Abstract
The plant hormone cytokinin (CK) is an important developmental regulator, promoting morphogenesis and delaying differentiation and senescence. From developmental processes, to growth, to stress tolerance, CKs are central in plant life. CKs are also known to mediate plant immunity and disease resistance, and several classes of microbes can also produce CKs, affecting the interaction with their plant hosts. While host species and genotype can be a driving force in shaping the plant microbiome, how plant developmental hormones such as CK can shape the microbiome is largely uninvestigated. Here, we examined the relationship between CK and the phyllosphere microbiome, finding that CK acts as a selective force in microbiome assembly, increasing richness, and promoting the presence of Firmicutes. CK-mediated immunity was found to partially depend on the microbial community, and bacilli isolated from previously described CK-rich plant genotypes, which overexpress a CK biosynthesis gene or have increased CK sensitivity, induced plant immunity, and promoted disease resistance. Using a biomimetic system, we investigated the relationship between the leaf microstructure, which is differentially patterned upon changes in CK content or signaling, and the growth of different phyllosphere microbes. We found that leaf structures derived from CK-rich plant genotypes support bacilli in the biomimetic system. CK was able to promote the growth, swarming, and biofilm formation of immunity inducing bacillus isolates in vitro. Overall, our results indicate that host genotype and hormonal profiles can act as a strong selective force in microbiome assembly, underlying differential immunity profiles, and pathogen resistance as a result.
Subject terms: Microbiome, Plant ecology
Introduction
The plant microbiome performs various beneficial functions, including promoting plant health, and agricultural sustainability [1]. The microbial community associated with above ground parts of the plant (phyllosphere) is dynamic, and influences ecological functions. Several phyllosphere studies have provided insights into factors that may play a key role in shaping phyllosphere community structure, such as host genotypes, leaf age, and seasonal variation [2]. It is apparent that these microbial communities do not signify random processes, but rather, reflect selection pressures that result, at least to some extent, in predictable microbial communities, with some dominant groups originating from the phyla Proteobacteria, followed by Actinobacteria, Bacteroidetes, and Firmicutes [3]. A recent study showed that the effect of the host plant genotype in shaping the diversity of phyllosphere microbiota declines over time, which suggests habitat selection of microbial communities under short host timescales [4].
The phylloshpere microbiome is comprised of epiphytes and endophytes. Leaf surfaces can host a dense population of epiphytes, estimated to reach 107 bacteria per cm2 of leaf surface [5]. Leaf surfaces are considered challenging ecosystems for bacterial colonization, due to ultraviolet radiation exposure, low water and nutrient availability, and day-night temperature fluctuations [6]. The epiphytic microbiome was found to be more phylogenetically diverse than the endophytic microbiome [7]. In recent years, the phyllosphere microbial community has attracted increasing attention; however, the processes that are responsible for determining the composition of phyllosphere microbiota, and how they contribute to plant health, remain mostly unresolved.
The plant developmental hormone cytokinin (CK) plays important roles in varied processes throughout plant life, including stem-cell support, vascular differentiation, chloroplast biogenesis, seed development, growth and branching of root, shoot and inflorescence, leaf senescence, nutrient balance, and stress tolerance [8]. CKs are often viewed as “juvenility” factors, promoting morphogenesis and delaying plant differentiation and/or senescence [9]. In a previous study aimed at identifying the role of cytokinin (CK) in plant pathogen interactions, we found that tomato plants overexpressing the CK biosynthesis gene ISOPENTENYL TRANSFERASE7 (IPT7), resulting in elevated endogenous levels of CK [10, 11], or plants having increased CK sensitivity [12], have significantly lower disease symptoms when infected with two pathogens: the gray mold necrotrophic fungus Botrytis cinerea, and the powdery mildew biotrophic fungus Oidium neolycopersici. Concurrently, plants with decreased endogenous levels of CK due to overexpression of a gene encoding CK OXIDASE (CKX3) [11], had significantly increased disease levels [13].
Manipulating the CK pathway in tomato results in alterations to leaf development and structure [9]. Overexpression of the CK biosynthesis gene AtIPT7 in tomato leaves leads to the formation of highly complex leaves bearing increased numbers of higher order leaflets, and conversely, decreasing CK levels by overexpression of the CK oxidase/degradation gene CKX, results in reduced leaf complexity [11, 12]. The CLAU gene promotes differentiation by negatively affecting CK signaling, resulting in heightened CK signals, and increased leaf complexity, in the clausa mutant [12]. Furthermore, altered levels of CK have been reported to affect the cell wall structure [14]. Thus, differential levels of CK, or CK-mediated signaling, can result in alterations to leaf structure, likely affecting the leaf microenvironment colonized by the microbial community.
Environmental factors, as well as host genotype, determine the abundance and diversity of the phyllosphere microflora [15]. Much work has been done to investigate various environmental impacts on phyllosphere microbiome formation (reviewed in [16]). Host genotype also influences microbiome formation. For instance, cuticle and ethylene (ET) signaling mutants exhibited altered phyllosphere bacterial communities, when compared to wild-type Arabidopsis plants [17]. The phylloplane of Arabidopsis plants grown under similar conditions and inoculated initially with the same synthetic bacterial community (SynCom), showed that plant host states likely act in shaping the composition of the leaf microbiota [17].
The epiphytic microbiome can confer disease resistance [18], though interestingly, it was found to possess less direct inhibitory activity against plant pathogens than the endophytic microbiome [7]. A recent study demonstrated that plants mutated in pattern triggered immunity and vesicle trafficking pathways harbor a restructured microbiome, resulting in dysbiosis in the phyllosphere [19]. This defines host genetics as a major driver of the leaf bacterial community, and associates the phyllosphere microbiota to plant health. Defense response pathways such as salicylic acid (SA) and jasmonic acid (JA) signaling were also found to affect microbial diversity in leaves of Arabidopsis [20].
In tomato, the roles of hormones in shaping the phyllopshere bacterial community remain mostly uninvestigated. In this work, we examined how CK shapes bacterial communities in the tomato phyllosphere. We found that alterations in endogenous CK levels or sensitivity, as well as external application of CK, resulted in predictable and reproducible changes to the phyllobiome. Increased CK, or CK-mediated signaling, supported Firmicutes in the phyllosphere. We found that phyllosphere isolated bacilli promote plant immunity, and disease resistance. Using a synthetic leaf system and synthetic bacterial communities, we were able to demonstrate that the effect of CK on leaf structure is one of the mechanisms by which differential colonization is supported.
Materials and methods
Plant materials
Plant genotypes and sample collection
During the winter or summer of 2018, tomato leaf samples were collected from a net house (summer), or roofed net house (winter) in ARO, Volcani Institute, Rishon Lesion, Israel. The net houses both have a 2 mm nylon mesh net. Genotypes used, all in the cv. M82 background, were as follows: M82 background line; pBLS>>IPT7, which contains elevated endogenous levels of CK- referred to hereinafter as “pBLS>>IPT “or “IPT”; clausa, which has increased CK sensitivity coupled with decreased CK content, referred to hereinafter as “clausa” or “clau”; and the CK depleted pFIL>>CKX3, referred to hereinafter as “pFIL>>CKX” or “CKX”. In tomato, the Arabidopsis FIL (filamentous flower) promoter drives expression throughout leaf primordia, starting from initiation (first plastochron), in initiating leaflets, and the abaxial side of the leaves. No expression of the FIL promoter was detected in the shoot apical meristem, rib meristem, or stems. The BLS promoter drives expression later in leaf development, in primordia from about the fourth plastocrhon stage, and in young leaves [21]. Plants overexpressing IPT have been shown to contain increased levels of cytokinin many times, in various plant species [22–24]. Strongly increasing CK levels throughout the plant also led in some cases to phenotypes that were not agriculturally compatible, such as reduced apical dominance, increased lateralization, and late flowering. Therefore, tissue specific promoters were used to express IPT [11, 25, 26]. This eliminated the undesired effects, and allowed plants to be viable and fertile, though mild effects of increased CK were occasionally observed in the non-targeted organs as well.
Plant overexpressing CKX have been shown to contain reduced levels of cytokinin many times, in various plant species. CKX3 overexpression was specifically shown to cause reduction in CKs in several works [27, 28]. Reducing cytokinin levels with CKX overexpression led to stunting in Arabidopsis when expressed from the strong 35 S promoter [29], but overexpression of CKX3 in tomato had relatively minimal phenotypes under optimal conditions [30].
The lines we used, which overexpress Arabidopsis AtIPT7 or AtCKX3 from the leaf specific promoters pFIL and pBLS, in tomato cv M82, were previously characterized [11]. The transgenic plants have normal early development and are viable [11]. pFIL>>IPT7 is mostly infertile, which is why we used pBLS>>IPT7, that has a milder phenotype, due to the expression being later in development, and normal fertility [11]. pFIL>>CKX3 is viable and fertile. We used pFIL>>CKX3 because pBLS>>CKX3 lacks any observable phenotype, and therefore, was not suitable for our biomimetic platform.
CK content was previously analyzed in the tomato clausa mutant. clausa is highly CK sensitive, displaying meristematic and leaf phenotypes similar to those of overexpression of IPT, coupled with a significant reduction in the content of many CK compounds [12].
16 S rRNA amplification, amplicon sequencing and bioinformatic analysis
To examine whether endogenous CK levels affect tomato phyllosphere composition, phyllosphere microbial DNA was extracted from altered-CK S. lycopersicum genotypes in the cv. M82 background. To determine the impact of exogenous CK on tomato (cv. M82) phyllosphere microorganisms, 100 µM of different CK compounds: 6-BAP (6-Benzylaminopurine; Sigma–Aldrich), kinetin (6‐furfurylaminopurine riboside; Sigma–Aldrich) and trans‐zeatin (6‐[4‐hydroxy‐3‐methyl but‐2‐enylamino] purine; Sigma–Aldrich) were used to treat 4‐week‐old plants. All solutions were prepared from a stock in 10 mM NaOH and diluted into an aqueous solution to the desired concentration, with the addition of Tween 20 (100 µl L−1) [13]. The CK solution (10 mL) was sprayed on the leaves of M82, twice a week for two weeks. For mock, M82 plants were treated with the aforementioned solution of NaOH with Tween 20.
For DNA isolation, five leaflet samples per genotype/treatment were collected from the middle lateral leaflets of leaves 5–6 of 10 different plants per sample, using ethanol-sterilized forceps. Twenty mL of 0.1 M potassium phosphate buffer at pH 8 were added to the tubes. The samples were sonicated in a water bath (47 kHz ± 6%) for 2 min and vortexed for 30 s; this step was repeated twice. The pellet of microbes was obtained after centrifugation at 12,000 g for 20 min at 4 °C. Sonication was used in order to minimize undesirable amplification of chloroplast rRNA genes and contamination by endophytic bacteria. The pellet of microbes was re-suspended in the potassium phosphate buffer. Total DNA from tomato phyllosphere microorganisms was isolated using modified protocols described by Yang et al. [31] and Tian et al. [32], and used as a template for 16 S rRNA PCR amplification. 16 S rRNA amplicons were generated with the following primers:
CS1_515F ACACTGACGACATGGTTCTACAGTGCCAGCMGCCGCGGT; CS2_806R TACGGTAGCAGAGACTTGGTCTGGACTACHVGGGTWTCT.
Amplicon sequencing was conducted at the UIC core facility, using Illumina MiSeq sequencing. QIIME 1.9 [33] was used for basic bioinformatics analysis: read merging, primer trimming, quality trimming, length trimming, chimera removal, clustering of sequences, annotation of clusters, generation of a biological observation matrix (BIOM; sample-by-taxon abundance table). Taxonomy for the operational taxonomic units (OTUs) was assigned using BLAST against the Silva database [34] (silva_132_16S.97). Alpha and beta-diversity, and Shannon index, were performed with QIIME 1.9 as well the workflow script core_diversity_analysis.py. The sequence data generated in this study was deposited to the Sequence Read Archive (SRA) at NCBI under Bioproject PRJNA729221.
Bacterial isolates, identification, and treatments
Epiphytic bacterial isolation
For epiphytic bacteria isolation, tomato leaves (1 g, similar sizes from five individual plants) were placed in 5 mL phosphate‐buffered saline (pH 7.4) and kept on a 100 rpm shaker for 30 min [35]. A 50 µL fraction of the resulting suspension was spread onto different bacterial media such as LB (Luria-Bertani) medium, nutrient agar medium, and YPGA (Yeast extract: 7 g L−1, Peptone: 7 g L−1, Glucose: 7 g L−1, Agar: 15 g L−1). The plates were incubated at 28 °C for 24–48 h. All isolated bacterial colonies were purified on LB medium based on their different morphology. The isolated bacterial strains were suspended in 30% glycerol in cryogenic tubes and kept at −80 °C for long-term storage.
