Abstract
Sporothrix (Ophiostomatales) comprises species that are pathogenic to humans and other mammals as well as environmental fungi. Developments in molecular phylogeny have changed our perceptions about the epidemiology, host-association, and virulence of Sporothrix. The classical agent of sporotrichosis, Sporothrix schenckii, now comprises several species nested in a clinical clade with S. brasiliensis, S. globosa, and S. luriei. To gain a more precise view of outbreaks dynamics, structure, and origin of genetic variation within and among populations of Sporothrix, we applied three sets of discriminatory AFLP markers (#3 EcoRI-GA/MseI-TT, #5 EcoRI-GA/MseI-AG, and #6 EcoRI-TA/MseI-AA) and mating-type analysis to a large collection of human, animal and environmental isolates spanning the major endemic areas. A total of 451 polymorphic loci were amplified in vitro from 188 samples, and revealed high polymorphism information content (PIC = 0.1765–0.2253), marker index (MI = 0.0001–0.0002), effective multiplex ratio (E = 15.1720–23.5591), resolving power (Rp = 26.1075–40.2795), discriminating power (D = 0.9766–0.9879), expected heterozygosity (H = 0.1957–0.2588), and mean heterozygosity (Havp = 0.000007–0.000009), demonstrating the effectiveness of AFLP markers to speciate Sporothrix. Analysis using the program structure indicated three genetic clusters matching S. brasiliensis (population 1), S. schenckii (population 2), and S. globosa (population 3), with the presence of patterns of admixture amongst all populations. AMOVA revealed highly structured clusters (PhiPT = 0.458–0.484, P < 0.0001), with roughly equivalent genetic variability within (46–48 %) and between (52–54 %) populations. Heterothallism was the exclusive mating strategy, and the distributions of MAT1-1 or MAT1-2 idiomorphs were not significantly skewed (1:1 ratio) for S. schenckii (χ2 = 2.522; P = 0.1122), supporting random mating. In contrast, skewed distributions were found for S. globosa (χ2 = 9.529; P = 0.0020) with a predominance of MAT1-1 isolates, and regional differences were highlighted for S. brasiliensis with the overwhelming occurrence of MAT1-2 in Rio de Janeiro (χ2 = 14.222; P = 0.0002) and Pernambuco (χ2 = 7.364; P = 0.0067), in comparison to a higher prevalence of MAT1-1 in the Rio Grande do Sul (χ2 = 7.364; P = 0.0067). Epidemiological trends reveal the geographic expansion of cat-transmitted sporotrichosis due to S. brasiliensis via founder effect. These data support Rio de Janeiro as the centre of origin that has led to the spread of this disease to other regions in Brazil. Our ability to reconstruct the source, spread, and evolution of the ongoing outbreaks from molecular data provides high-quality information for decision-making aimed at mitigating the progression of the disease. Other uses include surveillance, rapid diagnosis, case connectivity, and guiding access to appropriate antifungal treatment.
Key words: AFLP, AMOVA, Linkage disequilibrium, Mating-type, Ophiostomatales, Sporotrichosis, Zoonosis
Introduction
Sporothrix (Ascomycota) is embedded in the plant-associated order, Ophiostomatales, and comprises at least 53 reported species (Rodrigues et al. 2020b). This largely saprotrophic genus is frequently associated with decaying wood, insects, and soil; however, several members have emerged in recent years with the ability to cause infections in mammalian hosts. Sporothrix develops a filamentous form in the environment (25–30 °C) however undergoes a dimorphic switch to a yeast phase at elevated temperatures (35–37 °C). Infections follow two main routes of acquisition, one of which involves animal (zoonotic) transmission (e.g., cat-to-cat and cat-to-human), and the other involves exposure to infected decaying plant material (i.e., classic sapronosis). The resulting disease is called sporotrichosis, a subacute or chronic infection of the skin, subcutaneous tissues, and adjacent lymphatics, which usually suppurate, ulcerate, and drain (Rippon 1988, Orofino-Costa et al. 2017, Queiroz-Telles et al. 2017).
In humans, cutaneous and subcutaneous lesions develop at the inoculation site, and fungal spreading typically occurs through the lymphatics in the course of the first 2–3 wk (Rodrigues et al. 2017). As a polymorphic disease, clinical presentation varies from fixed localized cutaneous lesions, lymphocutaneous lesions to severe, disseminated sporotrichosis (Al-Tawfiq & Wools 1998, Silva-Vergara et al. 2012). In cats, Sporothrix meets a highly susceptible host population, and the most common clinical symptoms include multiple skin nodules and ulcers, often related to nasal mucosa lesions and respiratory signs (Schubach et al. 2004, Gremião et al. 2015, Seyedmousavi et al. 2018). Feline sporotrichosis can progress to severe forms that are difficult to treat and may lead to the death of infected animals (Schubach et al. 2003, Pereira et al. 2010, Rodrigues et al. 2018).
Benjamin R. Schenck first described human sporotrichosis in 1898, and the etiologic agent was two years later assigned to the genus Sporothrix by Hektoen and Perkins (Hektoen & Perkins 1900). Several years lapsed before animal sporotrichosis was described in naturally infected rats in Brazil (Lutz & Splendore 1907). Thus, for over a century, human and animal sporotrichosis was attributed to a sole etiologic agent, the classical Sporothrix schenckii. However, the application of molecular tools has led to the description of four cryptic species recognized in clinical practice (Marimon et al. 2006, Marimon et al. 2007). The classical species, S. schenckii, now comprises S. brasiliensis, S. schenckii s. str., S. globosa, and S. luriei (Marimon et al. 2007, Rodrigues et al. 2013b, Zhou et al. 2014). Outside this clinical clade, pathogenicity to mammals is rarely observed, and only a few reports have appeared in the literature of infections by members of the S. pallida and S. stenoceras complexes (de Beer et al. 2016, Makri et al. 2020) or by relatives in the genus Ophiostoma, e.g., O. piceae (Bommer et al. 2009).
Sporotrichosis has a worldwide occurrence, but endemic areas are located in tropical and subtropical regions (Pappas et al. 2000) with high occurrences reported in South Africa, India, Australia, China, Japan, the USA, and Mexico (Chakrabarti et al. 2015). In South America, endemic areas include Argentina, Brazil, Colombia, Peru, Uruguay, and Venezuela (Rodrigues et al. 2020b). In Brazil, the South/Southeast axis has been the epicentre of zoonotic sporotrichosis, and cats are the main vectors of disease transmission to humans and other animals (Rodrigues et al. 2013b; Gremião et al. 2017). Thousands of Sporothrix infections persist for many months in infected cats, leading to sustained transmission of sporotrichosis by cat-to-cat and cat-to-human contact patterns (Rodrigues et al. 2016b; Macêdo-Sales et al. 2018). The predominant etiologic agent in cats is S. brasiliensis, which is known to be the most virulent Sporothrix species (Arrillaga-Moncrieff et al. 2009).
Until recently, the occurrence of S. brasiliensis was geographically restricted to Brazil's South and Southeast regions (Rodrigues et al. 2013a; Maschio-Lima et al. 2021). However, a geographic expansion of sporotrichosis driven by S. brasiliensis has occurred in Brazil and neighbouring countries such as Argentina and Paraguay (García Duarte et al. 2017, Córdoba et al. 2018, Etchecopaz et al. 2019, Aldama et al. 2020). In addition, suspected cases have been reported in Bolivia, Colombia, and Panama (Rios et al. 2018, PAHO 2019).
Here we sought to identify aspects that underpin the emergence of sporotrichosis in South America. To gain insights into the trends in the epidemiology and genetic diversity of clinical isolates, we explored Brazilian isolates of Sporothrix collected over 70 years to determine the genetic diversity, population structure, and recognize different genotypes associated with Sporothrix species. In addition, a well-characterized collection of isolates from Argentina, Austria, Brazil, Chile, Italy, Japan, Mexico, Peru, Spain, South Africa, the USA, Uruguay, and Venezuela was used to evaluate our hypotheses in a wider international context.
Material and methods
Fungal isolates and DNA extraction
We included 188 Sporothrix isolates previously identified by species-specific PCR and phylogenetic analysis of the calmodulin, ITS1/2+5.8S, and β-tubulin loci as S. brasiliensis (n = 72), S. schenckii (n = 67), S. globosa (n = 34), S. luriei (n = 1), S. mexicana (n = 4), S. pallida (n = 3), S. chilensis (n = 2), S. brunneoviolacea (n = 2), S. dimorphospora (n = 2), and S. stenoceras (n = 1), and representing both clinical and environmental clades (Marimon et al. 2008, Arrillaga-Moncrieff et al. 2009, Madrid et al. 2010, Rodrigues et al. 2014, Zhou et al. 2014, Rodrigues et al. 2015, Zhang et al. 2015, Moussa et al. 2017). These isolates were recovered from Argentina, Austria, Brazil, Chile, Italy, Japan, Mexico, Peru, Spain, South Africa, the USA, Uruguay, and Venezuela (Supplementary Table S1) and are deposited in the Laboratory of Emerging Fungal Pathogens culture collection at the Federal University of São Paulo (UNIFESP), São Paulo, Brazil.
Filamentous colonies were grown on Sabouraud Dextrose Agar (SDA) at 25 °C and co-cultured every fourteen days (Brilhante et al. 2015). DNA extraction was performed from a 14-d-old monosporic culture using the FastDNA kit (MP Biomedicals, Solon, OH, USA) as previously described (Rodrigues et al. 2014). The genomic DNA concentration and purity were analysed by spectrophotometry (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA, USA). We considered a good quality DNA extraction when the OD 260/280 ratio was between 1.8–2.0, and an amplicon was detected by PCR using the primers ITS1 and ITS4 (White et al. 1990), indicating that the sample was free of PCR inhibitors (Table 1).
Table 1.
Primers used in this study for genetic characterization of Sporothrix species.
| Target | Primer | Sequence (5’→3’) | Reference |
|---|---|---|---|
| ITS1/2+5.8S | ITS1 | TCCGTAGGTGAACCTTGCGG | White et al. (1990) |
| ITS4 | TCCTCCGCTTATTGATATGC | White et al. (1990) | |
| MAT1-1 | SPMAT1-1F | GATCCCTACAAAAGCAAATGGACCATG | de Carvalho et al. (2021) |
| SPMAT1-1R | CTGCAATTGGGTTGTGCCTGATG | de Carvalho et al. (2021) | |
| MAT1-2 | SPMAT1-2F | CCAATTTCCTCTTCCACTATTCGTCGC | de Carvalho et al. (2021) |
| SPMAT1-2R | GCTTGATATCCACGGCCATCTTG | de Carvalho et al. (2021) | |
| ATP9-COX2 | 975–8038F | GCTAGAAATCCTTCTTTAAGAGGAC | Kawasaki et al. (2012) |
| 975–9194R | CCTTCCATTTGAGGTGTAGC | Kawasaki et al. (2012) |
AFLP fingerprinting
To perform the AFLP, we followed the protocol of Vos et al. 1995 with modifications described by de Carvalho et al. 2020. Briefly, Sporothrix genomic DNA (200 ng) was digested using EcoRI (GˆAATTC) and MseI (TˆTAA) restriction enzymes (New England Biolabs, Ipswich, MA) and ligated to EcoRI and MseI adapters simultaneously. A pre-selective PCR was carried out with EcoRI+0 and MseI+0 primers (Vos et al. 1995). Fluorescent AFLP was performed with 6-carboxyfluorescein (FAM; blue) EcoRI primer with two bases selection (5′-GAC TGC GTA CCA ATT CNN-3′) and unlabelled MseI primer with two bases selection (5′-GAT GAG TCC TGA GTA ANN-3′). Three combinations were chosen for genotyping Sporothrix isolates (#3 EcoRI-GA/MseI-TT, #5 EcoRI-GA/MseI-AG, and #6 EcoRI-TA/MseI-AA). AFLP fragments were resolved by capillary electrophoresis with an ABI3730xl Genetic Analyzer alongside a GeneScan LIZ600 internal size standard (35–600 bp; Applied Biosystems. Foster City, CA, USA) at the Human Genome and Stem Cell Research Centre Core Facility (University of São Paulo, São Paulo, Brazil) under previously described conditions (de Carvalho et al. 2020). To evaluate the ability to reproduce results accurately, electropherograms are representative of two independent assays.
Bioinformatics analysis
The raw data were imported into BioNumerics v. 7.6 software (Applied Maths, Sint-Martens-Latem, Belgium). To reduce scoring errors, each electropherogram was carefully examined to exclude low confidence peaks, setting the minimum threshold at 100 relative fluorescence units (RFU), and considering only peaks with sizes in the range of 50 and 500 base pairs. AFLP fragments were converted to the dominant presence (1) or absence (0) at probable fragment positions.
Distance-based techniques were used to assess relationships among Sporothrix isolates and taxa. The band-based Jaccard similarity coefficient was used to compute pairwise genetic distances (Jaccard 1912) combined with a "Fuzzy logic" option. Dendrograms were inferred using the unweighted pair group mean arithmetic method (UPGMA). To evaluate the consistency of a given cluster, we employed the cophenetic correlation coefficient and its standard deviation.
The congruence index (Icong), as defined by de Vienne and colleagues, was used to assess the presence of topological correspondence among AFLP dendrograms and the associated confidence level (de Vienne et al. 2007), based on maximum agreement subtrees (MAST). In addition, to calculate the congruence between the three AFLP experiments, we calculated the Pearson product-moment correlation coefficient (Pearson correlation) (Schober et al. 2018).
Dimensionality reduction methods such as principal component analysis (PCA) and multidimensional scaling (MDS) were used to create three-dimensional plots with the isolates dispersed according to their similarity. Automated fragment matching was implemented on all fingerprint profiles within the comparison, considering a minimum profiling of 5 %, with the optimization and position tolerances for picking fragments established for 0.10 %. Default settings were applied for PCA and MDS, subtracting the average for characters. In addition, the Self-Organizing Map (SOM), a typical artificial neural network algorithm in the unsupervised learning category, was utilized to categorize AFLP data in a two-dimensional space (map) according to their similarity (Kohonen 2001). The size of the Kohonen map was set to 100 (i.e., the neural network nodes in each direction).
The evolutionary relationships among all the genotypes of Sporothrix species were investigated using AFLP-derived Minimum Spanning Trees (MSTs). All figures were exported and treated using Corel Draw X8.
Genetic diversity analysis and linkage disequilibrium
To assess the potential of the three selective primer combinations evaluated here, the following descriptive genetic parameters for dominant markers were calculated: polymorphic information content (PIC) (Botstein et al. 1980), expected heterozygosity (H) (Liu 1998), effective multiplex ratio (E) (Powell et al. 1996), arithmetic mean heterozygosity (Havp) (Powell et al. 1996), marker index (MI) (Powell et al. 1996, Varshney et al. 2007), discriminating power (D) (Tessier et al. 1999), and resolving power (Rp) (Prevost & Wilkinson 1999).
Linkage disequilibrium (LD) was estimated by the standardized disequilibrium coefficient (D'), as well as squared allele-frequency correlations (r2) between pairs of polymorphic loci using the software package TASSEL v. 5.0 (Bradbury et al. 2007). A two-sided Fisher's Exact test determined P-values for each r2 estimate, and loci were considered to be in significant LD when P < 0.001 (Slatkin 2008). Mapping positions were not available for the AFLP markers.
Structure analysis
Analysis of AFLP data in structure v. 2.3.4 (Pritchard et al. 2000) was performed using the admixture model, allowing alpha to be inferred and assuming correlated allele frequencies, using a burn-in period of 10 000 Markov chain Monte Carlo (MCMC) replications followed by 10 000 sampling replications, with 20 independent runs performed for K values one to twenty. We evaluated the posterior distribution of alpha to ensure all chains for K values 2–20 converged. All data were analysed using the method of Evanno and colleagues as implemented in StructureHARVESTER (v. 0.6.94) (Evanno et al. 2005, Earl & vonHoldt 2012) to determine the optimal number of clusters (K). Consensus population distributions were obtained with CLUMPP (v. 1.1.2) (Jakobsson & Rosenberg 2007), using the greedy algorithm over 104 replicates. Final plots were generated using ggplot2 (Wickham 2016) in R (The R Core Team 2014).
Recombination analysis
A split network (Neighbor-Net) was created using the program SplitsTree v. 5.0.0 alpha to examine the relationships among Sporothrix species on AFLP profiles (Huson & Bryant 2006). We used the Hamming distances method (Hamming 1950) with the Neighbor-Net algorithm (Bryant & Moulton 2004) adapted for binary sequences (Huson & Kloepper 2005) for the construction of networks.
Analysis of molecular variance (AMOVA)
The AFLP data was converted into a binary matrix of presence/absence of each allele for each individual, and employed for further analysis of molecular variance using GenAlex v. 6.5 (Peakall & Smouse 2006, Peakall & Smouse 2012). The genetic differentiation among populations was determined using PhiPT (ΦPT). This measure allows intra-individual variation to be suppressed and is ideal for comparing binary data with 9 999 permutations (Teixeira et al. 2014).
