Abstract
Introduction
Intrinsically disordered proteins (IDPs) and regions (IDRs) lack stable three-dimensional structure making drug discovery challenging. A validated therapeutic target for diseases such as prostate cancer is the androgen receptor (AR) which has a disordered amino-terminal domain (NTD) that contains all of its transcriptional activity. Drug discovery against the AR-NTD is of intense interest as a potential treatment for disease such as advanced prostate cancer that is driven by truncated constitutively active splice variants of AR that lack the C-terminal ligand-binding domain (LBD).
Areas covered
This article presents an overview of the relevance of AR and its intrinsically disordered NTD as a drug target. AR structure and approaches to blocking AR transcriptional activity are discussed. The discovery of small molecules, including the libraries used, proven binders to the AR-NTD, and site of interaction of these small molecules in the AR-NTD are presented along with discussion of the Phase I clinical trial.
Expert opinion
The lack of drugs in the clinic that directly bind IDPs/IDRs reflects the difficulty of targeting these proteins and obtaining specificity. However, it may also point to an inappropriateness of too closely borrowing concepts and resources from drug discovery to folded proteins.
Keywords: androgen receptor, clinical trial, drug discovery, EPI-002, intrinsically disordered protein, mechanism of action, N-terminal domain inhibitor, prostate cancer, ralaniten, sintokamides
1. Introduction
1.1. Androgens
Androgens such as testosterone and dihydrotestosterone (DHT) are abundantly produced in the mature male predominantly in the testes with some contribution from the adrenal glands. The biological activity of androgen is mediated by the androgen receptor (AR) which is a ligand-activated transcription factor that belongs to the steroid receptor family. In the male, androgens and functional AR are essential for sexual differentiation, maintenance of spermatogenesis, and male gonadotropin regulation. Androgen-dependent tissue such as the prostate relies on functional androgen signaling. When levels of androgen are reduced in a mature male, the prostate will involute with apoptosis of prostate luminal epithelial cells. It is this dependency of prostate tissue on androgens that underlies the rationale of androgen deprivation therapy (ADT) as a systemic approach for advanced prostate cancer. ADT and inhibition of AR transcriptional activity has been the focus of therapeutics for prostate cancer for the past six decades. In addition to prostate cancer, the androgen axis plays a role in other pathologies such as alopecia, polycystic ovarian syndrome, spinal bulbar muscular atrophy (SBMA), androgen insensitivity syndrome, and some breast cancers.
1.2. Androgen receptor
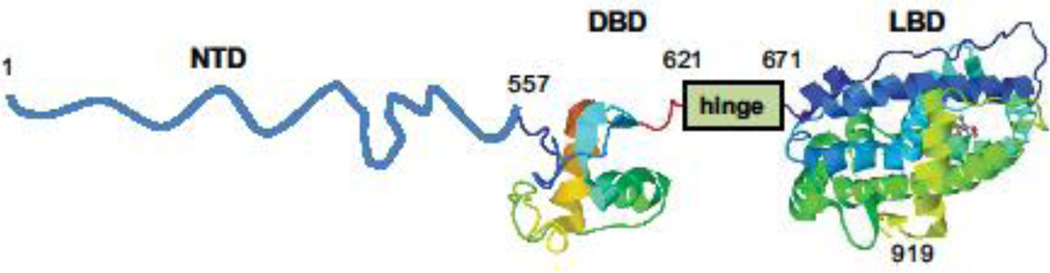
Full-length AR (NR3C4) ranges in size due to polymorphic variation of several repeats regions in its N-terminal domain (NTD) [1]. Most literature refers to a 910 or 919 amino acid full-length AR that is comprised of an intrinsically disordered NTD (547 to 556 residues), a folded DNA-binding domain (DBD; 65 residues), a disordered hinge region (49 residues) and folded C-terminal ligand-binding domain (LBD; 249 residues) that contains the ligand-binding pocket (Fig 1).
Fig 1.
Structure of the androgen receptor. N-terminal domain (NTD) is intrinsically disordered. DNA-binding domain (DBD) is an ordered and folded domain. Hinge region is disordered. Ligand-binding domain (LBD) is an ordered and folded domain.
1.2.1. AR-DBD and Hinge Region
The AR is a transcription factor which interacts with DNA through its structured DNA-binding domain (DBD) that has a resolved crystal structure [2]. This region contains a P-box and a D-box in which two zinc fingers are essential for AR transcriptional activity. Drug design against AR-DBD would be useful to block the transcriptional activity of full-length AR and truncated AR splice variants (AR-Vs) that lack AR-LBD as described below. Due to the high sequence similarity of this domain (77–80%) with other members of the steroid receptor family, finding a drug with specificity to AR-DBD may be difficult but efforts are underway [3]. AR-DBD is linked to its LBD through an unstructured hinge region which plays a role in nuclear translocation but other functions have also been noted and are regulated by phosphorylation, acetylation, methylation and ubiquitination [4].
1.2.2. AR-LBD and Antiandrogens
The C-terminus LBD is comprised of 11 α-helices which encompass a ligand-binding pocket. When androgen binds, there is a shift in conformation to reposition helix 12 over the ligand-binding pocket to create the AF-2 surface for interaction with coactivators [5,6]. In addition to binding androgen, the AR-LBD is the direct or indirect target for all currently FDA-approved drugs against the androgen axis. Drugs indirectly targeting the AR to reduce the levels of androgen that bind to the AR-LBD include LHRH analogues and CYP17 inhibitors (abiraterone) that blocks steroidogenesis. Due to the three-dimensional structure of AR-LBD and resolved crystal structure [7], structure-based drug discovery is possible and thus there are an abundance of drugs that have been developed that directly bind to this domain. These drugs include both agonists (selective AR modifiers or SARMs) as well as antagonists that are called, “antiandrogens”. There are steroidal and non-steroidal antiandrogens. For prostate cancer, the non-steroidal antiandrogens (stem name “lutamide”) are the most frequently used and include flutamide, nilutamide, bicalutamide, enzalutamide, apalutamide, and darolutamide. The mechanism of action for antiandrogens is to compete with androgens for the AR-LBD and induce an AR conformation that is not transcriptionally active. DHT is the most relevant androgen for prostate cancer and also the most potent with a binding affinity in the low nM range. This means that for an antiandrogen to be efficacious it must have very strong affinity to be able to compete with DHT for the ligand-binding pocket in AR-LBD. For these reasons, ADT to reduce levels of androgen (DHT) is continued while patients receive newer and more potent second generation non-steroidal antiandrogens. Unfortunately, resistance to these therapies will develop by mechanisms that predominantly involve: 1) gain-of-function mutations in the AR-LBD that result in promiscuous transactivation by other steroids or yielding a receptor where the antiandrogen behaves as an agonist; or 2) expression of truncated constitutively active AR-Vs such as AR-V7 that lack the AR-LBD. Hence antiandrogens have lost their binding site on AR-Vs and thereby have no effect. AR is unique from other steroid hormone receptors in that no transcriptional activity is attributed to its LBD {activation function-2 (AF-2) has no identified transactivation unit-2 (tau-2)}, but rather all transcriptional activity resides in it NTD [8–12].
