Summary
Microbial rhodopsins are photoreceptive membrane proteins showing various light-dependent biological activities. Styrene-maleic acid (SMA) copolymers spontaneously form nanoscale lipid particles containing membrane proteins and associated lipids without detergent, and can be used to characterize membrane molecules. Here, we provide a protocol to functionally express a thermally stable rhodopsin, Rubrobacter xylanophilus rhodopsin, and an unstable rhodopsin, Halobacterium salinarum sensory rhodopsin I, in Escherichia coli. We then describe the preparation of SMA and the extraction and purification of rhodopsin molecules using SMA.
For complete details on the use and execution of this protocol, please refer to Ueta et al. (2020).
Subject areas: Cell Membrane, Protein Biochemistry, Protein expression and purification, Biotechnology and bioengineering
Graphical abstract

Highlights
-
•
Functional expression of microbial rhodopsins in Escherichia coli cells
-
•
Preparation of styrene-maleic acid (SMA) copolymer
-
•
Extraction and purification of microbial rhodopsins using SMA
-
•
Applicability of SMA for biophysical analysis of microbial rhodopsins in membrane
Microbial rhodopsins are photoreceptive membrane proteins showing various light-dependent biological activities. Styrene-maleic acid (SMA) copolymers spontaneously form nanoscale lipid particles containing membrane proteins and associated lipids without detergent, and can be used to characterize membrane molecules. Here, we provide a protocol to functionally express a thermally stable rhodopsin, Rubrobacter xylanophilus rhodopsin, and an unstable rhodopsin, Halobacterium salinarum sensory rhodopsin I, in Escherichia coli. We then describe the preparation of SMA and the extraction and purification of rhodopsin molecules using SMA.
Before you begin
Microbial rhodopsins are photoreceptive membrane proteins that consist of a 7-transmembrane α-helical domain and retinal as a chromophore (Ernst et al., 2014; Kojima et al., 2020a). Since the activities of microbial rhodopsins can be precisely controlled by visible light and monitored by spectroscopic measurements with high spatiotemporal resolution, rhodopsins have been studied as models both for membrane proteins and for photoreceptive proteins (Ernst et al., 2014; Kojima et al., 2020a). Styrene-maleic acid (SMA) copolymers are widely applied for various membrane proteins such as potassium channels, ATP-binding cassette (ABC) transporters, cytochrome bc1 and cytochrome b6f complexes (Dorr et al., 2014, 2016; Gulati et al., 2014; Swainsbury et al., 2018). The protocols described herein provide useful information for expressing and purifying various microbial rhodopsins using an Escherichia coli recombinant protein expression system and SMA copolymers.
Expression of histidine-tagged microbial rhodopsins (RxR and HsSRI) in E. coli cells
Timing: 3 days
-
1.Preparation of LB medium
-
a.Dissolve Bacto Tryptone, Bacto Yeast Extract and NaCl for Milli-Q water according to materials and equipment section.
-
b.Autoclave the medium at 120°C for 20 min under the pressure of 0.2 MPa.
-
c.Cool the medium to room temperature (ca. 20°C).
-
d.Add filter-sterilized ampicillin (final concentration = 50 μg/mL) to the medium.
-
a.
-
2.Expression of Rubrobacter xylanophilus rhodopsin (RxR) in E. coli cells
-
a.Add the expression plasmid of RxR inserted into the pET21a plasmid vector (100 ng) to a 100 μL of E. coli competent cells.
-
b.Incubate the cells on ice for 30 min.
-
c.Incubate the cells in a heat block incubator at 42°C for 1 min.
-
d.Sow the cells on the LB/Agar plate by using a spreader.
-
e.Incubate the cells in an incubator at 37°C for 12–15 h.
-
f.Transfer more than 10 fresh colonies of E. coli cells (BL21 (DE3)) harboring the expression plasmid of RxR to 50 mL LB medium in a 200 mL Erlenmeyer flask.
-
g.Grow the cells in a shaking incubator (180 rpm) at 37°C for 12–15 h.
-
h.Transfer the growth medium to 1.0 L LB medium in a 3 L Erlenmeyer flask.
