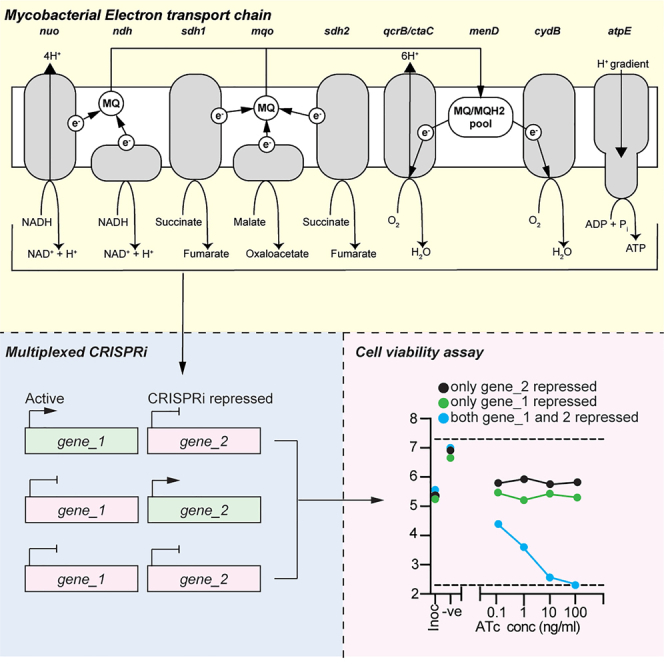
Summary
Mycobacterium tuberculosis remains a leading cause of infectious disease morbidity and mortality for which new drug combination therapies are needed. Combinations of respiratory inhibitors can have synergistic or synthetic lethal interactions with sterilizing activity, suggesting that regimens with multiple bioenergetic inhibitors could shorten treatment times. However, realizing this potential requires an understanding of which combinations of respiratory complexes, when inhibited, have the strongest consequences on bacterial growth and viability. Here we have used multiplex CRISPR interference (CRISPRi) and Mycobacterium smegmatis as a physiological and molecular model for mycobacterial respiration to identify interactions between respiratory complexes. In this study, we identified synthetic lethal and synergistic interactions between respiratory complexes and demonstrated how the engineering of CRISPRi-guide sequences can be used to further explore networks of interacting gene pairs. These results provide fundamental insights into the functions of and interactions between bioenergetic complexes and the utility of CRISPRi in designing drug combinations.
Subject areas: Tropical medicine, Drugs, Microbiology
Graphical abstract
Highlights
-
•
We identified interactions between respiratory complexes using multiplexed CRISPRi
-
•
CRISPRi sgRNAs with reduced efficacy expanded the network of interacting complexes
-
•
Strength of lethal interactions is influenced by gene pair and metabolic state
Tropical medicine; Drugs; Microbiology
Introduction
Mycobacterium tuberculosis remains a significant health problem (WHO, 2018). Although a curable disease requiring a combination of four drugs for six months, drug-resistant strains of M. tuberculosis require long treatment times with often highly toxic drugs. In addition to new drugs, rational approaches to the design of combination therapies are needed to shorten the length of treatment and prevent the emergence of drug resistance.
To generate sufficient energy to support aerobic growth, mycobacterial respiration couples the oxidation of electron donors from organic carbon catabolism (including NADH, succinate and malate) to the reduction of oxygen as a terminal electron acceptor (Cook et al., 2014, Cook et al., 2017). Mycobacteria encode several primary dehydrogenases that can donate electrons to the electron transport chain (ETC) via the electron carrier menaquinone (Cook et al., 2014, Cook et al., 2017). This includes two classes of NADH:menaquinone oxidoreductases that couple the oxidation of NADH to the reduction of menaquinone i.e., a proton-pumping type I NADH dehydrogenase (NDH-1) and a non-proton pumping type II NADH dehydrogenase (NDH-2); two succinate dehydrogenase enzymes that couple the oxidation of succinate to the reduction of menaquinone i.e., SDH-1 and SDH-2 and a malate:quinone oxidoreductase (MQO) that couples the oxidation of malate to the reduction of menaquinone (Cook et al., 2014; Cook et al., 2017; Hartman et al., 2014). Under aerobic conditions, a supercomplex of cytochrome bc1 (Complex III; qcrBCD) and a aa3-type cytochrome c oxidase (Complex IV; ctaBCDE) couples the transfer of electrons from reduced menaquinol to cytochrome c to proton translocation and the generation of a proton motive force (PMF) (Wiseman et al., 2018; Gong et al., 2018). An alternative cytochrome bd-type menaquinol oxidase (cydAB) functions as a purported higher affinity terminal oxidase under low oxygen conditions (Kalia et al., 2017). The generated proton gradient is used to drive ATP synthesis via the F1Fo-ATP synthase (atpBEFHAGDC).
Therapeutic targeting of mycobacterial respiration holds significant clinical promise, as highlighted by the FDA approval of the inhibitors bedaquiline (inhibitor of F1Fo-ATP synthase) (Andries et al., 2005; Cox and Laessig, 2014), delamanid (Stover et al., 2000), and the clinical testing of Q203 (inhibitor of Complex III) (Pethe et al., 2013; de Jager et al., 2020). Combinations of respiratory inhibitors can also have synergistic or synthetic lethal interactions against both replicating and non-replicating cells. For example, genetic or chemical inhibition of the terminal oxidase, cytochrome bd has synthetic lethal interactions in M. tuberculosis with the bacteriostatic inhibitor Q203 resulting in rapid cell death against both replicating and non-replicating cells in vitro and in murine infection models (Kalia et al., 2017; Lee et al., 2021). New regimens with multiple respiratory inhibitors have the potential to rapidly kill M. tuberculosis. However, realizing this potential requires an understanding of which combinations of respiratory complexes, when inhibited, have the strongest consequences on bacterial growth and viability.
To address this lack in knowledge we have utilized a combination of mycobacterial CRISPR interference (CRISPRi) and phenotypic assays of bacterial growth and viability to identify respiratory complexes that when simultaneously inhibited result in cell death (Rock et al., 2017; McNeil and Cook, 2019; de Wet et al., 2020). These results demonstrate that the simultaneous transcriptional inhibition of the supercomplex III-IV (i.e., qcrB) in combination with cydB or ndh2 produces a synthetically lethal interaction that results in cell death. However, the strength of synthetic lethal interactions is influenced by the specific gene pair being targeted and mycobacterial metabolism. We also demonstrate the bactericidal consequences of inhibiting ndh2 or atpE and how sgRNA engineering to produce hypomorphic bacteriostatic sgRNAs can further resolve networks of interacting gene pairs. These results provide fundamental insights into the functional interactions between respiratory complexes, the clinical promise of targeting these processes and the general utility of CRISPRi in designing new combination therapies.
