Abstract
Expression of the CD4 gene is tightly controlled throughout thymopoiesis. The downregulation of CD4 gene expression in CD4− CD8− and CD4− CD8+ T lymphocytes is controlled by a transcriptional silencer located in the first intron of the CD4 locus. Here, we determine that the c-Myb transcription factor binds to a functional site in the CD4 silencer. As c-Myb is also required for CD4 promoter function, these data indicate that depending on the context, c-Myb plays both positive and negative roles in the control of CD4 gene expression. Interestingly, a second CD4 silencer-binding factor, HES-1, binds to c-Myb in vivo and induces it to become a transcriptional repressor. We propose that the recruitment of HES-1 and c-Myb to the silencer leads to the formation of a multifactor complex that induces silencer function and repression of CD4 gene expression.
T-cell development is controlled by the ordered regulation of genes involved in the progression of the thymocyte through each stage of maturation. One of the most important genes expressed at specific stages of T-cell development is that encoding the coreceptor CD4. The earliest committed T-cell precursor cells do not express either CD4 or the coreceptor CD8 and are referred to as double-negative (DN) thymocytes. Expression of CD4 and CD8 is first seen in T cells that have undergone successful rearrangement of the T-cell receptor (TCR) β genes. This double-positive (DP) population subsequently completes rearrangement of the TCR α chain gene and undergoes the selection process to ensure a properly restricted T-cell repertoire (6). CD4 expression is maintained in mature T cells that survive selection and recognize antigen bound to major histocompatibility complex class II (5, 15). These cells downregulate expression of CD8 and become committed helper T cells (TH). Those cells that survive selection and recognize antigen bound to major histocompatibility complex class I will downregulate expression of CD4, maintain expression of CD8, and become committed cytotoxic T cells (TC) (42, 46). Thus, the activation or downregulation of CD4 gene expression defines the different stages of developing T cells. We have sought to understand how CD4 gene expression is linked to thymocyte development by identifying factors that bind to and mediate the function of the CD4 transcriptional control regions. As these trans-acting factors are likely to be responsive to the T-cell selection process, their study will help delineate the signaling pathways that drive repertoire selection.
Expression of the CD4 gene is controlled by four elements: a promoter, a thymocyte enhancer which functions early in development, a mature enhancer which begins function in mature post-selection T cells, and a transcriptional silencer (1, 8–10, 33, 34, 39, 40, 42, 43, 47, 49, and M. Adlam and G. Siu, unpublished). The silencer is the critical element that represses CD4 expression in DN thymocytes and as the DP thymocyte matures into the CD8 single-positive (SP) TC cell (41, 43). There are three factor-binding sites in the CD4 silencer, which we refer to as S1, S2, and S3, all of which are important in mediating silencer function (10). We have previously determined that the Notch pathway intermediate HES-1 binds to the silencer site S1 and silencer-associated factor (SAF) binds to S3; both are important in mediating silencer function (22, 23). Interestingly, SAF is located in the cytoplasm of DP and CD4 SP T cells and in the nucleus of DN and CD8 SP T cells. Its active transport to the nucleus is DP thymocyte specific, is controlled by lck signaling via the Mek1 pathway, and induces the downregulation of CD4 expression, the initiating step of CD8 SP development (W. W. S. Kim, N. de Souza, and G. Siu, submitted for publication). It is thus likely that SAF is critical for transmitting signals from the TCR complex to the CD4 silencer during the repertoire selection process. The factor(s) that binds to the S2 site of the silencer is unknown.
The c-Myb transcription factor plays a role in the control of transcription of many genes that are critical for early hematopoiesis (25); mouse genetic studies have also demonstrated a role for c-Myb in early thymopoiesis (2, 4). Consensus c-Myb-binding sites have been found in the transcriptional control elements of genes important in later stages of thymopoiesis, indicating that c-Myb may also play a role in late T-cell development as well (12, 17, 19, 28, 44). We and others have previously determined that c-Myb induces CD4 expression by binding to a consensus c-Myb site in the CD4 promoter (28, 44). Here, using molecular and transgenic approaches, we determine that c-Myb binds to the S2 functional site of the CD4 silencer and that its binding is important for mediating silencer function. In addition, we demonstrate that HES-1 binds to c-Myb in vivo and induces it to become a transcriptional repressor. Our data thus indicate that c-Myb can function as either a transcriptional repressor at the CD4 silencer or as a transcriptional activator at the CD4 promoter, depending on the context of its binding site within the transcriptional control element, and indicate that HES-1 and c-Myb form a multifactor complex that mediates CD4 silencer function.
MATERIALS AND METHODS
Electrophoretic mobility shift assays and UV cross-links and immunoprecipitations.
Nuclear extracts were purified from the CD4+ CD8− TH clone D10 or the CD8 SP TC clone L3 using a modified Dignam protocol (8), and electrophoretic mobility shift assay (EMSA) analyses were conducted using oligonucleotide probes (Gibco BRL) as described previously (10). The S2L probe contains sequences extending from position 121 to 185 in the CD4 silencer (10). Briefly, the EMSA reaction included reaction buffer (10 mM HEPES [pH 7.9], 50 mM NaCl, 5 mM Tris-HCl [pH 7.5], 25 mM EDTA, 1 mM dithiothreitol, and 10% glycerol), 1 mM spermidine, and 1 μg of deoxyinosine-deoxycytosine or 100 ng of herring sperm DNA. For the competition EMSAs, nonradioactive oligonucleotides were added to the binding mix simultaneously with the radioactive probe in 100- or 300-fold molar excess and incubated at room temperature for 15 min. Reactions were then resolved on a nondenaturing 4% polyacrylamide gel and run at 150 V for 2 h in glycine buffer (190 mM glycine, 25 mM Tris-HCl [pH 8.5], 1 mM EDTA). For UV cross-linking, a 5× EMSA binding reaction was performed using a bromodeoxyuridine-substituted S2 probe and exposed for 15 min to short-wave UV light using a Stratalinker (Stratagene). Immunoprecipitations of the UV cross-linking reactions with the BP2 and BP7 c-Myb-specific antisera were conducted as previously described (44). BP2 and BP7 were kindly provided by Joseph Lipsick. Briefly, cross-linked reactions were first incubated in the presence of normal rabbit serum for 4 to 6 h and precipitated using GammaBind Plus Sepharose beads (Amersham Pharmacia Biotech). This preincubation was used to remove all nonspecific interactions between the nuclear extracts and the antiserum. Supernatants were then incubated overnight at 4°C in the presence of specific antiserum (BP2 or BP7). Following precipitation with GammaBind Plus Sepharose beads, boiled immunoprecipitates were resolved on a sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) gel. Protein-DNA complexes were visualized using autoradiography and were sized in reference to 14C-labeled protein markers (Sigma). To estimate relative intensities of the bands in the in vivo immunoprecipitation experiment, mean channel intensities were determined for 49-pixel boxes encompassing representative portions of each band, and relative ratios were determined. Multiple exposures of the autoradiograms were analyzed to ensure that the exposure was within the linear range. 35S-methionine labeling of the DN thymoma S49 and coupling of the antisera to Sepharose beads were conducted as described previously (3).
Immunodepletion and Western analysis.
For the immunodepletion of T-cell extracts, 250 μg of nuclear extract was incubated with 2 μg of the mouse monoclonal anti-Myb antibody 1.1 (Upstate Biotechnology). Extract-antibody solutions were incubated overnight in the presence of GammaBind Plus Sepharose beads at 4°C, and immunoprecipitates were pelleted by centrifugation. Protein concentrations of the resulting supernatants were determined using a Bradford assay (Bio-Rad), and equivalent amounts of protein from treated and mock-treated extracts were used in EMSA reactions. Treated and mock-treated extracts were assessed for c-Myb content by Western blot analysis. Briefly, extracts were resolved on SDS–8% PAGE gels and transferred to nitrocellulose (36). Membranes were incubated overnight in the presence of anti-c-Myb polyclonal antibody M-19 (Santa Cruz Biotechnology, Inc.) and were developed using a chemiluminescent detection system (Boehringer Mannheim). c-Myb null embryonic stem (ES) cells were provided by Michael Mucenski (27).