-
i.Grow the cells in a shaking incubator (120 rpm) at 37°C until the optical density at 660 nm reaches 1.4–1.6.Note: In general, it takes 2.5–3.0 h to reach this OD.
-
j.Add isopropyl β-D-1-thiogalactopyranoside (IPTG, final concentration = 1 mM) and all-trans retinal (final concentration = 10 μM).
-
k.Grow the cells in a shaking incubator (120 rpm) at 37°C for 3 h to induce protein production.
-
l.Transfer the growth medium to the 50 mL centrifuge bottles (IWAKI, Japan).
-
m.Centrifuge the bottles (5,535×g) for 10 min at 4°C.
-
n.Dispose the supernatant and keep the pellet. Troubleshooting 1
-
o.Store the cells at −20°C until use.Note: The cells are recommended to be used within a year.
-
a.
-
3.Expression of Halobacterium salinarum sensory rhodopsin I (HsSRI) in E. coli cells
-
a.Add the expression plasmid of HsSRI inserted into the pET21c plasmid vector (100 ng) to a 100 μL of E. coli competent cells.
-
b.Incubate the cells on ice for 30 min.
-
c.Incubate the cells in a heat block incubator at 42°C for 1 min.
-
d.The cells were plated on the LB/Agar plate and incubate the plate in an incubator at 37°C for 12–15 h.
-
e.Transfer more than 10 fresh colonies of E. coli cells (BL21 (DE3)) harboring the expression plasmid of HsSRI to 50 mL LB medium in a 200 mL Erlenmeyer flask.
-
f.Grow the cells in a shaking incubator (180 rpm) at 37°C for 12–15 h.
-
g.Transfer the growth medium to 1.9 L LB medium in a 2 L round-bottom flask.Note: Preincubate the 1.9 L LB medium at 30°C for 1 h in an incubator before this step.
-
h.Grow the cells in a shaking incubator (160 rpm) at 30°C until the optical density at 660 nm reaches 0.3–0.4.Note: In general, it takes 2.5–3.0 h to reach this OD.
-
i.Cool the round-bottom flask on ice for 15 min.
-
j.Add IPTG (final concentration = 1 mM) and all-trans retinal (final concentration = 10 μM).
-
k.Grow the cells in a shaking incubator (160 rpm) at 18°C for 12–14 h to induce protein production.
-
l.Transfer the growth medium to the centrifuge bottles.
-
m.Centrifuge the bottles (5,535×g) for 10 min at 4°C.
-
n.Dispose the supernatant and keep the pellet. Troubleshooting 1
-
o.Store the cells at −20°C until use.Note: The cells are recommended to be used within a year.
-
a.
Preparation of SMA
Timing: 4 days
-
4.
Preparation of SMA
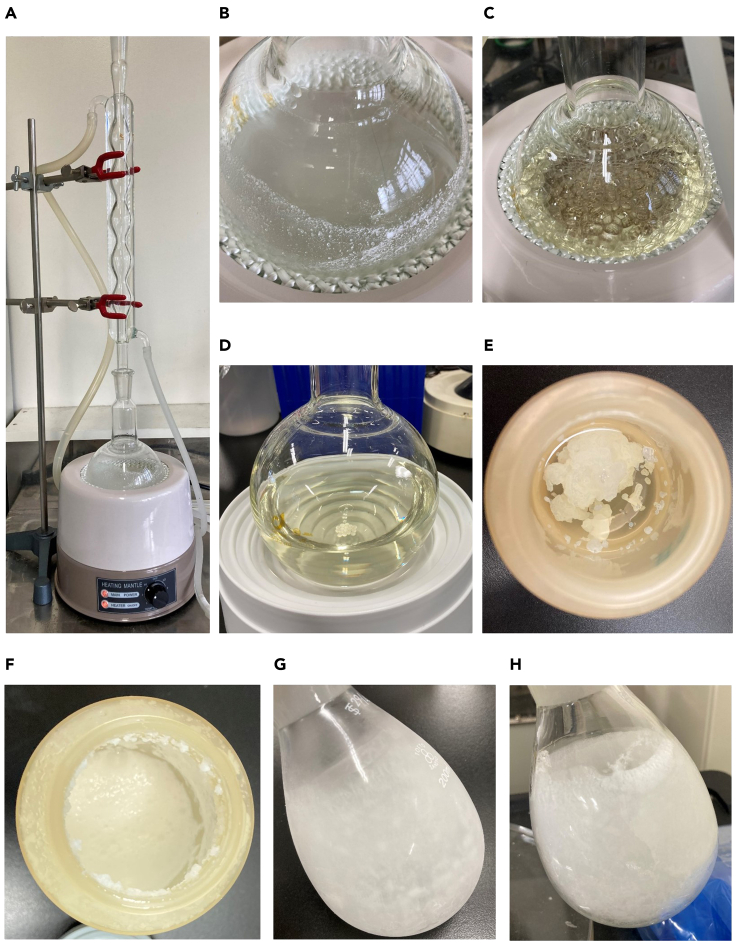
