Summary
A library of well-characterized human induced pluripotent stem cell (hiPSC) lines from clinically healthy human subjects could serve as a useful resource of normal controls for in vitro human development, disease modeling, genotype-phenotype association studies, and drug response evaluation. We report generation and extensive characterization of a gender-balanced, racially/ethnically diverse library of hiPSC lines from 40 clinically healthy human individuals who range in age from 22 to 61 years. The hiPSCs match the karyotype and short tandem repeat identities of their parental fibroblasts, and have a transcription profile characteristic of pluripotent stem cells. We provide whole-genome sequencing data for one hiPSC clone from each individual, genomic ancestry determination, and analysis of mendelian disease genes and risks. We document similar transcriptomic profiles, single-cell RNA-sequencing-derived cell clusters, and physiology of cardiomyocytes differentiated from multiple independent hiPSC lines. This extensive characterization makes this hiPSC library a valuable resource for many studies on human biology.
Keywords: induced pluripotent stem cells, clinically well-characterized healthy subjects, racially/ethnically diverse, whole-genome sequencing, genetic variants of mild common disease phenotypes or more severe disease with incomplete penetrance, age- and sex-independent gene signature, cardiomyocytes, similar physiology and transcriptomes
Highlights
-
•
A library of induced pluripotent stem cells from 40 healthy human subjects
-
•
Racially/ethnically diverse subjects of clinically well-characterized health
-
•
Whole-genome sequencing identifies variants of mild common phenotypes or incomplete penetrance
-
•
Similar physiology of cardiomyocytes from independent hiPSC clones and individuals
In this resource, Schaniel et al. report the generation and characterization of a gender-balanced, racially/ethnically diverse library of hiPSC lines from 40 clinically well-characterized healthy human individuals. The authors provide whole-genome sequencing data, ancestry determination, and analysis of mendelian disease genes and risks. Furthermore, similar physiology of cardiomyocytes differentiated from independent hiPSC clones derived from two individuals is documented.
Introduction
Since their groundbreaking discovery (Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007), human induced pluripotent stem cells (hiPSCs) and cells differentiated from them have become a powerful system to model in vitro human phenotypes, disease etiology and mechanisms, genotype-phenotype correlations, and drug responses. However, such studies are often hampered by the small number of hiPSC lines or respective controls used for comparative analysis, reported somatic variability of derived hiPSCs, and heterogeneity of differentiated cells (International Stem Cell Initiative et al., 2011; Dubois et al., 2011; Fusaki et al., 2009; Nazor et al., 2012; Witty et al., 2014). In recent years, several groups reported on the establishment of various hiPSC libraries with a wide range in the number of hiPSC lines (Carcamo-Orive et al., 2017; Kilpinen et al., 2017; Panopoulos et al., 2017; Park et al., 2017; Streeter et al., 2017). These libraries included mostly disease-specific hiPSCs and controls. The control hiPSC lines were derived from subjects without the specific diseases studied, either relatives or non-related individuals (Carcamo-Orive et al., 2017; Panopoulos et al., 2017), who were self-declared healthy, or individuals with no medical disease history (Kilpinen et al., 2017; Panopoulos et al., 2017; Rouhani et al., 2014). However, in these studies, a clinical health evaluation was not explicitly performed and no official documentation of health status reported. Nevertheless, such libraries have been used for the study of how genetic variants associated with complex genomic traits and phenotypes drive molecular and physiological variation in hiPSCs and their differentiated cells (Carcamo-Orive et al., 2017; D'Antonio-Chronowska et al., 2019; DeBoever et al., 2017; Karch et al., 2019; Kaserman et al., 2020; Kilpinen et al., 2017; Panopoulos et al., 2017; Park et al., 2017; Rouhani et al., 2014). Larger-scale comparative and effective disease modeling, drug discovery and evaluation, and genotype-phenotype association studies suffer from the limited availability and inclusion of hiPSC lines from clinically screened, healthy individuals of various racial and ethnic backgrounds and age. Here, as part of the NIH-Common Fund Library of Integrated Network-Based Cellular Signatures (LINCS) program (Keenan et al., 2018), we report the creation of a hiPSC library from 40 selected individuals of diverse racial/ethnic backgrounds and ages who passed a rigorous clinical health evaluation. We provide the clinical characteristics of each of the participants from whom hiPSCs were derived, cytogenetics reports, short tandem repeat (STR) authentication, and pluripotency analyses for all 40 hiPSC lines, as well as whole-genome sequencing data, genomic ancestry determination, and mendelian disease gene and risk assessment.
Several studies have suggested an impact of donor cell source, cellular heterogeneity of established hiPSCs, as well as sex on cellular differentiation and physiological behavior (D'Antonio-Chronowska et al., 2019; Hu et al., 2016; Pianezzi et al., 2020; Sanchez-Freire et al., 2014). Variability in measured physiological parameters might be affected by the lack of cellular homogeneity in differentiated hiPSCs. It remains unclear, however, whether cells differentiated from multiple hiPSC clones from the same healthy subject will behave physiologically the same. Therefore, we studied the characteristics of ventricular and atrial cardiomyocytes (CMs) differentiated from independent hiPSC clones from the same individual and from hiPSC clones from different individuals.
In summary, our diverse hiPSCs library from 40 clinically well-characterized healthy human individuals contributes a valuable resource to the scientific community for a broad variety of biomedical and pharmacological research.