Generation of reporter constructs.
The mutation of the CD4 promoter P1 site to generate S2/Pro was created with the primer TGGCGGGGGGCACATCCCACAACTG using the dut− ung− method as described previously (38). Mutant promoter fragments were subsequently cloned into the pGL2 luciferase reporter vector. Mutations of the CD4 silencer S2 region were generated from the Δ1Δ3 silencer template using an overlap extension PCR as described previously (24). The following primers (Gibco BRL, Sigma/Genosys) were used: 5′ GGG CAC ATC CCA TTT TTT GGC TAG AGT GGG 3′ and 5′ CCC ACT CTA GCC AAA AAA TGG GAT GTG CCC 3′. The external primers used were either T7 or M13R. PCR products were subcloned into pCR 2.1-TOPO vector (Invitrogen). DNA sequencing analysis and restriction enzyme digests confirmed each mutation. Mutant silencers were subcloned into the pTG construct, which contains the CD4 transcriptional control elements and the human HLA-B7 gene as a marker (10).
Generation of transgenic mice.
Generation of transgenic mice using this DNA was carried out using previously described methods (18). Prior to injection, the transgenic DNA insert was excised from the vector DNA and separated across a sucrose gradient as previously described (10). Purified insert DNA was dialyzed against transgenic injection buffer (5 mM Tris [pH 7.5], 0.1 mM EDTA) and injected at a concentration of 5 to 10 μg/ml (18). Transgenic founder mice were identified by the staining of peripheral lymphocytes as described below and by PCR analysis of genomic DNA. Multiple expressing founders for each construct were generated and analyzed.
Flow cytometry.
All analyses were performed on 3- to 6-week-old littermates housed in the pathogen-free Animal Facility of the Herbert W. Irving Cancer Center at Columbia University. The following monoclonal antibody reagents were obtained from Pharmingen to identify peripheral T cells using previously described protocols (36): allophycocyanin-conjugated RM4-5 (anti-CD4) and peridinin chlorophyll-A protein-conjugated 53-6.7 (anti-CD8α). The transgenic marker was stained with a phycoerythrin-conjugated ME-1 (anti-HLA-B7) antibody. Peripheral blood lymphocytes were stained with α-CD4, α-CD8, and α-ME-1. T cells were identified based on their expression of CD4 or CD8 and then assessed for their expression of HLA-B7. Representative progeny from all founder mice were analyzed; typical results from one founder are shown. Analyses were performed using the FACSCalibur flow cytometer and CellQuest software (Becton Dickinson) at the Flow Cytometry Facility of the Herbert W. Irving Cancer Center at Columbia University.
Transient transfection of T-cell lines.
The CD4+ CD8− TH clone D10 was transfected using previously described methods (22, 38). Briefly, test and control plasmids were cotransfected into cells by the DEAE-dextran method; the test plasmid contained the experimental CD4 promoter subcloned upstream of the luciferase gene in the pGL2 vector, and the transfection control plasmid contained the Renilla luciferase gene under the control of the herpes simplex virus 1 thymidine kinase promoter (pRL-TK; Promega). The total amount of DNA added to each transfection point was kept constant with the addition of the pGL2 vector. Cells were harvested after 48 h, and extracts were prepared for the Dual Luciferase assay as recommended by the manufacturer (Promega). Renilla and firefly luciferase levels were measured using a TD 20/20 Luminometer (Turner Designs). Results shown are averaged for 3 to 7 experiments per data point.
RESULTS
Characterization of the S2-binding factor.
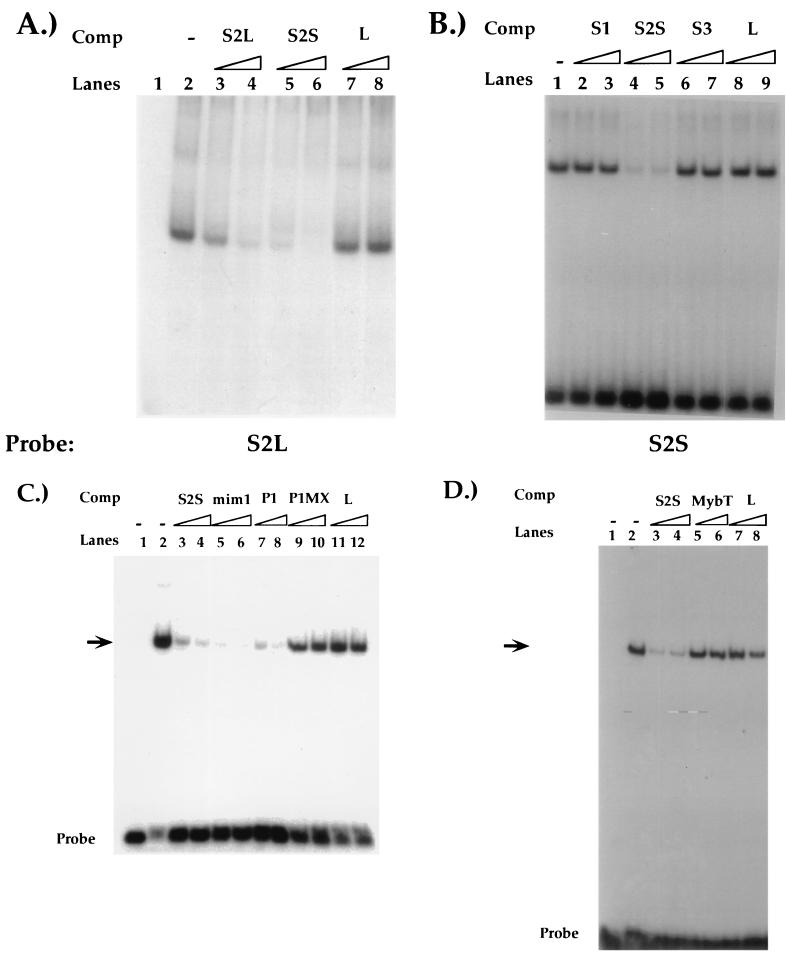
The CD4 silencer contains three factor-binding sites, referred to as S1, S2, and S3, that were originally defined by DNase footprinting analyses (10). As discussed above, HES-1 and the novel transcription factor SAF bind to S1 and S3, respectively (22, 23). To characterize the S2-binding factor further, we conducted EMSAs with oligonucleotides encompassing the S2 region (Fig. 1 and 2). The S2L probe encompasses the complete S2 footprint as well as an additional 40 bp that flank the site. Incubation of this probe with nuclear extracts from either CD4 SP TH- or CD8 SP TC-cell clones resulted in the formation of a single complex (Fig. 2A and data not shown). We have been unable to detect other complexes with this probe using a variety of different binding conditions, suggesting that this represents the sole factor-DNA complex in the S2 region (data not shown). Molar excesses of unlabeled probe but not nonspecific oligonucleotide inhibited complex formation, indicating that the factor(s) that forms this complex binds specifically to the S2L probe (Fig. 2A, lanes 3, 4, 7, and 8). Interestingly, a smaller 27-bp probe encompassing only the S2 footprint (the S2S probe) (Fig. 1A) also competes for complex formation, indicating that the factor(s) that binds to S2 binds within this region (Fig. 2A, lanes 5 and 6).
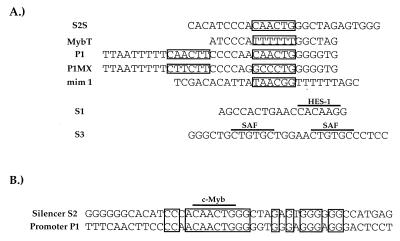
FIG. 1.
(A) Sequences of oligonucleotides used in the competition EMSA analyses. Boxed regions indicate consensus c-Myb recognition sequences within each oligonucleotide. The HES-1 and SAF DNA recognition sequences in S1 and S3, respectively, are overlined. (B) The DNA sequence identities between the P1 and S2 regions are indicated by boxed nucleotides. Consensus c-Myb recognition sequences are overlined.
FIG. 2.