Results
Transcriptional repression of respiratory complexes in M. smegmatis has both bacteriostatic and bactericidal consequences
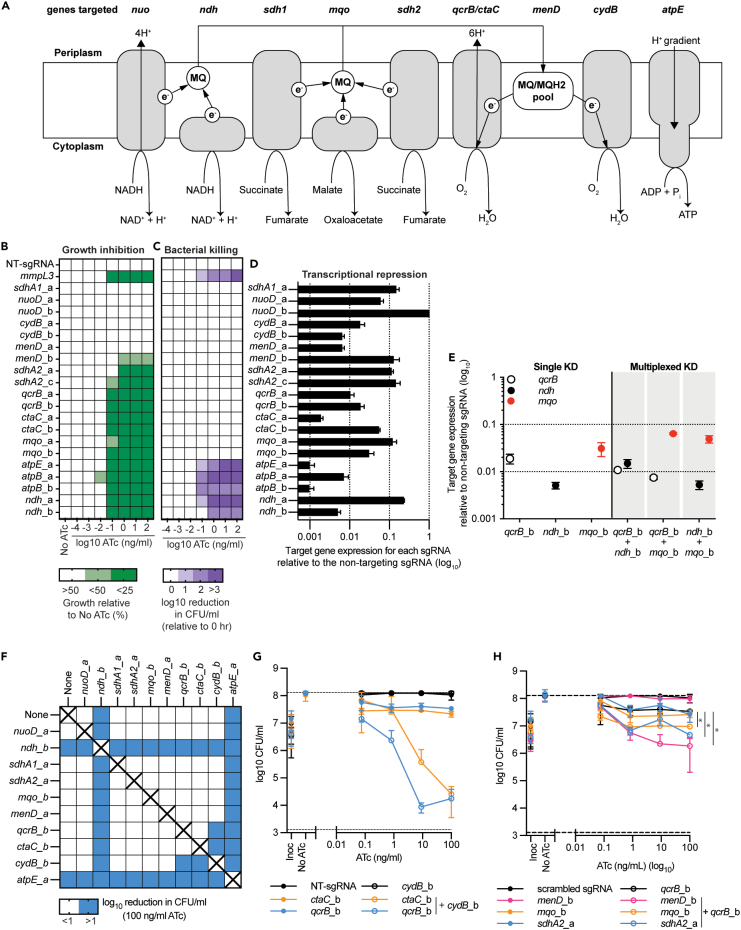
Using Mycobacterium smegmatis as physiological model for mycobacterial respiration and ATP synthesis, we investigated the consequences of transcriptionally inhibiting genes involved in mycobacterial respiration (Figure 1A). Transcriptional repression was achieved using mycobacterial CRISPRi that uses 20–25 nucleotide sequences with complementarity to the non-template strand of target genes, termed sgRNA, to guide a deactivated Cas9 nuclease from Streptococcus thermophilus CRISPR1 (dCas9) to bind target genes (Rock et al., 2017; McNeil et al., 2020; McNeil and Cook, 2019). Target-bound sgRNA-dCas9 impedes the progression of RNA polymerase, thereby inhibiting transcription. Both dCas9 and the sgRNA are expressed from a single integrative plasmid under the control of anhydrotetracycline (ATc)-inducible promoters (Rock et al., 2017). Two sgRNAs per target gene were individually cloned into the CRISPRi plasmid (i.e., pLJR962). Consequences of transcriptional repression on bacterial growth and viability were determined when grown in HdeB-minimal media using succinate as a sole, non-fermentable, carbon source in a 96-well plate format. Targeting nuoD (i.e., complex I), sdhA1 (i.e., SDH1, complex II), cydB (i.e., terminal oxidase) or menD (i.e., menaquione biosynthesis) had no effect on cell growth or viability (Figures 1B and 1C). A single sgRNA targeting menD (i.e., menD_b) impaired growth, but had no effect on CFU/ml (Figures 1B and 1C). With the exception of menD, these results are consistent with previously published deletion mutants and high-throughput transposon mutagenesis, confirming that these genes are dispensable for growth under the tested conditions (Pecsi et al., 2014; Kana et al., 2001). Transcriptional inhibition of ndh2, sdhA2 (i.e., SDH2, Complex II), mqo, qcrB (i.e., Complex III), ctaC (i.e., complex IV), atpE, or atpB (ATP synthase) resulted in impaired bacterial growth (Figure 1B). The transcriptional inhibition of sdhA2, mqo, qcrB, and ctaC had bacteriostatic effects on cell viability with no reduction in detectable CFU (Figure 1C). The transcriptional inhibition of ndh, atpB, and atpE had bactericidal consequences on cell viability, with >1 log reduction in CFU/ml (Figure 1C). For atpB and atpE, this is consistent with our previously published work (McNeil et al., 2020). In contrast to M. tuberculosis, M. smegmatis has only a single gene encoding NDH-2, yet the bactericidal phenotype of inhibiting ndh2 in this study is consistent with the CRISPRi targeting of ndh in M. tuberculosis (McNeil et al., 2021). Transcriptional analysis demonstrated that all sgRNAs, with the exception of nuoD_b, repressed target gene expression at least 10-fold relative to a non-targeting sgRNA control, with an absence of correlation between transcriptional repression and phenotype (Figure 1D). This suggests that variation in CRISRPi transcriptional repression does not influence the observed phenotype. In conclusion, CRISRPi is able to identify essential genes involved in mycobacterial respiration that have either a bacteriostatic or bactericidal consequence on cell viability.
Figure 1.
Individual and simultaneous transcriptional repression of respiratory complexes in M. smegmatis
(A) Schematic of the genes targeted in this study and their functions in the mycobacterial electron transport chain. MQ: menaquinone, e−: electron, H+: proton. Arrowheads denote the direction of the catalyzed reaction, electron transfer or proton movement.
(B) Growth of M. smegmatis expressing sgRNAs from a starting OD600 of 0.005 in 96 well plates in a total volume of 100 μL with differing levels of ATc. Results are the mean ± standard deviation of four replicates, expressed relative to a no-ATc control. Increasing shades of green represent increasing levels of growth inhibition. White is representative of no growth impairment.
(C) Viability of M. smegmatis expressing sgRNAs as described for (B). Increasing shades of purple represent increasing levels of reduction in CFU/ml. White is representative of no reduction in CFU/ml. For B and C, results are mean ± standard deviation of four replicates with either non-targeting (NT) or mmpL3 targeting sgRNA that were used as negative and positive-bactericidal controls, respectively.
(D) mRNA levels of target genes following CRISPRi induction of stated sgRNA when induced with 100 ng/mL ATc and grown in LBT. Bar length represents the level of mRNA for each target gene with the stated sgRNA relative to a non-targeting sgRNA. 1 is representative of no change, whereas 0.1, 0.01, and 0.001 is representative of a 1, 2, and 3 log10 reduction in detectable mRNA, respectively. Results are the mean ± standard deviation of three technical replicates. (B–D) Labels a and b represent the guides a and b targeting each genetic target.
(E) Transcriptional analysis of qcrB, ndh, and mqo in either single or multiplexed knockdown strains. Results of targeted genes are expressed relative to a non-targeting control in the presence of 100 ng/mL ATc and are the mean ± SD of three technical replicates.
(F) Log10 reduction (0–26 h) in CFU/ml for single and multiplex strains in the presence of 100 ng/mL ATc. Blue shading represents a gene pair that when simultaneously repressed has at least a one-log reduction in CFU/ml. sgRNAs paired with “none” denote results from strains that express a single sgRNA. Combinations marked with “X” were not tested.
(G–H) CFU/ml plots of selected single and multiplexed KD against increasing concentrations of ATc. A strain expressing a non-targeting (NT) sgRNA is used as a negative control. (G) Expression of sgRNAs qcrB_b and ctaC alone or in combination with sgRNAs cydB_b. (H) Expression of sgRNAs menD_b, mqo_b, and sdhA2_a alone or in combination with sgRNA qcrB_b. For G and H, results are the mean ± SD of at least four biological replicates. ∗ denotes statistically significant difference with a p value of <0.05 between the average CFU/ml at 100 ng/mL for the multiplex sgRNA compared to the qcrB_b sgRNA as determined by an unpaired t test.
Simultaneous inhibition of mycobacterial respiratory complexes identifies interacting gene pairs
To identify synergistic or synthetically lethal interactions between mycobacterial respiratory complexes we constructed multiplexed CRISPRi plasmids that simultaneously repress the expression of two target genes. Transcriptional analysis of multiplexed strains demonstrated that target genes are repressed to comparable levels as observed in the single KD strains (Figure 1E). Phenotypic analysis demonstrated that all multiplexed combinations with at least a single essential gene repressed mycobacterial growth (Figures S1–S6). Analysis of cell viability demonstrated that the simultaneous transcriptional repression of qcrB or ctaC with cydB resulted in bacterial killing (Figures 1F and 1G), whereas repression of individual complexes had a bacteriostatic or non-inhibitory phenotype (Figures 1F and 1G). Although there were no detectable reductions in CFU/ml, multiplexed strains of qcrB or ctaC with sdhA2, menD, or mqo exhibited a growth defect that required at least five days of incubation before visible colonies could be counted, in contrast to the typical three days of incubation required of other strains. These multiplexed interactions also had an additive or synergistic interaction with a stronger bacteriostatic phenotype than the single sgRNAs alone (i.e., the single knockdowns had a 0.5–1 log increase in CFU/ml) (Figures 1H and S3). The bactericidal phenotypes of ndh and atpE prevented the observation of interactions with other respiratory complexes (Figures 1F, S7A, and S7B). In conclusion, multiplex CRISPRi identifies interactions between mycobacterial respiratory components.
