Specific populations of immune cells in the adult develop from the embryonic yolk sac, independent of hematopoietic stem cells. Using the human pluripotent stem cell model, this study has mapped human yolk sac hematopoiesis, providing access to these developmentally restricted progenitors.
Abstract
In the mouse, the first hematopoietic cells are generated in the yolk sac from the primitive, erythro-myeloid progenitor (EMP) and lymphoid programs that are specified before the emergence of hematopoietic stem cells. While many of the yolk sac–derived populations are transient, specific immune cell progeny seed developing tissues, where they function into adult life. To access the human equivalent of these lineages, we modeled yolk sac hematopoietic development using pluripotent stem cell differentiation. Here, we show that the combination of Activin A, BMP4, and FGF2 induces a population of KDR+CD235a/b+ mesoderm that gives rise to the spectrum of erythroid, myeloid, and T lymphoid lineages characteristic of the mouse yolk sac hematopoietic programs, including the Vδ2+ subset of γ/δ T cells that develops early in the human embryo. Through clonal analyses, we identified a multipotent hematopoietic progenitor with erythroid, myeloid, and T lymphoid potential, suggesting that the yolk sac EMP and lymphoid lineages may develop from a common progenitor.
Introduction
Hematopoiesis in the mouse is initiated in the yolk sac by distinct programs that collectively produce a broad range of lineages, independent of hematopoietic stem cells (HSCs; Ghosn et al., 2019). The primitive program, the first to develop, has limited potential and gives rise to a transient population of progenitors on embryonic day (E)7.0, the majority of which are committed to the erythroid lineage. In addition to erythroid cells, primitive hematopoiesis also generates macrophages and megakaryocytes (Palis et al., 1999; Tober et al., 2007; Wong et al., 1986). Following the onset of primitive hematopoiesis, a second program, the erythro-myeloid progenitor (EMP) program, emerges at E8.25 and generates an expanded set of lineages that includes granulocytes and mast cells in addition to the erythroid, macrophage, and megakaryocyte lineages (Palis et al., 1999; Tober et al., 2007; Wong et al., 1986). EMPs can be identified and isolated based on the coexpression of CD41, Kit, and CD16/32 (FcγRII/III), markers not expressed on primitive hematopoietic progenitors (Ferkowicz et al., 2003; McGrath et al., 2015; Mikkola et al., 2003). Clonal analyses have demonstrated that EMP hematopoiesis contains CD41+Kit+CD16/32+ multipotent progenitors (MPPs) that are able to generate the spectrum of lineages produced at this stage (McGrath et al., 2015). The recent demonstration that this population can also give rise to natural killer (NK) cells (Dege et al., 2020) suggests that the potential of this program extends beyond the erythroid, myeloid, and megakaryocyte fates. Although the primitive and EMP programs share common lineages, there are differences in some of the end-stage cells generated. For example, primitive erythroid cells are larger than their EMP counterparts and display a globin expression pattern characterized by the predominance of the embryonic εγ and β H1 globins (Palis et al., 1999; Wong et al., 1986). In contrast, EMP-derived erythroid cells predominantly express the adult form of β-globin, β-major along with a low level of β H1 globin (McGrath et al., 2015, 2011; Palis et al., 1999; Wong et al., 1986).
In addition to the erythroid, myeloid, and megakaryocyte lineages, there is compelling evidence that the yolk sac also generates lymphoid cells independent of HSCs. Progenitors with T lymphoid potential have been detected in the yolk sac as early as E9.0 by culture in fetal thymic organs or with OP9 stromal cells (Huang and Auerbach, 1993; Yoshimoto et al., 2012). Stromal cell-based cultures have also identified B cell progenitors in the yolk sac at the same stage of development (Yoshimoto et al., 2011). Despite the presence of EMPs and lymphoid progenitors in the E9.0 yolk sac, a clonal relationship between these lineages has not been established. MPPs able to generate T cells, B cells, macrophages, and granulocytes have been identified by the expression of CD45, Kit, Il7ra, and Flt3 in E11.5 liver at a stage when few HSCs are present (Böiers et al., 2013). The observation that the E9.5 yolk sac contains a low frequency of CD45+Kit+Il7ra+ cells with a similar molecular profile raises the possibility that these progenitors are specified in the yolk sac.
Yolk sac hematopoiesis was long thought to function solely to support the developing embryo before the generation of HSCs by the definitive program. The view, however, has been challenged by a series of lineage tracing experiments in the mouse which have demonstrated that populations of tissue-resident macrophages in the adult, including microglia, Kupffer cells, and alveolar macrophages, develop from HSC-independent yolk sac–derived progenitors (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015; Schulz et al., 2012). These macrophage progenitors seed the developing organs and generate the tissue-resident populations that maintain themselves throughout adult life (Mass et al., 2016). More recent lineage tracing studies have shown that the contribution of the yolk sac hematopoietic programs to the adult is broader than first appreciated and includes subpopulations of mast cells (Gentek et al., 2018a; Li et al., 2018) and T cells (Gentek et al., 2018b). The T cells traced from yolk sac progenitors in this study represent a unique subset of γδ T cells, known as dendritic epidermal T cells (DETCs), that are distinguished from all other T cells by their expression of an invariant Vγ3 T cell receptor (Havran et al., 1989). These Vγ3+ cells are the first TCR-expressing lymphocyte detected in the thymus and are found as early as E14.0 (Havran and Allison, 1988). In utero depletion, parabiosis, and transplantation studies have shown that HSCs do not generate this lineage, indicating that DETCs are generated by embryonic-restricted progenitors and maintain themselves throughout life (Gentek et al., 2018b; Havran and Allison, 1990).
Given the limited access to human tissue at early stages of development, the structure of the human embryonic hematopoietic system is far less well understood than that of the mouse. Studies of the human yolk sac at 5 wk have identified large, nucleated erythroblasts that express embryonic (HBE; ε) globin indicative of human primitive hematopoiesis (Peschle et al., 1985). Analyses of the colony-forming progenitor potential of the yolk sac and liver at this stage demonstrated the presence of granulocyte and erythroid progenitors. The cells within these erythroid colonies expressed a combination of the embryonic (HBE; ε) and fetal (HBG1/2; Aγ and Gγ) globins, suggesting that they originate from the human equivalent of EMP-derived progenitors. Similar globin expression patterns were detected in erythroblasts present in the liver at 6 wk (Migliaccio et al., 1986; Peschle et al., 1985, 1984). The detection of rare MPPs with erythroid, macrophage, and granulocyte potential in the yolk sac at Carnegie stage 12 (approximately day 30; Bian et al., 2020) has provided further evidence of an EMP program in the human. Although there are no detailed analyses of lymphoid progenitors in the human yolk sac, studies on the developing liver and thymus have shown that a subset of γδ T cells, characterized by the expression of the Vδ2 gene, is present at 5 wk and is the dominant T lymphoid population before 12 wk (Haynes and Heinly, 1995; McVay and Carding, 1996). The proportion of Vδ2+ T cells decreases in these tissues beyond this time as these cells are supplanted by a second wave made up of Vδ1+ T cells detected in the fetal blood by 40 wk (Dimova et al., 2015). The early emergence of the Vδ2+ population suggests that it may be uniquely specified from the HSC-independent progenitors in the yolk sac, similar to the Vγ3+ T lymphoid lineage in the mouse. The recent finding that fetal, but not postnatal, progenitors efficiently generate Vδ2+ T cells following culture with OP9-DL1 stromal cells is consistent with this interpretation (Tieppo et al., 2020).
Owing to the scarcity of human yolk sac, efforts to characterize the earliest stages of human embryonic hematopoiesis, such as the relationship of the different hematopoietic programs and the pathways that regulate their specification, have turned to the human pluripotent stem cell (hPSC) system to model these developmental steps (Ditadi et al., 2017). Findings from these studies have shown that hematopoietic differentiation recapitulates the temporal organization of mouse yolk sac hematopoietic development (Kennedy et al., 2007; Zambidis et al., 2005), validating this approach for the study of human embryonic hematopoiesis. Following on these studies, our group has shown that appropriately staged inhibition of Wnt signaling induces a population of KDR+CD235a+ mesoderm from hPSCs that displays erythroid and myeloid potential indicative of yolk sac hematopoiesis (Sturgeon et al., 2014). This mesoderm, however, did not give rise to the T lymphoid lineage, suggesting that it does not recapitulate the full developmental potential of the mouse yolk sac.
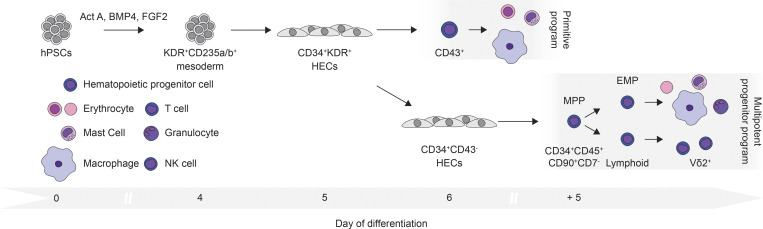
As the progenitors of tissue-resident immune cells are not found after birth, hPSCs represent an unlimited source of these cell types for further study and for future clinical applications, including novel cell-based therapies. To model yolk sac development from hPSCs, we used the combination of Activin A, BMP4, and FGF2 to induce KDR+CD235a/b+ mesoderm that gives rise to the human equivalent of the primitive and EMP hematopoietic programs. In addition to erythroid and myeloid lineage, we show that the human EMP program gives rise to γδ and αβ T cells, including the Vδ2+ lineage. Through clonal analyses, we demonstrate that the EMP-derived population contains a CD34+CD45+CD90+CD7− multipotent hematopoietic progenitor capable of generating erythroid, myeloid, NK, and T cell progeny. Analyses of the mouse EMP program revealed that it has similar potential and generates T cells as well as bipotent progenitors and MPPs that can give rise myeloid, erythroid, and T cell progeny in vitro. Together, these findings demonstrate that the early stages of human hematopoietic development recapitulate many aspects of hematopoiesis in the mouse yolk sac and provide evidence that the EMP and lymphoid yolk sac lineages develop from a common MPP.
Results
Specification of primitive hematopoiesis
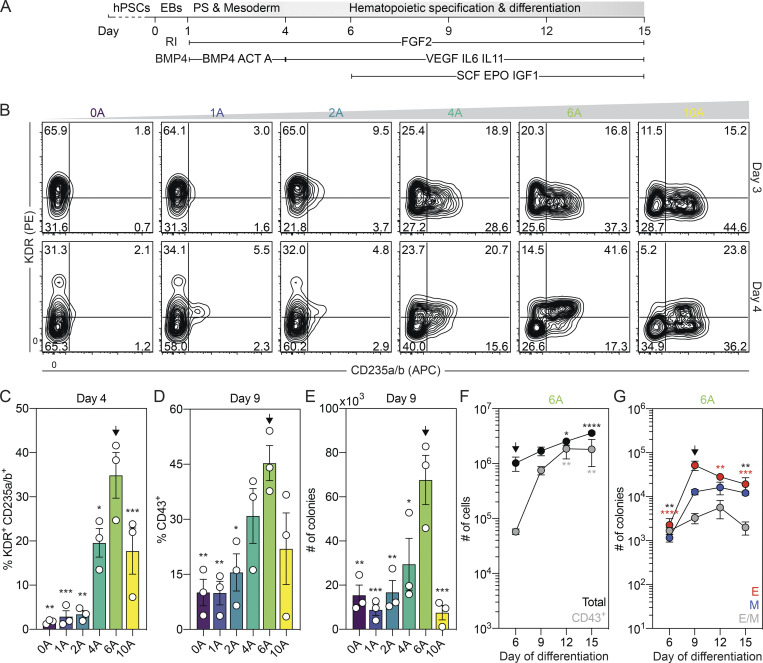
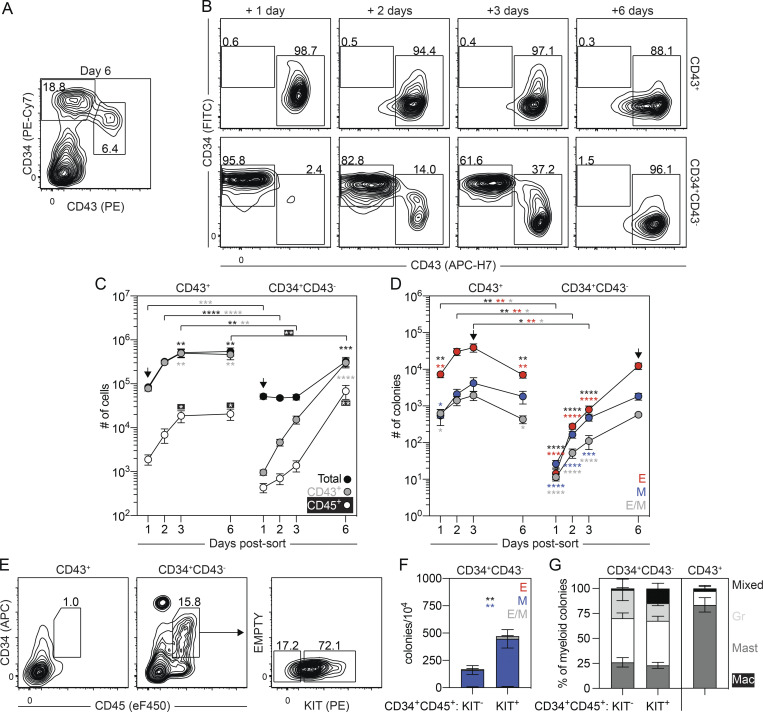
In an effort to generate hPSC-derived CD235a+ (hereafter referred to as CD235a/b, as the HIR2 clone used recognizes both proteins) mesoderm that gives rise to the spectrum of erythroid, myeloid, and lymphoid lineages found in the mouse yolk sac, we induced this population with combinations of Activin A, BMP4, and FGF2 without Wnt inhibition (Fig. 1 A), similar to the approach used by our group to generate CD235a/b+ cardiogenic mesoderm (Lee et al., 2017). For these studies, mesoderm was generated through the addition of different amounts of Activin A (0 to 10 ng/ml) together with a single concentration of BMP4 (10 ng/ml) and FGF2 (5 ng/ml) between days 1 and 4 of differentiation. As observed in our previous study, we found that the induction of CD235a/b+ mesoderm was dependent on Activin/Nodal signaling, as the size of this population (KDR+ and KDR−) measured on either day 3 or 4 of differentiation increased as the concentration of Activin A increased up to 6 ng/ml (Fig. 1 B). Additionally, we observed an increase in the proportion of KDR+CD235a/b+ cells between days 3 and 4 of differentiation in cultures induced with either 6 or 10 ng/ml of Activin A (Fig. 1, B and C). To determine if the mesoderm induced under these conditions displayed hematopoietic potential, we cultured the embryoid bodies (EBs) for an additional 5 d in the presence of vascular endothelial growth factor (VEGF), FGF2, and hematopoietic cytokines and then assayed the cultures for the presence of CD43+ hematopoietic cells and hematopoietic colony-forming progenitors. Cultures induced with 6 ng/ml of Activin A contained the highest proportion of CD43+ cells and the highest number of colony-forming progenitors (Fig. 1, D and E; and Fig. S1 A). Further analyses of this population showed that CD43+ cells were present by day 6 of differentiation and increased in number over the next 6 d (Fig. 1 F and Fig. S1 B), indicative of the expansion and maturation of hematopoietic progenitors. The number of colony-forming progenitors also increased between days 6 and 9 of differentiation and then decreased over the next 6 d (Fig. 1 G). While some colonies of mast cells and macrophages were detected, small erythroid colonies dominated the cultures (Fig. 1 G; and Fig. S1, C and D). Large and burst erythroid colonies were also observed at much lower frequencies (Fig. S1 D). Quantitative RT-PCR (RT-qPCR) analyses showed that both the small erythroid (Fig. S1 E) and large (data not shown) colonies predominantly expressed the embryonic globin HBE, indicating that they represent the human primitive erythroid lineage.
