Highlights
-
•
122 (20.4%) of 599 carbapenem-resistant Enterobacterales (CRE) isolates carried blaKPC genes.
-
•
Predominance of ST15 K. pneumoniae, whereas E. coli presented more diverse sequence types.
-
•
blaKPC-bearing plasmids were diverse in size.
-
•
Three different models of genetic context of blaKPC-2.
-
•
Hypothesis of circulation of resistant bacteria and transmission among hospitals.
Keywords: Carbapenem resistance, KPC, Enterobacterales, Vietnam
Abstract
Objectives
: The incidence of carbapenem resistance among nosocomial Gram-negative bacteria in Vietnam is high and increasing, including among Enterobacterales. In this study, we assessed the presence of one of the main carbapenemase genes, blaKPC, among carbapenem-resistant Enterobacterales (CRE) from four large hospitals in Hanoi, Vietnam, between 2010 and 2015, and described their key molecular characteristics.
Methods
: KPC-producing Enterobacterales were detected using conventional PCR and were further analysed using S1 nuclease pulsed-field gel electrophoresis (S1-PFGE), Southern blotting and whole-genome sequencing (WGS) for sequence typing and genetic characterisation.
Results
: blaKPC genes were detected in 122 (20.4%) of 599 CRE isolates. blaKPC-carrying plasmids were diverse in size. Klebsiella pneumoniae harbouring blaKPC genes belonged to ST15 and ST11, whereas KPC-producing Escherichia coli showed more diverse sequence types including ST3580, ST448, ST709 and ST405. Genotypic relationships supported the hypothesis of circulation of a population of ‘resident’ resistant bacteria in one hospital through the years and of transmission among these hospitals via patient transfer. WGS results revealed co-carriage of several other antimicrobial resistance genes and three different genetic contexts of blaKPC-2. Among these, the combination of ISEcp1–blaCTX-M and ISKpn27–blaKPC–ΔISKpn6 on the same plasmid is reported for the first time.
Conclusion
: We describe the dissemination of blaKPC-expressing Enterobacterales in four large hospitals in Hanoi, Vietnam, since 2010, which may have started earlier, along with their resistance patterns, sequence types, genotypic relationship, plasmid sizes and genetic context, thereby contributing to the overall picture of the antimicrobial resistance situation in Enterobacterales in Vietnam.
1. Introduction
The incidence of carbapenem resistance among nosocomial pathogens in Vietnam is high and increasing, especially among Escherichia coli and Gram-negative ‘ESKAPE’ organisms (Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.). Data from a nationwide hospital surveillance network in 2016–2017 showed that rates of carbapenem resistance among K. pneumoniae, E. coli and Enterobacter spp. were 29%, 11% and 27%, respectively [1]. A recent study from Vietnam including more than 2200 patients admitted to 12 hospitals throughout the country during 2017 and 2018 reported that 52% of patients were colonised with carbapenem-resistant Enterobacterales (CRE) [2]. Since the first NDM-1-producing K. pneumoniae in Vietnam was isolated from the urinary tract of a 62-year-old hospitalised patient in 2010 [3], most class A, B and D carbapenemases in Enterobacterales have been reported from Vietnam [4], [5], [6]. In the southern part of Vietnam, carbapenem-resistant K. pneumoniae clinical isolates producing various carbapenemases such as KPC-2, NDM-1, NDM-4 and OXA-48 have been described [6,7]. Studies from three hospitals in Hanoi, in the northern part of Vietnam, detected diverse variants of carbapenemase genes such as KPC-2, KPC-3, KPC-4, NDM-1, IMP-4, IMP-79, VIM-1 and OXA-48 in Enterobacterales [4,5,8,9]. Data on the molecular characteristics of carbapenemases from carbapenemase-producing Enterobacterales (CPE) in clinical isolates in Vietnam, a lower-middle income country with a high and increasing burden of antimicrobial resistance and hospital-acquired infections, are still sparse. Here we present the phenotypic and molecular characteristics of KPC-producing CPE isolates from four major hospitals in Hanoi between 2010 and 2015 in order to gain a better understanding of the circulation of CPE in Vietnam and to compare this with local, regional and global data to add to the current knowledge base. Our results will contribute to outline a larger picture of CPE in Vietnam and will serve as important scientific information for government action plans on antibiotic resistance control.
2. Materials and methods
2.1. Study sites and sample collection
We prospectively collected CRE isolates from four large hospitals including Saint Paul (A), Thanh Nhan (B), Viet Duc (C) and 108 Military Central Hospital (D) located in the centre of Hanoi, Vietnam. All four are public hospitals; three are general hospitals (A, B and D) and C is a specialised hospital for surgery. A and B are city hospitals with a 600-bed capacity, while C and D are central hospitals with a greater than 1000-bed capacity. Demographic and basic clinical information of patients whose specimens were positive for CRE were collected from clinical notes and included age, sex, date of admission, clinical diagnosis, origin of collected sample, date of sample collection and culture results. Treatment and clinical outcome data were not available for this study.
CRE isolated from clinical specimens were tested for antimicrobial susceptibility at the four sites by the disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines [10,11]. Microbiology laboratories in the four hospitals were requested to collect and send all bacterial isolates resistant to at least one carbapenem to the National Institute of Hygiene and Epidemiology for further characterisation (n = 599, including 179 isolates from hospital A, 87 from hospital B, 95 from hospital C and 238 from hospital D).