Results
Recruitment, health evaluation, and characterization of individuals in the Mount Sinai library of hiPSC lines derived from diverse, clinically healthy subjects
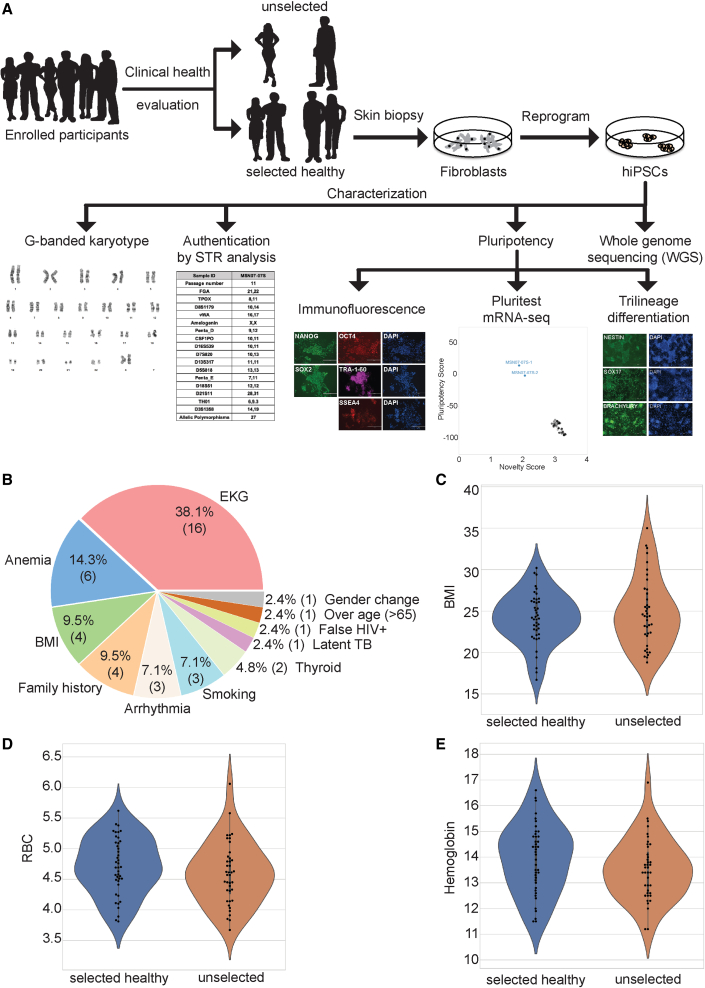
Potential healthy volunteers were recruited through Institutional Review Board (IRB)-approved advertisements, and were pre-screened for potential inclusion in the study. Ninety-six men and women who satisfied the initial pre-screening consented (documents S1 and S2), and their sex, age, and race/ethnicity were recorded through an enrollment questionnaire (Figure 1A). Eighty-five underwent a formal and thorough evaluation by a screening study physician. Formal screening involved a full medical history; measurement of weight, height, waist and hip circumference, heart rate, blood pressure, respiratory rate, and oxygen saturation; a physical examination; and an electrocardiogram (ECG). Blood was drawn for analysis of clinically relevant parameters. A pregnancy test was included for female participants. All blood draws and pregnancy tests were sent to a certified clinical laboratory for analysis. The more than 20 exclusion criteria included abnormal ECG, family history of any cardiovascular disorder excluding hypertension in any first- or second-degree relative at age <50 years, family history of non-ischemic cardiomyopathy in any first- or second-degree relative at any age, prior organ transplantation, HIV-positive status, history of myopathy, obesity, renal impairment, autoimmune disease, abnormal blood test results including brain natriuretic peptide, body mass index of ≥30 kg/m2, lifetime smoking of >2 pack-years, or abnormality on physical examination. Data were entered onto a clinical report form (document S3) and a final assessment of study eligibility was then made by consensus of two senior study physicians (J.C.K. and D.T.) and the screening study physician. If eligibility was approved, the subject then underwent skin biopsy and formally became one of the included study subjects. Of the 85 subjects who were screened, 42 (48.3%) were deemed eligible for final inclusion. Reasons for a screened subject's exclusion from further participation are summarized in Figure 1B. Of the 42 eligible individuals, a skin biopsy was scheduled and performed on 40. Table S1 lists their clinical parameters. The 40 healthy male (22) and female (18) individuals ranged in age from 22 to 61 years.
Figure 1.
Description of subject selection for the Mount Sinai clinically healthy hiPSC library
(A) Flow chart for subject selection and hiPSC generation and characterization. See Table S1 for the summary characterizations performed for all 40 selected, clinically healthy subjects and derived hiPSCs.
(B) Pie diagram of reasons for excluding screened participants from the final group of eligible subjects. The percentage as well as the number of participants (in parentheses) excluded for each specific main reason is presented.
(C–E) Violin plots of clinical selection criteria of (C) BMI (kg/m2), (D) red blood cell counts (RBC; ×106/μL), and (E) hemoglobin level (g/dL) of healthy/selected and excluded participants. See document S3 for the clinical report form and Table S1 for the clinical characteristics of all clinically healthy/selected individuals.
See also Table S1. Clinical characteristics of all 40 selected, clinically healthy subjects, related to Figure 1, Table S2. Summary characterization of all 40 selected, clinically healthy subjects and derived iPSCs, related to Figures 1, 2 and S1, and Tables S1, S3, S4, and S6, Table S3. Cytogenetics of 40 iPSC lines, related to Figures 1 and 2, Table S4. RNA-seq read counts, related to Figures 1–3, S2, and S4B, Table S5. PCA projections, standard deviation, variance, and percentage of variance of each principal component, and PCA variable loadings of hiPSC- and hiPSC-derived CM RNA-seq data, related to Figures 3B and S4B, Table S6. Ancestry analysis, related to Figure 1, Table S7. Pathogenic disease variants, related to Figures 1 and 3A, and Tables S1 and S2.
These observations demonstrate that an inclusive health evaluation and strict selection criteria will produce more stringent definition of health status than simply relying on a participant’s self-declaration or medical history.
Generation, authentication, and characterization of hiPSC clones
We derived fibroblast lines from skin biopsy samples taken from the 40 eligible healthy subjects. We used integration-free reprogramming (mRNA with microRNA boost [R], Warren et al., 2010; and/or Sendai virus [S], Fusaki et al., 2009) to generate hiPSCs and establish multiple clones from all 40 fibroblast lines. Our present resource consists of one hiPSC clone per individual plus two additional clones each for two hiPSC lines (Table S2). All data, as well as linked associated metadata, can also be found on the searchable LINCS data portal (Koleti et al., 2018; Stathias et al., 2020). All hiPSCs in our library match the karyotype of the parental fibroblast line. In all cases except one, the karyotype was normal (Figure 2A; Table S3). The abnormal karyotype, a t(1;17)(p34;q23) translocation, was observed for MSN24. We analyzed the karyotypes of five independent MSN24 hiPSC clones. All carried the t(1;17)(p34;q23) translocation. It is not uncommon that a karyotype is abnormal (International Stem Cell Initiative et al., 2011; Mayshar et al., 2010; Peterson and Loring, 2014; Taapken et al., 2011); however, a frequency of 100% abnormal hiPSC clones is very unusual. Thus, we karyotyped the parental fibroblasts. Surprisingly, the parental fibroblasts already carried this abnormality. Hence, this may represent a case of a healthy individual who is the carrier of a balanced translocation for which no report exists. Of 84 hiPSC clones we karyotyped, 26 (in addition to the five analyzed MSN24 hiPSC clones) showed an abnormal karyotype (31%). This is consistent with published reports of abnormal karyotypes in up to one-third of derived human embryonic stem cells (ESCs) and hiPSCs (International Stem Cell Initiative et al., 2011; Mayshar et al., 2010; Peterson and Loring, 2014; Taapken et al., 2011). All hiPSC clones in our library are negative for Sendai virus (Tables S2 and S4) and are authenticated by STR analysis to match the profile of their respective parental fibroblast line (STR table available at dbGaP under accession dbGaP: phs002088.v2.p1). This is an important and necessary quality control measure to ensure the origin and authenticity of the derived hiPSC clones.
