Abstract
Objective:
To compare the early responses to joint injury in conventional and germ-free mice.
Design:
Post-traumatic osteoarthritis (PTOA) was induced using a non-invasive anterior cruciate ligament rupture model in 20-week old germ-free (GF) and conventional C57BL/6 mice. Injury was induced in the left knees of n=8 GF and n=10 conventional mice. To examine the effects of injury, n=5 GF and n=9 conventional naïve control mice were used. Mice were euthanized seven days post-injury, followed by synovial fluid recovery for global metabolomic profiling and analysis of epiphyseal trabecular bone by micro-computed tomography (μCT). Global metabolomic profiling assessed metabolic differences in the joint response to injury between GF and conventional mice. Magnitude of trabecular bone volume loss measured using μCT assessed early OA progression in GF and conventional mice.
Results:
μCT found that GF mice had significantly less trabecular bone loss compared to conventional mice, indicating that the GF status was protective against early OA changes in bone structure. Global metabolomic profiling showed that conventional mice had greater variability in their metabolic response to injury, and a more distinct joint metabolome compared to their corresponding controls. Furthermore, differences in the response to injury in GF compared to conventional mice were linked to mouse metabolic pathways that regulate inflammation associated with the innate immune system.
Conclusions:
These results suggest that the gut microbiota promote the development of PTOA during the acute phase following joint trauma possibly through the regulation of the innate immune system.
Introduction
Osteoarthritis (OA) is a disease of the whole joint with low-grade articular inflammation driving disease progression[1-4]. Inflammation is associated with age, traumatic injury, and obesity; all of which are OA risk factors[2, 5]. Joint injury triggers an acute inflammatory response immediately after injury, and further perpetuation of inflammation can result in post-traumatic OA (PTOA) via catabolism in the joint[6]. Inflammation post-injury, therefore, is an important contributing factor in OA development. A better understanding of immune responses in this context may lead to new pharmacological targets for prevention and treatment.
The perpetual inflammatory response that persists after joint injury includes innate immune activity. Joint injury results in production of endogenous signals termed damage associated molecular patterns (DAMPs) that signal to innate immune cells. In OA, DAMPs include matrix components from cartilage degradation, plasma proteins from vascular leakage, and intracellular alarmins from stressed or necrotic cells[7-10]. Macrophages, neutrophils, and dendritic cells with pattern recognition receptors recognize DAMPs and induce NF-kβ- mediated production of proinflammatory cytokines, such as IL-6[7-10].
In addition to DAMPs, the immune system is modulated by antigens from microorganisms living in and on the body (microbiome)[11]. Recent studies suggest the gut microbiome may play a role in OA pathogenesis[12, 13] such that innate cells initially activated by joint injury are further and perpetually activated by microbial antigens. For example, increased abundance of Streptococcus species in the gut is associated with increased knee pain in OA driven by increased local inflammation in the joint[14]. Moreover, the gut microbiome-produced metabolites, hippurate and trigonelline, distinguish OA progressors from non-progressors[15]. Finally, microbial DNA has been reported in human and mouse cartilage, suggesting that molecules of microbial origin help drive OA-associated inflammation[16]. In a recent study, germ-free (GF) mice (mice lacking a microbiome) had less severe histological OA eight weeks after surgical destabilization of the medial meniscus[17]. However, joint injury induced by surgery requires opening the joint cavity for surgical destabilization, which does not occur in human joint injuries. Finally, in a model of obesity, altering the gut microbiome with antibiotics resulted in protection against cartilage loss in a mechanical overload model of PTOA[18].
Non-invasive PTOA mouse models simulate human injury and avoid some confounding factors from surgical models. Furthermore, non-invasive models allow examination of early timepoints during the acute phase after joint trauma. The acute response to injury is important because it provides a unique opportunity for intervention when patients often seek medical attention. Thus, the objective of this study was to compare the acute responses after joint injury in gnotobiotic (i.e. germ free) and conventional mice. We hypothesize that injured gnotobiotic mice exhibit less severe OA than conventional mice due to the absence of immune activity driven by the gut microbiome.
Materials and Methods
Joint Injury Model and Experimental Design
We utilized a non-invasive injury model to induce PTOA in C57BL/6 mice[19, 20]. This model results in ACL rupture that closely mimics ACL tears in humans by producing a highly reproducible joint injury and stimulates degeneration of bone and cartilage[19, 20]. Typically, this model first displays changes in subchondral trabecular bone at 7 days with subsequent cartilage pathology detected by 14-56 days post-injury[21]. All mouse studies were approved by the Montana State University IRB (protocol: 2018-02).
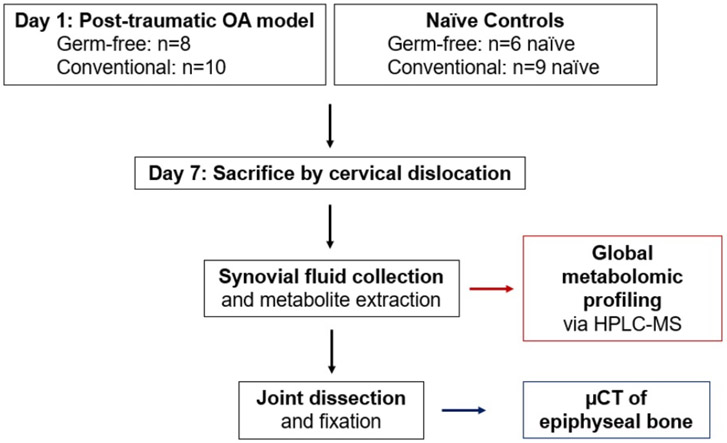
Mice were anesthetized using isoflurane inhalation and PTOA was induced in conventional and germ-free (GF) mice using a tibial compression system by applying a single compressive overload to the left leg with a target force of 12N at a loading rate of 130 mm/s[22]. After injury, mice were given a single dose of sustained-release buprenorphine (1.0 mg/kg) subcutaneously to alleviate pain associated with ACL rupture. GF mice were maintained in GF-conditions after injury. Naïve uninjured controls without buprenorphine treatment were used for both conventional and GF mice. Mice were euthanized by cervical dislocation seven days post-injury for synovial fluid (SF) collection and whole knee μCT (Figure 1).
Figure 1:
Experimental design to study the role of the microbiome in post-traumatic osteoarthritis. 20-week old C57/BL6 mice were used in this study. Mice were raised in either gnotobiotic (i.e. germ-free) or conventional conditions. On Day 1, mice were subjected to a post-traumatic OA model by single compressive overload of the joint. At Day 7, mice were sacrificed before synovial fluid harvest and joint fixation. Synovial fluid was profiled by global metabolomic analysis, and the epiphyseal bone was analyzed by μCT.
