Abstract
In an attempt to identify transcription factors which activate sterol-regulatory element-binding protein 1c (SREBP-1c) transcription, we screened an expression cDNA library from adipose tissue of SREBP-1 knockout mice using a reporter gene containing the 2.6-kb mouse SREBP-1 gene promoter. We cloned and identified the oxysterol receptors liver X receptor (LXRα) and LXRβ as strong activators of the mouse SREBP-1c promoter. In the transfection studies, expression of either LXRα or -β activated the SREBP-1c promoter-luciferase gene in a dose-dependent manner. Deletion and mutation studies, as well as gel mobility shift assays, located an LXR response element complex consisting of two new LXR-binding motifs which showed high similarity to an LXR response element recently found in the ABC1 gene promoter, a reverse cholesterol transporter. Addition of an LXR ligand, 22(R)-hydroxycholesterol, increased the promoter activity. Coexpression of retinoid X receptor (RXR), a heterodimeric partner, and its ligand 9-cis-retinoic acid also synergistically activated the SREBP-1c promoter. In HepG2 cells, SREBP-1c mRNA and precursor protein levels were induced by treatment with 22(R)-hydroxycholesterol and 9-cis-retinoic acid, confirming that endogenous LXR-RXR activation can induce endogenous SREBP-1c expression. The activation of SREBP-1c by LXR is associated with a slight increase in nuclear SREBP-1c, resulting in activation of the gene for fatty acid synthase, one of its downstream genes, as measured by the luciferase assay. These data demonstrate that LXR-RXR can modify the expression of genes for lipogenic enzymes by regulating SREBP-1c expression, providing a novel link between fatty acid and cholesterol metabolism.
Sterol-regulatory element (SRE)-binding proteins (SREBPs) are transcription factors which belong to the basic helix-loop-helix leucine zipper family (3–5). In contrast to other members of this family, SREBPs are synthesized as precursor proteins which remain bound to the endoplasmic reticulum and the nuclear envelope in the presence of sufficient sterol concentrations. Upon sterol deprivation, the precursor protein undergoes a sequential two-step cleavage process to release the NH2-terminal portion (28). This mature SREBP then enters the nucleus and activates the transcription of genes involved in cholesterol and fatty acid synthesis by binding to SREs or to palindromic sequences called E boxes within their promoter regions (20, 39). Three forms of SREBP have been characterized: SREBP-1a and -1c (also known as ADD1) (14, 38, 43) and SREBP-2. It has been shown that all of the cultured cells analyzed to date express primarily SREBP-2 and the SREBP-1a isoform, whereas most organs, including the liver and adipose tissue, express predominantly SREBP-2 and the SREBP-1c isoform (36). SREBP-1a is a stronger activator than SREBP-1c due to its longer transactivation domain and has a wider range of target genes involved in both cholesterol and fatty acid synthesis (30, 31).
Lipogenic enzymes, which are involved in energy storage through synthesis of fatty acids and triglycerides, are coordinately regulated at the transcriptional level during different metabolic states (9, 11). Recent in vivo studies demonstrated that SREBP-1c plays a crucial role in the dietary regulation of most hepatic lipogenic genes, whereas SREBP-2 is actively involved in the transcription of cholesterogenic enzymes (13). These include studies of the effects of the absence or overexpression of SREBP-1 on hepatic lipogenic gene expression (30, 31, 33), as well as physiological changes of SREBP-1c protein in normal mice after dietary manipulation such as placement on high-carbohydrate diets, polyunsaturated fatty acid-enriched diets, and fasting-refeeding regimens (12, 17, 37, 40, 41). The similar coordinated changes in SREBP-1c and lipogenic gene expression upon fasting and refeeding were also observed in adipose tissue (18). In fat tissue, SREBP-1c (ADD1) appears to be involved in adipocyte differentiation and insulin resistance (19, 35). Recent studies suggest that insulin or insulin-facilitated glucose uptake mediates lipogenesis through SREBP-1c induction (7, 8, 10, 21, 34). Previous reports on the regulation of SREBP-1c have all demonstrated the induction to be at the mRNA level. Up-regulation of hepatic SREBP-1 mRNA was observed in the livers of rodents on a fasting-refeeding regimen or a chronic high-carbohydrate diet and in primary hepatocytes with a high-glucose medium (8, 10, 12, 41). In contrast, down-regulation of SREBP-1c was observed in livers from fasted rodents, from insulin-depleted diabetic rats with streptozotocin treatment, and from mice on a diet containing polyunsaturated fatty acids (12, 17, 34, 37, 40, 41).
In contrast to SREBP-2, which mediates sterol regulation completely at the cleavage level through interaction with SREBP cleavage-activating protein (SCAP) and site 1 protease, SREBP-1c controls the transcriptional regulation of lipogenic enzymes by self-regulating the nuclear concentration of its mature form, which is highly correlated to its precursor and mRNA levels. The initial analysis of the mouse SREBP-1c promoter identified two functional regions (1). One region is the SRE complex, which confers a response to SREBPs. Upstream of this region is an oxysterol-inducible region, the details of which remain unknown. To investigate the relationship between the promoter sequence and physiological regulation of SREBP-1c, we sought to identify transcription factors that might regulate the SREBP-1c promoter. In the screening of the SREBP-1c gene activators, we cloned and identified liver X receptors (LXRα [NR1H3] and LXRβ [NR1H2]) as strong activators of SREBP-1c gene promoter.
LXRs belong to a subclass of nuclear hormone receptors that form obligate heterodimers with retinoid X receptors (RXRs) and are bound and activated by oxysterols (16, 22, 24, 27). LXRs regulate the expression of cholesterol 7α-hydroxylase, the rate-limiting enzyme of the classic bile acid biosynthetic pathway (22, 25, 27); cholesterol ester transfer protein, which translocates cholesterol ester between lipoprotein fractions (23); and ATP-binding cassette transporter 1 (ABC1), which modulates the reverse cholesterol transport from peripheral tissues and cholesterol absorption in the intestine (26). Thus, LXR-RXR heterodimers serve as key regulators of cholesterol homeostasis. Identification of LXR-RXR as an activator of the SREBP-1c promoter reveals a new aspect of cholesterol and fatty acid metabolism.
MATERIALS AND METHODS
Materials and general methods.
We obtained cholesterol, 22(R)-hydroxycholesterol [22(R)-HC], 25-hydroxycholesterol (25-HC), and 9-cis-retinoic acid (9CRA) from Sigma; Redivue [α-32P]dCTP (6,000 Ci/mmol) from Amersham Pharmacia; restriction enzymes from New England BioLabs; and polyclonal antibodies against mouse LXRα and -β (C19, sc-6063x) and RXRα (D-20, sc-553x) from Santa Cruz Biotechnology, Inc.
Standard molecular biology techniques were used. DNA sequencing was performed with the PRISM dye terminator cycle sequencing kit and an ABI PRISM 310 genetic analyzer (Applied Biosystems).
Plasmids.
