Abstract
Deltaproteobacteria, now proposed to be the phyla Desulfobacterota, Myxococcota, and SAR324, are ubiquitous in marine environments and play essential roles in global carbon, sulfur, and nutrient cycling. Despite their importance, our understanding of these bacteria is biased towards cultured organisms. Here we address this gap by compiling a genomic catalog of 1 792 genomes, including 402 newly reconstructed and characterized metagenome-assembled genomes (MAGs) from coastal and deep-sea sediments. Phylogenomic analyses reveal that many of these novel MAGs are uncultured representatives of Myxococcota and Desulfobacterota that are understudied. To better characterize Deltaproteobacteria diversity, metabolism, and ecology, we clustered ~1 500 genomes based on the presence/absence patterns of their protein families. Protein content analysis coupled with large-scale metabolic reconstructions separates eight genomic clusters of Deltaproteobacteria with unique metabolic profiles. While these eight clusters largely correspond to phylogeny, there are exceptions where more distantly related organisms appear to have similar ecological roles and closely related organisms have distinct protein content. Our analyses have identified previously unrecognized roles in the cycling of methylamines and denitrification among uncultured Deltaproteobacteria. This new view of Deltaproteobacteria diversity expands our understanding of these dominant bacteria and highlights metabolic abilities across diverse taxa.
Subject terms: Microbial ecology, Environmental microbiology, Metagenomics
Introduction
Deltaproteobacteria are globally distributed, metabolically and phylogenetically diverse bacteria with numerous cultured representatives. These bacteria have historically been a class within the Proteobacteria phylum. Recently, it was proposed that Deltaproteobacteria be reclassified into three phyla, the Desulfobacterota, Myxococcota, and SAR324 [1]. Desulfobacterota are known for their ability to respire sulfate utilizing protein complexes sulfate adenylyltransferase (Sat), adenylyl sulfate reductase (Apr), and dissimilatory sulfite reductase (Dsr), and have been identified from the environment based on the presence of the dsrA gene [2, 3]. However, these organisms have a variety of other metabolic abilities, including sulfite and thiosulfate disproportionation [4], mercury methylation [5, 6], aliphatic and aromatic hydrocarbon degradation [7], nitrogen fixation [8, 9], organohalide respiration [10], and dissimilatory iron reduction (mainly within Desulfuromonadia) [11, 12]. The Myxococcota exhibit complex social behavior, gliding motility, and sporulation, and are known for their predominantly aerobic, predatory lifestyle with the ability to produce a variety of secondary metabolites [13]. Finally, SAR324 lacks cultured representatives and is predicted to be metabolically flexible, encoding genes for short-chain alkane and sulfur oxidation, carbon fixation, and the utilization of oxygen and nitrite or nitrate as electron acceptors [14, 15]. Desulfobacterota and Myxococcota account for a large proportion of microbial communities in soils [16], subterranean environments [17], wetlands [18], and marine sediments [19], while SAR324 is ubiquitous in freshwater and marine water columns [14, 15].
16S rRNA gene analyses have revealed numerous populations of Desulfobacterota and Myxococcota that are phylogenetically distinct from characterized cultures [20]. Our current understanding about the ecophysiology of these uncultured organisms has been advanced through the use of omics approaches such as single-cell genomics, de novo metagenomic assembly, and metatranscriptomics [15, 21]. These methods have been especially crucial for examining these bacteria in extreme environments, such as hydrothermal vents, where conditions are difficult to recreate in a laboratory setting. In recent years, omics methods have resulted in a rapid expansion of new genomes reconstructed from diverse environments [22, 23]. However, the metabolic potential of Desulfobacterota, Myxococcota, and SAR324 has not yet been systematically described in the context of this new diversity.
To address this research need, we compiled a comprehensive collection of 1 559 Desulfobacterota, Myxococcota, and SAR324 genomes, including well-studied cultured organisms as well as genomes reconstructed from the environment. Of these, 346 MAGs are newly reconstructed metagenome-assembled genomes (MAGs) from estuary and deep-sea hydrothermal sediments obtained in this study, and 56 were obtained in a previously published study from the same deep-sea site (Dombrowski et al., 2018). Using this expanded genomic database, we define eight ecologically distinct metabolic groups based on protein content. Each of these protein-level groupings reveals distinct potential ecological roles across these taxa and provides initial insights into the lifestyle of a large number of uncultured bacteria.
Materials and methods
A total of 346 Desulfobacterota and Myxococcota MAGs were reconstructed in this study from marine sediments in hydrothermal vents of Guaymas Basin (GB), Gulf of California (314/346 MAGs) and a coastal site in Mesquite Bay (MB), Texas (32/346 MAGs). 56 GB MAGs have been published previously [24] as part of a larger analysis. The methods used to produce the 346 MAGs reconstructed in this study and analyze the total set of 1 390 reference genomes are described below.
Sampling of marine sediments
GB samples were collected from sediments in the Gulf of California, Mexico (27°N 0.388, 111°W 24.560). Samples were collected during two Alvin dives in 2008 and 2009 (dives 4 486 and 4 573) from a depth of approximately 2000 m using polycarbonate cores (45–60 cm in length, 6.25 cm interior diameter). These were subsampled into cm layers under N2 gas in the ship’s laboratory and immediately frozen at −80 °C. Twenty-seven subsamples from different depths yielded sufficient DNA for metagenomic sequencing. Details on geochemical characteristics are provided in Supplementary Table 1. MB samples were collected in July 2016, in Mesquite Bay, Mission-Aransas National Estuarine Research Reserve, Texas, (28°N 0.147, −96°W 0.8455) using a PVC sediment core. The sediment core was stored on ice and then immediately subsampled into four (1D–4D) 3 cm sections spanning 3–15 cm (3–6 cm, 6–9 cm, 9–12 cm, and 12–15 cm) and stored at −80 °C until processing. Oxygen profiles were taken at the site (Supplementary Table 1).
Metagenomic sequencing and assembly
Whole community DNA from ≥10 g of sediment was extracted from GB samples using the DNeasy PowerSoil kit (Qiagen, Germantown, Maryland, USA) following the manufacturer’s instructions for each of sixteen samples. DNA concentrations were quantified using a QUBIT 2.0 fluorometer (Thermo-Fisher, Singapore) and metagenomic sequencing was performed at the Michigan State University RTSF Genomics Core. Libraries were prepared using the Illumina TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, California, USA) on a Perkin Elmer Sciclone G3 robot (PerkinElmer, Waltham, Massachusetts, USA) following the manufacturer’s recommendations. Completed libraries were quality controlled and quantified using a combination of Qubit dsDNA HS and Advanced Analytical Fragment Analyzer High Sensitivity DNA assays (Advanced Analytical Technologies, Ankeny, Iowa, USA) The libraries were divided into 4 pools, each with 4 libraries combined in equimolar amounts. Pools were quantified using the Kapa Biosystems Illumina Library Quantification qPCR kit (Kapa Biosystems, Wilmington, Massachusetts, USA). Each pool was loaded onto 2 lanes of an Illumina HiSeq 4 000 flow cell (8 lanes total) and sequencing was performed in a 2 × 150 bp paired end format using HiSeq 4 000 SBS reagents. Base calling was completed by Illumina Real Time Analysis v2.7.7 and the output was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.19.1. An additional round of sequencing was completed on these library pools for improved resolution during genomic reconstruction. Sequences were trimmed and quality controlled using Sickle v1.33 [25] and assembly was performed using IDBA-UD v1.0.9 [26].
Whole community DNA from ≥10 g of sediment was extracted from MB samples using a DNeasy PowerSoil kit (Qiagen) following the manufacturer’s instructions. DNA concentrations were quantified using a QUBIT 2.0 fluorometer (Thermo-Fisher) and metagenomic sequencing was performed at the Michigan State University RTSF Genomics Core. Libraries were prepared using the Illumina TruSeq Nano DNA Library Preparation kit on a PerkinElmer Sciclone NGS robot following the manufacturer’s protocols. Completed libraries were quality controlled and quantified using a combination of Qubit dsDNA HS and Caliper LabChipGX HS (Caliper Life Sciences, Hopkinton, Massachusetts, USA) DNA assays. All libraries were pooled in equimolar quantities and the final pool was quantified using the Kapa Biosystems Illumina Library Quantification qPCR Assay kit. The pool was loaded onto 2 lanes of an Illumina HiSeq 4 000 flow cell and sequencing was performed in a 2 × 150 bp paired end format using HiSeq 4 000 SBS reagents. Base calling was completed by Illumina Real Time Analysis v2.7.6 and output demultiplexed and converted to FASTQ format with Illumina Bcl2fastq v2.18.0. Adapters were trimmed from FASTQ reads with CutAdapt v1.13 [27] for TruSeq adapters and quality controlled using Sickle v1.33 [25]. Assembly was performed with MegaHit v1.1.1 [28] (–12 –k-list 21,33,55,77,99,121 –min-count 2 –verbose -t 4 –memory 500000000000). Read mapping was performed with the BWA aligner v.0.7.12 BWA-MEM algorithm and Samtools v.0.1.19 [29, 30].
Genome binning
Binning of individual GB assemblies (scaffolds >2 000 bp) was performed from dives 4 486 and 4 573 using Concoct v0.4.0 [31] and Metabat v2.12.1 [32]. Concoct was used with default settings and Metabat was run with the following parameters: –minCVSum 0 –saveCls -d -v –minCV 0.1 -m 2000. Consensus MAGs produced from these two binning tools were determined using DAS Tool v1.0 [33] with default settings. CheckM v1.0.11 [34] was used to determine MAG completeness and contamination (Supplementary Table 2). MAGs were only analyzed further if they were more than 50% complete and had less than 10% contamination. There were 314 MAGs identified as Deltaproteobacteria from these GB samples and included in this analysis (Supplementary Table 3).
Binning of MB assembled fragments was performed with MetaBat v2.12.1 [30], Maxbin v2.2.4 [35], and Anvi’o v3 [36] using contigs ≥2 500 bp. Consensus MAGs were determined with DASTool [33] v1.0 (–search_engine diamond) and MAG completeness and contamination were determined with CheckM v1.0.11 [34]. For subsequent analyses, only MAGs with completeness greater than 50% and contamination less than 10% were used. There were 32 MAGs identified as Deltaproteobacteria and included in this analysis. All 346 MAGs are described in Supplementary Table 2.