Figure 2.
Characterization of two representative hiPSC lines/clones
(A) G-banded karyotypes for female hiPSC clone MSN07-07S and male hiPSC clone MS34-05S. See Table S3 for the cytogenetics of all 40 fibroblast lines and one associated derived hiPSC clone.
(B) Immunocytochemistry of pluripotency markers, NANOG, OCT4, SOX2, TRA-1-60, and SSEA4 in representative hiPSC clones, MSN07-07S and MS34-05S. DAPI is used to stain nuclei. Bar, 400 μm. See Figure S1 for the immunocytochemistry of one representative hiPSC clone derived from each of the 40 fibroblast lines.
(C) PluriTest summary plot of RNA-seq-based transcriptomic analyses performed in duplicate for one hiPSC clone per clinically healthy individual (colored circles). As a reference, the PluriTest results of transcriptomic data of fibroblasts from 66 individuals (Hagai et al., 2018) are plotted in black/gray. See Figure S2 for the PluriTest plots of one representative hiPSC clone derived from each of the 40 fibroblast lines. See also Figures S1–S3 and Table S2. Summary characterization of all 40 selected, clinically healthy subjects and derived iPSCs, related to Figures 1, 2 and S1, and Tables S1, S3, S4, and S6, Table S3. Cytogenetics of 40 iPSC lines, related to Figures 1 and 2, Table S4. RNA-seq read counts, related to Figures 1–3, S2, and S4B.
One karyotypically normal (with the exception of MSN24) and authenticated hiPSC clone generated from each of the 40 clinically healthy individuals was further characterized by immunocytochemistry for expression of NANOG, OCT4, SOX2, TRA-1-60, and SSEA4 (Figures 2B and S1). We also conducted a PluriTest assay based on RNA sequencing (RNA-seq) (Table S4) (Panopoulos et al., 2017) as an additional measure of the pluripotent status (Figures 2C and S2; Table S4). We included the pluripotency and novelty scores retrieved from fibroblasts of 66 individuals (Hagai et al., 2018), from which Kilpinen et al. (2017)derived hiPSC lines, as comparative reference. We next compared the gene expression profiles of the 40 hiPSC lines with 77 randomly chosen hiPSC lines described by Kilpinen and colleagues as well as these same 66 human fibroblast lines. Pairwise Pearson correlation and principal component analysis (PCA) demonstrate that the two sets of hiPSC lines are similar to each other and very distinct from the profiles of the fibroblast lines (Figure 3; Tables S5A–S5C). Lastly, we assessed pluripotency of a selection of hiPSC lines/clones by in vitro tri-lineage differentiation (Figure S3). All assayed hiPSC lines/clones differentiated efficiently to cells representing (neuro)ectoderm (NESTIN+), endoderm (SOX17+) and mesoderm (BRACHYURY+) demonstrating their pluripotency.
Figure 3.
Comparative hiPSC transcriptome analysis
(A) Pairwise Pearson correlation was determined between RNA-seq-based gene expression profiles of the 40 hiPSC samples (in duplicates) from this study with 77 randomly selected hiPSC samples (Kilpinen et al., 2017) and 66 human fibroblast samples (Hagai et al., 2018). Hierarchical clustering documents close correlation of the two hiPSC sample sets with clear separation from the fibroblast lines.
(B) PCA of all three combined sets identifies principal component 1 (PC1) with ∼60% of the total variance to not significantly distinguish between the two hiPSC sets but to clearly separate them from the fibroblast lines. See also Table S4. RNA-seq read counts, related to Figures 1–3, S2, and S4B, Table S5. PCA projections, standard deviation, variance, and percentage of variance of each principal component, and PCA variable loadings of hiPSC- and hiPSC-derived CM RNA-seq data, related to Figures 3B and S4B, Table S6. Ancestry analysis, related to Figure 1.
We performed genetic ancestry determination using whole-genome sequencing information from all 40 hiPSC lines (Table S6). This analysis provided confirmation of the authenticity of the hiPSC lines. In addition, it reflected the diverse nature of the subjects’ racial and ethnic backgrounds. The selected individuals have origins in East Africa, West Africa, East Asia, the central India subcontinent, the southern India subcontinent, eastern Mediterranean, northeast Europe, northern and central Europe, southwest Europe, and the Anatolia/Caucasus/Iranian Plateau region.
These data support our conclusion that we established a gender-balanced, racially/ethnically diverse library of well-characterized hiPSCs from 40 clinically healthy individuals who range in age from 22 to 61 years.
Clinically healthy individuals can be carriers of genetic variants with disease risks
Although all 40 individuals represented in the hiPSC library were determined to be clinically healthy, we next assessed whether they carry potential genetic disease risks. To assess this, we interrogated the whole-genome sequencing data by performing annotation of gene variants associated with both dominant and recessive disorders (variant call format files are provided along with the whole-genome sequencing data in dbGaP under accession dbGaP: phs002088.v2.p1). All variants called with non-conflicting interpretations of at least likely pathogenic in Clinvar (Landrum et al., 2014) with defined assertion criteria were annotated using Annovar. For nine individuals (22.5%), zero pathogenic or likely pathogenic variants were detected. Two subjects (5%) carried one likely pathogenic variant, seven individuals (17.5%) carried one to two pathogenic variants, three subjects (7.5%) carried one pathogenic/likely pathogenic variant, and 19 (47.5%) were carriers of a combination of pathogenic and/or likely pathogenic and/or pathogenic/likely pathogenic variants (Table S7). The maximum of pathogenic and/or likely pathogenic variants per subjects detected was six. Variants were reviewed by a board-certified medical geneticist (S.I.B.). Variants associated with dominant diseases are noted in Figure 4A. These variants are consistent with the history of coming from a healthy adult as they can result in mild common phenotypes or disorders with later onset and incomplete penetrance. These findings highlight that clinically well-characterized healthy human individuals can harbor genetic variants with disease risks, a fact that may not be surprising but should to be considered when using control hiPSCs, including those from our library, as controls in specific disease modeling and/or drug evaluation/toxicity studies where certain variants might affect the results and their interpretation.
