Abstract
Several members of the kinesin superfamily are known to play a prominent role in the motor-driven transport processes that occur in mitotic cells. Here we describe a new mitotic human kinesin-like protein, RB6K (Rabkinesin 6), distantly related to MKLP-1. Expression of RB6K is regulated during the cell cycle at both the mRNA and protein level and, similar to cyclin B, shows a maximum during M phase. Isolation of the RB6K promoter allowed identification of a CDE-CHR element and promoter activity was shown to be maximal during M phase. Immunofluorescence microscopy using antibodies raised against RB6K showed a weak signal in interphase Golgi but a 10-fold higher signal in prophase nuclei. During M phase, the newly synthesized RB6K does not colocalise with Rab6. In later stages of mitosis RB6K localized to the spindle midzone and appeared on the midbodies during cytokinesis. The functional significance of this localization during M phase was revealed by antibody microinjection studies which resulted exclusively in binucleate cells, showing a complete failure of cytokinesis. These results substantiate a crucial role for RB6K in late anaphase B and/or cytokinesis, clearly distinct from the role of MKLP-1.
Recently, we described the isolation of a new human cDNA (GenBank accession no. AF070672) encoding a protein that has all the characteristics of a kinesin-like protein (KLP) (11). Its sequence is 86% identical to murine RB6K (Rabkinesin 6) (GenBank accession no. Y09632), and because the differences in amino acids were randomly scattered throughout the sequence in structural rather than functional regions, we assumed it to be the human homolog. Murine RB6K was identified as a Golgi-localized KLP that, upon interaction with GTP-bound forms of Rab6, may be involved in retrograde vesicular traffic between the Golgi apparatus and the endoplasmic reticulum (6, 34). The human RB6K showed differential levels of expression in cytokine-stimulated human umbilical vein endothelial cells (HUVEC) (11). Downregulation of human RB6K upon cytokine stimulation was a late response but did not correlate with changes in Golgi architecture. Comparable to the gene encoding RB6K, GSPT1 and RGS5, two genes involved in cell cycle initiation, were found to be repressed, whereas the antiproliverative BTG1 gene was upregulated by tumor necrosis factor alpha (11). Serum starvation, leading to entry of the cells into G0, also resulted in downregulation of RB6K. In a recent work, the sequence of the human gene RAB6KIFL, encoding RB6K, was described (15). The gene is located on chromosome 5 in a region containing several cell cycle genes, including cdc25. In adult tissue the mRNA for RB6K is almost exclusively expressed in tissues with high proliferation rates (thymus, bone marrow, and testis), which suggests a role in the cell cycle. Sequence conservation within the extensive KLP family is mainly restricted to the catalytic motor domain, which typically comprises approximately 350 amino acids and is involved in microtubule binding and ATP hydrolysis (2, 10; L. Greene and S. Henikoff, http://howard.fhcrc.org/kinesin/). The phylogenetic tree of kinesins shows that both human and mouse RB6K are related to the MKLP-1 subfamily, based on homology within the motor domain (52% homology; 34% identity), whereas the C termini lack any discernible homology (website of Greene and Henikoff). MKLP-1 has been reported to play a role in assembly of the mitotic spindle and separation of spindle poles during anaphase B based on its ability to cross-link and slide antiparallel microtubuli past each other in vitro (22). Like MKLP-1, RB6K has been shown to be able to bind microtubules both with its N and C terminus (6). Knowledge on cargo binding and control of motoractivity of KLPs is still rather limited. Interestingly, regulation at the level of expression has been suggested for MKLP-1, but no supporting data were provided (16). Intracellular levels of CENP-E were reported to be regulated by steady synthesis throughout the cell cycle, combined with stabilization during the S and G2 phase and rapid proteolytic degradation at the end of mitosis (3, 27). As cells proceed through the cell cycle, activity of different members of the KLP family is required. The interphase microtubule network supports motor-driven transport of, e.g., vesicles and organelles by members of the KHC, Unc-104, and KRP85/95 subfamilies. Motility events during M phase include centrosome separation, chromosome positioning on the metaphase plate, chromosome movement, force generation in the mitotic spindle, and, finally, cytokinesis. In these processes, members of the BimC, C-terminal, MKLP-1, and chromokinesin subfamilies have been implicated (2, 10). Regulation of KLP expression and function could therefore be an important means by which cells generate and regulate the required transport capacity. The study presented here was undertaken to establish a possible role of the novel human KLP RB6K in mitosis. We provide evidence that RB6K is maximally expressed during M phase, specifically localizes to the spindle midzone and midbody, and plays an obligatory role during cytokinesis.
MATERIALS AND METHODS
Cell culture and immunofluorescence.
HeLa cells were cultured in Iscove's modified minimal medium, supplemented with penicillin (100 U/ml), streptomycin (100 U/ml) and 10% (vol/vol) fetal bovine serum. EC-RF24 cells were cultured as described (7). The medium was composed of equal parts of medium 199 and RPMI 1640, supplemented with 20% (vol/vol) heat-inactivated, pooled human serum; 2mM glutamine; and a 1:100 dilution of antibiotic-antimycotic mix (final concentrations: penicillin, 100 U/ml; streptomycin 100 U/ml; amphotericin B [Fungizone], 2.5 mg/ml, [Life Technologies]).
Primary endothelial cells or EC-RF24 cells were grown on gelatin-coated coverslips for at least 24 h. Cells were then washed with serum-free medium and fixed for 10 min with methanol at room temperature. After fixation, cells were washed twice with phosphate-buffered saline (PBS) and incubated for 1 h with rabbit anti-murine RB6K antibodies (6) in PBS supplemented with 1% (wt/vol) bovine serum albumin (BSA) (Organon Teknika, Boxtel, The Netherlands). Then, the coverslips were washed twice with PBS and incubated for 1 h with affinity-purified Cy3-conjugated goat anti-rabbit antibodies (Jackson Immunoresearch, West Grove, Pa.) in PBS with 1% (wt/vol) BSA. Nuclei were counterstained for 10 min in a dilution (100 ng/ml) of Hoechst 33258 in PBS. After washing three times in PBS, the coverslips were mounted in Mowiol (Calbiochem, La Jolla, Calif.) and viewed with a Zeiss AxioplanII microscope. Confocal laser scanning microscopy (CLSM) was performed using a Leica CLSM (Leica Microsystems, Heidelberg, Germany) equipped with an argon-krypton laser. Images were collected with a fixed setting for the laser power (excitation; 568 nm; detection, 580 nm; dichroid mirror and a 610-nm long-pass filter). A 40/1.30 NPL Fluotar objective was used together with a pinhole setting, giving an optical thickness of approximately 1 μm, and images were adjusted to the full dynamic range of the system (8 bit). The amount of Cy3 fluorescence as found in either the nucleus or the Golgi for individual cells in the confocal stack was determined with the Leica Qwin image analysis software (Leica Cambridge). The values for each cell were corrected for background (the amount of fluorescence outside of the cells) and photomultiplier settings.
In situ hybridization.
EC-RF24 cells were grown on gelatin-coated glass slides for at least 24 h. Cells were washed with serum-free medium and fixed in 4% (wt/vol) paraformaldehyde in PBS. Slides were dehydrated by passing them successively through 30, 50, 75, 85, 95, and 100% ethanol. A riboprobe was synthesized by in vitro transcription of a cDNA fragment spanning RB6K nucleotides 1190 to 1426 cloned in pGEM-4Z. The construct was linearized, and RNA was synthesized with T7 RNA polymerase (Stratagene, La Jolla, Calif.), according to the manufacturer's instructions, using 35S-UTP (Amersham, Amersham, United Kingdom) as label.
