Abstract
Aerobic methanotrophy is strongly controlled by copper, and methanotrophs are known to use different mechanisms for copper uptake. Some methanotrophs secrete a modified polypeptide—methanobactin—while others utilize a surface-bound protein (MopE) and a secreted form of it (MopE*) for copper collection. As different methanotrophs have different means of sequestering copper, competition for copper significantly impacts methanotrophic activity. Herein, we show that Methylomicrobium album BG8, Methylocystis sp. strain Rockwell, and Methylococcus capsulatus Bath, all lacking genes for methanobactin biosynthesis, are not limited for copper by multiple forms of methanobactin. Interestingly, Mm. album BG8 and Methylocystis sp. strain Rockwell were found to have genes similar to mbnT that encodes for a TonB-dependent transporter required for methanobactin uptake. Data indicate that these methanotrophs “steal” methanobactin and such “theft” enhances the ability of these strains to degrade methylmercury, a potent neurotoxin. Further, when mbnT was deleted in Mm. album BG8, methylmercury degradation in the presence of methanobactin was indistinguishable from when MB was not added. Mc. capsulatus Bath lacks anything similar to mbnT and was unable to degrade methylmercury either in the presence or absence of methanobactin. Rather, Mc. capsulatus Bath appears to rely on MopE/MopE* for copper collection. Finally, not only does Mm. album BG8 steal methanobactin, it synthesizes a novel chalkophore, suggesting that some methanotrophs utilize both competition and cheating strategies for copper collection. Through a better understanding of these strategies, methanotrophic communities may be more effectively manipulated to reduce methane emissions and also enhance mercury detoxification in situ.
Subject terms: Soil microbiology, Biogeochemistry, Bacterial genetics
Introduction
Aerobic methanotrophs play a critical role in the biogeochemical cycling of carbon [1, 2]. More specifically, these intriguing microbes consume substantial amounts of methane generated via methanogenesis [3], and thus may be invaluable means or “levers” to control not only future emissions of methane, but also remove methane from the atmosphere [4–8].
Any application of aerobic methanotrophy, however, requires a detailed understanding of their metabolism, particularly their need for trace metals. Specifically, expression and activity of alternative forms of methane monooxygenase (MMO, responsible for the conversion of methane to methanol), are controlled by copper, or the canonical “copper-switch” [9]. There are two forms of MMO—a cytoplasmic or soluble methane monooxygenase (sMMO) and a membrane-bound or particulate methane monooxygenase (pMMO). sMMO—a soluble di-iron-containing enzyme [10]—is only expressed under copper limitation [11, 12]. Expression and activity of pMMO—a copper and iron-containing enzyme [13, 14]—increases with increasing copper [11, 12]. The two forms of MMO have widely different properties, e.g., cells expressing sMMO have high methane turnover but poor affinity for methane while pMMO-expressing cells have lower turnover but greater affinity [15]. Given that methane oxidation is critical for methanotrophic growth, copper sequestration is also very important to these microbes, and methanotrophs have been found to have multiple mechanisms of copper uptake.
The first well-characterized copper-binding compound or chalkophore—methanobactin (MB)—is secreted by some methanotrophs of the Methylocystaceae family within the Alphaproteobacteria, e.g., Methylosinus trichosporium OB3b and Methylocystis sp. strain SB2 [16–18]. MB is a modified polypeptide containing two heterocyclic rings with associated thioamide groups that are responsible for copper binding with extremely high affinity [19]. To date, two forms of MB have been described—Group I MBs with two oxazolone rings and an internal disulfide bridge (e.g., MB from Ms. trichosporium OB3b) and Group II MBs that contain one oxazolone ring and one pyrazinedione or imidazolone ring as well as a sulfate group (e.g., MB from Methylocystis sp. strain SB2 [17, 19]). The gene encoding the polypeptide precursor of MB has been identified (mbnA [11]) as have several genes involved in ring formation (mbnBCN [20, 21]) and MB uptake (mbnT, encoding a TonB-dependent transporter [22]). Not all methanotrophs, however, can produce MB. Rather, methanotrophs of the Methylococcaceae family of the Gammaproteobacteria rely on an outer membrane protein (MopE) and a secreted form of this molecule (MopE*) for copper sequestration [23–27]. Finally, some Methylocystaceae methanotrophs lack both MB and the MopE/MopE* systems for copper uptake, suggesting that they collect copper by some unknown system(s) [28].
Given the importance of copper in methanotrophy, this raises several intriguing questions. First, do methanotrophs that express MB have a competitive advantage for copper sequestration? Competition between methanotrophs for copper is likely, with such competition affecting overall methanotrophic activity. Second, given that MB is secreted into the environment to collect copper, can copper-MB complexes be “stolen” by other microbes? Such a phenomenon would require non-MB-expressing methanotrophs to have the uptake system identified for MB, i.e., MbnT [22, 29]. Such “theft” would not be unprecedented, as many microbes have been found to act as “cheaters” where they steal siderophores produced by others to collect iron [30–33]. Further, it has been found that methanotrophs that produce and take up MB are able to degrade the potent neurotoxin methylmercury (MeHg) [34]. If some methanotrophs act as MB-cheaters, does such theft enable these microbes to degrade MeHg? Herein we examine several methanotrophs unable to produce MB to determine: (1) if copper requirements of these methanotrophs can be met either through MB theft and/or competition (i.e., expression of some other copper uptake system) and (2) if MB theft enables these microbes to degrade MeHg.
Materials and methods
Identification of putative MbnTs in Mm. album BG8, Methylocystis sp. strain Rockwell, and Mc. capsulatus Bath
Predicted MbnT amino acid sequences of Ms. trichosporium OB3b (ADVE02_v2_13651) and Methylocystis sp. strain SB2 (MSB2v1_460017) were used to search for putative mbnT genes in the genomes of Mm. album BG8, Methylocystis sp. strain Rockwell, and Mc. capsulatus Bath using tblastn or blastp ([35]).
Growth conditions
Initial inocula of Mm. album BG8 and Mc. capsulatus Bath were grown in nitrate mineral salts (NMS) while Methylocystis sp. strain Rockwell was grown in ammonium mineral salts (AMS) media [36], all with 1 µM copper (as CuCl2). Mm. album BG8 and Methylocystis sp. strain Rockwell were grown at 30 °C, and Mc. capsulatus Bath at 45 °C in 250-mL sidearm Erlenmeyer flasks while shaken at 220 rpm in the dark. CH4 was supplemented at a CH4-to-air ratio of 1:2. MBs from Ms. trichosporium OB3b (OB3b-MB) and Methylocystis sp. strain SB2 (SB2-MB) were purified as described previously [37]. Cu-MB stocks were freshly prepared by adding CuCl2 and either OB3b-MB or SB2-MB at 1:5 molar ratio and incubating in the dark at 30 °C for 1 h [38]. Cu-triethylenetetramine (TRIEN, a strong abiotic chelator of copper [39]) was prepared by adding CuCl2 and TRIEN at 1:5 molar ratio. To investigate the effect of copper chelation on the methanotrophic growth, cultures grown with 1 µM copper were washed with fresh NMS or AMS media and then transferred to four different conditions: 0 μM Cu, 1 μM Cu, 1 μM Cu + 5 μM OB3b-MB, and 1 μM Cu + 5 μM SB2-MB. Mm. album BG8 and Methylocystis sp. strain Rockwell were also grown in the presence of TRIEN to determine if these strains produce novel chalkophores in the presence of an abiotic competitive ligand. After reaching the stationary phase, all cultures were washed with fresh NMS or AMS media and used to inoculate a new set of flasks for a second growth cycle. Methanotrophic growth in the presence of varying amounts of copper and copper-chelating agents was non-invasively monitored by measuring the optical density at 600 nm (OD600) in sidearm flasks using a Genesys 20 Visible spectrophotometer (Spectronic Unicam, Waltham, MA). All conditions were run in biological triplicates.
