Summary
Hepatic stellate cells (HSCs) play a central role in the progression of liver fibrosis by producing extracellular matrices. The development of drugs to suppress liver fibrosis has been hampered by the lack of human quiescent HSCs (qHSCs) and an appropriate in vitro model that faithfully recapitulates HSC activation. In the present study, we developed a culture system to generate qHSC-like cells from human-induced pluripotent stem cells (hiPSCs) that can be converted into activated HSCs in culture. To monitor the activation process, a red fluorescent protein (RFP) gene was inserted in hiPSCs downstream of the activation marker gene actin alpha 2 smooth muscle (ACTA2). Using qHSC-like cells derived from RFP reporter iPSCs, we screened a repurposing chemical library and identified therapeutic candidates that prevent liver fibrosis. Hence, hiPSC-derived qHSC-like cells will be a useful tool to study the mechanism of HSC activation and to identify therapeutic agents.
Keywords: liver fibrosis, hepatic stellate cells, induced pluripotent stem cells, drug screening
Graphical abstract

Highlights
-
•
Generation of quiescent hepatic stellate cell (qHSC)-like cells from human-induced pluripotent stem cells (hiPSCs)
-
•
hiPSC-derived qHSC-like cells can be converted into activated HSCs in vitro
-
•
Monitoring of the HSC activation process using ACTA2-RFP reporter system
-
•
High-throughput drug screening model using reporter iPSC-derived qHSC-like cells
Kido and colleagues developed a culture system to generate qHSC-like cells from human iPSCs, which can be converted into activated HSCs in vitro. The qHSC-like cells derived from ACTA2-RFP reporter iPSCs express RFP fluorescence after conversion into the activated state. Using this reporter system, they screened a drug repositioning library and identified therapeutic agents that prevent liver fibrosis.
Introduction
The liver is a central organ for homeostasis and consists of parenchymal hepatocytes and non-parenchymal cells, such as hepatic stellate cells (HSCs), liver sinusoidal endothelial cells, cholangiocytes, and Kupffer cells. Regardless of etiology, chronic liver injury induces fibrosis that often proceeds to cirrhosis and hepatocellular carcinoma, indicating that prevention and/or resolution of fibrosis is a promising therapeutic target. HSCs are liver-specific mesenchymal cells that are localized in the perisinusoidal space known as the space of Disse (Friedman, 2008). In a healthy liver, they are quiescent and store vitamin A but convert to activated HSCs (aHSCs) in response to liver injury (Tsuchida and Friedman, 2017). Although aHSCs support the reconstruction of liver structure and the improvement of liver function after liver injury by producing growth factors and extracellular matrices, continuous HSC activation results in cirrhosis and hepatocellular carcinoma (Friedman, 2008). Thus, the inhibition of HSC activation should be an effective means to prevent chronic liver diseases (Mederacke et al., 2013; Osawa et al., 2015). To develop such therapeutic agents, it is necessary to recapitulate and monitor the HSC activation process in vitro. However, it is practically impossible to obtain enough quiescent HSCs (qHSCs) from human healthy livers, and commercially available primary HSCs and HSC lines are already activated and do not exhibit the quiescent characteristics of HSCs (Herrmann et al., 2007; Xu et al., 2005). Human-induced pluripotent stem cells (hiPSCs) have been used as an alternative cell source to develop disease models, and there are reports that show the generation of HSCs from hiPSCs. However, these hiPSC-derived HSCs are activated and exhibit limited characteristics of HSCs (Coll et al., 2018; Koui et al., 2017; Miyoshi et al., 2019). Hence, there are no practically available human qHSCs for the development of therapeutic agents for liver fibrosis. Furthermore, there is no convenient method for the quantitative assessment of HSC activation.
To overcome these challenges, we developed a protocol to generate qHSC-like cells from hiPSCs, which can be activated in vitro to produce extracellular matrices. As actin alpha 2 smooth muscle (ACTA2) encodes α smooth muscle actin, a protein that is highly inducible in the activation of HSCs, we developed a reporter hiPSC line by inserting red fluorescent protein (RFP) at the ACTA2 locus to monitor the activation processes of HSCs. Using the qHSC-like cells derived from reporter ACTA2-RFP iPSCs, we established an in vitro HSC activation model and demonstrated an application of the system by identifying chemical compounds that prevent liver fibrosis.
Results
Generation of hiPSC-derived qHSC-like cells
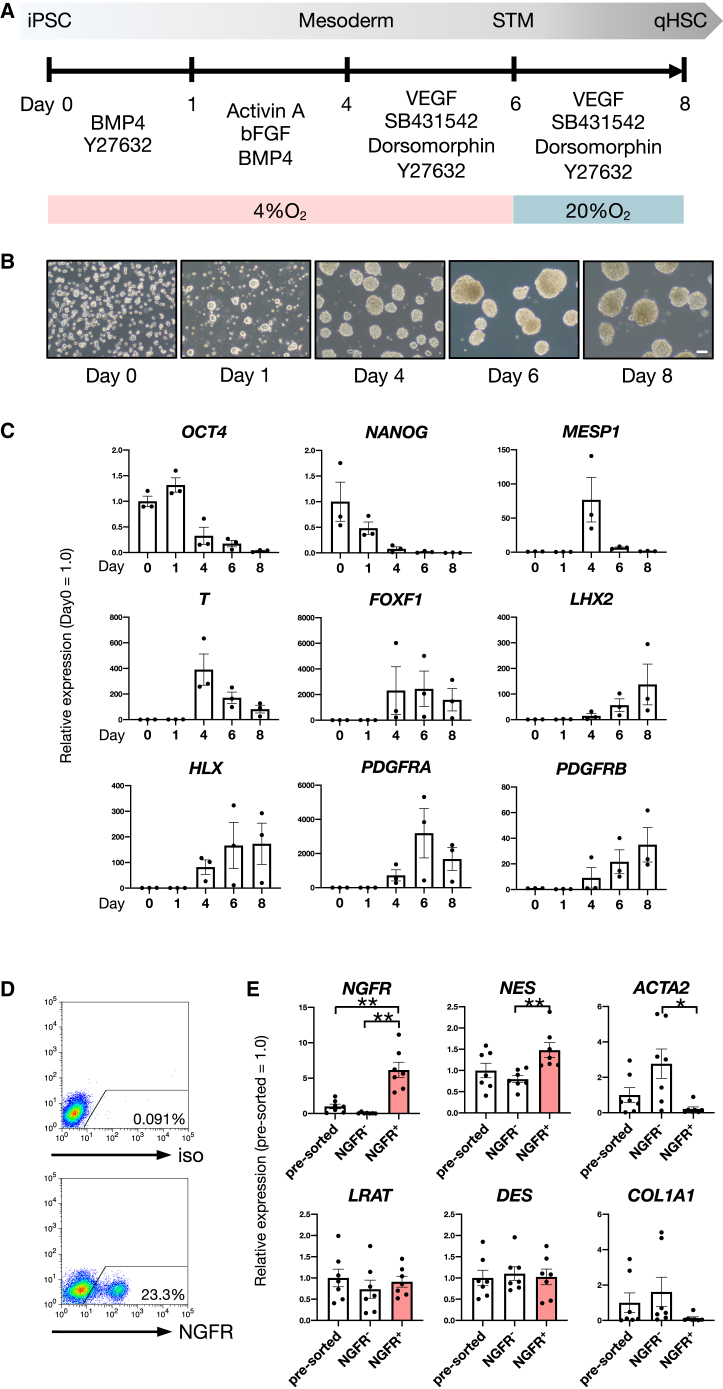
We established a method to generate qHSC-like cells from hiPSCs by mimicking the HSC developmental process. As HSCs were reported to derive from submesothelial cells showing mesenchymal characteristics (Asahina et al., 2011), we developed a differentiation system for qHSC-like cells in three-dimensional culture through mesodermal induction of hiPSCs (Figure 1A). Embryoid bodies (EBs) were formed and gradually became larger in culture (Figure 1B). Because the aHSC-specific markers, ACTA2 and collagen type I alpha 1 chain (COL1A1), were increased in a 4% O2 culture condition compared with 20% O2, we increased the oxygen concentration at day 6 (Figures 1A and S1A). We found that expression of the pluripotency marker genes, octamer-binding protein 4 (OCT4) and nanog homeobox (NANOG), was decreased, whereas expression of the mesodermal marker genes, mesoderm posterior bHLH transcription factor 1 (MESP1) and T-box transcription factor T (T), was increased along with mesodermal differentiation (Figure 1C). The septum transversum mesenchyme (STM) and HSC marker genes, forkhead box F1 (FOXF1), LIM homeobox 2 (LHX2), H2.0 like homeobox (HLX), platelet-derived growth factor receptor alpha (PDGFRA), and platelet-derived growth factor receptor beta (PDGFRB), were upregulated during differentiation (Figure 1C). The expression of the low-affinity nerve growth factor receptor (NGFR), which is a specific cell surface molecule of HSCs, was detected by flow cytometric (FCM) analysis (Figure 1D). NGFR+ cells derived from hiPSCs were separated from NGFR− cells using a cell sorter. NGFR and nestin (NES) were enriched in NGFR+ cells compared with pre-sorted or NGFR− cells (Figure 1E). Importantly, we found that expression of the aHSC-specific marker genes, ACTA2 and COL1A1, were hardly detected in NGFR+ cells (Figure 1E). Moreover, we applied our differentiation protocol to two additional hiPSC lines (FF-1 and FF-2), and using FCM analysis, we found that NGFR+ populations were clearly detected in both cell lines (Figure S1B), indicating that this protocol is applicable to various hiPSC lines.
