Abstract
Metastasis is the main cause of cervical cancer lethality, but to date, no effective treatment has been developed to block metastasis. Circular RNAs (circRNAs) were recently found to be involved in cancer metastasis. In this study, we identified a downregulated circRNA derived from the host gene Gli1 (hsa_circ_0005358) in cervical cancer tissues, which was expressed at lower levels in tissues with extracervical metastasis than in those without extracervical metastasis. Upregulation of hsa_circ_0005358 significantly suppressed the migration and invasion of cervical cancer cells in vitro, and downregulation of hsa_circ_0005358 had the opposite effect. A mouse model revealed that cervical cancer cells overexpressing hsa_circ_0005358 possessed weaker metastatic potential in vivo. RNA-pull-down assay, mass spectrometry, and RNA immunoprecipitation validated the findings that hsa_circ_0005358 functions via its 215–224 sequence, which interacts with polypyrimidine tract-binding protein 1 (PTBP1). RNA-sequencing profiling revealed that CUB-domain-containing protein 1 (CDCP1) is a common target for hsa_circ_0005358 and PTBP1. We further confirmed that hsa_circ_0005358 sequestered PTBP1, preventing it from stabilizing CDCP1 mRNA, reducing CDCP1 protein translation and ultimately suppressing cancer metastasis. Our findings reveal the function of hsa_circ_0005358 in tumor metastasis, which may be applied to a potential therapeutic approach for patients with metastatic cervical cancer.
Keywords: circular RNA, hsa_circ_0005358, PTBP1, CDCP1, metastasis, cervical cancer
Graphical abstract

Cen et al. provide evidence of circRNA hsa_circ_0005358 modulating metastatic behavior of cervical cancer by serving as a protein decoy. This offers a potential therapeutic approach for advanced cervical cancer patients.
Introduction
Cervical cancer ranks as the fourth most common malignant tumor in females, and squamous cell carcinoma (SCC) accounts for 80% of all cases.1 Approximately 90% of deaths caused by cervical cancer occur in developing countries,2 where women lack access to cancer screening and human papillomavirus vaccination, and many patients are first diagnosed at an advanced stage with pelvic and/or lymph node metastasis.3,4 Unfortunately, current treatments, namely, radiotherapy and chemoradiotherapy, yield unsatisfactory results for advanced cervical cancer.3 The 5-year survival rate of cervical cancer patients at an advanced stage was reported to be only 17.8% (SEER:2011–2017, All Races). Exploring the mechanism of tumor metastasis will advance the development of new strategies to block the metastasis of cervical cancer.
Metastasis is one of the main causes of cancer-related deaths,5 including cervical cancer deaths. Various mechanisms associated with tumor metastasis have been demonstrated, such as the epithelial-to-mesenchymal transition (EMT),6 the cancer stem cell theory,7,8 a remodeled tumor microenvironment,9 and others.10,11 Recently, the involvement of circular RNAs (circRNAs) in tumor metastasis has attracted considerable attention.12 circRNAs are non-coding RNAs characterized by a covalently closed loop without a 5′ end cap or 3′ poly(A) tail.13 The distribution of circRNAs reveals their functional specificity in diverse tissues and different diseases.14 Recent studies have shown that the same circRNA affects metastasis differently in different tumors. For example, the classic circRNA CDR1as promotes metastasis of lung squamous carcinoma and can be inhibited by miR-671-5p.15 In contrast, the deletion of CDR1as facilitates the invasion of melanoma in an IGF2BP3-mediated manner.16 These studies suggest a complicated role for circRNAs in cancer metastasis. Moreover, the competitive endogenous RNA (ceRNA) network has been recognized as a major mechanism by which circRNAs function,17 yet the capacity of circRNAs to affect molecules in addition to microRNAs is likely underestimated. Therefore, it is necessary to investigate circRNA involvement in cervical cancer metastasis to discover a potential therapeutic approach for cervical cancer.
In this study, we identified a circRNA derived from the Gli1 gene (circBase: hsa_circ_0005358) that is significantly downregulated in squamous cervical cancer tissues compared with normal cervical tissues. Gene function experiments showed that hsa_circ_0005358 functioned as a suppressor of the migration and invasion of cervical cancer in vitro and in vivo. Mechanistically, hsa_circ_0005358 bound directly to an RNA-binding protein (RBP), polypyrimidine tract-binding protein 1 (PTBP1), at sequence 215–224 and served as a decoy to intervene in the regulation by PTBP1 of CUB-domain-containing protein 1 (CDCP1), specifically blocking the metastasis-promoting ability of CDCP1. Our findings provide new evidence that circRNAs modulate tumor metastasis and suggest that hsa_circ_0005358 might be a potential agent that blocks the metastasis of cervical cancer.
Results
Identification and characteristics of hsa_circ_0005358
In our previous study,18 circRNA sequencing profiling (GEO: GSE147009) was performed with a set of 12 cervical samples, including 6 normal cervical tissues and 6 cervical SCC tissues. The results of the sequencing yielded a total of 257 circRNAs that were significantly downregulated in cervical cancer tissues (the threshold values were |log2FC (fold change)| ≥ 2 and p < 0.05) (Figure 1A). We focused on the 15 most differentially downregulated circRNAs in SCC (Table S1). Of these circRNAs, we excluded circRNAs expressed at insufficiently low levels and those of intron origin, and amplified putative back-splice junctions of the remaining 9 candidates using divergent primers and polymerase chain reaction (PCR) (Figure S1A). Considering that hsa_circ_0005358 was one of the most significantly downregulated circRNAs in cervical cancer tissues and showed the greatest specificity among the divergent primers, we chose hsa_circ_0005358 for further investigation.
Figure 1.
Identification and characteristics of hsa_circ_0005358
(A) The heatmap of the differentially downregulated circRNAs in six human squamous cervical cancer tissues compared with six normal cervical tissues. The line indicates hsa_circ_0005005358. (B) Schematic illustration of the generation of hsa_circ_0005358. hsa_circ_0005358 is back-spliced from the Gli1 gene. The existence of hsa_circ_0005358 was validated by Sanger sequencing. The black arrow indicates the splicing site of hsa_circ_0005358. (C) Linear and back-splicing products were amplified using convergent and divergent primers of gDNA and cDNA to validate the existence of hsa_circ_0005358 in SiHa and CaSki cells. (D) Full-length hsa_circ_0005358 was amplified using two pairs of different primers and PCR. (E) Northern blot analysis using a probe targeting the specific junction of hsa_circ_0005358 verified the endogenous existence of hsa_circ_0005358 in SiHa and CaSki cells. (F) The expression of hsa_circ_0005358 and Gli1 mRNA in SiHa and CaSki cells was detected by qRT-PCR analysis after RNase R treatment (Student's t test). (G) A fluorescence in situ hybridization (FISH) assay was conducted to determine the subcellular localization of hsa_circ_0005358. Scale bar, 10 μm. (H) The distribution of hsa_circ_0005358, GAPDH, and U1 in the cytoplasmic and nuclear fractions of SiHa (top) and CaSki (bottom) cells (Student's t test). The data were represented as means ± SEM, n = 3. ∗∗∗∗p < 0.0001.
