Abstract
Cryptosporidium is one of the most important genera of intestinal zoonotic pathogens that cause diarrhea in both humans and animals. Rodents are common and important hosts or carriers of pathogens with public health importance, and rodents play an important role in the ecology of zoonotic transmission. The overall worldwide prevalence of Cryptosporidium spp. in rodents is 19.8% (4589/23142). Twenty-five known Cryptosporidium species and 43 genotypes have been identified, and C. parvum is the dominant species in rodents worldwide. Rodents transfer pathogens to humans by the direct route or by serving as intermediate hosts transmitting the pathogens to other animals. We review the epidemiology, diversity, and transmission routes of Cryptosporidium spp. in rodents. The main purpose of this review is to highlight Cryptosporidium infection in rodents and its transmission, associated risk factors, and prevention; in addition, we assess the public health and ecological significance of Cryptosporidium infections from the One Health perspective.
Keywords: Cryptosporidium, Public health, Ecological significance, Rodents
Highlights
-
•
Review of the epidemiology and diversity of Cryptosporidium in rodents.
-
•
The overall worldwide prevalence is 19.8% (4589/23142), C. parvum is the dominant species.
-
•
Public health and ecological significance of rodent-borne Cryptosporidium at “One Health” perspective.
1. Introduction
Cryptosporidium spp. are foodborne and waterborne parasites with zoonotic potential. The parasites cause watery diarrhea in both humans and animals (domestic animals, mammals, rodents, birds, fish, marsupials, reptiles, and amphibians). Drinking source water, coastal water, recreational use water, wastewater, and market vegetables have been found to contain Cryptosporidium spp. in field investigations [1,5,9,11,12]. At least 44 valid Cryptosporidium species and approximately 120 genotypes have been described to date. Cryptosporidiosis is a global parasitic disease that usually presents as self-limiting diarrhea, abdominal pain, low-grade fever, nausea, vomiting, and weight loss [1,5]. The condition can be fatal in immunosuppressed individuals (e.g., persons infected with HIV/AIDS) [1]. The current treatment for cryptosporidiosis, nitazoxanide (NTZ), has only moderate clinical efficacy, and no vaccines are available [1,5].
The life cycle of Cryptosporidium involves several developmental stages (schizogamy, gametogenesis, and spore stages). The oocysts are ingested by susceptible hosts through contaminated food or water, after which they invade the epithelial cells lining the gastrointestinal gland mucosa and replicate intracellularly [11,53]. Fresh oocysts are excreted from the host with the feces and can cause infections in other susceptible hosts by contaminating food or water [11,12,53]. The rigid spore wall may play a vital role in the survival of the parasite in hostile environments, thus being responsible for large waterborne as well as foodborne outbreaks of the disease [4,53,[92], [93], [94]].
Rodents are an abundant and diversified order of mammals [92,99]. Since the Middle Ages, it has been recognized that rodents can contribute to human disease [9,11,98,99]. In modern times, rodents are also recognized as carriers of many pathogens with public health importance. Almost 10% of the global rodent population is either a carrier or reservoir of pathogens with public health importance [11,98]. Rodents have high population densities and live close to the ground, so they are frequently infected with Cryptosporidium spp. While much progress has been made in Cryptosporidium research, no retrospective analyses have been done on the epidemiology, diversity, or transmission routes of this parasite in rodents. This review aims to explore the current situation for cryptosporidiosis in rodents and to assess the potential risks posed to human and animal populations.
2. Molecular characteristics of Cryptosporidium in rodents
2.1. Prevalence of Cryptosporidium in rodents
To date, Cryptosporidium infection in rodents has been documented in 19 countries, with the prevalence ranging from 0.7% to 100%. The overall average infection rate is 19.8% (4589/23142). Regarding the geographical distribution, the prevalence across different regions ranges from 2.2% to 28.0%. Europe has a higher prevalence (28.0%) (1860/6638) than other documented regions. The overall prevalence values in Asia, North America, South America, Africa, and Oceania are 18.6% (1394/7510), 15.2% (1265/8299), 7.3% (11/150), 2.2% (3/135), and 13.7% (56/410), respectively (Table 1).
Table 1.
Cryptosporidium species and genotypes identified in rodents in the world.
| Factors | Positive/total samples (%) | Zoonotic Cryptosporidium spp. Positive no. | ||
|---|---|---|---|---|
| Location | Asia | China | 16.8% (1010/6010) | C. parvum (189); C. muris (21); C. tyzzeri (13); C. andersoni (42); C. suis (1); C. ubiquitum (125); C. occultus (7); C. erinacei (1); C. canis (1); C. viatorum (36) |
| Korea | 34% (64/188) | C. muris (13); C. ubiquitum (9); Chipmunk genotype I (8) | ||
| Philippines | 26.3% (51/194) | C. muris (3); C. suis (5); C. scrofarum (4); C. occultus (1) | ||
| Japan | 25.9% (171/661) | C. parvum (1); C. muris (4); C.meleagridis (1); C. ubiquitum (14) | ||
| Malaysia | 30.9% (69/223) | C. parvum (12); C. muris (7); C. meleagridis (1) | ||
| Iran | 12.4% (29/234) | C. parvum (3); C. muris (21) | ||
| Subtotal | 18.6% (1394/7510) | C. parvum (205); C. muris (69); C. meleagridis (2); C. tyzzeri (13); C. andersoni (42); C. suis (6); C. scrofarum (4); C. ubiquitum (148); C. occultus (8); C. erinacei (1); C. canis (1); C. viatorum (36); Chipmunk genotype I (8) | ||
| Europe | Poland | 50.3% (863/1715) | C. parvum (687); C. ubiquitum (1) | |
| Finland | 0.7% (2/272) | – | ||
| Czech Republic | 15.5% (219/1409) | C. parvum (3); C. muris (4); C. tyzzeri (44); C. ditrichi (21); C. andersoni (4); C. ubiquitum (1); C. occultus (3) | ||
| Slovak Republic | 19.4% (97/499) | C. parvum (15); C. muris (1); C. ditrichi (5); C. suis (4); C. scrofarum (28); C. hominis (2); C. ubiquitum (4) | ||
| Spain | 25.1% (156/621) | C. parvum (108); C. muris (18); C. meleagridis (1); C. tyzzeri (2) | ||
| Italy | 27.7% (36/130) | Chipmunk genotype I (2) | ||
| England | 27.0% (427/1580) | C. parvum (263); C. muris (73) | ||
| Unkonw | 14.6% (69/412) | C. parvum (2); C. muris (2); C. tyzzeri (2); C. ditrichi (22) | ||
| Subtotal | 28.0% (1860/6638) | C. parvum (1078); C. muris (98); C. meleagridis (1); C. tyzzeri (48); C. ditrichi (48); C. andersoni (4); C. suis (4); C. scrofarum (28); C. hominis (2); C. ubiquitum (6); C. occultus (3); Chipmunk genotype I (2) | ||
| North America | America | 15.2% (1265/8299) | C. parvum (510); C. ubiquitum (19); Muskrat genotype I (24); Muskrat genotype II (6); Skunk genotype (4) | |
| South America | Brazil | 7.3% (11/150) | C. parvum (8) | |
| Africa | Kenya | 100% (1/1) | – | |
| Nigeria | 1.5% (2/134) | C. andersoni (1) | ||
| Subtotal | 2.2% (3/135) | C. andersoni (1); C. ubiquitum (19); Muskrat genotype I (24); Muskrat genotype II (6); Skunk genotype (4) | ||
| Oceania | New Zealand | 20.0% (5/25) | – | |
| Australia | 13.2% (51/385) | C. tyzzeri (6); C. viatorum (3) | ||
| Subtotal | 13.7% (56/410) | C. tyzzeri (6); C. viatorum (3) | ||
| Rodent type | Wild | 20.5% (3848/18804) | C. parvum (1617); C. muris (145); C. meleagridis (3); C. tyzzeri (66); C. ditrichi (48); C. andersoni (3); C. suis (10); C. scrofarum (32); C. hominis (2); C. ubiquitum (37); C. occultus (7); C. erinacei (1); C. canis (1); C. viatorum (39); Muskrat genotype I (24); Muskrat genotype II (6); Skunk genotype (4); Chipmunk genotype I (10) | |
| Farm | 14.5% (354/2439) | C. parvum (169); C. muris (10); C. ubiquitum (85); C. occultus (4) | ||
| Pet | 27.0% (373/1381) | C. parvum (15); C. muris (11); C. andersoni (42); C. ubiquitum (50) | ||
| Lab | 2.7% (14/518) | C. muris (1); C. tyzzeri (1); C. andersoni (2); C. ubiquitum (1) | ||
| Total | 19.8% (4589/23142) | C. parvum (1801); C. muris (167); C. meleagridis (3); C. tyzzeri (67); C. ditrichi (48); C. andersoni (47); C. suis (10); C. scrofarum (32); C. hominis (2); C. ubiquitum (173); C. occultus (11); C. erinacei (1); C. canis (1); C. viatorum (39); Muskrat genotype I (24); Muskrat genotype II (6); Skunk genotype (4); Chipmunk genotype I (10) | ||
Note: “–” indicates no Zoonotic Cryptosporidium spp.