Escherichia coli used for mutant construction in Mm. album BG8 was grown in Luria broth medium (Dot Scientific, Burton, MI). Kanamycin was used for maintaining E. coli and Mm. album BG8 containing mutant construct plasmids (25 and 10 μg mL−1, respectively). Nalidixic acid (15 μg mL−1) was used to remove residual E. coli strain S17-1 after conjugation with Mm. album BG8.
RNA extraction and cDNA synthesis
Total RNA from cultures at the end of the second growth cycle was extracted as described previously [11, 40]. Removal of DNA was confirmed by the absence of products from polymerase chain reaction (PCR) with the universal primers 27F and 1492R targeting 16S rRNA gene (Table S1). cDNA was synthesized from DNA-free RNA samples using Superscript III reverse transcriptase kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Genes involved in methane oxidation (pmoA and mmoX), methanol oxidation (mxaF), putative MB uptake (Metal_1282 in Mm. album BG8, hereafter called mbnT-BG8; MSPATv1_230027 and MSPATv1_550006 in Methylocystis sp. strain SB2, hereafter called mbnT1-Rockwell and mbnT2-Rockwell; MCA1957 in Mc. capsulatus Bath, hereafter called mbnT-Bath; and mopE in Mc. capsulatus Bath (MCA2589)), and copper storage (csp3 in Mm. album BG8 (Metal_0689) and MSPATv1_280020 in Methylocystis sp. strain Rockwell, hereafter called csp1) were quantified by RT-qPCR. Expression of 16s rRNA genes was used as an internal reference. Primer sets used for qPCR of these select genes are shown in Table S1 and calibration curves generated as shown in Figs. S1–S3. qPCR reactions were performed as described previously [40]. Threshold cycle (CT) values were imported from CFX Manager Software (Bio-Rad, Hercules, CA) to calculate relative gene expression levels with 16S rRNA as the internal standard by the comparative threshold amplification cycle method [41]. Measurements were performed for at least biological duplicates for each condition.
Construction of mbnT mutant of Mm. album BG8
The gene most similar to any mbnT in Mm. album BG8 (Metal_1282) was knocked out via markerless mutagenesis as described previously [20]. Primers used in this study are shown in Table S1. Deletion of the putative MB uptake gene in Mm. album BG8 ΔmbnT was confirmed by checking for kanamycin sensitivity as well as via PCR and sequencing (Fig. S4).
Metal analysis
Copper uptake by Mm. album BG8 wild type and ΔmbnT strains, Methylocystis sp. strain Rockwell, and Mc. capsulatus Bath at the end of the second growth cycle was determined as described previously [38]. Triplicate biological samples were analyzed for every condition.
Immunoblotting
Monoclonal antibody (10B10) to MB from Ms. trichosporium OB3b was produced and purified as described earlier [22, 42]. 1,4-phenylene diisothiocyanate-derivatized polyvinylidene difluoride membranes were modified as previously described [43], with changes as detailed in the supplementary material. A detailed method of immunoblotting is provided in the supplementary material.
Mm. album BG8 chalkophore purification and characterization
To isolate a putative novel chalkophore produced by Mm. album BG8, cultures grown in the presence of 1 μM copper and 5 μM TRIEN were first centrifuged at 4300 × g for 10 min. Supernatant was collected and filtered through a 0.2-μm PES filter unit (Thermo Scientific, Waltham MA). A reversed-phase C18 Sep-Pak cartridge (Waters Corp., Milford MA) was sequentially conditioned with 3 mL methanol, 3 mL 60% acetonitrile, 3 mL methanol, and 6 mL H2O, and then loaded with the filtered spent medium. The chalkophore bound to the column was washed with 6 mL H2O, then eluted with 60% acetonitrile until a yellow band was collected. The eluant was frozen at −80 °C and lyophilized for chalkophore concentration and removal of acetonitrile (FreeZone 6 Freeze Dry System, Labconco, Kansas City MO). For larger scale production and isolation of the chalkophore from Mm. album BG8, cells were cultured in a 15 L Solida fermenter (Solida Biotechnology, Munich Germany) using the culture conditions described above. The chalkophore was isolated from the spent media and purified as described for MB from Methylocystis sp. strain SB2 [44] except the chalkophore from Mm. album BG8 was eluted from the Targa C18 column in the 65-75% methanol: H2O fraction. UV-visible spectroscopy was recorded on a Cary 50 (Agilent, Santa Clara, CA). Matrix-assisted laser desorption/ionization mass spectroscopy (MALDI-MS) was performed on a Shimadzu AXIMA Confidence MALDI TOF Mass Spectrometer (Shimadzu Corp., Kyoto, Japan) using a mixture of 2,5-dihydroxybenzoic acid and 2-hydroxy-5-methoxybenzoic acid (SuperDHB) in a 1:1 matrix to sample mixture. Electrospray ionization (ESI)MS/MS was performed on an Agilent LC using a Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer with an HCD fragmentation cell and an Agilent 1260 Infinity Capillary Pump with an Agilent Zorbax SB-C18, 0.5 mm × 150 mm, 5 micron, part#5064-8256 using 0.1% formic acid/water and 0.1% formic acid/acetonitrile.
Methylmercury degradation assays
MeHg degradation by Mm. album BG8 wild type and ΔmbnT, Methylocystis sp. strain Rockwell, and Mc. capsulatus Bath was measured in biological triplicate samples as described earlier [34, 45–47]. Experiments were performed in the absence/presence of MB from either Ms. trichosporium OB3b or Methylocystis sp. strain SB2. The final concentrations of cells, MeHg, and MB (if added) were 108 cells mL−1, 5 nM, and 45 µM, respectively [46].
Statistical analyses
Data were analyzed using the Tukey’s honestly significant difference test or Student’s t-test. Potential outliers of biological triplicates were determined using the Grubbs’ outlier test. Microbial growth was fitted to a logistic curve using the R package growthcurver [48] to calculate growth rates. All statistical analyses were performed using R version 3.4.4 [49].
Results
Identification of putative MbnTs in Mm. album BG8, Methylocystis sp. strain Rockwell, and Mc. capsulatus Bath
In Mm. album BG8, one gene encoding for a putative TonB-dependent receptor was most similar to both OB3b-MbnT and SB2-MbnT (Metal_1282; Table S2). Similarity was much greater, however, to OB3b-MbnT (E value = 2 × 10−145, identity = 36%) than SB2-MbnT (E value = 1 × 10−12, identity = 28%). For Methylocystis sp. strain Rockwell, several genes encoding for putative TonB-dependent transporters similar to OB3b-MbnT were found in the genome of Methylocystis sp. strain Rockwell, of which MSPATv1_230027 exhibited the highest similarity (E value = 1 × 10−136, identity = 36%; Table S2). Two genes encoding for putative TonB-dependent transporters highly similar to SB2-MbnT (MSPATv1_550006 and MSPATv1_50173; E value = 0.0 for both) were found in the genome of Methylocystis sp. strain Rockwell. The identification of MSPATv1_550006 to SB2 MbnT, however, was much higher than that found for MSPATv1_50173, i.e., 65% vs. 42%. For Mc. capsulatus Bath, no TonB-dependent transporter was found to have significant similarity to either OB3b-MbnT or SB2-MbnT, the closest being MCA1957 to OB3b-MbnT (E value = 2 × 10−6, identity = 21%) and MCA2074 to SB2-MbnT (E value of 9 × 10−13, identity = 28%). Expression of genes with high similarity to known mbnT genes, i.e., Metal_1282 (mbnT-BG8), MSPATv1_230027 (mbnT1-Rockwell), MSPATv1_550006 (mbnT2-Rockwell), and MCA1957 (mbnT-Bath) was monitored under different growth conditions as described below.