The genomic structure indicates that hsa_circ_0005358 is generated from the human Gli1 gene (chr12:57,861,115–57,862,007) (Figure 1B). Divergent primers were designed to detect the specific junction of hsa_circ_0005358, distinguishing it from its linear mRNA counterpart. We performed PCR using complementary DNA (cDNA) and genomic DNA (gDNA) of two cervical cancer cell lines, SiHa and CaSki. Without a specific circular junction, gDNA cannot amplify PCR products when divergent primers are used (Figure 1C). We also designed two pairs of primers to amplify the full-length hsa_circ_0005358 and validated the amplified product by DNA gel electrophoresis (Figure 1D). The Sanger sequencing results (Table S2) confirmed the full-length sequence of hsa_circ_0005358 with the intron region of the host gene spliced out. Endogenous expression of hsa_circ_0005358 was also detected in SiHa and CaSki cells using a probe targeting specific junctions in a northern blot assay (Figure 1E). Moreover, after RNase R exonuclease treatment, the mRNA expression of its host gene, Gli1, was significantly decreased, and hsa_circ_0005358 was resistant to RNase R digestion due to the lack of a 5′ end cap and 3′ poly(A) tail (Figure 1F), suggesting that hsa_circ_0005358 is a stable RNA with a circular structure. RNA fluorescence in situ hybridization (FISH) (Figure 1G) and subcellular fractionation assays (Figure 1H) revealed that hsa_circ_0005358 was mainly located in the nucleus, but also in the cytoplasm, of SiHa and CaSki cells. RNA in situ hybridization assays showed that cancer cells, but not stromal cells, expressed hsa_circ_0005358 in cervical cancer tissues (Figure S1B). Taken together, the findings indicate that hsa_circ_0005358 is a stable circRNA in cervical cancer cells.
hsa_circ_0005358 acts as a suppressor of metastatic behaviors of cervical cancer in vitro and in vivo
To explore the biological function of hsa_circ_0005358, we constructed a plasmid to overexpress hsa_circ_0005358. Northern blot analysis confirmed that this overexpression vector did not produce linear products (Figure S2A). qRT-PCR confirmed the overexpression efficiency in SiHa and CaSki cells (Figure S2B). Transwell assays were performed to detect the migration and invasion ability of cells. After hsa_circ_0005358 was overexpressed, the migration and invasion rates of both SiHa and CaSki cells were significantly decreased compared with the rate of the control cells (Figures 2A and 2B). Wound healing assays showed that the overexpression of hsa_circ_0005358 resulted in slower gap closure (Figure 2C). In addition, CCK-8 assays showed no significant differences in proliferation between cells with and without hsa_circ_0005358 overexpression (Figure 2D), suggesting that decreased migration and invasion of cells overexpressing hsa_circ_0005358 does not result from a lower rate of cellular proliferation.
Figure 2.
hsa_circ_0005358 acts as a suppressor of the metastatic behavior of cervical cancer in vitro and in vivo
(A and B) The migration (A) and invasion (B) abilities of SiHa and CaSki cells overexpressing hsa_circ_0005358 or vector as determined by transwell assay (Student's t test). Scale bar, 100 μm. (C) The migration capability of SiHa and CaSki cells overexpressing hsa_circ_0005358 or vector as determined by wound healing assay (Student’s t test). Scale bar, 100 μm. (D) The proliferation of SiHa and CaSki cells overexpressing hsa_circ_0005358 or vector as determined by CCK-8 assay (Student’s t test). (E and F) Representative images (E) and luminescence quantification (F) of lung metastases of transplanted tumors at the endpoints (Student's t test). (G) H&E staining of lung metastases. The arrows indicate classic morphology of lung metastases. Scale bars, 100 and 200 μm. (H) The number of metastatic lesions in five randomly scanned regions of each image was calculated to evaluate the metastasis of both groups (Student's t test). (I) Relative levels of hsa_circ_0005358 expression in SCC and normal cervical tissues as determined by qRT-PCR analysis, 18s was used as internal control (ANOVA). The data are represented as means ± SEM, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
In addition, we designed two small interfering RNAs (siRNAs) targeting the junction sites of hsa_circ_0005358 to knock down hsa_circ_0005358 expression in the C-4I cervical cell line, which expresses a higher level of hsa_circ_0005358 than both SiHa and CaSki cells (Figure S2C). The expression of hsa_circ_0005358, but not its linear counterpart, was significantly downregulated by both siRNAs (Figure S2D). In contrast to hsa_circ_0005358 overexpression, hsa_circ_0005358 knockdown significantly promoted cell migration and invasion rates (Figure S2E) and accelerated wound healing speed (Figure S2F) without affecting the proliferation (Figure S2G) of C-4I cells.
To confirm the suppressive effect of hsa_circ_0005358 on cancer metastasis in vivo, we injected SiHa cells stably overexpressing hsa_circ_0005358 or the control vector into female SCID mice via the caudal vein (n = 5/group, 2 × 106 cells injected per mouse), and monitored tumor metastasis once a week by luminescence using an in vivo imaging technique. We found that hsa_circ_0005358 overexpression significantly suppressed the metastasis of SiHa cells compared with the control group cells 4 weeks after injection (Figures 2E and 2F). Then, the mice were all sacrificed under anesthesia 4 weeks after injection, and the lungs were removed for further examination. Histology with H&E staining showed classic morphology of lung metastases (Figure 2G). The calculation of metastatic nodular numbers observed at 10× magnification revealed that the number of metastatic nodules in the hsa_circ_0005358 overexpression group was significantly less than that in the control group (Figure 2H). Moreover, we detected the expression level of hsa_circ_0005358 in 55 pathologically confirmed normal cervical tissue and 60 cervical SCC tissue samples (Table S3). Significant loss of hsa_circ_0005358 was observed in cervical cancer tissues compared with normal cervical tissues, and tumor samples with extracervical invasion or node metastasis (FIGO stage II or more) showed a significantly greater reduction in hsa_circ_0005358 expression than samples of tumors limited to the cervix (FIGO stage I) (Figure 2I). Together, these results suggest that hsa_circ_0005358 is a suppressor of cervical cancer metastasis.