1.2.3. Intrinsically disordered AR-NTD
AR-NTD is largely unstructured and described as having limited stable secondary structure which can be induced by interactions with binding partners to increase α-helical content and thereby conforms to a molten-globule-like conformation referred to as ‘collapsed disordered’ [13–15] (Fig 2 A). This domain is the most abundantly post-translationally modified of the AR and acts as a hub for interactions with many other proteins (Fig 2B). Most importantly is interaction of this domain with the basal transcriptional machinery that is necessary for its transcriptional activity. Within AR-NTD is AF-1 which is estimated to have 13% helical secondary structure but this can increase upon interaction with a binding partner [13,14]. AF-1 is comprised of two transactivation units 1 and 5 (Tau-1 and Tau-5). Tau-1 is comprised of amino acid residues 101–370 of which a large number are acidic amino acids. Tau-5 is comprised amino acid residues 360–485 and is not acidic. Interestingly AR-NTD harbors several repeat regions for glutamine (polyglutamine tract or polyQ), proline, alanine, and glycine.
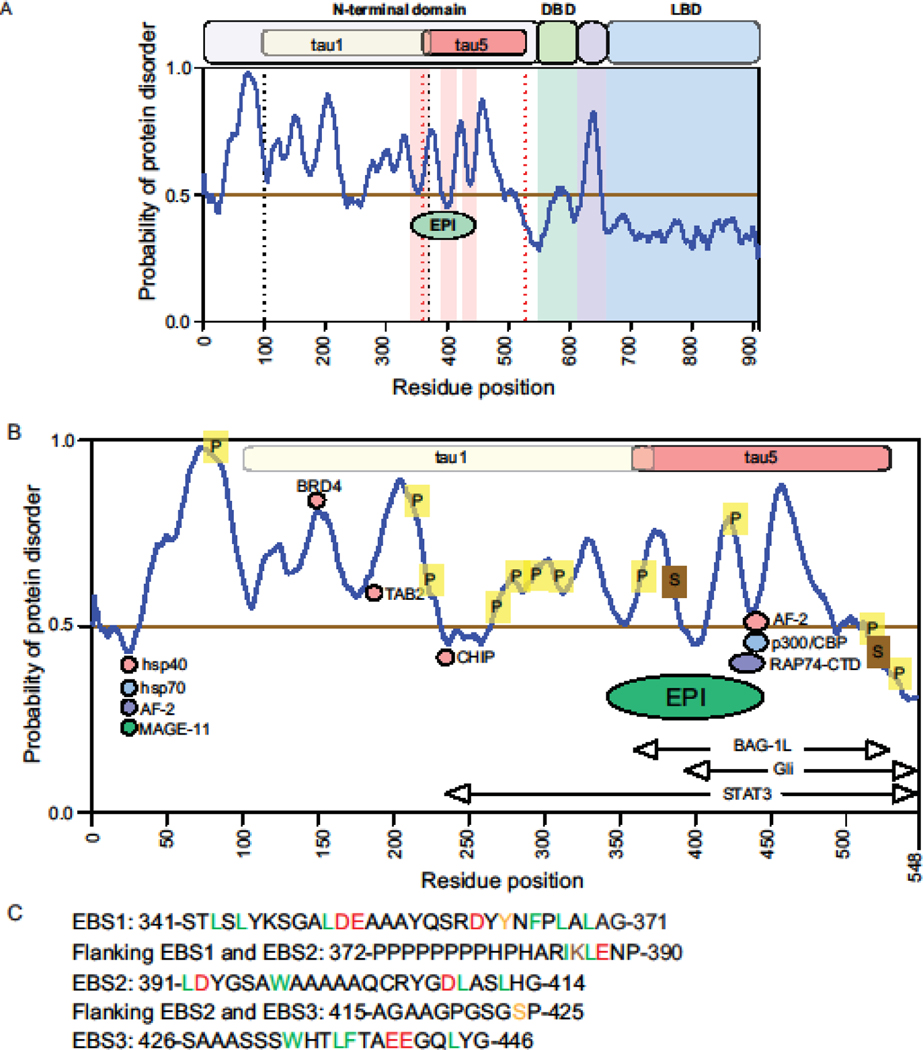
Fig 2.
Ronn plot of the androgen receptor with the EPI-002 binding site (EBS1–3). A. The probability of protein disorder across the amino acid residues of androgen receptor. A probability score below 0.5 is considered ordered and above 0.5 as disordered. Transactivation units (Tau) 1 and 5 are shown within the N-terminal domain. The EPI-002 binding site is depicted with the regions of interaction shown in light red. DBD, DNA-binding domain; Hinge region in purple; LBD, ligand-binding domain. B. Regions of posttranslational modification and interactions with some other proteins on the N-terminal domain in context to EPI-002 binding site. Within the EPI-binding site are regions for phosphorylation (P) and sumolyation (S). Protein interactions shown include chaperones hsp40 and hsp90; AF-2, activation function-2 in the AR LBD for N/C interaction; RAP74 of the basal transcriptional machinery; p300/CBP, BRD4, TAB2, CHIP, MAGE-11, BAG1L, Gli, and STAT3. C. Residues in the three EPI-002 binding sites (EBS) and the flanking residues. EBS1, EPI-002 binding site 1; EBS2, EPI-002 binding site 2; EBS3, EPI-002 binding site 3.
The variable length polyQ tract is the most studied of these repeat regions. Its length impacts AR solubility and transcriptional activity. ARs with short polyQ tracts tend to have increased transcriptional activity whereas a longer repeat region has less activity. PolyQ tracts longer than 37 residues tend to form cytotoxic fibrillar aggregates associated with SBMA. Helicity of the polyQtract is stabilized by H-bonds between the side chains of glutamine and the main carbonyl groups and helical structure of the tract correlates with its length [16]. Changes in conformation would presumably impact AR-NTD interactions with other proteins and thereby modulate its transcriptional activity in addition to its solubility and propensity to form fibrils. Androgens cause release of chaperone proteins Hsp40 and Hsp70 from the AR NTD to enable transcriptional activity but unfortunately also increase the propensity of aggregation. These chaperones bind to the 23FQNLF27 motif within the NTD with μM affinity [17]. In the absence of androgen or ligand, the apo-receptor is in a complex with chaperone proteins that facilitate AR’s solubility and ability to readily bind ligands [18]. Upon binding androgen, the 23FQNLF27 motif interacts as an α-helix with its AR C-terminal LBD (called N/C interaction) to mediate transcriptional activity [19]. The shift in conformation that occurs upon binding of androgen in AR-LBD results in dynamic changes in interactions with chaperones that reduce its solubility and improve its affinity for DNA thereby enabling nuclear translocation and transactivation. Application of a small molecule that binds Hsp70, JG-98, stabilizes interaction of AR-NTD with Hsp70 to promote AR degradation and reduce aggregation [17]. Thus the AR-NTD is a drug-target for multiple indications including SBMA as well as prostate cancer and other pathologies driven by AR.
