Abstract
The Fusarium oxysporum species complex (FOSC) is a group of closely related plant pathogens long-considered strictly clonal, as sexual stages have never been recorded. Several studies have questioned whether recombination occurs in FOSC, and if it occurs its nature and frequency are unknown. We analysed 410 assembled genomes to answer whether FOSC diversified by occasional sexual reproduction interspersed with numerous cycles of asexual reproduction akin to a model of predominant clonal evolution (PCE). We tested the hypothesis that sexual reproduction occurred in the evolutionary history of FOSC by examining the distribution of idiomorphs at the mating locus, phylogenetic conflict and independent measures of recombination from genome-wide SNPs and genes. A phylogenomic dataset of 40 single copy orthologs was used to define structure a priori within FOSC based on genealogical concordance. Recombination within FOSC was tested using the pairwise homoplasy index and divergence ages were estimated by molecular dating. We called SNPs from assembled genomes using a k-mer approach and tested for significant linkage disequilibrium as an indication of PCE. We clone-corrected and tested whether SNPs were randomly associated as an indication of recombination. Our analyses provide evidence for sexual or parasexual reproduction within, but not between, clades of FOSC that diversified from a most recent common ancestor about 500 000 years ago. There was no evidence of substructure based on geography or host that might indicate how clades diversified. Competing evolutionary hypotheses for FOSC are discussed in the context of our results.
Key words: Ascomycota, Clonal reproduction, Index of association, Phylogenomic networks, Phylogenomics, Population genomics, Sexual reproduction, Taxonomic boundaries
Introduction
Fungi possess different reproductive strategies to increase their survival. Most fungi can reproduce mitotically, which facilitates rapid proliferation and maintains successful genotypes. Many fungi intersperse extended bouts of asexual reproduction with occasional sexual reproduction (Tsai et al. 2008, Tibayrenc & Ayala 2012, Taylor et al. 2015, Nieuwenhuis & James 2016) resulting in life cycles with many more asexual than sexual generations, termed predominant clonal evolution (PCE) (Tibayrenc & Ayala 2021).
The PCE model assumes clonal evolution is dominant and genetic recombination is rare, but not absent. Clonality is evidenced by statistically significant linkage disequilibrium and the presence of identical or near identical multilocus genotypes over space and time (Tibayrenc & Ayala 2012, Tibayrenc & Ayala 2014, Tibayrenc & Ayala 2017). Very few, if any, fungal species have forgone sexuality entirely, and most exchange genetic material through sexual and/or parasexual reproduction, hybridisation and/or horizontal gene transfer (Taylor et al. 2015, Nieuwenhuis & James 2016, Steenkamp et al. 2018). The occurrence of genetic recombination events is not always clear in fungal plant pathogens, especially where asexual stages dominate the life cycle (Taylor et al. 2015, Drenth et al. 2019).
Virulent fungal genotypes benefit from asexual recombination, which preserves favourable combinations of alleles over multiple generations, and avoids recombinational load. Asexual populations in general evolve through mutation and non-recombinant sharing of DNA (for example, Ma et al. 2010). Asexual populations may show low levels of recombination through inbreeding, i.e. haploid selfing (Billiard et al. 2012), with no or scarce evidence of sexual recombination (Tibayrenc & Ayala 2012, Tibayrenc & Ayala 2017). Geographic isolation, disruption of life cycles, or other mechanisms that structure populations, may restrain recombination. Evidence for reduced recombination in fungal populations is not evidence for the absence of sex in fungal populations.
The Fusarium oxysporum species complex (FOSC) is a ubiquitous and cosmopolitan group of fungi (Summerell 2019). FOSC includes economically and medically important pathogens (Dean et al. 2012) and mycotoxigenic fungi (Munkvold et al. 2019) that impact agriculture, horticulture, and human and animal health (O'Donnell et al. 2004, O'Donnell et al. 2016).
FOSC has been cited as an example in Fungi of evolution by restrained recombination (Tibayrenc & Ayala 2012). FOSC were thought to have evolved as independent clonal lineages (Gordon & Martyn 1997, Koenig et al. 1997). More recently, competing taxonomic hypotheses have treated FOSC as either a complex of more than 10 closely-related, asexual taxa (Lombard et al. 2019, Maryani et al. 2019), or fewer than five phylogenetic groups (Laurence et al. 2014, Brankovics et al. 2017, Achari et al. 2020). Each of these hypotheses depends on gene selection, taxon sampling and species criteria.
Studies on genetic diversity and plasticity indicate that FOSC is comprised of clonal populations that have exchanged DNA through homologous recombination of mitochondrial genomes and horizontal sharing of nuclear chromosomes. O'Donnell et al. (2004) showed that opposite mating types were present in populations of FOSC in the United States and questioned whether MAT genes were functional. Ma et al. (2010) showed that F. oxysporum shared accessory chromosomes through horizontal chromosome transfer. Laurence et al. (2015) and van Dam & Rep (2017) used phylogenetic incongruence of genes and mobile elements to demonstrate the natural occurrence of horizontal gene transfer in FOSC. Vlaardingerbroek et al. (2016) found horizontal transfer of chromosomes in FOSC as well as homologous recombination among the core chromosomes. Brankovics et al. (2017) and Achari et al. (2020) concluded there was evidence of recombination in mitochondria of FOSC.
We used 410 publicly available genomes of FOSC to resolve the current ambiguity around speciation in FOSC. Phylogenomics and population genomics were used to test the hypothesis that F. oxysporum diversified by sexual reproduction under a model of predominant clonal evolution. Evidence of sexual reproduction was sought from (i) near-equal frequencies of different MAT-idiomorphs among phylogroups of FOSC, (ii) phylogenetic incongruence and reticulation of single-copy orthologs, (iii) random associations of SNPs in clone-corrected data, and (iv) past activity of repeat-induced point mutation (RIP) as a signature of meiosis. Evidence of PCE was sought from statistically significant linkage disequilibrium in non-clone-corrected data. The occurrence, contribution and mechanisms of sexual recombination in Fusarium is fundamental to understanding their diversity and a key component in developing strategies for disease management.
Methods
Genome sampling and annotation
We downloaded 490 nucleotide assemblies of FOSC publicly available on GenBank (including genomes from these studies: Ma et al. 2010, Thatcher et al. 2012, Guo et al. 2014, Kitts et al. 2016, Pu et al. 2016, van Dam et al. 2016, Williams et al. 2016, Singh et al. 2017, van Dam & Rep 2017, Armitage et al. 2018, Ayhan et al. 2018, Lv et al. 2018, Urbaniak et al. 2018, Asai et al. 2019, Gebru et al. 2019, Henry et al. 2019, Seo et al. 2019, Taylor et al. 2019, Urbaniak et al. 2019, Achari et al. 2020, Batson et al. 2020, Fokkens et al. 2020, Henry et al. 2020, Hudson et al. 2020, Kanapin et al. 2020, Khayi et al. 2020, Kim et al. 2020, Krasnov et al. 2020, Li et al. 2020, Srivastava et al. 2020, Thangavelu et al. 2020, Wang et al. 2020, Yu et al. 2020, Zhang et al. 2020, Henry et al. 2021, Chang & Cook unpublished). Genes were predicted in Augustus using a model of F. graminearum (Stanke & Morgenstern 2005). Seqtk (available at: https://github.com/lh3/seqtk) was used to exclude assemblies that had fewer than 3 700 genes longer than 600 amino acids; 410 assemblies met our criteria for inclusion (Table S1).
Identification of orthologs and MAT idiomorphs
We used OrthoFinder v. 1.0.6 (Emms & Kelly 2019) with a Diamond search (Buchfink et al. 2015) to identify single copy orthologs in annotated genomes of F. oxysporum. MAT genes annotated from KT876066 (MAT1-1-1) and KT883580 (MAT1-2-1) were searched for in orthogroup outputs from OrthoFinder, and PopART (Leigh & Bryant 2015) was used to visualise networks of haplotypes.