hsa_circ_0005358 directly interacts with PTBP1
A bioinformatic analysis (online tool: RBP map) of hsa_circ_0005358 sequences predicted several specific sequences that can bind to the corresponding RNA recognition motif (RRM) of different RBPs (Table S4), suggesting that hsa_circ_0005358 may potentially interact with RBPs. To explore the protein partners of hsa_circ_0005358, we performed RNA-pull-down assays. A synthesized biotin-labeled probe targeting the specific junction of hsa_circ_0005358 was used to capture hsa_circ_0005358, and the result was displayed as an obvious band at approximately 55–70 kDa in the hsa_circ_0005358 probe lane (Figure 3A). Mass spectrometry (MS) results confirmed that the major component of the differential band was PTBP1 (Table S5), an upregulated gene in cervical cancer.19 Pull-down assays with western blot analysis (Figures 3B and S3A) and RNA-immunoprecipitation (RIP) assays (Figures 3C and S3B) revealed an interaction between hsa_circ_0005358 and PTBP1. RNA-pull-down assays (Figure 3D) and RIP assays (Figure 3E) showed a stronger interaction between hsa_circ_0005358 and PTBP1 when hsa_circ_0005358 was overexpressed. When hsa_circ_0005358 was knocked down in C-4I cells, the interaction got weaker (Figures S3C and S3D). In addition, the upregulation or downregulation of hsa_circ_0005358 expression did not affect PTBP1 expression (Figure S3E), and PTBP1 expression did not affect hsa_circ_0005358 expression (Figures S3F–S3G). Moreover, we found that PTBP1 facilitated the metastatic behavior of cervical cancer cells in vitro (Figures S3H–S3L), which was consistent with the results in other studies.19,20 Therefore, we speculated that hsa_circ_0005358 acts as a decoy for PTBP1 and compromises the metastasis-promoting property of PTBP1.
Figure 3.
hsa_circ_0005358 directly interacts with PTBP1
(A) A probe targeting the junction of hsa_circ_0005358 was used to capture the hsa_circ_0005358-protein complex. The pull-down products were verified by silver staining and mass spectrum identification. PTBP1 was identified as the most likely candidate protein interacting with hsa_circ_0005358. (B) Pull-down western blot (WB) assay confirmed that hsa_circ_0005358 interacted with PTBP1 in SiHa and CaSki cell lines. (C) RIP assays using anti-PTBP1 antibodies confirmed that PTBP1 precipitated hsa_circ_0005358 in SiHa and CaSki cell lysates (Student's t test). (D and E) The interaction between hsa_circ_0005358 and PTBP1 in SiHa and CaSki cells overexpressing hsa_circ_0005358 as determined by pull-down WB (D) and RIP (E) assays (Student's t test). (F) Schematic illustration of different binding sites of hsa_circ_0005358 on PTBP1. Mut1 to Mut5 represent different mutant binding sites. Mut6 represents hsa_circ_0005358 with all potential binding sites mutated. (G and H) The migration (G) and invasion (H) abilities of SiHa and CaSki cells transfected plasmids with different mutants as determined by transwell assays (ANOVA). Scale bar, 100 μm. (I) Schematic illustrating mutated hsa_circ_0005358 at sequence 215–224, named Mut-hsa_circ_0005358. (J and K) The interaction between hsa_circ_0005358 and PTBP1 in SiHa and CaSki cells overexpressing Mut-hsa_circ_0005358 shown by pull-down assays (J) and RIP assays (K) (ANOVA). (L and M) The migration (L) and invasion (M) abilities of SiHa and CaSki cells overexpressing hsa_circ_0005358 or Mut-hsa_circ_0005358 as determined by transwell assays (ANOVA). Scale bar, 100 μm. The data are represented as means ± SEM, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Using bioinformatic analysis, we also predicted five potential binding sites between hsa_circ_0005358 and PTBP1, and then constructed five mutant circRNA plasmids (Mut1–Mut5) specific to each binding site and another plasmid with all the potential mutant binding sites (Mut6) (Figure 3F). None of the mutations affected the circular structure of hsa_circ_0005358. As shown in Figures 3G and 3H, the overexpression of Mut1, Mut2, and Mut3 plasmids significantly suppressed the migration and invasion of SiHa and CaSki cells as the overexpression of wild-type hsa_circ_0005358 did, but Mut4, Mut5, or Mut6 lost such function, suggesting that the sequence where Mut4 and Mut5 were located, i.e., sequence 215–224, is necessary for hsa_circ_0005358 function. We named the hsa_circ_0005358 plasmid with the sequence 215–224 mutation Mut-hsa_circ_0005358 (Figure 3I). RNA-pull-down assays showed that PTBP1 was less enriched in SiHa and CaSki cells overexpressing Mut-hsa_circ_0005358 than in cells overexpressing wild-type hsa_circ_0005358 (Figure 3J). RIP assays followed by qRT-PCR revealed that Mut-hsa_circ_0005358 could not be captured by PTBP1 (Figure 3K). The cells overexpressing Mut-hsa_circ_0005358 presented migration and invasion abilities similar to those of control cells (Figures 3L and 3M). Together, our results suggest that hsa_circ_0005358 functions via sequence 215–224 directly binding to PTBP1.