2. Drugs Targeting Intrinsically Disordered Proteins and Regions
2.1. Characteristics of Intrinsically Disordered Proteins/Regions for Drug Development
Intrinsically disordered proteins (IDPs) or regions (IDRs) lack stable structure in isolation. This property of structural plasticity permits an IDP/R to exist as multiple and changing conformations that vary depending upon its interacting binding partners and or the protein’s environment. The lack of a stable binding site together with presumed flat large protein-protein interaction areas reflect the challenges in developing drugs to these targets and contrast with the long-term success of drug development to folded proteins with three-dimensional structures that is set in the “lock-and-key” model. Uncertainties to the feasibility of achieving specificity with small molecule inhibitors or activators and the laborious drug discovery approaches needed are probably the main reasons that only two drugs that directly bind to an IDP/IDR have reached clinical trials (ClinicalTrials.gov Identifiers: NCT02606123 and NCT00509132).
The disordered states and structures of IDP/Rs are dictated by the protein sequence and thus “intrinsic” to the encoding sequence. Amino acids that are predominantly found in these regions include those with charged groups and a low content of hydrophobic amino acid residues. Hence IDRs tend to have a high net charge and low hydrophobicity [20–22]. Cysteines form covalent bonds as sulfide bridges to stabilize protein structure when the environment is oxidizing, but under reducing conditions the bridges are broken and usually the protein will be disordered and inactivated [23,24]. Prolines also disrupt protein structure. The presence of aromatic residues within IDRs may reveal a molecular recognition region (MoRF). MoRF are defined as “short, interaction-prone segments of protein disorder that undergo disorder-to-order transitions upon specific binding, representing a specific class of intrinsically disordered regions that exhibit molecular recognition and binding functions” [25]. MoRF regions are of substantial interest in drug development due to their potential for intervention [26].
2.2. Targeting a folded binding partner versus direct binding to IDR
Proteins that are transcription factors such AR are enriched with IDRs. The ability for AR to regulate gene expression depends on reversible interactions with other proteins. Compared to folded/structured regions, IDRs have the advantage of high-specificity, poor-affinity interactions (micromolar to millimolar binding strength) which facilitate reversibility required for signaling [27]. Interaction of IDRs and their partners may shift the ensemble to a different conformation which may yield secondary structure. The most well-known example is the interaction of the disordered IDR of p53 with its ordered partner MDM2. Upon interaction, p53 becomes folded [28]. Drug development to p53 has been predominantly directed to the ordered binding partner MDM2’s site of interaction with p53. There are very few small molecules reported that directly interact with an IDR or IDP [29]. The IDR of c-Myc interacts with the IDR of its binding partner Max to form a folded complex that binds to target genes to regulate their expression. Because c-Myc is implied in so many cancers, there is substantial interest in developing drugs against this protein [30]. Some drug development approaches are directed to blocking binding of the folded c-Myc/Max complex to DNA as this avenue may be easier than directly targeting c-Myc IDR. For example, Omomyc miniprotein was designed to act as a dominant negative of Myc with mutated residues that are similar to the residues in the bHLH domain of Max [31]. Omomyc inhibits Myc transcriptional activity by multiple mechanisms including binding E boxes on DNA to inhibit binding of Myc/Max heterodimers and decreasing levels of Myc protein [32,33]. Cellular penetration and in vivo efficacy of Omomyc have been recently shown in preclinical models thereby supporting the potential of this approach for clinical development [34]. The small molecules that directly bind to c-Myc IDR that have been discovered bind to a short linear region of amino acids residues and these compounds can bind multiple sites independently [35]. An example is 10074-A4 that binds to multiple conformations of c-Myc on a short amino acid segment of approximately 10 residues [35]. None of these small molecules that directly bind to c-Myc IDR have reached clinical testing presumably due to lack of specificity inherent with the mechanism of binding. There are several recent excellent reviews on drug discovery to IDR/Ps [36–39], here we focus on AR-NTD. The first small molecule that directly binds to an IDR to be tested in clinical trials is the prodrug of ralaniten (EPI-002), called ralaniten-acetate (EPI-506), that directly binds AR-NTD [40–43].
3. Discovery of drugs that directly bind to AR-NTD IDR
3.1. AR-NTD
Interest in developing drugs to the AR-NTD predominantly comes from the discovery of constitutively active AR-Vs that lack LBD that are associated with resistance mechanisms in lethal castration-resistance prostate cancer (CRPC) [44–47]. Also, unlike its related steroid hormone receptors with transcriptional activity within AF-2 in their LBDs, all of the transcriptional activity of AR resides in its NTD [48]. Thus an inhibitor to the AR-NTD should block the transcriptional activities of all AR species. AR-NTD has little sequence similarity (<15%) to its most closely related steroid hormone receptors and thereby considered a drug target that could be highly specific.
AR is a hub for interactions with more than 170 different proteins with many of these interactions occurring with AR-NTD [49]. One of the most critical protein-protein interactions with AR-NTD required for transcriptional activity is with the basal transcriptional machinery that includes RAP74, the subunit of TFIIF that aids in recruitment of the initiation complex and binds to RNA polymerase II [50–52]. The region of AR-NTD (residues 423–446) essential for interaction with RAP74 lies within Tau-5 [53,54] and may contain a MoRF with aromatic residues W433, Y445 and F437. The affinity for interaction between this region of the AR-NTD and RAP74 is improved from the millimolar to micromolar range (KD=1749 μM to KD=702 μM) with phosphorylation of serine 424 of AR-NTD [54]. These data stress the importance of post-translational modifications within this region [54,55] and are consistent with the general observation for high specificity and poor affinity for protein-protein interactions with IDRs. An inhibitor with an IC50 in the higher μM range may have therapeutic value to block this weak interaction with RAP74 and AR-NTD unlike the antiandrogens that compete with DHT in the low nM range.