Luciferase gene constructs containing 2.6-kb and 550-bp fragments of the mouse SREBP-1c promoter (pBP1c2600-Luc and pBP1c550-Luc, respectively) were prepared as described previously (1). Human RXRα expression plasmid pRXR was a kind gift from D. J. Mangelsdorf. Other SREBP-1 promoter-luciferase constructs were produced by PCR using pBP1c550-Luc as a DNA template and subcloning the PCR products into the pGL2 basic or pGL2 promoter vector (Promega). The PCR primers used were as follows: 5′ primers BP1c408-Luc (5′-GGCCAGGAGTGGGTAAA-3′), BP1c357-Luc (5′-GTGTGCGAACGAACCAG-3′), BP1c296-Luc (5′-GGATTCCGGACCCAGGCT-3′), BP1c249-Luc (5′-GGGTTGGGACGACAGTGA-3′), BP1c197-Luc (5′-GGCGCAGACGCGGTTA-3′), BP1c148-Luc (5′-GGGAGAAACCCGAGCT-3′), BP1c186-Luc (5′-GGACGCCCGCTAGTAACC-3′), BP1cLXREa-Luc (5′-GACAGTGACCGCCAGTAACCC-3′), and BP1cLXREaM-Luc (5′-GACAGAGTCCGCCAGAATCCC-3′) and 3′ primers BP1c41-Luc (5′-TAAGAGCTCGGTACCTCCCCTAGGGC-3′), BP1c149-Luc (5′-GGTGCTCTGAATGGGGC-3′), BP1cLXREb-Luc (5′-CCGGGGTTACTAGCGGGC-3′), and BP1cLXREbM-Luc (5′- CCGGGGATTCTAGCGGGC-3′). The primers were tailed with an SmaI site (5′ primers) and a KpnI site (3′ primers).
Preparation of expression library and screening of transactivator of the SREBP-1c promoter.
For preparation of the cytomegalovirus (CMV) promoter-expression vector, the SmaI site of CMV7 (2) was converted to a NotI site using a NotI linker and designated CMV7-NotI. Total RNA was extracted from adipose tissues from three SREBP-1 knockout mice (33) using Trizol (Life Technologies, Inc.). Poly(A)+ RNA was prepared from total RNA by two cycles of chromatography on oligo(dT) (mRNA Purification Kit; Amersham Pharmacia Biotech). A size-fractionated, directional cDNA library with SalI and NotI cohesive ends at the 5′ and 3′ termini, respectively, was constructed from 5 μg of poly(A)+ RNA using a Superscript Plasmid Kit (Life Technologies, Inc.). Size-fractionated cDNA (>1.0 kb) was ligated with expression vector pCMV7-NotI using the protocol and reagents supplied with the Superscript kit. The ligation reaction of cDNAs and the vectors digested with SalI and NotI was transformed into electrocompetent DH5α cells and aliquoted into subpools containing approximately 1,500 clones. The transcriptional activity of each pool (1.5 to 2.0 μg of total DNA) for the SREBP-1c promoter was assayed by transfection into human embryonic kidney (HEK) 293 cells with pBP1c2600-Luc (0.25 μg) and control β-galactosidase reference plasmid pCMV-βgal (0.1 μg; Promega) as described below.
Transfections and luciferase assays.
HEK 293 and HepG2 cells were grown at 37°C in an atmosphere of 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 25 mM glucose, 100 U of penicillin per ml, and 100 μg of streptomycin sulfate per ml supplemented with 10% fetal calf serum (FCS). Transfection studies were carried out with cells plated on 12-well plates as described previously (1). The indicated amount of each expression plasmid was transfected simultaneously with a luciferase reporter plasmid (0.25 μg) and pCMV-βgal or pSV-βgal (0.1 to 0.4 μg). The total amount of DNA in each transfection was adjusted to 1.5 μg/well with pCMV7-NotI vector DNA. Ligands for LXR and RXR were dissolved in ethanol and added to cells after transfection in DMEM with 10% FCS. For suppressed conditions, DMEM with 10% FCS, cholesterol (10 μg/ml), and 25-HC (1 μg/ml) was used to suppress endogenous SREBP activity. The amount of luciferase activity in transfectants was measured and normalized to the amount of β-galactosidase activity as measured by standard kits (Promega).
Gel mobility shift assays.
Gel mobility shift assays were done as previously described (1). Briefly, nuclear extracts were prepared from HEK 293 cells as described previously (31). Double-stranded oligonucleotides used in gel mobility shift assays were prepared by annealing both strands of each LXR response element (LXRE) of the LXRE complex in the SREBP-1c promoter (LXREa [5′-CAGTGACCGCCAGTAACCCCAGC-3′] and LXREb [5′-GGACGCCCGCTAGTAACCCCGGC-3′]), labeled with [α-32P]dCTP by Klenow enzyme, and purified on Sephadex G50 columns. The labeled probes (3,000 to 10,000 cpm) were incubated with nuclear extracts (3 μg) in a mixture (20 μl) containing 10 mM Tris-HCl (pH 7.6), 50 mM KCl, 0.05 mM EDTA, 2.5 mM MgCl2, 8.5% glycerol, 1 mM dithiothreitol, poly(dI-dC) at 0.5 μg/ml, 0.1% Triton X-100, and nonfat milk at 1 mg/ml for 30 min on ice. The DNA-protein complexes were resolved on a 4.6% polyacrylamide gel at 140 V for 1 h at 4°C. Gels were dried and exposed to a BAS2000 filter with BAStation software (Fuji Photo Film Co., Ltd.). For competition experiments, at least a 100-fold molar excess of unlabeled DNA relative to labeled DNA was added to the reaction mixture before addition of the labeled probe. In the supershift experiments, the gel shift reactions were first incubated for 1 h with 2 to 10 μg of polyclonal antibodies against LXRα and -β or RXRα on ice.
Nuclear extract preparation and immunoblot analysis.
Nuclear extracts from HepG2 cells were prepared as described previously (33). The samples of 30 μg of nuclear protein were subjected to immunoblot analysis with mouse immunoglobulin G (IgG) against human SREBP1 (30) or rabbit IgG against human SREBP1c and SREBP2 (32, 33), followed by horseradish peroxidase-linked mouse or rabbit IgG and the ECL kit (Amersham Pharmacia Biotech).
Total RNA preparation and blot hybridization with cDNA probes.
Total RNA was extracted from mouse livers, adipose tissues, and HepG2 cells using TRIZOL Reagent (Life Technologies, Inc.). Equal aliquots of total RNA from mice in each group were pooled (total, 10 μg), subjected to formalin-denatured agarose electrophoresis, and transferred to nylon membrane (Hybond N; Amersham Pharmacia Biotech). Blot hybridization was performed with the cDNA probes labeled with [α-32P]CTP (6,000 Ci/mmol) using the Megaprime DNA Labeling System (Amersham Pharmacia Biotech). The cDNA probes for mouse LXRs were prepared by digesting the cloned full-length LXR cDNAs (CMV-LXRα and -β) with SalI and with EcoRI and NotI, respectively. The cDNA probes for human SREBP1, human SREBP2, human fatty acid synthase (FAS), and human acidic ribosomal phosphoprotein P0 (36B4) were prepared as previously described (33). The resulting bands were visualized by exposure to BAS2000 filters with BAStation software (Fuji Photo Film Co., Ltd.).