In total, there were six experimental groups for comparisons: GF injured (GF-inj; n=8), GF contralateral (GF-cl; n=8), GF naïve (GF-ctrl; n=5), conventional injured (Conv-inj; n=10), conventional contralateral (Conv-cl; n=10), and conventional naïve (Conv-ctrl; n=9).
Germ-free status was monitored and confirmed using standard cultivation and molecular biology techniques[23]. Briefly, liquid ‘traps’ were established inside hermetically sealed and air purified (HEPA filtered) husbandry isolators containing sterilized drinking water and chow pellets (i.e. the same water and chow that mice were drinking and eating). Traps were monitored daily for any signs of microbial growth. Samples from traps were collected along with stool pellets from mice for aerobic and anaerobic cultivation on non-selective media (Luria broth and Mueller-Hinton broth agar plates) following 48 hours incubation at 37°C. In addition, bulk DNA was isolated from mouse pellets (DNeasy Powersoil Kit, Qiagen, Inc) and attempts were made to amplify a 16S rRNA locus using broad-range (universal) primers[24]. No positive results (i.e. growth on agar plates or amplification of PCR) were observed.
For injury of GF mice, two cages of animals were removed from the isolator inside a sealed cage with a HEPA-filtered lid and place immediately inside a biosafety cabinet with laminar flow and containing the compressive overload device. All surfaces were contact sterilized ahead of time and the cage was doused repeatedly with disinfectant (Ecolab® Exspor™). Following injury mice were placed back inside the cages and moved back inside husbandry isolators. The isolator environment and mice were checked repeatedly for any indication of breach in sterility as described above.
Synovial Fluid Collection
Immediately after euthanasia, SF was recovered using the calcium sodium alginate compound (CSAC) method as previously described[25]. Joint cavities were accessed and the patellar tendon was retracted to place a 3mm Melgisorb (Tendra, REF# 250600; Goteborg, Sweden) wound dressing on the cartilage surfaces for SF absorption. The Melgisorb wound dressing was submerged in 35 μL of Alginate Lyase in H2O (1 unit/mL concentration; derived from Flavobacterium, Sigma-Aldrich A1603-100MG), vortexed, and digested at 34°C for 30 minutes. After digestion, 15 μL of 1.0M sodium citrate (C6H5Na3O7) was added to chelate the Ca2+ ions and reduce the viscosity. Sample volumes were recorded prior to freezing at −80°C until metabolite extraction and analysis.
Metabolite Extraction
Metabolites were extracted from mouse SF following our previously established protocol with minor modifications[26, 27]. SF samples were thawed, centrifuged at 4°C at 500xg for 5 minutes, and supernatant was collected for vacuum concentration for ~2 hr. Metabolites were extracted from the dried pellet by re-suspension in 50:50 water: acetonitrile at −20°C for 30 min. Following extraction, the sample was vortexed for 3 min and centrifuged at 16100xg for 5 min at 4°C. Proteins were precipitated by adding 250 μL of acetone and shaken for 3 min followed by overnight refrigeration at 4°C. The mixture was then centrifuged for 5 min at 16100xg for 5 min and supernatant was collected for vacuum concentration prior to re-suspension in 50:50 water:acetonitrile for mass spectrometry analysis. All solvents were either HPLC or mass-spectrometry grade.
Global Metabolomic Profiling
Metabolite extracts were analyzed using established methods[28]. Samples were analyzed on an Agilent 1290 UPLC system (Agilent, Santa Clara, CA, USA) coupled to an Agilent 6538 Q-TOF mass spectrometer (Agilent Santa Clara, CA, USA) in positive mode. The chromatographic run used a Cogent Diamond Hydride HILIC 150 x 2.1 mm column (MicroSolv, Eatontown, NJ, USA) in normal phase with our established elution methods[27]. Spectra were analyzed as previously described[27].
μCT Analysis of Subchondral Bone
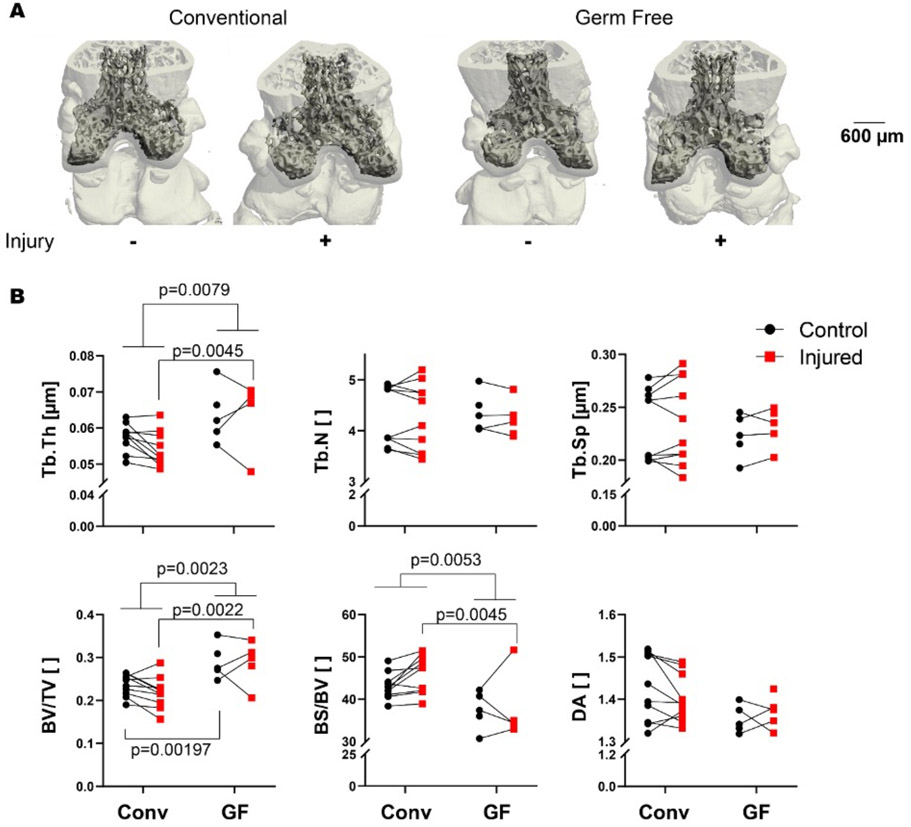
Following dissection, whole knee joints from the same animal were fixed in 4% paraformaldehyde for 24-48 hours and preserved in 70% ethanol. Joints were individually imaged using micro-computed tomography (SkyScan 1173, Bruker, Belgium) to quantify trabecular bone microstructure of the distal femoral epiphysis. During imaging, bones were embedded in 1.5% agarose, and were scanned with a 8.1 μm nominal voxel size (x-ray tube potential = 40 kVp, current = 200 μA, integration time = 1300 ms) according to the JBMR guidelines for μCT analysis of rodent bone structure[29]. Transverse images were exported as bitmap image files, then converted to raw files using ImageJ and uploaded to another μCT system for analysis (SCANCO μCT 35, Brüttisellen, Switzerland). The trabecular bone volume of interest was contoured by manually drawing on transverse images, and included the full epiphyseal region, excluding cortical bone and the growth plate (Figure 2). Trabecular bone volume fraction (BV/TV), trabecular thickness (Tb.Th), and other microstructural outcomes were determined using the SCANCO analysis software for paired statistical analysis.
