Abstract
The insulin/insulin-like growth factor signalling pathway has been hypothesized as a major determinant of life-history profiles that vary adaptively in natural populations. In Drosophila melanogaster, multiple components of this pathway vary predictably with latitude; this includes foxo, a conserved gene that regulates insulin signalling and has pleiotropic effects on a variety of fitness-associated traits. We hypothesized that allelic variation at foxo contributes to genetic variance for size-related traits that vary adaptively with latitude. We first examined patterns of variation among natural populations along a latitudinal transect in the eastern United States and show that thorax length, wing area, wing loading, and starvation tolerance exhibit significant latitudinal clines for both males and females but that development time does not vary predictably with latitude. We then generated recombinant outbred populations and show that naturally occurring allelic variation at foxo, which exhibits stronger clinality than expected, is associated with the same traits that vary with latitude in the natural populations. Our results suggest that allelic variation at foxo contributes to adaptive patterns of life-history variation in natural populations of this genetic model.
Keywords: body size, cline, foxo, genetic architecture, starvation tolerance
1 |. INTRODUCTION
Elucidating the mechanistic basis of adaptive differentiation for complex traits in natural populations remains a fundamental goal in evolutionary biology. Fitness traits often exhibit a highly polygenic architecture (e.g., Arnegard et al., 2014; McCown et al., 2014; Savolainen et al., 2013). The likelihood of effectively mapping complex traits to causative polymorphism may depend on architecture of these quantitative traits (Barton et al., 2017; Boyle et al., 2017; Rockman, 2012; Roff, 2007; Wellenreuther & Hansson, 2016). Many empirical advances in understanding the mechanistic basis of adaptation in sexual, outbred populations include both a clear identification of traits that drive local adaptation as well as an apparently simple genetic architecture, with at least one locus that demonstrates a strong statistical association between allelic and phenotypic variation (e.g., Colosimo et al., 2005; Comeault et al., 2015; van’t Hof et al., 2016; Jones et al., 2018; Lamichhaney et al., 2016). In such examples, alleles segregating at identified candidate loci can then be directly examined for functional differences that affect performance and fitness (e.g., Chakraborty & Fry, 2016; Cheviron et al., 2012; Laurie & Stam, 1988; Manceau et al., 2011).
Body size is a trait commonly associated with fitness in a variety of taxa (Blanckenhorn, 2000; Bonnet et al., 2017; Brown et al., 1993) including Drosophila melanogaster (e.g., Promislow et al., 1998; Reeve et al., 2001; reviewed in Flatt, 2020). Size often varies predictably across environmental gradients such as those associated with latitude (Ashton, 2002; Blanckenhorn & Demont, 2004; Huey et al., 2000; James et al., 1995; Stillwell et al., 2007); such clines suggest that body size is affected by spatially varying selection and reflects local adaptation (Partridge & Coyne, 1997; Stillwell, 2010). While size-related traits are in general highly polygenic (e.g., Boyle et al., 2017), individual loci can have large effects on size (e.g., Sutter et al., 2007). In particular, multiple components of the insulin/insulin-like growth factor signalling pathway (IIS) can regulate size (e.g., Colombani et al., 2005; Sutter et al., 2007); the forkhead box-O transcription factor gene foxo is a major regulator of IIS and impacts size as well as a variety of other traits associated with fitness (Fielenbach & Antebi, 2008; Hwangbo et al., 2004; Kramer et al., 2003, 2008; Libina et al., 2003; Mattila et al., 2009). Thus, the analysis of variation in body size offers an excellent system in which to examine the translation between genotype, phenotype and fitness in natural populations.
In Drosophila melanogaster, body size increases with increasing latitude on multiple continents (Coyne & Beecham, 1987; James et al., 1995, 1997; Karan et al., 1998). Such latitudinal patterns are mirrored by altitudinal clines where size increases with increasing altitude (Fabian et al., 2015; Lack et al., 2016). These parallel and replicated patterns suggest that patterns of size variation are adaptive and associated with thermally mediated selection (e.g., Partridge et al., 1994; Stillwell, 2010). However, it remains unknown why small body size is associated with higher fitness at low latitudes and large size with higher fitness at high latitudes. De Jong and Bochdonavits (2003) hypothesized that adaptive patterns of size variation are driven primarily by one or more components of the IIS pathway; the simple prediction is that any causative variants would also exhibit pronounced and replicated allele frequency clines. Analysis of PoolSeq data has shown that multiple IIS genes (e.g., Pi3K, foxo, InR) are segregating for many alleles that vary predictably with latitude in D. melanogaster (Bergland et al., 2014; Fabian et al., 2012; Kapun et al., 2016; Kolaczkowski et al., 2011; Machado et al., 2021). The question is whether these clinal alleles are distinct with respect to gene function and, at least in part, underlie the observed patterns of local adaptation in size (Paaby et al., 2014).
