Abstract
Objective
Gastric bypass surgery results in long‐term weight loss. Small studies have examined protein changes during rapid weight loss (up to 1 or 2 years post surgery). This study tested whether short‐term changes were maintained after 12 years.
Methods
A 12‐year follow‐up, protein‐wide association study of 1,297 SomaLogic aptamer‐based plasma proteins compared short‐ (2‐year) and long‐term (12‐year) protein changes in 234 individuals who had gastric bypass surgery with 144 nonintervened individuals with severe obesity.
Results
There were 51 replicated 12‐year protein changes that differed between the surgery and nonsurgery groups. Adjusting for change in BMI, only 12 proteins remained significant, suggesting that BMI change was the primary reason for most protein changes and not non‐BMI‐related surgical effects. Protein changes were related to BMI changes during both weight‐loss and weight‐regain periods. The significant proteins were associated primarily with lipid, uric acid, or resting energy expenditure clinical variables and metabolic pathways. Eight protein changes were associated with 12‐year diabetes remission, including apolipoprotein M, sex hormone binding globulin, and adiponectin (p < 3.5 × 10−5).
Conclusions
This study showed that most short‐term postsurgical changes in proteins were maintained at 12 years. Systemic protection pathways, including inflammation, complement, lipid, and adipocyte pathways, were related to the long‐term benefits of gastric bypass surgery.
Study Importance.
What is already known?
-
►
Plasma protein amounts change 1 to 2 years after bariatric surgery.
-
►
Many of these proteins are in known obesity‐related metabolic pathways and have been associated with improved clinical risk factors for disease.
What does this study add?
-
►
Most short‐term postsurgical plasma protein changes were maintained up to 12 years post surgery and were related to weight change rather than gastric bypass surgery, per se.
-
►
Long‐term protein changes were related to improvements in clinical disease risk factors primarily involving lipid pathways rather than diabetic pathways.
-
►
Inflammation, complement, and adipocyte function pathways were other important pathways that were overexpressed by the significantly changed proteins.
How might these results change the direction of research or the focus of clinical practice?
-
►
Beneficial protein changes identified after gastric bypass surgery are also likely to be induced by nonsurgical weight loss.
-
►
Identification of plasma proteins involved with improved clinical risk factors may, in the future, allow a more effective targeting of treatments to reduce adverse clinical outcomes of obesity.
INTRODUCTION
Bariatric surgery results in significant, sustained weight loss in most patients. The surgery also improves most cardiovascular risk factors, including blood pressure, diabetes, dyslipidemia, fatty liver disease, inflammation, and health‐related quality of life (1, 2, 3, 4). Total mortality and cardiovascular mortality are reduced by 40% and 56%, respectively (5). Whereas voluntary, nonsurgical weight loss is difficult to maintain over the long term (6), surgical weight loss is generally maintained long term and, as such, it may provide an efficient way of identifying proteins and protein pathways contributing to improved health outcomes induced by bariatric surgery or weight loss in general.
Short‐term studies of 1 or 2 years (i.e., the nadir of maximum weight loss) (7) have suggested plasma proteins significantly change after bariatric surgery (8, 9, 10, 11), but it is unknown which protein changes are durable in the long term. Long‐term assessment is important because short‐term postsurgical changes in many proteins may be due to the invasive nature of the surgery itself and the dramatic first year of postsurgical weight loss, which is expected to affect many related physiological systems. However, once weight has stabilized and modest weight regain progresses, the underlying short‐term benefits of the protein changes may disappear if these proteins return to near presurgical levels. Identification of proteins that remain changed long term may help identify biological pathways responsible for disease reduction. Therefore, a proteome‐wide association study (PWAS) was conducted to test for significant differences in 12‐year changes in 1,297 plasma protein levels between 234 individuals with severe obesity who had gastric bypass surgery and a nonintervened group of 144 individuals with severe obesity. Protein changes were also related to significant changes in disease risk factors.
METHODS
A longitudinal, controlled study of the risks and benefits of Roux‐en‐Y gastric bypass surgery was begun in 2000. After a baseline exam (exam 1), additional exams were conducted at the University of Utah at approximately 2 years (exam 2), 6 years (exam 3), and 12 years (exam 4) (3, 12, 13). Follow‐up of major clinical variables was more than 90% at exam 4, although only 67% of individuals had their blood drawn at the University of Utah and could provide a sample for proteomic measurements. Of the 67% returning individuals, a subset of the individuals was selected who either had gastric bypass surgery after the baseline exam or who never had bariatric surgery during follow‐up. The nonsurgery group did not have a study‐related intervention during the follow‐up period, and even though mean weight did not change, there was a wide variation in weight change in this group over the 12 years. An initial subset of 203 individuals (137 in the surgery group and 66 in the nonsurgery group) was used as a discovery set, and a second subset of 175 individuals (97 in the surgery group and 78 in the nonsurgery group) was used for replication. In addition to plasma protein measurements at baseline and 12 years, proteins were measured at the 2‐year follow‐up (exam 2) for 204 of the 234 individuals who had gastric bypass surgery. Proteins from exam 3 were not measured. All individuals provided written informed consent, and the study was approved by the University of Utah Institutional Review Board.
Clinical measurements
The clinical measurements shown in Table 1 were measured or calculated as previously described (3, 12, 13, 14). Fat‐free mass and fat mass (FM) were measured by bioimpedance. Resting energy expenditure (REE) was measured after an overnight fast by indirect calorimetry (TrueOne 2400, ParvoMedics, Sandy, Utah) (12). Clinical biochemistries were obtained after an overnight fast. Diabetes remission at 12 years in individuals with baseline diabetes, diabetes incidence in individuals without baseline diabetes, and 10‐year risk of coronary heart disease (CHD) as assessed by the Framingham Risk Score (FRS) were derived. To ensure that the FRS represented change in CHD risk only due to changes in the clinical variables and not age, age at exam 4 was used in both the baseline and 12‐year follow‐up risk equations. Diabetes was defined as a fasting glucose of 126 mg/dL or greater or being on antidiabetic medication. Blood pressure, lipids, and diabetes‐related variables that are affected by antihypertensive, antidiabetic, or lipid medications were adjusted prior to analysis, as described previously (3). Briefly, individuals taking medications for each condition had their values changed to the sex‐specific means of untreated individuals with the condition. Analysis with and without medication adjustment was performed.
TABLE 1.
