Abstract
In the current study, novel four electrode-based impedimetric biosensors have been fabricated using photolithography techniques and utilized to evaluate the cytotoxicity of tamoxifen on cervical cancer cell lines. The cell impedance was measured employing the electric cell-substrate impedance sensing (ECIS) method over the frequency range of 100 Hz to 1 MHz. The results obtained from impedimetric biosensors indicate that tamoxifen caused a significant reduction in the number of HeLa cells on the electrode surfaces in a dose-dependent manner. Next, the impedance values recorded by the fabricated biosensors have been compared with the results obtained from the different conventional techniques such as 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), live-dead cell assay, and flow cytometric analysis to estimate the cytotoxicity of tamoxifen. The impedimetric cytotoxicity of tamoxifen over the growth and proliferation of HeLa cells correlates well with the traditional methods. In addition, the IC50 values obtained from impedimetric data and MTT assay are comparable, signifying that the ECIS technique can be an alternative method to assess the cytotoxicity of different novel drugs. The working principle of the biosensor has been examined by scanning electron microscopy, indicating the detachment of cells from gold surfaces in a dose-dependent manner, signifying the decrease in impedance at higher drug doses.
In the current study, novel four electrode-based impedimetric biosensors have been fabricated using photolithography techniques and utilized to evaluate the cytotoxicity of tamoxifen on cervical cancer cell lines.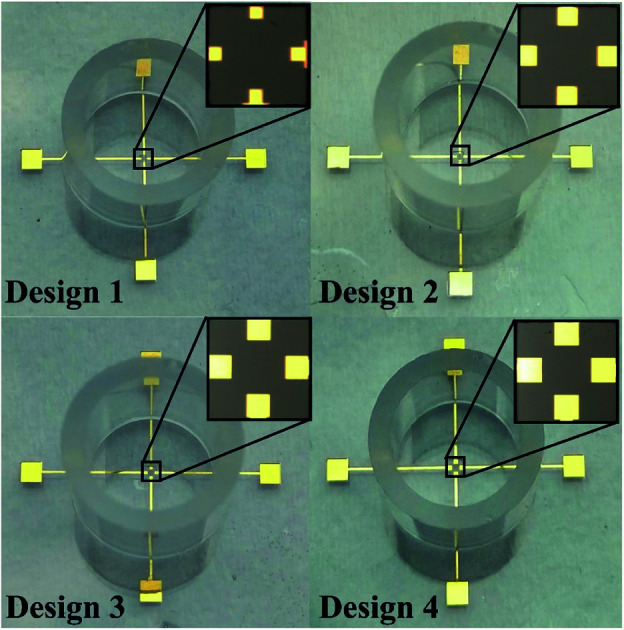
1. Introduction
Cervical cancer is known as the fourth most deadly form of malignancy affecting females globally.1 It has tremendously high morbidity involving around 530 000 new cases along with 270 000 deaths yearly, making it a serious health issue worldwide.2–4 In addition, the mortality of cervical cancer has been found to be 18 times higher in underdeveloped or developing countries.4 The most common histological subtypes of cervical cancer involve squamous cell carcinoma (70%) and adenocarcinoma (25%) and the remaining 5% of cervical cancer includes adenosquamous carcinoma and small cell cancer.5,6 Despite the advancements in vaccine program over the last decade, the overall situation has not improved considerably due to lack of availability of vaccine and organized screening in underdeveloped or developing countries.1,4 In the early stages of the disease, cervical cancer is managed by radical hysterectomy followed by primary chemoradiotherapy, tailored chemoradiotherapy, or neoadjuvant chemotherapy depending upon the stage of the disease.7 Chemotherapeutic drugs like cisplatin, gemcitabine, vinorelbine, paclitaxel, camptothecins, hydroxyurea, and fluorouracil or a regimen, including the combination of the drugs, have been regularly the choice.8 Therefore, there is a constant search for new chemotherapeutic drugs, which may help in the management of cervical cancer with fewer side effects.
The uterine cervix is a part of the female genital tract that is hormonally regulated, and hence anti-estrogenic agents may be a valued addition to the neoadjuvant chemotherapeutic regime.9 Tamoxifen, a non-steroidal anti-estrogenic drug, has already been successfully used in adjuvant therapy for breast cancer.10 However, before the use of tamoxifen in cervical cancer therapy, it is essential to evaluate the cytotoxic effects of the particular drug. The conventional techniques used to study the cytotoxicity are time consuming, destructive, costly, and do not provide the real time data. Thus, impedimetric biosensors are getting tremendous interest for the non-destructive analysis of the cellular parameters and interfacial properties related to cell–cell interactions,11 cell–drug interactions,12 interactions between molecules such as antigen–antibody,13 protein–ligand,14 receptor–ligand,15 protein–DNA,16 DNA–ligand,17 drug–DNA,18 biomolecular recognition,19 and biomolecular interactions20 by measuring their electrical properties. These biosensors have provided new insights into drug development, cytotoxic evaluation, cancer biology, and pathology.21 The technique used to study the live cell is known as electric cell-substrate impedance sensing (ECIS) that helps to determine the variations in the electrical properties of cells.22–24 ECIS is an in vitro methodology that senses real-time cellular activities via electrodes.25 An alternating current (AC) potential of different frequencies is applied to an electrochemical cell for the electrical impedance measurement. This real-time cell sensing technology makes it more efficient than microscopic imaging or observation.26–28 The particular technique has been utilized in the successful monitoring of cellular properties like changes during cell adhesion, growth, differentiation, and death.29–32
In the present study, we have evaluated the cytotoxicity of tamoxifen using a novel microfabricated four electrode-based impedimetric biosensors wherein, the ECIS technique has been employed to understand the responses of HeLa cells upon exposure to various doses of tamoxifen without studying its side effect. To the best of our knowledge, this impedance monitoring of cytotoxic effects of tamoxifen on HeLa cells has not been studied to date.