Figure 2:
Germ-free mice have increased epiphyseal bone mass compared with conventional controls in both injured and contralateral knees. (A) Representative μCT reconstructions of conventional and germ-free femurs. (B) Microstructural parameters of femoral epiphysis quantified by μCT. Injured joints plotted in red, and contralateral joints plotted in black. Joints from the same mouse are connected with a line. Germ free mice had greater trabecular thickness (Tb.Th, p=0.0079), and injury induced thicker trabeculae in GF mice (p=0.0045). Germ free mice also had greater volume fraction than conventional mice independent of injury, as well as in both control and injured knees (all p<0.0022). Numbers of trabeculae, trabecular spacing, and degree of anisotropy (DA) were similar between GF and conventional mice.
Statistical Analyses
Before statistical analyses, median metabolite intensities were calculated for each metabolite across each experimental group. Metabolite features (m/z values) with a median intensity of zero across all experimental groups were removed from further analyses. Any remaining metabolites with intensity of zero (i.e. non-detected) were replaced with one-half the minimum peak intensity for statistical analyses.
Statistical analyses were performed in MetaboAnalyst and GraphPad Prism (GraphPad Prism Software, La Jolla, CA, USA) for metabolomics data and μCT data, respectively[30]. Metabolomics data were log transformed (log2) and standardized (mean centered divided by standard deviation) in MetaboAnalyst prior to statistical analyses[30]. All statistical tests used a false discovery rate (FDR)-corrected a priori significance level of 0.05.
The metabolomic profiles were analyzed using multivariate statistical methods to determine between-group changes and patterns of co-regulated metabolites. Unsupervised statistical methods, hierarchical cluster analysis (HCA) with Euclidean distances, and principal component analysis (PCA) were used to examine variation within the overall dataset. PCA was visualized with scatterplot projections onto the principal axes to examine sample to sample variation.
Differentially regulated metabolites were identified using two-tailed Student’s t-tests, analysis of variance (ANOVA) F-tests, and volcano plot analysis. While Student’s t-tests and ANOVA identify metabolites that were significantly different by FDR-corrected p-values, volcano plot assesses significance and magnitude of change simultaneously (FDR-corrected p-value<0.05 and fold-change greater than twofold).
Specific metabolite features of interest were matched to putative metabolite identities using the MetaboAnalyst “MS Peaks to Pathways” function[30, 31]. This generates a compound match list, which matches m/z values to possible metabolite identities, while simultaneously determining enriched metabolic pathways within the relevant group of metabolite features. Raw metabolomics data is found in Supplemental Figure 1.
μCT data were analyzed in GraphPad Prism 8.4.3. Each of the microstructural parameters for the epiphyseal bone (trabecular thickness Tb.Th, trabecular number Tb.N, trabecular spacing Tb.Sp, bone volume fraction BV/TV, bone surface per bone volume BS/BV, and the degree of anisotropy DA) were analyzed using mixed effects analysis of variance (ANOVA). Each mouse was treated as a random effect. Fixed effects were mouse status (conventional or GF), joint (control or injured), and the interaction between mouse status and joint. Post hoc comparisons between either control or injured joints across mouse status (conventional or GF) were assessed with Sidak’s multiple comparisons tests. Statistical significance was defined using an a priori significance level of 0.05.
Results
Less evidence of OA-related injury-induced bone loss in germ-free mice
Consistent with prior results [32], GF mice (mean±SD: 29.0±2.1, 95% CI: 27.2-30.7 g) had greater mass than conventional mice (25.5±3.1, 23.4-27.6 g, Table 1). We found no evidence that GF mice became colonized by any microorganism up to eight days following injury. μCT analysis showed that GF mice had greater trabecular bone volume than conventional mice, with 23% greater trabecular bone volume fraction (BV/TV, mean difference of 0.0543, 95% confidence interval of difference (CI) [0.00798-0.100])) and 11% greater trabecular thickness (Tb.Th, mean difference of 0.00879, 95% CI [0.00270-0.0149], Figure 2). GF mice had thicker trabeculae independent of injury (overall ANOVA, p=0.0079) and injured GF mice had thicker trabeculae than conventional mice (mean difference 0.0108, 95% CI [0.00324-0.0184], p=0.0045). Bone volume fraction was also greater in GF mice than conventional independent of injury status (mean difference of 0.0629, 95% CI [0.0267-0.991], p=0.0023) consistent with prior studies [33]. Both the contralateral and injured knees of GF mice had greater BV/TV than conventional mice (p=0.00197 and p=0.0022, respectively. Mean difference for contralateral 0.0543, 95% CI [0.00798-0.100]. Mean difference for injured 0.0716 [0.0253-0.118]). There were no differences in trabecular number (TB.N) or trabecular separation (Tb.Sp) between GF and conventional mice.
Table 1.
Mouse information from experimental cohorts. This study used both conventional (Conv) and germ-free (GF) mice from both sexes at 20 weeks of age. Two groups of mice were used, injured and naïve.
| Injured Group | Naïve Group | ||||
|---|---|---|---|---|---|
| Mouse | Sex | Weight (g) | Mouse | Sex | Weight (g) |
| A, GF | F | 28.3 | AC, GF | M | 25.6 |
| C, GF | F | 29.7 | AD, GF | M | 27.2 |
| E, GF | F | 29.5 | AE, GF | M | 28.7 |
| F, GF | F | 27.2 | AF, GF | M | 29.1 |
| G, GF | F | 29.8 | AG, GF | M | 28.4 |
| H, GF | F | 25.7 | S, Conv | F | 24.0 |
| B, GF | M | 28.6 | T, Conv | F | 21.9 |
| D, GF | M | 33.0 | U, Conv | F | 24.3 |
| N, Conv | F | 22.5 | V, Conv | F | 22.6 |
| O, Conv | F | 23.6 | W, Conv | F | 25.6 |
| P, Conv | F | 25.9 | X, Conv | M | 28.9 |
| Q, Conv | F | 24.1 | Y, Conv | M | 30.7 |
| R, Conv | F | 21.7 | Z, Conv | M | 29.6 |
| I, Conv | M | 25.5 | AB, Conv | M | 30.8 |
| J, Conv | M | 27.2 | |||
| K, Conv | M | 28.6 | |||
| L, Conv | M | 29.2 | |||
| M, Conv | M | 30.9 | |||
On average, injured joints of conventional mice had a 7.4% lower BV/TV compared to their contralateral limbs, although this difference was not statistically significant (Figure 2B, p = 0.061). Injured joints of conventional mice also had 5.9% lower Tb.Th (p = 0.016) and 7.0% greater BS/BV (Figure 2B, p = 0.015) than contralateral joints. In contrast, no significant differences in trabecular bone microstructure were observed between injured and contralateral joints from GF mice.
