Summary
Efficient translation of human induced pluripotent stem cells (hiPSCs) requires scalable cell manufacturing strategies for optimal self-renewal and functional differentiation. Traditional manual cell culture is variable and labor intensive, posing challenges for high-throughput applications. Here, we established a robotic platform and automated all essential steps of hiPSC culture and differentiation under chemically defined conditions. This approach allowed rapid and standardized manufacturing of billions of hiPSCs that can be produced in parallel from up to 90 different patient- and disease-specific cell lines. Moreover, we established automated multi-lineage differentiation and generated functional neurons, cardiomyocytes, and hepatocytes. To validate our approach, we compared robotic and manual cell culture operations and performed comprehensive molecular and cellular characterizations (e.g., single-cell transcriptomics, mass cytometry, metabolism, electrophysiology) to benchmark industrial-scale cell culture operations toward building an integrated platform for efficient cell manufacturing for disease modeling, drug screening, and cell therapy.
Keywords: iPS cell, robotic cell culture, cell differentiation, high-throughput, biomanufacturing, single-cell transcriptomics, mass cytometry, standardization, CEPT cocktail
Graphical abstract

Highlights
-
•
Robotic cell culture platform facilitates standardized maintenance and rapid expansion of multiple hiPSC lines in parallel
-
•
Automated multi-lineage differentiation and reliable generation of functional cell types
-
•
Detailed comparison of manual versus automated cell culture
-
•
Robotic scalable biomanufacturing compatible with 2D and 3D cell culture formats
In this article, Singeç and colleagues show standardized culture and differentiation of hPSCs into neurons, cardiomyocytes, and hepatocytes by using a robotic cell culture platform. Industrial-scale biomanufacturing enables the production of large numbers of human cells, which are qualitatively similar to manually cultured cells as demonstrated by detailed molecular and functional characterizations.
Introduction
Human pluripotent stem cells (hPSCs) are characterized by extensive self-renewal capacity and differentiation into all somatic cell types, enabling novel approaches to model, diagnose, and treat human diseases (Kimbrel and Lanza, 2020; Sato et al., 2019; Sharma et al., 2020). However, several important challenges remain to be addressed for their efficient and safe utilization. These challenges include technical and biological variability, lack of standardization, laborious differentiation protocols, limited methods for scale up, and inefficient manufacturing of functional cell types representing the diversity of human tissues. Since isolation of the first human embryonic stem cells (hESCs) (Thomson et al., 1998), significant progress has been made in improving cell culture conditions, including the development of new reagents, coating substrates, medium formulations, and passaging tools (Chen et al., 2011; Kuo et al., 2020; Ludwig et al., 2006; Rodin et al., 2014). Despite these advances, manual cell culture of hPSCs remains time consuming, laborious, and subject to human bias or error (e.g., risk of contamination, medium change at different intervals). Other inherent challenges are due to variability in handling cells and reagents across laboratories, use of different reprogramming methods, and cell-line-to-cell-line variability (Cahan and Daley, 2013; Niepel et al., 2019; Osafune et al., 2008; Panopoulos et al., 2017).
Automated cell culture has several practical and scientific characteristics designed to improve quality control, increase productivity, implement standard operating procedures (SOPs), and develop commercial cellular products (Aijaz et al., 2018; Daniszewski et al., 2018). These advantages ensure scale-up of cell manufacturing, standardization of liquid handling, control of incubation times, minimization of batch-to-batch variability, reduction of human error, and seamless documentation of operations. Automated cell reprogramming by using liquid handlers can increase efficiency and reproducibility of new induced pluripotent stem cell (iPSC) line generation (Paull et al., 2015). Previous studies used various two- and three-dimensional (2D, 3D) systems to either automate or scale-up some aspects of hPSC culture (Archibald et al., 2016; Hookway et al., 2016; Konagaya et al., 2015; Liu et al., 2014; McLaren et al., 2013; Rigamonti et al., 2016; Schwedhelm et al., 2019; Soares et al., 2014a; Thomas et al., 2009). However, a comprehensive automation strategy for biomanufacturing of hPSCs under flexible scale-up and scale-down conditions and compatibility with 2D and 3D culture (e.g., embryoid bodies, neurospheres, monolayer differentiation) has not been established so far. Here we present and characterize a versatile robotic cell culture platform that can be utilized for scale-up and multi-lineage differentiation of human induced pluripotent stem cells (hiPSCs). We performed a functional analysis of neurons, cardiomyocytes, and hepatocytes and demonstrate their utility for high-throughput screening and Zika virus experiments. We envision that automation will help to overcome technical and economic challenges and leverage the full translational potential of hiPSCs.
Results
Automated and scalable culture of hPSCs
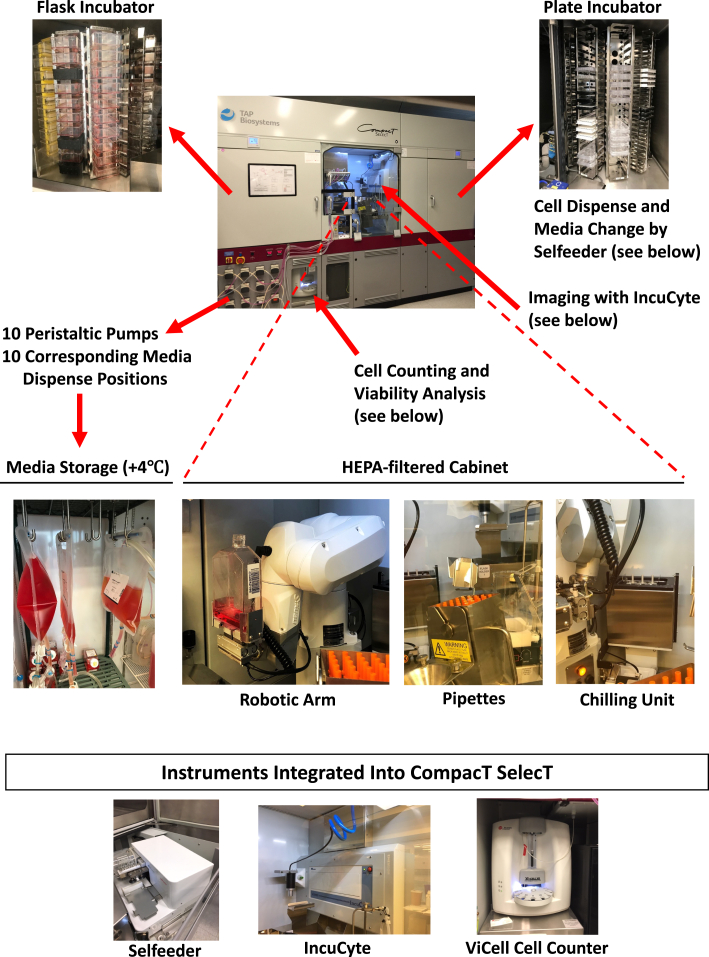
The CompacT SelecT (CTST) platform is a modular robotic system that integrates a full range of cell culture procedures under sterile conditions that mimic the manual cell culture process (Figure 1). These procedures include automated handling of different cell culture vessels, pipetting large and small volumes at adjustable speeds, cell counting, cell viability analysis, cell density assessment, microscopic imaging, cell passaging, cell harvest, and medium changes. Moreover, two independent incubator carousels (humidified 37°C, 5% CO2) enable culturing cells in various cell culture vessels (T75 and T175 flasks and 6-, 24-, 96-, or 384-well formats). Notably, the CTST system has the capacity to simultaneously culture up to 280 assay-ready plates and up to 90 different hiPSC lines in large T175 flasks (Figures 1 and 2A, Video S1, and full movie: https://youtu.be/-GSsTSO-WCM). Moreover, as CTST is handling different cell lines and protocols, scientists may remotely access, control, and monitor ongoing experiments without the need to physically enter the laboratory. Hence, the system allows non-stop cell culture operations with minimal manual intervention.