Unincorporated nucleotides were removed on a G-50 Sephadex spin column. The riboprobe was added to and stored in hybridization mixture, consisting of 40% (vol/vol) formamide, 8% (wt/vol) dextrane sulfate, 0.8× Denhardt's solution, 0.5 mg of yeast tRNA per ml, 4 mM EDTA, 16 mM Tris-HCl (pH 8.0), and 0.24 mol of NaCl per liter. In situ hybridization was performed as described (35) with minor modifications. The slides with fixed cells were rehydrated and treated for 10 min with 0.25% (wt/vol) acetic anhydride in 0.1 M triethanolamnine (pH 8.0). After washing and dehydration, approximately 0.1 μCi per cm2 of fixed cells was added, and hybridization was performed overnight at 50°C under a coverslip in a moist chamber.
After hybridization, coverslips were removed in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10 mM dithiothreitol at 50°C (30 to 60 min), followed by a high-stringency wash for 30 min at 65°C in 50% (vt/vol) formamide–2× SSC–10 mM dithiothreitol. RNase A (20 μg/ml) digestion was performed for 30 min at 37°C in 10 mM Tris-HCl (pH 8.0)–5 mM EDTA–500 mM NaCl. The high stringency wash was repeated and was followed by washing for 15 min in 2× SSC. After dehydration, slides were prepared for autoradiography by dipping in a 1:0.4 (vt/vol) dilution of Ilford G5 photographic emulsion (Ilford, Paramus, N.I.) with 2% glycerol. After an exposure of 4 weeks, slides were developed in Kodak D19 and fixed in Kodak UNIFIX (Kodak-Pathe, Paris, France). Finally, developed slides were counterstained with hematoxylin and eosin.
Cell cycle arrest, synchronization, and flow cytometry.
Cells were arrested in different stages of the cell cycle by addition of one of the following drugs to the medium: hydroxyurea (2 mM) or nocodazole (0.17 mM). Twenty hours after addition of the drug, cells were harvested for analysis. To synchronize cells, near-confluent cultures were blocked at the G1/early S stage by addition of hydroxyurea to the medium to a final concentration of 2 mM. After 20 h, cells were released from the block by washing them three times with serum-free medium and growing them on complete medium. For monitoring synchrony by flow cytometry, cells were briefly trypsinized, pelleted by centrifugation at 200 × g, and resuspended in 2 ml of PBS. While gently mixing on a vortex, ethanol was added to a final concentration of 75%. Shortly before flow cytometry, propidium iodide staining was performed. Cells were centrifuged at 200 × g and carefully resuspended in 0.25 ml of PBS. The actual staining was performed during 30 min at 37°C in 1 ml of PBS containing propidium iodide (0.025 mg/ml), 0.01% (wt/vol) saponin, and RNase A (1 mg/ml). Subsequently, cell cycle distribution of the cells was determined by analyzing their DNA content on a Becton Dickinson FACSVantage SE flow cytometer. Data were analyzed using WinMDI 2.8 software (J. Trotter, Scripps Research Institute, La Jolla, Calif.).
RNA isolation and Northern blot analysis.
RNA was isolated from synchronized cultures and analyzed by Northern blotting as described previously (11). As probes we used agarose-purified restriction fragments containing RB6K cDNA nucleotides 1712 to 2972 or cyclin B as an insert of approximately 1.5 kb from IMAGE clone 549825 (17). The fragments were labeled to high specific radioactivity using the random primers DNA labeling system (Life Technologies) and [α-32P]dATP (Redivue; Amersham). Unincorporated nucleotides were removed by the Qiaquick nucleotide removal kit (Qiagen, Hilden, Germany). Radioactivity was quantified using a STORM device and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
Cell lysates and immunoblotting.
Cells were washed in PBS and lysed in a buffer containing 150 mM NaCl, 10 mmol of EDTA per liter, 1% (vol/vol) Triton X-100, 25 mM Tris-HCl (pH 8.0), and a 1:10 dilution of a protease and phosphatase inhibitor cocktail (catalog no. P8340; Sigma, St. Louis, Mo.). Insoluble material was pelleted by centrifugation at 15,000 × g for 5 min. Protein content of the lysate was measured using a micro-BCA protein assay (Pierce, Rockford, III.). Fifteen micrograms of total protein was used for electrophoresis on an 8% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel under reducing conditions (14) and subsequently transferred to a 0.45-μm-pore-size nitrocellulose membrane (Schleicher and Schuell, Dassel, Germany). Filters were blocked by incubation with 2% (wt/vol) BSA in Tris-buffered saline (TBS) and incubated with affinity-purified rabbit immunoglobulins (Ig) raised against RB6K (6) diluted 1:1,000 in TBS containing 0.4% BSA. After three washes in TBS, the blot was developed using the ProtoBlot II AP system (Promega, Madison, Wis.), according to the manufacturer's instructions. As a control for equal loading the filters were reprobed using antibodies against α-tubulin (Cedarlane, Hornby, Ontario, Canada) and an ECL Western blotting detection system (Amersham Pharmacia Biotech, Uppsala, Sweden).
Microinjection.
Forty hours before microinjection, HeLa cells were seeded onto microinjection grids. Twenty hours before injection, cells were arrested in G1/S by addition of 2 mM hydroxyurea. Needles were pulled using a PB-7 micropipette puller (Narishage Co., Tokyo, Japan) and back-filled with affinity-purified polyclonal antibody preparations (6) containing Ig (1.5 mg/ml) in 0.5× PBS. Cells were released from the block by transferring the grids to fresh medium and injected into the cytoplasm with the appropriate Ig preparation. After injection, cells were washed and incubated in complete medium for 20 h to allow at least one passage through the cell cycle. Subsequently, cells were washed in PBS, fixed in methanol for 15 min at room temperature, and prepared for inspection by immunofluorescence, using Cy3-conjugated anti-rabbit Ig antibodies to detect injected cells.
RB6K-EGFP, RB6K 5′UTR, and RB6K-5′ upstream sequences: isolation, constructs, and functional analyses.
The 5′ untranslated region (5′ UTR) of RB6K was extended by 5′ rapid amplification of cDNA ends (5′-RACE) using human placenta Marathon-Ready cDNA and the Marathon cDNA amplification kit (Clontech, Palo Alto, Calif.). The nested gene-specific primers used, corresponded to nucleotides 121 to 145 and 151 to 175 of the RB6K cDNA (GenBank accession no. AF070672). PCR-amplification products were cloned in the pGEM-T vector (Promega) for sequence analysis.
pRB6K-EGFP encodes RB6K with its C terminus fused in frame to EGFP cDNA. A fragment of RB6K cDNA with an XmaI site replacing the stop codon was generated by PCR and cloned in frame with EGFP in the pEGFP-N2 expression vector (Clontech). Genomic sequences 5′ upstream of the RB6K cDNA were PCR-amplified from the human PAC clone DJ0309D19 (GB:AC004826) using Advantage cDNA polymerase mix (Clontech) and synthetic oligonucleotides 5′ATCACCAGTGACCGGGGTACC3′ and 5′ATGGAAGATCTCCGAAGACGTGCCACTTGCTCC3′ as the forward and reverse primer, respectively. The amplified fragment was cloned in the pGL3-Basic luciferase reporter plasmid (Promega) using KpnI and BglII sites. Thus, the resulting plasmid, referred to as pGL3 RB6K-5′, comprises 7 nucleotides of the 5′UTR and 1,342 nucleotides upstream of the originally published RB6K cDNA.
Transfection of EC-RF24 cells (7) or HeLa cells with RB6K constructs were performed using Superfect (Qiagen), essentially according to the manufacturer's instructions. pRB6K-EGFP-transfected cells were analyzed after fixation in 4% (wt/vol) paraformaldehyde in PBS and mounted for microscopy as mentioned under “Cell culture and immunofluorescence” above. Cells transfected with pGL3RB6K-5′ were analyzed for luciferase activity, using the luciferase assay system (Promega). Transfection efficiencies in these experiments were monitored by measuring β-galactosidase activity expressed from the cotransfected pSV-β-Galactosidase vector (Promega) using the Galacto-light chemiluminiscent reporter assay (Tropix, Bedford, Mass.). Both assays were performed on a Luminat LB 9501 illuminometer.