It is difficult to explain the discrepancies in the prevalence of Cryptosporidium spp. among studies because prevalence is affected by many factors, including the host species composition, host species gender, host species age, season, the geographical distribution of the sample population, the sample size, and the ecological conditions. However, according to current data, the major rodent species vary among regions; the prevalence of Cryptosporidium and the major Cryptosporidium species in different species of rodents are also different, and the types of rodents studied may influenced the differences in prevalence (Table 3, Supplementary Table S1). However, the reported prevalence of Cryptosporidium in Africa and Oceania is possibly related to the relative lack of research data.
Table 3.
The prevalence of Cryptosporidium and Cryptosporidium spp. in the rodents.
| Animal species | Locations total samples no. | Positive/total samples (%) | Cryptosporidium species/genotypes positive no. | gp60 subtypes |
|---|---|---|---|---|
| Brown Rat (Rattus norvegicus) | Japan (206); England (511); China (491); Iran (106); Nigeria (134); Czech Republic (−) | 17.2% (249/1448) | C. parvum (164); C. ubiquitum (1); C. ratti (37); rat genotype IV (35); C. occultus (5); C. tyzzeri (1); rat genotype III (1); rat genotype V (5); C. muris (4); C. andersoni (4); rat genotype II (1); C. meleagridis (1); C. ryanae (1); | IIdA15G1 |
| House Rat (Rattus rattus) | Japan (346); New Zealand (8); Spain (102); Iran (40); Australia (85) | 26.3% (153/581) | C. parvum (1); rat genotype II/III (10); C. muris (14); C. meleagridis (1); C. ratti (1); C. sp. 1 (1) | – |
| House mouse (Mus musculus) | England (715); New Zealand (17); USA (303); Spain (78); Iran (63); China (31); Czech Republic (45) | 22.0% (276/1252) | C. muris (61); C. parvum (60); C. tyzzeri (51); C. sp. 2 (1); rat genotype II (2); rat genotype III (2) | Ixa; Ixb |
| Yellow-necked mouse (Apodemus flavicollis) | Poland (331); Spain (2); Belgium (2); Czech Republic (274); Finland (2); France (16); Germany (10); Serbia (14); Slovakia (35); Slovak Republic (196) | 26.2% (66/252) | C. parvum (70); C. ditrichi (43); C. apodemi (11); apodemus genotype I (8); C. tyzzeri (1); apodemus genotype II (4); C. microti (2); C. muris (3); C. scrofarum (5); C. environment (3); C. suis (4) | IXaA8; XVIIIa; XVIIa; IIaA16GlRlb; IIaA18G3R1; IIiA10 |
| Myod glareolus (Clethrionomys glareolus) | Poland (836); Finland (141); Spain (49); England (123); USA (301) | 30.7% (445/1450) | C. parvum (442); C. muris (2) | – |
| Common Vole (Microtus arvalis) | Poland (274); Czech Republic (328); Slovak Republic (75) | 42.8% (290/677) | C. parvum (203); C. alticolis (7); C. microti (46); vole genotype II (1); vole genotype III (1); vole genotype IV (3); vole genotype V (2); vole genotype VI (1); vole genotype VII (6); C. scrofarum (4); C. environment (4); muscrat genotype I (3) | IIaA18G3R1; IIaA10G1R1 |
| Common field vole (Microtus agrestis) | Finland (131) | 0.8% (1/131) | – | – |
| Wood mice (Apodemus sylvaticus) | England (230); Spain (278); Czech Republic (25); France (4); Netherlands (6); Serbia (3); Slovakia (8) | 30.0% (166/554) | C. parvum (132); C. muris (18); C. ditrichi (3); C. tyzzeri (1); C. apodemi (2) | IXcA6 |
| Algerian mouse (Mus spretus) | Spain (22) | 27.3% (6/22) | C. parvum (1); C. muris (5) | – |
| Apodemus speciosus | Japan (33) | 12.1% (4/33) | C. muris (2); C. muris novel genotype (2) | – |
| White-footed mice (Peromyscus sp.) | USA (2706) | 8.2% (222/2706) | C. parvum (165) | – |
| Red-backed vole (Myodes gapperi) | USA (5) | 80.0% (4/5) | – | – |
| Prairie vole (Microtus pennsylvanicus) | USA (307) | 5.2% (16/307) | C. parvum (13) | – |
| Striped field mouse (Apodemus agrarius) | Slovak Republic (107) | 31.8% (34/107) | C. scrofarum (19); C. environment (2); muscrat genotype II (3); C. parvum (8); C. hominis (1) | IbA10G2; IIcA5G3a; IIaA18G3R1; IIiA10 |
| Muskrat (Ondatra zibethicus) | Poland (9); USA (353); Spain (90) | 26.3% (119/452) | C. parvum (85); C. muris (5); muskrat genotype I (24); muskrat genotype II (6) | – |
| California Ground Squirrels (Spermophilus beecheyi) | USA (1162) | 12.8% (149/1162) | C. parvum (149) | – |
| Eastern Gray Squirrel (Sciurus carolinensis) | USA (106) | 17.0% (18/106) | C. parvum (6) | – |
| Red squirrels (Tamiasciurus hudsonicus) | USA (80); China (333) | 9.4% (39/413) | C. parvum (9); rat genotype II (8); ferret genotype (13); chipmunk genotype III (5); C. ratti (4); | – |
| Eurasian Red Squirrel (Sciurus vulgaris) | Italia (70); USA (2) | 25.0% (18/72) | ferret genotype (15); chipmunk genotype I (2) | – |
| Eastern chipmunks (Tamias striatus) | USA (268); China (20) | 23.0% (66/288) | C. parvum (38); ferret genotype (3); ferret genotype + C. parvum (1); C. muris + C. parvum + chipmunk genotype III (1); chipmunk genotype II (28); chipmunk genotype IV (−) | – |
| Woodchuck (Marmota monax) | USA (38); Czech Republic (−) | 7.9% (3/38) | C. parvum (2); C. andersoni (1) | – |
| American Beaver (Castor canadensis) | USA (170) | 4.1% (7/170) | C. parvum (2) | – |
| Eurasian Beaver (Castor fiber) | Poland (82); Slovak Republic (19) | 19.8% (20/101) | C. parvum (16) | – |
| North American Porcupine (Erethizon dorsatum) | USA (18) | 11.1% (2/18) | C. parvum (2) | |
| Capybara (Hydrochoerus hydrochaeris) | Brazil (145) | 5.5% (8/145) | C. parvum (8) | – |
| Root rat (Tachyoryctes splendens) | Kenya (−) | – | C. proliferans | – |
| Striped field mouse (A. agrarius) | Latvia (11); Lithuania (3); Romania (2); Serbia (4); Slovakia (33); Slovak Republic (72) | 16.8% (21/125) | apodemus genotype II (5); C. ditrichi (2); C. apodemi (12); C. parvum (1); C. hominis (1) | XVIIIa; IIaA16G1R1b; IbA10G2 |
| Guinea pig (Cavia porcellus) | China (350); Australia (29); Italia (60); Brazil (5); England (1) | 50.8% (226/445) | C.rairi (159); C. homai (39); C. muris (1); C. parvum (1) | – |
| Chinchillas (Chinchilla lanigera) | China (420); Japan (63) | 11.0% (53/483) | C. ubiquitum (49); C. parvum (3); chipmunk genotype V (1); C. varanii (−) | – |
| Djungarian Hamster (Phodopus sungorus) | China (226) | 38.9% (88/226) | hamster genotype (31), C. andersoni (40); C. muris (8); C. parvum (4); C. andersoni + C. parvum (2); C. muris + C. parvum (2); | – |
| Chipmunk (Tamias) | China (4) | 75.0% (3/4) | ferret genotype (2); chipmunk genotype V (1) | – |
| Pteromys volans (Siberian flyingsquirrel) | China (1) | 100% (1/1) | C. ubiquitum (1) | XIIi |
| Cricetid rodents | USA (586); Czech Republic (493) | 33.2% (362/1089) | – | – |
| Nutria (Myocastor coypus) | Czech Republic (120); Slovak Republic (30) | 8.0% (12/150) | C. parvum (1); C. ubiquitum (5); C. myocastoris (5) | IIa; XXIIb; XXIIa; XIId; |
| Red-bellied treesquirrels (Callosciurus erythraeus) | China (287); USA (302) | 23.1% (136/589) | C. parvum (1); C. wrairi (1); rat genotype II (2); C. rubeyi (2); squirrel genotypes I (2); squirrel genotypes II (1); squirrel genotypes III (2); C. ubiquitum (19); skunk genotype (4); deer mouse genotype III (5) | – |
| Bamboo rat (Rhizomys sinensis) | China (1960) | 15.8% (309/1960) | C. parvum (158); C. occultus (4); C. ubiquitum (85); bamboo rat genotype I (54); bamboo rat genotype II (1); bamboo rat genotype III (5); C. muris (1) | IIpA9; IIpA6; IIoA15G1; IIoA13G1 |
| Porcupine (Hystrix hodgsoni) | China (147) | 6.8% (10/147) | C. tyzzeri (3); rat genotype III (1) | – |