Figure 1.
Specification of yolk sac–like hematopoietic mesoderm from hPSCs. (A) Schematic of hematopoietic differentiation from hPSCs through the addition of Activin A (ACT A), BMP4, and FGF2 followed by culture in the presence of VEGF, FGF2, and hematopoietic cytokines. PS, primitive streak; RI, ROCK inhibitor. (B) Representative flow cytometric analysis of KDR and CD235a/b expression on days 3 and 4 of differentiation in cultures induced with different concentrations of Activin A (A; ng/ml). (C) Quantification of the proportion of KDR+CD235a/b+ cells generated on day 4 of differentiation for a given concentration of Activin A (A; ng/ml; n = 3). ANOVA. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus cultures induced with 6 ng/ml of Activin A (arrow). (D) Quantification of the proportion of CD43+ hematopoietic cells on day 9 of differentiation for a given concentration of Activin A (A; ng/ml; n = 3). ANOVA. *, P < 0.05; **, P < 0.01 versus cultures induced with 6 ng/ml of Activin A (arrow). (E) Colony-forming progenitor number on day 9 of differentiation for a given concentration of Activin A (A; ng/ml) based on an input of 500,000 H1 hESCs (n = 3). ANOVA. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus cultures induced with 6 ng/ml of Activin A (arrow). Counts represent the sum of the erythroid, myeloid, and mixed erythromyeloid colonies. (F) Quantification of the number of total (black) and CD43+ (gray) cells generated from 500,000 input H1 hESCs in 6 ng/ml Activin A–induced cultures at the indicated days (n = 4). ANOVA. *, P < 0.05; **, P < 0.01; and ****, P < 0.0001 versus the indicated population on day 6 of differentiation (arrow). (G) Colony-forming progenitor numbers generated between days 6 and 15 of differentiation in cultures induced with 6 ng/ml Activin A based on an input of 500,000 H1 hESCs (n = 3). ANOVA. **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus the indicated lineage on day 9 of differentiation (arrow; black = all colonies). Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythro-myeloid (gray).
Figure S1.
Characterization of hematopoietic differentiation from hPSCs. Related to Fig. 1. (A) Representative flow cytometric analysis of CD43 expression on day 9 of differentiation in cultures induced with different concentrations of Activin A (A; ng/ml). (B) Representative flow cytometric analysis of CD43 expression between days 6 and 15 of differentiation. (C) Distribution of myeloid progenitors generated between days 6 and 15 of differentiation (n = 3). Colonies: Mac, macrophage (white); Mast, mast cell (dark gray); mixed, mixed lineage myeloid (black). (D) Distribution of erythroid progenitors generated between days 6 and 15 of differentiation (n = 3). ANOVA. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ##, P < 0.01; and ####, P < 0.0001 versus small and large erythroid colony morphologies, respectively. Representative brightfield images of the different erythroid colony morphologies that were observed. Scale bar, 50 µm. Colonies: circle = small, square = large, and hexagon = burst morphology. (E) RT-qPCR analysis of the percentage of HBE, HBG, and HBB globin expression in small erythroid colonies (n = 3). ANOVA. ****, P < 0.001 versus the indicated globin gene.
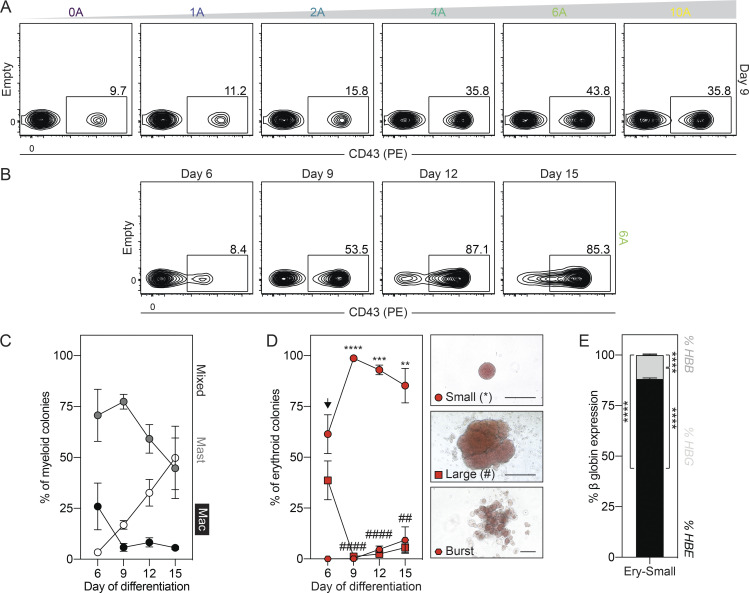
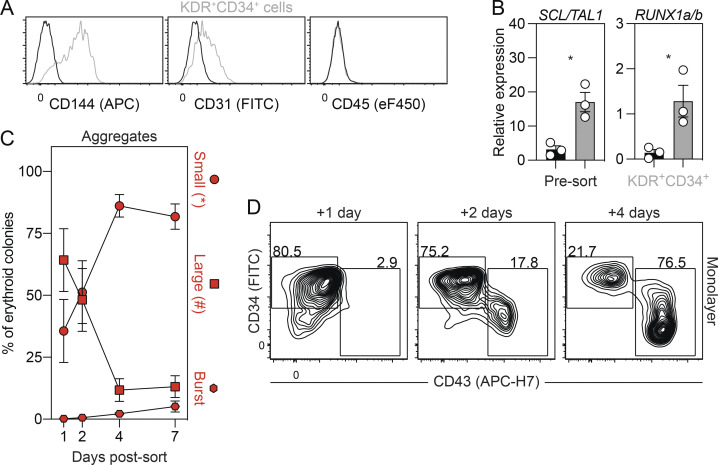
The human primitive program transitions through a hemogenic endothelial cell (HEC) intermediate
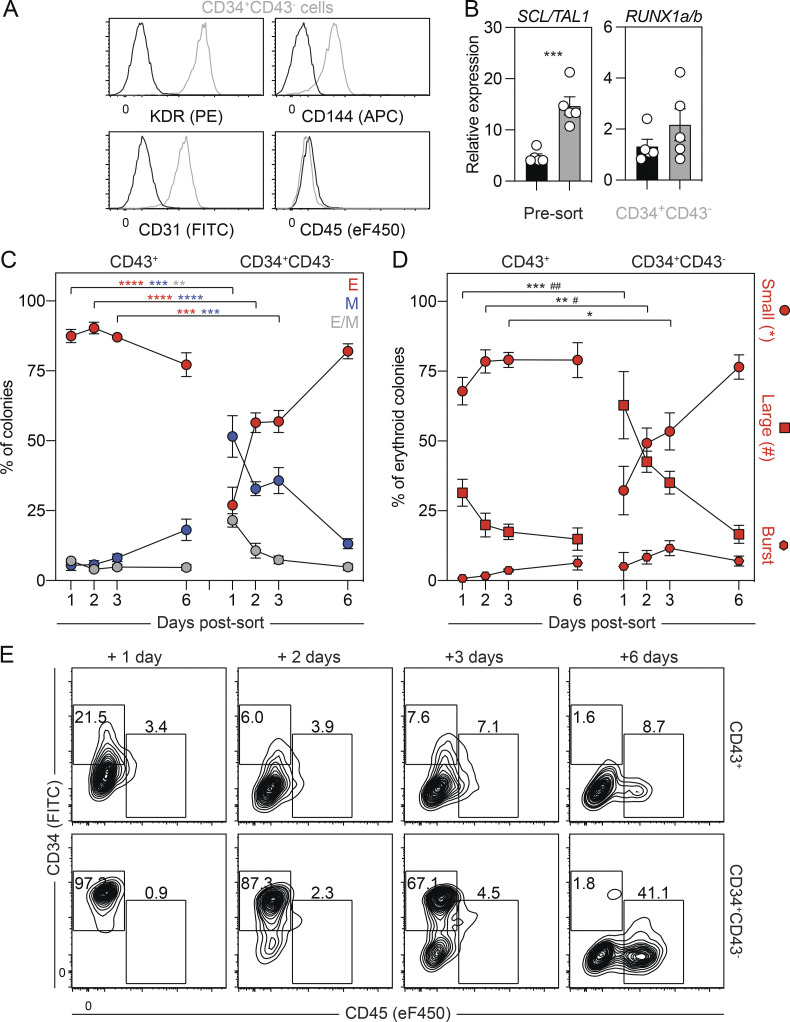
As studies in the mouse have shown that primitive hematopoiesis transitions through a HEC intermediate (Lancrin et al., 2009; Stefanska et al., 2017), we were next interested in determining if the same holds true in the human. To address this question, we monitored the cultures at different days for the emergence of a population that expressed HEC markers before the development of the first CD43+ primitive hematopoietic cells. As shown in Fig. 2 A and Fig. S2 A, a CD34+KDR+CD144+CD31+CD45− vascular/HEC population was detected on day 5 of differentiation, 1 d before the appearance of CD43+ cells. RT-qPCR analyses showed that two transcription factors associated with HECs, SCL/TAL1 and RUNX1a/b, were expressed in the day 5 KDR+CD34+ population (Fig. S2 B), supporting the interpretation that it contains HECs. To formally test this, we isolated the day 5 KDR+CD34+ population by FACS and cultured the cells as aggregates in the presence of VEGF, FGF2, and hematopoietic cytokines for 7 d. Within 1 d, we detected a small population of CD43+ hematopoietic cells that became increasingly abundant over the duration of the culture, demonstrating that the day 5 KDR+CD34+ population has hematopoietic potential (Fig. 2, B and C). Analyses of colony-forming potential revealed a transient wave of progenitors in the cultures that peaked in number on day 4 of culture (Fig. 2 D). The majority of these progenitors gave rise to small erythroid colonies characteristic of the primitive erythroid lineage (Fig. 2 D and Fig. S2 C). When plated on a thin layer of Matrigel, the day 5 KDR+CD34+ cells formed an adherent monolayer. Within 24 h, round, nonadherent CD43+ hematopoietic cells appeared in the cultures (Fig. 2 E and Fig. S2 D), indicative of the endothelial-to-hematopoietic transition, characteristic of hematopoietic commitment of HECs. Collectively, these results provide strong evidence that the human primitive program transitions though an HEC intermediate before the onset of hematopoiesis.
Figure 2.
The human primitive program transitions through a HEC intermediate. (A) Representative flow cytometric analysis of CD43, KDR, and CD34 expression between days 4 and 6 of differentiation, including the gating used to isolate the day 5 KDR+CD34+ population. (B) Representative flow cytometric analysis of CD34 and CD43 expression in cultures generated from FACS-isolated day 5 KDR+CD34+ cells. (C) Quantification of the number of total (black) and CD43+ (gray) cells generated from 62,500 day 5 KDR+CD34+ cells over 7 d of aggregation culture (n = 4–5). ANOVA. *, P < 0.05; **, P < 0.01; and ****, P < 0.0001 versus the indicated population after 1 d of culture (arrow). (D) Colony-forming progenitor numbers generated from 62,500 day 5 KDR+CD34+ cells over 7 d of aggregation culture (n = 4–5). ANOVA. *, P < 0.05; **, P < 0.01 versus the indicated lineage after 4 d of culture (arrow; black = all colonies). Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythro-myeloid (gray). (E) Representative phase-contrast images of monolayer cultures derived from the day 5 KDR+CD34+ population over 4 d of culture. Scale bar, 50 µm.
Figure S2.
Characterization of the day 5 KDR+CD34+ population. Related to Fig. 2. (A) Representative flow cytometric analysis of CD144, CD31, and CD45 expression in the KDR+CD34+ population (gray) on day 5 of differentiation. Black line = unstained cells. (B) RT-qPCR analysis of SCL/TAL1 and RUNX1a/b expression in KDR+CD34+ (gray) and unsorted (black) cells on day 5 of differentiation (n = 3). t test. *, P < 0.05. (C) Distribution of erythroid progenitors generated over 7 d of culture in aggregates derived from day 5 KDR+CD34+ cells (n = 4–5). Colonies: circle = small, square = large, and hexagon = burst morphology. (D) Representative flow cytometric analysis of CD34 and CD43 expression in monolayer cultures generated from FACS-isolated day 5 KDR+CD34+ cells.