hsa_circ_0005358 regulates CDCP1 expression by binding to PTBP1
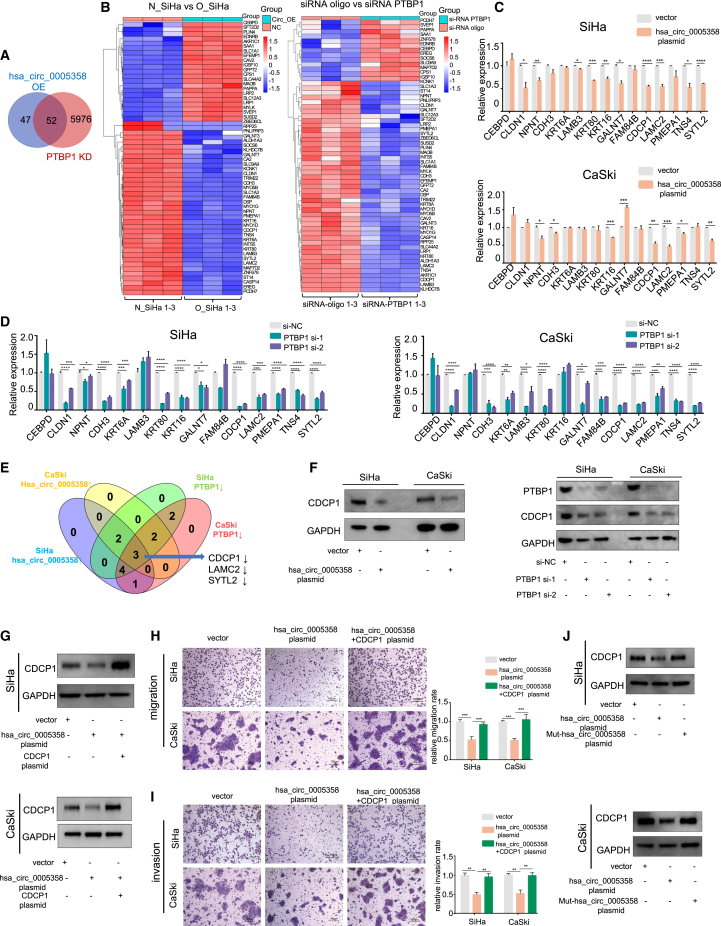
Since hsa_circ_0005358 served as a decoy and acted against PTBP1, we conducted two RNA-sequencing (RNA-seq) analyses with SiHa cells with and without overexpressed hsa_circ_0005358 (RNA-seq-1, N_SiHa versus O_SiHa) and SiHa cells with and without silenced PTBP1 (RNA-seq-2, siRNA_oligo versus siRNA_PTBP1). By applying filtered thresholds of |log2 FC| ≥ 1.2 and p < 0.05, RNA-seq analyses yielded 99 differentially expressed transcripts for RNA-seq-1 and 6,027 differentially expressed transcripts for RNA-seq-2. The overlap of RNA-seq-1 and RNA-seq-2 was analyzed and displayed in clustered heatmaps (Figures 4A and 4B). Of the overlapping genes, 38 showed a similar expression trend in cells with hsa_circ_0005358 overexpression and PTBP1 silencing. We further reduced the number of candidates according to the reported function of these genes and selected 15 genes for further verification in SiHa and CaSki cells by qRT-PCR (Figures 4C and 4D). The expression of three mRNAs, laminin subunit γ2 (LAMC2), CDCP1, and synaptotagmin-like 2 (SYTL2), showed consistent change trends in both SiHa and CaSki cells with hsa_circ_0005358 overexpression or PTBP1 silencing (Figure 4E). Next, we detected the expression of three proteins and found that the expression of the CDCP1 protein was more dramatically decreased in both SiHa and CaSki cells with hsa_circ_0005358 overexpression or PTBP1 silencing (Figure 4F) than that of the other two proteins (Figures S4A and S4B). The results suggested that hsa_circ_0005358 and PTBP1 commonly regulate CDCP1 expression.
Figure 4.
hsa_circ_0005358 regulates CDCP1 expression by binding to PTBP1
(A and B) Venn diagram (A) and clustered heatmaps (B) showing the overlap of differentially expressed mRNAs in SiHa cells with hsa_circ_0005358 that either is or is not overexpressed, and with PTBP1 that either is or is not knocked down. (C and D) Fifteen candidates in RNA-seq in SiHa and CaSki cells with hsa_circ_0005358 overexpression (C) or PTBP1 knockdown (D) as validated by qRT-PCR (Student's t test, ANOVA). (E) Venn diagram illustrating the overlap of RNA-seq results as verified by qRT-PCR in SiHa and CaSki cells. (F) The protein levels of CDCP1 in SiHa and CaSki cells with hsa_circ_0005358 overexpression or PTBP1 knockdown as measured by western blot analysis. (G) The protein levels of CDCP1 in SiHa and CaSki cells with hsa_circ_0005358 overexpressed or hsa_circ_0005358 overexpressed plus the CDCP1 plasmid as validated by western blot analysis. (H and I) The migration (H) and invasion (I) abilities of SiHa and CaSki cells with hsa_circ_0005358 overexpression or hsa_circ_0005358 overexpression plus CDCP1 plasmids as assessed by transwell assay (ANOVA). Scale bar, 100 μm. (J) The protein levels of CDCP1 in SiHa and CaSki cells overexpressing wild-type hsa_circ_0005358 or Mut-hsa_circ_0005358 as evaluated by western blot analysis. The data are represented as means ± SEM, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We then investigated CDCP1 function in cervical cells and found that forced CDCP1 overexpression significantly promoted the migration and invasion of SiHa and CaSki cells (Figures S4C–S4E), and downregulation of CDCP1 had the opposite effect (Figures S4F–S4H). Moreover, normalized hsa_circ_0005358 expression was negatively correlated with normalized CDCP1 mRNA expression in cervical cancer tissues (Pearson r = −0.7342, p < 0.0001, Figure S4I).
We simultaneously upregulated hsa_circ_0005358 and CDCP1 in SiHa and CaSki cells and found that hsa_circ_0005358 overexpression alone led to reduced migration and invasion rates in both cell lines, and these effects were neutralized by CDCP1 upregulation (Figures 4G–4I). In contrast, we found that hsa_circ_0005358 knockdown significantly increased the expression of CDCP1 (Figure S5A) and promoted the migration and invasion rates of C-4I cells, and these effects were reversed by CDCP1 knockdown (Figures S5B and S5C). We also simultaneously downregulated PTBP1 and upregulated CDCP1 in SiHa and CaSki cells and found that downregulated PTBP1 inhibited the migration and invasion rates of both cell lines, and these effects were neutralized by upregulated CDCP1 (Figures S5D–S5F). These results suggest that overexpression of CDCP1 reverses cellular migration and invasion abilities induced by hsa_circ_0005358 upregulation or PTBP1 downregulation.
Furthermore, we wondered whether hsa_circ_0005358 regulated CDCP1 expression in a PTBP1-dependent manner. The overexpression of Mut-hsa_circ_0005358, which has been shown not to interact with PTBP1, was associated with CDCP1 expression level similar to that of the vector group (Figure 4J). Combined with the phenotype of Mut-hsa_circ_0005358, as shown in Figures 3L and 3M, these results suggest that upregulated hsa_circ_0005358 inhibits CDCP1 expression by binding to PTBP1, thus leading to the attenuation of the aggressive metastatic behavior of cervical cancer cells.
hsa_circ_0005358 blocks PTBP1 from binding to and stabilizing CDCP1 mRNA
Considering that PTBP1 knockdown inhibited CDCP1 mRNA expression in our RNA-seq analysis and that a Clip-seq database (POSTAR2: PTBP1) implied that PTBP1 bound to CDCP1 mRNA directly, we deduced that the binding of PTBP1 to CDCP1 mRNA probably enhanced CDCP1 mRNA stability. Pull-down assays and RIP assays validated that PTBP1 bound to CDCP1 mRNA in SiHa and CaSki cells (Figures 5A and 5B).
