Abstract
We have isolated and microsequenced Snu17p, a novel yeast protein with a predicted molecular mass of 17 kDa that contains an RNA recognition motif. We demonstrate that Snu17p binds specifically to the U2 small nuclear ribonucleoprotein (snRNP) and that it is part of the spliceosome, since the pre-mRNA and the lariat-exon 2 are specifically coprecipitated with Snu17p. Although the SNU17 gene is not essential, its knockout leads to a slow-growth phenotype and to a pre-mRNA splicing defect in vivo. In addition, the first step of splicing is dramatically decreased in extracts prepared from the snu17 deletion (snu17Δ) mutant. This defect is efficiently reversed by the addition of recombinant Snu17p. To investigate the step of spliceosome assembly at which Snu17p acts, we have used nondenaturing gel electrophoresis. In Snu17p-deficient extracts, the spliceosome runs as a single slowly migrating complex. In wild-type extracts, usually at least two distinct complexes are observed: the prespliceosome, or B complex, containing the U2 but not the U1 snRNP, and the catalytically active spliceosome, or A complex, containing the U2, U6, and U5 snRNPs. Northern blot analysis and affinity purification of the snu17Δ spliceosome showed that it contains the U1, U2, U6, U5, and U4 snRNPs. The unexpected stabilization of the U1 snRNP and the lack of dissociation of the U4 snRNP suggest that loss of Snu17p inhibits the progression of spliceosome assembly prior to U1 snRNP release and after [U4/U6.U5] tri-snRNP addition.
Nuclear pre-mRNA splicing occurs via a two-step transesterification reaction that is catalyzed by a multicomponent complex termed the spliceosome (25). The spliceosome is built by the ordered association of the U1, U2, U4, U5, and U6 small nuclear ribonucleoprotein particles (snRNPs) and extrinsic protein factors with the pre-mRNA (reviewed in references 19, 25, 40, and 46). Each individual snRNP contains a unique set of particle-specific proteins and seven common Sm proteins, while the U6 snRNP contains a unique set of Sm-like (Lsm) proteins. The U5, U4, and U6 snRNPs form a trimeric complex in which the U4 and U6 snRNAs are extensively base paired.
The spliceosome is a highly dynamic molecular machine, forming anew on each pre-mRNA substrate in an ordered manner (4, 40). First, the U1 snRNP binds the pre-mRNA by interacting with the 5′ splice site (34, 38), leading to the formation of commitment complexes in yeast (33, 38) and the E complex in mammals (24). The U2 snRNP, which interacts with the pre-mRNA branchpoint region, is then added in an ATP-dependent step, resulting in the formation of the prespliceosome (reviewed in reference 25). Finally, U4, U5, and U6 snRNPs, preassembled in a single particle, join the prespliceosome to form the mature spliceosome, in which the pre-mRNA splicing reaction occurs. Several rearrangements then take place that activate the spliceosome for the two catalytic steps of splicing. For example, U1 and U4 are destabilized and the spliceosome is activated for catalysis (4). The first splicing reaction creates two intermediates, a cleaved upstream exon and an intron-downstream exon in a branched “lariat” configuration. The second splicing reaction results in the ligation of the exons and excision of the lariat-intron. The spliceosome then disassembles, and the products of the reaction are released.
As mentioned above, at the molecular level the assembly pathway involves rearrangements of RNA-RNA interactions, many of which must be disrupted to allow the formation of others (27, 40). After the 5′ splice site is recognized by U1 through base pairing (37, 39, 49), this interaction must be replaced by a mutually exclusive interaction with U6 (15, 18, 21, 36, 44). Similarly, the base-pairing interaction between U4 and U6 must be disrupted to allow the U2-U6 interaction, which comprises a major element of the spliceosome's catalytic core (23). These rearrangements are thought to be critical for activating the spliceosome for the catalytic steps of splicing. Notably, the DEAD-box ATPase Prp28p was identified as a potential mediator of the U1-to-U6 switch, perhaps by unwinding either the U1-5′ splice site duplex or the U4-U6 duplex (41). In addition, a mutation in U4 snRNA, which blocks U4-U6 unwinding as well as U1 release, can be suppressed by a specific mutation in the U5 snRNP protein Prp8p (20).
These and other data indicate that snRNP proteins play a key role in triggering the activation of the spliceosome and in spliceosome assembly (8). Some of these proteins were identified genetically in yeast, while others were characterized biochemically in various systems. Using these tools, the yeast U1 snRNP and the [U4/U5.U6] tri-snRNP recently have been extensively characterized (11, 12, 26, 32, 42). In contrast, our knowledge of the yeast U2 snRNP is less comprehensive, since a distinct U2 snRNP equivalent to the human U2 snRNP could only be partially purified from yeast (6). This led to the identification of a novel essential splicing factor, Rse1p/Snu154p, the yeast ortholog of the human protein SAP130, which is one of the subunits of the heteromeric U2 snRNP-associated splicing factor SF3b (6, 11). The human U2 snRNP contains nine characterized specific proteins besides the seven common Sm proteins (2). Sequence analysis revealed that homologs of all these proteins exist in yeast (5, 35, 43). Yib9p and Lea1p are the orthologs of metazoan U2B" and U2A′, respectively (5, 43). Cus2p is also a yeast U2 snRNP-associated protein but apparently does not have a human counterpart that associates with the human U2 snRNP (48). Based on several criteria, the PRP9, PRP11, and PRP21 gene products are homologs of the mammalian SF3a subunits; Hsh49p, Cus1p, and ySAP155, together with Rse1p (Snu154p), are homologs of the mammalian SF3b subunits (14, 28, 30, 31, 45, 47).
Although considerable progress has been made in identifying the critical RNA-RNA and RNA-protein interactions required for splicing, little is understood about the factors mediating the structural dynamics. While characterizing the yeast [U4/U6.U5] tri-snRNP proteins, we identified several U1 and U2 snRNP-specific proteins in our tri-snRNP preparations, which probably were due to the small amounts of copurifying U1 and U2 snRNPs (11). Here, we describe the characterization of one of these proteins with a predicted molecular mass of 17 kDa encoded by open reading frame YIR005w. This protein, named Snu17p, contains an RNA recognition motif (RRM). We show that Snu17p is specifically associated with the U2 snRNP and is an intrinsic component of prespliceosomes and spliceosomes, even after the first catalytic step of splicing has occurred. Deletion of the SNU17 gene leads to a slow-growth phenotype and to a splicing defect in vivo and in vitro. We demonstrate that the in vitro splicing defect is due to the lack of Snu17p since it can be specifically reversed by addition of recombinant protein. Analysis of spliceosomal complexes shows that in extracts lacking Snu17p an unusual complex accumulates that contains U1, U2, U4, U5, and U6 snRNPs. Thus, deletion of Snu17p traps the prespliceosome and allows the [U4/U6.U5] tri-snRNP to bind to this intermediate. However, progression to a more mature form of the spliceosome is inhibited.
MATERIALS AND METHODS
Yeast strains and plasmids.