Hydrolyze styrene-maleic anhydride (SMAnh) copolymers (SMA® 2000, in which the ratio of styrene and maleic anhydride is ca. 2:1, Cray Valley Co., USA) into the water-soluble amphiphilic membrane-active acid derivative SMA according to a previous study (Lee et al., 2016). As an alternative to SMA® 2000, use XIRAN 2000 in which the ratio of styrene and maleic anhydride is ca. 2:1 (Polyscope Polymers).-
a.Add 25 g SMAnh copolymer to 250 mL 1 M NaOH and 0.5 g anti-bumping granules in a 500 mL round-bottom flask and mix until the copolymer is completely resuspended.
-
b.Rest the flask on a heating mantle and set up a reflux apparatus with a water supply (Figures 1A and 1B).
-
c.Heat and reflux the solution for 2 h (Figure 1C).
-
d.Cool the solution to room temperature (ca. 20°C) (Figure 1D).Note: Ensure no solid SMAnh remained.
-
e.Divide the solution into two equal aliquots in 250 mL polypropylene centrifuge bottles.Note: Do not transfer more than 150 ml of polymer since it is needed to add ca. 100 mL of Milli-Q water in step g.
- f.
-
g.Add Milli-Q water to the precipitated polymer and fill the centrifuge bottles to the maximum permitted volume (ca. 250 mL).
-
h.Centrifuge the bottles (11,000×g) for 15 min at 25°C.
-
i.Pour off the remaining supernatant (Figure 1F).
 CRITICAL: Do not disturb the precipitate. Gently discard the supernatant using plastic pipettes.
CRITICAL: Do not disturb the precipitate. Gently discard the supernatant using plastic pipettes. -
j.Add Milli-Q water to the precipitated polymer at the maximum permitted volume (ca. 250 mL).
-
k.Mix the bottles well by vigorous shaking up and down for several minutes to completely resuspend the precipitate.
-
l.Centrifuge the bottles (11,000×g) for 15 min at 25°C and pour off the remaining supernatant
 CRITICAL: Do not disturb the precipitates. Gently discard the supernatant using plastic pipettes.
CRITICAL: Do not disturb the precipitates. Gently discard the supernatant using plastic pipettes. -
m.Repeat steps j – l two more times.
-
n.Solubilize the polymer in 125 mL 0.6 M NaOH by adding a few drops of NaOH at a time. Stir the suspension with a magnetic stirrer until the pellet has completely dissolved.
-
o.Check the pH using a pH meter.
-
p.Add HCl or NaOH to adjust the pH to 8.0.
-
q.Transfer the solution into a 1 L round-bottom flask.
-
r.Freeze the polymer for more than 18 h at −20°C (Figure 1G).
-
s.Cover the flask with a poly-net protective cover after the step r has been completed.
-
t.Place the flask in a freeze dryer and freeze-dry the polymer (Figure 1H).
-
u.Store the polymer in a sealed vessel at room temperature until use.Note: The dried polymer can be stored for up to 12 months without degradation.
-
a.
Figure 1.