Figure 5.
hsa_circ_0005358 blocks PTBP1 from binding to and stabilizing CDCP1 mRNA
(A) RNA-pull-down assay followed by western blot analysis was performed to determine the interaction between CDCP1 mRNA and PTBP1 protein in SiHa and CaSki cells. (B) RIP assays were used to measure the recruitment of PTBP1 protein to CDCP1 mRNA (Student's t test). (C) SiHa and CaSki cells with PTBP1 knockdown or overexpression were treated with actinomycin-D (5 μg/mL). Total RNA was extracted at different time points and CDCP1 mRNA was analyzed by qRT-PCR and normalized to GAPDH (Student's t test, ANOVA). (D) Western blot analysis was used to detect CDCP1 protein levels when PTBP1 was knocked down or overexpressed in SiHa and CaSki cells. (E and F) The interaction between PTBP1 protein and CDCP1 mRNA was measured using pull-down assays (E) and RIP assays (F) in which SiHa and CaSki cells were transfected with mock vector, hsa_circ_0005358 or Mut-hsa_circ_0005358 (ANOVA). (G) SiHa and CaSki cells overexpressing hsa_circ_0005358, Mut-hsa_circ_0005358, or hsa_circ_0005358 plus PTBP1 were treated with actinomycin-D (5 μg/mL). Total RNA was extracted at different time points and CDCP1 mRNA was analyzed by qRT-PCR and normalized to GAPDH (ANOVA). (H) Western blot analysis was used to determine the CDCP1 protein levels in SiHa and CaSki cells overexpressing hsa_circ_0005358, Mut-hsa_circ_0005358, or hsa_circ_0005358 plus PTBP1. The data are represented as means ± SEM, n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Actinomycin-D (Act-D), a classical transcription inhibitor, has been used in previous studies21,22 to determine the endogenous mRNA decay rate. By inhibiting RNA polymerase Ⅱ, Act-D can globally rapidly stop the transcription of cellular RNA. In this study, Act-D was used to examine the decay rate of CDCP1 mRNA levels in SiHa and CaSki cells. The qRT-PCR assay showed that PTBP1 knockdown accelerated endogenous CDCP1 mRNA degradation and reduced CDCP1 protein levels in SiHa and CaSki cells compared with control cells, and PTBP1 upregulation had the opposite effect (Figures 5C and 5D). The results suggest that PTBP1 binds to and stabilizes CDCP1 mRNA and consequently elevates CDCP1 protein expression.
Pull-down assays and RIP assays showed that the overexpression of hsa_circ_0005358, but not that of Mut-hsa_circ_005358, reduced PTBP1 recruitment by CDCP1 mRNA (Figure 5E) and decreased CDCP1 mRNA levels enriched by PTBP1 antibody (Figure 5F), indicating that hsa_circ_0005358 blocks PTBP1 from binding to CDCP1 mRNA. Next we examined CDCP1 mRNA expression under different conditions after Act-D treatment. As shown in Figures 5G and 5H, the overexpression of hsa_circ_0005358, but not that of Mut-hsa_circ_0005358, accelerated CDCP1 mRNA decay and decelerated protein synthesis, which was also reversed by co-transfection of hsa_circ_0005358 and PTBP1. Taken together, these results suggest that hsa_circ_0005358 prevents PTBP1 from binding to and stabilizing CDCP1 mRNA and thus decreases CDCP1 protein levels.
Discussion
Pelvic and lymph node metastasis is the main biological behavior and cause of lethality of cervical cancer.2 Accumulating evidence has revealed that circRNAs play roles in tumor metastasis in different ways.23, 24, 25, 26, 27 In our previous RNA-seq data, we found hsa_circ_0005358 to be significantly downregulated in squamous cervical cancer compared with normal cervical tissues. In this study, we further investigated the function of hsa_circ_0005358 and found that hsa_circ_0005358 served as a tumor suppressor for cervical cancer metastasis in vitro and in vivo. Moreover, the analysis of the data of 60 cervical cancer samples showed significantly more decreased hsa_circ_0005358 expression in tumor tissues with extracervical invasion or node metastasis than in those without extracervical involvement. Mechanistically, hsa_circ_0005358 bound to the PTBP1 protein and blocked the interaction between the PTBP1 protein and the CDCP1 mRNA, leading to the decay of CDCP1 mRNA and subsequent metastasis suppression. Taken together, our results suggest that the restoration of hsa_circ_0005358 expression may be a potential approach to blocking cervical cancer metastasis.
Initially, research on circRNA biological functions focused on their role in sponging microRNAs.28,29 However, recent studies demonstrated that some circRNAs may also bind to regulatory RBPs via specific binding sites, acting as protein sponges, decoys, or scaffolds to regulate downstream targets.26,30,31 Our bioinformatic analysis revealed the sequence of hsa_circ_0005358 containing RBP motifs. Accordingly, we conducted an RNA-pull-down assay combined with MS analysis and found that hsa_circ_0005358 bound directly to the PTBP1 protein. In our previous study based on RNA-seq, we found that PTBP1 was overexpressed in cervical cancer,19 implying that PTBP1 serves as an oncogene in cervical cancer. Here, our gene function experiments confirmed the role of PTBP1 in facilitating the metastatic behavior of cervical cancer cells in vitro. A recent study showed that specifically designed oligonucleotides can bind to PTBP1 like a decoy,32 compromising the original function of PTBP1 without changing PTBP1 protein expression. In the current study, we did not find that hsa_circ_0005358 regulated the expression of PTBP1 in cervical cancer cells, suggesting that inhibition of the oncogenic properties of PTBP1 is due to hsa_circ_0005358 binding to PTBP1.
It has been revealed that the RRM of PTBP1 has a preference for CU-rich sequences,33,34 which are abundantly scattered across the hsa_circ_0005358 sequence. Thus, we mutated five sites in the hsa_circ_0005358 sequence, each of which were predicted to bind possibly to PTBP1. We found that hsa_circ_0005358 with mutated sequence 215–224 no longer had the capacity to capture PTBP1 or inhibit the migration and invasion of cervical cancer cells, suggesting that sequence 215–224 is critical for the interaction between hsa_circ_0005358 and PTBP1.