Growth of Mm. album BG8 wild type and ΔmbnT mutant in the presence of varying amounts of copper, MB, and TRIEN
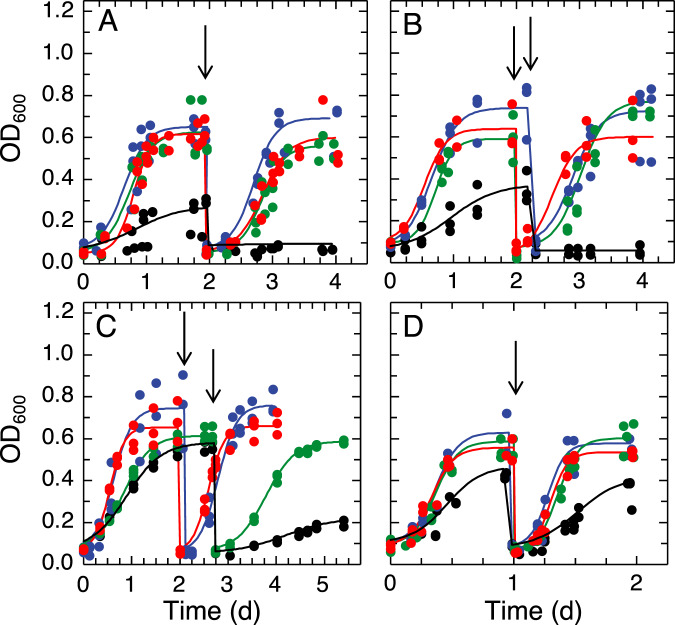
Growth of Mm. album BG8 wild type was strongly dependent on the availability of copper as described previously [50]. Growth clearly occurred in the presence of 1 µM copper, but was significantly reduced with no added copper in the first growth cycle (final OD600 of 0.60 ± 0.03 vs. 0.24 ± 0.10; p = 0.018) (Fig. 1A, S5A). Growth was abolished when this culture was transferred a second time to copper-free medium, indicating that original growth was likely due to the transfer of a small amount of copper with the initial inoculum. Such a result is not unexpected as Mm. album BG8 can only express pMMO that requires copper for its activity.
Fig. 1. Methanotrophic growth in the presence of varying amounts of copper and methanobactin.
Growth of A Mm. album BG8 wild type, B Mm. album BG8 ΔmbnT, C Methylocystis sp. strain Rockwell, and D Mc. capsulatus Bath with 0 μM Cu (black), 1 μM Cu (blue), 1 μM Cu, and 5 μM OB3b-MB (green), and 1 μM Cu and 5 μM SB2-MB (red). Solid lines indicate data fitted to a logistic growth curve using growthcurver [48], and arrows indicate the beginning of second growth cycle.
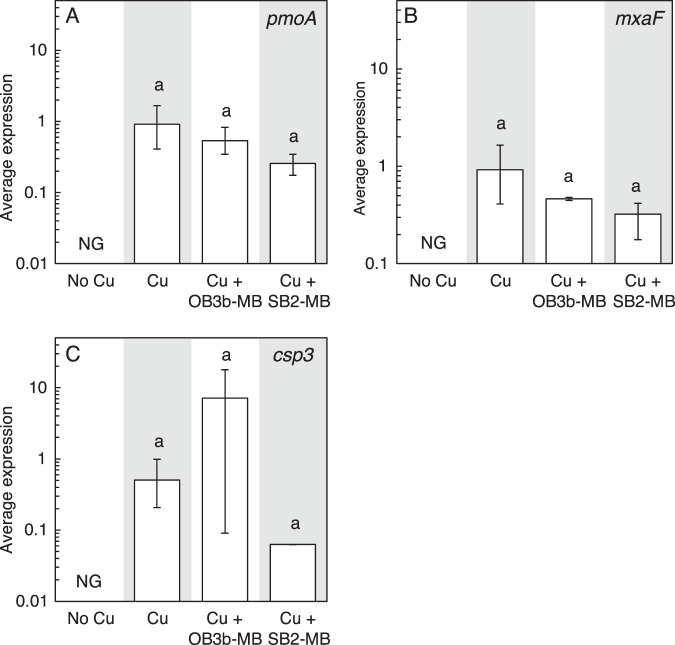
The addition of either 5 µM OB3b-MB (a Group I MB) or SB2-MB (a Group II MB) in the presence of 1 µM copper did not affect the growth of Mm. album BG8 as compared to growth in the presence of 1 µM copper only, indicating that neither form of MB inhibited copper uptake (Fig. 1A, S5A). This was confirmed by measuring copper associated with biomass at the end of the second growth cycle—no significant difference was found for cultures of Mm. album BG8 grown with copper and either OB3b-MB or SB2-MB (Fig. 2A). Expression of various genes involved either in copper storage (csp3), carbon oxidation (pmoA and mxaF), or putative MB uptake (mbnT-BG8) was not significantly affected by the addition of either type of MB (Fig. 3). Growth of the Mm. album BG8 ΔmbnT mutant was comparable to that of wild type under all conditions tested (Fig. 1B, S5B). Copper uptake by Mm. album BG8 ΔmbnT was also not affected by the addition of either form of MB, nor was expression of various genes involved in methane oxidation or copper storage (Figs. 2B, 4).
Fig. 2. Copper associated with methanotrophic biomass in the presence of varying amounts of copper and methanobactin.
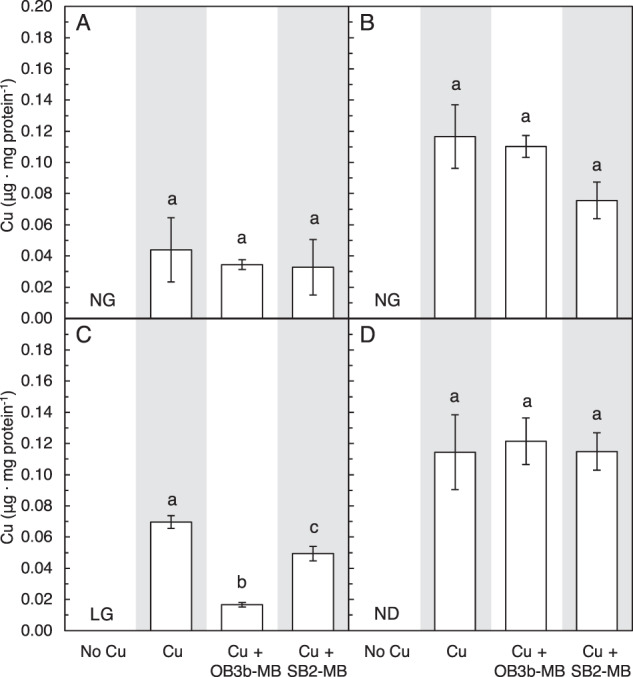
Copper associated with A Mm. album BG8 wild type, B Mm. album BG8 ΔmbnT, C Methylocystis sp. strain Rockwell, and D Mc. capsulatus Bath biomass at the end of the second growth cycle with 0 μM Cu, 1 μM Cu, and 5 μM OB3b-MB, 1 μM Cu and 5 μM SB2-MB. Error bar indicates the standard deviation of biological triplicate samples. Letter over bars indicates no significant differences determined by Tukey’s honestly significant difference test (p < 0.05). No detected (ND) copper associated with biomass and no growth (NG) is indicated. Low growth (LG) indicates insufficient biomass for metal analysis.
Fig. 3. Gene expression in Mm. album BG8 wild type.