Figure 4.
Genetic variants with disease risks and global gene expression independence of age and sex
(A) Subjects with genetic disease variants associated with a dominant presentation.
(B) Pairwise correlation was determined between RNA-seq raw read counts of duplicate samples of all 40 hiPSC lines, followed by hierarchical clustering. The resulting dendrogram documents high similarity between duplicate samples and hiPSC lines. No influence of age or sex was found on gene expression.
hiPSC gene signature is independent of age or sex of individuals from whom hiPSCs are derived
To assess whether age or sex of included individuals could segregate derived hiPSC lines based on global gene expression, we determined pairwise correlations between bulk mRNA sequencing raw read counts of duplicate samples of all 40 lines followed by hierarchical clustering. The resulting dendrogram shows a high similarity both between duplicate samples and across the 40 lines (Figure 4B). Similar to pluripotency marker expression (Figures 2B and S1) and PluriTest scores (Figures 2C and S2), we observed no significant influence of age or sex on global gene expression. These results indicate that global gene expression, within the wide age range we studied, is independent of age and sex of clinically well-characterized healthy human subjects from whom hiPSC lines are derived.
Similar functional characteristics of CMs derived from multiple hiPSC lines
A recent report determined that that sex affects cardiac ventricular and atrial differentiation outcomes, whereas inherited genetic variation does not (D'Antonio-Chronowska et al., 2019). It remains unknown, however, whether hiPSCs derived from different healthy individuals will exhibit comparable molecular and physiological characteristics when differentiated using similar protocols. We sought to answer this question by performing both molecular and physiological analysis on CMs. For this, we differentiated several age-matched, male and female hiPSC lines into cardiac mesoderm (kinase insert domain receptor [KDR]+/platelet-derived growth factor receptor alpha [PDGFRA]+) and ventricular CMs (signal regulatory protein alpha [SIRPA]+/CD90−) (Dubois et al., 2011; Kattman et al., 2011) followed by purification using metabolic selection (Tohyama et al., 2013). The vast majority of cells in our differentiation cultures post selection were beating. We then measured calcium transients and compared several features between the different hiPSC lines (Figure S4A; Table S8A). We also conducted bulk and single-cell RNA-seq experiments to interrogate global transcriptomes at the population and single-cell levels (Figures S4B–S4G). High-level comparisons of both the physiological and RNA-seq data failed to reveal obvious differences between CMs differentiated from the various hiPSC lines (Figures S4A–S4G) aside from some heterogeneity in relative cluster distribution (Figures S4D and S4E).
For more in-depth comparison, we also performed experiments in CMs differentiated from multiple hiPSC clones from two individuals. We differentiated three independent, karyotypically normal hiPSC clones derived from two age-matched males into atrial and ventricular cardiac mesoderm (KDR+/PDGFRA+) and CMs (SIRPA+/CD90−) (Devalla et al., 2015; Dubois et al., 2011; Kattman et al., 2011; Lee et al., 2017; Zhang et al., 2011) (Figure 5). As expected, we observed some variability in the differentiation efficiency between both experimental replicates (separate differentiations) and between the two hiPSC lines, both for cardiac mesoderm and differentiated CMs (Figure 5). Similar variability was present in both atrial and ventricular differentiations. We again measured intracellular calcium waveforms in preparations purified by metabolic selection (Tohyama et al., 2013) (Figure 6; Table S8B). As expected, atrial preparations tended to exhibit shorter calcium waveforms and faster beating rates than ventricular preparations (Figure 6A). We quantified various metrics (see supplemental experimental procedures) and performed PCA on the collected results (Figure 6B; Table S8B). The results suggest that clear differences in physiological metrics can be seen between atrial and ventricular preparations, but not between cells from the two donors, or between the different clones tested. An examination of selected metrics (beating rate and calcium transient duration) is consistent with this impression, as significant differences were observed between atrial and ventricular cells, but were difficult to detect between donors or between clones (Figures 6C–6E). Overall, these results suggest that CMs differentiated from hiPSCs originally derived from different clinically healthy individuals, or independent clones from the same individual, exhibit similar characteristics, at least when differentiated under well-controlled conditions.
Figure 5.
Atrial and ventricular differentiation of hiPSCs
(A) Representative flow cytometry plots of KDR and platelet-derived growth factor receptor (PDGFR) expression on cells at day 5 of atrial CM (ACM) and ventricular CM (VCM) differentiation from two racially/ethnically distinct, age-matched, male, clinically healthy subjects (MSN14 and MSN25, respectively) (top).
(B) Percentage of cells expressing both KDR and PDGFR across three independent hiPSC clones each established from subjects MSN14 (dark gray) and MSN25 (light gray). Symbols (circles, squares, and triangles) represent independent differentiations. Data are represented as the mean ± standard deviation (SD).
(C) Representative flow cytometry plots of SIRPA and CD90 expression at day 20 of atrial (ACM, right) and ventricular (VCM, left) differentiation from hiPSC clone MSN14-01 before and after lactate selection.
(D) Percentage of cells expressing SIRPA but not CD90 across the three independent hiPSC clones established from the same two racially/ethnically distinct, age-matched, male, clinically healthy subjects as in (A) before (light gray) and after (dark gray) lactate selection. Individual dots represent independent differentiations. Data are represented as the mean ± SD. The red dotted line indicates 95% purity, which was the cutoff set for a differentiated line to be used for the subsequent physiology assays.
Figure 6.
Calcium transient analysis of hiPSC-derived CMs from two racially/ethnically distinct, age-matched males
(A) Representative calcium transient analysis traces of ventricular (red trace) and atrial (blue trace) hiPSC-derived CMs. hiPSC-CMs were lactate purified at day 20 of differentiation and analyzed at day 30 of differentiation.
(B) PCA of multiple features measured from calcium transients recorded from atrial (blue) and ventricular (red) cell types differentiated from independent hiPSC clones established from each of two racially/ethnically distinct, age-matched, male, clinically healthy subjects (MSN14, orange; MSN25, blue) at day 30 of differentiation after lactate selection at day 20. The data are accrued from two to three independent differentiations for each hiPSC line and clone.
(C) Comparison of calcium transient duration (left) and beat frequency (right) between VCM (light gray) and ACM (dark gray) differentiated from three hiPSC clones derived from two patients (MSN14, orange; MSN25, blue). The data are accrued from two to three independent differentiations. ∗p ≤ 0.01 from unpaired Mann-Whitney test.