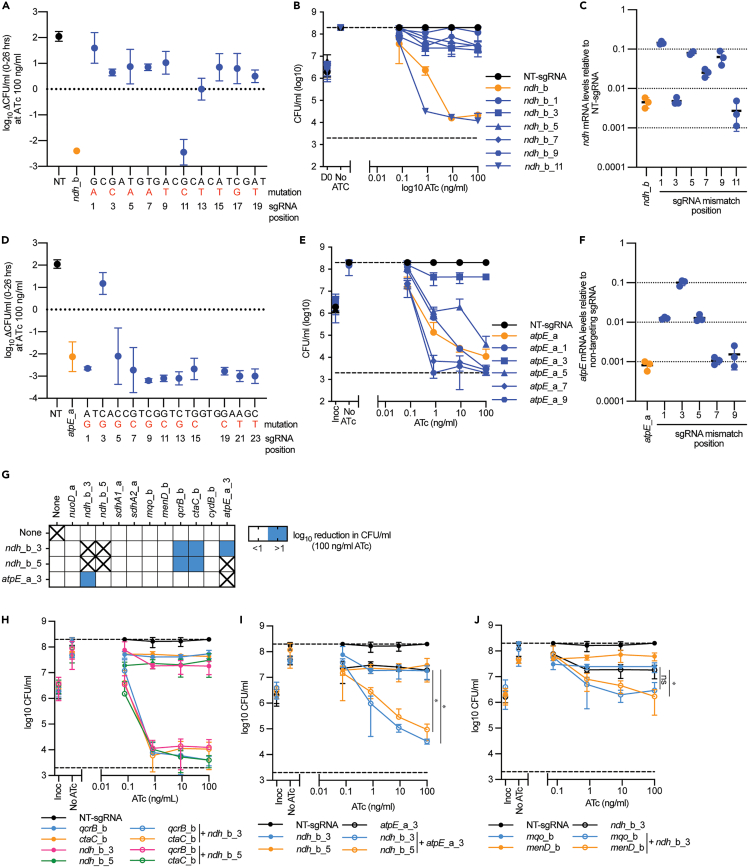
Mutagenesis identifies suboptimal sgRNAs targeting ndh and atpE
The bactericidal phenotypes of ndh and atpE prevented the detection of genetic interactions (Figures 1F, S7A, and S7B). We hypothesized that suboptimal sgRNAs with a growth inhibitory but non-lethal phenotype would allow for the investigation of interactions between ndh/atpE and other respiratory complexes. To identify suboptimal sgRNAs we (I) mutated every second nucleotide in the sgRNAs ndh_b and atpE_a and (II) selected sgRNAs with lower predicted PAM scores targeting ndh and atpB/E. Phenotypic analysis demonstrated that mismatched and alternative PAM targeting sgRNAs were active and retained their inhibitory effects on bacterial growth (Figures S8A–S8D). The majority of the mismatched ndh_b sgRNAs abolished the ndh_b bactericidal phenotype and had a bacteriostatic phenotype with no reduction in CFU/ml (Figures 2A and 2B). A mismatch at position 11 of ndh_b retained the bactericidal activity (Figures 2A and 2B). Conversely, the majority of mismatched atpE_a sgRNAs retained the bactericidal phenotype of atpE_a (Figures 2D and 2E). However, the one exception was a mismatch at position three that had a bacteriostatic phenotype with no reduction in CFU/ml (Figures 2D and 2E). Similarly, the majority of sgRNAs with weaker PAM scores resulted in a bacteriostatic phenotype when targeting ndh; however, they were bactericidal when targeting atpE or atpB (Figures S8E and S8F).
Figure 2.
Mutagenesis of sgRNAs targeting ndh and atpE identifies suboptimal sgRNAs with unique synthetic lethal interactions
(A) log10 Reduction in CFU/ml (CFU/ml at 0 h −26hrs) in the presence of 100 ng/mL ATc for strains expressing a non-targeting (NT), ndh_b, or a ndh_b mismatched sgRNA. Mismatch base pair is highlighted in red, with the sgRNA position noted below.
(B) CFU/ml plots for strains expressing a non-targeting (NT), ndh_b, or selected ndh_b mismatched sgRNA against increasing concentrations of ATc. Mismatched sgRNAs are noted by the position of their mutation in the parental sgRNA.
(C) mRNA levels of ndh_b or selected ndh_b mismatched sgRNAs following CRISPRi induction with 100 ng/mL. mRNA levels are expressed relative to a non-targeting sgRNA. Results are the mean ± standard deviation of three technical replicates.
(D) log10 Reduction in CFU/ml (CFU/ml at 0 h −26hrs) in the presence of 100 ng/mL ATc for strains expressing either a non-targeting (NT), atpE_a, or a atpE_a mismatched sgRNA. Mismatch base pair is highlighted in red, with the sgRNA position noted below.
(E) CFU/ml plots for strains expressing a non-targeting (NT), atpE_a, or selected atpE_a mismatched sgRNA against increasing concentrations of ATc. Mismatched sgRNAs are noted by the position of their mutation in the parental sgRNA.
(F) mRNA levels of atpE_a or selected atpE_a mismatched sgRNAs following CRISPRi induction with 100 ng/mL. mRNA levels are expressed relative to a non-targeting sgRNA. Results are the mean ± standard deviation of three technical replicates. All phenotypic results (A, B, D, and E) are the mean ± SD of at least four biological replicates.
(G) Log10 reduction (0–26 h) in CFU/ml for single and multiplex strains in the presence of 100 ng/mL ATc. Blue shading represents a gene pair that when simultaneously repressed has at least a one-log reduction in CFU/ml. sgRNAs paired with “none” denote results from strains that express a single sgRNA. Combinations marked with “X” were not tested.
(H–J) CFU/ml plots of selected single and multiplexed KD against increasing concentrations of ATc. A strain expressing a non-targeting (NT) sgRNA is used as a negative control. (H) Expression of sgRNAs qcrB_b and ctaC alone or in combination with sgRNAs ndh_b_3 or ndh_b_5. (I) Expression of sgRNA atpE_a_3 alone or in combination with sgRNAs ndh_b_3 or ndh_b_5.
(J) Expression of sgRNAs mqo_b and menD_b alone or in combination with sgRNA ndh_b_3. For H-J, results are the mean ± SD of at least four biological replicates. ∗ denotes statistically significant difference with a p value of <0.05 between the average CFU/ml at 100 ng/mL for the multiplex sgRNA compared to the (I) atpE_a_3 or (J) ndh_b_3 targeting sgRNA as determined by an unpaired t test.
Transcriptional profiling of suboptimal sgRNAs demonstrated that for the ndh mismatches, the majority of bacteriostatic sgRNAs had reduced levels of repression compared to the bactericidal parental sgRNA or ndh_b_11 (Figure 2C). Interestingly, the bacteriostatic ndh_b_3 had a comparable level of repression to the parental sgRNA and ndh_b_11 (Figure 2C). For the atpE mismatches, the single bacteriostatic sgRNA repressed atpE expression by only 10-fold, whereas bactericidal mismatched sgRNAs repressed atpE expression by at least 100-fold and the parental sgRNA repressed expression by approximately 1000-fold (Figure 2F). In conclusion, mutations in sgRNA targeting sequences or weaker predicted PAM sequences reduce the level of transcriptional repression and can generate a hypomorphic phenotype.
Suboptimal sgRNAs targeting ndh and atpE identify interacting gene pairs
To identify interactions with atpE and ndh, the suboptimal bacteriostatic sgRNAs atpE_a_3, ndh_b_3, and ndh_b_5 were multiplexed into CRISPRi plasmids with other respiratory components. Phenotypic analysis of these interactions demonstrated that suboptimal ndh targeting sgRNAs had synthetic lethal interactions with qcrB and ctaC achieving a >2 log reduction in CFU/ml at 1 ng/mL ATc (Figures 2G and 2H). Interestingly, the hypomorphic ndh and atpE targeting sgRNA also had a, albeit weaker, synthetically lethal interaction between themselves, achieving an approximately 1 log reduction in CFU/ml at the maximum ATc concentration (i.e., 100 ng/mL) (Figures 2G and 2I). The simultaneous repression of menD or mqo with the transcriptionally stronger ndh hypomorphic sgRNA (i.e., mismatch at position three, but not five) had an additive or synergistic interaction with a stronger bacteriostatic phenotype than the single sgRNAs alone (i.e., the single knockdowns had a 0.5–1 log increase in CFU/ml) (Figure 2J). In conclusion, the construction of suboptimal sgRNA with bacteriostatic phenotypes allows the identification of interactions associated with the inhibition of ndh and atpE.