Phylogenetic analyses
Phylosift v1.0.1 [37] was used to extract 37 single-copy, protein-coding marker genes for the phylogenetic placement of the assembled metagenomic bins. A set of 1 391 reference genomes were downloaded from NCBI in February of 2019 for comparison with GB and MB genomes (Supplementary Table 2) [38, 39]. 402 reconstructed genomic bins were used as input for Phylosift v1.0.1 [37] using the ‘phylosift search’ followed by the ‘phylosift align’ mode. One MAG (MB3D Bin 30) did not contain most of these markers and thus was not included in the phylogeny. The concatenated protein alignments of 37 universal marker genes were combined for all genomes of interest and trimmed using TrimAl v3 [40] using the automated1 setting. A phylogenetic tree was generated using a maximum likelihood-based approach using RAxML v8.2.10 [41], called as raxmlHPC-PTHREADS-AVX -f a -m PROTGAMMAAUTO -N autoMRE. This run converged on the LG substitution matrix and the GAMMA model of rate heterogeneity.
All 402 MAGs were assessed for average amino acid identity (AAI) using CompareM (v0.0.23, function aai_wf; https://github.com/dparks1134/CompareM). Publicly available genomes branching closely to MAGs on the 37 marker gene tree were included in the AAI analysis to create an AAI matrix (Supplementary Table 4).
16S rRNA gene sequences were extracted from reconstructed genomes using barrnap v.3 (https://github.com/tseemann/barrnap) using the following parameters: –lencutoff 0.2 –reject 0.3 –evalue 1e-05. Sequences were uploaded to ARB v2.5b [42] and aligned with the reference arb database SILVA_132_SSURef_NR99_13_12_17_opt.arb.gz. The alignment was checked and manually refined, exported from ARB v2.5b, and trimmed using BMGE v.1.12 [43]. This analysis produced an alignment of 149 sequences: 55 16S rRNA gene sequences from reconstructed genomes and 94 from the reference database. Using this alignment, a maximum likelihood tree was created with RaxML v.8.2.4 [41] as: raxmlHPC-PTHREADS-AVX -f a -m GTRGAMMA -N autoMRE. Trees were uploaded to iTOL [44] for visual annotation of groups such as color coding and heatmaps. Further annotations were completed in Inkscape (https://inkscape.org/) and the annotated 16S rRNA gene phylogeny is shown in Supplementary Fig. 1. MAGs were also classified using the Genome Taxonomy Database (GTDB-Tk v0.3.3) using database version r95 (Supplementary Table 2) [45].
Hierarchical clustering of genomes based on protein content
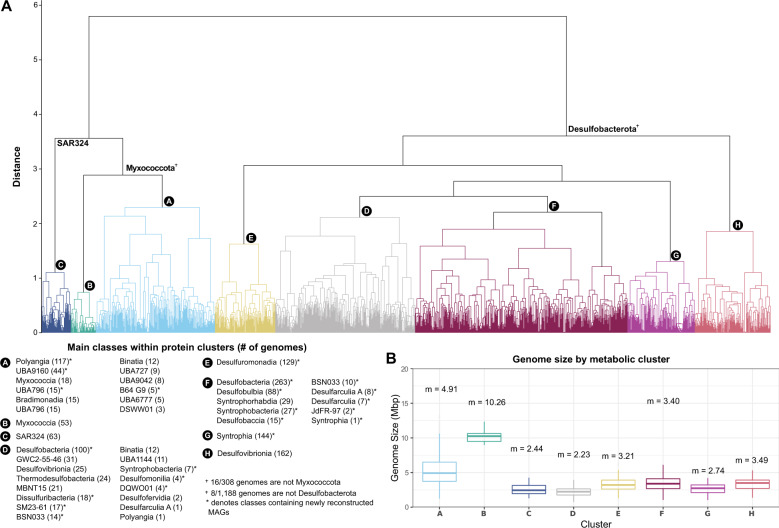
We conducted an unsupervised clustering of high-quality (>90% complete, <5% contamination, 598 genomes) and medium-quality (>50% complete, <10% contamination, 961 genomes) genomes [46]. This includes 402 newly reconstructed MAGs plus 1 157 publicly available reference genomes. These genomes (1 559 total) were scanned against the Pfam v3.0 database to obtain a protein presence/absence profile with MEBS (mebs.pl using the -comp option). The mapping file of all Pfams searched is shown in Supplementary Table 5. We hierarchically clustered the 1 559 genomes with MEBS (mebs_clust.py) using Jaccard distance, Ward variance minimization, and a maximum distance threshold of 0.4 (options –distance –method and –cutoff respectively). A threshold of 0.4 was chosen using the following methods; first, clustering results were examined at various maximum distance thresholds. At a maximum distance threshold of 0.5 or 0.3, five and 16 clusters are produced, respectively. These clustering results are consistent with taxonomy (shown in Supplementary Table 2). At a threshold of 0.5, the clusters are broad with more organisms encompassed in one cluster, and thus some metabolic distinctions are missed. At a threshold of 0.3, the number of clusters is too numerous to resolve broad metabolic distinctions. For these reasons, a maximum distance threshold of 0.4 was chosen and this produced eight genomic clusters, A-H (hereafter genomic clusters throughout the manuscript, Figs. 1–3). To further characterize the protein composition of each cluster, the conserved protein domains (present in at least 95% of genomes within a cluster, Supplementary Table 6) were identified using the script parse_pangenome_matrix.pl within the GET_HOMOLOGUES package [47].
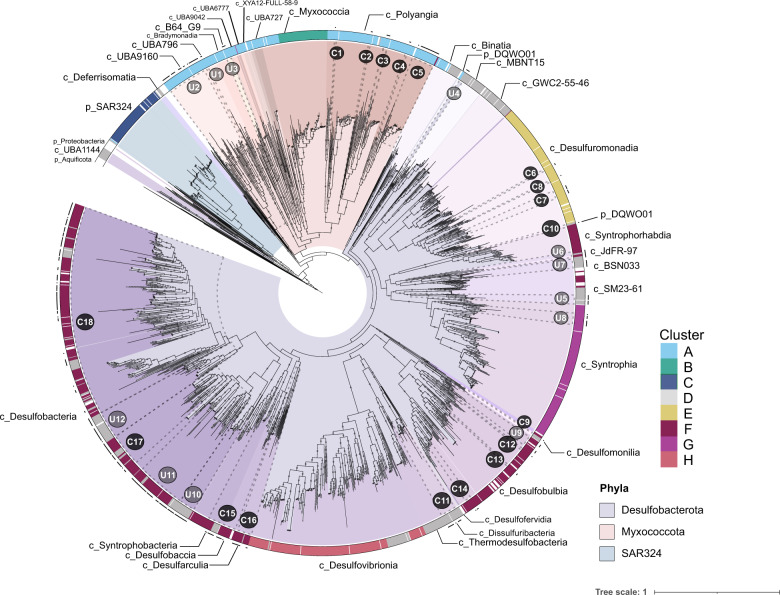
Fig. 1. Phylogeny and proposed metabolic groups of Deltaproteobacteria (Desulfobacterota, Myxococcota, and SAR324).
This phylogeny was generated using 1 793 genomes including 401 MAGs described in this study (black dots in the outer ring), 1 283 publicly available Deltaproteobacteria genomes (Supplementary Table 2), and 109 non-Deltaproteobacteria (mainly Proteobacteria and Aquificota) genomes in order to root the tree. MAGs reconstructed in this study are labeled as C (C1–C18) if they are associated with any known cultured representative and U (U1–U12) if they are related to only uncultured representatives. Genomic clusters based on protein content (cluster A–H) are shown in the outer ring of the phylogeny and their color codes can be cross-referenced with Figs. 2–4. Labels that begin with “p_” are phyla, and labels that are labeled with “c_” are classes, according to GTDB-Tk v1.5.0. The phylogeny was generated using 37 conserved marker proteins (mostly ribosomal proteins extracted using PhyloSift; see methods) using RAxML (v8.2.12) under the GAMMA model of rate heterogeneity, LG likelihood, and a maximum of 1 000 bootstrap replicates (option -N autoMRE). Bootstraps are shown in gray circles and are based on 1 000 replicated trees.
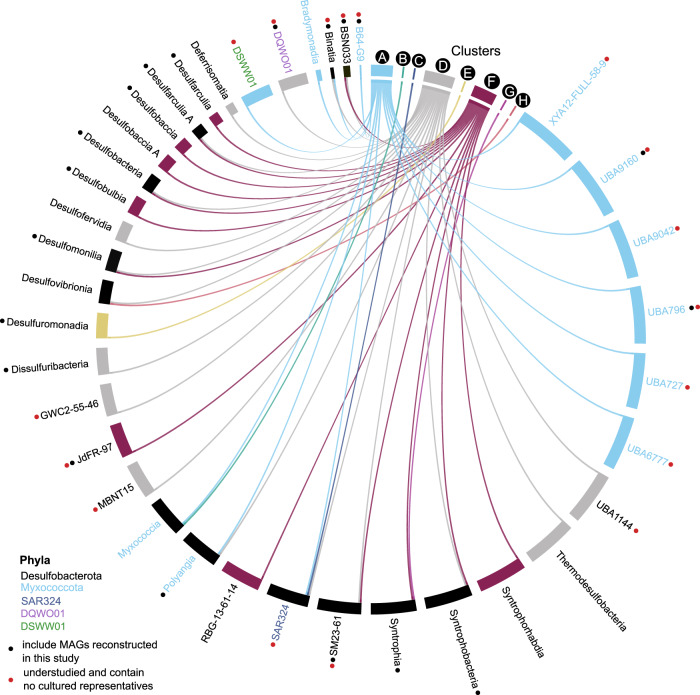
Fig. 3. Distribution of taxonomic lineages within eight genomic clusters.
Circos visualization map showing the relationship between metabolically related groups (genomic clusters based on their protein content similarity: A–H) and their corresponding taxonomy. The connections are drawn from clusters A-H to classes of Desulfobacterota, Myxococcota, or SAR324. Those with red dots are groups that contain MAGs reconstructed in this study and have no cultivated members (See U groups in Fig. 1). Black dots signify classes with newly reconstructed MAGs from cultured groups. Classes are colored according to their corresponding phylum.
To visualize the genomic clusters and to cross-reference taxonomy with metabolically coherent groups, a Circos diagram was generated online (http://mkweb.bcgsc.ca/tableviewer/, Fig. 3). The input file was created by mapping classes of Desulfobacterota, Myxococcota, and SAR324 to the cluster(s) in which they occur, resulting in a presence/absence table of classes in each cluster. Options “row with col size” and “col with row size” were selected and ribbon caps, tick labels/marks, and contribution tracks were turned off. The Circos diagram was downloaded and edited in Inkscape (https://inkscape.org/) to modify class labels and colors.