RESULTS
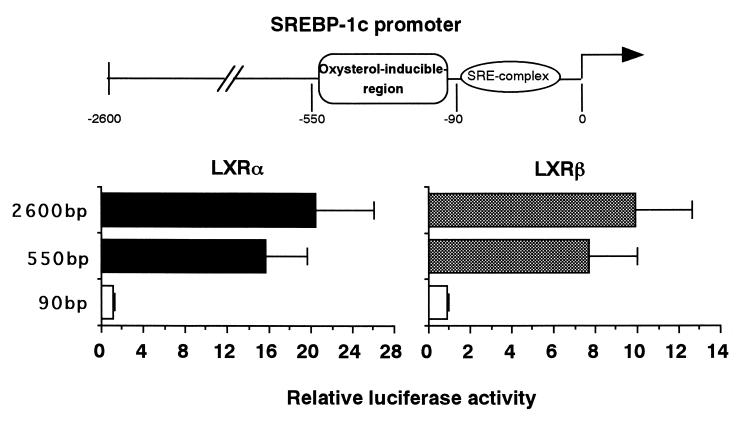
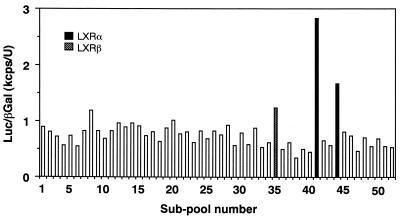
Our previous promoter analysis of the SREBP-1c gene 5′ flanking region identified an SRE complex which confers an autoloop response to nuclear SREBPs and another upstream region which was induced by the presence of oxysterols (1; see Fig. 4). The presence of two regions with opposing sterol regulation makes SREBP-1c gene expression very complex. The initial goal of this study was to isolate transcription factors that activate SREBP-1c expression. We used an expression cloning strategy of placing the luciferase gene containing 2.6 kb of the 5′ flanking region of the mouse SREBP-1c promoter in reporter gene assays. We previously observed that adipose tissues from SREBP-1 knockout mice highly expressed the aberrant SREBP-1c mRNA from the intact promoter of the disrupted SREBP-1c gene (33). Therefore, we assumed that these tissues might also abundantly express the transcription factors we were seeking. From mRNAs of these tissues, we constructed an expression cDNA library under the CMV promoter. Screening was performed using luciferase assays after the cotransfection of subpools of the library and the SREBP-1c promoter luciferase reporter gene into 293 cells. Figure 1 shows a representative result of the initial screening of subpools of the library. We screened roughly 210,000 clones and picked up six positive subpools of 1,500 clones that exhibited two- to threefold increases in luciferase activity of the SREBP-1c promoter reporter compared to background levels. After several rounds of screening, six single clones were isolated from these subpools. Each of these isolated clones increased the luciferase activity 10- to 20-fold above the background. DNA sequencing revealed that three of these six clones were LXRα and the remaining three were LXRβ. No other clones were identified in this system. Both LXRα and -β clones contained full-length cDNAs, were assumed to express their complete respective proteins in the assays, and were designated pCMV-LXRα and -β, respectively.
FIG. 4.
Activation of SREBP-1c promoter by LXR via the oxysterol-inducible region in the promoter. The previously identified oxysterol-inducible region and the SRE complex in the SREBP-1c promoter are illustrated at the top. pBP-1c2600-Luc and two shorter versions, pBP1c550- and pBP1c90-Luc, were prepared and used for transfection into 293 cells as described in Materials and Methods. Luciferase activity was measured and normalized to β-galactosidase activity. Fold induction of luciferase activity (mean ± standard deviation) by LXRs is shown.
FIG. 1.
Screening of an SREBP-1 knockout mouse adipose tissue expression library to identify cDNA clones which activate the SREBP-1c promoter. An expression cDNA library from the adipose tissues of SREBP-1-null mice was constructed as described in Materials and Methods. A luciferase reporter gene containing the mouse SREBP-1c promoter (2.6 kb), pBP1c2600-Luc, and each subpool containing approximately 1,500 cDNA clones were cotransfected into HEK 293 cells with pSV-βgal as a reference plasmid. Luciferase activity was measured and normalized by β-galactosidase activity. After several rounds of screening, six positive single clones were isolated from the library (three LXRα and three LXRβ). A representative result from the initial screening is shown.
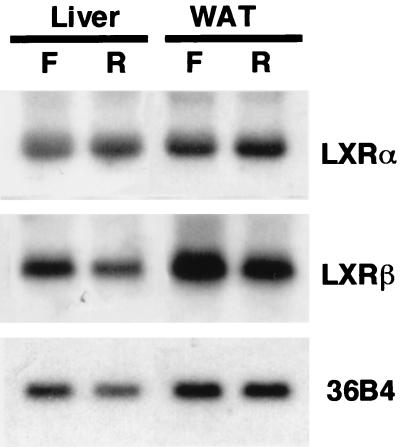
From a standpoint of nutritional regulation of these genes, we examined the expression of LXRα and -β in lipogenic organs, a location where the expression of SREBP-1c is suppressed by fasting and highly upregulated by refeeding. As shown in Fig. 2, both LXRα and -β were highly expressed in both livers and adipose tissues; however, the mRNA levels were not markedly changed by these dietary manipulations. Thus, expressions of LXRs in the lipogenic organs were not subjected to the nutritional change at fasting and refeeding, unlike SREBP-1c and peroxisome proliferator-activated receptor α.
FIG. 2.
Northern blot analysis of LXRs from livers and white adipose tissues (WAT) from mice fasted and refed. Total RNA was prepared and pooled (10 μg) equally from either livers or adipose tissues of normal mice fasted (F) or refed (R) after fasting as described previously (41), blotted to a nylon membrane, and hybridized with the indicated cDNA probes. A cDNA probe for 36B4 (acidic ribosomal phosphoprotein P0) was used as a loading control.