Global metabolomic profile of injury response – conventional injured vs. conventional naïve
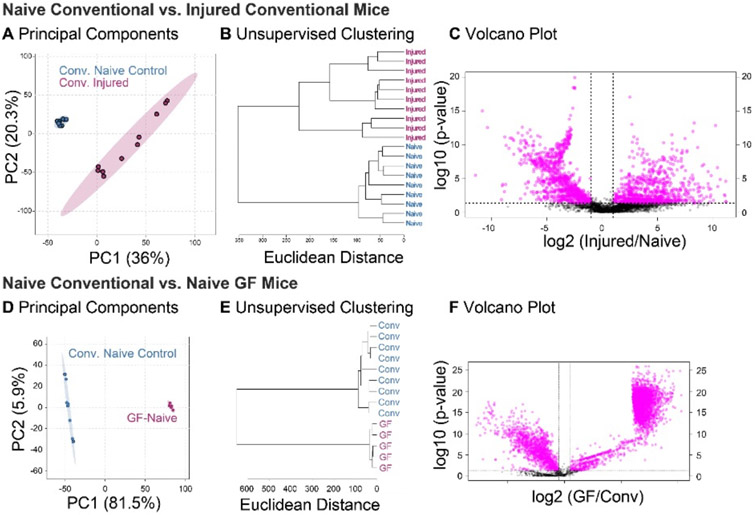
To assess the joint response to injury and gain insight into the metabolomic profile of PTOA, conventional injured mice were compared to conventional naïve mice (Conv-inj vs. conv-ctrl). A total of 4636 metabolite features were detected between conventional injured and naïve cohorts, with 2051 metabolite features identified as significantly different by Student’s T-test (FDR-adjusted p-value<0.05). Global differences between conventional injured and naïve mice were assessed using unsupervised PCA and HCA. Both PCA and HCA showed distinct clustering of SF samples within their respective cohorts (Figure 3A-B).
Figure 3:
Global metabolomic profiles of synovial fluid are driven by both injury and germ-free status. (A) Principal components analysis (PCA) finds complete separation between injured and naïve metabolomic profiles of conventional mice, with 36% and 20.3% of the variance associated with PC1 and PC2, respectively. (B) Conventional injured and naïve mice separate into two distinct clusters by unsupervised hierarchical cluster analysis. (C) 1038 metabolite features were significantly downregulated, and 971 metabolite features were significantly upregulated in Conv-inj compared to Conv-ctrl mice. (D) PCA finds clear separation between synovial fluid metabolomic profiles of germ-free and conventional mice, with 81.5% and 5.9% of the variance associated with principal components (PC) 1 and 2, respectively. (E) Unsupervised hierarchical clustering analysis finds distinct clusters between conventional and germ-free mice. (F) Volcano plot analysis found 4357 metabolites significantly different between germ-free and conventional mice. 1279 metabolites were significantly downregulated, and 3078 metabolites were significantly upregulated in naïve GF compared to naïve conventional mice.
Volcano plot analysis was used to identify specific metabolite features that were significantly upregulated or downregulated in conventional injured compared to naïve mice. 1038 metabolite features were significantly downregulated, and 971 metabolite features significantly upregulated in conventional injury compared to naïve controls (Figure 3C). Injury resulted in up- and down-regulation of several pathways of interest in conventional mice (Table 2).
Table 2.
Pathways altered in injured conventional mice. Metabolite features identified by volcano plot analysis as significantly upregulated or downregulated in injured conventional mice compared with conventional naïve were mapped to KEGG metabolic pathways. Pathways are reported with directionality (upregulated or downregulated); the total number of metabolites in the pathway, total number of metabolite features detected in samples in that pathway, total number of significant metabolite features (significant by both FDR-corrected p-value and fold change), and p-value for enrichment. Most significant pathways are reported up until arbitrary p-value cut-off for upregulated (p-value < 0.05) and downregulated pathways (p-value < 0.05). Full enrichment analyses are reported in Supplemental Tables 4 and 5.
| Pathway | Up/Down | Total metabolites in pathway |
Total pathway hits |
Significant pathway hits |
P-value |
|---|---|---|---|---|---|
| Porphyrin and chlorophyll metabolism | ↑ | 27 | 6 | 5 | 0.008626 |
| Primary bile acid biosynthesis | ↑ | 46 | 3 | 3 | 0.013698 |
| Steroid biosynthesis | ↑ | 41 | 6 | 4 | 0.022535 |
| Citrate cycle (TCA cycle) | ↑ | 16 | 4 | 3 | 0.025269 |
| Alanine, aspartate and glutamate metabolism | ↑ | 28 | 4 | 3 | 0.025269 |
| alpha-Linolenic acid metabolism | ↑ | 12 | 2 | 2 | 0.0321 |
| Fatty acid degradation | ↑ | 36 | 7 | 4 | 0.035143 |
| Valine, leucine and isoleucine degradation | ↑ | 35 | 5 | 3 | 0.041607 |
| Cysteine and methionine metabolism | ↓ | 33 | 5 | 5 | 0.008523 |
| Inositol phosphate metabolism | ↓ | 21 | 5 | 5 | 0.008523 |
| Phosphatidylinositol signaling system | ↓ | 17 | 4 | 4 | 0.012936 |
| Purine metabolism | ↓ | 66 | 9 | 7 | 0.013601 |
| N-Glycan biosynthesis | ↓ | 38 | 7 | 5 | 0.029627 |
| Citrate cycle (TCA cycle) | ↓ | 16 | 4 | 3 | 0.045722 |
| Tyrosine metabolism | ↓ | 42 | 6 | 4 | 0.046601 |
| Glyoxylate and dicarboxylate metabolism | ↓ | 31 | 6 | 4 | 0.046601 |
| Porphyrin and chlorophyll metabolism | ↓ | 27 | 6 | 4 | 0.046601 |
| Taurine and hypotaurine metabolism | ↓ | 8 | 2 | 2 | 0.048977 |
| Glycosphingolipid biosynthesis | ↓ | 15 | 2 | 2 | 0.048977 |
| Pyruvate metabolism | ↓ | 19 | 2 | 2 | 0.048977 |
Germ-free mice had distinct metabolomic profiles compared to conventional mice, injured and uninjured
Global metabolomic profiling of mouse SF detected 13,488 metabolite features across all experimental groups. 4,689 metabolite features were identified as significant by ANOVA (FDR-adjusted p-values<0.05). Unsupervised analyses including PCA and HCA found metabolomic variation between mouse SF cohorts, while FDR-corrected t-tests, and volcano plot analyses identified differentially regulated metabolites.