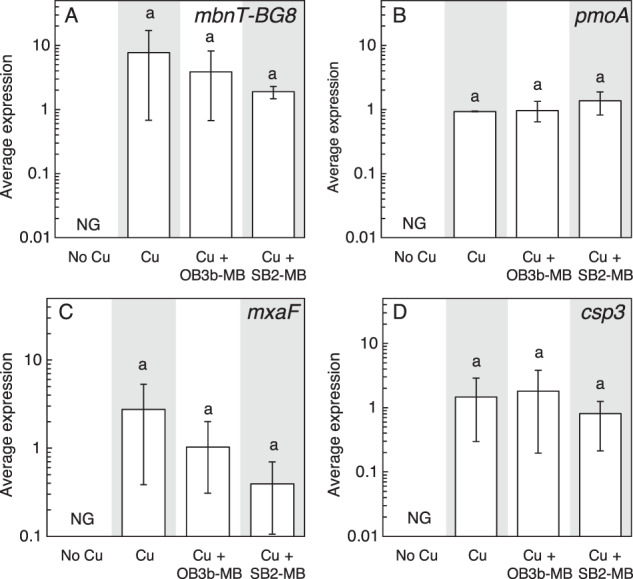
RT-qPCR of A mbnT-BG8, B pmoA, C mxaF, and D csp3 in Mm. album BG8 wild type grown with or without 1 μM Cu and 5 μM MB. Error bar indicates the range of biological duplicate or triplicate samples. Letter over bars indicates no significant differences determined by Tukey’s honestly significant difference test (p < 0.05). No growth is indicated as NG.
Fig. 4. Gene expression in Mm. album BG8 ΔmbnT.
RT-qPCR of A pmoA, B mxaF, and C csp3 in Mm. album BG8 ΔmbnT grown with or without 1 μM Cu and 5 μM MB. Error bar indicates the range of biological duplicate or triplicate samples. Letter over bars indicates no significant differences determined by Tukey’s honestly significant difference test (p < 0.05). No growth is indicated as NG.
The addition of 5 µM TRIEN in the presence of 1 µM copper significantly inhibited the growth of Mm. album BG8 wild type as compared to growth in the presence of copper alone (Figs. S5A and S6A). The addition of either form of MB did not improve the growth of Mm. album BG8 wild type in the presence of copper and TRIEN (Figs. S5A and S6A). Expression of various genes involved in methane/methanol oxidation (pmoA, mxaF) or copper storage (csp3) was not significantly affected in Mm. album BG8 grown in the presence of TRIEN, copper, and/or either form of MB (Fig. S7), nor was copper uptake (Fig. S8A). The addition of OB3b-MB in conjunction with TRIEN did reduce the growth of Mm. album BG8 ΔmbnT as compared to the presence of copper alone or copper plus TRIEN (Figs. S5B and S6B). Expression of various genes involved in methane/methanol oxidation (pmoA, mxaF) or copper storage (csp3) was not significantly affected in Mm. album BG8 ΔmbnT (Fig. S9), nor was copper uptake when the mutant was grown in the presence of TRIEN with or without either form of MB, although the mutant collected more copper in the presence of SB2-MB vs. OB3b-MB (Fig. S8B).
Growth of Methylocystis sp. strain Rockwell in the presence of varying amounts of copper, MB and TRIEN
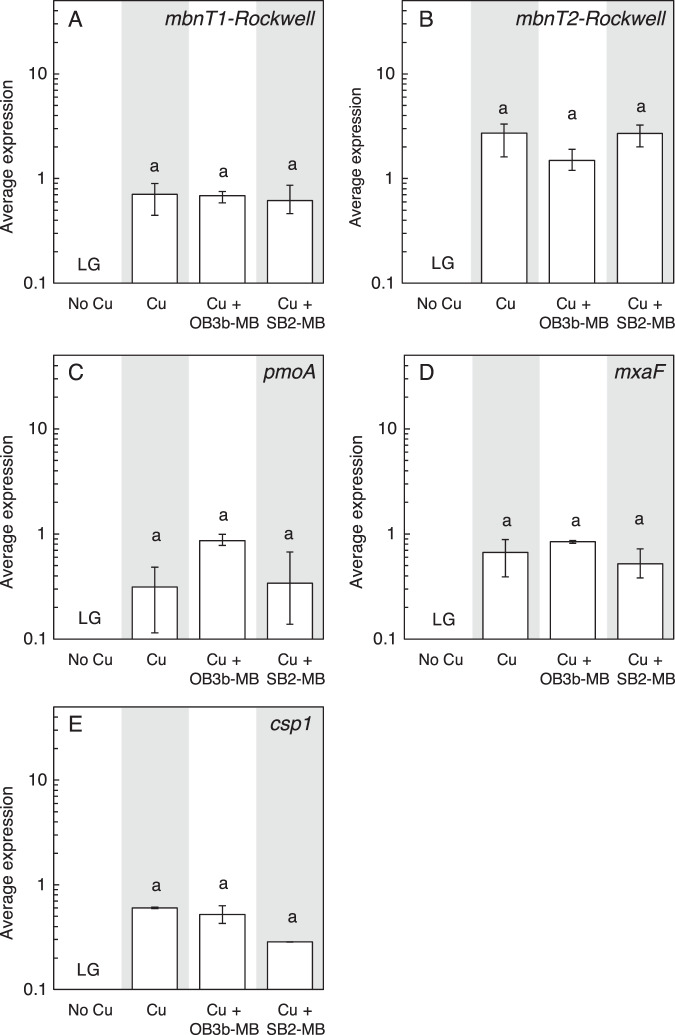
Similar to Mm. album BG8, Methylocystis sp. strain Rockwell cannot express sMMO, and its growth was inhibited in the absence of copper as compared to the presence of 1 µM copper (Fig. 1C, S5C). Addition of SB2-MB in the presence of copper did not affect the growth of Methylocystis sp. strain Rockwell, whereas OB3b-MB significantly reduced growth (Fig. 1C, S5C). Expression of various genes involved in carbon assimilation (pmoA, mxaF), copper storage (csp1), or putative MB uptake (mbnT1-Rockwell, mbnT2-Rockwell) in Methylocystis sp. strain Rockwell was not affected by the addition of either form of MB (Fig. 5). Overall, 5 µM TRIEN significantly inhibited the growth of Methylocystis sp. strain Rockwell in the presence of 1 µM copper, which was resolved only in the presence of 5 µM SB2-MB (Figs. S5C and S6C). Copper uptake by Methylocystis sp. strain Rockwell was significantly reduced in the presence of OB3b-MB, but not in the presence of SB2-MB, regardless if TRIEN was present or not (Fig. 2C, S8C). Expression of mbnT1-Rockwell and csp1 of Methylocystis sp. strain Rockwell increased in the presence of copper, TRIEN, and OB3b-MB as compared to that under 1 µM copper while no significant difference was observed for pmoA, mbnT2-Rockwell, or mxaF when comparing these two conditions (Fig. S10).
Fig. 5. Gene expression in Methylocystis sp. Rockwell.
RT-qPCR of A mbnT1-Rockwell, B mbnT2-Rockwell, C pmoA, D mxaF, and E csp1 in Methylocystis sp. Rockwell grown with or without 1 μM Cu and 5 μM MB. Error bar indicates the range of biological duplicate or triplicate samples. Letter over bars indicates no significant differences determined by Tukey’s honestly significant difference test (p < 0.05). Low growth is indicated as LG.