2.2. Antimicrobial susceptibility testing and detection of antimicrobial resistance genes
Minimum inhibitory concentrations (MICs) were determined centrally by the agar dilution method for imipenem, meropenem, cefotaxime, ceftazidime and ciprofloxacin and by microdilution for colistin (Sigma-Aldrich) according to CLSI and European Committee on Antimicrobial Susceptibility testing (EUCAST) guidelines [11,12]. Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as control strains.
Collected isolates were screened for four common carbapenemase genes, including blaKPC, blaNDM-1, blaIMP and blaOXA-48, as well as three other common β-lactamase-encoding genes (blaTEM, blaSHV and blaCTX-M) as described previously [4]. Resulting amplicons were sequenced using conventional Sanger sequencing.
2.3. Multilocus sequence typing (MLST)
MLST was done using PubMLST for all blaKPC-positive isolates (n = 122) [13]. Briefly, seven housekeeping genes were amplified by PCR, sequenced and compared with the sequences submitted to the MLST database to determine the sequence type (ST).
2.4. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting
blaKPC-positive isolates were further analysed using S1-PFGE with XbaI-digested Salmonella enterica serovar Braenderup H9812 as a reference molecular weight marker on a CHEF-DR III platform (Bio-Rad, Hercules, CA, USA), followed by membrane transfer and hybridisation with labelled probes in an HL-2000 HybriLinker Hybridization Oven (UPV, Germany). Autoradiograms were visualised according to standard Southern blotting protocols [14].
2.5. Whole-genome sequencing (WGS) and characterisation of the genetic environment of blaKPC genes
A subset of blaKPC-carrying CPE isolates (n = 69) was selected for further analysis including representatives of different hospitals, collection years, departments and sample types. Genomic DNA libraries of selected strains were prepared for WGS using a Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA, USA) according to the manufacturer's instructions. Then, 300-bp paired-end sequencing was performed on an Illumina MiSeq platform (MiSeq Reagent Kit v3; 600 cycles). Raw sequence reads were de novo assembled into contigs using SPAdes v.3.9.0 with pre-defined Kmers set. Antimicrobial resistance genes were identified using ResFinder v.2.1, and MLST profiles were generated using the platform of the Center for Genomic Epidemiology, Technical University of Denmark, coupled with the PubMLST.org database.
Plasmids were identified and typed using PlasmidFinder v.1.3. Structures of genetic contexts surrounding blaKPC genes were mapped using ISfinder, BLASTN v.2.6.0 and genoPlotR v.0.8.3.
Phylogenetic trees based on the core genome single nucleotide polymorphisms (SNPs) were constructed from WGS data of the 60 blaKPC-carrying K. pneumoniae and 9 blaKPC-carrying E. coli isolates using Parsnp 1.2 and IQ-TREE 2.0 [15,16].
2.6. Statistical methods
Isolates and patient data were analysed in Microsoft Excel 2017 (Microsoft Corp., Redmond, WA, USA) using descriptive statistics as appropriate.
3. Results
3.1. Distribution of blaKPC-positive carbapenemase-producing Enterobacterales (CPE) isolates among hospitals and their phenotypes
During the study period, 599 CRE were collected in four hospitals, comprising 64 isolates in 2010, 97 isolates in 2011, 64 isolates in 2012, 68 isolates in 2013, 149 isolates in 2014 and 157 isolates in 2015. Among these, K. pneumoniae (n = 305) and E. coli (n = 186) accounted for the majority of isolates; other bacteria included other Klebsiella spp. (n = 54), Enterobacter spp. (n = 29) and Citrobacter spp. (n = 25).
Of the 599 CRE isolates, 122 (20.4%) were positive for blaKPC, including 13 E. coli (10.7%) and 109 K. pneumoniae (89.3%). No blaKPC gene was detected in the other bacteria. Hospital A contributed the largest number of blaKPC-positive isolates (n = 101, accounting for 82.8%), followed by hospital B (n = 9; 7.4%), hospital D (n = 8; 6.6%) and hospital C (n = 4; 3.3%) (Table 1). The first detected blaKPC-positive bacterium was K. pneumoniae, isolated from bronchial fluid of a ventilated patient in the intensive care unit (ICU) of hospital B on 16 January 2010.
Table 1.
Distribution of blaKPC-positive Enterobacterales isolates among hospitals in Hanoi, Vietnam, from 2010–2015.
| Hospital | No. of isolates | Total [no. (%)] | |||||
|---|---|---|---|---|---|---|---|
| 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | ||
| A | 5 | 30 | 18 | 18 | 30 | 0 | 101 (82.8) |
| B | 6 | 0 | 0 | 2 | 1 | 0 | 9 (7.4) |
| C | 2 | 0 | 0 | 0 | 1 | 1 | 4 (3.3) |
| D | 0 | 0 | 0 | 0 | 2 | 6 | 8 (6.6) |
| Total | 13 | 30 | 18 | 20 | 34 | 7 | 122 (100) |
From the clinical information collected, the clinical characteristics of blaKPC-positive isolates (n = 122) were revealed after de-duplication. The highest proportion was from the neonatal ICU (n = 97; 79.5%), followed by the ICU (n = 15; 12.3%), tuberculosis (TB) and lung diseases (n = 4; 3.3%) and four other departments (Table 2). The most dominant sample type among blaKPC-positive isolates was bronchial fluid (n = 92; 75.4%). Other types of sample included blood (n = 18; 14.8%), sputum (n = 6; 4.9%), urine (n = 4; 3.3%), pleural fluid (n = 1; 0.8%) and abdominal fluid (n = 1; 0.8%).
Table 2.