Statistical analysis
To ascertain the degree of concordance between AFLP typing and species-specific PCR (Rodrigues et al. 2015) or phylogenetic-based identification (Rodrigues et al. 2014), we calculated Cohen's kappa coefficient (κ) and its 95 % confidence interval (CI). Kappa values were read as follows: 0.00–0.20, poor agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, good agreement; 0.81–1.00, very good agreement (Altman 1991). A P-value ≤ 0.05 was considered significant. All statistical calculations were performed with MedCalc Statistical Software v. 20.013 (MedCalc Software, Ostend, Belgium; http://www.medcalc.org; 2021). We calculated Simpson’s diversity (Simpson 1949) and Shannon’s diversity (Shannon 1948) for each organism/genetic group with the relative abundances estimated with frequency data.
Characterization of the mating-type idiomorphs and mitochondrial DNA typing.
A duplex PCR using primers targeting the MAT1-1 or MAT1-2 region was used to determine the mating-types idiomorphs, as described before (de Carvalho et al. 2021). Approximately 50 ng of genomic DNA was used for PCR with two sets of oligonucleotide primers: SPMAT1-1F and SPMAT1-1R, which amplify a 673 bp fragment from the α box region of the MAT1-1 idiomorph, or SPMAT1-2F and SPMAT1-2R, which amplify a 291 bp fragment from the HMG domain gene, present in the MAT1-2 idiomorph (Table 1). Mitochondrial DNA (mtDNA) typing in Sporothrix was performed using primers 975–8038F and 975–9194R (Table 1) that selectively amplify an intergenic locus between the ATP9 and COX2 genes (Kawasaki et al. 2012). Amplicons were resolved using 1.2 % agarose gel in 1× TBE buffer at 100 V for 1 h at room temperature in the presence of GelRed (Biotium, Hayward, CA, USA) (Sambrook & Russell 2001). DNA fragments were detected by UV illumination with the L-Pix Touch imaging system (Loccus Biotecnologia, São Paulo, Brazil). Fragment size was based on comparison with a 100 bp GeneRuler DNA Ladder (Thermo Fisher Scientific, Waltham, MA, USA).
Results
Identification of medically relevant Sporothrix
We conducted a retrospective molecular epidemiological study using a temporally and geographically diverse collection of Sporothrix isolates (n = 188). Species-specific PCRs were used to speciate Sporothrix isolates confirming 72 S. brasiliensis, 67 S. schenckii, and 34 S. globosa. The remaining Sporothrix isolates (n = 15), mainly in the environmental clade, were classified based on molecular phylogenetics analysis of the calmodulin gene and included as members of the S. pallida and S. stenoceras complexes, S. brunneoviolacea, and S. dimorphospora (Supplementary Table S1).
Mitochondrial DNA typing revealed that the distribution of genotypes of S. brasiliensis and S. globosa remained as expected, carrying the genotypes 1 157 bp or 557 bp, respectively (Supplementary Fig. S1). For S. schenckii, we observed a distribution of 46 isolates with the mtDNA genotype 557 bp, primarily present in South America after the 1990 decade, and 18 isolates with the mtDNA genotype 1 157 bp, widely distributed before the 1990 decade. Only three S. schenckii isolates did not present any amplicon (i.e., Ss192, Ss194, and Ss214).
AFLP markers for Sporothrix
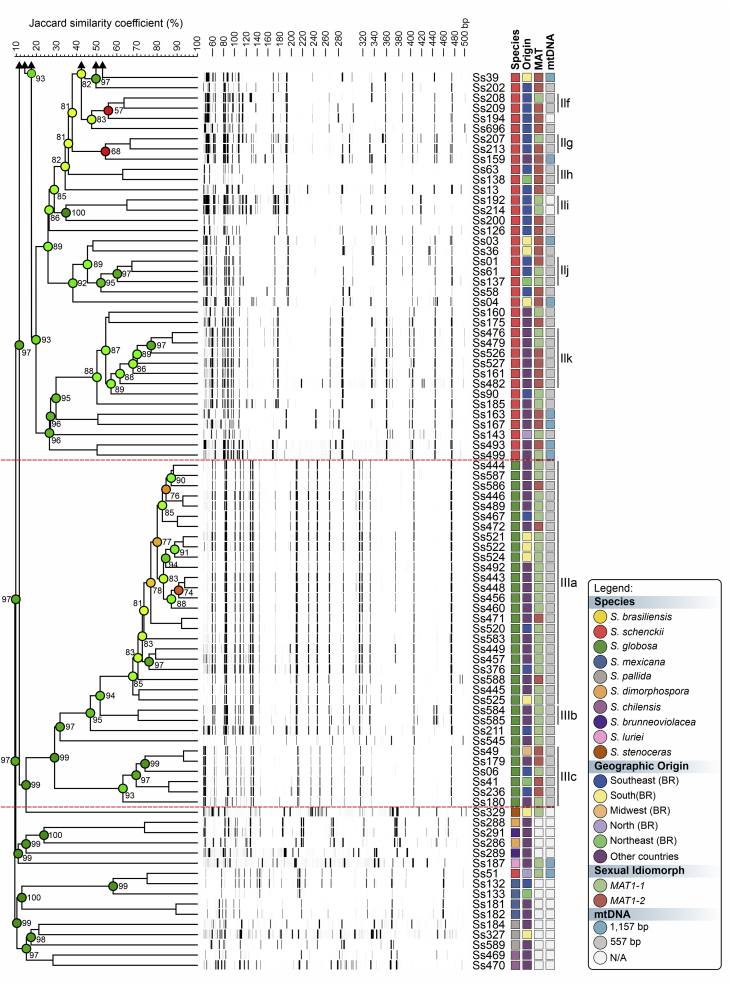
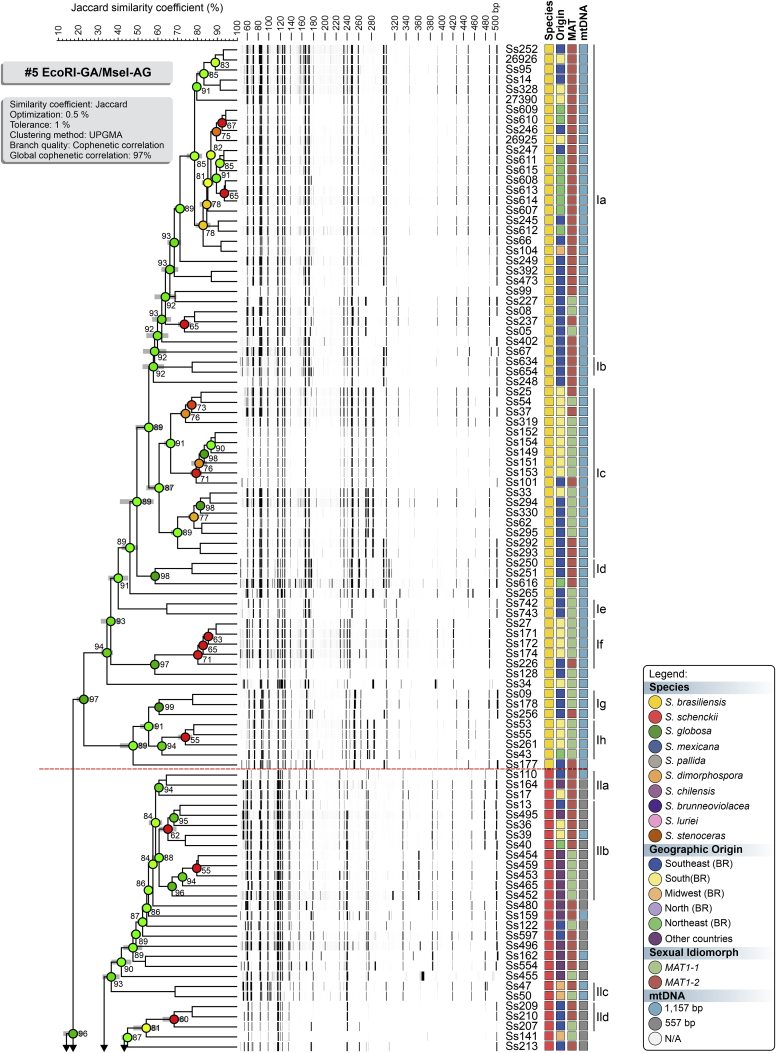
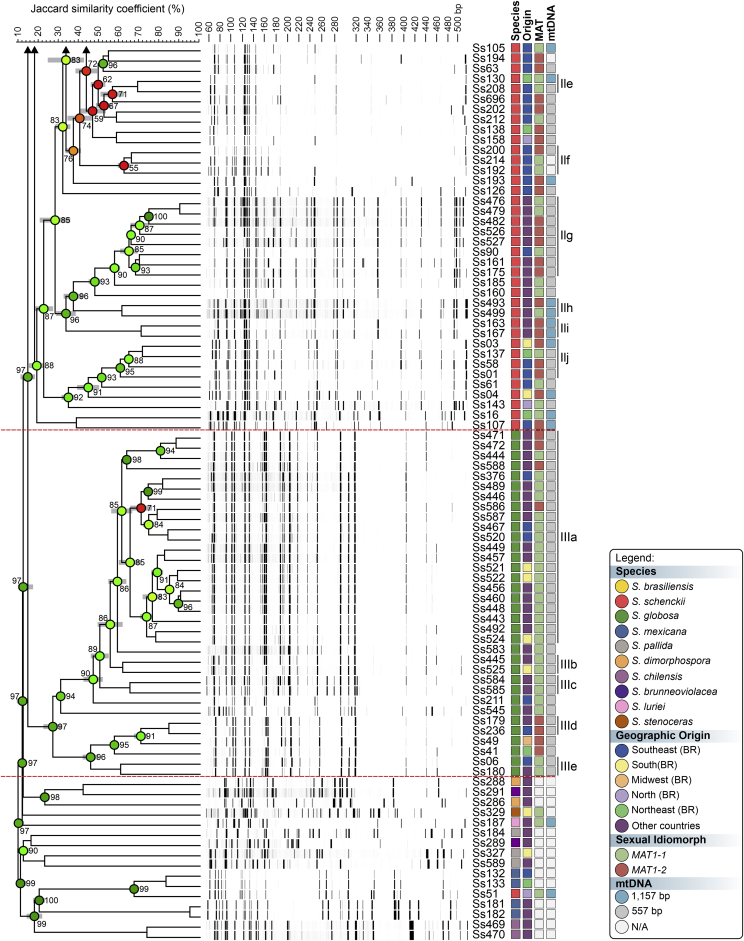
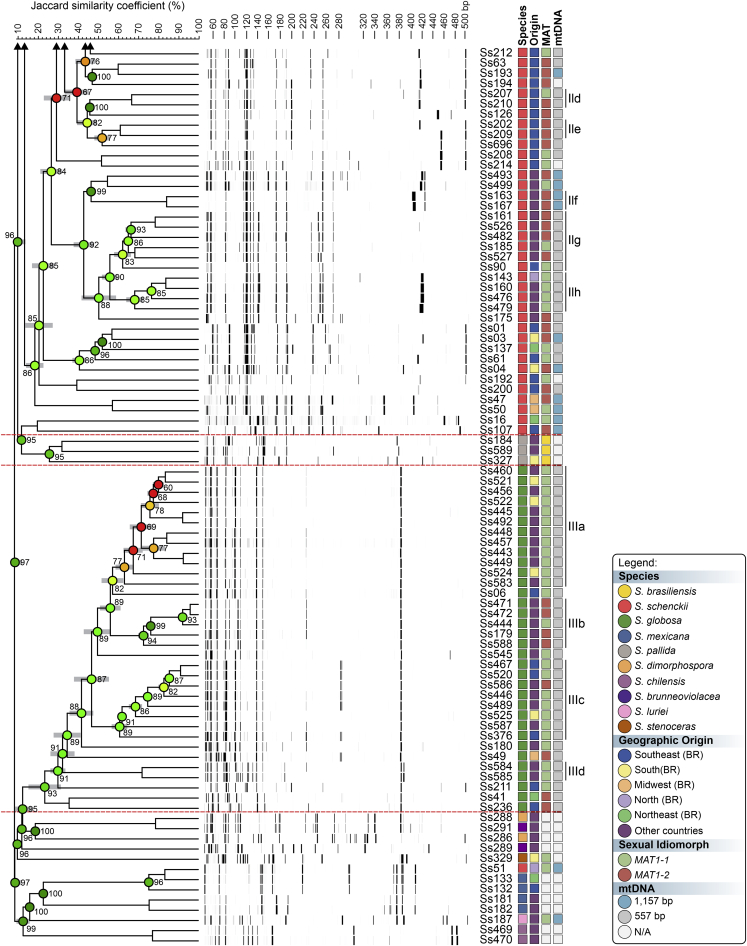
To evaluate the hypothesis of diversity in Sporothrix species, we employed three AFLP markers to investigate genetic variation in these agents (#3 EcoRI-GA/MseI-TT, #5 EcoRI-GA/MseI-AG, and #6 EcoRI-TA/MseI-AA). A total of 451 scorable fragments were amplified in the range of 50 to 500 bp, combining the three selective primers. From this total, 153, 160, and 138 were considered polymorphic fragments of the combinations #3, #5, and #6, respectively. A cut-off of 60.000 % ± 5.00 % was established to identify subclades that generated similarity levels ranging from 58.040 % ± 5.14 % to 91.320 % ± 0.30 % (Supplementary Table S2). Typical AFLP dendrograms established on Jaccard's similarity coefficient for Sporothrix species are shown in Fig. 1, Fig. 2, Fig. 3. Clustering analysis shows three well-supported clades (I–III) with a significant global cophenetic correlation coefficient (97 %) for all markers supporting a great degree of confidence in the association obtained for 188 isolates of Sporothrix (Supplementary Table S2). This clustering pattern agrees with the broadly applied calmodulin-based phylogeny (Marimon et al. 2006).
Fig. 1.
The UPGMA dendrogram, based on AFLP fingerprint, generated with a total of four selective bases (#3 EcoRI-GA/MseI-TT) for 188 Sporothrix isolates originated worldwide. The dendrogram shows cophenetic correlation values (circles are represented by colour ranges between green-yellow-orange-red according to decreasing cophenetic correlation) for a given clade and its standard deviation (grey bar). For pairwise genetic distances calculation, the Jaccard similarity coefficient was used. The cophenetic correlation of the dendrogram is 97 %. Further information about isolate sources can be found in Supplementary Table S1.
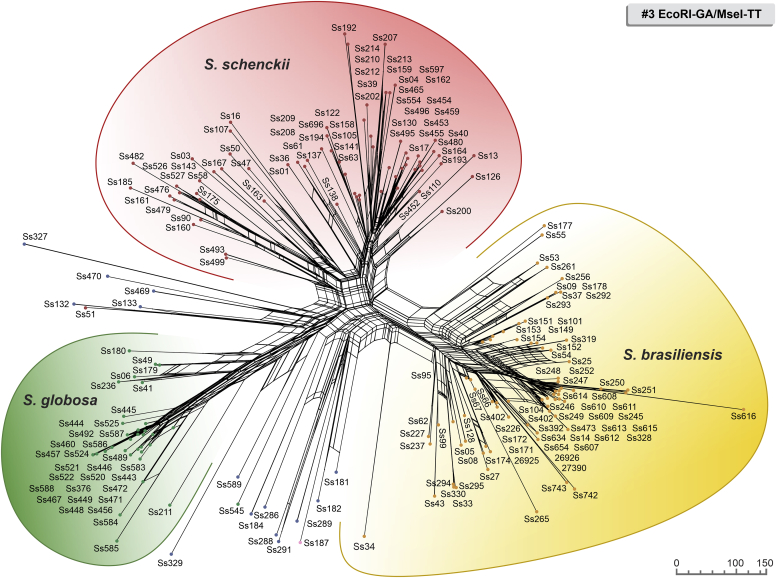
Fig. 2.
The UPGMA dendrogram, based on AFLP fingerprint, generated with a total of four selective bases (#5 EcoRI-GA/MseI-AG) for 188 Sporothrix isolates originated worldwide. The dendrogram shows cophenetic correlation values (circles are represented by colour ranges between green-yellow-orange-red according to decreasing cophenetic correlation) for a given clade and its standard deviation (grey bar). For pairwise genetic distances calculation, the Jaccard similarity coefficient was used. The cophenetic correlation of the dendrogram is 97 %. Further information about isolate sources can be found in Supplementary Table S1.