(D) Comparison of calcium transient duration and frequency between two patient lines (MSN14 and MSN25) for atrial cells alone. The data are accrued from two to three independent differentiations for each hiPSC line/clone. ∗p ≤ 0.01 from unpaired Mann-Whitney test was considered significant; n.s. (p > 0.01), non-significant.
(E) Comparison of calcium transient duration and frequency between two patient lines from ventricular cells alone. The data are accrued from two to three independent differentiations for each hiPSC line/clone. ∗p ≤ 0.01 from unpaired Mann-Whitney test; n.s. (p > 0.01), non-significant.
Discussion
Previous efforts to create hiPSC libraries have focused on diseases and associated controls that included unaffected relatives or non-related individuals, self-declared healthy subjects, or persons with no medical disease record (Carcamo-Orive et al., 2017; Kilpinen et al., 2017; Panopoulos et al., 2017; Park et al., 2017; Streeter et al., 2017). To our knowledge, no gender-balanced, racially/ethnically diverse hiPSC library of well-characterized, clinically screened, healthy individuals exists. We have created such a library consisting of well-characterized hiPSC lines that were established using integration-free reprogramming methods from 40 healthy male and female individuals of diverse racial/ethnic backgrounds who passed a rigorous health evaluation. As was done for other studies that generated control hiPSCs (Carcamo-Orive et al., 2017; Kilpinen et al., 2017; Panopoulos et al., 2017; Park et al., 2017; Streeter et al., 2017), our subjects had to complete a health questionnaire and had their medical histories evaluated. However, and in contrast to these other studies, the individuals who were selected for a skin biopsy in our study underwent a detailed health screen conducted by an internal medicine physician, and their results were evaluated by a three-member clinical panel. This screen included the measurement of general parameters, analysis of clinically relevant parameters present in the blood, a physical examination of the respiratory and gastrointestinal/abdominal systems, a neurological examination, and a cardiac examination, including an ECG.
While all our hiPSC lines were distinct from human fibroblast lines in the RNA-seq-based PluriTest assay (Figures 2D and S2), pluripotency and novelty scores of certain hiPSC lines and even of certain duplicate samples (Table S4) were inexplicably different and beyond the threshold set by Panopoulos et al. (2017), who first applied the RNA-seq-based PluriTest. However, comparative whole-transcriptome analysis of the 40 hiPSC lines with 77 randomly chosen hiPSC lines (Kilpinen et al., 2017) and 66 human fibroblast lines (Hagai et al., 2018) and PCA clearly showed that the two sets of hiPSC lines are similar to each other and very distinct from the fibroblasts (Figure 3). This finding together with expression of five pluripotency markers for all hiPSC lines and clones (Figures 2 and S1) and demonstrated in vitro tri-lineage differentiation potential of a selection of lines/clones (Figure S3) provides confidence in the pluripotency of our hiPSC library.
It is well established that race and ethnicity, which are used as a way of categorizing people of shared ancestry and physical traits (Sankar et al., 2004), as well as sex (Soldin and Mattison, 2009), are contributing factors to interindividual differences in drug exposure and/or response (Ramamoorthy et al., 2015; Wilson et al., 2001). These are important factors to consider during drug development and evaluation of drug responses as they imply diverse risk-benefit ratios in specific populations. Therefore, any hiPSC library, if it is not a library of lines derived from individuals with a disorder that is preferentially associated with a particular racial/ethnic group, should be gender balanced and racially/ethnically diverse. Our hiPSC library with 18 females and 22 males fulfills these criteria.
While our hiPSC library was derived from well-characterized clinically healthy individuals, whole-genome sequencing analysis revealed that several individuals were carriers of genetic variants associated with recessive and dominant pathogenic disease risk. This is important and useful information, particularly when using the lines as controls for modeling a particular disease/disorder, or for evaluating the response to drugs. Several expression quantitative trait loci (eQTL) hiPSC studies have shown that genetic variations account for many gene expression differences between lines (Carcamo-Orive et al., 2017; DeBoever et al., 2017; Kilpinen et al., 2017; Panopoulos et al., 2017; Rouhani et al., 2014). While we did not investigate eQTL on global gene expression in hiPSCs, we found that age and sex had no impact (Figure 4B). This is in agreement with Kilpinen et al. (2017), who showed that neither sex nor cell passage number substantially influenced gene and protein expression.
If hiPSCs from healthy individuals were to be used as controls for any study, then ideally the molecular and physiological functions of differentiated cells should be similar. We tested this possibility using multiple age-matched, male and female hiPSC lines differentiated into CMs. Physiological function and molecular profiles were assessed using calcium transient measurements and RNA-seq, respectively. Although small differences between groups could be seen when dozens of samples from each hiPSC line were assessed (e.g., Figure S4), which might be attributed to differences in proportions of cell sub-populations, there was always substantial overlap between the lines in all the measurements performed.
Detailed analyses of three independent clones each from two hiPSC lines under conditions where the atrial and ventricular CMs showed clearly observable differences did not reveal significant differences between either the independent clones or between the two lines (Figures 4 and 5). This may have practical and financial implications, especially for large-scale studies, where use of a single hiPSC clone per subject rather than multiple clones would curtail costs and reduce potential technical variability. Although our observations suggest that there are unlikely to be large differences between multiple hiPSC lines from healthy individuals, the general applicability and drawing firm generalized conclusions will require studies with tens or hundreds of independent hiPSC lines conducted under standardized conditions. Additionally, these conclusions from CMs may not be applicable to other cell types or organoids.
The possibility of differences between lines should be considered when planning to test drug responses across cell types differentiated from hiPSCs derived from a range of racially/ethnically diverse, clinically well-characterized healthy male and female individuals. One could potentially expect different responses to the drugs investigated based on racial/ethnic background as well as genetic variants (pharmacogenomics). In this context, it is noteworthy that a recent drug screening report of 10 control human pluripotent stem cell lines in an engineered heart tissue format found that, while there was great variability in baseline contractile parameters across the different lines, the variability appeared less relevant for drug screening (Mannhardt et al., 2020). A number of constraints precluded us from testing every hiPSC line in our library for each of the conditions and assays. Hence, further studies will be required to unequivocally establish all of the conclusions of this study. These limitations notwithstanding, we provide a gender-balanced, racially/ethnically diverse library of hiPSCs from 40 clinically well-characterized healthy human individuals ranging in age from 22 to 61 years. The library is accompanied by comprehensive quality control characterization of karyotype, STR-matching to each individual (authentication), pluripotency analysis, whole-genome sequencing, ancestry determination, and disease gene and risk analysis. This hiPSC library can be useful to investigators looking for appropriate controls to study normal human development or to investigate drug responses or specific diseases.