Clinical and genotypic characteristics of blaKPC-positive Enterobacterales isolates (n = 122) among hospitals (A–D) in Hanoi, Vietnam, from 2010–2015.
| Characteristic | No. of isolates | Total [no. (%)] | ||||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| Department | Neonatal ICU | 95 | 2 | 0 | 0 | 97 (79.5) |
| ICU | 6 | 7 | 2 | 0 | 15 (12.3) | |
| TB and lung diseases | 0 | 0 | 0 | 4 | 4 (3.3) | |
| Urology | 0 | 0 | 2 | 0 | 2 (1.6) | |
| Neurology | 0 | 0 | 0 | 2 | 2 (1.6) | |
| Cardiology | 0 | 0 | 0 | 1 | 1 (0.8) | |
| Thoracic surgery | 0 | 0 | 0 | 1 | 1 (0.8) | |
| Sample type | Bronchial fluid | 86 | 5 | 1 | 0 | 92 (75.4) |
| Blood | 15 | 1 | 0 | 2 | 18 (14.8) | |
| Sputum | 0 | 3 | 0 | 3 | 6 (4.9) | |
| Urine | 0 | 0 | 2 | 2 | 4 (3.3) | |
| Pleural fluid | 0 | 0 | 0 | 1 | 1 (0.8) | |
| Abdominal fluid | 0 | 0 | 1 | 0 | 1 (0.8) | |
| Carbapenemase-encoding gene | blaNDM-1 | 8 | 1 | 1 | 0 | 10 (8.2) |
| blaOXA-48 | 1 | 0 | 0 | 0 | 1 (0.8) | |
| blaIMP | 0 | 0 | 0 | 0 | 0 (0.0) | |
| ESBL-encoding gene | blaTEM | 99 | 7 | 4 | 6 | 116 (95.1) |
| blaSHV | 90 | 9 | 2 | 8 | 109 (89.3) | |
| blaCTX-M | 33 | 8 | 2 | 2 | 45 (36.9) | |
ICU, intensive care unit; TB, tuberculosis; ESBL, extended-spectrum β-lactamase.
Based on sequencing results, 67 (97.1%) of a selected subset of 69 blaKPC-positive isolates harboured blaKPC-2, whereas the other 2 isolates harboured blaKPC-12 and blaKPC-14, both from hospital D. No blaIMP gene was detected and one isolate harboured blaOXA-48. Ten isolates (six K. pneumoniae and four E. coli) co-carried blaKPC and blaNDM-1 genes (Table 2). Almost all blaKPC-harbouring strains carried blaTEM and blaSHV genes [116 (95.1%) and 109 (89.3%), respectively], whilst blaCTX-M genes were less common among these isolates [45 (36.9%)].
Results of MIC testing (Table 3) revealed that 110 (90.2%) and 98 (80.3%) isolates were resistant to third-generation cephalosporins (ceftazidime and cefotaxime, respectively) and 111 (91.0%) isolates were resistant to fluoroquinolones (ciprofloxacin). Moreover, 80 (65.6%) and 79 (64.8%) isolates were resistant to carbapenems (imipenem and meropenem, respectively) and 29 (23.8%) were resistant to colistin. All E. coli were susceptible to meropenem and colistin. No significant difference was detected in MICs among isolates carrying only blaKPC genes and those co-carrying blaKPC and other β-lactamase genes.
Table 3.
Minimum inhibitory concentrations (MICs) of blaKPC-positive Enterobacterales.
| Species | Hospital (no. of isolates) | Resistant phenotype MIC in μg/mL (no. of isolates) | |||||
|---|---|---|---|---|---|---|---|
| IPM | MEM | CAZ | CTX | CIP | COL | ||
| Klebsiella pneumoniae (n = 109) | A (n = 90) | ≥8 (63) | ≥8 (69) | ≥16 (83) | ≥16 (75) | ≥16 (83) | ≥8 (24) |
| B (n = 9) | ≥16 (4) | ≥16 (3) | ≥64 (7) | ≥16 (5) | ≥8 (7) | ≥4 (2) | |
| C (n = 2) | >64 (1) | >64 (1) | ≥256 (1) | ≥512 (1) | ≥128 (1) | ≥8 (1) | |
| D (n = 8) | ≥4 (4) | ≥4 (6) | ≥64 (8) | ≥128 (7) | ≥64 (8) | ≥16 (2) | |
| Escherichia coli (n = 13) | A (n = 11) | >64 (6) | – | ≥128 (9) | ≥8 (8) | ≥32 (10) | – |
| C (n = 2) | ≥4 (2) | – | >512 (2) | ≥512 (2) | ≥16 (2) | – | |
| Total [no. (%) of isolates] | n = 122 | 80 (65.6) | 79 (64.8) | 110 (90.2) | 98 (80.3) | 111 (91.0) | 29 (23.8) |
MIC, minimum inhibitory concentration; IPM, imipenem; MEM, meropenem; CAZ, ceftazidime; CTX, cefotaxime; CIP, ciprofloxacin; COL, colistin.
