Abstract
Transcription of yeast class III genes involves the formation of a transcription initiation complex that comprises RNA polymerase III (Pol III) and the general transcription factors TFIIIB and TFIIIC. Using a genetic screen for positive regulators able to compensate for a deficiency in a promoter element of the SNR6 gene, we isolated the NHP6A and NHP6B genes. Here we show that the high-mobility-group proteins NHP6A and NHP6B are required for the efficient transcription of the SNR6 gene both in vivo and in vitro. The transcripts of wild-type and promoter-defective SNR6 genes decreased or became undetectable in an nhp6AΔ nhp6BΔ double-mutant strain, and the protection over the TATA box of the wild-type SNR6 gene was lost in nhp6AΔ nhp6BΔ cells at 37°C. In vitro, NHP6B specifically stimulated the transcription of SNR6 templates up to fivefold in transcription assays using either cell nuclear extracts from nhp6AΔ nhp6BΔ cells or reconstituted transcription systems. Finally, NHP6B activated SNR6 transcription in a TFIIIC-independent assay. These results indicate that besides the general transcription factors TFIIIB and TFIIIC, additional auxilliary factors are required for the optimal transcription of at least some specific Pol III genes.
Transcription of small genes by RNA polymerase III (Pol III) in yeast involves a multistep assembly of transcription factors into a preinitiation complex which recruits RNA Pol III (for a review, see reference 35). The A and B blocks found in most Pol III promoters are first recognized by a multisubunit complex called Pol III transcription factor C (TFIIIC). TFIIIC, one of the largest and most complex transcription factors known, has a molecular mass of about 600 kDa and is composed of six subunits. It acts as an assembly factor to direct the binding of the initiation factor TFIIIB to an upstream gene position. Once assembled into a highly stable protein-DNA complex at Pol III promoters, TFIIIB can direct multiple rounds of transcription by Pol III in vitro in the absence of TFIIIC (17, 18). TFIIIB is composed of three loosely associated polypeptides, the TATA-binding protein (19), a general transcription factor used by all eukaryotic and archeal RNA polymerases (14, 27); B" or TFIIIB90, which displays little homology to other known proteins except for a putative SANT domain (1, 20, 28, 29); and Brf1 or TFIIIB70, which shows 44% similarity to TFIIB in its N-terminal 320 residues (3, 7, 21).
In addition to these basal factors, there are hints that additional components exist which influence transcription efficiency or accuracy. A protein called TFIIIE, which has yet to be characterized, is able to stimulate transcription under certain conditions (9, 29). TFIIIE has been suggested to act by facilitating TFIIIB recruitment, by inducing conformational rearrangements of TFIIIB, or by stabilizing transcription complexes. A partially purified B" fraction was found to direct a more efficient and more accurate transcription initiation than the recombinant TFIIIB90 protein (6, 29), but the factors postulated to influence start site selection and transcription efficiency remain to be identified. Among the potential candidates, factors belonging to the class of chromatin proteins might play a role in adjusting Pol III transcription to the cell physiology, but this hypothesis has not been explored so far.
In this paper we report the first characterization of yeast Pol III gene-specific activating factors. Using a screen for multicopy suppressors of a mutation affecting an extragenic promoter element of the SNR6 Pol III gene, we isolated the NHP6A and NHP6B genes. Both genes encode proteins with DNA-binding domains similar to those of the HMG1 and HMG2 proteins. NHP6A and NHP6B were found to increase specifically the transcription efficiency of wild-type and mutant SNR6 genes in vivo and in vitro.
MATERIALS AND METHODS
Yeast strains.
The Saccharomyces cerevisiae strains used for this study are derived from YPH500α (31) and Y865 (8). MCM260 is a derivative of YPH500α. It corresponds to strain FTY115 (22) with the snr6Δ2 allele at the chromosomal locus, but it is rescued at 30°C by the 2μm plasmid pRS425-snr6Δ2 instead of the centromeric plasmid pRS314-U6 for FTY115 (the snr6Δ2 allele has a 2-bp deletion in its B block, which strongly reduces its functionality in vitro and in vivo). YPH500α was used as a tester strain to monitor the effects of NHP6A or NHP6B overexpression on the transcription of the SNR6 genes. The wild-type Y865 and the nhp6AΔ nhp6BΔ double mutant Y869 have been described (8).
Isolation of high-copy-number suppressors of snr6Δ2.
A yeast genomic DNA library carried in the multicopy, URA3-marked vector pFL44 (32) was transformed into MCM260, and transformants were directly selected at 37°C on a medium lacking uracil. Of 60,000 transformants, 16 colonies were identified for growth at 37°C. To ensure that the ability to grow at 37°C was due to the presence of the genomic clone in pFL44, the thermoresistant colonies were streaked on 5-fluoroorotic acid (5-FOA) plates and tested again for growth at 37°C. All colonies tested became thermosensitive. The plasmids were then rescued into Escherichia coli and retested for suppressor activity by transformation into MCM260; all plasmids restored thermoresistance. The plasmids were finally sequenced and identified by comparison with sequences in the GenBank data base.
Plasmids.
pRS425-snr6Δ2 was obtained by subcloning the SNR6 sequences of pB6Δ238-239 (4) into the LEU2-marked 2μm plasmid pRS425 (31) using the BamHI and HindIII sites. All the plasmids used for the in vitro transcription assays are derived from the Bluescript SK vector (Stratagene) and contain the region of the SNR6 gene spanning bp −140 to +314 relative to the SNR6 transcription start site, except for the Aup-ΔB construct, which harbors a truncated fragment from −120 to +122 lacking the B block (4). These fragments were mutated as described previously (4). To study the transcriptional activity of wild-type and mutated SNR6 genes in yeast, we used YEp352-derived plasmids (15), harboring SNR6 genes with a 59-pb DNA fragment inserted in their transcribed sequences and the same mutations as those described above. Their construction has been described previously (22).