(A) Characterization of the S2-binding factor. EMSAs using a 3′-extended radioactive S2L probe and CD4 SP TH D10 nuclear extracts. Reactions were performed in the absence of competitor (Comp) oligonucleotides (lane 2) or in the presence of excess S2L (lanes 3 and 4), S2S (lanes 5 and 6), or nonspecific (L, lanes 7 and 8) oligonucleotides. Lane 1, probe only. (B) EMSAs using the radioactive S2 probe and CD4 SP TH D10 nuclear extracts. Reactions were performed in the absence of competitor oligonucleotides (lane 1) or in the presence of excess S1 (lanes 2 and 3), S2S (lanes 4 and 5), S3 (lanes 6 and 7), or nonspecific (L, lanes 8 and 9) oligonucleotides. Sequences of the S2S probe and competitors are listed in Fig. 1. (C) Reactions were performed in the absence of competitor oligonucleotides (lane 2) or in the presence of excess S2S (lanes 3 and 4), mim-1 (lanes 5 and 6), P1 (lanes 7 and 8), P1MX (lanes 9 and 10), or nonspecific (L, lanes 11 and 12) oligonucleotides. Lane 1, probe only. (D) Reactions were performed in the absence of competitor oligonucleotides (lane 2) or in the presence of excess S2S (lanes 3 and 4), MybT (lanes 5 and 6), or nonspecific (L, lanes 7 and 8) oligonucleotides. Unlabeled oligonucleotides were used at 100- and 300-fold molar excesses. Sequences of competitor oligonucleotides are shown in Fig. 1. Arrows indicate S2-specific binding complex; free probe is indicated. Lane 1, probe only.
We have previously demonstrated that there is functional redundancy among the three factor-binding sites in that silencer function is abrogated only when S2 is deleted in combination with S1 or S3 (10). One explanation is that a common factor is binding to more than one of these sites. To test this, we performed EMSAs using the S2S probe and competitor oligonucleotides that encode the other functional sites of the silencer (Fig. 1 and 2B). As described above, we detected a single major complex forming in EMSAs with the S2S probe by using nuclear extracts from both CD4 SP and CD8 SP T-cell clones (Fig. 2B and data not shown). Although molar excesses of nonradioactive S2S oligonucleotide competed for complex formation, similar molar excesses of unlabeled S1 or S3 oligonucleotides did not, indicating that the S2-binding factor does not recognize the S1 or S3 regions of the silencer (Fig. 2B, lanes 2 through 7). These data support the hypothesis that the factor binding S2 is distinct from the S1-binding protein HES-1 and the S3-binding protein SAF.
The S2-binding factor has the same sequence specificity as c-Myb.
Proteins of the Myb family, including c-Myb, bind as monomers to the sequence YAAC(T/G)G (25). Sequence analysis of the S2 region revealed a consensus c-Myb recognition sequence (Fig. 1B). Indeed, the putative c-Myb recognition site in S2 is almost identical in sequence to a previously defined c-Myb in the CD4 promoter (Fig. 1A). These observations led to the hypothesis that c-Myb could be binding to S2 and mediating silencer function. To test this hypothesis, we conducted cold competition EMSA experiments with the S2 probe and T-cell nuclear extracts (Fig. 2C and D). As in previous experiments, the S2S probe bound a single complex that was competed away specifically by addition of excess unlabeled S2S to the reaction but not by similar addition of a nonspecific oligonucleotide (Fig. 2C, lanes 3, 4, 11, and 12). Molar excesses of an unlabeled P1 oligonucleotide containing the CD4 promoter c-Myb site also compete for S2 factor binding (Fig. 1A and 2C, lanes 7 and 8); mutation of the c-Myb recognition sequences in P1 abrogates its ability to compete for S2 complex formation (the P1MX probe; Fig. 1A and 2C, lanes 9 and 10). In addition, molar excesses of a competitor oligonucleotide containing a known c-Myb recognition site from the mim-1 promoter also compete for S2 complex formation (the mim-1 probe; Fig. 1A and 2C, lanes 5 and 6). The mim-1 consensus Myb site is completely different in sequence from the putative c-Myb site in S2; due to degeneracy within the consensus sequence, the mim-1 site differs in sequence within the c-Myb site itself at two of six nucleotides (Fig. 1A) (30). Nonetheless, the mim-1 oligonucleotide competes as efficiently for S2 complex formation as the S2S oligonucleotide itself.
To confirm that the c-Myb consensus sequence itself in the S2 region is required for factor binding, we determined the effect of mutating this site on S2-binding complex formation. If c-Myb is indeed binding to S2, one can predict that the formation of the S2-binding complex would be dependent on an intact c-Myb recognition sequence. We therefore conducted cold-competition EMSA experiments with T-cell extracts and S2S oligonucleotides containing mutations in the c-Myb recognition site (Fig. 2D and data not shown). The MybT mutation contains insertions in the core c-Myb recognition site of S2; molar excesses of this oligonucleotide do not compete for complex formation, indicating that the S2-binding factor recognizes the consensus c-Myb recognition site in S2. In addition, the S2-binding complex could not be detected in EMSAs when the MybT oligonucleotide was used as a radiolabeled probe (data not shown). Taken together, these data provide strong evidence that the S2-binding protein has the same sequence specificity as c-Myb and support the hypothesis that c-Myb binds to the CD4 silencer.
The S2-binding factor shares antigenic epitopes with c-Myb.
To determine if the endogenous factor binding to S2 shares antigenic epitopes with c-Myb, we conducted UV cross-linking and immunoprecipitation analyses (Fig. 3). An EMSA reaction with the S2S probe and T-cell nuclear extract was exposed to UV light, inducing the formation of covalent bonds between the DNA probe and bound nuclear proteins. The products of the binding reaction were then resolved on an SDS-PAGE gel, and the cross-linked protein-DNA complexes were visualized with autoradiography. As can be seen in Fig. 3A, UV cross-linking of the S2S probe with T-cell nuclear extracts results in a 96-kDa protein-DNA complex. By subtracting the molecular mass of the DNA probe, we determined the apparent molecular mass of the protein binding to the S2S probe to be 75 kDa, which is similar to the molecular mass of c-Myb (25). Occasionally, we could also detect an approximately 84-kDa protein-DNA complex in this experiment (Fig. 3 and data not shown). The predicted molecular mass of the factor that would make up this complex would be 63 kDa. Although the identity of this factor is unknown, this complex is not detected reproducibly; in addition, there is no known Myb-like factor of this molecular mass. It is also possible that this 63-kDa protein is not a member of the Myb family. However, we are unable to detect more than one factor binding to the S2 region (see above), and depletion experiments indicate that all S2-binding factors share antigenic epitopes with c-Myb (see below). It is thus likely that this complex represents a degradation product of c-Myb.
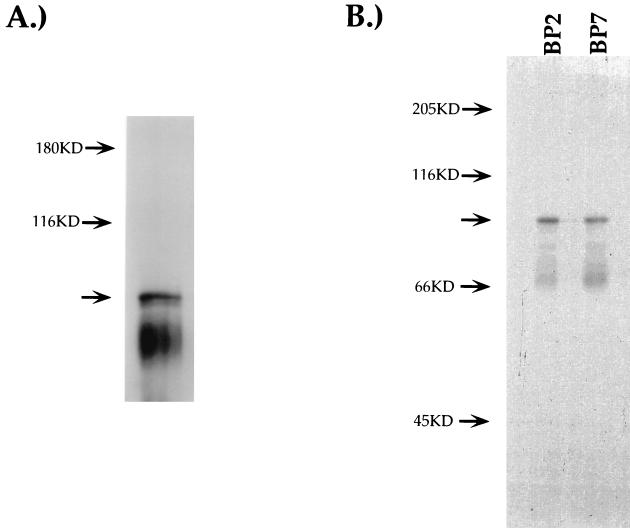
FIG. 3.
c-Myb binding to S2. EMSA reactions using a radioactive S2S probe and CD4 SP TH D10 nuclear extracts. (A) EMSA reactions were exposed to UV light for 15 min, resolved on SDS–10% PAGE gels, and visualized by autoradiography. (B) EMSA reactions were treated with UV light as for panel A and subsequently were immunoprecipitated using either the BP2 or BP7 anti-Myb antiserum. Immunoprecipitates were resolved on SDS–10% PAGE gels and visualized by autoradiography. Arrows indicate putative c-Myb DNA complexes.