Figure 1.
Overview of the automated CTST system
Features and components of CTST, including flask incubator, plate incubator, storage of large volumes of medium, cell counting, viability analysis, microscopic imaging, and a sterile HEPA-filtered cabinet housing a robotic arm, various pipettes, and a chilling unit to store temperature-sensitive reagents such as recombinant proteins.
Figure 2.
Characterization of hiPSCs (LiPSC-GR1.1) cultured by CTST
(A) Characteristics and advantages of automated cell culture.
(B) Representative hiPSCs growing in densely packed colonies at 3 days post-passaging. Scale bar, 500 μm.
(C) Colony of hiPSCs showing typical morphological features of human pluripotent cells at 3 days post-passaging. Scale bar, 250 μm.
(D) hiPSCs immunostained for pluripotency-associated markers OCT4 and NANOG. Scale bar, 100 μm.
(E) Long-term robotically cultured hiPSCs maintain a normal karyotype (passage 40).
(F) Seahorse XF glycolysis stress test profile comparison of glycolytic function in hiPSCs maintained by automated or manual cell culture. Cells were treated with serial injections of metabolic modulators (glucose, oligomycin, 2-deoxyglucose [2-DG]).
(G) Seahorse XF mitochondrial stress test profile comparison of mitochondrial function in hiPSCs maintained by automated or manual cell culture. Cells were treated with serial injections of metabolic modulators (oligomycin, FCCP, and rotenone/antimycin A [Rot/AA]).
(H) Comparison of medium change intervals during automated and manual cell culture of hiPSCs.
(I–O) Supernatants of cultures maintained manually or robotically were measured daily (Vi-Cell MetaFLEX Bioanalyte Analyzer). Boxplots show the variation of spent medium from hiPSC cultures. (I) pO2, (J) pH, (K) cLac, (L) cGlu, (M) cCa+, (N) cNa+, (O) cK+.
(P) Image-based analysis comparing cell growth in hiPSC cultures expanded manually and robotically.
(Q) Automated cell expansion strategy showing massive scale-up in only 12 days. Data expressed as mean ± SD, n > 3 biological replicates using two independent cell lines (B–P). p = 0.0001 in (I), unpaired t test.
To establish standardized high-throughput protocols for CTST, we focused on culturing hPSCs under feeder-free conditions using Essential 8 (E8) medium, recombinant vitronectin (VTN-N) as coating substrate, and EDTA for cell passaging. Use of EDTA for non-enzymatic cell dissociation was critical to minimize cellular stress and skip a manual intervention step (offline centrifugation and removal of enzymatic cell dissociation reagents). Under these chemically defined conditions, we were able to robustly culture, expand, and cryopreserve various hESC and hiPSC lines over the last 5 years (Figure S1A and Table S1). hPSCs maintained typical characteristics, such as growth in densely packed colonies, high nucleus-to-cytoplasm ratio, expression of pluripotency-associated markers OCT4 and NANOG, and normal karyotypes (Figures 2B–2E and S2A–S2C). Energy production in hPSCs depends on high glycolytic rates (Gu et al., 2016; Zhang et al., 2016), and live-cell metabolic analysis (Seahorse XF analyzer) confirmed expected metabolic profiles in hESCs and hiPSCs when cultured manually or robotically (Figures 2F, 2G, S2D, and S2E).
Because suboptimal conditions such as overgrowing cells in high-cell-density cultures can lead to cellular stress and impaired quality of hPSCs (Horiguchi et al., 2018; Jacobs et al., 2016; Paull et al., 2015), we sought to directly compare manual with automated cell cultures. Medium change intervals can be precisely controlled and documented by CTST, whereas manual cell culture is typically investigator dependent and variable. To monitor manual cell culture, we maintained hPSCs in live-cell imaging systems (IncuCyte), which enable the monitoring of cell growth and daily interventions by investigators. By tracking the online use of our IncuCyte instruments, we were able to capture the typical variability of medium change intervals in our laboratory (Figure 2H), which is likely to be representative for most laboratories culturing hPSCs. In contrast, medium change intervals were tightly controlled by using CTST (Figure 2H). To assess the consequences of variable medium change intervals, we measured the spent media of cultures maintained either manually or robotically. Indeed, culturing hPSCs by CTST resulted in less deviation from the mean in several measured endpoints such as oxygen concentration, pH fluctuations, lactate levels, glucose concentration, and ionic milieu (calcium, sodium, potassium) (Figures 2I–2O and S2F–S2L).
Process automation is of particular importance to produce large quantities of cells in a standardized fashion for high-throughput applications. One additional challenge for cell manufacturing is the fact that hPSCs are sensitive to environmental perturbations, and poor cell survival can be a limiting factor (Archibald et al., 2016; Soares et al., 2014a; Watanabe et al., 2007). Taking advantage of the newly developed CEPT small molecule, which promotes viability and cytoprotection during routine cell passaging (Chen et al., 2021), we aimed at optimizing the expansion of hPSCs. Combining CTST with the CEPT cocktail enabled consistent cell passaging and cell growth (Figures 2P and S2M). Robotic cell passaging was robust and predictable, resulting in minimal cell death, and cultures were devoid of cellular debris at 24 h post-passaging in the presence of CEPT (Figure S1B). The efficiency of this approach enabled rapid scale-up and production of large quantities of hPSCs. For instance, using the WA09 cell line and starting with one T175 flask containing 5.25 million cells and passaging at 70% to 80% confluency (∼42 million cells per flask) in a 1:6 ratio every 3 days, we were able to generate a total of 9.07 billion hPSCs in 12 days (Figure 2Q). To our knowledge, such dramatic scale-up in a short period of time has not been reported previously and should be invaluable for biobanking of hPSCs or CryoPause, an approach to increase experimental reproducibility by using the same batch of cryopreserved cells (Wong et al., 2017). Furthermore, since CTST can operate in a virtually non-stop fashion and handle large flasks or assay-ready plates, we compared these features with typical manual cell culture performed during a typical 8 h workday. This comparison demonstrated enormous advantages of robotic cell culture for biomanufacturing large quantities of pluripotent and differentiated cells (Table S2).
Similar molecular signatures of hPSCs cultured manually or robotically
Manual cell culture is the most widely used approach in the stem cell field. In parallel to our automated platform and depending on experimental needs, we continue to carry out significant amounts of cell culture work manually. To perform a side-by-side comparison of cultures maintained manually versus robotically, we performed a single-cell analysis, including RNA sequencing (RNA-seq) and mass cytometry. Deriving detailed information at single-cell resolution can aid in defining cell type identities and cellular heterogeneity (Quadrato et al., 2017; Veres et al., 2019). We randomly selected hESCs (WA09) and hiPSCs (LiPSC-GR1.1) that were cultured either manually or robotically by different investigators in our laboratory, and samples were processed for RNA-seq using the 10× Genomics platform. Single-cell transcriptome libraries of 18,817 cells derived from manual (5,573 cells for WA09; 4,835 cells for LiPSC-GR1.1) and automated (4,485 cells for WA09; 3,922 cells for LiPSC-GR1.1) cultures were analyzed for differential gene expression and comparison between both culture conditions. t-Distributed stochastic neighbor embedding (t-SNE) projection demonstrated that hiPSCs and hESCs cultured either manually or robotically showed a highly similar distribution (Figures 3A–3D). Thus, cells cultured by CTST substantially mirrored manually cultured hESCs and hiPSCs. Of 32,894 transcripts analyzed, there were only 98 differentially expressed genes among manually and robotically cultured hiPSCs (Figure 3B and Table S3). Similarly, there were only 15 differentially expressed genes in the hESC line (Figure 3D and Table S3). A total of only five genes (SFRP1, SLIRP, HNRNPAB, APOE, and COPS9) were downregulated comparing automated with manual cell cultures (Figure 3D and Table S3). Together, it was striking to see that the transcriptomic profiles of manually and robotically cultured cells were largely overlapping (Figure 3E).