Preparation of SMA copolymer
(A and B) SMAnh copolymer is added to 250 mL 1 M NaOH and 0.5 g anti-bumping granules in a round-bottom flask. The flask is put on a heating mantle and attached to a reflux apparatus with a water supply.
(C) The reaction mixture is heated for 2 h.
(D) The reaction mixture is cooled down after the heat treatment.
(E) The polymer is precipitated by adding HCl.
(F) The precipitated polymer after centrifugation.
(G) The polymer frozen for 18 h at −20°C.
(H) Freeze-dried polymer.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli BL21 (DE3) | Invitrogen | #C600003 |
| Chemicals, peptides, and recombinant proteins | ||
| Bacto Tryptone | Becton, Dickinson and Company | #211705 |
| Bacto Yeast Extract | Becton, Dickinson and Company | #212750 |
| All-trans retinal | Sigma-Aldrich | #R2500 |
| SMA® 2000 | Cray Valley Co. | SMA® 2000 |
| Ampicillin sodium | Fujifilm Wako Pure Chemical Co. | #014-23302 |
| Agar | Nacalai Tesque | #01028-85 |
| Isopropyl β-D-1-thiogalactopyranoside | Fujifilm Wako Pure Chemical Co. | #097-05014 |
| Tris(hydroxymethyl)aminomethane | Fujifilm Wako Pure Chemical Co. | #011-16381 |
| Glycerol | Fujifilm Wako Pure Chemical Co. | #075-00611 |
| Imidazole | Fujifilm Wako Pure Chemical Co. | #095-00015 |
| Recombinant DNA | ||
| Expression plasmid of RxR | (Kojima et al., 2020b) | n/a |
| Expression plasmid of HsSRI | (Kitajima-Ihara et al., 2008) | n/a |
| pET21a(+) vector | Novagen | #69740-3 |
| pET21c(+) vector | Novagen | #69742-3 |
| Others | ||
| 50 mL centrifuge bottle | IWAKI | #2344-050 |
| Anti-bumping granules | Fujifilm Wako Pure Chemical Co. | #021-07195 |
| 500 mL round-bottom flask | Climbing Co.,Ltd. | #CL0070-16-10 |
| Ultrasonic disruptor | TOMMY SEIKO Co., Ltd. | #UD-2119 |
| HisTrap FF column | Cytiva | #17-5319-01 |
| Chromatography system | GE Healthcare | ÄKTA prime plus, ÄKTA purifier 10 |
| UV-vis spectrophotometer | Shimadzu | UV-2450 |
| Amicon Ultra filter (30,000 Mw cut-off) | Merck Millipore | #UFC903024 |
| Dialysis membrane Size 8 (MWCO: 14,000) | Fujifilm Wako Pure Chemical Co. | #046-30911 |
Alternatives: Use XIRAN 2000 in which the ratio of styrene and maleic anhydride is ca. 2:1 (Polyscope Polymers, #9011-13-6) as an alternative to SMA® 2000.
Materials and equipment
| LB medium | Final concentration | Amount |
|---|---|---|
| Bacto Tryptone | 1 % (w/v) | 10 g |
| Bacto Yeast Extract | 0.5 % (w/v) | 5 g |
| NaCl | 1 % (w/v) | 10 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
Sterilize by autoclaving at 120°C for 20 min.
Add filter-sterilized ampicillin (final concentration = 50 μg/mL) before use.
The medium can be stored at room temperature for up to two days.
| LB/agar plate | Final concentration | Amount |
|---|---|---|
| Bacto Tryptone | 1 % (w/v) | 10 g |
| Bacto Yeast Extract | 0.5 % (w/v) | 5 g |
| NaCl | 1 % (w/v) | 10 g |
| Agar | 1.5 % (w/v) | 15 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
Sterilize by autoclaving at 120°C for 20 min.
After adding filter-sterilized ampicillin (final concentration = 50 μg/mL), pour the medium to polystyrene dishes (diameter 10 cm). Use after the agar hardened.