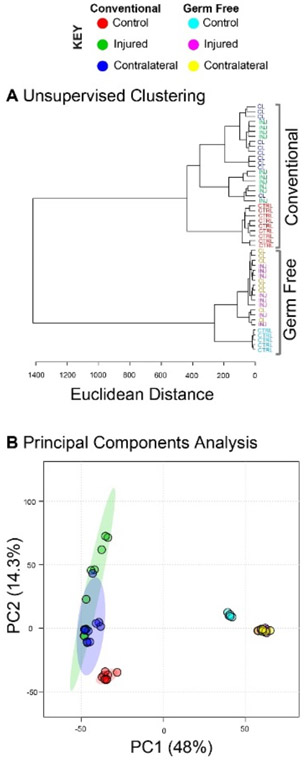
PCA and HCA revealed two large clusters that corresponded to GF and conventional mice, confirming that the presence of the gut microbiome influences the overall joint metabolome (Figure 4A-B). Conventional mice displayed greater overall variability as shown by increased distance between samples in PCA ordination space and increased branch lengths in HCA dendrogram (Figure 4A). Similarly, comparing synovial fluid of naïve, uninjured mice (Figure 4), there was dramatically less variability in the GF mice. Unsupervised clustering resulted in complete separation of GF and conventional samples (Figure 4B). Taken together, these results showed that the presence of a gut microbiome produces a distinct joint metabolome with greater variability between individuals.
Figure 4:
Global metabolomic profiles are significantly different between germ-free and conventional mice. The total of 13,488 metabolites were subjected to both unsupervised hierarchical clustering analysis and principal components analysis. (A) Unsupervised hierarchical clustering analysis finds distinct clusters between the metabolomic profiles of germ-free and conventional mouse synovial fluid. Furthermore, within both groups the naïve controls were distinct from both the contralateral and injured knees of injured mice. (B) Principal components analysis indicates that both injury and germ-free status drive differences in global metabolomic profiles.
After injury, the metabolomes of both GF and conventional mice shifted markedly from their naïve, uninjured counterparts (Figure 4). Furthermore, there was considerable overlap between injured and contralateral samples in both conventional and GF mice, suggesting injury produces a systemic response that influences both joint metabolomes.
Metabolomic profiles also suggested that GF mice were less sensitive to injury than conventional mice. In HCA branch lengths between dendrogram clusters represent differences in global metabolomic profiles, or dissimilarity. Within conventional mice, naïve individuals had a dissimilarity (Euclidean distance) of approximately 450 from injured mice (Figure 4A). The same comparison in GF mice yielded a dissimilarity of only 275, suggesting that the lack of a microbiome results in decreased metabolomic plasticity following injury.
Joint metabolome in germ-free and conventional naïve mice
To confirm that GF mice have a distinct joint metabolome than conventional mice in uninjured control conditions, a pairwise analysis of naïve experimental groups was conducted (GF-ctrl vs. Conv-ctrl). A total of 12,306 metabolite features were detected between GF and conventional naïve cohorts, with 4382 features identified as significantly different by Student’s T-test (FDR-adjusted p-value<0.05).
Global differences between GF and conventional naïve mice were assessed with unsupervised PCA and HCA. Unsupervised analyses showed clustering of SF samples within their respective cohorts, with greater variability within the C57-ctrl cohort than GF-ctrl (Figure 3D-E). Specific differences were identified using volcano plot analysis. Volcano plot analysis identified 3078 metabolite features as significantly upregulated and 1279 metabolite features as significantly downregulated in GF-ctrl compared to Conventional mice (Figure 3F).
Metabolic response to injury in germ-free injured and conventional mice
To assess the distinct response to injury in GF compared to conventional mice, pairwise analyses were conducted using only injured experimental groups (GF-inj vs. Conv-inj). A total of 12,075 metabolite features were detected between GF and conventional injured cohorts, with 3552 features identified as significantly different by Student’s T-test (FDR-adjusted p-value<0.05).
Global differences between GF and conventional injured mice were assessed with unsupervised PCA and HCA. Both PCA and HCA showed distinct clustering of SF samples within their respective cohorts (Figure 5A-B). However, PCA and HCA once again showed greater variability within the Conv -inj mice cohort as shown by less tight clustering of samples compared to GF-inj samples (Figure 5A-B).
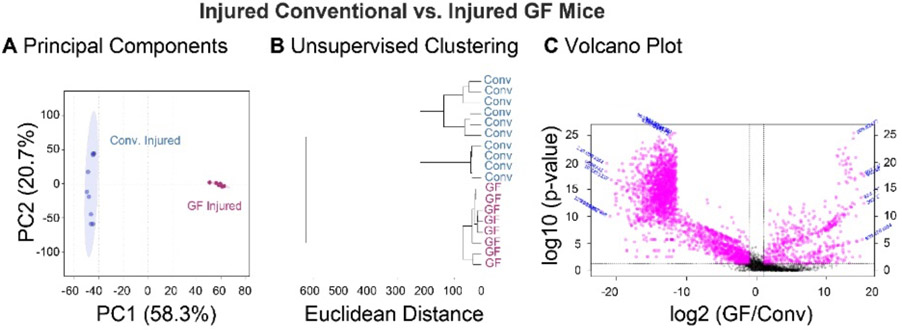
Figure 5:
Global metabolomic profiles of synovial fluid are driven by gut microbiota. Synovial fluid metabolites compared between injured germ free and conventional mice. (A) Principal components analysis finds complete separation between germ-free and conventional metabolomic profiles, with 58.3% and 20.7% of the variance associated with PC1 and PC2, respectively. (B) Germ free and conventional mice separate into two distinct clusters by unsupervised hierarchical clustering analysis. (C) 2728 metabolite features were significantly downregulated, and 578 metabolite features were significantly upregulated in GF-inj compared to Conv-inj mice.