Phylogenetic analyses and tests for recombination with single copy orthologs
We used phylogenetic concordance of 40 single copy orthologs (48 868 amino acids) identified by OrthoFinder to assign structure a posteriori from 410 genomes of FOSC and test for recombination among and within the assigned groups. Analysed genes occurred on 10 of 11 core chromosomes in a chromosome-level assembly of F. oxysporum (Table S2, Fokkens et al. 2020). Single copy orthologs were aligned using default settings with MUSCLE (Edgar 2004), trimmed using ClipKIT (Steenwyk et al. 2020) and concatenated with FASconCAT-G (Kück & Longo 2014). The most likely tree was searched in IQ-TREE v. 2 (Minh et al. 2020b) with a model test for each partition (command -spp -m TEST), 10 000 ultrafast bootstraps (Minh et al. 2013), and genealogical concordance factors calculated from gene trees for each locus and applied to the concatenated topology (Minh et al. 2020a). We considered a group well-supported if it was recovered by at least eight out of 40 single copy orthologs (20 % genealogical concordance factor, from here referred to as phylogroups).
We visualised the discordance of gene tree topologies using DensiTree v. 2.2.5 (Bouckaert 2010). We visualised putative recombination events from the aligned, single-copy orthologs as a neighbour net in SplitsTree v. 4.14.8, and tested recombination by calculating the pairwise homoplasy index (PHI) for the entire dataset and each phylogroup (Huson & Kloepper 2005).
Dating analyses with subsampled genomes and loci
The divergence ages of phylogroups in F. oxysporum were estimated to determine whether reproduction has occurred on a long or short geological time scale. We subsampled the phylogroups to reduce potential tree space, including 118 of the 410 genomes, and two outgroup taxa, F. fujikuroi and F. verticillioides. We used 11 of the 40 single-copy orthologs (11 999 amino acids) that were congruent with the concatenated topology, which was constrained for divergence analyses in BEAST v. 2.5.2 (Bouckaert et al. 2019). The most recent common ancestor of F. oxysporum was calibrated with a mean age of 5 million years, and a log normal distribution sampled ages between 0.198–26.5 mya (median distribution 2.29 mya). The sampling age covers time estimated for a clock-like rate of speciation (Hedges et al. 2015), and has a wide sampling space based on age estimates for Fusarium in studies of Sordariomycetes (van der Nest et al. 2015).
Identification of SNPs and tests for recombination
We used kSNP v. 3.1.2 (Gardner et al. 2015) with strict settings to identify SNPs across all genomes and in each of the phylogroups. kSNP used kmers to search for SNPs at sites present in coding regions annotated by Augustus (mean size of 24.4 million base pairs) of all genomes (min_frac = 1.0) over 101 homologous base pairs for the entire dataset (k = 101) and 31 homologous base pairs in each phylogroup (k = 31). These settings ensured each SNP site was homologous in all genomes, with either 101 or 31 flanking base pairs.
We visualised evolutionary relationships using SNP data from across the whole dataset and in each phylogroup using SplitsTree. The index of association (Brown et al. 1980) implemented in the R package poppr (Kamvar et al. 2014, R Core Team 2014) was used to test for evidence of clonality in phylogroups (where significant linkage disequilibrium is expected due to linkage among loci) and whether there was evidence of recombination in clone-corrected SNP data. We pruned SNP loci that were under linkage disequilibrium using PLINK (Chang et al. 2015), with a window size set to the number of SNPs, and a strict r2 threshold of 0.999. Haploid VCF files were imported into R with vcfR (Knaus & Grünwald 2017), and the bitwise.ia and samp.ia functions in poppr were used to determine the standardised index of association (rbarD) as a measure of linkage disequilibrium (Agapow and Burt, 2001). Genomes were clone-corrected based on their genetic distance across 40 loci using SplitsTree with a cut-off of <0.00002.
Analyses of repeat-induced point mutation
We searched for evidence of past RIP activity in genome assemblies with the highest N50 value from each phylogroup as an indication of meiosis in FOSC. We used the REPET pipeline (available at: http://urgi.versailles.inra.fr/index.php/urgi/Tools/REPET Bao & Eddy 2002, Quesneville et al. 2003, Edgar & Myers 2005, Flutre et al. 2011) to identify and annotate TE families, then searched the most dominant retrotransposon family for RIP-like mutations. We aligned TE sequences using MAFFT (Katoh & Standley 2013) and used RIPCAL (Hane & Oliver 2008) to quantify RIP-like mutations using the alignment-based method and majority consensus options. Only TE copies that were longer than half of the total alignment length were considered in the analyses.
Application of taxonomic names to phylogroups
We applied taxonomic names based on a phylogenetic species hypothesis of the TEF and RPB2 genes, aligned to the dataset of Lombard et al. (2019). The most likely tree was searched for in IQ-TREE v. 2 (Minh et al. 2020b) with a model test for each partition (command -spp -m TEST), and 10 000 UltraFast Bootstraps and aLRT values calculated for each node.
Results
Analyses of single copy orthologs
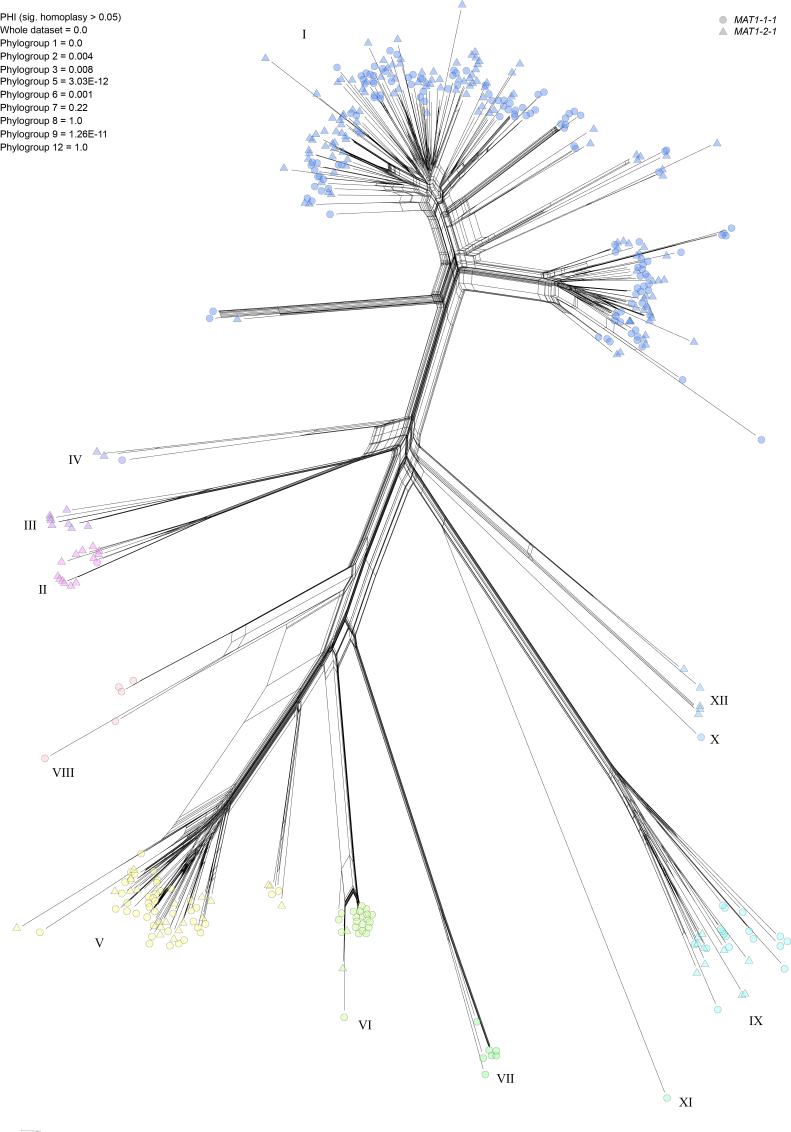
Genealogical concordance (20 % of 40 loci) from a maximum likelihood analysis (Fig. S1) recovered 12 phylogroups, congruent with relationships from a neighbour net analysis in SplitsTree (Fig. 2). Phylogenetic analyses and PHI tests provided evidence of recombination within F. oxysporum (Table 1). Single copy orthologs were not concordant in phylogenetic analyses based on genealogical concordance factors (Figs 1, S1) and visualisation of incongruence between topologies in DensiTree (Fig. S2), which indicates either incomplete lineage sorting of the 40 selected genes, or recombination between selected genomes. The reticulate neighbour net for the entire dataset was evidence of recombination, with homoplasy and incomplete lineage sorting as alternative explanations of reticulation. PHI test values < 0.05 supported recombination across the entire dataset (PHI = 0.0) as well as within most phylogroups, with the exceptions of phylogroups 7, 8 and 12. Phylogroups 2, 3 and 6 had less support for recombination based on their higher PHI scores than phylogroups 1, 5 and 9.