Characterization of dissimilatory sulfite reductases
DsrA and DsrB proteins were identified in the 402 MAGs reconstructed in this study (from GB and MB) using KAAS (KEGG Automatic Annotation Server) [48] and HMMER 3.1b2 [49]. A reference database of DsrAB sequences was manually curated using the following methods; first, 1 092 reference DsrA sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov/) using the query: (dsrA) AND “d-proteobacteria”[porgn:__txid28221] NOT “PRJNA362212” on October 10th, 2019. This was repeated for DsrB for a total of 1 307 DsrB reference sequences. Archaea DsrA sequences were obtained from NCBI using the query: “dsrA AND pyrobaculum” and then sorted by RefSeq only results. The query “dsrA AND vulcanisaeta” was also used, and these searches were repeated for DsrB. Finally, homologous sequences of DSRs were extracted from the 402 MAGs and used as a query against the non-redundant NCBI database using blastp with default settings [50]. The resulting DSRs with >90% sequence identity were included in the final database containing 1 505 DsrA and 1 695 DsrB sequences. DsrA and DsrB sequences were aligned separately using MAFFT v7.271 (default parameters) [51]. Alignments were masked in Geneious 8.1.9 (https://www.geneious.com) (at least 50% gaps) and manually refined. Trees were constructed using IQ-TREE v1.3.11.1 [52] with the ultrafast bootstrapping option -bb 1 000 and model LG + R7 (Fig. 4, Supplementary Fig. 2).
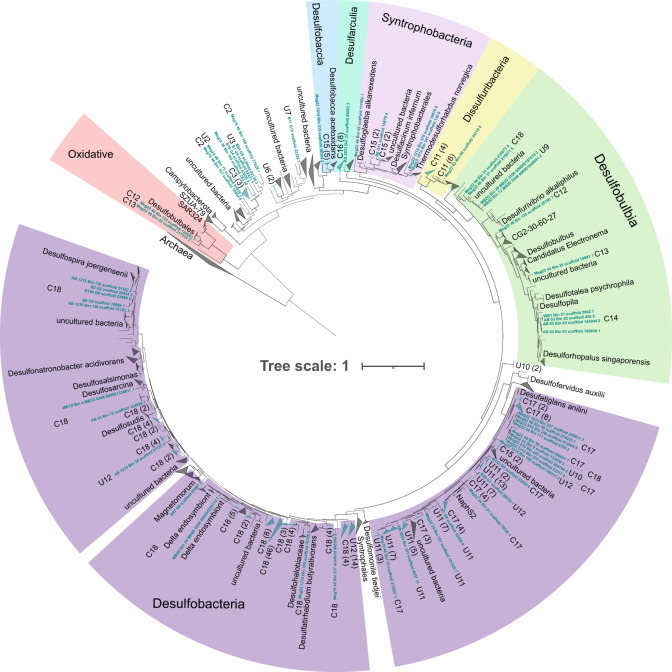
Fig. 4. Dissimilatory sulfite reductase (DsrA) phylogenetic tree showing the diversity of MAGs involved in dissimilatory sulfur cycling using the dsr system.
The tree includes 226 DsrA proteins identified in the 402 MAGs obtained in this study. These sequences are highlighted in blue text and labeled according to the 30 groups described in Fig. 1 (U and C for uncultured and cultured, respectively). The tree also contains 337 reference DsrA sequences from a manually curated database that includes representatives from public databases (see methods). The phylogeny was inferred using IQ-TREE (v.1.6.11, model LG + R7) with the ultrafast bootstrapping option -bb 1000. Numbers shown in parentheses are the number of protein sequences in a collapsed group belonging to the 402 MAGs.
Characterization of hydrogenases
Hydrogenases were identified in the 402 MAGs by using a set of 3 261 hydrogenase reference sequences [53], consisting of NiFe-, FeFe-, and Fe-hydrogenase catalytic subunits [54]. Hydrogenases were identified using DIAMOND v0.9.24.125 [55] against the reference hydrogenase database and then filtered to ensure an alignment length cutoff >40 amino acid residues and a sequence identity >50%. Putative hydrogenase sequences identified in these searches were then uploaded to the HydDB webserver [54] to identify and remove non-hydrogenases. This resulted in a total of 297 hydrogenases identified in the 402 MAGs that were concatenated with the reference set of hydrogenases (Supplementary Table 7). Finally, 4 005 hydrogenase sequences were aligned with MAFFT v7.271 [51] using default parameters and the –anysymbol option to allow for “U” as selenocysteine in reference sequences. The alignment was trimmed using TrimAl v1.4.22 [40] with the -automated1 option and manually refined in Geneious 8.1.9 (https://www.geneious.com). The final alignment was used to construct a phylogenetic tree using IQ-TREE v1.3.11.1 [52] with the ultrafast bootstrapping option -bb 1 000 and model LG + R10 (Supplementary Figs. 3 and 4).
Functional characterization of genomes
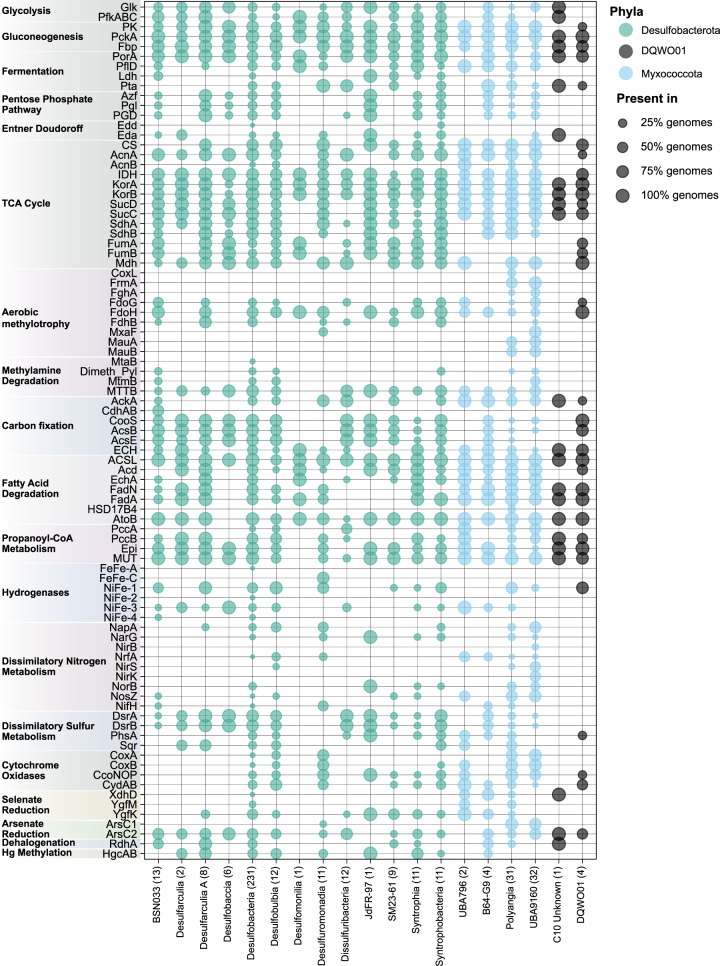
Gene prediction for the 402 MAGs was performed using Prodigal v2.6.2 (default settings) [56]. Predicted genes of MAGs were further characterized using KAAS [48] and dbCAN v8 [57]. For KAAS, protein sequences of each individual genome were uploaded to the KAAS webserver using the ‘Complete or Draft Genome’ setting (parameters: GHOSTX, custom genome dataset, BBH assignment method). MAGs and reference genomes were also annotated using custom databases searched using DIAMOND v2.0.4.142 (Supplementary Table 8) [55] and hmmsearch (Supplementary Table 9) [49]. DIAMOND searches were conducted using a custom database of genes related to central metabolic processes with manually curated cut-offs [58]. MAGs and reference genomes were annotated using the KEGG-based annotation program METABOLIC v1.3 (Supplementary Table 10) [59]. A subset of verified marker genes present in the 402 MAGs was compiled into Supplementary Table 11 using KAAS, DIAMOND, HMMER, and METABOLIC output. Genes encoding carbohydrate-degrading enzymes described in the Carbohydrate-Active enZYmes (CAZYmes) database were identified in MAGs with version 2 of the dbCAN meta server (http://bcb.unl.edu/dbCAN2/blast.php) retaining only those hits that were detected in at least two databases using the HMMER, DIAMOND, and Hotpep tools [60]. Protein localization of CAZYmes and peptidases were predicted with Psort v3.0 using the command-line options (-n -terse) (Supplementary Table 12) [61]. Anaerobic hydrocarbon degradation genes were identified in MAGs using a DIAMOND database (makedb and blastp options) of reference sequences from the anaerobic hydrocarbon degradation database AnHyDeg [62] and confirmed through phylogenetic analysis (Supplementary Table 13 and Supplementary Fig. 5). Reductive dehalogenases were identified in the 402 MAGs using METABOLIC v1.3 and confirmed through phylogenetic analysis (Supplementary Table 14 and Supplementary Fig. 6). Mercury methylation genes (hgcAB) were identified in the 402 MAGs using publicly available hmm profiles of hgcA and hgcB [63] and the search-custom-markers function in metabolisHMM [64]. Sulfide:quinone oxidoreductases were identified in the 402 MAGs using DIAMOND blastp (Supplementary Table 15), and hits were confirmed with phylogeny (Supplementary Fig. 7). Finally, MAGs and reference genomes were analyzed using the built-in function of MEBS, which evaluates the likelihood of a given genome to perform metabolic reactions involved in major cycles (S, N, Fe, and O) based on informative entropy scores [65]. We also utilized the custom function of MEBS to search MAGs and publicly available genomes for the presence of pfam domains involved in methylamine degradation (see Supplementary Table 11). Because trimethylamine methyltransferase activity is dependent on pyrrolysine [66], pyrrolysine residues were identified in MAGs and reference genomes using a DIAMOND blastp search (v2.0.4.142 default settings, one maximum target sequence per query) [55] of 24 reference pyrrolysine biosynthesis genes (pylB) obtained from UniProt (Supplementary Table 16). The distribution of 88 biogeochemically important genes identified using these methods is shown in Fig. 5 and metabolic features within each genomic cluster (A–H) is shown in Fig. 6.
Fig. 5. Distribution of biogeochemically important genes among 402 newly reconstructed MAGs described in this study.