Figure 1.
Generation of hiPSC-derived qHSC-like cells
(A) Schematic representation of a qHSC differentiation culture system from hiPSCs.
(B) Phase-contrast images of the differentiating cells from hiPSCs. Scale bar, 100 μm.
(C) Expression levels of the pluripotency marker genes (OCT4 and NANOG), the mesodermal marker genes (MESP1 and T), and the STM and HSC marker genes (FOXF1, LHX2, HLX, PDGFRA, and PDGFRB) in the differentiating cells at each stage. The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). n = 3 in each group.
(D) FCM analysis of hiPSC-derived cells after 8 days of differentiation (lower). Positive gates were defined by the isotype control (upper).
(E) Expression levels of HSC marker genes (NGFR, LRAT, NES, and DES) and aHSC marker genes (ACTA2 and COL1A1) in NGFR+ cells compared with NGFR− cells and pre-sorted cells (pre-sorted). The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). n = 7 in each group. ∗p < 0.05, ∗∗p < 0.01.
See also Figure S1.
Characterization of hiPSC-derived qHSC-like cells
hiPSC-derived qHSC-like cells exhibited a stellate cell-like morphology (Figure 2A) and expressed neural cell adhesion molecule (NCAM) at the protein level (Figure 2B). These cells also exhibited the ability to store vitamin A (Figures 2C and 2D), which is a unique function of qHSCs. Notably, compared with commercially available primary human HSCs, the hiPSC-derived qHSC-like cells highly expressed HSC marker genes and did not express the aHSC marker genes, ACTA2 and COL1A1 (Figure 2E), indicating that the commercial primary human HSCs were already activated and lost their characteristics of qHSCs. In addition, to clearly show the quiescent phenotype of hiPSC-derived qHSC-like cells, we compared their gene expression profile with that of HSCs derived from hiPSCs using a previously reported protocol (Koui et al., 2017). We found that HSC marker genes were highly expressed in hiPSC-derived qHSC-like cells (Figure S2A), whereas by contrast, the levels of aHSC-specific markers were much higher in HSCs that were generated using the conventional differentiation protocol (Figure S2A). Collectively, these findings indicate that hiPSC-derived qHSC-like cells exhibit in vivo qHSC-like phenotypes.
Figure 2.
Characteristics of hiPSC-derived qHSC-like cells
(A) Phase-contrast image of hiPSC-derived qHSC-like cells. Scale bar, 100 μm.
(B) FCM analysis of NCAM in hiPSC-derived qHSC-like cells (right). Positive gates were defined based on the isotype control (left).
(C) FCM analysis of autofluorescence of intracellular vitamin A droplets in hiPSC-derived qHSC-like cells (iPS-qHSC) (lower). Human mesenchymal stem cells (hMSC) are used as a control (upper).
(D) Phase-contrast and fluorescence images of hiPSC-derived qHSC-like cells incubated with or without retinol (vitamin A). Arrowheads indicate droplets of vitamin A (blue). Scale bar, 100 μm.
(E) Expression levels of HSC marker genes (NGFR, LRAT, and NES) and aHSC marker genes (ACTA2 and COL1A1) in primary human HSCs (hHSC) and hiPSC-derived NGFR+ qHSC-like cells (iPS-qHSC). The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). hHSC n = 2, iPS-qHSC n = 7.
(F) Schematic representation of an in vitro HSC activation model using hiPSC-derived qHSC-like cells (iPS-qHSC) (upper). Expression levels of aHSC marker genes (ACTA2, COL1A1, COL1A2, and COL3A1) in the activation process of hiPSC-derived qHSC-like cells (lower). The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). Day 0 n = 7, day 1 n = 7, day 3 n = 4, day 5 n = 7, day 10 n = 3. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) Immunofluorescence staining for αSMA (green) in hiPSC-derived aHSCs. Scale bar, 100 μm.
(H) Secretion levels of pro-collagen I alpha 1 in the activation process of hiPSC-derived qHSC-like cells. The results are shown as the mean ± SEM of independent experiments. n = 3 in each group.
See also Figure S2.
In vitro HSC activation model
First, we evaluated the activation of freshly isolated mouse qHSCs in two-dimensional culture because it is well established that qHSCs are rapidly activated in two-dimensional culture (Olsen et al., 2011). As expected, we found that the expression of the aHSC marker genes, Acta2 and Col1a1, was upregulated in mouse HSCs after 5 days of two-dimensional culture on a collagen-coated dish (Figure S2B). Based on this result, we then established an in vitro HSC activation model using hiPSC-derived NGFR+ qHSC-like cells by transfer to two-dimensional culture, a stiff condition (Olsen et al., 2011) (Figure 2F). Although the results were variable between experiments, the expression level of the activation marker gene, ACTA2, was significantly increased at day 3 in two-dimensional culture (Figure 2F). Expression of COL1A1, collagen type I alpha 2 chain (COL1A2), and collagen type III alpha 1 chain (COL3A1) was gradually increased and reached the highest level at day 10 (Figure 2F). Immunocytochemical analysis showed a strong expression of alpha smooth muscle actin (αSMA) encoded by ACTA2, which is a specific marker of aHSC (Figure 2G). The production of pro-collagen I alpha 1 was also upregulated after 7 days of culture (Figure 2H). These results demonstrate that hiPSC-derived qHSC-like cells exhibit the potential to become aHSCs in a manner similar to that of freshly isolated primary qHSCs.