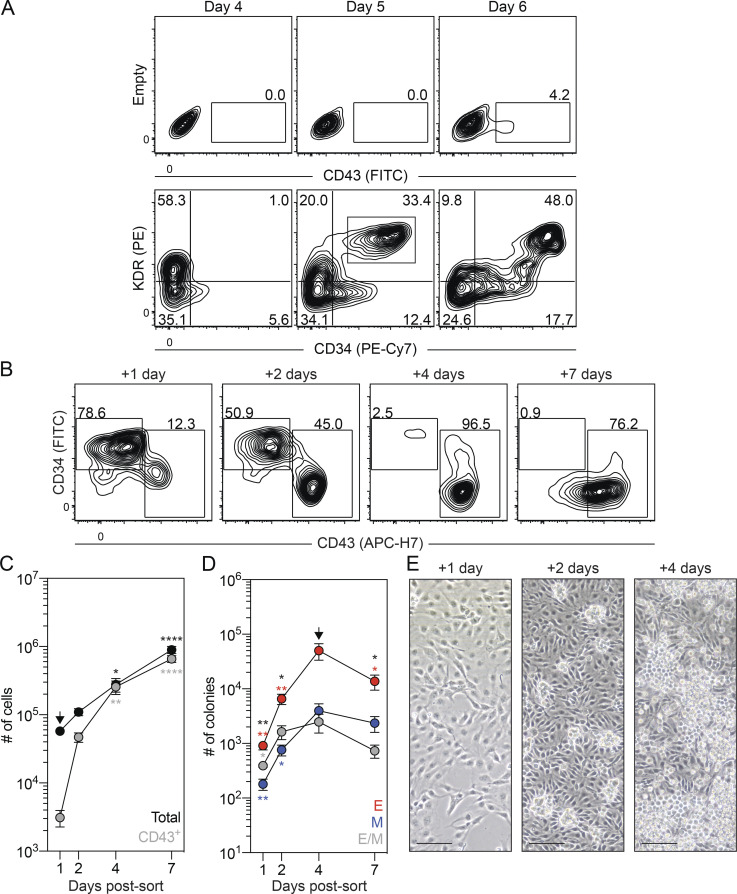
Identification of a second HEC population with EMP and lymphoid potential
In addition to the CD43+ hematopoietic cells, the day 6 cultures also contained a CD34+KDR+CD144+CD31+CD43−CD45− population that expressed SCL/TAL1 and RUNX1a/b (Fig. 3 A; and Fig. S3, A and B), suggesting that it also contains HECs. To characterize the potential of the day 6 CD34+CD43− cells, we isolated them by FACS and cultured the cells as aggregates, as described above. For comparison, we also isolated and cultured the CD43+ cells. The number of cells generated by the day 6 CD43+ population increased over the initial 3 d of culture and then remained stable (Fig. 3, B and C). Colony-forming analyses of day 6 CD43+-derived cells revealed a transient pattern of progenitor development almost identical to that generated by the day 5 KDR+CD34+ population (Fig. 3 D and Fig. S3 C). This pattern strongly suggests that the day 6 CD43+ population represents the emerging primitive hematopoietic program. Analyses of the myeloid compartment following 3 d of culture revealed the presence of macrophage and mast cell progenitors. The high proportion of mast cell progenitors indicates that this lineage is generated by the human primitive program (Fig. 3 G). The day 6 CD34+CD43− cells gave rise to a small CD43+ population within 1 d of culture that continued to increase in size over the next 5 d (Fig. 3, B and C). Colony-forming progenitors were detected within 24 h of culture, and their numbers continued to increase over the 6-d period (Fig. 3 D), a pattern distinct from that of the day 6 CD43+ population. At early stages, cultures derived from the day 6 CD34+CD43− population contained a more even distribution of erythroid and myeloid colony-forming progenitors than the erythroid bias observed in the day 6 CD43+-derived cultures (Fig. 3 D and Fig. S3 C). Within the first 3 d of culture, the day 6 CD34+CD43− population generated erythroid progenitors that gave rise to large and burst colonies in addition to the progenitors that formed the small colonies. Progenitors that formed the small erythroid colonies dominated the population by 6 d of culture and may well represent progeny of the progenitors that generated the large and burst colonies at the earlier stages (Fig. S3 D). Taken together, these patterns of hematopoietic development support the interpretation that the day 6 CD34+CD43− HECs give rise to hematopoietic progenitors that are distinct from those of the primitive program and may represent the human EMP program.
Figure 3.
Separation of the primitive and EMP programs on day 6 of differentiation. (A) Representative flow cytometric analysis of CD34 and CD43 expression on day 6 of differentiation, including the gating strategy used to isolate the day 6 CD43+ and CD34+CD43− populations. (B) Representative flow cytometric analysis of CD34 and CD43 expression in cultures generated from FACS-isolated day 6 CD43+ and CD34+CD43− cells. (C) Quantification of the number of total (black), CD43+ (gray), and CD45+ (white) cells generated from 62,500 day 6 CD43+ or CD34+CD43− isolated cells (n = 5). t test and ANOVA. **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus the stage-matched sample or versus after 1 d of culture within the same sample (arrow), as indicated. (D) Colony-forming progenitor numbers in populations generated from 62,500 day 6 CD43+ or CD34+CD43− cells (n = 5). t test and ANOVA. *, P < 0.05; **, P < 0.01; and ****, P < 0.0001 versus the stage-matched sample or versus after 1 d of culture within the same sample (arrow), as indicated (black = all colonies). Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythromyeloid (gray). (E) Representative flow cytometric analysis of CD34 and CD45 expression following 5 d of culture of day 6 CD43+ and CD34+CD43− cells, including the gating strategy used to isolate the KIT+ and KIT− CD34+CD45+ populations generated from the day 6 CD34+CD43− population. (F) Colony-forming progenitor frequency in the isolated CD34+CD45+ populations shown in E (n = 3). ANOVA. **, P < 0.01 versus the indicated lineage (black = all colonies). Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythro-myeloid (gray). (G) Distribution of myeloid progenitors observed in F and after 3 d of culture of the day 6 CD43+ population. Colonies: Mac, macrophage (white); Mast, mast cell (dark gray); Gr, granulocyte (light gray); mixed, mixed lineage myeloid (black).
Figure S3.
Characterization of the day 6 CD43+ and CD34+CD43− populations. Related to Fig. 3. (A) Representative flow cytometric analysis of KDR, CD144, CD31, and CD45 expression in the day 6 CD34+CD43− population (gray). Black line = unstained cells. (B) RT-qPCR analysis of SCL/TAL1 and RUNX1a/b expression in CD34+CD43− (gray) and unsorted (black) cells on day 6 of differentiation (n = 5). t test. ***, P < 0.001. (C) Distribution of lineages observed in Fig. 3 D (n = 5). ANOVA. **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus the stage-matched sample. Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythromyeloid (gray). (D) Distribution of erythroid progenitors observed in Fig. 3 D. ANOVA. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; #, P < 0.05; and ##, P < 0.01 versus small and large erythroid colony morphologies, respectively. Colonies: circle = small, square = large, and hexagon = burst morphology. (E) Representative flow cytometric analysis of CD34 and CD45 expression in cultures generated from FACS-isolated day 6 CD43+ and CD34+CD43− cells.
As CD45 is expressed on a subset of EMPs, but not primitive hematopoietic progenitors (McGrath et al., 2015; Mikkola et al., 2003), we next analyzed our hPSC-derived populations for the presence of this marker. Both the day 6 CD43+ and CD34+CD43− populations gave rise to few CD45+ cells after 3 d of culture. By day 6, both populations generated CD45+ cells; however, the number was significantly higher in the CD34+CD43−-derived than in the CD43+-derived population (Fig. 3 C and Fig. S3 E). The majority of the CD45+ cells in both groups did not express CD34 at day 6, suggesting that these populations represent maturing hematopoietic cells (Fig. S3 E). Analysis on day 5 showed a different picture, as it revealed the presence of a distinct CD34+CD45+ population in the CD34+CD43−-derived cultures that was not detected in the cultures generated from the CD43+ cells at any stage of development (Fig. 3 E and Fig. S3 E). The majority of the CD34+CD45+ cells expressed KIT, a marker found on EMPs but not on primitive hematopoietic progenitors in the mouse (Fig. 3 E; Ferkowicz et al., 2003; McGrath et al., 2015; Mikkola et al., 2003). Progenitor analyses showed that the frequency of colony-forming cells was significantly higher in the KIT+ than in the KIT− fraction of the CD34+CD45+ population (Fig. 3 F). Few erythroid-restricted progenitors were detected; however, both fractions gave rise to mixed-lineage colonies containing erythroid and myeloid cells. Primitive erythroid progenitors were not detected in either fraction. In addition to the macrophage and mast cell lineages, colonies of granulocytes developed from both KIT fractions (Fig. 3 G). The presence of granulocyte progenitors in the CD34+CD43−-derived population is consistent with observations in the mouse that EMP, but not the primitive program, generates this lineage (Palis et al., 1999). Taken together, these findings identify distinct populations of CD34+ progenitors that display developmental potential indicative of the human primitive and EMP hematopoietic programs.
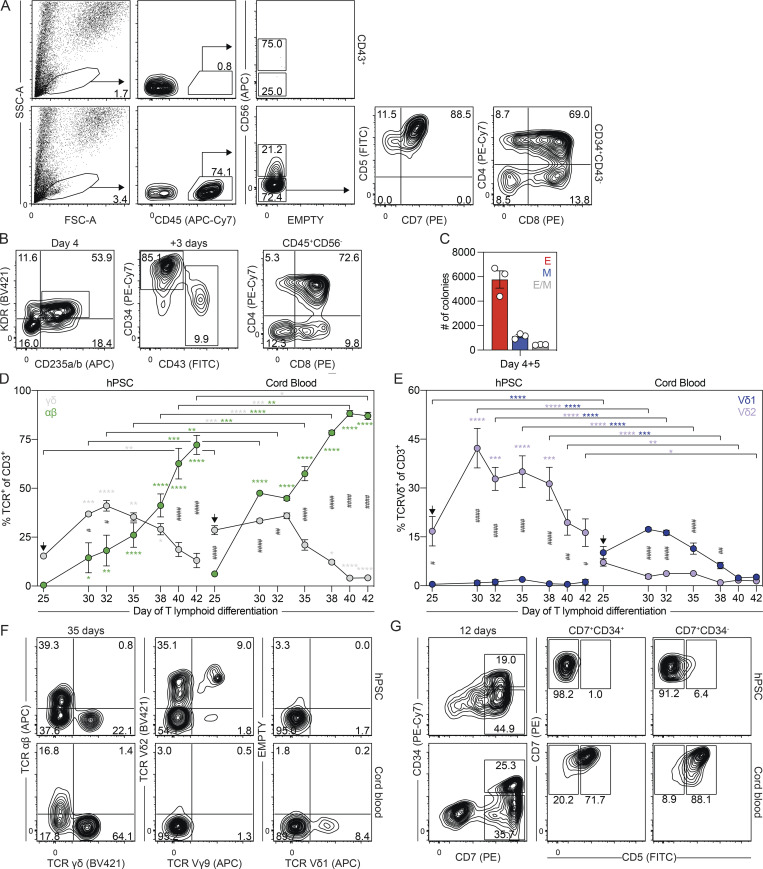
Given that the mouse yolk sac has lymphoid potential, we next were interested in determining if the day 6 CD34+CD43− population also has the capacity to generate lymphoid cells. To evaluate this, we cultured the cells with OP9-DL4 stromal cells to assay T lymphoid potential (Mohtashami et al., 2013; Schmitt and Zuniga-Pflucker, 2006). The day 6 CD43+ population was analyzed in parallel. As shown in Fig. 4 A, the CD34+CD43− population, but not the primitive CD43+ population, generated CD45+CD56−CD4+CD8+ T lymphoid progenitors following 1 mo of culture. To confirm that the T lymphoid lineage develops from KDR+CD235a/b+ mesoderm, we next isolated this population and cultured the cells as aggregates for 3 d before their transfer to OP9-DL4 cells. During this 3-d aggregation period, the day 4 KDR+CD235a/b+ cells gave rise to CD43+ and CD34+CD43− populations similar to those found in unmanipulated EBs on day 6 of differentiation (Fig. 4 B). As observed with the sorted day 6 CD34+CD43− cells, the KDR+CD235a/b+-derived cells generated CD45+CD56−CD4+CD8+ T lymphoid progenitors after 1 mo of culture (Fig. 4 B), indicating that this subset of mesoderm harbors T lymphoid potential. In addition to T lymphoid progenitors, this population also contained primitive erythroid and myeloid colony-forming progenitors (Fig. 4 C), demonstrating that the KDR+CD235a/b+ mesoderm gives rise to the spectrum of the hematopoietic lineages found in the mouse yolk sac.
Figure 4.
The day 6 CD34+CD43− population give rise a unique T lymphoid lineage. (A) Representative flow cytometric analysis of CD45, CD56, CD5, CD7, CD4, and CD8 expression on cells derived from the day 6 CD43+ or CD34+CD43− populations following 30 d of culture with OP9-DL4 cells. FSC-A, forward scatter area; SSC-A, side scatter area. (B) Gating strategy used for the FACS-based isolation of day 4 KDR+CD235a/b+ cells, which give rise to CD43+ and CD34+CD43− populations when cultured as aggregates for 3 d. Transfer of these cultures to OP9-DL4 cells generated T lymphoid progenitors marked by the presence of CD45+CD56−CD4+CD8+ cells after 30 d. (C) Colony-forming progenitor number after 5 d of culture of 62,500 day 4 KDR+CD235a/b+ cells (n = 3). Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythro-myeloid (gray). (D) Quantification of the frequency of γδ (gray) and αβ (green) CD3+ T cells between days 25 and 42 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− or cord blood CD34+ cells (n = 3). ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus the stage and population-matched sample or versus day 25 of T lymphoid differentiation (arrow), as indicated. #, P < 0.05; ##, P < 0.01; and ####, P < 0.0001 between stage-matched γδ and αβ T cells for the indicated sample. (E) Quantification of the frequency of Vδ1+ (blue) and Vδ2+ (purple) CD3+ T cells between days 25 and 42 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− or cord blood CD34+ cells (n = 3). ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus the stage- and population-matched sample or versus day 25 of T lymphoid differentiation (arrow), as indicated. #, P < 0.05; ##, P < 0.01; and ####, P < 0.0001 between stage-matched Vδ1+ and Vδ2+ T cells for the indicated sample. (F) Representative flow cytometric analysis of TCRγδ+, TCRαβ, Vδ1, Vδ2, and Vγ9 expression on day 35 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− or cord blood CD34+ cells. (G) Representative flow cytometric analysis of CD34, CD7, and CD5 expression on cells derived from day 6 CD34+CD43− or cord blood CD34+ cells following 12 d of culture with OP9-DL4 cells.