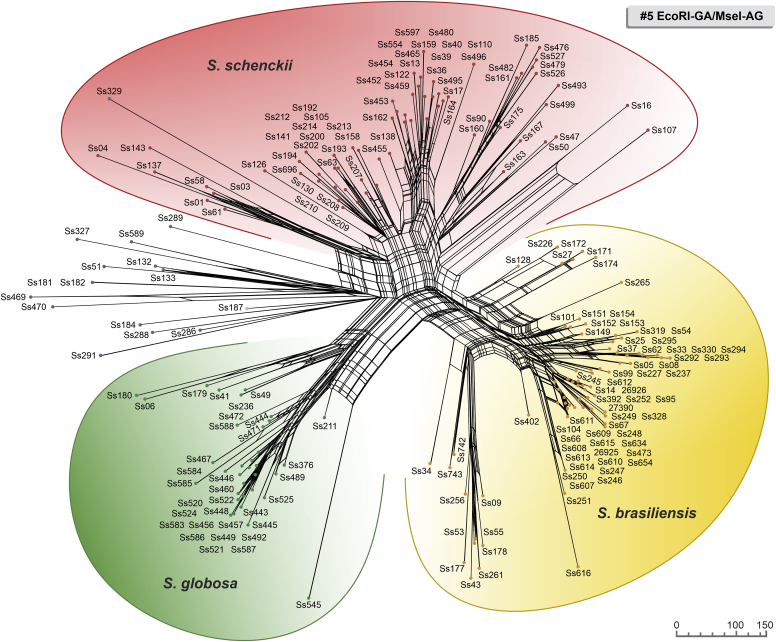
Fig. 3.
The UPGMA dendrogram, based on AFLP fingerprint, generated with a total of four selective bases (#6 EcoRI-TA/MseI-AA) for 188 Sporothrix isolates originated worldwide. The dendrogram shows cophenetic correlation values (circles are represented by colour ranges between green-yellow-orange-red according to decreasing cophenetic correlation) for a given clade and its standard deviation (grey bar). For pairwise genetic distances calculation, the Jaccard similarity coefficient was used. The cophenetic correlation of the dendrogram is 97 %. Further information about isolate sources can be found in Supplementary Table S1.
The first clade represents S. brasiliensis isolates (n = 72) and has global similarities ranging from 29.920 % ± 4.05 % to 46.913 % ± 4.43 %. Clade I was divided into subclades Ia to Ik in combination #3 (Fig. 1), Ia to Ih in combination #5 (Fig. 2), and Ia to Ii in combination #6 (Fig. 3). Isolates originating from the Northeast, represented by clade Ia in all combinations, had higher genetic similarity (i.e., lowest genetic diversity) than isolates from other Brazilian regions (e.g., Rio de Janeiro and Rio Grande do Sul). The second clade comprises S. schenckii isolates (n = 67) and presented global similarity levels ranging from 26.747 % ± 4.38 % to 31.766 % ± 5.33 %. The clade was divided into subclades IIa to IIk in combination #3 (Fig. 1), IIa to IIj in combination #5 (Fig. 2), and Ia to Ih in combination #6 (Fig. 3). The isolates from Venezuela remained grouped in the three combinations (e.g., Ss452, Ss453, Ss454, Ss455, Ss459, and Ss465), except for combination #5, where one isolate (Ss455) was distanced. The remaining isolates followed a similar pattern in all combinations, except for isolate Ss51, which clustered with S. mexicana isolates. In previous studies, isolate Ss51 formed a cluster with isolates Ss16 and Ss107 (Rodrigues et al. 2014). The S. globosa clade (III, n = 34) showed global similarity values ranging from 38.780 % ± 4.29 % to 42.007 % ± 5.14 %. Clade III was subdivided into clades IIIa to IIIc in combination #3 (Fig. 1), IIIa to IIIe in combination #5 (Fig. 2), and IIIa to IIId in combination #6 (Fig. 3). Interestingly, two Japanese isolates (i.e., Ss584 and Ss585) remained grouped in a subclade separately in all combinations, suggesting differentiation of Asian isolates. The remaining isolates (non-clinical species) clustered in the environmental clade, including representative members of the S. pallida or S. stenoceras species complexes, S. brunneoviolacea, and S. dimorphospora.
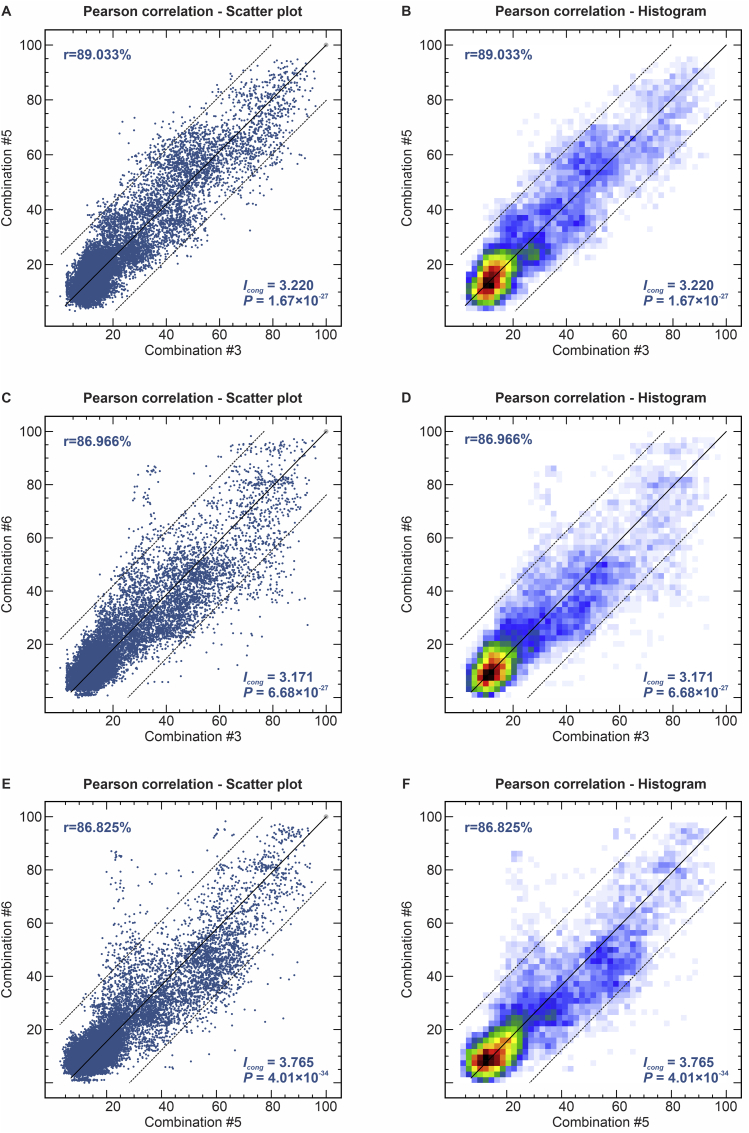
To determine the level of concordance of the results of the species-specific PCR and any AFLP assay, we calculated the kappa (κ) statistic and its 95 % confidence interval (CI). A very good agreement was observed for S. brasiliensis (κ = 1.0, 95 % CI 1.000–1.000), S. schenckii (κ = 0.98 ± 0.01, 95 % CI 0.965–1.000), and S. globosa (κ = 1.0, 95 % CI 1.000–1.000). To evaluate the existence of topological congruence between any two dendrograms, we used the congruence index (Icong) (de Vienne et al. 2007) and the Pearson product-moment correlation coefficient (Pearson correlation). Pairwise comparisons revealed a similar and consistent clustering pattern, as evidenced by the excellent congruence index values and their significant associated P-values (Fig. 4), as well as a strong positive correlation for the Pearson product-moment correlation coefficient (Fig. 4). Therefore, the dendrograms were more congruent than expected by chance and in full agreement with species-specific PCR, supporting the use of AFLP markers to speciate medically relevant Sporothrix and accessing genetic diversity.
Fig. 4.
The correlation between AFLP experiments evaluated for 188 Sporothrix isolates. (A) and (B): combination #3 EcoRI-GA/MseI-TT vs. combination #5 EcoRI-GA/MseI-AG; (C) and (D): combination #3 EcoRI-GA/MseI-TT vs. combination #6 EcoRI-TA/MseI-AA; (E) and (F): combination #5 EcoRI-GA/MseI-AG vs. combination #6 EcoRI-TA/MseI-AA. A similarity plot for two experiments was assessed using the Pearson correlation coefficient (scatter plot) to plot each pair of similarity values as one dot, and the Pearson correlation coefficient (histogram) representing the average the number of dots in each area. A multi-colour scale ranges continuously from white over blue, green, yellow, orange, and red to black. Icong = Congruence index.
The highest number of fragments were noted for combination #3 (EcoRI-GA/MseI-TT) and varied per species between 15–47 for S. brasiliensis (median = 26; CV = 18.12 %), 11–32 for S. schenckii (median = 21; CV = 20.09 %), and 17–38 for S. globosa (median = 23; CV = 17.61 %). The averages of fragments for combination #5 (EcoRI-GA/MseI-AG) varied between 15–39 for S. brasiliensis (median = 24; CV = 15.18 %), 12–33 for S. schenckii (median = 24; CV = 24.47 %), and 16–30 for S. globosa (median = 22; CV = 13.25 %). The lowest number of fragments was observed for combination #6 (EcoRI-TA/MseI-AA) and varied between 9–23 for S. brasiliensis (median = 14; CV = 19.30 %), 8–25 for S. schenckii (median = 15; CV = 19.06 %), and 8–19 for S. globosa (median = 10.5; CV = 24.05 %) (Supplementary Table S3). The characteristics of marker attributes for different AFLP primer combinations are given in Table 2.
Table 2.
Polymorphic statistics calculated for three combinations of selective primers for Sporothrix spp.
|
#3 EcoRI-GA/MseI-TT | ||||||||
|
Species (n) |
Fragments |
H |
PIC |
E |
Havp |
MI |
D |
Rp |
| S. brasiliensis (72) | 100 | 0.3781 | 0.3066 | 25.3194 | 0.0001 | 0.0013 | 0.9359 | 21.0278 |
| S. schenckii (67) | 115 | 0.3008 | 0.2555 | 21.2089 | 0.0000 | 0.0008 | 0.9660 | 29.2537 |
| S. globosa (34) | 71 | 0.4478 | 0.3475 | 24.0294 | 0.0002 | 0.0045 | 0.8855 | 14.7647 |
| Clinical clade (173) | 149 | 0.2662 | 0.2308 | 23.5606 | 0.0000 | 0.0002 | 0.9750 | 38.9132 |
| S. mexicana (4) | 36 | 0.4996 | 0.3748 | 18.5000 | 0.0035 | 0.0642 | 0.7377 | 28.0000 |
| S. pallida (3) | 49 | 0.4998 | 0.3749 | 25.0000 | 0.0034 | 0.0850 | 0.7414 | 29.3333 |
| S. chilensis (2) | 31 | 0.4370 | 0.3415 | 21.0000 | 0.0070 | 0.1480 | 0.5447 | 20.0000 |
| S. brunneoviolacea (2) | 36 | 0.4753 | 0.3623 | 22.0000 | 0.0066 | 0.1452 | 0.6299 | 28.0000 |
| S. dimorphospora (2) | 30 | 0.4061 | 0.3236 | 21.5000 | 0.0068 | 0.1455 | 0.4898 | 17.0000 |
| Overall (186) |
153 |
0.2588 |
0.2253 |
23.3709 |
0.000009 |
0.0002 |
0.9766 |
39.9032 |
|
#5 EcoRI-GA/MseI-AG | ||||||||
|
Species (n) |
Fragments |
H |
PIC |
E |
Havp |
MI |
D |
Rp |
| S. brasiliensis (72) | 84 | 0.4100 | 0.3259 | 24.1806 | 0.0001 | 0.0016 | 0.9172 | 20.1389 |
| S. schenckii (67) | 110 | 0.3323 | 0.2771 | 23.1492 | 0.0000 | 0.0010 | 0.9557 | 27.5820 |
| S. globosa (34) | 67 | 0.4438 | 0.3453 | 22.2647 | 0.0002 | 0.0043 | 0.8897 | 15.7059 |
| Clinical clade (173) | 143 | 0.2742 | 0.2366 | 23.4566 | 0.0000 | 0.0002 | 0.9730 | 38.832 |
| S. mexicana (4) | 39 | 0.4815 | 0.3656 | 23.2500 | 0.0031 | 0.0718 | 0.6462 | 31.5000 |
| S. pallida (3) | 50 | 0.4992 | 0.3746 | 24.0000 | 0.0033 | 0.0799 | 0.7713 | 31.3333 |
| S. chilensis (2) | 34 | 0.1107 | 0.1046 | 32.0000 | 0.0016 | 0.0521 | 0.1150 | 4.0000 |
| S. brunneoviolacea (2) | 54 | 0.4957 | 0.3728 | 29.5000 | 0.0046 | 0.1354 | 0.7039 | 49.0000 |
| S. dimorphospora (2) | 30 | 0.4550 | 0.3515 | 19.5000 | 0.0076 | 0.1479 | 0.5814 | 21.0000 |
| Overall (186) |
160 |
0.2511 |
0.2195 |
23.5591 |
0.000008 |
0.0001 |
0.9783 |
40.2795 |
|
#6 EcoRI-TA/MseI-AA | ||||||||
|
Species (n) |
Fragments |
H |
PIC |
E |
Havp |
MI |
D |
Rp |
| S. brasiliensis (72) | 75 | 0.3103 | 0.2622 | 14.4028 | 0.0001 | 0.0008 | 0.9632 | 14.5278 |
| S. schenckii (67) | 100 | 0.2903 | 0.2482 | 17.6268 | 0.0000 | 0.0007 | 0.9689 | 22.0895 |
| S. globosa (34) | 43 | 0.3912 | 0.3147 | 11.4706 | 0.0003 | 0.0031 | 0.9290 | 8.9412 |
| Clinical clade (173) | 126 | 0.2108 | 0.1886 | 15.0924 | 0.0000 | 0.0001 | 0.9856 | 25.3872 |
| S. mexicana (4) | 20 | 0.4688 | 0.3589 | 12.5000 | 0.0059 | 0.0732 | 0.6123 | 11.0000 |
| S. pallida (3) | 28 | 0.4955 | 0.3727 | 15.3333 | 0.0059 | 0.0904 | 0.7031 | 14.0000 |
| S. chilensis (2) | 14 | 0.0000 | 0.0000 | 14.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| S. brunneoviolacea (2) | 42 | 0.4898 | 0.3698 | 24.0000 | 0.0058 | 0.1399 | 0.6764 | 36.0000 |
| S. dimorphospora (2) | 33 | 0.4224 | 0.3332 | 23.0000 | 0.0064 | 0.1472 | 0.5175 | 20.0000 |
| Overall (186) | 138 | 0.1957 | 0.1765 | 15.1720 | 0.000007 | 0.0001 | 0.9879 | 26.1075 |
D: discriminating power; E: effective multiplex ratio; H: expected heterozygosity; Havp: mean heterozygosity; MI: marker index; PIC: polymorphism information content; Rp: resolving power. The clinical clade included S. brasiliensis (n = 72), S. schenckii (n = 67), and S. globosa (n = 34). S. luriei (n = 1) and S. stenoceras (n = 1) were not included in the analysis. A comprehensive view polymorphic statistics is presented in Supplementary Table S4.
The PIC values demonstrated the excellent capability of each primer pair combinations to detect intra and interspecific polymorphisms revealing average polymorphism in S. brasiliensis (PIC = 0.2622–0.3259), S. schenckii (PIC = 0.2482–0.2771), and S. globosa (PIC = 0.3147–0.3475). The highest overall PIC value was observed for primer combination #3 (PIC = 0.2253), and the lowest was recorded for primer combination #6 (PIC = 0.1765), indicating good diversity among the studied Sporothrix. In general, S. brasiliensis, S. schenckii, and S. globosa showed equivalent levels of polymorphic information content (Table 2). Discriminating power (D) is considered as the probability of two random individuals present a different pattern of bands, and all markers demonstrated overall high discriminating power (D = 0.9766–0.9879), especially among medically relevant species (Table 2).
Marker index (MI) was calculated as the product of the effective multiplex ratio (E), and the average expected heterozygosity (Havp) for polymorphic markers and was used to estimate the overall utility of each marker system. Comparable overall MI values (MI = 0.0001–0.0002) were obtained for all combinations. The resolving power (Rp), which is the ability of each primer combination to detect the level of variation among individuals, was found to be higher in primer combination #5 (Rp = 40.2795) and lower for primer combination #6 (Rp = 26.1075) (Table 2). A moderate positive correlation was observed between PIC and MI values (Pearson correlation = 0.6039, r2 = 0.3647) or Rp and MI values (Pearson correlation = 0.608, r2 = 0.3697), only for combination #6.
We also calculated the expected heterozygosity (H), which is described as the likelihood that an isolate is heterozygous for the locus in the population. The expected heterozygosity corresponds to Nei’s unbiased gene diversity (HS), as it has been modified for dominant markers based on the assumptions of Hardy-Weinberg equilibrium and the Lynch-Milligan model (Lynch & Milligan 1994). The whole average expected heterozygosity for Sporothrix species ranged between 0.1957–0.2588 (Table 2). Considering the previous report of clonality among members of the clinical clade, the high combined expected heterozygosity for S. brasiliensis (H = 0.3103–0.4100) and S. globosa (H = 0.3912–0.4478) was surprising when compared to S. schenckii isolates (H = 0.2903–0.3323), confirming that the three markers can uncover cryptic diversity in medically relevant Sporothrix.