Figure 3.
Single-cell RNA-seq and comparison of manual and automated cell culture
(A and C) t-SNE plots illustrating (A) hiPSCs (LiPSC-GR1.1) and (C) hESCs (WA09) maintained either manually or robotically show a high degree of transcriptomic similarity.
(B and D) Venn diagrams showing overlap of expressed genes in (B) hiPSCs (LiPSC GR1.1) and (D) hESCs (WA09).
(E) Direct comparison of transcriptomes of hiPSCs (LiPSC-GR1.1) and hESCs (WA09) cultured manually and robotically. Data from n = 5,573, 4,835, 4,485, and 3,922 single cells obtained from n = 4 independent experiments using two independent cell lines for hESC manual, hiPSC manual, hESC auto, and hiPSC auto, respectively (A–E). Single-cell RNA-seq data were analyzed in the Seurat R package.
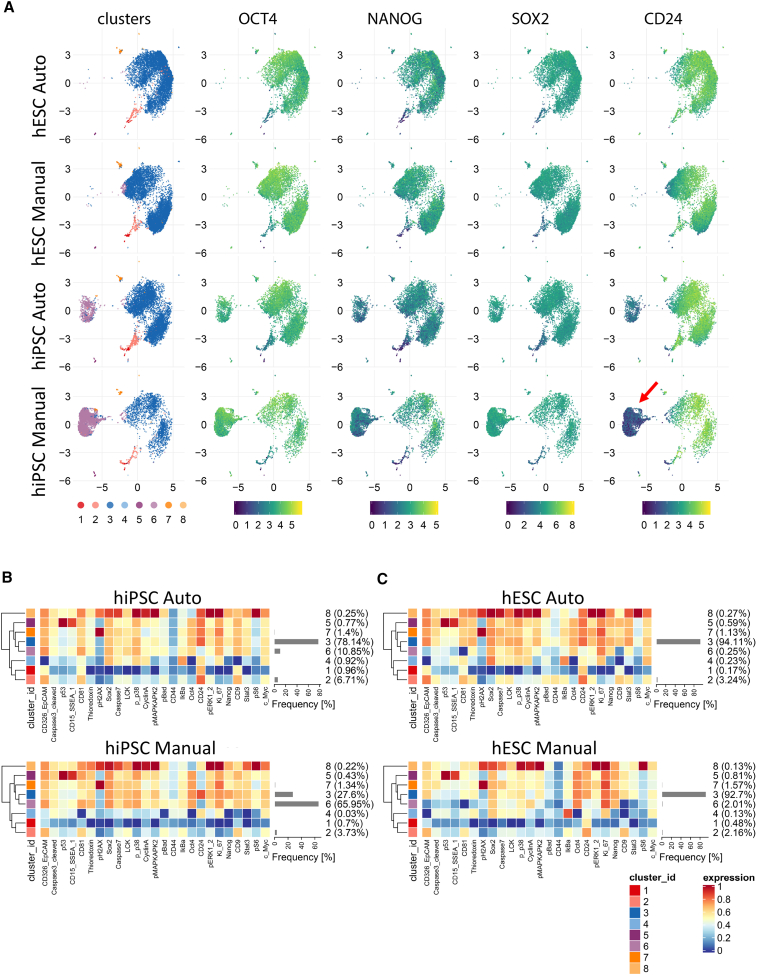
Cytometry time-of-flight (CyTOF) is a new technology that allows the simultaneous analysis of more than 30 proteins in single cells by using metal-conjugated antibodies (Qin et al., 2020; Zunder et al., 2015). We used a panel of 25 cell-surface cluster-of-differentiation (CD) antigens and intracellular proteins, including phosphorylated proteins (Table S4), to carefully compare markers of cell health and pluripotency in hPSCs cultured either manually or robotically. The expression of pluripotency-associated transcription factors OCT4, NANOG, and SOX2 showed, again, strikingly similar expression levels across different samples (Figures 4A–4C). A total of 96,861 cells derived from manual (11,898 cells from WA09; 19,217 cells from LiPSC-GR1.1) and automated (32,889 hESCs; 32,857 hiPSCs) cell culture experiments were subjected to single-cell mass cytometry. An analysis of an additional 22 proteins covering diverse cellular mechanisms confirmed the predominant similarity of cultures maintained either manually or by automation (Figures 4B and 4C). Expression of the cell-surface marker and sialoglycoprotein CD24 is regulated during cell reprogramming, and its expression may indicate a more differentiated state compared with naive pluripotency (Shakiba et al., 2015). The hiPSC line displayed a population of cells (cluster 6) that lacked CD24 expression and could be distinguished from the main cluster (cluster 3, Figure 4B). Interestingly, cluster 6 cells were more abundant in manually cultured hiPSC samples (Figure 4B). However, the hESC line (WA09) showed only a negligible percentage of CD24-negative cells in both automated and manual cell culture (Figure 4C).
Figure 4.
Mass cytometry of hiPSCs and hESCs and comparison of manual and automated cell culture
(A) Uniform manifold approximation and projection (UMAP) plots showing subpopulations of cells within each group organized into eight clusters identified by FlowSOM and ConsensusClusterPlus algorithms. Cluster 6 was prominent in hiPSCs (LiPSC-GR1.1) when cultured manually and its representation was mitigated by automated culture. Core pluripotency markers OCT4, NANOG, and SOX2 were expressed at similar levels across clusters. Surface-antigen CD24 was expressed at a considerably higher level in cluster 6 in hiPSCs cultured manually (red arrow).
(B) Heatmaps comparing protein expression levels for each analyzed marker in individual clusters and the abundance of the clusters within the hiPSC populations (LiPSC-GR1.1) cultured manually or by automation. Manual culture led to a large proportion of CD24-negative cells, 66% versus 11% in automated culture.
(C) Heatmaps of protein expression levels and cluster abundances in hESCs (WA09) after manual and automated cell culture. The abundance of the major cluster 3 was similar in both culture conditions, and CD24-negative cluster 6 was represented at a negligible level. UMAP plots were constructed from 8,000 single cells per sample (n = 4 independent experiments) obtained from two independent cell lines (A–C). CyTOF data were analyzed using a modified CyTOF workflow (Robinson et al., 2017).