PTBP1 has been demonstrated to be a crucial regulator of mRNA metabolism and is involved in splicing regulation, internal ribosomal entry site (IRES)-mediated translation initiation, mRNA stability, and other functions.35, 36, 37 To verify that hsa_circ_0005358 works as a decoy of PTBP1, we investigated the activity of hsa_circ_0005358 in modulating the target gene of PTBP1. The Venn graph of the two RNA-seq datasets revealed that CDCP1 was a candidate commonly controlled by both hsa_circ_0005358 and PTBP1. Previous studies revealed the association of CDCP1 upregulation with metastasis in various cancers,38,39 including cervical cancer.40 Consistently, in this study, we confirmed that CDCP1 functions as an oncogene to promote the metastatic behavior of cervical cancer cells. The cell phenotypes acquired through hsa_circ_0005358 overexpression or knockdown can be reversed by enforced overexpression or inhibition of CDCP1, respectively, suggesting that CDCP1 functions via hsa_circ_0005358. Importantly, overexpressed mutant hsa_circ_0005358 did not bind to PTBP1, alter CDCP1 expression, or affect cell phenotypes. Furthermore, we verified that hsa_circ_0005358 blocked the interaction between PTPB1 and CDCP1 mRNA, thus accelerating CDCP1 mRNA decay and leading to decreased expression of the CDCP1 protein. These findings demonstrated that hsa_circ_0005358 modulates the expression of CDCP1 mRNA and protein by binding to PTBP1.
To evaluate the function of hsa_circ_0005358 in promoting cervical cancer metastasis in vivo, we employed an animal model and injected cervical cancer cells stably overexpressing hsa_circ_0005358 into mice via the caudal vein, as described in previous reports.41,42 In this model, cancer cells are directly injected into the blood circulation, which does not truly reflect the characteristics of cancer cell metastasis. To overcome this shortcoming of the animal model, we detected the level of hsa_circ_0005358 expression in cancer tissues with metastasis and without metastasis, and the results supported that cervical cancer tissues with higher hsa_circ_0005358 expression levels exhibit weakened metastatic potential.
In summary, we found that the restoration of circRNA hsa_circ_0005358 in cervical cancer, which normally interacts with PTBP1 via sequence 215–224, impedes PTBP1 binding to and stabilization of CDCP1 mRNA, resulting in CDCP1 downregulation and cancer metastasis inhibition. Our findings reveal a new mechanism of tumor metastasis that may indicate a potential therapeutic approach for patients with metastatic cervical cancer.
Materials and methods
Clinical specimens
Pathologically confirmed cervical cancer and normal cervical epithelium tissues were collected from September 2015 to December 2020 at Women's Hospital, School of Medicine, Zhejiang University. All samples and clinical data were obtained with the approval of the Hospital Ethical Committee. Normal cervical tissues were collected from patients who underwent hysterectomy for benign gynecological diseases. Squamous cervical cancer tissues were obtained from patients who received radical hysterectomy treatment. In total, 55 normal cervical tissues and 60 squamous cervical cancer tissues were subjected to qRT-PCR analysis. All samples were stored at −80°C until use.
RNA isolation and real-time quantitative reverse-transcription PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was applied to extract total RNA according to the manufacturer's instructions. The cDNA was synthesized using the Prime Script RT Master Mix kit (Takara, Japan). To quantify the amount of circRNAs and mRNAs, qRT-PCR was performed using TB Green Premix Ex Taq II (Takara, Japan), and 18s and GAPDH were used as internal controls. Relative expression levels of circRNAs and mRNAs were calculated using the 2−ΔΔCt method. The primers used in this study are listed in Table S6.
PCR and agarose gel electrophoresis
The 2×Taq MasterMix (CWbiotech, China) was used to perform PCR according to the manufacturer's instructions. The PCR products were subject to agarose gel electrophoresis.
RNase R treatment
Total RNA was treated with RNase R (Epicentre, Madison, WI, USA) following the manufacturer's instructions. The relative expression levels of hsa_circ_0005358 and Gli1 mRNA were analyzed by qRT-PCR.
Northern blot
The probe targeting the back-splice junction of hsa_circ_0005358 was synthesized by Sangon Biotech (Shanghai, China). The northern blot assay was conducted as previously described.18 The probe used in this study is listed in Table S6.
Subcellular fractionation assay
The nuclear and cytoplasmic fractions of cells were separated using the PARIS kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions.
RNA fluorescence in situ hybridization
The FISH kit was purchased from RiboBio (Guangzhou, China). The specific probe targeting the back-splice junction of hsa_circ_0005358 was labeled by Cy3, and the probe sequences are listed in Table S6. Cells were seeded and infused to 70% beforehand. After fixation and permeabilization, the cells were incubated with prehybridization buffer at 37°C for 30 min and then hybridized overnight with probe added. The cells were then washed and dyed with DAPI. Representative images of RNA FISH were captured by a confocal microscope.
RNA in situ hybridization assay
hsa_circ_0005358 was detected in cervical cancer tissues using the BaseScope kit (Advanced Cell Diagnostics, Newark, CA, USA) according to the manufacturer's instructions.
Cell culture
Human cervical cancer cell lines SiHa and C-4I were purchased from American Type Culture Collection (Manassas, MA, USA). Human cervical CaSki was purchased from Cell Resource Center, Shanghai Institute of Life Sciences, Chinese Academy of Sciences (China). The cells were cultured as described.18
circRNA overexpression construction and stable transfection
The lentivirus-containing hsa_circ_0005358 overexpression plasmid and mock vector (pcD-ciR) were designed and synthesized by Geneseed Biotech (Guangdong, China). After 48 h of transfection, the cells were selected using puromycin for 2 weeks to construct stable cell lines.
Plasmids and oligonucleotide construction and transfection
Expression plasmids for PTBP1 and CDCP1 were designed and synthesized by GenePharma Biotech (Shanghai, China). siRNAs targeting hsa_circ_0005358 and PTBP1 were designed by GenePharma Biotech (Shanghai, China), and siRNAs targeting CDCP1 were designed by Guannan Biosciences, Inc. (Hangzhou, China). Sequences of all oligonucleotides are listed in Table S6. Cells were transfected using X-treme GENE (Roche, Basel, Switzerland) according to the manufacturer's instructions.
Cell migration and invasion assay
Cell invasion abilities were assessed using transwell chambers coated with diluted Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The lower chambers were filled with 500 μL medium with 18% FBS. Treated cells were suspended in Opti-Mem (Thermo Scientific, Waltham, MA, USA) and seeded into the upper chamber. After being cultured at 37°C for 8 h (SiHa) or 24 h (CaSki and C-4I), the cells were fixed and stained using crystal violet for 20 min at room temperature. The images of invaded cells were taken with a light microscope. Cell migration abilities were measured using the same methods without Matrigel added to the upper chambers.