To determine if the 75-kDa S2-binding factor shares antigenic epitopes with c-Myb, we used antisera directed against different domains of c-Myb in UV cross-linking–immunoprecipitation experiments (Fig. 3B). This approach has been used successfully to characterize c-Myb binding to other transcriptional control elements (44). The UV cross-linking experiment described above was repeated and subjected to immunoprecipitation with different antisera. The BP7 antiserum is specific for the c-Myb transactivation domain, whereas the BP2 antiserum is specific for the DNA-binding domain (7). As seen in Fig. 3B, a 96-kDa protein-DNA complex was precipitated using either antiserum, confirming that the S2-binding factor shares antigenic epitopes with c-Myb.
Depletion of c-Myb leads to loss of S2-binding activity.
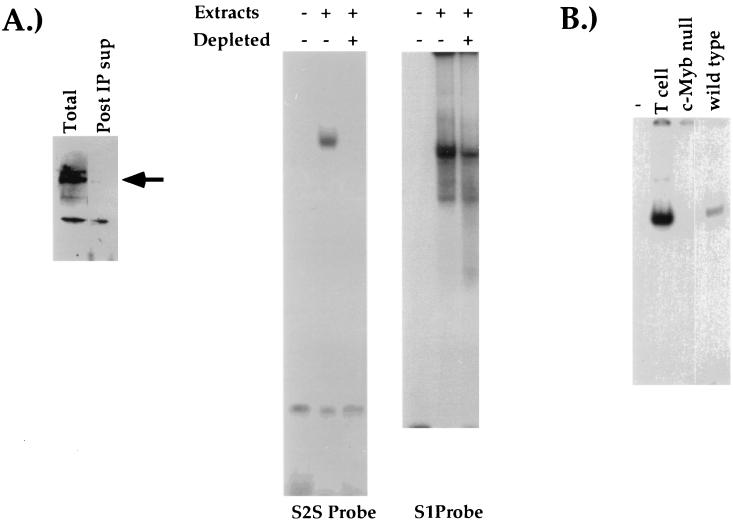
To demonstrate further that c-Myb is necessary for formation of the S2-binding complex, we used immunodepletion to generate T-cell nuclear extracts that lacked c-Myb. Based on Western analyses, c-Myb is expressed in cell lines representing each of the four major T-cell developmental stages (data not shown). If c-Myb is the S2-binding factor, then we would expect to observe a loss of S2-binding activity in extracts from which c-Myb had been depleted. Nuclear extracts from DP AKR1G1 thymoma cells were depleted of c-Myb by immunoprecipitation with the anti-c-Myb antibody 1.1, and the depletion was confirmed by Western blot analysis (Fig. 4A, left panel). We could not detect S2-binding activity in EMSA reactions with the depleted extracts (Fig. 4A, center panel); in contrast, we could still detect the binding of HES-1 to the S1 probe (Fig. 4A, right panel). Thus, depleting T-cell extracts of c-Myb also leads to a specific loss of S2-binding activity.
FIG. 4.
c-Myb is necessary for formation of S2-binding complex. (A) Western blot of AKR1G1 DP nuclear extracts either untreated or depleted of c-Myb (left panel). The arrow indicates c-Myb. AKR1G1 DP nuclear extracts were either mock-treated or depleted of c-Myb and used in EMSA reactions with either the S2S (center panel) or the S1 (right panel) radioactive probes. (B) EMSA reactions using a radiolabeled S2 probe and extracts from the CD4 SP TH D10 clone, c-Myb null ES cells, or wild-type ES cells. See Materials and Methods for details. Post IP sup, postimmunoprecipitation supernatant.
We also sought to determine the effect of genetically disrupting c-Myb expression on S2-binding activity. Wild-type ES cells or ES cells carrying a null mutation in both alleles of c-Myb (2) were grown in culture, and whole-cell extracts were prepared. If c-Myb binds to S2, we predicted that we would not be able to detect factor binding to the S2 probe in extracts from c-Myb null ES cells in comparison with extracts from wild-type ES cells. As can be seen in Fig. 4B, EMSAs with extracts from wild-type ES cells and the S2S probe resulted in the formation of a single S2-binding complex, whereas EMSAs performed with extracts from c-Myb null ES cells did not. Taken together, our biochemical experiments provide strong evidence that endogenous c-Myb binds to S2 of the CD4 silencer.
The c-Myb recognition site is essential for silencer function.
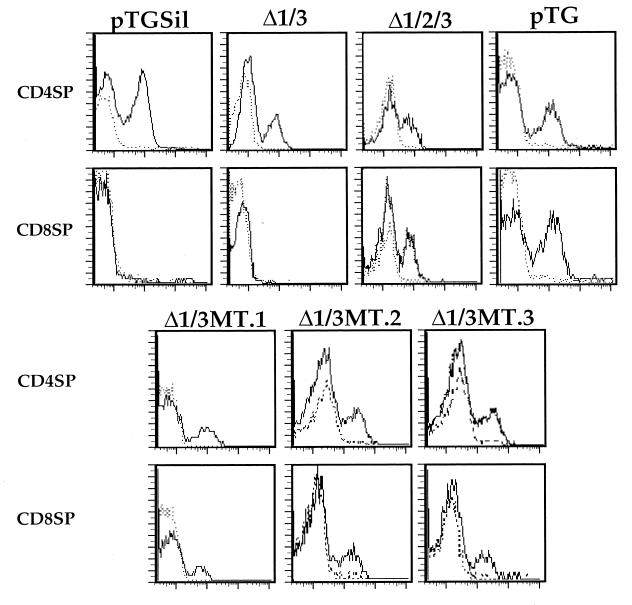
If the binding of c-Myb to S2 is important for silencer function, we could predict that the site-specific mutation of the c-Myb recognition site in S2 in the appropriate context would lead to abrogation of silencer function. To test this, we generated a mutation of the c-Myb site in the S2 region and tested this mutant silencer in our transgenic assay (10). We utilized reporter constructs that contain a cell surface marker gene under the transcriptional control of the CD4 promoter and enhancers as well as different mutations of the CD4 silencer (10). As we have reported previously, mice that are transgenic with a construct that contains the unmutated CD4 silencer (pTGSil) express the marker gene in CD4 SP but not CD8 SP T cells, whereas mice transgenic with constructs that do not contain the silencer (pTG) express the marker gene in both mature T-cell subsets (10) (Fig. 5). Deletion of any one of the three sites does not affect silencer function; function is abrogated only when S2 is deleted in combination with either S1 or S3 or both. Thus, a mutated CD4 silencer with S1 and S3 deleted (Δ1/3) still functions, whereas a silencer with all three sites deleted (Δ1/2/3) does not (10) (Fig. 5). To determine if the c-Myb-binding site within the S2 region is the critical functional site within the S2-footprinted region, we generated a mutation in the consensus c-Myb site in the Δ1/3 silencer and cloned this mutant silencer into the pTG construct (Δ1/3MT). The S1 and S3 deletions are identical to those in the Δ1/3 mutated silencer tested previously, whereas the mutation introduced into the c-Myb recognition site has been shown in our biochemical experiments to abrogate S2 factor binding (Fig. 2D). As can be seen in Fig. 5, mice transgenic with the Δ1/3MT construct express the marker in both CD4 SP and CD8 SP T cells, indicating that silencer function has been broken. These data indicate that inhibiting the binding of c-Myb to S2 abrogates silencer function.
FIG. 5.
The c-Myb-binding site in S2 is critical for CD4 silencer function. CD8 SP and CD4 SP T cells from pTG, pTGΔ1Δ3, pTGΔ1Δ2Δ3, pTGMT, or pTG transgenic mouse lines were gated on and analyzed for HLA-B7 expression. The presence of CD8+ HLA-B7+ T cells in the pTG, pTGΔ1Δ2Δ3, and pTGMT mice indicates a loss of silencer function in these constructs. Solid and dashed lines indicate staining with the anti-marker antibody and the isotype-matched control, respectively. Multiple founders for each construct were generated and analyzed; typical results are shown.