Growth of Mc. capsulatus Bath in the presence of varying amounts of copper and methanobactin
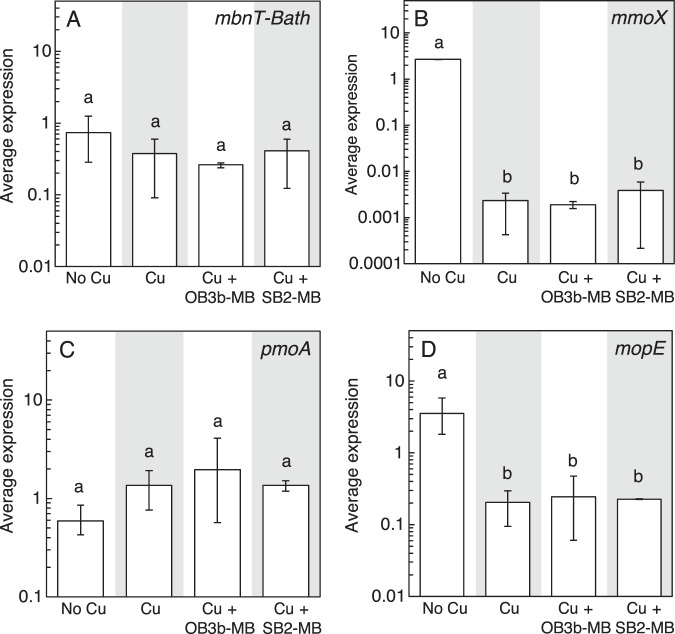
Mc. capsulatus Bath grows in both the presence and absence of copper as it can express both forms of MMO (Fig. 1D, S5D). Growth was faster and more extensive in the presence of copper, indicating that, as found earlier [51], Mc. capsulatus Bath has greater carbon conversion efficiency under pMMO-expressing conditions. Addition of either OB3b-MB or SB2-MB in the presence of copper did not affect growth (Fig. 1D, S5D). Copper uptake by Mc. capsulatus Bath was also not affected by the presence of either form of MB (Fig. 2D). Expression of various genes by Mc. capsulatus Bath was not affected by the addition of MB including a putative MB uptake system (mbnT-Bath; Fig. 6). Only the presence/absence of copper had any significant effect on gene expression, and then only on mmoX (encoding for a subunit of the sMMO) and mopE (encoding for a copper uptake protein). Activity of sMMO was also not affected by the presence of either form of MB, i.e., activity via the naphthalene assay was only evident in the absence of copper (Fig. S11).
Fig. 6. Gene expression in Mc. capsulatus Bath.
RT-qPCR of A mbnT-Bath, B mmoX, C pmoA, and D mopE in Mc. capsulatus Bath grown with or without 1 μM Cu and 5 μM MB. Error bar indicates the range of biological duplicate or triplicate samples. Letter over bars indicates no significant differences determined by Tukey’s honestly significant difference test (p < 0.05).
Localization of MB via Immunoblotting in Mm. album BG8
To determine if methanotrophs can take up foreign MB, immunoblotting assays were first performed. Monoclonal antibodies were successfully raised to OB3b-MB, but repeated attempts to generate high-affinity antibodies in rats and mice were unsuccessful for SB2-MB (data not shown). Control immunoblots showed successful monoclonal antibody hybridization to OB3b-MB, but not to lysozyme or E. coli cell extracts (Fig. S12). Monoclonal OB3b-MB antibody (10B10), however, cross-hybridized with cell extracts of Mm. album BG8 grown in the presence of 1 µM copper and absence of OB3b-MB over two growth cycles. Greater hybridization to Mm. album BG8 cell extract was observed in the presence of 1 µM copper + 5 µM OB3b-MB than in the absence of OB3b-MB (Fig. S12), but very little hybridization was observed in the spent medium or wash buffer when Mm. album BG8 was grown in the presence of OB3b-MB (Fig. S12). These data suggest that Mm. album BG8 produces some compound analogous to OB3b-MB, but this methanotroph also takes up OB3b-MB as evidenced by greater hybridization signal in the cell extract and low signal in the spent medium and wash buffer when Mm. album BG8 was grown in the presence of OB3b-MB. Due to the evidence of cross-hybridization of monoclonal OB3b-MB antibodies in Mm. album BG8 and the inability to raise monoclonal antibodies to SB2-MB, these experiments were not replicated in other methanotrophs.
Evidence of a novel chalkophore from Mm. album BG8
Given that neither form of MB had any measurable effect on Mm. album BG8 wild type or the ΔmbnT mutant and the monoclonal OB3b-MB antibody cross-hybridized to cell extracts of Mm. album BG8, the possibility that Mm. album BG8 makes some copper-binding compound was investigated further. Earlier efforts indicated that Mm. album BG8 does secrete some sort of chalkophore, but under standard growth conditions produces very little of it, making characterization difficult [52]. Mm. album BG8 was grown in the presence of TRIEN, a strong abiotic chelator of copper, to determine if copper limitation could induce the production of this chalkophore. When Mm. album BG8 was grown in the presence of 1 μM copper + 5 μM TRIEN, growth was visibly reduced (Figs. S5A and S6A) and the spent medium became yellow (Fig. 7A). Such coloration was not observed when Mm. album BG8 was grown in the presence of copper, indicating that Mm. album BG8 secretes some yellowish substance when copper availability is reduced through the addition of TRIEN. This putative chalkophore was found to have a molecular mass of 649.95 (Fig. S13A) or 653.29 Da (Fig. S13B) as determined by MALDI-TOF or ESI-MS, respectively. Following the addition of CuCl2 the molecular mass shifted to 711.35 (Fig. S13A) and 713.35 Da (Fig. S13C) as determined by MALDI-TOF or ESI-MS, indicating that this substance indeed binds copper (i.e., is a chalkophore), but likely loses 2 or 3H+ after doing so (Fig. S13). The UV-VIS spectrum of the isolated chalkophore did not have the characteristic peaks present in MBs (i.e., at ~340 and 394 nm), but did exhibit distinct absorption maxima at 396 and 402 nm with a molar extinction coefficient of 1.6 mM−1 cm−1 at 402 nm (Fig. 7B, C, S14A). A discrete isosbestic point at 340 nm at mole ratios of copper:chalkophore between 0 and 0.19 was evident (Fig. S14A). Absorption maxima shifted to 404 nm at copper:chalkophore ratios between 0.2 and 0.45 (Fig. 7, S14B) with a discrete isosbestic point at 398 and to 390 nm at Cu to chalkophore ratios above 0.6 (Fig. 7, S14C).
Fig. 7. Isolation and characterization of a novel copper-binding compound produced by Mm. album BG8.
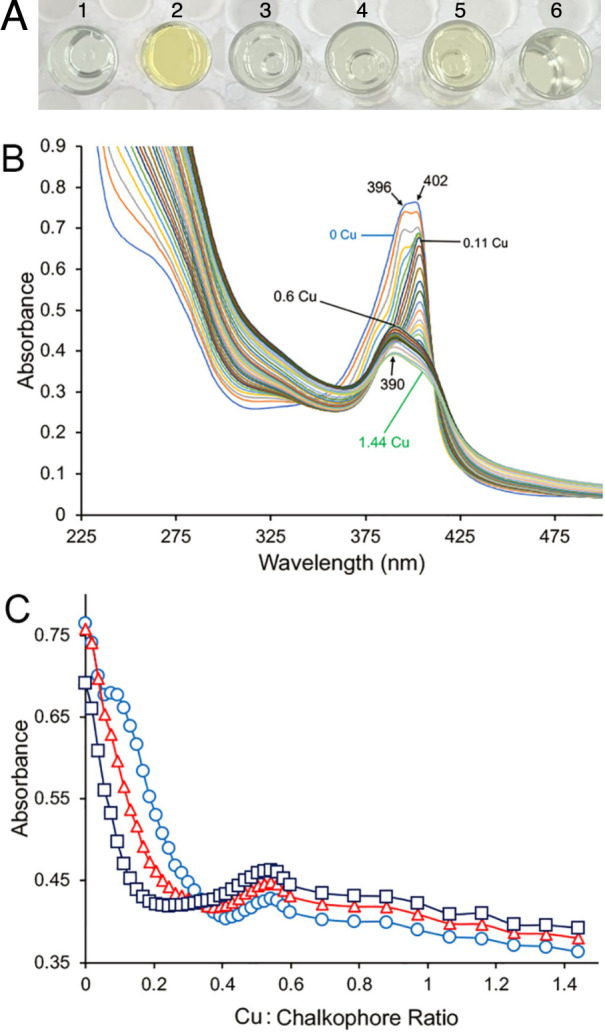
A Filtered spent medium of Mm. album BG8 grown in the presence of (1) 1 μM copper and (2) 1 μM copper and 5 μM TRIEN, with abiotic controls (3) NMS, (4) NMS, 1 μM copper, and 5 μM TRIEN, (5) NMS and 5 μM OB3b-MB, (6) NMS and 5 μM SB2-MB. B UV-Visible absorption spectra of 535 nmol of the chalkophore isolated from Mm. album BG8 and following the addition of copper (as CuCl2) initially in 10 nmol increments (up to 320 nmol) and then in 50 nmol increments (for an additional 450 nmol copper, or 770 nmol copper in total). C Absorbance changes at 402 nm (○), 396 nm (Δ), and 390 nm (□) following copper addition. Numbers in B refer to mole ratio of copper to Mm. album BG8 chalkophore.