Discovery and replication participant subset characteristics and winsorized variable means and standard deviations (SD)
| Variable | Discovery | Replication | |||||
|---|---|---|---|---|---|---|---|
| Groups | Surgery (n = 137) | Nonsurgery (n = 66) | All (n = 203) | Surgery (n = 97) | Nonsurgery (n = 78) | All (n = 175) | |
| Gender (M) | 20% | 12% | 17% | 12% | 11% | 12% | |
| Age (exam 1, y) | 46.4 ± 10.5 | 47.1 ± 11 | 46.6 ± 10.9 | 40.1 ± 10 | 43.9 ± 12.3 | 41.8 ± 11.2 | |
| BMI (exam 1, kg/m2) | 46.4 ± 7.1 | 44.6 ± 6.4 | 45.8 ± 6.9 | 46.1 ± 6.4 | 45.3 ± 7.5 | 45.8 ± 6.9 | |
| BMI (exam 2, kg/m2; n = 204) | 29.7 ± 5.5 (n = 137) | NA | NA | 34.3 ± 9.1 (n = 67) | NA | NA | |
| BMI (exam 4, kg/m2) | 34.6 ± 8.0 | 44.8 ± 7.5 | 37.9 ± 9.1 | 35.0 ± 8.8 | 45.0 ± 8.9 | 39.6 ± 10.2 | |
| Diabetes (exam 1, %) | 34 | 38 | 35 | 8 | 32 | 19 | |
| Diabetes (exam 4, %) | 17 | 38 | 24 | 7 | 54 | 28 | |
| Changes in clinical variables (exam 4‐exam 1) | |||||||
| FFM (kg) | M ± SD | −13.4 ± 6.7 | −2.4 ± 6.5 | −9.9 ± 8.4 | −13.5 ± 7.0 | −4.9 ± 7.4 | −9.8 ± 8.4 |
| n | 127 | 59 | 186 | 91 | 70 | 161 | |
| FM (kg) | M ± SD | −22.2 ± 11.5 | −2.1 ± 10.3 | −15.8 ± 14.5 | −21.0 ± 11.6 | −1.2 ± 10.1 | −12.4 ± 14.7 |
| n | 127 | 59 | 186 | 91 | 70 | 161 | |
| BMI (kg/m2) | M ± SD | −11.5 ± 5.4 | −0.2 ± 5.6 | −7.8 ± 7.6 | −11.0 ± 5.5 | −0.2 ± 5.3 | −6.2 ± 7.6 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| REE (kcal) | M ± SD | −550 ± 238 | −301 ± 215 | −478 ± 257 | −569 ± 260 | −325 ± 249 | −452 ± 282 |
| n | 79 | 32 | 111 | 50 | 46 | 96 | |
| AST (U/l) | M ± SD | −0.5 ± 8.1 | −2.4 ± 8.5 | −1.1 ± 8.3 | −0.5 ± 10.0 | −4.0 ± 7.8 | −2.1 ± 9.2 |
| n | 136 | 66 | 202 | 96 | 77 | 173 | |
| ALT (U/l) | M ± SD | −5.0 ± 11.3 | −2.7 ± 13.1 | −4.3 ± 11.9 | −4.4 ± 15.0 | −5.4 ± 12.8 | −4.8 ± 14.0 |
| n | 137 | 66 | 203 | 97 | 77 | 174 | |
| Uric acid (mg/dL) | M ± SD | −0.7 ± 1.2 | 0.1 ± 1.3 | −0.4 ± 1.3 | −0.8 ± 1.3 | −0.2 ± 1.2 | −0.6 ± 1.3 |
| n | 137 | 65 | 202 | 96 | 77 | 173 | |
| SBP (mmHg) | M ± SD | −4.6 ± 19.1 | 9.3 ± 21.5 | −0.1 ± 20.9 | −2.0 ± 21.5 | 8.8 ± 20.3 | 2.8 ± 21.6 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| DBP (mmHg) | M ± SD | −1.4 ± 13.9 | 5.5 ± 14.0 | 0.8 ± 14.3 | 1.1 ± 14.9 | 7.0 ± 14.4 | 3.7 ± 14.9 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| Glucose (mg/dL) | M ± SD | −13.3 ± 20.7 | −2.5 ± 23.9 | −9.8 ± 22.3 | −7.5 ± 14.0 | −1.9 ± 25.9 | −5.0 ± 20.3 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| Insulin (μU/mL) | M ± SD | −10.8 ± 11.8 | −6.7 ± 13.9 | −9.5 ± 12.6 | −11.3 ± 13.0 | −6.2 ± 12.8 | −9.1 ± 13.1 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| HOMA‐IR | M ± SD | −3.1 ± 3.4 | −2.0 ± 4.1 | −2.8 ± 3.7 | −2.9 ± 3.5 | −1.5 ± 3.9 | −2.3 ± 3.7 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| HOMA‐B | M ± SD | −71 ± 147 | −69 ± 177 | −70 ± 157 | −100 ± 179 | −38 ± 166 | −73 ± 175 |
| n | 133 | 63 | 196 | 96 | 76 | 172 | |
| HbA1c (%) | M ± SD | 0.01 ± 0.93 | 0.25 ± 1.00 | 0.09 ± 0.96 | 0.07 ± 0.76 | 0.44 ± 1.22 | 0.23 ± 1.01 |
| n | 136 | 66 | 202 | 97 | 78 | 175 | |
| TG (mg/dL) | M ± SD | −70.6 ± 59.8 | −24.8 ± 56.8 | −55.7 ± 62.5 | −73.0 ± 67.3 | −33.9 ± 69.7 | −55.6 ± 70.9 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| LDL‐C (mg/dL) | M ± SD | −2.9 ± 29.5 | 26.4 ± 29.7 | 6.6 ± 32.5 | −3.4 ± 30.6 | 27.8 ± 28.4 | 10.5 ± 33.4 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| HDL‐C (mg/dL) | M ± SD | 13.7 ± 12.6 | 2.2 ± 9.4 | 10.0 ± 12.8 | 15.6 ± 13.8 | 2.3 ± 9.0 | 9.7 ± 13.6 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| FRS (%) | M ± SD | −2.3 ± 3 | −0.8 ± 2 | −1.8 ± 3 | −1.0 ± 2 | −0.8 ± 2 | −0.9 ± 2 |
| n | 137 | 66 | 203 | 97 | 78 | 175 | |
| DMINC | % | 0 | 0 | 0 | 3 | 21 | 24 |
| n | 91 | 41 | 132 | 88 | 51 | 139 | |
| DMREM | % | 23 | 0 | 23 | 4 | 5 | 9 |
| n | 46 | 25 | 71 | 8 | 25 | 33 | |
Exam 1: baseline exam; exam 2: exam at 2 years; exam 4: exam at 12 years. Only 204 of the 234 participants who had gastric bypass surgery had available exam 2 proteomic measurements.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; DBP, diastolic blood pressure; DMINC, diabetes incidence at 12 years; DMREM, diabetes remission at 12 years; FFM, fat‐free mass; FM, fat mass; FRS, Framingham Risk Score for 10‐year cardiovascular disease incidence; HbA1c, hemoglobin A1c; HDL‐C, high‐density lipoprotein cholesterol; HOMA‐B, homeostatic assessment of insulin secretion; HOMA‐IR, homeostatic assessment of insulin resistance; LDL‐C, low‐density lipoprotein cholesterol; NA, not measured at exam 2; REE, resting energy expenditure; SBP, systolic blood pressure; TG, triglycerides.
Plasma aliquots were stored at −80°C. Prior to assay, the plasma was thawed, and approximately 70 μL was used for the proteomics assay. Proteins were measured by a Slow Off‐rate Modified Aptamer (SOMAmer)‐based protein array (SomaLogic, Boulder, Colorado) (15). A total of 1,297 proteins were measured and are referred to by their gene names. Normalization and calibration of protein levels were done using SomaScan proprietary software (SomaLogic). Sample data were normalized to remove hybridization variation within a run, followed by median normalization across all samples to remove other assay biases within the run and, finally, calibration to remove assay differences between runs. Calibrator coefficients of variation on each plate were calculated; at least 50% of SOMAmer reagents had to have coefficients of variation less than 0.1, and 95% had to have coefficients of variation below 0.2. Any flagged samples by SomaScan software were removed from the analysis. Protein abundance was calculated as log of the relative fluorescence units.