Marker genes are shown on the y-axis and taxonomic classes (according to GTDB-tk taxonomy) are shown on the x-axis. Numbers in parentheses show the number of genomes in each group. General categories of metabolism are shown on the left. For more detailed information about these pathways and a full description of the marker genes present in the 402 MAGs see Supplementary Table 11.
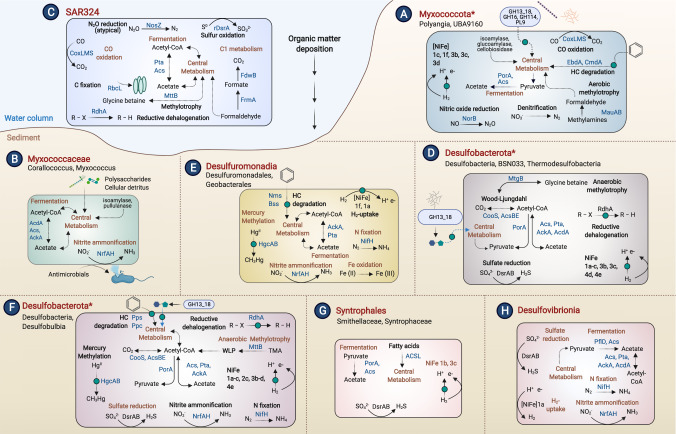
Fig. 6. Metabolic features of eight genomic clusters based on the protein content of 1 559 Desulfobacterota, Myxococcota, and SAR324 genomes.
For those clusters that contain MAGs reconstructed in this study (A, D, E, F, G), the metabolic features of the novel MAGs are highlighted. Predominant taxa for each cluster are shown next to each cluster letter label. Common metabolic pathways (present in >50% of genomes in a cluster) are shown in dark orange font inside the cells. Pathways labeled in black indicate the presence in <50% of genomes. Asterisks in cluster names (A, D, F) signify the presence of some genomes in that cluster that are not part of the listed taxa name (shown in Supplementary Table 2). Curved lines indicate transformations that are carried out either in the periplasm or are membrane-bound and straight arrows are cytoplasmic reactions. Proteins searched and identified in the 402 MAGs only are indicated with green circles. Dashed lines signify metabolic pathways with several steps that are only shown as one transformation. Central metabolism includes glycolysis, gluconeogenesis, and the TCA cycle at some degree of completion among genomes (see Supplementary Table 8). Outer blue circles are extracellular CAZYmes. This figure was created using BioRender.com.
Results
Phylogeny highlights understudied and novel organisms
To understand the taxonomic relationships of Desulfobacterota, Myxococcota, and SAR324 (formerly Deltaproteobacteria), we constructed a phylogenetic tree using 37 single-copy marker genes (primarily ribosomal proteins) for 401 newly reconstructed MAGs and 1 391 publicly available reference genomes (1 792 genomes total, Fig. 1, Supplementary Table 2) [37]. One MAG (MB3D Bin 30) was not included in the 37 protein phylogeny due to a low number of markers in this genome. All MAGs and reference genomes in the 37 marker phylogeny were annotated using GTDB-tk v1.5.0. We also manually examined branches from the resulting tree to identify the placement of the 402 MAGs reconstructed in this study and their relationship to cultured and uncultured organisms. We designated 18 groups containing MAGs related to cultured representatives (C1-C18, 229 MAGs) and 12 groups of MAGs without closely related, cultured representatives (U1-U12, 173 MAGs). These groups are labeled in Fig. 1 and GTDB-tk taxonomic classifications are provided in Supplementary Table 2.
Newly reconstructed MAGs belong to Myxococcota and Desulfobacterota and fall within the understudied classes of these phyla. For example, we reconstructed 22 MAGs belonging to the Desulfobacterota class BSN033, which is composed of uncultured lineages from an aquifer system in Rifle, Colorado [22] and a large-scale genome reconstruction effort from public data [23]. Within the Myxococcota, we reconstructed 34 novel MAGs in the classes UBA9160 and UBA796 (UBA for Uncultivated Bacteria and Archaea), lineages also known from the large-scale genome reconstruction effort [23]. These classes are all uncultured and only recently discovered, and thus they lack defined biogeochemical or ecological roles. Though not represented in the MAGs, the publicly available genomes also span other uncultivated lineages of the phyla Desulfobacterota, Myxococcota, and SAR324. For example, GWC2-55-46, MBNT15, and Binatia are all uncultured Desulfobacterota organisms reconstructed from a variety of environments including an aquifer system [22], grassland soil [67], and permafrost [68]. Myxococcota has several other UBA-named classes (Fig. 1) lacking cultured representatives [1]. Many of the novel uncultured branches within Desulfobacterota and Myxococcota are derived from large sequencing projects where reconstructed genomes have not been characterized in detail.
Although recent phylogenetic changes to Deltaproteobacteria largely match our phylogeny, there are some inconsistencies. For example, GTDB-tk classifies U4 and C10 MAGs as a novel phylum, DQWO01. In our phylogeny, U4 branches between the Desulfobacterota classes Binatia and MBNT15, while C10 branches close to a Desulfobacterota genome in an unknown class, between Desulfuromonadia and Syntrorhabdia. Furthermore, these genomes branch distantly from each other. In addition, previously described Desulfobacterota classes UBA1144 (Candidatus Dadabacteria) [17] and Deferrisomatia [69] branch outside of the Desulfobacterota in our phylogeny.
Protein clustering of Deltaproteobacteria
To understand protein content relatedness of Deltaproteobacteria (Desulfobacterota, Myxococcota, and SAR324), we hierarchically clustered proteins coded by 1 559 high- and medium-quality genomes based on the presence/absence of 17 935 protein family domains (see methods). This large-scale metabolic analysis grouped genomes with similar protein content into eight clusters (hereafter referred to as genomic clusters A-H throughout the manuscript). The distribution of these genomic clusters is largely consistent with the phylogeny (Fig. 1). To understand how these groups of metabolically related genomes are unique, we examined their metabolic pathways by comparing their predicted proteins to a variety of functional databases (see methods). From this, we inferred their metabolic and ecological capabilities (Fig. 6) and examined the predicted proteins that are conserved in the eight clusters (A-H).
Cluster A Myxococcota have overlooked metabolic roles
Nearly all Myxococcota in this analysis (255 genomes, U1, U2, and C1-C5 MAGs) are within genomic cluster A (Fig. 2, light blue). This cluster spans the cultivated Myxococcota classes Polyangia, Myxococcia, and Bradymonadia, several newly established UBA classes reconstructed from the environment [23], and 21 non-Myxococcota genomes. Cultured organisms in cluster A Myxococcota (e.g., Sorangium cellulosum and Anaeromyxobacter dehalogenans) have complex social behavior, are often predatory, adjust gene expression patterns under varying environmental conditions [70], and have versatile metabolisms, including dehalorespiration, denitrification, and facultative anaerobic respiration [71]. This versatility is broadly reflected in cluster A genomes in their relatively large genome sizes (average genome size of 4.91 Mb, Fig. 2). Many cluster A Myxococcota contain genes for acetate fermentation (pyruvate ferredoxin-oxidoreductase alpha subunit porA, acetate kinase ack, acetyl-CoA synthetase acs, and phosphate acetyltransferase pta) [72], the degradation of complex organic compounds (cbhA [73], ramA [74], and xynB [75]), and a variety of terminal oxidases (coxAB [76], ccoNOP [77], and cydAB [78], Supplementary Table 10). 95% of cluster A genomes contain cytochrome c554 (PF13435), which can act as an electron transfer protein from hydroxylamine oxidoreductase (HAO) and can act as a nitric oxide reductase (Supplementary Table 6) [79]. In addition, organisms primarily within the classes Myxococcia, Polyangia, UBA6777, UBA9042, UBA9160, and cluster A Binatia (in the Desulfobacterota phylum), encode genes for nitric oxide and nitrous oxide reduction (norB and nosZ), nitrogen fixation (nifH), and nitrate and nitrite reduction (napA, narG and nrfA) (Supplementary Tables 8 and 10) [80].
Fig. 2. Overview of eight genomic clusters identified through protein clustering.
(A) Dendrogram of genomic groups (labeled A–H) based on their protein content similarity (determined using Jaccard distance and Ward variance minimization, see methods). Genomic groups include 1559 Desulfobacterota, Myxococcota, and SAR324 genomes that were clustered based on their protein family content using a set of 17 395 pfam domains (see methods). General taxonomic representatives are provided in the dendrogram and more detailed information about the classes present within clusters is provided in the lower-left box labeled “Classes within protein clusters”. Numbers in parentheses in the lower-left box show the number of genomes within that class. Classes that contain MAGs reconstructed in this study are shown with an asterisk. (B) Box plot distribution of genome size in Mbp of all genomes included in the eight metabolic clusters shown in panel (A). Each box plot shows the mean genome size of that cluster. The upper boundary of a box plot represents the 75% quantile, and the lower boundary is the 25% quantile. The middle line in each box plot represents the median. Upper whiskers are the largest observation less than or equal to the upper hinge +1.5 * the interquartile range (IQR), while lower whiskers are the smallest observation greater than or equal to the lower hinge −1.5 * IQR.
Newly reconstructed cluster A MAGs from U2 (UBA9160, 32 MAGs/44 genomes) and C5 (Polyangiales, genus SG8-38, 22 MAGs/24 genomes) appear unique in their shared abilities in denitrification, aromatic hydrocarbon degradation, aerobic methylotrophy, and hydrogenotrophic respiration (Fig. 5). Four U2 MAGs encode all genes for complete denitrification from nitrate to N2 (narGH, napAB, nirK, nirS, norBC, and nosZ), and several C5 and U2 MAGs encode genes for nitric oxide and nitrous oxide reduction (nitric oxide reductase norBC, nitrous oxide reductase nosZ, Supplementary Table 10). U2 MAGs putatively encode both subunits of the anaerobic hydrocarbon degradation genes ethylbenzene dehydrogenase (ebdAB) and cymene dehydrogenase (cmdAB, Supplementary Fig. 5), and these genes are known to occur in denitrifiers [81, 82]. U2 and C5 genomes also uniquely encode methylamine dehydrogenase (mauAB) for the oxidation of methylamine to formaldehyde, which was not identified in any other MAGs reconstructed in this study and was found in only 10 uncultured representatives from cluster A. Furthermore, these organisms both encode group 1f NiFe-hydrogenases (Supplementary Table 7 and Supplementary Fig. 3), which is thought to support aerobic hydrogenotrophic respiration [54]. Interestingly, C5 Polyangiales also appear to have the capacity to use carbon monoxide (CO) as an electron donor for aerobic respiration (6 C5 MAGs encode coxL). This trait is relatively rare in the phyla studied here and is largely limited to SAR324. Finally, we identified genes for organohalide respiration (rdh) in 8 of 69 Myxococcota MAGs (Supplementary Fig. 6), and some Myxococcota encode 2-haloacid dehalogenase (Supplementary Table 10) [83]. The predicted ecological role of Cluster A Myxococcota as a nitrogen cycling, heterotrophic group is depicted in Fig. 6.