Gene expression profiles of qHSCs and aHSCs
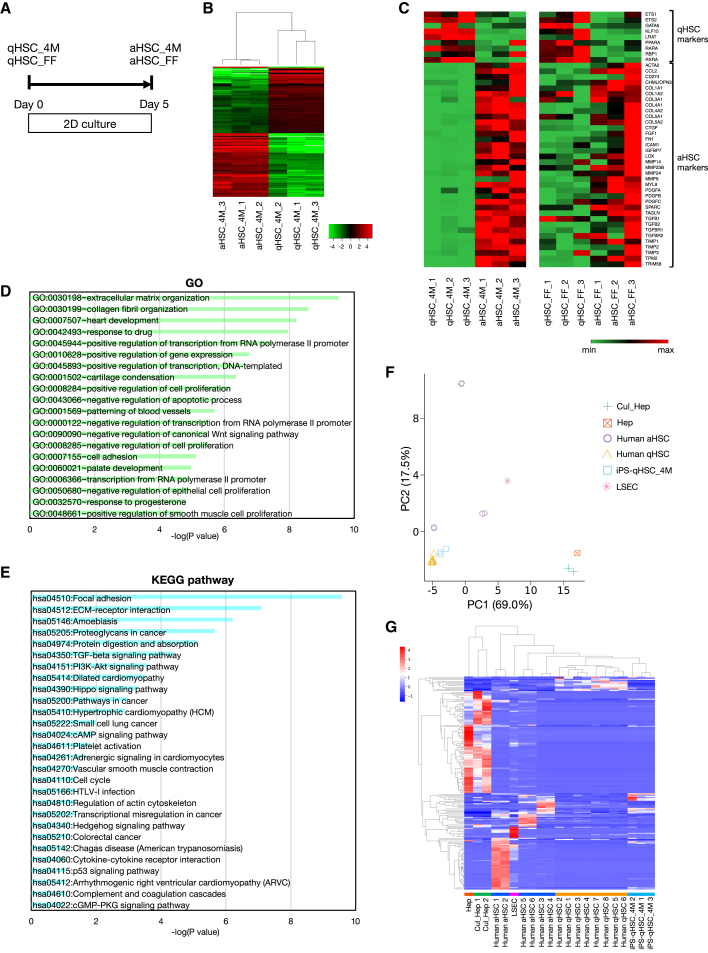
To further characterize our hiPSC-derived qHSC-like cells and aHSCs, we performed RNA-sequencing (RNA-seq) analysis on hiPSC-derived HSCs prepared from two iPSC lines (Figure 3A). Clustering analysis showed that iPSC-derived qHSC-like cells and aHSCs were clustered into different groups (Figures 3B and S3A; Table S1). The expression patterns of known qHSC and aHSC markers are shown in Figure 3C. We found that qHSC marker genes were specifically expressed in hiPSC-derived qHSC-like cells, whereas aHSC marker genes, such as those encoding collagens, matrix metalloproteinases, and members of the transforming growth factor β (TGFβ) family, were enriched in hiPSC-derived aHSCs. In addition, gene ontology (GO) enrichment analysis and pathway analysis showed that liver fibrosis-related gene clusters involved in such processes and pathways, like extracellular matrix (ECM) organization, collagen fibril organization, TGFβ signaling pathway, ECM-receptor interaction, and cellmatrix adhesion, were significantly changed in the process of activating hiPSC-derived qHSC-like cells (Figures 3D, 3E, S3B, and S3C; Table S2). Then, we compared gene expression profiles of hiPSC-derived qHSC-like cells with those of primary human qHSCs and aHSCs. Gene expression data of human qHSCs, aHSCs (Liu et al., 2020) (GSE141100), sinusoidal endothelial cells, hepatocytes (Shahani et al., 2014) (GSE43984), and cultured hepatocytes (Koui et al., 2017) (GSE98710) were extracted from a publicly available database and further processed for comparison. Principal component analysis (PCA) demonstrated that gene expression patterns of hiPSC-derived qHSC-like cells were similar to those of human qHSCs but distinct from other liver cells (Figure 3F). Although human aHSCs revealed diverse gene expression profiles between donors, PCA represented that there was a clear difference in gene expression profile between hiPSC-derived qHSC-like cells and primary human aHSCs. Clustering analysis demonstrated that expression profiles of hiPSC-derived qHSC-like cells were similar to those of human qHSCs and can be distinguished from other hepatic cells including aHSCs (Figure 3G). Although it has been reported that the expression profiles are different between in vivo and in vitro aHSCs (De Minicis et al., 2007), our results demonstrate that hiPSC-derived qHSC-like cells exhibit a qHSC-type gene expression profile and partially recapitulate the activation process of HSCs.
Figure 3.
Gene expression profiles of qHSC-like cells and aHSCs
(A) Schematic representation of the activation culture system of hiPSC-derived qHSC-like cells. qHSC-like cells (qHSC) and aHSCs were prepared from two iPS cell lines (4M: TkDN4-M and FF: FF-1).
(B) Hierarchical clustering analysis of 4M iPSC-derived qHSC-like cells and aHSCs (qHSC n = 3, aHSC n = 3). Expression of 276 high fold-change genes between qHSC-like cells and aHSCs is depicted by color.
(C) Comparison of gene expression levels of qHSC and aHSC marker genes in 4M iPSC-derived HSCs (qHSC_4M n = 3, aHSC_4M n = 3) and FF iPSC-derived HSCs (qHSC_FF n = 3, aHSC_FF n = 3). Gene expression is depicted by color.
(D) GO analysis of 276 high fold-change genes between 4M iPSC-derived qHSC-like cells and aHSCs. p values of the top 20 terms are indicated.
(E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of 276 high fold-change genes between 4M iPSC-derived qHSC-like cells and aHSCs. p values of enriched pathways are indicated.
(F) PCA of hiPSC-derived qHSC-like cells (iPS-qHSC_4M), primary human qHSCs (human qHSC, GSE141100), primary human aHSCs (human aHSC, GSE141100), hepatocytes (Hep, GSE43984), cultured hepatocytes (Cul_Hep, GSE98710), and liver sinusoidal endothelial cells (LSEC, GSE43984) based on log(TPM). Raw data were extracted from GEO and further processed as described in Supplemental experiment procedures.
(G) Cluster map of hiPSC-derived qHSC-like cells (iPS-qHSC_4M), primary human qHSCs (Human qHSC, GSE141100), primary human aHSCs (Human aHSC, GSE141100), hepatocytes (Hep, GSE43984), cultured hepatocytes (Cul_Hep, GSE98710), and liver sinusoidal endothelial cells (LSEC, GSE43984) based on Z-scores. The y axis indicates DEG.