No such coloration in the spent medium was observed for Methylocystis sp. strain Rockwell under any condition (data not shown). Attempts to identify novel chalkophore(s) in Mc. capsulatus Bath were not pursued as this strain has already been shown to produce membrane-bound and secreted copper-binding polypeptides (i.e., MopE and MopE*, respectively) [23–26].
Effect of MB on methylmercury degradation by Mm. album BG8 wild type, ΔmbnT mutant, Methylocystis sp. strain Rockwell, and Mc. capsulatus Bath
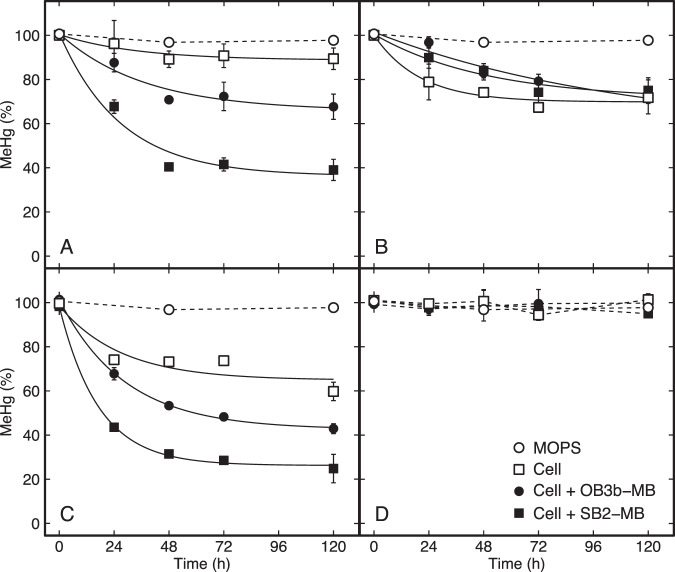
Given the uncertainty of the immunoblot data and evidence that Mm. album BG8 produces a competitive chalkophore, to determine if MB theft occurs between methanotrophs, demethylation of MeHg by various strains was monitored in the absence or presence of OB3b-MB and SB2-MB (Fig. 8). This was done as earlier work has shown methanotrophs expressing and taking up MB can degrade significant amounts of MeHg [34]. That is, MB appears to serve as a device to deliver MeHg inside the cell where it is degraded, but not by the well-known organomercurial lyase as these microbes lack merB. Rather, data suggest that MeHg degradation may be carried out by the periplasmic methanol dehydrogenase that all methanotrophs possess [34]. If Mm. album BG8, Methylocystis sp. strain Rockwell, and/or Mc. capsulatus Bath can take up MB, one would expect that these methanotrophs would be able to degrade MeHg in the presence of MB but not in its absence. Relatively little MeHg degradation was observed in Mm. album BG8 in the absence of MB (~10%), but this increased in the presence of both OB3b-MB and SB2-MB (32 and 61%, respectively; Fig. 8). In the absence of either MB, MeHg degradation was observed in Methylocystis sp. strain Rockwell (40%), and degradation increased in the presence of both OB3b-MB and SB2-MB (57 and 75%, respectively). Interestingly, under no condition was MeHg degradation observed in Mc. capsulatus Bath, nor was the degradation of MeHg by the ∆mbnT mutant of Mm. album BG8 significantly different in the presence or absence of either form of MB (Fig. 8).
Fig. 8. Degradation of methylmercury (MeHg) by different methanotrophs.
A Mm. album BG8 wild type, B Mm. album BG8 ΔmbnT, C Methylocystis sp. strain Rockwell, and D Mc. capsulatus Bath in MOPS buffer (5 mM). Degradation was fitted to an exponential decay model with stabilization over time (solid line). The total added MeHg, methanobactin (MB), and cell concentrations were 5 nM, 45 μM, and 108 cells mL−1 at t = 0 h. Error bars represent the standard deviation of at least biological duplicates. Where error bars are not visible, symbol size is greater than the measured standard deviation.
Discussion
It is well-known that microorganisms have active “social lives”, i.e., microbes exhibit a range of behaviors ranging from cooperation, competition, and cheating [53–55]. An example is the sharing of siderophores amongst microorganisms to meet iron requirements. Iron availability commonly limits microbial growth due to the insolubility of Fe(III), and many microbes produce siderophores for iron solubilization and collection [56, 57]. Given that these compounds are secreted, they can be considered “public goods”, i.e., they are costly for an individual microorganism to make, but can be utilized by other microbes for iron collection [53–55]. A challenge that then arises is that microbes can and do develop cheating strategies, i.e., some microorganisms with the inability to produce siderophores steal them to meet their needs and such cheating strategies likely play important roles in the diversification and evolution of microbial communities [30–33].
Previous studies suggested that methanotrophs do not utilize a public good for copper collection, i.e., it was shown earlier that Ms. trichosporium OB3b outcompetes Mm. album BG8 for copper, and thus predominates in mixed cultures [58]. This conclusion, however, appears to be overstated as the Mm. album BG8 was unequivocally present in large numbers in these experiments, suggesting that they have some mechanism(s) to collect copper in the presence of MB-expressing methanotrophs. Although Mm. album BG8 does not have genes for MB production, it does express something akin to MB, i.e., it secretes a copper-binding compound, especially under copper-limiting conditions, that appears to compete with MBs for copper. Further, the Mm. album BG8 ΔmbnT mutant exhibited a wild-type phenotype, indicating that the chalkophore expressed by Mm. album BG8 is effective in competing for copper in the presence of MB. Nonetheless, multiple data sets indicate that Mm. album BG8, in addition to being able to effectively compete with MB by producing a novel chalkophore, also engages in MB theft. First, although TRIEN affected growth of both Mm. album BG8 wild type and ΔmbnT mutant, growth of Mm. album BG8 wild type was not further affected by the concurrent addition of OB3b-MB, but that of the ΔmbnT mutant was. Second, MB enhanced MeHg demethylation in Mm. album BG8 wild type, but not the ∆mbnT mutant.