Primer extension analysis was performed according to standard methods (26), using 5 μg of total RNA isolated from ECRF-24 cells. Ten picomoles of a Cy5-5′-labeled synthetic oligonucleotide, 5′CCGAAGACGTGCCACTTGCTCCTCCTGGGATACTGGC3′, was used to initiate first-strand synthesis, and resulting products were analyzed on a high-resolution gel on an ALF-express automatic sequence analyzer (Amersham Pharmacia Biotech).
RESULTS
RB6K is a mitotic kinesin-like protein.
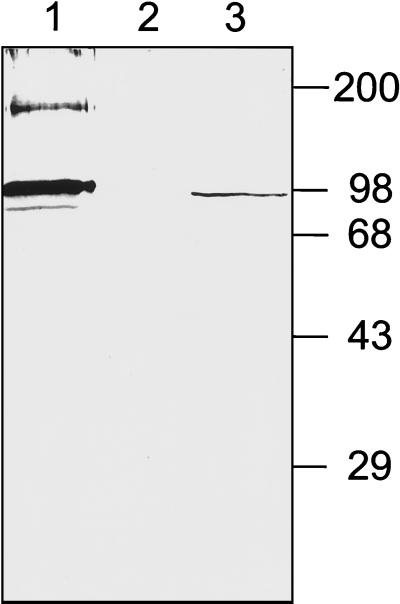
As the observation that RB6K is repressed by cytokine incubation was initially made in HUVEC, we chose to study its subcellular localization in either HUVEC or EC-RF24, a cell line derived from HUVEC and previously characterized as a reliable model for studying endothelial -cell functions (7). The affinity-purified anti-mouse RB6K antibodies that we used in this study were previously described (6). The reactivity of these antibodies with human RB6K and specificity of the antibodies in lysates of the cell types that we used in this study were tested on a Western blot. For this purpose we performed in vitro transcription-translation of human RB6K cDNA and Triton X-100 extracts of EC-RF24 cells were made. Samples were electrophoresed under reducing conditions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto nitrocellulose. Immunostaining with anti-mouse RB6K antibodies detected a major band of approximately 98 kDa, the expected size for human RB6K (SwissProt accession no. O95235) (Fig. 1). In the samples of the in vitro transcription-translation material some bands with slightly lower molecular weights were observed, probably resulting from leaky scanning for translation initiation sites. An additional band at approximately 200 kDa is probably a remainder of the dimer conformation.
FIG. 1.
Reactivity of affinity-purified anti-mouseRB6K polyclonal antibodies. Samples were electrophoresed under reducing conditions on an 8% (wt/vol) polyacrylamide gel and transferred to a Protran membrane, which was subsequently probed with anti-RB6K antibodies. Lane 1, product of coupled in vitro transcription-translation: reticulocyte lysate programmed with hRB6K cDNA cloned in pGEM4Z; lane 2, control, reticulocyte lysate without plasmid added; lane 3, lysate of EC-RF24 cells. Molecular weight markers (in thousands) are indicated at the right side of the figure.
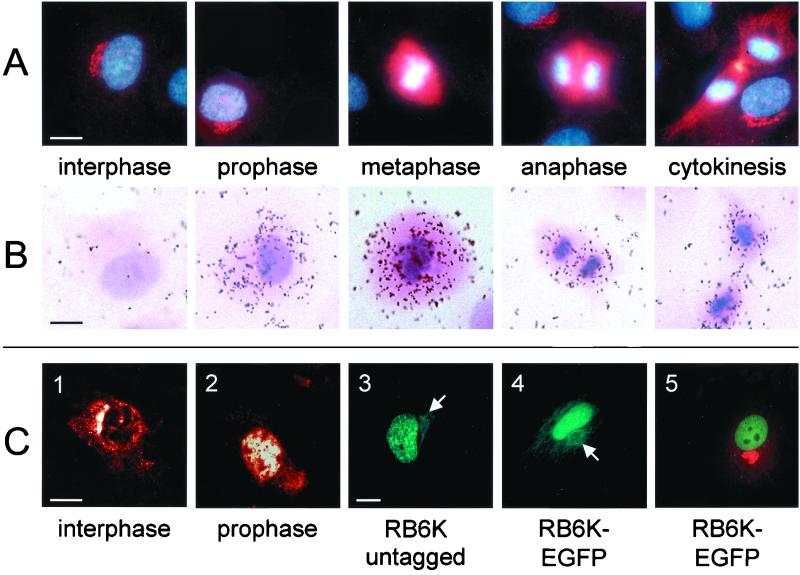
Next, we investigated expression of RB6K by immunofluorescence microscopy. The vast majority of the cells showed a perinuclear stain that has previously been localized to the medial Golgi compartment by colocalization with CTR 433 (6). Remarkably, in a minor part of the cell population different staining patterns with a relatively high fluorescence intensity were observed. Costaining of DNA with the Hoechst 33258 dye revealed that these patterns were associated with various stages of mitosis (Fig. 2A). A higher resolution localization during cell cycle progression is shown and discussed in relation to Fig. 6 (vide infra). Strikingly, the appearance of fluorescence signal in prophase nuclei was apparently accompanied neither by loss of signal from the Golgi apparatus nor by loss of integrity of the Golgi structure, prompting us to make a quantitative assessment of the fluorescence at these sites, using CLSM. As RB6K fluorescence in nuclei of prophase cells and in Golgi structures in interphase cells was spatially well defined due to compartmentalization (cf. Fig. 2C panels 1 and 2), we chose to use these compartments for further quantitative analysis. The mean integral fluorescence (standard deviation) measured in prophase nuclei, 1,244 (322) arbitrary units, was ∼1 order of magnitude higher than that in interphase Golgi, 117 (60) arbitrary units. To further substantiate the nuclear targeting of RB6K, transfection experiments were performed. Expression from a constitutive promoter of native, untagged human RB6K resulted in the accumulation of the excess protein in the nucleus as detected by anti-RB6K antibodies and a fluorescein isothiocyanate (FITC) conjugate. (Fig. 2C, panel 3). As a second approach, independent of the use of antibodies, we transfected EC-RF24 cells with a construct in which the C -terminus of RB6K, devoid of its stop codon, was fused in -frame to enhanced green fluorescent protein (EGFP) in the pEGFP-N2 vector. Again, the EGFP-tagged RB6K localized predominantly to the nucleus in all transfected cells (Fig. 2C, panels 4 and 5). Thus, nuclear targeting of endogenous RB6K in prophase cells was confirmed by two independent approaches. Typically, the nuclei contain several nucleoli, indicating that the cells are indeed in G1 phase (Fig. 2C, panels 3 and 5). Not only do these observations confirm targeting of RB6K to the nucleus, they also imply that when expression is driven from a constitutive promoter, entrance of (GFP-tagged) RB6K into the nucleus is independent from cell cycle progression. Thus, it is conceivable that cell cycle-induced increase of synthesis of the endogenous RB6K in prophase cells saturates the Golgi localization machinery and leads to the observed nuclear localization.
FIG. 2.