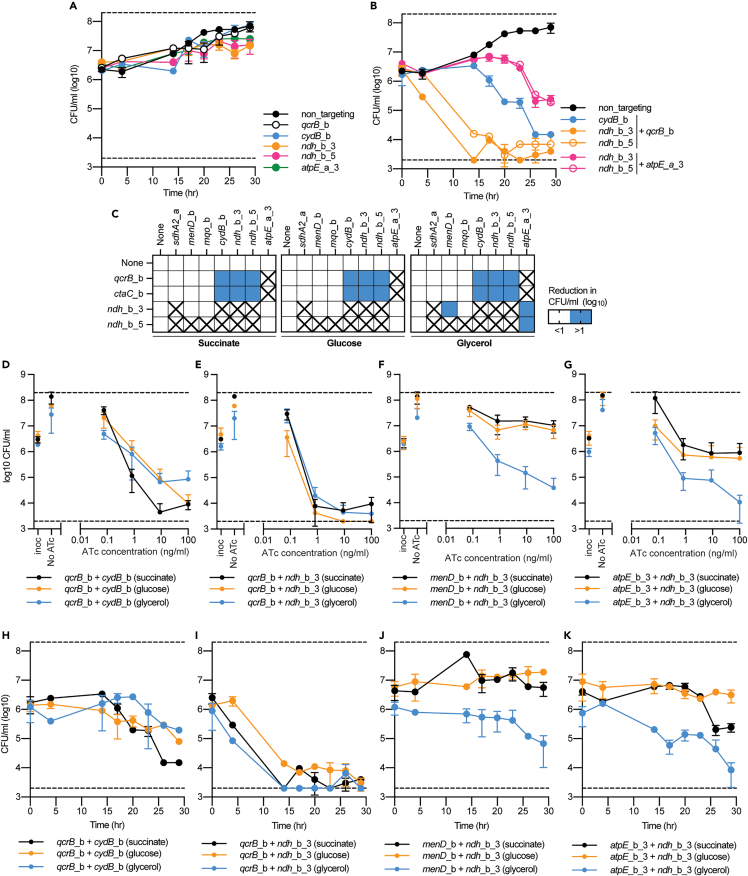
Rate and magnitude of killing by simultaneous target inhibition is influenced by gene pair and metabolic state
To validate synthetic lethal interactions and to investigate possible variations in the rate or magnitude of killing, time kill experiments were performed. Time kill experiments reproduced the synthetic lethal interactions between qcrB and cydB and qcrB with the ndh hypomorphs, in addition to the weaker interaction between the ndh and atpE hypomorphs (Figures 3A and 3B). The synthetic lethal interaction between qcrB and ndh hypomorphs had a greater than two-log reduction within 14 h, whereas the qcrB + cydB interaction was slower and reached a one-log and two-log reduction in CFU/ml at 20 and 26 h, respectively (Figures 3A and 3B). The interaction between the ndh and atpE hypomorph was the weakest and reached a one log reduction in CFU/ml at 26 h (Figures 3A and 3B).
Figure 3.
Rate and magnitude of killing by synthetic lethal interactions is influenced by gene pair and growth on alternative carbon sources
(A–B) Time kill assays of either (A) single or (B) multiplexed gene pairs when grown in the presence of 100 ng/mL ATc with succinate as a sole carbon source. CFUs were taken at stated time points. For A-B, data is the mean ± SD of biological duplicates from a representative experiment.
(C) Log10 reduction (0–26 h) in CFU/ml for single and multiplex strains in the presence of 100 ng/mL ATc. Blue shading represents a gene pair that when simultaneously repressed has at least a 1 log greater reduction in CFU/ml. sgRNAs paired with “none” denote results from strains that express a single sgRNA. Boxes marked with an “X”, denoted gene pairs that were not analyzed on alternative carbon sources because they failed to show a synergistic or synthetic lethal interaction when previously tested (see Figures 1 and 2).
(D–G) CFU/ml plots of selected single and multiplexed KD against increasing concentrations of ATc. A strain expressing a non-targeting (NT) sgRNA is used as a negative control. Black, orange, and blue lines represent results when grown in succinate, glucose or glycerol, respectively. (D-G) Reduction in CFU/ml for M. smegmatis strains expressing the sgRNA (D-E) qcrB_b in combination with sgRNAs (D) cydB_b or (E) ndh_b_3, or (F-G) expression of the sgRNA ndh_b_3 in combination with (F) menD_b or (G) atpE_a_3. For D-G, results are the mean ± SD of at least four biological replicates.
(H–K) Kill curves for M. smegmatis strains expressing the sgRNA (H-I) qcrB_b in combination with sgRNAs (D) cydB_b or (E) ndh_b_3, or (F and G) expression of the sgRNA ndh_b_3 in combination with (F) menD_b or (G) atpE_a_3. For H-K, results are the mean ± SD two biological replicates from a representative experiment. Black, orange, and blue lines represent results when grown in succinate, glucose, or glycerol, respectively. For H-K, the succinate kill curve is the same as presented in panel 3B.
Our experimental set up utilized succinate as a sole non-fermentable carbon source, which forces mycobacterial respiration to use the TCA cycle and ETC (i.e., oxidative phosphorylation) to generate ATP. We sought to determine whether the synthetic lethal interactions would be rescued or amplified when grown on alternative carbon sources such as glucose or glycerol (i.e., fermentable carbon sources) that allow ATP to be generated via glycolysis and provide opportunities for metabolic rerouting to overcome a dysfunctional TCA cycle (de Carvalho et al., 2010; Mackenzie et al., 2020; Koul et al., 2014). The synthetic lethal interactions between qcrB-cydB and qcrB-ndh_b_3 were conserved on all carbon sources (Figures 3C–3E). The magnitude of killing by the qcrB-cydB interaction was greater when grown in succinate as a sole carbon source, with a 1 log increase in killing at 10 ng/mL ATc (Figure 3D). There was no variation in the magnitude of killing by the qcrB-ndh_b_3 interaction across different carbon sources (Figure 3E). Interestingly, the ndh_b_3 interaction with either menD or atpE_b_3 became bactericidal when grown with glycerol as a sole carbon, with a >1 log reduction in CFU/ml at 100 ng/mL ATc (Figures 3C, 3F and 3G). Time kill experiments validated the consistency of killing by the qcrB-ndh_b_3 interaction across different carbon sources, with a greater than two-log reduction in CFU/ml reach by 14 h (Figure 3I). The qcrB-cydB interaction, although present across all carbon sources, was stronger when grown in succinate with a two-log reduction in CFU/ml at 26 h, compared to a one-log reduction when grown on glucose or glycerol (Figure 3H). Consistent with ATc titration results, the ndh_b_3 interaction with menD was bactericidal when grown on glycerol as a sole carbon source with a one-log reduction at 30 h, compared to no reduction in CFU/ml when grown on succinate or glucose (Figure 3J). Similarly, the ndh_b_3 interaction with atpE_b_3 had the greatest level of killing when grown on glycerol as a sole carbon source with a >two-log reduction in CFU/ml at 30 h compared to the one-log or less than one-log reduction when grown on succinate or glucose, respectively (Figure 3K). In conclusion, the rate and magnitude of killing resulting from the simultaneous inhibition of respiratory components is influenced by both the genes pairs that are targeted and mycobacterial metabolic state.
Discussion
New drugs and drug regimens are urgently needed to combat M. tuberculosis. Mycobacterial respiration and metabolism have emerged as a promising area for the development of new drugs and regimens. This is highlighted by the approval of the ATP synthase inhibitor BDQ and the cytochrome-bc1 inhibitor, Q203, being advanced into clinical trials (de Jager et al., 2020; Cox and Laessig, 2014). There is also evidence that combinations of respiratory inhibitors have the potential for synergistic or synthetically lethal interactions with sterilizing activity, supporting the development of new regimens involving multiple respiratory inhibitors (Berube et al., 2019; Kalia et al., 2017; Lamprecht et al., 2016). Herein, we have used mycobacterial CRISPRi to identify the cellular consequences of individually and simultaneously inhibiting the major respiratory components of oxidative phosphorylation in mycobacteria. We demonstrate that in M. smegmatis, the majority of respiratory components are essential and there is a network of interactions centered around the inhibition of the CIII-CIV supercomplex.