2. Materials and methods
2.1. Chemicals and reagents
The microscopic slides were acquired from Merck, India. SU-8 was procured from MicroChem, USA. Polydimethylsiloxane (PDMS) has been obtained from Dow Corning, Inc., India. All other chemicals or reagents unless mentioned otherwise were of analytical grade.
2.2. Design of the impedimetric biosensor
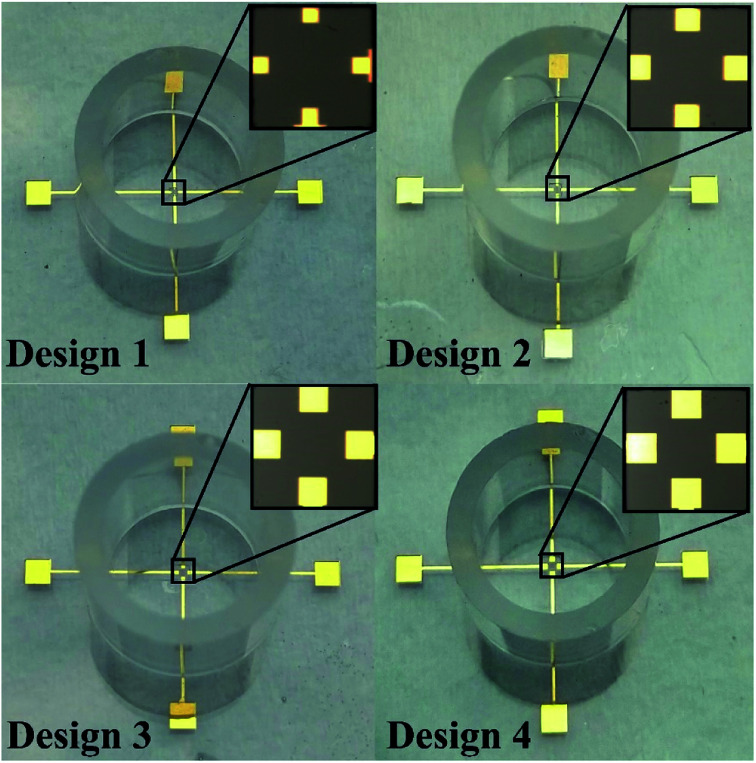
The present design provides novel four electrode-based biosensors for measuring bioimpedance to understand the cytotoxic activity of tamoxifen on HeLa cells. The areas of all the electrodes, such as the sensing electrode (SE), working electrode (WE), counter electrode (CE), and the reference electrode (RE), have been kept identical for all the designs.33 Pradhan et al. reported a design with a minimum working electrode area of 2500 μm2, providing uniform current density along with lower noises in the final impedance data.34 Thus in the present study, the lower electrode area has been selected as 2500 μm2. Preventing cross-contamination remains a significant challenge that was solved by allocating the SE, RE, and CE with a minimum space of 100 μm from the working electrode.35,36 The dimensions of four electrode based novel designs to increase the sensitivity are described in Table 1. A coating of SU-8 of 50 μm thickness was carried out over the connecting width to remove noises in the final impedance data due to interconnections.
Dimensions of different design of sensors.
| Devices | Electrode dimensions (μm2) |
|---|---|
| Design 1 | WE-50 × 50, SE-50 × 50, RE-50 × 50, CE-50 × 50 |
| Design 2 | WE-100 × 100, SE-100 × 100, RE-100 × 100, CE-100 × 100 |
| Design 3 | WE-150 × 150, SE-150 × 150, RE-150 × 150, CE-150 × 150 |
| Design 4 | WE-200 × 200, SE-200 × 200, RE-200 × 200, CE-200 × 200 |
2.3. Fabrication of the biosensor
The four electrode-based biosensors have been fabricated over the microscopic glass slides by using the photolithography technique. The glass slides were cleaned thoroughly by using piranha solution (H2SO4 : H2O2 (5 : 1)), followed by the deposition of Ti (100 Å) and Au (200 nm) at 80 °C (substrate heating at 120 °C) and ∼10−5 torr (base pressure ∼ 10−6 torr) utilizing the thermal evaporation technique (Vacuum Technologies (P) Ltd, India). Next, patterns of the electrodes and the contact pads were demarcated over the deposited Ti/Au film lithographically by coating the deposited slide with HPR-504, exposing photoresist to ultraviolet ray for 6.5 s, developing with HPRD 429 for 35 s and rinsing with DI water for 30 s. The unwanted Ti and Au layers were selectively removed by wet etching using standard Ti and Au etchants for 4–5 min and then the photoresist covering the active electrode area, leads, and its contact pads was stripped off by using acetone at 65 °C for 15 minutes and then thoroughly rinsed with DI water. Subsequently, spin coating of SU8, a photosensitive polymer, was carried out, and a lithographical pattern of SU8 coating was acquired over the metal interconnections. Using PDMS (elastomer: curing agent (10 : 1)) as adhesive, the cloning cylinders were immobilized around the biosensors to aid as a cell culture chamber, as presented in Fig. 1.
Fig. 1. Microphotograph of different designs of four electrode-based devices.
2.4. Cell culture
HeLa cell line was procured from National Centre for Cell Sciences, Pune. The cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Sigma Aldrich), supplemented with 1% penicillin–streptomycin (Sigma Aldrich), 10% fetal bovine serum (Sigma Aldrich), and 3.75 g L−1 sodium bicarbonate. The cells were incubated at 37 °C, 5% CO2 under humidified condition.