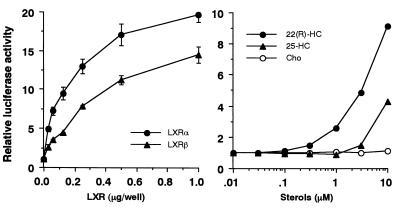
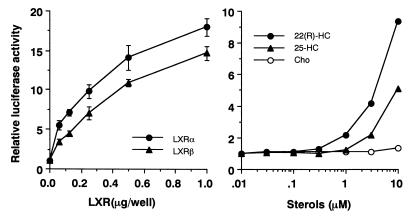
Studies were next focused on the effects of LXRα and -β on the SREBP-1c promoter. As an initial step, we tested the dose-dependent effects of LXRα and -β on the transcriptional activity of the SREBP-1c promoter (Fig. 3, left panel). Expression of both LXRα and -β resulted in a dose-dependent increase in the luciferase activity of the 2.6-kb SREBP-1c promoter in the range of 0.03 to 1 μg of DNA. SREBP-1c promoter activity was also induced by addition of 22(R)-HC, a well-established ligand for LXRs, to the medium in transfection studies with HEK 293 cells (Fig. 3, right panel). A marked dose-dependent increase in the luciferase activities was observed at 22(R)-HC concentrations between 1 and 10 μM, a dose range commonly used for activation of LXR. These data suggest that either the endogenous LXRα or -β, or both, in the transfected cells was activated by 22(R)-HC and was able to induce SREBP-1c promoter activity. 25-HC was a significant, but much weaker, activator, whereas cholesterol was essentially inactive. To evaluate the effect of free cholesterol, different doses of cholesterol were added to cells cultured in delipidated serum and the luciferase activity was measured. Up to 10 μM cholesterol did not activate the SREBP-1c promoter; however, 30 μM cholesterol slightly increased the luciferase activity (data not shown). These results are consistent with the previously reported affinities of these oxysterols for LXRs (15, 16, 22).
FIG. 3.
Dose-dependent activation of SREBP-1c promoter by LXRs (left panel) and their ligands (right panel). (Left panel) pBP1c2600-Luc was cotransfected into HEK 293 cells with a positive clone expressing LXR from the screening (pCMV-LXRα or pCMV-LXRβ), an empty vector (CMV-7) used as a control, or pSV-βgal, which was used as a reference plasmid. (Right panel) Ligands for LXR or ethanol (used as a control) were added to the cells after transfection of pBP1c2600-Luc and pSV-βgal in medium with 10% fetal bovine serum 24 h prior to the assay. After the incubation, luciferase activity was measured and normalized to β-galactosidase activity. The fold induction of luciferase activity by LXRs or their ligands (means ± standard deviations in the left panel [n = 3]; means in the right panel [n = 2]) compared to that of controls is shown. Cho, cholesterol.
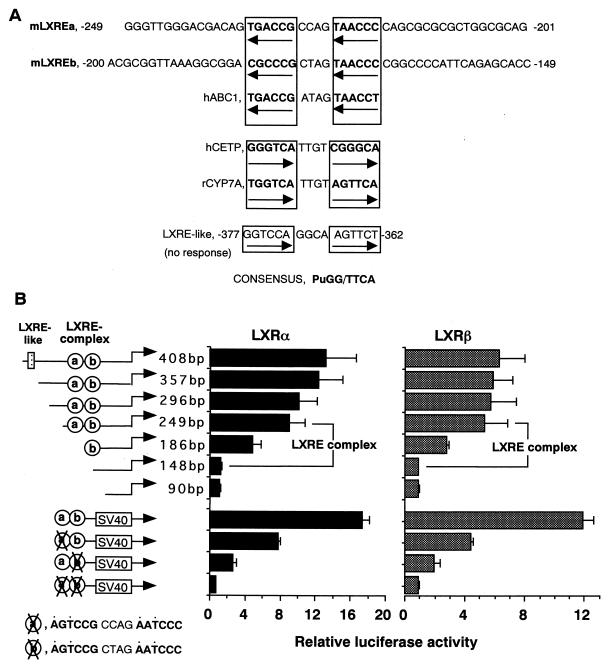
To identify the element in the SREBP-1c promoter that was responsible for LXR activation, we compared the LXR activation of the luciferase constructs with different sizes of the SREBP-1c promoter. Our previous study identified the sterol-inducible region between bp −550 and −90 (1; Fig. 4). Since LXR has been shown to be activated by sterols (15, 16, 22), 25-HC, and cholesterol, compounds commonly used for a sterol-suppressive condition, it was reasonable to assume that this region could be an LXR activation site. Initial luciferase assays with BP1c2600-Luc, BP1c550-Luc, and BP1c90-Luc demonstrated that LXR activation was observed with BP1c2600 and BP1c550, but not with BP1c90, confirming the presence of an LXR activation site between bp −550 and −90 (Fig. 4). Figure 5 shows luciferase activities after LXRα or -β coexpression in sequential-deletion-containing SREBP-1c promoter constructs assembled to identify the LXR activation sites. In the sterol-inducible region, we found a sequence at bp −377 to −362 which was similar to the LXRE (GGTCCAGGCAAGTTCT) (1). However, as shown in Fig. 5B, deletion of this sequence did not affect the LXR activation of the SREBP-1c promoter, indicating that this sequence was not involved. In contrast, considerable reduction in both LXRα and -β activation was observed when two regions, bp −249 to −187 and bp −186 to −148 were deleted. Loss of both regions essentially abolished the LXR activation, suggesting that these regions are crucial. As depicted in Fig. 5A, each candidate region contains a DR-4 sequence which was similar to the inverted LXRE. While this report was being prepared; a detailed promoter analysis of the ABC1 gene containing a new LXR-binding sequence in an inverted orientation was reported (6; Fig. 5A). LXREa, an upstream element of the two LXREs in the SREBP-1c promoter closely matched this new sequence (9 out of 10 bases in the two direct repeats), and the other (LXREb) also showed significant similarity (7 out of 10 bases).
FIG. 5.
Identification of LXREs by deletional and mutational analysis of the oxysterol-inducible region in the SREBP1c-promoter. (A) DNA sequencing of the oxysterol-inducible region of the SREBP1c promoter identified two elements, which were designated mLXREa and -b. These elements are highly similar to the LXRE of the human ABC1 promoter. (B) Sequential deletion and mutation analysis of the oxysterol-inducible region was performed with reporter genes with the indicated sizes and positions of the promoter. HEK 293 cells were transfected with each reporter plasmid, pCMV-LXR, and reference plasmid pSV-βgal and thereafter cultured for 24 h in medium with 10% fetal bovine serum. Luciferase activity was measured and normalized to β-galactosidase activity. Fold induction of luciferase activity by LXRs (means ± standard deviations) is shown. SV40, simian virus 40.