Cluster D and F Desulfobacterota differ in their genome sizes and sulfate reduction capacity
Most Desulfobacterota diversity is encompassed within-cluster D (185 reference genomes, 114 MAGs) and cluster F (247 reference genomes, 198 MAGs), and these organisms are closely phylogenetically related (Fig. 1). For example, representatives from the same class, such as Desulfobacteria, Syntrophobacteria, and BSN033 (see Supplementary Table 2), are present in both genomic clusters. Despite the taxonomic similarities between cluster D and F, several factors appear to contribute to their separation; first, genomes within-cluster D have smaller median genome sizes (2.2 Mbp) compared to genomes in cluster F (median size of 3.4 Mbp, Fig. 2, panel B). We also cannot discount the possibility that differences in genome quality could contribute to the split of genomes between these clusters (Supplementary Fig. 8). Cluster D organisms have a lower median genome completeness compared to cluster F (80% in cluster D and 88% in cluster F) and fewer high-quality genomes (83 genomes >90% completeness and <10% contamination in cluster D, 170 genomes >90% completeness and <10% contamination in cluster F).
However, differences in genome size also appear to reflect different metabolic capabilities. For example, cluster F has the second-highest median sulfur cycling MEBS score compared to all other clusters (Supplementary Fig. 9), and 95% of cluster F genomes contain a heterodisulfide reductase subunit (PF02662, Supplementary Table 6), a protein domain that is conserved among sulfur cycling microorganisms [65]. In contrast, cluster D has the third-highest sulfur cycling MEBS score and does not contain this conserved domain in 95% of genomes. When examining the completeness of sulfur cycling pathways, cluster D is predicted to have fewer genes for sulfur cycling (aprAB, dsrABC, qmoABC, Supplementary Table 17). For example, 42.5% of cluster D organisms are predicted to have dsrAB genes for sulfite reduction, compared to 93.2% of cluster F. Desulfobacterota classes in cluster D that generally lack these genes include UBA1144, Binatia, BSN033, Desulfomonilia_A, Defferisomatia, GWC2-55-46, and MBNT15. Many of the organisms that lack genes for sulfate reduction in cluster D also lack the carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS) complex for acetate oxidation and carbon fixation through the Wood-Ljungdahl pathway (WLP, Supplementary Tables 10 and 18). In contrast, this protein complex is predicted to be widespread in cluster F organisms. Cluster D MAGs also encode fewer hydrogenases compared to cluster F (47/114 cluster D MAGs, 136/198 cluster F MAGs) indicating that a hydrogenotrophic lifestyle is less widespread in these organisms. Overall, the lower genome size of cluster D organisms is in agreement with fewer genes present in these organisms for the above-mentioned pathways compared to genomes in cluster F.
Newly reconstructed Desulfobacterota MAGs in clusters D and F appear to be obligate anaerobes as they largely lack heme-copper oxidases. Instead, most sedimentary MAGs and public genomes encode cytochrome bd ubiquinol oxidase (cydAB), which is expressed under microoxic conditions primarily to detoxify O2 [84, 85]. Oxygen-sensitive group 1b NiFe hydrogenases are widespread in Desulfobacterota classes Desulfatiglandales and Desulfobacterales (Supplementary Table 7 and Supplementary Fig. 3), and thus these organisms are predicted to perform hydrogenotrophic respiration using a variety of terminal electron acceptors such as sulfate, nitrate, and metals [53]. Additional support for anaerobic lifestyles is evident in the distribution of anaerobic hydrocarbon degradation genes in these organisms; MAGs are predicted to encode putative anaerobic benzene carboxylase abcA, acetophenone carboxylase (apc γ and δ subunits), phenylphosphate carboxylase (ppcAB), and phenylphosphate synthase (ppsA, Supplementary Table 13 and Supplementary Fig. 5). These genes are primarily distributed within the class BSN033, and orders Desulfobacterales (containing known hydrocarbon-degrading organisms such as Desulfococcus oleovorans [81, 86] and Desulfosarcina sp. BuS5 [87] and Desulfatiglandales within the class Desulfobacteria. Additionally, MAGs from cluster F likely contribute to methylmercury production in marine sediments, as 108/198 encode the Hg-methylating gene acetyl-CoA synthase/corrinoid iron-sulfur protein/putative mercury methyltransferase (hgcAB) [88]. In contrast, few cluster D MAGs are predicted to encode this gene (9/114). Cluster F MAGs also uniquely encode putative genes for trimethylamine metabolism, where organisms predominantly in the order Desulfobacterales encode the pyrrolysine biosynthesis gene pylB (Supplementary Table 16) and trimethylamine methyltransferase (mttB, PF06253) [66, 89]. Finally, Desulfobacterota MAGs are predicted to encode reductive dehalogenases (rdhA genes) for dehalorespiration (22/114 cluster D and 77/198 cluster F MAGs) [90]. Most of these sequences branch separately from known rdhA sequences (Supplementary Fig. 6), suggesting they could utilize different substrates than known reductive dehalogenases.
Cluster E is composed of genomes involved in iron and manganese cycling
Genomes in cluster E are within the Desulfuromonadia class, including 118 publicly available genomes known to be involved in iron and manganese cycling, as well as 11 MAGs (C6-C8). Desulfuromonadia is made up of two orders, the Desulfuromonadales and Geobacterales, which are known to mediate dissimilatory Fe (III) and Mn (IV) reduction and include cultured representatives such as Desulfuromonas acetoxidans and Geobacter sulfurreducens [91, 92]. Desulfuromonadia genomes have distinct protein content compared to other Desulfobacterota classes. We identified unique protein domains in 95% of the cluster E genomes including those involved in two-component signal transduction, chemotaxis, nitrogen fixation, metal tolerance and homeostasis, and extracellular domains involved in small molecule recognition (Supplementary Table 6). These characteristics align with previously published literature, which highlight many of these features as unique and crucial to Geobacter physiology [93]. Desulfuromonadia genomes have the highest potential (according to MEBS scores) for iron cycling when compared to other clusters (Supplementary Fig. 9), further highlighting their unique role in this cycle. Eleven newly reconstructed MAGs in cluster E (C6-C8) add diversity to families BM103 and Geopsychrobacteraceae, and largely seem to mirror patterns identified broadly in Desulfuromonadia. These MAGs are related to cultured representatives (Desulfuromusa kysingii and Desulfuromonas acetoxidans) known to couple acetate oxidation to the reduction of elemental sulfur [94, 95]. Sulfur respiration in Desulfuromonadales MAGs may be sustained by electron donors such as formate or acetate with polysulfide as an acceptor (polysulfide reductase, PF03916). C6-C8 organisms also encode group 1f NiFe hydrogenase, (7/11 MAGs, Supplementary Table 7 and Supplementary Fig. 3) which has been implicated in aerotolerance in G. sulfurreducens [96]. Finally, these C6-C8 MAGs encode genes for nitrogen fixation (nifDKH) and nitrate reduction (napAB).
Genomic cluster G is uniquely composed of the class Syntrophia
Cluster G is made up of 10 MAGs reconstructed in this study (U8) and 134 publicly available genomes in the class Syntrophia (within Desulfobacterota). Only eight bacteria within-cluster G are cultured organisms, including Syntrophus aciditrophicus and Syntrophus gentianae. Syntrophia are predicted to lack pathways present in other Desulfobacterota classes (e.g., genes associated with central metabolism, sulfur, nitrogen, and iron cycling) and have the lowest potential for nitrogen, oxygen, and iron cycling compared to other genomic clusters (Supplementary Fig. 9). Also, Syntrophia has fewer complete KEGG modules for central metabolic pathways (Supplementary Table 10). Ninety-five percent of Syntrophia organisms encode a benzoyl-CoA reductase domain (PF01869), indicating these organisms may be able to anaerobically oxidize aromatic compounds to CO2. This trait is shared with cultured representatives, which degrade benzoate in syntrophic association with hydrogen-using microorganisms [97]. In addition, several Syntrophia families (UBA5619, UBA2251, UBA2185, Smithellaceae, Fen-1087, and CG2-30-49-12) encode dsrAB, dsrD, and dsrKMJOP, but lack adenylyl sulfate reductase subunits A and B (aprBA), sulfate adenylyltransferase (sat), and quinone-interacting membrane-bound oxidoreductase subunits A, B, and C (qmoABC). This indicates these organisms may not conserve energy via sulfate reduction, as has been identified in Firmicutes [98]. U8 Syntrophales may also utilize acetate as a source of carbon and energy via the conversion of acetate to acetyl-CoA (acs, present in 6/10 MAGs and porA, present in 10/10 MAGs). This may then be fed into gluconeogenesis since genes involved in fermentation were absent (Supplementary Table 10). The identification of putative anaerobic benzene carboxylase gene abcA in 2 U8 MAGs suggests these organisms may degrade benzene. U8 MAGs also encode genes for formate oxidation (9/10 encode formate dehydrogenase, fdoH) and hydrogenotrophic metabolism (two U8 MAGs encode the anaerobic respiratory group 1b and electron-bifurcating group 3c NiFe-hydrogenases). Overall, the phylogenetic placement of U8 MAGs with Syntrophia and their limited metabolic abilities suggest that these organisms may be involved in syntrophic interactions, providing fermentation products to methanogens or anaerobic methane-oxidizing archaea, while benefiting from the removal of hydrogen and formate from the environment.
Genomic clusters of Myxoccocaceae, SAR324, and Desulfovibrionia
Genomes in clusters B, C, and H are composed of publicly available representatives from Myxococcaceae, SAR324, and Desulfovibrionia (53, 63, and 185 genomes respectively). Cluster C Myxococcaceae have the highest average genome completeness of those analyzed in this study (98.8% complete and 2.6% average genome contamination). The clustering of almost all SAR324 genomes into cluster B is consistent with the unique protein content of this lineage and their particle-associated lifestyle in the water column [14, 15]. Our analyses support previous findings [14, 15] that these organisms mediate sulfur-dependent chemolithoautotrophy (sulfide-quinone oxidoreductase sqr, RuBisCO rbcL) (Supplementary Tables 8 and 10). In addition, we identified atypical nitrous-oxide reductase nosZ genes associated with low oxygen environments and non-denitrifying organisms [99], as well as a potential nonpyrrolysine methyltransferase (mttB, PF06253) since they lack the pyrrolysine biosynthesis gene pylB (Supplementary Table 16) [66]. Finally, Desulfovibrionia have the highest sulfur cycling MEBS score (Supplementary Fig. 9) and numerous genes for dissimilatory sulfate reduction. They also encode genes for nitrogen fixation, nitrite reduction, and hydrogen utilization (Supplementary Table 10).