Taxonomic identification of bacterial isolates
The genomic DNA of the purified bacterial isolates was used to amplify 16 S rRNA using bacterial primers 27 f (5′-AGAGTTTGATCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) [36]. PCR was performed using a Thermal cycler: 94 °C for 3 min (1 cycle); 94 °C for 1 min, 55 °C for 45 s and 72 °C for 1.5 min (15 cycles); and 72 °C for 10 min (1 cycle). PCR products (approx. 1500 bp) were purified using a PCR purification kit (Zymo Research), following the protocol of the manufacturer, and were sent to an external company for sequencing (Hylabs, Israel). The obtained chromatograms were visually inspected and sequences obtained were compared with those from the EZTaxon database, aligned using the Clustal W software. Phylogenetic trees were inferred using the neighbor-joining method in the MEGA X program [37]. The identified sequences were submitted to the NCBI Gene Bank database. Accession numbers and details of bacterial isolates used in this study are provided in Table 1.
Table 1.
Bacterial isolates used in this work.
| ID | Accession | Species | Source | Assays |
|---|---|---|---|---|
| R2E | MZ148745 | Bacillus pumilus | pBLS»IPT isolate | Plant immunity, synthetic leaf replica, in vitro growth |
| 4C | MZ148746 | Bacillus megaterium | pBLS»IPT isolate | SynCom, plant immunity, synthetic leaf replica, in vitro growth |
| R3C | MZ148747 | Ralstonia pickettii | pBLS»IPT isolate | SynCom, plant immunity, synthetic leaf replica, in vitro growth |
| *IN68 | – | Pseudomonas aeruginosa | Wheat phyllopshere isolate obtained from Jonathan Friedman, Hebrew University of Jerusalem | SynCom, plant immunity, synthetic leaf replica, in vitro growth |
| *Bs SB491 | SB491 “legacy” strain [101, 102] | Bacillus subtilis | Jonathan Friedman, Hebrew University of Jerusalem | SynCom, plant immunity, synthetic leaf replica, in vitro growth |
| *Ea | ATCC 13048 | Enterobacter aerogenes | Jonathan Friedman, Hebrew University of Jerusalem [103] | SynCom |
*Used after initial verification of ability to survive on tomato leaves in a three week period.
Sterile plant growth and SynCom experiments
Seeds of various genotypes viz., M82, altered CK genotypes pBLS>>IPT7, clausa and pFIL>>CKX3 were sterilized using 100% ethanol followed by 1.5% sodium hypochlorite (NaOCl), and washed with sterile distilled water three times to remove any excess bleach. Sterilized seeds were germinated in sterile vented magenta boxes (Duchefa) on half strength MS medium (Duchefa M0225). After three weeks, plants were inoculated with SynComs, at a 1:1:1:1 ratio of each bacterium. Two SynComs were created: one comprised a Pseudomonas isolate (IN68), a Ralstonia isolate (R3C), an Enterobacter isolate (Ea), and a B. megaterium isolate (4C), for a 1:3 Gram-positive to Gram-negative bacterial ratio (community #1). The second community comprised a Pseudomonas isolate (IN68), a Ralstonia isolate (R3C), and two different bacillus isolates (4C and SB491), for a 1:1 Gram-positive to Gram-negative bacterial ratio (community #2). This solution was then diluted to OD600 = 0.01. Plants were inoculated by spraying 200 µL of bacterial suspension with a sterile spray bottle. The synthetic communities were allowed to develop for three weeks, and were then re-isolated from leaves of each sterile plant, and plated for colony forming unit (CFU) count. Briefly, the third youngest leaf from at least three plants from three separate boxes was harvested. Leaves were washed in 100 mM phosphate buffer (pH 7) containing 0.2% Silwett by shaking for 15 min, and ten-fold serial dilutions were plated on NB media [17].
Biomimetic leaf experiments
Synthetic leaf replica preparation
Synthetic polydimethylsiloxane (PDMS) leaf replicas were prepared as detailed in Zhang et al. [38], with modifications described in Kumari et al. [39]. Initially, negative replicas were prepared by molding the PDMS directly onto the leaf surface. Negative replicas are a topographical mirror image of the leaf structure. Positive replicas, which are a copy of the leaf structure, were then obtained by molding the PDMS on the existing negative replicas. We used the negative replicas to prepare agar plates which copy the leaf structure, and the positive replicas for imaging and validation of the ability to copy the leaf micro-structure. For preparation of a PDMS negative replica of the leaf surface, the Sylgard 184 polymer kit was used. The Pre-polymer and curing agent were vigorously mixed at a 10 to 1 w/w ratio of polymer/curing agent respectively, and then subjected to vacuum for 1 h to remove air bubbles. Natural leaves of each genotype: M82, pBLS>>IPT, clausa, and pFIL>>CKX, were individually taped to a separate petri dish and the polymer solution was poured onto the leaf. Vacuum was applied for 2 h to ensure full coverage and uniform distribution on the surface microstructure. The covered leaf was kept at room temperature overnight to cure the polymer. The leaf was then peeled off the cured polymer, leaving the PDMS template of the leaf surface microstructure mirror image the negative replica.
To generate a positive replica, which structurally simulates the natural leaf, the negative replica was functionalized to turn the polymer hydrophilic, and prevent adherence of the two layers. For functionalization, a BD-20 AC laboratory CORONA treater was used for 30 s to activate the surface of the negative replica. The negative replica was then immediately placed in a desiccator with 100 μL of Trichloro (1H,1H,2H,2Hperfluorooctyl) silane, for 3 h. The negative replica was kept at room temperature wrapped in an airtight container until further use. The negative replica could be used dozens of times for months, with reproducible results.
To generate a positive PDMS replica, the negative replica was placed in a petri dish. Liquid polymer solution (identical to the one used to form the negative replica) was poured onto the negative replica, vacuumed for 2 h, and cured at RT overnight. The negative replica was removed from the newly formed polymer layer, which represents the synthetic positive replica.
For all synthetic replica experiments, five leaflets from the fifth leaf of five different plants of each genotype underwent the replica preparation process and were used in the replica mono-culture and community assays.
To generate agar plates containing an imprint which structurally simulates the natural leaf, desired liquid media with 0.8% agar was poured into a petri dish, let to sit for 10 min, and when the media was cooled but not yet hardened, an autoclaved negative PDMS replica was placed on the petri dish. Once the agar hardened, the PDMS negative was gently removed.
Bacterial growth on synthetic leaf replicas
To examine on-replica growth of each of the individual bacterial isolates, bacteria were uniformly sprayed onto the leaf replicas at OD600 adjusted to 0.01. Preference or aversion to the leaf structure was assessed by quantifying the CFU growing on the leaf structure, and dividing it by the CFU growing on an equal area in the surrounding structure-less agar.
We examined the growth and dispersal of combinations of two bacteria on the leaf replicas using the same methodology. Pairwise communities (“PC”) comprising each one Gram-positive (G(+)):Bs SB491, R2E, or 4C, in combination with one Gram-negative (G(−)) bacterium, R. pickettii R3C, in equal ratios (OD600 = 0.01 of each), were spray inoculated onto agar replicas of leaves of the altered CK genotypes. Preference or aversion to the leaf structure was assessed by quantifying the CFU growing on the leaf structure and dividing it by the CFU growing on an equal area in the surrounding structure-less agar, after 24 h.
Tissue staining and imaging
Leaves of indicated genotypes were individually cleared in an increasing ethanol gradient (30–70%) for three days. Epidermal peels were generated using two forceps and applying gentle force in opposing directions, and, to improve visibility of epidermal cells, stained with 1% Toluidine Blue O (sigma) for 2 min, then washed in water. Images were analyzed using a Nikon SMZ-25 stereomicroscope equipped with a Nikon-D2 camera and NIS Elements v. 5.11 software. ImageJ software was used for analysis and quantification of the cell area and perimeter of epidermis and mesophyll cells, and for counting non-glandular trichomes per mm2 adaxial leaf.
In vitro bacterial growth assays
Bacterial growth curves
To check the direct effect of CK on growth of isolated bacteria, in mono-culture or community, we inoculated bacteria B. pumilus R2E, B. megaterium 4 C, R. pickettii R3C, P. aeruginosa IN68, B. subtilis Bs SB491, as well as two additional Gram-positive bacilli, B. aryabhattai R2A or B. subtilis R1D (also isolated from tomato leaves), into nutrient broth (NB) media or minimal media (containing the following amounts per L: Dextrose, 1 g; K2HPO4, 7 g; KH2PO4, 3 g; (NH4)2SO4, 1 g; MgSO4.7H20, 100 mg; agar, 15 g; and pH adjusted to 7.0 ± 0.2), at a starting OD600 = 0.01, from overnight monocultures. For monoculture growth curves, bacteria were grown from OD600 = 0.01 in 96 well plates (with and without 6-BAP, 100 µM), with OD measured every 2 h for 24 h. Adenine (100 µM, dissolved in 1 M HCl) was used as a structurally similar negative control to 6-BAP, as is customary [40]. For community assays, we constructed two communities: one comprised a Pseudomonas isolate (IN68), a Ralstonia isolate (R3C), and a B. megaterium isolate (4C) (community #1). The second community comprised a Pseudomonas isolate (IN68), a Ralstonia isolate (R3C), and two different bacillus isolates (4C and R2E) (community #2). Communities were grown in 100 mL flasks with or without the addition of CK, 100 µM, and samples were removed every 2 h and plated in 10-fold serial dilutions in triplicate, to assess CFU count.
Biofilm formation, swimming and swarming assays
The effect of CK on bacterial growth parameters such as biofilm formation and motility assays (swim and swarm) of the phyllosphere derived microbes, was assessed using several assays. For biofilm formation, an overnight grown culture of isolated bacteria was diluted to an OD600 of 0.1, and 1 μL culture was added to 99 μL NB broth in the absence and presence of 6-BAP (100 μM) in a 96-well plastic titer plate. The plate was allowed to stand at 28 °C for 16–24 h. After the incubation time, the cultures were carefully removed from each well using a pipette. Sterile water was used to gently rinse each well, and the remaining adherent cell matrices were stained with a 1% crystal violet (CV) solution for 30 min. The wells were washed with distilled water twice after incubation, and the CV attached to the biofilm was dissolved in DMSO. To quantify the amount of biofilm adherent dye, absorbance was measured at 595 nm [41]. For swimming and swarming assays, the overnight grown bacterial cells (OD600 = 0.01, 1 uL) were gently inoculated in the center of the solidified NB agar plates (0.3% agar for swim and 0.5% agar for swarm) supplemented with CK, and incubated at 28 °C for 24 h. The swimming and swarming traits were examined after 24 h, by measuring the diameter of the bacterial zone. Similar plates without the addition of CK in the respective media served as a control [41].
Data analysis
All experimental data is presented as minimum to maximum values with median, in boxplots or floating bars, or as average ± SEM, with all points displayed. For microbiome analyses, differences between two groups were analyzed for statistical significance using a Mann–Whitney test, or a two-tailed t test, with Welch’s correction where applicable (unequal variances). Differences among three groups or more were analyzed for statistical significance with a Kruskal–Wallis ANOVA, with Dunn’s multiple comparisons post-hoc test. For all other analyses, differences between two groups were analyzed for statistical significance using a two-tailed t test, with Welch’s correction where applicable (unequal variances), and differences among three groups or more were analyzed for statistical significance with a one-way ANOVA. Regular ANOVA was used for groups with equal variances, and Welch’s ANOVA for groups with unequal variances. When a significant result for a group in an ANOVA was returned, significance in differences between the means of different samples in the group were assessed using a post-hoc test. The Tukey test was employed for samples with equal variances, when the mean of each sample was compared to the mean of every other sample. The Bonferroni test was employed for samples with equal variances, when the mean of each sample was compared to the mean of a control sample. The Dunnett test was employed for samples with unequal variances. All statistical analyses were conducted using Prism8.