2.5. Cell viability assay
The cell viability on treatment with the chemotherapeutic tamoxifen was measured by MTT assay.37 For this assay, approximately 104 cells per well in a 96-well plate were seeded and incubated overnight. Next, cells were treated with various doses of the drug by considering the IC50 value of tamoxifen as 25.25 ± 0.52 μM (0, 12.5 μM, 25 μM, and 37.5 μM) and incubated for 24 hours to understand the drug–cell interaction before and after IC50 value. After this, the drug was removed, and 10 μL MTT dye dissolved in 100 μL DMEM was added to cells for 30 min. The purple-colored formazan crystals formed after 30 minutes were dissolved in DMSO, and the absorbance was measured at 570 nm (A570) by using Cytation 3 microplate reader (Biotek).38 The background absorbance was measured at 690 nm (A690), and the cell viability was calculated using the eqn (1).39
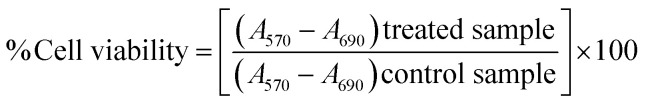 |
1 |
2.6. Live/dead assay
Cells were stained with acridine orange (AO) and ethidium bromide (EB) to detect apoptosis. Cells were treated with different drug concentrations for 24 hours, after which the drug-containing media was removed, and cells were washed with phosphate buffered saline (PBS). Cells were stained with a mixture of AO–EB and incubated for 15 min at 37 °C to allow uptake of the dye, and excess dye was removed by a PBS wash.40 Phase-contrast and fluorescence images were captured with an EVOS fluorescence microscope (EVOS® FL Color, AMEFC 4300 inverted microscope).
2.7. Flow cytometry studies
Flow cytometric analysis of the HeLa cells was carried out to study the cell death in G1 phase. The control and treated HeLa cells after 24 h trypsinized, collected, and washed to remove residual media. Cells were fixed with 70% ethanol by incubating them overnight at 20 °C. After fixation, cells were stained with propidium iodide solution (containing 40 mg mL−1 PI, 100 mg mL−1 RNase, 0.1% (v/v) Triton X-100 dissolved in PBS) and incubated at 37 °C for 1 hour. Around 10 000 cells were acquired using the appropriate channel in the Amnis FlowSight (Imaging Flow Cytometer).41 The obtained data were analyzed using the Amnis IDEAS software.
2.8. Cell morphology analysis
Scanning electron microscopy (SEM) imaging was carried out to analyze the cell morphology on gold surfaces. After 24 h of drug treatment, cells were fixed with 2% glutaraldehyde (10–15 min), followed by fixation with ethanol gradient (20%, 40%, 60%, 80%). Cells were air-dried at room temperature and observed under Zeiss EVO18 LaB6 filament SEM at 5 kV EHT.42
2.9. Experimental procedures
The ECIS devices were decontaminated by using 75% ethanol, nitrogen-drying, and treatment with ultraviolet radiation, all procedures for a while of 15 min. The cell-seeding media, DMEM (1 mL) was then loaded in the biosensor and incubated for 20 min at 37 °C to note the value of the contextual impedance (ZNo cell). Subsequently, HeLa cells (1 mL, 1 × 106 cells) were supplemented in the cell culture chamber of the biosensor. Then, the different doses of tamoxifen (0, 12.5, 25, and 37.5 μM) were added to the chamber of the biosensor after half an hour of cell insemination to observe the cytotoxicity of the drug.
2.10. Impedimetric studies
The electrical impedance measurement of the cell sample was carried out by using the electrochemical work station (M204, Metrohm Autolab). After 24 h of treatment with different dosages of tamoxifen, the impedance of HeLa cells was recorded by applying an AC voltage of 10 mV over a frequency range of 100 Hz to 1 MHz in a logarithmic scale. The final impedance used to generate a Bode plot is the mean value of three successive measurements of impedance value. The acquired impedance values have been analyzed by using ZsimpWin software.43 The sensitivity of the ECIS devices has been computed by using eqn (2) extracted from previous literature.30,44
| Sensitivity (f) = (|ZCell(f)| − ZNo cell(|(f)|)QCell−1 | 2 |
where Qcell is the maximum cell density (about 106 cells per cm2), and f is sensing frequency.
3. Results and discussion
3.1. Impedimetric studies
The ECIS techniques can be classified into two, three and four electrodes configurations in order to study the in vitro bio impedance and cellular behavior. In two electrode system, WE and CE are present while in three electrode system a specialized RE is present along with WE and CE. On the other hand, the four-electrode system possesses an extra SE for accurate impedance measurement by minimizing the noise level. Generally, the AC current is applied to WE and collected by CE, while the RE provides a constant potential. As SE is separated from the WE in the four-electrode configuration, it provides the ability to increase signal to noise ratio. Thus in the current study four electrode techniques has been used to measure the cytotoxicity effects of tamoxifen on cervical cancer cells.
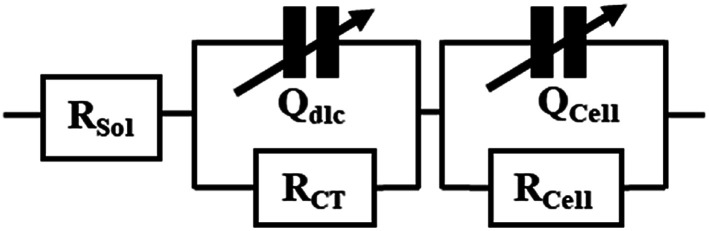
After the addition of different doses of tamoxifen (12.5 μM, 25 μM, and 37.5 μM) into the inoculations of cells in the device, the impedance values were measured after 24 h. The equivalent circuit used to fit the impedance data has been extracted from the previous literature,30,45 and described in Fig. 2. RS denotes solution resistance; Qdlc symbolizes double-layer capacitance. RCT signifies the charge transfer resistance, while QCell and RCell are the membrane capacitance and intracellular resistance of HeLa cells cultured on electrodes.
Fig. 2. Equivalent circuit of ECIS devices.