Statistical methods
Outliers were removed at four standard deviations above or below the mean value of each protein. Within‐individual changes in protein levels (protein at exam 4 − protein at exam 1) were calculated for all 1,297 proteins. Clinical data were winsorized at 5% to 95% quantiles.
β coefficients from linear regression models were used to test for protein changes versus surgery status (surgery group compared with nonsurgery group) or versus BMI changes (exam 4 − exam 1) after adjusting for age, sex, and baseline BMI, first in the discovery batch and then in the replication batch. Additional regression models were run after adding change in BMI as a covariate when comparing the surgery and nonsurgery groups. All statistical tests were two‐sided tests and were adjusted for multiple comparisons using the Bonferroni correction.
A functional enrichment analysis was performed in R using the fgsea (Korotkevich G et al, bioRxiv doi:10.1101/060012) and clusterProfiler (16) packages (R Foundation, Vienna, Austria) for gene set enrichment analysis (GSEA). The analysis was performed using pathway annotation from human Reactome (version 73; June 2020 release), WikiPathways (July 2020 release), and Kyoto Encyclopedia of Genes and Genomes (KEGG; July 7, 2020, release) databases. The full SomaScan panel of proteins was used as the background set of proteins. GSEA results were filtered for redundant entries based on semantic similarity between protein groups. UniProt keyword (October 15, 2019, release) annotations were used for classification of proteins. Significantly overrepresented diseases were assessed using the Genetic Association Database (17). Clustering in heat maps was performed using the complete linkage Euclidean distance method.
RESULTS
Baseline and 12‐year change variables are presented for the discovery and replication subsets in Table 1. For the gastric bypass surgery group, unadjusted protein means at baseline, 2 years, and 12 years for all 1,297 proteins are color coded for those that increased or decreased over 12 years (Supporting Information Table S1). The unadjusted protein means at baseline and at the 12‐year follow‐up in the comparison nonsurgery group are listed in Supporting Information Table S2.
Long‐term effect of surgery and weight loss on the proteomic profile
A total of 58 adjusted protein changes differed between the gastric bypass surgery and nonsurgery groups in the discovery set at a Bonferroni significance level (p < 0.05/1,297 = 3.85 × 10−5), among which 51 were replicated (p < 0.05/58 = 8.6 × 10−4; Figure 1A, Supporting Information Tables S3‐S5 for discovery, replication, and combined analysis, respectively).
FIGURE 1.
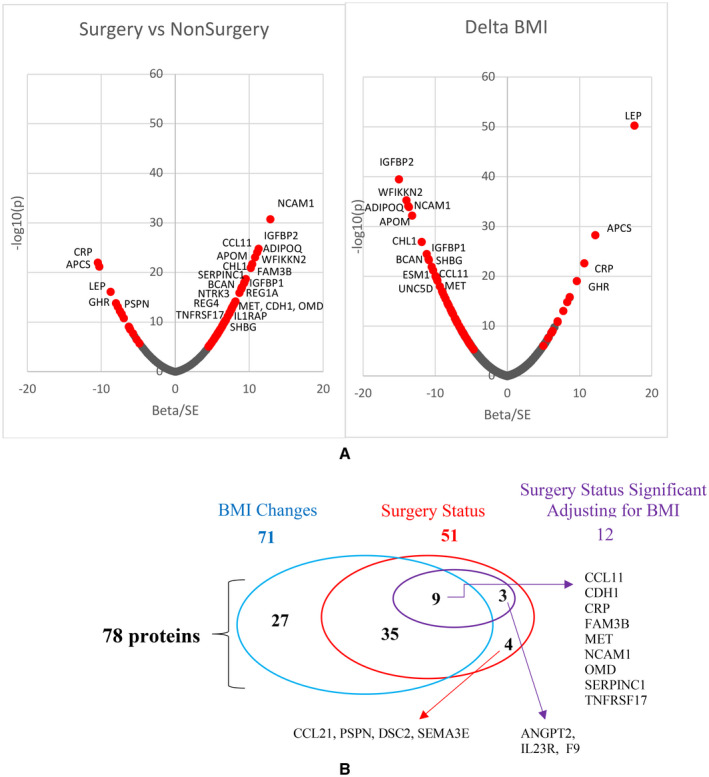
(A) Plot of 12‐year protein changes’ normalized β values (β/SE β) vs. nominal −log10 p values. Left plot from the surgery vs. nonsurgery analysis of the combined discovery and replication samples. Right plot from the Δ BMI (exam 4 − exam 1) analysis. Red color identifies the 78 replicated and Bonferroni‐corrected significant protein changes. (B) Overlap of 71 significant 12‐year protein changes associated with BMI change (blue oval) and 51 associated with gastric bypass surgical status without correcting for change in BMI in the model (red oval). Twelve of the fifty‐one proteins were still significantly associated with surgery status after correcting for changes in BMI (purple oval). Four proteins were no longer associated with surgery status after adjusting for BMI changes even though they were not found to be associated with BMI changes
Because the 12‐year change in weight varied within both the surgery and nonsurgery groups, additional PWAS was conducted to find the association of 12‐year changes in proteins with the quantitative change in BMI, adjusting for age, sex, and baseline BMI. In the discovery set, 12‐year changes in levels of 99 proteins were associated with changes in BMI (p < 3.85 × 10−5). Of these 99 protein changes, 71 were replicated (p < 0.05/99 = 5.1 × 10−4; Figure 1A). Sixty of the seventy‐one identified proteins increased as BMI decreased over the 12 years, including insulinlike growth factor binding protein 2 (IGFBP2); apolipoprotein M (APOM); and adiponectin, C1Q and collagen domain containing (ADIPOQ); whereas 11 proteins decreased, including leptin (LEP), C‐reactive protein (CRP), growth hormone receptor (GHR), afamin (AFM), and myeloperoxidase (MPO; Figure 1A).
Combining the significant results from the two PWAS analyses (Figure 1B), 78 long‐term protein changes were associated with change in BMI (n = 71) or between surgery and nonsurgery groups (n = 51). All but seven protein changes that were significantly different between surgery groups were also associated with change in BMI (Figure 1B). Therefore, the protein change differences between groups were further adjusted for change in BMI to see whether some proteins changed independently of BMI change. Twelve of the fifty‐one replicated protein changes that were significantly associated with surgery/nonsurgery status remained significantly associated with surgery status even after further adjustment for BMI change (Figure 1B).
Short‐ versus long‐term effect of surgery and weight loss on the proteomic profile
Short‐term protein change associations with BMI change were maintained in the long term (Figure 2A, Supporting Information Tables S6 and S7). There was a strong correlation of the 12‐year follow‐up versus the 2‐year follow‐up standardized β coefficients (β/SE) associated with the p values shown in Figure 2A for the 71 proteins (r = 0.93).
FIGURE 2.