hPSC-derived progenitors give rise to Vδ2+ T cells
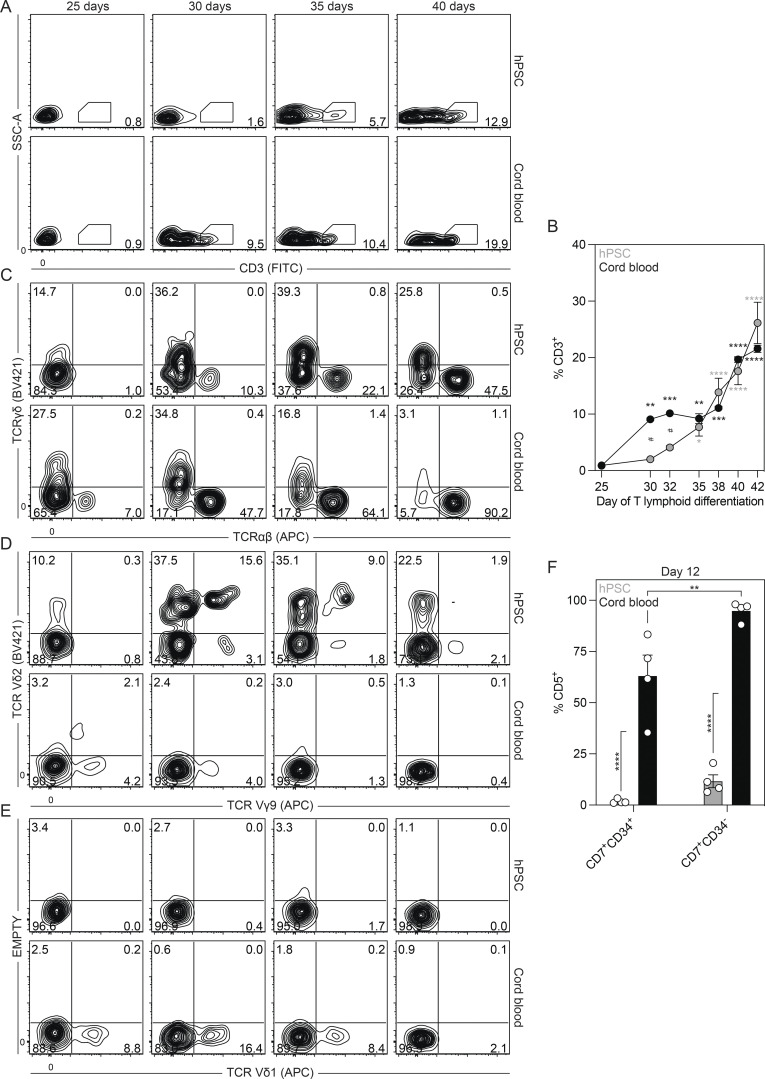
To further characterize the T lymphoid lineage generated by the day 6 CD34+CD43− progenitors, we analyzed the population for the presence αβ and γδ T cells, including those that express Vδ2 and Vδ1. For comparison, we analyzed the T lymphoid potential of CD34+ cord blood progenitors, representative of definitive hematopoiesis. Kinetic studies showed that CD3+ T cells emerged between the third and fourth weeks of culture (Fig. S4, A and B). γδ T cells predominated the TCR-expressing population at early stages of differentiation and preceded the emergence of αβ T cells in both the hPSC- and cord blood–derived cultures (Fig. 4 D and Fig. S4 C). Analyses of Vδ2 and Vδ1 expression showed that the hPSC-derived day 6 CD34+CD43− population gives rise to Vδ2+ but not Vδ1+ T cells (Fig. 4, E and F; and Fig. S4, D and E). A fraction of the hPSC-derived cells also expressed Vγ9 (Fig. 4 F and Fig. S4 D), an observation consistent with the presence of Vγ9+Vδ2+ T cells in the human fetus at early gestational stages (Dimova et al., 2015; McVay and Carding, 1996). In contrast, while the cord blood progenitors generated few Vδ2+ T cells, Vδ1+ T cells were abundant (Fig. 4, E and F; and Fig. S4, D and E). These observations suggest that human Vδ2+ and Vδ1+ T cells develop from yolk sac and definitive progenitors, respectively. To determine if the T cell lineages generated from hPSCs and cord blood differ at earlier stages of development, we compared the CD7+ progenitors generated after 12 d of culture, as the expression of this marker represents one of the earliest stages of T lymphoid commitment. Analyses of the cord blood–derived cells showed that the CD34+ and CD34− fractions of the CD7+ population coexpressed CD5, consistent with the expected pattern of T lymphopoiesis. In contrast, the hPSC-derived CD7+CD34+ population did not express CD5 at this stage or any other stage that was analyzed. A small proportion of the CD7+CD34− population expressed CD5 at this time (Fig. 4 G and Fig. S4 F). Collectively, these findings show that KDR+CD235a/b+ mesoderm induced with Activin A, BMP4, and FGF2 gives rise to the primitive, EMP, and lymphoid yolk sac hematopoietic programs, largely reflecting the hematopoietic potential of the mouse yolk sac. These studies also provide evidence that the hPSC-derived T cells differ from those generated from postnatal progenitors.
Figure S4.
Characterization of the embryonic T cell lineage. Related to Fig. 4. (A) Representative flow cytometric analysis of CD3 expression between days 25 and 40 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− or cord blood CD34+ cells. SSC-A, side scatter area. (B) Quantification of the frequency of CD3+ cells between days 25 and 42 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− (gray) or cord blood CD34+ (black) cells (n = 3). ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus day 25 of T lymphoid differentiation for the indicated population and #, P < 0.05 versus the stage-matched sample. (C) Representative flow cytometric analysis of TCRγδ and TCRαβ expression between days 25 and 40 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− or cord blood CD34+ cells. (D) Representative flow cytometric analysis of Vγ9 and Vδ2 expression between days 25 and 40 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− cells or cord blood CD34+ cells. (E) Representative flow cytometric analysis of Vδ1 expression between days 25 and 40 of T lymphoid differentiation in cultures derived from day 6 CD34+CD43− or cord blood CD34+ cells. (F) Quantification of frequency CD5+ cells in the CD34+CD7+ and CD34−CD7+ populations derived from day 6 CD34+CD43− (gray) or cord blood CD34+ (black) cells after 12 d of culture with OP9-DL4 cells (n = 4). ANOVA. **, P < 0.01; ****, P < 0.0001 versus the indicated sample.
The EMP and lymphoid lineages develop from a multipotent hematopoietic progenitor
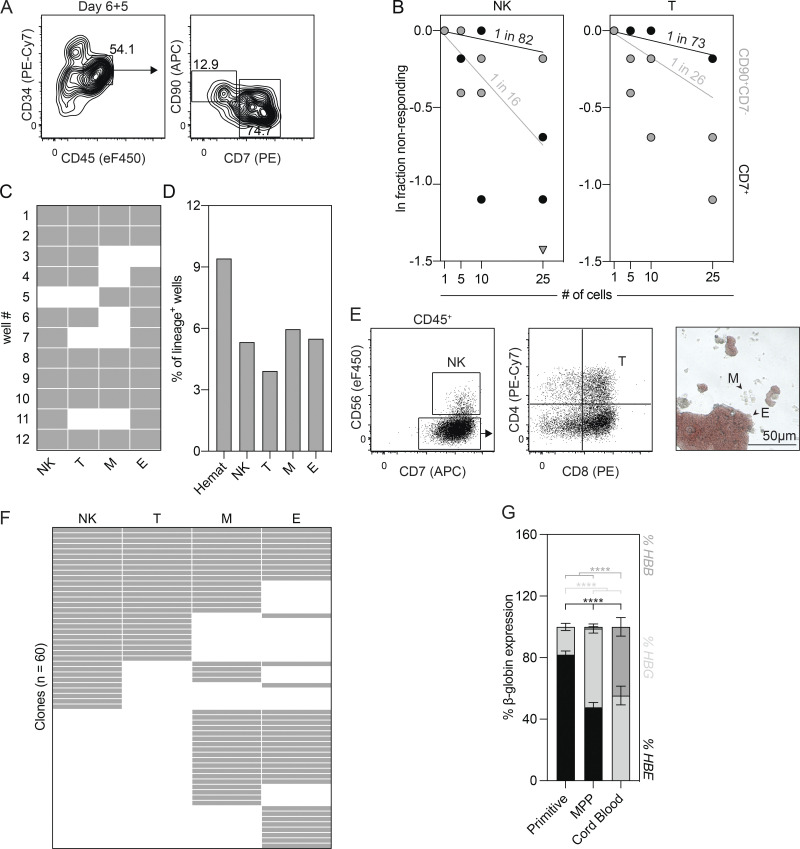
The close temporal association between the EMP and lymphoid hematopoietic programs in the mouse yolk sac and the demonstration that the hPSC-derived day 6 CD34+CD43− population contains erythroid, myeloid, and T lymphoid potential raised the possibility that these lineages develop from a common multipotent hematopoietic progenitor. To search for this progenitor, we cultured the day 6 CD34+CD43− cells with OP9-DL4 cells for 5 d to initiate the endothelial-to-hematopoietic transition and then analyzed cultures for the presence of cells that express markers of hematopoietic progenitors. Through this analysis, we identified distinct CD34+CD45+CD90+CD7− (CD90+CD7−) and CD34+CD45+CD7+ (CD7+) populations (Fig. 5 A) that displayed lymphoid potential. Limiting dilution analyses revealed that the CD90+CD7− population had a higher frequency of both NK (1 in 16 cells) and T lymphoid (1 in 26 cells) progenitors than the CD7+ population (NK, 1 in 82 cells; T lymphoid, 1 in 73 cells; Fig. 5 B). Given this, we continued to evaluate the CD90+CD7− population and next asked if it contains erythroid and myeloid potential. To test this, we deposited 25 CD90+CD7− cells onto OP9-DL4 cells by FACS and cultured them for 4 d. Thereafter, 30% of the cells were transferred to methylcellulose to measure erythroid and myeloid colony-forming progenitors, and the remaining 70% of cells were maintained on the OP9-DL4 cells to evaluate lymphoid potential. As shown in Fig. 5 C, all of the cultures displayed hematopoietic activity, and 50% of them contained all four lineages, indicating that these conditions broadly support multilineage hematopoietic differentiation and therefore should be appropriate for expanding clones.
Figure 5.
The day 6 CD34+CD43− population gives rise to multipotent hematopoietic progenitors. (A) Representative flow cytometric analysis of CD34, CD45, CD90, and CD7 expression on cells generated from the day 6 CD34+CD43− populations following 5 d of culture with OP9-DL4 cells. (B) Limiting dilution analysis of NK cell (CD45+CD7+CD56+) and T lymphoid (CD45+CD7+CD56−CD4+CD8+) progenitor frequencies in the CD34+CD45+CD90+CD7− and CD34+CD45+CD90−CD7+ populations (n = 2). (C) Summary of cultures initiated with 25 CD34+CD45+CD90+CD7− cells that gave rise to NK cell (NK), T lymphoid (T), myeloid (M), and erythroid (E) progeny. Gray = lineage positive. (D) Summary of the cultures initiated with one CD34+CD45+CD90+CD7− cell that gave rise to a hematopoietic clone (Hemat). (E) Representative flow cytometric analysis of CD56, CD7, CD4, and CD8 expression and brightfield image identifying the NK cell (NK), T lymphoid (T), myeloid (M), and erythroid (E) lineages generated from a single CD34+CD45+CD90+CD7− cell. (F) Summary of the NK cell (NK), T lymphoid (T), myeloid (M), and erythroid (E) lineage potential of cells that gave rise to a hematopoietic clone (n = 60). Gray = lineage positive. (G) RT-qPCR analysis of the percentage of HBE, HBG, and HBB globin expression in erythroid colonies generated from the hPSC-derived primitive, MPP, and cord blood progenitors (n = 5–35). ANOVA. ****, P < 0.0001 versus the indicated sample.
To determine if the CD90+CD7− population contains multipotent hematopoietic progenitors, we deposited single cells onto OP9-DL4 cells by FACS and cultured them for 4 d to initiate clonal expansion. The emerging clones were then assayed for erythroid, myeloid, and lymphoid (NK and T) potential, as described above. From 637 sorted cells, 60 (9.4%) produced a hematopoietic clone (Fig. 5 D), and, of these, 10 (16.7%) contained NK cell, T lymphoid, erythroid, and myeloid progeny (Fig. 5, E and F), demonstrating the existence of MPPs within the CD90+CD7− population. The remaining 50 clones were largely restricted to the lymphoid (20%) or erythromyeloid (25%) fates (Fig. 5 F). We next compared the globin expression of the erythroid colonies generated from each clone with erythroid colonies derived from hPSC-derived primitive and cord blood hematopoietic progenitors. The erythroid colonies generated from the CD90+CD7− progenitors expressed lower levels of the embryonic globin HBE and higher levels of the fetal globin HBG than the primitive erythroid colonies. In contrast, the cord blood–derived colonies did not express HBE but did express both HBG and the adult globin, HBB (Fig. 5 G). This intermediate pattern is consistent with the globin expression profile observed in erythroblasts in the human fetal liver between 5 and 6 wk of gestation, a stage that likely contains EMPs (Migliaccio et al., 1986; Peschle et al., 1985, 1984). Taken together, these findings demonstrate that the day 6 CD34+CD43− HECs differentiate to give rise to a CD34+CD45+CD90+CD7− population that contains multipotent hematopoietic progenitors with erythro-myeloid and lymphoid potential.