3.2. Small molecule libraries that generated “hits” to the AR-NTD
Starting in 2003, several available small molecule libraries were tested empirically over the next 17 years by the Sadar lab against a series of assays developed to discover small molecules that specifically inhibited the AR NTD. These libraries included the NCI Diversity set (2,100 compounds), the 50k microformat Diverset from Chembridge (50,000 compounds), and natural compounds libraries of marine sponge and invertebrates extracts (Professor RJ Andersen, Vancouver, BC). Over 52,000 compounds from the NCI or Chembridge libraries were tested with little to no hits. Fortunately the natural compounds libraries provided approximately 30 hits from unique extracts which were further fractionated to purification and isolation of the active compounds. Three of these compounds (niphatenones, sintokamides, and ralaniten) that directly interact with the AR-NTD have been published. The remaining active scaffolds and extracts continue to be optimized and characterized.
3.3. Niphatenones
Niphatenones were isolated from extracts of the marine sponge Niphates digitalis collected in Dominica [56]. Synthetic analogues of niphatenone were compared to the natural products for activity against AR activity and found to be more potent and that the Michael acceptor enone functionality is not required for activity [56]. An IC50 value of approximately 6 μM was measured for inhibiting transcriptional activity of full-length AR [57]. Niphatenone binds covalently to activation function-1 (AF1) region of the AR-NTD and inhibits the transcriptional activity of AR-Vs without affecting the transcriptional activity of the closely related progesterone receptor [57]. Unfortunately, niphatenone decreased glucocorticoid receptor (GR) transcriptional activity by a mechanism involving covalent binding to GR AF-1 [57]. Further development of these compounds was halted upon discovery of alkylation with glutathione [57].
3.4. Sintokamides
In 2008, the first of a series of reports was published that chlorinated peptides, sintokamides A to E, isolated from the marine sponge Dysidea sp. collected in Indonesia were potent inhibitors of AR-NTD [58]. Evidence that sintokamide A (SINT1) binds AR AF-1 region to specifically inhibit transactivation of AR NTD was provided [59]. Transcriptional activities of both full-length AR and AR-Vs were blocked. In vivo studies using human prostate cancer xenografts grown in castrated male mice revealed regression of tumors and reduced expression of an AR-regulated gene, prostate-specific antigen (PSA) [59]. Combination experiments of SINT1 with ralaniten (described below) had additive effects on AR transcriptional activities which implies that SINT1 binds to a site on AF-1 that is unique from ralaniten [59]. Based upon differences between the two compounds in blocking AR-NTD interaction with STAT3, it was proposed that SINT1 interacts possibly with Tau-1 rather than Tau-5 [59].
3.5. EPI-067 and Ralaniten analogues
3.5.1. Discovery and structure
The original EPI compound isolated from the marine sponge Geodia lindgreni collected in Papau New Guinea was EPI-067 [60,61]. This is not a natural compound and most likely of industrial origin based upon its structural resemblance to Bisphenol A Diglycidic Ether (BADGE). BADGE is a harmless metabolite of bisphenol A with no estrogenic or androgenic effects [62–65] and it cannot be converted by biological systems back to the estrogenic bisphenol A [63]. BADGE and its chlorohydrins are not carcinogenic nor genotoxic at high daily doses [66]. Structure activity relationship studies yielded EPI-002, as single stereoisomer of the mixture called EPI-001. These compounds were demonstrated to not be generally reactive as shown at physiological pH in vitro [59], in vivo using an radioactive imaging agent [67], and from patient clinical samples [43,68].
3.5.2. Mechanism of action
EPI analogues inhibit the transcriptional activities of full-length AR in response to androgen and AR-Vs. Inhibition of AR transcriptional activity by EPI is specific and it has no effect on the activities of related human steroid receptors. EPI analogues do not bind to the AR-LBD and consistent with this they do not compete with androgen in a competitive ligand-binding assay [40]. Upon binding androgen, the full-length AR translocates to the nucleus. Similarly, in the absence of androgens, some antiandrogens also cause the full-length AR to translocate to the nucleus and bind to DNA binding sites of target genes. EPI analogues do not induce AR nuclear translocation in the absence of androgen [40]. EPI blocks AR binding to its binding sites (androgen response elements) in the promoters and enhancers of target genes to decrease expression of these genes in response to androgens [40,41]. N/C interaction of AR is required for androgen-dependent transactivation of AR and is inhibited by EPI. CREB-binding protein (CBP) interacts with AR-NTD and is essential for AR transcriptional activity. EPI blocks this interaction with CBP as well as interaction with RAP74 [40]. Efficacy of EPI as a therapeutic for prostate cancer was provided using prostate cancer cells maintained in vitro as well as cell line and patient-derived xenografts, and the Herschberger assay [40].
3.5.3. Tau-5 Binding site for EPI
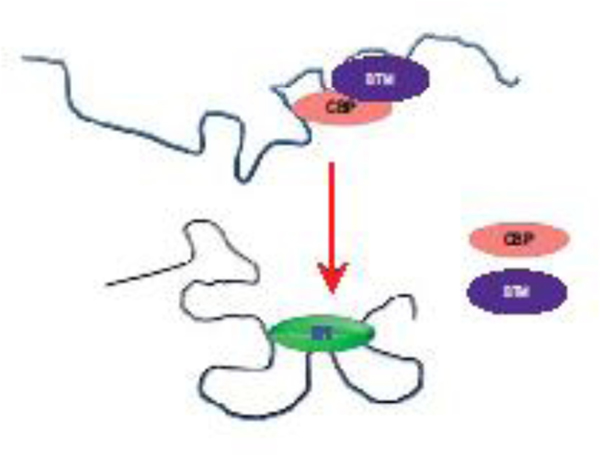
Direct interaction of the EPI analogues with the AF-1 region of AR was first shown with recombinant protein in a cell-free assay by fluorescence emission spectroscopy [40] and then later using Click-chemistry probes [41]. Due to the sensitivity of IDR conformations on their environment and protein-protein interactions, proof that EPI bound endogenous AR in living cells was important to ensure that studies with the recombinant protein in cell-free assays were not artifactual. To do this, the first demonstration of direct binding of a small molecule to an endogenous IDR in cells using both Click-chemistry probes and radiolabelled analogues was reported [41,67]. In vivo, a radiolabelled EPI analogue was injected into mice carrying both an AR-positive xenograft and an AR-negative xenograft. Only AR-positive tumors accumulated the radioactive compound thereby reaffirming the specificity of EPI to AR as well as providing proof-of-concept of the potential to image tumors using EPI compounds that bind to the AR-NTD [67]. While these studies provided evidence of EPI specifically binding to AF-1, it is the elegant NMR data from Dr. Salvatella’s group that revealed the amino acid residues required for this interaction [42]. EPI-001 and its stereoisomers were shown by NMR to directly bind to three regions within Tau-5 of AF-1 (Fig 2C). This is the region where RAP74 interacts [54] thereby supporting earlier studies showing EPI blocked this interaction [40]. EPI did not bind linear amino acid sequence as seen with small molecules discovered for c-Myc but rather EPI binding required all three regions to bind and did not bind independently to the individual regions [42]. All three regions within residues 354–448 must simultaneously be present for binding to occur. This suggests that EPI binds to a conformation that had a pocket, or that it induced formation of a conformation that created a pocket (Fig 3). The result however, is that binding of EPI to Tau-5 of AR-NTD yields a transcriptionally dead conformation that cannot interact with either RAP74 of the basal transcriptional machinery, or CBP [40] which are necessary for transcriptional activity [53,69]. Binding was specific and EPI did not interact with other regions of AR AF-1. It is suggested that EPI-001 interacts with an ensemble of conformations of AF-1 where the regions of sequence adopt a partially folded structure [42].