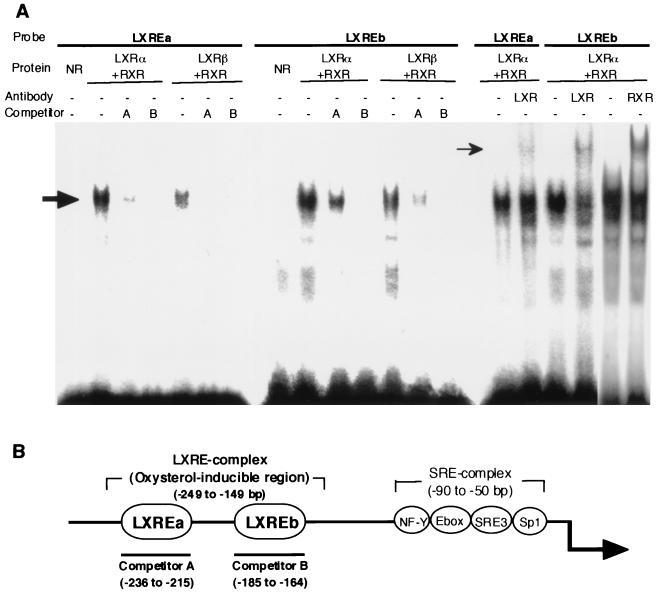
To show direct binding of LXRs to LXREa or LXREb, gel mobility shift assays were performed (Fig. 6). A labeled LXREa or LXREb probe was shifted by incubation of nuclear extracts from cells in which LXRα or -β and RXR were overexpressed. Coexpression of RXR was required for the binding of LXRs to LXREs (data not shown). Preincubation of the reaction mixture with antibodies against LXRα and -β or RXRα caused a supershift of the LXR-RXR DNA complex, confirming the specific binding of LXR-RXR to these LXREs, although the shifts caused by these antibodies were partial, as previously described (23). Competition assays suggest that LXREb has a slightly higher affinity for LXRs than does LXREa.
FIG. 6.
Binding of LXR to LXREs in the SREBP1c promoter as measured by gel mobility shift assay. (A) LXREa and LXREb in the SREBP-1c promoter were labeled and incubated with nuclear extracts from transfected 293 cells in which LXRα or -β and RXR were overexpressed. A molar excess of unlabeled LXREa or -b fragment and antibody against LXRα and -β or RXRα were added to the incubations in competition and supershift assays, respectively. The DNA-protein complexes were resolved in a 4.8% polyacrylamide gel. The shifted and supershifted LXRE probes are indicated by thick and thin arrows, respectively. NR, nuclear extract from mock-transfected cells used as a negative control. (B) LXREa and -b in the oxysterol-inducible region of the SREBP-1c promoter are shown.
Based upon these data, we tentatively designated the region between bp −249 and bp −149 containing the two LXR-binding sites an LXRE complex (Fig. 6B). An LXRE complex enhancer construct (pLXRE-Luc) in which the LXRE complex was inserted upstream of simian virus 40 promoter luciferase gene (pGL2 promoter vector) was prepared and tested for activation by LXR. Dose dependency and the effect of 22(R)-HC were essentially the same as those observed in the 2.6-kb SREBP-1c promoter construct (Fig. 7), indicating that this region basically represents the whole response of the SREBP-1c promoter to LXR. Mutation analysis was also performed on these two LXREs using the LXRE complex enhancer construct (pLXRE-Luc) (Fig. 5B). Mutation of either LXREa or LXREb significantly reduced the induction of luciferase activity by LXR expression. Mutation of both LXREs essentially abolished the effect of LXR. These results confirmed the role of both LXREs in LXR activation of the SREBP-1c promoter, although LXREb seemed relatively more important.
FIG. 7.
Dose-dependent activation of the LXRE complex enhancer in the SREBP-1c promoter by LXRs (left panel) and their ligands (right panel). The LXRE complex (LXREa and -b, bp −249 to −148) in the SREBP-1c promoter was fused to a luciferase reporter plasmid which contained a simian virus 40 promoter (pGL2 promoter vector; Fig. 5) to estimate LXRE enhancer activity (pLXRE-Luc). (Left panel) pLXRE-Luc was cotransfected into HEK 293 cells with pCMV-LXRα, pCMV-LXRβ, or an empty vector (CMV-7, used as a control) and pSV-βgal, which was used as a reference plasmid. (Right panel) Ligands for LXR or ethanol (used as a control) was added to the cells after transfection of pLXRE-Luc and pSV-βgal 24 h prior to the assay. After the incubation, luciferase activity was measured and normalized to β-galactosidase activity. Fold induction of luciferase activity by LXRs or their ligands (means ± standard deviations in the left panel [n = 3]; means in the right panel [n = 2]) compared to that of the control is shown. Cho, cholesterol.
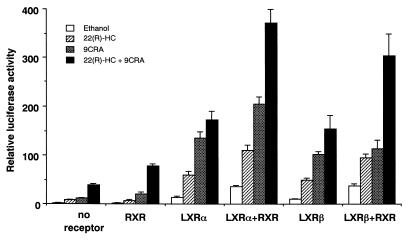
Since LXRs are obligated to heterodimerize with RXR in order to activate their target genes, the effects of coexpression of RXR and its ligand on LXR activation of LXRE-Luc were investigated (Fig. 8). Addition of 22(R)-HC or 9CRA to the medium caused some increase in this enhancer activity, and both ligands caused synergistic induction, presumably due to endogenous expression of LXR and RXR in 293 cells (Fig. 8, no receptor). RXR expression alone showed only slight additional effects on the activities of the respective ligands. LXRα or -β expression alone caused an ∼15-fold increase in the enhancer activity, which was consistent with the original result obtained with BP1c2600Luc. LXRα (or -β) expression with 22(R)-HC synergistically induced activation of the promoter 60-fold (50-fold), with further induction by RXR expression and 9CRA up to 170-fold (155-fold). There was no significant difference in transcriptional activity between LXRα and LXRβ. Finally, LXRα (or -β)-RXR coexpression caused a further additional effect, with a maximal effect of 370-fold (300-fold) induction in the presence of both 22(R)-HC and 9CRA. These data demonstrate that LXR and RXR and their ligands activate the SREBP-1c promoter activity in a synergistic fashion through the LXRE enhancer complex.
FIG. 8.
Synergistic activation of the LXRE complex in the SREBP-1c promoter by LXRs, RXR, and their ligands. The luciferase reporter construct containing the LXRE complex in the mouse SREBP-1c promoter (pLXRE-Luc) was cotransfected into HEK 293 cells with pCMV-LXRα or -β, pRXR, or both and pSV-βgal. After the transfection, the cells were incubated in the absence or presence of the ligand 22(R)-HC, 9CRA, or both for 24 h. Luciferase activity was measured and normalized to β-galactosidase activity. Fold induction (means ± standard deviations) compared to that of the control (an empty expression vector with no addition of ligands) is shown.