Results
The effect of CK on microbial community formation
Endogenous CK supports Gram-positive bacteria in the pyllosphere
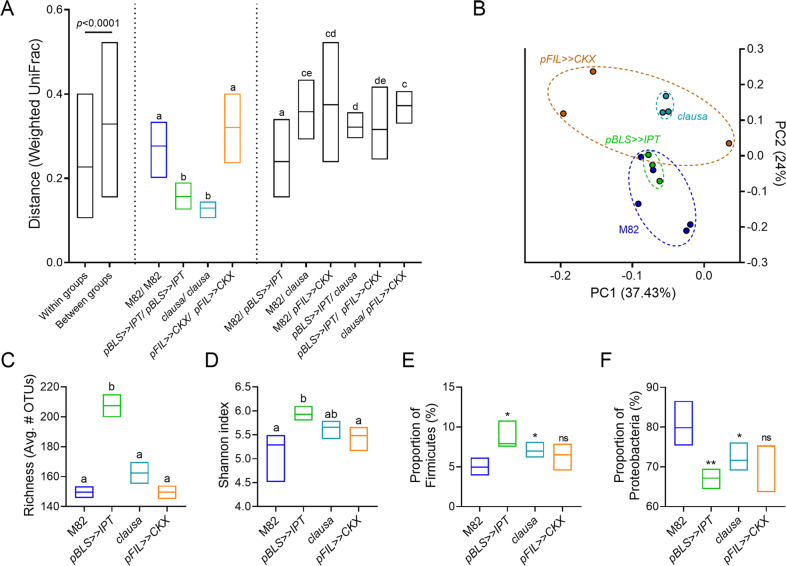
To assess the effect of altered CK profiles on phyllosphere microbial communities, microbial DNA was prepared from the phyllosphere of randomly interspersed tomato plants from the M82 background line, and the altered CK genotypes pBLS>>IPT, clausa and pFIL>>CKX, grown in a net house in the winter of 2018. Details on the previously described CK content and plant development of these genotypes is provided in the “Methods”. When assessing community structure between the samples using the weighted UniFrac distances, we observed a significant clustering of the samples based on their genotype, demonstrating that the distance among biological replicates is significantly smaller within groups then between groups (Fig. 1A, B). Interestingly, the CK rich pBLS>>IPT and CK hypersensitive clausa exhibited significantly less variation in community composition between the replicates compared to the WT M82, and the CK deficient pFIL>>CKX, observed through the lower distance between the biological replicates of each group. The higher homogeneity in community composition between the pBLS>>IPT, and clausa samples, suggests that these genotypes elicit a higher constraint on community composition compared to WT, and pFIL>>CKX. Furthermore, pBLS>>IPT exhibited a significantly higher species richness (alpha diversity) (Fig. 1C), and Shannon index (Fig. 1D). These results, combined with the higher similarity between the replicates, suggest that this genotype promotes a diverse but defined bacterial community. The CK rich pBLS>>IPT and CK hypersensitive clausa also possessed a higher average proportion of Firmicutes, (Fig. 1E) and a lower average proportion of proteobacteria (Fig. 1F), in the bacterial community.
Fig. 1. Endogenous cytokinin supports Gram-positive bacteria in the pylloshpere.
16 S rRNA sequencing of the bacterial phyllosphere of randomly interspersed plants grown in a net house in the winter of 2018, N = 3–5 for each genotype: wild type M82 (background line), High-CK content (pBLS»IPT), increased CK sensitivity coupled with decreased CK content (clausa), and decreased CK content (pFIL»CKX). A Weighted UniFrac beta diversity. Distance is significantly smaller within groups then between groups (p < 0.0001). B Principal coordinates analysis of distance between all individual samples in the weighted UniFrac beta diversity calculations. C Species richness- alpha diversity. D Shannon index. E Proportion of Firmicutes in the bacterial community of indicated genotypes. F Proportion of Proteobacteria in the bacterial community of indicated genotypes. Floating bars encompass minimum to maximum values, line indicates mean. Different letters indicate statistical significance between samples, and asterisks indicate statistical significance from WT M82, in a two-tailed t test with Welch’s correction. (*p < 0.05; **p < 0.01; NS non significant).
The experiment was repeated with the same genotypes in the summer of 2018 (Fig. S1). In the summer, the alpha diversity and Shannon index decreased, and the weighted UniFrac diversity between some of the samples increased, however, the significantly higher similarity within the pBLS>>IPT and clausa samples, and their significantly higher proportion of Firmicutes, was retained (Fig. S1).
Exogenous CK supports Gram-positive bacteria in the pylloshpere
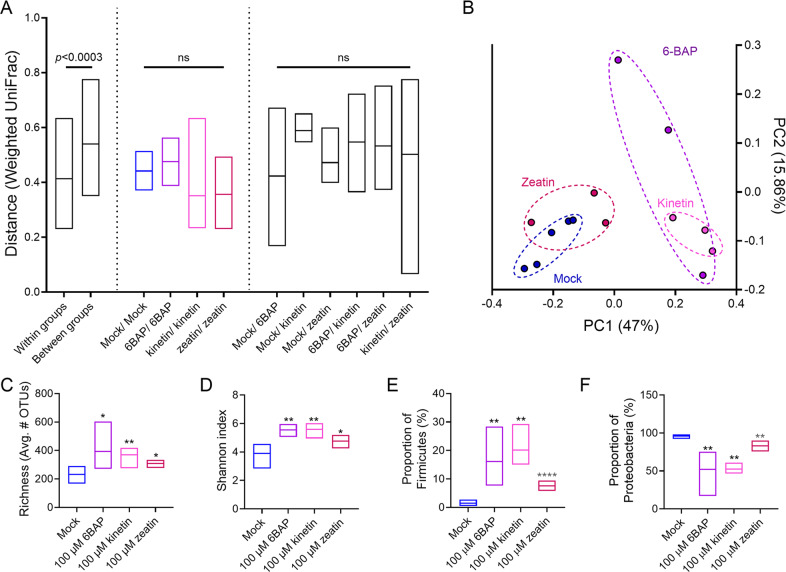
Since we observed that high endogenous CK content can affect phylloshpere microbial community composition, we proceeded to examine whether exogenous CK treatment could have a similar effect. Microbial DNA was prepared from the bacterial phyllosphere of randomly interspersed WT M82 plants treated with CK compounds (100 µM of 6-BAP, kinetin, or zeatin), grown in a net house in the winter of 2018. Weighted UniFrac beta diversity indicates that the distance is significantly smaller within groups than between groups (Fig. 2A, B). While the treatment with exogenous CK compounds did not significantly increase the similarity between the replicates when compared with the untreated mock, as observed with the CK rich or CK hypersensitive genotypes (Fig. 1), it significantly increased the community richness (Fig. 2C) and Shannon index (Fig. 2D), compared to the mock treated plants. Similar to the results observed in the CK altered genotypes, where high CK content or sensitivity promoted an increase in the relative content of Firmicutes and a decrease in the relative content of proteobacteria, the average proportion of Firmicutes in the bacterial community (Fig. 2E) increased upon exogenous CK treatment, while the average proportion of proteobacteria in the bacterial community decreased upon exogenous CK treatment (Fig. 2F).
Fig. 2. Exogenous cytokinin supports Gram-positive bacteria in the pylloshpere.
16 S rRNA sequencing of the bacterial phyllosphere of randomly WT M82 plants treated with 100 µM of the indicated cytokinin (CK) compounds (6-Benzylaminopurine (6-BAP), kinetin, and trans‐zeatin), grown in a net house in the winter of 2018, N = 3–5 for each genotype. A Weighted UniFrac beta diversity. Distance is significantly smaller within groups then between groups (p < 0.0003). B Principal coordinates analysis of distance between all individual samples in the weighted UniFrac beta diversity calculations. C Species richness- alpha diversity. D Shannon index. E Proportion of Firmicutes in the bacterial community. F Proportion of Proteobacteria in the bacterial community. (*p < 0.05; **p < 0.01; ***p < 0.001; NS non significant). Floating bars encompass minimum to maximum values, line indicates mean. Asterisks indicate statistical significance from mock treatment in a two-tailed t test with Welch’s correction.
Endogenous CK promotes Gram-positive bacteria in the phyllosphere under sterile conditions
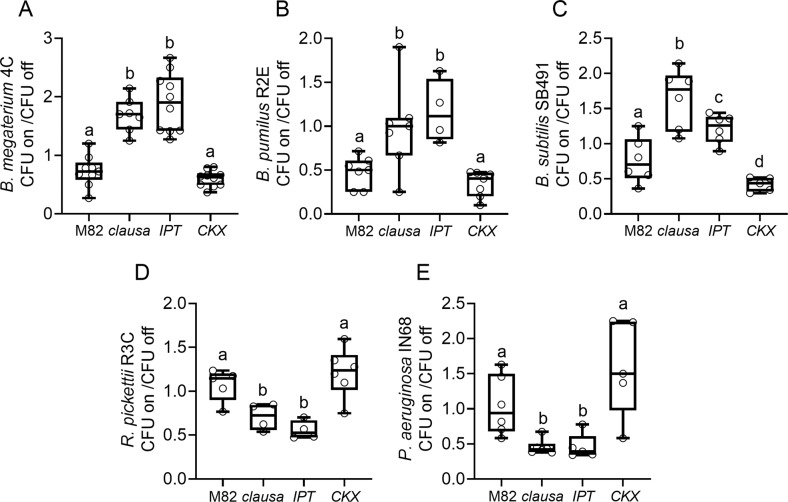
Next, we examined whether genotypes with high CK content or sensitivity could also support Firmicutes in a synthetic community (SynCom). Plants of altered CK genotypes were grown in sterile conditions, and then inoculated with synthetic communities containing morphologically identifiable bacteria of representative OTUs. The bacilli we included in these communities were isolated from the phyllosphere of CK-rich plant genotypes (see details in the “Materials”). We constructed two different communities as detailed in the “Methodology”. Plants were inoculated with either community at a 1:1:1:1 ratio of each bacterium, and community composition was examined three weeks after inoculation. The results of the sterile inoculations essentially confirmed the 16 S rRNA sequencing results, with the high CK pBLS>>IPT, and the CK hypersensitive mutant clausa, supporting more Gram-positive bacilli than the WT M82, or the CK deficient pFIL>>CKX (Fig. 3A, B). Total bacterial CFU on the plants was also increased on pBLS>>IPT in both communities, and on clausa in community #1 (Fig. 3C). The synthetic community results confirm that CK-rich plant genotypes are better able to support bacilli.
Fig. 3. Endogenous cytokinin supports Gram-positive bacteria in the phyllosphere under sterile conditions.
