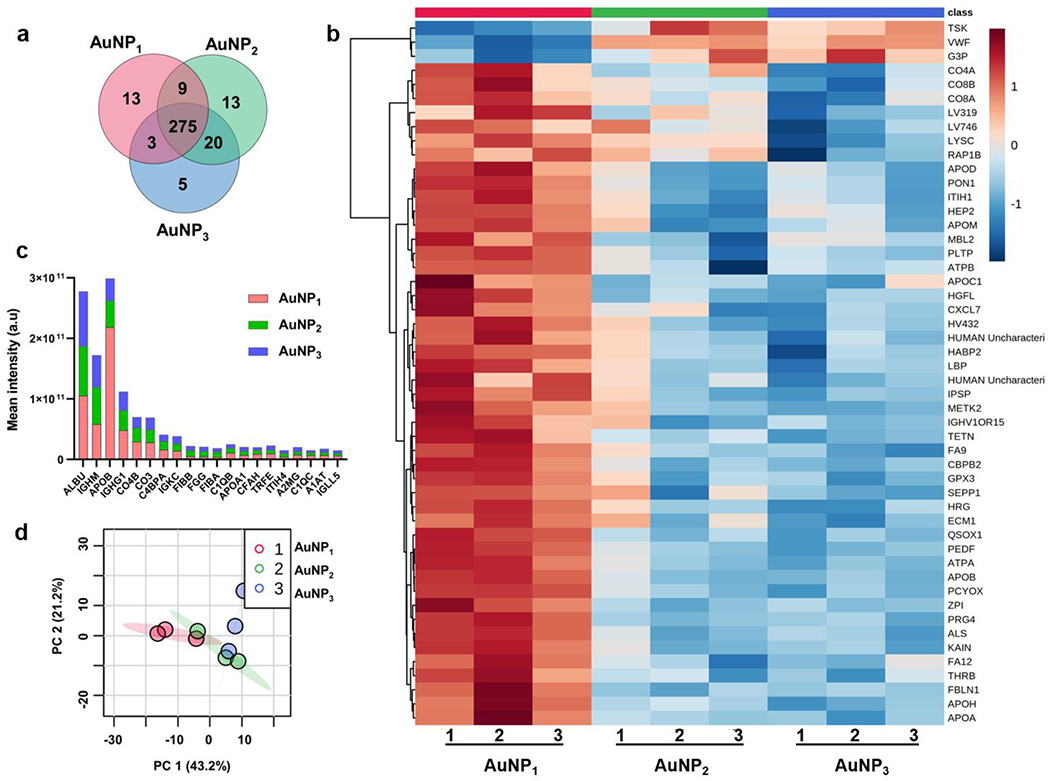
Proteomic analysis of the plasma coronal proteins acquired by the three types of AuNPs. a) Venn diagram illustrating the distribution of coronal proteins associated with different AuNPs. b) Hierarchical clustering of the three types of AuNPs; AuNP1 (red), AuNP2 (green), and AuNP3 (blue). Vertical clustering displays similarities between sample groups, while horizontal clusters reveal the relative abundances of the 50 most significant different proteins ranked by p-values. White indicates no change, while red and blue indicate increased and decreased abundances, respectively. Ward’s minimum variance method algorithm was used to generate the hierarchical cluster analysis. c) Total mean intensity comparison of top proteins contributing more than 1% of the total protein abundance. d) Principal component analysis (PCA) score plot for different AuNPs showing variations in the AuNP groups. The data is from at least three independent biological replicates.
Table 2.
Top-10 proteins identified in the AuNP-protein coronae.
| AuNP1 | AuNP2 | AuNP3 |
|---|---|---|
| Apolipoprotein B-100 | Serum albumin | Serum albumin |
| Serum albumin | Immunoglobulin heavy constant mu | Immunoglobulin heavy constant mu |
| Immunoglobulin heavy constant mu | Apolipoprotein B-100 | Apolipoprotein B-100 |
| HUMAN Immunoglobulin heavy constant gamma 1 | Immunoglobulin heavy constant gamma 1 | Immunoglobulin heavy constant gamma 1 |
| Complement C4-B | Complement C4-B | Complement C3 |
| Complement C3 | Complement C3 | Complement C4-B |
| C4b-bindinq protein alpha chain | C4b-binding protein alpha chain | Immunoglobulin kappa constant |
| Immunoglobulin kappa constant | Immunoglobulin kappa constant | C4b-binding protein alpha chain |
| Complement C1q subcomponent subunit B | Fibrinogen beta chain | Alpha-2-macroglobulin |
| Serotransferrin | Fibrinogen gamma chain | Fibrinogen beta chain |
The common proteins comprising the bulk of the AuNP-corona (top-50 significantly different proteins ranked by p-values from one-way ANOVA) were summarized in the heatmap with hierarchical clustering showing log transformed normalized protein intensities based on their abundance on the NP surfaces (Figures 2b&S2). There were distinct variations in the abundance of the coronal proteins across the AuNP species, where a large cluster of proteins had higher relative intensities for spiky AuNP1 and decreased as the morphology changed to spherical AuNP3. The unique coronal proteins (Table 3) provided fingerprints specific to the morphologies of the AuNPs. PCA was performed in MetaboAnalyst to determine significant variations among the AuNPs using the MS/MS label-free quantification (LFQ) intensity as the principal component. Visual analysis based on the first two principal components showed a large qualitative difference (43.2% variance in the principal component 1) in the proteins adsorbed onto AuNP1 than AuNP3. In contrast, the AuNP2-corona shared some similarities with both the AuNP1-corona and AuNP3-corona (Figure 2d).
Table 3.