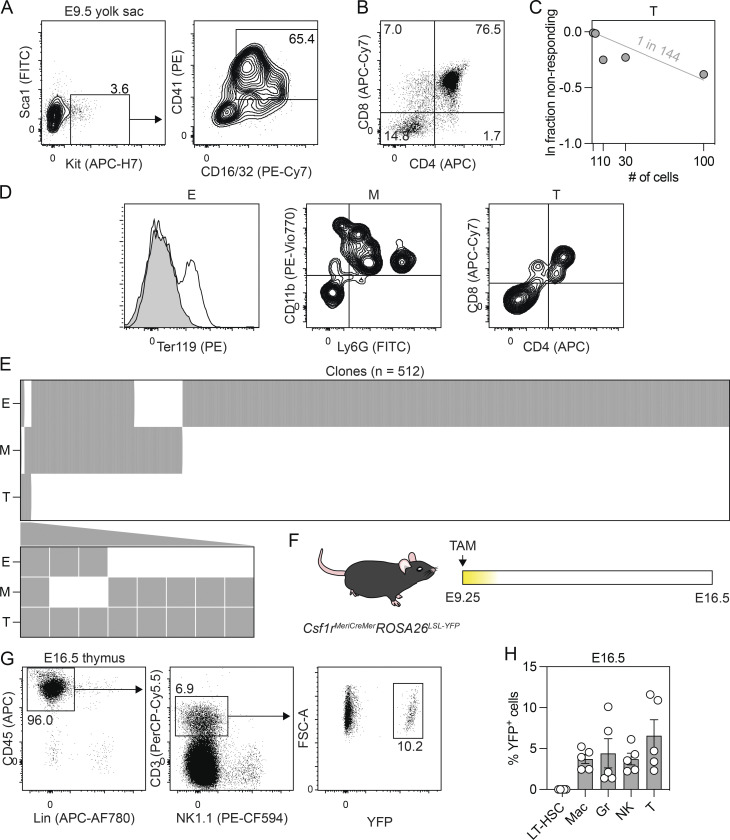
The mouse yolk sac CD41+Kit+CD16/32+ EMP population contains T lymphoid progenitors
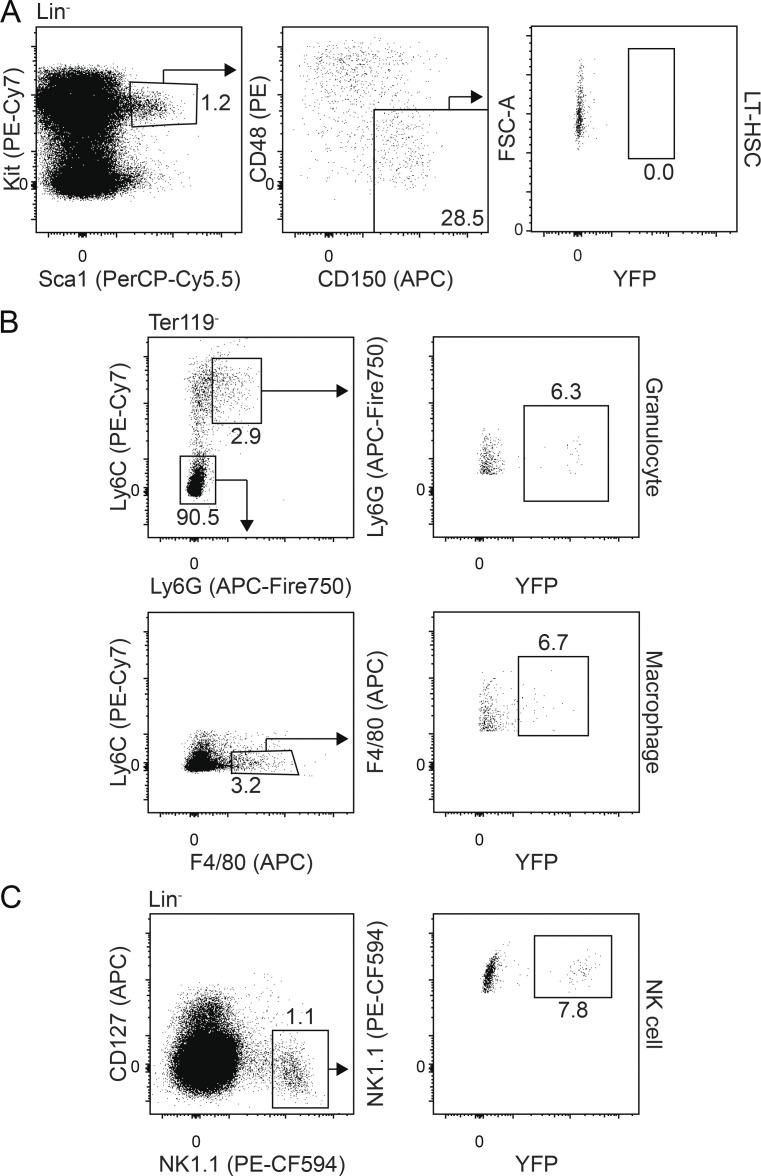
Given our demonstration of a multipotent hematopoietic progenitor in what we consider to be the human equivalent of EMP hematopoiesis, we were next interested in determining if the mouse CD41+Kit+CD16/32+ population (Fig. 6 A) found in the yolk sac at E9.5 has similar potential. As shown in Fig. 6 B, these progenitors are able to generate CD45+CD4+CD8+ T lymphoid progenitors following 11 d of culture with OP9-DL1 cells, indicating that the potential of this population is not restricted to erythroid, myeloid, and NK cell fates. Limiting dilution analyses revealed that the frequency of T lymphoid progenitors in the CD41+Kit+CD16/32+ population was 1 in 144 cells (Fig. 6 C). To determine if the population contains multipotent hematopoietic progenitors, we deposited single CD41+Kit+CD16/32+ cells onto OP9-DL1 cells by FACS and cultured them in a broad spectrum of cytokines known to support multilineage hematopoietic differentiation. Developing clones were analyzed at 11 d of culture by flow cytometry for the presence of erythroid (Ter119+), myeloid (CD45+CD11b+ and/or Ly6G+), and T lymphoid (CD45+CD4+CD8+) cells (Fig. 6 D). From 859 sorted CD41+Kit+CD16/32+ progenitors, 512 (59.6%) produced a hematopoietic clone, and, of these, 8 (1.5%) contained T lymphoid cells and at least one other lineage. Five of the eight clones had myeloid and T lymphoid cells, two had erythroid and T lymphoid cells, and one contained all three lineages. The remaining 506 clones were largely lineage restricted (erythroid, 77.0%; myeloid, 6.8%); however, a small proportion gave rise to both the erythroid and myeloid (14.6%) fates (Fig. 6 E). To evaluate the potential of the EMP/MPP progenitors in vivo, we traced the fate of Csf1r-expressing cells at E9.25 using an inducible Csf1rMeriCreMerRosa26LSL-YFP model (Fig. 6 F; Qian et al., 2011; Schulz et al., 2012). With this approach, hematopoietic progenitors can be labeled by the administration of tamoxifen at specific developmental stages, and their progeny can be identified and monitored by the expression of YFP. For this study, we initiated labeling at E9.25 and monitored YFP expression to verify that immunophenotypic HSCs were not labeled. As shown in Fig. 6 G and Fig. S5, YFP+ macrophages (F4/80+), granulocytes (Ly6G+Ly6C+), and NK cells (NK1.1+) were detected in the liver at E16.5. Phenotypic long-term HSCs (Lin−Kit+Sca1−CD150+CD48−) were not labeled, supporting the interpretation that these lineages are HSC independent. Analyses of the E16.5 thymus revealed the presence of YFP+ T cells (CD3+NK1.1+; Fig. 6 H), a finding that is consistent with that reported by Gentek et al. (2018b) and one that provides additional evidence that the earliest T cells originate from yolk sac progenitors, independent of HSCs.
Figure 6.
The mouse CD41+Kit+CD16/32+ EMP population displays T lymphoid potential. (A) Gating strategy used for the FACS-based isolation of Sca1−CD41+Kit+CD16/32+ EMPs from the E9.5 yolk sac. (B) Representative flow cytometric analysis of CD4 and CD8 expression on CD45+ cells derived from the E9.5 yolk sac CD41+Kit+CD16/32+ population following 11 d of culture with OP9-DL1 cells. (C) Limiting dilution analysis of T lymphoid (CD45+CD4+CD8+) progenitor frequency in the E9.5 yolk sac CD41+Kit+CD16/32+ population (data collated from two to five embryos). (D) Representative flow cytometric analysis of Ter119, CD11b, Ly6G, CD4, and CD8 expression identifying the erythroid (E), myeloid (M), and T lymphoid (T) lineages generated from clonal cultures. (E) Summary of the T lymphoid (T), myeloid (M), and erythroid (E) lineage potential of all cells that gave rise to a hematopoietic clone (n = 512). Cells that gave rise to a T lymphoid clone are shown below. Gray = lineage positive. (F) Schematic of the lineage-tracing approach used to track the contribution of Csf1r-expressing cells to the E16.5 fetus following the administration of a single dose of tamoxifen (TAM) at E9.25. (G) Representative flow cytometric analysis of CD45, CD3, NK1.1, and YFP expression on lineage-negative cells isolated from the E16.5 thymus. FSC-A, forward scatter area. (H) Quantification of the frequency of YFP+ cells in the E16.5 liver and thymus. Liver: LT-HSC = Lin−(Ter119, Gr1, CD19, NK1.1, CD3)Sca1+Kit+CD150+CD48−; Mac = macrophage (Ter119−Ly6C−F4/80+); Gr = granulocyte (Ter119−Ly6C+Ly6G+); and NK = NK cell (Ter119−Gr1−CD3−CD19−CD127−NK1.1+). Thymus: T cell = CD45+Lin−(Ter119, Gr1, CD19, NK1.1) CD3+ (n = 5).
Figure S5.
Characterization of the E9.25 Csf1r+-traced YFP+ populations in the E16.5 liver. Related to Fig. 6. (A) Representative flow cytometric analysis of Kit, Sca1, CD48, CD150, and YFP expression on lineage-negative cells (Ter119−Gr1−CD19−NK1.1−CD3−). FSC-A, forward scatter area. (B) Representative flow cytometric analysis of Ly6C, Ly6G, F4/80, and YFP expression on Ter119− cells. (C) Representative flow cytometric analysis of CD127, NK1.1, and YFP expression on lineage-negative cells (Ter119−Gr1−CD3−CD19−).
Yolk sac hematopoietic differentiation from induced PSCs (iPSCs)
To demonstrate that the protocol described here is applicable to other hPSC lines, we tested it on the CHOPWT10 iPSC line (Maguire et al., 2016). Given our understanding that hPSC lines differ in their responsiveness to cytokines, we first titrated Activin A (0 to 10 ng/ml) in the presence of a single concentration of BMP4 (10 ng/ml) and FGF2 (5 ng/ml) between days 1 and 4 of differentiation to optimize the induction of KDR+CD235a/b+ mesoderm. Cultures induced with either 2 ng/ml or 4 ng/ml of Activin A generated the highest proportion of KDR+CD235a/b+ mesoderm at day 4 of differentiation (Fig. 7, A and B). These cultures also contained the highest proportion of CD43+ cells when differentiated in the presence of VEGF, FGF2, and hematopoietic cytokines for an additional 5 d (Fig. 7, A and C). Further analysis of cultures induced with 2 ng/ml of Activin A showed that CD43+ cells were present from day 6 of differentiation and increased in number thereafter (Fig. 7, D and E). Finally, we isolated the day 4 KDR+CD235a/b+ population to evaluate whether the mesoderm generated with 2 ng/ml of Activin A gives rise to the broad set of lineages indicative of yolk sac hematopoiesis. Consistent with our studies using the H1 human embryonic stem cell line (hESCs), the iPSC-derived KDR+CD235a/b+ mesoderm gave rise to progenitors with primitive erythroid potential when assayed in methylcellulose and T lymphoid progenitors when cultured with OP9-DL4 cells (Fig. 7, F and G). Taken together, these findings demonstrate that the yolk sac hematopoietic programs can be specified from different hPSC lines through the appropriate optimization of mesoderm induction.
Figure 7.
Specification of yolk sac–like hematopoietic mesoderm from CHOPWT10 iPSCs. (A) Representative flow cytometric analysis of KDR and CD235a/b expression on day 4 of differentiation and CD43 expression on day 9 of differentiation in cultures induced with different concentrations of Activin A (A; ng/ml). (B) Quantification of the proportion of KDR+CD235a/b+ cells generated on day 4 of differentiation for a given concentration of Activin A (A; ng/ml; n = 3). ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 versus the indicated sample. (C) Quantification of the proportion of CD43+ hematopoietic cells on day 9 of differentiation for a given concentration of Activin A (A; ng/ml; n = 3). (D) Representative flow cytometric analysis of CD43 expression between days 6 and 12 of differentiation in cultures induced with 2 ng/ml of Activin A. (E) Quantification of the number of total and CD43+ cells generated from 500,000 CHOPWT10 iPSCs between days 6 and 12 of differentiation (n = 3). ANOVA. *, P < 0.05; **, P < 0.01; and ****, P < 0.0001 versus the indicated population after 1 d of culture (arrow). (F) Colony-forming progenitor number after 5 d of culture of 62,500 day 4 KDR+CD235a/b+ cells (n = 3). Colonies: E, erythroid (red); M, myeloid (blue); E/M, mixed erythromyeloid (gray). (G) Representative flow cytometric analysis of CD4 and CD8 expression on CD45+CD56− cells derived from the day 4 KDR+CD235a/b+ population following 30 d of culture with OP9-DL4 cells.
KDR+CD235a/b+ mesoderm is detected in vivo
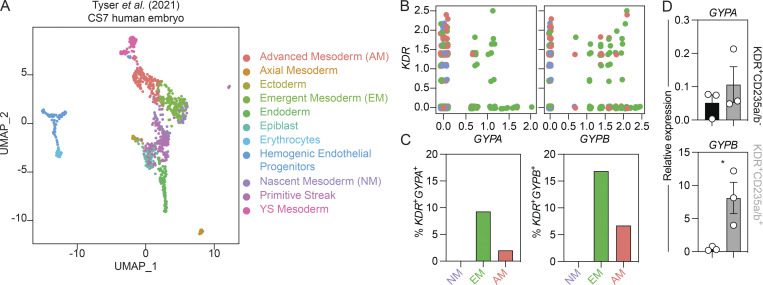
The presence of CD235a/b (encoded by GYPA and GYPB) on the KDR+ mesoderm that gives rise to yolk sac hematopoiesis suggests that the expression of these glycophorins marks a subpopulation of early mesoderm. To determine if CD235a/b is also found on KDR+ mesoderm in vivo, we analyzed single-cell RNA-sequencing data from a human gastrulating embryo (Carnegie stage 7; ∼16-19 d after fertilization) for the expression of these markers (Fig. 8 A; Tyser et al., 2021). As shown in Fig. 8, B and C, a fraction of KDR+ cells in emergent and advanced mesoderm coexpress GYPA and/or GYPB. These analyses also showed that GYPB is expressed at higher levels than GYPA (Fig. 8 B), suggesting that it is the predominant glycophorin expressed in KDR+ mesoderm. This difference in gene expression is consistent with our finding that the level of GYPB expression is substantially higher (80-fold) than GYPA in hPSC-derived day 4 KDR+CD235a/b+ cells (Fig. 8 D). Together, these findings show that GYPB is expressed in a subpopulation of KDR+ mesoderm in vivo.
Figure 8.
Glycophorin expression in mesoderm in vivo. (A) Uniform Manifold Approximation and Projection (UMAP) plot of the human embryo at Carnegie stage 7 (CS7) colored by cluster identity. Data made available by Tyser et al. (2021). (B) Single-cell RNA-sequencing analysis of KDR+GYPA+ and KDR+GYPB+ cells in selected populations of mesoderm in the CS7 human embryo. Mesoderm: NM, nascent (purple); EM, emergent (green); AM, advanced (salmon). (C) Quantification of the number of KDR+GYPA+ and KDR+GYPB+ cells shown in B. Mesoderm: NM, nascent (purple); EM, emergent (green); AM, advanced (salmon). (D) RT-qPCR analysis of GYPA and GYPB expression in KDR+CD235a/b+ (gray) and KDR+CD235a/b− (black) cells on day 4 of hPSC differentiation (n = 3). t test. *, P < 0.05.