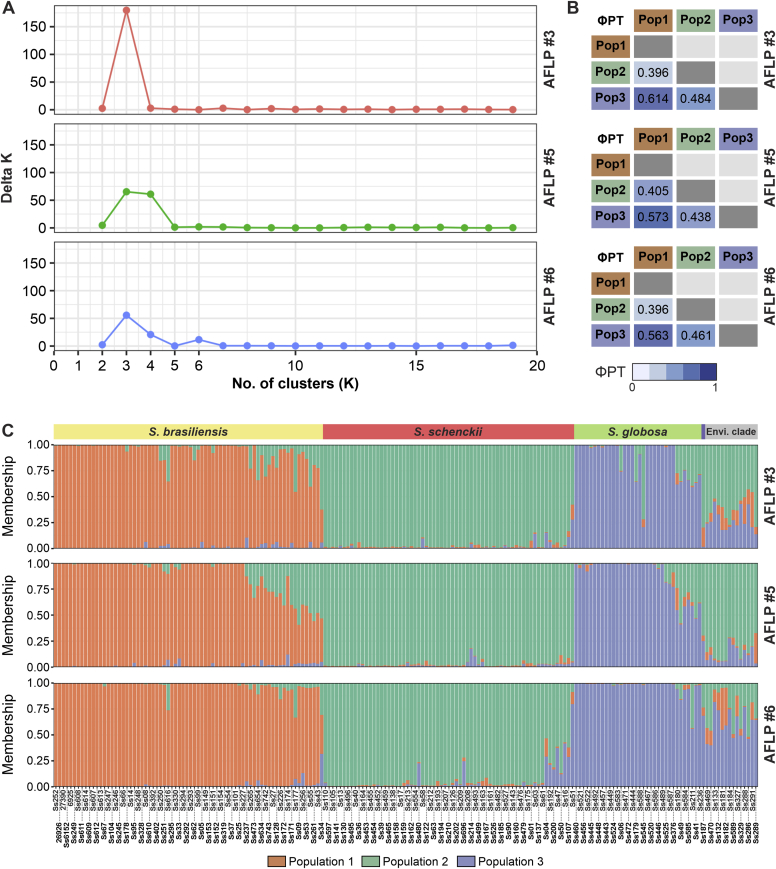
Structure analysis
The Delta K plot revealed the maximum peak at K = 3 (Fig. 5A), supporting the division into three genetic clusters as the most probable number of genetically distinct populations with a great signal of admixture (Supplementary Figs S2–S4). We found a strong correlation between population structure and Sporothrix species (r2 = 0.995, P < 0.00001). For K = 3, S. brasiliensis isolates clustered with population 1, S. schenckii clustered with population 2, and S. globosa isolates correspond to population 3. In contrast, members of the environmental clade show some degree of admixture, as we might expect, given their low relatedness – but the small sample number likely explains why they were not consistently partitioned into their own groups (Fig. 5C).
Fig. 5.
Population structure of clinical and environmental Sporothrix isolates. (A) Structure HARVESTER results. The most plausible number of genetic clusters (K) within the complete data set of 188 individuals based on the method depicted by Evanno et al. (Evanno et al. 2005). Population genetic structure of the estimated ΔK value determined the maximum value at K = 3. (B) PhiPT values for all pairwise comparisons among the S. brasiliensis (population 1), S. schenckii (population 2), and S. globosa (population 3). (C) Bayesian cluster analyses with STRUCTURE (k = 3) of 188 Sporothrix isolates based on AFLP combinations #3 EcoRI-GA/MseI-TT, #5 EcoRI-GA/MseI-AG, and #6 EcoRI-TA/MseI-AA. Each vertical bar represents one individual and its probabilities of being assigned to clusters. Further information about isolate sources can be found in Supplementary Table S1.
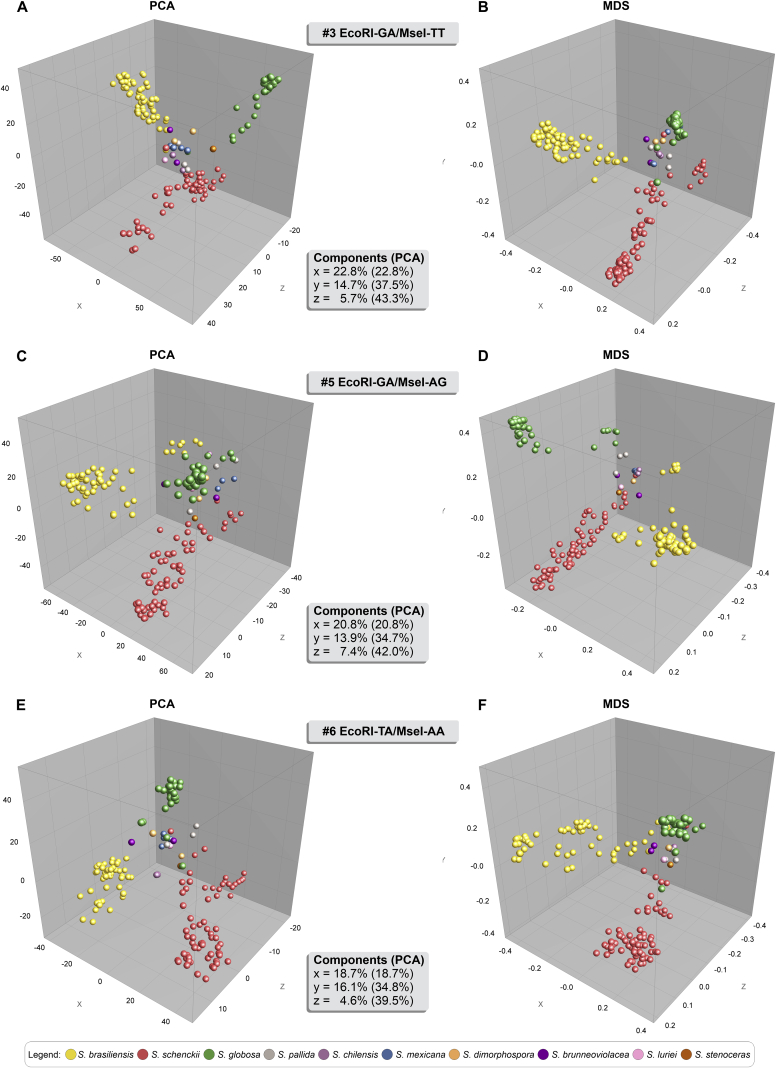
The AFLP markers were used to generate pairwise genetic distance matrices using Jaccard's similarity coefficient, which were then subjected to a PCA utilizing BioNumerics. Fig. 6 depicts the PCA plots for combinations #3, #5, and #6, and the distribution of 188 Sporothrix isolates among the three coordinates illustrated a similar trend to cluster analysis. Combination #3 revealed the highest cumulative percentage explained, with 43.3 % of the variation described by the first three components (coordinates X, Y, and Z), indicating a robust genetic structure. The dimensioning technique revealed an excellent level of intraspecific clustering and a significant genetic distance between any two taxa (interspecific variation). The structure evidenced by PCA supports the separation of S. brasiliensis, S. schenckii, and S. globosa, consistent with the higher level of intraspecific variability shown in dendrogram analysis. Although PCA is explicitly a non-parametric data summary, the dispersion of sample projections along an axis was diagnostic of the samples being admixed among populations at the axis ends (McVean 2009). Therefore, PCA was a robust analysis to reveal outliers in S. brasiliensis (e.g., Ss34, Ss128, and Ss265), S. schenckii (e.g., Ss16, Ss51, Ss107), and S. globosa populations (e.g., Ss41, Ss49, and Ss211).
Fig. 6.
Principal component analysis (PCA) and Multidimensional scaling (MDS) analysis of the #3 EcoRI-GA/MseI-TT (156 loci), #5 EcoRI-GA/MseI-AG (163 loci), and #6 EcoRI-TA/MseI-AA (142 loci) informative AFLP markers plotted in three-dimensional space coloured according to the genetic groups. (A) PCA, and (B) MDS based on combination #3 EcoRI-GA/MseI-TT (n = 188). (C) PCA, and (D) MDS based on combination #5 EcoRI-GA/MseI-AG (n = 188). (E) PCA, and (F) MDS based on combination #6 EcoRI-TA/MseI-AA (n = 188). PCAs and MDS were created in the software BioNumerics v. 7.6.
We performed an independent dimensioning analysis for S. brasiliensis from the South, Southeast and Northeast regions using the three AFLP combinations (Supplementary Fig. S5). The cumulative percentages were 37.5 %, 47.5 % and 46 % for combinations #3, #5, and #6, respectively. We observed that isolates originating from Rio Grande do Sul remained distinct from Rio de Janeiro in all combinations. Only one isolate belonging to Rio Grande do Sul clustered with isolates from Rio de Janeiro. Isolates from Pernambuco represent a recent expansion of S. brasiliensis to Northeast Brazil. These isolates followed the same clustering pattern of isolates from Rio de Janeiro, indicating a possible dissemination route within the country (Supplementary Fig. S5).
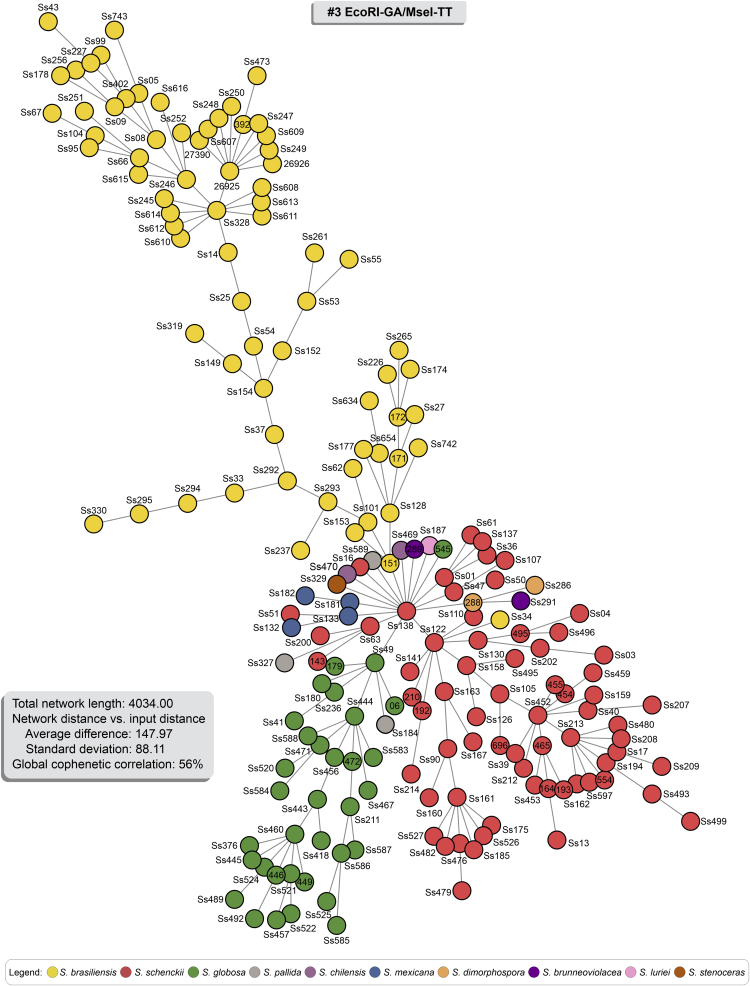
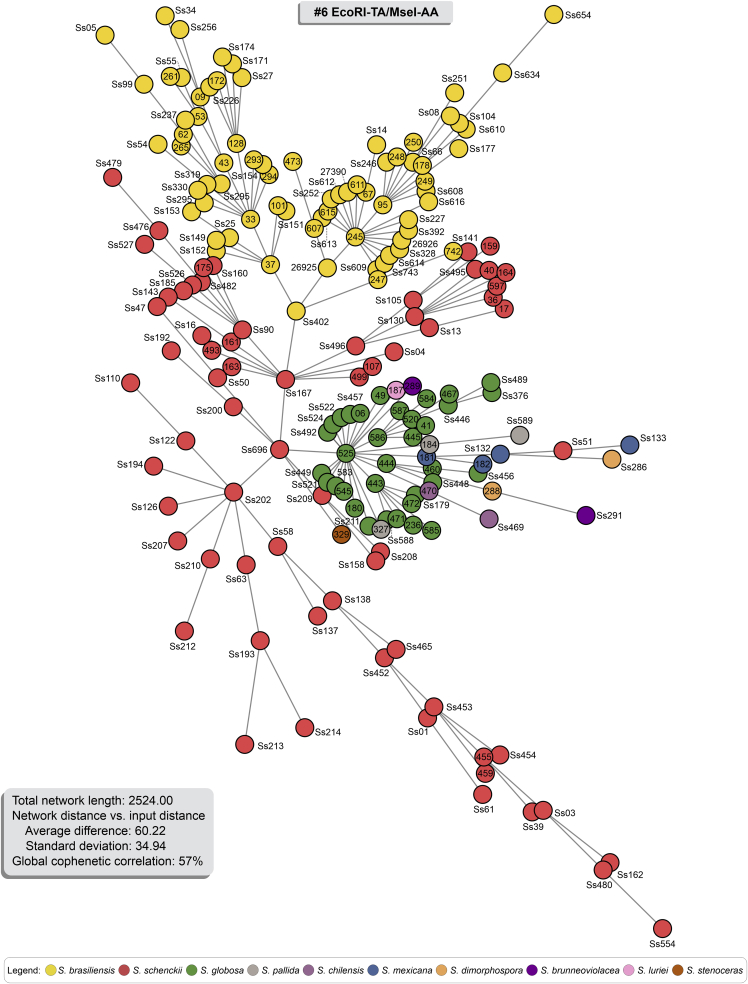
The AFLP-derived MSTs in Fig. 7, Fig. 8, Fig. 9 effectively confirm the genetic structure of members of the clinical clade in Sporothrix in dimensioning analysis, with most isolates having a single genotype. However, similar to the dendrogram, a few isolates in S. brasiliensis (e.g., Ss34) and S. schenckii (e.g., Ss51) were more randomly distributed in the minimum spanning tree analysis, notably for combination #3 (Fig. 7). A MST is a tree that connects all samples (individual sequences) to minimize all tree branches' summed distance. Thus, MSTs provide an indication of evolutionary directionality, which may be used to understand pathogen transmission (Salipante & Hall 2011). Accordingly, the oldest isolates in our collection assumed a more internal position in our MSTs analysis (e.g., Ss05, Ss14, Ss128). In contrast, those isolates recovered more recently in outbreaks taking place in Northeast Brazil were usually placed in terminal nodes (e.g., Ss607, Ss608, Ss609, Ss610, Ss611, Ss612, Ss613, Ss614, Ss615, and Ss616). Associated with the low diversity (Havp) of the Northeast isolates (See cluster Ia in all combinations; Supplementary Table S4) suggest the direction of migration (southeast-northeast), producing a founder effect and explaining the expansion dynamics of S. brasiliensis outbreaks.
Fig. 7.
Minimum Spanning Trees (MSTs) showing the genetic relationship between 188 Sporothrix isolates using for combination #3 EcoRI-GA/MseI-TT (Total network length 4 034.00). Isolates were colour-coded according to their genetic groups. Therefore, the distance between genotypes in the diagram does not reflect any relationship between genotypes' genetic distance.
Fig. 8.
Minimum Spanning Trees (MSTs) showing the genetic relationship between 188 Sporothrix isolates using for combination #5 EcoRI-GA/MseI-AG (Total network length 3 907.00). Isolates were colour-coded according to their genetic groups. Therefore, the distance between genotypes in the diagram does not reflect any relationship between genotypes' genetic distance.
Fig. 9.
Minimum Spanning Trees (MSTs) showing the genetic relationship between 188 Sporothrix isolates using for combination #6 EcoRI-TA/MseI-AA (Total network length 2 524.00). Isolates were colour-coded according to their genetic groups. Therefore, the distance between genotypes in the diagram does not reflect any relationship between genotypes' genetic distance.
In all combinations it was possible to notice S. brasiliensis, S. schenckii, and S. globosa coming from a single ancestor, which is consistent with many conserved fragments observed. Furthermore, the overall high similarity of > 70 % between the fingerprints supports the monophyletic origin of these isolates (Supplementary Table S2).