| Asian house rat (Rattus tanezumi) | China (79) | 50.6% (40/79) | rat genotype IV (24); rat genotype III (8); C. occultus (1); C. erinacei (1); C. parvum (3); C. muris (3) | – |
| Edward's long-tailed rat (leopoldamys edwardsi) | China (38) | 55.3% (21/38) | C. viatorum (11); rat genotype IV (8); rat genotype III (2) | XVcA2G1a (4); XVcA2G1b (1); XVdA3 (1) |
| Muridae | China (10) | 40.0% (4/10) | rat genotype III (2); rat genotype IV (2) | – |
| Brandt vole (Microtus brandti) | China (678) | 18.7% (127/678) | C. suis, C. environmental; muskrat genotype II; novel genotype of Brandt's vole | – |
| Spermophilus ground squirrel | USA (−) | – | C. rubeyi (−) | – |
| White-footed Mouse (Peromyscus maniculatus) | USA (1071) | 6.9% (74/1071) | – | – |
| Yellow-bellied Marmot (Marmota flaviventris) | USA (224) | 14.7% (33/224) | C. parvum (33) | – |
| Australian Mice (Mus domesticus) | Australia (250) | 7.6% (19/250) | C. tyzzeri (6); mouse genotype II (11) | – |
| Indian mole rat (Bandicota bengalensis) | Iran (25) | 36.0% (9/25) | C. muris (9) | – |
| Asian chipmunk (Eutamias sibiricus) | Czech Republic (−) | – | C. muris (−) | – |
| Qinghai vole (Lasiopodomys fuscus) | China (90) | 8.9% (8/90) | C. parvum (3); C. canis (1); C. ubiquitum (1); Qinghai vole genotype (3) | – |
| Swamp rats | Australia (21) | 14.3% (3/21) | C. viatorum (3) | XVbA2G1 |
| Prairie vole (Meadow vole) | USA (10) | 10.0% (1/10) | Vole genotype I (1) | – |
| Deer mouse | USA (177) | 32.2% (57/177) | Deer mouse I (13); Deer mouse II (3); Deer mouse III (20); Deer mouse IV (21); | – |
| Laboratory rats | China (355); Czech Republic (−) | 0.6% (2/355) | C. ubiquitum (1); Cryptosporidium spp. (1); C. muris (−) | |
| Wild rats | Philippines (194); Japan (14); China (228); Poland (266); Malaysia (223); Korea (188) | 33.0% (387/1173) | C. muris (25); C. scrofarum (4); C. ratti (1); rat genotypes II (19); rat genotypes III (23); rat genotypes IV (6); C. occultus (5); C. ubiquitum (10); Naruko genotype (1); C. viatorum (25); C. parvum (13); C. meleagridis (1); chipmunk genotype I (8); bear genotype (14). | XVaA6; XVaA3g; XVaA3h; XVcA2G1; IIaA15G2R1; IIaA17G2R1; IIaA16G3R1 |
Note: “-” indicates unknown information.
2.2. Cryptosporidium species distributions in rodents
To date, 25 known Cryptosporidium species (C. parvum, C. hominis, C. muris, C. tyzzeri, C. andersoni, C. meleagridis, C. suis, C. ditrichi, C. apodemi, C. scrofarum, C. alticolis, C. microti, C. myocastoris, C. ubiquitum, C. occultus, C. homai, C. wrairi, C. varanii, C. erinacei, C. canis, C. viatorum, C. proliferans, C. rubeyi, C. ratti, and C. ryanae) and 43 genotypes (rat genotype II-V, bamboo rat genotype I-III, mouse genotype II, muskrat genotype I-II, skunk genotype, hamster genotype, ferret genotype, chipmunk genotype I-V, vole genotype I-VII, apodemus genotype I-II, muskrat genotype I-II, squirrel genotypes I-III, deer mouse genotype I-IV, Cryptosporidium sp. 1, Cryptosporidium sp. 2, Naruko genotype, Qinghai vole genotype, C. muris novel genotype, and the C. environment isolate) have been identified in rodents (Table 2).
Table 2.
Recognized Cryptosporidium spp. in the rodents.
| Species name | Type host(s) | Major host(s) | Reports in humans | Reports in rodents Positive no. | gp60 | Reference |
|---|---|---|---|---|---|---|
| C. hominis* | Human (Homo sapiens) | Humans | Most common reported | Apodemus agrarius (1) | IbA10G2 | [64,87] |
| C. parvum* | Cattle (Bos taurus) | Ruminants; humans | Second most common reported | Rattus norvegicus (164); Rattus rattus (1); Mus musculus (60); Apodemus flavicollis (70); Clethrionomys glareolus (442); Microtus arvalis (203); Apodemus sylvaticus (132); Mus spretus (1); Peromyscus sp. (165); Microtus pennsylvanicus (13); A. agrarius (13); Ondatra zibethicus (8); Spermophilus beecheyi (149); Sciurus carolinensis (6); Tamiasciurus hudsonicus (9); Tamias striatus (38); Marmota monax (2); Castor canadensis (2); Castor fiber (16); Erethizon dorsatum (2); Hydrochoerus hydrochaeris (8); Cavia porcellus (1); Chinchilla lanigera (3); Phodopus sungorus (4); Myocastor coypus (1); Rhizomys sinensis (158); Rattus tanezumi (3); Marmota flaviventris (33); Wild rat (13); Qinghai vole (3) | IIaA15G2R1; IIaA16G2R1; IIaA17G2R1; IIaA18G1R1b; IIaA18G3R1; IIdA15G1; IIiA10; IIpA9; IIpA6; IIoA15G1; IIoA13G1 | [5,37,80,87,24,64] |
| C. meleagridis | Turkey (Meleagris gallopavo) | Birds, humans | Commonly reported | R. norvegicus (1); R. rattus (1); Wild rat (1) | – | [38,65,80] |
| C. ubiquitum | Cattle (B. taurus) | Ruminants, rodents, primates | Commonly reported | R. norvegicus (1); C. lanigera (49); Siberian flyingsquirrel (1); M. coypus (5); Callosciurus erythraeus (19); Wild rat (10); Laboratory rats (1); Qinghai vole (1) | XIIa; XIId; XIIi | [5,11,40,42,43,46] |
| C. viatorum | Human (Homo sapiens) | Rodents | Many reported | leopoldamys edwardsi (11); swamp rats (3); Wild rats (25) | XVbA2G1; XVaA6; XVaA3g; XVaA3h; XVcA2G1; XVcA2G1a; XVcA2G1b; XVdA3 | [52,66,91] |
| C.muris | House mouse (Mus musculus) | Rodents | Commonly reported | R. norvegicus (4); R. rattus (14); M. musculus (61); A. flavicollis (3); C. glareolus (2); M. spretus (5); A. sylvaticus (18); O. zibethicus (5); T. striatus (1); P. sungorus (8); R. sinensis (1); R. tanezumi (3); Bandicota bengalensis (9); Eutamias sibiricus (9); Wild rat (25); Laboratory rats (−);Apodemus speciosus (2); C. porcellus (1); | – | [7,8,20,27,52,59,63,87] |
| C. canis | Dog (Canis familiaris) | Dogs | Commonly reported | Qinghai vole (1) | – | [89] |
| C. tyzzeri | Mouse (Mus musculus) | Rodents | Some reported | R. norvegicus (1); M. musculus (51); A. sylvaticus (1); Hystrix hodgsoni (3); Mus domesticus (6); | IXa; IXb; IXc | [5,37,75] |
| C. andersoni | Cattle (B. taurus) | Cattle | Some reported c | R. norvegicus (4); M. monax (1); P. sungorus (40) | – | [40,47,70,78] |
| C. erinacei | European hedgehog (Erinaceus europaeus) | Hedgehogs, horses | Some reported | R. tanezumi (1) | – | [5,52] |
| C. suis | Pig (Sus scrofa) | Pig | Some reported | A. flavicollis (4); Microtus brandti (−) | – | [87] |
| C. ditrichi | Yellow-necked mouse (Apodemus flavicollis) | Rodents | Two reported | A. flavicollis (43); A. sylvaticus (4); A. agrarius (2) | – | [37,64] |
| C. occultus | Brown rat (Rattus norvegicus) | Rodents | Two reported | R. norvegicus (5); R. sinensis (4); R. tanezumi (1); Wild rats (5) | – | [5,46,49,50,52,49] |
| C. scrofarum | Pig (S. scrofa) | Pig | One reported | A. flavicollis (5); M. arvalis (4); A. agrarius (19) | – | [61,87] |
| C. ryanae | Cattle (B. taurus) | Cattle | None reported | R. norvegicus (1) | – | [13] |
| C wrairi | Guinea pig (Cavia porcellus) | Rodents | None reported | C. porcellus (159); C. erythraeus (1); | – | [40,45,47,82] |
| C. homai | Guinea pig (Cavia porcellus) | Rodents | None reported | C. porcellus (39) | – | [40,82] |
| C. apodemi | Striped field mouse (A. agrarius) | Rodents | None reported | A. flavicollis (11);A. agrarius (12); A. sylvaticus (2) | – | [37,64] |
| C. alticolis | Common vole (M. arvalis) | Rodents | None reported | M. arvalis (7) | – | [35] |
| C. microti | Common vole (M. arvalis) | Rodents | None reported | M. arvalis (46); A. flavicollis (2); | – | [35,37] |
| C. myocastoris | Nutria (Myocastor coypus) | Rodents | None reported | M. coypus (5) | – | [42] |
| C. proliferans | East African mole rat (Tachyoryctes splendes) | Rodents | None reported | Tachyoryctes splendens (−) | – | [46] |
| C varanii | numerous reptiles | Reptiles | None reported | C. lanigera (−) | – | [53] |