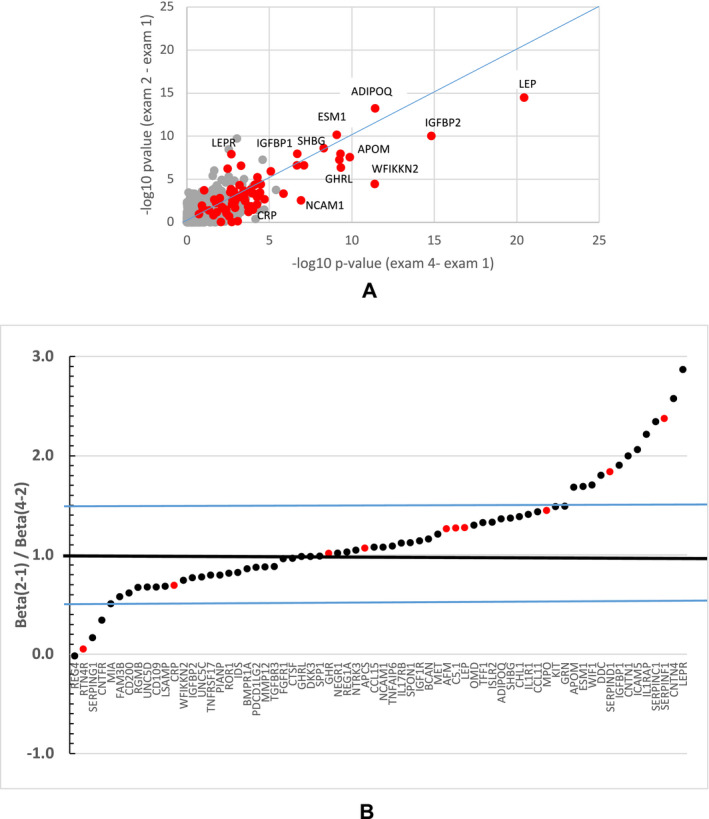
(A) Comparison of −log10 p values from the association of short‐term (exam 2 – exam 1) vs. long‐term (exam 4 − exam 1) protein change associations with BMI change for all 1,297 proteins in the combined discovery and replication groups. Red dots indicate the 71 replicated proteins for BMI change. (B) Patterns of protein changes per unit of BMI change after substantial weight loss (exam 2 − exam 1) and after some weight regain (exam 4 − exam 2) obtained from the surgery group (N = 204 who have measurements at all three time points) for the 78 proteins in Figure 1. Ratio of β coefficients derived from the regression of the protein change on BMI change for the 2‐year follow‐up (exam 2 − exam 1; β1‐2) divided by the β coefficient using changes from the subsequent 10 years of follow‐up (exam 4 − exam 2, β2‐4). Ratios greater than one suggest greater protein change per unit of BMI change during the weight‐loss period compared with the weight‐gain period. Ratios less than one suggest greater protein changes per unit of BMI change in the weight‐gain period compared with the weight‐loss period. Proteins shown in red increased with increasing BMI and proteins in black decreased with increasing BMI. Blue lines represent ±50% increase or decrease in the ratio. FCN2, with a ratio of 5.0, is not shown to improve the readability of the figure. Serpin family A member 4 (SERPINA4) and cadherin 1 (CDH1) had greater changes during weight loss but also had negative ratios of −7.0 and −4.8, respectively, indicating that the protein change association with BMI change did not reverse from the weight‐loss to the weight‐gain period, as did the other proteins (points not shown). The β coefficients used in the ratios can be found in Supporting Information Table S6
All protein changes except regenerating family member 4 (REG4) had the same direction of association with BMI change during weight‐loss (exam 2 ‐ exam 1) and weight‐regain (exam 4 ‐ exam 2) time periods (Supporting Information Table S6). If the protein change was positively associated with BMI change from exam 1 to 2 (both decreased over time), it was positively associated from exam 2 to 4 (both increased). If the protein change was inversely associated from exam 1 to 2 (BMI decreased but the protein increased), the protein change was inversely associated with BMI change from exam 2 to 4 (BMI increased but the protein decreased). Therefore, protein changes consistently reflected both BMI decreases and BMI increases.
In addition to the consistent direction of the association of protein changes with BMI change, the amount of protein change per unit of BMI change was assessed to see whether the period of rapid weight loss had a differing effect on a protein than the period of slow weight regain. All but 16 proteins had a protein change per unit of BMI change during weight regain that was within 50% (0.5 ≤ ratio of β coefficients ≤ 1.5) of the change per unit of BMI during weight loss (Figure 2B, Supporting Information Table S6). Proteins that changed more per unit of BMI change during weight loss than weight regain included leptin receptor (LEPR), insulinlike growth factor binding protein 1 (IGFBP1), APOM, and three serpin proteins. Only four proteins had greater than a 50% change per unit of BMI change during the weight‐regain period than the weight‐loss period.
Individuals who regained less than 10% of their baseline presurgical weight from exam 2 to exam 4 were compared with individuals who regained more than 10% of their baseline weight (18). Six proteins were significantly associated with weight regain versus weight maintenance (p < 0.05/1,297; Supporting Information Figure S1, Supporting Information Table S8). LEP and amyloid P component, serum (APCS) were higher at exam 4 in those who regained more than 10% of their baseline weight, whereas IGFBP2; WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 (WFIKKN2); HtrA serine peptidase 2 (HTRA2); and sex hormone binding globulin (SHBG) were lower. The clinical characteristics of the two groups are presented in Supporting Information Table S9.
Clinical variable associations with the proteome
A heat map of the regression β‐coefficients and the cluster patterns of the 71 protein and 20 clinical variable 12‐year changes showed the strongest protein associations with lipids (58 proteins with high‐density lipoprotein cholesterol [HDL‐C], 47 with low‐density lipoprotein cholesterol [LDL‐C], and 44 with triglycerides [TG]), REE (36 proteins), and uric acid (23 proteins; Figure 3, Supporting Information Table S10). Surprisingly few associations were seen with variables in the glucose/insulin pathways (fasting glucose with 8; insulin, homeostatic assessment of insulin resistance, and homeostatic assessment of insulin secretion with 0; hemoglobin A1c [HbA1c] with 4, which included ADIPOQ and SHBG; or blood pressure [systolic blood pressure with 4, which included APOM, and diastolic blood pressure with 1, which included AFM]). Eight protein changes were associated with 12‐year remission of diabetes (including APOM, ADIPOQ, and SHBG), and eight proteins increased when BMI decreased and were associated with a decreased 10‐year risk of CHD (Supporting Information Table S10).
FIGURE 3.