Unique proteins identified in the AuNP-protein coronae.
| AUNP1 | AUNP2 | AUNP3 |
|---|---|---|
| Immunoglobulin kappa variable 2D-29 | Immunoglobulin lambda variable 1-44 | Transmembrane protein 131 |
| Coagulation factor VII | Immunoglobulin heavy variable 3-48 | Vitamin K-dependent protein Z |
| Immunoglobulin heavy variable 1-8 | Matrix Gla protein | Trypsin-3 |
| Immunoglobulin lambda-like polypeptide 1 | Collagen alpha-3(VI) | Thrombospondin-4 |
| Insulin-like growth factor-binding protein 3 | Junction plakoglobin | Immunoglobulin kappa variable 3-7 |
| Insulin-like growth factor-binding protein 6 | Erythrocyte band 7 integral membrane protein | |
| Insulin-like growth factor-binding protein 5 | Transgelin-2 | |
| Structural maintenance of chromosomes protein 1B | Myosin light polypeptide 6 | |
| Titin | Ras-related protein Rab-10 | |
| Collectin-11 | Desmocollin-1 | |
| Golgi-associated plant pathogenesis-related protein 1 | Microtubule-associated protein RP/EB family member 2 | |
| Fetuin-B | Retinoic acid receptor responder protein 2 | |
| Immunoglobulin heavy variable 3/OR16-13 | IgGFc-binding protein |
Coronal proteins with significant changes between AuNP1, AuNP2, and AuNP3 can be identified from their corresponding Variable Importance of Projection (VIP) score ranks from multivariate PLS-DA analysis for component 1 (normalized intensities) (Figure 3a,b&Supplementary Excel file). The quantitative abundance in the coronal protein association with AuNP1 and AuNP3 was analyzed using a volcano plot with a fold change threshold of 1.5 to distinguish the major outliners by comparing the size of the fold change to the statistical significance level with a P-value threshold of 0.05 (Figure 3c&Supplementary Excel file). The horizontal axis plotted the fold change between the two groups (on a log scale), while the vertical axis represented the P-value for a t-test (one-way ANOVA) of differences between samples (on a negative log scale). The resulting volcano plot for the fold change differences between coronal protein abundance found 88 proteins to increasingly bind AuNP1 and 4 proteins to be increasingly associated with AuNP3 within the given threshold. The top enriched proteins on AuNP1 included Ras-related protein Rap-1b, prenylcysteine oxidase 1, selenoprotein P, immunoglobulin lambda variable 3-19, cholesteryl ester transfer protein, apolipoprotein(b), sulfhydryl oxidase 1, phospholipid transfer protein, and coagulation factor IX, whereas the top coronal proteins enriched in AuNP3 were von Willebrand factor, glyceraldehyde-3-phosphate dehydrogenase, Tsukushin, and C-reactive protein. The fold change variations in the coronal protein association with AuNP1 vs. AuNP2 as well as AuNP2 vs. AuNP3 were also analyzed using a volcano plot (Figure S3&Supplementary Excel file). The fold change analysis found 50 proteins to increasingly bind AuNP1 and 11 proteins to be increasingly associated with AuNP2 within the given threshold. Comparing the AuNP2 and AuNP3 fold change values, only 5 coronal proteins were bound more to AuNP2. Largely, the common coronal proteins had very similar relative abundances for both AuNP2 and AuNP3. These results clearly showed that the protein corona evolved according to the changing morphology of the AuNPs.
Figure 3.
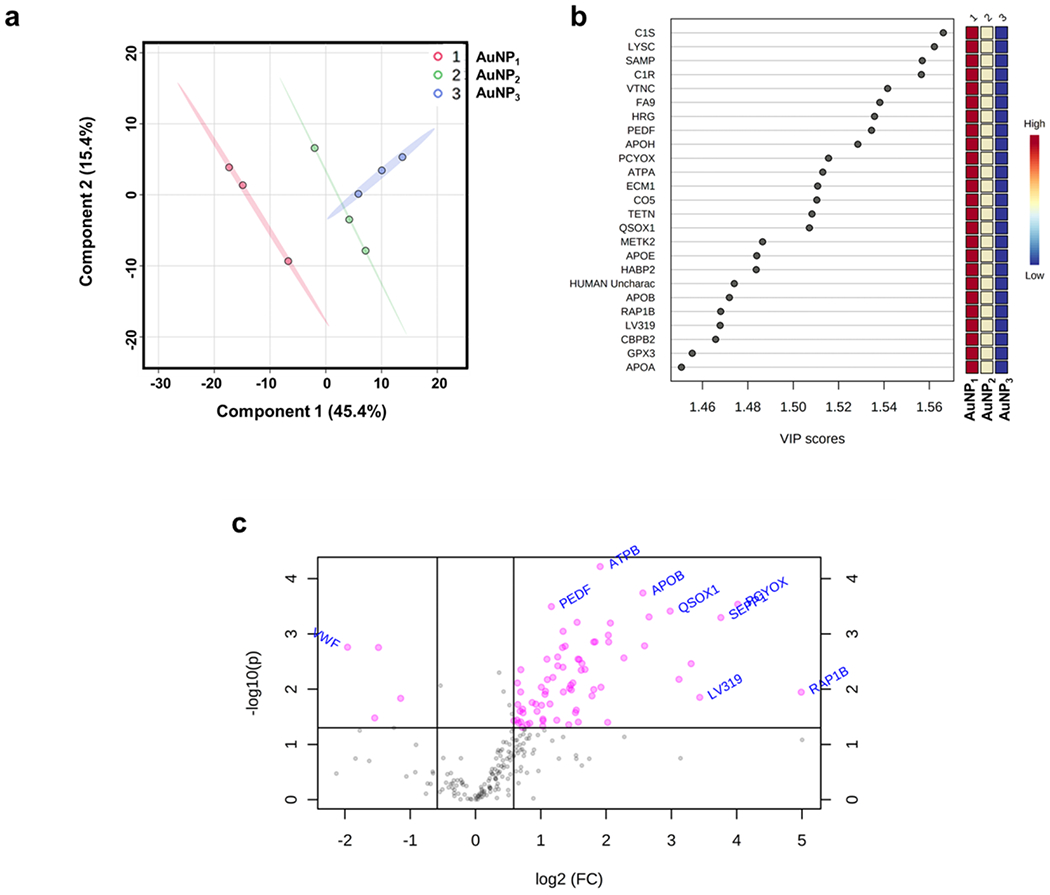
a) Partial least squares-discriminant analyses (PLS-DA) PC1/PC2 score plots revealing a clear variation in coronal protein intensities between AuNP1 and AuNP3. b) Variable Importance of Projection (VIP) scores for the top-25 significantly enriched coronal proteins for the AuNP series (1-AuNP1, 2-AuNP2 and 3-AuNP3) for component 1. The VIP score plots show the differences in the mean intensity of coronal proteins between different AuNP samples as shown by the color chart on the right indicating high variations between AuNP1, AuNP2 and AuNP3 mean intensities. The values of the VIP plot indicate significance of the proteins for the PLS-DA models based on relative abundance. c) Volcano plot for the fold-change variation (fold change threshold 1.5) of the most varied coronal proteins among the AuNP1 and AuNP3 samples indicating the most significantly enriched proteins.