See also Figure S3.
Quantitative assessment of HSC activation
To establish a method to quantitatively assess HSC activation in vitro, we searched for a critical marker of aHSCs. We examined the expression of the HSC activation markers, Acta2, Col1a1, Col1a2, and Col3a1, in qHSCs and aHSCs isolated from normal and injured mouse livers. The expression of Col1a1, Col1a2, and Col3a1 was detected in qHSCs and upregulated in aHSCs, whereas that of Acta2 was not detected in qHSCs (Figure S4A). These results were consistent with the activation of iPSC-derived qHSC-like cells. Among the HSC activation marker genes, we found that the expression of ACTA2 was undetectable in hiPSC-derived qHSC-like cells and induced immediately after conversion from the quiescent to the activated state (Figure 2F). These results suggest that qHSCs can be clearly distinguished from aHSCs by the expression of ACTA2. Accordingly, we generated an ACTA2-RFP reporter hiPSC line by inserting the RFP reporter gene at the ACTA2 locus using the CRISPR-Cas9 system (Figure 4A). A positive hiPSC clone (clone #25) containing donor DNA was identified by PCR and sequencing following CRISPR-Cas9-mediated gene edition (Figures S4B and S4C). We also confirmed that the Cas9 vector was not integrated into the genomic DNA of cells from the ACTA2-RFP reporter hiPSC line (clone #25) (Figure S4D) and that these ACTA2-RFP reporter hiPSCs had alkaline phosphatase activity (Figure S4E) and expressed OCT3/4 (Figure S4F). We then induced the NGFR+ qHSC-like cells from ACTA2-RFP reporter hiPSCs and cultured them under activation conditions. Expectedly, although ACTA2-RFP reporter hiPSCs and NGFR+ qHSC-like cells did not express RFP (Figures 4B and S4G), we found that RFP fluorescence gradually increased along with HSC activation (Figures 4B and S4H). Thus, the qHSC-like cells derived from reporter hiPSCs express RFP fluorescence after conversion into the activated state. In addition, ACTA2-RFP reporter iPSC-derived NGFR+ qHSC-like cells can be cryopreserved without phenotypic changes (Figure S4I). Next, we determined whether ACTA2-RFP reporter iPSC-derived NGFR+ qHSC-like cells can be used for the identification of therapeutic agents for liver fibrosis including late-stage cirrhosis. We tested two well-known inhibitors of HSC activation, A83-01 and ICG-001, both of which inhibit TGFβ receptor 1 and the binding of β-catenin and CREB binding protein (CBP), respectively (Aimaiti et al., 2019; Akcora et al., 2018). We found that ACTA2 and COL1A1 expression was completely inhibited by A83-01 and ICG-001 (Figure 4C). Consistently, RFP fluorescence was also severely suppressed by treatment of either inhibitor (Figure 4D), indicating that a reporter iPSC line is a useful tool for monitoring the activation process of HSCs and for drug discovery.
Figure 4.
Quantitative assessment of HSC activation
(A) Schematic representation of generating ACTA2-RFP reporter iPSCs. The structure of the donor vector and the wild-type human ACTA2 locus are shown. The RFP reporter gene was inserted at the ACTA2 locus using CRISPR-Cas9 system. Primers 1 to 4 (p1–4) were used for PCR genotyping.
(B) Schematic representation of a differentiation and activation culture system for ACTA2-RFP reporter iPSCs (upper). Phase-contrast and fluorescent images of RFP (red) in ACTA2-RFP reporter iPSCs (day 8), ACTA2-RFP reporter iPSC-derived qHSC-like cells (iPS-qHSC) cultured under the two-dimensional condition for several days (day 1, day 3, and day 5) (lower). Scale bars, 100 μm.
(C) Expression levels of activation marker genes (ACTA2 and COL1A1) in hiPSC-derived qHSC-like cells after 5 days of two-dimensional culture. hiPSC-derived qHSC-like cells were cultured with or without (W/O) TGFβ1, A83-01, or ICG-001 from day 1. The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). n = 6 in each group. ∗p < 0.05.
(D) Fluorescence images of RFP (red) in hiPSC-derived qHSC-like cells after 5 days of two-dimensional culture. Nuclei were counter-stained with Hoechst 33342 (blue). Scale bar, 500 μm.
See also Figure S4.
Screening of therapeutic agents for liver fibrosis
To identify therapeutic agents for liver fibrosis including late-stage liver cirrhosis, we developed a high-throughput screening system using the qHSC-like cells derived from ACTA2-RFP reporter iPSCs (Figure 5A). The screening system was optimized and validated in a 384-well plate. We developed an automatic evaluation system for HSC activation (RFP-positive area) and cell viability (Hoechst33342-positive cells) using image processing software (Figure 5B). To validate the screening system, a Z'-factor was calculated and determined to be 0.515, indicating that the system can be used for screening compounds (Zhang et al., 1999) (Figure 5C). In addition, the screening assay was verified by inhibiting HSC activation using A83-01 and ICG-001. We found that the RFP area was significantly decreased in the presence of either inhibitor (Figure 5D). These results demonstrate that this screening system is useful to evaluate the inhibitory effect of compounds on HSC activation. Thus, using a drug repositioning library, we then screened for therapeutic agents of liver fibrosis (Figure 5E). First, based on an initial analysis of the inhibitory effects of 1,453 compounds in a validated compound library on HSC activation (RFP-positive area-3XSD, n = 1 of each compound), we identified 218 compounds (Figures 5E–5G). Second, we verified the inhibitory effect of each compound on HSC activation (n = 3 for each compound) and excluded cytotoxic compounds based on the number of cells in each well (<1,000 cells/well), resulting in a shortlist of 16 compounds that significantly inhibit HSC activation without affecting cell viability (Figure 5E). Finally, we evaluated these 16 compounds on freshly isolated qHSCs from normal mouse livers and found that the activated HSC marker genes, Acta2, Col1a1, Col3a1, and Col1a2, were inhibited in the presence of compounds B2, F2, H2, A3, and B3 (Figures 5H and S5A). Thus, using our screening system, we identified five compounds as candidates for therapeutic agents of liver fibrosis. Among these five compounds, we further excluded molecules that would not be suitable for oral administration and concluded with two candidates including artemisinin. Artemisinin is a longstanding antimalarial drug with a well-known antifibrotic effect (Cui and Su, 2009). Hence, we performed animal experiments using the artemisinin derivative artesunate. We administrated artesunate to carbon tetrachloride (CCl4)-induced liver fibrosis model mice (Figure 5I). After 2 weeks of administration, we found that liver/body weight and Acta2 expression level were significantly improved (Figures 5J and 5K), indicating that artesunate has a therapeutic effect on mouse liver fibrosis. These findings are consistent with those of previous studies reporting on the antifibrotic activity of artesunate in animal models (Kong et al., 2019; Lv et al., 2018). Moreover, the expression of the aHSC marker genes ACTA2, COL1A1, COL1A2, and COL3A1 in cells of the human HSC line LX2 was inhibited by artesunate in a dose-dependent manner (Figure S5B). All of these findings demonstrate that our drug screening system based on ACTA2-RFP reporter iPSC-derived qHSC-like cells will be a useful tool for the identification of novel therapeutic agents for liver fibrosis.