| C. rubeyi | Spermophilus ground squirrel | Rodents | None reported | C. erythraeus (2); Spermophilus ground squirrel (−) | – | [55,57] |
| C. ratti | Brown rats (Rattus norvegicus) | Rodents | None reported | R. norvegicus (37); R. rattus (1); T. hudsonicus (4); Wild rats (1) | – | [4] |
| Chipmunk genotype I | Peromyscus spp. | Rodents | Many reported | Sciurus vulgaris (2); Wild rat (8) | – | [32,40,44,47,48,76] |
| Skunk genotype | Mephitis mephitis | Fox, human | Many reported | C. erythraeus (4) | – | [77] |
| Muskrat genotype I | O. zibethicus | Rodents | One reported | O. zibethicus (24) | – | [29,54] |
| Muskrat genotype II | O. zibethicus | Rodents | One reported | O. zibethicus(6); Microtus brandti (−) | – | [29,54] |
| rat genotype II | R. tanezumi | Rodents | None reported | R. norvegicus (1); R. rattus (10); M. musculus (2); C. erythraeus (2); T. hudsonicus (8); Wild rat (19) | – | [29,43,45] |
| rat genotype III | R. tanezumi | Rodents | None reported | R. norvegicus (1); R. rattus (10); M. musculus (2); R. tanezumi (8); leopoldamys edwardsi (2); Muridae (2); Wild rat (23); Hystrix hodgsoni (1) | – | [29,43,45] |
| rat genotype IV | R. tanezumi | Rodents | None reported | R. norvegicus (35); R. tanezumi (24); leopoldamys edwardsi (8); Muridae (2); Wild rats (6) | – | [29,43,45] |
| rat genotype V | R. tanezumi | Rodents | None reported | R. norvegicus (5) | – | [29,43,45] |
| Bamboo rat genotypeI | R. sinensis | Rodents | None reported | R. sinensis (54) | – | [49,50] |
| Bamboo rat genotype II | R. sinensis | Rodents | None reported | R. sinensis (1) | – | [49,50] |
| Bamboo rat genotype III | R. sinensis | Rodents | None reported | R. sinensis (5) | – | [49,50] |
| chipmunk genotype II | T. striatus | Rodents | None reported | T. striatus (28) | [50,54] | |
| chipmunk genotype III | T. striatus | Rodents | None reported | T. hudsonicus (5); T. striatus (1) | – | [50,54] |
| chipmunk genotype IV | Eutamias sibiricus | Rodents | None reported | T. striatus (−) | – | [50,54] |
| chipmunk genotype V | T. striatus | Rodents | None reported | T. striatus (1); C. lanigera (1) | – | [50,54] |
| Mouse genotype II | M. domesticus | Rodents | None reported | M. domesticus (11) | – | [47,77] |
| Muskrat genotypes I | O. zibethicus | Rodents | None reported | M. arvalis (1); O. zibethicus (24) | – | [29,54] |
| Muskrat genotypes II | O. zibethicus | Rodents | None reported | Microtus brandti(−); O. zibethicus (6) | – | [29,54] |
| Vole genotypes I | M. pennsylvanicus | Rodents | None reported | Meadow vole (1) | – | [35] |
| Vole genotypes II | M. arvalis | Rodents | None reported | M. arvalis (1) | – | [35] |
| Vole genotypes III | M. arvalis | Rodents | None reported | M. arvalis (1) | – | [35] |
| Vole genotypes IV | M. arvalis | Rodents | None reported | M. arvalis (3) | – | [35] |
| Vole genotypes V | M. arvalis | Rodents | None reported | M. arvalis (2) | – | [35] |
| Vole genotypes VI | M. arvalis | Rodents | None reported | M. arvalis (1) | – | [35] |
| Vole genotypes VII | M. arvalis | Rodents | None reported | M. arvalis (6) | – | [35] |
| Apodemus genotypes I | Apodemus spp. | Rodents | None reported | A. flavicollis (8) | XVIIa | [37] |
| Apodemus genotypes II | Apodemus spp. | Rodents; water | None reported | A. flavicollis (4); A. agrarius (5) | XVIIIa | [37] |
| squirrel genotype I | S. beecheyi | Sciuridae spp. | None reported | C. erythraeus (2) | – | [77] |
| squirrel genotype II | S. beecheyi | Sciuridae spp. | None reported | C. erythraeus (1) | – | [77] |
| squirrel genotype III | S. beecheyi | Sciuridae spp. | None reported | C. erythraeus (2) | – | [77] |
| Deer mouse genotype I | Peromyscus spp. | Rodents; water | None reported | Deer mouse (13) | – | [77] |
| Deer mouse genotype II | Peromyscus spp. | Rodents; water | None reported | Deer mouse (3) | – | [77] |
| Deer mouse genotype III | Peromyscus spp. | Rodents; water | None reported | C. erythraeus (5); Deer mouse (20) | – | [77] |
| Deer mouse genotype IV | Peromyscus spp. | Rodents; water | None reported | Deer mouse (21) | – | [77] |
| Cryptosporidium sp. 1 | environmental sample; water | – | None reported | R. rattus (1) | – | [38] |
| Cryptosporidium sp. 2 | – | – | None reported | M. musculus (1) | – | [5] |
| Qinghai vole genotype | Microtus fuscus | Vole | None reported | Qinghai vole (3) | – | [89] |
| Novel genotype of Brandt's vole | Brandt's vole | Vole | None reported | Brandt vole (−) | – | [54] |
| Novel genotype | Apodemus spp. | Vole | None reported | Wild rat (14); A. speciosus (2) | – | [48] |
| Naruko genotype | Wild rats | Rodents | None reported | Wild rats (1) | – | [79] |
Note: “–” indicates unknown information; “*” indicates Cryptosporidium spp. that caused the outbreak of human cryptosporidiosis.
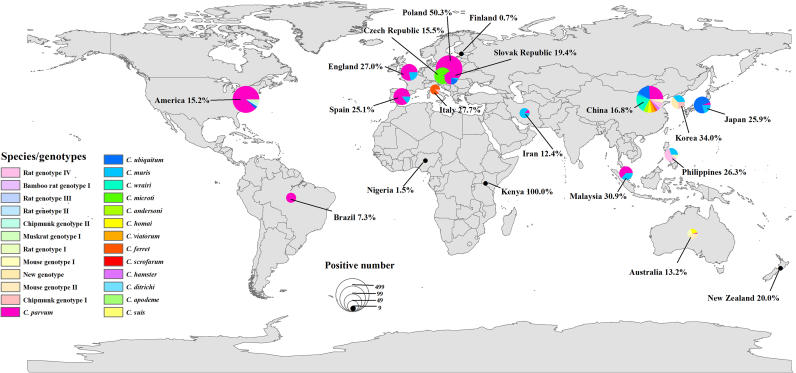
Cryptosporidium species and dominant distributions differ by region. In Asia, 19 species and 15 genotypes have been identified; C. parvum is the dominant species. In Europe, 15 species and 16 genotypes have been identified, and C. parvum is the dominant species. In North America, three species and four genotypes have been identified, and C. parvum is the dominant species. In South America, only C. parvum has been detected. In Africa, two species and one genotype have been identified, and C. andersoni is the dominant species. In Oceania, four species and five genotypes have been identified, and C. homai is the dominant species (Fig. 1). The species and genotypes of Cryptosporidium infecting rodents in Asia and Europe are very diverse; this may be due to the comparatively large number of studies and the species richness of rodents studied. C. parvum has been shown to be common in rodents throughout the world; the species is similarly prevalent in humans and cattle.
Fig. 1.
Prevalence and geographic distribution of Cryptosporidium spp. in rodents. The figure was originally designed by the authors using ArcGIS 10.2 software. The original vector diagram imported in ArcGIS was adapted from Natural Earth (http://www.naturalearthdata.com).
2.3. Cryptosporidium in rodent types
Cryptosporidium infections have been documented 54 rodent species. According to the descriptions of the environment of the sample collection sites and details concerning rodents' living habitats listed in 86 epidemiological articles, the types of rodents can be divided into wild, domestic pet, farm, and laboratory animals. The overall prevalence rates in wild, pet, farm, and laboratory animals are 20.5% (3848/18804), 27.0% (373/1381), 14.5% (354/2439), and 2.7% (14/518), respectively (Table 1).
The dominant species of Cryptosporidium are different in various animal types. In wild rodents, 21 species and 32 genotypes have been identified, and C. parvum is the dominant species. In pet rodents, 11 species and two genotypes have been identified, and C. wrairi is the dominant species. In farm rodents, five species and six genotypes have been identified, and C. parvum is the dominant species. In laboratory rodents, five species and one genotype have been identified, and C. homai is the dominant species. Wild and farm rodents have zoonotic potential. Pet rodents are often in close contact with humans, and the zoonotic species C. parvum, C. muris, C. andersoni, and C. ubiquitum have frequently been detected in pet rodents. Therefore, pet rodents pose a certain zoonotic risk that cannot be considered negligible.