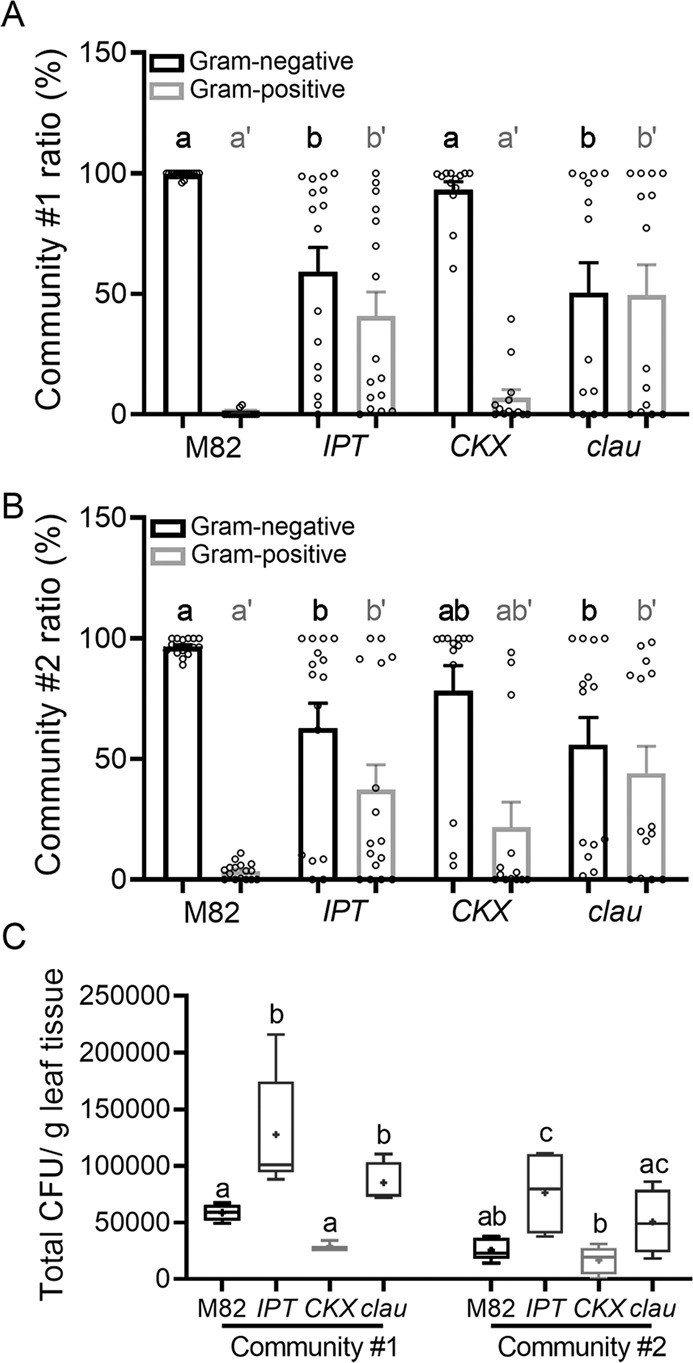
Plants of indicated genotypes: M82 (WT), IPT (pBLS»IPT), CKX (pFIL»CKX) and clau (clausa), were grown in sterile conditions for three weeks, and then inoculated with bacterial communities containing morphologically identifiable bacteria. Firmicutes included in these communities were isolated from the phyllosphere of the CK rich genotype pBLS»IPT. Plants were inoculated with each community at a 1:1:1:1 ratio of each bacterium. Community #1 included one Firmicute (25%), and community #2 included 2 Firmicutes (50%), as detailed in the “Methodology”. Community composition was examined three weeks after inoculation, by preparing epiphyte bacteria and plating them for colony forming unit (CFU) count. A Proportion of Gram-negative (black) and Gram-positive (gray) bacteria found on indicated genotypes inoculated with community #1. Experiment was conducted four times, with 3–4 plants of each genotype in each experiment, average ± SEM, N = 14. Bar graph displays all points. Letters (lower case for Gram-negative, tagged lower case for Gram-positive) indicate statistical significance in a one-way ANOVA with a Dunnett post-hoc test, p < 0.028). B Proportion of Gram-negative (black) and Gram-positive (gray) bacteria found on indicated genotypes inoculated with community #2. Experiment was conducted four times, with 3–4 plants of each genotype in each experiment, average ± SEM, N = 14. Bar graph displays all points. Letters (lower case for Gram-negative, tagged lower case for Gram-positive) indicate statistical significance in a one-way ANOVA with a Dunnett post-hoc test, p < 0.03). C Total CFU found in the phyllosphere of indicated genotypes three weeks after inoculation. Box plots indicate inner quartile ranges (box), outer quartile ranges (whiskers), median (line), mean (“+”). Letters indicate statistical significance in a one-way ANOVA with a Tukey post-hoc test, p < 0.026.
The role of the bacterial community in CK-mediated immunity and disease resistance
CK-mediated immunity is bacterial community dependent
Several previous works by us and others have investigated CK-mediated immunity, primarily against bacterial and fungal pathogens that attack aerial plant tissues [13, 42, 43]. Recently, the crucial role of immunity promoting gram positive bacilli in the phyllosphere has been confirmed [19]. However, possible roles of the microbial community in CK-mediated immunity have not yet been elucidated. Thus, we next examined CK-mediated immunity in sterile and non-sterile conditions.
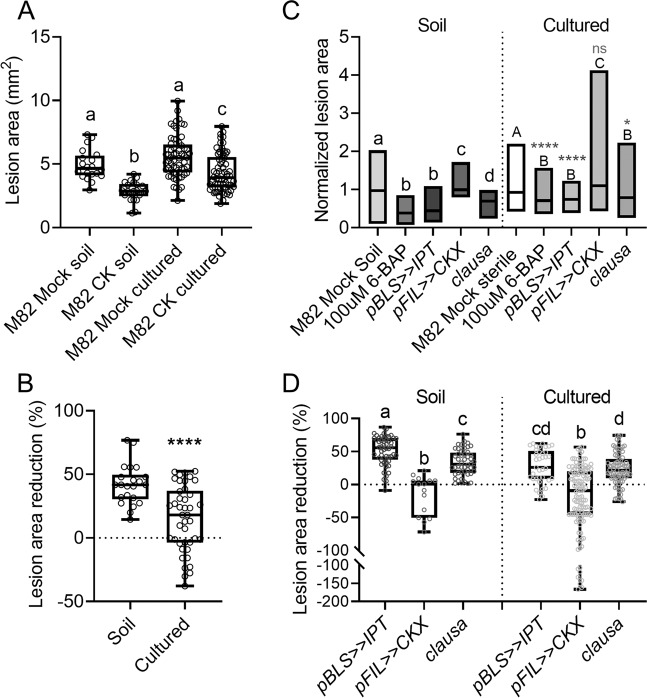
CK reduced B. cinerea disease in both natural (soil) and sterile (culture) conditions (Fig. 4A). However, the reduction in sterile conditions was significantly smaller (Fig. 4A). Disease was reduced by close to 50% in soil-grown plants, and by only ~20% in sterile grown plants (Fig. 4B), indicating that the microbial community likely influences the degree of CK-mediated disease reduction. As we previously reported, pBLS>>IPT and clausa have reduced disease levels when inoculated with B. cinerea [13]. In agreement with the sterile CK-treatment disease results, we found that under sterile conditions, these genotypes display a reduction in the level of increased immunity they possess when soil-grown (Fig. 4C, D). Similar disease levels were achieved with B. cinerea in sterile and cultured conditions (Fig. 4A), however, disease reduction following CK treatment or in altered CK genotypes was significantly lower under sterile conditions (Fig. 4C, D), indicating the bacterial community is required to achieve the full extent of CK-mediated disease reduction, and/or, that CK mediates the growth of a disease-suppressive microbiome.
Fig. 4. Cytokinin mediated immunity is bacterial community dependent.
CK-mediated immunity was examined using increased cytokinin (CK) genotypes and exogenous CK treatment, on identical aged plants, in sterile (cultured) and natural (soil) conditions, using B. cinerea infection. S. lycopersicum cv. M82 plants were treated with 100 µM 6BAP (CK) and inoculated with 10 µL of B. cinerea spore solution (106 spores mL−1) after 24 h. The lesion area was measured five days after B. cinerea inoculation using ImageJ. A Lesion area observed in M82 soil grown and cultured plants, with and without CK. Box plots indicate inner quartile ranges (box), outer quartile ranges (whiskers), median (line). All points shown. Letters indicate statistical significance in a one way ANOVA with a Dunnett post hoc test, N > 22, p < 0.0001. B Lesion area reduction with CK treatment, in soil and cultured conditions. Box plots indicate inner quartile ranges (box), outer quartile ranges (whiskers), median (line). All points shown. Asterisks indicate statistical significance in an unpaired t test with Welch’s correction, N > 22, p < 0.0001. C Normalized disease levels in genotypes with altered CK levels/signaling, in soil and sterile conditions. Values were normalized to allow comparison of the percent of disease reduction under both conditions. Floating bars encompass minimum to maximum values, line indicates median. Lower case letters indicate statistical significance in soil conditions, and uppercase letters indicate statistical significance in sterile conditions, in a one-way ANOVA with a Dunnett post hoc test, N > 15, p < 0.0048. Asterisks indicate statistical significance of each “cultured” sample from its counterpart “soil” sample. D Lesion area reduction in genotypes with altered CK levels/signaling, in soil and cultured conditions. Box plots indicate inner quartile ranges (box), outer quartile ranges (whiskers), median (line). All points shown. Letters indicate statistical significance in an unpaired t test with Welch’s correction, N > 15, p < 0.028.
Gram-positive bacilli isolated from the phyllosphere of high CK genotypes promote disease resistance
Since we found that CK-mediated immunity is partly phyllobiome dependent, we next tested whether our phyllosphere microbial isolates could themselves promote plant immunity and disease resistance. We treated WT M82 plants with the bacterial isolates, and separately challenged with different pathogens three days after bacterial treatment. Plants treated with any of the bacilli isolates all displayed increased resistance to B. cinerea (Fig. S2A), X. euvesicatoria (Fig. S2B) and O. neolycopersici (Fig. S2C). P. aeruginosa was also able to promote disease resistance against X. euvesicatoria and O. neolycopersici, suggesting that it is able to prime defenses against biotrophic pathogens, while our bacilli isolates likely prime defenses against both nectrotrophic and biotrophic pathogens.
Activity against a wide range of pathogens with different lifestyles suggests that the primary mode of action is priming plant defenses. To verify this, we examined immunity parameters and defense gene expression following individual treatment with different bacterial isolates: B. pumilus R2E, B. megaterium 4C, R. pickettii R3C, P. putida IN68, and B. subtilis SB491. WT M82 plants were pre-treated with indicated bacteria (OD600 = 0.1), and challenged with wounding, or the immunity elicitors EIX and flg22, three days after bacterial treatment. Plants pre-treated with either of the three bacillus isolates, 4C, R2E, or SB491, displayed a significant increase in ethylene production in response to wounding (Fig. S3A) and EIX elicitation (Fig. S3B), and in ROS production by flg22 elicitation (Figure S3C). The Gram-negative Ralstonia and Pseudomonas isolates did not effect similar immunity increases. Defense gene expression was assayed by qRT-PCR on cDNA samples prepared from plants pre-treated with the bacterial isolates 48 h prior to RNA preparation (Fig. S3D–K). Assayed genes were normalized to a geometric mean of the expression of three housekeeping genes: SlRPL8 (Solyc10g006580), Slcyclophilin (Solyc01g111170) and SlActin (Solyc03g078400). We examined the expression of genes related to the SA, ET, and JA pathways, as well as PRR genes. The SA pathway was activated by the pseudomonas and two of the bacillus isolates (Fig. S3D), verified by PR1a induction [44]. ET is thought to signal induced resistance, in synergy with JA, during colonization by beneficial microorganisms [44, 45]. Ethylene related genes were activated by all isolates (Fig. S3E–G), though only the bacillus isolates activated SlACO-1 (Fig. S3G), an ACC oxidase [44], the upregulation of which indicates activation of the ET signaling pathway. The ethylene-responsive factor 1 (ERF1) which is rapidly elicited by ET or JA [46], was activated by all isolates (Fig. S3F). Chitinase (Chi) and glucanase (Gluc), which are regulated by JA/ET and activate induced resistance against necrotrophic pathogens [47], were activated by all isolates except the pseudomonas (Fig. S3H–I). Interesting differential induction was observed with the PRRs, with LeEIX1 activated only by bacilli (Fig. S3J), and SlFLS2 activated only by the Gram-negative isolates (Fig. S3K). Gene accessions, primers, and primer efficiencies are detailed in Supplementary Table 1.
Overall, our results indicate that one mode of action of the phyllosphere bacterial isolates in disease prevention is via host immunity activation. To examine whether they can also function through direct inhibition bio-control against the pathogens, we conducted on-plate biocontrol confrontation assays between our bacterial isolates, and B. cinerea or X. euvesicatoria. We found no effect of the bacterial isolates on the growth of either pathogen (Fig. S4).
The role of leaf structure in phyllosphere community formation
Altered CK content or signaling results in altered leaf structure
We found that CK treatment or endogenous alterations in CK levels or response affect tomato immunity [13], and microbial community formation (Figs. 1–3). We observed that the microbial community likely mediates part of the CK protective effect (Figs. 4 and S2, S3). Leaf wax mutants have been shown to possess altered leaf structure, and host distinct microbial communities in Arabidopsis [48]. Alterations in CK levels have both chemical and structural effects on the plant. We proceeded to investigate whether genotype-dependent alterations in leaf structure could affect the leaf microbial community.