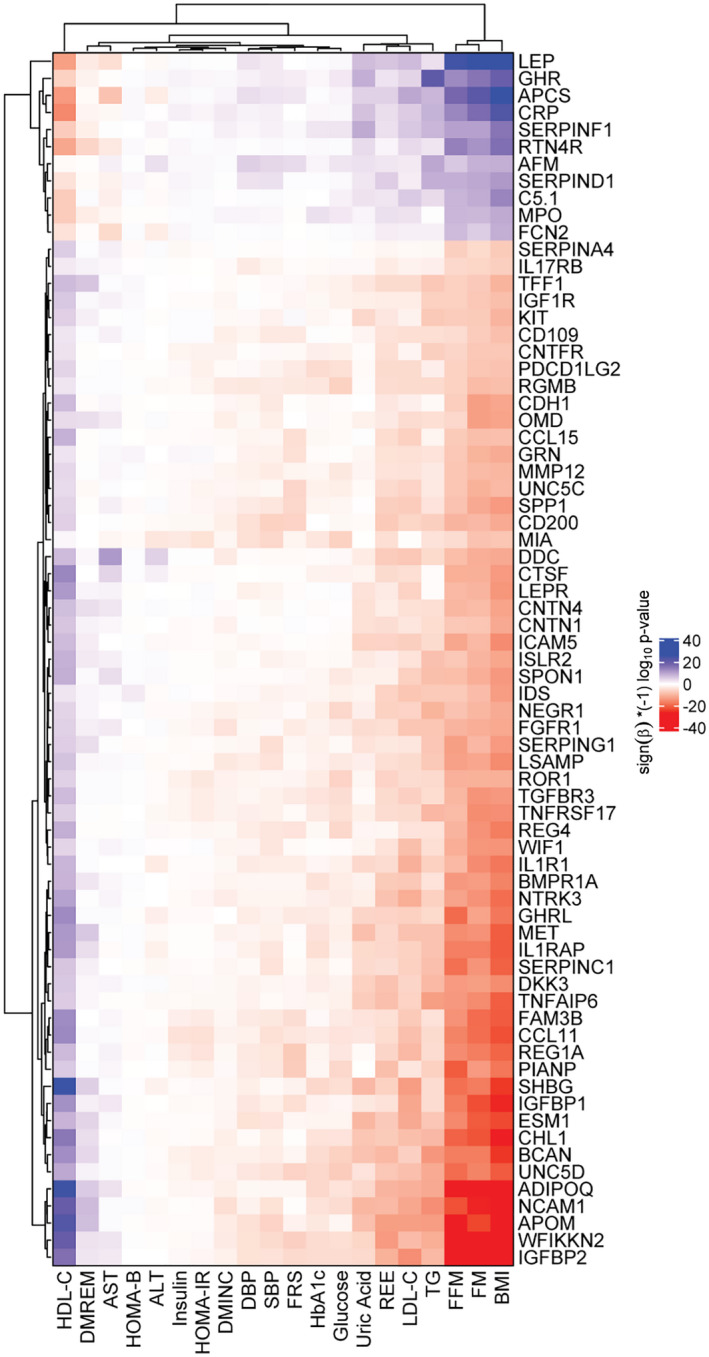
Heat map of clinical variable associations with the 71 protein changes that were associated with change in BMI from a regression model using combined surgery and nonsurgery groups. Colors represent (sign[β] × [−1]log10[p value]). Significance level is Bonferroni‐corrected p value (p ≤ 0.05/71/20 = 3.5 × 10−5) for 71 proteins and 20 clinical variables. Red indicates high significance with negative β (the protein increases with a decrease in the clinical variable), and blue indicates high significance with positive β
Because so few glucose metabolism‐related proteins were found among the protein changes that were related to BMI change, a PWAS analysis was done for all 1,297 proteins in the surgical group (Supporting Information Figure S2, Supporting Information Table S11). Longitudinal changes in glucose, insulin, homeostatic assessment of insulin resistance, homeostatic assessment of insulin secretion, HbA1c, and blood pressure still did not show highly significant associations with protein changes. The strongest associations of non‐BMI‐related protein changes were with liver function (particularly protein cluster 3; Supporting Information Figure S2). HDL‐C change was associated with ADIPOQ, APOM, WFIKKN2, ghrelin and obestatin prepropeptide (GHRL), SHBG, sonic hedgehog signaling molecule (SHH), and complement C3 (C3). Lower TG were associated with lower AFM, GHR, apolipoprotein A1 (APOA1), apolipoprotein E (APOE), cysteine rich with EGF like domains 1 (CRELD1), and crystallin zeta like 1 (CRYZL1) and with higher tissue inhibitor of metallopeptidase inhibitor 2 (TIMP2). All significant proteins remained significant after repeating the analyses without adjusting the clinical variables for medication use.
Pathway analysis
Overexpressed pathways using GSEA on all 78 proteins in Figure 1 were identified using the SomaScan panel (SomaLogic) of proteins as background (Table 2). Pathways overrepresented by the proteins altered after weight loss through bariatric surgery involved inflammation, cellular growth, and apoptosis. These pathways were represented by the interleukins, mitogen‐activated protein kinase (MAPK) signaling proteins, immune/complement system proteins, adipogenesis, and insulinlike growth‐related proteins. CRP, representing acute‐phase inflammation, was limited to the complement pathway. In addition to identifying overexpressed pathways, there were six diseases that were overrepresented by the significant protein changes (Supporting Information Table S12). These diseases or conditions included bone mineral density, obesity/weight, metabolic syndrome, and diabetes.
TABLE 2.
Top differential pathways identified by GSEA of genes identified to be associated with quantitative BMI change or surgery group status differences
| Pathway | Overlap (BMI change; surgery group) | p value (BMI change; surgery group) | Proteins from BMI change or surgery group comparisons in each pathway |
|---|---|---|---|
| Signaling by interleukins | 128/201; 135/201 | 7.91e‐05; 4.97e‐04 | CCL11, IL1R1, IL23R, CNTFR, IL17RB |
| Adipogenesis | 16/31; 17/31 | 2.35e‐03; 9.33e‐03 | LEP, ADIPOQ, CNTFR |
| Cell adhesion molecules | 24/41; 17/41 | 4.67e‐03; 1.63e‐02 | NCAM1, CDH1, CNTN1, NEGR1, PDCD1LG2 |
| Cytokine signaling in immune system | 194/314; 189/314 | 5.19e‐03; 1.87e‐02 | NCAM1, CCL11, MET, GHR, IL1R1, TNFRSF17, PSPN, KIT, IL23R, CNTFR, IL17RB |
| MAPK family signaling cascades | 71/98; 60/98 | 6.36e‐03; 1.73e‐02 | NCAM1, MET, PSPN, KIT |
| Regulation of IGF transport and uptake by IGFBPs | 26/57; 25/57 | 6.83e‐03; 2.41e‐02 | IGFBP2, IGFBP1, SERPINC1, SPP1, SERPIND1 |
| Synthesis of GPI‐anchored proteins | 7/12; 7/12 | 8.81e‐03; 2.52e‐02 | LSAMP, CNTN4, NEGR1, CD109 |
| Complement cascade | 12/33; 16/33 | 1.44e‐02; 3.38e‐03 | CRP, C5a, SERPING1, FCN2, C5, C5b/6 complex |
| IL4 and IL13 signaling | 43/64; 45/64 | 1.76e‐02; 1.45e‐03 | CCL11, IL23R |
Protein examples for each pathway are shown only if the protein was among the 78 Bonferroni significant proteins according to Figure 1.
Abbreviations: GPI, glucose‐6‐phosphate isomerase; GSEA, gene set enrichment analysis; IGF, insulinlike growth factor; IGFBP, IGF binding protein; IL, interleukin; MAPK, mitogen‐activated protein kinase.
Pathways were also defined using only the surgery subset of individuals and GSEA (Figure 4). The pathways with the greatest number of significant clinical variable associations with protein changes involved BMI and HDL‐C. HDL‐C was overexpressed primarily in the inflammation (interleukin signaling) and the immune (cytokine signaling) pathways.
FIGURE 4.