A yeast strain expressing the Snu17p protein with a C-terminal protein A tag was created using a PCR-based strategy as previously described (11, 29). The SNU17 3′ untranslated region (3′-UTR) was amplified from yeast genomic DNA using oligonucleotides B1 and B2. This construct was ligated to the protA-TRP1 cassette from plasmid pBS1173, cleaved with XhoI, and filled in with Klenow fragment. The DNA cassette encoding the protein A tag and carrying the TRP1 marker gene and the SNU17 3′-UTR was then amplified by PCR from the ligation mixture using oligonucleotide B3, which is homologous to the 3′-end of the SNU17 coding region and eliminates the stop codon, and oligonucleotide B2. The PCR product was transformed into strain TR2 (MATa trp1-Δ1 his3-Δ ura3-52 lys2-801 ade2-101) to give rise to strain AGY9.
The SNU17 gene was deleted by a PCR-based strategy, replacing the entire coding region with the HIS3-marker gene. First, the 5′- and 3′-UTRs were amplified from yeast genomic DNA using primers SNU17 5′-up and SNU17 5′-low as well as SNU17 3′-up and SNU17 3′-low. The HIS3 marker gene was amplified using primers HIS3 up BsrGI and HIS3 low AatII, which are homologous to the 5′-UTR and 3′-UTR of SNU17, respectively. The 5′-UTR product was then fused by PCR to the HIS3 gene using primers SNU17 5′-up and HIS3 low AatII. The resulting product was purified and then fused by PCR to the 3′-UTR product using primers SNU17 5′-up and SNU17 3′-low. The final 5′-UTR–HIS3–3′-UTR cassette was then transformed into strain TR1. Integrants were selected on medium lacking histidine, giving rise to strain AGY17. These cells were then sporulated, and tetrads were dissected. All four spores were viable, although the two spores containing the HIS3 marker grew slower. Thus, a haploid snu17Δ strain was isolated (AGY21) (trp1-Δ1 his3-Δ ura3-52 lys2-801 ade2-101 snu17Δ::HIS3).
Expression and purification of GST-Snu17p fusion protein.
The coding region of SNU17 was amplified by PCR from yeast genomic DNA using oligonucleotides SNU17 up BamHI and SNU17 low XhoI. The PCR product was cut with BamHI and XhoI and ligated into vector pGEX-4T1 (Amersham-Pharmacia) between the respective restriction sites. This plasmid was transformed into Escherichia coli strain BL21. Glutathione S-transferase (GST)–Snu17p fusion protein was overexpressed and purified as specified by the manufacturer (Amersham-Pharmacia). It was subsequently dialyzed against buffer D150 (20 mM HEPES-KOH [pH 7.9], 150 mM KCl, 8% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol) and stored in small aliquots at −80°C.
Immunoprecipitation, Northern analysis, and primer extension.
Splicing extracts were prepared from strain AGY9, essentially as described previously (22). A 20-μl volume of immunoglobulin G (IgG)-agarose (Sigma) was washed with buffer NET-2 75 (50 mM Tris-HCl [pH 7.5], 75 mM NaCl, 0.05% NP-40) and used to precipitate Snu17p-ProtA from 50 μl of extract for 1.5 h at 4°C. The beads were then washed three times with 1.5 ml of NET-2 75. Parallel precipitations were washed with NET-2 buffers containing 150, 225, 300, or 375 mM NaCl. As control, extract from the isogenic strain TR2 was precipitated with IgG beads. The beads were then incubated with proteinase K for 15 min at 37°C. The copreciptated snRNAs were subsequently extracted with phenol-chloroform-isoamyl alcohol (50:49:1) and separated by electrophoresis on a denaturing gel containing 6 M urea and 8% polyacrylamide. After the gels were blotted to a nylon membrane (Quiagen), which was UV irradiated for 3 min, Northern analysis with uniformly radiolabeled DNA probes specific for the U1, U2, U4, U5, and U6 snRNAs was performed. In vivo accumulation of unspliced pre-U3A and pre-U3B RNA transcripts was assayed by primer extension, as described previously (11).
In vitro splicing complementation with recombinant Snu17p, precipitation of splicing-reaction products, nondenaturing gels, and affinity purification of spliceosomes.
Splicing reactions were performed as described previously (22), generally using actin pre-mRNA transcribed in the presence of [α-32P]UTP. When splicing extracts were complemented with recombinant GST-Snu17p protein, 50 ng of protein (1.2 pmol) in 0.5 μl of buffer D150 per 5 μl of reaction mixture was used. As a control, only 0.5 μl of buffer D150 was added.
Precipitation of Snu17p-protA and associated pre-mRNA or derived mRNA species from in vitro splicing-reaction mixtures was performed as previously described (11), using the AGY9 extract and radiolabeled transcripts of actin or U3 pre-mRNA. A 30-μl volume of extract was incubated with 150,000 cpm of 32P-labeled pre-mRNA (specific activity, 10,000 cpm/fmol) for 40 min under splicing conditions. We kept 1/15 of the mixture for analysis of the splicing reaction, while the remaining splicing mixture was precipitated using 20 μl of IgG-agarose (Sigma) in NET-2 150 buffer for 1.5 h at 4°C. Coprecipitated pre-mRNA and splicing products were extracted from the washed IgG-agarose beads and analyzed by denaturing gel electrophoresis followed by autoradiography. Nondenaturing gel electrophoresis was performed as described previously (7), except that the EDTA concentration used to block spliceosome assembly in A1 was 8 mM instead of 5 mM. Oligonucleotide d1, used to destroy U6 snRNA, is described in reference 10. For affinity purification of spliceosomes, actin pre-mRNA was transcribed in the presence of biotin-16-UTP (Boehringer) with UTP/biotin-16-UTP ratio of 20:1. Standard splicing-reaction mixtures (125 μl) containing 50 μl extract and 640 fmol of biotinylated actin pre-mRNA were incubated for 20 min at 23°C, treated with 2 μg of heparin per μl of extract, and then sedimented on a 2.4-ml 10 to 30% glycerol gradient in buffer D containing 100 mM KCl. The gradients were run for 3 h at 55,000 rpm in a Beckman TL-100 ultracentrifuge. After fractionation of the gradient into 15 fractions of 160 μl, four-fifths of each fraction was used for affinity purification by addition of 15 μl of streptavidin agarose in 450 μl of NET-2 buffer containing 100 mM NaCl, for 1.5 h at 4°C. The precipitates were washed extensively with NET-2 100, and RNAs were extracted from the beads and analysed by Northern blot analysis.
Oligonucleotides used in this work.