Here, we examine patterns of variation among natural populations for two measures of body size (thorax length, wing area and their ratio) as well as two additional complex traits associated with fitness and correlated with size, development time and starvation tolerance. We evaluate patterns of clinality for SNPs at the foxo locus and test the functional significance of naturally occurring allelic variation; we specifically assess whether the inferred effects of allelic variation at foxo are consistent with patterns of trait variation among natural populations. If size in D. melanogaster is highly polygenic with no loci of major effect, then alternative alleles segregating at foxo should be effectively and functionally equivalent in their phenotypic effects in laboratory- or field-based functional assays. However, if foxo is a strong determinant of body size in this species, then differences among natural alleles may be of sufficient magnitude to be detectable in association studies. If allelic variation at foxo substantially contributes to phenotypic variation and local adaptation, then the effects of foxo alleles on phenotype should be concordant with patterns observed in natural populations.
2 |. MATERIALS AND METHODS
2.1 |. Identification of SNPs/alleles that vary predictably with latitude
Fabian et al. (2012) identified a series of SNPs in foxo that exhibited high FST in pooled sequencing of natural populations derived from Florida (low latitude), Pennsylvania (mid-latitude), and Maine, USA (high latitude). From this analysis, which suggested that foxo might be a clinal outlier in D. melanogaster populations in eastern North America, we selected candidate foxo alleles to test whether allelic variation at this locus might be of functional significance and contribute to phenotypic clines for traits linked to foxo function. The selection of the foxo alleles was based on: (i) High FST in the Fabian et al. (2012) data, (ii) preliminary indication that latitudinal differences in allele frequencies were concordant with seasonal differences in allele frequencies (following the rationale and methods in Paaby et al., 2014), and (iii) alleles being at intermediate frequency and present in sufficient numbers in the Drosophila Genetic Reference Panel (DGRP, Mackay et al., 2012) so that we could establish multiple, independent sets of lines for functional analysis (see Constructing recombinant outbred populations below). The selected foxo allele, which satisfied the three criteria described above, was actually a haplotype defined by two SNPs (synonymous A/G at position 3R:9892517, D. melanogaster reference genome v.5.0, and intronic T/G at position 3R:9894559) spanning approximately 2 kb within the foxo gene (see Figure 1, Table S2). These sites were observed to be in perfect linkage disequilibrium (r2 =1) in the DGRP, such that they covary and are not independent. Further details regarding these polymorphic sites and the landscape of linkage disequilibrium at foxo are provided in our previous publication (Durmaz et al., 2019).
FIGURE 1.