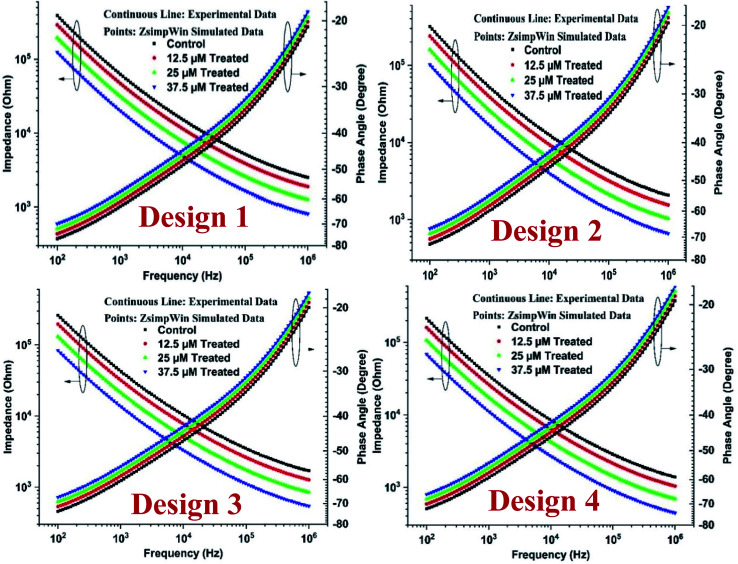
The Bode plot has been described as in Fig. 3, and it is apparent from the figures that the impedance value reduces with the rise of drug doses. The highest assessment of impedance was observed for the control sample because cells do not act as conductors and thus offer a lot of resistance to the flow of current, thereby increasing impedance value. Also, the obtained results demonstrate that the impedance magnitude is maximum for Design 1 and progressively drops with the escalations in dimensions of the electrode.
Fig. 3. Impedimetric cytotoxicity of tamoxifen on HeLa cells for all designs.
The impedance magnitude declines progressively with the surge in frequency. It is well known that at higher frequencies, the current permeates the cell instead of moving around the extra-cellular spaces, as seen in lower frequencies. However, there is a rise in the phase angle value of up to 1 MHz. The differences found in curves of control and drug-treated samples are because of the increased drug–cell interactions, which leads to the death of cells and decrease in impedance for higher drug doses as compared to previous literature.30,43 The relative standard deviations (RSD) for different designs for control and treated sample was obtained below 10%, signifying the higher reproducibility of the fabricated ECIS devices.29,30
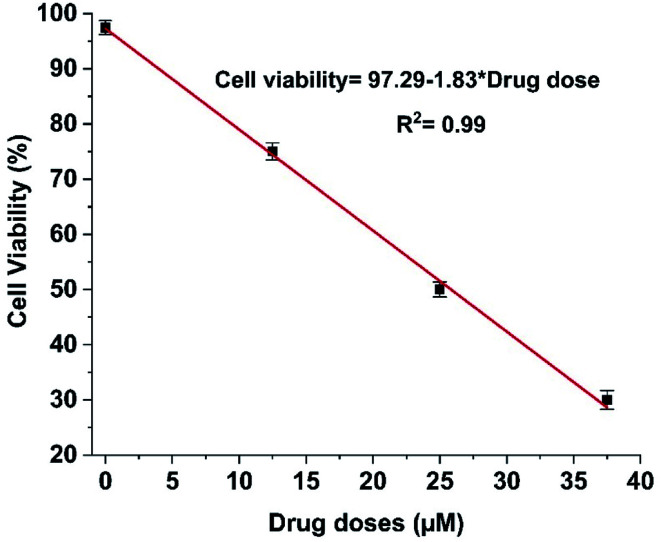
The IC50 value of tamoxifen in HeLa cells measured via MTT assay was 25.25 ± 0.52 μM. The cell viability percentage was computed from impedance using eqn (3)29 and deciphered in Fig. 4.
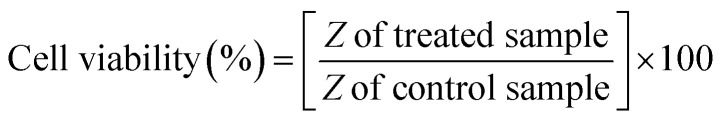 |
3 |
Fig. 4. Impedimetric cell viability calibration curve for tamoxifen on HeLa cells.
The IC50 value of tamoxifen obtained from magnitude, as described in Fig. 4, is 25.84 ± 0.12 μM. Thus, the IC50 value obtained from impedance data is comparable with IC50 value acquired from MTT assay, which infers that the fabricated ECIS devices can be used as an alternative method for cytotoxic evaluation of anti-cancer drugs.
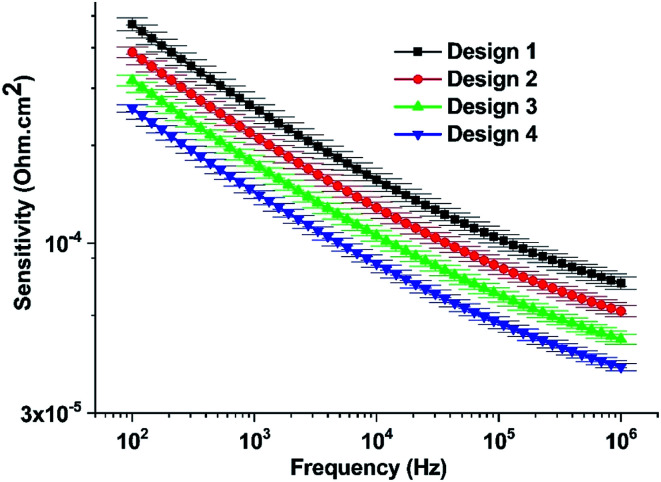
The sensitivity of four electrode-based biosensors was determined using eqn (2), and the corresponding plot has been denoted in Fig. 5. It can be apparent from the figure that Design 1 has maximum sensitivity and varies inversely to the working electrode area. The higher sensitivity of Design 1 may be attributed to the higher cell-binding ability of the smaller working electrode area. The sensitivity results obtained here for all the devices are comparable to previous literature.29,30
Fig. 5. The sensitivity of the biosensors for different designs.