Fig. 2.
SplitsTree neighbour network based on 40 concatenated genes. Tests for the pairwise homoplasy index calculated in SplitsTree are provided for the entire dataset and alignments of individual phylogroups. Genotypes are coloured by phylogroup. SplitsTree networks show all putative evolutionary relationships between tips and reticulation is an indication of recombination.
Table 1.
Summary of datasets, distribution of MAT idiomorphs, dating and phylogenetic analyses in all phylogroups.
| Phylogroup | Genomes | Taxonomic names | MAT1-1 | MAT1-2 | χ2 | P-value | PHI | Mean age at MRCA | Distribution | Monophyletic TEF/RPB2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 256 | F. fabacearum, F. gossypinum, F. glycines, F. cugenangense, F. elaeidis, F. callistephi, F. carminascens, F. duoseptatum, F. tardichlamydosporum | 119 | 137 | 1.266 | 0.2606 | 0 | 142 000 | Australia, China, Ethiopia, Greece, Israel, Japan, Netherlands, Russia, Spain, Taiwan, UK, USA | Paraphyletic with Phylo4 |
| 2 | 13 | F. oxysporum | 1 | 12 | 9.308 | 0.0023 | 0.004 | 46 000 | Australia, Italy, Spain | Yes |
| 3 | 8 | NA | 0 | 8 | 8 | 0.0047 | 0.008 | 62 000 | Australia, Ethiopia, India | Yes |
| 4 | 3 | NA | 1 | 2 | 0.333 | 0.5637 | NA | 23 000 | Ethiopia, India, Japan | No |
| 5 | 61 | F. nirenbergiae, F. curvatum | 42 | 19 | 8.672 | 0.0032 | 3.03E-12 | 109 000 | Australia, Canada, China, France, Greece, Morocco, Netherlands, South Korea, Space, Spain, Switzerland, UK, USA | No |
| 6 | 23 | F. languescens | 21 | 2 | 15.696 | 0.0001 | 0.001 | 51 000 | Australia, France, India, Netherlands, Spain, USA | Yes |
| 7 | 7 | F. contaminatum, F. veterinarium, F. pharetrum | 7 | 0 | 7 | 0.0082 | 0.22 | 74 000 | Australia, Space, Ukraine, USA | Yes |
| 8 | 5 | F. triseptatum | 5 | 0 | 5 | 0.0253 | 1.0 | 102 000 | Australia, Spain, USA | Yes |
| 9 | 27 | F. odoratissimum | 17 | 10 | 1.286 | 0.2568 | 1.25E-11 | 79 000 | Australia, China, India, Malaysia, UK | Yes |
| 10 | 1 | NA | 1 | 0 | NA | NA | NA | NA | USA | NA |
| 11 | 1 | NA | 1 | 0 | NA | NA | NA | NA | Spain | NA |
| 12 | 5 | F. libertatis | 0 | 5 | 5 | 0.0253 | 1.0 | 89 000 | Australia | No |
| All | 410 | NA | 215 | 195 | 0.976 | 0.3233 | 0 | 382 000 | NA | NA |
Taxonomic names based on phylogenetic species concept in Fig. S5.
χ2 = Chi-square value relative to the expected 1:1 ratio from a random mating population.
P-value = Two-tailed P-value obtained from Chi-square test.
PHI = Pairwise Homoplasy Index over 40 loci (sig. < 0.05).
NA = not applicable.MRCA = Most recent common ancestor (calibrated to 5 million years at MRCA of F. oxysporum).
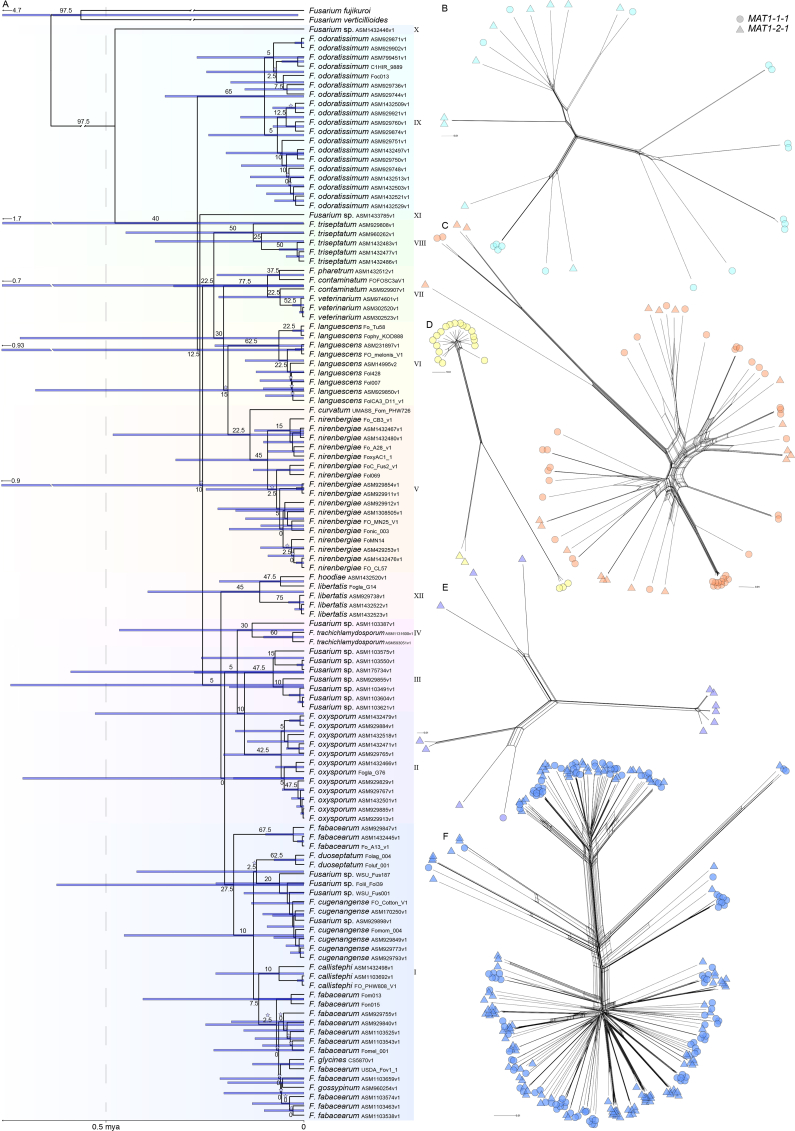
Fig. 1.