Self-organizing maps based on AFLP fingerprints (character data and similarity matrix) were used and are plotted according to species identification. A self-organizing map (SOM) is a category of an artificial neural network trained using an unsupervised learning algorithm and usually applied to explore input spaces, for which there is no previous information, and is, therefore, a method to make dimensionality reduction (Kitani et al. 2010). In Fig. 10, SOMs contain areas of high distance and regions of high similarity, and samples are organized into 2-dimensional genetic distance maps, in which samples with low genetic distance form clusters (represented by black blocks). The relative genetic distance between neighbouring groups (black blocks) is indicated by the intensity of white lines separating the clusters, with closely related clusters separated by dark lines and more distantly related isolates separated by increasingly lighter lines. Therefore, in S. brasiliensis, those isolated from recent outbreaks appear in cells separated by faint lines, indicating little diversification from the outbreak's beginning (founder effect). The older the isolates are, the more robust and intense the lines separate individual isolates, showing the intraspecific diversity inherent to S. brasiliensis. Sporothrix schenckii isolates are as different from each other as distinct from other species, demonstrating marked intraspecific diversity. Sporothrix globosa isolates are roughly similar to each other, and we highlight the formation of two groups, one belonging to a global population recovered from Argentina, Brazil, Chile, Venezuela, Mexico, and Spain. The Japanese isolates represent the second group (Asian cluster). A limitation of our study was the absence of S. globosa isolates from other regions of Asia, such as India and China. It is essential to highlight that all the isolates belonging to the same species remain close to each other on the self-organizing maps, but bright solid lines were observed separating clusters interspecifically (Fig. 10).
Fig. 10.
The distribution of the studied AFLP genotypes of 188 Sporothrix isolates originated worldwide, using self-organizing mapping (SOM). The dimensioning analyses were performed using BioNumerics v. 7.6 to determine the consistency of the differentiation of the populations defined by the cluster analysis. (A) and (B) show the SOM for combination #3 EcoRI-GA/MseI-TT (n = 188) using character data (binary matrix) and similarity matrix, respectively. (C) and (D) show the SOM for combination #5 EcoRI-GA/MseI-AG (n = 188) using character data (binary matrix) and similarity matrix, respectively. (E) and (F) show the SOM for combination #6 EcoRI-TA/MseI-AA (n = 188) using character data (binary matrix) and similarity matrix, respectively. The lighter and thicker the line (white, grey) between black blocks, the more distant are those samples contained in the black block from the adjacent black block. Isolates were colour-coded according to their genetic groups.
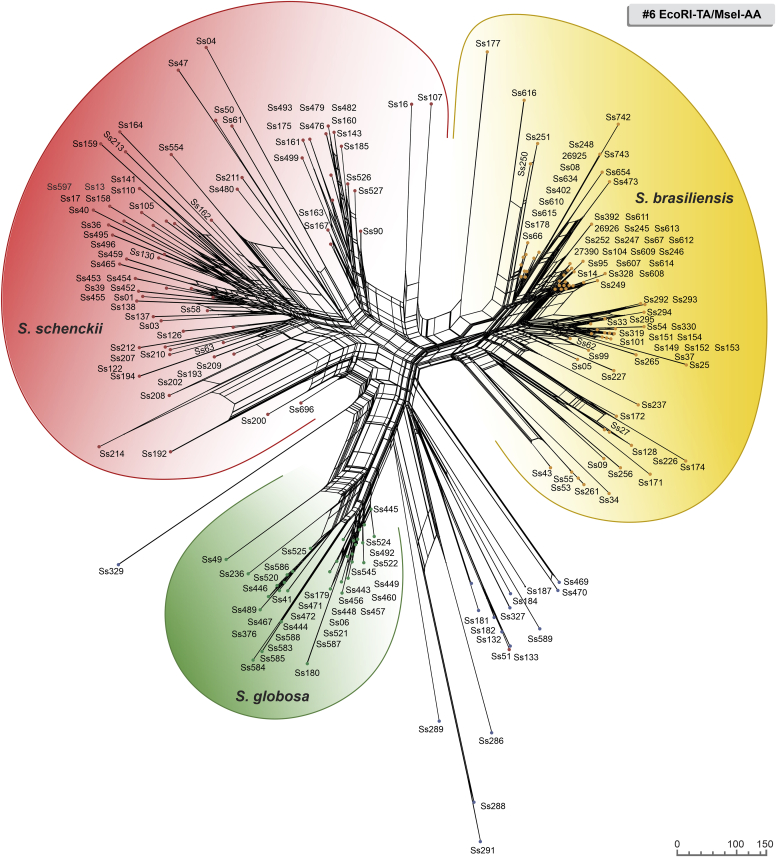
Phylogenetic networks represent conflicting and incompatible signals in the AFLP data set to reveal recombination or hybridization events. Relationships among Sporothrix are depicted in Fig. 11, Fig. 12, Fig. 13 by the Neighbor-Net results obtained from Hamming Distances method. All members of the clinical clade are differentiated from each other in all markers. Large splits/reticulations provided molecular evidence of recombination in Sporothrix in Neighbor-Net, confirming previous results based on DNA sequencing (Rodrigues et al. 2014). However, in S. brasiliensis, we noticed that the isolates recovered from recent epidemics in the Northeast of Brazil. (i.e., 2017) have fewer splits and shorter branches. The discrete recombination events detected occurred with isolates recovered in the long-lasting outbreak of cat-transmitted sporotrichosis in Rio de Janeiro, suggesting proximity between these samples (Fig. 11, Fig. 12, Fig. 13).
Fig. 11.
Neighbor-Net network showing genetic relationships based on AFLPs #3 EcoRI-GA/MseI-TT among Sporothrix species (scale equals genetic distance). Analysis was performed using SplitsTree v. 5.0.0_alpha (Huson & Bryant 2006) for binary sequences (Huson & Kloepper 2005), and the original input consisted of 188 standard character sequences. Method: Neighbor-net. Weights: NNet2004. Original input: 188-character sequences. Length: 156 characters. No. of splits: 604 cyclic. Split’s network: 3 990 nodes and 7 374 edges.
Fig. 12.
Neighbor-Net network showing genetic relationships based on AFLPs #5 EcoRI-GA/MseI-AG among Sporothrix species (scale equals genetic distance). Analysis was performed using SplitsTree v. 5.0.0_alpha (Huson & Bryant 2006) for binary sequences (Huson & Kloepper 2005), and the original input consisted of 188 standard character sequences. Method: Neighbor-net. Weights: NNet2004. Original input: 188-character sequences. Length: 163 characters. No. of splits: 585, cyclic. Split’s network: 3 733 nodes and 6 879 edges.
Fig. 13.
Neighbor-Net network showing genetic relationships based on AFLPs #6 EcoRI-TA/MseI-AA among Sporothrix species (scale equals genetic distance). Analysis was performed using SplitsTree v. 5.0.0_alpha (Huson & Bryant 2006) for binary sequences (Huson & Kloepper 2005), and the original input consisted of 188 standard character sequences. Method: Neighbor-net. Weights: NNet2004. Original input: 188-character sequences. Length: 142 characters. No. of splits: 660, cyclic. Split’s network: 6 498 nodes and 12 334 edges.
AMOVA
We used AMOVA as a statistical framework for hypothesis testing of different patterns of population structure, including 173 individuals of the three genetic populations in Sporothrix (S. brasiliensis, n = 72, population 1; S. schenckii, n = 67, population 2, S. globosa, n = 34, population 3). The AMOVA results for the population analysis are shown in Table 3. AMOVAs performed for combination #3 showed that 48 % of the total genetic variance was triggered by variability among populations, whereas 52 % was driven by variability within populations (PhiPT = 0.484, P < 0.0001). A similar trend was observed for combinations #5 and #6 with 46 % of total variation among genetic populations and 54 % within populations (PhiPT #5 = 0.461, PhiPT #6 = 0.458, P < 0.0001) (Supplementary Fig. S6). The hierarchical analysis of molecular variance demonstrated significant genetic differentiation among populations and within populations (P < 0.0001), indicating that the total genetic variation was almost evenly split between intrapopulation and interpopulation variations, irrespective of the marker used. This pattern could have resulted from substantial differentiation among the populations and relatively frequent gene flow among S. brasiliensis, S. schenckii, and S. globosa populations represented by the presence of frequent patterns of admixture in the three populations evaluated (e.g., Ss34, Ss51, Ss49). Pairwise PhiPT comparison revealed that all three populations differed significantly from one other. The lowest PhiPT values were found between the S. brasiliensis and S. schenckii populations and ranged from 0.396 to 0.405. PhiPT values between the S. schenckii and S. globosa populations ranged from 0.438 to 0.484. The largest range of PhiPT was found between the S. brasiliensis and S. globosa populations (PhiPT = 0.563–0.614) (Fig. 5B).
Table 3.
Analysis of molecular variance (AMOVA) shows the partitioning of genetic variation within and between Sporothrix species populations.
| Marker | Source of variation | df | SS | MS | Est. var. | % | P-value |
|---|---|---|---|---|---|---|---|
| AFLP #3 | Among Population | 2 | 961.086 | 480.543 | 8.540 | 48 % | 0.0001 |
| Within Population | 170 | 1 546.232 | 9.095 | 9.095 | 52 % | 0.0001 | |
| Total marker #3 |
172 |
2 507.318 |
17.636 |
100 % |
|||
| AFLP #5 | Among Population | 2 | 879.705 | 439.852 | 7.803 | 46 % | 0.0001 |
| Within Population | 170 | 1 551.786 | 9.128 | 9.128 | 54 % | 0.0001 | |
| Total marker #5 |
172 |
2 431.491 |
16.931 |
100 % |
|||
| AFLP #6 | Among Population | 2 | 611.680 | 305.840 | 5.424 | 46 % | 0.0001 |
| Within Population | 170 | 1 092.991 | 6.429 | 6.429 | 54 % | 0.0001 | |
| Total marker #6 | 172 | 1 704.671 | 11.853 | 100 % |
df = degree of freedom, SS = sum of squares, MS mean squares, Est. var. = estimate of variance, % = percentage of total variation, P-value is based on 9 999 permutations.
We performed an in-depth analysis of S. brasiliensis isolates by dividing them into three geographic populations, namely: population S matching the southern region (isolates from Rio Grande do Sul and Paraná), population SE covering isolates from the southeast region (Rio de Janeiro, São Paulo, Minas Gerais, and Espírito Santo), while population NE corresponds to isolates from northeast Brazil (Pernambuco & Ceará). The most significant PhiPT values were found between the South and Northeast isolates (PhiPT = 0.184–0.193). Moreover, the lowest values were found in pairwise comparisons between the Southeast and Northeast, demonstrating the genetic proximity of these isolates (PhiPT = 0.030–0.103). AMOVAs indicated that only 7–10 % of the total genetic variance was triggered by variability among geographic populations, whereas 90–93 % was driven by variability within populations (PhiPT = 0.071–0.104, P < 0.0001) (Supplementary Fig. S7, Supplementary Table S5).
Linkage disequilibrium (LD)
Squared allele frequency correlations (r2) were estimated in the complete set (n = 173) using 149 (combination #3), 143 (combination #5), and 126 (combination #6) AFLP markers (Supplementary Figs S8–10). Only around 11.3 % out of 11 026 (combination #3), 10.51 % out of 10 153 (combination #5), and 6.60 % out of 7 875 (combination #6) possible genome-wide marker pairs were in LD at P < 0.001, indicating that the LD level remained low in the Sporothrix isolates included in this study. Splitting the Sporothrix species eliminated some of the observed LD levels, though residual patterns remain. In S. brasiliensis (n = 72), a total of 383 out of 11 211 (3.41 %) genome-wide marker pairs were in LD at P < 0.001, and the strongest LD (r2 = 1) was observed for 14 marker pairs (0.12 %). In S. schenckii (n = 67), a total of 389 out of 17 500 (2.22 %) genome-wide marker pairs were in LD at P < 0.001, and the strongest LD (r2 = 1) was observed for 16 marker pairs (0.09 %). In S. globosa (n = 34), a total of 145 out of 5 599 (2.58 %) genome-wide marker pairs were in LD at P < 0.001, and the strongest LD (r2 = 1) was observed for 89 marker pairs (1.58 %) (Table 4).
Table 4.
Linkage disequilibrium (LD) analysis in the complete Sporothrix database (n = 173) and within members of the clinical clade.
|
#3 EcoRI-GA/MseI-TT |
||||
|
LD characteristic |
Clinical clade (n = 173) |
S. brasiliensis (n = 72) |
S. schenckii (n = 67) |
S. globosa (n = 34) |
| Total number of markers | 149 | 100 | 115 | 71 |
| Number of markers pairs | 11 026 | 4 950 | 6 555 | 2 485 |
| Number of markers pairs at 0 ≤ P ≤ 0.001 | 1 246 (11.3 %) | 114 (2.30 %) | 190 (2.89 %) | 112 (4.50 %) |
| Number of markers pairs at 0.001 ≤ P ≤ 0.01 | 442 (4 %) | 132 (2.66 %) | 141 (2.15 %) | 52 (2.09 %) |
| Number of markers pairs at r2 = 1 | 3 (0.02 %) | 5 (0.10 %) | 14 (0.21 %) | 31 (1.24 %) |
| Mean r2 | 0.0379 | 0.0472 | 0.0436 | 0.1666 |
| Mean D′ |
0.6729 |
0.6931 |
0.6737 |
0.7983 |
|
#5 EcoRI-GA/MseI-AG |
||||
|
LD characteristic |
Clinical clade (n = 173) |
S. brasiliensis (n = 72) |
S. schenckii (n = 67) |
S. globosa (n = 34) |
| Total number of markers | 143 | 84 | 110 | 67 |
| Number of markers pairs | 10 153 | 3 486 | 5 995 | 2 211 |
| Number of markers pairs at 0 ≤ P ≤ 0.001 | 1 067 (10.51 %) | 188 (5.39 %) | 145 (2.41 %) | 22 (0.99 %) |
| Number of markers pairs at 0.001 ≤ P ≤ 0.01 | 420 (4.13 %) | 134 (3.84 %) | 139 (2.31 %) | 102 (4.61 %) |
| Number of markers pairs at r2 = 1 | 0 | 8 (0.22 %) | 0 | 43 (1.94 %) |
| Mean r2 | 0.0370 | 0.0697 | 0.0453 | 0.1247 |
| Mean D′ |
0.6299 |
0.6959 |
0.6193 |
0.7527 |
|
#6 EcoRI-TA/MseI-AA |
||||
|
LD characteristic |
Clinical clade (n = 173) |
S. brasiliensis (n = 72) |
S. schenckii (n = 67) |
S. globosa (n = 34) |
| Total number of markers | 126 | 75 | 100 | 43 |
| Number of markers pairs | 7 875 | 2 775 | 4 950 | 903 |
| Number of markers pairs at 0 ≤ P ≤ 0.001 | 520 (6.60 %) | 81 (2.91 %) | 54 (1.09 %) | 11 (1.21 %) |
| Number of markers pairs at 0.001 ≤ P ≤ 0.01 | 298 (3.78 %) | 59 (2.12 %) | 91 (1.83 %) | 22 (2.43 %) |
| Number of markers pairs at r2 = 1 | 0 | 1 (0.03 %) | 2 (0.04 %) | 15 (1.66 %) |
| Mean r2 | 0.0271 | 0.0543 | 0.0322 | 0.1167 |
| Mean D′ | 0.7111 | 0.7084 | 0.6968 | 0.7505 |
Mating-type
A mating type-specific duplex PCR assay was used to successfully amplify either the MAT1-1 or the MAT1-2 region among 173 medically relevant Sporothrix isolates. The MAT1-1 region was observed in 83 isolates (47.97 %), while the MAT1-2 region was observed among 90 isolates (52.02 %). Thus, molecular data suggest that heterothallism (self-sterility) is the universal mating strategy amongst Sporothrix species. The distribution of each sexual idiomorph within Sporothrix is presented in Table 5. The distributions of the MAT1-1 or MAT1-2 idiomorph were not significantly skewed (1:1 ratio) for S. brasiliensis s. str. (χ2 = 2.000; P = 0.1573) and S. schenckii (χ2 = 2.522; P = 0.1122), but for S. globosa (χ2 = 9.529; P = 0.0020), supporting the presence of random mating within each species. However, regional partition highlighted a biased distribution of S. brasiliensis, such as Rio de Janeiro (χ2 = 14.222; P = 0.0002), Rio Grande do Sul (χ2 = 7.364; P = 0.0067) and Northeast Brazil (χ2 = 7.364; P = 0.0067) with an overwhelming presence of a single idiomorph. The dominance of MAT 1-2 in Northeast Brazil suggests a close connection with isolates from the Rio de Janeiro epidemic (Supplementary Table S6).
Table 5.
Chi-square value and P-value calculated for Sporothrix isolates based on the distribution of mating-type alleles.
| Species | No. of isolates | No. of isolates by mating-type |
χ2 | P-value | |
|---|---|---|---|---|---|
| MAT 1-1 (%) | MAT 1-2 (%) | ||||
| S. brasiliensis | 72 | 30 (41.66) | 42 (58.33) | 2.000 | 0.1573 |
| S. brasiliensis (RJ) | 18 | 1 (5.55) | 17 (94.44) | 14.222 | 0.0002 |
| S. brasiliensis (RS) | 11 | 10 (90.90) | 1 (9.09) | 7.364 | 0.0067 |
| S. brasiliensis (NE) | 11 | 1 (9.09) | 10 (90.90) | 7.364 | 0.0067 |
| S. schenckii | 67 | 27 (40.29) | 40 (59.70) | 2.522 | 0.1122 |
| S. globosa | 34 | 26 (76.47) | 8 (23.52) | 9.529 | 0.0020 |
| Overall | 173 | 83 (47.97) | 90 (52.02) | 0.283 | 0.5946 |
RJ: Rio de Janeiro; RS: Rio Grande do Sul; NE: Northeast Brazil (Pernambuco and Ceará States).