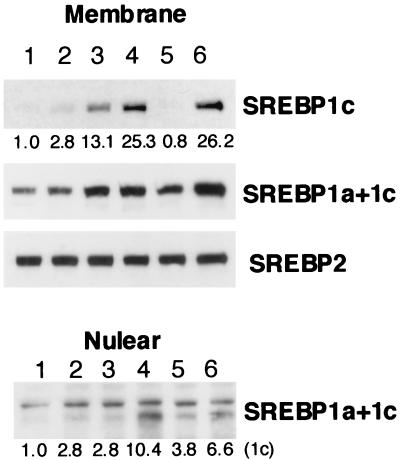
To evaluate the consequences of LXR-RXR activation, the effects of LXR and/or RXR ligands on endogenous SREBP-1c expression were estimated in HepG2 cells. As shown in Fig. 9, SREBP-1c precursor protein, as estimated by immunoblot analysis of cell membranes with mouse SREBP-1c-specific antibody, was barely detectable in HepG2 cells but was slightly induced by addition of the LXR ligand 22(R)-HC. Interestingly, the RXR ligand 9CRA induced the SREBP-1c protein to a greater extent than did 22(R)-HC and combination of the two ligands had an additive effect. LXR-RXR induction of SREBP-1, as demonstrated by an SREBP-1 antibody that detects both SREBP-1a and -1c, was less marked, suggesting that SREBP-1a was not regulated by those ligands. SREBP-2 precursor protein was not regulated by LXR either. The data suggest that LXR-RXR activation is specific to SREBP-1c. Nuclear SREBP-1c protein was detected in nuclear extracts from the HepG2 cells treated with both 22(R)-HC and 9CRA, demonstrating that induced SREBP-1c expression by activation of LXR-RXR could result in the cleavage and movement of the mature form of SREBP-1c protein to the nucleus.
FIG. 9.
Induction of SREBP-1c protein by LXR-RXR ligands in HepG2 cells as measured by immunoblot analysis. HepG2 cells were incubated with the indicated ligands for LXR or RXR for 24 h. In some groups, 1 μg of 25-HC per ml and 10 μg of cholesterol per ml were also added to the medium to suppress sterol-regulatory cleavage activity of SREBPs (sterol-suppressed condition). After the incubation, nuclear extracts and membranes were prepared from the cells and aliquots (30 μg) of protein were subjected to immunoblot analysis using antibodies against SREBP-1c, SREBP-1a and -1c, and SREBP-2. The bands were visualized with the ECL system. The values below the bands refer to relative fold changes in protein levels. Lanes: 1, ethanol; 2, 22(R)-HC; 3, 9CRA; 4, 22(R)-HC plus 9CRA; 5, ethanol in a sterol-suppressed condition; 6, 22(R)-HC plus 9CRA in a sterol-suppressed condition.
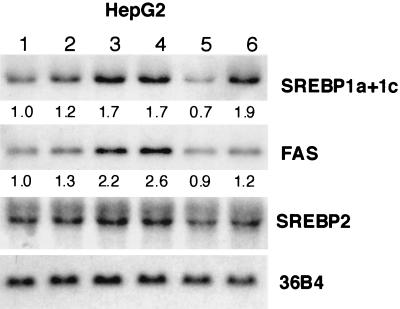
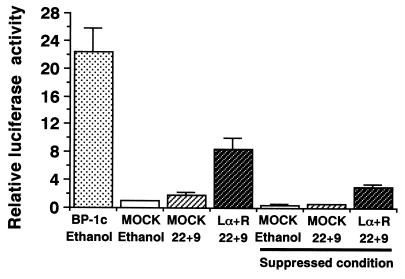
Northern blot analysis demonstrated that addition of both 22(R)-HC and 9CRA induced mRNAs of the SREBP-1 gene and the gene for FAS, a representative SREBP-1 target lipogenic gene, but not SREBP-2 gene mRNA (Fig. 10). HepG2 cells were transfected with the FAS promoter-luciferase gene to evaluate the transcriptional activity of LXR-induced nuclear SREBP-1c. As shown in Fig. 11, addition of 22(R)-HC and 9CRA to the medium caused only a marginal increase in the FAS-Luc activity. However, coexpression of LXRα and -β with these ligands markedly increased the luciferase activity up to one-third of the level of activation caused by by direct overexpression of nuclear SREBP-1c (CMV-SREBP1c). This induction was significant even in the presence of 25-HC and cholesterol, a condition that suppresses endogenous SREBP-1a and -2 activities by inhibiting the sterol-regulatory cleavage of SREBPs. These data demonstrate that LXR-RXR induction of SREBP-1c can potentially increase its target gene expression regardless of sterol regulation.
FIG. 10.
SREBP-1, -2, and FAS mRNA levels in HepG2 cells treated with LXR or RXR ligands as measured by Northern blot analysis. HepG2 cells were incubated with the indicated ligands for LXR or RXR for 24 h. In some groups, 25-HC at 1 μg/ml and cholesterol at 10 μg/ml were added to the medium to suppress the sterol-regulatory cleavage activity of SREBPs (sterol-suppressed condition). Total RNA was prepared and pooled (10 μg) equally from HepG2 cells, blotted to a nylon membrane, and then hybridized with the indicated cDNA probes. The values below the bands refer to relative fold changes in mRNA levels normalized relative to the 36B4 signal. Lanes: 1, ethanol; 2, 22(R)-HC; 3, 9CRA; 4, 22(R)-HC plus 9CRA; 5, ethanol in a sterol-suppressed condition; 6, 22(R)-HC plus 9CRA in a sterol-suppressed condition. A cDNA probe for 36B4 (acidic ribosomal phosphoprotein P0) was used as a loading control.
FIG. 11.
Activation of the FAS promoter by LXRs and their ligands. A luciferase reporter construct containing the rat FAS promoter (31) and reference plasmid pSV-βgal with empty plasmid pCMV7 or expression plasmids pCMV-LXR and pRXR were transfected into HepG2 cells. After the transfection, ligands for LXR and RXR or ethanol (used as a negative control) was added to the cells, followed by a 24-h incubation in the medium with 10% fetal bovine serum. In some sets, 25-HC at 1 μg/ml and cholesterol at 10 μg/ml were also added to the medium to suppress the sterol-regulatory cleavage activity of SREBPs (sterol-suppressed condition). Luciferase activity was measured and normalized to β-galactosidase activity. Fold induction (means ± standard deviations) by LXRs is shown. pCMV-nSREBP-1c, an expression plasmid of human nuclear SREBP-1c was used as a positive control for FAS promoter activation (BP-1c). Lα+R, LXRα plus RXR; 22+9, 22(R)-HC plus 9CRA.
DISCUSSION
The current studies clearly demonstrate that LXR-RXR activates SREBP-1c expression by binding to newly identified LXR-binding sites in the SREBP-1c promoter. In our expression cloning system from mouse adipose tissue with a reporter gene containing the native SREBP-1c promoter (2.6 kb), we could isolate only LXRα and -β, suggesting that LXRs are abundant and potent activators of the SREBP-1c promoter. The LXRE complex, composed of two inverted DR4-type LXREs at approximately a 30-bp interval. Both binding sites contributed to activation of the SREBP-1c promoter. The promoters of the LXR target genes which have been analyzed to date contain only a single LXRE (6, 22, 23). The presence of two copies of LXREs in the LXRE complex could make the response of the SREBP-1c gene to LXR-RXR strong.