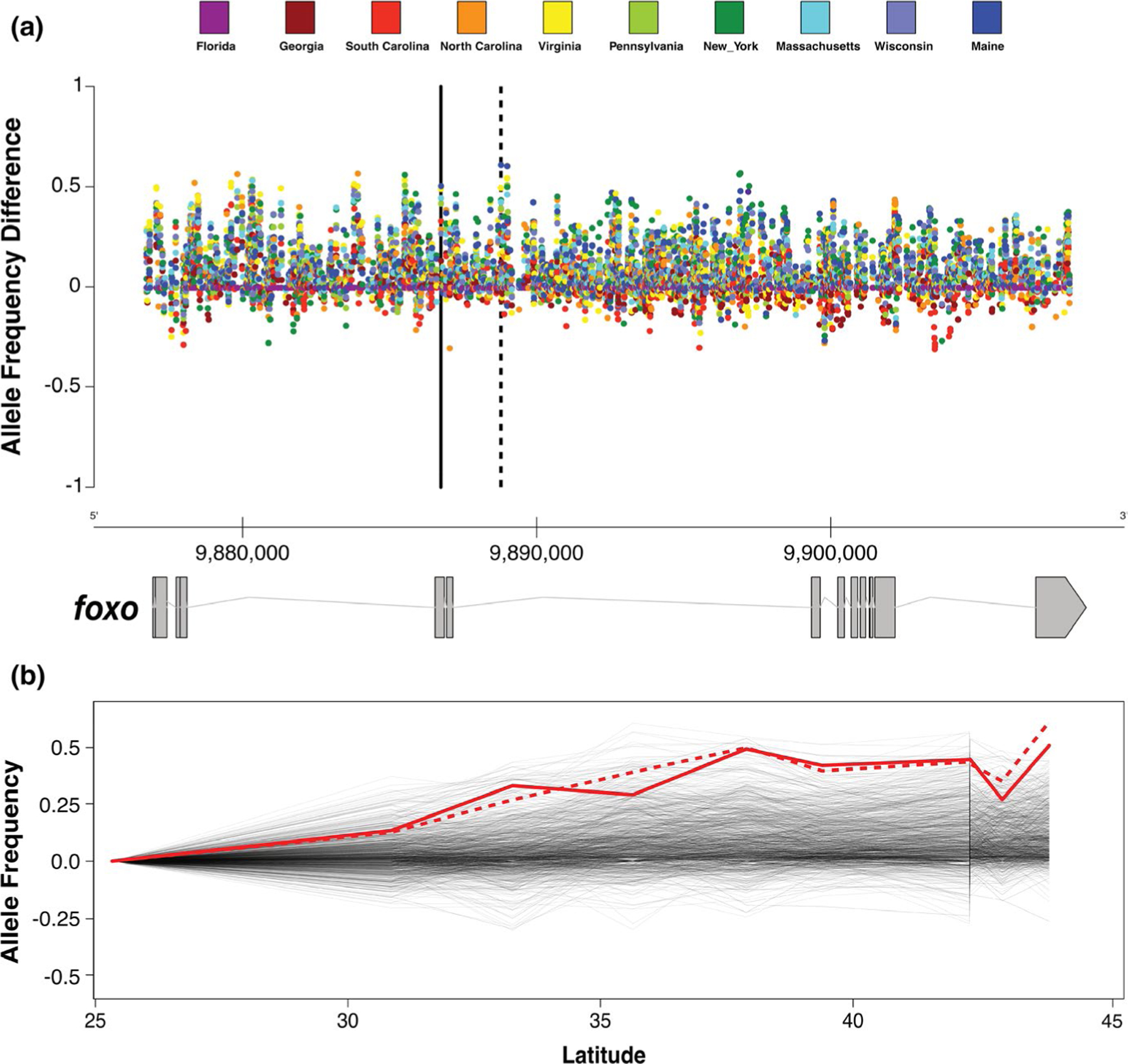
Allele frequency changes for foxo-associated SNPs in 10 populations sampled from the eastern U.S (population specifics given in Table S1). Both plots show allele frequency differences conditioned to increase from south to north, with frequencies in Florida being set to zero. (a) Allele frequency differences for a given population (colour coded) compared to the reference (Florida) for all SNPs according to their genomic position. The foxo candidate SNPs are denoted by two vertical black lines (solid: 3R: 9,892,517; dashed: 3R: 9,894,449; D. melanogaster reference genome v6). (b) Shows how allele frequencies change with latitude. The two foxo candidate SNPs are shown in red (solid: 3R: 9,892,517; dashed: 3R: 9,894,449) [Colour figure can be viewed at wileyonlinelibrary.com]
To examine clinal patterns associated with the foxo locus in general (i.e., foxo compared to the genome as a whole) and the focal low- and high-latitude foxo haplotypes in particular (i.e., the focal foxo SNPs as compared to other SNPs across the foxo locus), we analysed patterns of allele frequency variation in 10 populations collected at different latitudes in the eastern U.S. and sequenced as pools (Figure 1; Table S1; Bergland et al., 2014; Kapun et al., 2016; Machado et al., 2021). We restricted our analyses to high-confidence SNPs that were polymorphic in the DGRP data set (Langley et al., 2012; Mackay et al., 2012) and isolated a total of 1372 SNPs located inside or within 2 kbp up- and downstream of the annotated foxo gene. To provide a null genomic context for allele frequency differentiation at foxo, we isolated 20,000 SNPs located in short introns (<60 bp) and at least 100 kbp distance from the breakpoints of common cosmopolitan inversions (Corbett-Detig et al., 2012) that are consistent with patterns of neutrality (Clemente & Vogl, 2012; Parsch et al., 2010). For each of these neutral and foxo-associated SNPs, we tested for significant correlations between latitude and allele frequencies using generalized linear models (GLM) with a binomial error structure of the form: yi = L + εi, where yi is the allele frequency of the ith SNP, L is the continuous factor “Latitude” and εi is the binomial error of the ith SNP (Figure 2a,b). We further assessed whether allele frequency changes of the two candidate SNPs that constitute our focal haplotype were more clinal than neutral SNPs or other SNPs located within or in the proximity of foxo (Figure 2c,d). To this end, we compared the −log10(p)-values from the GLMs for each of the two focal SNPs to distributions of −log10(p)-values from GLMs of either neutral SNPs or noncandidate SNPs associated with foxo. We subsequently calculated empirical cumulative distribution functions (ECDF; with total area = 1) based on −log10(p)-values from all neutral or noncandidate foxo SNPs in R (R Development Core Team, 2009). To test if the significance values associated with the two focal SNPs were greater than the 95 percentiles of each distribution, we integrated over the area under each ECDF with values larger than the significance of each candidate SNP and subtracted the integral value from 1, which represents the total area of the ECDF.