Phylogenetic trends in Sporothrix
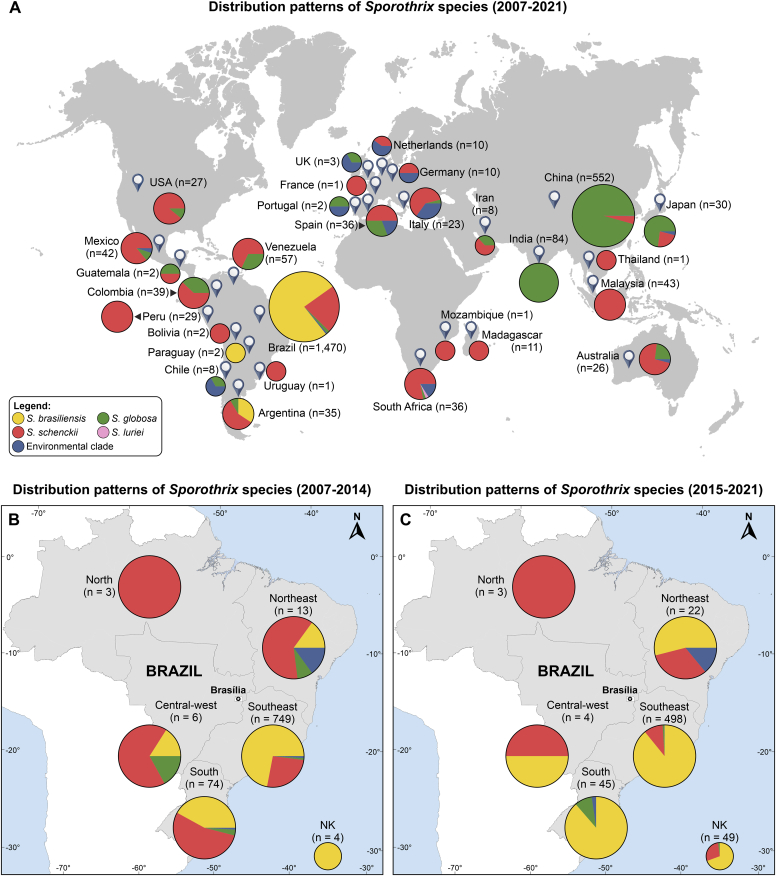
We explored distribution patterns by combining our dataset (n = 188) with data from 2 394 Sporothrix isolates reported in the literature using molecular methods (e.g., multilocus sequence analysis, DNA fingerprint, species-specific PCR, and whole-genome sequencing). The search strategy is available in Supplementary Table S7. Despite its high transmissibility in epizootic and zoonotic episodes, S. brasiliensis shows an endemic geographic distribution to date. However, molecular trends revealed that S. brasiliensis is widely distributed in Brazil and is rapidly spreading to different countries in South America, especially those bordering Brazilian Southern states such as Argentina and Paraguay. In Brazil, most molecular siblings occur in sympatry, as exemplified by S. brasiliensis, S. schenckii, and S. globosa, with a clear overlapping distribution (Fig. 14).
Fig. 14.
Distribution patterns of 2 582 medically relevant Sporothrix spp. isolates based on molecular characterization. (A) Distribution patterns observed worldwide show that most endemic areas are in (sub)tropical areas, and Brazil and China are the main areas to carry out molecular identification. The sizes of circumferences are roughly proportional to the number of strains included. (B, C) Temporal distribution patterns were observed in Brazil (n = 1 470). Codes reported within the pies denote Sporothrix species. Further information about isolate sources can be found in supplementary files (see Search strategy, Supplementary Table S7).
The source of isolation revealed an overwhelming occurrence of clinical isolates (68 %, n = 1 768/2 582) followed by animals (e.g., cats, dogs, and armadillos; 28 %, n = 716/2 582), and from the environment (e.g., soil and decaying wood; 3 %, n = 77/2 582). The source of isolation was unknown for 21 isolates (1 %). Most molecularly characterized isolates are from Brazil (56.66 %, n = 1 470), followed by China (21 37 %, n = 552), India (3.25 %, n = 84), Venezuela (2.20 %, n = 57), Malaysia (1.66 %, n = 43), Mexico (1.62 %, n = 42), Colombia (1.51 %, n = 39), South Africa (1.39 %, n = 36), Spain (1.39 %, n = 36), Argentina (1.35 %, n = 35), and Japan (1.16 %, n = 30). These data emphasize the urgency to increase genetic surveillance efforts in endemic areas (Fig. 14A).
In Brazil, a striking difference in the geographical occurrence of each phylogenetic species was observed, especially if we consider a temporal variable (Fig. 14B and C). The south (Simpson Index = 0.7896; Shannon Index = 0.5928) and southeast regions (Simpson Index = 0.8042; Shannon Index = 0.5507) corresponds to most S. brasiliensis infections and present the highest levels of diversity, with all medically relevant species being reported. In the central-west region, cases are mainly due to S. schenckii, followed by S. brasiliensis (Simpson Index = 0.3333; Shannon Index = 1.0000). Sporothrix schenckii was the prevalent agent in Northeast Brazil in the past (2007–2014) but is currently losing space to infections driven by S. brasiliensis (Simpson Index = 0.3896; Shannon Index = 1.3950). No diversity was found for the North Brazil, as only three isolates were recovered from this region (Fig. 14B and C). A comparative Simpson and Shannon diversity indexes are presented in Supplementary Table S8.
Discussion
Our study provides a comprehensive view of genetic diversity and population structure for the mammalian pathogen Sporothrix in Brazil and globally. Many evolutionary processes, such as mutation, local adaptation, migration (gene flow), genetic drift, and natural selection, clearly depend on a species’ past and present population structure (Chen et al. 2017). Consequently, our approach is relevant towards defining evolutionary relationships in order to gain insights into the epidemiology of emerging sporotrichosis in humans and animals. To obtain a robust view of the genetic basis of ongoing outbreaks, three sets of highly discriminatory AFLP markers were applied in a vast collection of isolates spanning the major endemic areas in order to dissect both deep and fine-scale genetic structures.
Our AFLP dendrograms for Sporothrix species are compatible with the evolutionary history of the etiological agents of sporotrichosis, as determined by multilocus sequencing analysis of proteins-encoding genes such as calmodulin, β-tubulin, elongation factor 1-α, or phylogenomic analyses (Marimon et al. 2006, Marimon et al. 2007, Huang et al. 2016, Rodrigues et al. 2016a, New et al. 2019). Likewise, convergence between fingerprints and DNA-sequencing methods has already been demonstrated for Paracoccidioides (Roberto et al. 2021), Sporothrix (de Carvalho et al. 2020), Fusarium (Al-Hatmi et al. 2016), or Candida auris using AFLP (Vatanshenassan et al. 2020), short tandem repeat typing (de Groot et al. 2020), and whole-genome sequencing (Lockhart et al. 2017). Moreover, phylogenetic studies of Sporothrix suggest that S. brasiliensis, S. schenckii, and S. globosa are closely related taxonomic entities (Rodrigues et al. 2013a; de Beer et al. 2016), and this clustering profile was easily recognized in our AFLP dendrograms. Therefore, we highlight that the extensive in silico screening of selective bases was fundamental to guarantee the efficacy of these markers (de Carvalho et al. 2020), avoiding, for example, splitting monophyletic clades into paraphyletic groups as previously reported for S. brasiliensis and S. schenckii using the combination EcoRI-AA/MseI-G (Zhang et al. 2015).
Pairwise comparisons of our dendrograms were more congruent than expected by chance, supported by the Icong value and a positive Pearson correlation, confirming that different markers reveal compatible evolutionary histories. In each case, S. brasiliensis and S. schenckii are sister species, and this clade is sister to S. globosa. This indicates that members of the clinical clade share a more recent common ancestor than they do with members of the environmental clade, confirming previously reported evolutionary analyses (Marimon et al. 2006, Marimon et al. 2007, de Beer et al. 2016).
Our AFLP technique revealed significant polymorphisms among closely related isolates formerly thought to be clonal (Rodrigues et al. 2013b, Rodrigues et al. 2014, Rangel-Gamboa et al. 2016, Moussa et al. 2017, Rudramurthy et al. 2021), which may help resolve local epidemiological patterns as well as broader changes within populations over time and in response to selection pressures imposed by the environment and host dynamics (McDonald & Linde 2002). In addition, all Sporothrix species in the clinical clade showed high diversity, suggesting that these lineages have high fitness that may have favoured its dispersion, allowing the survival and adaptation to varied geographic conditions worldwide, as population heterogeneity might pool together individuals that contribute disproportionately to the spread of infection or enhanced virulence (Ekroth et al. 2021).
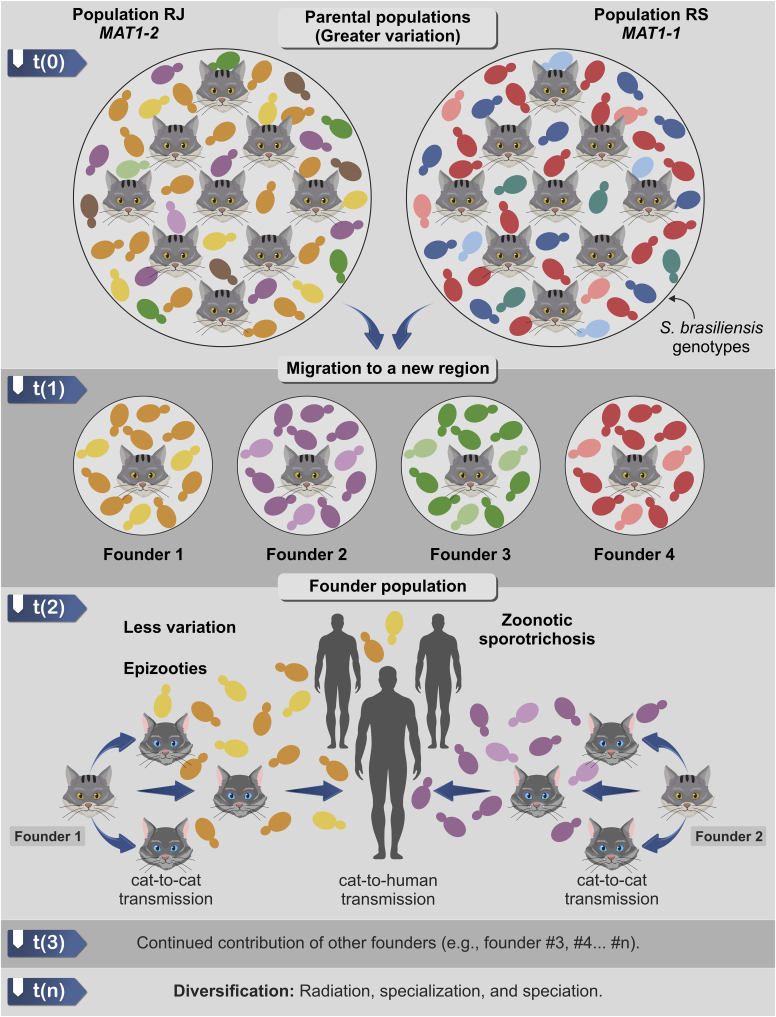
The combination of genetic diversity and population structure reveals a plethora of S. brasiliensis genotypes circulating in the Brazilian South/Southeast axis. A geographic analysis of the isolates indicates that unidirectional migration from the Southeast to Northeast Brazil is the most parsimonious explanation for the observed genetic patterns. Temporal trends reveal that S. brasiliensis was detected earlier than recent reports in the Northeast region (e.g., Ss43, Ss44, Ss244, since 1997) (Rodrigues et al. 2014, Rodrigues et al. 2016b, Rodrigues et al. 2020b). However, recent outbreaks are not related to these specific isolates but rather to the genotypes circulating in Rio de Janeiro. Thus, the introduction was probably a recent phenomenon, suggestive of a founder effect, given the low diversification of isolates in the Northeast compared to the epicentre in Rio de Janeiro (RJ). Indeed, the samples from the parental population, such as RJ, have significantly high levels of genetic diversity over small geographic distances, as RJ genotypes were hitchhiking through most of the interspecific clades (e.g., AFLP#3, clades Ia–Ik). Small, local outbreak events explain the genetic drift hypothesis after an initial migration of a small number of diseased cats into a new region. Indeed, proof for a founder effect is rarely accompanied by evidence for fast population growth, but this illustrates that founders rarely colonize a newly accessible environment and must compete with an already existing group (Böhme et al. 2007, Allcock & Strugnell 2012). However, contact patterns (e.g., scratches and bites) shown in the epizootic and zoonotic transmission chain are effective drivers of the emergence of the disease (Fig. 15). On the other hand, weak genetic differentiation among isolates from different outbreak areas resulted in their grouping into a single genetic cluster using structure.
Fig. 15.
The founder effect in Sporothrix brasiliensis. The parental population of S. brasiliensis represented by the epicentre of sporotrichosis outbreaks (in Rio de Janeiro or Rio Grande do Sul, Brazil) could give rise to different founder populations. Population genetics analysis revealed that parental populations of S. brasiliensis possess more significant genetic variation. Migration of new founder individual t(1), i.e., a diseased cat to a new region leads to direct animal horizontal transmission (cat-to-cat) and zoonotic transmission (cat-to-human), generating a founder population, which is composed of a fraction of genotypes circulating in the parental population (i.e., less variation). The absence of sanitary barriers and public police health to mitigate the epidemic leads to the continued contribution of other founders, accelerating the pace of diversification of the founder population. Such diversification via host jumps is usually followed by radiation, specialization, and speciation.
A different perspective was proposed by Eudes Filho et al. (Eudes Filho et al. 2020), which suggests that the S. brasiliensis population in the city of Brasília is extremely unlikely to be derived from the one in Rio de Janeiro, as the latter has a shallow genetic diversity, suggesting an extreme bottleneck. However, such a conclusion was founded on sequencing only two isolates from Brasília (A001 and A005) and four isolates from Rio de Janeiro (s15677, s28606, s34180, and s48605). Here we show that isolates from Rio de Janeiro are highly diverse and are scattered across dendrograms, suggesting a large contribution by the Rio de Janeiro epidemic in seeding the ongoing outbreaks taking places in other states, such as those bordering Rio de Janeiro (e.g., São Paulo, Espírito Santo, and Minas Gerais), or even those areas spanning over 2 000 km from the epicentre, such as Pernambuco, Rio Grande do Norte, and Ceará (Zhang et al. 2015, do Monte Alves et al. 2020, de Oliveira Bento et al. 2021). Therefore, combining a broader sampling and markers designed to reveal genetic variation in a temporal and spatial setting, we likely captured most of the diversity in S. brasiliensis. Additional sampling and genotyping efforts may well uncover new, rare genotypes, but substantial changes are unlikely.
Temporal and spatial information revealed the evolutionary potential to jump to new hosts, as recently witnessed in epizootic events taking place in Brazil that are caused by S. brasiliensis. Such diversification via host jumps may be followed by specialization, and speciation (Thines 2019) if gene-pools remain distinct. In an evolutionary setting, host jumps are expected to initiate following a suboptimal interaction of a pathogen with a new host and progress by adaptation to infecting the new host with higher efficacy (Thines 2019). If this holds true, it is expected that the archetypical S. brasiliensis would show lower fitness in animal infection. A low virulence profile has been observed in some plesiomorphic lineages of S. brasiliensis (e.g., Ss34, Ss67, Ss104, Ss99), which have little capacity for infection and dissemination in a murine host (Fernandes et al. 2013, Sasaki et al. 2014). Subsequently, adaptation following the initial host jump occurs when the new host can be subsequently colonized with similar efficiency to that of the original pathogen on the original host (Thines 2019). Likewise, for the host-adapted lineage of S. brasiliensis, we would expect a greater fitness in animal infection compared to the ancestral relative. In support of this argument, it has been shown that isolates where admixture analysis revealed evidence of shared ancestry (e.g., Ss174, Ss226, Ss252, Ss265) have a greater capacity for infection, dissemination, leading to impaired development and death of infected animals (Della Terra et al. 2017). This suggests that hybridization between two closely related species could lead to the emergence of genotypes that combine the virulence traits of each species (Maxwell et al. 2018).
A framework of evolutionary epidemiology theory assumes that selection for horizontal transmission is maximum at the onset of an epidemic but declines thereafter as the epidemic depletes the pool of susceptible hosts (Berngruber et al. 2013). Observational epidemiological studies indicate that severe, atypical, and refractory cases of sporotrichosis are repeatedly observed during S. brasiliensis epizootic and zoonotic episodes (Almeida-Paes et al. 2014, Montenegro et al. 2014, Sanchotene et al. 2015, Boechat et al. 2018, Nepomuceno Araújo et al. 2021), suggesting that we are perhaps at an early stage of the sporotrichosis epidemic. This pattern is likely supported by continuous founder events, leading to predominantly clonal outbreaks in a naïve host population, followed shortly by genetic expansion and diversification during geographical range expansion (Fig. 15).