Automated embryoid body formation
Cell differentiation is a dynamic process with cells progressing through developmental states, which can be recapitulated in vitro by spontaneous or controlled differentiation when appropriate factors and morphogens are administered at defined time points. Spontaneous differentiation of hPSCs by embryoid body (EB) formation is a widely used assay for pluripotency assessment (i.e., capacity to differentiate into ectoderm, mesoderm, and endoderm), toxicity testing, organoid formation, and other developmental studies (Guo et al., 2019; Lancaster et al., 2013; Osafune et al., 2008; Tsankov et al., 2015). Hence, developing defined protocols for automated large-scale production of EBs is of great relevance. Typically, in manual cell culture work EBs are maintained as free-floating 3D structures in ultra-low attachment six-well plates. Although the CTST system can culture cells and change medium in different plate formats (6, 24, 96, or 384 wells), T175 flasks would represent the largest vessel for EB production in this context. To our knowledge, T175 flasks are currently not available in an ultra-low attachment version. However, we found that rinsing regular T175 flasks with a commercially available anti-adherence solution (STEMCELL Technologies) was sufficient to prevent unwanted cell attachment and, in combination with the CEPT cocktail, enabled highly efficient formation of free-floating EBs (Figures S1C and S1D). Again, enzyme-free passaging with EDTA, which obviates an offline centrifugation step, was ideal for fully automated EB production. As expected, EB formation from hESCs and hiPSCs and comparison of manual and automated cell culture by using the standardized ScoreCard method (Tsankov et al., 2015) showed multi-lineage differentiation potential (Figure S1E). Of note, a difference in spontaneous endoderm differentiation was observed when comparing hiPSCs (LiPSC-GR1.1) to hESCs (WA09). This difference is likely due to the absence of endoderm-promoting factors in Essential 6 (E6) medium, which is known to favor differentiation into ectoderm (Lippmann et al., 2014). Indeed, in adherent cultures using a commercial kit for directed differentiation, both cell lines efficiently produced endodermal cells (Figures 5 and S3).
Figure 5.
Controlled multi-lineage differentiation of hPSCs by using CTST
(A) Western blot of hiPSCs (LiPSC-GR1.1) and hESCs (WA09) before (OCT4) and after differentiation into ectoderm (PAX6) at day 7, mesoderm (Brachyury) at day 5, and endoderm (SOX17) at day 5. Tubulin was used as loading control.
(B) Immunocytochemical analysis of hiPSC (LiPSC-GR1.1)-derived ectoderm (PAX6) at day 7, endoderm (SOX17) at day 5, and mesoderm (Brachyury) at day 5. Cultures were differentiated by CTST. Scale bar, 200 μm.
(C) Single-cell RNA-seq of pluripotent and differentiated cultures (LiPSC-GR1.1).
(D) Heatmap showing the highly expressed genes for pluripotent cells (LiPSC-GR1.1) and differentiated cultures representing ectoderm (day 7), endoderm (day 5), and mesoderm (day 5).
(E) Comparison of undifferentiated and differentiated hESCs (WA09) and hiPSCs (LiPSC-GR1.1) shows that gene expression signatures are similar. Data are from n = 4 biological replicates using two independent cell lines (A and B). Data are from n = 19,759 or 16,582 single cells obtained from n = 4 independent experiments using two independent cell lines (C–E). Cell counts for hiPSCs: 4,772 pluripotent, 6,457 ectoderm, 4,160 endoderm, and 4,370 mesoderm. Cell counts for hESCs: 3,627 pluripotent, 5,062 ectoderm, 4,267 endoderm, and 3,626 mesoderm. Single-cell RNA-seq data were analyzed in the Seurat R package.
Controlled multi-lineage differentiation in monolayer cultures
While spontaneous EB differentiation is useful for certain applications, directed differentiation under adherent monolayer conditions is highly desirable for developing scalable protocols for different lineages. Hence, we established automated protocols for directed differentiation into the three embryonic germ layers. For neural differentiation, hPSCs were cultured in E6 medium containing the bone morphogenetic protein (BMP) pathway inhibitor LDN-193189 (100 nM) and the transforming growth factor (TGF) β pathway inhibitor A83-01 (2 μM). Simultaneous inhibition of these pathways is typically referred to as dual-SMAD inhibition (dSMADi) (Chambers et al., 2009; Singec et al., 2016). For mesodermal and endodermal differentiation, we utilized standardized kits from a commercial vendor (experimental procedures). Stock solutions of different reagents can be stored in the chilling unit of the CTST (Figure 1), and the robotic arm can add fresh reagents during daily medium changes. By using these protocols, we were able to efficiently generate cultures with ectodermal (PAX6), mesodermal (Brachyury), and endodermal (SOX17) precursors as demonstrated by western blotting and immunocytochemistry (Figures 5A, 5B, and S3A). To confirm efficient automated multi-lineage differentiation, we performed single-cell RNA-seq analysis of lineage-committed precursor cells derived from either hiPSCs (Figures 5C and 5D) or hESCs (Figures S3B and S3C). We analyzed a total of 19,759 cells for the hiPSC line and a total of 16,582 cells for the hESC line. For both independently tested cell lines, comparison of transcriptomes by unsupervised clustering revealed distinct signatures for pluripotent, ectodermal, mesodermal, and endodermal cells (Figures 5C and S3B). Similarly, a heatmap analysis for typical lineage-specific markers demonstrated distinct molecular signatures for pluripotent and differentiated germ layer cells (Figures 5D and S3C). Comparison of cultures generated either manually or robotically, showed similar quantitative polymerase chain reaction (qPCR) expression profiles for ectodermal, mesodermal, and endodermal markers (Figure S4). Interestingly, some genes, such as NES, TUBB3, HES4, MAP2, Brachyury, VIM, NODAL, and ABCA4, were expressed at higher levels when cultures were differentiated robotically versus manually (Figure S4). Last, comparing automated hiPSCs and hESCs cultures with each other revealed a high degree of similarity among pluripotent and lineage-committed progeny (Figure 5E). Together, the robotic cell differentiation protocols established here generated primary embryonic germ layers with high efficiency and reproducibility.
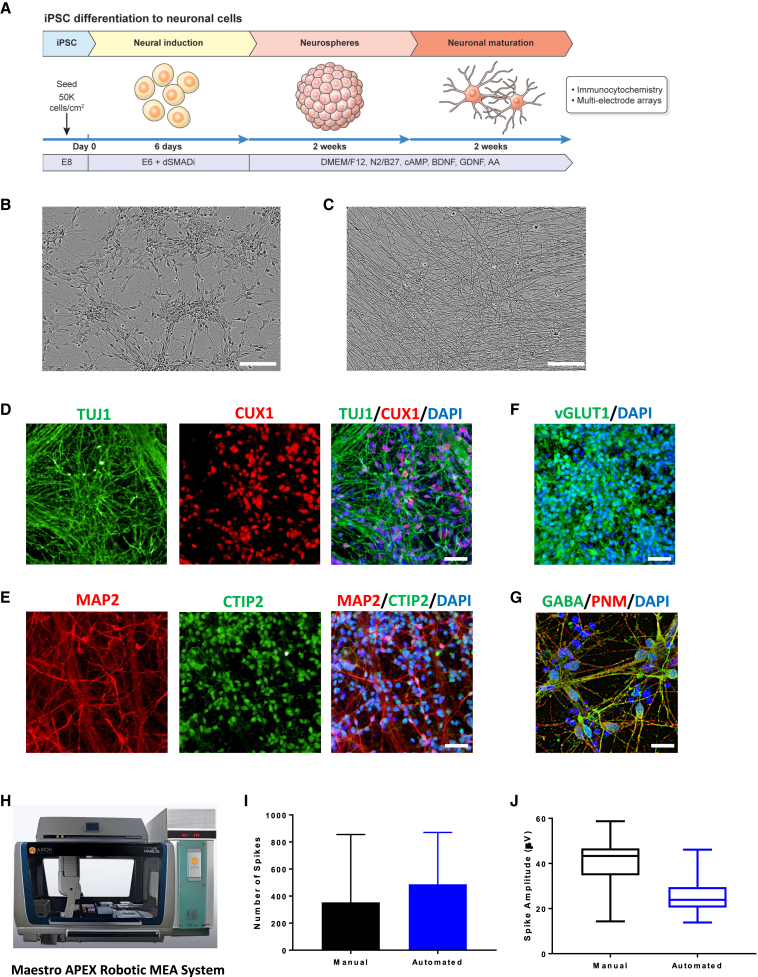
Scalable production of functional human neurons
The translation of hiPSCs depends on controlled and scalable differentiation into diverse cellular phenotypes that can be used for disease modeling, drug screening, and cell therapies. We asked if executing complex multi-step protocols over several weeks could be performed by using robotic cell culture in a fully automated “touch-and-go” fashion. Hence, we developed a cost-efficient differentiation protocol that utilizes the dSMADi strategy followed by culturing cells as neurospheres and then replating them for further maturation and analysis (Figures 6A–6C). Most neuronal cells (>90%) generated by using this simple protocol (Figure 6A) expressed neuronal markers β-III-tubulin (TUJ1) and microtubule-associated protein 2 (MAP2) at day 30 (Figures 6D, 6E, and S5A). Expression of transcription factors CUX1 (a marker for cortical layers 2/3) and CTIP2 (a marker for cortical layers 5/6) indicated the generation of specific forebrain neurons (Figures 6D and 6E) that were generated at higher numbers using automated versus manual differentiation (Figure S5A). Moreover, specific antibodies against vesicular glutamate transporter 1 (vGLUT1) and γ-aminobutyric acid (GABA) suggested that cultures contained a mixed population of cells, with the majority (>80%) representing glutamatergic neurons (Figures 6F, 6G, and S5A).