FIGURE 2.
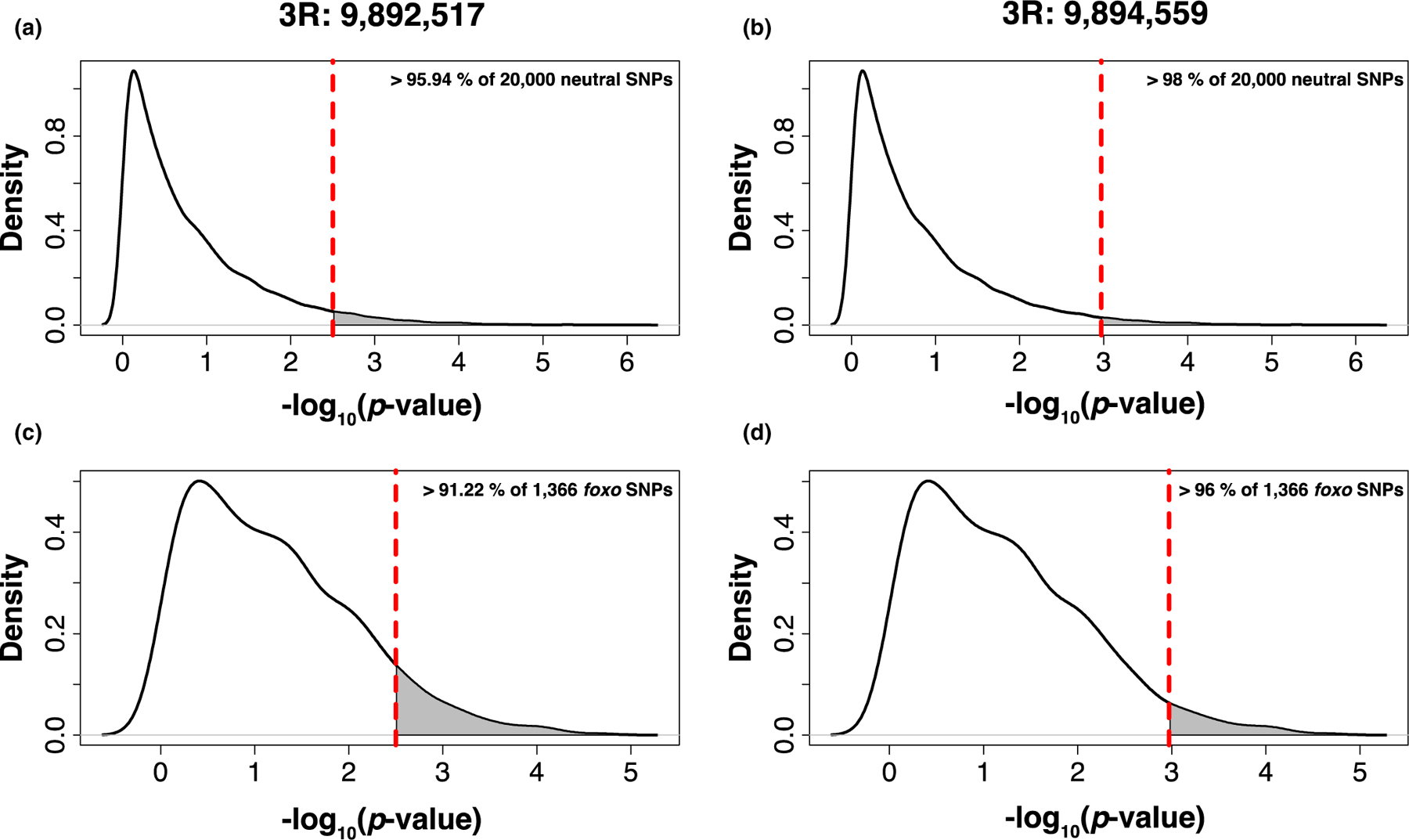
Empirical cumulative density functions (ECDF; total area = 1) calculated from the distribution of −log10 (p-values) for generalized linear models that test for associations between allele frequencies and latitude in 20,000 neutrally evolving SNPs (a,b) and 1372 noncandidate SNPs located inside or within 2 kbp distance to foxo. The vertical dashed lines indicate the significance values of the two candidate SNPs 3R: 9,892,517 (a,c) and 3R: 9,894,449 (b,d). The grey areas limited by the dashed line indicate the percentiles of neutral or noncandidate foxo SNPs with significance values larger than the candidates [Colour figure can be viewed at wileyonlinelibrary.com]
To investigate and visualize the relative patterns of allele frequency change for the two focal SNPs with respect to all other SNPs located inside or in close proximity to foxo, we conditioned the alleles to have lower frequencies in Florida compared to the population in Maine for each foxo-associated SNP. Allele frequencies in Florida were set to zero and we then calculated the allele frequency differences relative to Florida for all other populations (Figure 1).
2.2 |. Constructing recombinant outbred populations
Based on the combination of the results of Fabian et al. (2012) and the DrosRTEC (Drosophila Real-Time Evolution Consortium) sequencing effort (Machado et al., 2021), we identified individual lines in the DGRP (Mackay et al., 2012) that were homozygous for the candidate foxo allele that was at high frequency in high-latitude populations [hereafter, the high-latitude (HL) allele], and lines that were fixed for the foxo allele that was at high frequency in low-latitude samples [hereafter, the low-latitude (LL) allele]. Scripts for this filtering of the DGRP based on nucleotide state and locus are provided in Files S4 and S5. Two biological replicates were established using 18 independent (different and nonoverlapping) lines per cage per foxo allele; thus, each cage was constructed with a completely distinct and independent set of inbred lines (see Table S2). These biological replicates