The plate can be stored at 4°C for up to one month.
| Retinal stock solution | Final concentration | Amount |
|---|---|---|
| All-trans retinal | 10 mM | 284 mg |
| Ethanol | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
The medium can be stored at −20 or −80°C for six months in brown bottles.
| IPTG stock solution | Final concentration | Amount |
|---|---|---|
| Isopropyl β-D-1-thiogalactopyranoside | 1 M | 2.38 g |
| Milli-Q water | n/a | Up to 10 mL |
| Total | n/a | 10 mL |
Sterilize the solution using a membrane filter (0.22 μm pore size).
The medium can be stored at −20°C for six months.
| Buffer A | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| NaCl | 1 M | 58.44 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer B | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| NaCl | 4 M | 233.76 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer AN | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| Glycerol | 10 % (v/v) | 100 mL |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer BN | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.5) | 50 mM | 50 mL |
| Glycerol | 10 % (v/v) | 100 mL |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer AW | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| Glycerol | 10 % (v/v) | 100 mL |
| Imidazole | 20 mM | 1.36 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer BW | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| NaCl | 500 mM | 29.22 g |
| Glycerol | 10 % (v/v) | 100 mL |
| Imidazole | 20 mM | 1.36 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer AE | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| Glycerol | 10 % (v/v) | 100 mL |
| Imidazole | 1 M | 68.08 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
| Buffer BE | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl (pH 8.0) | 50 mM | 50 mL |
| NaCl | 500 mM | 29.22 g |
| Glycerol | 10 % (v/v) | 100 mL |
| Imidazole | 1 M | 68.08 g |
| Milli-Q water | n/a | Up to 1000 mL |
| Total | n/a | 1000 mL |
The medium can be stored at 4°C for up to six months.
Step-by-step method details
Extraction of rhodopsin molecules with SMA
Timing: 4–7 h
-
1.Preparation of the E. coli membrane fraction
-
a.Suspend the crude E. coli membranes obtained from 1 L culture medium (Figure 2A) in 50–100 mL Buffer A for RxR or Buffer B for HsSRI using plastic pipettes.Note: When you use the frozen membranes, place it at room temperature for 1–2 h until the pellet is thawed.
-
b.Disrupt the cells in a 100 mL beaker using an ultrasonic disruptor (60 pulses/min for 30 min, Duty: 50, Output control: 7, UD-2119, TOMMY SEIKO Co., Ltd., Japan) (Figure 2B). Troubleshooting 2
 CRITICAL: Cool the samples in ice-cold water during the ultrasonication.
CRITICAL: Cool the samples in ice-cold water during the ultrasonication. -
c.Transfer the suspension to 30 mL centrifuge tubes.
-
d.Centrifuge the tubes (5,000×g) for 10 min at 4°C and collect the supernatants into clean30 mL centrifuge tubes.
-
e.Centrifuge the tubes (40,000×g) for 60 min at 4°C and pour off the remaining supernatants.
-
f.Collect the precipitates as the membrane fraction (Figure 2C).
-
a.
-
2.Extraction of rhodopsin molecules with SMA
-
a.Suspend the collected membranes with ca. 10 mL Buffer AN for RxR or Buffer BN for HsSRI using plastic pipettes.
-
b.Homogenize the suspension using a glass-Teflon Dounce homogenizer (Labo-Stirrer LR 41A, Yamato Co., Japan).
-
c.Transfer the suspension to a 10 mL beaker.
-
d.Add SMA (final concentration = 5 % w/v) to the suspension.
 CRITICAL: Add the polymer to a buffer above pH 7.4 containing a low salt concentration (below 500 mM) for the solubilization because the polymer forms aggregates under low pH and high salt conditions.Note: If you need to change the lipid composition of SMA lipid particles, add the exogenous lipids in addition to SMA.
CRITICAL: Add the polymer to a buffer above pH 7.4 containing a low salt concentration (below 500 mM) for the solubilization because the polymer forms aggregates under low pH and high salt conditions.Note: If you need to change the lipid composition of SMA lipid particles, add the exogenous lipids in addition to SMA. -
e.Stir the suspension (RxR: for 2 h at room temperature (23°C–28°C), HsSRI for 30 min at 4°C) with a magnetic stirrer (Figure 2D).Note: Since RxR and HsSRI are not significantly denatured by room light, it is not needed to perform this step in the dark. However, when you solubilize other unstable rhodopsins, we recommend to perform the steps after the solubilization in the dark.