There was no evident relationship observed for the coronal proteins of the three types of AuNPs in terms of their enrichment with grand average of hydropathy (GRAVY) index, molecular weight, or isoelectric point (pI) (Figure S4&Supplementary Excel file). Generally, proteins with a GRAVY score less than 0 are hydrophilic in nature and more likely to possess a globular structure, while a GRAVY score above 0 indicates hydrophobicity. The isoelectric point (pI) refers to the pH value at which a protein is electrically neutral and least soluble. Here, both sets of total proteins for AuNP1 and AuNP3 shared similar GRAVY indices (i.e., hydrophobicity) and pI (i.e., charge) distributions indicating that majority of coronal proteins associated with the AuNPs tended to entail GRAVY values ranging from −1.03 to 0.67, including a few extreme values for Proteoglycan 4 (−1.03), Cell division cycle 5-like protein (−0.96), Histidine-rich glycoprotein (−0.9567) and Tetraspanin (0.675). The pI ranged from 4.3 to 11.36 and assumed a bimodal distribution, with one centered at ~pI 5-6 and another within pI 8-9 due to the presence of large numbers of immunoglobulin side chains. The proteins with the outmost pI values included Platelet glycoprotein Ib beta chain (9.67), Immunoglobulin heavy variable 4-30-2 (9.7), Angiogenin (9.73), and Histone H4 (11.36).
To derive the relative composition differences of the protein coronae pertaining to the AuNPs and to understand their biological implications, a protein network analysis was performed using the STRING resource (string-db.org) for all identified coronal proteins across the AuNP series (Figure S5). A total of 173 out of the 275 coronal proteins were identified in the StringDB (excluding immunoglobulin side chains) with 27 KEGG pathways significantly enriched (Supplementary Excel file), including complement and coagulation pathway, focal adhesion, platelet activation, cholesterol metabolism, and phagosome. For the differentially bound coronal proteins between AuNP1 and AuNP3, the network analysis assigned 4 major protein groups, based on k-mean clustering nodes derived from the protein-protein interaction records obtained from cellular/biological Gene Ontology (Figure 4&Supplementary Excel file). The most significantly associated proteins identified in the coronal formation were mainly involved in blood coagulation intrinsic pathways, opsonization, complement and coagulation cascade, and cholesterol metabolism pathways. The combined interaction score of protein-protein interaction was imported from StringDB to Cytoscape and the interaction fold change network was created (Figure S6&Supplementary Excel file) indicating a relative enrichment of apolipoproteins including Apolipoprotein C1 (APOC1), Apolipoprotein B (APOB), Cholesteryl ester transfer protein (CETP), Apolipoprotein A (LPA), Apolipoprotein M (APOM) and others for spiky AuNP1 along the protein network. Apolipoproteins, especially that of Apolipoproteins J and A1,44 have been featured as prominent binders for stealth NPs in the protein corona literature, while Apolipoprotein E has been associated with enhanced cellular uptake of NPs.45, 46 Although there were no large variations among the number of immunoglobulin side chains identified across different AuNPs, there was a significantly greater amount of immunoglobulin bound to AuNP1 and AuNP2 than AuNP3 (Figure S7&Supplementary Excel file). It is known that the enrichment of immunoglobulins and complement proteins can result in higher opsonization and immune recognition by macrophages, which in turn can affect the retention, transport, and clearance of nanoparticles.21, 47, 48 This suggests a differential recognition pattern of the AuNPs by the immune system as well as a gradual release of immunoglobulin would occur as AuNPs evolve from spiky to spherical in morphology.
Figure 4.
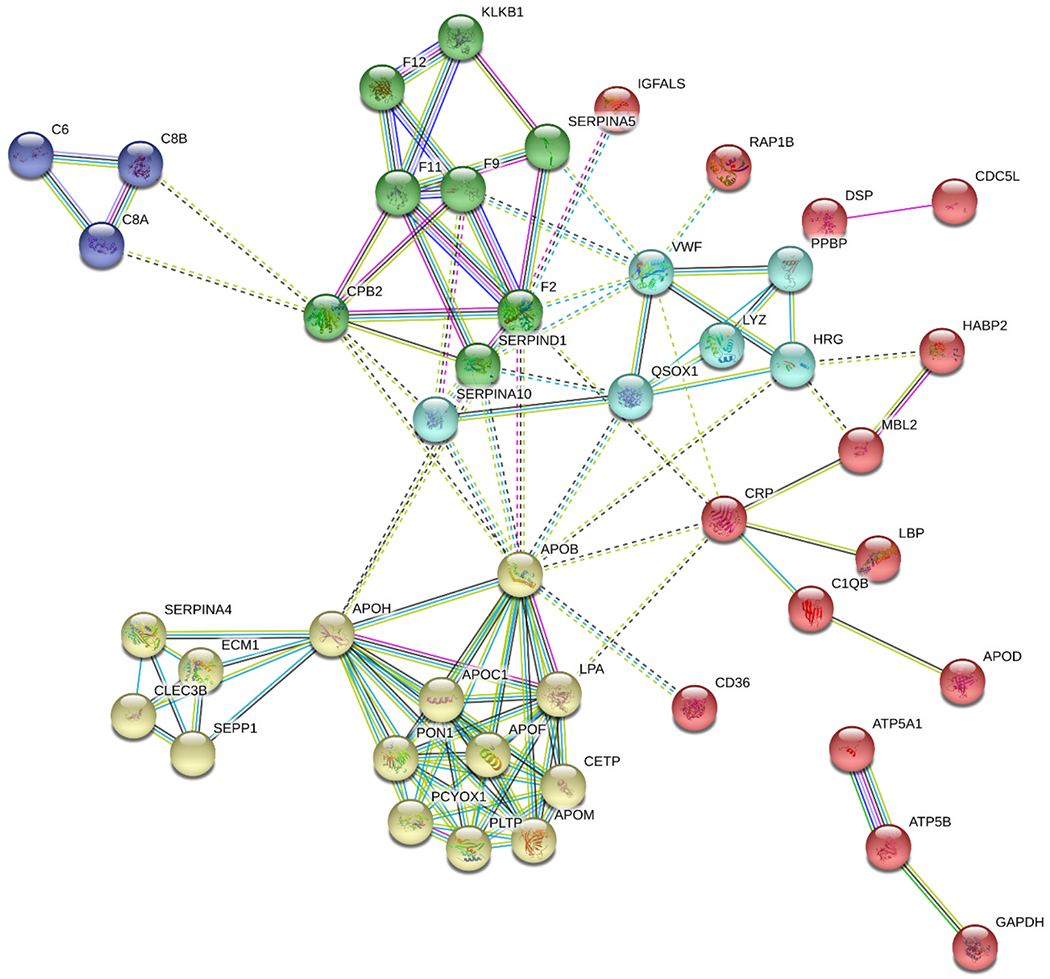
Protein interaction network for the most significantly bound coronal proteins differentially associated with AuNP1 & AuNP3 (proteins with a fold change cut-off below 0.5 and above 1.5, and with a P-value < 0.05). The most significantly associated proteins identified in the corona formation were mainly involved in blood coagulation intrinsic pathways, opsonization, complement and coagulation cascades, and cholesterol metabolism pathways.