were then also replicated as technical replicates, resulting in a total of eight experimental population cages (Table S2). Each population cage was individually founded using 10 individuals of each sex, from age- and density-controlled cohorts, from each of the 18 inbred DGRP founding lines associated with each specific cage.
After establishment, each of the eight population cages was cultured at a population size of ~2000 adults on standard cornmeal-molasses medium for eight discrete generations of outcrossing under conditions of constant temperature (25°C) and photoperiod (12L:12D). Thus, at the end of the experimental period, we generated replicate population cages in which the focal foxo allele was homozygous and fixed for either the high- or low-latitude allele and the genomic background was randomized across the 18 inbred lines used to find each respective cage (Behrman, Howick, et al., 2018; Paaby et al., 2014). To test for potential genome-wide patterns of genetic differentiation among the recombinant outbred populations (ROP) fixed for either the high or low-latitude haplotypes, we used available genome sequence data for the inbred lines used to found experimental populations (Table S2) for sets A and B (representing the two biological replicates) to calculate SNP-wise FST based on the method of Weir and Cockerham (1984). This was done to evaluate whether any other position in the genome was highly differentiated among our experimental populations and could thus confound interpretation of results (Figure 3; Files S4 and S5). Note that our experimental approach makes two assumptions: (i) De novo mutational input at the foxo locus is insufficient to meaningfully affect experimental outcomes, and (ii) genetic drift and/or selection did not result in random fixation of any additional alleles in the construction of these populations. After eight generations of density-controlled culture under standardized environmental conditions, we established replicate density-controlled vial cultures (30 ± 10 eggs/vial) for subsequent phenotyping of the high-latitude and low-latitude foxo alleles.
FIGURE 3.