A. BEAST time-tree estimate with ages sampled from 11 concatenated, protein-coding genes, calibrated to a mean age of 5 million years at the most recent common ancestor of the Fusarium oxysporum species complex. The BEAST search was constrained to a maximum likelihood topology from 40 concatenated, protein-coding genes in IQ-TREE, from which genealogical concordance factors are provided above nodes. Blue stars indicate nodes that were not supported by UltraFast Bootstraps (<95 %, 10 000 replicates). Taxon names are based on a phylogenetic species hypothesis shown in Fig. S5. B–E. SplitsTree neighbour networks based on SNPs called in each phylogroup. B. Phylogroup 9 (471 669 SNPs), C. Phylogroup 5 (625 191 SNPs), D. Phylogroup 6 (70319 SNPs), E. Phylogroup 2 (197 928 SNPs), F. Phylogroup 1 (1 630 980 SNPs).
The BEAST analyses recovered a mean age for the most recent common ancestor of FOSC as 382 000 years ago (maximum sampled age 1.47 million years ago), with sampled ages calibrated to a mean of 5 million years (Fig. 1). All phylogroups were sampled with a mean age lower than 200 000 years, with the maximum sampled ages of phylogroup 1 the oldest at 547 000 years ago (mean 142 000).
MAT idiomorphs
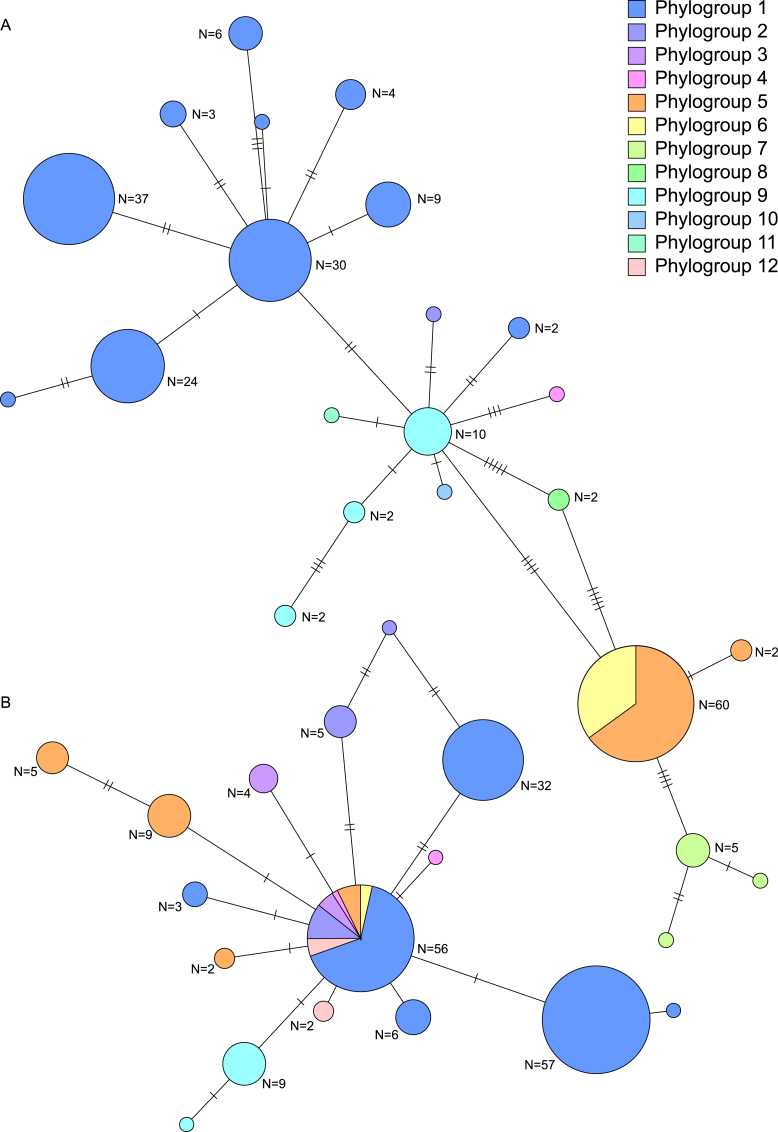
The frequencies of MAT1-1-1 (52 %) and MAT1-2-1 (48 %) across the entire dataset were not significantly different from a 1:1 ratio (Table 1). Phylogroups with PHI test values <0.05 had similar frequencies of MAT1-1-1 and MAT1-2-1, which supports recombination in those groups, i.e. phylogroup 1 with MAT1-1-1 (46 %) and MAT1-2-1 (54 %), phylogroup 5 with MAT1-1-1 (69 %) and MAT1-2-1 (31 %), and phylogroup 9 with MAT1-1-1 (63 %) and MAT1-2-1 (37 %). Phylogroups with lower support for recombination based on high PHI test values had a high frequency of one copy of a MAT locus, e.g. phylogroup 2 with MAT1-1-1 (8 %) and MAT1-2-1 (92 %), phylogroup 3 with MAT1-1-1 (0 %) and MAT1-2-1 (100 %), phylogroup 6 with MAT1-1-1 (91 %) and MAT1-2-1 (9 %), phylogroup 7 with MAT1-1-1 (100 %) and MAT1-2-1 (0 %), phylogroup 8 with MAT1-1-1 (100 %) and MAT1-2-1 (0 %), and phylogroup 12 with MAT1-1-1 (0 %) and MAT1-2-1 (100 %). Haplotypes of MAT1-2-1 were admixed within phylogroups, and sequences of MAT1-1-1 and MAT1-2-1 varied in phylogroups, although MAT1-2-1 showed more sequence conservation (Fig. 3). MAT1-1-1 had greater variability and fewer shared genotypes in different phylogroups than MAT1-2-1.
Fig. 3.
Haplotype networks from alignments of (A) MAT1-1-1 and (B) MAT1-2-1. Haplotypes are coloured by whether they are present in different phylogroups.
Analyses of SNP data
Up to 56 % of called SNPs were filtered from phylogroups based on their r2 pairwise frequencies across all loci (Table 2). The rbarD values, which approach zero in randomly recombining populations, were high (>0.01) for all phylogroups in non-clone-corrected data (Table 2). The non-clone-corrected rbarD values in phylogroups 1, 5 and 9 were an order of magnitude smaller than other phylogroups and suggested incomplete linkage between sites and recombination. Clone-corrected datasets reduced the standardised index of association by an order of magnitude, which supported recombination in all examined phylogroups except 6 and 8. rbarD values did not change by an order of magnitude when phylogroup 9 and the entire dataset were clone-corrected, which may indicate near-clones were not removed with the criteria for clone-correction.
Table 2.
Tests for linkage disequilibrium based on the standardised index of association, which approaches zero in recombining populations.
| Phylogroup | Non-clone-corrected |
Clone-corrected |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genomes | No. called SNPs1 | LD corrected SNPs2 | % SNPs removed under LD | rbarD3 | Genomes | No. called SNPs1 | LD corrected SNPs2 | % SNPs removed under LD | rbarD3 | |
| 1 | 256 | 1 630 980 | 714 955 | 56 | 0.014 | 61 | 802 433 | 413 923 | 48 | 0.002 |
| 2 | 13 | 197 928 | 140 357 | 29 | 0.111 | 9 | 183 604 | 136 284 | 26 | 0.057 |
| 3 | 8 | 150 897 | 102 449 | 32 | 0.155 | 4 | 120 558 | 95 211 | 21 | 0.018 |
| 4 | 3 | 99 029 | 93 548 | 6 | 0.245 | NA | NA | NA | NA | NA |
| 5 | 61 | 625 191 | 352 735 | 44 | 0.025 | 28 | 401 258 | 238 426 | 41 | 0.008 |
| 6 | 23 | 70 319 | 54 140 | 23 | 0.455 | 9 | 63 419 | 54 160 | 15 | 0.316 |
| 7 | 7 | 141 561 | 120 891 | 15 | 0.228 | 4 | 136 590 | 119 677 | 12 | 0.004 |
| 8 | 5 | 117 541 | 100 285 | 15 | 0.326 | 4 | 114 316 | 99 302 | 13 | 0.220 |
| 9 | 27 | 471 669 | 310 211 | 34 | 0.026 | 17 | 440 908 | 296 782 | 33 | 0.021 |
| 10 | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 11 | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 12 | 5 | 144 481 | 124 829 | 14 | 0.337 | 3 | 132 108 | 119 599 | 9 | 0.021 |
| All | 410 | 4 664 221 | 4 628 213 | 1 | 0.0344 | 136 | 748 225 | 377 645 | 50 | 0.0384 |
Number of SNPs from a k-mer search (k = 31 for phylogroups, k = 101 for all) of coding sequences within each phylogroup using kSNP.