Experimental procedures
Subject recruitment and health screening
Subjects of diverse racial/ethnic background (as per NIH standards described in NOT-OD-15-089) were recruited and consented under Mount Sinai Institutional Review Board-approved protocol (HS# 14-00530) (documents S1 and S2). Consented subjects were evaluated for their health status at the Mount Sinai Clinical Research Unit by internal medicine physicians. Screen involved completion of a health questionnaire, clinical history, and measurement of weight, height, waist and hip circumference, heart rate, blood pressure, respiratory rate, and oxygen saturation. Blood was drawn for analysis of clinically relevant parameters (determination of glucose levels was not limited to a specific state [fasting/non-fasting] or time of sampling), and subjects underwent respiratory, gastrointestinal/abdominal, neurological, and cardiovascular examinations, including an ECG (document S3). A pregnancy test was included for female participants. Subjects with no clinical history of disease, normal physical examination, normal ECG, and evaluated parameters within the normal range (document S3) were classified as healthy by a medical panel consisting of two internal medicine physicians and an interventional cardiologist. Clinical health information remained within Mount Sinai’s protected electronic medical records system.
Clinical data of all evaluated subjects were extracted from the individual’s electronic medical record and downloaded manually. Subjects’ age, sex, race/ethnicity, and selected healthy/passed or unselected/unpassed status were transcribed to a database and healthy subjects were de-identified with a unique sample ID (MSNxx, where xx is a two-digit number ranging from 01 to 40) (Tables S1 and S2). The primary reason for exclusion of 42 individuals from the final pool of eligible healthy subjects was plotted as a pie chart (Figures 1B–1E). To visualize the distribution of eligible healthy subjects versus excluded subjects, we plotted violin swarm plots of (1) body mass index (BMI) with median value for healthy subjects of 24.15 versus unselected of 24.4 kg/m2, (2) hemoglobin with median value for healthy of 14.5 versus excluded of 13.5 g/dL, (3) red blood cells with median value for healthy of 4.7 versus unselected of 4.6 × 106/μL. All analyses were done using Python programming.
Biopsy punch, derivation, and culture of fibroblasts
Our detailed standard operating procedure (SOP) can be found in supplementary experimental procedures. Briefly, a skin sample was taken from each of the 40 clinically healthy subjects using a 3-mm sterile disposable biopsy punch (Integra Miltex, catalog number [Cat#] 98PUN3-11) during a second visit at the Clinical Research Unit. Each sample was cut into pieces and placed into gelatin-coated dishes in DMEM, 20% FBS, penicillin/streptomycin, non-essential amino acids, 2 mM L-glutamine, 2 mM sodium pyruvate (all from Thermo Fisher Scientific), and 100 μM 2-mercaptoethanol (MP Biomedicals, Cat# 194705) in a humidified incubator at 5% CO2, 37°C. Fibroblasts were harvested using TrypLE Express (Thermo Fisher Scientific, Cat# 12605010) and passaged at a 1:4 split ratio. Fibroblast lines were cryopreserved in 40% DMEM, 50% FBS and 10% DMSO (Millipore Sigma, Cat# D2438).
Reprogramming fibroblasts to hiPSCs
Mycoplasma-free fibroblasts at passage number 3–5 were reprogrammed using the mRNA reprogramming kit (Stemgent, Cat # 00-0071) in combination with the microRNA Booster Kit (Stemgent, Cat# 00-0073) and/or the CytoTune-iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific, Cat# A16517) according to our SOPs (see supplementary experimental procedures).
Human iPSC culture
Human iPSCs were cultured on Matrigel (Corning, Cat# 254248)-coated plates in mTeSR (STEMCELL Technologies, Cat# 05850) in a humidified incubator at 5% CO2, 37°C. Cells were passaged using ReLeSR (STEMCELL Technologies, Cat# 05872) according to the manufacturer’s instructions and grown for 16–24 h in mTeSR supplemented with 2 μM Thiazovivin (Millipore Sigma, Cat# 420220).
Human iPSC-CM differentiation
Human iPSCs were maintained in E8 medium and passaged every 4 days onto Matrigel-coated plates before differentiation. On day 0 (start of differentiation), hiPSCs were treated with 1 mg/mL Collagenase B (Roche, Cat# 11088807001) for 1 h, or until cells dissociated from plates, to generate embryoid bodies (EBs). Cells were collected and centrifuged at 300 relative centrifugal force (rcf) for 3 min, and resuspended as small clusters of 50–100 cells by gentle pipetting in CM differentiation medium composed of RPMI 1640 (Thermo Fisher Scientific, Cat# 11875085) containing 2 mM/L L-glutamine (Thermo Fisher Scientific, Cat# 25030149), 4 × 10−4 M monothioglycerol (Millipore Sigma, Cat# M6145), and 50 μg/mL ascorbic acid (Millipore Sigma, Cat# A4403). Differentiation medium was supplemented with 3-ng/mL BMP4 (Biotechne-R&D Systems) and 3 μM Thiazovivin (Millipore Sigma, Cat# 420220), and EBs were cultured in 6-cm dishes (USA Scientific, Cat# 8609-0160) in a humidified incubator at 37°C in 5% CO2, 5% O2, and 90% N2. On day 1, medium was changed to differentiation medium supplemented with 20 ng/mL BMP4 (Biotechne-R&D Systems), 20 ng/mL Activin A (Biotechne-R&D Systems), 5ng/mL bFGF (Biotechne-R&D Systems), and 1 μM Thiazovivin (Millipore Sigma, Cat# 420220). On day 3, EBs were harvested and washed once with DMEM (Gibco). Medium was changed to differentiation medium supplemented with 5 ng/mL VEGF (Biotechne-R&D Systems) and 5 μM XAV939 (Reprocell-Stemgent, Cat# 04-0046). To generate atrial CMs, retinoic acid (RA) was added to the differentiation medium at 0.5 μM (Devalla et al., 2015; Lee et al., 2017). On day 5, medium was changed to CM differentiation medium supplemented with 5 ng/mL VEGF (Biotechne-R&D Systems). After day 8, medium was changed every 3–4 days to differentiation medium without supplements (Dubois et al., 2011).