Dependence of AuNPs-induced endothelial leakiness on their protein coronae
Towards nanomedicinal applications of the dynamic protein corona, HUVEC cell monolayers were treated with different AuNPs of 30 nm (100 μM of Au) and AuNP-coronae, where the occurrence of NanoEL was detected after 30 min of treatment. Specifically, paracellular gaps of ~10 μm were evident upon exposure to the AuNPs with and without their corona derivatives, whereas the control cells retained their confluent monolayers (Figure 5a). Semi-quantitative analysis (gap area%) of the acquired confocal fluorescence images was carried out using trainable Weka segmentation plugin in ImageJ software. The analysis revealed that NanoEL was dependent on the surface morphology of the AuNPs as well as their protein coronae (Figure 5b). Specifically, the morphologically most rugged AuNP1 induced a maximum extent of endothelial leakiness, closely followed by AuNP2 and then by the morphologically smoothest AuNP3. The extent of endothelial leakiness induced by AuNP3 was notably less than for the other two morphologies, indicating a minimal effect of AuNP3 on the integrity of the HUVEC endothelial monolayer. Semi-quantitative image analysis indicated marked reductions in the extent of NanoEL due to the protein corona formation in AuNP1 and AuNP2, where the gap area percentage decreased by 47% (1.9 ± 0.7% to 1.0 ± 0.3%) for AuNP1 and by 53% (from 1.9 ± 0.6% to 0.9 ± 0.3%) for AuNP2, respectively.
Figure 5.
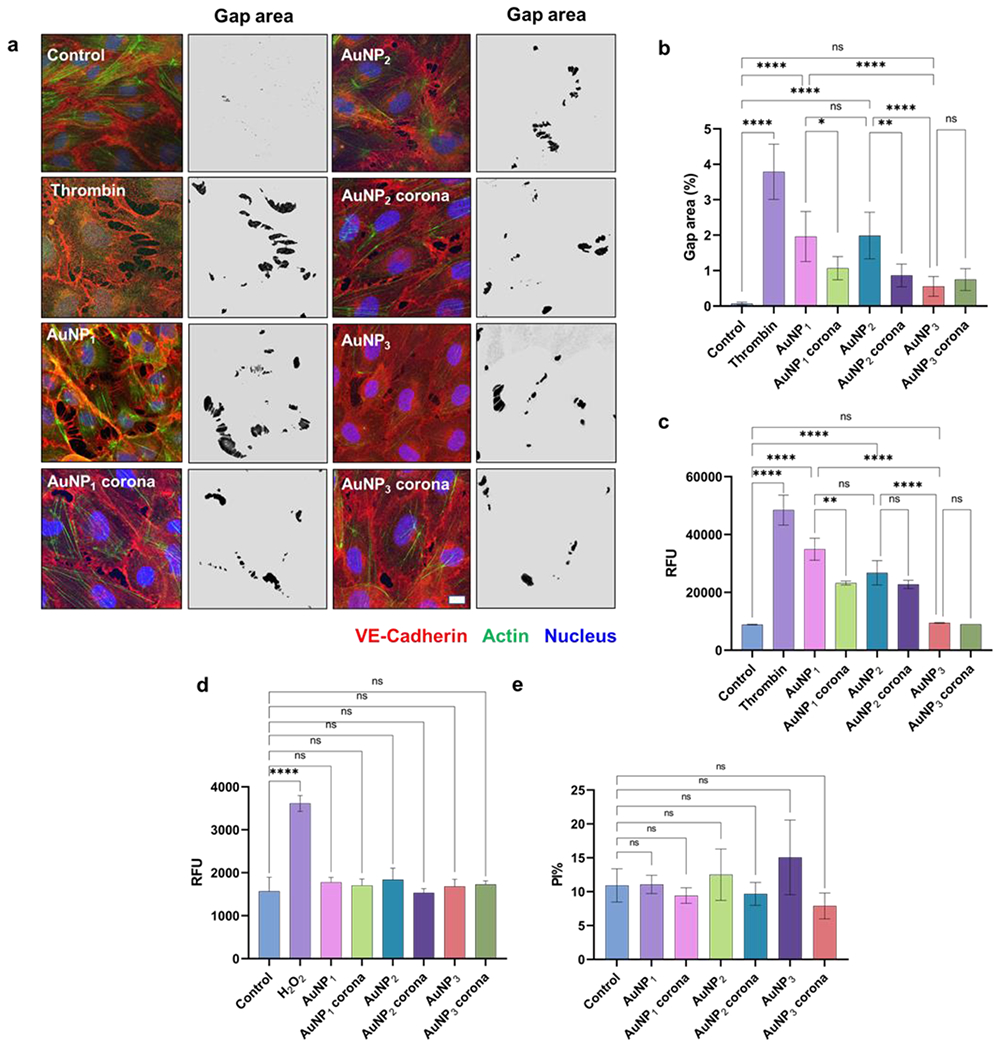
AuNP-induced endothelial leakage and influence of their protein corona on HUVEC cell monolayers. a) Confocal imaging and gap area analysis of cell control, thrombin, AuNP1-3, and AuNP1-3 coronae. b) Semi-quantitative image analysis of gap area% (shown in black in the images) highlighted on a grey background derived from the trainable Weka segmentation plugin in ImageJ software. HUVEC cells exposed to the AuNPs and their coronae evaluated by c) a transwell assay, d) ROS production, and e) a viability assay. Scale bars: 10 μm. *P < 0.05, **P < 0.001, ***P < 0.0005 and ****P < 0.0001.