Volcano plot analysis was used to identify specific metabolite features that were significantly upregulated or downregulated in GF compared to conventional injured mice. 2728 metabolite features were significantly downregulated, and 578 metabolite features were significantly upregulated in GF-inj compared to Conv -inj mice (Figure 5C). Metabolite features that were significantly downregulated in GF-inj compared to Conv -inj mice mapped to arachidonic acid metabolism, the TCA cycle, porphyrin metabolism, arginine and proline metabolism, and pyruvate metabolism (Table 3). Metabolite features significantly upregulated in GF-inj compared to Conv -inj mapped to valine, leucine, and isoleucine degradation, porphyrin metabolism, glycerophospholipid metabolism, arachidonic acid metabolism, fatty acid degradation, N-glycan biosynthesis, propanoate metabolism, and pantothenate and Coenzyme A (CoA) biosynthesis (Table 3).
Table 3.
Pathways altered in injured Germ-Free mice. Metabolite features identified by volcano plot analysis as significantly upregulated or downregulated in injured Germ Free mice compared with injured conventional were mapped to KEGG metabolic pathways. Pathways are reported with directionality (upregulated or downregulated); the total number of metabolites in the pathway, total number of metabolite features detected in samples in that pathway, total number of significant metabolite features (significant by both FDR-corrected p-value and fold change), and p-value for enrichment. Most significant pathways are reported up until arbitrary p-value cut-off for upregulated (p-value < 0.05) and downregulated pathways (p-value < 0.1). Full enrichment analyses are reported in Supplemental Tables 2 and 3.
| Pathway | Up/Down | Total metabolites in pathway |
Total pathway hits |
Significant pathway hits |
P-value |
|---|---|---|---|---|---|
| Valine, leucine and isoleucine degradation | ↑ | 40 | 16 | 8 | 0.012306 |
| Porphyrin metabolism | ↑ | 30 | 12 | 5 | 0.020232 |
| Glycerophospholipid metabolism | ↑ | 36 | 9 | 4 | 0.022792 |
| Arachidonic acid metabolism | ↑ | 36 | 13 | 5 | 0.023064 |
| Fatty acid degradation | ↑ | 39 | 7 | 3 | 0.033277 |
| N-Glycan biosynthesis | ↑ | 41 | 3 | 2 | 0.036671 |
| Propanoate metabolism | ↑ | 23 | 8 | 3 | 0.040544 |
| Pantothenate and CoA biosynthesis | ↑ | 19 | 8 | 3 | 0.040544 |
| Arachidonic acid metabolism | ↓ | 36 | 13 | 13 | 0.062431 |
| Citrate cycle (TCA cycle) | ↓ | 20 | 12 | 12 | 0.07051 |
| Porphyrin metabolism | ↓ | 30 | 12 | 12 | 0.07051 |
| Arginine and proline metabolism | ↓ | 38 | 10 | 10 | 0.091788 |
| Pyruvate metabolism | ↓ | 22 | 10 | 10 | 0.091788 |
Discussion
The goal of this study was to establish new mouse modeling techniques to experimentally evaluate host response to acute OA-associated injury in the absence of a microbiome. Since we found no evidence that GF mice became colonized by microorganisms following injury, this study provides direct proof-of-principle that such experimental manipulations can be achieved in a sterile setting. Although technically challenging[34] gnotobiotic (i.e. germ free) mice have provided great insight into interactions between bacterial and mammalian metabolism[35] and similar approaches can be used to further examine gnotobiotic, or defined microbiome, impacts on mammalian host response following joint injury.
Inflammation plays a role in the development of OA in humans[1-4] as well as in the development of OA risk factors, including age, obesity, and joint trauma. After joint trauma there is an initial acute inflammatory response followed by low-grade chronic inflammation[6]. Initial inflammatory responses from DAMPs promote a state of stress[36, 37], and it is increasingly clear that other endogenous signals from the microbiome further exacerbate pro-inflammatory responses[11, 38].
Bacteria in the mammalian gut play key roles in many diseases associated with chronic inflammation, including type 2 diabetes, rheumatoid arthritis, and ankylosing spondylitis[39-42]. Given that chronic inflammation is now an established contributor to OA progression, a better understanding of the impact of the overall gut microbiome and that of individual members as activators (pro-inflammatory) and regulators (anti-inflammatory) of OA-related inflammation promises to yield new targets for prevention and treatment. Furthermore, because joint trauma increases OA risk and because these patients typically only seek care shortly after injury, determining how the gut microbiome influences PTOA over a longer recovery period is critically important.
It is well-known that immunity in GF mice is markedly different from conventional counterparts with observed deficits in overall numbers and distribution of leukocytes[43]. These deficits lead to altered acute responses to stress in other injury-induced models[44], including lymph node trafficking. Similarly, increased bone loss is associated with inflammatory diseases via an imbalance in bone resorption and formation [45].
The results of this study provide a global metabolomic profile of mouse early PTOA, identifying many pathways affected by injury. Consistent with our previous work analyzing metabolic phenotypes of OA human SF [28], the metabolic phenotype of early PTOA was associated with perturbations in structural deterioration pathways (N-glycan biosynthesis), amino acid metabolism, fatty acid and lipid metabolism (fatty acid degradation, glycosphingolipid biosynthesis), energy metabolism (pyruvate metabolism, TCA cycle), and cell signaling pathways (phosphatidylinositol signaling system, inositol phosphate metabolism). These results further support the involvement of these metabolic shifts in OA pathogenesis.
In our study, synovial fluid metabolomes between injured and naïve mice were more distinct from one another in conventional compared to GF mice. This was further supported by μCT data. Collectively these data suggest that the GF phenotype is protective against the development of OA and the lack of a microbiome may slow the progression of disease. Metabolomic analyses also showed greater variability within conventional versus GF mice, which is consistent with known differences in immune cell activation/maturation and in turn, responses to a diverse community of microorganisms.
Moreover, the observed alterations in joint metabolomic profiles are consistent with different inflammatory responses in conventional versus GF mice. For example, fatty acid degradation, pantothenate and CoA biosynthesis, and arachidonic acid metabolism identified in volcano plots are known to play key roles in inflammation[46, 47]. Cellular fatty acid degradation is typically accomplished by beta oxidation, which is an important anti-inflammatory mediator of innate immunity[48]. Interestingly, this pathway was significantly upregulated in GF versus conventional mouse joint metabolome. While these results suggest that the absence of a microbiome promotes anti-inflammatory processes via fatty acid degradation, other studies report that short-chain fatty acids (SCFAs) produced by the microbiome also suppress immune functions[49]. Therefore, the effect of SCFAs may not be sufficient to counterbalance the skewed immune responses of GF mice.