Figure 6.
Robotic scalable production of hiPSC (LiPSC-GR1.1)-derived human neurons
(A) Neuronal differentiation strategy established for automated cell culture.
(B) Phase-contrast image showing a typical neuronal culture (day 30). Scale bar, 200 μm.
(C) Neurons develop a dense network of neurites upon maturation (day 50). Scale bar, 200 μm.
(D–F) hiPSC-derived cortical neurons (day 40) immunostained for (D) TUJ1 and CUX1, (E) MAP2 and CTIP2, and (F) vGLUT. Scale bar, 50 μm.
(G) hiPSC-derived neuronal cells (day 40) showing immunoreactivity for inhibitory neurotransmitter GABA. Scale bar, 20 μm.
(H) Robotic MEA platform used for high-throughput electrophysiology and functional cell characterization.
(I and J) Comparison of (I) spontaneous neuronal spikes and (J) spike amplitudes in hiPSC-derived cultures after 6 weeks of manual or robotic cell differentiation as measured by MEA. Representative data are expressed as mean ± SD, n > 3 biological replicates (B–G, I, J).
p > 0.5, unpaired t test.
To demonstrate neuronal activity, we conducted electrophysiological analysis using the robotic Maestro APEX multi-electrode array (MEA) platform (Figure 6H). At day 30, cultures were dissociated into single cells and 140,000 neurons were seeded into a well with 16 electrodes in the presence of the CEPT cocktail (applied for 24 h to improve viability). At day 7 post-plating, an analysis of extracellular field potentials revealed spontaneous activity in hiPSC-derived neuronal cultures generated manually or robotically (Figure 6I). Similar spike shapes and amplitudes were also detected 2 weeks later in both groups (Figures 6J, S5B, and S5C). Moreover, comparing cultures generated either manually or robotically by qPCR, both cultures expressed typical neuronal markers, SATB2, CUX1, GAD1, SLC6A1, BCL11B, and SLC17A7 (Figure S5D).
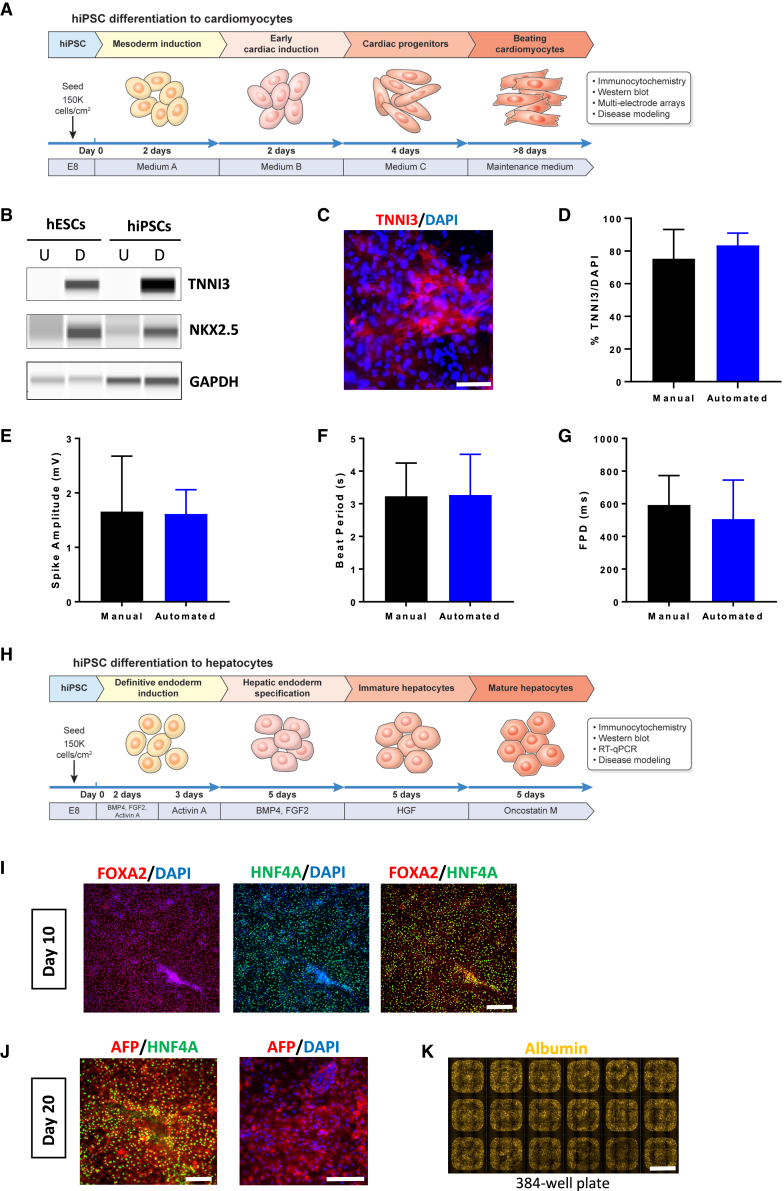
Standardized production of functional cardiomyocytes and hepatocytes
Derivation of large quantities of hiPSC-derived cardiomyocytes and hepatocytes is important for drug development, toxicology, and regenerative medicine (Kimbrel and Lanza, 2020; Sharma et al., 2020). To generate cardiomyocytes, we adopted a kit-based protocol for automated differentiation of hPSCs (Figure 7A). A western blot analysis of differentiated hESCs and hiPSCs demonstrated strong induction of TNNI3 (cardiac troponin) and transcription factor NKX2.5 at day 14 (Figure 7B). Immunocytochemistry and flow cytometry showed that 80% to 90% of cells expressed cardiomyocyte-specific markers TNNI3 and ACTC1 (α-cardiac actin) irrespective of manual or robotic differentiation (Figures 7C, 7D, S6A, and S6B). Moreover, a comparison of manually and robotically differentiated cultures (day 24) indicated similar expression levels of typical cardiomyocyte markers as measured by qPCR (Figure S6H). A functional analysis confirmed that cardiomyocytes were active and spontaneously beating as measured by MEA (Figures 7E and S6C). An analysis of field potentials documented spontaneous cardiomyocyte activity (Figures 7G and S6E). Beat-to-beat variance analysis showed that cardiomyocytes exhibited regular and consistent beat intervals, confirming the presence of non-arrhythmic cardiomyocytes, field potential durations, and conduction velocities that were comparable in cultures generated by manual and automated differentiation (Figures 7F, 7G, and S6D–S6G).
Figure 7.
Characterization of cardiomyocytes and hepatocytes derived by automated cell culture
(A) Overview of cardiomyocyte differentiation protocol.