We were unable to identify any novel chalkophore produced by Methylocystis sp. strain Rockwell, but it does appear to act as a cheater by taking up MB—preferentially Group II MB—to meet its copper requirements as growth and copper uptake was inhibited in the presence of OB3b-MB (a group I MB), but not SB2-MB (a Group II MB). This conclusion is supported by finding that the addition of TRIEN inhibited the growth of Methylocystis sp. strain Rockwell, but concurrent addition of SB2-MB relieved such inhibition. Although we could not directly determine SB2-MB uptake via immunoblots as we were unsuccessful in raising monoclonal antibodies to SB2-MB, MeHg degradation data also indicate MB can be taken up by Methylocystis sp. strain Rockwell. On the other hand, Mc. capsulatus Bath does not appear to steal MB as its genome had no genes with high similarity to mbnT and the addition of MB did not enable this methanotroph to degrade MeHg. Rather, Mc. capsulatus Bath appears to have an effective strategy to compete for copper in the presence of MB (i.e., MopE/MopE*), as the addition of MB had no effect on its growth, copper uptake, and gene expression. Thus, MB may serve as a sort of public good to some methanotrophs, but is not of benefit to all methanotrophs. Given that methanotrophs use a variety of strategies to collect copper, these interactions likely are significant in structuring methanotrophic communities in situ.
While herein, we report methanotrophic interactions based on competition for copper, including MB theft, interspecies interactions have been documented earlier for methanotrophs, e.g., recognition of and response to acyl-homoserine lactone receptor/transcription factors and uptake of foreign MB by species that can make MB. Such interactions, however were within species of the same family [59, 60]. Here we show interactions not only between members of the same class of methanotrophs (i.e., Alphaproteobacteria), but also between members of different classes (i.e., Alpha- vs. Gammaproteobacteria methanotrophs), indicating methanotrophic interactions can be phylogenetically far-ranging. It may be that uptake of MB from the environment by non-MB-producing methanotrophs not only enhances their ability to collect copper, but also gives them an advantage by acting as a MB sink, thereby placing MB-producing methanotrophs at a disadvantage. Such a finding lends support to the hypothesis that as competition for resources becomes more local, the influence of species relatedness for cooperation is reduced, thus decreasing altruism and allowing cheating to become more pronounced [53–55]. That is, as methanotrophs of different phylogenies co-habitate, kin recognition/discrimination becomes less effective for these microbes and MB can be more readily stolen. These findings raise the intriguing question as what prevents MB stealers from overwhelming the population? As suggested for siderophores, it may be that MB production and distribution to non-MB-producing methanotrophs provides both direct and indirect fitness benefits to MB-producers that outweigh the costs of MB synthesis and loss. The magnitude and distribution of such benefits, however, are likely to be highly dependent on environmental conditions (e.g., copper availability) and population density as suggested for siderophores [53].
Finally, the finding that the heterologous uptake or theft of MB enables MB non-producers to detoxify a highly toxic organic form of mercury suggests that methanotrophic-mediated MeHg detoxification may be more widespread than previously thought in the natural environment. Examining this in more detail will likely be very informative and will serve as key inputs for metabolic and reactive transport models that can be used to better predict net MeHg production and Hg biogeochemical cycling in the environment.
Supplementary information
Acknowledgements
This work was supported in part by grants from Helmholtz Zentrum München, the Iowa State Presidential Interdisciplinary Research Seed Grant Program, the National Science Foundation (Grants 1724430 and 1724744), and the US Department of Energy (Grants DE-SC0018059 and DE-SC0020174). Oak Ridge National Laboratory is managed by UT-Battelle LLC under contract no. DE-AC05-00OR22725 with the US Department of Energy.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Christina S. Kang-Yun, Xujun Liang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01062-1.
References
- 1.Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 2.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Mol Biol Rev. 1996;60:439–71. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhury TR, Dick RP. Ecology of aerobic methanotrophs in controlling methane fluxes from wetlands. Appl Soil Ecol. 2013;65:8–22. [Google Scholar]
- 4.Cai Y, Zheng Y, Bodelier PLE, Conrad R, Jia Z. Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat Commun. 2016;7:11728. doi: 10.1038/ncomms11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tveit AT, Hestnes AG, Robinson SL, Schintlmeister A, Dedysh SN, Jehmlich N, et al. Widespread soil bacterium that oxidizes atmospheric methane. Proc Natl Acad Sci USA. 2019;116:8515–24. doi: 10.1073/pnas.1817812116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratscher J, Vollmers J, Wiegand S, Dumont MG, Kaster A-K. Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing upland soil cluster α: Illuminating the USCα methanotrophs from soil. Environ Microbiol. 2018;20:1016–29. doi: 10.1111/1462-2920.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon S, Carey JN, Semrau JD. Feasibility of atmospheric methane removal using methanotrophic biotrickling filters. Appl Microbiol Biotechnol. 2009;83:949–56. doi: 10.1007/s00253-009-1977-9. [DOI] [PubMed] [Google Scholar]
- 8.Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci USA. 2008;105:10203–8. doi: 10.1073/pnas.0702643105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanley SH, Prior SD, Leak DJ, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane mono-oxygenase in methane-oxidizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–92. [Google Scholar]
- 10.Lipscomb JD. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–99. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 11.Semrau JD, Jagadevan S, DiSpirito AA, Khalifa A, Scanlan J, Bergman BH, et al. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ Microbiol. 2013;15:3077–86. doi: 10.1111/1462-2920.12150. [DOI] [PubMed] [Google Scholar]
- 12.Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han J-I, et al. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol. 2003;185:5755–64. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinho M, Choi D, DiSpirito AA, Antholine WE, Semrau JD, Münck E. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: Evidence for a diiron center. J Am Chem Soc. 2007;129:15783–5. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao L, Caldararu O, Rosenzweig AC, Ryde U. Quantum refinement does not support dinuclear copper sites in crystal structures of particulate methane monooxygenase. Angew Chem Int Ed. 2018;57:162–6. doi: 10.1002/anie.201708977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S-W, Keeney DR, Lim D-H, Dispirito AA, Semrau JD. Mixed pollutant degradation by Methylosinus trichosporium OB3b expressing either soluble or particulate methane monooxygenase: Can the tortoise beat the hare? Appl Environ Microbiol. 2006;72:7503–9. doi: 10.1128/AEM.01604-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, et al. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–5. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 17.DiSpirito AA, Semrau JD, Murrell JC, Gallagher WH, Dennison C, Vuilleumier S. Methanobactin and the link between copper and bacterial methane oxidation. Microbiol Mol Biol Rev. 2016;80:387–409. doi: 10.1128/MMBR.00058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semrau JD, DiSpirito AA, Obulisamy PK, Kang-Yun CS. Methanobactin from methanotrophs: genetics, structure, function and potential applications. FEMS Microbiol Lett. 2020;367:fnaa045. doi: 10.1093/femsle/fnaa045. [DOI] [PubMed] [Google Scholar]