Discussion
There is now compelling evidence that yolk sac hematopoiesis gives rise to populations of myeloid (macrophage and mast cell) and lymphoid cells (DETCs) that seed specific sites in developing tissues, where they maintain themselves and function throughout adult life (Gentek et al., 2018a, 2018b; Ginhoux et al., 2010; Gomez Perdiguero et al., 2015; Li et al., 2018; Schulz et al., 2012). As these lineages are not readily generated from HSCs, access to them for detailed studies on their development and function and for potential therapeutic applications relies on our ability to generate them from hPSCs. In this study, we show that the combination of Activin A, BMP4, and FGF2 induces KDR+CD235a/b+ mesoderm from hPSCs that displays the capacity to generate the blood cell lineages indicative of human yolk sac primitive and EMP/lymphoid hematopoiesis. As the primitive and EMP/lymphoid lineages are generated, they transition through HEC intermediates, indicating that all stages of hematopoietic development share this developmental progression. Through clonal analyses, we identified a CD34+CD45+CD90+CD7− multipotent hematopoietic progenitor with erythroid, myeloid, NK cell, and T lymphoid potential that we propose defines an MPP yolk sac hematopoietic program that encompasses both EMP and lymphoid hematopoiesis. While the relationship between the different progenitors within the CD34+CD45+CD90+CD7− population is currently unknown, we speculate that they establish a developmental hierarchy, with the EMP and lymphoid progenitors representing progeny of the MPP (Fig. 9). Our analyses of the mouse yolk sac CD41+Kit+CD16/32+ population showing that, in addition to EMPs, it also contains T cell progenitors, suggest that an MPP yolk sac program may be a unifying concept across species.
Figure 9.
A model of human yolk sac hematopoietic development. In this model, Activin A (Act A), BMP4, and FGF2 promote the generation of KDR+CD235a/b+ mesoderm on day 4 of differentiation. Within 24 h, the mesoderm gives rise to HECs that, in turn, generate CD43+ primitive hematopoietic progenitors with erythroid, mast cell, and macrophage lineage potential. Coincident with the emergence of these primitive hematopoietic progenitors on day 6 of differentiation, a second population of HECs gives rise to CD34+CD45+CD90+CD7− multipotent hematopoietic progenitors (MPPs). The MPP program represents the unification of the EMP and lymphoid programs with its erythroid, mast cell, macrophage, NK cell, and αβ and γδ T lymphoid lineage potential. Of the γδ T cells, the Vδ2+ lineage was preferentially generated.
The approach to modeling human yolk sac hematopoiesis in this study builds on our previous work which showed that the expression of CD235a/b identifies a subset of KDR+ mesoderm that gives rise to primitive, but not definitive hematopoiesis (Sturgeon et al., 2014). The CD235a/b+ mesoderm in that study, induced through the inhibition of the WNT pathway in the context of BMP and FGF2 signaling, did not display T cell potential. Our demonstration that the KDR+CD235a/b+ mesoderm induced with Activin A, BMP4, and FGF2 does give rise to T lymphoid lineage cells stands in contrast to those findings and highlights important differences in the capacity of these signaling pathways to induce seemingly similar mesoderm populations with different developmental potential. The protocol used here is the same as the one that we developed for the induction of CD235a/b+ mesoderm fated to the ventricular cardiomyocyte lineage (Lee et al., 2017). As was the case in that earlier study, we found that the induction of CD235a/b+ mesoderm is exquisitely sensitive to the concentration of Activin A used, suggesting tight developmental control of this mesoderm by the Activin/Nodal pathway. Our demonstration that the H1 and CHOPWT10 hPSCs require slightly different concentrations of Activin A underscores the sensitivity of this mesoderm to Activin/Nodal signaling and the need to establish optimal concentrations of this factor for the efficient generation of KDR+CD235a/b+ mesoderm from different hPSC lines. The observation that ventricular cardiomyocytes, the first cardiomyocyte lineage to develop, and yolk sac hematopoiesis originate from CD235a/b+ mesoderm suggests that expression of these glycophorins marks an early emerging mesoderm population. Our analysis of single-cell RNA-sequencing data of a gastrulating human embryo (Tyser et al., 2021), which showed that KDR+GYPB+ cells are present in advanced and emergent, but not nascent mesoderm, provides evidence that, as in the hPSC differentiation model, GYPB defines a subpopulation of KDR+ mesoderm in vivo.
The contribution of yolk sac progenitors to populations of tissue-resident macrophages in the adult has been well documented by lineage-tracing studies in the mouse (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015; Schulz et al., 2012). As most studies have traced the fate of Csf1r-expressing cells from E8.5, a primitive versus EMP origin of these populations is difficult to determine because both programs are present at this time and can be labeled. Our ability to separate the primitive and MPP hematopoietic populations will enable us to characterize the derivative macrophages with respect to molecular profile, proliferative ability, and engraftment potential. Access to developmentally relevant tissue-resident macrophage progenitors from hPSCs will provide a source of these cells for studies on their role in different diseases, including those of the brain and liver (Krenkel and Tacke, 2017; Salter and Stevens, 2017), and for developing novel cell-based therapies to treat them.
In addition to tissue-resident macrophage progenitors, MPP hematopoiesis also represents a source of Vδ2+ T cells. Our demonstration that progenitors derived from hPSCs, but not cord blood, can generate this subset of T cells suggests that their development is restricted to the HSC-independent yolk sac hematopoiesis. The observations that they represent the predominant γδ T cell population early in fetal life (Haynes and Heinly, 1995; McVay and Carding, 1996), that they are not efficiently reconstituted in pediatric recipients who underwent CD34+ hematopoietic cell transplantation (Airoldi et al., 2015), and that fetal, but not postnatal progenitors efficiently generate these T cells when cultured with OP9-DL1 cells (Tieppo et al., 2020) support this notion. A yolk sac origin of Vδ2+ T cells in the human would recapitulate the pattern of Vγ3+ DETC development in the mouse (Gentek et al., 2018b; Havran and Allison, 1988). Vδ2+ T cells isolated from adults have potent antimicrobial activity and display the capacity to kill certain types of cancers, highlighting possible therapeutic applications (Hoeres et al., 2018; Lawand et al., 2017). Access to these cells from hPSCs provides an unprecedented opportunity to further explore this therapeutic potential.
The concept of an MPP is not inconsistent with many of the observations on hematopoietic lineage specification in the early embryo. As MPPs can only be identified with sensitive assays that support multilineage hematopoiesis, these progenitors may not have been detected in previous studies, or they may have been characterized as lymphoid or erythro-myeloid restricted, depending on the conditions used in the experiment (Bian et al., 2020; McGrath et al., 2015; Yoshimoto et al., 2011, 2012). Alternatively, some of these progenitors, such as the CD45+Kit+Il7ra+Flt3+ cells identified in the fetal liver, may represent progeny of the MPPs that have migrated from the yolk sac (Böiers et al., 2013). Evidence for such migration has been provided by lineage-tracing studies that showed that EMPs from the yolk sac could be detected in the liver at E12.5 (Gomez Perdiguero et al., 2015). Similar migration may also take place in the human, as erythroid progenitors that give rise to colonies that express both embryonic (HBE; ε) and fetal (HBG1/2; Aγ and Gγ) globins, a pattern characteristic of EMP-derived progenitors, have been identified in the liver at 5 wk (Peschle et al., 1985). The concept that MPPs and their progeny colonize the liver has important implications for studies on human fetal liver hematopoiesis. For instance, given the phenotype of the MPPs described here, these progenitors would contaminate CD34+CD45+CD90+CD7− populations enriched for HSCs (Laurenti and Gottgens, 2018). Additionally, it is also possible that hematologic diseases that appear soon after birth, including acute leukemias, arise from MPP and not definitive hematopoiesis (Cazzola et al., 2021).
In summary, our study provides a roadmap of human hematopoietic development that we propose represents the equivalent of human yolk sac hematopoiesis. The temporal emergence and lineage potential of the primitive and MPP programs characterized here recapitulate the development of the primitive and EMP and lymphoid programs in the mouse yolk sac, demonstrating that many aspects of embryonic hematopoiesis are conserved. The advances in this study provide access to the human yolk sac–derived hematopoietic lineages that persist and function in the adult. With this access, it is now possible to further investigate their role in normal tissue homeostasis and their contribution to disease. Evidence for a role in adult disease is provided by the findings that a mutation introduced into the yolk sac hematopoietic progenitors of microglia led to late-onset neurodegenerative disease in mice (Mass et al., 2017) and by the work of Daniels et al. (2020) which showed that Vδ2+ T cells can develop into an aggressive form of cutaneous lymphoma in humans. In addition to providing models to study their role in disease, access to the yolk sac–derived hematopoietic populations provides an opportunity to develop new cell-based strategies to replace and regenerate these cell types in damaged or degenerating tissues.
Materials and methods
hPSC culture
The H1 hESCs (Thomson et al., 1998) and CHOPWT10 induced hPSC line (Maguire et al., 2016) were used in this study. hPSCs were maintained on irradiated mouse embryonic fibroblasts in hESC media containing DMEM/F12 (CellGro) with penicillin/streptomycin (1%; Thermo Fisher Scientific), L-glutamine (2 mM; Thermo Fisher Scientific), nonessential amino acids (1×; Thermo Fisher Scientific), 2-ME (55 µM; Thermo Fisher Scientific), and KnockOut serum replacement (20%; Thermo Fisher Scientific) on 0.1% gelatin (MilliporeSigma)-coated tissue culture plates. Media supplemented with FGF2 (H1 15 ng/ml and CHOPWT10 10 ng/ml; R&D Systems) was changed daily for 7 d before differentiation. hPSCs were routinely tested for mycoplasma. hPSCs were maintained in normoxic conditions (37°C, 5% CO2). The hPSC studies were subject to approval by the Stem Cell Oversight Committee (Canadian Institutes of Health Research).
Hematopoietic differentiation from hPSCs
hPSC differentiation to the hematopoietic lineage was performed in StemPro-34 media supplemented with penicillin/streptomycin (1%; Thermo Fisher Scientific), L-glutamine (2 mM; Thermo Fisher Scientific), ascorbic acid (50 µg/ml; MilliporeSigma), transferrin (150 µg/ml; Roche), monothioglycerol (50 µg/ml; MilliporeSigma), and other stage-specific factors. Differentiation cultures were maintained in hypoxic conditions (37°C, 5% CO2, 5% O2) unless otherwise indicated. On day 0, hPSC cultures at 80–90% confluency were treated with TrypLE for 3 min at 37°C. Thereafter, 80–90% of the TrypLE was aspirated, and the cultures were incubated at 37°C for an additional 2 min. Small clusters of hPSCs (<5 cells per cluster) were generated by gentle pipetting and transferred to 4 ml of StemPro-34 media (Gibco) containing ROCK inhibitor Y-27632 (10 µM) and BMP4 (1 ng/ml) at 500,000 cells/ml. EBs were generated in 60-mm Petri dishes by culture on an orbital shaker (H1, 70 revolutions per minute; CHOPWT10, 60 revolutions per minute) for 18 h. On day 1 of differentiation, the EBs were collected by centrifugation at 40 relative centrifugal force for 5 min and cultured in StemPro-34 media supplemented with BMP4 (10 ng/ml), FGF2 (5 ng/ml), and Activin A (H1, 6 ng/ml; CHOPWT10, 2 ng/ml). Cultures were maintained under static conditions in 5% poly(2-hydroxyethyl methacrylate; MilliporeSigma)-treated tissue culture plates for the duration of the differentiation. On day 4 of differentiation, the EBs were collected by centrifugation at 150 relative centrifugal force for 5 min and cultured in StemPro-34 media supplemented with FGF2 (5 ng/ml), VEGF (15 ng/ml), IL-6 (10 ng/ml), and IL-11 (10 ng/ml). From day 6 of differentiation onward, cultures were maintained in StemPro-34 media containing FGF2 (5 ng/ml), VEGF (15 ng/ml), IL-6 (10 ng/ml), and IL-11 (10 ng/ml), stem cell factor (SCF; 100 ng/ml), IGF1 (50 ng/ml), and erythropoietin (EPO; 4 U/ml). Thereafter, the media were replenished every 2 d for the duration of the differentiation. All recombinant factors are human and were purchased from R&D Systems.
Flow cytometry and cell sorting
EBs before day 9 of differentiation, monolayer cultures, and cultures aggregated after cell sorting were dissociated with trypsin (Corning) for 5 min at 37°C. From day 9 of differentiation onward, EB cultures were dissociated with trypsin for 5 min at 37°C before incubation in collagenase type II (0.2%; Worthington Biochemical) for 1 h at 37°C. Cells were stained at a concentration ≤5,000,000 cells/ml for 30 min at 4°C in the dark. For flow cytometry, cells were stained in IMDM (Gibco) supplemented with penicillin/streptomycin (1%), FCS (2%; Wisent), and DNase I (MilliporeSigma). For FACS, cells were stained, sorted, and collected in StemPro-34 media supplemented with penicillin/streptomycin (1%), L-glutamine (2 mM), and DNase I. Cells were sorted through a 100-µm nozzle using the Influx (BD), FACSAriaII (BD), FACSAriaIII (BD), and MoFlo (Beckman Coulter Life Sciences) cell sorters at the SickKids/University Health Network Flow Cytometry Facility. Thereafter, the sorted cells were counted and resuspended at 250,000 cells/ml in StemPro-34 media supplemented with recombinant factors appropriate for the day of differentiation. To aggregate the sorted cells, 250 µl of the single-cell suspension was transferred to individual wells of 5% poly(2-hydroxyethyl methacrylate)-treated 24-well tissue culture plates. 750 µl of supplemented StemPro-34 media was added 1 d after sorting. To generate monolayer cultures, 80 µl of the single-cell suspension was spotted on individual wells of 25% Matrigel-coated 12-well tissue culture plates in supplemented StemPro-34 media. After 1 d, 1 ml of supplemented StemPro-34 media was added.
To access hematopoietic cells from OP9-DL4 cocultures, human cells were isolated by magnetic-activated cell sorting. The cultures were dissociated with trypsin for 5 min at 37°C and stained with the Mouse Cell Depletion Cocktail (1:5, Miltenyi) at 100,000,000 cells/ml for 15 min at 4°C in the dark. Following magnetic-activated cell sorting on an MS column (Miltenyi Biotec), the flow-through fraction was stained with fluorescence-conjugated antibodies in calcium- and magnesium-free PBS (CellGro) supplemented with 2% FCS and DNase I for 30 min at 4°C in the dark. Cells were sorted and collected in calcium- and magnesium-free PBS supplemented with 2% FCS and DNase I or deposited into wells of 96-well tissue culture plates containing 150 µl of supplemented culture media and stromal cells where indicated.