Figure 5.
Screening of therapeutic agents for liver fibrosis
(A) Schematic representation of high-throughput drug screening system using the ACTA2-RFP reporter iPSCs-derived qHSC-like cells.
(B) Representative fluorescent images of RFP (red) and Hoechst33342 (blue) in ACTA2-RFP reporter iPSC-derived HSCs after 5 days of culture (left). RFP-positive area (green) and Hoechst33342-positive area (yellow) were detected by image processing software (right). Scale bars, 200 μm.
(C) Values of RFP-positive area in ACTA2-RFP reporter iPSC-derived HSCs cultured for 5 days (green dots, n = 307) and control iPSC-derived HSCs cultured for 5 days (pink dots, n = 8). Results are shown as total RFP-positive area in each well. CV (%) of RFP-positive area in ACTA2-RFP reporter iPSC-derived HSCs and Zʹ-factor of this screening system were calculated.
(D) Values of RFP-positive area in ACTA2-RFP reporter iPSC-derived HSCs cultured with A83-01 (yellow dots) and ICG-001 (blue dots) for 5 days. Control n = 148, A83-01 n = 72, ICG-001 n = 73. ∗∗∗p < 0.001.
(E) The process of a three-step screening to identify therapeutic agents for liver fibrosis.
(F) Representative results of first screening. Values of RFP-positive area in ACTA2-RFP reporter iPSC-derived HSCs treated with the compound were plotted (red dots, n = 1 of each compound). Green dots indicate the RFP-positive areas of ACTA2-RFP reporter iPSC-derived HSCs treated without the compound (control, n = 29). Blue dashed line indicates the average of the controls. Blue line indicates the average - 3XSD of the controls.
(G) Representative images of RFP signals in each well. The values of RFP-positive area are shown under each image. Scale bar, 500 μm.
(H) Expression levels of activation marker genes (Acta2 and Col1a1) in primary mouse HSCs cultured with compounds. The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). n = 3 in each group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(I) Schematic representation of CCl4-induced liver fibrosis model. The mice were intraperitoneally injected with CCl4 twice a week for 8 weeks. Then, artesunate was administrated for 2 weeks.
(J) Average of liver/body weight after PBS or artesunate (AS) administration. The results are shown as the mean ± SEM of independent experiments. n = 7 in each group. ∗∗p < 0.01.
(K) Expression levels of Acta2 in whole livers after PBS or AS administration. The results are shown as the mean ± SEM of independent experiments (each experiment contains two technical replicates). n = 7 in each group. ∗p < 0.05.
See also Figure S5.
Discussion
In this study, we developed a culture system to generate qHSC-like cells from hiPSCs that can be converted into aHSCs in culture. We characterized the hiPSC-derived NGFR+ cells by gene and protein expression analyses and functional assays. The definition of “quiescence” of HSCs is a sensitive issue. In fact, there are many studies on the markers of qHSC and aHSC (Cassiman et al., 2001; Coll et al., 2015; El Taghdouini et al., 2015; Passino et al., 2007; Ramachandran et al., 2019; Trim et al., 2000; Tschaharganeh et al., 2014). Several comprehensive gene expression analyses of human HSCs and immunostaining analyses of human tissues demonstrated that NGFR and NES expressions were detected in both qHSCs and aHSCs, and their expressions were elevated in injured liver in humans (Cassiman et al., 2001; Coll et al., 2015; El Taghdouini et al., 2015; Passino et al., 2007; Ramachandran et al., 2019; Trim et al., 2000; Tschaharganeh et al., 2014). We reanalyzed the single-cell RNA-seq dataset to examine expression changes of NGFR and NES genes in HSCs in uninjured and cirrhotic livers and found that both qHSCs and aHSCs expressed these genes in vivo (Ramachandran et al., 2019). NGFR and NES expression in hiPSC-derived NGFR+ cells was also confirmed by qPCR and RNA-sequencing experiments, indicating that hiPSC-derived NGFR+ cells exhibit HSC-like phenotypes.
Among several qHSC makers, LRAT has been identified as a reliable and specific marker for qHSCs in healthy human livers and reported that its expression was decreased along with HSC activation (Mederacke et al., 2013; Nagatsuma et al., 2009). Similarly, LRAT expression was detected in hiPSC-derived NGFR+ cells and dramatically decreased after induction of activation (Figure 3C). LRAT is known to catalyze the esterification of retinol for storage and is necessary for vitamin A storage in qHSCs. We demonstrated that hiPSC-derived NGFR+ cells stored vitamin A (Figures 2B–2D), suggesting that hiPSC-derived NGFR+ cells exhibit a qHSC-like phenotype. Moreover, as shown in Figures 2E and 2F, we also showed that the aHSC-specific markers, ACTA2 and COL1A1, were hardly detected in hiPSC-derived NGFR+ cells. Therefore, we defined hiPSC-derived NGFR+ cells as qHSC-like cells in this study. However, because NGFR and NES expressions were not upregulated in the process of iPSC-derived qHSC-like cell activation, iPSC-derived qHSC-like cells were not exactly the same as primary qHSCs. Moreover, we identified the 23 differentially expressed genes between human qHSCs and iPSC-derived qHSC-like cells in Figure 3G (11 upregulated and 12 downregulated genes in iPSC-derived qHSC-like cells, listed in Table S3). Among the 23 genes, IGFBP3 and TGM2 were reported to regulate the character of the HSCs. Although the expression of IGFBP3 and TGM2 was upregulated in the process of the HSC activation (Mannaerts et al., 2013; Wen et al., 2017; Yaqoob et al., 2020), our analysis of publicly available RNA-seq data demonstrated that they were highly expressed in the human qHSCs compared with the human aHSCs (Liu et al., 2020), indicating that the expression of IGFBP3 and TGM2 is controversial.
HSCs play essential roles in the progression of liver fibrosis by changing their characteristics from a quiescent to an activated state, leading to the production of ECMs. However, existing drug screening efforts using human HSCs have been hampered by the lack of qHSCs. Although there are several approaches to address the preservation of the quiescent state of HSCs in vitro (Guvendiren et al., 2014; Olsen et al., 2011), the quiescent phenotype of HSCs freshly isolated from the body cannot be maintained in culture. Similarly, human HSC lines such as LX1 and LX2 (Xu et al., 2005) express aHSC marker genes and have lost the quiescent phenotype. In this study, to overcome the shortage of qHSCs needed for drug discovery, we generated qHSC-like cells from human iPSCs. As the HSCs generated by our current protocol are capable of being activated, they are superior to primary human HSCs and hiPSC-derived HSCs using previously established protocols (Coll et al., 2018; Koui et al., 2017; Miyoshi et al., 2019). In concordance with their activation, qHSC-like cells dramatically changed their gene expression profile into the activated state. Thus, hiPSC-derived qHSC-like cells can be converted to aHSCs, and these phenotypic changes are partially similar to the activation process during liver fibrosis in vivo.