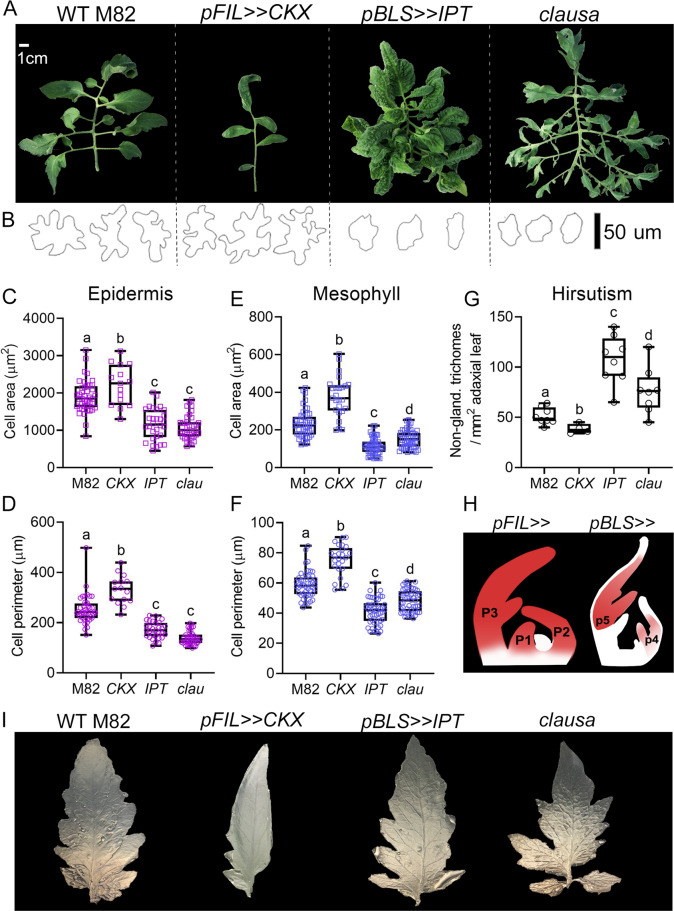
Altered CK levels cause dramatic alterations in leaf structure (Fig. 5 and S5), likely creating different micro-structural niches that could explain the support of different OTUs in the pyhllosphere. We quantified some of the altered structural features of leaf surfaces of altered CK genotypes, finding that high CK (pBLS>>IPT), or increased CK sensitivity (clausa) result in smaller epidermal and mesophyll cells with reduced patterning (Fig. 5B–F), and increased amounts of non-glandular trichomes (Fig. 5G). Reduced CK levels (pFIL>>CKX) result in larger cells, as was previously reported in Arabidopsis [49], and decreased amounts of non-glandular trichomes. These features could create different micro-structural niches and affect microbe placement. Expression domains of the FIL and BLS promoters are provided in Fig. 5H. Quantification of plant development and representative images of the altered CK content transgenic lines used in this study are provided in Fig. S6. The clausa mutant was previously compared to WT M82 in detail [12, 50].
Fig. 5. Leaf physical properties of indicated genotypes.
A Image of a mature fifth leaf of indicated genotypes, bar = 1 cm. B Outline of abaxial epidermal cells of indicated genotypes, bar = 50 µM. C, D Average area and perimeter of abaxial epidermal cells in indicated genotypes. E–F Average area and perimeter of mesophyll cells in indicated genotypes. G Average number of non-glandular trichome hairs on adaxial leaf surface in indicated genotypes. Cell size and trichome numbers were counted using ImageJ. H Cartoon depicting the expression domain of the FIL and BLS promoters, adapted with permission from Shani et al. [104]. I Representative images of recapitulation in agar using prepared polydimethylsiloxane (PDMS) negative replicas of the left hand second lateral leaflet from each indicated genotype. See also Supplementary Fig. S8. C–G Box plots are shown with inner quartile ranges (box), outer quartile ranges (whiskers), median (line), all points indicated. A–F Letters indicate significance in a one-way ANOVA with a Dunnett post hoc test, N > 15, p < 0.0005. G Letters indicate significance in an unpaired two-tailed t test, N = 8, p < 0.02.
Synthetic leaf replicas can recapitulate the leaf structure of altered CK genotypes
To isolate the effect of the structural component(s) on microbial community formation, we used biomimetics. Biomimetics is the use of chemistry and material sciences to mimic natural systems, and has been used previously to recapitulate the microstructure of leaves [51, 52]. We created replicas of tomato adaxial leaf surfaces using the synthetic polymer polydimethylsiloxane (PDMS), and leaflets from leaves of six-week-old S. lycopersicum cv. M82 plants of the different CK altered genotypes. The process of preparing leaf replicas is detailed in Fig. S7. We used the PDMS replicas to recapitulate the leaf structures in agar. The leaf microstructure was accurately re-produced in the PDMS (Fig. S8), and in the agar (Fig. 5I). We proceeded to examine bacterial growth and dispersion on our synthetic agar leaves.
Bacilli isolates display increased CFU growth on the leaf structure of high CK genotypes
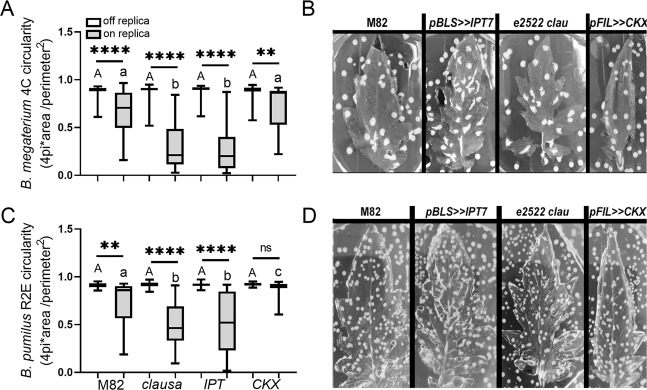
We first examined the growth of each of the individual bacterial isolates on the different structures. We found that our bacillus isolates B. megaterium 4C (Fig. 6A), B. pumilus R2E (Fig. 6B), and B. subtilis SB491 (Fig. 6C), showed a significant preference for the structures derived from high CK content or sensitivity genotypes, with the ratio between CFU growth on the structure/CFU growth off the structure being higher on strcutures mimicking high CK content or sensitivity genotypes. We also examined growth of the Gram-negative isolates on the leaf replicas. R. pickettii R3C (Fig. 6D), and P. aeruginosa IN68 (Fig. 6E), displayed an aversion for the structures derived from the high CK genotypes, preferring to grow on the structures derived from M82 or pFIL>>CKX.
Fig. 6. Preference of Gram-positive bacteria and aversion of Gram-negative bacteria to synthetic leaf structures derived from high cytokinin content/signaling genotypes.
Indicated bacteria (OD600 = 0.01) were spray inoculated onto agar replicas of leaves of the indicated genotypes. Bacterial growth was quantified after 24 h. Preference or aversion to the leaf structure was assessed by quantifying the colony forming units (CFU) growing on the leaf structure, and dividing it by the CFU growing on an equal area in the surrounding structure-less agar. Box plots are shown with inner quartile ranges (box), outer quartile ranges (whiskers), median (line), depicting results from three independent experiments, all points shown. A B. megaterium 4C. Letters indicate significance in a one-way ANOVA with a Dunnett post hoc test, N = 9, p < 0.0001. B B. pumilus R2E. Letters indicate significance in a one-way ANOVA with a Tukey post hoc test, N = 7, p < 0.04. C B. subtilis SB491. Letters indicate significance in a one-way ANOVA with a Tukey post hoc test, N = 6, p < 0.0016. D R. pickettii R3C. Letters indicate significance in a one-way ANOVA with a Tukey post hoc test, N = 6, p < 0.029. E P. aeruginosa IN68. Letters indicate significance in an unpaired two-tailed t test, N = 7, p < 0.019.
Interestingly, the colony shape and size were also dependent on the leaf structure. The bacillus isolates B. megaterium 4C (Fig. 7A, B), and B. pumilus R2E (Fig. 7C, D), had less circular shapes on the structures derived from high CK content or sensitivity genotypes. Growth on the surrounding agar area that did not contain the leaf structure was similar in all cases. Circularity was not strongly affected in B. subtilis SB491, which has wavy colony margins, making circularity quantification less meaningful- except in the case of pFIL>>CKX, where circularity increased on the replica, likely due to the colonies being very small, and thus less patterned (Fig. S9A, B). Colony circularity was not affected by the presence of the replica structure in the case of R. pickettii R3C (Fig. S9C, D), and P. aeruginosa IN68 (Fig. S9E, F).
Fig. 7. Gram-positive bacteria exhibit decreased circularity on synthetic leaf structures derived from high cytokinin content/signaling genotypes.
Indicated bacteria (OD600 = 0.01) were spray inoculated onto agar replicas of leaves of the indicated genotypes. Colony circularity of indicated bacteria was measured after 24 h using ImageJ. A, C Box plots are shown with inner quartile ranges (box), outer quartile ranges (whiskers), median (line), depicting results from three independent experiments. Asterisks denote significant differences in colony circularity on and off the synthetic leaf replica derived from each genotype. Upper case letters denote differences in colony circularity on the structure-less agar (“off replica”). Lower case letters indicate statistically significant differences in colony circularity on the synthetic leaf replicas (“on replica”) derived from different genotypes. Asterisks and different letters represent statistically significant differences found in a one-way ANOVA with a Dunnett post hoc test. A N > 50, p < 0.0012. C N > 20, p < 0.017. (**p < 0.01, ****p < 0.0001, NS non significant). B, D Images depicting the colony morphology of B. megaterium 4C (B) and B. pumilus R2E (D) growing on the different genotype- derived synthetic leaf structures.
Colonies of the bacillus isolates B. megaterium 4C grew larger (Fig. S10A) on the structures derived from high CK content or sensitivity genotypes. Colony size of B. pumilus R2E was not affected (Fig. S10B). B. subtilis SB491 colony size was significantly affected, with colonies growing larger on the replica area on structures derived from high CK genotypes, and significantly smaller on the structure derived from the CK deficient genotype (Fig. S10C). R. pickettii R3C colony size was significantly reduced on the structures derived from the high CK genotypes (Fig. S10D), while P. aeruginosa IN68 colony size was mostly unaffected by the replica structure (Fig. S10E). Growth on the surrounding agar area that did not contain the leaf structure was similar in most cases.
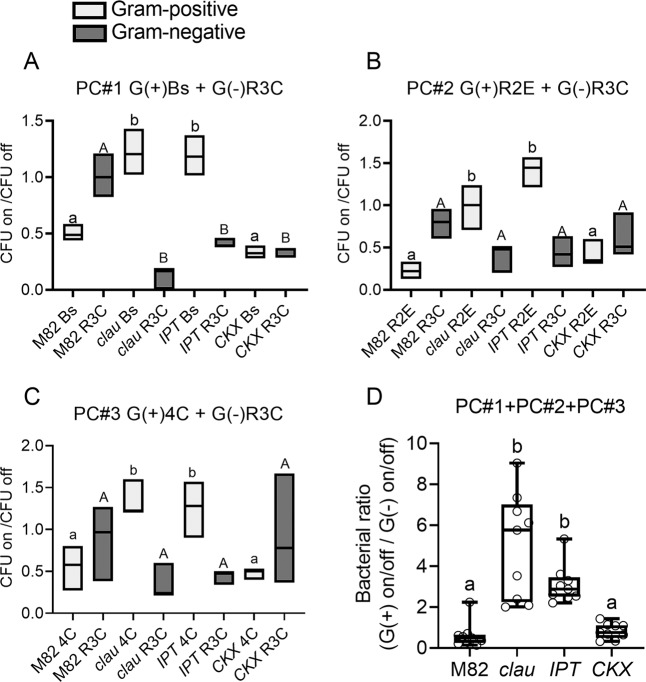
We next examined the growth and dispersal of combination of two bacteria on the leaf replicas, using the same methodology, with pairwise communities (“PC”), comprising each one Gram-positive (G(+)) and one Gram-negative (G(–)) bacterium (see “Methodology” for further details). In all communities, the changing Gram-positive bacillus isolate showed a clear preference for the high CK genotype derived structures (Fig. 8A–C). The ratio between Gram-positive and Gram-negative was calculated for the three pairwise communities, demonstrating a clear advantage for Gram-positive bacilli isolates on the clausa and pBLS>>IPT derived leaf replicas (Fig. 8D). Representative pictures of the PCs growing on the replicas are provided in Fig. S11.
Fig. 8. Preference of Gram-positive bacteria to leaf replica structures derived from high cytokinin content/signaling genotypes, in a pairwise community context.