Expression of RB6K protein and mRNA during the cell cycle in nonsynchronously growing cells. (A) Immunofluorescence of RB6K. Cells were fixed with methanol and incubated with anti-RB6K polyclonal antibodies followed by Cy3-conjugated goat anti-rabbit Ig (red). DNA was stained using Hoechst 33258 (blue). (B) In situ hybridization of paraformaldehyde-fixed EC-RF24 cells with an RB6K antisense riboprobe. (C) Details of subcellular localization. Panels 1 and 2, 1-μm-thick CLSM sections of RB6K immunofluorescence in interphase (1) and prophase (2) cells; panel 3, fluorescence of EC-RF24 cells transiently transfected with RB6K cDNA and stained with anti-RB6K polyclonal antibodies followed by incubation with FITC-conjugated goat anti-rabbit Ig (green); panel 4, fluorescence of EC-RF24 cells transiently transfected with RB6K-EGFP cDNA (in panels 3 and 4, arrows indicate localization of RB6K to the microtubuli); panel 5, fluorescence of EC-RF24 cells transiently transfected with RB6K-EGFP cDNA (the Golgi apparatus is stained using antibodies against Giantin and a Cy3 conjugate [red]). Bars: A, B, and C, panels 1 and 2, 15 μm; C, panels 3 to 5, 20 μm.
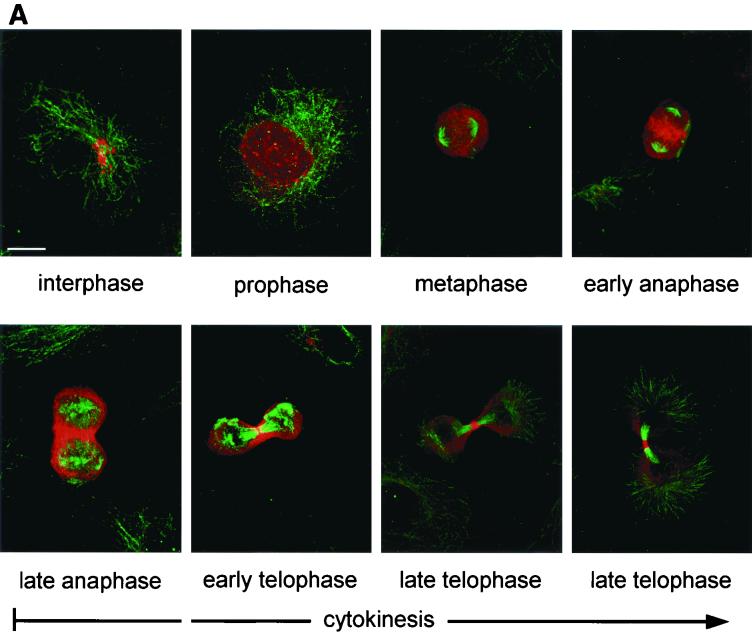
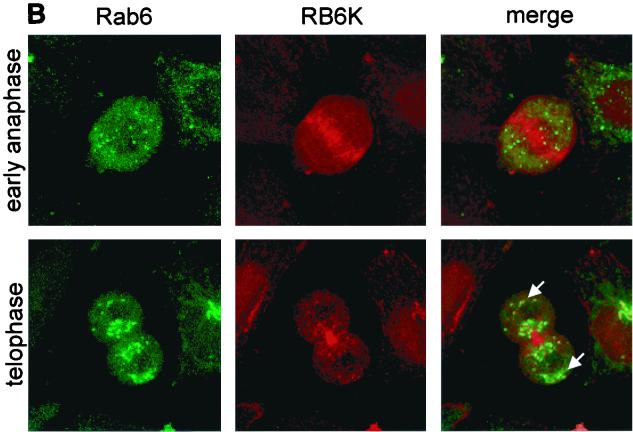
FIG. 6.
(A) Subcellular localization of RB6K during the cell cycle. Endogenous RB6K was detected in EC-RF24 cells using an affinity-purified anti-RB6K preparation and Cy3-conjugated goat anti-rabbit Ig (red). Microtubuli were stained using anti-α-tubulin antibodies and a FITC-labeled conjugate (green). Shown is a series of the subsequent stages of the cell cycle as brightest point projections of confocal sections taken every 1.0 μm. Bar, 20 μm. (B) Subcellular localization of RB6K during the cell cycle. Endogenous RB6K was detected in HeLa cells stably expressing Rab6-GFP (35) using an affinity-purified anti-RB6K preparation and Texas red-conjugated goat anti-rabbit Ig (red).
In cells showing overexpression of either native or GFP-tagged RB6K, fluorescence also localized to the microtubuli, which then obtained an abnormal, bundled appearance (Fig. 2C, panels 3 and 4 [arrows]). Cells constitutively overexpressing RB6K-EGFP did not enter mitosis and died within 48 h after transfection. Essentially the same observations were made using the native, untagged, construct, whereas cells transfected with the nonfused EGFP construct did maintain their proliferative capacity (data not shown). These results suggest that the constitutive overexpression of RB6K, either native or as a moiety of an EGFP-tagged fusion protein, is detrimental to the cells. No fluorescence of de novo-synthesized RB6K on the Golgi was observed, neither for wild-type nor for EGFP-tagged RB6K, which may be due to high levels of fluorescence of RB6K(-EGFP), in the nucleus and on the microtubules, that hamper detection of the signal of a much smaller pool of the protein localizing to the Golgi. Integrity of the Golgi during RB6K overexpression was substantiated by costaining with the Golgi-specific marker Giantin (Fig. 2C, panel 5 [red stain]).
RB6K expression is upregulated in mitosis.
In a culture of growing cells, at any time point, a minor fraction of the population will be in mitosis. In fact, the relative number of cells in a certain stage of the cell cycle can be considered to reflect the time span in which individual cells pass that particular stage of the cell cycle. As the duration of M phase is short relative to interphase, mitotic cells are underrepresented in nonsynchronized cultures, hampering analysis of expression regulation around M phase. Here, we used two different approaches to further study cell cycle dependency of RB6K expression. First, in situ hybridization of nonsynchronized EC-RF24 cells with an antisense RB6K riboprobe was performed to examine mRNA expression related to different stages of the cell cycle and, more specifically, throughout different stages of mitosis. Indeed, expression of RB6K mRNA was almost exclusively observed in cells that could be characterized as mitotic cells, based on hematoxylin and eosin counterstaining that was applied after visualizing the in situ hybridization signals by autoradiography (Fig. 2B). Control hybridizations run in parallel with an antisense probe for the constitutively expressed vWF mRNA showed no changes in mRNA signal during the cell cycle, thus indicating that the observed changes for RB6K were indeed cell cycle-dependent and excluding that the typical change in morphology of mitotic cells, due to partial detachment from the matrix, had influenced interpretation (data not shown).
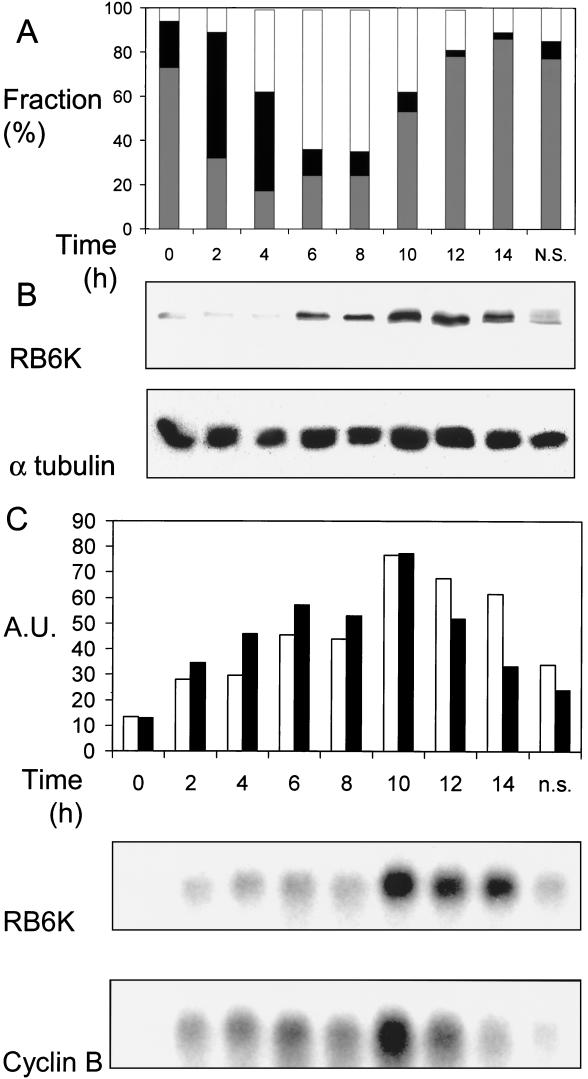
Second, EC-RF24 cells were synchronized by a hydroxyurea block and subsequent release in fresh medium. With 2-hour intervals, cells were harvested for further analysis. For each timepoint, three parameters were analyzed. First, synchrony of the cell population was monitored by flow cytometric analysis of propidium iodide-stained cells. Second, lysates were prepared and used for making Western blots that were subsequently stained with anti-RB6K antiserum. Third, RNA was isolated and analyzed on Northern blot by hybridization with an hRB6K probe and, as a control, a cyclin B probe.