Fig 3.
A model proposing that direct binding of EPI to Tau-5 in the AR-NTD creates a conformation that prevents interactions with CBP and the basal transcriptional machinery (BTM).
The single stereoisomer of EPI-001, EPI-002, was assigned the generic name Ralaniten by the USAN council and a new stem class “-aniten” created based upon its novel mechanism of action that distinguishes it from the C-terminal LBD nonsteroidal antiandrogens with the stem name “lutamide”. Since the original discovery of EPI-067, approximately 500 analogues of this compound have been tested by the Sadar Lab for potential clinical development as a therapeutic and also as an imaging agent. In 2015, the prodrug of ralaniten, ralaniten-acetate also known as EPI-506, entered first-in-human clinical trials, thereby being the first drug that directly binds to an IDR to ever be tested in clinical trials. Trodusquemine/MSI-1436 has also been tested in clinical trials but it first binds to a folded region and then to an IDR of PTP1B [70]. Trodusquemine originated from a natural compound isolated from the dogfish shark, Squalus acanthias [71].
3.6. First-in-human clinical trials for ralaniten/EPI-002 that directly binds to an IDR
EPI-002, ralaniten, is a first-in-class drug and the first drug to be tested in the clinic that directly binds an IDR (Clinical trial information: NCT02606123). November 2015, the first of 28 heavily pretreated CRPC patients was dosed with ralaniten acetate. These patients had previously failed abiraterone (a CYP17 inhibitor) and/or the nonsteroidal antiandrogen enzalutamide. This Phase 1 clinical trial showed signs of efficacy evidenced by reduction of serum PSA and stable disease in some patients receiving higher doses in spite of not achieving steady-state Cmin concentrations of what would be required for optimal therapeutic concentrations based upon in vitro data (10μg/mL or 25μM). The highest dosed patients that received 3,600 mg/daily had trough levels of approximately 200ng/mL (0.5μM) which is 50X lower than the 25μM required for optimal activity of ralaniten in vitro and 48 to 58-fold lower than steady-state Cmin for enzalutamide and its active metabolite respectively [72]. Several patients remained on ralaniten for more than one year with stable disease. The drug was considered “well-tolerated” but due to poor pharmacokinetics (PK) there was excessive pill burden. Analysis of the metabolism of ralaniten acetate using the plasma obtained from these patients showed that ralaniten was both oxidized and glucuronidated [43]. The next generation analogue of ralaniten, EPI-7386, has improved PK and better metabolic stability. It is expected to enter Phase I clinical trials in 2020 (73).
4.0. Conclusion
Drug discovery has focused upon folded proteins with pockets or clefts that can be targeted with drugs to modulate the protein’s function and downstream pathway. Unfortunately many proteins involved in diseases are IDPs or have IDRs that act as hubs to interact with many binding partners. These disordered structures can have multiple and changing conformations thereby creating a challenge to discover drugs that would be specific with durable therapeutic effects. Full-length AR has been a validated drug target for pathologies involving the androgen axis for more than 50 years. All FDA-approved drugs to AR target the folded AR-LBD. The major resistance mechanism to these drugs in advanced prostate cancer is the expression of constitutively active AR-Vs that lack the folded LBD and are thought to continue to drive tumor growth. Theoretically the intrinsically disordered AR-NTD provides an attractive new target since a drug that binds to specific regions in this domain should be efficacious against all AR species including full-length AR, AR-Vs and any AR with gain-of-function mutations in the LBD. To date, most approaches have been exploring drugs that target interacting binding partners of AR-NTD, but now there has been success with finding drugs that directly bind to AF-1 and Tau-5 that have reached clinical trials with promising results.
5.0. Expert Opinion
IDPs and IDRs have the ability to change conformation to interact with many different binding partners and tend to be enriched in proteins with molecular functions in transcription and cell cycle that are dependent on reversible interactions with other proteins. These qualities of multiple and changing conformations of IDPs/IDRs along with the presumed large flat interaction binding sites for protein-protein interactions have been viewed by established drug developers as being insurmountable hurdles to achieve specificity with small molecules. An approach that improves the success rate in finding specific drugs has been to target a folded binding protein rather than the IDR itself.
The AR-NTD is an IDR and essential for the transcriptional activities of full-length AR and truncated constitutively active AR-Vs. The discovery of AR-Vs that lack the folded LBD as a resistance mechanism that drives lethal CRPC has emphasized the need to find drugs that target this domain. A number of approaches have been proposed and have shown some success in preclinical studies but none have moved towards clinical testing with the exception of ralaniten. Most studies have been targeting proteins that interact with the AR-NTD. The first in vivo proof-of-concept that sequestering AR-NTD interacting proteins yields a therapeutic response for CRPC was provided in preclinical studies using decoys [74,75]. Since then targeting individual interacting binding partners of AR-NTD are now being examined such as: hsp40/70 to induce degradation of AR-Vs and reduce aggregation of full-length AR with extended polyQ tracts [7,76–78]; BRD4 [79]; BAG1L [80,81]; and steroid receptor coactivators-1 and 3 [82]. The shortfalls of drugs that do not directly bind the AR-NTD, but rather an interacting protein, are predominantly a lack of specificity to blocking AR function since these interacting partners are not unique for AR and interact with many other proteins. Thus the quest to find drugs that directly bind to AR-NTD are of substantial interest.