The following oligonucleotides were used: B1 (5′-ATAATTTTTTCTTTCATC-3′), B2 (5′-GCTACTGCCGTGCAAAAA-3′), B3 (5′-AAGAACAATGCTGAGAAACTCATTTTGGCTAAAAAGGACCAACCACCTCCGCTGGAGCTCAAAAC-3′), SNU17 5′-up (5′-GAAGAGCTGAAGCAATGA-3′), SNU17 5′-low (5′-TTCTAGATGTACACCGCCGAATTGCACGCTGCACTT-3′), SNU17 3′-up (5′-TCGATACGACGTCCCGCCGAACAATGCTGAGAAACTC-3′), SNU17 3′-low (5′-AGCTTTAGCTGGAGGCGT-3′), HIS3 up BsrGI (5′-GGCGGTGTACATCTAGAAAGAGCTTGGTG-3′), HIS3 low AatII (5′-GGCGGGACGTCGTATCGAGTTCAAGAGAA-3′), SNU17 up BamHI (5′-GGCGGGGATCCAACAAAATTCAGCAAATC-3′), and SNU17 low XhoI (5′-GGCGGCTCGAGTTAATTTGGTTGGTCCTT-3′).
RESULTS
Snu17p contains an RRM.
We have recently isolated the yeast [U4/U6.U5] tri-snRNP using two affinity chromatography steps (11). Subsequently, by glycerol gradient centrifugation we could partially separate the [U4/U6.U5] tri-snRNP from contaminating U1 and U2 snRNPs. Thirty-four protein bands with molecular masses ranging from ca. 10 to 200 kDa and comigrating with the U4, U5, and U6 snRNAs were identified. Peptides generated proteolytically from these bands were analyzed by mass spectrometry and database searching. Twenty-eight distinct proteins were shown to be specifically associated with the [U4/U6.U5] tri-snRNP. Two of the remaining proteins were identified as U2-specific proteins, Hsh49p (14) and Rse1p (6, 11). Both are subunits of the heteromeric, U2 snRNP-associated splicing factor SF3b. In addition to the proteins of the [U4/U6.U5] tri-snRNP and these two U2 snRNP proteins, we identified a novel protein which we named Snu17p. This protein contains an RNA recognition motif or RRM (Fig. 1) (3).
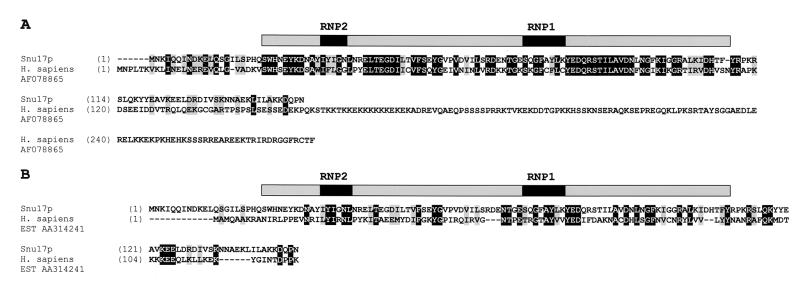
FIG. 1.
Sequence alignment of S. cerevisiae Snu17p with homologous proteins. (A) Snu17p is aligned with a homolog from H. sapiens (AF078865). Identical residues are boxed in black, and similar residues are boxed in grey. The amino acid positions are indicated by numbers. (B) Snu17p is aligned with an H. sapiens cDNA product from EST186153 AA314241. The position of the Snu17p RRM is indicated above the sequences by a shaded box.
A BLAST search in the protein database (1) with Snu17p identified homologous proteins, which could potentially be orthologs, in Drosophila melanogaster (accession number AAP53845), Arabidopsis thaliana (AL133292), Caenorhabditis elegans (U23450), Schizosaccharomyces pombe (AL033388), Plasmodium falciparum (AL034559), and Homo sapiens (AF078865); these proteins exhibited a high degree of homology to Snu17p (50% identity in a region spanning the first 120 amino acids [Fig. 1A and data not shown]). All of these proteins share an RRM which spans positions 21 to 109 of Snu17p (3) (Fig. 1A and data not shown). All of these homologs, with the exception of the Drosophila protein, are much longer at their C terminus than is Snu17p. However, homology to Snu17p is not confined solely to their RRMs, since additional short regions of homology were also detected in the flanking N-terminal and C-terminal regions (Fig. 1A). Since all of these proteins had alignment scores higher than 100, they may be possible orthologs of Snu17p. Several other homologous proteins with lower alignment scores (ca. 60 to 40) emerged during the search, but since their homology to Snu17p is restricted exclusively to their RRMs, they are most probably putative RNA binding proteins, functionally unrelated to Snu17p. A TBLASTN search in the human expressed sequence tag (EST) database revealed that beside the ESTs encoding the human protein shown in Fig. 1A (alignment score, 124), several more ESTs exist that encode a product homologous to Snu17p. Interestingly, among these is EST AA314241 (alignment score, 53), which encodes a 14-kDa protein, named p14, that is associated with both U2 and U11-U12 snRNPs (C. L. Will, C. Schneider, A. M. MacMillan, N. Katopodis, G. Neubauer, M. Wilm, R. Lührmann, and C. C. Query, submitted for publication). Although this protein shows only 36% identity and 53% similarity to Snu17p in a region spanning 96 amino acids from positions 30 to 125, it is similar in length (Fig. 1B). Since similar length may be an important parameter to conclude whether two proteins are genuine orthologs and, most importantly, considering that there is experimental evidence that p14 is associated with U2 snRNPs in HeLa cells (Will et al., submitted), p14, and not the protein shown in Fig. 1A, may be the bona fide human counterpart of Snu17p.
Snu17p is a U2 snRNP protein.
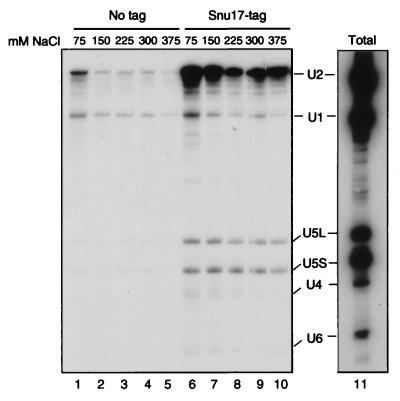
To investigate whether Snu17p is stably associated with one of the snRNP particles, we added a cassette coding for two IgG binding units of Staphylococcus aureus protein A (29) downstream of the SNU17 coding sequence in vivo. Splicing extracts were prepared and used in immunoprecipitation experiments. Immunoprecipitation reactions were performed with extracts containing protein A-tagged Snu17p (Snu17p-protA) and control extracts obtained from an isogenic strain containing untagged protein and then washed at increasing salt concentrations. Subsequently, the precipitated snRNAs were identified by Northern blot analysis. The U2 snRNA coprecipitated with Snu17p in a specific manner (Fig. 2). The amount of coprecipitated U2 snRNP only slightly decreased, increasing the NaCl concentration from 75 to 375 mM KCl. Only background U1 snRNA was coprecipitated (Fig. 2, compare with the control extract [No tag] and with the total RNA [lane 11]). Also, very small amounts of U5L, U5S, U4, and U6 snRNAs were coprecipitated at all salt concentrations tested. This is most probably due to the interaction of the U2 snRNP with the tri-snRNP (11, 16). This experiment demonstrates that Snu17p is stably and specifically associated with the U2 snRNP even at high salt concentrations. Therefore, we have named this protein Snu17p, for the “snurp” protein of 17 kDa.
FIG. 2.