We also generated an ACTA2-RFP reporter iPSC line by CRISPR-Cas9-mediated gene editing. The qHSC-like cells derived from ACTA2-RFP reporter iPSCs can be used to quantitatively monitor the HSC activation process. Using this model, we demonstrated that A83-01 and ICG-001 inhibited HSC activation, and furthermore, by screening a drug repositioning library, we identified compounds that inhibit HSC activation, among which we found the antimalarial artemisinin (Cui and Su, 2009). Artemisinin and its derivatives were previously shown to inhibit HSC activation in vitro and in vivo by regulating the PDGFRB/extracellular signal-regulated kinase (ERK) and farnesoid X receptor-sphingosine 1 phosphate receptor 2 (FXR-S1PR2) signaling pathways (Chen et al., 2016a; Kong et al., 2019; Lv et al., 2018; Xu et al., 2016, 2017). Furthermore, histopathological changes of cirrhotic livers were previously examined by histological analysis, and it was clearly shown that the artemisinin derivative artesunate improved liver fibrosis (Chen et al., 2016b; Lai et al., 2015; Shen et al., 2021). Similar to the histological findings of these previous studies, we also showed the effect of artesunate on liver fibrosis, reporting improved liver/body weight and decreased Acta2 expression in CCl4-induced liver fibrosis model mice. We are currently performing animal experiments using the other candidate compound that was identified by our drug screening system using hiPSC-derived qHSC-like cells to confirm its effectiveness on liver fibrosis treatment.
In conclusion, our drug screening assay using hiPSC-derived qHSC-like cells is an effective means to identify novel therapeutic agents for liver fibrosis. This screening approach can be readily applied to robustly screen for other types of therapeutic agents, such as peptides, antibodies, and oligonucleotides.
Experimental procedures
Maintenance of human iPSCs
Three human iPSC lines (TkDN4-M, FF-1, and FF-2) were used in this study. TkDN4-M was provided by Dr. Otsu (Institute of Medical Science, The University of Tokyo) (Takayama et al., 2010) and maintained on Vitronectin (Thermo Fisher Scientific)-coated dishes in StemFlex Medium (Thermo Fisher Scientific). FF-1 and FF-2 were provided by FUJIFILM Cellular Dynamics, Inc, and maintained on GFR Matrigel (Corning)-coated dishes in StemFlex Medium.
Differentiation of qHSC-like cells from human iPSCs
Human qHSC-like cells were induced from iPSCs in a three-dimensional EB culture system. To form EBs, TkDN4-M iPSCs were dissociated into single cells and seeded on Ultra-Low Attachment plates (Corning) at a density of 10,000 cells/cm2. FF-1 and FF-2 iPSCs were dissociated into small cell clusters and seeded on Ultra-Low Attachment plates. Cells were cultured in Stempro-34 SFM medium (Thermo Fisher Scientific) supplemented with Y27632 (10 μM) (Wako Pure Chemical Industries, Ltd.) and bone morphogenetic protein 4 (BMP4) (2 ng/mL) (PeproTech) (day 0 to day 1), Activin A (TkDN4-M: 5 ng/mL, FF-1: 1.25 ng/mL, and FF-2: 5 ng/mL) (PeproTech), basic fibroblast growth factor (bFGF) (5 ng/mL) (Thermo Fisher Scientific), and BMP4 (TkDN4-M: 30 ng/mL, FF-1: 120 ng/mL, and FF-2: 180 ng/mL) (day 1 to day 4), vascular endothelial growth factor (VEGF) (10 ng/mL) (PeproTech), SB431542 (5.4 μM) (Tocris), Dorsomorphin dihydrochloride (0.5 μM) (Tocris), and Y27632 (10 μM) (day 4 to day 8). EBs were maintained in a 5% CO2, 4% O2 environment from day 0 to day 6 and in a 5% CO2, ambient O2 environment from day 6 to day 8. After 8 days of culture, EBs were collected and dissociated in 1xTripLE select (Thermo Fisher Scientific)/1 mM EDTA solution for 15 min at 37°C. Cells were blocked by FcR blocking reagent (Miltenyi Biotech) for 20 min and incubated with APC-conjugated anti-NGFR antibody (Miltenyi Biotech, 130-113-418) and PE-conjugated anti-NCAM antibody (BioLegend, 304605) for 30 min on ice. NGFR+ qHSC-like cells were isolated using a MoFlo XDP cell sorter (Beckman Coulter).
Activation of qHSC-like cells in vitro
hiPSC-derived qHSC-like cells were seeded on Cellmatrix Type I-C (Nitta gelatin)-coated plates at a density of 10,000 cells/cm2 in Stempro-34 SFM medium supplemented with VEGF (10 ng/mL), SB431542 (5.4 μM), Dorsomorphin dihydrochloride (0.5 μM), and Y27632 (10 μM). After 24 h of culture, medium was changed to Stempro-34 SFM medium. The cells were cultured for 5 days in 5% CO2, ambient O2 environment.
Drug screening system
Validated compound library (1,453 compounds) was provided by Drug Discovery Initiative, The University of Tokyo. qHSC-like cells derived from ACTA2-RFP reporter iPSCs were seeded on collagen-coated 384-well plates (Corning) at a density of 1,000 cells/well in Stempro-34 SFM medium supplemented with VEGF (10 ng/mL) and Y27632 (10 μM). After 24 h of culture, 1,453 validated compounds were dissolved in Stempro-34 SFM medium and added into each well at a final concentration of 1 μM. After 4 days of incubation with each compound, Hoechst 33342 was added into each well to visualize nuclei in live cells, and Hoechst 33342 and RFP fluorescence were imaged by the FV3000 confocal laser scanning microscope (OLYMPUS). The cell numbers were quantitated based on Hoechst 33342 fluorescence signal. The levels of HSC activation were evaluated by a total area of RFP fluorescence signal in each well. Image processing and quantification were performed by cellSens imaging analyzer (OLYMPUS).
Mice
C57BL/6J mice were purchased from CLEA Japan, Inc (Tokyo, Japan). Mice were maintained in a temperature-controlled room with 12 h light/dark cycles. All mouse experiments were performed according to the guidelines of the institutional Animal Care and Use Committee of the University of Tokyo.
Mouse liver fibrosis model
Adult male C57BL/6J mice at 6 to 8 weeks of age were used for experiments. The mice were randomly assigned to the control group and administrated group. Liver fibrosis was induced by intraperitoneal CCl4 administration (1 mL/kg body weight diluted in corn oil) (Sigma) twice a week for 8 weeks (17 injections). To evaluate the antifibrotic effects of the hit compounds, the mice were subjected to daily intraperitoneal injection with or without 100 mg/kg artesunate (Sigma) dissolved in PBS for 2 weeks. The mice were killed 24 h after the final injection.
Isolation of qHSCs and aHSCs from mouse livers
qHSCs or aHSCs were isolated from normal livers or chronically injured livers of male mice, respectively. For a chronic liver injury model, C57BL/6J mice were administrated 300 mg/L thioacetamide (TAA) in drinking water for 8 weeks or fed a diet containing 0.1% 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) (F-4643; Bio-Serv) for 5 weeks (Katsumata et al., 2017). A cell suspension of hepatocytes and non-parenchymal cells (NPCs) was prepared from livers by a two-step collagenase perfusion method (Okabe et al., 2009). After isolation of the NPC fraction by centrifugal separation, qHSCs and aHSCs were identified based on the vitamin A autofluorescence signal detected by a violet laser at 405 nm and sorted by MoFlo XDP cell sorter.