The importance of LXR for expression of SREBP-1 has already been recognized in a previous report on LXRα knockout mice (25). In the LXRα knockout mice, induction of cholesterol 7α-hydroxylase was impaired, resulting in hepatic accumulation of cholesterol when dietary cholesterol was consumed. One intriguing observation in the livers of these mice was a considerable reduction of SREBP-1 mRNA and protein levels, leading to a decrease in some lipogenic enzyme mRNA levels. Our current data on activation of SREBP-1c expression by LXRs can explain this observation. Furthermore, the LXRα-deficient mouse data suggest that LXRα is an important regulator of SREBP-1c expression in the liver because LXRβ cannot entirely compensate for the absence of LXRα. It would be interesting to see a tissue survey of SREBP-1c expression in LXRα- and -β-deficient mice, as well as mice doubly deficient in both LXRs, to estimate the contribution of these two factors to SREBP-1c expression in different tissues.
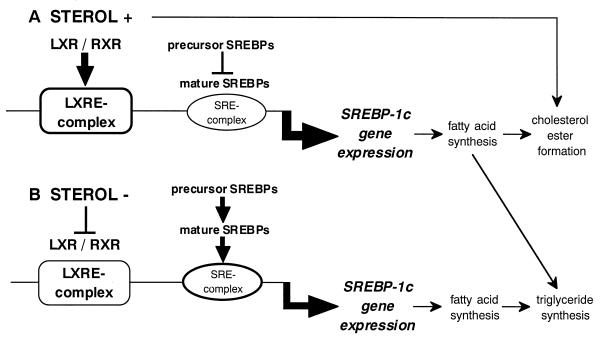
It is intriguing to speculate about the physiological significance of SREBP-1c induction by LXR. Recent studies identifying the genes for 7α-cholesterol hydroxylase (25), cholesterol ester transfer protein (23), and ABC1 (6, 26) as target genes of LXRs suggest that LXR-RXR seems to play an important role in cellular adaptation to the accumulation of excess cholesterol by enhancing catabolism of cholesterol into bile acid (23, 25) or by promoting cholesterol efflux (26, 29). SREBP-1c is a transcription factor for lipogenesis, and its induction should cause an increase in the synthesis of fatty acids. These fatty acids can then provide a source for acyl residues to produce cholesterol esters from cholesterol and protect cells from the cytotoxic effect of free cholesterol accumulation (Fig. 12A). On the other hand, in cells with a cholesterol overload leading to activation of the cholesterol efflux system, cholesterol synthesis is negatively regulated through suppression of cleavage of SREBPs by a sterol-sensing and regulation system consisting of SCAP and site 1 protease. This reduction in SREBP cleavage activity should counterbalance the effect of LXR induction of the SREBP-1c precursor protein. However, our recent findings suggest that cleavage of SREBP-1c is not as tightly regulated by sterols as is that of SREBP-2 in liver cell lines, suggesting that nuclear SREBP-1c has a chance to liberate its nuclear active form into the nucleus even in a state of cholesterol accumulation (10). This is supported by our current finding that transfection of LXR-RXR into HepG2 cells with addition of 22(R)-HC increased the amount of endogenous nuclear SREBP-1c and the resultant luciferase activity of its downstream gene even in a sterol-suppressed condition (Fig. 11). Taken together, these data allow us to hypothesize that LXR-RXR activation of SREBP-1c might have a physiologic function situations of excess cellular cholesterol accumulation (Fig. 12A). While this would result in an overall decrease in the cleavage of SREBPs, the lack of complete control of sterols over the SREBP-1c precursor, combined with the ability of LXRs to simulate SREBP-1c expression, would ensure that a certain amount of nuclear SREBP-1c was available to stimulate the production of fatty acids, presumably to protect cells from the deposition of free cholesterol to a toxic level. On the other hand, when cells are depleted of cholesterol, activation of the cleavage system will increase nuclear SREBPs, including SREBP-1c, to maintain lipogenesis even in the absence of LXR activation (Fig. 12B). As illustrated in Fig. 12, this multiple-complex system for SREBP-1c promoter activity links SREBP-1c expression, and thus lipogenesis, partially to sterol regulation but also to some levels of control irrespective of sterols. Considering that the list of SREBP-1 target genes seems to be expanding, it is also possible that LXR activation of SREBP-1c could activate unknown target genes to cause as yet unidentified physiological consequences.
FIG. 12.
Dual regulation of SREBP-1c promoter by LXR-RXR and SREBPs. In the presence of excess amount of sterols, presumably a concomitant increase in oxysterol levels activates the SREBP-1c promoter through the LXRE complex while the cleavage activity of SREBPs is suppressed (A). When cells are deprived of sterols, increased nuclear SREBPs activate the SREBP-1c promoter through the SRE complex (B). The dual activation of the SREBP-1c promoter through LXRE and SRE complexes ensures the maintenance of lipogenesis irrespective of cellular sterol levels.
Oxysterol receptors are involved in cholesterol catabolism (activation of 7α-cholesterol hydroxylase for bile acid synthesis and ABC1 for the reverse cholesterol transporter [6, 25, 26]), with the ligands 22(R)-HC, 24(S)-HC, and 24,25(S)-epoxycholesterol demonstrating the highest affinities for LXRs (15, 16, 22). In contrast, 25-HC is the most potent known suppressor of SREBP-2 in the regulation of cholesterol synthesis (42). The cause for different affinities of oxysterols in the regulation of cholesterol catabolism and synthesis in response to the same cholesterol accumulation is currently unknown. Further investigation to seek out other ligands for LXR could lead to the discovery of a common and real physiological signaling molecule for cholesterol metabolism. Meanwhile, our current study suggests that the expression level of LXRs, in addition to their ligand concentrations, can be another potential method for the regulation of LXR target genes.
It is noteworthy that in some experiments, RXR and its ligand exhibited an effect similar to or even higher than those of LXRs and their ligands on SREBP-1c activation. Since RXR forms heterodimers with different nuclear receptors such as retinoic acid receptor, peroxisome proliferator-activated receptors, farnesoid X receptor, and some orphan receptors, the effect of RXR activation should be diverse. Therefore, it was also possible that an alternative heterodimerizing nuclear receptor might be involved in SREBP-1c activation. Tissue specificity of copartners for RXR heterodimers might be crucial for the physiological consequences of RXR manipulation. SR=BP-1c seems to be involved in energy disturbances causing fatty liver, remnant particle production, and insulin resistance and may therefore be a therapeutic target for these fuel metabolism disorders. Together with recent reports on the activation of ABC1 as a reverse cholesterol transporter, LXR-RXR could be a useful tool with clinical application in the control of cholesterol and fuel metabolic disorders.
The current studies raise the possibility of a novel link between lipogenesis and sterol regulation through SREBP-1c and LXR-RXR. Its physiological relevance should be tested in different tissues in vivo. Investigation of the involvement of the LXRE complex in the nutritional regulation of SREBP-1c should be investigated. It might open up a new aspect of the roles of oxysterols and a novel link between lipogenesis and sterol regulation through SREBP-1c and LXR-RXR.
ACKNOWLEDGMENTS
We thank N. Emoto and A. Amemiya for great help in construction of the expression library.