Pantothenate, also known as vitamin B5, and arachidonic acid (AA) were other known mediators of the innate immunity associated with the GF joint metabolome. One study found that B5 had pro-inflammatory effects during Mycobacterium tuberculosis infection[50], while another study found the overall vitamin B complex (B1, B2, B3, B5, B6, B12) suppressed local inflammation after peripheral nerve transection[51]. Furthermore, B5 is a precursor to CoA, which plays a role in regulating the innate immune system[52]. AA is released from the lipid bilayer in response to injury or stress and initiates a series of signaling cascades that can trigger inflammation[53]. While our results suggest these pathways help explain observed differences in response to joint injury in the context of a microbiome, neither have received much attention with respect to OA development.
An important difference between previously published approaches and ours is the use of the non-invasive PTOA mouse model, which has been well-established to initiate rapid loss of trabecular bone as early as day seven post-injury[21]. The present results show smaller changes than prior studies which may reflect the older mice (20 week) used in this study [54, 55]. Microstructural differences between injured and contralateral joints observed in conventional mice were not observed in GF mice, supporting the conclusion that immune deficits in mice lacking a microbiome protected them against rapid trabecular bone loss that is typical in this PTOA mouse model.
This work has important limitations. First, GF mice are raised in isolators, which can alter behavior[56]. Thus, differences observed between conventional and GF mice may result from additional factors besides the lack of a microbiome, such as differences in activity levels or body composition that could potentially confound our results. Second, while the CSAC method of rodent synovial fluid collection is established[25], this method can be challenging. In preliminary studies, this method consistently yielded thousands of metabolites compared with blank and neat controls indicating that synovial fluid metabolites can pe profiled using the CSAC method. Third, per IACUC requirements, injured mice received pain medication after injury. While pharmacokinetics studies[57] suggest that plasma concentrations of sustained-release buprenorphine diminish before the 7-day endpoint, the observed effects of the GF response to injury might have been greater without buprenorphine. Fourth, while all mice limped after injury, ACL tears were not confirmed through laxity testing which would provide further insight into these data. Furthermore, microCT analysis was only performed on the injured and contralateral legs of injured mice. This potentially explains the smaller changes in epiphyseal bone parameters compared to previous studies[21]. Finally, a priori power calculation was not possible because there have been no prior joint injury studies in GF mice. Therefore this study utilized available mice and it is not clear if non-significant findings represent true negatives or result from low power.
In conclusion, to our knowledge this study is the first to investigate the role of the microbiome in the acute phase after joint injury. These results demonstrate that the gut microbiome drives key injury responses, specifically via perturbations in the innate inflammatory response to injury. Thus, microbiota may be a potential therapeutic target during the acute phase after joint trauma to abrogate the innate immune response and thereby diminish some of the first events that result in PTOA.
Supplementary Material
Acknowledgements
We thank Dr. Tim Griffin for showing us the CSAC method, and we acknowledge Dr. Ladean McKittrick for his assistance with μCT imaging. We thank the Montana State University Proteomics, Metabolomics, and Mass Spectrometry Facility for assistance with sample analysis. This facility is supported in part by funding from the Murdock Charitable Trust and NIH P20GM103474 of the IDEA program and AG007996. This research is funded by the NSF (CMMI 1554708 to RKJ), NIH (R01AR073964 to RKJ, R01AR075013 to BAC), Murdock Charitable Trust (FSU-2017207 to AKH), and Montana Space Grant Consortium for student funding.
Role of the funding source
The funding sources played no role in the design or execution of this study.
Footnotes
Conflict of interest
The authors have no conflicts of interests to disclose. Dr. June owns stock in Beartooth Biotech which was not involved in this study.
References
- 1.Liu-Bryan R. Synovium and the innate inflammatory network in osteoarthritis progression. Curr Rheumatol Rep 2013; 15: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol 2014; 28: 5–15. [DOI] [PubMed] [Google Scholar]
- 3.Houard X, Goldring MB, Berenbaum F. Homeostatic mechanisms in articular cartilage and role of inflammation in osteoarthritis. Curr Rheumatol Rep 2013; 15: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu-Bryan R, Terkeltaub R. Emerging regulators of the inflammatory process in osteoarthritis. Nat Rev Rheumatol 2015; 11: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldring MB, Otero M. Inflammation in osteoarthritis. Curr Opin Rheumatol 2011; 23: 471–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberthal J, Sambamurthy N, Scanzello CR. Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthritis Cartilage 2015; 23: 1825–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecil DL, Appleton CT, Polewski MD, Mort JS, Schmidt AM, Bendele A, et al. The pattern recognition receptor CD36 is a chondrocyte hypertrophy marker associated with suppression of catabolic responses and promotion of repair responses to inflammatory stimuli. J Immunol 2009; 182: 5024–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin C, Frayssinet P, Pelker R, Cwirka D, Hu B, Vignery A, et al. NLRP3 inflammasome plays a critical role in the pathogenesis of hydroxyapatite-associated arthropathy. Proc Natl Acad Sci U S A 2011; 108: 14867–14872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu-Bryan R, Terkeltaub R. The growing array of innate inflammatory ignition switches in osteoarthritis. Arthritis Rheum 2012; 64: 2055–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zreiqat H, Belluoccio D, Smith MM, Wilson R, Rowley LA, Jones K, et al. S100A8 and S100A9 in experimental osteoarthritis. Arthritis Res Ther 2010; 12: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 2009; 22: 240–273, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favazzo LJ, Hendesi H, Villani DA, Soniwala S, Dar QA, Schott EM, et al. The gut microbiome-joint connection: implications in osteoarthritis. Curr Opin Rheumatol 2020; 32: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins KH, Paul HA, Reimer RA, Seerattan RA, Hart DA, Herzog W. Relationship between inflammation, the gut microbiota, and metabolic osteoarthritis development: studies in a rat model. Osteoarthritis Cartilage 2015; 23: 1989–1998. [DOI] [PubMed] [Google Scholar]