The tetO-NHP6A and tetO-NHP6B constructs were generated as follows. The entire coding sequences of the NHP6A and NHP6B genes were amplified by PCR using as a template the genomic sequences harbored by the pFL44 plasmids isolated by our screen. After digestion with BamHI and HpaI, the PCR products were cloned into the BamHI-HpaI sites of pCM183 (13), behind the tetracycline operator tetO.
Proteins.
Recombinant NHP6B was produced in E. coli strain BL21(DE3) (hupA::Cm hupB::Km) as previously described (36). Crude yeast extracts were prepared from the Y869 yeast strain as previously described (4), except for the DEAE-Sephadex column purification stage, which was omitted. The recombinant TBPm3, TFIIIB70, and TFIIIB90 were a gift from Giorgio Dieci (10). The purified fractions containing Pol III or TFIIIC were obtained as previously described (16).
RNA analysis.
The multicopy plasmids YEp352 harboring the various SNR6 constructs were introduced into YPH500α, Y865, and Y869. Yeast transformation procedures, RNA extraction, and Northern blot analysis were performed as previously described (4), using body-labeled DNA fragments encompassing the SNR6, SNR31, and tDNAHis-KL coding sequences. Alternatively, the primers 5′-TGTTGCTATAAGCACGAAGCTCTAACCACT-3′ and 5′-GTCAGGCTCTTACCAGCTTAA-3′ were phosphorylated by T4 polynucleotide kinase and used to detect the tRNAIle(UAU) and 5S RNA, respectively. Quantifications were performed using PhosphorImager software (Amersham Pharmacia).
Chromatin analysis by MNase.
Cultures of strains Y865 (wild type) and Y869 (double mutant nhp6AΔ nhp6BΔ) were grown in yeast extract-peptone-dextrose (YPD) at 30°C to 1 × 107 to 2 × 107 cells/ml. Cells were harvested and converted to spheroplasts using Zymolyase. For chromatin analysis at 37°C, cultures were grown in YPD at 30°C to 1 × 107 to 2 × 107 cells/ml, incubated at 37°C for 4 h, and spheroplasted at 37°C. Chromatin and genomic DNA were prepared and digested with micrococcal nuclease (MNase) for 5 min at 37°C, and the cutting sites were mapped by indirect end labeling from the PstI site as previously described (22).
In vitro transcription assays.
In vitro transcription reactions were performed using 150 ng of either Bluescript SK-derived plasmids, harboring SNR6 genes, or the KS-tDNAIle(TAT)199 plasmid (10), the pSIRT plasmid (23), or the pFL44-t(His)K plasmid (24), harboring the I(TAT)LR1, the 5S DNA, and the tDNAHis-KL genes, respectively. The templates were incubated at 25°C for 40 min in 40-μl reaction mixtures (20 mM HEPES buffer [pH 7.5], 0.1 mM EDTA, 5% glycerol, 5 mM MgCl2, 5 mM dithiothreitol, 8 U of RNasin (Promega), 0.6 mM each ATP, CTP and GTP, 0.03 mM UTP, and 10 μCi of [α-32P]UTP) with or without 10 to 200 ng of NHP6B recombinant protein. Transcription reaction mixtures with purified components contained 50 ng of purified Pol III (16) and recombinant TBPm3 (40 ng), TFIIIB70 (80 ng), and TFIIIB90 (80 ng) (10), with or without 100 ng of purified TFIIIC (16). Transcription reactions with cell extracts prepared from the Y869 strain contain 20 μg of proteins.
RESULTS
Identification of high-copy-number suppressors of the temperature-sensitive snr6Δ2 gene.
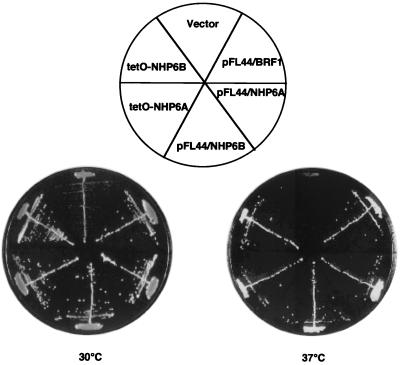
SNR6 is a Pol III gene which encodes the yeast U6 RNA, the catalytic part of the spliceosome. This gene has unusual promoter elements. Its A block, at position +21 relative to the transcription start site, is degenerate compared with the consensus tRNA gene element, its B block is located downstream of the transcribed sequence, 202 bp away from the A block, and it has a TATA box at position −30 (2). A mutant snr6Δ2 strain in which the chromosomal SNR6 gene has been inactivated by a 2-bp deletion in the B block is not viable but could be rescued by the same snr6Δ2 allele if the allele is harbored by a multicopy plasmid. This new snr6Δ2 strain, MCM260, grew slowly at 30°C and failed to form colonies at 37°C (see Fig. 1). We reasoned that by searching multicopy suppressors of the snr6Δ2 allele, which is impaired only in a gene-distal promoter element, we should select for genes involved in the transcriptional activation of SNR6.
FIG. 1.
Overexpression of the BRF1, NHP6A, or NHP6B genes rescues the thermosensitivity of snr6Δ2. MCM260 transformants containing either 2μm plasmids with the BRF1 (pFL44/BRF1), NHP6A (pFL44/NHP6A), or NHP6B (pFL44/NHP6B) genes, or the tetO-NHP6A or tetO-NHP6B constructs, or an empty vector (vector), were streaked on YPD plates and grown at 30 or 37°C for 3 days.