Judging from the founder effect perspective, the phylogeographic patterns described here demonstrate the ability of S. brasiliensis to spread rapidly at local and regional scales, perhaps vectored by the feline host. The centre of origin of S. brasiliensis is Rio de Janeiro, based on greater genetic diversity and historical evidence (Schubach et al. 2001, Schubach et al. 2002) and geographical distribution (Rodrigues et al. 2013b). The ubiquitous presence of S. brasiliensis in epizootic events suggests that the most plausible mode of expansion of this pathogens range occurs with the migration of a small number of diseased cats into a new region. As Brazil borders ten countries in South America, it is characterized as the third largest land border globally; thus, increased biosecurity restrictions should be taken to minimize the risk of pathogen movement to neighbouring countries (via epizooty). Continued clinical and environmental sampling within and beyond the regions evaluated in this study and implementing an ecoepidemiological analysis would be required to fully understand the progression of Sporothrix.
The results of our study indicate strong signals of genetic introgression among S. brasiliensis, S. schenckii, and S. globosa with the presence of putative hybrids in these populations. Therefore, a focus on whole-genome sequencing with a higher depth of coverage would further increase the number of alleles that can be detected and provide a clearer view of the genetic diversity of these pathogens. The Chinese S. globosa population was recently divided into eight distinct clustering groups with high-resolution AFLP markers (Zhao et al. 2017) and three genetic clusters using ten microsatellites (Gong et al. 2019), which were not related to the clinical manifestations. On the other hand, in India, AFLP results exhibited low genetic diversity, and likewise, no correlation was observed between genotypes and clinical presentation or geographic distribution (Rudramurthy et al. 2021). The typical lack of hierarchical phylogenetic structuring is expected in an outbreak scenario where numerous variants can be found before they finally go extinct (Eppinger et al. 2014). Our analyses suggest that the S. brasiliensis circulating in Rio de Janeiro and Rio Grande Sul are different. This can be explained by clustering profile, neural networks, recombination, dimensioning analyses, and skewed mating-type idiomorph distribution. The fact that these variants were found throughout Brazil and even in neighbouring countries indicates the speed and extent of feline dispersal. This epizootic/zoonotic mediated distribution pattern was not limited to the onset of the outbreak, as phylogeographic patterns show a shallow population structure even within the two largest Brazilian subclades (i.e., Rio de Janeiro and Rio Grande do Sul). The mobility of the human and animal populations may continue to complicate eradication efforts in the long term (Eppinger et al. 2014).
Detection of recombination based on linkage disequilibrium (LD) pattern is influenced by diverse selection pressures (Stukenbrock & Dutheil 2018). Here, we used LD analysis to determine if Sporothrix populations are predominantly clonal or sexual. Meiotic recombination, a significant driver of rapid pathogen evolution, results in the free exchange of alleles in a panmictic sexual population (i.e., LD among loci is unexpected), which produces their random association. On the other hand, one practical consequence of the paucity or absence of sex are populations that are in significant LD (i.e., considerable LD is expected due to linkage among loci) (Little & Ebert 2000, Schurko et al. 2009, Úbeda et al. 2011, Tibayrenc & Ayala 2012, Ruggiero et al. 2018). Therefore, the low levels of LD found for Sporothrix species provide robust estimates of recombination, supporting that alleles may recombine freely into new genotypes during the process of reproduction (Hosid et al. 2010).
Molecular data suggest that members of Sporothrix are heterothallic fungi with a single mating-type locus that produces two alleles, MAT1-1 and MAT1-2, in agreement with previous reports (Teixeira et al. 2015, de Carvalho et al. 2021). The sexual development in Sporothrix has been demonstrated for isolates embedded in the environmental clade; however, such observation seems to be complex-specific. For example, sexual development has been frequently observed among members of the S. stenoceras, S. gossypina, and S. candida complexes, leading to the development of ophiostoma-like structures, such as ephemeral asci and long-necked ophiostomatoid perithecia through which the ascospores are passively discharged (de Beer & Wingfield 2013). On the other hand, members of the S. pallida complex (except for S. palmiculminata) and members of the clinical clade (i.e., S. brasiliensis, S. schenckii, S. globosa, and S. luriei) has never been linked to sexual development under laboratory conditions (Rodrigues et al. 2020b). Recombination events reported in population genetic studies could support the hypothesis of a sexual cycle leading to diversification in these pathogens (de Carvalho et al. 2020). Confirming previous results, mating-type markers were vital to tracking the emergence of Sporothrix during outbreaks, as MAT1-2 isolates are frequently associated with the Rio de Janeiro epidemic, and MAT 1-1 isolates are commonly associated with the Rio Grande do Sul epidemic. A single isolate from Rio Grande do Sul (i.e., Ss328) was characterized as MAT1-2, and it did not group with the other South clade isolates of S. brasiliensis, indicating a possible introduction from Rio de Janeiro, or even an infection by S. brasiliensis in Rio de Janeiro that was diagnosed in Rio Grande do Sul.
Murine models of infection and observational epidemiological studies demonstrated that both mating-type idiomorphs are highly virulent to the warm-blooded hosts (Montenegro et al. 2014, Sanchotene et al. 2015, Della Terra et al. 2017, Macêdo-Sales et al. 2020, Maschio-Lima et al. 2021). The skewed MAT loci distribution observed here may be related to the scarcity of sexual reproduction and/or the contact patterns in the host population (cat-to-cat and cat-to-human transmission), or even a phenomenon of small populations (Valero et al. 2018). This paradoxical reproduction system has been observed in Sporothrix (Teixeira et al. 2015, Rocha et al. 2020, de Carvalho et al. 2021), Histoplasma (Rodrigues et al. 2020a), Paracoccidioides (Roberto et al. 2021), and Cryptococcus (Nielsen et al. 2005), with species prevalently clonal along with recombinant molecular siblings coexisting in the same geographical range.
Conclusions
Our approach highlights AFLP as a powerful and inexpensive tool to explore genetic diversity in medically relevant Sporothrix species. For the first time in S. brasiliensis, S. schenckii, and S. globosa, patterns of admixture were detected, which may explain the sudden emergence of genotypes with increased virulence traits. Our ability to reconstruct the source, spread, and evolution of the ongoing outbreaks from molecular data provides essential information for decision-making to mitigate the progression of the disease. Our analyses support the expansion of S. brasiliensis in Brazil and identify Rio de Janeiro as the most likely centre of origin. This region then appears to have vectored the infection to other country regions as seen by the occurrence of genetic founder effects. The detection of Sporothrix in frontier countries reinforces the need for biosecurity measures to contain further spread. These measures include surveillance, rapid diagnosis, follow-up of the cases, access to appropriate antifungal treatment, and education of the population about sporotrichosis. Moreover, our study highlights the urgency of improving the availability of molecular diagnosis to speciate Sporothrix and identify sources of epizootic and zoonotic pathogens that can more widely threaten animals and humans.
Acknowledgements
The authors acknowledge the financial support granted by the São Paulo Research Foundation (FAPESP 2017/27265-5 and FAPESP 2018/21460-3), National Council for Scientific and Technological Development (CNPq 433276/2018-5), and Coordination for the Improvement of Higher Education Personnel (CAPES 88887.159096/2017-00). AMR is a CNPq Research Productivity Fellow (CNPq 304902/2020-9). SAP is a CNPq Research Productivity Fellow (CNPq 312238/2020-7). MAB was funded by the Wellcome Trust (#206194). MCF was funded by the UK Medical Research Council and Wellcome Trust and is a Fellow in the CIFAR ‘Fungal Kingdoms’ program. For the purpose of Open Access, the author has applied a CC-BY public copyright license to any Author Accepted Manuscript version arising from this submission. The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2021.100129.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Al-Hatmi A.M., Hagen F., Menken S.B., et al. Global molecular epidemiology and genetic diversity of Fusarium, a significant emerging group of human opportunists from 1958 to 2015. Emerging Microbes & Infections. 2016;5 doi: 10.1038/emi.2016.126. e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A., Wools K.K. Disseminated sporotrichosis and Sporothrix schenckii fungemia as the initial presentation of Human Immunodeficiency Virus infection. Clinical Infectious Diseases. 1998;26:1403–1406. doi: 10.1086/516356. [DOI] [PubMed] [Google Scholar]
- Aldama A., Aldama J.G., Pereira J. [Value of molecular techniques and risk factors in the diagnosis and evolution of sporotrichosis. About 2 cases of Sporothrix brasiliensis and S. globosa] Anales de la Facultad de Ciencias Médicas (Asunción) 2020;53:177–184. [Google Scholar]
- Allcock A.L., Strugnell J.M. Southern Ocean diversity: new paradigms from molecular ecology. Trends in Ecology & Evolution. 2012;27:520–528. doi: 10.1016/j.tree.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Almeida-Paes R., de Oliveira M.M., Freitas D.F., et al. Sporotrichosis in Rio de Janeiro, Brazil: Sporothrix brasiliensis is associated with atypical clinical presentations. PLoS Neglected Tropical Diseases. 2014;8 doi: 10.1371/journal.pntd.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman D.G. Chapman and Hall; London: 1991. Practical statistics for medical research; p. 624. [Google Scholar]
- Arrillaga-Moncrieff I., Capilla J., Mayayo E., et al. Different virulence levels of the species of Sporothrix in a murine model. Clinical Microbiology and Infection. 2009;15:651–655. doi: 10.1111/j.1469-0691.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- Berngruber T.W., Froissart R., Choisy M., et al. Evolution of virulence in emerging epidemics. PLoS Pathogens. 2013;9 doi: 10.1371/journal.ppat.1003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boechat J.S., Oliveira M.M.E., Almeida-Paes R., et al. Feline sporotrichosis: associations between clinical-epidemiological profiles and phenotypic-genotypic characteristics of the etiological agents in the Rio de Janeiro epizootic area. Memorias do Instituto Oswaldo Cruz. 2018;113:185–196. doi: 10.1590/0074-02760170407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme M.U., Schneeweiß N., Fritz U., et al. Small edge populations at risk: genetic diversity of the green lizard (Lacerta viridis viridis) in Germany and implications for conservation management. Conservation Genetics. 2007;8:555–563. [Google Scholar]
- Bommer M., Hütter M.-L., Stilgenbauer S., et al. Fatal Ophiostoma piceae infection in a patient with acute lymphoblastic leukaemia. Journal of Medical Microbiology. 2009;58:381–385. doi: 10.1099/jmm.0.005280-0. [DOI] [PubMed] [Google Scholar]
- Botstein D., White R.L., Skolnick M., et al. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Bradbury P.J., Zhang Z., Kroon D.E., et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Brilhante R.S., Silva N.F., Lima R.A., et al. Easy storage strategies for Sporothrix spp. strains. Biopreservation and Biobanking. 2015;13:131–134. doi: 10.1089/bio.2014.0071. [DOI] [PubMed] [Google Scholar]
- Bryant D., Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A., Bonifaz A., Gutierrez-Galhardo M.C., et al. Global epidemiology of sporotrichosis. Medical Mycology. 2015;53:3–14. doi: 10.1093/mmy/myu062. [DOI] [PubMed] [Google Scholar]
- Chen Y., Tong D., Wu C.I. A new formulation of random genetic drift and its application to the evolution of cell populations. Molecular Biology and Evolution. 2017;34:2057–2064. doi: 10.1093/molbev/msx161. [DOI] [PubMed] [Google Scholar]
- Córdoba S., Isla G., Szusz W., et al. Molecular identification and susceptibility profile of Sporothrix schenckii sensu lato isolated in Argentina. Mycoses. 2018;61:441–448. doi: 10.1111/myc.12760. [DOI] [PubMed] [Google Scholar]
- de Beer Z.W., Duong T.A., Wingfield M.J. The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Studies in Mycology. 2016;83:165–191. doi: 10.1016/j.simyco.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer Z.W., Wingfield M.J. In: The Ophiostomatoid Fungi: Expanding Frontiers. Seifert K.A., de Beer Z.W., andWingfield M.J., editors. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2013. Emerging lineages in the Ophiostomatales; pp. 21–46. [Google Scholar]
- de Carvalho J.A., Hagen F., Fisher M.C., et al. Genome-wide mapping using new AFLP markers to explore intraspecific variation among pathogenic Sporothrix species. PLoS Neglected Tropical Diseases. 2020;14 doi: 10.1371/journal.pntd.0008330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho J.A., Pinheiro B.G., Hagen F., et al. A new duplex PCR assay for the rapid screening of mating-type idiomorphs of pathogenic Sporothrix species. Fungal Biology. 2021;125:834–843. doi: 10.1016/j.funbio.2021.05.005. [DOI] [PubMed] [Google Scholar]
- de Groot T., Puts Y., Berrio I., et al. Development of Candida auris short tandem repeat typing and its application to a global collection of isolates. MBio. 2020;11 doi: 10.1128/mBio.02971-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Bento A., de Sena Costa A.S., Lima S.L., et al. The spread of cat-transmitted sporotrichosis due to Sporothrix brasiliensis in Brazil towards the Northeast region. PLoS Neglected Tropical Diseases. 2021;15 doi: 10.1371/journal.pntd.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne D.M., Giraud T., Martin O.C. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;23:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- Della Terra P.P., Rodrigues A.M., Fernandes G.F., et al. Exploring virulence and immunogenicity in the emerging pathogen Sporothrix brasiliensis. PLoS Neglected Tropical Diseases. 2017;11 doi: 10.1371/journal.pntd.0005903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Monte Alves M., Pipolo Milan E., da Silva-Rocha W.P., et al. Fatal pulmonary sporotrichosis caused by Sporothrix brasiliensis in Northeast Brazil. PLoS Neglected Tropical Diseases. 2020;14 doi: 10.1371/journal.pntd.0008141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl D.A., vonHoldt B.M. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Ekroth A.K.E., Gerth M., Stevens E.J., et al. Host genotype and genetic diversity shape the evolution of a novel bacterial infection. The ISME Journal. 2021;15:2146–2157. doi: 10.1038/s41396-021-00911-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinger M., Pearson T., Koenig S.S.K., et al. Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic. MBio. 2014;5 doi: 10.1128/mBio.01721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchecopaz A.N., Lanza N., Toscanini M.A., et al. Sporotrichosis caused by Sporothrix brasiliensis in Argentina: Case report, molecular identification and in vitro susceptibility pattern to antifungal drugs. Journal of Medical Mycology. 2019;30:100908. doi: 10.1016/j.mycmed.2019.100908. [DOI] [PubMed] [Google Scholar]
- Eudes Filho J., Santos I.B.D., Reis C.M.S., et al. A novel Sporothrix brasiliensis genomic variant in Midwestern Brazil: evidence for an older and wider sporotrichosis epidemic. Emerging Microbes & Infections. 2020;9:2515–2525. doi: 10.1080/22221751.2020.1847001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fernandes G.F., dos Santos P.O., Rodrigues A.M., et al. Characterization of virulence profile, protein secretion and immunogenicity of different Sporothrix schenckii sensu stricto isolates compared with S. globosa and S. brasiliensis species. Virulence. 2013;4:241–249. doi: 10.4161/viru.23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Duarte J.M., Wattiez Acosta V.R., Fornerón Viera P.M.L., et al. Sporotrichosis transmitted by domestic cat. A family case report. Revista del Nacional. 2017;9:67–76. [Google Scholar]
- Gong J., Zhang M., Wang Y., et al. Population structure and genetic diversity of Sporothrix globosa in China according to 10 novel microsatellite loci. Journal of Medical Microbiology. 2019;68:248–254. doi: 10.1099/jmm.0.000896. [DOI] [PubMed] [Google Scholar]
- Gremião I.D., Menezes R.C., Schubach T.M., et al. Feline sporotrichosis: epidemiological and clinical aspects. Medical Mycology. 2015;53:15–21. doi: 10.1093/mmy/myu061. [DOI] [PubMed] [Google Scholar]
- Gremião I.D., Miranda L.H., Reis E.G., et al. Zoonotic epidemic of sporotrichosis: Cat to human transmission. PLoS Pathogens. 2017;13 doi: 10.1371/journal.ppat.1006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming R.W. Error detecting and error correcting codes. The Bell System Technical Journal. 1950;29:147–160. [Google Scholar]
- Hektoen L., Perkins C.F. Refractory subcutaneous abscesses caused by Sporothrix schenckii: A new pathogenic fungus. Journal of Experimental Medicine. 1900;5:77–89. doi: 10.1084/jem.5.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosid E., Grishkan I., Yusim E., et al. The mode of reproduction in natural populations of ascomycetous fungus, Emericella nidulans, from Israel. Genetics Research. 2010;92:83–90. doi: 10.1017/S0016672310000066. [DOI] [PubMed] [Google Scholar]