Pairwise communities (“PC”) comprising each one Gram-positive (G(+)) and one Gram-negative (G(−)) bacterium, in equal ratios, at a combined OD600 of 0.01, were spray inoculated onto agar replicas of leaves of the indicated genotypes. Bacterial growth parameters were assessed after 24 h. Preference or aversion to the leaf structure was assessed by quantifying the colony forming unit (CFU) growing on the leaf structure, and dividing it by the CFU growing on an equal area in the surrounding structure-less agar. A–C Ratio of CFU on structure to CFU off structure in three communities of indicated bacteria. Floating bars depict minimum to maximum values, line indicates median. All graphs represent three independent replicates. Different letters represent statistically significant differences among samples, lower case letters for the Gram-positive bacterium (gray) and upper case letters for the Gram-negative bacterium (black), in a one-way ANOVA with a Dunnett (A) or Tukey (B) post hoc test, or in an unpaired t test (C). A B. subtilis Bs SB491 + R. pickettii R3C, p < 0.0004. B B. pumilus R2E + R. pickettii R3C, p < 0.0255. C B. megaterium 4C + R. pickettii R3C, p < 0.047. D Ratio between Gram-positive and Gram-negative was calculated for the three pairwise communities. Box plots are shown with inner quartile ranges (box), outer quartile ranges (whiskers), median (line), depicting results from three independent experiments, all point shown. Letters indicate significance in a one-way ANOVA with a Dunnett post hoc test, N = 9, p < 0.0072.
The role of leaf chemistry in phyllosphere community formation
Direct effect of CK on phyllosphere derived microbes
We observed a clear effect of the CK-derived leaf structure on the leaf phyllobiome (Figs. 6, 7 and S9–S11). In 16S rRNA sequencing experiments, exogenous application of CK on M82 plants supported increases in bacilli in the microbial community, without observable changes to the leaf structure (Fig. 2). Therefore, in addition to the structural effect, and although CK was found not to directly affect the growth of bacterial plant pathogens [42, 53], CK could directly affect the growth of phyllosphere isolates.
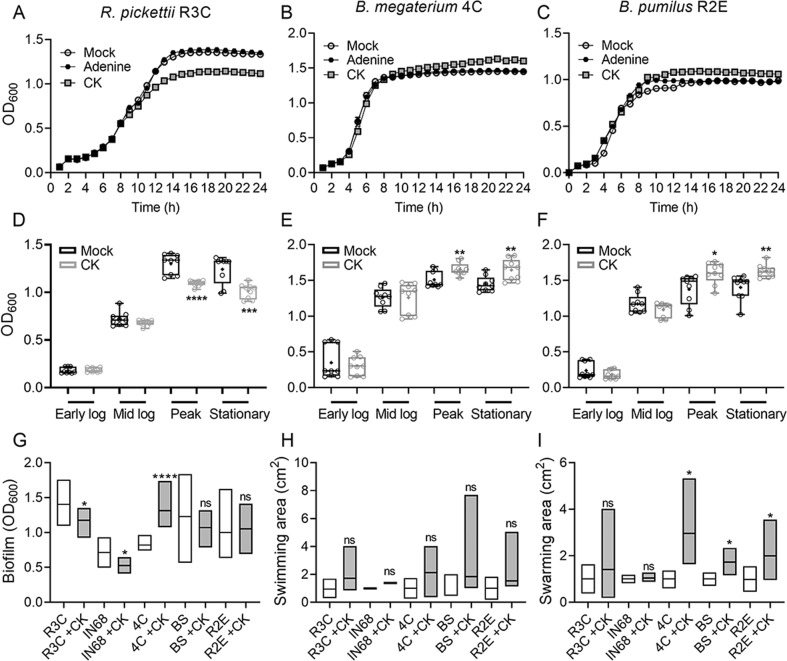
To test this, bacterial strains were grown in vitro with or without the addition of 6-BAP (CK) or adenine (100 µM), for 24 h. We found that CK directly inhibits the growth of R. pikettii R3C (Fig. 9A, D), and slightly but significantly enhances the growth of B. megaterium 4C (Fig. 9B, E), and B. pumilus R2E (Fig. 9C, F), when compared with mock and adenine controls. Analysis of growth with and without CK during log phase, peak, and stationary phase, indicates that CK mostly affects the peak of growth reached (Fig. 9D–F). CK also inhibited the growth of P. aeruginosa IN68 at all growth phases (Fig. S12A, D), and prevented the decline of B. subtilis (Bs, SB491) in the stationary phase (Fig. S12B, E). Two additional bacillus isolates we examined, B. aryabhattai R2A and B. subtilis R1D, were unaffected by CK (Fig. S12C, F), indicating that this effect is not common to all bacilli. The effect of CK on all isolates except R2E was retained in minimal media (Fig. S12G).
Fig. 9. Direct effect of cytokinin on phyllosphere derived microbes.
Bacterial strains were grown in nutrient broth with or without the addition of cytokinin (6-Benzylaminopurine (6-BAP), 100 µM) or the structural control adenine (100 µM) for 24 h. A–C growth curves; D–F analysis of growth with and without CK during log phase, peak, and stationary phase. A, D R. pikettii R3C; B, E B. megaterium 4 C; C, F B. pumilus R2E. Graphs represent three independent experiments, N = 9. G Biofilm formation with and without CK was assessed using crystal violet dye. H Bacterial swimming with and without CK was assessed in 0.3% agar. I Bacterial swarming with and without CK was assessed in 0.5% agar. D–F Box plots are shown with inner quartile ranges (box), outer quartile ranges (whiskers), median (line), mean (“+”), depicting results from three independent experiments, all points shown. Asterisks indicate significance in a one-way ANOVA with a Bonferroni post hoc test, N = 9. D: p < 0.0001; E: p < 0.0214; F: p < 0.012 (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). G–I Floating bars encompass minimum to maximum values, line indicates mean. Asterisks indicate statistical significance in a two-tailed t test with Welch’s correction. G N = 6, p < 0.047. H N = 6, no significant differences found. I N = 6, p < 0.038. (*p < 0.05, ****p < 0.0001, NS non significant).
We next examined the direct effect of CK on phyllosphere derived microbes in a community context, using synthetic created communities (“CC”) comprising 3 phyllosphere microbes. We found that the effect was similar to that observed in the monocultures, with CK promoting the growth of the bacilli 4C or R2E, and inhibiting the growth of the Gram-negative R3C and IN68 (Fig. S13). We repeated the assay with the same created communities, diluting the initial starting ratio of the bacilli in the community to 10%. We observed similar results, with CK similarly promoting the growth of the bacilli when they were diluted to a smaller proportion of the starting community (Fig. S14).
We also investigated a possible effect of CK on additional growth parameters of phyllosphere derived microbes. Examining the ability of phyllosphere isolates to form biofilms in the presence of CK, we found that this ability was reduced in the Gram-negative R3C and IN68, and strongly increased in the bacillus 4C (Fig. 9G). The ability to swim was not affected by CK in any of the isolates (Fig. 9H), however, the ability to swarm was significantly increased in all the bacilli (Fig. 9I).
We have demonstrated that CK-mediated changes to the leaf surface structure play a central role in the formation of microbial communities (Figs. 1–2, 6–8). Further, a direct effect of CK on particular bacterial isolates may also play a part in leaf bacterial colonization (Fig. 9).
Discussion
CK drives phyllopshere microbial content
The mechanisms by which plants influence the formation of the particular microbial communities they host are the subject of intense investigation. The plant hormone CK is a central developmental regulator with important roles in many aspects of plant life. The effect of plant hormones on the assembly of the plant microbiome is complex [54], and has been examined in a few works [55–57]. Here, we set out to investigate what roles CK may have in influencing plant microbial communities. The host genotype and the environment were reported to have varying degrees of influence on the plant microbiome. Many works have investigated whether the plant genotype can influence the composition of its microbiome, finding larger or smaller genotype-based effects in different cases [4, 44, 58, 59]. Niches available for microbial colonization will differ in different hosts. A recent work examining microbial samples from different compartment niches in several plant hosts, demonstrated that microbiome assembly is shaped predominantly by compartment niche and host species, with increased host selection pressure reducing bacterial diversity and network complexity. Interestingly, the strongest host effect was observed in the leaf [58]. Here, we have demonstrated that plant genotypes that result in increased CK content or sensitivity can have a strong effect on shaping the microbiome (Figs. 1, 2, S1). Genotypes with increased CK content or sensitivity demonstrated enrichment of Firmicutes (Figs. 1, 2, S1). Interestingly, high CK content or signaling increased species richness while reducing distance among different samples within those genotypes, with results consistent across different seasons/environments, despite overall changes to the microbiome. This indicates that CK-rich plant genotypes apply stronger, more consistent, selective forces on bacterial community formation. These selective forces also increased community richness, rather than reducing it. Pre-treating plants with exogenous CK also resulted in increases in Firmicutes (Fig. 2). Synthetic community experiments confirmed that CK-rich plant genotypes are better able to support bacilli (Fig. 3). CK content and/or signaling is therefore a determining selective factor in microbial diversity, and can predict microbial community composition to a certain extent, favoring Gram-positive bacteria, and bacilli in particular.
CK-mediated immunity is partly dependent on the microbiome
Not surprisingly, like in other eukaryotes, plant immunity can depend on the microbiome [60–62]. Induced systemic resistance (ISR) can trigger plant immunity and subsequent pathogen resistance following exposure to particular microbes, with reports of ISR being triggered by various phylogenetically distinct OTUs, in various plant species [45]. Inducing plant immunity is known to promote a disease protective microbiome [63–66], and microbiome transplant was shown to protect plants against disease in several cases [4, 62, 67]. The plant microbiome also responds to the presence of pathogens and pests [44, 68–70], with pathogen infection demonstrated to lead to differential recruitment of protective bacteria that can improve disease outcomes [68, 71, 72].
Plant hormones have central roles in plant immunity and disease resistance. Signaling pathways of the plant defense hormones SA, JA, and ET, were all found to affect microbial diversity in leaves [17, 20]. CK is also gaining traction as a defense hormone of late. Several previous works by us and others have investigated CK-mediated immunity [13, 42, 43]. We observed that CK-mediated immunity partially depends on the microbiome (Fig. 4), and that increased CK content or signaling genotypes, which are also disease resistant [13], promote the formation of a protective epiphytic microbiome. Bacilli isolates from high CK genotypes were found to prime plant defenses (Fig. S2, S3). Recently, the necessity of Gram-positive bacilli in the microbial community for protecting plants from disease has been re-affirmed [19]. Bacillius spp. have been reported to have disease resistance promoting activities (reviewed in [73]), and have been developed as biocontrol products [74]. Some bacilli can have direct antimicrobial activities, acting directly as antifungal or antibacterial agents [75, 76]. Other isolates indirectly suppress disease by the elicitation of ISR [77, 78]. The bacilli we isolated from pBLS>>IPT do not have direct antimicrobial activities as far as we found (Fig. S4), consistent with the notion that bacteria with direct activities against plant pathogens may preferentially colonize the endosphere [7], and suggesting that our isolates improve plant disease outcomes by inducing immunity in the host plant.
Since immune system activation influences the microbiome composition, and CK promotes plant immunity [13, 79, 80], it cannot be ruled out that the activated immune state upon CK treatment influences the recruitment of beneficial microbes. Notably, some CK-mediated immunity was retained in sterile conditions (Fig. 4), indicating that another mechanism, such as CK-mediated priming, we as well as others reported [13, 81], also promotes pathogen resistance in response to high CK levels. Given that CK influences microbial community composition and mediates disease resistance, the possibility that CK causes a priority effect by promoting the establishment of particular microbial OTUs that are beneficial in disease resistance arises.
CK influences microbial content through structural and chemical cues
To investigate how CK promotes the establishment of bacilli in the microbiome, we examined whether CK influences the bacterial community composition through colonizable structural niches preferentially formed in CK-rich plant genotypes, or by directly promoting the growth of disease suppressive bacteria. Our results demonstrate that both avenues are plausible. Disease suppressive bacteria were able to preferentially grow on structures derived from CK-rich plant genotypes (Figs. 6, 7 and S9, S10), with some isolates also being promoted by the addition of CK in vitro (Fig. 9, S12-S14). The growth profile of particular bacilli in swarming, but not swimming assays, was also altered with the addition of CK (Fig. 9). Swarming and swimming are both powered by rotating flagella [82]. Bacteria can use both these types of motility machinery to colonize environments. Increased swarming can underlie higher biofilm formation, which may play a role in microbial plant colonization, thereby exerting beneficial effects on plant health [83]. Taken together with the changes to colony morphology of the bacilli on CK-rich derived synthetic leaf structures, we conclude that CK may promote changes to the biofilm formation abilities of particular bacteria which, in a community context, can allow them to thrive, particularly on leaf microstructures present in genotypes with high CK content or sensitivity.