Cell culture of mouse qHSCs
Mouse qHSCs were isolated from livers of 6- to 12-month old C57BL/6J female mice. After isolation of the NPC fraction, NPCs were subjected to a density centrifugation containing 11% Histodenz (Sigma). The purity was >90% based on vitamin A content by UV excitation by FCM analysis (Matsuda et al., 2018). Quiescent HSC fraction was collected and cultured on Cellmatrix Type I-C coated plates in DMEM (Sigma) supplemented with 10% FBS, penicillin-streptomycin-glutamine, and MEM non-essential amino acids solution (Thermo Fisher Scientific).
Statistics
Data were presented as the mean ± SEM. The F-test was performed to evaluate equal variance in the data. Significant differences were determined by two-tailed Student’s t test or Welch’s t test depending on scedasticity. p < 0.05 was considered statistically significant.
Author contributions
Y.K. designed the study, performed experiments, analyzed data, and wrote the manuscript. T.K., Y.S., and A.M. designed the study and wrote the manuscript. M.H., Y.N., N.T., N.M., L.W., Y.K., and H.A. performed culture experiments and mouse experiments. Y.M. and T.Y. performed culture experiments and RNA-sequencing analysis. E.S. and R.N. analyzed publicly available RNA-sequencing data.
Conflicts of interest
Y.K., A.M., and T.K. have filed a patent based on this study (PCT/JP2020/6147). This study was supported in part by the research fundings from FUJIFILM Corporation, ROHTO Pharmaceutical Co., Ltd., and International Space Medical Co., Ltd.
Acknowledgments
We thank the members of Miyajima lab for helpful discussions and suggestions. We also thank the Olympus Bioimaging Center at the Institute for Quantitative Biosciences, The University of Tokyo for acquiring imaging data. This study was supported by JSPS KAKENHI (Grant Number 16K18975, 18K15773, 20J40236, and 21H02710), Japan Agency for Medical Research and Development Grant Number JP16bm0704007 and JP17fk0310111, the Platform Project for Supporting Drug Discovery and Life Science Research from AMED under Grant Number JP19am0101086 (support number 1455), the research fundings from FUJIFILM Corporation, ROHTO Pharmaceutical Co., Ltd., and International Space Medical Co., Ltd. The authors thank Enago (www.enago.jp) for the English language review.
Published: December 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.11.002.
Supplemental information
FPKM values are shown in the table.
Top 20 GO terms, top 20 enriched pathways, and p values are listed in the table.
Data and code availability
The accession number for the RNA-seq data reported in this paper is GSE155017.
References
- Aimaiti Y., Yusufukadier M., Li W., Tuerhongjiang T., Shadike A., Meiheriayi A., Abudusalamu A., Wang H., Tuerganaili A., Shao Y., et al. TGF-beta1 signaling activates hepatic stellate cells through Notch pathway. Cytotechnology. 2019;71:881–891. doi: 10.1007/s10616-019-00329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akcora B.O., Storm G., Bansal R. Inhibition of canonical WNT signaling pathway by beta-catenin/CBP inhibitor ICG-001 ameliorates liver fibrosis in vivo through suppression of stromal CXCL12. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:804–818. doi: 10.1016/j.bbadis.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Asahina K., Zhou B., Pu W.T., Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology. 2011;53:983–995. doi: 10.1002/hep.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiman D., Denef C., Desmet V.J., Roskams T. Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology. 2001;33:148–158. doi: 10.1053/jhep.2001.20793. [DOI] [PubMed] [Google Scholar]
- Chen Q., Chen L., Kong D., Shao J., Wu L., Zheng S. Dihydroartemisinin alleviates bile duct ligation-induced liver fibrosis and hepatic stellate cell activation by interfering with the PDGF-betaR/ERK signaling pathway. Int. Immunopharmacol. 2016;34:250–258. doi: 10.1016/j.intimp.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Chen Y.X., Lai L.N., Zhang H.Y., Bi Y.H., Meng L., Li X.J., Tian X.X., Wang L.M., Fan Y.M., Zhao Z.F., et al. Effect of artesunate supplementation on bacterial translocation and dysbiosis of gut microbiota in rats with liver cirrhosis. World J. Gastroenterol. 2016;22:2949–2959. doi: 10.3748/wjg.v22.i10.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., El Taghdouini A., Perea L., Mannaerts I., Vila-Casadesús M., Blaya D., Rodrigo-Torres D., Affò S., Morales-Ibanez O., Graupera I., et al. Integrative miRNA and gene expression profiling analysis of human quiescent hepatic stellate cells. Sci. Rep. 2015;5:11549. doi: 10.1038/srep11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Perea L., Boon R., Leite S.B., Vallverdu J., Mannaerts I., Smout A., El Taghdouini A., Blaya D., Rodrigo-Torres D., et al. Generation of hepatic stellate cells from human pluripotent stem cells enables in vitro modeling of liver fibrosis. Cell Stem Cell. 2018;23:101–113.e107. doi: 10.1016/j.stem.2018.05.027. [DOI] [PubMed] [Google Scholar]
- Cui L., Su X.Z. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev. Anti Infect. Ther. 2009;7:999–1013. doi: 10.1586/eri.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Minicis S., Seki E., Uchinami H., Kluwe J., Zhang Y., Brenner D.A., Schwabe R.F. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- El Taghdouini A., Sørensen A.L., Reiner A.H., Coll M., Verhulst S., Mannaerts I., Øie C.I., Smedsrød B., Najimi M., Sokal E., et al. Genome-wide analysis of DNA methylation and gene expression patterns in purified, uncultured human liver cells and activated hepatic stellate cells. Oncotarget. 2015;6:26729–26745. doi: 10.18632/oncotarget.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman S.L. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvendiren M., Perepelyuk M., Wells R.G., Burdick J.A. Hydrogels with differential and patterned mechanics to study stiffness-mediated myofibroblastic differentiation of hepatic stellate cells. J. Mech. Behav. Biomed. Mater. 2014;38:198–208. doi: 10.1016/j.jmbbm.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J., Gressner A.M., Weiskirchen R. Immortal hepatic stellate cell lines: useful tools to study hepatic stellate cell biology and function? J. Cell Mol. Med. 2007;11:704–722. doi: 10.1111/j.1582-4934.2007.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata L.W., Miyajima A., Itoh T. Portal fibroblasts marked by the surface antigen Thy1 contribute to fibrosis in mouse models of cholestatic liver injury. Hepatol. Commun. 2017;1:198–214. doi: 10.1002/hep4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z., Liu R., Cheng Y. Artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway. Biomed. Pharmacother. 2019;109:2043–2053. doi: 10.1016/j.biopha.2018.11.030. [DOI] [PubMed] [Google Scholar]