Dot plot from gene set enrichment analysis showing enriched pathways defined by the protein associations with clinical variables. Normalized enrichment score (x‐axis) indicates the distribution of pathway proteins using sign([β] × [−log10(p value)]) from the regression of protein changes with the clinical variable changes. The size of this core‐enriched set is visualized by the size of the dots. Pathway enrichment cutoff for plotting is p < 0.022
Finally, KEGG Brite pathway‐based Voronoi treemaps were also created for 950 out of the 1,297 panel proteins that could be assigned to a specific KEGG Brite level. Supporting Information Figure S3A‐S3C (which can be enlarged to see more detail in any section of the panels) shows the −log10 p value for categories, pathways within categories, and proteins within pathways, respectively, associated with BMI change. BMI change was strongly associated with the janus kinase and signal transducer and activator of transcription (JAK‐STAT) signaling pathway (panel B) within the signal transduction category (panel A). Changes in LEP, LEPR, GHR, and interleukin‐19 (IL19) were the primary signal transduction pathway proteins. Change in BMI was also related to the complement and coagulation cascades, with complement C7 (C7), complement factor B (CFB), and complement factor I (CFI) being the primary proteins. The peroxisome proliferator activated receptor (PPAR) signaling pathway was represented by ADIPOQ. In particular, the HDL‐C β coefficients showed pathway and protein associations very similar to those seen for BMI (Supporting Information Figure S3B‐S3C).
DISCUSSION
As bariatric surgery results in substantial long‐term weight loss, identifying the underlying reasons for risk factor improvements after this large weight loss may make it possible to develop better interventions for patients with severe obesity without the need for major surgery and bariatric surgery’s required lifelong lifestyle changes. The large sample size and long follow‐up period of this study allowed us to test whether the short‐term protein changes observed in this study and previous studies are maintained long term, which is essential prior to pursuit of efforts to mimic the positive effects of these protein changes.
One of the major findings of this study of 378 individuals is that protein changes following gastric bypass surgery were durable 12 years post surgery, despite some weight regain. The majority of the protein changes were significant within 2 years of surgery after weight loss, and the changes reversed direction after the first 2 years according to the amount of weight regain during the following 10 years. These important results support the clinical data that the health benefits of gastric bypass surgery occur soon after the surgery rather than following a significant period of time but, nevertheless, they are maintained in the long term.
Another important finding is that most of the changes in proteins were associated with change in BMI per se and not BMI‐independent pathways induced by the surgical rerouting of the intestine. Furthermore, most proteins changed approximately the same per unit of BMI change, regardless of whether weight was lost or weight was regained. These paired within‐person results strongly support the involvement of BMI change with these protein changes, confirming proteins previously implicated with obesity, including LEP, SHBG, ADIPOQ, GHRL, WFIKKN2, IGFBP1, and IGFBP2 (19, 20, 21). Larger protein changes during rapid weight loss compared with slower weight‐regain periods were seen for several proteins, suggesting either BMI‐independent or pleiotropic mechanisms involving these proteins were activated during rapid weight loss. Although many known obesity‐related proteins were confirmed to be related to long‐term weight loss, proteins not established as obesity‐related proteins were also identified, such as neural cell adhesion molecule 1 (NCAM1).
Finally, the majority of protein changes that were significantly associated with BMI change were related to changes in lipid levels as opposed to blood pressure, liver function, or glucose‐ and insulin‐associated variables. Prior studies have shown that HDL‐C had the largest and most consistent but, at the time unexplained, increase after gastric bypass surgery of all the clinical variables measured (3, 22). The current study expands the previous findings by suggesting APOM, ghrelin, and SHH (cholesterol transferase activity in the endoplasmic reticulum) were significantly related to HDL‐C and the lipoprotein pathway (Supporting Information Figure S2B) and that they may be part of the underlying reason for the sustained 12‐year postsurgical elevation of HDL‐C.
At least three of the eight proteins related to 12‐year remission of diabetes were known obesity‐ and diabetes‐related proteins, including SHBG, ADIPOQ, and possibly APOM. However, of the eight proteins associated with fasting glucose, only MIA SH3 domain containing (MIA), related to the secretion of lipoproteins, pre‐chylomicrons, and pre‐very‐low‐density lipoproteins from the endoplasm (UniProt Knowledgebase, accessed December 23, 2020), had functions considered to be in a diabetes pathway. Therefore, the proteins associated with diabetes remission do not seem to be directly related to plasma glucose or insulin changes.
Specific protein decreases after gastric bypass surgery
Of the 78 proteins identified, 11 proteins decreased in accordance with their known functions. Decreasing LEP is highly correlated with weight loss (23). Weight loss is known to reduce inflammation in accordance with the reduction in CRP, C5.1, and MPO observed in our study. As tissue growth is being opposed by reduced calorie intake and intestinal reabsorption of nutrients, there is less need for GHR, which decreased. AFM decreased, confirming previous findings of higher levels of AFM associated with increased weight, the metabolic syndrome (24), and diabetes incidence (25). APCS is thought to bind to and remove apoptotic cells and it was greatly decreased at 2 years after most of the large tissue losses during the first 2 years stabilized. Ficolin 2 (FCN2) is involved with innate immunity and the lectin complement pathway and it dramatically changed during weight loss. The complement system has been previously shown to be altered after gastric bypass surgery (8, 26).
Specific protein increases after gastric bypass surgery
Proteins with the most significant increases with decreasing BMI included IGFBP1, IGFBP2, ADIPOQ, and APOM. IGFBP1 and IGFBP2 are insulinlike growth factor binding proteins, and higher levels have been associated with reduced glucose intolerance, blood pressure, or nonalcoholic fatty liver disease in multiple cross‐sectional or short‐term intervention studies (8, 27, 28, 29, 30, 31, 32). ADIPOQ has multiple beneficial functions, including improvement in most components of the metabolic syndrome as well as reducing diabetes and atherosclerosis (33).
APOM is a component of HDL and it is thought to be responsible for the anti‐inflammatory effects of HDL. The higher levels of APOM after bariatric surgery correspond to the reduced inflammation present after weight loss (34), which is also supported by a significant association of APOM with remission of diabetes. Because denser HDL particles are the primary carriers of APOM, one might also expect a shift to more dense HDL particles with increasing APOM after surgery. However, the significant increase in cholesterol content in the HDL particle along with lower APOA1 (32) at follow‐up (Supporting Information Table S1) suggests that the particles were less dense.
Clinical variable associations with protein changes
HDL‐C was clearly the mostly strongly associated clinical variable associated with the 71 protein changes that were associated with BMI change over 12 years (Figure 3). HDL‐C increased by approximately 25% 2 years after surgery, an increase that was maintained throughout the 12‐year follow‐up period despite weight regain and that was larger than any other clinical change (3). HDL‐C has a strong positive association with SHBG even after adjusting for multiple cardiovascular risk factors (35). Polymorphisms in the SHBG gene have been associated with abundance of the SHBG protein, HDL‐C, diabetes, and CHD (35, 36, 37). SHBG was tightly clustered with IGFBP2, suggesting additional effects beyond HDL‐C (31). Additionally, decreases in AFM and APOE (associated with decreased TG) and increases in ADIPOQ and APOM (associated with increased HDL‐C) after surgery suggest that these proteins may be responsible for the sustained large increase in HDL‐C. ADIPOQ, APOM, and SHBG were also associated with 12‐year diabetes remission, but not with HbA1c, glucose, or insulin.
The direction of the clinical variable associations suggests that CHD risk should be lowered (38) 12 years after surgery. Seven of the eight protein increases that were related to decreased CHD risk had significant positive associations with HDL‐C and negative associations with LDL‐C. All eight protein increases were also associated with the decreased REE that accompanied BMI decreases.