Snu17p is a yeast U2 snRNP protein. DNA coding for a tandem repeat of S. aureus protein A was fused 3′ to SNU17. Extracts (50 μl) from the resulting strain (Snu17p-tag), as well as from the parental wild-type strain (No tag), were immunoprecipitated using IgG-agarose and then washed at various NaCl concentrations, as indicated above the lanes. The snRNA content of the precipitates was assayed by Northern blot analysis (snRNAs are indicated). Total refers to the total RNAs from 4 μl of extract (8% of the total input) before immunoprecipitation.
Snu17p is associated with the spliceosome during splicing in vitro.
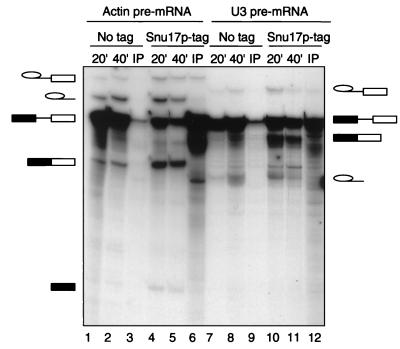
To learn more about Snu17p interactions, we investigated whether Snu17p is associated with the spliceosome during the course of a splicing reaction in vitro. We therefore used the Snu17p-protA extract and the untagged control extract to perform in vitro splicing with labeled actin and U3 pre-mRNAs (Fig. 3). Aliquots of the splicing-reaction mixtures were removed after 20 and 40 min and analyzed for splicing activity. After 40 min the remaining splicing-reaction mixture was immunoprecipitated using IgG-agarose beads (Fig. 3). Significant amounts of the pre-mRNA were coprecipitated from the extracts containing Snu17p-protA (Fig. 3, lanes 6 and 12) but not from the control extract containing untagged Snu17p (lanes 3 and 9). Using either of the two precursors, significant amounts of the lariat-exon 2 intermediate were also coprecipitated (lanes 6 and 12). This suggests that Snu17p, as part of the U2 snRNP, associates with the pre-mRNA in the prespliceosome. Moreover, Snu17p is also part of the catalytically active spliceosome, since it is associated with the lariat-exon 2 intermediate, which is formed during the first step of the pre-mRNA splicing reaction (lanes 6 and 12). Since Snu17p associates with spliceosomes assembled on at least two precursors, actin and U3 pre-mRNAs, this indicates that it is a general splicing factor.
FIG. 3.
Pre-mRNA and the lariat-intron intermediate coprecipitate with Snu17p under in vitro splicing conditions. Splicing reactions using either extracts containing Snu17p without tag (lanes 1 to 3 and 7 to 9) or protein-A tagged Snu17p (lanes 4 to 6 and 10 to 12) were incubated under splicing conditions with 32P-labeled actin pre-mRNA (lanes 1 to 6) or U3 pre-mRNA (lanes 7 to 12) for 20 or 40 min at 25°C. Then 93% of the 40-min reaction volume was precipitated with IgG agarose. For each time point, 7% of the total reaction mixtures (lanes 1, 2, 4, 5, 7, 8, 10, and 11) and the precipitate from the remaining 93% of the reaction mixtures (IP; lanes 3, 6, 9 and 12) were assayed for the presence of pre-mRNA, splicing intermediates, and products. Indicated on the left and on the right are the identities of the labeled RNA species: intron-lariat-exon 2 intermediate, excised lariat-intron, pre-mRNA, mature mRNA, and cleaved exon 1 intermediate.
Deletion of SNU17 confers a slow-growth phenotype.
In view of the results obtained above, we were interested in determining whether Snu17p is essential for cell viability. We performed a genetic knockout of the SNU17 gene in a diploid strain by replacing its coding sequence with the HIS3 marker gene. After sporulation and tetrad dissection of this strain at 25°C, we observed that all four spores were viable, demonstrating that this gene is not essential for vegetative cell growth (data not shown). However, we noticed that two spores in each tetrad grew slower (data not shown). These spores carried the HIS3 marker, indicating that the strains lacking SNU17 (snu17Δ) were viable but impaired for vegetative growth at 25°C. We then isolated haploid cells carrying snu17Δ and tested their growth ability at 17, 30, and 37°C. The slow-growth phenotype of these cells was not apparently exacerbated by increasing or lowering the temperature, compared to that of an isogenic wild-type strain grown at the same temperature. At each temperature, the snu17Δ::HIS3 cells grew about 1.3-fold slower than the wild-type cells did (data not shown). An identical phenotype was observed by Entian et al., who systematically studied S. cerevisiae genes of unknown function (9). After deletion of YIR005w ORF, which corresponds to SNU17, they observed a reduced growth rate on glucose and on several other fermentable carbon sources and alterations in the cell cycle, with an increased proportion of cells with 1n DNA content compared to the wild type (9). Although Snu17p is not essential for vegetative cell growth, we have shown that it is present in the spliceosome during the course of the splicing reaction at least prior to the second step of splicing (Fig. 3; also see above). It was therefore interesting to determine whether the deletion of SNU17 had an effect on splicing in vivo and in vitro.
Deletion of SNU17 leads to a pre-mRNA splicing defect in vivo and to inhibition of splicing prior to the first catalytic step in vitro.
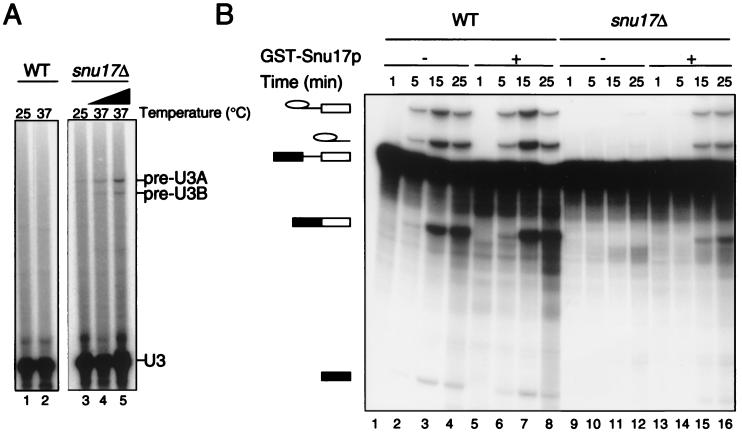
We determined whether SNU17 is required for pre-mRNA splicing in vivo by measuring the levels of unspliced U3A and U3B transcripts in cells grown at 25 and 37°C, respectively. Figure 4A shows the results of a primer extension analysis of U3 transcripts from wild-type and snu17Δ cells. A slight but higher than background accumulation of unspliced pre-U3A and pre-U3B transcripts was found (Fig. 4A, lane 3) (snu17Δ), in comparison to the parental control (lane 1). The splicing defect was exacerbated after the cells were switched for 2 and 4 h from 25 to 37°C (compare lane 2 with lanes 4 and 5). Thus, although deletion of SNU17 does not lead to a discernible temperature-sensitive phenotype, it does cause a splicing defect which is intensified by increasing the temperature.
FIG. 4.