3.2. Microscopic assessment of the cytotoxic effects of tamoxifen on HeLa cells
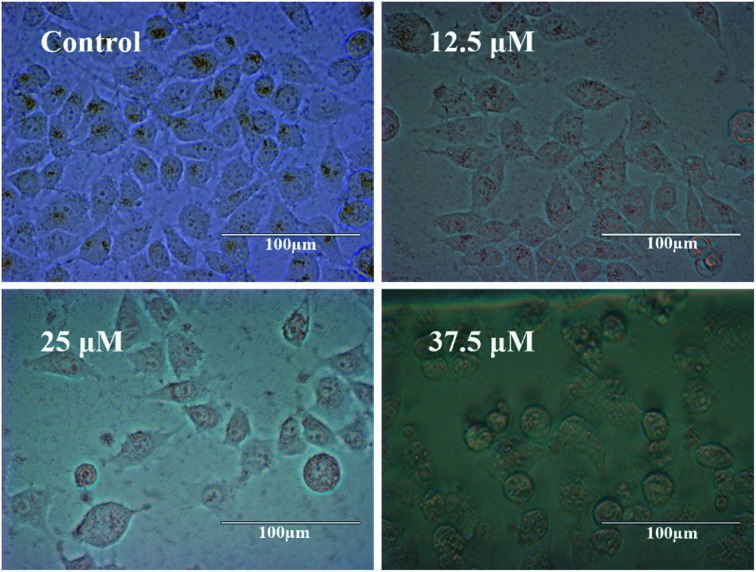
For observing the cytotoxicity of the chemotherapeutic drug tamoxifen, phase contrast imaging was done, and results have been shown in Fig. 6.
Fig. 6. Phase-contrast micrographs of HeLa cells treated with 0, 12.5, 25, and 37.5 μM of tamoxifen for 24 h (magnification 40×).
It can be inferred that with the increase of drug doses, the cell death increases, leading to a decrease in impedance as denoted in Fig. 3. Also, the percentage of living HeLa cells was calculated by using an automated cell counter (Invitrogen), and the obtained values for control and treated samples (12.5 μM, 25 μM, and 37.5 μM) are 98 ± 2, 69 ± 4, 45 ± 3, and 26 ± 5, respectively.
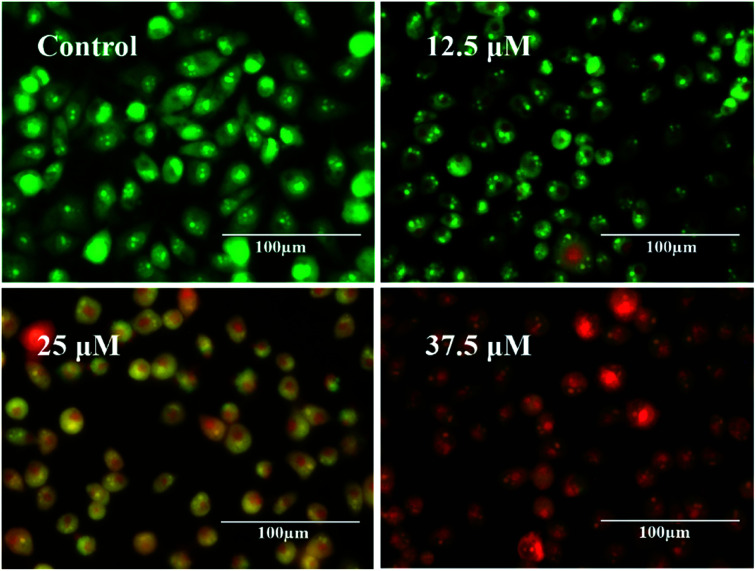
Next, fluorescence imaging was carried out for control and treated samples, and the acquired results have been indicated in Fig. 7. It can be evident that a large number of green cells are visible in control, indicating the presence of more live cells. In cells treated with 12.5 μM tamoxifen, green cells, along with some orange/red cells, are realized, indicating early apoptosis due to fragmentation of the nucleus.46 As the concentration of drug increases to 25 μM and 37.5 μM, more and more orange/red cells are observed, indicating higher cell death. Therefore, it can be anticipated that tamoxifen is exceptionally useful in destroying the cancerous HeLa cells due to its anti-estrogenic nature, which may be comparable to the other anti-cancer drugs for chemotherapeutic management of cervical cancer.
Fig. 7. Fluorescent micrographs of HeLa cell obtained from live/dead cell assay.
Further, the results obtained from live/dead cell assay can be compared qualitatively to impedimetric data. The live cells are stained green and found more in control data so that cellular impedance is higher in the untreated sample. The cell death is less in the case of lower drug doses as the IC50 of tamoxifen has been calculated to be 25.25 ± 0.52 μM signifying a lower impedance curve in the Bode plot. However, with the increase of drug concentration, the number of distorted orange/red dots increases indicating higher cell death, and the equivalent data curve of impedance decreases gradually.
3.3. Flow cytometry studies
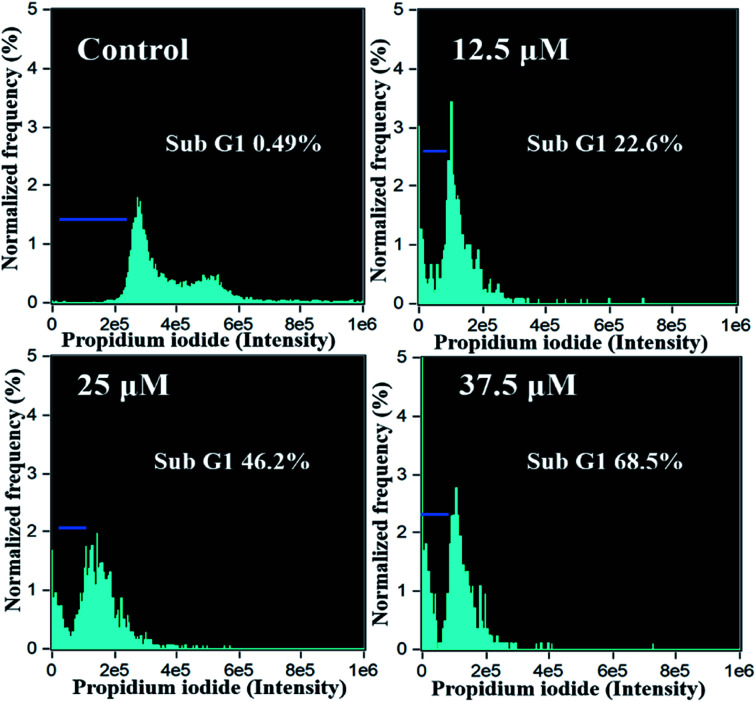
Flow cytometric analysis of the HeLa cells was carried out and depicted as in Fig. 8. It can be observed that the percentage of subG1 phase cells increases progressively with the increase of tamoxifen dose. There is approximately 46.2% and 68.5% relative increase in subG1 phase in cells treated with 25 and 37.5 μM of tamoxifen, respectively, compared to control value indicating higher cell death. Therefore, the accumulation of dead cells in the subG1 phase is dose-dependent, which is in good agreement with the impedimetric results obtained from fabricated biosensors.