(B) Western blot showing induction of cardiac troponin and NKX2.5 in undifferentiated (abbreviated as U) and differentiated (abbreviated as D) hESCs (WA09) and hiPSCs (LiPSC-GR1.1) at day 24. GAPDH was used as a loading control.
(C and D) (C) Immunocytochemistry and (D) quantification shows that hiPSC-derived cardiomyocytes express cardiac troponin (day 24). Scale bar, 75 μm.
(E) Comparison of spontaneous spike amplitudes in hiPSC-derived cardiomyocytes differentiated manually or robotically (day 24).
(F and G) Comparison of (F) beat periods and (G) field potential duration in cardiomyocyte cultures (LiPSC-GR1.1) differentiated manually or robotically and measured by MEA (day 24).
(H) Overview of hepatocyte differentiation protocol.
(I) Immunocytochemistry at day 10 shows most hiPSC (LiPSC-GR1.1)-derived cells express FOXA2 and HNF4A. Scale bar, 200 μm.
(J) hiPSCs (LiPSC-GR1.1) differentiated into hepatocytes express α-fetoprotein (AFP) and HNF4A (day 20). Scale bar, 200 μm.
(K) Immunocytochemistry showing albumin-expressing hepatocytes robotically differentiated in a 384-well plate. Representative overview of 18 whole wells containing hepatocytes. Scale bar, 2 mm. Representative data are expressed as mean ± SD, n > 3 biological replicates.
p > 0.5, unpaired t test.
Next, we established an automated protocol for hepatocyte differentiation using the CTST platform. A 20-day protocol (Mallanna and Duncan, 2013) was adopted to generate human hepatocytes entirely in scaled-down 384-well plates compatible with high-throughput screening (Figure 7H). Immunocytochemical analysis at day 10 showed that >80% of cells, differentiated manually and robotically, expressed endodermal markers FOXA2 and HNF4A (Figure 7I and S7A–S7C). Comparison of cultures at day 10, generated either manually or robotically, showed similar expression levels for FOXA2, GATA4, and GATA6 as measured by qPCR analysis (Figure S7F). By day 20, hepatocytes expressed HNF4A, α-fetoprotein (AFP), and albumin (Figures 7J, 7K, S7D, and S7E). qPCR showed that cultures differentiated manually or robotically exhibited similar levels of gene expression for HNF4A, AFP, Albumin, APOA1, SLC10A1, ASGR1, CYP3A4, CYP2D6, and CYP3A7 (Figure S7G).
Zika virus infection of robotically generated cardiomyocytes and hepatocytes
To demonstrate the utility of robotically differentiated cells, we performed translationally relevant assays. Human cellular models provide unique opportunities to better understand Zika virus (ZIKV) pathobiology (Qian et al., 2016; Tang et al., 2016; Zhou et al., 2017). Accordingly, we found that robotically generated cardiomyocytes and hepatocytes were susceptible to ZIKV infection after viral exposure for 24 h (Figures S8A and S8B). Moreover, since intrauterine ZIKV infections can lead to microcephaly in the developing human embryo by selectively damaging neural stem cells (NSCs) (Martinot et al., 2018), in a separate study we robotically generated NSCs sufficient for 184 plates (384-well format) and performed systematic genome-wide knockdown screens to identify host factors that can protect from ZIKV infection (data not shown). Collectively, these studies showed that the automated production of different neural and non-neural cell types can be established under standardized scale-up and scale-down conditions (Table S5) enabling high-throughput genetic and chemical screens.
Discussion
Cell reprogramming has allowed the generation of thousands of new hiPSC lines over the last decade. The ever-increasing number of new cell lines, including concerted efforts to generate, biobank, and distribute large numbers of cell lines derived from ethnically diverse individuals and patients with genetic diseases (Soares et al., 2014b), reinforces the need for implementing high-throughput cell culture methods that can be used as cost-efficient, standardized, and safe SOPs. It would be ideal if the production and quality testing of new hiPSC lines could be performed after employing the same reprogramming method (e.g., Sendai virus, episomal plasmids), consistently using the same chemically defined media and reagents and performing the same cell culture practices.
Currently, the culture and differentiation of hiPSC lines pose significant technical and scientific challenges for basic and translational research. Uniform and standardized processing of multiple cell lines and manufacturing various lineage-specific cell types in parallel are particularly cumbersome and inefficient for large-scale projects. Relying on a small number of cell lines for modeling human diseases and studying gene effects (e.g., population genetics) may lead to underpowered results (Cahan and Daley, 2013; Sharma et al., 2020). Another challenge is continuous passage of self-renewing hPSCs, while cell differentiation experiments are initiated in parallel. Therefore, to increase experimental reproducibility, the production of large cell quantities at a given passage number and establishing an original batch (CryoPause) was recommended (Wong et al., 2017). Automation can help to overcome these challenges, reduce the burden of manual hiPSC culture, and contribute to improving overall experimental reproducibility. Our daily experience using the CTST over the last 5 years convinced us of the advantages and versatility of automated cell culture. High-quality hPSCs can be expanded, cryopreserved, differentiated, and utilized on demand in large flasks or assay-ready microplates. In contrast with previous studies that also used the CTST system (Table S6), we were able to automate and characterize all essential steps of hiPSC culture, including massive cell expansion (Figure 2Q) and controlled multi-lineage differentiation yielding functional cell types. Systematic cell characterization experiments using complementary methods demonstrated that cells cultured manually or robotically were qualitatively similar, further supporting the notion that industrial-scale culture of hiPSCs is feasible and not limited by the availability, work schedule, and manual labor of specially trained scientists.
The combined use of E8 medium, VTN-N coating, enzyme-free cell passaging, and the CEPT cocktail was optimal for automated cell culture. Previous studies also reported improved culture of hPSCs in E8 medium compared with traditional feeder-based methods (Wang et al., 2013; Wong et al., 2017). In general, spontaneous cell differentiation and contamination with unwanted cells might be a challenge when culturing large quantities of hPSCs in a high-throughput fashion. The advantage of using CTST is that cells can be expanded as adherent cultures, while other 3D methods and suspension cultures (e.g., bioreactors, stirring tanks) will make metabolite and oxygen exchange less controlled, expose cells to shear stress, and lead to the merging of free-floating spheres (Hookway et al., 2016; Liu et al., 2014; Schwedhelm et al., 2019; Singec et al., 2006). However, spontaneous differentiation may also occur in adherent cultures after repeated enzymatic passaging (Barbaric et al., 2014; Garitaonandia et al., 2015; Wang et al., 2013), which can be avoided by using enzyme-free approaches such as EDTA. Based on our experience with growing cell lines in E8 medium over several years, spontaneous differentiation has not been a limiting factor for automated cell culture described here. Indeed, it is possible that the use of E8 medium, EDTA, and the cytoprotective CEPT cocktail may help to minimize the risk of spontaneous differentiation. Future work and data sharing across different laboratories using automated cell culture will help to further establish this notion. Last, all experiments in this study were carried out using the CTST system in a preclinical research setting (BSL-2). As other robotic cell culture systems are becoming available (e.g., Celltrio), the next critical step toward the development of clinical-grade cellular products should be the establishment and testing of automated systems that are compatible with good manufacturing practice (GMP) guidelines.
Experimental procedures
Detailed descriptions of experimental procedures can be found in the supplemental information.
Automated and manual cell culture
All hESC and hiPSC lines (Table S1) were maintained under feeder-free conditions in E8 medium (Thermo Fisher) and VTN-N-coated (Thermo Fisher) microplates or T175 flasks as described in the supplemental information.