A transwell assay was performed to further quantify the AuNPs-induced leakiness in HUVEC monolayers, where AuNP1 caused the maximum extent and AuNP3 the least extent of endothelial leakage (Figure 5c) corroborating the observations by confocal fluorescence microscopy (Figure 5a&b). While a significant decrease in the relative fluorescence (indicating endothelial leakiness) was observed for AuNP1 corona compared to bare AuNP1, little variations were seen in paracellular leakiness due to the formation of a protein corona for both AuNP2 and AuNP3. Furthermore, there was no significant increase in either ROS production or cytotoxicity in the HUVEC cells exposed to the AuNPs and their respective protein coronae (Figure 5d&e), indicating a non-cytotoxic nature of the NanoEL phenomenon associated with AuNPs.
The role of phosphorylation of vascular endothelial (VE)-cadherin tyrosine residues in promoting its internalization and resulting in higher vascular permeability has been established in recent literature.49, 50 Here, molecular regulation of AuNPs-induced NanoEL through the activation of VE-cadherin signalling pathways was analyzed by western blotting of treated HUVEC cells to identify the phosphorylation of two pivotal residues of VE-cadherin, tyrosine 658 (Y658) and tyrosine 731 (Y731), along with α-tubulin (Figure 6). The amount of phosphorylation is directly related to the degree of the NanoEL effect.51 Specifically, AuNP1 exerted the highest amount of phosphorylation of both Y658 and Y731 residues, whereas AuNP3 induced the least amount of phosphorylation of both residues. In comparison, the protein corona formation on the spiky AuNP1 markedly alleviated the amount of phosphorylation of both resides, whereas AuNP2 corona formation gave rise to a significant reduction in the phosphorylation of only the Y731 reside. In comparison, there were no major variations in the phosphorylation of the residues for the cells exposed to AuNP3 with or without their corona. Furthermore, there were no significant differences in the expression of α-tubulin among the groups indicating minimal cytoskeletal reorganization associated with the AuNPs.
Figure 6.
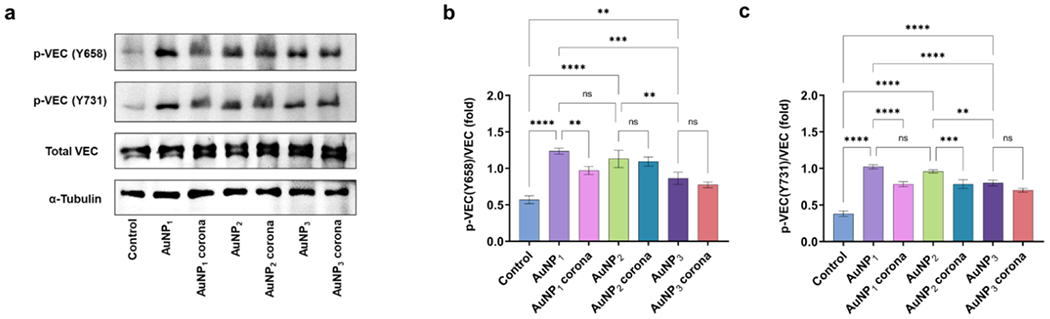
Western blot and corresponding semi-quantitative analysis of activation of VE-cadherin (VEC) signalling pathways for NanoEL induced by different AuNPs in HUVEC cells. An increased phosphorylation of VE-cadherin at tyrosine 658 and 731 sites was observed for AuNP1 which significantly reduced due the protein corona formation. Data are mean ± SD, biologically independent samples (n=3), analyzed via one-way ANOVA, * represents P < 0.01 and **,## represents P < 0.001, between the compared groups.
In addition to the in vitro assays described above, an ex vivo assay was further performed using swine vessels to gauge the NanoEL effect on vascular endothelium (Figure 7). Specifically, AuNP1 and AuNP2 elicited a comparable level of endothelial leakiness as indicated by the Evans blue dye (EBD) penetration, whereas AuNP3 induced the smallest extent of NanoEL than AuNP1 and AuNP2. The degree of vessel leakiness was significantly reduced due to the corona formation on AuNP1 and AuNP2. A 5-fold reduction in EBD fluorescence was observed for spiky AuNP1 and mid-spiky AuNP2 in the presence of their protein coronae, whereas no reduction in EBD fluorescence was detected for spherical AuNP3 with or without a protein corona. These results corroborated the in vitro observations for HUVEC cells (Figures 5&6) implicating an attenuating effect of the protein corona on NanoEL, a new finding which may be exploited for controlling the permeability of NPs in the vasculature for future nanomedicine.
Figure 7.