- 14.Boer CG, Radjabzadeh D, Medina-Gomez C, Garmaeva S, Schiphof D, Arp P, et al. Intestinal microbiome composition and its relation to joint pain and inflammation. Nat Commun 2019; 10: 4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeser RF, Pathmasiri W, Sumner SJ, McRitchie S, Beavers D, Saxena P, et al. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthritis Cartilage 2016; 24: 1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn CM, Velasco C, Rivas A, Andrews M, Garman C, Jacob PB, et al. Identification of cartilage microbial DNA signatures and associations with knee and hip osteoarthritis. Arthritis Rheumatol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulici V, Kelley KL, Azcarate-Peril MA, Cleveland RJ, Sartor RB, Schwartz TA, et al. Osteoarthritis induced by destabilization of the medial meniscus is reduced in germ-free mice. Osteoarthritis Cartilage 2018; 26: 1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guss JD, Ziemian SN, Luna M, Sandoval TN, Holyoak DT, Guisado GG, et al. The effects of metabolic syndrome, obesity, and the gut microbiome on load-induced osteoarthritis. Osteoarthritis Cartilage 2019; 27: 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christiansen BA, Guilak F, Lockwood KA, Olson SA, Pitsillides AA, Sandell LJ, et al. Non-invasive mouse models of post-traumatic osteoarthritis. Osteoarthritis Cartilage 2015; 23: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little CB, Hunter DJ. Post-traumatic osteoarthritis: from mouse models to clinical trials. Nat Rev Rheumatol 2013; 9: 485–497. [DOI] [PubMed] [Google Scholar]
- 21.Christiansen BA, Anderson MJ, Lee CA, Williams JC, Yik JH, Haudenschild DR. Musculoskeletal changes following non-invasive knee injury using a novel mouse model of post-traumatic osteoarthritis. Osteoarthritis Cartilage 2012; 20: 773–782. [DOI] [PubMed] [Google Scholar]
- 22.Lockwood KA, Chu BT, Anderson MJ, Haudenschild DR, Christiansen BA. Comparison of loading rate-dependent injury modes in a murine model of post-traumatic osteoarthritis. J Orthop Res 2014; 32: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontaine CA, Skorupski AM, Vowles CJ, Anderson NE, Poe SA, Eaton KA. How free of germs is germ-free? Detection of bacterial contamination in a germ free mouse unit. Gut Microbes 2015; 6: 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002; 148: 257–266. [DOI] [PubMed] [Google Scholar]
- 25.Seifer DR, Furman BD, Guilak F, Olson SA, Brooks SC 3rd, Kraus VB. Novel synovial fluid recovery method allows for quantification of a marker of arthritis in mice. Osteoarthritis Cartilage 2008; 16: 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Hwang J, Xuan J, Jung YH, Cha HS, Kim KH. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One 2014; 9: e97501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson AK, Rawle RA, Adams E, Greenwood MC, Bothner B, June RK. Application of global metabolomic profiling of synovial fluid for osteoarthritis biomarkers. Biochem Biophys Res Commun 2018; 499: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson AK, Rawle RA, Wallace CW, Brooks EG, Adams E, Greenwood MC, et al. Characterization of synovial fluid metabolomic phenotypes of cartilage morphological changes associated with osteoarthritis. Osteoarthritis Cartilage 2019; 27: 1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 2010; 25: 1468–1486. [DOI] [PubMed] [Google Scholar]
- 30.Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics 2016; 55: 14 10 11–14 10 91. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013; 9: e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabay O, Vicenty J, Smith D, Tiffany L, Ascher J, Curry T, et al. Using a Model of Germ-Free Animals to Study the Impact of Gut Microbiome in Research: A Step by Step Sterility Setting and Management. Methods Protoc 2020; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sjogren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res 2012; 27: 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osbelt L, Thiemann S, Smit N, Lesker TR, Schroter M, Galvez EJC, et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog 2020; 16: e1008448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 2009; 106: 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piccinini AM, Midwood KS. DAMPening inflammation by modulating TLR signalling. Mediators Inflamm 2010; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg JH, Rai V, Dilisio MF, Agrawal DK. Damage-associated molecular patterns in the pathogenesis of osteoarthritis: potentially novel therapeutic targets. Mol Cell Biochem 2017; 434: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015; 163: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018; 175: 947–961 e917. [DOI] [PubMed] [Google Scholar]
- 40.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018; 67: 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minter MR, Zhang C, Leone V, Ringus DL, Zhang X, Oyler-Castrillo P, et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer's disease. Sci Rep 2016; 6: 30028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32: 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005; 122: 107–118. [DOI] [PubMed] [Google Scholar]
- 44.van de Wouw M, Lyte JM, Boehme M, Sichetti M, Moloney G, Goodson MS, et al. The role of the microbiota in acute stress-induced myeloid immune cell trafficking. Brain Behav Immun 2020; 84: 209–217. [DOI] [PubMed] [Google Scholar]
- 45.Hardy R, Cooper MS. Bone loss in inflammatory disorders. J Endocrinol 2009; 201: 309–320. [DOI] [PubMed] [Google Scholar]
- 46.Nitto T, Onodera K. Linkage between coenzyme a metabolism and inflammation: roles of pantetheinase. J Pharmacol Sci 2013; 123: 1–8. [DOI] [PubMed] [Google Scholar]
- 47.Innes JK, Calder PC. Omega-6 fatty acids and inflammation. Prostaglandins Leukot Essent Fatty Acids 2018; 132: 41–48. [DOI] [PubMed] [Google Scholar]
- 48.Namgaladze D, Brune B. Macrophage fatty acid oxidation and its roles in macrophage polarization and fatty acid-induced inflammation. Biochim Biophys Acta 2016; 1861: 1796–1807. [DOI] [PubMed] [Google Scholar]
- 49.Sivaprakasam S, Prasad PD, Singh N. Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol Ther 2016; 164: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He W, Hu S, Du X, Wen Q, Zhong XP, Zhou X, et al. Vitamin B5 Reduces Bacterial Growth via Regulating Innate Immunity and Adaptive Immunity in Mice Infected with Mycobacterium tuberculosis. Front Immunol 2018; 9: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehmedah A, Nedeljkovic P, Dacic S, Repac J, Draskovic Pavlovic B, Vucevic D, et al. Vitamin B Complex Treatment Attenuates Local Inflammation after Peripheral Nerve Injury. Molecules 2019; 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung S, Kim MK, Choi BY. The long-term relationship between dietary pantothenic acid (vitamin B5) intake and C-reactive protein concentration in adults aged 40 years and older. Nutr Metab Cardiovasc Dis 2017; 27: 806–816. [DOI] [PubMed] [Google Scholar]
- 53.Dhall S, Wijesinghe DS, Karim ZA, Castro A, Vemana HP, Khasawneh FT, et al. Arachidonic acid-derived signaling lipids and functions in impaired healing. Wound Repair Regen 2015; 23: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khorasani MS, Diko S, Hsia AW, Anderson MJ, Genetos DC, Haudenschild DR, et al. Effect of alendronate on post-traumatic osteoarthritis induced by anterior cruciate ligament rupture in mice. Arthritis Res Ther 2015; 17: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson MJ, Diko S, Baehr LM, Baar K, Bodine SC, Christiansen BA. Contribution of mechanical unloading to trabecular bone loss following non-invasive knee injury in mice. J Orthop Res 2016; 34: 1680–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niimi K, Takahashi E. New system to examine the activity and water and food intake of germ-free mice in a sealed positive-pressure cage. Heliyon 2019; 5: e02176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kendall LV, Hansen RJ, Dorsey K, Kang S, Lunghofer PJ, Gustafson DL. Pharmacokinetics of sustained-release analgesics in mice. J Am Assoc Lab Anim Sci 2014; 53: 478–484. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.