Pathways
The gene set and Voronoi treemap pathway analyses replicated several pathways associated with weight loss and obesity (20, 32). Major pathways associated with BMI were related to lipid metabolism, adipose tissue functionality, inflammation, cell adhesion, immunity, and coagulation. Reduced inflammation appeared to be a primary benefit of protein changes associated with weight loss and gastric bypass surgery, as suggested by the overexpression of protein changes in the pathways involving signaling by interleukins and cytokine signaling in the immune system. These pathways coincided with the overexpressed HDL‐C pathways, supporting the anti‐inflammatory functions of elevated HDL‐C. The cytokine signaling pathway contains several interleukins and adipocytokines, and adipose tissue colonization by proinflammatory macrophages and macrophage‐derived factors such as tumor necrosis factor α (TNF‐α) and IL‐β are hallmarks of obesity (39, 40).
Voronoi treemaps implicated the protein kinase AMP‐activated catalytic subunit α 2 (AMPK) pathway, which regulates cellular energy stores and preservation of ATP and which is activated by leptin and adiponectin (41, 42), both of which had significant changes in this study. The pathway is involved with inhibition of gluconeogenesis and lipid and protein synthesis. Another pathway, JAK‐STAT, can be activated by cytokines (43) and leptin, and functions include regulating cellular division and inflammation processes and proper adipose function. A third pathway, the phosphatidylinositol‐4,5‐bisphosphate 3‐kinase (PIK3)‐AKT‐RAS pathway, influences cell survival and growth, is involved with lipid phosphorylation, and has been associated with diabetes (44).
Proteomic studies with 12‐year follow‐up or even 5‐year follow‐up have not been reported, to our knowledge, but many of the proteins identified replicated those found in a short‐term weight‐gain and weight‐loss study (20). In that study, coagulation, complement, and inflammation proteins were in primary pathways involved with weight gain and were reversed with weight loss, similar to our long‐term study. Findings in a study of 47 individuals (19 of whom were followed at least 2 years) who had gastric bypass surgery were also similar, with apolipoproteins in particular (including APOM) having the most significant changes, along with complement and inflammation pathways (8). A very low‐calorie diet intervention study of 43 individuals found strong apolipoprotein associations at 1 year (19).
CONCLUSION
Long‐term weight loss after gastric bypass surgery resulted in sustained protein changes despite some weight regain occurring over the 12 years. Most protein changes found associated with BMI change in the discovery sample were replicated in a second sample, although an external long‐term replication cohort would have been preferred. The reversal of direction of protein changes after weight loss when weight was partially regained suggests the findings were robust.
Most protein changes were related to change in BMI rather than non‐weight‐related long‐term surgical effects, suggesting that the beneficial changes of these proteins might be obtained by other methods of weight loss besides bariatric surgery. The direction of the protein and clinical variable changes was consistent with the health benefits and decreased mortality observed after bariatric surgery. These BMI‐associated protein changes were associated with lipid‐related proteins (primarily HDL‐C), greater diabetes remission, and pathways that control inflammation, energy conservation, cell replication and apoptosis, and adipocyte function. The non‐BMI‐related protein changes were related to coagulation and organismal injury pathways.
Mechanisms responsible for sustained higher HDL‐C after gastric bypass surgery; the nature of HDL‐C’s anti‐inflammatory functions; the related protein increases of APOM, SHH, and GHRL in the lipoprotein pathway; and decreases in C3 in multiple pathways need to be clarified in future studies. Known obesity‐related proteins that change 1 to 2 years after bariatric surgery remain changed 12 years after surgery, including LEP, ADIPOQ, SHBG, IGFBP1, IGFBP2, WFIKKN2, and AFM.O
CONFLICT OF INTEREST
The authors declared no conflict of interest.
AUTHOR CONTRIBUTIONS
NAY, KS, and SCH designed the study; RE and HS performed the proteomics assays; NAY and SCH performed the data analysis and wrote the manuscript; RE and FS performed the analyses of the protein pathways; TDA, RDM, SCS, and SCH recruited the patients and collected the clinical data and blood; and all authors edited the final manuscript.
Supporting information
Fig S1
Fig S2
Fig S3
Table S1‐S12
Yousri NA, Engelke R, Sarwath H, et al. Proteome‐wide associations with short‐ and long‐term weight loss and regain after Roux‐en‐Y gastric bypass surgery. Obesity (Silver Spring). 2022;30:129–141. doi: 10.1002/oby.23303
Funding information
This work was supported by a grant (DK‐55006) from the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH, a United States Public Health Service research grant (MO1‐RR00064) from the National Center for Research Resources, Biomedical Research Program funds from Intermountain Healthcare, and grant #NPRP11S‐0114‐180299 from the Qatar National Research Fund (a member of Qatar Foundation). The findings herein are solely the responsibility of the authors.
REFERENCES
- 1. Pou KM, Massaro JM, Hoffmann U, et al. Visceral and subcutaneous adipose tissue volumes are cross‐sectionally related to markers of inflammation and oxidative stress: the Framingham Heart Study. Circulation. 2007;116:1234‐1241. [DOI] [PubMed] [Google Scholar]
- 2. Bays H, Kothari SN, Azagury DE, et al. Lipids and bariatric procedures Part 2 of 2: scientific statement from the American Society for Metabolic and Bariatric Surgery (ASMBS), the National Lipid Association (NLA), and Obesity Medicine Association (OMA). Surg Obes Relat Dis. 2016;12:468‐495. [DOI] [PubMed] [Google Scholar]
- 3. Adams TD, Davidson LE, Litwin SE, et al. Weight and metabolic outcomes 12 years after gastric bypass. N Engl J Med. 2017;377:1143‐1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolotkin RL, Kim J, Davidson LE, Crosby RD, Hunt SC, Adams TD. 12‐year trajectory of health‐related quality of life in gastric bypass patients versus comparison groups. Surg Obes Relat Dis. 2018;14:1359‐1365. [DOI] [PubMed] [Google Scholar]
- 5. Adams TD, Gress RE, Smith SC, et al. Long‐term mortality after gastric bypass surgery. N Engl J Med. 2007;357:753‐761. [DOI] [PubMed] [Google Scholar]
- 6. Wing RR, Phelan S. Long‐term weight loss maintenance. Am J Clin Nutr. 2005;82(1 suppl):222S‐225S. [DOI] [PubMed] [Google Scholar]
- 7. Zhou K, Wolski K, Malin SK, et al. Impact of weight loss trajectory following randomization to bariatric surgery on long‐term diabetes glycemic and cardiometabolic parameters. Endocr Pract. 2019;25:572‐579. [DOI] [PubMed] [Google Scholar]
- 8. Wewer Albrechtsen NJ, Geyer PE, Doll S, et al. Plasma proteome profiling reveals dynamics of inflammatory and lipid homeostasis markers after Roux‐en‐Y gastric bypass surgery. Cell Syst. 2018;7:601‐612.e3. [DOI] [PubMed] [Google Scholar]