Arrest of cultures by incubation with hydroxyurea, an inhibitor of DNA replication, caused an efficient, reversible arrest at late G1/early S phase. Upon release, the degree of synchronization was sufficient to allow reliable analysis of regulation of RB6K mRNA and protein in the subsequent S and G2/M phases. Two hours after release, the majority of the cells were in S phase, and they subsequently shifted to G2/M phase at 6 h. The net distribution over the phases did not change during the next 2-h period, which we interpret as a reflection of the duration of the G2 phase. Then, at 10 h after release, a strong shift from G2/M to G1/G0 occurred, indicating that at this point a maximum in the number of M-phase cells was reached. At 14 h, 86% of the cells again entered the G1 phase which lasted at least another 7 h, leading to loss of synchronization (Fig. 3A). Changes in RB6K protein levels after release from the G1/early S stage were assessed by analyzing equal amounts of total cellular protein, isolated at 2-h intervals, on a Western blot with an affinity-purified anti-RB6K preparation.
FIG. 3.
Expression of RB6K mRNA and protein in synchronized cultures. (A) Synchrony of cultures. Cells were synchronized as described in Materials and Methods. During progression through the cell cycle, cultures were harvested at 2-h intervals and the distribution of the cells over the stages of the cell cycle was determined by analyzing DNA contents using flow cytometry. The x axis shows time in hours after release from the hydroxyurea block. Bars: white, G2/M population; black, S population; grey, G1/G0 population. (B) RB6K protein expression. In parallel with the experiment shown in panel A, cell extracts were prepared and total protein was determined. Equal amounts were electrophoresed on an 8% (wt/vol) polyacrylamide gel under denaturing conditions and transferred to a Protran membrane, which was subsequently probed with anti-RB6K antibodies. As a control for equal loading the blot was reprobed with antibodies against α-tubulin. (C) RB6K mRNA levels and comparison to cyclin B. In parallel with the experiment shown in panel A, RNA was isolated. Ten micrograms of total RNA was denatured, electrophoresed, and blotted to Hybond-N filters. Filters were hybridized with radiolabeled probes for RB6K and cyclin B. After autoradiography and quantification, blots were stripped and reprobed for 28S RNA to allow normalization of the signals. The x axis shows time in hours after release from the hydroxyurea block. Bars: white, RB6K; black, cyclin B. n.s., nonsynchronized.
Combination of the data from the Western blot with flow cytometric analyses shows that a slight increase of the RB6K protein level occurred at 6 and 8 h after release when the majority of the cells passed through G2. Then, at 10 h after release, the protein level showed a further increase when cells passed M phase. In early G1, at 12 and 14 h after release, a gradual decrease of protein level was observed. During the subsequent G1 phase there is a further decline of RB6K to a level comparable to that at the time of release from the hydroxyurea block, as can be inferred from the control nonsynchronized cultures that contain mainly cells randomly distributed over the G1 phase (Fig. 3A and B). The level of RB6K mRNA was measured at intervals of 2 h after release from the hydroxyurea block by probing a Northern blot of total RNA, isolated at the respective time points, with an RB6K cDNA probe. In order to show the validity of synchronization of EC-RF24 cultures and subsequent mRNA quantification as a method to assess cell cycle dependency of RB6K expression, we hybridized identical blots with a cyclin B cDNA probe. Using a comparable approach for synchronized HeLa cells, a fourfold increase of the cyclin B mRNA in G2/M populations has been reported (23). Obviously, the levels of RB6K and cyclin B mRNA showed nearly identical temporal changes in the synchronized cultures. After 20 h of hydroxyurea incubation, signal levels of both mRNAs were at a minimum. Upon release from the block, the levels of both mRNAs increased, showed a maximum at 10 h after release, and subsequently started to decrease at 12 and 14 h after release, the decline for RB6K mRNA being somewhat slower than that for cyclin B mRNA (Fig. 3C). Thus, the maximal mRNA levels for both RB6K and cyclin B occur at the time point (10 h) when the synchronized cultures are maximally enriched in cells that are in or near M phase as can be deduced from the large flux of cells from the G2/M to the G1/G0 population that was observed in flow cytometry.
Structure and promoter activity of the 5′ upstream region of the RB6K gene.
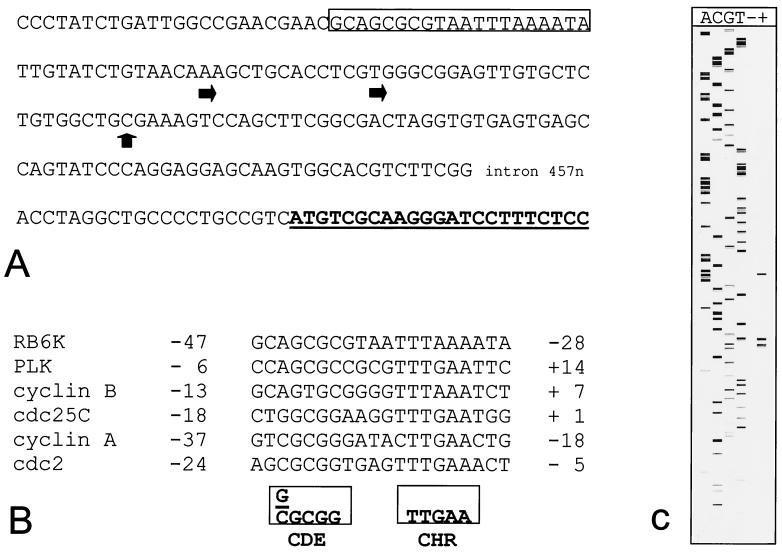
To investigate whether an increased transcriptional activity of the RB6K promoter contributes to the observed increase in RB6K mRNA levels in M phase, we isolated the 5′ upstream sequences of the RB6K gene. The 5′UTR of the RB6K cDNA (GenBank accession no. AF070672) comprised only 27 nucleotides and was therefore unlikely to represent the complete 5′UTR of the transcript. Therefore, we completed the cDNA by 5′-RACE. The longest transcript we obtained extended the 5′UTR with 64 nucleotides. BLAST searches on the high-throughput genomic sequence database revealed that RB6K cDNA sequences matched to a contig from PAC DJ0309D19, (GenBank accession no. AC004826). This contig comprises 17.3 kb upstream of our cDNA; therefore, it is conceivable that it contains RB6K promoter sequences.
By analysis of RNA from EC-RF24 cells using a primer extension assay, two potential transcription start sites were mapped respectively 25 and 38 nucleotides upstream of the longest transcript that we isolated in 5′-RACE (Fig. 4A and C). These initiation sites will be referred to as position 1 and −13, respectively.
FIG. 4.