A yeast genomic DNA library in a 2μm multicopy plasmid (32) was transformed into MCM260, and transformants were selected for growth at 37°C. Of 60,000 transformants, 16 plasmids that reproducibly allowed growth at 37°C were isolated. The yeast genomic DNA inserts harbored by these plasmids were identified by sequence analysis and fell into seven classes. One of the largest classes of suppressor plasmids had four members and contained the wild-type SNR6 gene. The BRF1 gene encoding the TFIIIB70 subunit of TFIIIB was found in three plasmids and defined a second class. Two classes of inserts present in six plasmids contained either the NHP6A or the NHP6B gene and were selected for further study. The other three classes of plasmids contained uncharacterized yeast genes, whose analysis will be reported elsewhere.
The BRF1 gene was found in some genomic inserts in the absence of any other open reading frame, so its identification as a suppressor gene was immediate. To confirm the suppressor activity of the NHP6A and NHP6B genes, their coding sequences were cloned into a yeast expression vector under the control of the tetracycline operator (tetO). The tetO-NHP6A and tetO-NHP6B constructs improved the growth rate of MCM260 transformants at 30°C and suppressed their thermosensitive phenotype at 37°C, confirming the identification of NHP6A and NHP6B as suppressors of the snr6Δ2 allele (Fig. 1).
In vivo transcript levels of mutant SNR6 genes are increased by overexpression of NHP6A or NHP6B.
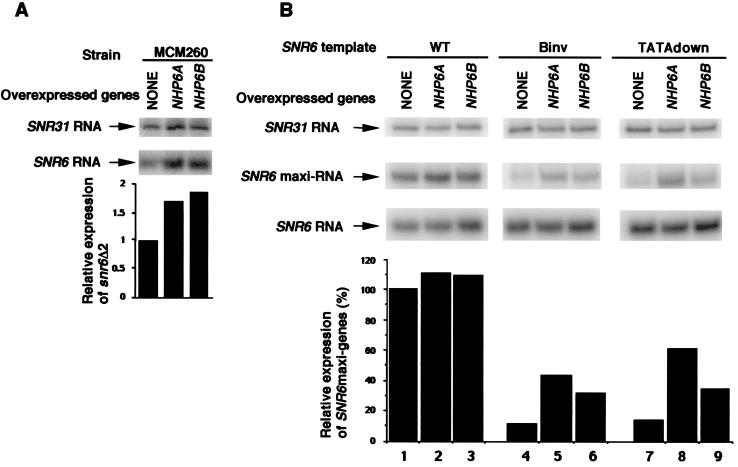
To confirm that the suppression of snr6Δ2 thermosensitivity was due to an effect on SNR6 expression, the steady-state levels of SNR6 transcripts in the MCM260 strain at 30°C, in the presence or the absence of the tetO-NHP6A and tetO-NHP6B constructs, were analyzed by Northern blotting (Fig. 2A). Equivalent amounts of RNA, as determined by measuring of optical density, were loaded in each lane of the gel, so as to normalize the levels of SNR6 transcripts in relation to rRNA. We also noticed that the quantities of RNA derived from SNR31, a Pol II-transcribed gene, showed no correlation with the levels of NHP6A and NHP6B proteins, and we used them systematically to correct for variations in RNA loading and to quantify more precisely the amounts of SNR6 transcripts. As shown in Fig. 2A, the overexpression of NHP6A or NHP6B increased significantly the level of SNR6 transcripts (derived from both the plasmid and chromosomal snr6Δ2 genes).
FIG. 2.
The transcription of mutant SNR6 genes is increased by overexpression of NHP6A or NHP6B. (A) The tetO-NHP6A and tetO-NHP6B constructs were introduced into the MCM260 strain, which contains only the snr6Δ2 allele. Transcripts derived from the snr6Δ2 genes were quantified in Northern blots by PhosphorImager analysis, using SNR31 transcripts as internal controls. (B) The Northern blot illustrates the transcriptional activation of SNR6 constructs harboring a 59-bp insert in the transcribed region and different mutations. These genes are borne by multicopy plasmids and generate transcripts (SNR6 maxi-RNA) easily distinguishable from the SNR6 RNA produced from the wild-type, chromosomal SNR6 gene. The steady-state levels of transcripts derived from the SNR6 maxigene constructs were analyzed in the wild-type strain YPH500α, with or without the overexpression of NHP6A or NHP6B, and quantified using SNR31 transcripts as internal controls. The transcription level of the wild-type (WT) maxigene construct without the overexpression of NHP6A or NHP6B (lane 1) was arbitrarily assigned the value 100%.
To test whether the effect of NHP6A or NHP6B overexpression was restricted to the snr6Δ2 mutation or could also affect other promoter-defective alleles of SNR6, we analyzed the expression of two other mutant SNR6 genes at 30°C in wild-type cells overexpressing NHP6A or NHP6B. We used another SNR6 mutant affected in the B-block element, the Binv mutant, whose B block and surrounding sequences have been inverted, and an SNR6 gene whose TFIIIC-binding sequences have been left intact but whose TATA box (5′-TATAAAT-3′) has been replaced by an unrelated sequence (5′-GTGCACG-3′), the TATAdown mutant, and a wild-type SNR6 gene as a control. A 59-bp DNA fragment was introduced into the transcribed regions of these genes, at position +73, to distinguish the corresponding transcripts from the endogenous, wild-type SNR6 RNA (22). These SNR6 maxigene constructs were inserted into the multicopy YEp352 plasmid and introduced into the wild-type strain YPH500α. As previously described (22), in the absence of NHP6A and NHP6B overexpression, the level of transcripts derived from the Binv maxigene represented about 15% of the level produced from the wild-type construct, whereas the TATAdown construct generated fewer RNA molecules than the wild-type maxigene and produced transcripts which were slightly shorter, in agreement with the proposed role of the TATA box on the selection of the transcription start site (Fig. 2B, lanes 1, 4, and 7). The overexpression of NHP6A or NHP6B was found to increase the steady-state levels of the transcripts derived from the TATAdown and Binv constructs (lanes 4 to 9), whereas it had no effect on the transcript level of the wild-type construct (lanes 1 to 3). Similarly, the overexpression of NHP6A or NHP6B had no effect on the abundance of the transcripts derived from the wild-type, chromosomal SNR6 gene. In conclusion, it seems that the overexpression of NHP6A or NHP6B increased the steady-state levels only of transcripts derived from SNR6 genes that are impaired in their promoter elements, either TFIIIC- or TFIIIB-binding sequences: the snr6Δ2 allele, the TATAdown construct, and the Binv construct. These observations confirmed that the suppressor activity of NHP6A and NHP6B was due to a direct effect on snr6Δ2 expression and implied that wild-type levels of NHP6A and NHP6B were not a limiting factor at 30°C for transcription of the wild-type SNR6 gene.