Snu17p is required for efficient splicing in vivo and in vitro. (A) RNA was extracted from wild-type (WT) and snu17Δ cells grown at 25°C (lanes 1 and 3) and after the switch to 37°C for 2 h (lane 4) and 4 h (lanes 2 and 5). Primer extension analysis was performed to measure the levels of unspliced pre-U3A and pre-U3B transcripts, indicated on the right. (B) Splicing reactions were performed at 25°C using the wild-type (lanes 1 to 8) and snu17Δ (lanes 9 to 16) extracts for the time indicated above each lane. To restore splicing, 1.2 pmol of recombinant GST-Snu17p was added to the reaction mixtures prior to addition of the actin precursor (lanes 5 to 8 and 13 to 16). Indicated on the left is the identity of the 32P-labeled RNA species (from top to bottom): intron-lariat-exon 2 intermediate, excised lariat-intron, pre-mRNA, mature mRNA, and cleaved exon 1 intermediate.
To study whether the deletion of SNU17 has an influence on the splicing reaction, we prepared splicing extract from the snu17Δ strain and compared its ability to carry out splicing in vitro with that of a wild-type extract. We performed in vitro splicing reactions at 25°C and withdrew aliquots of the reaction mixtures at 1, 5, 15, and 25 min. (Figure 4B). The deletion of SNU17 led to a dramatic decrease in splicing prior to the first step (Fig. 4B, compare lanes 9 to 12 with lanes 1 to 4). To determine whether this reduction was due specifically to the lack of Snu17p, we tested whether the splicing activity of the extract lacking Snu17p could be restored by adding recombinant GST-Snu17p to the splicing-reaction mixture. We therefore performed splicing in the presence of GST-Snu17p, which was preincubated with the wild-type and snu17Δ extracts prior to the addition of the pre-mRNA. With the snu17Δ extract, the addition of recombinant Snu17p efficiently reversed the splicing defect (Fig. 4B, lanes 13 to 16). Addition of recombinant Snu17p to the wild-type extract did not have any detectable effect (lanes 5 to 8). We conclude that the in vitro splicing defect is due to the specific loss of Snu17p and not to an indirect effect, such as loss of another factor in addition to Snu17p or instability and degradation of the U2 snRNA.
Deletion of SNU17 leads to accumulation of an unusual spliceosomal complex.
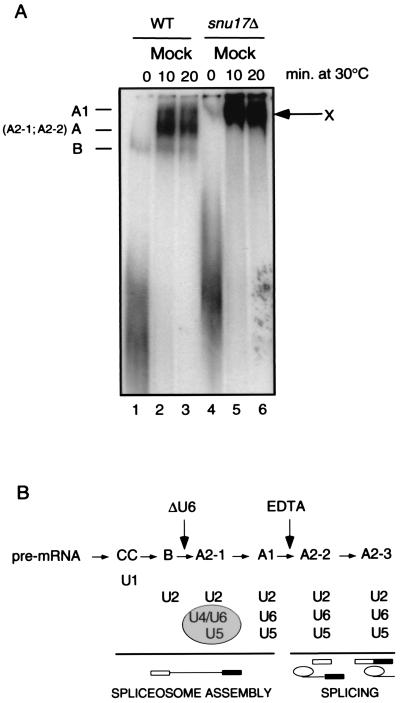
To investigate the step of spliceosome assembly at which Snu17p acts, we separated spliceosomal complexes by electrophoresis on nondenaturing polyacrylamide gels. Spliceosomal complexes were assembled at 30°C on 32P-labeled actin pre-mRNA, using wild-type and snu17Δ extracts (Fig. 5A). Interestingly, the lack of SNU17 led to the formation of a single, slowly migrating complex, which we named complex X (Fig. 5A, lanes 5 and 6). This complex had already accumulated after 10 min of incubation. Under the same conditions, at least two distinct complexes were formed in wild-type extracts: the B and A complexes. A third complex, A1, was usually less abundant (lanes 2 and 3). Previous analysis of the B complex (7) showed that it contains the U2 snRNP but not the U1 snRNP, which is only loosely bound and is lost from complex B due to heparin treatment (7, 17). The A complex (named A2-1 and A2-2 at early and later incubation times, respectively) and the A1 complex usually contain the U2 and [U4/U6.U5] snRNPs or the U2, U6, and U5 snRNPs, respectively (Fig. 5B gives a summary).
FIG. 5.
Deletion of SNU17 leads to accumulation of an unusual spliceosomal complex. (A) Spliceosomes were assembled on a 32P-labeled actin pre-mRNA by incubation in mock-treated wild-type (WT) and snu17Δ extracts at 30°C. Aliquots were withdrawn after 0, 10, and 20 min and treated with a buffer containing heparin. Nondenaturing gels were run for 5 h (7). The identities of the complexes are indicated on the left. The unusual, slow-migrating complex seen only in snu17Δ extracts is designated complex X. (B) Yeast spliceosome assembly, as in reference 7. RNase H-mediated digestion of U6 snRNA (ΔU6) blocks spliceosome assembly at the B complex stage, whereas addition of 5 to 8 mM EDTA blocks spliceosome assembly at the A1 complex stage.
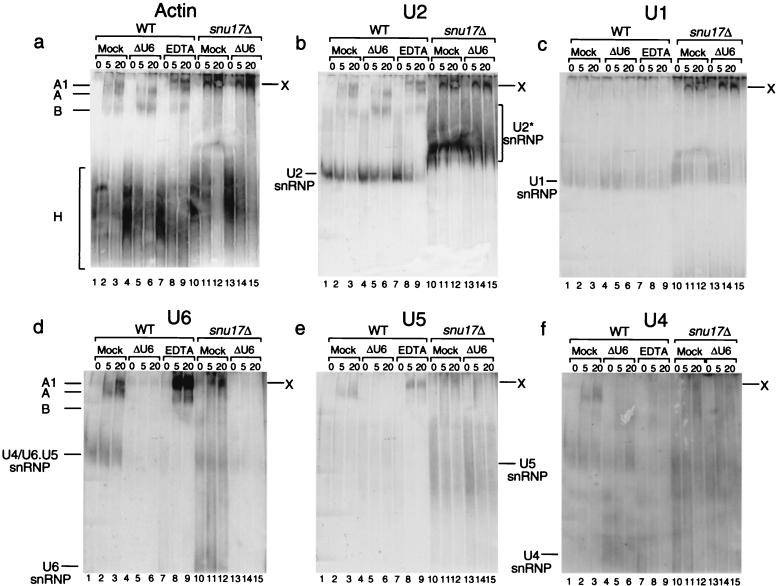
We noticed that complex X exhibits migration behavior similar to that of complex A1. To analyze whether the deletion of SNU17 leads to accumulation of complex A1, we studied the RNA composition of complex X by Northern blot analysis (Fig. 6). Spliceosomal complexes were assembled on unlabeled actin precursor at 30°C. After 0, 5, and 20 min, aliquots of the reaction mixture were incubated on ice and a solution containing heparin was added. The complexes were separated on a native gel, blotted, and then hybridized with probes for the actin precursor and subsequently for the U1, U2, U4, U5, and U6 snRNAs (Fig. 6a to f). Hybridization with the actin and the U2 probes revealed the position and composition of complexes B, A, and A1 in the wild-type extract (Fig. 6a and b, lanes 2 and 3). To increase the signal of the A1 complex in this experiment, we added EDTA to the splicing-reaction mixture. As shown previously (7), addition of EDTA blocked spliceosome assembly, leading to accumulation of complex A1, which, as anticipated, contained the U2 snRNP but not the U1 snRNP (Fig. 6b and c, lanes 8 and 9, respectively). Surprisingly, analysis of complex X showed that it contained not only the U2 snRNP but also the U1 snRNP (Fig. 6b and c, lanes 11 and 12, respectively). In addition, we noticed that in extracts deficient in Snu17p the U2 snRNP (U2* snRNP) ran aberrantly (Fig. 6b, lanes 10 to 15), forming a diffuse, slower-migrating band (compare with the wild-type U2 snRNP), indicating that its structure is somehow compromised.