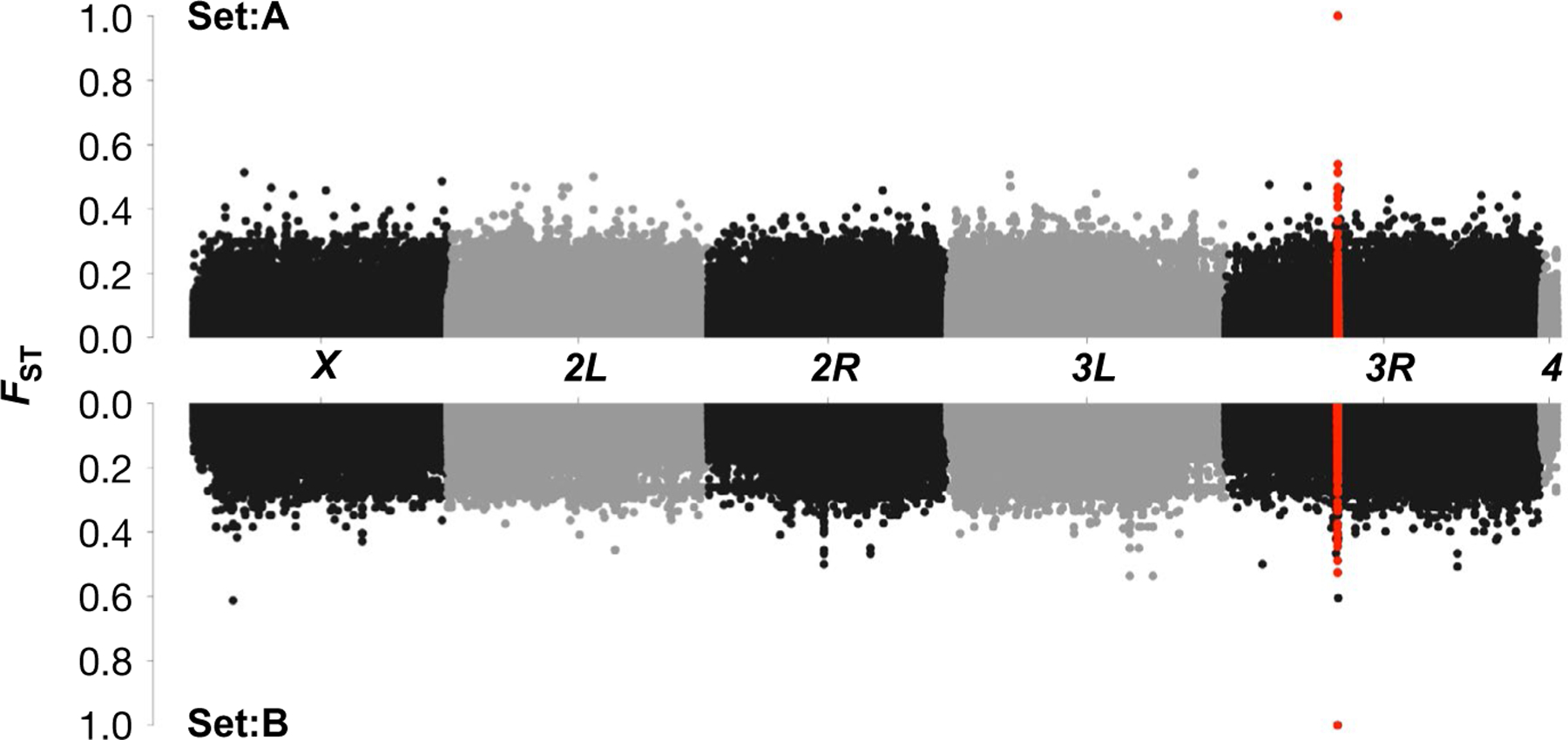
FST Manhattan plots for the biological replicates that were constructed from independent sets of inbred lines from the DGRP panel. FST values for all SNPs at the foxo locus are highlighted in red. The analyses show that only the two focal foxo SNPs are fixed (FST =1) for alternative alleles in the low- and high-latitude population cages, and that this is consistent for both of the biological replicates (Set A, top; Set B, bottom); all other foxo SNPs shown in red do not reach fixation at FST = 1. Note that the two focal, fixed foxo SNPs are so close to each other that they appear as a single red dot at FST = 1, both in Set A and Set B. The construction of the recombinant outbred population cages, using 18 inbred lines that are fixed for the SNPs of interest, is thus not confounded by fixed differences between experimental populations at other positions in the genome [Colour figure can be viewed at wileyonlinelibrary.com]
2.3 |. Isofemale lines from natural populations
Thirty isofemale lines were randomly selected from each of six natural populations along the east coast of the U.S. to serve as a latitudinal comparison to the recombinant foxo populations [described in Rajpurohit et al., 2017, 2018: Homestead, FL (HFL), Jacksonville, FL (JFL), Charlottesville, VA (CVA), Media, PA (MPA), Lancaster, MA (LMA), and Bowdoin, ME (BME)]. Individual lines from each population were maintained on a 21-day culture regime under the same environmental conditions as the foxo recombinant cages (25°C, 12L:12D). Prior to phenotyping, each isofemale line was cultured for two generations at low density (30 ± 10 eggs/vial) at 25°C, 12L:12D; in the third generation, freshly eclosed flies were collected in daily cohorts and used in the phenotypic assays described below.