- Koui Y., Kido T., Ito T., Oyama H., Chen S.W., Katou Y., Shirahige K., Miyajima A. An in vitro human liver model by iPSC-derived parenchymal and non-parenchymal cells. Stem Cell Rep. 2017;9:490–498. doi: 10.1016/j.stemcr.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L., Chen Y., Tian X., Li X., Zhang X., Lei J., Bi Y., Fang B., Song X. Artesunate alleviates hepatic fibrosis induced by multiple pathogenic factors and inflammation through the inhibition of LPS/TLR4/NF-κB signaling pathway in rats. Eur. J. Pharmacol. 2015;765:234–241. doi: 10.1016/j.ejphar.2015.08.040. [DOI] [PubMed] [Google Scholar]
- Liu X., Rosenthal S.B., Meshgin N., Baglieri J., Musallam S.G., Diggle K., Lam K., Wu R., Pan S.Q., Chen Y., et al. Primary alcohol-activated human and mouse hepatic stellate cells share similarities in gene-expression profiles. Hepatol. Commun. 2020;4:606–626. doi: 10.1002/hep4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Bai R., Wang L., Gao J., Zhang H. Artesunate may inhibit liver fibrosis via the FAK/Akt/beta-catenin pathway in LX-2 cells. BMC Pharmacol. Toxicol. 2018;19:64. doi: 10.1186/s40360-018-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannaerts I., Schroyen B., Verhulst S., Van Lommel L., Schuit F., Nyssen M., van Grunsven L.A. Gene expression profiling of early hepatic stellate cell activation reveals a role for Igfbp3 in cell migration. PLoS One. 2013;8:e84071. doi: 10.1371/journal.pone.0084071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Tsurusaki S., Miyata N., Saijou E., Okochi H., Miyajima A., Tanaka M. Oncostatin M causes liver fibrosis by regulating cooperation between hepatic stellate cells and macrophages in mice. Hepatology. 2018;67:296–312. doi: 10.1002/hep.29421. [DOI] [PubMed] [Google Scholar]
- Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H., Pradere J.P., Schwabe R.F. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi M., Kakinuma S., Kamiya A., Tsunoda T., Tsuchiya J., Sato A., Kaneko S., Nitta S., Kawai-Kitahata F., Murakawa M., et al. LIM homeobox 2 promotes interaction between human iPS-derived hepatic progenitors and iPS-derived hepatic stellate-like cells. Sci. Rep. 2019;9:2072. doi: 10.1038/s41598-018-37430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatsuma K., Hayashi Y., Hano H., Sagara H., Murakami K., Saito M., Masaki T., Lu T., Tanaka M., Enzan H., et al. Lecithin: retinol acyltransferase protein is distributed in both hepatic stellate cells and endothelial cells of normal rodent and human liver. Liver Int. 2009;29:47–54. doi: 10.1111/j.1478-3231.2008.01773.x. [DOI] [PubMed] [Google Scholar]
- Okabe M., Tsukahara Y., Tanaka M., Suzuki K., Saito S., Kamiya Y., Tsujimura T., Nakamura K., Miyajima A. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- Olsen A.L., Bloomer S.A., Chan E.P., Gaça M.D., Georges P.C., Sackey B., Uemura M., Janmey P.A., Wells R.G. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa Y., Oboki K., Imamura J., Kojika E., Hayashi Y., Hishima T., Saibara T., Shibasaki F., Kohara M., Kimura K. Inhibition of cyclic adenosine monophosphate (cAMP)-response element-binding protein (CREB)-binding protein (CBP)/beta-Catenin reduces liver fibrosis in mice. EBioMedicine. 2015;2:1751–1758. doi: 10.1016/j.ebiom.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passino M.A., Adams R.A., Sikorski S.L., Akassoglou K. Regulation of hepatic stellate cell differentiation by the neurotrophin receptor p75NTR. Science. 2007;315:1853–1856. doi: 10.1126/science.1137603. [DOI] [PubMed] [Google Scholar]
- Ramachandran P., Dobie R., Wilson-Kanamori J.R., Dora E.F., Henderson B.E.P., Luu N.T., Portman J.R., Matchett K.P., Brice M., Marwick J.A., et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahani T., Covens K., Lavend'homme R., Jazouli N., Sokal E., Peerlinck K., Jacquemin M. Human liver sinusoidal endothelial cells but not hepatocytes contain factor VIII. J. Thromb. Haemost. 2014;12:36–42. doi: 10.1111/jth.12412. [DOI] [PubMed] [Google Scholar]
- Shen S., Luo J., Ye J. Artesunate alleviates schistosomiasis-induced liver fibrosis by downregulation of mitochondrial complex I subunit NDUFB8 and complex Ⅲ subunit UQCRC2 in hepatic stellate cells. Acta Trop. 2021;214:105781. doi: 10.1016/j.actatropica.2020.105781. [DOI] [PubMed] [Google Scholar]
- Takayama N., Nishimura S., Nakamura S., Shimizu T., Ohnishi R., Endo H., Yamaguchi T., Otsu M., Nishimura K., Nakanishi M., et al. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J. Exp. Med. 2010;207:2817–2830. doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trim N., Morgan S., Evans M., Issa R., Fine D., Afford S., Wilkins B., Iredale J. Hepatic stellate cells express the low affinity nerve growth factor receptor p75 and undergo apoptosis in response to nerve growth factor stimulation. Am. J. Pathol. 2000;156:1235–1243. doi: 10.1016/s0002-9440(10)64994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh D.F., Xue W., Calvisi D.F., Evert M., Michurina T.V., Dow L.E., Banito A., Katz S.F., Kastenhuber E.R., Weissmueller S., et al. p53-dependent Nestin regulation links tumor suppression to cellular plasticity in liver cancer. Cell. 2014;158:579–592. doi: 10.1016/j.cell.2014.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- Wen Z., Ji X., Tang J., Lin G., Xiao L., Liang C., Wang M., Su F., Ferrandon D., Li Z. Positive feedback regulation between transglutaminase 2 and Toll-like receptor 4 signaling in hepatic stellate cells correlates with liver fibrosis post Schistosoma japonicum infection. Front. Immunol. 2017;8:1808. doi: 10.3389/fimmu.2017.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Hui A.Y., Albanis E., Arthur M.J., O'Byrne S.M., Blaner W.S., Mukherjee P., Friedman S.L., Eng F.J. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Lu C., Zhang F., Shao J., Yao S., Zheng S. Dihydroartemisinin counteracts fibrotic portal hypertension via farnesoid X receptor-dependent inhibition of hepatic stellate cell contraction. FEBS J. 2017;284:114–133. doi: 10.1111/febs.13956. [DOI] [PubMed] [Google Scholar]
- Xu W., Lu C., Zhang F., Shao J., Zheng S. Dihydroartemisinin restricts hepatic stellate cell contraction via an FXR-S1PR2-dependent mechanism. IUBMB Life. 2016;68:376–387. doi: 10.1002/iub.1492. [DOI] [PubMed] [Google Scholar]
- Yaqoob U., Luo F., Greuter T., Jalan Sakrikar N., Sehrawat T.S., Lu J., Hu X., Gao J., Kostallari E., Chen J., et al. GIPC-regulated IGFBP-3 promotes HSC migration in vitro and portal hypertension in vivo through a β1-Integrin pathway. Cell Mol. Gastroenterol. Hepatol. 2020;10:545–559. doi: 10.1016/j.jcmgh.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FPKM values are shown in the table.
Top 20 GO terms, top 20 enriched pathways, and p values are listed in the table.
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GSE155017.