3. Public health of zoonotic Cryptosporidium spp
3.1. Zoonotic Cryptosporidium spp. in humans
3.1.1. C. hominis
Although humans are the major host species for C. hominis, the species has been reported in a number of wildlife hosts, including rodents. To date, C. hominis has only been detected in wild striped field mice (Apodemus agrarius) in Slovakia [64,87]. Subtyping of C. hominis at the gp60 locus identified nine subtype families (Ia to Ij); only the subtype IbA10G2 of C. hominis has been reported in wild striped field mice [64]. The subtype IbA10G2 of C. hominis has been identified in humans, cattle, marsupials, and the European hedgehog. This is the main subtype associated with outbreaks of cryptosporidiosis caused by C. hominis [64]. Although the source of C. hominis in wild rodents is unclear, these animals can clearly serve as potential reservoirs for this pathogen.
3.1.2. C. parvum
C. parvum is one of the two most common Cryptosporidium species causing human cryptosporidiosis. The parasite infects a broad range of hosts, including various bovids, camelids, equids, canids, non-human primates, and marine mammals [94]. C. parvum is the dominant species in rodents; at least 20 species of rodents such as rats, mice, voles, squirrels, Rhizomys sinensis, and Chinchilla lanigera are known to be positive for C. parvum [8,11,24,28,37,44,64,68,80,87]. C. parvum has been detected in wild rodents, pet rodents, and farm rodents in 14 countries [37,80,87,24,64]. Pet rodents and farm rodents have close contact with humans. Therefore, pet rodents and farm rodents are potential reservoirs of C. parvum and thus may play an important role in the ecology of the zoonosis.
Subtyping of C. parvum at the gp60 locus identified more than 20 subtype families. Several studies that identified C. parvum in rodents have conducted typing at the gp60 locus; a variety of C. parvum subtypes including IIaA15G2R1, IIaA16G2R1, IIaA17G2R1, IIaA18G1R1b, IIaA18G3R1, IIdA15G1, IIiA10, IIpA9, IIpA6, IIoA15G1, and IIoA13G1 have been reported from rodents [37,80,87,24,64]. The IIa, IId, IIi, and IIo subtypes were previously reported in humans [87]. C. parvum IIp is genetically related to the IId and IIo subtypes; they have a broad host range and the potential for human infection. Rodents are frequently infected with IIdA15G1, and the most prevalent subtype family IId in rodents is also commonly found in cattle and other livestock.
3.1.3. C. meleagridis
C. meleagridis is a common cause of cryptosporidiosis in avian hosts. This species is the third most common species involved in human cryptosporidiosis [38,80]. To date, only four cases of C. meleagridis have been reported in wild rodents. Japan, USA, Spain, and Malaysia have reported the presence of C. meleagridis in R. norvegicus, Peromyscus spp., Rattus rattus, and in one unidentified wild rodent species. These findings may indicate a possible role of rodents in the mechanical transmission of this pathogen [38,65,80].
3.1.4. C. viatorum
C. viatorum has been frequently identified in human and urban wastewater; however, there were no reports in any animal species other than humans prior to recent studies reporting its occurrence in wild rodents [52,66,91]. Rodent species include Edward's long-tailed rat (55.3%, 21/38) and wild rats (12.0%, 25/228) in China and swamp rats in (14.3%, 3/21) in Australia [52,66,91]. To date, nine subtypes of C. viatorum (XVaA3a–XVaA3g, XVaA6, and XVbA2G1) have been identified globally. Subtypes XVbA2G1, XVaA6, XVaA3g, XVaA3h, XVcA2G1, XVcA2G1a, XVcA2G1b, and XVdA3 have been found in rodents. Interestingly, subtypes XVaA3a to XVaA3f were identified only in humans. XVaA6 was isolated from wastewater, and thus the presence of the XVa subtype family in rodents suggests that wild rats may have the potential for zoonotic transmission and must be considered a potential threat to human health.
3.1.5. C. ubiquitum
C. ubiquitum is considered an emerging zoonotic pathogen. The species has a broad host range that includes primates, carnivores, ruminants, and various rodents [46,94]. There are increasing research reports of C. ubiquitum being detected in rodents, and the host range has been expanding to include rodents such as wild squirrels, chipmunks, field mice, brown rats, Myocastor coypus, pet C. lanigera, farm bamboo rats, and laboratory mice [40,42,43,46,53,62,67,74,77,81]. In the USA, a study showed that transmission of C. ubiquitum to humans from rodents was likely to come from drinking untreated water contaminated by wildlife urine or feces [11].
Subtyping of C. ubiquitum at the gp60 locus identified nine subtype families (XIIa–XIIi). Subtypes XIIa and XIId have been found in pet C. lanigera in China (1.8%, 5/280 and 6.7%, 28/420, respectively). Subtype XIID has also been found in pet C. lanigera in Japan (20.6%, 13/63), M. coypus in the Czech Republic (5.8%, 7/120), M. coypus in the Slovak Republic (16.7%, 5/30), and C. lanigera in Japan (no data); subtype XIIi has been detected in Siberian flying squirrels in China (100%, 1/1) [40,42,46,53]. Subtype XIId appears to be common in rodents, occurring in C. ubiquitum isolates examined in many countries [42]. Subtypes XIIa and XIId have broadly specific and zoonotic potential, but novel subtype XIIi has unknown human infective potential.
3.2. Zoonotic Cryptosporidium spp. in rodents
A number of other zoonotic Cryptosporidium spp. have been identified in rodents, and several have been identified in humans. Of these, C. muris and C. andersoni are gastric parasites. C. muris has a wide host range that includes various mammals (rodents, canids, felids, suids, equids, NHPs, and marsupials) and birds, but C. andersoni primarily infects cattle, while the human infectivity is controversial. C. tyzzeri mostly infects domestic mice and small rodents, and it has been found in several non-specific hosts such as humans, pandas, black leopards, voles, snakes, and horses, among others [94]. Subtyping of C. tyzzeri at the gp60 locus identified three subtype families (IXa–IXc) that coevolved with hosts, each type having different natural host specificities [75]. Subtype IXa was restricted to the house mouse subspecies; subtype IXb was restricted to Mus musculus domesticus, and subtype IXc was detected only in A. sylvaticus.
C. suis and C. scrofarum are potential zoonotic species that are commonly detected in pigs. These species have been detected in rodents in Slovakia, China, and the Philippines; rodents may not be natural hosts for C. scrofarum and may have been infected as a result of mechanical transmission. C. occultus has a wide host range, mostly parasitizing rats genetically related to bamboo rats [46,49,50,52]. C. canis is the predominant species responsible for companion animal cryptosporidiosis, and the species has recently been reported in humans. C. ditrichi seems to be quite restricted to Apodemus spp. in Europe. The chipmunk genotype I has been identified in many rodents and in water [32,40,44,47,48,76]; this genotype is considered an emerging human pathogen. In C. erinacei, the skunk and muskrat genotypes I and II have also been reported in a few human cases of cryptosporidiosis.
3.3. Other Cryptosporidium species identified in rodents
Most rodent species and genotypes are host-specific or have narrow host ranges. Specific associations include C. myocastoris in nutrias [42]; C. proliferans in moles [46]; C. alticolis, C. microti, and vole genotypes I–VII in voles [29,35,37,54]; C. apodemi in mice [37,64]; C. homai and C. wrairi in guinea pigs [40,45,47,82]; C. rubeyi and squirrel genotypes I–III in ground squirrels [55,77]; C. ratti and mouse genotype II in rats [29,[43], [44], [45]]; hamster genotype in pet hamsters [40,47]; bamboo rat genotypes I–III in bamboo rats [49,50], and apodemus genotypes I and II in Apodemus spp. [37,48]. However, with increasing research effort, the host species ranges of these genotypes (rat genotypes II–V, ferret genotype, chipmunk genotypes II–V, muskrat genotypes I and II, and deer mouse genotypes I–IV) have gradually been extended [13,32,40,44,45,47,52,54,61,65,70,73,77].
C. varanii and C. scrofarum have mostly been isolated from reptiles and pigs; the presence in rodents may have been the result of mechanical transmission [53,61,87]. Cryptosporidium environmental has been found in wild Apodemus spp., suggesting that the environment plays an important role in transmission dynamics of the parasites [87]. Future studies aiming to characterize Cryptosporidium in environmental samples from areas with rodents are needed. The potential of these Cryptosporidium species and genotypes to cause disease in humans or animals is unknown, but C. ratti, rat genotype III, rat genotype IV, apodemus genotype II, and Cryptosporidium sp. 1 have been detected in streams in the USA and in raw sewage water in the UK, China, Japan, and USA [13,29,37,94]. To date, these species and genotypes have not been reported in humans, suggesting that they are unlikely to be of public health significance. However, more research is needed to confirm this.