Small molecule inhibitors proven to directly bind to the AR-NTD have all been isolated from natural compounds libraries and are: sintokamides that bind AF-1 possibly within Tau-1 [59], naphatenones that bind within AF-1 [57], and EPI-001/ralaniten that binds Tau-5 [40–42]. Also the bispecific antibody, 3E10-AR441, binds within Tau-1 at residues 299–315 [83]. Interestingly, deletions of small regions of Tau-1 (approximately 100 amino acid residues) do not eliminate AR transcriptional activity thereby suggesting its activity is not attributed to a single small structural element [84]. This suggests that a small molecule inhibitor, or antibody, directed to Tau-1 would need to impact the conformation across more than just a small discrete region. With the removal of the AR-LBD, the location of transcriptional activity shifts in AR-NTD with a loss of Tau-1 activity and induced use of Tau-5 activity [84]. Thus the AR-LBD plays a determinant role in the functioning of Tau-1 versus Tau-5. Theoretically this has implications for finding a small molecule inhibitor that blocks both full-length AR in response to ligand (Tau-1) and truncated AR-Vs that lack LBD (Tau-5). What is known so far, is that ralaniten binds residues 341–446 predominantly of Tau-5, including the core unit 435WHTLF439, but also has some overlap into Tau-1 (approximately 30 residues). Ralaniten inhibits the transcriptional activities of both full-length AR and constitutively active truncated ARs including AR-V7 that lacks AR-LBD [40,41,85]. Whether ralaniten is a better inhibitor of truncated AR-Vs compared to full-length AR has not been examined and studies to check this are warranted. It is possible that therapy combining a Tau-1 inhibitor with a Tau-5 inhibitor may be superior to individual monotherapies to a discrete Tau region as been shown with sintokamide and ralaniten [59]. Hence the discovery of additional small molecule inhibitors that directly bind to the AR-NTD are urgently needed.
To expedite success in finding drugs that directly bind to IDPs/IDRs may include different choices of small molecule libraries for screening compared to what have been predominantly used for drug discovery to folded proteins. To date, the only two drugs to reach clinical trials that directly bind to an IDR are from natural compound libraries and all small molecules that directly bind AR-NTD IDR were discovered in natural compound libraries. While EPI-067 (ralaniten analogue) was discovered in a marine sponge extract, it is not a natural compound, but such a chemical compound would never be included in a chemical library due to its chlorohydrin moiety, even though it has been proven to highly specific and not be generally reactive (i.e., does not form adducts with glutathione) [41,57].
Discovering drugs that directly bind AR-NTD IDR is labor intensive and slow. It is not high throughput screening and the assays are highly specialized and require unique expertise. This means most grant funding mechanisms of 3 or 5 years will probably not be adequate to ensure success for drug discovery projects against this target. An example is the empirical testing of each compound against an IDP/IDR, rather than having leads from virtual docking for a resolved protein to begin to narrow down potential drugs that might bind the target of interest. Although there are reports of rational drug design and druggable cavities for IDPs, these promising approaches are still structure-based and thereby currently limited [86,87]. Application of various different approaches including NMR and single particle cryo electron microscopy may yield structural information of IDP/Rs and structural ensembles to begin to facilitate drug discovery against these difficult drug targets [88,89 ]. The technical hurdles of aggregation, sensitivity to proteolytic cleavage, and insolubility of recombinant IDP/IDR also limits the abilities to complete many assays that are routinely done in drug discovery for folded proteins. Going forward the future of drug development will have to move towards targeting IDPs/IDRs in order to maximize and improve therapies to combat complex diseases such as cancer.
Article highlights.
Transcriptionally active androgen receptor (AR) drives the growth of most prostate cancer
Truncated constitutively active splice variants of AR (AR-Vs) cause resistance to current FDA-approved drugs that target full-length AR
AR N-terminal domain is intrinsically disordered and essential for transcriptional activity of full-length AR and AR-Vs and therefore an important, but difficult drug target
All drugs proven to directly bind the intrinsically disordered AR N-terminal domain were identified from screening natural compound libraries
Ralaniten/EPI-002 is the first drug that directly binds to an intrinsically disordered protein to be tested in clinical trials
Acknowledgments
Funding:
This work was supported by the National Cancer Institute via a National Institutes of Health grant (2R01CA105304).
Footnotes
Declaration of Interest:
MD Sadar is a paid consultant of ESSA Pharma Inc. She has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures:
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Annotated Bibliography
- 1.Claessens F1, Denayer S, Van Tilborgh N, et al. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal. 2008;6:e008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaffer PL, Jivan A, Dollins DE, et al. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A. 2004;101:4758–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalal K, Roshan-Moniri M, Sharma A, et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem. 2014;289:26417–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinckemalie L, Vanderschueren D, Boonen S, et al. The hinge region in androgen receptor control. Mol Cell Endocrinol. 2012;358:1–8 [DOI] [PubMed] [Google Scholar]
- 5.Heery DM, Kalkhoven E, Hoare S, et al. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6 [DOI] [PubMed] [Google Scholar]
- 6.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–91 [DOI] [PubMed] [Google Scholar]
- 7.Matias PM, Donner P, Coelho R, et al. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations. J Biol Chem. 2000;275:26164–71 [DOI] [PubMed] [Google Scholar]
- 8.Jenster G, van der Korput HA, van Vroonhoven C, et al. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–404 [DOI] [PubMed] [Google Scholar]
- 9.Simental JA, Sar M, Lane MV, et al. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–8 [PubMed] [Google Scholar]
- 10.Palvimo JJ, Kallio PJ, Ikonen T, et al. Dominant negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol Endocrinol. 1993;7:1399–407 [DOI] [PubMed] [Google Scholar]
- 11.He B, Kemppainen JA, Voegel JJ, et al. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J Biol Chem. 1999;274:37219–25 [DOI] [PubMed] [Google Scholar]
- 12.Slagsvold T, Kraus I, Bentzen T, et al. Mutational analysis of the androgen receptor AF-2 (activation function 2) core domain reveals functional and mechanistic differences of conserved residues compared with other nuclear receptors. Mol Endocrinol. 2000;14:1603–17 [DOI] [PubMed] [Google Scholar]
- 13.Reid J, Kelly SM, Watt K, et al. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. 2002;277:20079–86 [DOI] [PubMed] [Google Scholar]
- 14.Kumar R, Betney R, Li J, et al. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–13 [DOI] [PubMed] [Google Scholar]
- 15.Lavery DN, McEwan IJ. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry. 2008;47:3360–9 [DOI] [PubMed] [Google Scholar]