RB6K 5′ upstream sequence. (A) The sequence shown corresponds to nucleotides 102787 to 103454 from GenBank accession no. AC004826. Indicated are transcription initiation sites (horizontal arrows), the longest cDNA isolated by 5′-RACE (vertical arrow), the open reading frame (in boldface type and underlined), and the CDE-CHR element (box). (B) Alignment of CDE-CHR element sequences from cell cycle-regulated genes. The consensus sequence as originally described for cdc2, cyclin A, and cdc25C (35) is shown in boxes. (C) Primer extension mapping of the transcription initiation site. Sequence analysis and primer extention assay were performed using a Cy5-labeled oligonucleotide primer. The reaction products were analyzed on an ALF-express sequence analyzer. Lanes A, C, G, and T contain sequence ladder. Lanes − and + contain primer extension reaction mixtures using 5 μg of total RNA isolated from EC-RF24 cells without (−) and with (+) reverse transcriptase added.
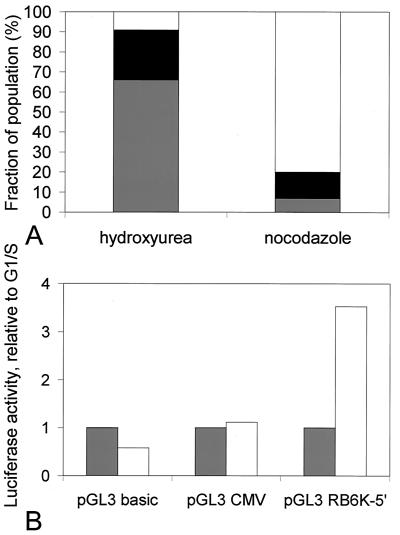
To test the upstream sequences for cell cycle-dependent promoter activity, we placed a fragment containing nucleotides −1253 to +96 into a luciferase reporter vector, pGL3-basic, and transfected the construct, pGL3RB6K-5′, into HeLa cells. As controls, the pGL3-basic vector or the pGL3 vector containing the strong, constitutive cytomegalovirus promoter was transfected. In all transfections, a pSV-β-galactosidase vector was cotransfected to allow for normalization. Twenty-four hours after transfection, cells were treated for 20 h with hydroxyurea or nocodazole to arrest them in late G1/early S phase and M phase, respectively. Cells were then either lysed and used for determination of luciferase activity or prepared for flow-cytometric analysis of the DNA content. As the half-life of luciferase protein is approximately 3 h (31), this approach allowed us to specifically compare the promoter activities of G1/S- and M-phase cells. First, the luciferase activity measured in extracts of pGL3RB6K-5′-transfected cells was typically found to be increased by almost 3 orders of magnitude compared to the pGL3-transfected controls. Second, as shown in Fig. 5, the luciferase expression driven by the cytomegalovirus promoter did not vary between M-phase-and G1/S-phase-arrested cells, whereas expression driven by the RB6K promoter in M-phase-arrested cells showed a 3.5-fold increase compared to G1/S-phase cells. Taken together, these observations show that a sequence spanning nucleotides −1253 to +96 relative to the RB6K transcription start not only provides promoter activity but also confers cell cycle-dependent variation to the expression of the luciferase reporter gene and is therefore likely to contain sequences responsible for the observed cell cycle-dependent expression of RB6K in cultured cells (Fig. 4B).
FIG. 5.
Cell cycle dependency of activity of the RB6K promoter. (A) HeLa cells were transiently cotransfected with the pGL3RB6K-5′- and pSV-β-galactosidase plasmids. Control cotransfections were performed with the pGL3 basic and pGL3CMV plasmids. Twenty-four hours after transfection, cultures were cell cycle arrested for 20 h as described in Materials and Methods. Cells were then harvested, and the distribution of the cells over the stages of the cell cycle was determined by flow cytometry. Bars: white, G2/M population; black, S population; grey, G1/G0 population. (B) In paralel with A, cells were lysed and luciferase and β-galactosidase activities were measured in the lysates. Luciferase values were normalized for β-galactosidase activities and are given relative to luciferase activity measured in G1/S (hydroxyurea)-arrested cells. Bars: grey, G1/S (hydroxyurea)-arrested cells; white, G2/M (nocodazole)-arrested cells.
RB6K plays a crucial role in cytokinesis.
To further delineate the mechanistic function of RB6K, a detailed confocal imaging was performed of cells, at various stages of mitosis, costained with an anti-RB6K antibody (red) and an antitubulin antibody (green). It was observed that a dominant nuclear staining appears at prophase, while in metaphase the staining becomes dispersed throughout the cytoplasm surrounding the mitotic spindle. In the subsequent stages of anaphase and telophase, concomittant with the onset of cytokinesis, RB6K accumulates throughout the equatorial region of the cell, thus colocalizing with the spindle midzone and the area of the contracting cleavage furrow. In the end stage of cytokinesis, RB6K becomes sharply concentrated on the midbodies (Fig. 6A). RB6K was previously shown to interact with the GTPase Rab6 in interphase cells (6). Figure 6B shows HeLa cells stably expressing Rab6-GFP (34) in early anaphase where RB6K (red) becomes concentrated in the equatorial zone, whereas Rab6-GFP (green) is dispersed throughout the cytoplasm, localizing to dispersed Golgi fragments (28). Cells in telophase show a clear concentration of RB6K on the forming cytoplasmic bridge, and only a small fraction of RB6K colocalizes with Rab6 at the centrosomes of the two daughter cells (Fig. 6B [arrows]), where the Golgi stacks are being re-formed. Rab6 also localizes adjacent to the cytoplasmic bridge (Fig. 6B) as do other Golgi markers, such as mannosidase II or galactosyl-transferase (data not shown). From the above data, it can be inferred that most of the RB6K in M-phase cells shows a behavior that is likely not compatible with a role related to the Golgi apparatus.
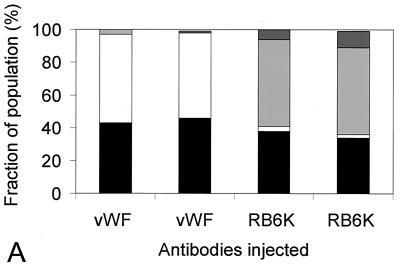
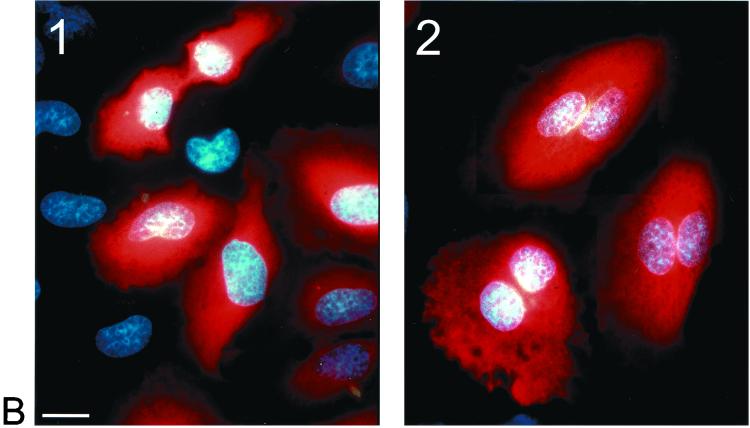
To test the functional significance of the observed subcellular localization of RB6K, we performed microinjection of synchronized cell cultures with anti-RB6K antibodies. For these studies we used HeLa cells as the passage of these cells through the cell cycle after control injections with buffer or inert antibody preparations was less affected than that of EC-RF24 cells. Control immunofluorescence microscopy showed that the RB6K expression patterns in HeLa cells during the cell cycle were identical to those observed in EC-RF24 cells (Fig. 6B). Cells were grown on grids to facilitate evaluation of the effects of injection. Twenty hours after seeding, near-confluent cultures were incubated for 20 h with 2 mM hydroxyurea. Cells were then washed to completely remove the hydroxyurea, released in fresh medium, and injected in the cytoplasm with an affinity-purified polyclonal anti-RB6K antibody preparation. As a control, cells were injected with identical amounts of affinity-purified polyclonal antibodies against von Willebrand factor, a secretory protein that does not have a role in the cell cycle. The injections were either clustered (all cells in one grid, approximately 100 cells) or scattered (9 cells per grid), allowing the examination of the phenotype and a possible effect on mitosis, respectively. Following microinjection, the cells were grown for another 20 h, in principle allowing at least one passage through the cell cycle. The grids were then fixed and the microinjected cells were detected by staining the injected antibodies with a Cy3-coupled anti-rabbit Ig preparation. Using immunofluorescence microscopy the grids were then screened for the following categories of injected cells: (i) single cells that were mononucleate and therefore had not passed mitosis, (ii) pairs of cells that were mononucleate, indicating successful completion of mitosis, and (iii) any phenotype indicative for arrest in M phase. Figure 7A shows that injection of anti-RB6K antibodies specifically and reproducibly results in cells showing a double nucleus and an increased cell size (Fig. 7B). This phenotype shows that injection of anti-RB6K antibodies results in a failure of cytokinesis. Mitosis is not disturbed by the microinjected anti-RB6K antibodies before completion of anaphase A, as can be inferred from the presence of two apparently normal nuclei. RB6K-injected cells that fail to undergo cytokinesis typically show close spacing of the two nuclei and an apparent lack of a functional cleavage furrow, suggesting that the RB6K antibodies interfere with processes during anaphase B or cytokinesis that are essential for completion of M phase.