Fig. 8. Flow cytometric analysis of HeLa cells treated with tamoxifen with different drug doses (0, 12.5, 25, 37.5 μM).
3.4. Cell morphology analysis
The cell morphology in the presence of different drug doses was studied by SEM and was described as in Fig. 9. Throughout cell growth and proliferation on the surface of the gold electrode, the impedance values of the control sample are high as the cells cover the electrode fully, indicating a tight attachment of the cell membrane with the electrode. At lower concentrations (12.5 μM) of the drug, cells are also in normal expanded morphology; however, at higher drug concentrations (37.5 μM) a large number of cells are rounded up and detached from the gold surface due to cell death as described in previous literature.31,47–51 Kustermann et al. reported that a rise in impedance reveals cell proliferation however, a reduction can be reflected as an insignia of cytotoxicity leading to death and detachment from the electrode surface.47 Similarly, Wei et al. also reported that with the increase of drug doses of doxorubicin, the cell death enhances which leads to the detachment of cells from the planer electrode areas.51
Fig. 9. SEM image analysis of HeLa cells treated with tamoxifen with different drug doses (0, 12.5, 25, 37.5 μM).
Also, RCell, as described in Fig. 2, has been calculated to understand the detachment of cells from the electrode as the percentage of RCell directly relates to the percentage of live cells present on the electrode surface.52 The percentage of RCell calculated for all the designs with respect to control value are 70 ± 2, 50 ± 4, 30 ± 3 for 12.5 μM, 25 μM, 37.5 μM treated samples, respectively. Thus, fewer cells are available on the electrode surfaces at higher drug doses, and most of the cells die and detach from the electrode surface, as noted in previous literature.31,53
3.5. Comparison of impedimetric data with conventional data
It can be evident from the above-performed experiments that cell death is directly proportional to drug doses. Cell death is significantly increased with the highest drug concentration, which is successfully reinforced by impedimetric studies. The impedance values are comparable for the cells treated with control and 12.5 μM of the drug, as the IC50 of tamoxifen is 25.25 ± 0.52 μM. The potential cytotoxic effect of tamoxifen is more prominent in treating drugs beyond this IC50 concentration, which involves obliteration of the spreading of HeLa cells in a dose-dependent manner necrosis/apoptosis of the cells. Literature suggests that the drug tamoxifen has been regularly used in all stages of breast cancer, restraining cellular chemoinvasion and proliferation.54,55 However, the cytotoxicity of tamoxifen against the cervical cancer cell line is being investigated for the first time as tamoxifen has an untoward association with gynecological cancers.56–58 The current study shows the restraining activity of tamoxifen over the growth and proliferation of HeLa cells by using impedimetric biosensors, which correlate well with the results obtained from different conventional techniques. However, the present four electrode based impidimetric method is advantageous in comparison to different conventional techniques as it is cost-effective, non-destructive, rapid, and provides the real time monitoring capability. Microscopic investigation of the sensors reveals apparent cell death near the electrode surfaces of the biosensors at higher drug doses indicating decreased impedance values. The dead cells detach themselves from the electrode surface of the impedimetric biosensor, as shown in SEM images, thereby exposing lesser cells on the electrode surface for impedance measurement. Therefore, it may be assumed that the biosensor's mechanism is dependent on the growth of HeLa cells on the surface of the electrodes and the detachment of apoptotic cells from the electrodes as described in previous literature.30,31
4. Conclusions
The current study demonstrates the cytotoxic evaluation of tamoxifen through the impedimetric studies further verified by MTT assay, live/dead assay, flow cytometric analysis, and detachment studies by SEM. In addition, the impedimetric assay has been compared with the different conventional techniques for the cytotoxic evaluation. The IC50 value obtained from impedimetric data is comparable to the IC50 value calculated from MTT assay. Moreover, the impedimetric biosensors indicated that tamoxifen causes significant cell death and detachment of HeLa cells from the electrode surface in a dose-dependent manner, as evident through the morphological analysis. Furthermore, the fabricated impedimetric biosensors provided an efficient platform to investigate the consequence of drug dosage on cellular behavior of the cancerous cell lines and its impedance characteristics by correlating the drug doses with impedance. Therefore, the development of biosensors based on impedance technique for determining cytotoxicity of drugs may be of great interest wherein anti-cancer different drugs can be tested for their cytotoxicity. In conclusion, this technique may be taken up for further investigation for cytotoxic evaluation of other anti-cancer drugs. Thereby, it may be safe to say that there is indeed a positive correlation of tamoxifen impedance values and other proven assays heralding the arrival of a novel method to evaluate the cytotoxicity of anti-cancerous drugs.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the Department of Biotechnology (No. BT/PR25095/NER/95/1011/2017), Government of India. R. P. and A. K. are thankful to the Ministry of Human Resource Development and S. J. is thankful to the Department of Biotechnology, Government of India, for the fellowship. Sincere thanks to Centre for Nanotechnology and Institute Instrumentation Centre, IIT Roorkee, for the various analytical facilities provided.