The expression of wild-type and mutant SNR6 genes is reduced in the absence of NHP6A and NHP6B.
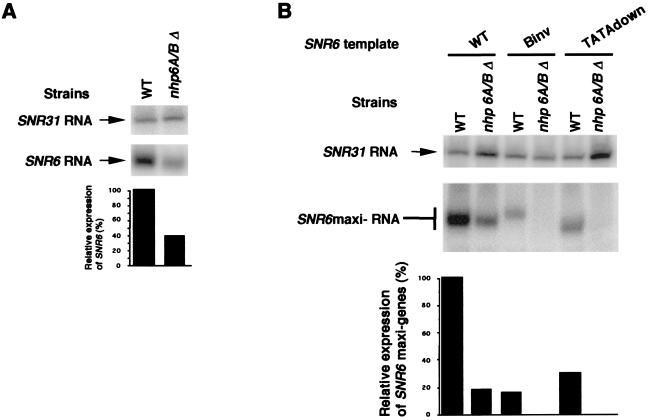
To assess the contribution of NHP6A and NHP6B to the expression of the wild-type SNR6 gene, the abundance of SNR6 transcripts was analyzed in wild-type and in double-mutant nhp6AΔ nhp6BΔ cells grown at 30°C. As shown in Fig. 3A, the transcript level of the wild-type SNR6 gene was specifically reduced 2.6-fold in the nhp6AΔ nhp6BΔ cells compared to that of other genes, such as SNR31. Likewise, the RNA levels derived from the SNR6 maxigene constructs were systematically reduced in the mutant nhp6AΔ nhp6BΔ strain, whether these SNR6 constructs were impaired in their promoter elements or not (Fig. 3B).
FIG. 3.
The transcriptional activity of wild-type and mutant SNR6 genes is reduced in the absence of NHP6A and NHP6B at 30°C. The transcription of the wild-type (WT), chromosomal SNR6 gene (A) and of SNR6 maxigene constructs either wild type or harboring different mutations (B) was monitored by Northern blotting in wild-type (WT, Y865 [8]) and nhp6AΔ nhp6BΔ mutant (Y869 [8]) cells. The steady-state levels of SNR6 RNA or maxi-RNA were quantified by PhosphorImager analysis, with SNR31 transcripts as internal controls. The transcription level of the wild-type maxigene construct in the wild-type strain was assigned the value 100%.
Protection over the TATA box of the wild-type SNR6 gene is dramatically altered in nhp6AΔ nhp6BΔ cells.
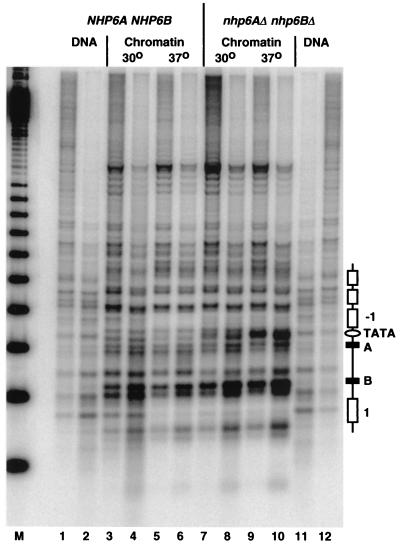
To investigate whether the absence of NHP6A and NHP6B would affect SNR6 chromatin structure, chromatin of the SNR6 locus in the wild-type and nhp6AΔ nhp6BΔ strains was analyzed by MNase digestions. MNase preferentially attacks linker DNA between the nucleosomes and leads to double-strand cut, but it may also introduce single-strand nicks on the nucleosome surface. Genomic chromatin and deproteinized DNA were digested with different amounts of MNase, and double-strand-cutting sites were displayed by indirect end labeling (Fig. 4). The bands in the DNA lanes represent the preferential cutting sites for MNase in deproteinized DNA. Some of these sites were protected in chromatin, whereas others remained or became accessible. Protected regions of 140 to 200 bp were interpreted as positioned nucleosomes (open boxes) (33). As previously described (22), the SNR6 chromatin structure in a wild-type strain at 30°C was characterized by the organization of the upstream and downstream regions into series of positioned nucleosomes, by a protection of the TATA box, and by hypersensitive sites around the A and B blocks (Fig. 4, lanes 1 to 4). Only minor changes were observed when the wild-type strain was grown at 37°C: the protection over nucleosome 1 was stronger, and the relative accessibility of the two sites between nucleosome 1 and the B block was slightly altered (lanes 3 to 6). In the nhp6AΔ nhp6BΔ cells, the nucleosomal organization in the upstream and downstream regions of the SNR6 locus was maintained at 30 and 37°C (lanes 7 to 10) and was similar to that of the wild-type cells. The relative sensitivities of the two sites around the A block and of the two sites between nucleosome 1 and the B block were similar to those of the wild-type strain grown at 37°C. The most dramatic change, however, which was induced by the absence of NHP6A and NHP6B, was observed at the TATA box: while the TATA box was completely protected in the wild-type cells at both temperatures (lanes 3 to 6), it was slightly accessible in the mutant at 30°C, as revealed by a weak band (lanes 7 and 8), and the protection appeared to be completely lost at 37°C, as indicated by a strong band (lanes 9 and 10). Loss of the footprint on the TATA box has been previously observed for the snr6Δ2 allele, whose transcriptional activity is crippled by a 2-bp deletion in the B block (22). These observations hinted that the occupancy of the A and B blocks by TFIIIC could be modified in the nhp6AΔ nhp6BΔ cells and strongly suggested that TFIIIB positioning on the TATA-box region was destabilized and lost in the absence of NHP6A and NHP6B at 37°C. The effects of NHP6A and NHP6B deletion on the chromosomal structure of the SNR6 locus indicate that NHP6A and NHP6B are acting at the level of SNR6 transcription complex formation and rule out the possibility that NHP6A and NHP6B are simply stabilizing SNR6 RNA.