Inhibition of menD produced a growth defect, but was not identified as an essential gene under the tested conditions. This is in contrast to previous TnSeq experiments in M. tuberculosis and CRISPRiseq studies in M. smegmatis where the menD is designated as an essential gene. These discrepancies maybe a combination of: (i) differences in experimental set up (growth in liquid vs selection on solid media), (ii) CRISPRi being unable to achieve sufficient levels of inhibition to produce a phenotype, or (iii) because following repression of menD residual menaquinone remains available and is sufficient to buffer the genetic depletion and support bacterial growth for a period of time (Donati et al., 2020; McNeil et al., 2021). Consistent with previous results, sdh1 and nuoD are non-essential (Pecsi et al., 2014; Vilcheze et al., 2018; Hartman et al., 2014; Beites et al., 2019). The inhibition of sdh2, qcrB, ctaC, and mqo produced an inhibitory phenotype with bacteriostatic consequences on cell viability. For qcrB, this is consistent with previous chemical inhibitors including Q203 and TB47 (Pethe et al., 2013; Lamprecht et al., 2016; Lu et al., 2019). The inhibition of ndh, atpE, and atpB had an inhibitory phenotype with bactericidal consequences. These results for atpE/B and ndh2 are consistent with our recent studies in M. tuberculosis (McNeil et al., 2020; McNeil et al., 2021). It is important to note, there are genetic redundancies in M. tuberculosis with it encoding two copies of ndh-2 (i.e., ndh and ndhA). In contrast to CRISPRi results (Bosch et al., 2021; McNeil et al., 2021), previous targeted deletion strategies have demonstrated that only when both copies of ndh (i.e., ndh and ndhA) in M. tuberculosis are deleted in the presence of fatty acids is an essential phenotype observed (Beites et al., 2019; Vilcheze et al., 2018). Resolving the discrepancies between these genetic approaches require additional work. Nevertheless, this suggests that chemical inhibitors of NDH-2 should have a bactericidal phenotype as has been observed for clofazimine and quinoline quinones (Heikal et al., 2016; Yano et al., 2011).
The combined results of this study validate the use of CRISPRi to identify favorable interactions between possible drug targets. For example, simultaneous transcriptional inhibition of cydB and qcrB, which had non-essential and bacteriostatic phenotypes, respectively, resulted in cell death. This synthetic lethal interaction is consistent with interactions between recently discovered chemical inhibitor of cydB (i.e., ND-011992) and the qcrB targeting compound Q203 (Kalia et al., 2017; Lee et al., 2021). Our multiplexed interactions also demonstrated that qcrB and ctaC, together which form the CIII-CIV supercomplex in mycobacteria (Wiseman et al., 2018; Gong et al., 2018), have additive or potentially synergistic interactions with sdhA2, mqo, and menD. Multiplexed interaction analysis of hypomorphic sgRNAs also identified a synthetic lethal interaction between qcrB/ctaC and ndh, as well as a synthetic lethal interaction between the inhibition of atpE and ndh. The rate and magnitude of killing by synthetic lethal interactions varied greatly, with ndh + qcrB being the strongest, followed by qcrB + cydB, and then the ndh + atpE hypomorph. Growth on alternative carbon sources also influenced synthetic lethal interactions, with the interaction between ndh + menD or atpE being greater when grown on glycerol, whereas the interaction between qcrB + cydB was stronger when grown with succinate as a sole carbon source. The retention of the qcrB + cydB interaction across diverse carbon sources is consistent with previous studies in M. tuberculosis (Kalia et al., 2019). Carbon source has previously been shown to influence the efficacy of inhibitors and the phenotype of deletion mutants targeting mycobacterial respiration and metabolism (Vilcheze et al., 2018; Koul et al., 2014; Beites et al., 2019). This phenotypic variation is widely attributed to the ability of mycobacteria to undergo metabolic remodeling, to either partially or completely compensate for the loss of individual metabolic pathways (Beites et al., 2019; Lee et al., 2019b; Puckett et al., 2014, 2017; Eoh and Rhee, 2014). Although this presents inherent challenges for the development of drugs that target mycobacterial respiration, identifying antibiotics or combinations thereof that retain their lethality irrespective of carbon source and metabolic state will have the greatest impact in a clinical setting. Combined, this work has significantly expanded the network of synthetic lethal interactions between mycobacterial respiratory components, demonstrated that the rate and magnitude of synthetic lethal interactions is highly variable and highlighted the potential for rapidly killing drug combinations that consist of multiple respiratory inhibitors.
This study also has wider implications on the understanding of CRISPR applications and the generation of suboptimal hypomorphic sgRNA sequences to probe interactions between genetic targets. Previous studies of Streptococcus pyogenes Cas9 demonstrated that as little as five bp similarity in the seed region (i.e., proximal to PAM sequence) is sufficient to facilitate sgRNA binding with disruption of this region perturbing sgRNA targeting (Cui et al., 2018). Conversely in this study, the transcriptional effects of sgRNA mutations in S. thermophilus Cas9 were variable, with mutations in the atpE sgRNA showing a positional effect (i.e., mutations closer to PAM had a weaker transcriptional repression), whereas mutations in the ndh2 sgRNA lacked a positional effect. Suboptimal sgRNAs targeting atpE suggest that a transcriptional repression between 10-100 fold is sufficient to prevent growth but not kill, whereas the above 100 fold repression results in bacterial killing. Conversely, for suboptimal sgRNAs targeting ndh even with no perturbations in transcriptional repression, cellular outcomes can be changed. This suggests that other factors, such as dCas9 retention on target sequences, are likely to be important factors when considering the efficacy of mutated sgRNAs. With a limited dataset of only two sgRNAs, the results of this current study cannot provide any general design principles for constructing hypomorphic sgRNAs when using this mycobacterial CRISPRi platform. However, the contrasting phenotypic consequences for suboptimal guides targeting ndh (i.e., majority bacteriostatic) and atpE (i.e., majority bactericidal) suggest that even with predictable effects of mutagenesis on sgRNA efficacy, as are available for S. pyogenes dCas9 (Hawkins et al., 2020), the effects on cellular phenotype are completely dependent on the gene being targeted and the relative level of transcriptional inhibition needed to produce a phenotype.
Several of our genetic interactions have been previously reported using chemical inhibitors. This includes: (1) the synthetic lethal interaction between qcrB (i.e., Q203) and cydB deletion (Kalia et al., 2017), and the synergistic killing between (2) ndh (i.e., clofazimine) and qcrB (i.e., Q203 and IMP) (Lamprecht et al., 2016), (3) menA and qcrB (i.e., IMP) (Berube et al., 2019), and (4) ndh (i.e., clofazimine) and atpE (i.e,. bedaquiline) (Lamprecht et al., 2016). The respiratory inhibitor C10 with a currently undefined mechanism of action is also synthetically lethal with Q203, suggesting that qcrB may have synthetic lethal interactions with other respiratory components outside of those investigated in this current study (Flentie et al., 2019). Furthermore, consistent with our lack of interaction between qcrB and atpE, Lamprecht et al. did not report synergistic killing between Q203 with bedaquiline (Lamprecht et al., 2016). For the interaction between clofazimine with either Q203 or bedaquiline, it has been proposed that Q203 and bedaquiline both deplete ATP levels while increasing carbon metabolism and TCA cycle activity (Lamprecht et al., 2016). The proposed increase in TCA cycle activity results in an accumulation of reducing equivalents that cause reductive stress and potentiate clofazimine’s ability to transfer electrons from NDH-2 to oxygen, resulting in rapid production of reactive oxygen species and bacterial killing (Lamprecht et al., 2016). Whether this potential mechanism of simultaneously starving mycobacteria of energy to limit their ability to utilize antioxidant defenses in the face of increasing oxidative stress is universal to all of our observed synthetic lethal interactions requires further investigation. It is also worth noting that previous studies have identified an interaction between chemical inhibition of menaquinone biosynthesis and chemical inhibition of either atpE (i.e., bedaquiline) or ndh (i.e., clofazimine) (Sukehka et al., 2017; Berube et al., 2019). We observed no interaction between menD + atpE, and the menD + ndh interaction was only observed when grown on glycerol. This may be because of an insufficient transcriptional repression of menD or because of metabolic buffering of residual menaquinone remains and is sufficient to support bacterial growth (McNeil et al., 2021). Alternatively, this lack of chemical validation in some cases may be because of potential off-target effects of chemical inhibitors. Furthermore, this CRISPRi platform will allow for wider investigations into how perturbations in mycobacterial metabolism influence antibiotic efficacy (Lee et al., 2019a).