Identification of novel dissimilatory sulfite reductases and sulfide:quinone oxidoreductases
Roughly half of the MAGs obtained in this study (208/402) contain both marker genes for sulfite reduction, dissimilatory sulfite reductase subunits A and B (dsrA and dsrB) (Fig. 4 and Supplementary Fig. 2) [100, 101]. Most of these MAGs (201 of 402) belong to clusters D and F Desulfobacterota and encode Dsr complexes that are related to those from environmental surveys. The Desulfobacterota Dsr complexes we identified are predominantly of the reductive-type, with the exception of 2 MAGs encoding oxidative-type Dsr complexes related to SAR324 (Fig. 4, C12 and C13) [14]. We identified dsrA in 477 and 466 dsrB in the 1 391 publicly available genomes. These organisms are mostly within clusters F and H, the Desulfobacterota and Desulfovibrionales. Desulfobacterota Dsr complexes in cluster F are distributed across a variety of classes, including BSN033, Desulfobacteria, Desulfobulbia, Syntrophia, and Syntrophobacteria. We found the deepest branching reductive Dsr complexes are derived from Desulfurella amilsii and D. acetivorans. Interestingly, these Dsr complexes form a sister clade with 3 Dsr protein sequences from Acidulodesulfobacterales [8] (now within phylum SZUA-79). In addition, deeply branching MAGs in the 37 marker gene tree (U1-U3, C1, Fig. 1) also appear to encode deeply branching Dsr complexes.
Furthermore, 95 sulfide:quinone oxidoreductase (SQR) sequences were identified in 79/402 MAGs reconstructed in this study (Supplementary Fig. 7). SQR is a key enzyme for maintaining sulfide homeostasis through the oxidation of sulfide to sulfur [102]. The SQR identified here belong to genomes from clusters A (17 MAGs), D (9 MAGs), and F (53 MAGs, Supplementary Table 15). Phylogenetic analyses indicate that most of these SQR sequences (88/95) belong to the membrane-bound type III SQRs, previously identified in Caldivirga maquilingensis [103]. We also identify eight type II and one group IV SQR sequences. Most group III SQR sequences have sequence homology to Desulfovibrio gigas and Desulfohalovibrio alkalitolerans. Despite this relationship, most SQRs we identified branch separately from known SQRs and form a unique group (Supplementary Fig. 7), highlighting their potential novelty.
Discussion
In this study, we compared the protein content (>17 000 protein domains) and metabolic capabilities of over 1 500 Desulfobacterota, Myxococcota, and SAR324 genomes (synthesized in Supplementary Table 19). This includes 402 new Desulfobacterota and Myxococcota MAGs reconstructed from hydrothermal vent and coastal bay sediments that are taxonomically related to understudied groups (i.e., MAGs in UBA classes). Our analyses identified eight genomic clusters that reflect distinct ecophysiologies. Many of these clusters (C: SAR324, B: Myxococcaceae, E: Desulfuromonadia, G: Syntrophia, and H: Desulfovibrionales) are highly consistent with phylogeny, indicating their genomic protein content is distinct and reflects conserved marker genes used for phylogenetic reconstructions (Fig. 3). The other clusters (A: Myxococcota, D: Desulfobacterota, and F: Desulfobacterota) are more broadly distributed throughout phylogeny (Figs. 1 and 3) and contain many distinct taxonomic classes with diverse metabolic abilities. Thus, for these organisms, it may be difficult to infer metabolism-based solely on phylogeny. Our work provides a broad view of the traits of these taxa and incorporates novel diversity within the reclassified Deltaproteobacteria [1].
We have reconstructed and analyzed 65 marine Myxococcota MAGs, which are understudied compared to terrestrial and soil Myxococcota. U2 UBA9160 and C5 Polyangia are predicted to have unique metabolisms, including shared abilities in complete or partial denitrification, methylamine degradation, and anaerobic hydrocarbon degradation, despite being phylogenetically distinct. Interestingly, we identified genes for complete denitrification (nitrate to N2) in four of 32 U2 MAGs; this is a rare trait within microorganisms, which more commonly rely on metabolic handoffs for this process [22]. Within Myxococcota, nitrogen cycling has largely been studied in Anaeromyxobacter dehalogenans [104, 105] and these pathways are not well characterized in other taxa within this phylum. Putative ethylbenzene (ebdAB) and cymene (cmdAB) dehydrogenase genes identified in U2 UBA9160 and C5 Polyangia are also unique features in the novel MAGs described here since previously studied anaerobic hydrocarbon-degrading Deltaproteobacteria generally do not possess denitrification genes [81]. Thus, the presence of these genes (edbAB and cmdAB) in these newly reconstructed putative denitrifiers may be a unique feature worth exploring in future studies. Methylamine degradation was identified as another unique feature of U2 and C5 and is understudied in Myxococcota. U2 and C5 encode methylamine dehydrogenases (mauAB) (Supplementary Table 10) which is thought to be conserved in Proteobacteria [106]. Thus, the marine Myxococcota reconstructed here potentially play a previously unrecognized role in competition with other methylotrophic degraders and have implications in global carbon and nitrogen cycling [107]. Finally, the Myxococcota reconstructed here are predicted to encode non-reductive dehalogenases (i.e., haloalkane dehalogenase), which are thought to be involved in the transformation of chlorinated natural organic matter [83], though the exact functions of these dehalogenases in the environment are largely unexplored.
We have also added a large number (333) of Desulfobacterota MAGs, a phylum known for sulfate reduction via dissimilatory sulfite reductases (DsrAB complexes) and associated enzymes [2, 108]. Our analyses confirm that dsr genes are widespread in the Desulfobacteria, Desulfarculia, Desulfobaccia, and Dissulfuribacteria classes. However, Desulfobacterota are metabolically versatile and are also capable of anaerobic hydrocarbon degradation, methylmercury production, hydrogen cycling, and reductive dehalogenation. The identification of putative methylmercury genes (hgcAB) in Desulfobacterota genomes reconstructed here supports previous findings that have identified these genes in Desulfobacterota from other marine systems [109, 110], and suggests they produce the neurotoxic and bioaccumulative compound methylmercury. We also identified over 200 hydrogenase sequences in 194 Desulfobacterota MAGs reconstructed in this study, in line with previous inferences that many of these organisms are hydrogenotrophs. In addition, uncultured Desulfobacterota contain putative reductive dehalogenases that appear phylogenetically distinct from characterized enzymes (Supplementary Fig. 6). Finally, Desulfobacterota MAGs encode putative type III SQRs related to D. gigas and D. alkalitolerans. SQR has previously been identified in Desulfurivibrio alkaliphilus, a chemolithotroph that appears to oxidize sulfide using sulfate reduction genes, and which lacks all genes for sulfide oxidation except SQR [101]. Also, previous bioinformatics analyses have identified SQRs as common in marine metagenomes, with type II being the dominant type in the open ocean [111]. Here we mainly identify type III SQRs, suggesting these could be dominant in deep sea Desulfobacterota. Also, phylogenetic analyses of SQR from the MAGs reconstructed in this study reveal that they form new branches within type III SQR, highlighting the potential novel role of these SQRs related to the maintenance of sulfide homeostasis or bioenergetics in deep-sea sediments. Molecular analyses or activity measurements [112] will be needed to confirm these genome-based metabolic predictions.
Most Desulfobacterota MAGs reconstructed here fall within genomic clusters D and F. These clusters raise questions about the role of genome size in differentiating the metabolic traits of phylogenetically related bacteria. Cluster D organisms have smaller genome sizes and fewer genes for sulfate reduction, carbon fixation, reductive dehalogenation, and other processes. Cluster F organisms have larger genome sizes compared to cluster D, which appears to confer greater metabolic versatility in these organisms and the ability to gain energy through the oxidation of a wider range of substrates. Genome quality could also play a role in this distinction, as cluster F has a higher median genome completeness compared to cluster D, and cluster F has more high-quality (>90% complete, <10% contamination) genomes (Supplementary Fig. 8). However, genomes reconstructed from understudied environments (i.e., aquifer sediment, hydrothermal vents) or large-scale reconstruction efforts almost entirely group together in cluster D, including Desulfobacterota classes UBA1144 (Dadabacteria), GWC2-55-46, Binatia, BSN033, and MBNT15. This suggests that the distinction between D and F could be due to differences in protein content as a result of adaptation to different environments, rather than simply an artifact of genome quality. The relationship between clusters D and F warrants further investigation as a potentially ecologically relevant and overlooked distinction.
According to GTDB-tk classification, five MAGs reconstructed in this study belong to a novel phylum, DQWO01. Our 37 marker gene phylogeny indicates that DQWO01 is related to Desulfobacterota, and in support of this, these genomes are within-cluster D Desulfobacterota (suggesting they have related protein content at the whole genome level). However, these organisms are undersampled and thus likely require additional genomes to better understand their relationship to Desulfobacterota. It is also possible that this GTDB-tk classification will change as the genomes reconstructed in this study and other metagenomic analyses are added to public databases, allowing these relationships to be further resolved.
We have significantly expanded the diversity of Desulfobacterota and Myxococcota. A comparison of the protein composition of over 1 500 genomes has revealed shared metabolic abilities between taxonomically distinct bacteria, as well as potential distinctions between closely related organisms. Our findings highlight that even within relatively well-studied microbial taxa, there are previously unrecognized metabolic pathways and uncultured lineages that are unexplored. This is especially true when examining understudied environments such as the deep sea. This work will be a resource for future analyses and provide a blueprint to aid in the exploration of the impressive metabolic versatility of these widespread sediment bacteria. Metagenomics has not only uncovered entirely new branches across the tree of life but has also enabled greater exploration of bacteria that have been studied for decades, like the Deltaproteobacteria. As a result, we are witnessing a dramatic reshuffling of taxonomy and a more comprehensive understanding of the genetic composition in these bacteria.