FIG. 4.
Protection over the TATA box of the SNR6 gene is dramatically altered in nhp6AΔ nhp6BΔ cells. Cultures of Y865 (NHP6A NHP6B) and Y869 (nhp6AΔ nhp6BΔ) were grown at 30°C (lanes 3 and 4 and lanes 7 and 8, respectively) and shifted to 37°C for 4 h (lanes 5 and 6 and lanes 9 and 10, respectively). Chromatin and genomic DNA were prepared and digested with different amounts of MNase. To display the cutting sites, the DNA was digested with PstI, fractionated on a 1% agarose gel, blotted to a nylon membrane, and hybridized to a probe close to the PstI site. Indicated are the nucleosome positions (white boxes), A and B blocks (black boxes), and TATA box (white oval) of the SNR6 gene. The marker (M) represents multiples of 256 bp and was hybridized separately. The protection of the TATA box (lanes 3 to 8) was lost when Y869 was shifted to 37°C (lanes 9 and 10).
In vitro transcription of SNR6 genes is stimulated by NHP6B.
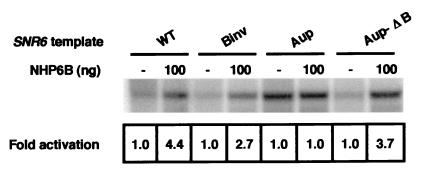
We next analyzed the effect of NHP6B on the in vitro transcription of several mutant SNR6 genes. The mutant template called Binv has been described above. The constructs Aup and Aup-ΔB have the SNR6 degenerate A block replaced by a consensus A block derived from the sequences of tRNA genes (4), and their B blocks have been either left intact (Aup) or deleted (Aup-ΔB). We used crude nuclear extracts so as to mimic as closely as possible the complexity of transcription processes occurring in the cell nucleus. The wild-type and mutant SNR6 templates were thus transcribed in a cell extract prepared from nhp6AΔ nhp6BΔ cells, with or without the addition of NHP6B (Fig. 5). NHP6B significantly stimulated the transcription of the wild-type and Binv templates. Interestingly, NHP6B strongly activated the transcription of the Aup-ΔB template but had no effect on the Aup construct, which remained the only SNR6 template insensitive to NHP6B activity. In all cases, in vitro as well as in vivo, the size of the transcripts was the same in the presence and in the absence of NHP6B, suggesting that the activation of the SNR6 genes by NHP6B did not alter the transcription start site.
FIG. 5.
NHP6B stimulates the transcription of wild-type and mutant SNR6 genes in vitro. NHP6B was added to in vitro transcription reaction mixtures containing cell extracts prepared from nhp6AΔ nhp6BΔ mutant cells (Y869 [8]) and different SNR6 templates, either wild type (WT) or mutated in the A or B block. The templates used were Bluescript derivatives harboring SNR6 wild-type or mutant genes. Reaction mixtures were incubated for 40 min at 25°C, and the transcription products were electrophoresed in a 6% polyacrylamide gel. The SNR6 transcripts were quantified by PhosphorImager analysis, and for each template, the basal level of SNR6 transcription in the absence of NHP6B was arbitrarily assigned the value of 1 unit.
NHP6B activates the transcription of SNR6 in a reconstituted transcription system.
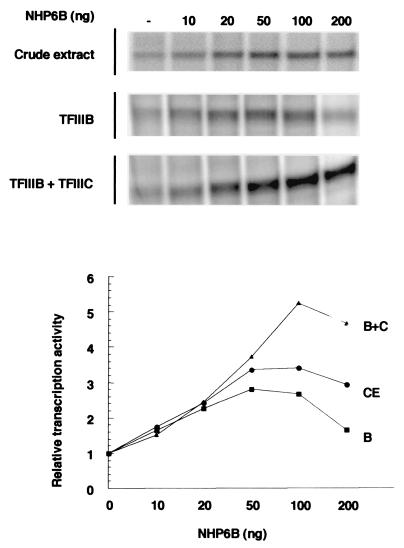
To get some insight into the molecular mechanisms underlying SNR6 transcriptional activation by NHP6A and NHP6B, we performed in vitro transcription reactions using purified fractions (TFIIIC and polymerase III) and recombinant proteins (TFIIIB70, TFIII90, and TBP). In contrast to many tRNA genes, naked DNA templates of SNR6 can be transcribed in vitro using only purified fractions of TFIIIB and Pol III. TFIIIC is not required for this reaction, although its presence increases the transcription efficiency. We first tested the effect of NHP6B on a transcription reaction involving the complete system with purified TFIIIC and recombinant TFIIIB. As shown in Fig. 6, a fivefold increase in SNR6 transcription was observed at optimal concentrations of NHP6B. This stimulation level was comparable to that observed, using the same concentration range, in yeast cell extracts. This result strongly suggested a direct stimulatory effect of NHP6B on the transcription system. Remarkably, as seen in Fig. 6, NHP6B was able to give a ca. threefold stimulation of SNR6 transcription directed by TFIIIB alone. This lower stimulation level might be due to the repression of transcription observed at the highest concentration of NHP6B, in the absence of TFIIIC.