2.4 |. Phenotype assays
In all assays, foxo ROPs were tested simultaneously in three replicates (experimental blocks, conducted on different days) in the same generation since founding of the experimental cages. For assessment of starvation resistance, virus infection of the cages precluded running three independent blocks, and a single block was included in the analysis. For the natural populations, all lines from all populations were assayed simultaneously for all phenotypes using discrete 1d cohorts for each phenotype.
2.4.1 |. Development time
For the foxo recombinant outbred populations, eggs were collected from each cage over a 3 h window using large Petri dishes containing standard medium supplemented with live yeast. The collected eggs were then counted and distributed in groups of 30 into three replicate vials per cage. For the natural populations, eggs from all isofemale lines were similarly collected over 3 h in small collection receptacles. Eggs were counted and distributed to new collection vials, with density also standardized at 30 eggs per vial. All experimental material was subsequently cultured under the standard conditions. Experimental vials were checked four times daily (9 a.m., 1 p.m., 5 p.m., 9 p.m.); eclosion events and sex were recorded.
2.4.2 |. Body size
For both the foxo cages as well as the natural populations, flies from the development assay were transferred to new vials, allowed to mate and age for five days post eclosion, then were preserved in 95% ethanol for subsequent size measurements. A total of 10 flies of each sex were randomly sampled and measured from each foxo ROP cage and five flies for each sex were measured for each isofemale line from the natural populations. Body size measurements (thorax length and wing area) were recorded using a Leica MZ9.5 microscope mounted with an Olympus DP73 camera with CellSens standard measuring software. Thorax length was measured as the longest length across the dorsal shield in lateral view; wing area was defined as a polygon using a standardized series of veinous landmarks. We also calculated and analysed the ratio of total wing area to thorax length, indicative of (the inverse of) “wing loading”, a variable thought to be an important determinant of flight ability (Azevedo et al., 1998; Gilchrist et al., 2000).
2.4.3 |. Starvation resistance
For the foxo recombinant outbred populations, embryos were collected from each cage in two replicate glass culture bottles and density was standardized at 150 ± 10 eggs per bottle. Isofemale lines from the natural populations were transferred into replicate vials and density standardized at 30 ± 10 embryos per vial. All culture was done under the standard conditions (25°C, 12L:12D). Upon eclosion, mixed sex daily cohorts were collected over 3 days and subsequently aged to 5 days. The flies were then separated by sex into replicate groups of 10 and placed into glass vials equipped with a small cotton ball saturated with 1 ml of water. Samples were placed in an incubator at 25°C, 12L:12D and mortality was recorded at four timepoints per day (9 a.m., 1 p.m., 5 p.m., and 9 p.m.) until all flies had died. The foxo cages were assayed in three replicates per cage population; all isofemale lines from each of the natural populations were also assayed.
2.5 |. Statistical analysis
For the natural populations, data were analysed separately by sex. All data were analysed using a mixed-effects ANOVA model, using population as a fixed factor and isofemale line nested within population as a random factor (estimated with restricted maximum likelihood). For all traits other than starvation tolerance, experimental block (N = 3) was also included as an additional effect. For the foxo recombinant outbred populations, we analysed data separately for both sexes with nested ANOVA models, using foxo allele, DGRP set nested in allelic state, and experimental replicate (population cage) nested in a combination of set and allele as factors.