FIG. 6.
RNA composition of complex X. Spliceosomes were assembled on unlabeled actin pre-mRNA. Splicing reactions were performed using wild-type (WT) or snu17Δ extracts at 30°C, either mock treated, treated to destroy U6, or treated with 8 mM EDTA as indicated above each lane. Aliquots were withdrawn after 0, 5, and 20 min, blocked on ice, and treated with a buffer containing heparin. Nondenaturing gels were run, blotted, and sequentially hybridized with probes for the actin precursor (Actin) (a) and subsequently for the U2 (b), U1 (c), U6 (d), U5 (e), and U4 (f) snRNAs. The complexes are indicated on the left. Complex X is an unusual slow-migrating complex seen only in snu17Δ extracts.
To determine whether the [U4/U6.U5] tri-snRNP is part of complex X, the blot was hybridized with the U6, U5, and U4 probes. U6 and U5 were identified in the wild-type A and A1 complexes (Fig. 6d and e, lanes 2 and 3 and lanes 8 and 9). As expected, U4 was identified only in the A but not in the A1 complexes (Fig. 6f, lanes 2 and 3 and lanes 8 and 9). As shown in Fig. 6d to f, lanes 11 and 12, complex X also contained the U6, U5, and U4 snRNPs. The controls show that in the wild-type extract, deletion of U6 by oligonucleotide-directed RNase H cleavage blocked the binding of the tri-snRNP and led to the accumulation of complex B, which, as expected, contained the substrate and the U2 snRNP but not the U1 snRNP (Fig. 6a to c, lanes 5 and 6). Digestion of U6 in wild-type extracts led to nearly complete disappearance of the U6, U5, and U4 signals (Fig. 6d to f, lanes 5 and 6), suggesting that, normally, deletion of U6 blocks the entry of the [U4/U6.U5] tri-snRNP. When the snu17Δ extract was treated to destroy U6, the latter completely disappeared from complex X (Fig. 6d, lanes 14 and 15). However, U5 still appeared to be present (Fig. 6e, lanes 14 and 15).
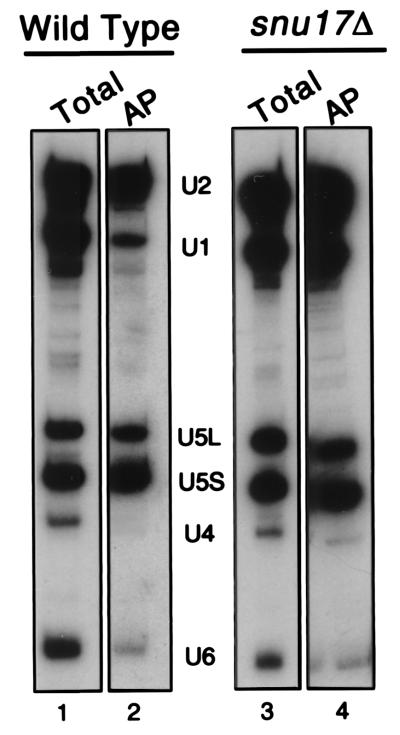
Since the U5 and U4 snRNP were never clearly detected by this method, as an independent assay to support the existence of the [U4/U6.U5] tri-snRNP in complex X, we have purified spliceosomes that were assembled onto biotinylated pre-mRNA in vitro. Splicing-reaction mixtures were incubated with biotinylated unlabeled substrate using either wild-type or snu17Δ extracts (Fig. 7). Spliceosomal complexes were then fractionated by centrifugation on a 10 to 30% glycerol gradient, and their RNA composition was analyzed following precipitation with streptavidin beads (Fig. 7, lanes 2 and 4). The snRNAs present in gradient-fractionated spliceosomes precipitated with streptavidin (the ca. 40S region of the gradient [lanes 2 and 4]), as well as the input of the total snRNPs before incubation and fractionation, were assayed by Northern blot analysis (lanes 1 and 3). The snu17Δ spliceosome contained large amounts of U1, U2, and U5 but smaller amounts of U4 and U6 snRNAs (lane 4). Note that the U4 and U6 snRNA were in general not well represented in both extracts. One reason may be that the U6 and U4 probes had a lower specific activity than the U1, U2, and U5 probes in some of our experiments. The snRNA composition of the wild-type spliceosome indicates that it contains in proportion slightly less U2 but much less U1 than the snu17Δ spliceosome (Fig. 7, compare lane 2 with lane 4). Importantly, the wild-type spliceosome has released U4, since the U4 snRNA is not detectable (lane 2). However, the snu17Δ spliceosome does contain detectable levels of U4 (lane 4). The controls show that the total snRNA contents of the wild-type and snu17Δ extracts before spliceosome assembly, gradient fractionation, and affinity purification are practically indistinguishable, including the lower levels of the U4 and U6 snRNAs (compare lane 1 with lane 3). In summary, the conclusion that complex X, the snu17Δ spliceosome, contains all of the snRNPs is supported and strengthened by this independent experiment. From the combination of these experiments, we conclude that complex X contains the U2, U6, U5, and U4 snRNPs and, unusually, a U1 snRNP that is stably associated with the complex even after treatment with heparin. The unexpected stabilization of the U1 snRNP and the lack of release of the U4 snRNP in spliceosomes lacking Snu17p suggest that deletion of Snu17p inhibits the progression of spliceosome assembly prior to U1 snRNP release and after [U4/U6.U5] tri-snRNP addition.
FIG. 7.
Deletion of SNU17 inhibits the dissociation of U1 and U4 from the spliceosome. Splicing-reaction mixtures were incubated with actin pre-mRNA that was transcribed in the presence of biotinylated UTP to incorporate an affinity label. The reaction mixtures were incubated for 20 min at 23°C. Then 5 μl was withdrawn and kept for analysis of total RNAs (Total; lanes 1 and 3). The remaining 120 μl was treated with heparin and then sedimented by centrifugation on a 10 to 30% glycerol gradient. rRNAs from yeast were run on a parallel gradient as sedimentation markers. Fifteen fractions (160 μl each) were collected, and four-fifths of each fraction was affinity purified with streptavidin-agarose (AP = affinity purification of a fraction of the 40S region of the gradient corresponding to fraction 15, the spliceosome [lanes 2 and 4]). After extensive washing, precipitated snRNAs were assayed by Northern blotting.