Cell culture medium analysis
Medium analyses were done using a Vi-Cell MetaFLEX Bioanalyte analyzer (Beckman). Spent cell culture medium was analyzed and evaluated for pH, pO2, pCO2, glucose, lactate, and electrolytes every 24 h.
Live-cell metabolic assays using the Seahorse XF analyzer
The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were analyzed using a Seahorse XF-96 analyzer (Agilent) according to the manufacturer's protocol. OCR and ECAR values were normalized to total cells per well.
MEA
Electrophysiology was performed on the Maestro APEX robotic platform (Axion Biosystems) as described in the supplemental information.
Mass CyTOF
Following single-cell dissociation, hESCs/iPSCs were stained and labeled for CyTOF analysis using the Maxpar Human ES/iPS Phenotyping Panel Kit (Fluidigm) and other lanthanide metal-labeled antibodies (Table S4).
Single-cell RNA library preparation and sequencing
hESCs, hiPSCs, and their derived cell types were single-cell dissociated, loaded on a Chromium Controller (10× Genomics) to generate single-cell gel bead-in-emulsions (GEMs) and barcoding. Libraries were sequenced on an Illumina HiSeq 3,000.
Analysis of single-cell RNA-seq
Details of the analysis procedure are described in the supplemental information.
Automated and manual differentiation into embryonic germ layers
Endoderm and mesoderm differentiations were induced using the TeSR-E8 optimized STEMdiff Definitive Endoderm Kit (STEMCELL Technologies) or STEMdiff Mesoderm Induction Medium (STEMCELL Technologies). Endoderm and mesoderm cells were analyzed on day 5. Ectoderm differentiation was induced using E6 medium supplemented with LDN-193189 (100 nM, Tocris) and A83-01 (2 μM, Tocris) and cells were analyzed on day 7. All automated and manual protocols were performed in parallel as described in the supplemental information.
Automated and manual neuronal differentiation
Neuronal differentiation is summarized (Figure 6A) and described in the supplemental information.
Automated and manual cardiomyocyte differentiation
Cardiomyocyte differentiation is summarized (Figure 7A) and described in the supplemental information.
Automated and manual hepatocyte differentiation
Hepatocyte differentiation was performed as summarized (Figure 7H) and described in the supplemental information.
Data and code availability
Raw sequencing data generated in this study can be found in the NCBI SRA database under the Bioproject accession number PRJNA657268.
Analysis code is available at https://github.com/cemalley/Tristan_methods.
Author contributions
C.A.T. and I.S. conceived the study and experiments. C.A.T., P.O., J.S., P.C., V.M.J., Y.G., Y.J., C.B., E.B., S.K.M., D.D., and M.J.I. performed experiments. C.A.T., P.O., J.S., C.M, P.C., V.M.J., J.B., S.K.M., M.J.I., T.C.V., S.M., A.S., and I.S. contributed to data analysis and discussions. C.T. and I.S. wrote the manuscript.
Conflict of interests
A.S. and I.S. are co-inventors on a US Department of Health and Human Services patent application covering CEPT and its use.
Acknowledgments
We thank all our colleagues at NCATS, DPI. We are grateful to Alan Hoofring and Ethan Tyler from the NIH Medical Arts Design Section for their technical expertise. We also acknowledge funding from the NIH Common Fund (Regenerative Medicine Program), the NIH HEAL Initiative and, in part, the intramural research program of NCATS.
Published: December 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.11.004.
Supplemental information
References
- Aijaz A., Li M., Smith D., Khong D., Leblon C., Fenton O.S., Olabisi R.M., Libutti S., Tischfield J., Maus M.V., et al. Biomanufacturing for clinically advanced cell therapies. Nat. Biomed. Eng. 2018;2:362–376. doi: 10.1038/s41551-018-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald P.R.T., Chandra A., Thomas D., Chose O., Massouridès E., Laâbi Y., Williams D.J. Comparability of automated human induced pluripotent stem cell culture: a pilot study. Bioproc. Biosyst. Eng. 2016;39:1847–1858. doi: 10.1007/s00449-016-1659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaric I., Biga V., Gokhale P.J., Jones M., Stavish D., Glen A., Coca D., Andrews P.W. Time-lapse analysis of human embryonic stem cells reveals multiple bottlenecks restricting colony formation and their relief upon culture adaptation. Stem Cell. Reports. 2014;3:142–155. doi: 10.1016/j.stemcr.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E., et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tristan C.A., Chen L., Jovanovic V.M., Malley C., Chu P.H., Ryu S., Deng T., Ormanoglu P., Tao D., et al. Springer US; 2021. A Versatile Polypharmacology Platform Promotes Cytoprotection and Viability of Human Pluripotent and Differentiated Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniszewski M., Crombie D.E., Henderson R., Liang H.H., Wong R.C.B., Hewitt A.W., Pébay A. Automated cell culture systems and their applications to human pluripotent stem cell studies. SLAS Technol. 2018;23:315–325. doi: 10.1177/2472630317712220. [DOI] [PubMed] [Google Scholar]
- Garitaonandia I., Amir H., Boscolo F.S., Wambua G.K., Schultheisz H.L., Sabatini K., Morey R., Waltz S., Wang Y.C., Tran H., et al. Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PLoS One. 2015;10:1–25. doi: 10.1371/journal.pone.0118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Gaeta X., Sahakyan A., Chan A.B., Hong C.S., Kim R., Braas D., Plath K., Lowry W.E., Christofk H.R. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell. 2016;19:476–490. doi: 10.1016/j.stem.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Tian L., Zhang J.Z., Kitani T., Paik D.T., Lee W.H., Wu J.C. Single-cell RNA sequencing of human embryonic stem cell differentiation delineates adverse effects of nicotine on embryonic development. Stem Cell. Reports. 2019;12:772–786. doi: 10.1016/j.stemcr.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookway T.A., Butts J.C., Lee E., Tang H., McDevitt T.C. Aggregate formation and suspension culture of human pluripotent stem cells and differentiated progeny. Methods. 2016;101:11–20. doi: 10.1016/j.ymeth.2015.11.027. [DOI] [PubMed] [Google Scholar]
- Horiguchi I., Urabe Y., Kimura K., Sakai Y. Effects of glucose, lactate and basic FGF as limiting factors on the expansion of human induced pluripotent stem cells. J. Biosci. Bioeng. 2018;125:111–115. doi: 10.1016/j.jbiosc.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Jacobs K., Zambelli F., Mertzanidou A., Smolders I., Geens M., Nguyen H.T., Barbé L., Sermon K., Spits C. Higher-density culture in human embryonic stem cells results in DNA damage and genome instability. Stem Cell Reports. 2016;6:330–341. doi: 10.1016/j.stemcr.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel E.A., Lanza R. Next-generation stem cells — ushering in a new era of cell-based therapies. Nat. Rev. Drug Discov. 2020;19:463–479. doi: 10.1038/s41573-020-0064-x. [DOI] [PubMed] [Google Scholar]
- Konagaya S., Ando T., Yamauchi T., Suemori H., Iwata H. Long-term maintenance of human induced pluripotent stem cells by automated cell culture system. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H.H., Gao X., DeKeyser J.M., Fetterman K.A., Pinheiro E.A., Weddle C.J., Fonoudi H., Orman M.V., Romero-Tejeda M., Jouni M., et al. Negligible-cost and weekend-free chemically defined human iPSC culture. Stem Cell. Reports. 2020;14:256–270. doi: 10.1016/j.stemcr.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann E.S., Estevez-Silva M.C., Ashton R.S. Defined human pluripotent stem cell culture enables highly efficient neuroepithelium derivation without small molecule inhibitors. Stem Cells. 2014;32:1032–1042. doi: 10.1002/stem.1622. [DOI] [PubMed] [Google Scholar]
- Liu N., Zang R., Yang S.T., Li Y. Stem cell engineering in bioreactors for large-scale bioprocessing. Eng. Life Sci. 2014;14:4–15. [Google Scholar]