FIG. 6.
NHP6B activates SNR6 transcription in a reconstituted system. NHP6B protein was added as indicated to in vitro transcription reaction mixtures containing either cell extracts (CE) prepared from nhp6AΔ nhp6BΔ mutant cells (Y869 [8]) or purified Pol III and recombinant TFIIIB (B) or Pol III, TFIIIB, and TFIIIC (B+C). A Bluescript-derived plasmid harboring the wild-type SNR6 gene was used as a template. Reaction mixtures were incubated for 40 min at 25°C, and the transcription products were electrophoresed in a 6% polyacrylamide gel. The SNR6 transcripts were quantified by PhosphorImager analysis, and the basal level of SNR6 transcription in the absence of NHP6B was arbitrarily assigned the value of 1 unit in each case.
NHP6A and NHP6B exert pleiotropic effects on the expression of Pol III genes.
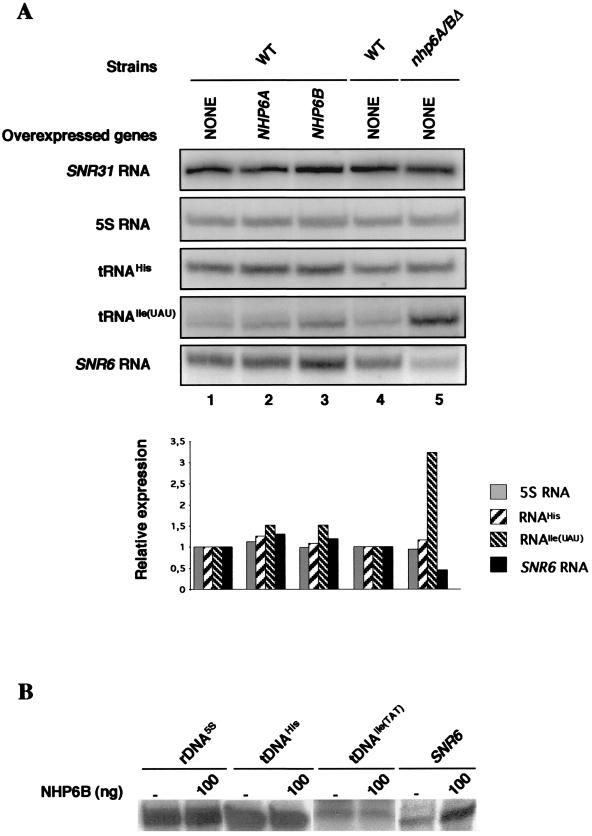
NHP6A and NHP6B are required for the induction of a subset of genes transcribed by Pol II (25). Since NHP6A and NHP6B are involved in the transcription of SNR6, we investigated whether these proteins could also influence the transcript levels of other Pol III genes. We analyzed by Northern blotting the levels of the 5S rRNA, tRNAIle(UAU), and tRNAHis in wild-type cells grown at 30°C, with or without the overexpression of NHP6A or NHP6B, and in mutant nhp6AΔ nhp6BΔ cells. The tRNAIle(UAU) genes were selected because they contain a canonical TATA sequence around position −30, like the SNR6 gene. Also, like SNR6, these tRNAIle(UAU) genes can be transcribed in vitro in the absence of TFIIIC (10). As shown in Fig. 7A, the levels of the 5S rRNA and of the tRNAHis were similar in all the strains analyzed whereas, unexpectedly, the abundance of the tRNAIle(UAU) strongly increased in the absence of NHP6A and NHP6B, in contrast to SNR6 transcript level that decreased (Fig. 7A, lane 5). tRNAIle(UAU) is encoded by two genes, I(TAT)LR1 and I(TAT)DR2, with identical transcribed sequences. To confirm these in vivo results, we investigated the effect of NHP6B on the in vitro transcription of 5S RNA, tRNAHis, and tRNAIle(UAU) genes using cell extracts prepared from nhp6AΔ nhp6BΔ cells, with or without the addition of NHP6B. As shown in Fig. 7B, NHP6B stimulated only the in vitro transcription of the SNR6 gene. Remarkably, under the same condition, the transcription of both tRNAIle(UAU) genes remained unaffected by the addition of NHP6B (Fig. 7B and data not shown).
FIG. 7.
NHP6A and NHP6B affect the transcript levels of several Pol III genes in vivo. (A) The steady-state levels of 5S RNA, tRNAHis, tRNAIle(UAU), and the transcripts derived from the SNR31 and SNR6 genes were analyzed by Northern blotting in the wild-type strain YPH500α (WT), with or without the overexpression of NHP6A or NHP6B (lanes 1 to 3), and in wild-type (lane 4, Y865 [8]) and nhp6AΔ nhp6BΔ mutant (lane 5, Y869 [8]) cells. The steady-state levels of the RNA were quantified by PhosphorImager analysis, and the quantity of each transcript was normalized with SNR31 transcripts as internal control. The relative RNA levels in the control wild-type strains (lanes 1 and 4) were arbitrarily assigned the value of 1 unit. (B) NHP6B was added to in vitro transcription reaction mixtures containing cell extracts prepared from nhp6AΔ nhp6BΔ mutant cells (Y869 [8]) and different templates containing either 5S rDNA, tDNAHis, tDNAIle(TAT), or the wild-type SNR6 gene as a control. The plasmids used are described in Materials and Methods. Reaction mixtures were incubated for 40 min at 25°C, and the transcription products were electrophoresed in a 6% polyacrylamide gel.