4. Ecological significance from a One Health perspective
Rodents are essential components of many terrestrial ecosystems. Their beneficial activities in nature are well known, as is their transmission potential of pathogens to humans [92,[98], [99], [100], [101]]. Rodent transfer of pathogens to humans occurs by direct contact with humans and animals or through contamination of human or animal food and water by rodent stools, hair, and urine [9,98,99]. Rodents live in close contact with human populations; farm animals, pets, and peri-urban rodents provide a nexus between wildlife communities and humans, exposing humans to some zoonoses circulating in these natural ecosystems [99]. Rodent-borne diseases are associated with the rodent population and human socioeconomic lifestyle factors [11]. Human migration, travel, trade, urbanization, and agricultural activities can be facilitating factors in transferring rodent-borne pathogens from one community to another.
4.1. Possible direct transmission of rodent-borne Cryptosporidium at the human–animal–environment interface
Data obtained thus far suggest that rodents may play a role in the transmission of zoonotic Cryptosporidium spp. The first pathway is the direct route in which humans come in contact with Cryptosporidium present in rodent excrement or in an environmental component (food, water, or soil) that is contaminated with rodent urine. Moreover, humans may consume food products or water that is contaminated with rodent feces. Wild rodents are commonly found in urban and rural areas, thus providing a link between rural and urban disease foci [99]. Rodents often occur in the streets of cities or are hidden in food stores, granaries, and kitchens in rural areas, and they inhabit farmland, forests, and other natural environments. Their fecal droppings may be left wherever they forage, thus contaminating human and animal feed stores and accommodations. This can contribute to many of the sporadic human cases of cryptosporidiosis in urban and rural areas [99,98].
C. ditrichi oocysts were detected in the feces of decoration workers in Sweden. Epidemiological data indicated that the workers shared the same room with wild mice when they were working and thus had contact with mouse feces [37]. Humans can be directly infected with Cryptosporidium through contact with rodent feces. Humans often live closely with pet rodents; feeding, handling of rodents, close contact with feces, or playing with rodents can directly cause humans to be infected with Cryptosporidium (Fig. 2). At the same time, pet rodents excrete feces during family activities and thus contaminate the home environment (food, water, and the ground). Humans can become infected with Cryptosporidium by contact with an environment that is contaminated with rodent excrement. Laboratory and farm rodents have close contact with the breeder, and thus the breeder may be directly infected with Cryptosporidium by contacting rodent feces.
Fig. 2.
Schematic diagram showing the ecological and public health significance of Cryptosporidium in rodents and the major routes of transmission. Possible direct, indirect, waterborne, and foodborne transmission of rodent-borne Cryptosporidium at the human–animal–environment interface.
4.2. Possible waterborne transmission of rodent-borne Cryptosporidium at the human–animal–environment interface
Cryptosporidium is one of the most prevalent waterborne parasitic infections. Cryptosporidium spores can be transported into water bodies and become waterborne pathogens [94,95]. C. ubiquitum has been found in untreated drinking water in the USA. C. viatorum, the chipmunk genotype I, and the muskrat genotypes I and II have also been found in water [4,11,29,32,54]. These species and genotypes have zoonotic potential, and they can be transmitted to humans through drinking water or recreational waters (Fig. 2). To date, C. parvum and C. hominis have been responsible for all typed waterborne outbreaks [94]. Although C. parvum and C. hominis have not been detected in surface water contaminated by rodents or in the watershed within the living range of rodents, this may be due to fewer relevant studies; it is still a public health problem that cannot be ignored. Rodents contributing to Cryptosporidium contamination in water may have major public health significance since rodents are generally infected with human-pathogenic species and genotypes.
4.3. Possible foodborne transmission of rodent-borne Cryptosporidium at the human–animal–environment interface
Foodborne illnesses are any infections or diseases caused by consuming contaminated foods or drinks. Almost all reported Cryptosporidium cases of foodborne diseases are caused by C. parvum [5,97]. Cryptosporidium foodborne illnesses commonly involve contaminated water supplies, fresh fruits, and vegetables [10,11,92]. Polluted waters may contaminate food through the process of irrigating crops and washing vegetables, thereby causing foodborne transmission [3] (Fig. 2). Rats within fields excrete feces ubiquitously, thereby increasing the risk for human foodborne illness, particularly if harvested products are to be consumed raw [3]. It is difficult to remove Cryptosporidium oocysts from the surfaces of vegetables and fruits using common cleaning methods. Humans may ingest Cryptosporidium under unknown circumstances. Unfortunately, there is little relevant research in this area, and thus more research is needed to better understand the potential health risks.
4.4. Possible indirect transmission of rodent-borne Cryptosporidium at the human–animal–environment interface
Rodent-borne pathogens can also be spread indirectly to humans. At least 25 species and 43 genotypes of Cryptosporidium have been detected in rodents; some are zoonotic and can cross the species barrier. In addition to humans, rodent-related zoonotic species and genotypes of Cryptosporidium are present among wildlife, livestock, farm captive animals, and companion animals (Fig. 2). Moreover, rodents can maintain pathogen transmission cycles in different environments. Rodents can serve as potential mediators of Cryptosporidium and can transmit the parasites to wildlife, livestock, farm captive animals, and companion animals.
The risk of Cryptosporidium spreading within a rodent colony is elevated in rodent farms (Cavia porcellus, R. sinensis, and Rattus tanezumi) and laboratory rats that achieve high rodent densities whereby collective fecal production by a cohort, social behaviors (grooming and licking), and close interactions can promote intraspecific transmission, fecal shedding, and environmental persistence of Cryptosporidium. Coprophagia, a common behavior in a variety of rodent species, is a significant route for autoinfection of fecal-orally transmitted Cryptosporidium and can amplify Cryptosporidium shedding and disease spread within a rodent colony [92,99]. Wild rats leave many small droppings wherever they forage (including watersheds) in wild areas. Wildlife and domestic animals can become infected with Cryptosporidium by ingesting contaminated food, water, and rodents (possible in carnivores) and thereby transfer the pathogens. At the same time, pet rodents excrete feces during family activities, possibly causing dogs or cats to be infected with Cryptosporidium. Ultimately, humans may be indirectly infected with Cryptosporidium through contact with wildlife, livestock, and companion animals or contaminated environments.
Wild rodents may spread Cryptosporidium on farms. This risk may be even greater on captive animal farms where wild rats can share the habitat with farm animals or travel through grazing land used by domestic animals where contact with livestock is more likely, thus providing ample opportunity for transmission of Cryptosporidium to the livestock. Infected wild rodents on livestock farms can potentially transmit pathogens to the livestock by contaminating animal feed and water sources with fecal pellets. Horizontal transmission by infected captive animals on farms can amplify Cryptosporidium shedding and disease spread. Additionally, rodenticides are used less often on farms, further increasing the possibility of Cryptosporidium spreading. Rodents may also acquire enteric microbes from livestock and amplify and mechanically vector the pathogens across agricultural landscapes, thereby facilitating the spread of disease [2]. Ultimately, breeders may be indirectly infected with Cryptosporidium through contact with farm animals and contaminated environments.
5. Conclusions
Cryptosporidium spp. are common in rodents. To date, 25 Cryptosporidium species and 43 genotypes have been identified in rodents, including the species that cause outbreaks of human cryptosporidiosis, C. parvum and C. hominis. C. parvum is the dominant species in rodents. The fact that zoonotic C. parvum, C. hominis, C. meleagridis, and C. ubiquitum, particularly subtypes IbA10G2 and IIdA15G1, have been found in rodents suggests that rodents infected with Cryptosporidium have significant zoonotic potential. For Cryptosporidium at the human–animal–environment interface, rodents can be a direct route or can become potential mediators in parasite transmission. There is no direct evidence with which to illustrate the transmission pathways of Cryptosporidium at the human–animal–environment interface. In addition to rodents, dogs, cats, wild animals, and livestock animals can be involved in the transmission cycle. Studies are required to investigate Cryptosporidium among the diverse human population, livestock, pet animals, and rodents in various ecosystems. Researchers should pursue a multidisciplinary One Health approach with contributions from zoologists, ecologists, veterinarians, and public health experts to understand rodent-related Cryptosporidium and possible transmission routes.
Search strategy
We conducted a systematic literature search from June 20, 2021 to June 26, 2021 through four databases: PubMed, Scopus, Science Direct, and Web of Science. The search included original field epidemiological articles in English for each of the 86 rodent-Cryptosporidium diseases individually, with no time limit of publication (Supplementary Table S1). The search terms included Cryptosporidium or Cryptosporidiosis and scientific names of animal species We screened the searches as “Title/Abstract” in PubMed, “Find articles with these terms” in Science Direct, “TITLE-ABS-KEY” in Scopus, and “Topic” in Web of Science.
Funding
This study was supported in part by the National Natural Science Foundation of China (U1904203), the Higher Education Teaching Reform Research and Practice Project of Henan Province in 2019 (2019SJGLX006Y), and the Leading talents of the Thousand Talents Program of Central China (19CZ0122). The sponsors played no role in the study design or in the collection, analysis, or interpretation of the data, in writing the report, or in the decision to submit the article for publication.
The following are the supplementary data related to this article.
Cryptosporidium species and genotypes identified in rodents in the world
Declaration of Competing Interest
The authors declare that no competing interests exist.