3.2. Antimicrobial resistance gene profile, sequence typing and genotypic relationship
WGS data revealed the resistance profile of blaKPC-harbouring isolates to a wide range of antibiotics (Table 4). All isolates carried genes conferring resistance to at least three different antibiotic categories, but with considerable variation in the resistance-conferring elements carried. Indeed, two resistance genes (fosA and dfrA) were present in all K. pneumoniae isolates but not all E. coli isolates. On the other hand, the mph(A) gene was observed in all E. coli but only a few K. pneumoniae isolates (Fig. 1, Fig. 2). Notably, one K. pneumoniae isolate carried resistance genes for all 10 investigated antibiotic categories. All 60 (100%) blaKPC-carrying K. pneumoniae isolates carried resistance genes against β-lactams, fosfomycin and trimethoprim. Resistance elements to aminoglycosides and quinolones were also detected in high proportions (≥95%) among K. pneumoniae isolates; however, few strains had genotypic resistance to macrolides. In E. coli, all nine (100%) blaKPC-carrying isolates carried genes encoding resistance to aminoglycosides, β-lactams, tetracyclines and macrolides. One E. coli isolate carried genes conferring resistance to quinolones.
Table 4.
Antimicrobial resistance gene profile of a selected subset of blaKPC-positive Enterobacterales.
| Species | Antibiotic categoriesa |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino-glycosides |
β-Lactams |
Fosfomycin |
Phenicols |
Rifampicin |
Sulfonamides |
Tetracyclines |
Trimethoprim |
Quinolones |
Macrolides |
||||||||||||
| + | – | + | – | + | – | + | – | + | – | + | – | + | – | + | – | + | – | + | – | ||
| Klebsiella pneumoniae (n = 60) | n | 57 | 3 | 60 | 0 | 60 | 0 | 5 | 55 | 16 | 44 | 23 | 37 | 15 | 45 | 60 | 0 | 59 | 1 | 6 | 54 |
| % | 95.0 | 5.0 | 100.0 | 0.0 | 100.0 | 0.0 | 8.3 | 91.7 | 26.7 | 73.3 | 38.3 | 61.7 | 25.0 | 75.0 | 100.0 | 0.0 | 98.3 | 1.7 | 10.0 | 90.0 | |
| Escherichia coli (n = 9) | n | 9 | 0 | 9 | 0 | 6 | 3 | 3 | 6 | 3 | 6 | 7 | 2 | 9 | 0 | 3 | 6 | 1 | 8 | 9 | 0 |
| % | 100.0 | 0.0 | 100.0 | 0.0 | 66.7 | 33.3 | 33.3 | 66.7 | 33.3 | 66.7 | 77.8 | 22.2 | 100.0 | 0.0 | 33.3 | 66.7 | 11.1 | 88.9 | 100.0 | 0.0 | |
+ indicates that the bacteria carried at least one resistance gene for the antibiotic group; – indicates that the bacteria did not carry any resistance gene for the antibiotic group.
Fig. 1.
Core genome phylogenetic tree of blaKPC-carrying Klebsiella pneumoniae isolates. Presence of resistance genes was colour coded by different antibiotic categories, while grey blocks showed no corresponding resistance genes were detected.
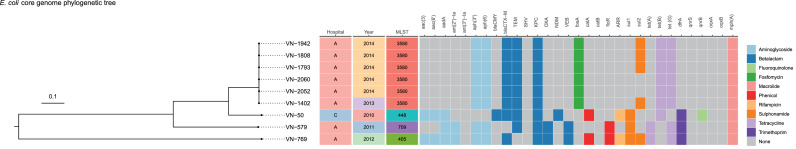
Fig. 2.
Core genome phylogenetic tree of blaKPC-carrying Escherichia coli isolates. Presence of resistance genes was colour coded by different antibiotic categories, while grey blocks showed no corresponding resistance genes were detected.
Klebsiella pneumoniae harbouring blaKPC genes belonged to ST15 and ST11 (Table 5). Notably, while ST15 was predominant, ST11 was observed only in two isolates, both from hospital D (Table 5; Fig. 1).
Table 5.
Key epidemiological information and sequence types (ST) of blaKPC carriers in the four hospitals (A–D)
| Strain ID | Species | Hospital | Collection year | Department | Sample type | MLST | Carbapenemase genes | |||
|---|---|---|---|---|---|---|---|---|---|---|
| KPC | OXA-48 | IMP | NDM-1 | |||||||
| 64 | Klebsiella pneumoniae | A | 2010 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 66 | K. pneumoniae | A | 2010 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 67 | K. pneumoniae | A | 2010 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 84 | K. pneumoniae | A | 2010 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 124 | K. pneumoniae | A | 2010 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 151 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 152 | K. pneumoniae | A | 2011 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 273 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 274 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 278 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 263 | K. pneumoniae | A | 2011 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 265 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 297 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 294 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 298 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 322 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 326 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 328 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 365 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 342 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 348 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | + |
| 389 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 408 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 410 | K. pneumoniae | A | 2011 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 447 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 451 | K. pneumoniae | A | 2011 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 452 | K. pneumoniae | A | 2011 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 454 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 455 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 457 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 437 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 529 | K. pneumoniae | A | 2011 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 580 | K. pneumoniae | A | 2011 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 676 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 681 | K. pneumoniae | A | 2012 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 683 | K. pneumoniae | A | 2012 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 686 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 724 | K. pneumoniae | A | 2012 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 725 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 729 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 732 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 847 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 869 | K. pneumoniae | A | 2012 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 914 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 980 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1040 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1045 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1048 | K. pneumoniae | A | 2012 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 1070 | K. pneumoniae | A | 2012 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1075 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1117 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1133 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1136 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1152 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1279 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1280 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1281 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1282 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1338 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1340 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1342 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1401 | K. pneumoniae | A | 2013 | Paediatrics | Blood | ST15 | KPC-2 | – | – | + |