DISCUSSION
We present genetic and biochemical evidence that the nonhistone chromatin proteins NHP6A and NHP6B are required for optimal transcription efficiency of wild-type and mutant SNR6 genes in vivo and in vitro. These observations suggest the potential of chromatin-associated proteins to act as positive or negative cofactors in Pol III transcription.
Reciprocal genetic interactions between SNR6 and the NHP6A and NHP6B genes.
Nonhistone proteins 6A and 6B (NHP6A and NHP6B) belong to a family of proteins characterized by the presence of one HMG box, a conserved domain of about 80 amino acids which mediates DNA binding (for a review, see reference 5). NHP6A and NHP6B are 96% similar and appear to be functionally redundant, as indicated by the absence of phenotype for the single nhp6AΔ or nhp6BΔ mutants (8). However, the double nhp6AΔ nhp6BΔ mutant grows slowly at 30°C and is nonviable at 38°C (8). The nhp6AΔ nhp6BΔ mutant shares many phenotypes with the pkc1Δ, slk1Δ, and slt2Δ mutants, and the NHP6A gene has been identified as a multicopy suppressor of the synthetic lethality of the slk1Δ and spa2Δ mutations, which has led to the suggestion that NHP6A and NHP6B function downstream of SLT2 to mediate its function in cell growth and morphogenesis (8). However, we have demonstrated here that the transcription of the SNR6 gene is strongly reduced in nhp6AΔ nhp6BΔ mutants and have found that the thermosensitivity of nhp6AΔ nhp6BΔ cells can be suppressed by the overexpression of either wild-type SNR6 or BRF1 (data not shown), which encodes the TFIIIB70 component of TFIIIB and whose overexpression was also found in our screen to suppress the thermosensitivity of snr6Δ2. Our data thus suggest that the thermosensitivity of the nhp6AΔ nhp6BΔ mutant could be primarily due to a defect in SNR6 transcription, which could in turn affect, via splicing defects, the expression of genes belonging to the SLT2 pathway. The chromatin analysis directly supports this hypothesis: the nhp6AΔ nhp6BΔ mutant revealed a loss of protection of the TATA box when the cells were shifted from 30°C to the nonpermissive temperature of 37°C.
Are NHP6A and NHP6B general regulators of Pol III transcription?
While NHP6 proteins increase the transcription efficiency of SNR6, the overexpression or deletion of the NHP6A and NHP6B genes did not affect the in vivo levels of 5S RNA or tRNAHis. Remarkably, the level of another tRNA species, tRNAIle(UAU), was found to be markedly increased in cells lacking NHP6 proteins. This observation raises the possibility that NHP6A and NHP6B could positively or negatively modulate the expression of a subset of Pol III genes. It should be noted, however, that we did not observe any effect of NHP6B on the transcription of tRNAIle(UAU) genes in vitro. NHP6 proteins might participate in the repression of the tRNAIle(UAU) genes only in the in vivo chromatin context or affect tRNAIle(UAU) levels in an indirect fashion. At this point, SNR6 remains the only Pol III gene whose transcription is unambiguously and directly influenced by NHP6A and NHP6B. It will be interesting, when the specific DNA microarrays are available, to test the influence of NHP6 gene deletion on the expression of all the Pol III genes. From the small set of genes examined, it appears that the presence of a canonical TATA sequence (TATAAATA) around position −30 is not a determinant for the stimulatory effect of NHP6. Both SNR6 and the tRNAIle(UAU) genes have this core promoter element. Furthermore, the TATAdown mutant SNR6 gene required NHP6A and NHP6B for detectable expression in vivo (Fig. 3). The only SNR6 mutant gene that was insensitive to the presence of NHP6B in vitro was the Aup template, which harbors an intact B block and a canonical A block (instead of the SNR6 degenerate A block). This mutant SNR6 gene was efficiently transcribed in the presence or absence of NHP6B. Therefore, the selective effect of NHP6A and NHP6B on the transcription of Pol III genes in vitro appears to be related to the strength of their A block, suggesting a role for NHP6A and NHP6B in transcription complex assembly.
Mechanisms of NHP6A and NHP6B transcriptional activation of SNR6.
The fact that NHP6B stimulated the transcription of SNR6 gene in a purified reconstituted system suggested a direct effect on transcription complex formation. In strong support of this conclusion, chromatin analysis with MNase revealed a dramatic loss of protection in the TATA region of the SNR6 gene in cells lacking NHP6 proteins. The protection of the TATA box is strictly linked to the transcriptional activity of the gene, and the extent of protection was found to correspond to TFIIIB footprinting on naked SNR6 DNA (22). NHP6A and NHP6B could favor TFIIIB assembly over the TATA region indirectly, by facilitating a TFIIIC-DNA interaction. In gel shift assays, we found that NHP6B interacted with TFIIIC-SNR6 DNA and TFIIIC-TFIIIB-DNA complexes to generate an upshifted complex. The specificity of this interaction, however, is uncertain because NHP6B by itself caused the formation of a ladder of protein-DNA complexes with SNR6 DNA (results not shown). The possibility that NHP6 proteins act at the level both of TFIIIC and TFIIIB DNA binding remains open inasmuch as NHP6 stimulated the TFIIIC-independent transcription of SNR6.