Acknowledgments
We thank Let Pub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
References
- 1.Tallant C., Huddleston P., Alshanberi A., Misra S. Acute, Severe Cryptosporidiosis in an Immunocompetent Pediatric Patient. Clin. Pract. 2016;6(2):837. doi: 10.4081/cp.2016.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donskow K., Bajer A., Bednarska M., Siñski E. Experimental transmission of Cryptosporidium parvum isolates from wild rodents and calves to laboratory bred common voles (Microtus arvalis) Acta Parasitol. 2005;50(1):19–24. [Google Scholar]
- 3.Kilonzo C. University of California; Davis: 2015. Epidemiology of Foodborne Pathogens in Wild Rodents in a Major Agricultural Region of the California Central Coast. [Google Scholar]
- 4.Feng Y., Alderisio K.A., Yang W., Blancero L.A., Kuhne W.G., Nadareski C.A. Cryptosporidium Genotypes in Wildlife from a New York Watershed. Appl. Environ. Microbiol. 2007;73(20):6475. doi: 10.1128/AEM.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Åberg R., Sjöman M., Hemminki K., Pirnes A., Räsänen S., Kalanti A., et al. Cryptosporidium parvum Caused a Large Outbreak Linked to Frisée Salad in Finland, 2012. Zoonoses Public Health. 2015;62(8):618–624. doi: 10.1111/zph.12190. [DOI] [PubMed] [Google Scholar]
- 7.Chalmers R.M., Sturdee A.P., Bull S.A., Miller A., Wright S.E. The prevalence of Cryptosporidium parvum and C. muris in Mus domesticus, Apodemus sylvaticus and Clethrionomys glareolus in an agricultural system. Parasitol. Res. 1997;83(5):478–482. doi: 10.1007/s004360050283. [DOI] [PubMed] [Google Scholar]
- 8.Torres J., Gracenea M., Gómez M.S., Arrizabalaga A., González-Moreno O. The occurrence of Cryptosporidium parvum and C. muris in wild rodents and insectivores in Spain. Vet. Parasitol. 2000;92(4):253–260. doi: 10.1016/s0304-4017(00)00331-9. [DOI] [PubMed] [Google Scholar]
- 9.Strand T.M., Lundkvist Å. Rat-borne diseases at the horizon. A systematic review on infectious agents carried by rats in Europe 1995-2016. Infect. Ecol. Epidemiol. 2019;9(1):1553461. doi: 10.1080/20008686.2018.1553461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rzezutka A., Nichols R.A., Connelly L., Kaupke A., Kozyra I., Cook N., et al. Cryptosporidium oocysts on fresh produce from areas of high livestock production in Poland. Int. J. Food Microbiol. 2010;139(1−2):96–101. doi: 10.1016/j.ijfoodmicro.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 11.Hamidi K. How do Rodents Play Role in Transmission of Foodborne Diseases, Nutr Food Sci. Int. J. 2018;6:1–4. [Google Scholar]
- 12.O’Hara S.P., Chen X.M. The cell biology of Cryptosporidium infection. Microbes Infect. 2011;13(8–9):721–730. doi: 10.1016/j.micinf.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ježková J., Prediger J., Holubová N., Sak B., Konečný R., Feng Y., et al. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitol. 2020;148(1):1–45. doi: 10.1017/S0031182020001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klesius P.H., Haynes T.B., Malo L.K. Infectivity of Cryptosporidium sp. isolated from wild mice for calves and mice. J. Am. Vet. Med. Assoc. 1986;189(2):192–193. [PubMed] [Google Scholar]
- 24.Siński E., Bednarska M., Bajer A. The role of wild rodents in ecology of Cryptosporidiosis in Poland. Folia Parasitol. 1998;45(2):173–174. doi: 10.1006/expr.1998.4215. [DOI] [PubMed] [Google Scholar]
- 27.Nakai Y., Hikosaka K., Sato M., Sasaki T., Kaneta Y., Okazaki N. Detection of Cryptosporidium muris Type Oocysts from Beef Cattle in a Farm and from Domestic and Wild Animals in and around the Farm (Parasitology) J. Vet. Med. Sci. 2004;66(8):983–984. doi: 10.1292/jvms.66.983. [DOI] [PubMed] [Google Scholar]
- 28.Perz J.F., Blancq S. Cryptosporidium parvum Infection Involving Novel in Wildlife from Lower New York State. Appl. Environ. Microbiol. 2001;67(3):1154–1162. doi: 10.1128/AEM.67.3.1154-1162.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L., Fayer R., Trout J.M., Ryan U.M., Schaefer F.W., Xiao L. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl. Environ. Microbiol. 2004;70(12):7574–7577. doi: 10.1128/AEM.70.12.7574-7577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kvác M., Hofmannová L., Bertolino S., Wauters L., Tosi G., Modrý D. Natural infection with two genotypes of Cryptosporidium in red squirrels (Sciurus vulgaris) in Italy. Folia Parasitol. 2008;55(2):95–99. [PubMed] [Google Scholar]
- 35.Horčičková M., Čondlová Š., Holubová N., Sak B., Květoňová D., Hlásková L., et al. Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. n. and Cryptosporidium microti sp. n. (Apicomplexa: Cryptosporidiidae) Parasitol. 2008;146(2):220–233. doi: 10.1017/S0031182018001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Čondlová Š., Horčičková M., Havrdová N., Sak B., Hlásková L., Perec-Matysiak A., et al. Diversity of Cryptosporidium spp. in Apodemus spp. in Europe. Eur. J. Protistol. 2019;69:1–13. doi: 10.1016/j.ejop.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García L.K., Martín A.A., Foronda P. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasit. Vectors. 2020;13(1):445. doi: 10.1186/s13071-020-04330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J., Wang W., Lin Y., Sun L., Li N., Guo Y., et al. Genetic characterizations of Cryptosporidium spp. from pet rodents indicate high zoonotic potential of pathogens from chinchillas. One Health. 2021;13 doi: 10.1016/j.onehlt.2021.100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ježková J., Limpouchová Z., Prediger J., Holubová N., Sak B., Konečný R., et al. Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus) Microorganisms. 2021;9(4):813. doi: 10.3390/microorganisms9040813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao W., Wang J., Ren G., Yang Z., Yang F., Zhang W., et al. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasit. Vectors. 2018;11(1):313. doi: 10.1186/s13071-018-2892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng L., Chai Y., Luo R., Yang L., Yao J., Zhong Z., et al. Occurrence and genetic characteristics of Cryptosporidium spp. and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-57896-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chai Y., Deng L., Liu H., Yao J., Zhong Z., Xiang L., et al. First detection of Cryptosporidium spp. in red-bellied tree squirrels (Callosciurus erythraeus) in China. Parasite. 2019;26:28. doi: 10.1051/parasite/2019029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F., Zhang Z., Hu S., Zhao W., Zhao J., Kváč M., et al. Common occurrence of divergent Cryptosporidium species and Cryptosporidium parvum subtypes in farmed bamboo rats (Rhizomys sinensis) Parasit. Vectors. 2020;13(1):149. doi: 10.1186/s13071-020-04021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv C., Zhang L., Wang R., Jian F., Zhang S., Ning C., et al. Cryptosporidium spp. in Wild, Laboratory, and Pet Rodents in China: prevalence and molecular characterization. Appl. Environ. Microbiol. 2009;75(24):7692–7699. doi: 10.1128/AEM.01386-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song J., Kim C.Y., Chang S.N., Abdelkader T.S., Han J., Kim T.H., et al. Detection and Molecular Characterization of Cryptosporidium spp. from Wild Rodents and Insectivores in South Korea. Korean J. Parasitol. 2015;53(6):737–743. doi: 10.3347/kjp.2015.53.6.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Z., Liu Q., Zhao W., Jiang X., Zhang Y., Zhao A., et al. Prevalence and diversity of Cryptosporidium spp. in bamboo rats (Rhizomys sinensis) in South Central China. Int. J. Parasitol. Parasites Wildl. 2019;9:312–316. doi: 10.1016/j.ijppaw.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F., Zhao W., Zhang C., Guo Y., Li N., Xiao L., et al. Cryptosporidium Species and C. parvum Subtypes in Farmed Bamboo Rats. Pathogens. 2020;9(12):1018. doi: 10.3390/pathogens9121018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao W., Zhou H., Huang Y., Xu L., Rao L., Wang S., et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: Molecular detection, species/genotype identification and implications for public health. Int. J. Parasitol. Parasites Wildl. 2019;9(C):317–321. doi: 10.1016/j.ijppaw.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melicherová J., Ilgová J., Kváč M., Sak B., Koudela B., Valigurová A. Life cycle of Cryptosporidium muris in two rodents with different responses to parasitization. Parasitol. 2014;141(2):287–303. doi: 10.1017/S0031182013001637. [DOI] [PubMed] [Google Scholar]