Context dependence of c-Myb function.
Our data indicate that c-Myb is playing a critical role in mediating silencer function and may therefore play a role in the repression of CD4 expression. As discussed above, we have previously shown that c-Myb also plays an important role in the induction of CD4 promoter function (44). Thus, c-Myb appears to play opposing roles in the control of CD4 expression: activation in CD4+ T cells, repression in CD4− T cells. The mechanism by which c-Myb mediates opposite functions in these closely related cell types is unknown. One possibility is that c-Myb is induced to assume different conformations when binding to its recognition sites in the different elements. In this model, the silencer site induces c-Myb to assume a conformation that leads it to become a transcriptional repressor, whereas the promoter sites induce c-Myb to become an activator. Thus, activating and repressing c-Myb-containing complexes may recognize different c-Myb consensus sequences.
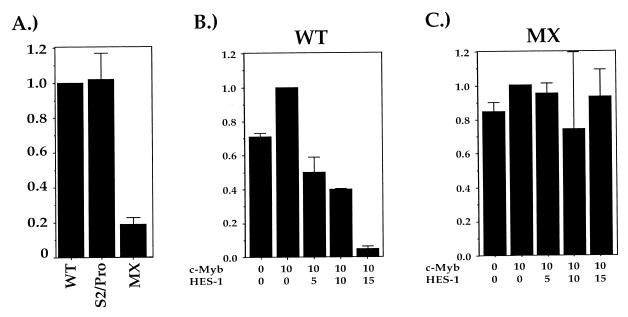
To determine if the differences in the silencer and promoter c-Myb recognition sites affect c-Myb function, we generated a mutation of the CD4 promoter that contains a replacement of its c-Myb recognition site with the silencer c-Myb site (the S2/Pro mutation). This mutation contains the CD4 silencer sequence from position 153 to 178 (10) substituting for the CD4 promoter sequence from position −100 to −75 (40) encompassing the defined functional c-Myb sites (28, 44); all spacing distances between the c-Myb site and the other factor-binding sites in the promoter were preserved (39). The S2/Pro mutant promoter was tested for function in transient transfection reporter assays in the CD4 SP TH clone D10. As we and others have reported earlier, the unmutated CD4 promoter functions at high levels in activated TH cells (28, 35, 37, 39, 44), whereas site-specific mutations of the c-Myb consensus sequence within the P1 promoter functional site (the MX mutation) lead to significant decreases in promoter activity (28, 44) (Fig. 6A). Interestingly, substitution of the silencer c-Myb recognition site into the c-Myb site in the CD4 promoter does not appreciably affect promoter function; we consistently obtain levels of reporter activity with the S2/Pro promoter construct comparable to that obtained with the wild-type CD4 promoter reporter construct (Fig. 6A). We can draw two conclusions from these data. First, the data further confirm that the S2 site is a functional c-Myb recognition site. Second, these data indicate that the c-Myb-binding sites in the silencer and the promoter are functionally equivalent, and sequence differences between these two sites do not result in differences in c-Myb function.
FIG. 6.
HES-1 modifies c-Myb function. (A) The c-Myb sites in the promoter and silencer are functionally interchangeable. Luciferase constructs containing the CD4 promoter with the c-Myb recognition site either intact (WT), mutated (MX), or substituted with the silencer c-Myb site (S2/Pro) were transfected into CD4 SP TH-clone D10 cells, and extracts from these cells were assayed for luciferase activity. Bars indicate percent of promoter activity when compared to that of the wild-type minimal CD4 promoter (100%). Data shown are compiled from at least three independent experiments with each construct. (B and C) HES-1 induces c-Myb to become a transcriptional repressor. The D10 CD4 SP TH clone was transfected with luciferase reporter constructs containing the CD4 promoter with either the c-Myb sites intact (B) or mutated (the MX mutation; panel C), a c-Myb expression vector, and increasing amounts of a HES-1 expression vector; error bars represent one standard deviation. Data are presented as fractions of the values obtained with the reporter construct and the c-Myb expression vector alone; typical values are 2 × 104 to 4 × 104 light units for the WT construct and 1 × 103 to 3 × 103 for the MX construct.
HES-1 binds to c-Myb in vivo.
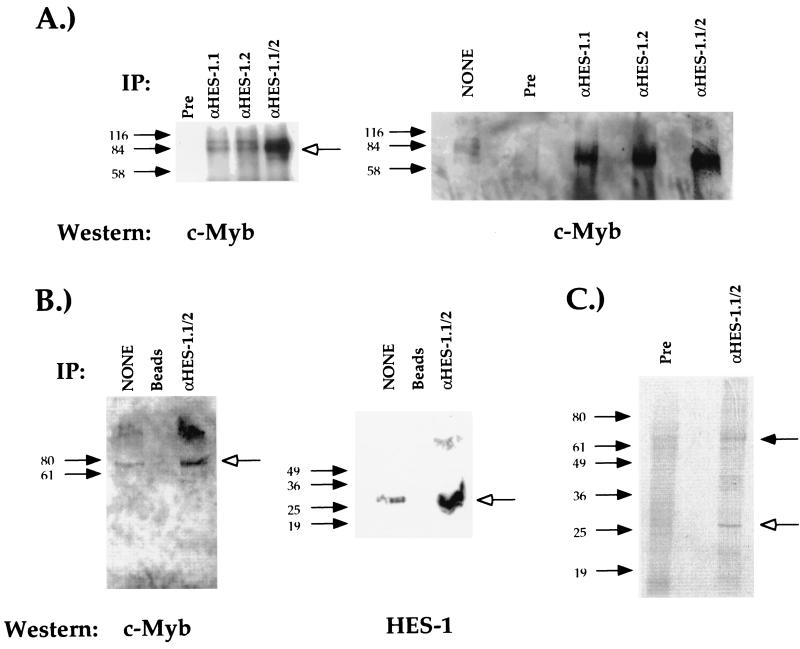
A second possible mechanism for the dual functionality of c-Myb is that the differential association of c-Myb with other DNA-binding transcription factors leads to different activating or repressing transcription factor complexes. In CD4+ T cells, the interaction of c-Myb with one factor would lead to its binding to the CD4 promoter and the induction of its function, whereas in CD4− T cells, c-Myb interacts with a second factor which leads to its binding to the silencer and repression of CD4 expression. One logical candidate for a c-Myb cofactor is HES-1, which binds to S1 of the CD4 silencer and is a known transcriptional repressor. Since the HES-1-binding site is next to the c-Myb-binding site, it is possible that the two factors may bind to each other directly and mediate silencer function as a multifactor complex. To test this, we conducted coimmunoprecipitation studies using nuclear extracts from the DP T-cell clone AKR1G1 and antibodies against HES-1 and c-Myb. In this experiment, endogenous HES-1 was immunoprecipitated from T-cell nuclear extracts with the HES-1 antiserum (22), the precipitate was resolved on an SDS-PAGE gel and transferred to nitrocellulose, and the membrane was subjected to Western blot analysis with the anti-c-Myb monoclonal antibody 1.1. As can be seen in Fig. 7A, we can detect a 75-kDa protein, which is the appropriate size for c-Myb. No complex is detected in lanes containing immunoprecipitates using the preimmune serum in place of the anti-HES-1 antiserum. These data indicate that the immunoprecipitation of HES-1 also brings down c-Myb, providing evidence that HES-1 can bind to c-Myb directly in vivo. To estimate the amount of c-Myb immunoprecipitated in this assay, we compared the intensities of the c-Myb band with that detectable in an input lane loaded with 20% of the T-cell nuclear extract used in the immunoprecipitation assay (Fig. 7A). Using densitometric analyses, we determined that 35% of the c-Myb in these extracts is complexed with HES-1, indicating that the amount of HES-1-complexed c-Myb is surprisingly high.