The following antibodies were used in the human studies: KDR-PE (3:20, clone 89106; R&D Systems), KDR-biotin (1:10, clone 89106; Novus Biologicals), CD235a/b-APC (1:100, clone HIR2/GA-R2; BD Pharmingen), CD34-PE-Cy7 (1:100, clone 4H11; Thermo Fisher Scientific), CD34-APC (1:100, clone 8G12; BD Pharmingen), CD34-FITC (1:100, clone 8G12; BD Pharmingen), CD43-PE (3:100, clone 1G10; BD Pharmingen), CD43-FITC (1:10, clone 1G10; BD Pharmingen), CD43-APC-H7 (1:100, clone 1G10; BD Pharmingen), CD45-eFluor 450 (1:50, clone HI30; Thermo Fisher Scientific), CD45-APC-Cy7 (3:100, clone 2D1; BD Pharmingen), CD45-BV605 (1:50, clone HI30; BioLegend), KIT-APC (1:50, clone 104D2; Thermo Fisher Scientific), KIT-PE (1:50, clone 104D2; BD Pharmingen), CD144-APC (3:100, clone BV9; BioLegend), CD31-FITC (3:20, clone WM59; BD Pharmingen), CD31-PE (3:100, clone WM59; BD Pharmingen), CD90-APC (1:1000, clone 5E10; BD Pharmingen), CD7-PE (7:100, clone M-T701; BD Pharmingen), CD7-APC (3:100, clone 124-1D1; Thermo Fisher Scientific), CD56-eFluor 450 (1:50, clone TULY56; Thermo Fisher Scientific), CD56-APC (3:100, clone B159; BD Pharmingen), CD5-FITC (1:10, clone UCHT2; BD Pharmingen), CD4-PE-Cy7 (1:50, clone SK3; Thermo Fisher Scientific), CD8-PE (1:20, clone HIT8a; BD Pharmingen), CD3-FITC (1:50, clone UCHT1; Thermo Fisher Scientific), TCRαβ-APC (3:100, clone IP26; BioLegend), TCRγδ-PE (5:100, clone IMMU510; Beckman Coulter Life Sciences), TCRγδ-BV421 (2:100, clone B1; BioLegend), TCR Vδ1-APC (1:50, clone REA173; Miltenyi Biotec), TCR Vδ2-BV421 (1:200, clone B6; BioLegend), and TCR Vγ9-APC (1:200, clone B3; BioLegend). Staining with biotin-conjugated antibodies was detected with streptavidin-BV421 (1:100; BioLegend) or streptavidin-PE-Cy7 (1:200; BD Pharmingen).
The following antibodies were used in the mouse studies: Sca1-FITC (1:50, clone REA422; Miltenyi Biotec), Sca1-PerCP-Cy5.5 (1:100, clone D7; Thermo Fisher Scientific), Kit-APC-H7 (1:200, clone 2B8; BD Pharmingen), Kit-PE-Cy7 (1:100, clone 2B8; Thermo Fisher Scientific), CD16/32-PE (1:100, clone REA377; Miltenyi Biotec), CD16/32-PE-Cy7 (1:20, clone 93; Thermo Fisher Scientific), CD48-PE (1:100, clone HM48-1; Thermo Fisher Scientific), CD150-APC (1:100, clone 9D1; Thermo Fisher Scientific), CD41-PE (1:50, clone MWReg30; BD Pharmingen), CD41-VioBlue (1:50, clone MWReag30; Miltenyi Biotec), Ter119-PE (1:50, clone TER-119; BD Pharmingen), Ter119-APC-eFluor 780 (1:100, clone TER-119; Thermo Fisher Scientific), CD45-BV605 (1:50, clone 30-F11; BD Pharmingen), CD45-VioBlue (1:50, clone REA737; Miltenyi Biotec), CD45-APC (1:100, clone 30-F11; Thermo Fisher Scientific), Ly6G-FITC (1:50, clone REA526; Miltenyi Biotec), Ly6G-APC-Fire750 (1:100, clone 1A8; BioLegend), Ly6C-PE-Cy7 (1:100, clone HK1.4; Thermo Fisher Scientific), Gr1-APC-eFluor 780 (1:100, clone RB6-8C5; Thermo Fisher Scientific), F4/80-APC (1:100, clone BM8; Thermo Fisher Scientific), CD11b-PE-Vio770 (1:50, clone REA592; Miltenyi Biotec), CD127-APC (1:100, clone A7R34; Thermo Fisher Scientific), NK1.1-PE-CF594 (1:100, clone PK136; BD Pharmingen), CD4-APC (1:200, clone RM4-5; BD Pharmingen), CD8a-APC-Vio770 (1:200, clone REA601; Miltenyi Biotec), CD3-PerCP-Cy5.5 (1:100, clone 145-2C11; Thermo Fisher Scientific), CD3-APC-eFluor 780 (1:100, clone 17A2; Thermo Fisher Scientific), and CD19-APC-eFluor 780 (1:100, clone eBio1D3; Thermo Fisher Scientific).
RT-qPCR
Total RNA was prepared with the RNAqueous RNA Isolation Kit (Thermo Fisher Scientific) that included treatment with RNase-free DNase. Reverse transcription to cDNA was performed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories). RT-qPCR was performed on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) with the QuantiFast SYBR Green PCR Kit (Qiagen). Gene expression was evaluated as ΔCt relative to TBP. For globin analyses, gene expression was evaluated as ΔCt relative to ACTB. Genomic DNA content was assessed using primers for the GAGEB1 promoter. Primer sequences are listed in Table 1.
Table 1. Primer sequences for RT-qPCR.
| Gene | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) |
|---|---|---|
| TBP | TGAGTTGCTCATACCGTGCTGCTA | CCCTCAAACCAACTTGTCAACAGC |
| ACTB | AAACTGGAACGGTGAAGGTGACAG | CAATGTGCAATCAAAGTCCTCGGC |
| GAGEB1 | CAACATACCTCATAGCATTATACAAGAC | CCCTATGTTCCTGGTTCTTCATATT |
| RUNX1a/b | CGTGCACATACATTAGTAGCACTACC | CCTCCACGAATCTTGCTTGCAGAGGT |
| SCL | CCGTGGATTCGCTTGAGTTA | GAAAGAAGAGGGAGCCAGAAG |
| HBE | TCTGGCTACTCACTTTGGCAAGGA | TCACAGGAACACCTGCAAACTGGA |
| HBG | TGGGAAATGTGCTGGTGACCGTTT | AAGCTCTGAATCATGGGCAGTGAG |
| HBB | ACTAAGCTCGCTTTCTTGCTGTCC | TCCAGATGCTCAAGGCCCTTCATA |
Hematopoietic colony-forming assay
Colony-forming progenitor number was quantified by plating 100–80,000 cells in methylcellulose (1%) containing plasma-derived serum (15%; Animal Technologies), protein-free hybridoma media II (5%; Invitrogen), L-glutamine (2 mM), ascorbic acid (25 µg/ml), transferrin (150 µg/ml), monothioglycerol (33 µg/ml), thrombopoietin (TPO; 50 ng/ml), IL-3 (50 ng/ml), IL-6 (10 ng/ml), IL-11 (5 ng/ml), SCF (100 ng/ml), EPO (4 U/ml), GM-CSF (1 ng/ml), M-CSF (10 ng/ml), IGF1 (25 ng/ml), VEGF (10 ng/ml), and FGF2 (10 ng/ml). Cultures were maintained in normoxic conditions (37°C, 5% CO2) for 9–14 d before quantification.
T lymphoid differentiation using OP9-DL4 cells
OP9 stromal cells that constitutively express Delta-like 4 (OP9-DL4; Mohtashami et al., 2013; Schmitt and Zuniga-Pflucker, 2006) were used to assay T lymphoid potential. Cocultures were maintained in normoxic conditions (37°C, 5% CO2). 1–250,000 cells were cultured with OP9-DL4 cells on 0.1% gelatin-treated tissue culture plates in αMEM (Gibco) supplemented with penicillin/streptomycin (1%), FCS (20%; HyClone), L-glutamine (2 mM), IL-7 (5 ng/ml), and FLT3L (5 ng/ml). SCF (30 ng/ml) was included at the start of the coculture and removed after 4–6 d. Cultures were transferred to new OP9-DL4 cells every 4–6 d by vigorous pipetting and passage through a 40-µm strainer. Cultures were analyzed by flow cytometry at the indicated stages and scored positive if >10 CD45+CD56−CD4+CD8+ events were observed.
Hematopoietic MPP cell assay
Hematopoietic cells were isolated from OP9-DL4 cocultures initiated with day 6 CD34+CD43− cells after 5 d. Single cells were sorted into the wells of 96-well plates, retaining index sorting information on an FACSAria II cell sorter. The perimeter wells were excluded from the sort. The cells were cultured in α-MEM supplemented with FCS (20%), penicillin/streptomycin (1%; Thermo Fisher Scientific), L-glutamine (2 mM; Thermo Fisher Scientific), SCF (30 ng/ml), IL-7 (5 ng/ml) and FLT3L (5 ng/ml). The cultures were maintained in normoxic conditions (37°C, 5% CO2). After 4 d, the cultures were dissociated with trypsin, and 30% of the culture was transferred to methylcellulose to evaluate erythro-myeloid potential, as described above. The cultures were scored after 14 d, and erythroid colonies were picked for β-globin expression analysis by RT-qPCR. As the OP9-DL4 cells express GFP, the colonies were visualized under fluorescence to confirm their human identity. The remaining fraction of the culture was seeded on new OP9-DL4 cells to evaluate NK and T cell potential, as described above. Cultures were dissociated with trypsin and transferred to fresh OP9-DL4 biweekly for another 12–14 d. OP9-DL4 cocultures were analyzed by flow cytometry. >10 CD45+-gated events (T cells, CD7+CD56−CD4+CD8+; NK cells, CD56+CD7+) were required to call a positive well.
Mouse tissue collection and processing
Animal experiments were performed in accordance with the institutional guidelines approved by the University of Rochester School of Medicine and Dentistry Institutional Animal Care and Use Committee and the Animal Care and Use Committee of Ospedale San Raffaele (IACUC 841) and communicated to the Ministry of Health and local authorities according to Italian law. Mice were housed with free access to food and water and maintained in a 12-h/12-h light/dark cycle. Pregnant CD-1 mice were purchased from Charles River Laboratories, and FVB-Tg(Csf1r-cre/Esr1*)1Jwp/J and B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J mice were purchased from The Jackson Laboratory. Mice were euthanized under CO2. Embryonic tissues were staged by somite pairs. E9.5 yolk sacs were dissected and incubated in 0.1% collagenase type I (Sigma-Aldrich) in calcium- and magnesium-free PBS (CellGro) supplemented with 5% FCS (HyClone) for 15 min. Single-cell suspensions were prepared thereafter by gentle pipetting. E16.5 thymi were dissected and incubated in 0.08% collagenase type I (STEMCELL Technologies) in PB2 made up of calcium- and magnesium-containing PBS (Thermo Fisher Scientific) supplemented with 0.3% BSA (Gemini Bioproducts) and 0.1% glucose (Invitrogen) for 30 min. Single-cell suspensions were prepared thereafter by gentle pipetting. E16.5 livers were dissected, and single-cell suspensions were prepared by gentle pipetting in PB2. For in vitro studies of lineage potential, cells were sorted through an 85-µm nozzle using the AriaII cell sorter at the Flow Cytometry Resource, Advanced Cytometry Technical Applications Laboratory (FRACTAL).
Mouse hematopoietic progenitor cell assay
OP9 stromal cells that constitutively express Delta-like 1 (OP9-DL1; Schmitt and Zuniga-Pflucker, 2002) were used to assay the hematopoietic potential of mouse yolk sac cells. Cocultures were maintained in normoxic conditions (37°C, 5% CO2). 500–10,000 cells were cultured with OP9-DL1 cells on tissue culture plates in α-MEM (Thermo Fisher Scientific) supplemented with penicillin/streptomycin (100 µg/ml; Lonza), L-glutamine (2 mM; Lonza), FCS (20%, HyClone), Scf (50 ng/ml), Tpo (50 ng/ml), Gm-csf (3 ng/ml), Il3 (30 ng/ml), Il-6 (5 ng/ml), Flt3l (10 ng/ml), Il-7 (5 ng/ml), and EPO (2 U/ml). All factors are mouse, with the exception of EPO (human). All factors were purchased from Miltenyi Biotec, except for Gm-csf and EPO, which were purchased from PeproTech. Cultures were transferred to new OP9-DL1 cells after 5 d by vigorous pipetting and passage through a 40-µm strainer. For clonal analyses, single cells were sorted into the wells of 96-well plates on an FACSAriaII cell sorter.
Mouse lineage tracing
FVB-Tg(Csf1r-cre/Esr1*)1Jwp/J and B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J mice were mated overnight, and the morning that the vaginal plug was detected was considered E0.3. To induce Cre, mice were administered 4-hydroxytamoxifen (75 mg/g; Sigma-Aldrich) and progesterone (37.5 mg/g; Sigma-Aldrich) by i.p. injection at E9.25.
Analysis of the single-cell RNA-sequencing dataset (Tyser et al., 2021)
Preprocessed count matrix and metadata containing the cell type annotation were retrieved from the study by Tyser et al. (2021). The data were accessed under the accession no. E-MTAB-9388 from ArrayExpress. Further filtering of low-quality cells (nFeature_RNA >2,000) and mitochondrial and cell cycle factors regression (vars.to.regress = c [“percent.mt”, “S.Score”, “G2M.Score”]) was performed with the SCTransform pipeline provided in the Seurat version 3.2.2 R package (Hafemeister and Satija, 2019; Macosko et al., 2015; Satija et al., 2015), as described in the tutorials (http://satijalab.org/seurat/). For clustering analysis, the top 25 principal components were chosen, and a resolution of 0.7 was applied. The cell type annotation provided by Tyser et al. was used to label cells in Uniform Manifold Approximation and Projection plots. KDR-, GYPA-, and GYPB-expressing cells were identified using a threshold of normalized expression value >0.
Quantification and statistical analyses
All data are presented as the mean ± SEM. Flow cytometry and cell sorting data were analyzed using FlowJo software (BD Biosciences). Statistical analyses were performed using GraphPad Prism 9 as indicated in the figure captions. Progenitor frequencies from limiting dilution analyses were calculated using extreme limiting dilution analysis (Hu and Smyth, 2009) in the R statistical computing environment.
Online supplemental material
Fig. S1 is related to Fig. 1 and shows (i) flow cytometric analysis of populations induced with different concentrations of Activin A at day 9 of differentiation and (ii) flow cytometric, colony-forming progenitor, and RT-PCR globin analyses of populations induced with 6 ng/ml of Activin A. Fig. S2 is related to Fig. 2 and shows (i) flow cytometric and RT-qPCR analyses of the day 5 KDR+CD34+ population, (ii) colony-forming progenitor analyses of populations generated from aggregates of day 5 KDR+CD34+ cells, and (iii) flow cytometric analysis of the populations generated from monolayers of the day 5 KDR+CD34+ cells. Fig. S3 is related to Fig. 3 and shows (i) flow cytometric and RT-qPCR analyses of the day 6 CD34+CD43− population and (ii) flow cytometric and colony-forming progenitor analyses of populations generated from the day 6 CD34+CD43− cells. Fig. S4 is related to Fig. 4 and shows flow cytometric analyses of hPSC- and cord blood–derived T cells. Fig. S5 is related to Fig. 6 and shows flow cytometric analysis of the hematopoietic populations analyzed in the Csf1r lineage-tracing study.