FIG. 7.
Effects of microinjection of anti-RB6K antibodies on the cell cycle. HeLa cells were synchronized and injected as described in Materials and Methods with either affinity-purified anti-RB6K antibodies or, as a negative control, anti-von Willebrand factor antibodies (vWF) at identical concentrations. Injected cells were visualized after 20 h by immunofluorescent staining of the injected antibodies. (A) Result of four independent injection procedures. In each procedure 9 cells per grid were injected with a total of approximately 100 cells (scattered protocol). In the injected population, frequencies of occurrence of the following events were counted and expressed as percentage of the total number of immunofluorescence-positive cells: (i) single cells, no signs of mitosis (black bars); (ii) doubled cells, after mitosis (white bars); (iii) cells containing double nuclei, failure of cytokinesis (light grey bars); (iv) cells in any stage of mitosis (dark grey bars). (B) Result of clustered injection with anti-von Willebrand factor polyclonal antibodies (panel 1) and affinity-purified anti-RB6K polyclonal antibodies (panel 2). Bar, 20 μm.
DISCUSSION
The novel human protein RB6K is shown to be a mitotic KLP that has a distinct function in the final stages of the cell cycle. Its expression is highly regulated to peak during M phase, with low intracellular mRNA and protein concentrations during G1 and S phase (Fig. 2 and 3). This is highly similar to the expression kinetics of cyclin B, a protein of which the expression has been extensively studied in relation to regulation of the cell cycle (23, 24). The observed fluctuations of the mRNA levels are at least in part based on cell cycle-dependent promoter regulation (Fig. 4 and 5). Similar to the cell cycle-regulated promoters of cdc25C, cyclin A, cdc2, and plk genes (18, 32, 37), the RB6K promoter contains a CDE-CHR element (Fig. 4B) that is known to induce repression of transcription during G1 (32, 37). In contrast to typical cell cycle-regulatory proteins of which the intracellular concentrations are regulated by ubiquitin-mediated proteolysis (12, 13, 36), RB6K protein shows a rather gradual decline that continues in early G1 (Fig. 3B). Indeed, no consensus sequences for ubiquitination were found, and no changes in molecular weight indicative of ubiquitination were observed (Fig. 3B and data not shown). The importance of correctly tuned de novo synthesis and proteolysis of RB6K is illustrated by the finding that constitutive overexpression of RB6K in interphase cells leads to cell death, probably caused by a bundling of interphase microtubuli (Fig. 2C, panels 3 and 4). This microtubule binding was not observed for the endogenous RB6K in interphase cells. Therefore, incorrectly regulated expression of RB6K might directly interfere with interphase microtubule function. The observed tight expression regulation of RB6K during the cell cycle represents a means by which the cell can adapt its transport capacity to the specific requirements of the various stages of the cell cycle.
Human RB6K showed 91% homology and 86% identity to mouse RB6K (11) and was therefore considered to be its human equivalent. Murine RB6K was initially isolated and characterized as a protein tightly binding the Golgi-localized GTPase Rab6. Overexpression in HeLa cells of GFP-tagged murine RB6K caused dispersion of the Golgi apparatus, suggesting a role for murine RB6K in Golgi dynamics (6). Like murine RB6K, the human RB6K is able to bind Rab6 in a two-hybrid assay (A. Echard and B. Goud, unpublished results) and is localized to the Golgi apparatus in interphase cells (Fig. 6A). Our study, however, also covered M phase, during which we found RB6K to be expressed at levels considerably higher than that in interphase cells. Concomitantly, RB6K no longer localizes exclusively to the Golgi but appears in the prophase nucleus. This localization is in good agreement with a PSORT-II prediction (60.9% nuclear versus 17.4% cytoplasmic). For MKLP-1, it was suggested that sequestration in the nucleus prevents undesirable binding to the interphase microtubuli and only after dispersal of the nuclear envelope after prophase the KLP is released and allowed to interact with the then-formed mitotic spindle (21). For RB6K, an explanation should concern both the function of RB6K in interphase Golgi and the function in M phase. The relatively small RB6K pool on the interphase Golgi is involved in retrograde transport and is regulated by the effector GTPase Rab6. However, Fig. 6B shows that localization of most of the M phase-induced RB6K is independent of Rab6, which seemingly contradicts a function related to the mitotic Golgi apparatus (28). Rather, taking into consideration the fact that several proteins that are critically involved in cytokinesis initially localize to the prophase nucleus (29, 30), it can be hypothesized that localization to the nucleus of the RB6K pool that is synthesized at the onset of M phase is instrumental in binding cargo and/or modulation of RB6K function by regulatory proteins. This hypothesis will require an unbiased search for proteins that interact with RB6K in various stages of the cell cycle.
Concomitant with the onset of cytokinesis during anaphase, RB6K concentrates in the equatorial zone of the cell. Consistent with this localization, effects of interference with RB6K function by injection of specific antibodies are restricted to cytokinesis. Phylogenetic analysis of the motor domain sequences has been used to classify most of the KLPs into 8 to 10 subfamilies that, in addition to having related motor domain sequences, usually have a related domain structure and show similarity with regard to motility behavior and cellular functions (2, 10; website of Greene and Henikoff). Detailed information on the function of members of the MKLP1 family has been derived from mutant analysis in the case of the Drosophila melanogaster gene pavarotti (1) and the C. elegans gene zen-4 (25) or from antibody microinjection studies of the MKLP-1 (CHO1 antigen) protein in mammalian cells (21). When antibodies raised against the CHO1 protein are injected in mammalian cells before onset of anaphase, cells are arrested in metaphase, showing partially impaired congression of chromosomes and a disorganized spindle, indicating that the antibodies interfere in a stage preceding anaphase A. Injection after onset of anaphase has little effect on completion of cell division (21). In contrast, RB6K antibody microinjection does not affect mitosis before anaphase B, indicating that its function temporally follows that of MKLP-1 rather than being redundant with it. As RB6K, like the other MKLP-1 family members, has the ability to cross-link antiparallel microtubules, our microinjection results might indicate interference with dynamics of midzone microtubules in anaphase B. It should be noted, however, that RB6K is only distantly related to MKLP-1, PAV-KLP, and ZEN-4, as sequence homology is confined to the motor domain.