-
f.Transfer the suspension to 30 mL centrifuge tubes.Centrifuge the tubes (103,900×g) for 30 min at 4°C and collect the supernatants as the solubilized fraction (Figure 2E). Troubleshooting 3
 CRITICAL: Perform the extraction steps of HsSRI at 4°C or on ice to prevent protein denaturation.
CRITICAL: Perform the extraction steps of HsSRI at 4°C or on ice to prevent protein denaturation.
-
a.
Figure 2.
Preparation of E. coli membrane fraction and solubilization of rhodopsins with SMA
(A) E. coli cells expressing RxR.
(B) Disruption of E. coli cells by ultrasonication. The beaker containing the sample is cooled in ice-cold water.
(C) Membrane fraction of E. coli cells expressing RxR.
(D) Membrane fraction of RxR is solubilized with SMA. The suspension is stirred using a magnetic stirrer.
(E) SMA-solubilized fraction of RxR collected after centrifugation.
(F) HisTrap FF column after the SMA-solubilized fraction containing RxR is loaded on the column.
Purification of rhodopsin molecules in SMA lipid particles
Timing: 2 days
-
3.Purification of rhodopsin molecules by affinity column chromatography
-
a.Wash a HisTrap FF column (5 mL, Cytiva) with 5 column volumes (CV) of Milli-Q water.Note: The flow rate was set at 1–3 mL/min in steps a – e.
-
b.Wash the column with 3 CV Buffer AN for RxR or Buffer BN for HsSRI.
-
c.Apply the lysate containing rhodopsins to the column (Figure 2F). Troubleshooting 4
-
d.Wash the column with 3 CV Buffer AW for RxR or Buffer BW for HsSRI.
-
e.Elute the column with a linear gradient of imidazole in Buffer AE for RxR or in Buffer BE for HsSRI using a chromatography system (ÄKTA prime plus, ÄKTA purifier 10, GE Healthcare).Note: The detector of the chromatography system is set to monitor the absorption at 541 nm for RxR and 535 nm for HsSRI.
-
f.Collect the red-colored fractions corresponding to each absorbance peak containing rhodopsins.
 CRITICAL: Perform the column chromatography steps of HsSRI at 4°C to prevent protein denaturation.
CRITICAL: Perform the column chromatography steps of HsSRI at 4°C to prevent protein denaturation. -
g.Measure absorption spectra of the collected samples using a UV-vis spectrophotometer (UV-2450, Shimadzu, Japan) to confirm the purity and amounts of rhodopsins.
-
a.
-
4.Remove imidazole from purified RxR samples
-
a.Buffer exchange and concentrate the samples with an Amicon Ultra filter (30,000 Mw cut-off; Millipore). Troubleshooting 5
-
b.Add more than a 100-fold volume of Buffer AN and gently mix the solution using a pipet.
-
c.Repeat steps a – b two more times.
 CRITICAL: Use a buffer above pH 7.4 for the purification because the polymer forms aggregates under low pH conditions, especially during the steps of concentrating the samples.
CRITICAL: Use a buffer above pH 7.4 for the purification because the polymer forms aggregates under low pH conditions, especially during the steps of concentrating the samples. -
d.Measure absorption spectra of the sample using a UV-vis spectrophotometer (UV-2450, Shimadzu, Japan) to estimate the concentration by using the absorbance at 541 nm with molar extinction coefficient of RxR (54,000 M−1 cm−1) (Kanehara et al., 2017).
-
a.
-
5.Remove imidazole from purified HsSRI samples
-
a.Place the samples into a dialysis membrane (Wako, Dialysis membrane Size 8, MWCO: 14,000).
-
b.Immerse the dialysis membrane in more than a 100-fold volume of Buffer BN for 4 h at 4°C.
-
c.Exchange Buffer BN to fresh Buffer BN and incubate for 4 h at 4°C.
-
d.Repeat step c two more times.