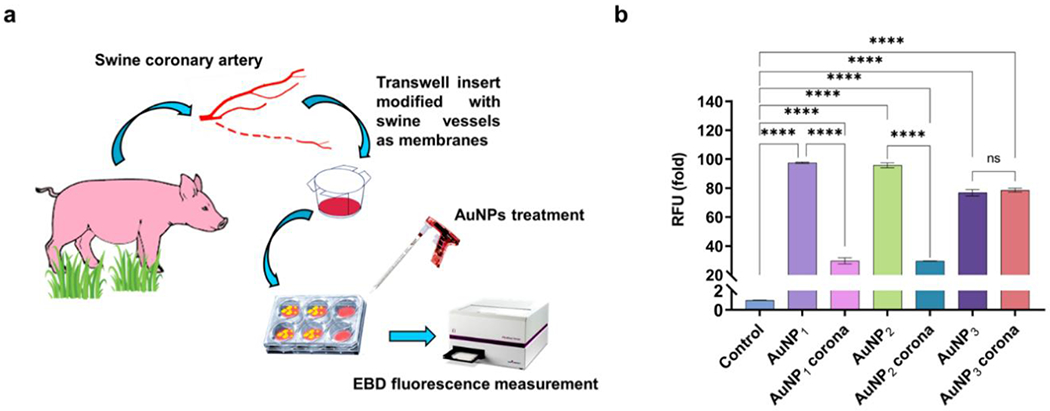
Quantitative measurement of AuNP-induced endothelial leakage in swine vessels. a) A scheme illustrating the ex vivo experimental setup. b) Migration of Evans blue dye (EBD) was used to quantify the effect of coronal formation on endothelial leakiness in swine vessels. The protein corona formation significantly reduced the leakiness irrespective of the AuNP species. A 5-fold reduction in the fluorescence readings were observed for AuNP1 corona and AuNP2 corona when compared to the bare AuNPs. ****P < 0.0001 compared with control.
Conclusion
The protein corona is a thermodynamic phenomenon associated with the transformation of NPs in aqueous environments. For well over a decade, the protein corona paradigm has shaped the field of nanomedicine from bench design to in vitro/in vivo deployment, with its impact further reaching the fields of nanotoxicology and environmental science. While research concerning the protein corona has been largely focused on the physicochemical properties (chemical composition, surface functionalization, charge, and shape) of the NPs, the environmental parameters (pH, temperature, salts), NP-biomolecular interactions (proteins, lipids, nucleic acids, ligands), the fate (biodistribution, opsonization) of the NPs and the response (toxicity, inflammation) of the biological host, few studies have considered a NP whose morphology evolves spontaneously driven by energy minimization that is a common feature to many nanomaterials post synthesis and during their shelf life.
Accordingly, the current study filled this knowledge void by looking into the human plasma protein corona of AuNPs undergoing the physical transitions from spiky to mid-spiky and spherical morphologies. For practicality involving LC-MS/MS, such temporal evolution of the AuNPs was instead mimicked by AuNPs assuming the initial states corresponding to the three morphologies. The proteomics study determined variations both qualitatively and quantitatively on the coronal compositions and abundance for the three types of AuNPs, revealing that each type of the AuNPs possessed biological fingerprints specific to their morphologies. Collectively, our physicochemical and biological network analyses of the coronal proteins implicated a dynamic binding-dissociation process of plasma proteins as the NPs contracted energetically from being rugged (i.e., AuNP1) to being smooth (AuNP3, via AuNP2) to accommodate increased adsorption of proteins which possessed compatible shapes and flexibilities and a more subdued immune sensitivity.
Rational design of NPs has previously enabled the control of NanoEL in different cellular and animal model systems,51 but little is known about the formation and effect of the protein corona on NanoEL. Furthermore, no data is available about the relationship between a dynamic protein corona and NanoEL. Using confocal fluorescence imaging, in vitro and ex vivo transwell assays, as well as molecular signalling pathway assays, we identified an alleviating effect of the protein corona on the extent of NanoEL, via the phosphorylation of VE-cadherin residues Y658 and Y731. While the spiky AuNP1 elicited the most significant leakiness in an HUVEC cell monolayer among the three types of AuNPs, the protein corona of AuNP1 reduced the phosphorylation of Y658 and Y751 by up to 3% and 22%, respectively. In contrast, the corona of the mid-spiky AuNP2 reduced the phosphorylation of Y731 but not Y658, by 21%, and the corona of the spherical AuNP3 exerted no major effect on NanoEL. Together, this study implicated the dynamic protein corona of AuNPs possessing a changing morphology and demonstrated their application for future nanomedicine harnessing the paracellular route.
Supplementary Material
Acknowledgements
This work was supported by Australian Research Council (CE140100036) and National Natural Science Foundation of China (21976145 and 21974110).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. Adsorption and total amounts of proteins bound to the AuNPs by a BCA assay (Figure S1). Hierarchical clustering heatmap for coronal proteins identified across the three AuNP groups (Figure S2). Volcano plot for the fold-change analysis of a) AuNP1 vs. AuNP2 and b) AuNP2 vs. AuNP3 (Figure S3). Coronal protein enrichment represented by GRAVY index, pI, and molecular weight for common proteins identified across the AuNP series (Figure S4). Compiled protein network for all StringDB identified coronal proteins across the three types of AuNPs (Figure S5). Protein interactive network for the differentially associated coronal proteins on AuNP1&AuNP3 (Figure S6). Mean intensity comparison of top-10 abundant immunoglobulin sidechains associated with coronal proteins of the AuNPs (Figure S7).
Competing financial interests: The authors declare no conflicting financial interests.