SNPs were filtered based on an r2 cutoff of 0.9999 calculated in PLINK.
Calculated from bitwise.ia across LD corrected SNPs in poppr.
Calculated from samp.ia with 1 000 repeats of 1 000 SNPs in poppr.
SplitsTree relationships based on SNP loci are shown for the entire dataset (Fig. S5) and recombinant phylogroups (Fig. 1). The neighbour net based on SNP data across the entire dataset was congruent with evolutionary hypotheses obtained with the 40 single-copy orthologs (compare Figs 2 and S3).
Analyses of RIP
RIPCAL analyses of the dominant retrotransposon families from a genome of each of the phylogroups indicated RIP activity in all phylogroups of FOSC (Fig. S4). The most frequent RIP type was CpT > TpT mutation followed by CpA > TpA mutation.
Application of taxonomic names
Based on a concatenated phylogeny of TEF and RPB2, the phylogroups were monophyletic, with the exception of phylogroups 4 and 12 (Table 1, Fig. S5). Species names can be applied to phylogroups 2 (F. oxysporum), 6 (F. languescens), 8 (F. triseptatum), 9 (F. odoratissimum) and 12 (F. libertatis). Phylogroup 5 included two monophyletic groups and two named species (F. nirenbergiae, F. curvatum). Phylogroup 7 was monophyletic with three named species (F. contaminatum, F. pharetrum and F. veterinarium), of which F. veterinarium applies to species isolated from the international space station and humans. Phylogroup 1 contained nine names applied by Lombard et al. (2019), and was similarly rich in names below species rank (Table S1). The TEF and RPB2 genes had congruent topologies and supported a hypothesis of restrained recombination in lineages of F. oxysporum.
Discussion
We have shown using gene and SNP data from 410 genomes that FOSC fits a model of predominant clonal evolution. The extant diversity of FOSC is the product of long-term sexual or parasexual reproduction, as demonstrated by (i) equal frequencies of mating-type idiomorphs within phylogroups, (ii) low phylogenetic concordance between 40 orthologous genes, (iii) significant recombination based on PHI, (iv) standardised indices of association that were evidence of recombination in clone-corrected datasets, and (v) past activity of RIP in TEs from all phylogroups. Asexual reproduction is frequent, evidenced by multiple near clones in all phylogroups and high rbarD values in non-clone-corrected phylogroups. We estimated the recent common ancestor for all lineages of F. oxysporum existed approximately 500 000 years ago. Independent lineages have likely diversified within the last 200 000 years, with episodic recombination within, but not between, restrained phylogroups.
FOSC shows signatures of restrained recombination, as evidenced by clustering in phylogenetic groups of house-keeping genes (shared multilocus genotypes in TEF and RPB2) and SNP sites under linkage disequilibrium in all phylogroups (Tibayrenc & Ayala 2012). Extant phylogroups of FOSC evolved from recombinant ancestors without subsequent outcrossing among sister lineages, and the diverse haplotype network of MAT1-1-1 suggests that mating is restricted within phylogroups. Based on the divergence age estimates of monophyletic phylogroups, and patterns of reticulation and edge lengths in SplitsTree analyses, phylogroups have not exchanged core genes by recombination since they shared a most recent common ancestor. We interpreted central reticulation in SplitsTree analyses as recombination, although incomplete lineage sorting and homoplasy are alternative explanations.
RIP is active during sexual reproduction (Hane et al. 2015), and has supported sexual reproduction in other Fungi, including taxa hypothesized to be clonal (Ikeda et al. 2002, Braumann et al. 2008, Crouch et al. 2008, Ropars et al. 2012). The activity of RIP in all phylogroups supported meiosis in FOSC, although signatures of RIP are not evidence of recent recombination. Hane et al. (2015) consider sexual reproduction the most likely explanation of RIP, and outlined possible alternatives to explain RIP in clonal species, namely loss of sexual reproduction, non-meiotic RIP and horizontal transfer from a meiotic donor.
Taylor et al. (2015) used Cryptococcus gattii as a case study (see Engelthaler et al. 2014) to show that clones under restrained recombination warranted names to improve communication between researchers. Clones under restrained recombination may be short-lived in an evolutionary sense (Drenth et al. 2019), but as agricultural pathogens they have long-lasting impact to humans. The assignment of (species) names to pathogens that have become successful clones under predominant clonal evolution can be accommodated under the current nomenclatural code.
Maryani et al. (2019) and Lombard et al. (2019) proposed taxonomic hypotheses that reflected restrained recombination in FOSC. Our phylogenomic and population genomic analyses partially support their taxonomies. We found congruent taxonomic groups (phylogroups) with sexual reproduction within these groups, but not between them, demonstrated by reticulation within but not among phylogroups in network analyses. Phylogroups 1 and 5 contain multiple species names. Networks based on over 600 000 SNPs revealed distinct clusters within phylogroups 1 and 5, despite recent evidence of recombination across them. We did not find population substructure based on host or geography in any of the phylogroups. Nor did we find pathogenicity on different hosts as a reliable indicator for taxonomic identification.
Successful asexual clones (genotypes) of FOSC are often collected during plant disease outbreaks. The sampling in our study may be biased by the collection of F. oxysporum during plant disease outbreaks, which results in under-sampling of non-pathogenic lineages. Recombination was most evident in phylogroups that were well-sampled, either based on a PHI test, or with rbarD values that approached zero in clone-corrected and non-clone-corrected datasets (e.g. phylogroups 1 and 5). In the more highly sampled phylogroups we detected near-equal frequencies of both mating types, a result that is most congruent with sexual reproduction. We hypothesise that with increased sampling, recombination will be supported in all lineages and that both mating types will be found in all lineages. The implication is that capacity for sexual/parasexual reproduction is maintained, despite predominant clonal evolution, as it is required for long-term adaptability of the species.
Our criteria to include single copy genes and SNPs that were present in all genomes analysed likely excluded loci on accessory chromosomes, which may be horizontally transferred between individuals without subsequent meiosis. A limitation of our study is that we did not test whether horizontal chromosome transfer, without recombination, occurs among phylogroups of FOSC.
Physical evidence of sexual recombination (ascomata) in FOSC has never been found. Leslie & Summerell (2006) reported structures that resembled sclerotia in cultures of F. oxysporum and hypothesised these were protoperithecia, similar in morphology to those of the F. fujikuroi species complex. Attempts to cross these isolates and produce sexual structures have been unsuccessful (Visser et al. 2005, and Summerell unpublished data). Parasexual reproduction is a competing hypothesis with sexual reproduction to explain recombination in FOSC in the absence of a meiotic stage, however, parasexual reproduction in nature may be hard to witness, may occur on host taxa that are less commonly investigated, and is a less parsimonious explanation for the occurrence of equal mating type frequencies.