- 19.Semrau JD, DiSpirito AA, Gu W, Yoon S. Metals and methanotrophy. Appl Environ Microbiol. 2018;84:e02289–17. doi: 10.1128/AEM.02289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu W, Baral BS, DiSpirito AA, Semrau JD. An aminotransferase is responsible for the deamination of the N-terminal leucine and required for formation of oxazolone ring A in methanobactin of Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2017;83:e02619–16. doi: 10.1128/AEM.02619-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenney GE, Dassama LMK, Pandelia M-E, Gizzi AS, Martinie RJ, Gao P, et al. The biosynthesis of methanobactin. Science. 2018;359:1411–6. doi: 10.1126/science.aap9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu W, Ul-Haque MF, Baral BS, Turpin EA, Bandow NL, Kremmer E, et al. A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2016;82:1917–23. doi: 10.1128/AEM.03884-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ve T, Mathisen K, Helland R, Karlsen OA, Fjellbirkeland A, Røhr ÅK, et al. The Methylococcus capsulatus (Bath) secreted protein, MopE*, binds both reduced and oxidized copper. PLoS One. 2012;7:e43146. doi: 10.1371/journal.pone.0043146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helland R, Fjellbirkeland A, Karlsen OA, Ve T, Lillehaug JR, Jensen HB. An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J Biol Chem. 2008;283:13897–904. doi: 10.1074/jbc.M800340200. [DOI] [PubMed] [Google Scholar]
- 25.Karlsen OA, Berven FS, Stafford GP, Larsen Ø, Murrell JC, Jensen HB, et al. The surface-associated and secreted MopE protein of Methylococcus capsulatus (Bath) responds to changes in the concentration of copper in the growth medium. Appl Environ Microbiol. 2003;69:2386–8. doi: 10.1128/AEM.69.4.2386-2388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fjellbirkeland A, Kruger PG, Bemanian V, Høgh BT, Murrell CJ, Jensen HB. The C-terminal part of the surface-associated protein MopE of the methanotroph Methylococcus capsulatus (Bath) is secreted into the growth medium. Arch Microbiol. 2001;176:197–203. doi: 10.1007/s002030100307. [DOI] [PubMed] [Google Scholar]
- 27.Berson O, Lidstrom ME. Cloning and characterization of corA, a gene encoding a copper-repressible polypeptide in the type I methanotroph, Methylomicrobium albus BG8. FEMS Microbiol Lett. 1997;148:169–74. doi: 10.1111/j.1574-6968.1997.tb10284.x. [DOI] [PubMed] [Google Scholar]
- 28.Kang CS, Dunfield PF, Semrau JD. The origin of aerobic methanotrophy within the Proteobacteria. FEMS Microbiol Lett. 2019;366:fnz096. doi: 10.1093/femsle/fnz096. [DOI] [PubMed] [Google Scholar]
- 29.Dassama LMK, Kenney GE, Ro SY, Zielazinski EL, Rosenzweig AC. Methanobactin transport machinery. Proc Natl Acad Sci USA. 2016;113:13027–32. doi: 10.1073/pnas.1603578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butaitė E, Baumgartner M, Wyder S, Kümmerli R. Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun. 2017;8:414. doi: 10.1038/s41467-017-00509-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan LL, Kanoh K, Kamino K. Effect of exogenous siderophores on iron uptake activity of marine bacteria under iron-limited conditions. Appl Environ Microbiol. 2001;67:1710–7. doi: 10.1128/AEM.67.4.1710-1717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champomier-Vergès M-C, Stintzi A, Meyer J-M. Acquisition of iron by the non-siderophore-producing Pseudomonas fragi. Microbiology. 1996;142:1191–9. doi: 10.1099/13500872-142-5-1191. [DOI] [PubMed] [Google Scholar]
- 33.Cordero OX, Ventouras L-A, DeLong EF, Polz MF. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA. 2012;109:20059–64. doi: 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X, Gu W, Zhao L, Haque MFU, DiSpirito AA, Semrau JD, et al. Methylmercury uptake and degradation by methanotrophs. Sci Adv. 2017;3:e1700041. doi: 10.1126/sciadv.1700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Whittenbury R, Phillips KC, Wilkinson JF. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970;61:205–18. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 37.Bandow NL, Gallagher WH, Behling L, Choi DW, Semrau JD, Hartsel SC, et al. Isolation of methanobactin from the spent media of methane-oxidizing bacteria. Meth Enzymol. 2011;495:259–69. doi: 10.1016/B978-0-12-386905-0.00017-6. [DOI] [PubMed] [Google Scholar]
- 38.Kalidass B, Ul-Haque MF, Baral BS, DiSpirito AA, Semrau JD. Competition between metals for binding to methanobactin enables expression of soluble methane monooxygenase in the presence of Copper. Appl Environ Microbiol. 2015;81:1024–31. doi: 10.1128/AEM.03151-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandow N, Gilles VS, Freesmeier B, Semrau JD, Krentz B, Gallagher W, et al. Spectral and copper binding properties of methanobactin from the facultative methanotroph Methylocystis strain SB2. J Inorg Biochem. 2012;110:72–82. doi: 10.1016/j.jinorgbio.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Gu W, Ul-Haque MF, DiSpirito AA, Semrau JD. Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol Lett. 2016;363:fnw129. doi: 10.1093/femsle/fnw129. [DOI] [PubMed] [Google Scholar]
- 41.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 42.Lichtmannegger J, Leitzinger C, Wimmer R, Schmitt S, Schulz S, Kabiri Y, et al. Methanobactin reverses acute liver failure in a rat model of Wilson disease. J Clin Investig. 2016;126:2721–35. doi: 10.1172/JCI85226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigues JD, Combrink J, Brandt WF. Derivatization of polyvinylidene difluoride membranes for the solid-phase sequence analysis of a phosphorylated sea urchin embryo histone H1 peptide. Anal Biochem. 1994;216:365–72. doi: 10.1006/abio.1994.1054. [DOI] [PubMed] [Google Scholar]
- 44.Dershwitz P, Bandow NL, Yang J, Semrau JD, McEllistrem MT, Heinze RA, et al. Oxygen generation via water splitting by a novel biogenic metal ion binding compound. Appl Environ Microbiol. 2021;87.e00286-21. [DOI] [PMC free article] [PubMed]
- 45.Yin X, Wang L, Zhang L, Chen H, Liang X, Lu X, et al. Synergistic effects of a chalkophore, methanobactin, on microbial methylation of mercury. Appl Environ Microbiol. 2020;86:e00122–20. doi: 10.1128/AEM.00122-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu X, Liu Y, Johs A, Zhao L, Wang T, Yang Z, et al. Anaerobic mercury methylation and demethylation by Geobacter bemidjiensis Bem. Environ Sci Technol. 2016;50:4366–73. doi: 10.1021/acs.est.6b00401. [DOI] [PubMed] [Google Scholar]
- 47.Lin H, Lu X, Liang L, Gu B. Thiol-facilitated cell export and desorption of methylmercury by anaerobic bacteria. Environ Sci Technol Lett. 2015;2:292–6. [Google Scholar]
- 48.Sprouffske K, Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016;17:172. doi: 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018.
- 50.Collins ML, Buchholz LA, Remsen CC. Effect of Copper on Methylomonas albus BG8. Appl Environ Microbiol. 1991;57:1261–4. doi: 10.1128/aem.57.4.1261-1264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leak DJ, Dalton H. Growth yields of methanotrophs 1. Effect of copper on the energetics of methane oxidation. Appl Microbiol Biotechnol. 1986;23:470–6. [Google Scholar]
- 52.Choi DW, Bandow NL, McEllistrem MT, Semrau JD, Antholine WE, Hartsel SC, et al. Spectral and thermodynamic properties of methanobactin from γ-proteobacterial methane oxidizing bacteria: a case for copper competition on a molecular level. J Inorg Biochem. 2010;104:1240–7. doi: 10.1016/j.jinorgbio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 53.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 54.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024–7. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 55.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu Rev Ecol Evol Syst. 2007;38:53–77. [Google Scholar]
- 56.Ahmed E, Holmström SJM. Siderophores in environmental research: roles and applications: siderophores in environmental research. Micro Biotechnol. 2014;7:196–208. doi: 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 58.Graham DW, Chaudhary JA, Hanson RS, Arnold RG. Factors affecting competition between type I and type II methanotrophs in two-organism, continuous-flow reactors. Micro Ecol. 1993;25:1–17. doi: 10.1007/BF00182126. [DOI] [PubMed] [Google Scholar]
- 59.Puri AW, Liu D, Schaefer AL, Yu Z, Pesesky MW, Greenberg EP, et al. Interspecies chemical signaling in a methane-oxidizing bacterial community. Appl Environ Microbiol. 2019;85:e02702–18. doi: 10.1128/AEM.02702-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ul-Haque MF, Kalidass B, Vorobev A, Baral BS, DiSpirito AA, Semrau JD. Methanobactin from Methylocystis sp. strain SB2 affects gene expression and methane monooxygenase activity in Methylosinus trichosporium OB3b. Appl Environ Microbiol. 2015;81:2466–73. doi: 10.1128/AEM.03981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.