Cell viability assay
Cell viability was detected with a Cell Counting Kit-8 (Dojindo, Japan).
Wound healing assay
Culture-Insert Wells (ibidi, Germany) were used for wound healing assay. The Culture-Inserts were first placed in a 24 well plate. Treated cells were seeded into the separated chambers of the ibidi Culture-Inserts. After cells had grown for approximately 24 h, the Culture-Inserts were removed to create a gap. The process of cell migration was monitored using a microscope at different time points. The distance between the gap was measured. The migration rate of the cells was expressed as relative gap closure.
RBP prediction
RBP target prediction was performed using the online database RBP Map (http://rbpmap.technion.ac.il/).
RNA-pull-down assay and protein MS analysis
Streptavidin-coupled magnetic beads (Thermo Scientific, Waltham, MA, USA) were incubated with biotin-labeled hsa_circ_0005358-specific probes for 30 min at room temperature, and the hsa_circ_0005358 antisense probe was used as the negative control. Approximately 1 × 106 cells were harvested and lysed on ice. Cell lysates were incubated with probe-bound beads at 4°C overnight. Bead-bound RNA complex was washed with washing buffer twice and extracted for further analysis. The sequences of all probes used in the pull-down assay are listed in Table S6. Technical support for protein MS analysis was supplied by OE Biotech Co. Ltd. (Shanghai, China).
RNA immunoprecipitation assay
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore, Germany) was used for RIP assay. In brief, 2 × 107 cells were harvested and lysed, and protease inhibitor and RNase inhibitor were added on ice. Five micrograms of PTBP1 or IgG antibodies was incubated with protein A/G magnetic beads for 30 min. Then the cell lysates were incubated with the antibody-bead complex at 4°C overnight. The beads were washed by wash buffer six times before the RNA was extracted and precipitated. The quality and abundance of hsa_circ_0005358 or CDCP1 mRNA were detected by qRT-PCR.
RNA-sequencing analysis
SiHa cells transfected with mock vector or hsa_circ_0005358 (n = 3) and siRNA-oligo or siRNA-PTBP1 (n = 3) were used for transcriptome sequencing analysis. The RNA-seq and further analysis were conducted by LC.Bio.Tech (Hangzhou, China) and OE Biotech Co. Ltd. (Shanghai, China). The RNA-seq raw data have been uploaded to Gene Expression Omnibus (GSE: 168549 and GSE: 168907).
Protein extraction and western blot
Cellular protein was extracted with RIPA buffer (Thermo Scientific, Waltham, MA, USA). Protein samples were subjected to 10% SDS-PAGE and then trans-blotted to polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). The membranes were probed with primary antibodies overnight at 4°C and then incubated with secondary antibodies for 1 h at room temperature. Antibodies against PTBP1, CDCP1, LAMC2, SYTL2, and GAPDH are listed in Table S6.
Actinomycin-D assay
Prepared SiHa and CaSki cells were treated with 5 μg/mL actinomycin-D (Sigma-Aldrich, St. Louis, MO, USA). RNA was extracted at the indicated time points (0,1, 2, 3, and 4 h), and qRT-PCR was performed to measure the stability of the mRNA.
Animal models
All in vivo experiments were performed at the animal research center of Zhejiang Chinese Medical University with the approval of the Institutional Animal Care and Use Committee (IACUC) of Zhejiang Chinese Medical University (ethic protocol IACUC-20181029-05). Ten 4-week-old female SCID mice were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (China) and randomly divided into two groups. SiHa cells (2 × 106) stably transfected with hsa_circ_0005358 plasmid or mock vector were injected into the caudal vein of each mouse. The cells were labeled with luciferase, and luminescence could be excited by intraperitoneal injection with substrate. In vivo bioluminescence imaging was utilized for luminescence measurement every week. At week 4 after injection, all mice were sacrificed under anesthesia after in vivo imaging, and the lungs were removed for further investigation.
Statistical analysis
All the experiments were performed in triplicate and repeated independently at least three times. The sample sizes and statistical methods are indicated in the corresponding figure legends. The data are represented as the mean ± SEM. All data plotting and statistical analyses were performed by GraphPad Prism 8 software. Differences were statistically significant at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Data avilability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper can be requested from the corresponding authors.
Acknowledgments
We thank Professor Xiaohang Yang, Professor Linyu Lu (School of Medicine, Zhejiang University, Hangzhou, China), and Professor Wanzhong Ge (Women's Hospital, School of Medicine, Zhejiang University, Hangzhou, China) for providing experimental guidance and Yu Huang (Zhejiang Chinese Medical University) for providing technical support for animal experiments.
This work was supported by the National Key Research and Development Program of China (2016YFC1302900), the Key Research and Development Plan in Zhejiang province (2020C03025), the Center for Uterine Cancer Diagnosis & Therapy Research of Zhejiang Province (JBZX-201803), the National Natural Science Foundation of China (82072855), and the Natural Science Foundation of Zhejiang Province (LQ17H160004).
Authors' contributions
W.L. and X.W. conceptualized and designed the study, revised the manuscript, and supervised and administered the study. Y.C. and T.Z. performed most of the experiments, analyzed the data, and wrote the manuscript. Y.Z., L.Z., J.Z., and L.W. collected and classified the human tissue samples and conducted the experiments. J.X., T.D., and X.X. performed most of the in vivo experiments and analyzed the data. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.11.020.
Contributor Information
Xinyu Wang, Email: wangxy@zju.edu.cn.
Weiguo Lu, Email: lbwg@zju.edu.cn.