Lactate metabolic selection
EBs were dissociated on day 20 of CM differentiation and replated on Matrigel-coated six-well plates at 1 × 106 cells/well in CM differentiation medium supplemented with 1 μM Thiazovivin (Millipore Sigma, Cat# 420220). Medium was removed the following day and replaced with CM differentiation medium. After 3 days, CM differentiation medium was replaced with lactate medium (stock solution, 1 M lactate, 1 M Na-HEPES in distilled water; working solution, 4 mM lactate in DMEM, no glucose) for 4 days. From days 5–8, lactate medium was titrated down in the following lactate to CM differentiation medium ratios: day 5, 3:1; day 6, 1:1; day 7: 1:3; day 8, 0:4 (Tohyama et al., 2013).
Data and software availability
All hiPSC RNA-seq data files have been deposited at the Gene Expression Omnibus (GEO) under accession number GEO: GSE156384. The bulk and single-cell RNA-seq data files of CMs differentiated form hiPSC lines have been deposited at GEO under accession numbers GEO: GSE174773 and GEO: GSE175761, respectively. The whole-genome sequencing data, enhanced carrier screen gene variant vcf files, subjects’ clinical examination parameters, and STR data have been deposited at the Database of Genotypes and Phenotypes (dbGaP) under accession number dbGaP: phs002088.v2.p1. These data are restricted to requestors from not-for-profit organizations and are available through controlled access requiring provision of the requester’s local IRB approval documentation as required by the Genomic Data Sharing (GDS) Extramural Institutional Certification agreement between Mount Sinai and NIH (NHGRI). Requesters must agree to make any results of studies using these data available to the larger scientific community.
Author contributions
C.S. recruited, enrolled, scheduled health evaluations and biopsies, and de-identified and coded samples. C.L., K.M., N.T., and H.W. conducted clinical health evaluations and biopsies. J.C.K. conceived the clinical health evaluation. D.T. and J.C.K. supervised clinical health evaluations, assessed clinical data, and made final determination on health pass/fail decisions. A.S.Y. performed clinical health data extraction, processing, and analysis. C.S., P.D., and T.R. established fibroblast lines. C.S., P.D., T.R., B.H., and S.L.D. generated hiPSCs. B.H. performed the tri-lineage differentiation. P.D., D.M.G., and N.C.D. differentiated hiPSCs to CMs and performed the physiology experiments. D.M.G., R.D., E.A.S., and N.C.D. analyzed the physiology experiments. C.S., P.D., T.R., and G.J. performed immunostaining and imaging. B.H. and G.J. isolated mRNA and genomic DNA. J.T. and V.N. supervised and interpreted karyotype analysis. K.C.M., C.S.L., H.W., and M.T. conducted clinical health evaluations. J.G. and M.A.M. read and interpreted ECG data. J.C.K. and D.T. supervised clinical health evaluations and made final determinations on subject eligibility. K.G.B. and R.S. supervised bulk and single-cell RNA-seq library preparations, sequencing, and quality control. Y.X. and J.H. performed RNA-seq data processing and computational analysis. B.M., M.M., M.M., and E.S. performed whole-genome sequencing data processing, ancestry, variant, and recessive disorder carrier mutation analysis, and interpretation. S.I.B. performed direct dominant genetic clinical risk analysis. J.G. provided quality assurance and control. D.V. and S.C.S. coordinated and integrated data into the LINCS data portal. C.S. and A.E. managed the project. C.S., M.B., E.A.S., N.C.D., and R.I. conceived, designed, and supervised the study. C.S. and R.I. wrote the manuscript with input from all authors.
Conflicts of interests
The authors declare no competing financial interests. B.M., M.M., J.T., M.M., and E.S. are current employees of Sema4 (Stamford, CT). N.C.T. is currently a medical doctor in private practice.
Acknowledgments
This work was supported by NIH Common Fund LINCS program awards U54HG008098 (R.I.) and U54HL127624 (S.C.S). B.M., M.M., J.T., M.M., and E.S. received salaries from Sema4. We thank the Mount Sinai Clinical Research Unit for providing examination rooms and nurses, and the Mount Sinai Genomics Core for RNA-seq.
Published: November 4, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.10.005.
Contributor Information
Christoph Schaniel, Email: christoph.schaniel@mssm.edu.
Nicole C. Dubois, Email: nicole.dubois@mssm.edu.
Ravi Iyengar, Email: ravi.iyengar@mssm.edu.
Supplemental information
References
- Carcamo-Orive I., Hoffman G.E., Cundiff P., Beckmann N.D., D'Souza S.L., Knowles J.W., Patel A., Papatsenko D., Abbasi F., Reaven G.M., et al. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell. 2017;20:518–532.e519. doi: 10.1016/j.stem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antonio-Chronowska A., Donovan M.K.R., Young Greenwald W.W., Nguyen J.P., Fujita K., Hashem S., Matsui H., Soncin F., Parast M., Ward M.C., et al. Association of human iPSC gene signatures and X chromosome dosage with two distinct cardiac differentiation trajectories. Stem Cell Rep. 2019;13:924–938. doi: 10.1016/j.stemcr.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C., Li H., Jakubosky D., Benaglio P., Reyna J., Olson K.M., Huang H., Biggs W., Sandoval E., D'Antonio M., et al. Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell. 2017;20:533–546 e537. doi: 10.1016/j.stem.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L., et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G., Gramolini A., Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagai T., Chen X., Miragaia R.J., Rostom R., Gomes T., Kunowska N., Henriksson J., Park J.E., Proserpio V., Donati G., et al. Gene expression variability across cells and species shapes innate immunity. Nature. 2018;563:197–202. doi: 10.1038/s41586-018-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Zhao M.T., Jahanbani F., Shao N.Y., Lee W.H., Chen H., Snyder M.P., Wu J.C. Effects of cellular origin on differentiation of human induced pluripotent stem cell-derived endothelial cells. JCI Insight. 2016;1:e85558. doi: 10.1172/jci.insight.85558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Stem Cell Initiative, Amps K., Andrews P.W., Anyfantis G., Armstrong L., Avery S., Baharvand H., Baker J., Baker D., Munoz M.B., et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat. Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch C.M., Kao A.W., Karydas A., Onanuga K., Martinez R., Argouarch A., Wang C., Huang C., Sohn P.D., Bowles K.R., et al. A comprehensive resource for induced pluripotent stem cells from patients with primary tauopathies. Stem Cell Rep. 2019;13:939–955. doi: 10.1016/j.stemcr.