FIG. 7.
c-Myb binds to HES-1 in vivo. (A) Nuclear extracts from the AKR1G1 DP thymoma were immunoprecipitated (IP) with either the anti-HES-1 (α-HES-1) or preimmune (Pre) serum, resolved on an SDS-PAGE gel, and blotted with an anti-c-Myb antibody. Open arrows indicate the position of specific complex. Two different antisera against HES-1 generated from two different rabbits were tested either separately (α-HES-1.1 and α-HES-1.2) or pooled (α-HES-1.1/2). The lane marked “none” in the left panel represents a direct loading of 20% of the nuclear extract used in the immunoprecipitations. Densitometric analyses (see Materials and Methods) indicate that the input band is 6.6× less intense than the c-Myb bands in the IP lanes, indicating that 35% of the c-Myb in the nuclear extract is being immunoprecipitated with the HES-1 antiserum. (B) Nuclear extracts from the AKR1G1 DP thymoma were immunoprecipitated using either the HES-1 pooled antisera coupled to Sepharose beads (α-HES-1.1/2) or beads alone (beads), loaded onto an SDS-PAGE gel, and blotted with either the antibody against c-Myb (left panel) or the pooled HES-1 antisera (right panel). The lanes marked “none” in the left and right panels represent a direct loading of 20 and 10% of the nuclear extract used in the immunoprecipitations, respectively. (C) Direct immunoprecipitation of both c-Myb and HES-1 with the pooled HES-1 antisera. The S49 T-cell lymphoma was grown in 35S-methionine, and whole-cell extracts were purified as described previously (3). The labeled extracts were then immunoprecipitated either with the pooled HES-1 antisera (α-HES-1.1/2) or with the pooled preimmune sera (Pre), and the immunoprecipitates were resolved on an SDS-PAGE gel and visualized by autoradiography. Filled and open arrows indicate protein species of the molecular masses of c-Myb and HES-1, respectively.
To confirm that we are able to immunoprecipitate both HES-1 and c-Myb, we used the pooled HES-1 antisera coupled to Sepharose beads to precipitate HES-1 in T-cell nuclear extracts. The immunoprecipitates were then resolved on an SDS-PAGE gel, transferred to nitrocellulose, and blotted with either the 1.1 monoclonal antibody against c-Myb (Fig. 7B, left panel) or the pooled HES-1 antisera (Fig. 7B, right panel). Similar to the results presented above, we can detect the immunoprecipitation of c-Myb with the pooled HES-1 antisera and not with the Sepharose beads alone (Fig. 7B, left panel). As expected, we can also detect the immunoprecipitation of HES-1 with the pooled HES-1 antisera and not with the Sepharose beads alone (Fig. 7B, right panel). These data indicate that the immunoprecipitation of HES-1 also precipitates c-Myb. To confirm these results, we conducted in vivo labeling of T-cell nuclear extracts with 35S-methionine and immunoprecipitated with either the preimmune serum (Fig. 7C, left lane) or the pooled HES-1 antisera (Fig. 7C, right lane). The immunoprecipitates were then resolved on an SDS-PAGE gel, and the products were identified by autoradiography. As can be seen in Fig. 7C, we can detect two major protein species in the anti-HES-1 immunoprecipitate. The slower-mobility complex has the apparent molecular mass of 75 kDa, which is the appropriate molecular mass for c-Myb (Fig. 7C and data not shown), whereas the faster-mobility complex has the apparent molecular mass of 27 kDa, which is the appropriate molecular mass for HES-1. Taken together, these experiments indicate that we are able to coimmunoprecipitate c-Myb with HES-1, further supporting the hypothesis that these two transcription factors bind to each other in vivo.
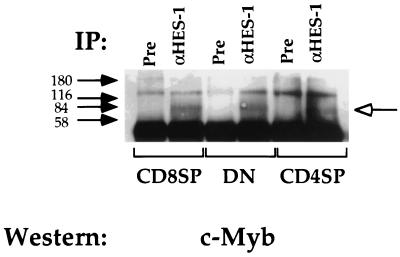
Although both HES-1 and c-Myb are expressed in all classes of T cells (22), it is possible that the interaction of HES-1 and c-Myb contributes to the specificity of silencer function and the repression of CD4 gene expression. In this model, either the subclass-specific modification of c-Myb or HES-1 or expression of a transcriptional coactivator allows for the interaction of these two factors and subsequent silencer function in DN and CD8 SP T cells. The lack of this modification or coactivator expression in DP and CD4 SP T cells results in the corresponding lack of silencer function. The prediction of this model is that we would be able to detect differences in the ability of HES-1 to interact with c-Myb in DN and CD8 SP T cells as opposed to DP and CD4 SP T cells. To test this, we conducted immunoprecipitation experiments with the pooled HES-1 antisera using extracts isolated from T cells of all developmental phenotypes (Fig. 8). We can immunoprecipitate c-Myb with the HES-1 antiserum in all T-cell extracts at approximately equivalent levels. These data indicate that the c-Myb–HES-1 interaction occurs in all T cells and thus is not likely to mediate the specificity of CD4 silencer function.
FIG. 8.
HES-1 and c-Myb interact in all T cells. In vivo immunoprecipitations (IP) using anti-HES-1 (α-HES-1) or preimmune (Pre) serum with the D10 CD4 SP TH clone (CD4SP), the L3 CD8 SP TC clone (CD8SP), and the S49 DN thymoma and the pooled antisera. The open arrow indicates protein species of the molecular weight of c-Myb.
HES-1 induces c-Myb-dependent repression of transcription.
Our biochemical data suggest that the HES-1–c-Myb complex binds to the silencer and mediates the repression of CD4 gene expression. We can thus make the prediction that the overexpression of HES-1 would lead to increased levels of HES-1 bound to c-Myb, which in turn could be recruited to a transcriptional control element by the binding of c-Myb in the complex to its recognition site, even in the absence of a consensus HES-1 site. According to this hypothesis, this would result in transcriptional repression. As discussed above, the CD4 promoter contains a c-Myb but not a HES-1 recognition site (38, 39). Should the HES-1–c-Myb complex function as a repressor, one can make the prediction that the overexpression of HES-1 should lead to the repression of CD4 promoter function dependent on the c-Myb site. To test these predictions, we conducted cotransfection reporter assays using eukaryotic expression constructs that contain the HES-1 and c-Myb cDNAs. The CD4 SP TH clone D10 was transfected with these constructs as well as with the wild-type and MX mutant CD4 promoter luciferase constructs described above (Fig. 6B and C). Transfection of the CD4 promoter luciferase construct alone leads to high levels of reporter activity (Fig. 6B, first lane). Cotransfection with the c-Myb expression vector leads to only a modest enhancement of promoter function (Fig. 6B); this is consistent with what we have reported previously (44) and is likely due to the high levels of endogenous c-Myb. Addition of increasing amounts of HES-1 expression vector to the transfection leads to the dose-dependent decrease in reporter activity (Fig. 6B). Similar results are obtained without the transfection of the c-Myb expression vector; this repression is likely due to the interaction of the overexpressed HES-1 with endogenous c-Myb (data not shown). These data indicate that the overexpression of HES-1 can indeed repress CD4 promoter function, even in the absence of its cognate binding site. We do not observe decreases in reporter activity when the MX variant of the CD4 promoter, which contains a site-specific mutation in the c-Myb-binding site, is used in the reporter construct (Fig. 6C). Similarly, overexpressed HES-1 does not repress simian virus 40 early promoter and enhancer function, indicating that it is not causing a general repression of transcription (data not shown). Thus, overexpression of HES-1 leads to repression of CD4 promoter function that is dependent on the ability of c-Myb to bind to its binding site. These observations fulfill the predictions listed above and indicate that the HES-1–c-Myb complex indeed functions as a transcriptional repressor.
DISCUSSION
Context-dependent c-Myb function in mediating CD4 gene expression.