O'Donnell et al. (2004) outlined other hypotheses competing with sexual reproduction and explained the presence of two mating loci in populations of FOSC as a remnant of past sexual reproduction and/or that the MAT genes have evolved a different function and no longer regulated mating. Tibayrenc & Ayala (2014) proposed that restrained recombination is a ubiquitous evolutionary strategy used to avoid recombinational load or break-up of favourable multilocus allele combinations, and is another valid hypothesis to explain a lack of recombination.
Asexual reproduction has evolutionary advantages for successful genotypes in the short-term, whereas long-term clonal genotypes decline via the accumulation of deleterious mutations (McDonald et al. 2016). Infrequent sexual/parasexual reproduction may lead to skewed mating type frequencies, as seen in MAT1-1-1 of FOSC, further reducing the potential for sexual reproduction. Despite fitting a model of predominant clonal evolution and the short-term success of clones, FOSC has maintained the capacity for sexual/parasexual reproduction. The signature of recombination in FOSC is clear and may guide species-rank taxonomic hypotheses for these fungi.
Data availability
All data and commands are available at https://drive.google.com/drive/folders/17EIfIiuLLR-1wx_hXK0d-UvNc4X5XhB7?usp=sharing
Acknowledgements
ARM acknowledges the University of Queensland Development Fellowships (UQFEL1718905) and support from the Department of the Environment and Energy under the Australian Biological Resources Study (grant number RG18-43). TAD and BDW acknowledge the Tree Protection Co-operative Programme (TPCP), the National Research Foundation of South Africa and the DST-NRF Centre of Excellence in Tree Health Biotechnology (CTHB). We thank Doug Cook for preliminary access to genomic data and comments that helped improve the manuscript. We thank two anonymous reviewers for their improvements to the final manuscript.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2021.100132.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Genome accession data, phylogroup, mating type and binomial applied based on a phylogenetic species hypothesis (Fig. S5) for all isolates used in study.
Location of single copy orthologs in the chromosome assembly of Fusarium callistephi (GCA_014154955).
Phylogram obtained from a maximum likelihood search of 40 concatenated, protein-coding genes in IQ-Tree, with a model test for each partition, and genealogical concordance factors provided above nodes. Genealogical concordance factors used to define phylogroups are in bold.
DensiTree visualisation of 40 single copy gene trees showing that the examined single copy orthologs were not concordant.
SplitsTree neighbour network based on 100 000 random SNPs out of 4.6 million called from kmers of 101 base pairs across 410 genomes of the Fusarium oxysporum species complex using kSNP. Genotypes are coloured by phylogroup.
Analyses of repeat-induced point mutation (RIP) for the highest quality genome (based on N50) in each phylogroup. Top panels display the distribution of the RIP-like mutations on alignments of TE copies. The bottom graphs show the frequency of different RIP-like mutations (orange lines indicate frequency of CpA > TpA mutations, purple lines indicate frequency of CpC > TpC mutation, green lines indicate frequency of CpG > TpG mutation, and turquoise lines indicate frequency of CpT > TpT mutation). Top row, phylogroups 1, 2, 3, 4; middle row, phylogroups 5, 6, 7, 8; bottom row, phylogroups 9, 10, 11, 12.
Phylogenetic hypothesis based on a maximum likelihood search of a concatenated alignment of TEF and RPB2 in IQ-Tree. Alignment is based on Lombard et al. (2019) and used to apply names in the Fusarium oxysporum species complex. aLRT and ultrafast bootstrap values from 10 000 replicates above nodes.
References
- Achari S.R., Kaur J., Dinh Q., et al. Phylogenetic relationship between Australian Fusarium oxysporum isolates and resolving the species complex using the multispecies coalescent model. BMC Genomics. 2020;21:248. doi: 10.1186/s12864-020-6640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agapow P.M., Burt A. Indices of multilocus linkage disequilibrium. Molecular Ecology Notes. 2001;1:101–102. [Google Scholar]
- Armitage A.D., Taylor A., Sobczyk M.K., et al. Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporumf. sp.cepae. Scientific Reports. 2018;8:13530. doi: 10.1038/s41598-018-30335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S., Ayukawa Y., Gan P., et al. High-quality draft genome sequence of Fusarium oxysporumf. sp.cubense strain 160527, a causal agent of Panama disease. Microbiology Resource Announcements. 2019;8 doi: 10.1128/MRA.00654-19. e00654-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayhan D.H., López-Díaz C., Di Pietro A., et al. Improved assembly of reference genome Fusarium oxysporumf. sp.lycopersici strain Fol4287. Microbiology Resource Announcements. 2018;7:e00910–e00918. doi: 10.1128/MRA.00910-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Research. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batson A.M., Fokkens L., Rep M., et al. Putative effector genes distinguish two pathogenicity groups of Fusarium oxysporumf. sp.spinaciae. Molecular Plant-Microbe Interactions. 2020;34:141–156. doi: 10.1094/MPMI-06-20-0145-R. [DOI] [PubMed] [Google Scholar]
- Billiard S., LoPez-Villavicencio M., Hood M.E., et al. Sex, outcrossing and mating types: unsolved questions in fungi and beyond. Journal of Evolutionary Biology. 2012;25:1020–1038. doi: 10.1111/j.1420-9101.2012.02495.x. [DOI] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R.R. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- Brankovics B., van Dam P., Rep M., et al. Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics. 2017;18:735. doi: 10.1186/s12864-017-4116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braumann I.M., van den Berg M., Kempken F. Repeat induced point mutation in two asexual fungi, Aspergillus niger and Penicillium chrysogenum. Current Genetics. 2008;53:287–297. doi: 10.1007/s00294-008-0185-y. [DOI] [PubMed] [Google Scholar]
- Brown A.H.D., Feldman M.W., Nevo E. Multilocus structure of natrual populations of Hordeum spontaneum. Genetics. 1980;96:523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C.A.M., et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4 doi: 10.1186/s13742-015-0047-8. s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch J.A., Glasheen B.M., Giunta M.A., et al. The evolution of transposon repeat-induced point mutation in the genome of Colletotrichum cereale: reconciling sex, recombination and homoplasy in an ''asexual" pathogen. Fungal Genetics and Biology. 2008;45:190–206. doi: 10.1016/j.fgb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Dean R., Van Kan J.A.L., Pretorius Z.A., et al. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth A., McTaggart A.R., Wingfield B.D. Fungal clones win the battle, but recombination wins the war. IMA Fungus. 2019;10:18. doi: 10.1186/s43008-019-0020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C., Myers E.W. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler D.M., Hicks N.D., Gillece J.D., et al. Cryptococcus gattii in North American Pacific Northwest: whole-population genome analysis provides insights into species evolution and dispersal. mBio. 2014;5 doi: 10.1128/mBio.01464-14. e01464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutre T., Duprat E., Feuillet C., et al. Considering transposable element diversification in de novo annotation approaches. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0016526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens L., Guo L., Dora S., et al. A chromosome-scale genome assembly for the Fusarium oxysporum strain Fo5176 to establish a model Arabidopsis-fungal pathosystem. G3 Genes|Genomes|Genetics. 2020;10:3549–3555. doi: 10.1534/g3.120.401375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S.N., Slezak T., Hall B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- Gebru S.T., Mammel M.K., Gangiredla J., et al. Draft genome sequences of 12 isolates from 3 Fusarium species recovered from moldy peanuts. Microbiology Resource Announcements. 2019;8 doi: 10.1128/MRA.01642-18. e01642-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon T.R., Martyn R.D. The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology. 1997;35:111–128. doi: 10.1146/annurev.phyto.35.1.111. [DOI] [PubMed] [Google Scholar]