- 9. Shah RV, Hwang S‐J, Yeri A, et al. Proteins altered by surgical weight loss highlight biomarkers of insulin resistance in the community. Arterioscler Thromb Vasc Biol. 2019;38:107‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodríguez‐Rivera C, Pérez‐García C, Muñoz‐Rodríguez JR, et al. Proteomic identification of biomarkers associated with eating control and bariatric surgery outcomes in patients with morbid obesity. World J Surg. 2019;43:744‐750. [DOI] [PubMed] [Google Scholar]
- 11. Doulamis IP, Konstantopoulos P, Tzani A, et al. Visceral white adipose tissue and serum proteomic alternations in metabolically healthy obese patients undergoing bariatric surgery. Cytokine. 2019;115:76‐83. [DOI] [PubMed] [Google Scholar]
- 12. Adams TD, Avelar E, Cloward T, et al. Design and rationale of the Utah obesity study. A study to assess morbidity following gastric bypass surgery. Contemp Clin Trials. 2005;26:534‐551. [DOI] [PubMed] [Google Scholar]
- 13. Adams TD, Pendleton RC, Strong MB, et al. Health outcomes of gastric bypass patients compared to nonsurgical, nonintervened severely obese. Obesity (Silver Spring). 2010;18:121‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson PW, Castelli WP, Kannel WB. Coronary risk prediction in adults (the Framingham Heart Study). Am J Cardiol. 1987;59:91G‐94G. [DOI] [PubMed] [Google Scholar]
- 15. Davies DR, Gelinas AD, Zhang C, et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A. 2012;109:19971‐19976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431‐432. [DOI] [PubMed] [Google Scholar]
- 18. Wing RR, Hill JO. Successful weight loss maintenance. Annu Rev Nutr. 2001;21:323‐341. [DOI] [PubMed] [Google Scholar]
- 19. Geyer PE, Wewer Albrechtsen NJ, Tyanova S, et al. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol Syst Biol. 2016;12:901. doi: 10.15252/msb.20167357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piening BD, Zhou W, Contrepois K, et al. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 2018;6:157‐170.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zaghlool SB, Sharma S, Molnar M, et al. Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nat Commun. 2021;12:1279. doi: 10.1038/s41467-021-21542-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sjöström L, Lindroos A‐K, Peltonen M, et al. Lifestyle, diabetes, and cardiovascular risk factors 10 years after bariatric surgery. N Engl J Med. 2004;351:2683‐2693. [DOI] [PubMed] [Google Scholar]
- 23. Rosenbaum M, Nicolson M, Hirsch J, Murphy E, Chu F, Leibel RL. Effects of weight change on plasma leptin concentrations and energy expenditure. J Clin Endocrinol Metab. 1997;82:3647‐3654. [DOI] [PubMed] [Google Scholar]
- 24. Dieplinger H, Dieplinger B. Afamin–A pleiotropic glycoprotein involved in various disease states. Clin Chim Acta. 2015;446:105‐110. doi 10.1016/j.cca.2015.04.010 [DOI] [PubMed] [Google Scholar]
- 25. Kollerits B, Lamina C, Huth C, et al. Plasma concentrations of afamin are associated with prevalent and incident type 2 diabetes: a pooled analysis in more than 20,000 individuals. Diabetes Care. 2017;40:1386‐1393. [DOI] [PubMed] [Google Scholar]
- 26. Carayol J, Chabert C, Di Cara A, et al. Protein quantitative trait locus study in obesity during weight‐loss identifies a leptin regulator. Nat Commun. 2017;8:2084. doi: 10.1038/s41467-017-02182-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sandhu MS, Heald AH, Gibson JM, Cruickshank JK, Dunger DB, Wareham NJ. Circulating concentrations of insulin‐like growth factor‐I and development of glucose intolerance: a prospective observational study. Lancet. 2002;359:1740‐1745. [DOI] [PubMed] [Google Scholar]
- 28. Rajwani A, Ezzat V, Smith J, et al. Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes. 2012;61:915‐924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cominetti O, Núñez Galindo A, Corthésy J, et al. Obesity shows preserved plasma proteome in large independent clinical cohorts. Sci Rep. 2018;8:16981. doi: 10.1038/s41598-018-35321-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choi H, Koh HWL, Zhou L, et al. Plasma protein and microRNA biomarkers of insulin resistance: a network‐based integrative ‐omics analysis. Front Physiol. 2019;10:379. doi: 10.3389/fphys.2019.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niu L, Geyer PE, Wewer Albrechtsen NJ, et al. Plasma proteome profiling discovers novel proteins associated with non‐alcoholic fatty liver disease. Mol Syst Biol. 2019;15:e8793. doi: 10.15252/msb.20188793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bruderer R, Muntel J, Müller S, et al. Analysis of 1508 plasma samples by capillary‐flow data‐independent acquisition profiles proteomics of weight loss and maintenance. Mol Cell Proteomics. 2019;18:1242‐1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hotta K, Funahashi T, Arita Y, et al. Plasma concentrations of a novel, adipose‐specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol. 2000;20:1595‐1599. [DOI] [PubMed] [Google Scholar]
- 34. Frej C, Mendez AJ, Ruiz M, et al. A shift in ApoM/S1P between HDL‐particles in women with type 1 diabetes mellitus is associated with impaired anti‐inflammatory effects of the ApoM/S1P complex. Arterioscler Thromb Vasc Biol. 2017;37:1194‐1205. [DOI] [PubMed] [Google Scholar]
- 35. Kurnaz‐Gomleksiz O, Akadam‐Teker B, Bugra Z, Omer B, Yilmaz‐Aydogan H. Genetic polymorphisms of the SHBG gene can be the effect on SHBG and HDL‐cholesterol levels in coronary heart disease: a case‐control study. Mol Biol Rep. 2019;46:4259‐4269. [DOI] [PubMed] [Google Scholar]
- 36. Perry JRB, Weedon MN, Langenberg C, et al. Genetic evidence that raised sex hormone binding globulin (SHBG) levels reduce the risk of type 2 diabetes. Hum Mol Genet. 2010;19:535‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ding EL, Song Y, Manson JE, et al. Sex hormone‐binding globulin and risk of type 2 diabetes in women and men. N Engl J Med. 2009;361:1152‐1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li T, Yang L, Zhao S, Zhang S. Correlation between apolipoprotein M and inflammatory factors in obese patients. Med Sci Monit. 2018;24:5698‐5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caër C, Rouault C, Le Roy T, et al. Immune cell‐derived cytokines contribute to obesity‐related inflammation, fibrogenesis and metabolic deregulation in human adipose tissue. Sci Rep. 2017;7:3000. doi: 10.1038/s41598-017-02660-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmidt FM, Weschenfelder J, Sander C, et al. Inflammatory cytokines in general and central obesity and modulating effects of physical activity. PLoS One. 2015;10:e0121971. doi: 10.1371/journal.pone.0121971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin TL, Alquier T, Asakura K, Furukawa N, Preitner F, Kahn BB. Diet‐induced obesity alters AMP kinase activity in hypothalamus and skeletal muscle. J Biol Chem. 2006;281:18933‐18941. [DOI] [PubMed] [Google Scholar]
- 42. Kubota N, Yano W, Kubota T, et al. Adiponectin stimulates AMP‐activated protein kinase in the hypothalamus and increases food intake. Cell Metab. 2007;6:55‐68. [DOI] [PubMed] [Google Scholar]
- 43. Richard AJ, Stephens JM. The role of JAK‐STAT signaling in adipose tissue function. Biochem Biophys Acta. 2014;1842:431‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang X, Liu G, Guo J, Su Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int J Biol Sci. 2018;14:1483‐1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Table S1‐S12