- 16.Escobedo A, Topal B, Kunze MBA, et al. Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor. Nat Commun. 2019;10:2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eftekharzadeh B, Banduseela VC, Chiesa G, et al. Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. Nat Commun. 2019;10:3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohen SP, Kralli A, Yamamoto KR. Hold ‘em and fold ‘em: chaperones and signal transduction. Science. 1995;268:1303–4 [DOI] [PubMed] [Google Scholar]
- 19.He B, Gampe RT Jr, Kole AJ, et al. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell. 2004;16:425–38 [DOI] [PubMed] [Google Scholar]
- 20.Hemmings HC Jr, Nairn AC, Aswad DW, et al. DARPP-32, a dopamine- and adenosine 3’:5’-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Purification and characterization of the phosphoprotein from bovine caudate nucleus. J Neurosci. 1984;4:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gast K, Damaschun H, Eckert K, et al. Prothymosin alpha: a biologically active protein with random coil conformation. Biochemistry. 1995;34:13211–8 [DOI] [PubMed] [Google Scholar]
- 22.Weinreb PH, Zhen W, Poon AW, et al. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–15 [DOI] [PubMed] [Google Scholar]
- 23.Fraga H, Graña-Montes R, Illa R, et al. Association between foldability and aggregation propensity in small disulfide-rich proteins. Antioxid Redox Signal. 2014;21:368–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mészáros B, Erdos G, Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y, LeGall T, Oldfield CJ, et al. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–42 [DOI] [PubMed] [Google Scholar]
- 26.Yan J, Dunker AK, Uversky VN, Kurgan L. Molecular recognition features (MoRFs) in three domains of life. Mol Biosyst. 2016;12:697–710. doi: 10.1039/c5mb00640f. [DOI] [PubMed] [Google Scholar]
- 27. Oldfield CJ, Uversky V, Dunker AK, et al. Intrinsically Disordered Proteins. Elsevier Inc. 2019 ** A comprehensive introduction to IDPs
- 28.Wang S, Zhao Y, Aguilar A, et al. Targeting the MDM2-p53 Protein-Protein Interaction for New Cancer Therapy: Progress and Challenges. Cold Spring Harb Perspect Med. 2017;7:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuertes G, Nevola L, Esteban-Martin S. Intrinsically Disordered Proteins. Elsevier Inc. 2019 ** A comprehensive review on drug discovery strategies to IDPs
- 30.Whitfield JR, Beaulieu ME, Soucek L. Strategies to Inhibit Myc and Their Clinical Applicability. Front Cell Dev Biol. 2017;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soucek L, Helmer-Citterich M, Sacco A, et al. Design and properties of a Myc derivative that efficiently homodimerizes. Oncogene. 1998;17:2463–72 [DOI] [PubMed] [Google Scholar]
- 32.Jung LA, Gebhardt A, Koelmel W, et al. OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors. Oncogene 2017;36:1911–1924. [DOI] [PubMed] [Google Scholar]
- 33.Demma MJ, Mapelli C, Sun A, et al. Omomyc Reveals New Mechanisms To Inhibit the MYC Oncogene. Mol Cell Biol. 2019;39(22). pii: e00248–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaulieu ME, Jauset T, Massó-Vallés D, et al. Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci Transl Med. 2019;11(484). pii: eaar5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammoudeh DI1, Follis AV, Prochownik EV, et al. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J Am Chem Soc. 2009;131:7390–401 [DOI] [PubMed] [Google Scholar]
- 36.Ruan H, Sun Q, Zhang W, et al. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov Today. 2019;24:217–227 [DOI] [PubMed] [Google Scholar]
- 37.Ambadipudi S, Zweckstetter M. Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin Drug Discov. 2016;11:65–77 [DOI] [PubMed] [Google Scholar]
- 38.Wójcik S, Birol M, Rhoades E, et al. Targeting the Intrinsically Disordered Proteome Using Small-Molecule Ligands. Methods Enzymol. 2018;611:703–734 [DOI] [PubMed] [Google Scholar]
- 39.Tsafou K, Tiwari PB, Forman-Kay JD, et al. Targeting Intrinsically Disordered Transcription Factors: Changing the Paradigm. J Mol Biol. 2018;430:2321–2341 [DOI] [PubMed] [Google Scholar]
- 40. Andersen RJ, Mawji NR, Wang J, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–46 **First report showing direct binding of a small molecule inhibitor an IDP/R.
- 41. Myung JK, Banuelos CA, Fernandez JG, et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013;123:2948–60 ** The first report to detect direct binding of a small molecule to an endogenous IDR in cells
- 42. De Mol E, Fenwick RB, Phang CT, et al. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem Biol. 2016;11:2499–505 ** The first NMR report for a small molecule binding to the IDR of the NTD of a steroid hormone receptor and defines the residues of AR-NTD that EPI binds.
- 43. Obst JK, Wang J, Jian K, et al. Revealing Metabolic Liabilities of Ralaniten To Enhance Novel Androgen Receptor Targeted Therapies. ACS Pharmacology & Translational Science, Articles ASAP. 2019;DOI: 10.1021/acsptsci.9b00065. ** Shows the metabolic liabilities of EPI-002/ralaniten that was tested in clinical trials to guide the chemistry of next generation analogues.
- 44. Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38 ** The first clinical study showing AR-V7 is associated with resistance to drugs that target the AR C-terminal ligand-binding domain in prostate cancer patients.
- 45.Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016;2:1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scher HI, Graf RP, Schreiber NA, et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018;4:1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Armstrong AJ, Halabi S, Luo J, et al. Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J Clin Oncol. 2019;37:1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jenster G, van der Korput HA, van Vroonhoven C, et al. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–404 ** Mapping of the function AR domains and its transactivation units (Taus).
- 49.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28:778–808 [DOI] [PubMed] [Google Scholar]
- 50.Flores O, Ha I, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990;265:5629–34 [PubMed] [Google Scholar]
- 51.Wang BQ, Burton ZF. Functional domains of human RAP74 including a masked polymerase binding domain.J Biol Chem. 1995;270:27035–44 [DOI] [PubMed] [Google Scholar]
- 52.Lei L, Ren D, Burton ZF. The RAP74 subunit of human transcription factor IIF has similar roles in initiation and elongation. Mol Cell Biol. 1999;19:8372–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid J, Murray I, Watt K, Betney R, McEwan IJ. The androgen receptor interacts with multiple regions of the large subunit of general transcription factor TFIIF. J Biol Chem. 2002. October 25;277(43):41247–53. [DOI] [PubMed] [Google Scholar]
- 54.De Mol E, Szulc E, Di Sanza C, et al. Regulation of Androgen Receptor Activity by Transient Interactions of Its Transactivation Domain with General Transcription Regulators. Structure. 2018;26:145–152.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tien AH, Sadar MD. Order within a Disordered Structure. Structure. 2018;26:4–6 [DOI] [PubMed] [Google Scholar]
- 56.Meimetis LG, Williams DE, Mawji NR, et al. Niphatenones, glycerol ethers from the sponge Niphates digitalis block androgen receptor transcriptional activity in prostate cancer cells: structure elucidation, synthesis, and biological activity. J Med Chem. 2012;55:503–14 [DOI] [PubMed] [Google Scholar]
- 57.Banuelos CA, Lal A, Tien AH, et al. Characterization of niphatenones that inhibit androgen receptor N-terminal domain. PLoS One. 2014;9:e107991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sadar MD, Williams DE, Mawji NR, et al. Sintokamides A to E, chlorinated peptides from the sponge Dysidea sp. that inhibit transactivation of the N-terminus of the androgen receptor in prostate cancer cells. Org Lett. 2008;10:4947–50 ** The first report of a small molecule inhibitor of the AR-NTD.