- Guo L., Han L., Yang L., et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporumf. sp.cubense causing banana vascular wilt disease. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0095543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane J.K., Oliver R.P. RIPCAL: a tool for alignment-based analysis of repeat-induced point mutations in fungal genomic sequences. BMC Bioinformatics. 2008;9:478. doi: 10.1186/1471-2105-9-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane J.K., Williams A.H., Taranto A.P., et al. In: Genetic transformation systems in fungi Volume 2. van den Berg M.A., Maruthachalam K., editors. Springer International Publishing; Denmark: 2015. Repeat-Induced Point Mutation: A fungal-specific, endogenous mutagenesis process; pp. 55–68. [Google Scholar]
- Hedges S.B., Marin J., Suleski M., et al. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry P., Kaur S., Pham Q.A.T., et al. Genomic differences between the new Fusarium oxysporumf. sp.apii (Foa) race 4 on celery, the less virulent Foa races 2 and 3, and the avirulent on celery f. sp.coriandrii. BMC Genomics. 2020;21:730. doi: 10.1186/s12864-020-07141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry P.M., Pincot D.D.A., Jenner B.N., et al. Horizontal chromosome transfer and independent evolution drive diversification in Fusarium oxysporumf. sp.fragariae. New Phytologist. 2021;230:327–340. doi: 10.1111/nph.17141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry P.M., Stueven M., Li S., et al. Genome sequence of a California isolate of Fusarium oxysporumf. sp.lycopersici Race 3, a fungus causing wilt disease on tomato. Microbiology Resource Announcements. 2019;8:e01713–e01718. doi: 10.1128/MRA.01713-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson O., Hudson D., Ji P., et al. Draft genome sequences of three Fusarium oxysporumf. sp.niveum isolates used in designing markers for race differentiation. Microbiology Resource Announcements. 2020;9:e01004–e01020. doi: 10.1128/MRA.01004-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Kloepper T.H. Computing recombination networks from binary sequences. Bioinformatics. 2005;21:ii159–ii165. doi: 10.1093/bioinformatics/bti1126. [DOI] [PubMed] [Google Scholar]
- Ikeda K-i, Nakayashiki H., Kataoka T., et al. Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Molecular Microbiology. 2002;45:1355–1364. doi: 10.1046/j.1365-2958.2002.03101.x. [DOI] [PubMed] [Google Scholar]
- Kamvar Z.N., Tabima J.F., Grünwald N.J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanapin A., Samsonova A., Rozhmina T., et al. The Genome Sequence of Five Highly Pathogenic Isolates of Fusarium oxysporumf. sp.lini. Molecular Plant-Microbe Interactions. 2020;33:1112–1115. doi: 10.1094/MPMI-05-20-0130-SC. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayi S., Khoulassa S., Gaboun F., et al. Draft genome sequence of Fusarium oxysporumf. sp.albedinis strain Foa 133, the causal agent of Bayoud disease on date palm. Microbiology Resource Announcements. 2020;9 doi: 10.1128/MRA.00462-20. e00462-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-S., Lohmar J.M., Busman M., et al. Identification and distribution of gene clusters required for synthesis of sphingolipid metabolism inhibitors in diverse species of the filamentous fungus Fusarium. BMC Genomics. 2020;21:510. doi: 10.1186/s12864-020-06896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P.A., Church D.M., Thibaud-Nissen F., et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Research. 2016;44:D73–D80. doi: 10.1093/nar/gkv1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus B.J., Grünwald N.J. vcfr: a package to manipulate and visualize variant call format data in R. Molecular Ecology Resources. 2017;17:44–53. doi: 10.1111/1755-0998.12549. [DOI] [PubMed] [Google Scholar]
- Koenig R.L., Ploetz R.C., Kistler H.C. Fusarium oxysporumf. sp.cubense consists of a small number of divergent and globally distributed clonal lineages. Phytopathology. 1997;87:915–923. doi: 10.1094/PHYTO.1997.87.9.915. [DOI] [PubMed] [Google Scholar]
- Krasnov G.S., Pushkova E.N., Novakovskiy R.O., et al. High-quality genome assembly of Fusarium oxysporumf. sp.lini. Frontiers in Genetics. 2020;11:959. doi: 10.3389/fgene.2020.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück P., Longo G.C. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Frontiers in Zoology. 2014;11:81. doi: 10.1186/s12983-014-0081-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence M.H., Summerell B.A., Burgess L.W., et al. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biology. 2014;118:374–384. doi: 10.1016/j.funbio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Laurence M.H., Summerell B.A., Liew E.C.Y. Fusarium oxysporumf. sp.canariensis: evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathology. 2015;64:1068–1075. [Google Scholar]
- Leigh J.W., Bryant D. popart: full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6:1110–1116. [Google Scholar]
- Leslie J.F., Summerell B.A. Blackwell Publishing Professional; USA: 2006. The Fusarium Laboratory Manual. [Google Scholar]
- Li J., Fokkens L., van Dam P., et al. Related mobile pathogenicity chromosomes in Fusarium oxysporum determine host range on cucurbits. Molecular Plant Pathology. 2020;21:761–776. doi: 10.1111/mpp.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard L., Sandoval-Denis M., Lamprecht S.C., et al. Epitypification of Fusarium oxysporum - clearing the taxonomic chaos. Persoonia. 2019;43:1–47. doi: 10.3767/persoonia.2019.43.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Yang Y., Liu X., et al. Draft genome sequence of FGL03-6, a race 1 strain of Fusarium oxysporumf. sp.conglutinans, the causal agent of cabbage Fusarium wilt. Genome Announcements. 2018;6 doi: 10.1128/genomeA.00191-18. e00191-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L.-J., van der Does H.C., Borkovich K.A., et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryani N., Lombard L., Poerba Y.S., et al. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporumf. sp.cubense in the Indonesian centre of origin. Studies in Mycology. 2019;92:155–194. doi: 10.1016/j.simyco.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.J., Rice D.P., Desai M.M. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. 2016;531:233–236. doi: 10.1038/nature17143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Hahn M.W., Lanfear R. New methods to calculate concordance factors for phylogenomic datasets. Molecular Biology and Evolution. 2020;37:2727–2733. doi: 10.1093/molbev/msaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Nguyen M.A.T., von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold G.P., Arias S., Taschl I., et al. In: Corn. Third Edition. Serna-Saldivar S.O., editor. AACC International Press; England: 2019. Chapter 9 – Mycotoxins in Corn: Occurrence, Impacts, and Management; pp. 235–287. [Google Scholar]
- Nieuwenhuis B.P.S., James T.Y. The frequency of sex in fungi. Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371:20150540. doi: 10.1098/rstb.2015.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Sutton D.A., Rinaldi M.G., et al. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. Journal of Clinical Microbiology. 2004;42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Sutton D.A., Wiederhold N., et al. Veterinary Fusarioses within the United States. Journal of Clinical Microbiology. 2016;54:2813–2819. doi: 10.1128/JCM.01607-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Z., Ino Y., Kimura Y., et al. Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Frontiers in Plant Science. 2016;7:31. doi: 10.3389/fpls.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A language and environment for statistical computing. [Google Scholar]
- Quesneville H., Nouaud D., Anxolabéhère D. Detection of new transposable element families in Drosophila melanogaster and Anopheles gambiae genomes. Journal of Molecular Evolution. 2003;57:S50–S59. doi: 10.1007/s00239-003-0007-2. [DOI] [PubMed] [Google Scholar]
- Ropars J., Dupont J., Fontanillas E., et al. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0049665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Pokhrel A., Coleman J.J. The genome sequence of five genotypes of Fusarium oxysporumf. sp.vasinfectum: A resource for studies on Fusarium wilt of cotton. Molecular Plant-Microbe Interactions. 2019;33:138–140. doi: 10.1094/MPMI-07-19-0197-A. [DOI] [PubMed] [Google Scholar]