Leaves possess many structural niches potentially habitable by microbes, and the leaf cuticle structure and trichome distribution were previously demonstrated to impact the composition of the bacterial phyllosphere community [48, 84, 85]. High CK content or sensitivity tomato genotypes have smaller epidermal cells, meaning more “depressions” on the leaf surface. Increased hairy trichomes could create additional sub-niches. CK altered genotypes likely also have altered cell wall structures [86], which would also affect the colonizable leaf microstructure. Furthermore, CK-rich plant genotypes possess rugose, curled leaves with increased marginal patterning, all of which could generate yet additional structural niches that preferentially support particular bacterial OTUs. We used a biomimetic system to confirm that structural elements can support particular bacterial isolates. Indeed, the morphology of bacilli isolate colonies on synthetic leaf structures derived from high CK content/signaling plants suggests that they are “hugging” the depressions of the venation pattern, which are deeper and more numerous in leaves of CK-rich plant genotypes (Figs. 6, 7 and S9, S10). The synthetic leaf imprints generated in agar did not indicate a bacillus preference to a particular area within the leaf structure other than the venation pattern. However, the agar system has the limitation of having equal nutrient distribution across the imprinted structure, which is likely different than the situation on the natural leaf, therefore, additional structural preferences cannot be ruled out.
CK may serve as a bidirectional signaling molecule in plant-microbe interactions
Many bacteria possess genes which could produce CKs [87–89]. Plant growth promoting B. cereus, B. subtilis, and P. fluorescens G20-18 were reported to synthesize cytokinins [90–93] and also promote disease resistance [91, 94]. Microbial CK can interact with plant hormone signaling pathways and promote disease resistance against pathogens [95], essentially mirroring the results we obtained when overexpressing the CK biosynthetic gene IPT, or exogenously providing CKs. Furthermore, the level of CKs within the plant is also affected by crosstalk with additional hormones [9], therefore, hormonal signaling networks operating within the framework of the interaction between the plant host and the microbiome, are likely also influenced by microbial produced auxins or additional hormones. CK signaling is perceived by membrane-located sensor HKs, and transmitted through a two-component system (TCS) in higher eukaryotes [96]. An analogous system was also identified in bacteria [97]. Thus, bacteria present in the microbiome can likely also sense the presence of plant CK, resulting in bacterial signaling cascades, the outcome of which could affect plant microbe interactions and phylloshpere colonization.
CKs play an important role in plant resource allocation, through their effects on the stimulation of invertase activity and carbohydrate metabolism [98, 99]. CK production by plant-associated microbes was thought to serve primarily as a plant growth promoting mechanism in the mutualistic host-microbe interaction [90, 93]. Our findings that CK may directly support the growth of particular isolates, suggest that microbial CKs may have a role in assisting plant colonization by their producers. Microbial produced CKs could promote plant metabolism, leading to increased vigor and underlying, at least in part, the observed plant-growth promoting effects reported for many microbiome isolates. CKs produced by the microbiome could also further support disease protective consortia, increasing the benefit for the host plant. Future work investigating the roles of microbial CKs in plant–microbe interactions should also focus on how plant physiology and development, as well as stress responses, are specifically affected by microbial CKs.
Conclusions
We show here that CK poses a dominant effect on microbial community content in the phyllosphere, promotes species richness, supports immunity inducing bacilli in the phyllosphere community through both structural and chemical routes, and activates plant immunity in part via the microbiome. CK can be viewed as a core component in a holistic or circular system, where CK from both the plant host and the colonizing microbial strains acts as a bidirectional signaling molecule, supporting a beneficial interaction and leading to increased vigor, preferential development, and disease resistance in the host, while simultaneously allowing the bacteria through their CK sensing system to identify preferred hosts for colonization. From an agricultural standpoint, single strain inoculation of particular bacilli isolated from high CK genotypes, or inclusion of these bacilli in multi-strain communities, could be agriculturally beneficial and have minimal negative effects [100]. Future work will investigate this further with different hosts and in different agricultural settings.
Supplementary information
Acknowledgements
The authors wish to thank Dror Minz, Stefan J Green and Jonathan Friedman for providing bacterial strains and for helpful discussions, and the Bar, Jami and Kleiman group members for continuous discussion and support.
Author contributions
MK and MB: Conceptualization. RG, ML-M, TS, EJ, MK, and MB: Methodology. RG, DE, ML-M, TS, and AS: Experimentation. RG, DE, ML-M, TS, AS, EJ, MK, and MB: Analysis. RG, EJ, MK, and MB: Manuscript.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Raw data is available through NCBI-SRA, Bioproject PRJNA729221.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01060-3.
References
- 1.Trivedi P, Trivedi C, Grinyer J, Anderson IC, Singh BK. Harnessing host-vector microbiome for sustainable plant disease management of phloem-limited bacteria. Front Plant Sci. 2016;7:1423. doi: 10.3389/fpls.2016.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leveau JH. A brief from the leaf: latest research to inform our understanding of the phyllosphere microbiome. Curr Opin Microbiol. 2019;49:41–49. doi: 10.1016/j.mib.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Carlström CI, Field CM, Bortfeld-Miller M, Müller B, Sunagawa S, Vorholt JA. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat Ecol Evol. 2019;3:1445–54. doi: 10.1038/s41559-019-0994-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morella NM, Weng FCH, Joubert PM, Jessica C, Lindow S, Koskella B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc Natl Acad Sci USA. 2020;117:1148–59. doi: 10.1073/pnas.1908600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remus-Emsermann MNP, Lücker S, Müller DB, Potthoff E, Daims H, Vorholt JA. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ Microbiol. 2014;16:2329–40. doi: 10.1111/1462-2920.12482. [DOI] [PubMed] [Google Scholar]
- 6.Schlechter RO, Miebach M, Remus-Emsermann MNP. Driving factors of epiphytic bacterial communities: a review. J Adv Res. 2019;19:57–65. doi: 10.1016/j.jare.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruisson S, Zufferey M, L’Haridon F, Trutmann E, Anand A, Dutartre A, et al. Endophytes and epiphytes from the grapevine leaf microbiome as potential biocontrol agents against phytopathogens. Front Microbiol. 2019;10:2726. [DOI] [PMC free article] [PubMed]
- 8.Wybouw B, De Rybel B. Cytokinin – a developing story. Trends Plant Sci. 2019;24:177–85. doi: 10.1016/j.tplants.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Shwartz I, Levy M, Ori N, Bar M Hormones in tomato leaf development. Dev Biol. 2016;419:132-142. [DOI] [PubMed]
- 10.Žižková E, Dobrev PI, Muhovski Y, Hošek P, Hoyerová K, Haisel D, et al. Tomato (Solanum lycopersicum L.) SlIPT3 and SlIPT4 isopentenyltransferases mediate salt stress response in tomato. BMC Plant Biol. 2015;15:1–20. doi: 10.1186/s12870-015-0415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shani E, Ben-Gera H, Shleizer-Burko S, Burko Y, Weiss D, Ori N. Cytokinin regulates compound leaf development in tomato. Plant Cell. 2010;22:3206–17. doi: 10.1105/tpc.110.078253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar M, Israeli A, Levy M, Ben Gera H, Jiménez-Gómez JM, Kouril S, et al. CLAUSA is a MYB transcription factor that promotes leaf differentiation by attenuating cytokinin signaling. Plant Cell. 2016;28:1602–15. doi: 10.1105/tpc.16.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta R, Pizarro L, Leibman-Markus M, Marash I, Bar M. Cytokinin response induces immunity and fungal pathogen resistance, and modulates trafficking of the PRR LeEIX2 in tomato. Mol Plant Pathol. 2020; 10.1111/mpp.12978. [DOI] [PMC free article] [PubMed]
- 14.Nafisi M, Fimognari L, Sakuragi Y. Interplays between the cell wall and phytohormones in interaction between plants and necrotrophic pathogens. Phytochemistry. 2015;112:63–71. doi: 10.1016/j.phytochem.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Brettell LE, Singh B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020;25:841–4. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Dastogeer KMG, Tumpa FH, Sultana A, Akter MA, Chakraborty A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr Plant Biol. 2020;23:100161.. [Google Scholar]
- 17.Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014;10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purahong W, Orrù L, Donati I, Perpetuini G, Cellini A, Lamontanara A, et al. Plant microbiome and its link to plant health: host species, organs and pseudomonas syringae pv. Actinidiae infection shaping bacterial phyllosphere communities of kiwifruit plants. Front Plant Sci. 2018;9:1563. [DOI] [PMC free article] [PubMed]
- 19.Chen T, Nomura K, Wang X, Sohrabi R, Xu J, Yao L, et al. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature. 2020;580:653–7. [DOI] [PMC free article] [PubMed]
- 20.Kniskern JM, Traw MB, Bergelson J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol Plant-Microbe Interact. 2007;20:1512–22. doi: 10.1094/MPMI-20-12-1512. [DOI] [PubMed] [Google Scholar]
- 21.Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103:6398–403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smigocki AC, Owens LD. Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc Natl Acad Sci. 1988;85:5131–5. doi: 10.1073/pnas.85.14.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redig P, Schmülling T, Van Onckelen H. Analysis of cytokinin metabolism in ipt transgenic tobacco by liquid chromatography-tandem mass spectrometry. Plant Physiol. 1996;112:141–8. doi: 10.1104/pp.112.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Márquez-López RE, Quintana-Escobar AO, Loyola-Vargas VM. Cytokinins, the Cinderella of plant growth regulators. Phytochem Rev. 2019;18:1387–408. [Google Scholar]
- 25.Smigocki A, Neal JW, McCanna I, Douglass L. Cytokinin-mediated insect resistance in Nicotiana plants transformed with the ipt gene. Plant Mol Biol. 1993;23:325–35. doi: 10.1007/BF00029008. [DOI] [PubMed] [Google Scholar]
- 26.Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23:69–80. doi: 10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid DE, Heckmann AB, Novák O, Kelly S, Stougaard J. CYTOKININ OXIDASE/DEHYDROGENASE3 maintains cytokinin homeostasis during root and nodule development in Lotus Japonicus. Plant Physiol. 2016;170:1060–74. doi: 10.1104/pp.15.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23:2169–83. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vercruyssen L, Gonzalez N, Werner T, Schmülling T, Inzé D. Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in arabidopsis. Plant Physiol. 2011;155:1339–52. doi: 10.1104/pp.110.167049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farber M, Attia Z, Weiss D. Cytokinin activity increases stomatal density and transpiration rate in tomato. J Exp Bot. 2016;67:6351–62. doi: 10.1093/jxb/erw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang CH, Crowley DE, Borneman J, Keen NT. Microbial phyllosphere populations are more complex than previously realized. Proc Natl Acad Sci USA. 2001;98:3889–94. doi: 10.1073/pnas.051633898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian X, Shi Y, Geng L, Chu H, Zhang J, Song F, et al. Template preparation affects 16S rRNA high-throughput sequencing analysis of phyllosphere microbial communities. Front Plant Sci. 2017;8:1623. doi: 10.3389/fpls.2017.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. 2017;261:169–76. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 35.Kuklinsky-Sobral J, Araújo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol. 2004;6:1244–51. doi: 10.1111/j.1462-2920.2004.00658.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Chen Y, Shi Q, Hou B, Yang Q. Identification of bacteria associated with periapical abscesses of primary teeth by sequence analysis of 16S rDNA clone libraries. Microb Pathog. 2020;141:103954. [DOI] [PubMed]
- 37.Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–61. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Luo Y, Pearlstein AJ, Aplin J, Liu Y, Bauchan GR, et al. Fabrication of biomimetically patterned surfaces and their application to probing plant–bacteria interactions. ACS Appl Mater Interfaces. 2014;6:12467–78. doi: 10.1021/am502384q. [DOI] [PubMed] [Google Scholar]
- 39.Kumari P, Ginzburg N, Sayas T, Saphier S, Bucki P, Miyara SB, et al. A biomimetic platform for studying root-environment interaction. Plant Soil. 2020;447:157–68. [Google Scholar]