- 59.Banuelos CA, Tavakoli I, Tien AH, et al. Sintokamide A Is a Novel Antagonist of Androgen Receptor That Uniquely Binds Activation Function-1 in Its Amino-terminal Domain. J Biol Chem. 2016;291:22231–22243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadar MD. Small molecule inhibitors targeting the “achilles’ heel” of androgen receptor activity. Cancer Res. 2011;71:1208–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersen RJ. Sponging off nature for new drug leads. Biochem Pharmacol. 2017;139:3–14 [DOI] [PubMed] [Google Scholar]
- 62.Völkel W, Colnot T, Csanády GA, et al. Metabolism and kinetics of bisphenol a in humans at low doses following oral administration. Chem Res Toxicol. 2002;15:1281–7 [DOI] [PubMed] [Google Scholar]
- 63.Poole A1, van Herwijnen P, Weideli H, et al. Review of the toxicology, human exposure and safety assessment for bisphenol A diglycidylether (BADGE). Food Addit Contam. 2004;21:905–19 [DOI] [PubMed] [Google Scholar]
- 64.Stroheker T1, Picard K, Lhuguenot JC, et al. Steroid activities comparison of natural and food wrap compounds in human breast cancer cell lines. Food Chem Toxicol. 2004;42:887–97 [DOI] [PubMed] [Google Scholar]
- 65.Tsai WT. Human health risk on environmental exposure to Bisphenol-A: a review. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2006;24:225–55 [DOI] [PubMed] [Google Scholar]
- 66.EFSA J. Opinion of the E.U. scientific panel on food additives flavourings, processing aids and materials in contact with food (AFC) on a request from the commission related to 2,2-bis(4-hydroxyphenyl)propane bis(2,3-epoxypropyl)ether (bisphenol A diglycidyl ether, BADGE) REF No 13510 and 39700 (EFSA-Q-2003–178) 86 (2004), pp. 1–40 [Google Scholar]
- 67. Imamura Y, Tien AH, Pan J, et al. An imaging agent to detect androgen receptor and its active splice variants in prostate cancer. JCI Insight. 2016;1(11) e87850 ** Development of an imaging agent that targets an IDR.
- 68.Wang L, Wu Y, Zhang W, et al. Widespread occurrence and distribution of bisphenol A diglycidyl ether (BADGE) and its derivatives in human urine from the United States and China. Environ Sci Technol. 2012;46:12968–76 [DOI] [PubMed] [Google Scholar]
- 69.Frønsdal K1, Engedal N, Slagsvold T, et al. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J Biol Chem. 1998;273:31853–9 [DOI] [PubMed] [Google Scholar]
- 70.Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss CM, Page R, Blackledge M, Muthuswamy SK, Peti W, Tonks NK. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol. 2014;10:558–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zasloff M, Williams JI, Chen Q, et al. A spermine-coupled cholesterol metabolite from the shark with potent appetite suppressant and antidiabetic properties. Int J Obes Relat Metab Disord. 2001;25:689–97 [DOI] [PubMed] [Google Scholar]
- 72.Ito Y, Sadar MD. Enzalutamide and blocking androgen receptor in advanced prostate cancer: lessons learnt from the history of drug development of antiandrogens. Res Rep Urol. 2018;10:23–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. https://www.essapharma.com/wp-content/uploads/2019/09/ESSA_ NonConfidential-Slide-Deck_090519-Read-Only.pdf.
- 74. Quayle SN, Mawji NR, Wang J, et al. Androgen receptor decoy molecules block the growth of prostate cancer. Proc Natl Acad Sci U S A. 2007;104:1331–6 ** Proof-of-concept that targeting the AR-NTD has in vivo efficacy.
- 75.Myung JK, Wang G, Chiu HH, et al. Inhibition of androgen receptor by decoy molecules delays progression to castration-recurrent prostate cancer. PLoS One. 2017;12:e0174134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C, Lou W, Yang JC, et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat Commun. 2018;9:4700. doi: 10.1038/s41467-018-07178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moses MA, Kim YS, Rivera-Marquez GM, et al.Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer. Cancer Res. 2018;78:4022–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong J, Wu Z, Wang D, et al. Hsp70 Binds to the Androgen Receptor N-terminal Domain and Modulates the Receptor Function in Prostate Cancer Cells. Mol Cancer Ther. 2019;18:39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cato L, Neeb A, Sharp A, et al. Development of Bag-1L as a therapeutic target in androgen receptor-dependent prostate cancer. Elife. 2017. August 10;6. pii: e27159. doi: 10.7554/eLife.27159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee II, Kuznik NC, Rottenberg J, et al. Bag-1L: a promising therapeutic target for androgen receptor-dependent prostate cancer. J Mol Endocrinol. 2019. pii: JME-19–0034.R1. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Lonard DM, Yu Y, et al. Small molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol Endocrinol. 2011;25:2041–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goicochea NL, Garnovskaya M, Blanton MG, et al. Development of cell-penetrating bispecific antibodies targeting the N-terminal domain of androgen receptor for prostate cancer therapy. Protein Eng Des Sel. 2017;30:785–793 [DOI] [PubMed] [Google Scholar]
- 84.Jenster G, van der Korput HA, Trapman J, et al. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–6 [DOI] [PubMed] [Google Scholar]
- 85.Yang YC, Banuelos CA, Mawji NR, et al. Targeting Androgen Receptor Activation Function-1 with EPI to Overcome Resistance Mechanisms in Castration-Resistant Prostate Cancer. Clin Cancer Res. 2016;22:4466–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Cao H, Liu Z. Binding cavities and druggability of intrinsically disordered proteins. Protein Sci. 2015;24:688–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu C, Niu X, Jin F, et al. Structure-based Inhibitor Design for the Intrinsically Disordered Protein c-Myc. Sci Rep. 2016;6:22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonomi M, Vendruscolo M. Determination of protein structural ensembles using cryo-electron microscopy. Curr Opin Struct Biol. 2019;56:37–45 [DOI] [PubMed] [Google Scholar]
- 89.Schneider R, Blackledge M, Jensen MR. Elucidating binding mechanisms and dynamics of intrinsically disordered protein complexes using NMR spectroscopy. Curr Opin Struct Biol. 2019;54:10–18 [DOI] [PubMed] [Google Scholar]