| 1407 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | + |
| 1403 | K. pneumoniae | A | 2013 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1406 | K. pneumoniae | A | 2013 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1408 | K. pneumoniae | A | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1503 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1508 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1512 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1577 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1587 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1591 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1592 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1663 | K. pneumoniae | A | 2014 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 1665 | K. pneumoniae | A | 2014 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 1758 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1768 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1797 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1838 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1847 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | + | – | – |
| 1853 | K. pneumoniae | A | 2014 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 1859 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1893 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1902 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1909 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | + |
| 2030 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 2054 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 2057 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 2059 | K. pneumoniae | A | 2014 | Paediatrics | Blood | ST15 | KPC-2 | – | – | – |
| 2060 | K. pneumoniae | A | 2014 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 13 | K. pneumoniae | B | 2010 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 26 | K. pneumoniae | B | 2010 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 25 | K. pneumoniae | B | 2010 | ICU | Sputum | ST15 | KPC-2 | – | – | – |
| 27 | K. pneumoniae | B | 2010 | ICU | Sputum | ST15 | KPC-2 | – | – | – |
| 32 | K. pneumoniae | B | 2010 | ICU | Blood | ST15 | KPC-2 | – | – | – |
| 33 | K. pneumoniae | B | 2010 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | + |
| 1203 | K. pneumoniae | B | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1202 | K. pneumoniae | B | 2013 | Paediatrics | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 1800 | K. pneumoniae | B | 2014 | ICU | Sputum | ST15 | KPC-2 | – | – | + |
| 1555 | K. pneumoniae | C | 2014 | ICU | Abdominal fluid | ST15 | KPC-2 | – | – | – |
| 2090 | K. pneumoniae | C | 2015 | ICU | Bronchial fluid | ST15 | KPC-2 | – | – | – |
| 01445 | K. pneumoniae | D | 2014 | Cardiology | Urine | ST15 | KPC-2 | – | – | – |
| 01478 | K. pneumoniae | D | 2014 | Neurology | Urine | ST15 | KPC-2 | – | – | – |
| 01512 | K. pneumoniae | D | 2015 | Thoracic Surgery | Blood | ST11 | KPC-12 | – | – | – |
| 01555 | K. pneumoniae | D | 2015 | TB and lung diseases | Sputum | ST15 | KPC-2 | – | – | – |
| 01557 | K. pneumoniae | D | 2015 | TB and lung diseases | Pleural fluid | ST15 | KPC-2 | – | – | – |
| 01556 | K. pneumoniae | D | 2015 | Neurology | Blood | ST11 | KPC-14 | – | – | – |
| 01567 | K. pneumoniae | D | 2015 | TB and lung diseases | Sputum | ST15 | KPC-2 | – | – | – |
| 01568 | K. pneumoniae | D | 2015 | TB and lung diseases | Sputum | ST15 | KPC-2 | – | – | – |
| 573 | Escherichia coli | A | 2011 | Paediatrics | Bronchial fluid | ST709 | KPC-2 | – | – | + |
| 579 | E. coli | A | 2011 | Paediatrics | Bronchial fluid | ST709 | KPC-2 | – | – | – |
| 769 | E. coli | A | 2012 | Paediatrics | Bronchial fluid | ST405 | KPC-2 | – | – | – |
| 774 | E. coli | A | 2012 | Paediatrics | Bronchial fluid | ST405 | KPC-2 | – | – | + |
| 1402 | E. coli | A | 2013 | ICU | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 1593 | E. coli | A | 2014 | Paediatrics | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 1942 | E. coli | A | 2014 | Paediatrics | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 1793 | E. coli | A | 2014 | Paediatrics | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 1808 | E. coli | A | 2014 | Paediatrics | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 2060 | E. coli | A | 2014 | Paediatrics | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 2052 | E. coli | A | 2014 | Paediatrics | Bronchial fluid | ST3580 | KPC-2 | – | – | – |
| 20 | E. coli | C | 2010 | Urology | Urine | ST448 | KPC-2 | – | – | + |
| 50 | E. coli | C | 2010 | Urological Surgery | Urine | ST448 | KPC-2 | – | – | + |
MLST, multilocus sequence typing; ICU, intensive care unit; TB, tuberculosis.
The core genome phylogenetic tree of 60 blaKPC-carrying K. pneumoniae isolates presented only three genotypic groups: two groups had ST15 and one group had ST11 (Fig. 1). The largest ST15 lineage contained all isolates from three hospitals (A, B and C) collected during 2010 and 2015. Interestingly, two resistance gene patterns were observed in this lineage originated from isolates from the three hospitals through the years. In contrast, strains collected from hospital D constituted two separate ST15 and ST11 groups, sharing a quite similar resistance gene profile.
KPC-producing E. coli had more diverse sequence types, including ST3580, ST448, ST709 and ST405 (Table 5), corresponding to four genotypic groups in its core genome phylogenetic tree (Fig. 2). The resistance profile was similar among E. coli strains belonging to the same sequence type.
3.3. Characterisation of blaKPC-carrying plasmids and genetic environment of blaKPC
Randomly selected blaKPC-positive isolates (n = 24) from the four hospitals were subsequently analysed by S1-PFGE and Southern blotting for blaKPC, showing that most (21/24) blaKPC genes were plasmid-borne (Fig. 3). Interestingly, the size of blaKPC-carrying plasmids in hospital A and B was similar (∼30 kb). In contrast, blaKPC-positive K. pneumoniae in hospitals C and D carried plasmids different in size: ∼170 kb and ∼55 kb, respectively. Two blaKPC-positive E. coli isolates and one K. pneumoniae isolate did not hybridise. IncFIB(K), IncN and IncFIIK were predominant plasmid types among blaKPC-carrying plasmids.