Combined, the results from this current study have significantly expanded the network of synthetic lethal interactions between mycobacterial respiratory components, the majority of which involve the inhibition of the cytochrome-bc1-aa3 supercomplex. Previous work highlighting the synthetic lethality of the interaction between the terminal oxidase cydB and qcrB has placed significant emphasis on the identification of cytochrome bd oxidase inhibitors to realize the full potential of Q203. These results expand the potential suite of drug targets that could be combined with Q203. In conclusion, this work supports the development of respiratory inhibitor combinations for the treatment of mycobacterial infections, energizes the discovery of small molecule inhibitors targeting other respiratory components, and highlights the power of CRISPRi in identifying unique combination therapies.
Limitations of the study
This current study uses a transcriptional approach to infer effects of potential chemical inhibitors. Although useful, there can be differences between their potential effects on targets. For example, transcriptional inhibition will affect both enzyme function and the formation of complexes, whereas chemical inhibition will typically only affect enzyme function. In addition, although M. smegmatis is widely regarded as a physiological and molecular model for mycobacterial respiration, there are genetic and physiological differences between M. smegmatis and pathogenic mycobacterial species, including M. tuberculosis. For this reason, interactions identified in M. smegmatis need to be validated in M. tuberculosis.
Data and code availability
-
•
This paper does not report original code
-
•
The data that support the findings of this study are available from the lead contact, Matthew McNeil (matthew.mcneil@otago.ac.nz).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli mc1061 | CGSC | 6649 |
| Mycobaterium smegmatis mc2155 | ATCC | 700084 |
| Chemicals, peptides, and recombinant proteins | ||
| Kanamycin sulfate | Merck | 60615 |
| Anhydrotetracycline | Merck | 37919-100MG-R |
| Critical commercial assays | ||
| SuperScript VILO | ThermoFisher | 11755050 |
| PowerUp SYBR Green Master Mix | ThermoFisher | A25780 |
| Oligonucleotides | ||
| All oligonucleotides used for plasmid construction or qPCR were ordered through IDT and are listed in supplemental materials | ||
| Experimental models: Organisms/strains | ||
| Escherichia coli mc1061 | CGSC | 6649 |
| Mycobaterium smegmatis mc2155 | ATCC | 700084 |
| Recombinant DNA | ||
| pLJR962 (CRISPRi plasmid for M. smegmatis | Addgene | 115162 |
Resource availability
Lead contact
Further information should be directed to lead contact, Matthew McNeil (matthew.mcneil@otago.ac.nz).
Materials availability
All requests for strains and plasmids constructed in this study should be directed to lead contact, Matthew McNeil (matthew.mcneil@otago.ac.nz).
Bacterial strains and growth conditions
Escherichia coli MC1061 and M. smegmatis mc2155 were grown in luria broth (LB) media or LB agar (1.5%) at 37°C and shaking at 200 rpm when required. For growth of M. smegmatis in liquid LB media tween-80 was added at 0.05% (LBT). M. smegmatis phenotypic experiments were performed in a minimal Hartmans de bont (HdeB) media (2 g (NH4)SO4, 1.08 g KH2PO4 and 2.99 g Na2HPO4 with 10 mL 100x trace metals per L (Berney et al., 2012)) supplemented with tyloxapol (0.05%) and either succinate (30 mM), glucose (20 mM) or glycerol (0.2%) as a sole carbon source. When necessary media was supplemented with Kanamycin at 50 μg/mL for E. coli and 25 μg/mL for M. smegmatis.
Construction and transformation of CRISPRi plasmids
A 20–25 bp sequence downstream of permissible PAM sequences targeting the non-template strand of target genes of interest were identified (Rock et al., 2017). Target sequences were ordered as oligos with GGGA and AAAC overhangs respectively (Table S1), annealed and cloned into pJLR962 using BsmB1 and golden gate cloning following protocols described in Table S3 (McNeil and Cook, 2019). Plasmids were cloned into E. coli and validated with sanger sequencing. CRISPRi plasmids expressing multiple sgRNA were constructed by cloning the promoter-sgRNA-dCas9 scaffold from single expression plasmids using the primer pair MMO120 + MMO121 (Table S2). PCR products were purified and cloned into target single sgRNA expression plasmids using Sap1 and golden gate cloning, following the protocol described in Table S3. Plasmids were cloned into E. coli and validated with sanger sequencing. Confirmed single and double sgRNA CRISPRi expression plasmids are listed in Tables S1–S4, respectively. Plasmids were electroporated into M. smegmatis mc2155 (McNeil et al., 2020; McNeil and Cook, 2019).
Electroporation of M. smegmatis mc2155
Cultures of M. smegmatis mc2155 were grown to an OD600 of 0.2–0.3 in LBT. 0.1 volumes of 2 M glycine were added to cultures and allowed to grow for an additional 3 h. Cultures were harvested, wash twice in 10 mL 10% glycerol and resuspended 5 mL 10% glycerol. 0.5–5 μg of DNA was added to 200 μL of resuspended cells, pulsed once (2.5 kV, 25 μF, 1000 Ω resistance) and allowed to recover in 1 mL LBT (in the absence of KAN) for approximately 3 h with shaking at 37°C. Cultures were harvested, diluted and plated onto LB agar supplemented with KAN. Plates were incubated at 37°C until colonies appeared, approximately 3–4 days.
Quantification of transcriptional knockdowns
To quantify the level of transcriptional inhibition, M. smegmatis strains expressing sgRNA were grown from a starting OD600 of 0.1 in HdeB-Succinate with 100 ng/mL ATc. Cultures were grown for 16 h at 37°C with shaking at 200 rpm. The volume of culture harvested was determined as follows (OD600 x volume of culture(ml) = 2.5). Pellets were resuspended in 1 mL TRIzol, bead beaten in a 2 mL tube with 200 μL of 0.1 mm Zirconia/silicon beads for 3 cycles of 30 s at 4800 rpm followed by 30 s on ice and frozen overnight at −20°C. Frozen samples were thawed, mixed with 0.2 mL chloroform and centrifuged in Invitrogen PhaseMaker tubes (Cat No: A33248) for 15 min at 12,000 g. The clear, upper aqueous phase was transferred to a clean 1.5 mL Eppendorf tube and mixed with an equal volume of ethanol. RNA was extracted using Zymo-RNA Clean and Concentrator (Cat No: R1019) and DNase treated using Invitrogen Turbo DNA-free kit (Cat No: AM1907) (McNeil and Cook, 2019). The removal of DNA was confirmed by PCR using 1 μL of extracted RNA as template and the primer combination MMO202 + MMO203. Extracted RNA, confirmed as DNA free, was converted into cDNA using 100 ng of RNA and Invitrogen SuperScript IV VILO Master Mix (Cat No: 11756050). qPCR reactions were performed in 384 well plates in a ViiA7 Thermocycler using Invitrogen PowerUp SYBR Green Master Mix (Cat No: A25780) with 1 μL of cDNA and 500 nM of fwd and rev primers as listed in Table S2. Signals were normalized to the housekeeping sigA transcript and quantified by the 2ΔΔCt method (Livak and Schmittgen, 2001). Error bars are the standard deviation of three technical replicates.