- Huang L., Gao W., Giosa D., et al. Whole-genome sequencing and in silico analysis of two strains of Sporothrix globosa. Genome Biology and Evolution. 2016;8:3292–3296. doi: 10.1093/gbe/evw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Kloepper T.H. Computing recombination networks from binary sequences. Bioinformatics. 2005;21:ii159–ii165. doi: 10.1093/bioinformatics/bti1126. [DOI] [PubMed] [Google Scholar]
- Jaccard P. The distribution of the flora in the Alpine. New Phytologist. 1912;11:37–50. [Google Scholar]
- Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Anzawa K., Mochizuki T., et al. New strain typing method with Sporothrix schenckii using mitochondrial DNA and polymerase chain reaction restriction fragment length polymorphism (PCR–RFLP) technique. Journal of Dermatology. 2012;39:362–365. doi: 10.1111/j.1346-8138.2011.01379.x. [DOI] [PubMed] [Google Scholar]
- Kitani E.C., Hernandez E.D.M., Thomaz C.E., et al. Eleventh Brazilian Symposium on Neural Networks. Institute of Electrical and Electronics Engineers; 2010. Visual Interpretation of Self Organizing Maps; pp. 37–42. [Google Scholar]
- Kohonen T. Self-Organizing Maps. Springer Series in Information Sciences. 2001;30:502. [Google Scholar]
- Little T.J., Ebert D. Sex, linkage disequilibrium and patterns of parasitism in three species of cyclically parthenogenetic Daphnia (Cladocera: Crustacea) Heredity. 2000;85:257–265. doi: 10.1046/j.1365-2540.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Liu B.H. CRC press; Boca Raton: 1998. Statistical genomics: linkage, mapping, and QTL analysis. [Google Scholar]
- Lockhart S.R., Etienne K.A., Vallabhaneni S., et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clinical Infectious Diseases. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz A., Splendore A. On a mycosis observed in men and mice: Contribution to the knowledge of the so-called sporotrichosis. Revista Médica de São Paulo. 1907;21:443–450. [Google Scholar]
- Lynch M., Milligan B.G. Analysis of population genetic structure with RAPD markers. Molecular Ecology. 1994;3:91–99. doi: 10.1111/j.1365-294x.1994.tb00109.x. [DOI] [PubMed] [Google Scholar]
- Macêdo-Sales P.A., Souto S.R.L.S., Destefani C.A., et al. Domestic feline contribution in the transmission of Sporothrix in Rio de Janeiro State, Brazil: a comparison between infected and non-infected populations. BMC Veterinary Research. 2018;14:19. doi: 10.1186/s12917-018-1340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macêdo-Sales P.A., Souza L.O.P., Della-Terra P.P., et al. Coinfection of domestic felines by distinct Sporothrix brasiliensis in the Brazilian sporotrichosis hyperendemic area. Fungal Genetics and Biology. 2020;140:103397. doi: 10.1016/j.fgb.2020.103397. [DOI] [PubMed] [Google Scholar]
- Madrid H., Gené J., Cano J., et al. Sporothrix brunneoviolacea and Sporothrix dimorphospora, two new members of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia. 2010;102:1193–1203. doi: 10.3852/09-320. [DOI] [PubMed] [Google Scholar]
- Makri N., Paterson G.K., Gregge F., et al. First case report of cutaneous sporotrichosis (Sporothrix species) in a cat in the UK. Journal of Feline Medicine and Surgery Open Reports. 2020;6 doi: 10.1177/2055116920906001. 2055116920906001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R., Cano J., Gené J., et al. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. Journal of Clinical Microbiology. 2007;45:3198–3206. doi: 10.1128/JCM.00808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R., Gené J., Cano J., et al. Sporothrix luriei: a rare fungus from clinical origin. Medical Mycology. 2008;46:621–625. doi: 10.1080/13693780801992837. [DOI] [PubMed] [Google Scholar]
- Marimon R., Gené J., Cano J., et al. Molecular phylogeny of Sporothrix schenckii. Journal of Clinical Microbiology. 2006;44:3251–3256. doi: 10.1128/JCM.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschio-Lima T., Marques M.D.R., Lemes T.H., et al. Clinical and epidemiological aspects of feline sporotrichosis caused by Sporothrix brasiliensis and in vitro antifungal susceptibility. Veterinary Research Communications. 2021;45:171–179. doi: 10.1007/s11259-021-09795-2. [DOI] [PubMed] [Google Scholar]
- Maxwell C.S., Sepulveda V.E., Turissini D.A., et al. Recent admixture between species of the fungal pathogen Histoplasma. Evolution Letters. 2018;2:210–220. doi: 10.1002/evl3.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B.A., Linde C. Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology. 2002;40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- McVean G. A genealogical interpretation of Principal Components Analysis. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro H., Rodrigues A.M., Galvão Dias M.A., et al. Feline sporotrichosis due to Sporothrix brasiliensis: an emerging animal infection in São Paulo, Brazil. BMC Veterinary Research. 2014;10:269. doi: 10.1186/s12917-014-0269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa T.A., Kadasa N.M., Al Zahrani H.S., et al. Origin and distribution of Sporothrix globosa causing sapronoses in Asia. Journal of Medical Microbiology. 2017;66:560–569. doi: 10.1099/jmm.0.000451. [DOI] [PubMed] [Google Scholar]
- Nepomuceno Araújo M., Nihei C.H., Rodrigues A.M., et al. Case Report: Invasive sinusitis due to Sporothrix brasiliensis in a renal transplant recipient. The American Journal of Tropical Medicine and Hygiene. 2021;105:1218–1221. doi: 10.4269/ajtmh.20-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New D., Beukers A.G., Kidd S.E., et al. Identification of multiple species and subpopulations among Australian clinical Sporothrix isolates using whole genome sequencing. Medical Mycology. 2019;57:905–908. doi: 10.1093/mmy/myy126. [DOI] [PubMed] [Google Scholar]
- Nielsen K., Marra R.E., Hagen F., et al. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics. 2005;171:975–983. doi: 10.1534/genetics.105.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orofino-Costa R.C., Macedo P.M., Rodrigues A.M., et al. Sporotrichosis: an update on epidemiology, etiopathogenesis, laboratory and clinical therapeutics. Anais Brasileiros de Dermatologia. 2017;92:606–620. doi: 10.1590/abd1806-4841.2017279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAHO . Pan American Health Organization; 2019. Sporothrix brasiliensis, an emerging fungal pathogen, notable for its zoonotic transmission and epidemic potential for human and animal health in the Americas; pp. 1–4. [Google Scholar]
- Pappas P.G., Tellez I., Deep A.E., et al. Sporotrichosis in Peru: Description of an area of hyperendemicity. Clinical Infectious Diseases. 2000;30:65–70. doi: 10.1086/313607. [DOI] [PubMed] [Google Scholar]
- Peakall R., Smouse P.E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R.O.D., Smouse P.E. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S.A., Passos S.R., Silva J.N., et al. Response to azolic antifungal agents for treating feline sporotrichosis. Veterinary Record. 2010;166:290–294. doi: 10.1136/vr.166.10.290. [DOI] [PubMed] [Google Scholar]
- Powell W., Morgante M., Andre C., et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding. 1996;2:225–238. [Google Scholar]
- Prevost A., Wilkinson M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical and Applied Genetics. 1999;98:107–112. [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz-Telles F., Fahal A.H., Falci D.R., et al. Neglected endemic mycoses. The Lancet Infectious Diseases. 2017;17:e367–e377. doi: 10.1016/S1473-3099(17)30306-7. [DOI] [PubMed] [Google Scholar]
- Rangel-Gamboa L., Martinez-Hernandez F., Maravilla P., et al. Update of phylogenetic and genetic diversity of Sporothrix schenckii sensu lato. Medical Mycology. 2016;54:248–255. doi: 10.1093/mmy/myv096. [DOI] [PubMed] [Google Scholar]
- Rios M.E., Suarez J.M.D., Moreno J., et al. Zoonotic sporotrichosis related to cat contact: first case report from Panama in Central America. Cureus. 2018;10 doi: 10.7759/cureus.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippon J.W. B. Saunders Company; Philadelphia, PA: 1988. Medical Mycology – The pathogenic fungi and the pathogenic actinomycetes W. [Google Scholar]
- Roberto T.N., De Carvalho J.A., Beale M.A., et al. Exploring genetic diversity, population structure, and phylogeography in Paracoccidioides species using AFLP markers. Studies in Mycology. 2021;100:100131. doi: 10.1016/j.simyco.2021.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha I.C.B., Terra P.P.D., Cardoso de Oliveira R., et al. Molecular-based assessment of diversity and population structure of Sporothrix spp. clinical isolates from Espirito Santo-Brazil. Mycoses. 2021 Apr;64(4):420–427. doi: 10.1111/myc.13230. Epub 2020 Dec 28. [DOI] [PubMed] [Google Scholar]
- Rodrigues A.M., Beale M.A., Hagen F., et al. The global epidemiology of emerging Histoplasma species in recent years. Studies in Mycology. 2020;97:100095. doi: 10.1016/j.simyco.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., Cruz Choappa R., Fernandes G.F., et al. Sporothrix chilensis sp. nov. (Ascomycota: Ophiostomatales), a soil-borne agent of human sporotrichosis with mild-pathogenic potential to mammals. Fungal Biology. 2016;120:246–264. doi: 10.1016/j.funbio.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Rodrigues A.M., de Hoog G.S., de Camargo Z.P. Molecular diagnosis of pathogenic Sporothrix species. PLoS Neglected Tropical Diseases. 2015;9 doi: 10.1371/journal.pntd.0004190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., de Hoog G.S., de Camargo Z.P. Sporothrix species causing outbreaks in animals and humans driven by animal-animal transmission. PLoS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., de Hoog G.S., de Camargo Z.P. In: Emerging and Epizootic Fungal Infections in Animals. Seyedmousavi S., de Hoog G.S., Guillot J., Verweij P.E., editors. Springer International Publishing; Cham: 2018. Feline Sporotrichosis; pp. 199–231. [Google Scholar]
- Rodrigues A.M., de Hoog G.S., Zhang Y., et al. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerging Microbes & Infections. 2014;3:e32. doi: 10.1038/emi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., de Hoog S., de Camargo Z.P. Emergence of pathogenicity in the Sporothrix schenckii complex. Medical Mycology. 2013;51:405–412. doi: 10.3109/13693786.2012.719648. [DOI] [PubMed] [Google Scholar]
- Rodrigues A.M., de Melo Teixeira M., de Hoog G.S., et al. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Neglected Tropical Diseases. 2013;7 doi: 10.1371/journal.pntd.0002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.M., Della Terra P.P., Gremiao I.D., et al. The threat of emerging and re-emerging pathogenic Sporothrix species. Mycopathologia. 2020;185:813–842. doi: 10.1007/s11046-020-00425-0. [DOI] [PubMed] [Google Scholar]
- Rodrigues A.M., Fernandes G.F., de Camargo Z.P. In: Emerging and re-emerging infectious diseases of livestock. Bayry J., editor. Springer; 2017. Sporotrichosis; pp. 391–421. [Google Scholar]
- Rudramurthy S.M., Shankarnarayan S.A., Hemashetter B.M., et al. Phenotypic and molecular characterisation of Sporothrix globosa of diverse origin from India. Brazilian Journal of Microbiology. 2021;52:91–100. doi: 10.1007/s42770-020-00346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero M.V., D'Alelio D., Ferrante M.I., et al. Clonal expansion behind a marine diatom bloom. The ISME Journal. 2018;12:463–472. doi: 10.1038/ismej.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salipante S.J., Hall B.G. Inadequacies of minimum spanning trees in molecular epidemiology. Journal of Clinical Microbiology. 2011;49:3568–3575. doi: 10.1128/JCM.00919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Cold Spring Harbor Laboratory, Cold Spring Harbor; N.Y: 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- Sanchotene K.O., Madrid I.M., Klafke G.B., et al. Sporothrix brasiliensis outbreaks and the rapid emergence of feline sporotrichosis. Mycoses. 2015;58:652–658. doi: 10.1111/myc.12414. [DOI] [PubMed] [Google Scholar]
- Sasaki A.A., Fernandes G.F., Rodrigues A.M., et al. Chromosomal polymorphism in the Sporothrix schenckii complex. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0086819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober P., Boer C., Schwarte L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesthesia & Analgesia. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- Schubach T.M., de Oliveira Schubach A., dos Reis R.S., et al. Sporothrix schenckii isolated from domestic cats with and without sporotrichosis in Rio de Janeiro, Brazil. Mycopathologia. 2002;153:83–86. doi: 10.1023/a:1014449621732. [DOI] [PubMed] [Google Scholar]
- Schubach T.M., Schubach A., Okamoto T., et al. Evaluation of an epidemic of sporotrichosis in cats: 347 cases (1998-2001) Journal of the American Veterinary Medical Association. 2004;224:1623–1629. doi: 10.2460/javma.2004.224.1623. [DOI] [PubMed] [Google Scholar]
- Schubach T.M., Schubach A., Okamoto T., et al. Haematogenous spread of Sporothrix schenckii in cats with naturally acquired sporotrichosis. Journal of Small Animal Practice. 2003;44:395–398. doi: 10.1111/j.1748-5827.2003.tb00174.x. [DOI] [PubMed] [Google Scholar]
- Schubach T.M., Valle A.C., Gutierrez-Galhardo M.C., et al. Isolation of Sporothrix schenckii from the nails of domestic cats (Felis catus) Medical Mycology. 2001;39:147–149. doi: 10.1080/mmy.39.1.147.149. [DOI] [PubMed] [Google Scholar]
- Schurko A.M., Neiman M., Logsdon J.M. Jr. Signs of sex: what we know and how we know it. Trends in Ecology & Evolution. 2009;24:208–217. doi: 10.1016/j.tree.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Seyedmousavi S., Bosco SdMG., de Hoog S., et al. Fungal infections in animals: a patchwork of different situations. Medical Mycology. 2018;56:165–187. doi: 10.1093/mmy/myx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- Silva-Vergara M.L., de Camargo Z.P., Silva P.F., et al. Disseminated Sporothrix brasiliensis infection with endocardial and ocular involvement in an HIV-infected patient. The American Journal of Tropical Medicine and Hygiene. 2012;86:477–480. doi: 10.4269/ajtmh.2012.11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. Measurement of Diversity. Nature. 1949;163 [Google Scholar]
- Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nature Reviews Genetics. 2008;9:477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock E.H., Dutheil J.Y. Fine-scale recombination maps of fungal plant pathogens reveal dynamic recombination landscapes and intragenic hotspots. Genetics. 2018;208:1209–1229. doi: 10.1534/genetics.117.300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira H., Rodríguez-Echeverría S., Nabais C. Genetic diversity and differentiation of Juniperus thurifera in Spain and Morocco as determined by SSR. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0088996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MdM., Rodrigues A.M., Tsui C.K.M., et al. Asexual propagation of a virulent clone complex in human and feline outbreak of sporotrichosis. Eukaryotic Cell. 2015;14:158–169. doi: 10.1128/EC.00153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier C., David J., This P., et al. Optimization of the choice of molecular markers for varietal identification in Vitis vinifera L. Theoretical and Applied Genetics. 1999;98:171–177. [Google Scholar]
- The R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A Language and Environment for Statistical Computing.http://wwwR-projectorg/ 2014. [Google Scholar]
- Thines M. An evolutionary framework for host shifts–jumping ships for survival. New Phytologist. 2019;224:605–617. doi: 10.1111/nph.16092. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3305–3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úbeda F., Haig D., Patten M.M. Stable linkage disequilibrium owing to sexual antagonism. Proceedings of the Royal Society B: Biological Sciences. 2011;278:855–862. doi: 10.1098/rspb.2010.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero C., Gago S., Monteiro M.C., et al. African histoplasmosis: new clinical and microbiological insights. Medical Mycology. 2018;56:51–59. doi: 10.1093/mmy/myx020. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., Chabane K., Hendre P.S., et al. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Science. 2007;173:638–649. [Google Scholar]
- Vatanshenassan M., Boekhout T., Mauder N., et al. Evaluation of microsatellite typing, ITS Sequencing, AFLP fingerprinting, MALDI-TOF MS, and fourier-transform infrared spectroscopy analysis of Candida auris. Journal of Fungi (Basel) 2020;6:146. doi: 10.3390/jof6030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S., et al. In: PCR Protocols: A Guide to Methods and Applications. Innis M., Gelfand D., Shinsky J., White T., editors. Academic Press; New York: 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Wickham H. Springer; Cham: XVI: 2016. ggplot2: Elegant Graphics for Data Analysis; p. 260. (Use R!). [Google Scholar]
- Zhang Y., Hagen F., Stielow B., et al. Phylogeography and evolutionary patterns in Sporothrix spanning more than 14,000 human and animal case reports. Persoonia. 2015;35:1–20. doi: 10.3767/003158515X687416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Cui Y., Zhen Y., et al. Genetic variation of Sporothrix globosa isolates from diverse geographic and clinical origins in China. Emerging Microbes & Infections. 2017;6:e88. doi: 10.1038/emi.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Rodrigues A.M., Feng P., et al. Global ITS diversity in the Sporothrix schenckii complex. Fungal Diversity. 2014;66:153–165. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.