- 40.Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, et al. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001;42:1017–23. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- 41.Singh A, Gupta R, Pandey R. Rice seed priming with picomolar rutin enhances rhizospheric bacillus subtilis CIM colonization and plant growth. PLoS ONE. 2016;11:e0146013. doi: 10.1371/journal.pone.0146013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta R, Leibman-Markus M, Pizarro L, Bar M. Cytokinin induces bacterial pathogen resistance in tomato. Plant Pathol. 2020 (in press). [DOI] [PMC free article] [PubMed]
- 43.Argueso CT, Ferreira FJ, Epple P, To JPC, Hutchison CE, Schaller GE, et al. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 2012;8:e1002448. [DOI] [PMC free article] [PubMed]
- 44.Teixeira PJP, Colaianni NR, Fitzpatrick CR, Dangl JL. Beyond pathogens: microbiota interactions with the plant immune system. Curr Opin Microbiol. 2019;49:7–17. doi: 10.1016/j.mib.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–75. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 46.Mehari ZH, Elad Y, Rav-David D, Graber ER, Meller, Harel Y. Induced systemic resistance in tomato (Solanum lycopersicum) against Botrytis cinerea by biochar amendment involves jasmonic acid signaling. Plant Soil. 2015;395:31–44. [Google Scholar]
- 47.Jayaraj J, Punja ZK. Combined expression of chitinase and lipid transfer protein genes in transgenic carrot plants enhances resistance to foliar fungal pathogens. Plant Cell Rep. 2007;26:1539–46. doi: 10.1007/s00299-007-0368-x. [DOI] [PubMed] [Google Scholar]
- 48.Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS ONE. 2013;8:78613. doi: 10.1371/journal.pone.0078613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–50. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bar M, Ben-Herzel O, Kohay H, Shtein I, Ori N. CLAUSA restricts tomato leaf morphogenesis and GOBLET expression. Plant J. 2015;83:888–902. doi: 10.1111/tpj.12936. [DOI] [PubMed] [Google Scholar]
- 51.Schulte AJ, Koch K, Spaeth M, Barthlott W. Biomimetic replicas: transfer of complex architectures with different optical properties from plant surfaces onto technical materials. Acta Biomater. 2009;5:1848–54. doi: 10.1016/j.actbio.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 52.Bhushan B. Biomimetics: lessons from nature-an overview. Philos Trans R Soc A Math Phys Eng Sci. 2009;367:1445–86. doi: 10.1098/rsta.2009.0011. [DOI] [PubMed] [Google Scholar]
- 53.Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, Dandekar T, et al. Integrated systems view on Networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell. 2012;24:1793–814. doi: 10.1105/tpc.112.098335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eichmann R, Richards L, Schäfer P. Hormones as go‐betweens in plant microbiome assembly. Plant J. 2020;105:518-541. [DOI] [PMC free article] [PubMed]
- 55.Liu F, Rice JH, Lopes V, Grewal P, Lebeis SL, Hewezi T, et al. Overexpression of strigolactone-associated genes exerts fine-tuning selection on soybean rhizosphere bacterial and fungal microbiome. Phytobiomes J. 2020;4:239–51. [Google Scholar]
- 56.Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science (80-) 2015;349:860–4. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y, Bonkowski M, Shen Y, Griffiths BS, Jiang Y, Wang X, et al. Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome. 2020;8:4. doi: 10.1186/s40168-019-0775-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong C, Zhu YG, Wang JT, Singh B, Han LL, Shen JP, et al. Host selection shapes crop microbiome assembly and network complexity. N. Phytol. 2021;229:1091–104. doi: 10.1111/nph.16890. [DOI] [PubMed] [Google Scholar]
- 59.Müller DB, Vogel C, Bai Y, Vorholt JA. The plant microbiota: systems-level insights and perspectives. Annu Rev Genet. 2016;50:211–34. doi: 10.1146/annurev-genet-120215-034952. [DOI] [PubMed] [Google Scholar]
- 60.Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, et al. Microbial interkingdom interactions in roots promote arabidopsis survival. Cell. 2018;175:973–983.e14. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berg M, Koskella B. Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr Biol. 2018;28:2487–2492.e3. doi: 10.1016/j.cub.2018.05.085. [DOI] [PubMed] [Google Scholar]
- 62.Choi K, Choi J, Lee PA, Roy N, Khan R, Lee HJ, et al. Alteration of bacterial wilt resistance in tomato plant by microbiota transplant. Front Plant Sci. 2020;11:1–12. doi: 10.3389/fpls.2020.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okon Levy N, Meller Harel Y, Haile ZM, Elad Y, Rav-David E, Jurkevitch E, et al. Induced resistance to foliar diseases by soil solarization and Trichoderma harzianum. Plant Pathol. 2015;64:365–74. [Google Scholar]
- 64.Levy NO, Elad Y, Katan J, Baker SC, Faull JL. Trichoderma and soil solarization induced microbial changes on plant surfaces. Bull OILB/SROP. 2006;29:21–6. [Google Scholar]
- 65.Innerebner G, Knief C, Vorholt JA. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol. 2011;77:3202–10. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel C, Bodenhausen N, Gruissem W, Vorholt JA. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. N. Phytol. 2016;212:192–207. doi: 10.1111/nph.14036. [DOI] [PubMed] [Google Scholar]
- 67.Morella NM, Zhang X, Koskella B. Tomato seed-associated bacteria confer protection of seedlings against foliar disease caused by pseudomonas syringae. Phytobiomes J. 2019;3:177–90. [Google Scholar]
- 68.Berendsen RL, Vismans G, Yu K, Song Y, De Jonge R, Burgman WP, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12:1496–507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dudenhöffer J, Scheu S, Jousset A. Systemic enrichment of antifungal traits in the rhizosphere microbiome after pathogen attack. J Ecol. 2016;104:1566–75. [Google Scholar]
- 70.Snelders NC, Rovenich H, Petti GC, Rocafort M, van den Berg GCM, Vorholt JA, et al. Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat Plants. 2020;6:1365–74. [DOI] [PubMed]
- 71.Saleem M, Meckes N, Pervaiz ZH, Traw MB. Microbial interactions in the phyllosphere increase plant performance under herbivore biotic stress. Front Microbiol. 2017;8:41. doi: 10.3389/fmicb.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song C, Zhu F, Carrión VJ, Cordovez V. Beyond plant microbiome composition: exploiting microbial functions and plant traits via integrated approaches. Front Bioeng Biotechnol. 2020;8:896. doi: 10.3389/fbioe.2020.00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miljaković D, Marinković J, Balešević-Tubić S. The significance of bacillus spp. In disease suppression and growth promotion of field and vegetable crops. Microorganisms. 2020;8:1–19. doi: 10.3390/microorganisms8071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albayrak ÇB. Bacillus species as biocontrol agents for fungal plant pathogens. Cham: Springer; 2019. p. 239–65.
- 75.Khan MS, Gao J, Chen X, Zhang M, Yang F, Du Y, et al. The endophytic bacteria bacillus velezensis Lle-9, isolated from lilium leucanthum, harbors antifungal activity and plant growth-promoting effects. J Microbiol Biotechnol. 2020;30:668–80. doi: 10.4014/jmb.1910.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monteiro L, De Lima Ramos Mariano R, Souto-Maior AM. Antagonism of Bacillus spp. against Xanthomonas campestris pv. campestris. Braz Arch Biol Technol. 2005;48:23–9. [Google Scholar]
- 77.Radhakrishnan R, Hashem A, Abd Allah EF. Bacillus: a biological tool for crop improvement through bio-molecular changes in adverse environments. Front Physiol. 2017;8:667. doi: 10.3389/fphys.2017.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Legein M, Smets W, Vandenheuvel D, Eilers T, Muyshondt B, Prinsen E, et al. Modes of action of microbial biocontrol in the phyllosphere. Front Microbiol. 2020;11:1619.. doi: 10.3389/fmicb.2020.01619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spallek T, Gan P, Kadota Y, Shirasu K. Same tune, different song—cytokinins as virulence factors in plant–pathogen interactions? Curr Opin Plant Biol. 2018;44:82–7. doi: 10.1016/j.pbi.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 80.Choi J, Huh SU, Kojima M, Sakakibara H, Paek K-HH, Hwang I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in arabidopsis. Dev Cell. 2010;19:284–95. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 81.Cortleven A, Leuendorf JE, Frank M, Pezzetta D, Bolt S, Schmülling T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019;42:998–1018. [DOI] [PubMed]
- 82.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–44. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Secchi E, Vitale A, Miño GL, Kantsler V, Eberl L, Rusconi R, et al. The effect of flow on swimming bacteria controls the initial colonization of curved surfaces. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-16620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. Phyllosphere bacterial communities of trichome-bearing and trichomeless Arabidopsis thaliana leaves. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol. 2012;101:551–60. doi: 10.1007/s10482-011-9669-8. [DOI] [PubMed] [Google Scholar]
- 85.Kusstatscher P, Wicaksono WA, Bergna A, Cernava T, Bergau N, Tissier A, et al. Trichomes form genotype-specific microbial hotspots in the phyllosphere of tomato. Environ Microbiomes. 2020;15:1–10. doi: 10.1186/s40793-020-00364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brenner WG, Ramireddy E, Heyl A, Schmülling T. Gene regulation by cytokinin in Arabidopsis. Front Plant Sci. 2012;3:8. doi: 10.3389/fpls.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kudoyarova GR, Melentiev AI, Martynenko EV, Timergalina LN, Arkhipova TN, Shendel GV, et al. Cytokinin producing bacteria stimulate amino acid deposition by wheat roots. Plant Physiol Biochem. 2014;83:285–91. doi: 10.1016/j.plaphy.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 88.Hussain A, Hasnain S. Cytokinin production by some bacteria: its impact on cell division in cucumber cotyledons. Afr J Microbiol Res. 2009;3:704–12. [Google Scholar]
- 89.Arkhipova TN, Veselov SU, Melentiev AI, Martynenko EV, Kudoyarova GR. Ability of bacterium Bacillus subtilis to produce cytokinins and to influence the growth and endogenous hormone content of lettuce plants. Plant Soil. 2005;272:201–9. [Google Scholar]
- 90.Pallai R, Hynes RK, Verma B, Nelson LM. Phytohormone production and colonization of canola (brassica napus l.) roots by pseudomonas fluorescens 6-8 under gnotobiotic conditions. Can J Microbiol. 2012;58:170–8. doi: 10.1139/w11-120. [DOI] [PubMed] [Google Scholar]
- 91.Großkinsky DK, Tafner R, Moreno MV, Stenglein SA, De Salamone IEG, Nelson LM, et al. Cytokinin production by Pseudomonas fluorescens G20-18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci Rep. 2016;6:24. doi: 10.1038/srep23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karadeniz A, Topcuoǧlu ŞF, Inan S. Auxin, gibberellin, cytokinin and abscisic acid production in some bacteria. World J Microbiol Biotechnol. 2006;22:1061–4. [Google Scholar]
- 93.García de Salamone IE, Hynes RK, Nelson LM. Cytokinin production by plant growth promoting rhizobacteria and selected mutants. Can J Microbiol. 2001;47:404–11. doi: 10.1139/w01-029. [DOI] [PubMed] [Google Scholar]
- 94.Tahir HAS, Gu Q, Wu H, Raza W, Hanif A, Wu L, et al. Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front Microbiol. 2017;8:171. doi: 10.3389/fmicb.2017.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akhtar SS, Mekureyaw MF, Pandey C, Roitsch T. Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front Plant Sci. 2020;10:1777.. doi: 10.3389/fpls.2019.01777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Naseem M, Sarukhanyan E, Dandekar T. LONELY-GUY knocks every door: crosskingdom microbial pathogenesis. Trends Plant Sci. 2015;20:781–3. doi: 10.1016/j.tplants.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 97.Wang FF, Cheng ST, Wu Y, Ren BZ, Qian W. A bacterial receptor PcrK senses the plant hormone cytokinin to promote adaptation to oxidative stress. Cell Rep. 2017;21:2940–51. doi: 10.1016/j.celrep.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 98.Roitsch T, Ehneß R. Regulation of source/sink relations by cytokinins. Plant Growth Regul. 2000;32:359–67.
- 99.Lara MEB, Garcia MCG, Fatima T, Ehneß R, Lee TK, Proels R, et al. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell. 2004;16:1276–87. doi: 10.1105/tpc.018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tosi M, Mitter EK, Gaiero J, Dunfield K. It takes three to tango: the importance of microbes, host plant, and soil management to elucidate manipulation strategies for the plant microbiome. Can J Microbiol. 2020;66:413–33. doi: 10.1139/cjm-2020-0085. [DOI] [PubMed] [Google Scholar]
- 101.Carlton BC. Transformation mapping of the genes controlling tryptophan biosynthesis in Bacillus subtilis. J Bacteriol. 1967;94:660–5. doi: 10.1128/jb.94.3.660-665.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, et al. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–95. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Friedman J, Higgins LM, Gore J. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol. 2017;1:109. doi: 10.1038/s41559-017-0109. [DOI] [PubMed] [Google Scholar]
- 104.Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, et al. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell. 2009;21:3078–92. doi: 10.1105/tpc.109.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information files. Raw data is available through NCBI-SRA, Bioproject PRJNA729221.