- Singh N.K., Blachowicz A., Romsdahl J., et al. Draft genome sequences of several fungal strains selected for exposure to microgravity at the International Space Station. Genome Announcements. 2017;5:e01602–e01616. doi: 10.1128/genomeA.01602-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S.K., Zeller K.A., Sobieraj J.H., et al. Genome resources of four distinct pathogenic races within Fusarium oxysporumf. sp.vasinfectum that cause vascular wilt disease of cotton. Phytopathology. 2020;111:593–596. doi: 10.1094/PHYTO-07-20-0298-A. [DOI] [PubMed] [Google Scholar]
- Stanke M., Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Research. 2005;33:W465–467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp E.T., Wingfield M.J., McTaggart A.R., et al. Fungal species and their boundaries matter – Definitions, mechanisms and practical implications. Fungal Biology Reviews. 2018;32:104–116. [Google Scholar]
- Steenwyk J.L., Buida T.J., Li Y., et al. ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biology. 2020;18 doi: 10.1371/journal.pbio.3001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerell B.A. Resolving Fusarium: current status of the genus. Annual Review of Phytopathology. 2019;57:323–339. doi: 10.1146/annurev-phyto-082718-100204. [DOI] [PubMed] [Google Scholar]
- Taylor A., Armitage A.D., Handy C., et al. Basal rot of Narcissus: understanding pathogenicity in Fusarium oxysporumf. sp.narcissi. Frontiers in Microbiology. 2019;10:2905. doi: 10.3389/fmicb.2019.02905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.W., Hann-Soden C., Branco S., et al. Clonal reproduction in fungi. Proceedings of the National Academy of Sciences of the USA. 2015;112:8901–8908. doi: 10.1073/pnas.1503159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavelu R., Edwin Raj E., Pushpakanth P., et al. Draft genome of Fusarium oxysporumf. sp.cubense strain tropical race-4 infecting cavendish (AAA) group of banana in India. Plant Disease. 2020;105:481–483. doi: 10.1094/PDIS-06-20-1170-A. [DOI] [PubMed] [Google Scholar]
- Thatcher L.F., Gardiner D.M., Kazan K., et al. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Molecular Plant Microbe Interactions. 2012;25:180–190. doi: 10.1094/MPMI-08-11-0212. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proceedings of the National Acadamy of Sciences of the USA. 2012;109:E3305–3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Cryptosporidium, Giardia, Cryptococcus, Pneumocystis genetic variability: cryptic biological species or clonal near-clades? PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1003908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Is predominant clonal evolution a common evolutionary adaptation to parasitism in pathogenic parasitic Protozoa, Fungi, bacteria, and viruses? Advances in Parasitology. 2017;97:243–325. doi: 10.1016/bs.apar.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M., Ayala F.J. Models in parasite and pathogen evolution: Genomic analysis reveals predominant clonality and progressive evolution at all evolutionary scales in parasitic protozoa, yeasts and bacteria. Advances in Parasitology. 2021;111:75–117. doi: 10.1016/bs.apar.2020.12.001. [DOI] [PubMed] [Google Scholar]
- Tsai I.J., Bensasson D., Burt A., et al. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proceedings of the National Academy of Sciences of the USA. 2008;105:4957. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam P., Fokkens L., Schmidt S.M., et al. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environmental Microbiology. 2016;18:4087–4102. doi: 10.1111/1462-2920.13445. [DOI] [PubMed] [Google Scholar]
- van Dam P., Rep M. The distribution of miniature impala elements and SIX genes in the Fusarium genus is suggestive of horizontal gene transfer. Journal of Molecular Evolution. 2017;85:14–25. doi: 10.1007/s00239-017-9801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Nest M.A., Steenkamp E.T., McTaggart A.R., et al. Saprophytic and pathogenic fungi in the Ceratocystidaceae differ in their ability to metabolize plant-derived sucrose. BMC Evolutionary Biology. 2015;15:273. doi: 10.1186/s12862-015-0550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., van Dam P., Zaborin A., et al. Genomic characterization and virulence potential of two Fusarium oxysporum isolates cultured from the International Space Station. mSystems. 2019;4 doi: 10.1128/mSystems.00345-18. e00345-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Massa G., Hummerick M., et al. Draft genome sequences of two Fusarium oxysporum isolates cultured from infected Zinnia hybrida plants grown on the International Space Station. Genome Announcements. 2018;6 doi: 10.1128/genomeA.00326-18. e00326-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M., Gordon T.R., Wingfield B.D., et al. Department of Genetics, University of Pretoria; South Africa: 2005. Chapter 2: Mating type genes and the reproductive potential of Fusarium oxysporumf. sp.cubense. PhD Dissertation. [Google Scholar]
- Vlaardingerbroek I., Beerens B., Rose L., et al. Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environmental Microbiology. 2016;18:3702–3713. doi: 10.1111/1462-2920.13281. [DOI] [PubMed] [Google Scholar]
- Wang B., Yu H., Jia Y., et al. Chromosome-scale genome assembly of Fusarium oxysporum strain Fo47, a fungal endophyte and biocontrol agent. Molecular Plant-Microbe Interactions. 2020;33:1108–1111. doi: 10.1094/MPMI-05-20-0116-A. [DOI] [PubMed] [Google Scholar]
- Williams A.H., Sharma M., Thatcher L.F., et al. Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporumformae speciales facilitates identification of candidate effectors. BMC Genomics. 2016;17:191. doi: 10.1186/s12864-016-2486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Ayhan D.H., Diener A.C., et al. Genome sequence of Fusarium oxysporumf. sp.matthiolae, a Brassicaceae pathogen. Molecular Plant-Microbe Interactions. 2020;33:569–572. doi: 10.1094/MPMI-11-19-0324-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang H., Turra D., et al. The genome of opportunistic fungal pathogen Fusarium oxysporum carries a unique set of lineage-specific chromosomes. Communications Biology. 2020;3:50. doi: 10.1038/s42003-020-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome accession data, phylogroup, mating type and binomial applied based on a phylogenetic species hypothesis (Fig. S5) for all isolates used in study.
Location of single copy orthologs in the chromosome assembly of Fusarium callistephi (GCA_014154955).
Phylogram obtained from a maximum likelihood search of 40 concatenated, protein-coding genes in IQ-Tree, with a model test for each partition, and genealogical concordance factors provided above nodes. Genealogical concordance factors used to define phylogroups are in bold.
DensiTree visualisation of 40 single copy gene trees showing that the examined single copy orthologs were not concordant.
SplitsTree neighbour network based on 100 000 random SNPs out of 4.6 million called from kmers of 101 base pairs across 410 genomes of the Fusarium oxysporum species complex using kSNP. Genotypes are coloured by phylogroup.
Analyses of repeat-induced point mutation (RIP) for the highest quality genome (based on N50) in each phylogroup. Top panels display the distribution of the RIP-like mutations on alignments of TE copies. The bottom graphs show the frequency of different RIP-like mutations (orange lines indicate frequency of CpA > TpA mutations, purple lines indicate frequency of CpC > TpC mutation, green lines indicate frequency of CpG > TpG mutation, and turquoise lines indicate frequency of CpT > TpT mutation). Top row, phylogroups 1, 2, 3, 4; middle row, phylogroups 5, 6, 7, 8; bottom row, phylogroups 9, 10, 11, 12.
Phylogenetic hypothesis based on a maximum likelihood search of a concatenated alignment of TEF and RPB2 in IQ-Tree. Alignment is based on Lombard et al. (2019) and used to apply names in the Fusarium oxysporum species complex. aLRT and ultrafast bootstrap values from 10 000 replicates above nodes.
Data Availability Statement
All data and commands are available at https://drive.google.com/drive/folders/17EIfIiuLLR-1wx_hXK0d-UvNc4X5XhB7?usp=sharing