Presently, the combined databases do not contain KLPs from other species that could be considered to be functional equivalents of human and mouse RB6K by showing even distant homology to the C-terminal 350 residues. Still, the phenotype resulting from microinjection of anti-RB6K antibodies is comparable to that of zen-4, polo, and pav mutants (1, 4, 25), suggesting that RB6K may fulfill a separate but comparable role in cytokinesis. In mammalian cells, Polo-like kinase has been shown to colocalize and interact with MKLP-1 in vivo and to phosphorylate MKLP-1 in vitro (16). ZEN-4 and PAV-KLP, show 50 and 58% overall homology to MKLP-1, including the C terminus that is supposed to be involved in cargo binding. In both cases, it was speculated that, in addition to a role in spindle function, the KLPs may also contribute to cytokinesis by transporting Polo-like kinase and Polo, respectively (1, 25). Indeed, several studies, both in Drosophila and in cultured cells (5, 8), have established that the spindle midzone provides stimuli for cytokinesis. Although the underlying molecular mechanisms are only partially understood, relocation of several proteins along microtubuli seems to be involved (5, 33). Based on these observations, we can speculate that RB6K is possibly involved in transport of one of the many essential components of the cleavage furrow that are currently emerging (4, 9, 19, 29, 30).
The failure of cytokinesis that we observed after anti-RB6K antibody microinjection is compatible with both a function for RB6K on the spindle midzone and a function more related to the cleavage furrow, since both processes are intimately linked, as evidenced by Giansanti et al. (8). These authors showed the cooperative interaction between the contractile ring and the spindle midzone, and if either of these stuctures is perturbed, the assembly of the other is disrupted. A more detailed understanding of RB6K function therefore requires knowledge of RB6K interacting proteins. At present, our data provide evidence for a tight cell cycle-regulated expression of RB6K and show that it is an essential, nonredundant component of the cell cycle that is required for successful completion of cytokinesis.
ACKNOWLEDGMENTS
We thank the members of the CMO department of the Academic Medical Center, University of Amsterdam, for their assistance with flow cytometric analyses.
This work was supported by Molecular Cardiology grant M93.007 from the Netherlands Heart Foundation, The Hague.
REFERENCES
- 1.Adams R R, Tavares A A M, Salzberg A, Bellen H J, Glover D M. pavarotti encodes a kinesin-like protein required to organise the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton N R, Goldstein L S B. Going mobile: microtubule motors and chromosome segregation. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown K D, Coulson R M R, Yen T J, Cleveland D W. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J Cell Biol. 1994;125:1303–1312. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmena M, Riparbelli M G, Minestrini G, Tavares A M, Adams R, Callaini G, Glover D M. Drosophila polo kinase is required for cytokinesis. J Cell Biol. 1998;143:659–671. doi: 10.1083/jcb.143.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao L G, Wang Y L. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echard A, Jollivet F, Martinez O, Lacapère J-J, Rouselet A, Janoueix-Lerosey I, Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 7.Fontijn R D, Hop C, Brinkman H-J, Slater R, Westerveld A, van Mourik J A, Pannekoek H. Maintenance of vascular endothelial cell-specific properties after immortalisation with an amphotrophic replication-deficient retrovirus containing human papillomavirus E6/E7 DNA. Exp Cell Res. 1995;216:199–207. doi: 10.1006/excr.1995.1025. [DOI] [PubMed] [Google Scholar]
- 8.Giansanti M G, Bonaccorsi S, Williams B, Williams E V, Santolamazza C, Goldberg M L, Gatti M. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glover D M, Hagan I M, Tavares A A M. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein L S B, Philp A V. The road less traveled: emerging principles of kinesin motor utilisation. Annu Rev Cell Dev Biol. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 11.Horrevoets A J G, Fontijn R D, van Zonneveld A J, de Vries C M, ten Cate J W, Pannekoek H. Vascular endothelial cells that are responsive to tumor necrosis factor-α in vitro are expressed in atherosclerotic lesions, including inhibtor of apoptosis protein-1, stannin and two novel genes. Blood. 1999;93:3418–3431. [PubMed] [Google Scholar]
- 12.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 13.Koepp D M, Harper J W, Elledge S J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Cleavage of structural proteins during tht assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Lai F, Fernald A A, Zhao N, Le Beau M M. cDNA cloning, expression pattern, genomic structure and chromosomal localisation of RAB6KIFL, a human kinesin-like gene. Gene. 2000;248:117–125. doi: 10.1016/s0378-1119(00)00135-9. [DOI] [PubMed] [Google Scholar]
- 16.Lee K S, Yuan Y-L O, Kuriyama R, Erikson R L. PLK is an M-phase specific protein kinase and interacts with a kinesin-like protein, CHO/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennon G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 18.Lucibello F C, Truss M, Zwicker J, Ehlert F, Beato M, Müller R. Periodic cdc25C transcription is mediated by a novel cell cycle-regulated repressor element (CDE) EMBO J. 1995;14:132–142. doi: 10.1002/j.1460-2075.1995.tb06983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 20.Moore J D, Endow S A. Kinesin proteins: a phylum of motors for microtubule-based mobility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- 21.Nislow C, Sellito C, Kuriyama R, McIntosh J R. A monoclonal antibody to a mitotic microtubule-associated protein blocks mitotic progression. J Cell Biol. 1990;111:511–522. doi: 10.1083/jcb.111.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nislow C, Lombillo V A, Kuriyama R, McIntosh J R. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- 23.Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 24.Pines J. Cyclin from sea urchins to HeLas: making the human cell cycle. Biochem Soc Trans. 1996;24:15–33. doi: 10.1042/bst0240015. [DOI] [PubMed] [Google Scholar]
- 25.Raich W B, Moran A N, Rothman J H, Hardin J. Cytokinesis and midzone microtubule organisation in Caenorhabditis elegans require the kinesin-like protein zen-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch F T, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- 27.Schaar B T, Chan G K T, Maddox P, Salmon E D, Yen T J. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shima D T, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: visualisation of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–927. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terada Y, Tatsuka M, Suzuki F, Yasuda Y, Fujita S, Otsu M. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J, Hayes L, Lloyd D. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene. 1991;103:171–177. doi: 10.1016/0378-1119(91)90270-l. [DOI] [PubMed] [Google Scholar]
- 32.Uchiumi T, Longo D L, Ferris D K. Cell cycle regulation of the human Polo-like kinase (PLK) promotor. J Biol Chem. 1997;272:9166–9174. doi: 10.1074/jbc.272.14.9166. [DOI] [PubMed] [Google Scholar]
- 33.Wheatly S P, O'Connel C B, Wang Y-L. Inhibition of chromosomal separation provides insights into cleavage furrow stimulation in cultured epithelial cells. Mol Biol Cell. 1998;9:2173–2148. doi: 10.1091/mbc.9.8.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White J, Johannes L, Mallard F, Girod A, Grill S, Reinsh S, Kellen P, Tzschaschel B, Echard A, Goud B, Stelzer E. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J Cell Biol. 1999;147:1–17. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkinson D, Green J. In situ hybridisation and the three-dimensional reconstruction of serial sections. In: Copp A J, Cockroft D L, editors. Postimplantation mammalian embryos: a practical approach. Oxford, United Kingdom: Oxford University Press; 1990. pp. 155–171. [Google Scholar]
- 36.Zachariae W. Progression into and out of mitosis. Curr Opin Cell Biol. 1999;11:708–716. doi: 10.1016/s0955-0674(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 37.Zwicker J, Lucibello F C, Wolfraim L A, Gross C, Truss M, Engeland K, Müller R. Cell cycle regulation of the cyclin A, cdc25C, and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]