Supplementary information
Acknowledgements
This work was funded by NSF Systems and Synthetic Biology award number 1817354 to B.J.B. and A.P.T. and Simons Foundation award 687165 to B.J.B. Alvin dives and sediment sampling in Guaymas Basin were supported by NSF Biological Oceanography (OCE-0647633 for sediments collected in 2008 and 2009, OCE-1357238 in 2018). C.G. was supported by an NHMRC EL2 Fellowship (APP1178715). We thank the Alvin teams for stellar performance over the years in Guaymas Basin. A portion of the Guaymas sediments were sequenced by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility (Contract No. DE-AC02-05CH11231 provided to ND). Thank you to Mirna Vazquez Rosas Landa for her technical support on the bubble plot (Fig. 5) implemented in the Rpackage rbims (https://github.com/mirnavazquez/RbiMs). Finally, many thanks to Augusto César Poot Hernandez, head of the Unidad de Bioinformática y Manejo de la Información of the Instituto de Fisiología Celular, UNAM, for his valuable input and the development of the source code used for the metabolic clustering analysis implemented in MEBS (https://github.com/valdeanda/mebs/).
Author contributions
BJB, APT, VDA, and MVL conceived the study. APT provided environmental samples. MVL, KWS, and IMR extracted DNA from environmental samples and performed metagenomic sequence assemblies and binning. CG provided metabolic databases and assistance with hydrogenase analyses. ND provided genomic data. MVL, VDA, and BJB performed phylogenomic analyses. MVL and VDA analyzed genomic data, carried out metabolic analyses and inferences. MVL, VDA, and BJB wrote the manuscript, and all authors edited and approved the manuscript.
Data availability
The final assembled and annotated genomic sequences of Deltaproteobacteria from deep-sea sediments have been deposited in NCBI under BioProject ID PRJNA688516. The NCBI accession numbers for the MAGs reconstructed in this study are provided in Supplementary Table 3, and geochemical data is provided in Supplementary Table 1.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised due to an error in figure 6.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marguerite V. Langwig, Valerie De Anda.
Change history
11/22/2021
A Correction to this paper has been published: 10.1038/s41396-021-01091-w
Contributor Information
Marguerite V. Langwig, Email: marguerite.langwig@gmail.com
Brett J. Baker, Email: acidophile@gmail.com
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01057-y.
References
- 1.Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, Wagner M, et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol. 2020;70:5972–6016. doi: 10.1099/ijsem.0.004213. [DOI] [PubMed] [Google Scholar]
- 2.Mußmann M, Ishii K, Rabus R, Amann R. Diversity and vertical distribution of cultured and uncultured Deltaproteobacteria in an intertidal mud flat of the Wadden Sea. Environ Microbiol. 2005;7:405–18. doi: 10.1111/j.1462-2920.2005.00708.x. [DOI] [PubMed] [Google Scholar]
- 3.Minz D, Flax JL, Green SJ, Muyzer G, Cohen Y, Wagner M, et al. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl Environ Microbiol. 1999;65:4666–71. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorokin DY, Yu, Sorokin D, Tourova TP, Henstra AM, Stams AJM, et al. Sulfidogenesis under extremely haloalkaline conditions by Desulfonatronospira thiodismutans gen. nov., sp. nov., and Desulfonatronospira delicata sp. nov. - a novel lineage of Deltaproteobacteria from hypersaline soda lakes. Microbiology. 2008;154:1444–53. doi: 10.1099/mic.0.2007/015628-0. [DOI] [PubMed] [Google Scholar]
- 5.Si Y, Zou Y, Liu X, Si X, Mao J. Mercury methylation coupled to iron reduction by dissimilatory iron-reducing bacteria. Chemosphere. 2015;122:206–12. doi: 10.1016/j.chemosphere.2014.11.054. [DOI] [PubMed] [Google Scholar]
- 6.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, et al. Mercury methylation by novel microorganisms from new environments. Environ Sci Technol. 2013;47:11810–20. doi: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 7.Bergmann F, Selesi D, Weinmaier T, Tischler P, Rattei T, Meckenstock RU. Genomic insights into the metabolic potential of the polycyclic aromatic hydrocarbon degrading sulfate-reducing Deltaproteobacterium N47. Environ Microbiol. 2011;13:1125–37. doi: 10.1111/j.1462-2920.2010.02391.x. [DOI] [PubMed] [Google Scholar]
- 8.Tan S, Liu J, Fang Y, Hedlund BP, Lian Z-H, Huang L-Y, et al. Insights into ecological role of a new deltaproteobacterial order Candidatus Acidulodesulfobacterales by metagenomics and metatranscriptomics. ISME J. 2019;13:2044–57. doi: 10.1038/s41396-019-0415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda Y, Itoh H, Shiratori Y, Isobe K, Otsuka S, Senoo K. Predominant but previously-overlooked prokaryotic drivers of reductive nitrogen transformation in paddy soils, revealed by metatranscriptomics. Microbes Environ. 2017;32:180–3. doi: 10.1264/jsme2.ME16179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Häggblom MM. Genome-guided identification of organohalide-respiring Deltaproteobacteria from the marine environment. MBio. 2018;9:e02471–18. doi: 10.1128/mBio.02471-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovley DR, Phillips EJ. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–80. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonergan DJ, Jenter HL, Coates JD, Phillips EJ, Schmidt TM, Lovley DR. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–8. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawid W. Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev. 2000;24:403–27. doi: 10.1111/j.1574-6976.2000.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 14.Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 15.Sheik CS, Jain S, Dick GJ. Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol. 2014;16:304–17. doi: 10.1111/1462-2920.12165. [DOI] [PubMed] [Google Scholar]
- 16.Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359:320–5. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 17.Hug LA, Thomas BC, Sharon I, Brown CT, Sharma R, Hettich RL, et al. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol. 2016;18:159–73. doi: 10.1111/1462-2920.12930. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Zhang J, Zhao L, Zhang X, Xie S. Spatial distribution of bacterial communities in high-altitude freshwater wetland sediment. Limnology. 2014;15:249–56. [Google Scholar]
- 19.Wang Y, Sheng H-F, He Y, Wu J-Y, Jiang Y-X, Tam NF-Y, et al. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microbiol. 2012;78:8264–71. doi: 10.1128/AEM.01821-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yilmaz P, Yarza P, Rapp JZ, Glöckner FO. Expanding the world of marine bacterial and archaeal clades. Front Microbiol. 2016;6:1524. doi: 10.3389/fmicb.2015.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jochum LM, Schreiber L, Marshall IPG, Jørgensen BB, Schramm A, Kjeldsen KU. Single-cell genomics reveals a diverse metabolic potential of uncultivated Desulfatiglans-related Deltaproteobacteria widely distributed in marine sediment. Front Microbiol. 2018;9:2038. doi: 10.3389/fmicb.2018.02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 24.Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun. 2018;9:4999. doi: 10.1038/s41467-018-07418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi N, Sickle FJ. A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). 2011. https://github.com/najoshi/sickle.
- 26.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 28.Li D, Luo R, Liu C-M, Leung C-M, Ting H-F, Sadakane K, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 32.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y-W, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–7. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 36.Eren AM, Esen ÖC, Quince C, Vineis JH, Sogin ML, Delmont TO. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, et al. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–5. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Criscuolo A, Gribaldo S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2019;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowers RM. The Genome Standards Consortium, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35:725–31. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016;10:761–77. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Søndergaard D, Pedersen CNS, Greening C. HydDB: A web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 56.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–51. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greening C. Greening lab metabolic marker gene databases. 10.26180/c.5230745.
- 59.Zhou Z, Tran P, Liu Y, Kieft K, Anantharaman K. METABOLIC: a scalable high-throughput metabolic and biogeochemical functional trait profiler based on microbial genomes. bioRxiv. 2019. Preprint at 10.1101/761643.
- 60.Terrapon N, Lombard V, Drula E, Coutinho PM, Henrissat B. The CAZy database/the carbohydrate-active enzyme (CAZy) database: principles and usage guidelines. In: Aoki-Kinoshita KF (ed). A Practical Guide to Using Glycomics Databases. (Springer Japan, Tokyo, 2017) pp 117–31.
- 61.Peabody MA, Laird MR, Vlasschaert C, Lo R, Brinkman FSL. PSORTdb: expanding the bacteria and archaea protein subcellular localization database to better reflect diversity in cell envelope structures. Nucleic Acids Res. 2016;44:D663–8. doi: 10.1093/nar/gkv1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callaghan AV, Wawrik B. AnHyDeg: a curated database of anaerobic hydrocarbon degradation genes. GitHub. 2016. https://github.com/AnaerobesRock/AnHyDeg.
- 63.McDaniel EA, Peterson BD, Stevens SLR, Tran PQ, Anantharaman K, McMahon KD. Expanded phylogenetic diversity and metabolic flexibility of mercury-methylating microorganisms. mSystems. 2020;5:e00299–20. doi: 10.1128/mSystems.00299-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDaniel EA, Anantharaman K, McMahon KD. metabolisHMM: Phylogenomic analysis for exploration of microbial phylogenies and metabolic pathways. bioRxiv. 2019. Preprint at 10.1101/2019.12.20.884627.