-
e.Concentrate the samples with an Amicon Ultra filter (30,000 Mw cut-off; Millipore) at 4°C. Troubleshooting 5
 CRITICAL: Use a buffer above pH 7.4 for the purification because the polymer forms aggregates under low pH conditions, especially during the steps of concentrating the samples.
CRITICAL: Use a buffer above pH 7.4 for the purification because the polymer forms aggregates under low pH conditions, especially during the steps of concentrating the samples. -
f.Measure absorption spectra of the sample using a UV-vis spectrophotometer (UV-2450, Shimadzu, Japan) to estimate the concentration by using the absorbance at 535 nm with molar extinction coefficient of HsSRI (63,000 M−1 cm−1) (Bogomolni and Spudich, 1982).
-
a.
Expected outcomes
Upon completion of this protocol, red-colored purified rhodopsins were obtained (Figure 3A). The yield of purified RxR was ca. 5 mg per 1 L culture medium. The sizes of SMA lipid particles containing RxR were estimated as ca. 54 nm by dynamic light scattering (DLS) measurements (Figure 3B). Absorption spectra of RxR and HsSRI samples are shown in Figure 3C. The absorbance observed in the visible region (around 540 nm for RxR and 535 nm for HsSRI) reflects the photoactive holoproteins in SMA lipid particles. The obtained samples are used for biophysical analysis of rhodopsins in a membrane environment with less light scattering (Ueta et al., 2020).
Figure 3.
Purified rhodopsin samples with SMA
(A) Photograph of purified RxR sample with SMA.
(B) Dynamic light scattering (DLS) patterns of purified RxR sample with SMA.
(C) Absorption spectra of purified RxR and HsSRI samples with SMA (left and right panels, respectively). Please see our previous study Ueta et al. (2020) for detailed methods of DLS and spectroscopic measurements.
Limitations
SMA polymers are more likely to aggregate under acidic or high salt conditions. Therefore, SMA-solubilized samples should be prepared and stored in alkaline (> 7.4) and low salt (< 500 mM NaCl) conditions to prevent aggregation. 10 % v/v glycerol should be added to the SMA-solubilized samples to prevent aggregation. SMA-solubilized samples are useful for biophysical analysis of membrane proteins including rhodopsins in alkaline and low salt conditions, but not in acidic or high salt conditions.
In addition, the protocol described here can be used to incorporate rhodopsin molecules in natural E. coli membrane lipids without adding exogenous lipids. If you would like to change the lipid composition of SMA lipid particles, please add the exogeneous lipids during the step of the addition of SMA (step 2d in Extraction of rhodopsin molecules with SMA) according to previous studies (Dorr et al., 2014; Knowles et al., 2009; Swainsbury et al., 2014).
Troubleshooting
Problem 1
The color of the collected E. coli cells is not reddish (steps 2f and 3e).
Potential solution
If you use colonies that have been stored at 4°C before the inoculation, the expression level may be low. Use colonies just after the incubation on the plate at 37°C (steps 2e and 3d).
Problem 2
The color of the E. coli membranes expressing rhodopsins is changed into yellow during the disruption process (step 1b).
Potential solution
When rhodopsins are denatured, the color of the E. coli membranes is changed into yellow due to the release of the retinal from the apoprotein. Because the ultrasonication generates heat, check whether the temperature of the samples is below 10°C to avoid thermal denaturation of rhodopsins. As an alternative, a freeze-thaw method is applicable.
Problem 3
Solubilization efficiency of rhodopsins from the E. coli membranes with SMA is low (step 2f).
Potential solution
To increase the solubilization efficiency, the E. coli membranes should be suspended well using an ultrasonic disruptor (step 1b) and a glass-Teflon Dounce homogenizer (step 2b). Repeat the above step until the suspension becomes less viscous and homogenous.
Problem 4
Waste samples, which are passed through a HisTrap FF column, show rhodopsin’s color (step 3c).
Potential solution
The situation indicates that the flow-through fraction contains unbound rhodopsins. For some rhodopsins including RxR, the binding affinity to the resin is low. Repeatedly apply the flow-through fraction to the column until rhodopsins are completely bound to the column.