We and others have shown that c-Myb can induce the CD4 promoter to function at high levels in CD4 SP TH cells (28, 44); here we present data indicating that c-Myb also binds to the CD4 silencer and helps mediate its function as well. We also demonstrate that c-Myb can function as a transcriptional repressor when bound to HES-1, a second CD4 silencer-binding factor. Our data thus suggest that in the appropriate contexts c-Myb can both activate and repress CD4 gene expression. Although c-Myb has been reported in separate systems to function as both an activator and a repressor, this is the first report of c-Myb playing both roles in the control of transcription of the same gene. Our data suggest that c-Myb function at either the promoter or the silencer is dependent on the other factors that bind to each element. We have demonstrated that c-Myb binds to HES-1 in vivo and forms a repressor complex; we propose that this multifactor complex binds to the CD4 silencer at S1 and S2, leading to the induction of silencer function and transcriptional repression of the CD4 gene. This HES-1–c-Myb complex may recruit other factors to the CD4 silencer as well. For example, HES-1 is known to recruit TLE, the mammalian homologue of groucho, as well as cooperate with Runt domain-containing proteins, such as Cbfa2-AML-PEBP2, to DNA to repress transcription (14, 26). It is interesting to note that Cbfa2-AML-PEBP2 is also capable of interacting with c-Myb to affect transcription (16, 51), and that there are consensus Cbfa2-AML-PEBP2 binding sites within footprinted regions of the CD4 silencer (10). It is thus possible that a multifactor repressor complex consisting of HES-1 and c-Myb, perhaps in conjunction with Cbfa2-AML-PEBP2 and TLE, may be critical for inducing silencer function and the downregulation of CD4 gene expression. At the CD4 promoter, however, c-Myb interacts with a different set of transcription factors. Although our experiments indicate that a significant amount of c-Myb in T cells is complexed with HES-1, there is still free c-Myb in these cells that may be capable of accessing the promoter alone. Previous studies have demonstrated that the MAZ and Elf-1 transcription factors mediate CD4 promoter function (11, 38); in addition, an unknown factor that displays subclass-specific properties binds to a third functional site (39). It is thus possible that the interaction of c-Myb with these other factors in the absence of HES-1 induces it to become a transcriptional activator.
Context-dependent transcription factor function has been previously demonstrated in dorsal-ventral pattern formation in Drosophila melanogaster development. In this system, the dorsoventral fate map of the embryo is determined by a concentration gradient of the maternal transcription factor Dorsal (32, 33, 45). The ability of Dorsal to mediate fate determination is dependent on its ability to function as both a transcriptional activator and a transcription repressor; Dorsal binds to the promoters of the mesoderm-determining genes snail and twist and induces their expression, whereas for the ectoderm-determining genes zerknullt and decapentaplegic, Dorsal binds to a ventral repression region and mediates transcriptional repression (31). Interestingly, Dorsal binding to its cognate recognition site alone leads to transcriptional activation (20, 21, 30, 47); transcriptional repression requires additional elements (30). For the ventral repression region of zerknullt, Dorsal binds as a multifactor complex with cut, dead ringer, and groucho, and it is this complete complex that mediates transcriptional repression (49). Thus, the context of the Dorsal-binding site determines whether or not Dorsal becomes a transcriptional activator or a transcriptional repressor. The similarities in mechanism of function between Dorsal in the control of dorsoventral fate and c-Myb in the control of CD4 gene expression suggest that context dependence is a general mechanism for modifying transcription factor function in complex developmental systems.
c-Myb and the mechanism of CD4 silencer function.
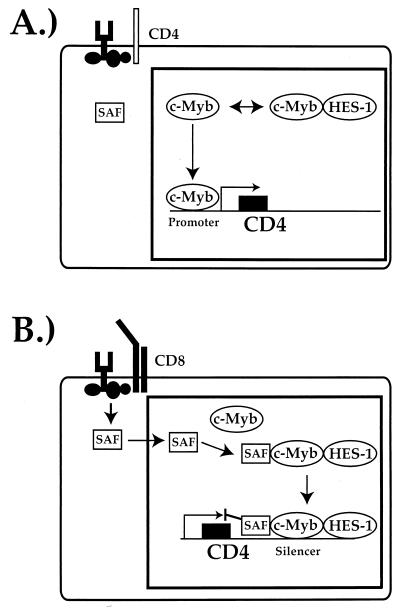
In controlling CD4-specific expression, the silencer must inhibit the transcriptional machinery from functioning (the mechanism of action), and it must do so in a subclass-specific manner (the specificity of action). These two functions may be completely distinct from each other and require different sets of factors, or there may be significant overlap. Nonetheless, in constructing models for the role of the CD4 silencer-binding factors in the control of CD4 gene expression, it is useful to consider these concepts separately. Because both HES-1 and c-Myb are expressed and their interaction in vivo can be demonstrated in all T-cell subclasses, it is less likely that these factors mediate the specificity of silencer function. We believe that the specificity of function of CD4 expression is mediated primarily by the S3-binding factor SAF. This novel homeodomain-like transcription factor is expressed in all T cells; however, SAF is present in the nucleus of T cells in which the silencer is functioning and which thus do not express CD4, such as DN and CD8 SP T cells (24; Kim et al., submitted). In contrast, in T cells in which the silencer is not functioning, such as CD4 SP and DP T cells, SAF is present in the cytoplasm (24; Kim et al., submitted). We believe that the transport of SAF across the nuclear membrane in developing T cells mediates the specificity of CD4 silencer function (Kim et al., submitted). However, our data demonstrating that the HES-1–c-Myb complex is a functional transcriptional repressor indicate that this complex is more likely to play an important role in mediating the actual repression mechanism. In our model, HES-1 and c-Myb bind together to form a repressor complex in DP T cells, but they are nonfunctional at the CD4 silencer due to the absence of SAF. Should the DP thymocyte develop into a CD4 SP T cell, SAF remains outside of the nucleus, and the HES-1–c-Myb complex is not recruited to the silencer; instead, uncomplexed c-Myb is recruited to the promoter, where in conjunction with other promoter-binding factors, it induces CD4 promoter function (Fig. 9A). Alternatively, should the DP T cell develop into a CD8 SP T cell, SAF is transported into the nucleus, where it recruits the HES-1–c-Myb complex to the silencer, thus leading to the induction of CD4 silencer function (Fig. 8B). This could be mediated either directly by the HES-1–c-Myb complex or by the recruitment of other transcriptional corepressors, such as Cbfa2-AML-PEBP2 and TLE. Further experiments in this system will enable us to address these questions directly.
FIG. 9.
A model for the mechanism of CD4 gene expression control by c-Myb and HES-1. (A) Induction of CD4 promoter function by c-Myb during CD4 SP TH-cell development. (B) Induction of CD4 silencer function by a HES-1–c-Myb complex during CD8 SP TC-cell development. See the text for details.
One interesting aspect of our data is that they may help explain the puzzling functional asymmetric redundancy that is observed in CD4 silencer function (10). We have shown that single deletions of the factor-binding sites in the CD4 silencer do not affect silencer function; silencer function is only abrogated when the c-Myb site is deleted in conjunction with either the HES-1- or the SAF-binding site. c-Myb itself cannot mediate silencer function, as the region around the c-Myb site alone does not function as a silencer in our transgenic assays (10). Because HES-1 binds to c-Myb in the absence of DNA, it is possible that the binding of one of these factors to the silencer helps recruit the other. Thus, the single deletion of either the HES-1 or the c-Myb site would not affect silencer function, since the presence of either factor would recruit the other to the silencer even in the absence of its binding site. In this model, in addition to its role in the repressor complex, c-Myb plays a central role in maintaining the transcription factor complex on the silencer itself.
ACKNOWLEDGMENTS
We thank Monica Mendelson for technical assistance, Nikhil deSouza for the 35S-labeled T-cell nuclear extracts, and Kathryn Calame, Alessandra Pernis, and Chris Schindler for critical review of the manuscript.
This work was funded by grants from the NIH (AI34925) and the Irma T. Hirschl-Monique Weill Caulier Trust to G.S. R.D.A. and H.K.K. were supported by NIH training grant no. T32AI07525.