Acknowledgments
We thank all members of the Keller laboratory and Norman Iscove for their advice and comments on this work. We also thank Paul D. Kingsley, Rose Guilfoyle, and Leah Vit of the Palis laboratory for technical assistance. We thank the University Health Network/SickKids Flow Cytometry Facility for their assistance with the cell-sorting experiments. We also thank Deborah French (Children’s Hospital of Philadelphia, Philadelphia, PA) for the CHOPWT10 iPSC line.
This work was supported by the Canadian Institutes of Health Research (FDN259937 [G.M. Keller]), the Fondazione Telethon (Italy; SR-TIGET Core Grant, Project C4 [A. Ditadi]), and National Heart, Lung, and Blood Institute, National Institutes of Health (R01 HL151777 [J. Palis]). M.H. Atkins was supported by a Canadian Institutes of Health Research Banting and Best Doctoral Research Award. R. Scarfò conducted this study as partial fulfillment for the PhD in Molecular Medicine, International PhD School, Vita-Salute San Raffaele University (Milan, Italy).
Author contributions: M.H. Atkins and G.M. Keller developed the project and wrote the manuscript. M.H. Atkins performed the hPSC experiments. R. Scarfò performed the mouse embryo ex vivo experiments. R. Scarfò and A. Ditadi analyzed the mouse embryo data. K.E. McGrath and J. Palis performed and supervised the mouse lineage tracing experiments. D. Yang, analyzed the single-cell RNA-sequencing data.
References
- Airoldi, I., Bertaina A., Prigione I., Zorzoli A., Pagliara D., Cocco C., Meazza R., Loiacono F., Lucarelli B., Bernardo M.E., et al. 2015. γδ T-cell reconstitution after HLA-haploidentical hematopoietic transplantation depleted of TCR-αβ+/CD19+ lymphocytes. Blood. 125:2349–2358. 10.1182/blood-2014-09-599423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, Z., Gong Y., Huang T., Lee C.Z.W., Bian L., Bai Z., Shi H., Zeng Y., Liu C., He J., et al. 2020. Deciphering human macrophage development at single-cell resolution. Nature. 582:571–576. 10.1038/s41586-020-2316-7 [DOI] [PubMed] [Google Scholar]
- Böiers, C., Carrelha J., Lutteropp M., Luc S., Green J.C., Azzoni E., Woll P.S., Mead A.J., Hultquist A., Swiers G., et al. 2013. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 13:535–548. 10.1016/j.stem.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Cazzola, A., Cazzaniga G., Biondi A., Meneveri R., Brunelli S., and Azzoni E.. 2021. Prenatal origin of pediatric leukemia: lessons from hematopoietic development. Front. Cell Dev. Biol. 8:618164. 10.3389/fcell.2020.618164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, J., Doukas P.G., Escala M.E.M., Ringbloom K.G., Shih D.J.H., Yang J., Tegtmeyer K., Park J., Thomas J.J., Selli M.E., et al. 2020. Cellular origins and genetic landscape of cutaneous γ δ T cell lymphomas. Nat. Commun. 11:1806. 10.1038/s41467-020-15572-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dege, C., Fegan K.H., Creamer J.P., Berrien-Elliott M.M., Luff S.A., Kim D., Wagner J.A., Kingsley P.D., McGrath K.E., Fehniger T.A., et al. 2020. Potently cytotoxic natural killer cells initially emerge from erythro-myeloid progenitors during mammalian development. Dev. Cell. 53:229–239.e7. 10.1016/j.devcel.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova, T., Brouwer M., Gosselin F., Tassignon J., Leo O., Donner C., Marchant A., and Vermijlen D.. 2015. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc. Natl. Acad. Sci. USA. 112:E556–E565. 10.1073/pnas.1412058112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditadi, A., Sturgeon C.M., and Keller G.. 2017. A view of human haematopoietic development from the Petri dish. Nat. Rev. Mol. Cell Biol. 18:56–67. 10.1038/nrm.2016.127 [DOI] [PubMed] [Google Scholar]
- Ferkowicz, M.J., Starr M., Xie X., Li W., Johnson S.A., Shelley W.C., Morrison P.R., and Yoder M.C.. 2003. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 130:4393–4403. 10.1242/dev.00632 [DOI] [PubMed] [Google Scholar]
- Gentek, R., Ghigo C., Hoeffel G., Bulle M.J., Msallam R., Gautier G., Launay P., Chen J., Ginhoux F., and Bajénoff M.. 2018a. Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity. 48:1160–1171.e5. 10.1016/j.immuni.2018.04.025 [DOI] [PubMed] [Google Scholar]
- Gentek, R., Ghigo C., Hoeffel G., Jorquera A., Msallam R., Wienert S., Klauschen F., Ginhoux F., and Bajénoff M.. 2018b. Epidermal γδ T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J. Exp. Med. 215:2994–3005. 10.1084/jem.20181206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn, E., Yoshimoto M., Nakauchi H., Weissman I.L., and Herzenberg L.A.. 2019. Hematopoietic stem cell-independent hematopoiesis and the origins of innate-like B lymphocytes. Development. 146:dev170571. 10.1242/dev.170571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero, E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., and Rodewald H.R.. 2015. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 518:547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister, C., and Satija R.. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20:296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havran, W.L., and Allison J.P.. 1988. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 335:443–445. 10.1038/335443a0 [DOI] [PubMed] [Google Scholar]
- Havran, W.L., and Allison J.P.. 1990. Origin of Thy-1+ dendritic epidermal cells of adult mice from fetal thymic precursors. Nature. 344:68–70. 10.1038/344068a0 [DOI] [PubMed] [Google Scholar]
- Havran, W.L., Grell S., Duwe G., Kimura J., Wilson A., Kruisbeek A.M., O’Brien R.L., Born W., Tigelaar R.E., and Allison J.P.. 1989. Limited diversity of T-cell receptor γ-chain expression of murine Thy-1+ dendritic epidermal cells revealed by V γ 3-specific monoclonal antibody. Proc. Natl. Acad. Sci. USA. 86:4185–4189. 10.1073/pnas.86.11.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, B.F., and Heinly C.S.. 1995. Early human T cell development: analysis of the human thymus at the time of initial entry of hematopoietic stem cells into the fetal thymic microenvironment. J. Exp. Med. 181:1445–1458. 10.1084/jem.181.4.1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeres, T., Smetak M., Pretscher D., and Wilhelm M.. 2018. Improving the efficiency of Vγ9Vδ2 T-cell immunotherapy in cancer. Front. Immunol. 9:800. 10.3389/fimmu.2018.00800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huang, H., and Auerbach R.. 1993. Identification and characterization of hematopoietic stem cells from the yolk sac of the early mouse embryo. Proc. Natl. Acad. Sci. USA. 90:10110–10114. 10.1073/pnas.90.21.10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, M., D’Souza S.L., Lynch-Kattman M., Schwantz S., and Keller G.. 2007. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 109:2679–2687. 10.1182/blood-2006-09-047704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenkel, O., and Tacke F.. 2017. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 17:306–321. 10.1038/nri.2017.11 [DOI] [PubMed] [Google Scholar]
- Lancrin, C., Sroczynska P., Stephenson C., Allen T., Kouskoff V., and Lacaud G.. 2009. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 457:892–895. 10.1038/nature07679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti, E., and Göttgens B.. 2018. From haematopoietic stem cells to complex differentiation landscapes. Nature. 553:418–426. 10.1038/nature25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawand, M., Déchanet-Merville J., and Dieu-Nosjean M.-C.. 2017. Key features of γδ T-cell subsets in human diseases and their immunotherapeutic implications. Front. Immunol. 8:761. 10.3389/fimmu.2017.00761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., Protze S.I., Laksman Z., Backx P.H., and Keller G.M.. 2017. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 21:179–194.e4. 10.1016/j.stem.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Li, Z., Liu S., Xu J., Zhang X., Han D., Liu J., Xia M., Yi L., Shen Q., Xu S., et al. 2018. Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity. 49:640–653.e5. 10.1016/j.immuni.2018.09.023 [DOI] [PubMed] [Google Scholar]
- Macosko, E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161:1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, J.A., Gagne A.L., Jobaliya C.D., Gandre-Babbe S., Gadue P., and French D.L.. 2016. Generation of human control iPS cell line CHOPWT10 from healthy adult peripheral blood mononuclear cells. Stem Cell Res. (Amst.). 16:338–341. 10.1016/j.scr.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Mass, E., Ballesteros I., Farlik M., Halbritter F., Günther P., Crozet L., Jacome-Galarza C.E., Händler K., Klughammer J., Kobayashi Y., et al. 2016. Specification of tissue-resident macrophages during organogenesis. Science. 353:aaf4238. 10.1126/science.aaf4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass, E., Jacome-Galarza C.E., Blank T., Lazarov T., Durham B.H., Ozkaya N., Pastore A., Schwabenland M., Chung Y.R., Rosenblum M.K., et al. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 549:389–393. 10.1038/nature23672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, K.E., Frame J.M., Fromm G.J., Koniski A.D., Kingsley P.D., Little J., Bulger M., and Palis J.. 2011. A transient definitive erythroid lineage with unique regulation of the β-globin locus in the mammalian embryo. Blood. 117:4600–4608. 10.1182/blood-2010-12-325357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath, K.E., Frame J.M., Fegan K.H., Bowen J.R., Conway S.J., Catherman S.C., Kingsley P.D., Koniski A.D., and Palis J.. 2015. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 11:1892–1904. 10.1016/j.celrep.2015.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVay, L.D., and Carding S.R.. 1996. Extrathymic origin of human γ δ T cells during fetal development. J. Immunol. 157:2873–2882. [PubMed] [Google Scholar]
- Migliaccio, G., Migliaccio A.R., Petti S., Mavilio F., Russo G., Lazzaro D., Testa U., Marinucci M., and Peschle C.. 1986. Human embryonic hemopoiesis. Kinetics of progenitors and precursors underlying the yolk sac----liver transition. J. Clin. Invest. 78:51–60. 10.1172/JCI112572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola, H.K., Fujiwara Y., Schlaeger T.M., Traver D., and Orkin S.H.. 2003. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 101:508–516. 10.1182/blood-2002-06-1699 [DOI] [PubMed] [Google Scholar]
- Mohtashami, M., Shah D.K., Kianizad K., Awong G., and Zúñiga-Pflücker J.C.. 2013. Induction of T-cell development by Delta-like 4-expressing fibroblasts. Int. Immunol. 25:601–611. 10.1093/intimm/dxt027 [DOI] [PubMed] [Google Scholar]
- Palis, J., Robertson S., Kennedy M., Wall C., and Keller G.. 1999. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 126:5073–5084. 10.1242/dev.126.22.5073 [DOI] [PubMed] [Google Scholar]
- Peschle, C., Migliaccio A.R., Migliaccio G., Petrini M., Calandrini M., Russo G., Mastroberardino G., Presta M., Gianni A.M., Comi P., et al. 1984. Embryonic----Fetal Hb switch in humans: studies on erythroid bursts generated by embryonic progenitors from yolk sac and liver. Proc. Natl. Acad. Sci. USA. 81:2416–2420. 10.1073/pnas.81.8.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschle, C., Mavilio F., Carè A., Migliaccio G., Migliaccio A.R., Salvo G., Samoggia P., Petti S., Guerriero R., Marinucci M., et al. 1985. Haemoglobin switching in human embryos: asynchrony of ζ----α and ε----γ-globin switches in primitive and definite erythropoietic lineage. Nature. 313:235–238. 10.1038/313235a0 [DOI] [PubMed] [Google Scholar]
- Qian, B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., and Pollard J.W.. 2011. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 475:222–225. 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter, M.W., and Stevens B.. 2017. Microglia emerge as central players in brain disease. Nat. Med. 23:1018–1027. 10.1038/nm.4397 [DOI] [PubMed] [Google Scholar]
- Satija, R., Farrell J.A., Gennert D., Schier A.F., and Regev A.. 2015. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33:495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, T.M., and Zúñiga-Pflücker J.C.. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. 10.1016/S1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- Schmitt, T.M., and Zúñiga-Pflücker J.C.. 2006. T-cell development, doing it in a dish. Immunol. Rev. 209:95–102. 10.1111/j.0105-2896.2006.00353.x [DOI] [PubMed] [Google Scholar]
- Schulz, C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Stefanska, M., Batta K., Patel R., Florkowska M., Kouskoff V., and Lacaud G.. 2017. Primitive erythrocytes are generated from hemogenic endothelial cells. Sci. Rep. 7:6401. 10.1038/s41598-017-06627-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon, C.M., Ditadi A., Awong G., Kennedy M., and Keller G.. 2014. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 32:554–561. 10.1038/nbt.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., and Jones J.M.. 1998. Embryonic stem cell lines derived from human blastocysts. Science. 282:1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Tieppo, P., Papadopoulou M., Gatti D., McGovern N., Chan J.K.Y., Gosselin F., Goetgeluk G., Weening K., Ma L., Dauby N., et al. 2020. The human fetal thymus generates invariant effector γδ T cells. J. Exp. Med. 217:e20190580. 10.1084/jem.20190580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tober, J., Koniski A., McGrath K.E., Vemishetti R., Emerson R., de Mesy-Bentley K.K., Waugh R., and Palis J.. 2007. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 109:1433–1441. 10.1182/blood-2006-06-031898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyser, R.C.V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., and Srinivas S.. 2021. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature. 600:285–289. 10.1038/s41586-021-04158-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, P.M., Chung S.W., Chui D.H., and Eaves C.J.. 1986. Properties of the earliest clonogenic hemopoietic precursors to appear in the developing murine yolk sac. Proc. Natl. Acad. Sci. USA. 83:3851–3854. 10.1073/pnas.83.11.3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, M., Montecino-Rodriguez E., Ferkowicz M.J., Porayette P., Shelley W.C., Conway S.J., Dorshkind K., and Yoder M.C.. 2011. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc. Natl. Acad. Sci. USA. 108:1468–1473. 10.1073/pnas.1015841108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, M., Porayette P., Glosson N.L., Conway S.J., Carlesso N., Cardoso A.A., Kaplan M.H., and Yoder M.C.. 2012. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 119:5706–5714. 10.1182/blood-2011-12-397489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambidis, E.T., Peault B., Park T.S., Bunz F., and Civin C.I.. 2005. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 106:860–870. 10.1182/blood-2004-11-4522 [DOI] [PMC free article] [PubMed] [Google Scholar]