References
- Guan Y.-H. Tian M. Liu X.-Y. Wang Y.-N. J. Cell. Physiol. 2019;234:16475–16484. doi: 10.1002/jcp.28316. [DOI] [PubMed] [Google Scholar]
- Koivusalo R. Hietanen S. Cancer Biol. Ther. 2004;3:1177–1183. doi: 10.4161/cbt.3.11.1340. [DOI] [PubMed] [Google Scholar]
- Hausen H. z. J. Natl. Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- Small Jr W. Bacon M. A. Bajaj A. Chuang L. T. Fisher B. J. Harkenrider M. M. Jhingran A. Kitchener H. C. Mileshkin L. R. Viswanathan A. N. Gaffney D. K. Cancer. 2017;123:2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- Cohen P. A. Jhingran A. Oaknin A. Denny L. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- Small Jr W. Bacon M. A. Bajaj A. Chuang L. T. Fisher B. J. Harkenrider M. M. Jhingran A. Kitchener H. C. Mileshkin L. R. Viswanathan A. N. Cancer. 2017;123:2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- Waggoner S. E. Lancet. 2003;361:2217–2225. doi: 10.1016/S0140-6736(03)13778-6. [DOI] [PubMed] [Google Scholar]
- Savarese A. Cognetti F. Crit. Rev. Oncol. Hematol. 2003;48:323–327. doi: 10.1016/S1040-8428(03)00131-8. [DOI] [PubMed] [Google Scholar]
- Grenman S. Shapira A. Carey T. E. Gynecol. Oncol. 1988;30:228–238. doi: 10.1016/0090-8258(88)90029-7. [DOI] [PubMed] [Google Scholar]
- Karimi M. Z. Behtash N. Sekhavat L. Dehghan A. Asian Pac. J. Cancer Prev. 2009;10:595–598. [PubMed] [Google Scholar]
- Shukla V. C. Kuang T. R. Senthilvelan A. Higuita-Castro N. Duarte-Sanmiguel S. Ghadiali S. N. Gallego-Perez D. Trends Biotechnol. 2018;36:549–561. doi: 10.1016/j.tibtech.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Pan Y. Hu N. Wei X. Gong L. Zhang B. Wan H. Wang P. Biosens. Bioelectron. 2019;130:344–351. doi: 10.1016/j.bios.2018.09.046. [DOI] [PubMed] [Google Scholar]
- Kumar S. Ashish Kumar S. Augustine S. Yadav S. Yadav B. K. Chauhan R. P. Dewan A. K. Malhotra B. D. Biosens. Bioelectron. 2018;102:247–255. doi: 10.1016/j.bios.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Rong Y. Padron A. V. Hagerty K. J. Nelson N. Chi S. Keyhani N. O. Katz J. Datta S. P. A. Gomes C. McLamore E. S. Analyst. 2018;143:2066–2075. doi: 10.1039/C8AN00065D. [DOI] [PubMed] [Google Scholar]
- Frense D. Kang S. Schieke K. Reich P. Barthel A. Pliquett U. Nacke T. Brian C. Beckmann D. J. Phys.: Conf. Ser. 2013;434:012091. doi: 10.1088/1742-6596/434/1/012091. [DOI] [Google Scholar]
- Tersch C. Lisdat F. Electrochim. Acta. 2011;56:7673–7679. doi: 10.1016/j.electacta.2011.06.063. [DOI] [Google Scholar]
- Witte C. Lisdat F. Electroanalysis. 2011;23:339–346. doi: 10.1002/elan.201000410. [DOI] [Google Scholar]
- Erdem A., Eksin E. and Kesici E., Biosensors and Nanotechnology: Applications in Health Care Diagnostics., John Wiley & Sons, 2017, pp. 349–365, 10.1002/9781119065036.ch15 [DOI] [Google Scholar]
- Sadik O. A. Aluoch A. O. Zhou A. Biosens. Bioelectron. 2009;24:2749–2765. doi: 10.1016/j.bios.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Katz E. Willner I. Electroanalysis. 2003;15:913–947. doi: 10.1002/elan.200390114. [DOI] [Google Scholar]
- Gupta N. Renugopalakrishnan V. Liepmann D. Paulmurugan R. Malhotra B. D. Biosens. Bioelectron. 2019;141:111435. doi: 10.1016/j.bios.2019.111435. [DOI] [PubMed] [Google Scholar]
- Ceriotti L. Ponti J. Broggi F. Kob A. Drechsler S. Thedinga E. Colpo P. Sabbioni E. Ehret R. Rossi F. Sens. Actuators, B. 2007;123:769–778. doi: 10.1016/j.snb.2006.10.024. [DOI] [Google Scholar]
- Daza P. Olmo A. Canete D. Yufera A. Sens. Actuators, B. 2013;176:605–610. doi: 10.1016/j.snb.2012.09.083. [DOI] [Google Scholar]
- Xiao C. Luong J. H. Biotechnol. Prog. 2003;19:1000–1005. doi: 10.1021/bp025733x. [DOI] [PubMed] [Google Scholar]
- Gu A. Y. Kho D. T. Johnson R. H. Graham E. S. O’Carroll S. J. Biosensors. 2018;8:90. doi: 10.3390/bios8040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiquei H. R., Nordin A. N., Ibrahimy M. I., Arifin M. A., Sulong N. H., Mel M. and Voiculescu I., International Conference on Computer and Communication Engineering (ICCCE'10), IEEE, 2010 [Google Scholar]
- Huang L. Xie L. Boyd J. M. Li X.-F. Analyst. 2008;133:643–648. doi: 10.1039/B714384B. [DOI] [PubMed] [Google Scholar]
- Wegener J. Keese C. R. Giaever I. Exp. Cell Res. 2000;259:158–166. doi: 10.1006/excr.2000.4919. [DOI] [PubMed] [Google Scholar]