DISCUSSION
In this work, we have presented the isolation and characterization of a novel yeast U2 snRNP protein, Snu17p. Using affinity purification experiments performed during the course of the splicing reaction, we showed that Snu17p coprecipitates unspliced precursor and spliced lariat-exon 2 intermediate. This demonstrates that Snu17p is a component of the spliceosome at least until prior to the second catalytic step of splicing. In addition, extracts prepared from the knockout strain exhibited a dramatic reduction in splicing prior to the first catalytic step. Notably, the splicing inhibition was reversed by addition of recombinant Snu17p (Fig. 4B). This indicates that, similar to the in vivo situation, Snu17p is required in vitro to sustain efficient splicing and that the splicing reduction is directly due to the lack of Snu17p. To exclude the possibility that deletion of Snu17p inhibits splicing due to an indirect effect, such as reduced levels of the U2 snRNA, we have analyzed the U2 snRNP lacking Snu17p by glycerol gradient centrifugation followed by Northern blot analysis. These experiments showed that the level of U2 snRNPs appears normal, if not increased, in the knockout strain (data not shown). In addition, the sedimentation behavior of the mutant U2 snRNP is indistinguishable from that of the wild type (data not shown). An alternative but highly likely indirect effect which would explain the reversion of the splicing defect after addition of recombinant Snu17p is an impaired structure or conformation of the U2 snRNP lacking Snu17p (Fig. 6b, U2* snRNP). A partially inactive U2 snRNP due to the presence of an altered structure could be reversed to normal after the addition of recombinant Snu17p, and as a result, pre-mRNA splicing may also be restored. This would explain why the deletion of Snu17p has a less drastic effect in vivo than in vitro. In vivo, an aberrant conformation of the U2 snRNP due to the lack of Snu17p may be stabilized and compensated by other factors which are found in an in vivo environment (physiological salt, pH, temperature, etc.). In vitro, the conformation of the weakened U2* snRNP may not be sufficiently stabilized to support a splicing reaction under the limiting assay conditions used in the test tube.
When the products of a splicing reaction performed in snu17Δ extract are analyzed on a non-denaturing gel, the spliceosome runs as a single slowly migrating complex, complex X, which contains the U2, U6, U5, and U4 snRNPs and a heparin-resistant U1 snRNP. A possible explanation for the formation of complex X is, again, misfolding of the U2 snRNP in the absence of Snu17p. Figure 6b shows that the migration range of the U2 snRNP lacking Snu17p (U2* snRNP) is considerably larger than that of the wild-type U2 snRNP. It may be possible that the unexpected stabilization of the U1 snRNP in the spliceosome after deletion of SNU17 is due to an indirect effect on the U2 snRNP, rather than to a direct role of Snu17p in the U1–pre-mRNA interaction. If, for example, Snu17p contributes to the correct positioning of the U2 snRNP at the branchpoint, deletion of Snu17p may lead to an aberrant conformation of the U2–pre-mRNA structure which is unable to displace U1. This aberrant U1.U2–pre-mRNA complex (a pseudo-prespliceosome) would nevertheless be capable of integrating the [U4/U6.U5] tri-snRNP; however, further rearrangements would be inhibited or greatly slowed such that the complex does not progress to a catalytic form. Whether the aberrant conformation of the U2* snRNP is reversed to normal after the addition of recombinant Snu17p has not been analyzed. However, addition of recombinant Snu17p to snu17Δ extracts, prior to addition of pre-mRNA, efficiently reverses the splicing defect (Fig. 4B). When the reconstitution experiment with recombinant Snu17p is performed and the reaction product is analyzed on a nondenaturing gel, complex X is replaced, at least in part, by a faster-migrating complex, which must be the active form of the spliceosome since splicing is restored (Fig. 4B and data not shown). Only further experiments may clarify this point.
It is interesting that the phenotype of the Snu17p deletion strain is unique. In extracts prepared from strains lacking other U2 proteins, like LEA1 and/or Y1B9/U2B", spliceosome assembly is blocked prior to the addition of the U2 snRNP (5). In addition, strains lacking LEA1 and/or Y1B9/U2B" contain reduced levels of U2 snRNA, while the U2 snRNA present in extracts lacking Snu17p seems to be even more abundant than the U2 snRNA found in the wild type (Fig. 6b). Since we used the same amounts of wild-type and snu17Δ extracts in our experiments (ca. 25 μg of proteins/2 μl of extract), we can conclude that the levels of U2 snRNP are even increased upon deletion of SNU17. The reason for this increase remains to be determined.
A surprising feature of complex X is the apparent lack of stepwise assembly. Even after a very short (5-min [Fig. 6]) incubation of pre-mRNA in snu17Δ extracts, only a single complex is detected. The formation of complex X is slow, appearing first after 2 min of incubation (data not shown). This complex already contains the U1 and U2 snRNPs, with only very small amounts of U4-U6 and U5 snRNPs detectable. Most probably, it is very difficult, if not impossible, to resolve the U1.U2 complex from the U1.U2.[U4/U6.U5] or U1.U2-U6.U5 complexes in these low-resolution gels. Moreover, we do not know what factors govern the mobility of these complexes in electrophoresis. Likewise, the fact that complex X does not run faster in the gel even after digestion of U6 (Fig. 6a to c, lanes 14 and 15) may also not be surprising. On digestion of U6, the tri-snRNP does not enter the wild-type spliceosome, and complex B, the prespliceosome containing only the U2 snRNP, accumulates. After digestion of U6, complex X apparently still runs in the same position. This may be because the resulting complex contains U1 in addition to U2 and also some U5 (Fig. 6e, lanes 14 and 15). Loss of a snRNP does not always lead to a faster-migrating behavior of splicing complexes. For example, the yeast complex A2-1, which contains U2, U4-U6, and U5 (Fig. 5B), runs faster in nondenaturing gels than does complex A1, which contains only U2, U6, and U5. This is presumably due to conformational changes that are coupled with the dissociation of U4 during the transition from the A2-1 to the A1 complex (7). We do not know which type of conformational rearrangement occurs in complex X. However, our results provide good evidence that the [U4/U6.U5] tri-snRNP joins the U1.U2 complex. Although the U1 snRNP appears to be hyperstabilized, the tri-snRNP nevertheless is able to bind. Complex X appears to be similar to the “U1-hyper” splicing intermediate observed by Staley and Guthrie (41), where U1 and U2 remain bound in the spliceosome along with U4-U6 and U5 only when the U1:5′ splice site interaction is hyperstabilized, leading to a concomitant block of U1 and U4 dissociation. Meanwhile, Kuhn et al. showed that U4-U6 unwinding and U1 release are both inhibited by the mutant U4 snRNA U4-cs1 (20). Similar to these mutations, loss of Snu17p traps U1 in the spliceosome, still allowing the tri-snRNP to bind but blocking the release of U4. These data also provide evidence that the [U4/U6.U5] tri-snRNP is bound to pre-mRNA at the prespliceosome step of assembly. This was also observed by Staley and Guthrie using a cold-sensitive mutant of Prp28p, prp28-1. These authors showed that at 15°C prp28-1 extracts trapped the prespliceosome and allowed the [U4/U6.U5] snRNP to bind, although weakly, to this intermediate (41).