- 65.De Anda V, Zapata-Peñasco I, Poot-Hernandez AC, Eguiarte LE, Contreras-Moreira B, Souza V. MEBS, a software platform to evaluate large (meta)genomic collections according to their metabolic machinery: unraveling the sulfur cycle. Gigascience. 2017;6:1–17. doi: 10.1093/gigascience/gix096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ticak T, Kountz DJ, Girosky KE, Krzycki JA, Ferguson DJ., Jr A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc Natl Acad Sci. 2014;111:E4668–76. doi: 10.1073/pnas.1409642111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diamond S, Andeer PF, Li Z, Crits-Christoph A, Burstein D, Anantharaman K, et al. Mediterranean grassland soil C-N compound turnover is dependent on rainfall and depth and is mediated by genomically divergent microorganisms. Nat Microbiol. 2019;4:1356–67. doi: 10.1038/s41564-019-0449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woodcroft BJ, Singleton CM, Boyd JA, Evans PN, Emerson JB, Zayed AAF, et al. Genome-centric view of carbon processing in thawing permafrost. Nature. 2018;560:49–54. doi: 10.1038/s41586-018-0338-1. [DOI] [PubMed] [Google Scholar]
- 69.Slobodkina GB, Reysenbach A-L, Panteleeva AN, Kostrikina NA, Wagner ID, Bonch-Osmolovskaya EA, et al. Deferrisoma camini gen. nov., sp. nov., a moderately thermophilic, dissimilatory iron(III)-reducing bacterium from a deep-sea hydrothermal vent that forms a distinct phylogenetic branch in the Deltaproteobacteria. Int J Syst Evol Microbiol. 2012;62:2463–8. doi: 10.1099/ijs.0.038372-0. [DOI] [PubMed] [Google Scholar]
- 70.Han K, Li Z-F, Peng R, Zhu L-P, Zhou T, Wang L-G, et al. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci Rep. 2013;3:1–7. doi: 10.1038/srep02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanford RA, Cole JR, Tiedje JM. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl Environ Microbiol. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castaño-Cerezo S, Pastor JM, Renilla S, Bernal V, Iborra JL, Cánovas M. An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Micro Cell Fact. 2009;8:54. doi: 10.1186/1475-2859-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meinke A, Gilkes NR, Kwan E, Kilburn DG, Warren RA, Miller RC, Jr, et al. CbhA) from the cellulolytic bacterium Cellulomonas fimi is a beta-1,4-exocellobiohydrolase analogous to Trichoderma reesei CBH II. Mol Microbiol. 1994;12:413–22. doi: 10.1111/j.1365-2958.1994.tb01030.x. [DOI] [PubMed] [Google Scholar]
- 74.Zverlov VV, Hertel C, Bronnenmeier K, Hroch A, Kellermann J, Schwarz WH. The thermostable alpha-L-rhamnosidase RamA of Clostridium stercorarium: biochemical characterization and primary structure of a bacterial alpha-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol Microbiol. 2000;35:173–9. doi: 10.1046/j.1365-2958.2000.01691.x. [DOI] [PubMed] [Google Scholar]
- 75.Galinier A, Josef Deutscher, Martin-Verstraete I. Phosphorylation of either Crh or HPr mediates binding of CcpA to the Bacillus subtilis xyn cre and catabolite repression of the xyn operon. Edited by IB Holland. J Mol Biol. 1999; 286: 307–14. [DOI] [PubMed]
- 76.Schmetterer G, Valladares A, Pils D, Steinbach S, Pacher M, Muro-Pastor AM, et al. The coxBAC operon encodes a cytochrome c oxidase required for heterotrophic growth in the cyanobacterium Anabaena variabilis strain ATCC 29413. J Bacteriol. 2001;183:6429–34. doi: 10.1128/JB.183.21.6429-6434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ducluzeau A-L, Ouchane S, Nitschke W. The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol Biol Evol. 2008;25:1158–66. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 78.Green GN, Fang H, Lin RJ, Newton G, Mather M, Georgiou CD, et al. The nucleotide sequence of the cyd locus encoding the two subunits of the cytochrome d terminal oxidase complex of Escherichia coli. J Biol Chem. 1988;263:13138–43. [PubMed] [Google Scholar]
- 79.Upadhyay AK, Hooper AB, Hendrich MP. NO reductase activity of the tetraheme cytochrome C554 of Nitrosomonas europaea. J Am Chem Soc. 2006;128:4330–7. doi: 10.1021/ja055183+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 81.Davidova IA, Marks CR, Suflita JM. Anaerobic hydrocarbon-degrading Deltaproteobacteria. In: McGenity TJ (ed). Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes. (Springer International Publishing, Cham, 2019) pp 207–43.
- 82.Strijkstra A, Trautwein K, Jarling R, Wöhlbrand L, Dörries M, Reinhardt R, et al. Anaerobic activation of p-cymene in denitrifying betaproteobacteria: methyl group hydroxylation versus addition to fumarate. Appl Environ Microbiol. 2014;80:7592–603. doi: 10.1128/AEM.02385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Temme HR, Carlson A, Novak PJ. Presence, diversity, and enrichment of respiratory reductive dehalogenase and non-respiratory hydrolytic and oxidative dehalogenase genes in terrestrial environments. Front Microbiol. 2019;10:1–14. doi: 10.3389/fmicb.2019.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lemos RS, Gomes CM, Santana M, LeGall J, Xavier AV, Teixeira M. The ‘strict’ anaerobe Desulfovibrio gigas contains a membrane-bound oxygen-reducing respiratory chain. FEBS Lett. 2001;496:40–43. doi: 10.1016/s0014-5793(01)02399-7. [DOI] [PubMed] [Google Scholar]
- 86.Aeckersberg F, Rainey FA, Widdel F. Growth, natural relationships, cellular fatty acids and metabolic adaptation of sulfate-reducing bacteria that utilize long-chain alkanes under anoxic conditions. Arch Microbiol. 1998;170:361–9. doi: 10.1007/s002030050654. [DOI] [PubMed] [Google Scholar]
- 87.Kniemeyer O, Musat F, Sievert SM, Knittel K, Wilkes H, Blumenberg M, et al. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria. Nature. 2007;449:898–901. doi: 10.1038/nature06200. [DOI] [PubMed] [Google Scholar]
- 88.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Jr, Smith SD, et al. The genetic basis for bacterial mercury methylation. Science. 2013;339:1332–5. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 89.Krzycki JA. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr Opin Chem Biol. 2004;8:484–91. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 90.Cole JR, Fathepure BZ, Tiedje JM. Tetrachloroethene and 3-chlorobenzoate dechlorination activities are co-induced in Desulfomonile tiedjei DCB-1. Biodegradation. 1995;6:167–72. doi: 10.1007/BF00695347. [DOI] [PubMed] [Google Scholar]
- 91.Caccavo F, Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol. 1994;60:3752–9. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roden EE, Lovley DR. Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl Environ Microbiol. 1993;59:734–42. doi: 10.1128/aem.59.3.734-742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, et al. Geobacter: the microbe electric’s physiology, ecology, and practical applications. Adv Micro Physiol. 2011;59:1–100. doi: 10.1016/B978-0-12-387661-4.00004-5. [DOI] [PubMed] [Google Scholar]
- 94.Liesack W, Finster K. Phylogenetic analysis of five strains of gram-negative, obligately anaerobic, sulfur-reducing bacteria and description of Desulfuromusa gen. nov., including Desulfuromusa kysingii sp. nov., Desulfuromusa bakii sp. nov., and Desulfuromusa succinoxidans sp. nov. Int J Syst Bacteriol. 1994;44:753–8. [Google Scholar]
- 95.Pfennig N, Biebl H. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch Microbiol. 1976;110:3–12. doi: 10.1007/BF00416962. [DOI] [PubMed] [Google Scholar]
- 96.Tremblay P-L, Lovley DR. Role of the NiFe hydrogenase Hya in oxidative stress defense in Geobacter sulfurreducens. J Bacteriol. 2012;194:2248–53. doi: 10.1128/JB.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, et al. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104:7600–5. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imachi H, Sekiguchi Y, Kamagata Y, Loy A, Qiu Y-L, Hugenholtz P, et al. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol. 2006;72:2080–91. doi: 10.1128/AEM.72.3.2080-2091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bertagnolli AD, Konstantinidis KT, Stewart FJ. Non-denitrifier nitrous oxide reductases dominate marine biomes. Environ Microbiol Rep. 2020;12:681–92. doi: 10.1111/1758-2229.12879. [DOI] [PubMed] [Google Scholar]
- 100.Wasmund K, Mußmann M, Loy A. The life sulfuric: microbial ecology of sulfur cycling in marine sediments. Environ Microbiol Rep. 2017;9:323–44. doi: 10.1111/1758-2229.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thorup C, Schramm A, Findlay AJ, Finster KW, Schreiber L. Disguised as a sulfate reducer: growth of the Deltaproteobacterium Desulfurivibrio alkaliphilus by sulfide oxidation with nitrate. MBio. 2017;8:e00671–17. doi: 10.1128/mBio.00671-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marcia M, Ermler U, Peng G, Michel H. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins. 2010;78:1073–83. doi: 10.1002/prot.22665. [DOI] [PubMed] [Google Scholar]
- 103.Lencina AM, Ding Z, Schurig-Briccio LA, Gennis RB. Characterization of the type III sulfide:quinone oxidoreductase from Caldivirga maquilingensis and its membrane binding. BBA-Bioenerg. 2013;1827:266–75. doi: 10.1016/j.bbabio.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Onley JR, Ahsan S, Sanford RA, Löffler FE. Denitrification by Anaeromyxobacter dehalogenans, a common soil bacterium lacking the nitrite reductase genes nirS and nirK. Appl Environ Microbiol. 2018;84:e01985–17. doi: 10.1128/AEM.01985-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Masuda Y, Yamanaka H, Xu Z-X, Shiratori Y, Aono T, Amachi S, et al. Diazotrophic Anaeromyxobacter isolates from soils. Appl Environ Microbiol. 2020;86:e01985–17. doi: 10.1128/AEM.00956-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–22. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 107.Taubert M, Grob C, Howat AM, Burns OJ, Pratscher J, Jehmlich N, et al. Methylamine as a nitrogen source for microorganisms from a coastal marine environment. Environ Microbiol. 2017;19:2246–57. doi: 10.1111/1462-2920.13709. [DOI] [PubMed] [Google Scholar]
- 108.Kaneko R, Hayashi T, Tanahashi M, Naganuma T. Phylogenetic diversity and distribution of dissimilatory sulfite reductase genes from deep-sea sediment cores. Mar Biotechnol. 2007;9:429–36. doi: 10.1007/s10126-007-9003-7. [DOI] [PubMed] [Google Scholar]
- 109.Capo E, Bravo AG, Soerensen AL, Bertilsson S, Pinhassi J, Feng C, et al. Deltaproteobacteria and spirochaetes-like bacteria are abundant putative mercury methylators in oxygen-deficient water and marine particles in the Baltic Sea. Front Microbiol. 2020;11:574080. doi: 10.3389/fmicb.2020.574080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Villar E, Cabrol L, Heimbürger-Boavida L-E. Widespread microbial mercury methylation genes in the global ocean. Env Microbiol Rep. 2020;12:277–87. doi: 10.1111/1758-2229.12829. [DOI] [PubMed] [Google Scholar]
- 111.Xia Y, Lü C, Hou N, Xin Y, Liu J, Liu H, et al. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J. 2017;11:2754–66. doi: 10.1038/ismej.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Landgraf P, Antileo ER, Schuman EM, Dieterich DC. BONCAT: metabolic labeling, click chemistry, and affinity purification of newly synthesized proteomes. Methods Mol Biol. 2015;1266:199–215. doi: 10.1007/978-1-4939-2272-7_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final assembled and annotated genomic sequences of Deltaproteobacteria from deep-sea sediments have been deposited in NCBI under BioProject ID PRJNA688516. The NCBI accession numbers for the MAGs reconstructed in this study are provided in Supplementary Table 3, and geochemical data is provided in Supplementary Table 1.