Fig. 3.
S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting of plasmids carrying blaKPC from clinical isolates. (A) DNA fingerprint of S1-treated plasmid DNA of selected Enterobacterales from clinical isolates stained with ethidium bromide. (B) Autoradiogram of gel A showing plasmids carrying the blaKPC gene. M, Salmonella Braenderup H9812 (molecular weight marker); lanes 1 and 4–8, Klebsiella pneumoniae isolates from hospital A; lanes 2 and 3, Escherichia coli isolates from hospital A; lanes 9–13, 23 and 24, K. pneumoniae isolates from hospital B; lane 14, K. pneumoniae from hospital C; lanes 15–19, K. pneumoniae isolates from hospital D; lanes 20 and 21, K. pneumoniae from hospital C; lane 22, E. coli isolates from hospital C.
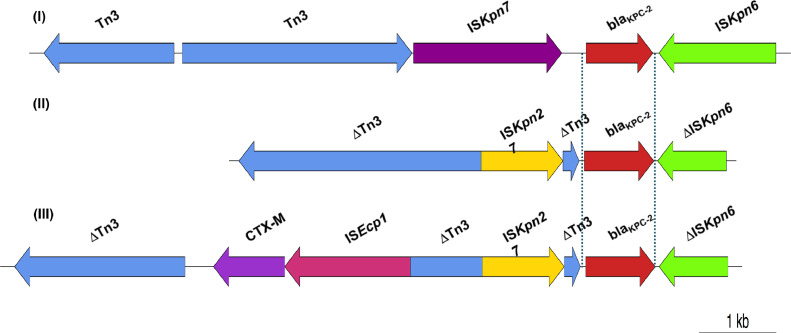
Analysis of the genetic environment of the blaKPC-2 gene revealed various genetic contexts of blaKPC-2 (Fig. 4) in different plasmids. Tn4401b isoform harbouring the blaKPC-2 gene was present in several E. coli and K. pneumoniae isolates carrying 55-bp or 30-bp plasmids with no deletion between ISKpn7 and blaKPC-2 (Model I). Two different structures of the blaKPC-2 gene environment were observed among K. pneumoniae isolates. The first variant possessed ISKpn27 upstream and a truncated ISKpn6 downstream of the blaKPC-2 gene (Model II), whereas the other had another mobile element containing ISEcp1–blaCTX-M inserted into a Tn3 transposon at 950 bp upstream of ISKpn27 (Model III).
Fig. 4.
Gene context models of blaKPC-2 in Escherichia coli and Klebsiella pneumoniae isolates. (I) Model observed in K. pneumoniae and E. coli isolates carrying 55-bp or 30-bp plasmids. (II) Model observed in K. pneumoniae isolates carrying 55-bp or 170-bp plasmids. (III) Model observed in K. pneumoniae isolates carrying 55-bp plasmids.
4. Discussion
Since their first detection in the USA [17], blaKPC-carrying plasmids have rapidly spread across countries and continents and the blaKPC-2 gene was recently described in several hospitals in Vietnam [4,[6], [7], [8], [9]]. Here we report the widespread distribution of blaKPC-producing Enterobacterales from samples collected from patients admitted to four large hospitals between 2010 and 2015 in Hanoi, Vietnam, with the first isolate detected in early 2010. It is possible there was already undetected circulation prior to 2010. blaKPC genes were detected in 122 (20.4%) of 599 CRE isolates. Co-expression of blaKPC and multiple other β-lactamase-encoding genes was found here as reported previously [6], [7], [8].
The most dominant sample type was bronchial fluid usually collected from mechanically ventilated patients, which are commonly found to be associated with nosocomial infections [18]. Genes conferring resistance to different antibiotic categories were also observed in other CRE isolated in Vietnam [5,8]. This shows the increasing number of diverse resistant strains in hospital settings in Vietnam, which pose a great challenge for doctors in efficient antibiotic selection.
The predominance of ST15 in the four hospitals was similar to other published data in Vietnam and Asian countries including China, which shares a >1000 km border and trading and tourism activities with Vietnam [6,8,19,20]. ST15 K. pneumoniae has been reported worldwide as a clone carrying multiple carbapenemase genes, which was also observed in other studies in Vietnam [8,21]. Carbapenemase-producing ST15 K. pneumoniae was first reported in Vietnam in samples collected between 2014–2015 [6]; however, in our study they were found in samples collected since 2010, facilitating the hypothesis of their presence in Vietnam hospitals prior to 2010. All blaKPC-carrying K. pneumoniae isolated from hospital A throughout the study period belonged to ST15, suggesting that they were resident flora. Although ST11 was reported among carbapenem-resistant K. pneumoniae in Vietnam previously [22,23], it was detected in only two isolates in the current study. This sequence type has also been associated with nosocomial outbreaks in several countries, especially in China [21,24,25]. Isolates from hospitals A, B and C belonged to the same lineage, supporting the hypothesis of exchange of bacteria and plasmids between these hospitals via patient transfer. Hospital D served a different targeted patient population and did not transfer patients with the other hospitals, which might explain the finding that resistant strains collected here evolved in their own ways forming two separate lineages.
Plasmid types carrying blaKPC genes found in this study were diverse and similar to those in many countries such as the UK, USA and China [26], [27], [28]. Four plasmid sizes were detected in this study that differed from the 150-kb blaKPC-carrying plasmid observed previously in Vietnam [6]. Notably, blaKPC-carrying plasmids isolated from hospital D all had the same size (∼55 kb) but belonged to two different ST groups (ST11 and ST15). This evidence supports the independent existence of blaKPC-carrying plasmids and, together with the diversity of sizes of blaKPC-carrying plasmids, shows the possibility of plasmid transmission across bacterial strains and species.