- Pradhan R. Mandal M. Mitra A. Das S. Sens. Actuators, B. 2014;193:478–483. doi: 10.1016/j.snb.2013.12.003. [DOI] [Google Scholar]
- Pradhan R. Rajput S. Mandal M. Mitra A. Das S. Biosens. Bioelectron. 2014;55:44–50. doi: 10.1016/j.bios.2013.11.060. [DOI] [PubMed] [Google Scholar]
- Anh-Nguyen T. Tiberius B. Pliquett U. Urban G. A. Sens. Actuators, A. 2016;241:231–237. doi: 10.1016/j.sna.2016.02.035. [DOI] [Google Scholar]
- Lei K. F. Wu Z.-M. Huang C.-H. Biosens. Bioelectron. 2015;74:878–885. doi: 10.1016/j.bios.2015.07.060. [DOI] [PubMed] [Google Scholar]
- Abdur Rahman A. R. Price D. T. Bhansali S. Sens. Actuators, B. 2007;127:89–96. doi: 10.1016/j.snb.2007.07.038. [DOI] [Google Scholar]
- Pradhan R. Mitra A. Das S. Electroanalysis. 2012;24:2405–2414. doi: 10.1002/elan.201200455. [DOI] [Google Scholar]
- Lind R. Connolly P. Wilkinson C. Thomson R. Sens. Actuators, B. 1991;3:23–30. doi: 10.1016/0925-4005(91)85004-3. [DOI] [Google Scholar]
- Breckenridge L. Wilson R. Connolly P. Curtis A. Dow J. Blackshaw S. Wilkinson C. J. Neurosci. Res. 1995;42:266–276. doi: 10.1002/jnr.490420215. [DOI] [PubMed] [Google Scholar]
- Tran T. B. Cho S. Min J. Biosens. Bioelectron. 2013;50:453–459. doi: 10.1016/j.bios.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Stockert J. C. Blazquez-Castro A. Canete M. Horobin R. W. Villanueva A. Acta Histochem. 2012;114:785–796. doi: 10.1016/j.acthis.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Kalkal A. Pradhan R. Kadian S. Manik G. Packirisamy G. ACS Appl. Bio Mater. 2020;3:4922–4932. doi: 10.1021/acsabm.0c00427. [DOI] [PubMed] [Google Scholar]
- Liu K. Liu P.-c. Liu R. Wu X. Med. Sci. Monit. Basic Res. 2015;21:15–20. doi: 10.12659/MSMBR.893327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi C. Nicoletti I. Nat. Protoc. 2006;1:1458–1461. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- Matai I. Sachdev A. Gopinath P. ACS Appl. Mater. Interfaces. 2015;7:11423–11435. doi: 10.1021/acsami.5b02095. [DOI] [PubMed] [Google Scholar]
- Pradhan R. Rajput S. Mandal M. Mitra A. Das S. RSC Adv. 2014;4:9432–9438. doi: 10.1039/C3RA45090B. [DOI] [Google Scholar]
- Wang L. Wang H. Wang L. Mitchelson K. Yu Z. Cheng J. Biosens. Bioelectron. 2008;24:14–21. doi: 10.1016/j.bios.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Pradhan R. Mandal M. Mitra A. Das S. IEEE Sens. J. 2014;14:1476–1481. [Google Scholar]
- Ogle A. E., Ethidium Bromide/Acridine Orange Viability Staining Method, 2010 [Google Scholar]
- Kustermann S. Boess F. Buness A. Schmitz M. Watzele M. Weiser T. Singer T. Suter L. Roth A. Toxicol. in Vitro. 2013;27:1589–1595. doi: 10.1016/j.tiv.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Atienza J. M. Yu N. Kirstein S. L. Xi B. Wang X. Xu X. Abassi Y. A. Assay Drug Dev. Technol. 2006;4:597–607. doi: 10.1089/adt.2006.4.597. [DOI] [PubMed] [Google Scholar]
- Park I. Nguyen T. Park J. Yoo A. Y. Park J. K. Cho S. J. Electrochem. Soc. 2018;165:B55–B59. doi: 10.1149/2.1201802jes. [DOI] [Google Scholar]
- Yeon J. H. Park J.-K. Anal. Biochem. 2005;341:308–315. doi: 10.1016/j.ab.2005.03.047. [DOI] [PubMed] [Google Scholar]
- Wei L. Wang S.-F. Gu H.-W. Li X. Biomed. Res. 2017;28:199–202. [Google Scholar]
- Senbiao Luo L. Z. Dai Y. Lu Y. Liu Q. Sens. Mater. 2018;30:1977–1987. [Google Scholar]
- Adcock A. F. Agbai C. O. Yang L. J. Anal. Sci. Technol. 2018;9:17. doi: 10.1186/s40543-018-0149-x. [DOI] [Google Scholar]
- Wickerham L. Breast Cancer Res. Treat. 2002;75(suppl. 1):S7–S12. doi: 10.1023/A:1020353530963. [DOI] [PubMed] [Google Scholar]; ; discussion S33
- Karimi Zarchi M. Behtash N. Sekhavat L. Dehghan A. Asian Pac. J. Cancer Prev. 2009;10:595–598. [PubMed] [Google Scholar]
- Fornander T. Rutqvist L. E. Wilking N. Ann. N. Y. Acad. Sci. 1991;622:469–476. doi: 10.1111/j.1749-6632.1991.tb37890.x. [DOI] [PubMed] [Google Scholar]
- Deligdisch L. Kalir T. Cohen C. J. de Latour M. Le Bouedec G. Penault-Llorca F. Gynecol. Oncol. 2000;78:181–186. doi: 10.1006/gyno.2000.5859. [DOI] [PubMed] [Google Scholar]
- Curtis R. E. Boice Jr J. D. Shriner D. A. Hankey B. F. Fraumeni Jr J. F. J. Natl. Cancer Inst. 1996;88:832–834. doi: 10.1093/jnci/88.12.832. [DOI] [PubMed] [Google Scholar]