CRISPRi phenotypic assessment of essentiality and viability
To determine the consequences of targeted gene repression on bacterial growth, phenotypic assays were performed as follows (Figure S9). M. smegmatis strains containing sgRNA expression plasmids were initially grown overnight in 5 mL HdeB-succinate + Kan. Cultures were diluted to an OD600 of 0.01 in HdeB-succinate + Kan and 50 μL was used to inoculate a 96 well assay plate at a starting OD600 of 0.005. 96 well assay plates were prepared as follows. Fifty μl of HdeB-succinate + Kan was added to all wells of column 2–11 of a 96 well plate except row H. 55 μL of HdeB-succinate + Kan containing the starting concentration of ATc (i.e. 200 ng/mL ATc) was added to row H. ATc was diluted along the vertical axis of the 96 well plate, transferring 5 μL between Rows, up to Row B. Row A was used as a no ATc control. Columns 1 and 12 contained 100 μL of media as contamination and background controls. Inoculated plates were grown at 37°C with shaking at 200 rpm for 26 h. OD600 was measured using a Varioskan-LUX microplate reader. The minimal inhibitory concentration (MIC) of ATc was determined relative to the growth of the no-ATc control, using a non-linear fitting of data to the Gompertz equation (Lamert and Pearson, 2000). Targeted genes classified as essential for growth generated an MIC for ATc, whilst non-essential genes failed to generate an MIC. To determine the consequences of targeted gene repression on bacterial viability, colony forming units were determined at 0 and 26 h. Briefly, at 0 h a 4-point ten-fold dilution of the 0.01 diluted culture was performed in PBS and 5 μL of each dilution was spotted onto to LBA. At 26 h, approximately 50 μL of culture was removed from the 96 well assay plate used in essentiality screens after OD600 was determined. A 4-point ten-fold dilution was performed in PBS and 5 μL of each dilution was spotted onto to LBA. Plates were incubated at 37°C for 3–4 days, at which points colonies were counted. Essential genes and gene pairs with bacteriostatic consequences resulted in no change in CFU/ml relative to 0 h, whilst bactericidal consequences produced at least a 1 log reduction in CFU/ml relative to 26 h. For cultures assessing essentiality and viability with alternative carbon sources, experiments were performed as described above, with the exception that succinate was substituted for alternative carbon sources throughout and used at stated concentrations.
Time kills assays
Time kill assays were performed by growing M. smegmatis strains containing sgRNA expressing plasmids overnight in 5 mL HdeB-succinate + Kan. Cultures were diluted to an OD600 of 0.1 in HdeB-succinate + Kan and 500 μL was used to inoculate into a final volume of 10 mL HdeB-succinate + Kan in a 100 mL flask, to give a starting OD600 of 0.005. Flasks were incubated at 37°C with shaking at 200 rpm. Samples were taken for OD600 and CFU at stated time points. OD600 was measured in a Jenway 6300 spectrophotometer. CFUs were determined as described above for viability assays in a 96 well plate. For time kill experiments with alternative carbon sources, experiments were performed as described above, with the exception that succinate was substituted for alternative carbon sources throughout and used at stated concentrations.
Quantification and statistical analysis
Statistical analysis was used to evaluate if the additive interactions between either multiplexed sgRNAs i.e. (i) qcrB with the sdhA2/mqo/menD knockdown (ii) atpE_a_3 with ndh_b_3/5 or (iii) ndh_3_B with mqo/menD were significantly different from the single knockdowns at 100 ng/mL ATc. Specifically, an unpaired t-test was used with a p value of <0.05 deemed as being statistically significant. All analysis were performed in Graphpad-prism.
Acknowledgments
This research was financially supported by the Maurice Wilkins Centre for Molecular Biodiscovery, the Marsden Fund (Royal Society of New Zealand) (grant number UOO1807) and the Health Research Council of New Zealand (grant number 20/459).
Author contributions
MBM and GMC conceived the idea. MBM, HWR, JT, and CYC performed the experiments. MBM, HWR, and GMC interpreted the data. MBM wrote the manuscript with input from all other authors.
Declaration of interests
We have no conflicts of interest to declare.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103573.
Supplemental information
Data and code availability
The data that support the findings of this study are available from the lead contact, Matthew McNeil (matthew.mcneil@otago.ac.nz).
References
- Andries K., Verhasselt P., Guillemont J., Gohlmann H.W.H., Neefs J.-M., Winkler H., Gestel J.V., Timmerman P., Zhu M., Lee E., et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- Beites T., O'brien K., Tiwari D., Engelhart C.A., Walters S., Andrews J., Yang H.J., Sutphen M.L., Weiner D.M., Dayao E.K., et al. Plasticity of the Mycobacterium tuberculosis respiratory chain and its impact on tuberculosis drug development. Nat. Commun. 2019;10:4970. doi: 10.1038/s41467-019-12956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney M., Weimar M.R., Heikal A., Cook G.M. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol Microbiol. 2012;84:664–681. doi: 10.1111/j.1365-2958.2012.08053.x. [DOI] [PubMed] [Google Scholar]
- Berube B.J., Russell D., Castro L., Choi S.R., Narayanasamy P., Parish T. Novel mena inhibitors are bactericidal against Mycobacterium tuberculosis and synergize with electron transport chain inhibitors. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02661-18. e02661-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B., Dejesus M.A., Poulton N.C., Zhang W., Engelhart C.A., Zaveri A., Lavalette S., Ruecker N., Trujillo C., Wallach J.B., et al. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell. 2021;184:4579–4592.e24. doi: 10.1016/j.cell.2021.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G.M., Hards K., Dunn E., Heikal A., Nakatani Y., Greening C., Crick D.C., Fontes F.L., Pethe K., Hasenoehrl E., Berney M. Oxidative phosphorylation as a target space for tuberculosis: Success, caution, and future directions. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.tbtb2-0014-2016. TBTB2–0014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G.M., Hards K., Vilchèze C., Hartman T., Berney M. Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2014;2:1–20. doi: 10.1128/microbiolspec.MGM2-0015-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E.M., Laessig K. FDA approval of bedaquiline — the benefit–risk balance for drug-resistant tuberculosis. N. Engl. J. Med. 2014;371:689–691. doi: 10.1056/NEJMp1314385. [DOI] [PubMed] [Google Scholar]
- Cui L., Vigouroux A., Rousset F., Varet H., Khanna V., Bikard D. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018;9:1912. doi: 10.1038/s41467-018-04209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho L.P., Fischer S.M., Marrero J., Nathan C., Ehrt S., Rhee K.Y. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- de Jager V., Dawson R., van Niekerk C., Hutchings J., Kim J., Vanker N., van der Merwe L., Choi J., Nam K., Diacon A. Telacebec (Q203), a new antituberculosis agent. N. Engl. J. Med. 2020;382:1280–1281. doi: 10.1056/NEJMc1913327. [DOI] [PubMed] [Google Scholar]
- de Wet T.J., Winkler K.R., Mhlanga M., Mizrahi V., Warner D.F. Arrayed CRISPRi and quantitative imaging describe the morphotypic landscape of essential mycobacterial genes. Elife. 2020;9:e60083. doi: 10.7554/eLife.60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati S., Kuntz M., Pahl V., Farke N., Beuter D., Glatter T., Gomes-Filho J.V., Randau L., Wang C.Y., Link H. Multi-omics analysis of CRISPRi-knockdowns identifies mechanisms that buffer decreases of enzymes in E. coli metabolism. Cell Syst. 2020;12:56–67. doi: 10.1016/j.cels.2020.10.011. [DOI] [PubMed] [Google Scholar]
- Eoh H., Rhee K.Y. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc. Natl. Acad. Sci. U S A. 2014;111:4976–4981. doi: 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentie K., Harrison G.A., Tukenmez H., Livny J., Good J.A.D., Sarkar S., Zhu D.X., Kinsella R.L., Weiss L.A., Solomon S.D., et al. Chemical disarming of isoniazid resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 2019;116:10510–10517. doi: 10.1073/pnas.1818009116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H., Li J., Xu A., Tang Y., Ji W., Gao R., Wang S., yu L., Tian C., Li J., et al. An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science. 2018;362:eaat8923. doi: 10.1126/science.aat8923. [DOI] [PubMed] [Google Scholar]