- Ludwig T.E., Levenstein M.E., Jones J.M., Berggren W.T., Mitchen E.R., Frane J.L., Crandall L.J., Daigh C.A., Conard K.R., Piekarczyk M.S., et al. Derivation of human embryonic stem cells in defined conditions. Nat. Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- Mallanna S.K., Duncan S.A. Differentiation of hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 2013;1:1G.4.1–1G.4.13. doi: 10.1002/9780470151808.sc01g04s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinot A.J., Abbink P., Afacan O., Prohl A.K., Bronson R., Hecht J.L., Borducchi E.N., Larocca R.A., Peterson R.L., Rinaldi W., et al. Fetal neuropathology in zika virus-infected pregnant female rhesus monkeys. Cell. 2018;173:1111–1122.e10. doi: 10.1016/j.cell.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren D., Gorba T., Marguerie De Rotrou A., Pillai G., Chappell C., Stacey A., Lingard S., Falk A., Smith A., Koch P., et al. Automated large-scale culture and medium-throughput chemical screen for modulators of proliferation and viability of human induced pluripotent stem cell-derived neuroepithelial-like stem cells. J. Biomol. Screen. 2013;18:258–268. doi: 10.1177/1087057112461446. [DOI] [PubMed] [Google Scholar]
- Niepel M., Hafner M., Mills C.E., Subramanian K., Williams E.H., Chung M., Gaudio B., Barrette A.M., Stern A.D., Hu B., et al. A multi-center study on the reproducibility of drug-response assays in mammalian cell lines. Cell Syst. 2019;9:35–48.e5. doi: 10.1016/j.cels.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-Gerald C.S., Sato Y., Cowan C.A., Chien K.R., Melton D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Panopoulos A.D., D’Antonio M., Benaglio P., Williams R., Hashem S.I., Schuldt B.M., DeBoever C., Arias A.D., Garcia M., Nelson B.C., et al. iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types. Stem Cell. Reports. 2017;8:1086–1100. doi: 10.1016/j.stemcr.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull D., Sevilla A., Zhou H., Hahn A.K., Kim H., Napolitano C., Tsankov A., Shang L., Krumholz K., Jagadeesan P., et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods. 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Sufi J., Vlckova P., Kyriakidou P., Acton S.E., Li V.S.W., Nitz M., Tape C.J. Cell-type-specific signaling networks in heterocellular organoids. Nat. Methods. 2020;17:335–342. doi: 10.1038/s41592-020-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Yang S.M., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P., et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti A., Repetti G.G., Sun C., Price F.D., Reny D.C., Rapino F., Weisinger K., Benkler C., Peterson Q.P., Davidow L.S., et al. Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system. Stem Cell Reports. 2016;6:993–1008. doi: 10.1016/j.stemcr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Nowicka M., Krieg C., Weber L.M., Hartmann F.J., Guglietta S., Becher B., Levesque M.P. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Research. 2017;6:1–53. doi: 10.12688/f1000research.11622.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Niaudet C., Simonson O.E., Salmela E., Hansson E.M., Domogatskaya A., Xiao Z., Damdimopoulou P., Sheikhi M., et al. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 2014;5:1–13. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
- Sato Y., Bando H., Di Piazza M., Gowing G., Herberts C., Jackman S., Leoni G., Libertini S., MacLachlan T., McBlane J.W., et al. Tumorigenicity assessment of cell therapy products: the need for global consensus and points to consider. Cytotherapy. 2019;21:1095–1111. doi: 10.1016/j.jcyt.2019.10.001. [DOI] [PubMed] [Google Scholar]
- Schwedhelm I., Zdzieblo D., Appelt-Menzel A., Berger C., Schmitz T., Schuldt B., Franke A., Müller F.J., Pless O., Schwarz T., et al. Automated real-time monitoring of human pluripotent stem cell aggregation in stirred tank reactors. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-48814-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba N., White C.A., Lipsitz Y.Y., Yachie-Kinoshita A., Tonge P.D., Hussein S.M.I., Puri M.C., Elbaz J., Morrissey-Scoot J., Li M., et al. CD24 tracks divergent pluripotent states in mouse and human cells. Nat. Commun. 2015;6:1–11. doi: 10.1038/ncomms8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Sances S., Workman M.J., Svendsen C.N. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell. 2020;26:309–329. doi: 10.1016/j.stem.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singec I., Knoth R., Meyer R.P., Maciaczyk J., Volk B., Nikkhah G., Frotscher M., Snyder E.Y. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat. Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- Singec I., Crain A.M., Hou J., Tobe B.T.D., Talantova M., Winquist A.A., Doctor K.S., Choy J., Huang X., La Monaca E., et al. Quantitative analysis of human pluripotency and neural specification by in-depth (phospho)proteomic profiling. Stem Cell. Reports. 2016;7:527–542. doi: 10.1016/j.stemcr.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares F.A.C., Chandra A., Thomas R.J., Pedersen R.A., Vallier L., Williams D.J. Investigating the feasibility of scale up and automation of human induced pluripotent stem cells cultured in aggregates in feeder free conditions. J. Biotechnol. 2014;173:53–58. doi: 10.1016/j.jbiotec.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares F.A.C., Sheldon M., Rao M., Mummery C., Vallier L. International coordination of large-scale human induced pluripotent stem cell initiatives: Wellcome Trust and ISSCR workshops white paper. Stem Cell. Reports. 2014;3:931–939. doi: 10.1016/j.stemcr.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M., et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.J., Anderson D., Chandra A., Smith N.M., Young L.E., Williams D., Denning C. Automated, scalable culture of human embryonic stem cells in feeder-free conditions. Biotechnol. Bioeng. 2009;102:1636–1644. doi: 10.1002/bit.22187. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tsankov A.M., Akopian V., Pop R., Chetty S., Gifford C.A., Daheron L., Tsankova N.M., Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H.R., Harb G., Poh Y.C., Sintov E., Gürtler M., Pagliuca F.W., et al. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chou B.K., Dowey S., He C., Gerecht S., Cheng L. Scalable expansion of human induced pluripotent stem cells in the defined xeno-free E8 medium under adherent and suspension culture conditions. Stem Cell Res. 2013;11:1103–1116. doi: 10.1016/j.scr.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S.I., Muguruma K., et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wong K.G., Ryan S.D., Ramnarine K., Rosen S.A., Mann S.E., Kulick A., De Stanchina E., Müller F.J., Kacmarczyk T.J., Zhang C., et al. CryoPause: a new method to immediately initiate experiments after cryopreservation of pluripotent stem cells. Stem Cell. Reports. 2017;9:355–365. doi: 10.1016/j.stemcr.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Badur M.G., Divakaruni A.S., Parker S.J., Jäger C., Hiller K., Murphy A.N., Metallo C.M. Distinct metabolic states can support self-renewal and lipogenesis in human pluripotent stem cells under different culture conditions. Cell Rep. 2016;16:1536–1547. doi: 10.1016/j.celrep.2016.06.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tan L., Cederquist G.Y., Fan Y., Hartley B.J., Mukherjee S., Tomishima M., Brennand K.J., Zhang Q., Schwartz R.E., et al. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell. 2017;21:274–283.e5. doi: 10.1016/j.stem.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Lujan E., Goltsev Y., Wernig M., Nolan G.P. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data generated in this study can be found in the NCBI SRA database under the Bioproject accession number PRJNA657268.
Analysis code is available at https://github.com/cemalley/Tristan_methods.