References
- (1).Walczyk D; Bombelli FB; Monopoli MP; Lynch I; Dawson KA, What the cell “sees” in bionanoscience. J. Am. Chem. Soc 2010, 132, 5761–5768. [DOI] [PubMed] [Google Scholar]
- (2).Cedervall T; Lynch I; Lindman S; Berggård T; Thulin E; Nilsson H; Dawson KA; Linse S, Understanding the Nanoparticle—Protein Corona Using Methods To Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 2050–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ke PC; Lin S; Parak WJ; Davis TP; Caruso F, A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. [DOI] [PubMed] [Google Scholar]
- (4).Monopoli MP; Walczyk D; Campbell A; Elia G; Lynch I; Baldelli Bombelli F; Dawson KA, Physical– chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc 2011, 133, 2525–2534. [DOI] [PubMed] [Google Scholar]
- (5).Norde W, Protein adsorption at solid surfaces: A thermodynamic approach. Pure Appl. Chem 1994, 66, 491–496. [Google Scholar]
- (6).Vroman L, Effect of Adsorbed Proteins on the Wettability of Hydrophilic and Hydrophobic Solids. Nature 1962, 196, 476–477. [DOI] [PubMed] [Google Scholar]
- (7).Monopoli MP; Aberg C; Salvati A; Dawson KA, Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat. Nanotechnol 2012, 7, 779–786. [DOI] [PubMed] [Google Scholar]
- (8).Lundqvist M; Stigler J; Cedervall T; Berggård T; Flanagan MB; Lynch I; Elia G; Dawson K, The Evolution of the Protein Corona around Nanoparticles: A Test Study. ACS Nano 2011, 5, 7503–7510. [DOI] [PubMed] [Google Scholar]
- (9).Xu M; Soliman MG; Sun X; Pelaz B; Feliu N; Parak WJ; Liu S, How entanglement of different physicochemical properties complicates the prediction of in vitro and in vivo interactions of gold nanoparticles. ACS nano 2018, 12, 10104–10113. [DOI] [PubMed] [Google Scholar]
- (10).Johnston BD; Kreyling WG; Pfeiffer C; Schäffler M; Sarioglu H; Ristig S; Hirn S; Haberl N; Thalhammer S; Hauck SM; Semmler-Behnke M; Epple M; Hühn J; Del Pino P; Parak WJ, Colloidal Stability and Surface Chemistry Are Key Factors for the Composition of the Protein Corona of Inorganic Gold Nanoparticles. Adv. Fund. Mater 2017, 27, 1701956. [Google Scholar]
- (11).Gagner JE; Lopez MD; Dordick JS; Siegel RW, Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials 2011, 32, 7241–7252. [DOI] [PubMed] [Google Scholar]
- (12).Maiorano G; Sabella S; Sorce B; Brunetti V; Malvindi MA; Cingolani R; Pompa PP, Effects of Cell Culture Media on the Dynamic Formation of Protein—Nanoparticle Complexes and Influence on the Cellular Response. ACS Nano 2010, 4, 7481–7491. [DOI] [PubMed] [Google Scholar]
- (13).Walkey CD; Olsen JB; Guo H; Emili A; Chan WC, Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc 2012, 134, 2139–2147. [DOI] [PubMed] [Google Scholar]
- (14).Lundqvist M; Stigler J; Elia G; Lynch I; Cedervall T; Dawson KA, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 14265–14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tenzer S; Docter D; Rosfa S; Wlodarski A; Kuharev J. r.; Rekik A; Knauer SK; Bantz C; Nawroth T; Bier C, Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS nano 2011, 5, 7155–7167. [DOI] [PubMed] [Google Scholar]
- (16).Docter D; Distler U; Storck W; Kuharev J; Wünsch D; Hahlbrock A; Knauer SK; Tenzer S; Stauber RH, Quantitative profiling of the protein coronas that form around nanoparticles. Nat. Protoc 2014, 9, 2030–2044. [DOI] [PubMed] [Google Scholar]
- (17).Gref R; Lück M; Quellec P; Marchand M; Dellacherie E; Harnisch S; Blunk T; Müller R, ‘Stealth’corona-core nanoparticles surface modified by polyethylene glycol (PEG): influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B Biointerfaces 2000, 18, 301–313. [DOI] [PubMed] [Google Scholar]
- (18).García-Álvarez R; Hadjidemetriou M; Sánchez-Iglesias A; Liz-Marzán LM; Kostarelos K, In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale 2018, 10, 1256–1264. [DOI] [PubMed] [Google Scholar]
- (19).Wang M; Gustafsson OJR; Pilkington EH; Kakinen A; Javed I; Faridi A; Davis TP; Ke PC, Nanoparticle—proteome in vitro and in vivo. J. Mater. Chem. B 2018, 6, 6026–6041. [DOI] [PubMed] [Google Scholar]
- (20).Caracciolo G; Farokhzad OC; Mahmoudi M, Biological Identity of Nanoparticles In Vivo: Clinical Implications of the Protein Corona. Trends Biotechnol. 2017, 35, 257–264. [DOI] [PubMed] [Google Scholar]
- (21).Hadjidemetriou M; Al-Ahmady Z; Mazza M; Collins RF; Dawson K; Kostarelos K, In Vivo Biomolecule Corona around Blood-Circulating, Clinically Used and Antibody-Targeted Lipid Bilayer Nanoscale Vesicles. ACS Nano 2015, 9, 8142–8156. [DOI] [PubMed] [Google Scholar]
- (22).Hadjidemetriou M; McAdam S; Garner G; Thackeray C; Knight D; Smith D; Al-Ahmady Z; Mazza M; Rogan J; Clamp A, The human in vivo biomolecule corona onto PEGylated liposomes: a proof-of-concept clinical study. Adv.Mater 2019, 31, 1803335. [DOI] [PubMed] [Google Scholar]
- (23).Casals E; Pfaller T; Duschl A; Oostingh GJ; Puntes V, Time evolution of the nanoparticle protein corona. ACS nano 2010, 4, 3623–3632. [DOI] [PubMed] [Google Scholar]
- (24).Dell’Orco D; Lundqvist M; Oslakovic C; Cedervall T; Linse S, Modeling the Time Evolution of the Nanoparticle-Protein Corona in a Body Fluid. PLoS One 2010, 5, e10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cheng X; Tian X; Wu A; Li J; Tian J; Chong Y; Chai Z; Zhao Y; Chen C; Ge C, Protein Corona Influences Cellular Uptake of Gold Nanoparticles by Phagocytic and Nonphagocytic Cells in a Size-Dependent Manner. ACS Applied Materials & Interfaces 2015, 7, 20568–20575. [DOI] [PubMed] [Google Scholar]