Regarding the gene context models of blaKPC-2, the Tn4401 isoform is endemic in many countries, whereas it is infrequently observed in China. Instead, ISKpn27–blaKPC–ΔISKpn6 within the Tn3 transposon frame accounted for the majority of isolates from China but not in other countries, which was also found in our study (Model II) [21,29]. This model was not only observed in Enterobacterales such as E. coli, K. pneumoniae and Citrobacter freundii but was also detected in P. aeruginosa [29], [30], [31], showing the possibility of plasmid transmission between different species and genera of bacteria. All isolates carrying this model in our study showed high resistance to carbapenems (MIC ≥ 8 μg/mL), except for one isolate maintaining susceptibility to imipenem.
Model III with the combination of ISEcp1–blaCTX-M and ISKpn27–blaKPC–ΔISKpn6 in the same plasmid (Fig. 4) has not been reported before. However, the co-existence of one plasmid carrying ISEcp1–blaCTX-M and one plasmid carrying ISKpn27–blaKPC–ΔISKpn6 in one isolate was reported from China in 2010 [32]. This suggests that a recombination event occurred bringing these two structures into one plasmid during the evolution of the isolate/mobile gene element. Interestingly, MIC results showed that this combination confers only weak resistance to carbapenems but still strong resistance to cefotaxime. Insertion of ISEcp1–blaCTX-M into the ISKpn27–blaKPC–ΔISKpn6 frame might affect the phenotype of carbapenem resistance.
This study has several limitations. We do not have full hospital denominators and only a limited amount of metadata were collected, and we do not know patient treatment outcomes. Therefore, we were unable to draw further conclusions on the epidemiological characteristics (such as burden of disease, distinction of community-acquired and hospital-acquired isolates, patient-to-patient and environmental persistence, commensal and pathogenic bacteria) of blaKPC-expressing Enterobacterales in these hospitals.
In conclusion, we describe the widespread presence of blaKPC-expressing Enterobacterales in four large hospitals in Hanoi, Vietnam, since 2010, which may have started earlier, along with their resistance patterns, sequence types, genotypic relationship, plasmid sizes and genetic context. The spread of these carbapenemase-producers adds an additional challenge to the treatment of diseases caused by these common bacteria with a very extensive expression of genes conferring additional resistances.
Our study also provides evidence for the likelihood of KPC-producer circulation among three of the four hospitals as well as the possibility of plasmid transmission across bacterial strains and species. Data from this study contribute to a more comprehensive picture of the antimicrobial resistance situation in hospitals in Hanoi in the context of overcrowding and lack of hospital infection control programmes.
Funding
This work was supported by a grant from the Newton Fund Vietnam [MOST: NHQT/SPDP/02.16], a Grant-in-Aid for the Research Program on Emerging and Re-emerging Infectious Diseases [JP20fk0108061 for KS; JP20fk0108093, JP20fk0108139 and JP20wm0225008 for MS] from the Japan Agency for Medical Research and Development (AMED), the Wellcome Trust of Great Britain, UK, and the IRD and LMI DRISA.
Ethical approval
The samples used in this study were taken from the Isolate Bank of the National Institute of Hygiene and Epidemiology (NIHE). This study is part of the main project ‘Assessing the impact and burden of antimicrobial resistance in Vietnam, genomic characterization and risk factors related to antimicrobial resistance of common bacteria in Vietnam’, which was approved by the institutional review board (IRB) of NIHE [IRB code IRB-VN01057-38/2016]. Individual informed consent was waived because of the retrospective nature of the work and because no personal identifiers were collected.
Declaration of Competing Interest
None declared.