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaserman J.E., Hurley K., Dodge M., Villacorta-Martin C., Vedaie M., Jean J.C., Liberti D.C., James M.F., Higgins M.I., Lee N.J., et al. A highly phenotyped open access repository of alpha-1 antitrypsin deficiency pluripotent stem cells. Stem Cell Rep. 2020;15:242–255. doi: 10.1016/j.stemcr.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Keenan A.B., Jenkins S.L., Jagodnik K.M., Koplev S., He E., Torre D., Wang Z., Dohlman A.B., Silverstein M.C., Lachmann A., et al. The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell Syst. 2018;6:13–24. doi: 10.1016/j.cels.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H., Goncalves A., Leha A., Afzal V., Alasoo K., Ashford S., Bala S., Bensaddek D., Casale F.P., Culley O.J., et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleti A., Terryn R., Stathias V., Chung C., Cooper D.J., Turner J.P., Vidovic D., Forlin M., Kelley T.T., D'Urso A., et al. Data portal for the library of integrated network-based cellular signatures (LINCS) program: integrated access to diverse large-scale cellular perturbation response data. Nucleic Acids Res. 2018;46:D558–D566. doi: 10.1093/nar/gkx1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Protze S.I., Laksman Z., Backx P.H., Keller G.M. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell. 2017;21:179–194.e174. doi: 10.1016/j.stem.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Mannhardt I., Saleem U., Mosqueira D., Loos M.F., Ulmer B.M., Lemoine M.D., Larsson C., Ameen C., de Korte T., Vlaming M.L.H., et al. Comparison of 10 control hPSC lines for drug screening in an engineered heart tissue format. Stem Cell Rep. 2020;15:983–998. doi: 10.1016/j.stemcr.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y., Ben-David U., Lavon N., Biancotti J.C., Yakir B., Clark A.T., Plath K., Lowry W.E., Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Nazor K.L., Altun G., Lynch C., Tran H., Harness J.V., Slavin I., Garitaonandia I., Muller F.J., Wang Y.C., Boscolo F.S., et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos A.D., D'Antonio M., Benaglio P., Williams R., Hashem S.I., Schuldt B.M., DeBoever C., Arias A.D., Garcia M., Nelson B.C., et al. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell Rep. 2017;8:1086–1100. doi: 10.1016/j.stemcr.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Park S., Gianotti-Sommer A., Molina-Estevez F.J., Vanuytsel K., Skvir N., Leung A., Rozelle S.S., Shaikho E.M., Weir I., Jiang Z., et al. A comprehensive, ethnically diverse library of sickle cell disease-specific induced pluripotent stem cells. Stem Cell Rep. 2017;8:1076–1085. doi: 10.1016/j.stemcr.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.E., Loring J.F. Genomic instability in pluripotent stem cells: implications for clinical applications. J. Biol. Chem. 2014;289:4578–4584. doi: 10.1074/jbc.R113.516419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianezzi E., Altomare C., Bolis S., Balbi C., Torre T., Rinaldi A., Camici G.G., Barile L., Vassalli G. Role of somatic cell sources in the maturation degree of human induced pluripotent stem cell-derived cardiomyocytes. Biochim. Biophys. Acta Mol. Cell Res. 2020;1867:118538. doi: 10.1016/j.bbamcr.2019.118538. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A., Pacanowski M.A., Bull J., Zhang L. Racial/ethnic differences in drug disposition and response: review of recently approved drugs. Clin. Pharmacol. Ther. 2015;97:263–273. doi: 10.1002/cpt.61. [DOI] [PubMed] [Google Scholar]
- Rouhani F., Kumasaka N., de Brito M.C., Bradley A., Vallier L., Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. doi: 10.1371/journal.pgen.1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Freire V., Lee A.S., Hu S., Abilez O.J., Liang P., Lan F., Huber B.C., Ong S.G., Hong W.X., Huang M., et al. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J. Am. Coll. Cardiol. 2014;64:436–448. doi: 10.1016/j.jacc.2014.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar P., Cho M.K., Condit C.M., Hunt L.M., Koenig B., Marshall P., Lee S.S., Spicer P. Genetic research and health disparities. JAMA. 2004;291:2985–2989. doi: 10.1001/jama.291.24.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldin O.P., Mattison D.R. Sex differences in pharmacokinetics and pharmacodynamics. Clin. Pharmacokinet. 2009;48:143–157. doi: 10.2165/00003088-200948030-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathias V., Turner J., Koleti A., Vidovic D., Cooper D., Fazel-Najafabadi M., Pilarczyk M., Terryn R., Chung C., Umeano A., et al. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020;48:D431–D439. doi: 10.1093/nar/gkz1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter I., Harrison P.W., Faulconbridge A., The HipSci Consortium, Flicek P., Parkinson H., Clarke L. The human-induced pluripotent stem cell initiative-data resources for cellular genetics. Nucleic Acids Res. 2017;45:D691–D697. doi: 10.1093/nar/gkw928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taapken S.M., Nisler B.S., Newton M.A., Sampsell-Barron T.L., Leonhard K.A., McIntire E.M., Montgomery K.D. Karyotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat. Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T., Hashimoto H., Suzuki T., Yamashita H., Satoh Y., et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.F., Weale M.E., Smith A.C., Gratrix F., Fletcher B., Thomas M.G., Bradman N., Goldstein D.B. Population genetic structure of variable drug response. Nat. Genet. 2001;29:265–269. doi: 10.1038/ng761. [DOI] [PubMed] [Google Scholar]
- Witty A.D., Mihic A., Tam R.Y., Fisher S.A., Mikryukov A., Shoichet M.S., Li R.K., Kattman S.J., Keller G. Generation of the epicardial lineage from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1026–1035. doi: 10.1038/nbt.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L., et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All hiPSC RNA-seq data files have been deposited at the Gene Expression Omnibus (GEO) under accession number GEO: GSE156384. The bulk and single-cell RNA-seq data files of CMs differentiated form hiPSC lines have been deposited at GEO under accession numbers GEO: GSE174773 and GEO: GSE175761, respectively. The whole-genome sequencing data, enhanced carrier screen gene variant vcf files, subjects’ clinical examination parameters, and STR data have been deposited at the Database of Genotypes and Phenotypes (dbGaP) under accession number dbGaP: phs002088.v2.p1. These data are restricted to requestors from not-for-profit organizations and are available through controlled access requiring provision of the requester’s local IRB approval documentation as required by the Genomic Data Sharing (GDS) Extramural Institutional Certification agreement between Mount Sinai and NIH (NHGRI). Requesters must agree to make any results of studies using these data available to the larger scientific community.