Snu17p contains an RRM spanning approximately 60% of its length. Since RRMs are found in a wide variety of proteins across species, this complicates the search for its genuine orthologs. In our search we found proteins homologous to Snu17p which are evolutionarily highly conserved not only in their central region, which contains an RRM, but also in the N-terminal part. However, since all but one are much longer than Snu17p in their C-terminal part, we cannot conclusively establish, without additional experimental information, whether they are functionally related to Snu17p. The p14 protein, the product of EST AA314241, may be the functional counterpart of Snu17p in the human spliceosome for reasons we find very convincing: experiments show that p14 is a human U2 snRNP-associated protein that is also found in the U11-U12 snRNP of the minor spliceosome (Will et al., submitted).
Since Snu17p is unequivocally a U2 snRNP protein and has an RRM, we performed initial immunoprecipitation experiments to determine whether it binds directly to the U2 snRNA. However, we could not reproducibly show that Snu17p binds directly to in vitro-transcribed U2 snRNA, to one of the other spliceosomal snRNAs, or to the pre-mRNA. This suggests that Snu17p binds the U2 snRNP via protein-protein interactions. Considering possible pre-mRNA contacts of Snu17p, all of the human SF3a components, as well as the SF3b subunits SAP49, SAP145, and SAP155, cross-link to the pre-mRNA close to the branch point (13). Although we have not analyzed whether Snu17p cross-links to the pre-mRNA, it is worth mentioning that in human splicing extracts, p14 can be cross-linked to the pre-mRNA at the branch point adenosine (Will et al., submitted). Since Snu17p may be functionally related to p14, it is conceivable that it also directly contacts the branch point sequence.
To summarize, we have identified a novel yeast U2 snRNP protein, Snu17p, which, despite its discernible in vitro phenotype, is not essential in vivo. However, its genetic deletion leads to a slow-growth phenotype and to a pre-mRNA splicing defect in vivo. Our in vitro studies suggest that Snu17p may play a role in the maturation of the spliceosome, since extracts lacking Snu17p accumulate an unusual splicing complex containing all five snRNPs: U1, U2, U4, U5, and U6. Loss of Snu17p, in contrast to other U2 snRNP proteins analyzed (5), allows not only the addition of the U2 snRNP to the prespliceosome but also the addition of the [U4/U6.U5] tri-snRNP, inhibiting the progression of spliceosome assembly before U1 release and after tri-snRNP addition. This phenotype is peculiar, and deletion of SNU17 can be used as a tool to isolate fully assembled spliceosomes stalled prior to the first catalytic step of splicing and to learn more about their protein composition.
ACKNOWLEDGMENTS
We thank Cindy L. Will for helpful discussions and communication of results prior to publication. For critical reading of the manuscript we are grateful to Klaus Hartmuth, Reinhard Rauhut, and Cindy L. Will. We also thank Marion Killian for expert technical assistance.
This work was supported by the Gottfried Wilhelm Leibniz Program and a grant from the Deutsche Forschungsgemeinschaft (SFB 397, A7) to P.F and R.L.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Behrens S E, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 4.Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 5.Caspary F, Séraphin B. The yeast U2A′/U2B complex is required for pre-spliceosome formation. EMBO J. 1998;17:6348–6358. doi: 10.1093/emboj/17.21.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caspary F, Shevchenko A, Wilm M, Séraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng S C, Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- 8.Collins C A, Guthrie C. The question remains: Is the spliceosome a ribozyme? Nat Struct Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- 9.Entian K D, Schuster T, Hegemann J H, Becher D, Feldmann H, Guldener U, Gotz R, Hansen M, Hollenberg C P, Jansen G, Kramer W, Klein S, Kotter P, Kricke J, Launhardt H, Mannhaupt G, Maierl A, Meyer P, Mewes W, Munder T, Niedenthal R K, Ramezani Rad M, Rohmer A, Romer A, Hinnen A, et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 10.Fabrizio P, McPheeters D S, Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989;3:2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- 11.Gottschalk A, Neubauer G, Banroques J, Mann M, Lührmann R, Fabrizio P. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 1999;18:4535–4548. doi: 10.1093/emboj/18.16.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot H V, Mann M, Séraphin B, Rosbash M, Lührmann R, Fabrizio P. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 13.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 14.Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 15.Kandels-Lewis S, Séraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 16.Konarska M M, Sharp P A. Association of U2, U4, U5, and U6 small nuclear ribonucleoproteins in a spliceosome-type complex in absence of precursor RNA. Proc Natl Acad Sci USA. 1988;85:5459–5462. doi: 10.1073/pnas.85.15.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konarska M M, Sharp P A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 18.Konforti B B, Koziolkiewicz M J, Konarska M M. Disruption of base pairing between the 5′ splice site and the 5′ end of U1 snRNA is required for spliceosome assembly. Cell. 1993;75:863–873. doi: 10.1016/0092-8674(93)90531-t. [DOI] [PubMed] [Google Scholar]
- 19.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn A N, Li Z, Brow D A. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 21.Lesser C F, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 22.Lin R J, Newman A J, Cheng S C, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
- 23.Madhani H D, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 24.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 25.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA by the spliceosome. In: Gesteland A, editor. RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 26.Neubauer G, Gottschalk A, Fabrizio P, Séraphin B, Lührmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nilsen T W. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 28.Pauling M H, McPheeters D S, Ares M., Jr Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol Cell Biol. 2000;20:2176–2185. doi: 10.1128/mcb.20.6.2176-2185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puig O, Rutz B, Luukkonen B G, Kandels-Lewis S, Bragado-Nilsson E, Séraphin B. New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast. 1998;14:1139–1146. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1139::AID-YEA306>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 30.Rain J C, Tartakoff A M, Kramer A, Legrain P. Essential domains of the PRP21 splicing factor are implicated in the binding to PRP9 and PRP11 proteins and are conserved through evolution. RNA. 1996;2:535–550. [PMC free article] [PubMed] [Google Scholar]
- 31.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 32.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 33.Rosbash M, Séraphin B. Who's on first? The U1 snRNP-5′ splice site interaction and splicing. Trends Biochem Sci. 1991;16:187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- 34.Ruby S W, Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988;242:1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- 35.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawa H, Abelson J. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc Natl Acad Sci USA. 1992;89:11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Séraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 39.Siliciano P G, Guthrie C. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 40.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 41.Staley J P, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 42.Stevens S W, Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc Natl Acad Sci USA. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang J, Abovich N, Rosbash M. Identification and characterization of a yeast gene encoding the U2 small nuclear ribonucleoprotein particle B" protein. Mol Cell Biol. 1996;16:2787–2795. doi: 10.1128/mcb.16.6.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wassarman D A, Steitz J A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 45.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 46.Will C L, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 47.Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan D, Perriman R, Igel H, Howe K J, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]