Problem 5
Purified rhodopsin samples form aggregates during the concentration step with an Amicon Ultra filter (steps 4a and 5e).
Potential solution
Purified rhodopsin samples are likely to form aggregates at high protein concentrations (e.g., 1 mg/mL). The protein concentration should be kept at low during the process.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yuki Sudo (sudo@okayama-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank Mr. Tetsuya Ueta, Dr. Tomoya Hino, Dr. Shingo Nagano and Dr. Mikihiro Shibata for sample preparation and invaluable discussion. This work was financially supported by JSPS KAKENHI grant numbers JP21K15054 to K.K. and JP20K21482, JP21H02446, and JP21H00404 to Y.S. This research was partially supported by CREST-JST (JPMJCR1656) to Y.S.
Author contributions
K.K. and Y.S. performed the research and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This study did not generate any unique datasets or code.
References
- Bogomolni R.A., Spudich J.L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc. Natl. Acad. Sci. U S A. 1982;79:6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr J.M., Koorengevel M.C., Schafer M., Prokofyev A.V., Scheidelaar S., van der Cruijsen E.A., Dafforn T.R., Baldus M., Killian J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. U S A. 2014;111:18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr J.M., Scheidelaar S., Koorengevel M.C., Dominguez J.J., Schafer M., van Walree C.A., Killian J.A. The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 2016;45:3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst O.P., Lodowski D.T., Elstner M., Hegemann P., Brown L.S., Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S., Jamshad M., Knowles T.J., Morrison K.A., Downing R., Cant N., Collins R., Koenderink J.B., Ford R.C., Overduin M., et al. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J. 2014;461:269–278. doi: 10.1042/BJ20131477. [DOI] [PubMed] [Google Scholar]
- Kanehara K., Yoshizawa S., Tsukamoto T., Sudo Y. A phylogenetically distinctive and extremely heat stable light-driven proton pump from the eubacterium Rubrobacter xylanophilus DSM 9941T. Sci. Rep. 2017;7:44427. doi: 10.1038/srep44427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima-Ihara T., Furutani Y., Suzuki D., Ihara K., Kandori H., Homma M., Sudo Y. Salinibacter sensory rhodopsin - sensory rhodopsin I-like protein from a eubacterium. J. Biol. Chem. 2008;283:23533–23541. doi: 10.1074/jbc.M802990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T.J., Finka R., Smith C., Lin Y.P., Dafforn T., Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- Kojima K., Shibukawa A., Sudo Y. The unlimited potential of microbial rhodopsins as optical tools. Biochemistry. 2020;59:218–229. doi: 10.1021/acs.biochem.9b00768. [DOI] [PubMed] [Google Scholar]
- Kojima K., Ueta T., Noji T., Saito K., Kanehara K., Yoshizawa S., Ishikita H., Sudo Y. Vectorial proton transport mechanism of RxR, a phylogenetically distinct and thermally stable microbial rhodopsin. Sci. Rep. 2020;10:282. doi: 10.1038/s41598-019-57122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Knowles T.J., Postis V.L., Jamshad M., Parslow R.A., Lin Y.P., Goldman A., Sridhar P., Overduin M., Muench S.P., et al. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- Swainsbury D.J., Scheidelaar S., van Grondelle R., Killian J.A., Jones M.R. Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew. Chem. Int. Ed. Engl. 2014;53:11803–11807. doi: 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainsbury D.J.K., Proctor M.S., Hitchcock A., Cartron M.L., Qian P., Martin E.C., Jackson P.J., Madsen J., Armes S.P., Hunter C.N. Probing the local lipid environment of the Rhodobacter sphaeroides cytochrome bc1 and Synechocystis sp. PCC 6803 cytochrome b6f complexes with styrene maleic acid. Biochim. Biophys. Acta Bioenerg. 2018;1859:215–225. doi: 10.1016/j.bbabio.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta T., Kojima K., Hino T., Shibata M., Nagano S., Sudo Y. Applicability of styrene-maleic acid copolymer for two microbial rhodopsins, RxR and HsSRI. Biophys. J. 2020;119:1760–1770. doi: 10.1016/j.bpj.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.