This study was supported by the Promotion of Fundamental Studies in Health Science of the Organization for Pharmaceutical Safety and Research and health sciences research grants (Research on Human Genome and Gene Therapy) from the Ministry of Health and Welfare.
ADDENDUM
During the manuscript review process, activation of SREBP-1c expression by LXRs was reported in studies using a pharmacological LXR agonist as well as mice deficient in LXRα, LXRβ, or both (25a, 28a). The researchers also studied the SREBP-1c promoter and found one of the LXREs that we identified in the current study. Their data, from a different approach to LXRs, and our present SREBP-1c promoter analysis data are basically consistent and confirm each other.
REFERENCES
- 1.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty A H, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Osuga J-I, Harada K, Gotoda T, Sato R, Kimura S, Ishibashi S, Yamada N. Promoter analysis of the mouse sterol regulatory element-binding protein (SREBP)-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S, Davis D L, Dahlback H, Jornvall H, Russell D W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Brown M S, Goldstein J L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 5.Brown M S, Ye J, Rawson R B, Goldstein J L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 6.Costet P, Luo Y, Wang N, Tall A R. Sterol-dependent transactivation of the human ABC1 promoter by LXR/RXR. J Biol Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 7.Foretz M, Guichard C, Ferre P, Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci USA. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Lièpvre X, Berthelier-Lubrano C, Spiegelman B, Kim J B, Ferré P, Foufelle F. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodridge A G. Dietary regulation of gene expression: enzymes involved in carbohydrate and lipid metabolism. Annu Rev Nutr. 1987;7:157–185. doi: 10.1146/annurev.nu.07.070187.001105. [DOI] [PubMed] [Google Scholar]
- 10.Hasty A H, Shimano H, Yahagi N, Amemiya-Kudo M, Perrey S, Yoshikawa T, Osuga J-I, Okazaki H, Tamura Y, Iizuka Y, Shionoiri F, Ohashi K, Harada K, Gotoda T, Nagai R, Ishibashi S, Yamada N. Sterol regulatory element-binding-1 protein is regulated by glucose at the transcriptional level. J Biol Chem. 2000;275:31069–31077. doi: 10.1074/jbc.M003335200. [DOI] [PubMed] [Google Scholar]
- 11.Hillgartner F B, Salati L M, Goodridge A G. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 12.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton J D, Shimomura I, Brown M S, Hammer R E, Goldstein J L, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Investig. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua X, Yokoyama C, Wu J, Briggs M R, Brown M S, Goldstein J L, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci USA. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janowski B A, Grogan M J, Jones S A, Wisely G B, Kliewer S A, Corey E J, Mangelsdorf D J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 17.Kim H J, Takahashi M, Ezaki O. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. A possible mechanism for down-regulation of lipogenic enzyme mRNAs. J Biol Chem. 1999;274:25892–25898. doi: 10.1074/jbc.274.36.25892. [DOI] [PubMed] [Google Scholar]
- 18.Kim J B, Sarraf P, Wright M, Yao K M, Mueller E, Solanes G, Lowell B B, Spiegelman B M. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Investig. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J B, Spiegelman B M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 20.Kim J B, Spotts G D, Halvorsen Y-D, Shih H-M, Ellenberger T, Towle H C, Spiegelman B M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo S-H, Towle H C. Glucose regulation of mouse S14 gene expression in hepatocytes: involvement of a novel transcription factor complex. J Biol Chem. 2000;275:5200–5207. doi: 10.1074/jbc.275.7.5200. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann J M, Kliewer S A, Moore L B, Smith-Oliver T A, Oliver B B, Su J L, Sundseth S S, Winegar D A, Blanchard D E, Spencer T A, Willson T M. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, Tall A R. Sterol upregulation of human CETP expression in vitro and in transgenic mice by an LXR element. J Clin Investig. 2000;105:513–520. doi: 10.1172/JCI8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peet D J, Turley S D, Ma W, Janowski B A, Lobaccaro J M, Hammer R E, Mangelsdorf D J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 25a.Repa J J, Liang G, Ou J, Bashmakov Y, Lobaccaro J A, Shimomura I, Shan B, Brown M S, Goldstein J L, Mangelsdorf D J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Repa J J, Turley S D, Lobaccaro J A, Medina J, Li L, Lustig K, Shan B, Heyman R A, Dietschy J M, Mangelsdorf D J. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 27.Russell D W. Nuclear orphan receptors control cholesterol catabolism. Cell. 1999;97:539–542. doi: 10.1016/s0092-8674(00)80763-1. [DOI] [PubMed] [Google Scholar]
- 28.Sakai J, Duncan E A, Rawson R B, Hua X, Brown M S, Goldstein J L. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 28a.Schultz J R, Tu H, Luk A, Repa J J, Medina G J C, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf D J, Lusting K D, Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz K, Lawn R M, Wade D P. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem Biophys Res Commun. 2000;274:794–802. doi: 10.1006/bbrc.2000.3243. [DOI] [PubMed] [Google Scholar]
- 30.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M, Goldstein J L. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Investig. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Investig. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimano H, Shimomura I, Hammer R E, Herz J, Goldstein J L, Brown M S, Horton J D. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Investig. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty A, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, Gotoda T, Ishibashi S, Yamada N. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 34.Shimomura I, Bashmakov Y, Ikemoto S, Horton J D, Brown M S, Goldstein J L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimomura I, Hammer R E, Richardson J A, Ikemoto S, Bashmakov Y, Goldstein J L, Brown M S. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Investig. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thewke D P, Panini S R, Sinensky M. Oleate potentiates oxysterol inhibition of transcription from sterol regulatory element-1-regulated promoters and maturation of sterol regulatory element-binding proteins. J Biol Chem. 1998;273:21402–21407. doi: 10.1074/jbc.273.33.21402. [DOI] [PubMed] [Google Scholar]
- 38.Tontonoz P, Kim J B, Graves R A, Spiegelman B M. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Briggs M, Hua X, Yokoyama C, Goldstein J, Brown M. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem. 1993;268:14497–14504. [PubMed] [Google Scholar]
- 40.Worgall T S, Sturley S L, Seo T, Osborne T F, Deckelbaum R J. Polyunsaturated fatty acids decrease expression of promoters with sterol regulatory elements by decreasing levels of mature sterol regulatory element-binding protein. J Biol Chem. 1998;273:25537–25540. doi: 10.1074/jbc.273.40.25537. [DOI] [PubMed] [Google Scholar]
- 41.Yahagi N, Shimano H, Hasty A, Amemiya-Kudo M, Okazaki H, Tamura Y, Iizuka Y, Shionoiri F, Ohashi K, Osuga J, Harada K, Gotoda T, Nagai R, Ishibashi S, Yamada N. A crucial role of sterol regulatory element-binding protein-1 in the regulation of lipogenic gene expression by polyunsaturated fatty acids. J Biol Chem. 1999;274:35840–35844. doi: 10.1074/jbc.274.50.35840. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Sato R, Goldstein J L, Brown M S. Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- 43.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]