The mode of action of NHP6 proteins is probably related to their DNA-binding properties. NHP6A and NHP6B belong to the subfamily of non-sequence-specific HMG box proteins. NHP6A was shown to bind linear DNA with little sequence specificity and to induce a large bend (26, 36). The abundance of NHP6A has been estimated to be ∼50,000 to ∼70,000 molecules per haploid cell, which would correspond to ∼1 molecule of NHP6A for every 1 to 2 nucleosomes (25). NHP6A and NHP6B also bind the TATA-box regions of Pol II genes and DNA molecules of random sequences with equivalent affinity in vitro (25, 26). On the other hand, a sequence-dependent binding of the NHP6A- and NHP6B-related HMG1 protein has recently been demonstrated on the BHLF-1 promoter (11). Therefore, NHP6A and NHP6B may contribute to stabilize bent DNA conformations within the preinitiation complexes in a specific or non-sequence-specific manner. This does not exclude the possibility that NHP6A/B could influence SNR6 transcription by also interacting with components of the preinitiation complex. Gal4(1–147)-NHP6A and Gal4(1–147)-NHP6B fusions were tested in the two-hybrid system (12) with fusions comprising the Gal4 activating domain and all the subunits of TFIIIB or TFIIIC. No interaction between NHP6A or NHP6B and any components of TFIIIC or TFIIIB could be detected in this way (data not shown). These negative results instead suggested that the major role of NHP6 proteins may reside in their DNA-binding and -bending properties. Paull et al. (25) previously reported that NHP6A promotes the formation of a Pol II preinitiation complex and suggested that NHP6A-induced structural changes in the TBP-TFIIA-DNA complex may facilitate TFIIB-DNA binding, which requires considerable DNA distortion. Similarly, TFIID-TFIIA-DNA complex formation was found to be enhanced by HMG2 (30). NHP6A and NHP6B could play a similar role for some Pol III genes. Interestingly, the C-terminal half of human TFIIB90 was found to contain an HMG1- and HMG2-related domain which is required for Pol III transcription (34). The presence of this domain, which is absent in yeast TFIIIB70, suggests that HMG boxes, here embedded in a Pol III common factor, could play a general role in the transcription of vertebrate Pol III genes.
ACKNOWLEDGMENTS
We are very grateful to Reid Johnson for his generous gift of the Y865 and Y869 yeast strains, of the RJ1963 and RJ1964 E. coli strains, and of the antibodies against NHP6A and NHP6B. We warmly thank Giorgio Dieci for providing us with the recombinant TBP, TFIIIB70, and TFIIIB90 purified proteins and with the KS-tDNAIle(TAT)199 and KS-tDNAIle(TAT)36 plasmids harboring the I(TAT)LR1 and I(TAT)DR2 genes; Emmanuel Favry for excellent technical assistance; and Olivier Lefebvre and Christine Conesa for stimulating discussions.
S.L. was supported by a Commissariat à l'Energie Atomique postdoctoral fellowship. F.T. and M.L. were supported by the Swiss National Science Foundation.
REFERENCES
- 1.Aasland R, Stewart A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 2.Brow D A, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 4.Burnol A F, Margottin F, Schultz P, Marsolier M C, Oudet P, Sentenac A. Basal promoter and enhancer element of yeast U6 snRNA gene. J Mol Biol. 1993;233:644–658. doi: 10.1006/jmbi.1993.1542. [DOI] [PubMed] [Google Scholar]
- 5.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 6.Chedin S, Ferri M L, Peyroche G, Andrau J C, Jourdain S, Lefebvre O, Werner M, Carles C, Sentenac A. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp Quant Biol. 1998;63:381–389. doi: 10.1101/sqb.1998.63.381. [DOI] [PubMed] [Google Scholar]
- 7.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 8.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dieci G, Duimio L, Coda-Zabetta F, Sprague K U, Ottonello S. A novel RNA polymerase III transcription factor fraction that is not required for template commitment. J Biol Chem. 1993;268:11199–11207. [PubMed] [Google Scholar]
- 10.Dieci G, Percudani R, Giuliodori S, Bottarelli L, Ottonello S. TFIIIC-independent in vitro transcription of yeast tRNA genes. J Mol Biol. 2000;299:601–613. doi: 10.1006/jmbi.2000.3783. [DOI] [PubMed] [Google Scholar]
- 11.Ellwood K B, Yen Y M, Johnson R C, Carey M. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol Cell Biol. 2000;20:4359–4370. doi: 10.1128/mcb.20.12.4359-4370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 15.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 16.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 17.Joazeiro C A, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 19.Kassavetis G A, Joazeiro C A, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 20.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-De-Leon A, Librizzi M, Puglia K, Willis I M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 22.Marsolier M C, Tanaka S, Livingstone-Zatchej M, Grunstein M, Thoma F, Sentenac A. Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes Dev. 1995;9:410–422. doi: 10.1101/gad.9.4.410. [DOI] [PubMed] [Google Scholar]
- 23.Musters W, Knol J, Maas P, Dekker A F, van Heerikhuizen H, Planta R J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro F, Thuriaux P. In vivo misreading by tRNA overdose. RNA. 2000;6:103–110. doi: 10.1017/s1355838200991714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paull T T, Carey M, Johnson R C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 26.Paull T T, Johnson R C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 27.Qureshi S A, Bell S D, Jackson S P. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 29.Ruth J, Conesa C, Dieci G, Lefebvre O, Dusterhoft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 30.Shykind B M, Kim J, Sharp P A. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- 33.Thoma F, Bergman L W, Simpson R T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White R J. RNA Polymerase III transcription. New York, N.Y: Springer-Verlag; 1998. [Google Scholar]
- 36.Yen Y M, Wong B, Johnson R C. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J Biol Chem. 1998;273:4424–4435. doi: 10.1074/jbc.273.8.4424. [DOI] [PubMed] [Google Scholar]