Supplemental information
and Tables S1–S6
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Canfell K., Kim J.J., Brisson M., Keane A., Simms K.T., Caruana M., Burger E.A., Martin D., Nguyen D.T.N., Benard E., et al. Mortality impact of achieving WHO cervical cancer elimination targets: a comparative modelling analysis in 78 low-income and lower-middle-income countries. Lancet. 2020;395:591–603. doi: 10.1016/S0140-6736(20)30157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbyn M., Weiderpass E., Bruni L., de Sanjose S., Saraiya M., Ferlay J., Bray F. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob. Health. 2020;8:e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao F., Qiao Y. Cervical cancer prevention in China: a key to cancer control. Lancet. 2019;393:969–970. doi: 10.1016/S0140-6736(18)32849-6. [DOI] [PubMed] [Google Scholar]
- 5.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 8.Lee S.Y., Jeong E.K., Ju M.K., Jeon H.M., Kim M.Y., Kim C.H., Park H.G., Han S.I., Kang H.S. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol. Cancer. 2017;16:10. doi: 10.1186/s12943-016-0577-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massague J., Obenauf A.C. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q., Zhang H., Jiang X., Qian C., Liu Z., Luo D. Factors involved in cancer metastasis: a better understanding to "seed and soil" hypothesis. Mol. Cancer. 2017;16:176. doi: 10.1186/s12943-017-0742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakhoum S.F., Ngo B., Laughney A.M., Cavallo J.A., Murphy C.J., Ly P., Shah P., Sriram R.K., Watkins T.B.K., Taunk N.K., et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu P., Mo Y., Peng M., Tang T., Zhong Y., Deng X., Xiong F., Guo C., Wu X., Li Y., et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen Lasse S., Andersen Maria S., Stagsted Lotte V.W., Ebbesen Karoline K., Hansen Thomas B., Jørgen K. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 14.Kristensen L.S., Hansen T.B., Veno M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison E.B., Porrello A., Bowman B.M., Belanger A.R., Yacovone G., Azam S.H., Windham I.A., Ghosh S.K., Wang M., McKenzie N., et al. A circle RNA regulatory axis promotes lung squamous metastasis via CDR1-mediated regulation of golgi trafficking. Cancer Res. 2020;80:4972–4985. doi: 10.1158/0008-5472.CAN-20-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanniford D., Ulloa-Morales A., Karz A., Berzoti-Coelho M.G., Moubarak R.S., Sanchez-Sendra B., Kloetgen A., Davalos V., Imig J., Wu P., et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70.e15. doi: 10.1016/j.ccell.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Zhao L., Yang S., Cen Y., Zhu T., Wang L., Xia L., Liu Y., Zou J., Xu J., et al. CircCDKN2B-AS1 interacts with IMP3 to stabilize hexokinase 2 mRNA and facilitate cervical squamous cell carcinoma aerobic glycolysis progression. J. Exp. Clin. Cancer Res. 2020;39:281. doi: 10.1186/s13046-020-01793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J., Liu H., Yang Y., Wang X., Liu P., Li Y., Meyers C., Banerjee N.S., Wang H.K., Cam M., et al. Genome-wide profiling of cervical RNA-binding proteins identifies human papillomavirus regulation of RNASEH2A expression by viral E7 and E2F1. mBio. 2019;10:e02687. doi: 10.1128/mBio.02687-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie R., Chen X., Chen Z., Huang M., Dong W., Gu P., Zhang J., Zhou Q., Dong W., Han J., et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31–44. doi: 10.1016/j.canlet.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 21.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S., et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Cui Y.H., Yang S., Wei J., Shea C.R., Zhong W., Wang F., Shah P., Kibriya M.G., Cui X., Ahsan H., et al. Autophagy of the m(6)A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nat. Commun. 2021;12:2183. doi: 10.1038/s41467-021-22469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M., et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Zhang J., Tian Y., Gao Y., Dong X., Chen W., Yuan X., Yin W., Xu J., Chen K., et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer. 2020;19:128. doi: 10.1186/s12943-020-01246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J., Lee E.E., Kim J., Yang R., Chamseddin B., Ni C., Gusho E., Xie Y., Chiang C.M., Buszczak M., et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 2019;10:2300. doi: 10.1038/s41467-019-10246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsitsipatis D., Grammatikakis I., Driscoll R.K., Yang X., Abdelmohsen K., Harris S.C., Yang J.H., Herman A.B., Chang M.W., Munk R., et al. AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 2021;49:1631–1646. doi: 10.1093/nar/gkaa1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen T.B., Kjems J., Damgaard C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 29.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W.Y., Cai Z.R., Liu J., Wang D.S., Ju H.Q., Xu R.H. Circular RNA: metabolism, functions and interactions with proteins. Mol. Cancer. 2020;19:172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 32.Denichenko P., Mogilevsky M., Clery A., Welte T., Biran J., Shimshon O., Barnabas G.D., Danan-Gotthold M., Kumar S., Yavin E., et al. Specific inhibition of splicing factor activity by decoy RNA oligonucleotides. Nat. Commun. 2019;10:1590. doi: 10.1038/s41467-019-09523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 34.Bielli P., Bordi M., Di Biasio V., Sette C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection. Nucleic Acids Res. 2014;42:12070–12081. doi: 10.1093/nar/gku922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgilis A., Klotz S., Hanley C.J., Herranz N., Weirich B., Morancho B., Leote A.C., D'Artista L., Gallage S., Seehawer M., et al. PTBP1-Mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell. 2018;34:85–102.e109. doi: 10.1016/j.ccell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z., Wang L., Welch J.D., Ma H., Zhou Y., Vaseghi H.R., Yu S., Wall J.B., Alimohamadi S., Zheng M., et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551:100–104. doi: 10.1038/nature24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David C.J., Chen M., Assanah M., Canoll P., Manley J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y., Davies C.M., Harrington B.S., Hellmers L., Sheng Y., Broomfield A., McGann T., Bastick K., Zhong L., Wu A., et al. CDCP1 enhances Wnt signaling in colorectal cancer promoting nuclear localization of beta-catenin and E-cadherin. Oncogene. 2020;39:219–233. doi: 10.1038/s41388-019-0983-3. [DOI] [PubMed] [Google Scholar]
- 39.Wright H.J., Hou J., Xu B., Cortez M., Potma E.O., Tromberg B.J., Razorenova O.V. CDCP1 drives triple-negative breast cancer metastasis through reduction of lipid-droplet abundance and stimulation of fatty acid oxidation. Proc. Natl. Acad. Sci. U S A. 2017;114:E6556–E6565. doi: 10.1073/pnas.1703791114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang L., Chen Y., Lai S., Guan H., Hu X., Liu J., Zhang H., Zhang Z., Zhou J. CUB domain-containing protein-1 promotes proliferation, migration and invasion in cervical cancer cells. Cancer Manag. Res. 2020;12:3759–3769. doi: 10.2147/CMAR.S240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong H., Zhu H., Zhao S., Wang K., Zhang N., Tian Y., Li Y., Wang Y., Lv X., Wei T., et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019;10:950. doi: 10.1038/s41419-019-2183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Ou J., Guo Y., Dai T., Li X., Liu J., Xia M., Liu L., He M. TBLR1 is a novel prognostic marker and promotes epithelial-mesenchymal transition in cervical cancer. Br. J. Cancer. 2014;111:112–124. doi: 10.1038/bjc.2014.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and Tables S1–S6