Editor: Stefania Stefani
References
- 1.Vu TVD, Choisy M, Do TTN, Nguyen VMH, Campbell JI, Le TH, et al. Antimicrobial susceptibility testing results from 13 hospitals in Vietnam: VINARES 2016–2017. Antimicrob Resist Infect Control. 2021;10:78. doi: 10.1186/s13756-021-00937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran DM, Larsson M, Olson L, Hoang NTB, Le NK, Khu DTK, et al. High prevalence of colonisation with carbapenem-resistant Enterobacteriaceae among patients admitted to Vietnamese hospitals: risk factors and burden of disease. J Infect. 2019;79:115–122. doi: 10.1016/j.jinf.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Hoang TH, Wertheim H, Minh NB, Duong TN, Anh DD, Phuong TTL, et al. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae strains containing New Delhi metallo-β-lactamase isolated from two patients in Vietnam. J Clin Microbiol. 2013;51:373–374. doi: 10.1128/JCM.02322-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran HH, Ehsani S, Shibayama K, Matsui M, Suzuki S, Nguyen MB, et al. Common isolation of New Delhi metallo-β-lactamase 1-producing Enterobacteriaceae in a large surgical hospital in Vietnam. Eur J Clin Microbiol Infect Dis. 2015;34:1247–1254. doi: 10.1007/s10096-015-2345-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi-Akiyama T, Ohmagari N, Phuong TT, Huy NQ, Anh NQ, Van Thanh D, et al. Epidemiology of Enterobacter cloacae strains producing a carbapenemase or metallo-β-lactamase in Vietnamese clinical settings in 2014–2017. J Med Microbiol. 2020;69:530–536. doi: 10.1099/jmm.0.001175. [DOI] [PubMed] [Google Scholar]
- 6.Tada T, Tsuchiya M, Shimada K, Nga TTT, Thu LTA, Phu TT, et al. Dissemination of carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA methylases (RmtB and RmtC) in Vietnam. BMC Infect Dis. 2017;17:467. doi: 10.1186/s12879-017-2570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoang CQ, Nguyen HD, Vu HQ, Nguyen AT, Pham BT, Tran TL, et al. Emergence of New Delhi metallo-β-lactamase (NDM) and Klebsiella pneumoniae carbapenemase (KPC) production by Escherichia coli and Klebsiella pneumoniae in Southern Vietnam and appropriate methods of detection: a cross-sectional study. Biomed Res Int. 2019;2019 doi: 10.1155/2019/9757625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund B, Hoang NTB, Tarnberg M, Le NK, Nilsson M, Khu DTK, et al. Molecular and phenotypic characterization of clinical isolates belonging to a KPC-2-producing strain of ST15 Klebsiella pneumoniae from a Vietnamese pediatric hospital. Antimicrob Resist Infect Control. 2019;8:156. doi: 10.1186/s13756-019-0613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund B, Hoang NTB, Tarnberg M, Le NK, Welander J, Nilsson M., et al. Colistin- and carbapenem-resistant Klebsiella pneumoniae carrying mcr-1 and blaOXA-48 isolated at a paediatric hospital in Vietnam. J Antimicrob Chemother. 2018;73:1100–1102. doi: 10.1093/jac/dkx491. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI) CLSI; Wayne, PA: 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24. [Google Scholar]
- 11.Clinical and Laboratory Standards Institute (CLSI) CLSI supplement M100. 28th ed. CLSI; Wayne, PA: 2018. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 12.European Committee on Antimicrobial Susceptibility testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0, 2018.
- 13.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Southern E. Southern blotting. Nat Protoc. 2006;1:518–525. doi: 10.1038/nprot.2006.73. [DOI] [PubMed] [Google Scholar]
- 15.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phu VD, Wertheim HF, Larsson M, Nadjm B, Dinh QD, Nilsson LE, et al. Burden of hospital acquired infections and antimicrobial use in Vietnamese adult intensive care units. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DAB, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. 2020;12:11. doi: 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Huang L, Liu C, Huang X, Zheng R, Lu Y, et al. Characterization of carbapenem-resistant Klebsiella pneumoniae ST15 clone coproducing KPC-2, CTX-M-15 and SHV-28 spread in an intensive care unit of a tertiary hospital. Infect Drug Resist. 2021;14:767–773. doi: 10.2147/IDR.S298515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Li F, Cui S, Mao L, Li X, Awan F, et al. Prevalence and distribution characteristics of blaKPC-2 and blaNDM-1 genes in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:2901–2910. doi: 10.2147/IDR.S253631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts L, Hoi LT, Khokhar F, Hoa NT, Van Giang T, Bui C, et al. A genomic epidemiology study of multidrug-resistant Escherichia coli, Klebsiella pneumoniae and Acinetobacter baumannii in two intensive care units in Hanoi, Vietnam. medRxiv. 2020 Dec 11 doi: 10.1101/2020.12.09.20246397. [DOI] [PubMed] [Google Scholar]
- 23.Chang J, Lee JY, Joo JY, Kim K, Park HY, Kim SH, et al. Emergence of NDM-4-producing Klebsiella pneumoniae in a Korean hospital due to a patient hospitalized in Vietnam and case review. J Infect Chemother. 2019;25:909–912. doi: 10.1016/j.jiac.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Yu F, Hu L, Zhong Q, Hang Y, Liu Y, Hu X, et al. Dissemination of Klebsiella pneumoniae ST11 isolates with carbapenem resistance in integrated and emergency intensive care units in a Chinese tertiary hospital. J Med Microbiol. 2019;68:882–889. doi: 10.1099/jmm.0.000981. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Yang Y, Chen G, Lin M, Chen Y, He R, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10:700–709. doi: 10.1080/22221751.2021.1906163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerqueira GC, Earl AM, Ernst CM, Grad YH, Dekker JP, Feldgarden M, et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A. 2017;114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Findlay J, Hopkins KL, Doumith M, Meunier D, Wiuff C, Hill R, et al. KPC enzymes in the UK: an analysis of the first 160 cases outside the North-West region. J Antimicrob Chemother. 2016;71:1199–1206. doi: 10.1093/jac/dkv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang LH, Wei DD, Wan LG, Yu Y, Deng Q, Liu Y. Diversity of the genetic environment of the blaKPC-2 gene among Klebsiella pneumoniae clinical isolates in a Chinese hospital. Microb Drug Resist. 2016;22:15–21. doi: 10.1089/mdr.2014.0281. [DOI] [PubMed] [Google Scholar]
- 29.Dai X, Zhou D, Xiong W, Feng J, Luo W, Luo G, et al. The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel ΔISEc33-associated blaKPC-2 gene cluster. Front Microbiol. 2016;7:310. doi: 10.3389/fmicb.2016.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J, Qiu Y, Yin Z, Chen W, Yang H, Yang W, et al. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob Chemother. 2015;70:2987–2991. doi: 10.1093/jac/dkv232. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Fang H, Feng J, Yin Z, Xie X, Zhu X, et al. Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front Microbiol. 2015;6:838. doi: 10.3389/fmicb.2015.00838. [DOI] [PMC free article] [PubMed] [Google Scholar]