- (26).Choi K; Riviere JE; Monteiro-Riviere NA, Protein corona modulation of hepatocyte uptake and molecular mechanisms of gold nanoparticle toxicity. Nanotoxicology 2017, 11, 64–75. [DOI] [PubMed] [Google Scholar]
- (27).Dobrovolskaia MA; Patri AK; Zheng J; Clogston JD; Ayub N; Aggarwal P; Neun BW; Hall JB; McNeil SE, Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomedicine: Nanotechnology, Biology and Medicine 2009, 5, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lacerda SHDP; Park JJ; Meuse C; Pristinski D; Becker ML; Karim A; Douglas JF, Interaction of gold nanoparticles with common human blood proteins. ACS nano 2010, 4, 365–379. [DOI] [PubMed] [Google Scholar]
- (29).Walkey CD; Olsen JB; Song F; Liu R; Guo H; Olsen DWH; Cohen Y; Emili A; Chan WC, Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS nano 2014, 8, 2439–2455. [DOI] [PubMed] [Google Scholar]
- (30).Molleman B; Hiemstra T, Size and shape dependency of the surface energy of metallic nanoparticles: unifying the atomic and thermodynamic approaches. Phys. Chem 2018, 20, 20575–20587. [DOI] [PubMed] [Google Scholar]
- (31).Takato Y; Benson ME; Sen S, Small nanoparticles, surface geometry and contact forces. Proc. Math Phys. Eng. Sci 2018, 474, 20170723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Vollath D; Fischer FD; Holec D, Surface energy of nanoparticles — influence of particle size and structure. Beilstein J. Nanotechnol 2018, 9, 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lee S; Chon H; Lee M; Choo J; Shin SY; Lee YH; Son SW; Oh CH, Surface-enhanced Raman scattering imaging of HER2 cancer markers overexpressed in single MCF7 cells using antibody conjugated hollow gold nanospheres. Biosens. Bioelectron 2009, 24, 2260–2263. [DOI] [PubMed] [Google Scholar]
- (34).Qian X; Peng X-H; Ansari DO; Yin-Goen Q; Chen GZ; Shin DM; Yang L; Young AN; Wang MD; Nie S, In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol 2008, 26, 83–90. [DOI] [PubMed] [Google Scholar]
- (35).Huang X; Jain PK; El-Sayed IH; El-Sayed MA, Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Lasers Med. Sci 2008, 23, 217–223. [DOI] [PubMed] [Google Scholar]
- (36).Butterworth K; Coulter J; Jain S; Forker J; McMahon S; Schettino G; Prise K; Currell F; Hirst D, Evaluation of cytotoxicity and radiation enhancement using 1.9 nm gold particles: potential application for cancer therapy. Nanotechnology 2010, 21, 295101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Duncan B; Kim C; Rotello VM, Gold nanoparticle platforms as drug and biomacromolecule delivery systems. J. Control. Release 2010, 148, 122–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Turkevich J; Stevenson PC; Hillier J, A study of the nucleation and growth processes in the synthesis of colloidal gold. Faraday Discuss. 1951, 11, 55–75. [Google Scholar]
- (39).Schneider CA; Rasband WS; Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wang M; Siddiqui G; Gustafsson OJR; Käkinen A; Javed I; Voelcker NH; Creek DJ; Ke PC; Davis TP, Plasma Proteome Association and Catalytic Activity of Stealth Polymer-Grafted Iron Oxide Nanoparticles. Small 2017, 13, 1701528. [DOI] [PubMed] [Google Scholar]
- (41).Siddiqui G; Srivastava A; Russell AS; Creek DJ, Multi-omics Based Identification of Specific Biochemical Changes Associated With PfKelch 13-Mutant Artemisinin-Resistant Plasmodium falciparum. J. Infect. Dis 2017, 215, 1435–1444. [DOI] [PubMed] [Google Scholar]
- (42).Szklarczyk D; Franceschini A; Wyder S; Forslund K; Heller D; Huerta-Cepas J; Simonovic M; Roth A; Santos A; Tsafou KP; Kuhn M; Bork P; Jensen LJ; von Mering C, STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Shannon P; Markiel A; Ozier O; Baliga NS; Wang JT; Ramage D; Amin N; Schwikowski B; Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schöttler S; Becker G; Winzen S; Steinbach T; Mohr K; Landfester K; Mailänder V; Wurm FR, Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nature Nanotechnology 2016, 11, 372–377. [DOI] [PubMed] [Google Scholar]
- (45).Mahmoudi M; Lynch I; Ejtehadi MR; Monopoli MP; Bombelli FB; Laurent S, Protein—Nanoparticle Interactions: Opportunities and Challenges. Chem. Rev 2011, 111, 5610–5637. [DOI] [PubMed] [Google Scholar]
- (46).Bertrand N; Grenier P; Mahmoudi M; Lima EM; Appel EA; Dormont F; Lim J-M; Karnik R; Langer R; Farokhzad OC, Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics. Nat. Commun 2017, 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Corbo C; Molinaro R; Parodi A; Toledano Furman NE; Salvatore F; Tasciotti E, The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 2015, 11, 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Tenzer S; Docter D; Kuharev J; Musyanovych A; Fetz V; Hecht R; Schlenk F; Fischer D; Kiouptsi K; Reinhardt C, Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol 2013, 8, 772–781. [DOI] [PubMed] [Google Scholar]
- (49).Setyawati M; Tay CY; Chia SL; Goh SL; Fang W; Neo MJ; Chong H; Tan S-M; Loo J; Ng KW; Xie J; Ong CN; Tan NS; Leong D, Titanium dioxide nanomaterials cause endothelial cell leakiness by disrupting the homophilic interaction of VE-cadherin. NATURE COMMS 2013. Nat. Commun 2013, 4, 1673–1684. [DOI] [PubMed] [Google Scholar]
- (50).Setyawati MI; Tay CY; Bay BH; Leong DT, Gold Nanoparticles Induced Endothelial Leakiness Depends on Particle Size and Endothelial Cell Origin. ACS Nano 2017, 11, 5020–5030. [DOI] [PubMed] [Google Scholar]
- (51).Tee JK; Yip LX; Tan ES; Santitewagun S; Prasath A; Ke PC; Ho HK; Leong DT, Nanoparticles’ interactions with vasculature in diseases. Chem. Soc. Rev 2019, 48, 5381–5407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