- 54.Feng S., Chang H., Wang Y., Huang C., Han S., He H. Molecular Characterization of Cryptosporidium spp. in Brandt's Vole in China. Front. Vet. Sci. 2020;7:300. doi: 10.3389/fvets.2020.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X., Pereira M.D., Larsen R., Xiao C., Phillips R., Striby K., et al. Cryptosporidium rubeyi n. sp. (Apicomplexa: Cryptosporidiidae) in multiple Spermophilus ground squirrel species. Int. J. Parasitol. Parasites Wildl. 2015;4(3):343–350. doi: 10.1016/j.ijppaw.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montecino L.D., Li X., Xiao C., Atwill E.R. Elevation and vegetation determine Cryptosporidium oocyst shedding by yellow-bellied marmots (Marmota flaviventris) in the Sierra Nevada Mountains. Int. J. Parasitol. Parasites Wildl. 2015;4(2):171–177. doi: 10.1016/j.ijppaw.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brar S.K., Singla L.D. Concurrent infection of Cryptosporidium and Giardia in synanthropic rodents: First report from Punjab, India. Indian J. Vet. Sci. Biotechnol. 2021;17(1):1–3. [Google Scholar]
- 61.Ng-Hublin J.S., Singleton G.R., Ryan U. Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect. Genet. Evol. 2013;16:5–12. doi: 10.1016/j.meegid.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 62.Murakoshi F., Fukuda Y., Matsubara R., Kato Y., Sato R., Sasaki T., et al. Detection and genotyping of Cryptosporidium spp. in large Japanese field mice, Apodemus speciosus. Vet. Parasitol. 2013;196(1−2):184–188. doi: 10.1016/j.vetpar.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Zhao Z., Wang R., Zhao W., Qi M., Zhao J., Zhang L., et al. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology. 2015;142(6):800–806. doi: 10.1017/S0031182014001929. [DOI] [PubMed] [Google Scholar]
- 64.Čondlová Š., Horčičková M., Sak B., Květoňová D., Hlásková L., Konečný R., et al. Cryptosporidium apodemi sp. n. and Cryptosporidium ditrichi sp. n. (Apicomplexa: Cryptosporidiidae) in Apodemus spp. Eur. J. Protistol. 2018;63:1–12. doi: 10.1016/j.ejop.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 65.Tan T.K., Low V.L., Ng W.H., Ibrahim J., Wang D., Tan C.H., et al. Occurrence of zoonotic Cryptosporidium and Giardia duodenalis species/genotypes in urban rodents. Parasitol. Int. 2019;69:110–113. doi: 10.1016/j.parint.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y.W., Zheng W.B., Zhang N.Z., Gui B.Z., Lv Q.Y., Yan J.Q., et al. Identification of Cryptosporidium viatorum XVa subtype family in two wild rat species in China. Parasit. Vectors. 2019;12(1):502. doi: 10.1186/s13071-019-3763-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matysiak A.P., Buńkowska G.K., Zaleśny G., Hildebrand J. Small rodents as reservoirs of Cryptosporidium spp. and Giardia spp. in south-western Poland. Ann. Agric. Environ. Med. 2015;22(1):1–5. doi: 10.5604/12321966.1141359. [DOI] [PubMed] [Google Scholar]
- 68.Bystrianska J., Papajová I., Šmiga Ľ., Šoltys J., Majláthová V., Majláth I., et al. First report on parasites of European beavers in the Slovak Republic. Parasitol. Res. 2020;120(1):355–358. doi: 10.1007/s00436-020-06943-6. [DOI] [PubMed] [Google Scholar]
- 70.Ayinmode A.B., Ogbonna N.F., Widmer G. Detection and molecular identification of Cryptosporidium species in laboratory rats (Rattus norvegicus) in Ibadan, Nigeria. Ann. Parasitol. 2017;63(2):105–109. doi: 10.17420/ap6302.92. [DOI] [PubMed] [Google Scholar]
- 73.Paparini A., Jackson B., Ward S., Young S., Ryan U. Multiple Cryptosporidium genotypes detected in wild black rats (Rattus rattus) from northern Australia. Exp. Parasitol. 2012;131(4):404–412. doi: 10.1016/j.exppara.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Qi M., Luo N., Wang H., Yu F., Wang R., Huang J., et al. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol. Int. 2015;64:339–341. doi: 10.1016/j.parint.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 75.Ren X., Zhao J., Zhang L., Ning C., Jian F., Wang R., et al. Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus) Exp. Parasitol. 2012;130(3):274–281. doi: 10.1016/j.exppara.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 76.Stenger B.L., Clark M.E., Kváč M., Khan E., Giddings C.W., Dyer N.W., et al. Highly divergent 18S rRNA gene paralogs in a Cryptosporidium genotype from eastern chipmunks (Tamias striatus) Infect. Genet. Evol. 2015;32:113–123. doi: 10.1016/j.meegid.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stenger B.L.S., Clark M.E., Kváč M., Khan E., Giddings C.W., Prediger J. North American tree squirrels and ground squirrels with overlapping ranges host different Cryptosporidium species and genotypes. Infect. Genet. Evol. 2015;36:287–293. doi: 10.1016/j.meegid.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 78.Ryan U., Xiao L., Read C., Zhou L., Lal A.A., Pavlasek I. Identification of Novel Cryptosporidium Genotypes from the Czech Republic. Appl. Environ. Microbiol. 2003;69(7):4302–4307. doi: 10.1128/AEM.69.7.4302-4307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hikosaka K., Nakai Y. A novel genotype of Cryptosporidium muris from large Japanese field mice, Apodemus speciosus. Parasitol. Res. 2005;97(5):373–379. doi: 10.1007/s00436-005-1459-7. [DOI] [PubMed] [Google Scholar]
- 80.Kimura A., Edagawa A., Okada K., Takimoto A., Yonesho S., Karanis P., et al. Detection and genotyping of Cryptosporidium from brown rats (Rattus norvegicus) captured in an urban area of Japan. Parasitol. Res. 2007;100(6):1417–1420. doi: 10.1007/s00436-007-0488-9. [DOI] [PubMed] [Google Scholar]
- 81.Koyama Y., Satoh M., Maekawa K., Hikosaka K., Nakai Y. Isolation of Cryptosporidium andersoni Kawatabi type in a slaughterhouse in the northern island of Japan. Vet. Parasitol. 2005;130(3–4):323–326. doi: 10.1016/j.vetpar.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 82.Zahedi A., Durmic Z., Gofton A.W., Kueh S., Austen J., Lawson M., et al. Cryptosporidium homai n. sp. (Apicomplexa: Cryptosporidiiae) from the guinea pig (Cavia porcellus) Vet. Parasitol. 2017;245:92–101. doi: 10.1016/j.vetpar.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 87.Danišová O., Valenčáková A., Stanko M., Luptáková L., Hatalová E., Čanády A. Rodents as a reservoir of infection caused by multiple zoonotic species/genotypes of C. parvum, C. hominis, C. suis, C. scrofarum, and the first evidence of C. muskrat genotypes I and II of rodents in Europe-Science Direct. Acta Trop. 2017;172:29–35. doi: 10.1016/j.actatropica.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X., Jian Y., Li X., Ma L., Karanis G., Karanis P. The first report of Cryptosporidium spp. in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in the Qinghai-Tibetan Plateau area, China. Parasitol. Res. 2018;117(5):1401–1407. doi: 10.1007/s00436-018-5827-5. [DOI] [PubMed] [Google Scholar]
- 91.Koehler A.V., Wang T., Haydon S.R., Gasser R.B. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus - An emerging zoonotic pathogen. Int. J. Parasitol. Parasites Wildl. 2018;7(1):18–26. doi: 10.1016/j.ijppaw.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kunz Mammal species of the world: a taxonomic and geographic reference. Mastozool. Neotropical. 2006;13(2):290–293. [Google Scholar]
- 93.Ranjbar-Bahadori S.H., Mostoophi A., Shemshadi B. Study on Cryptosporidium contamination in vegetable farms around Tehran. Trop. Biomed. 2013;30(2):193–198. [PubMed] [Google Scholar]
- 94.Zahedi A., Paparini A., Jian F., Robertson I., Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int. J. Parasitol. Parasites Wildl. 2016;5(1):88–109. doi: 10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rochelle P.A., Giovanni G. Springer Vienna; 2014. Cryptosporidium Oocysts in Drinking Water and Recreational Water; pp. 489–513. [Google Scholar]
- 97.Pönka A., Kotilainen H., Rimhanen-Finne R., Hokkanen P., Hänninen M.L., Kaarna A., et al. A foodborne outbreak due to Cryptosporidium parvum in Helsinki, November 2008. Euro Surveill. 2009;14(28):19269. doi: 10.2807/ese.14.28.19269-en. [DOI] [PubMed] [Google Scholar]
- 98.Han B.A., Schmidt J.P., Bowden S.E., Drake J.M. Rodent reservoirs of future zoonotic diseases. Proc. Natl. Acad. Sci. 2015;112:7039–7044. doi: 10.1073/pnas.1501598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meerburg B.G., Singleton G.R., Kijlstra A. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 2009;35:221–270. doi: 10.1080/10408410902989837. [DOI] [PubMed] [Google Scholar]
- 100.Khaghani R. The economic and health impact of rodent in urban zone and harbours and their control methods. Ann. Mil. Health Sci. Res. 2007;4:1071–1078. [Google Scholar]
- 101.Rabiee M.H., Mahmoudi A., Siahsarvie R., Kryštufek B., Mostafavi E. Rodent-borne diseases and their public health importance in Iran. PLoS Negl. Trop. Dis. 2018;12(4) doi: 10.1371/journal.pntd.0006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cryptosporidium species and genotypes identified in rodents in the world