REFERENCES
- 1.Adlam M, Duncan D D, Ng D K, Siu G. Positive selection induces CD4 promoter and enhancer function. Int Immunol. 1997;9:877–887. doi: 10.1093/intimm/9.6.877. [DOI] [PubMed] [Google Scholar]
- 2.Allen R D, Bender T P, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 4.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 5.Berg L J, Pullen A M, Fazekas de St. Groth B, Mathis D, Benoist C, Davis M M. Antigen/MHC-specific T cells are preferentially exported from the thymus in the presence of their MHC ligand. Cell. 1989;58:1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- 6.Bevan M J, Hogquist K A, Jameson S C. Selecting the T cell receptor repertoire. Science. 1994;264:796–797. doi: 10.1126/science.8171333. [DOI] [PubMed] [Google Scholar]
- 7.Boyle W J, Lipsick J S, Baluda M A. Antibodies to the evolutionarily conserved amino-terminal region of the v-Myb-encoded protein detect the c-Myb protein in widely divergent metazoan species. Proc Natl Acad Sci USA. 1986;83:4685–4689. doi: 10.1073/pnas.83.13.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam J D. Preparation of extracts from higher eukaryotes. In: Deutscher M P, editor. Methods in enzymology. Vol. 182. New York, N.Y: Academic Press, Inc.; 1990. pp. 194–203. [DOI] [PubMed] [Google Scholar]
- 9.Donda A, Schulz M, Burki K, De Libero G, Uematsu Y. Identification and characterization of a human CD4 silencer. Eur J Immunol. 1996;26:493–500. doi: 10.1002/eji.1830260232. [DOI] [PubMed] [Google Scholar]
- 10.Duncan D D, Adlam M, Siu G. Asymmetric redundancy in CD4 silencer function. Immunity. 1996;4:301–311. doi: 10.1016/s1074-7613(00)80438-0. [DOI] [PubMed] [Google Scholar]
- 11.Duncan D D, Stupakoff A, Hedrick S M, Marcu K B, Siu G. A Myc-associated zinc-finger protein (MAZ) binding site is one of four important functional regions in the CD4 promoter. Mol Cell Biol. 1995;15:3179–3186. doi: 10.1128/mcb.15.6.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ess K C, Whitaker T L, Cost G J, Witte D P, Hutton J J, Aronow B J. A central role for a single c-Myb binding site in a thymic locus control region. Mol Cell Biol. 1995;15:5707–5715. doi: 10.1128/mcb.15.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia A, LaMontagne K, Reavis D, Stober-Grasser U, Lipsick J S. Determinants of sequence-specific DNA-binding by p48v-myb. Oncogene. 1991;6:265–273. [PubMed] [Google Scholar]
- 14.Grbavec D, Lo R, Liu Y, Stifani S. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur J Biochem. 1998;258:339–349. doi: 10.1046/j.1432-1327.1998.2580339.x. [DOI] [PubMed] [Google Scholar]
- 15.Grusby M J, Johnson R S, Papaionnou V E, Glimcher L H. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Munain C, Krangel M S. c-Myb and core-binding factor/PEBP2 display functional synergy but bind independently to adjacent sites in the T-cell receptor delta enhancer. Mol Cell Biol. 1995;15:3090–3099. doi: 10.1128/mcb.15.6.3090. . (Erratum, 15:4654.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Munain C, Krangel M S. Regulation of the T-cell receptor d enhancer by functional cooperation between c-Myb and core-binding factors. Mol Cell Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. New York, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 19.Hsiang Y-H, Goldman J P, Raulet D H. The role of c-Myb or a related factor in regulating the T cell receptor γ gene enhancer. J Immunol. 1995;154:5195–5204. [PubMed] [Google Scholar]
- 20.Ip Y T, Kraut R, Levine M, Rushlow C A. The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell. 1991;64:439–446. doi: 10.1016/0092-8674(91)90651-e. [DOI] [PubMed] [Google Scholar]
- 21.Jiang J, Kosman D, Ip Y T, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 22.Kim H K, Siu G. The Notch pathway intermediate HES-1 silences CD4 gene expression. Mol Cell Biol. 1998;18:7166–7175. doi: 10.1128/mcb.18.12.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim W W S, Siu G. Subclass-specific nuclear localization of a novel CD4 silencer-binding factor. J Exp Med. 1999;190:281–291. doi: 10.1084/jem.190.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin H H, Robinson B H. Approaches to DNA mutagenesis: an overview. Anal Biochem. 1997;254:157–178. doi: 10.1006/abio.1997.2428. [DOI] [PubMed] [Google Scholar]
- 25.Lipsick J S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 26.McLarren K W, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfal. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 27.Mucenski M L, McLain K, Kier A B, Swerdlow S H, Schreiner C M, Miller T A, Pietryga D W, Scott J W J, Potter S S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama K, Yamamoto R, Ishii S, Nakauchi H. Binding of c-Myb to the core sequence of the CD4 promoter. Int Immunol. 1993;5:817–824. doi: 10.1093/intimm/5.8.817. [DOI] [PubMed] [Google Scholar]
- 29.Ness S A, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 30.Pan D, Courey A J. The same dorsal binding site mediates both activation and repression in a context-dependent manner. EMBO J. 1992;11:1837–1842. doi: 10.1002/j.1460-2075.1992.tb05235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray R P, Arora K, Nusslein-Volhard C, Gelbart W M. The control of cell fate along the dorsal-ventral axis of the Drosophila embryo. Development. 1991;113:35–54. doi: 10.1242/dev.113.1.35. [DOI] [PubMed] [Google Scholar]
- 32.Roth S, Stein D, Nusslein-Volhard C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- 33.Rushlow C A, Han K, Manley J L, Levine M. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell. 1989;59:1165–1177. doi: 10.1016/0092-8674(89)90772-1. [DOI] [PubMed] [Google Scholar]
- 34.Salmon P, Boyer O, Lores P, Jami J, Klatzmann D. Characterization of an intronless CD4 minigene expressed in mature CD4 and CD8 T cells, but not expressed in immature thymocytes. J Immunol. 1996;156:1873–1879. [PubMed] [Google Scholar]
- 35.Salmon P, Giovane A, Wasylyk B, Klatzmann D. Characterization of the human CD4 gene promoter: transcription from the CD4 gene core promoter is tissue-specific and is activated by Ets proteins. Proc Natl Acad Sci USA. 1993;90:7739–7743. doi: 10.1073/pnas.90.16.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual (2nd ed.). Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sarafova S D, Siu G. Control of CD4 gene expression: connecting signals to outcomes in T cell development. Braz J Med Biol Res. 1999;32:785–803. doi: 10.1590/s0100-879x1999000700001. [DOI] [PubMed] [Google Scholar]
- 38.Sarafova S D, Siu G. A potential role for Elf-1 in CD4 promoter function. J Biol Chem. 1999;274:16126–16134. doi: 10.1074/jbc.274.23.16126. [DOI] [PubMed] [Google Scholar]
- 39.Sarafova S D, Siu G. Precise orientation of factor-binding sites is required for CD4 promoter function. Nucleic Acids Res. 2000;28:2664–2671. doi: 10.1093/nar/28.14.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawada S, Littman D R. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawada S, Scarborough J D, Killeen N, Littman D R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 42.Sha W C, Nelson C A, Newberry R D, Kranz D M, Russell J H, Loh D. Positive and negative selection of an antigen receptor on T cells in transgenic mice. Nature. 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- 43.Siu G, Wurster A L, Duncan D D, Soliman T M, Hedrick S M. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siu G, Wurster A L, Lipsick J S, Hedrick S M. Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steward R. Relocalization of the dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell. 1989;59:1179–1188. doi: 10.1016/0092-8674(89)90773-3. [DOI] [PubMed] [Google Scholar]
- 46.Teh H S, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H. Thymic major histocompatibility complex antigens and the αβ T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–233. doi: 10.1038/335229a0. [DOI] [PubMed] [Google Scholar]
- 47.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 48.Uematsu Y, Donda A, De Libero G. Thymocytes control the CD4 gene differently from mature T-lymphocytes. Int Immunol. 1997;9:179–187. doi: 10.1093/intimm/9.1.179. [DOI] [PubMed] [Google Scholar]
- 49.Valentine S A, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey A J. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wurster A L, Siu G, Leiden J, Hedrick S M. Elf-1 binds to a critical element in a second CD4 enhancer. Mol Cell Biol. 1994;14:6452–6463. doi: 10.1128/mcb.14.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaiman A L, Lenz J. Transcriptional activation of a retrovirus enhancer by CBF (AML1) requires a second factor: evidence for cooperativity with c-Myb. J Virol. 1996;70:5618–5629. doi: 10.1128/jvi.70.8.5618-5629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]