- Hartman T., Weinrick B., Vilchèze C., Berney M., Tufariello J., Cook G.M., Jacobs W.R. Succinate dehydrogenase is the regulator of respiration in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004510. doi: 10.1371/journal.ppat.1004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J.S., Silvis M.R., Koo B.-M., Peters J.M., Osadnik H., Jost M., Hearne C.C., Weissman J.S., Todor H., Gross C.A. Mismatch-CRISPRi reveals the co-varying expression-fitness relationships of essential genes in Escherichia coli and Bacillus subtilis. Cell Syst. 2020;11:523–535. doi: 10.1016/j.cels.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikal A., Hards K., Cheung C.Y., Menorca A., Timmer M.S.M., Stocker B.L., Cook G.M. Activation of type II NADH dehydrogenase by Quinolinequinones mediates antitubercular cell death. J. Antimicrob. Chemother. 2016;71:2840–2847. doi: 10.1093/jac/dkw244. [DOI] [PubMed] [Google Scholar]
- Kalia N.P., Hasenoehrl E.J., AB Rahman N.B., Koh V.H., Ang M.L.T., Sajorda D.R., Hards K., Grüber G., Alonso S., Cook G.M., et al. Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc. Natl. Acad. Sci. U S A. 2017;114:7426–7431. doi: 10.1073/pnas.1706139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia N.P., Shi Lee B., AB Rahman N.B., Moraski G.C., Miller M.J., Pethe K. Carbon metabolism modulates the efficacy of drugs targeting the cytochrome bc1:aa3 in Mycobacterium tuberculosis. Sci. Rep. 2019;9:8608. doi: 10.1038/s41598-019-44887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana B.D., Weinstein E.A., Avarbock D., Dawes S.S., Rubin H., Mizrahi V. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 2001;183:7076–7086. doi: 10.1128/JB.183.24.7076-7086.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A., Vranckx L., Dhar N., Göhlmann H.W.H., Özdemir E., Neefs J.-M., Schulz M., LU P., Mørtz E., Mckinney J.D., et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 2014;5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamert R., Pearson J. Susceptibility testing- accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values. J. Appl. Microbiol. 2000;88:784–790. doi: 10.1046/j.1365-2672.2000.01017.x. [DOI] [PubMed] [Google Scholar]
- Lamprecht D.A., Finin P.M., Rahman M.A., Cumming B.M., Russell S.L., Jonnala S.R., Adamson J.H., Steyn A.J.C. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat. Commun. 2016;7:12393. doi: 10.1038/ncomms12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.S., Hards K., Engelhart C.A., Hasenoehrl E.J., Kalia N.P., Mackenzie J.S., Sviriaeva E., Chong S.M.S., Manimekalai M.S.S., Koh V.H., et al. Dual inhibition of the terminal oxidases eradicates antibiotic-tolerant Mycobacterium tuberculosis. EMBO Mol. Med. 2021;13:e13207. doi: 10.15252/emmm.202013207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.S., Kalia N.P., Jin X.E.F., Hasenoehrl E.J., Berney M., Pethe K. Inhibitors of energy metabolism interfere with antibiotic-induced death in mycobacteria. J. Biol. Chem. 2019;294:1936–1943. doi: 10.1074/jbc.RA118.005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.J., Lee S.K., Song N., Nathan T.O., Swarts B.M., Eum S.Y., Ehrt S., Cho S.N., Eoh H. Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis. Nat. Commun. 2019;10:2928. doi: 10.1038/s41467-019-10975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu X., Williams Z., Hards K., Tang J., Cheung C.Y., Aung H.L., Wang B., Liu Z., Hu X., Lenaerts A., et al. Pyrazolo[1,5- a]pyridine inhibitor of the respiratory cytochrome bcc complex for the treatment of drug-resistant tuberculosis. ACS Infect. Dis. 2019;5:239–249. doi: 10.1021/acsinfecdis.8b00225. [DOI] [PubMed] [Google Scholar]
- Mackenzie J.S., Lamprecht D.A., Asmal R., Adamson J.H., Borah K., Beste D.J.V., Lee B.S., Pethe K., Rousseau S., Krieger I., et al. Bedaquiline reprograms central metabolism to reveal glycolytic vulnerability in Mycobacterium tuberculosis. Nat. Commun. 2020;11:6092. doi: 10.1038/s41467-020-19959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M., Cook G. Utilization of CRISPR interference to validate MmpL3 as a drug target in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2019;63:e00629-19. doi: 10.1128/AAC.00629-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M., Ryburn H., Harold L.K., Tirados J., Cook G. Transcriptional inhibition of the F1F0-type ATP synthase has bactericidal consequences on the viability of mycobacteria. Antimicrob. Agents Chemother. 2020;64:e00492-20. doi: 10.1128/AAC.00492-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M.B., Keighley L.M., Cook J.R., Cheung C.Y., Cook G.M. Crispr interference identifies vulnerable cellular pathways with bactericidal phenotypes in Mycobacterium tuberculosis. Mol. Microbiol. 2021;116:1033–1043. doi: 10.1111/mmi.14790. [DOI] [PubMed] [Google Scholar]
- Pecsi I., Hards K.J., Ekanayaka N., Berney M., Hartman T., Jacobs W.R., Cook G.M. Essentiality of succinate dehydrogenase in Mycobacterium smegmatis. mBIO. 2014;5:1–14. doi: 10.1128/mBio.01093-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethe K., Bifani P., Jang J., Kang S., Park S., Ahn S., Jiricek J., Jung J., Jeon H.K., Cechetto J., et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 2013;19:1157–1160. doi: 10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- Puckett S., Trujillo C., Eoh H., Marrero J., Spencer J., Jackson M., Schnappinger D., Rhee K., Ehrt S. Inactivation of fructose-1,6-bisphosphate aldolase prevents optimal Co-catabolism of glycolytic and gluconeogenic carbon substrates in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004144. doi: 10.1371/journal.ppat.1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckett S., Trujillo C., Wang Z., Eoh H., Ioerger T.R., Krieger I., Sacchettini J., Schnappinger D., Rhee K.Y., Ehrt S. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U S A. 2017;114:E2225–E2232. doi: 10.1073/pnas.1617655114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.M., Hopkins F.F., Chavez A., Diallo M., Chase M.R., Gerrick E.R., Pritchard J.R., Church G.M., Rubin E.J., Sassetti C.M., et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2017;2:16274. doi: 10.1038/nmicrobiol.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C.K., Warrener P., Vandevanter D.R., Sherman D.R., Arain T.M., Langhorne M.H., Anderson S.W., Towell J.A., Yuan Y., Mcmurray D.N., et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–966. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- Sukehka P., Kumar P., Mittal N., Shao-Gang L., Singleton E., Russo R., Perryman A., Shrestha R., Awasthi D., Husain S., et al. A novel small-molecule inhibitor of the Mycobacterium tuberculosis demethylmenaquinone methyltransferase MenG is bactericidal to both growing and nutritionally deprived persister cells. MBio. 2017;8:1–15. doi: 10.1128/mBio.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcheze C., Weinrick B., Leung L.W., Jacobs W.R., Jr. Plasticity of Mycobacterium tuberculosis NADH dehydrogenases and their role in virulence. Proc. Natl. Acad. Sci. U S A. 2018;115:1599–1604. doi: 10.1073/pnas.1721545115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2018. Global tuberculosis report 2018. World Health Organization. https://apps.who.int/iris/handle/10665/274453. [Google Scholar]
- Wiseman B., Nitharwal R.G., Fedotovskaya O., Schafer J., Guo H., Kuang Q., Benlekbir S., Sjostrand D., Adelroth P., Rubinstein J.L., et al. Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis. Nat. Struct. Mol. Biol. 2018;25:1128–1136. doi: 10.1038/s41594-018-0160-3. [DOI] [PubMed] [Google Scholar]
- Yano T., Kassovska-Bratinova S., Shin Teh J., Winkler J., Sullivan K., Isaacs A., Schechter N.M., Rubin H. Reduction of clofazimine by mycobacterial type 2 NADH: Quinone oxidoreductase: A pathway for the generation of bactericidal levels of reactive oxygen species. J. Biol. Chem. 2011;286:10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code
-
•
The data that support the findings of this study are available from the lead contact, Matthew McNeil (matthew.mcneil@otago.ac.nz).
The data that support the findings of this study are available from the lead contact, Matthew McNeil (matthew.mcneil@otago.ac.nz).