3 |. RESULTS
3.1 |. Alleles at foxo exhibit steep latitudinal clines
By analysing genome-wide Pool-Seq data from 10 populations sampled along the U.S. east coast generated by the DrosRTEC consortium (Kapun et al., 2016; Machado et al., 2021), we show that numerous SNPs associated with foxo exhibit steep latitudinal clines and extensive differentiation as a function of geography (Figure 1). Notably and consistent with previous observations by Fabian et al. (2012), the two focal foxo candidate SNPs that are denoted by black and red lines in Figure 1a,b, respectively, exhibit strong patterns of allele frequency change across latitudes. In fact, clinal patterns of the two candidate SNPs were more pronounced than 95% of neutrally evolving SNPs located in short introns (>95.94% for 3R:9,892,517 and >98% for 3R:9,894,559, respectively; Figure 2a,b) and 91% of all noncandidate SNPs located within or close to foxo (>91.22% for 3R:9,892,517 and >96% for 3R:9,894,559, respectively; Figure 2c,d).
In establishing the recombinant outbred populations (ROPs) using the DGRP panel of inbred lines, the goal was to use candidate SNPs as markers of functional effects for naturally occurring alleles or haplotypes at this locus (Berhman et al., 2018; Paaby et al., 2014). The utility of this method is predicated on using a sufficient number of independent inbred lines such that no other position in the genome, other that the candidate site(s), is fixed or highly differentiated between experimental sets. In Figure 3, FST between experimental cages (foxo allele AT vs. foxo allele GG) is plotted as a function of chromosomal position for all SNPs segregating in the biological replicate sets A (Figure 3a) and B (Figure 3b). While there are multiple sites on each chromosome arm with FST > 0.4 between the sets of lines used to construct the alternative foxo allelic cages, only the candidate sites are fixed between the cages that comprise the allelic states.
3.2 |. Natural populations vary clinally for size-related traits
3.2.1 |. Development time
Development time did not vary predictably with latitude. For both males and females, significant variation was observed among lines and among populations (Table 1) but there was no major association with latitudinal origin (Table S3, Figures 4a and 5a). We did, however, observe distinct patterns of development time among the replicate experimental blocks, despite controlling for density and culture conditions. This suggests that this trait is affected by additional environmental variables, measurement or other experimental error, or a combination of the two. It should be noted, however, that in examination of the experimental blocks individually, no significant association between development time and latitudinal origin of the population was observed. This is in contrast to patterns of seasonal variation, which demonstrate predictable change in development time as a function of time of collection (Behrman et al., 2015).
TABLE 1.
Analyses of variance (ANOVA) for the assayed phenotypic traits among natural populations from across the latitudinal cline. The models shown below account for the random effect of line nested in population (estimated with restricted maximum likelihood, REML), but we do not report variance component estimates here since they are of little biological interest. Note that we did not use experimental blocks for measuring starvation tolerance. See Materials and Methods for further details
| Females |
Males |
|||||||
|---|---|---|---|---|---|---|---|---|
| F | dfnum | dfden | p | F | dfnum | dfden | p | |
| Developmental time | ||||||||
| Population | 6.75 | 5 | 133.3 | <.0001 | 6.42 | 5 | 132.1 | <.0001 |
| Block | 69.69 | 2 | 649.8 | <.0001 | 75.51 | 2 | 539.6 | <.0001 |
| Thorax length | ||||||||
| Population | 4.71 | 5 | 135.4 | .0005 | 3.28 | 5 | 140.9 | .0078 |
| Block | 0.05 | 2 | 363.6 | .96 | 1.96 | 2 | 365.8 | .14 |
| Wing area | ||||||||
| Population | 14.15 | 5 | 136.8 | <.0001 | 11.54 | 5 | 136.4 | <.0001 |
| Block | 2.72 | 2 | 386.6 | .067 | 4.71 | 2 | 394.9 | .0095 |
| Wing area/thorax length | ||||||||
| Population | 13.91 | 5 | 135.6 | <.0001 | 11.45 | 5 | 137.7 | <.0001 |
| Block | 3.69 | 2 | 365.3 | 0.026 | 3.27 | 2 | 372.1 | 0.039 |
| Starvation tolerance | ||||||||
| Population | 10.41 | 5 | 116.5 | <.0001 | 5.53 | 5 | 113.7 | .0001 |
FIGURE 4.
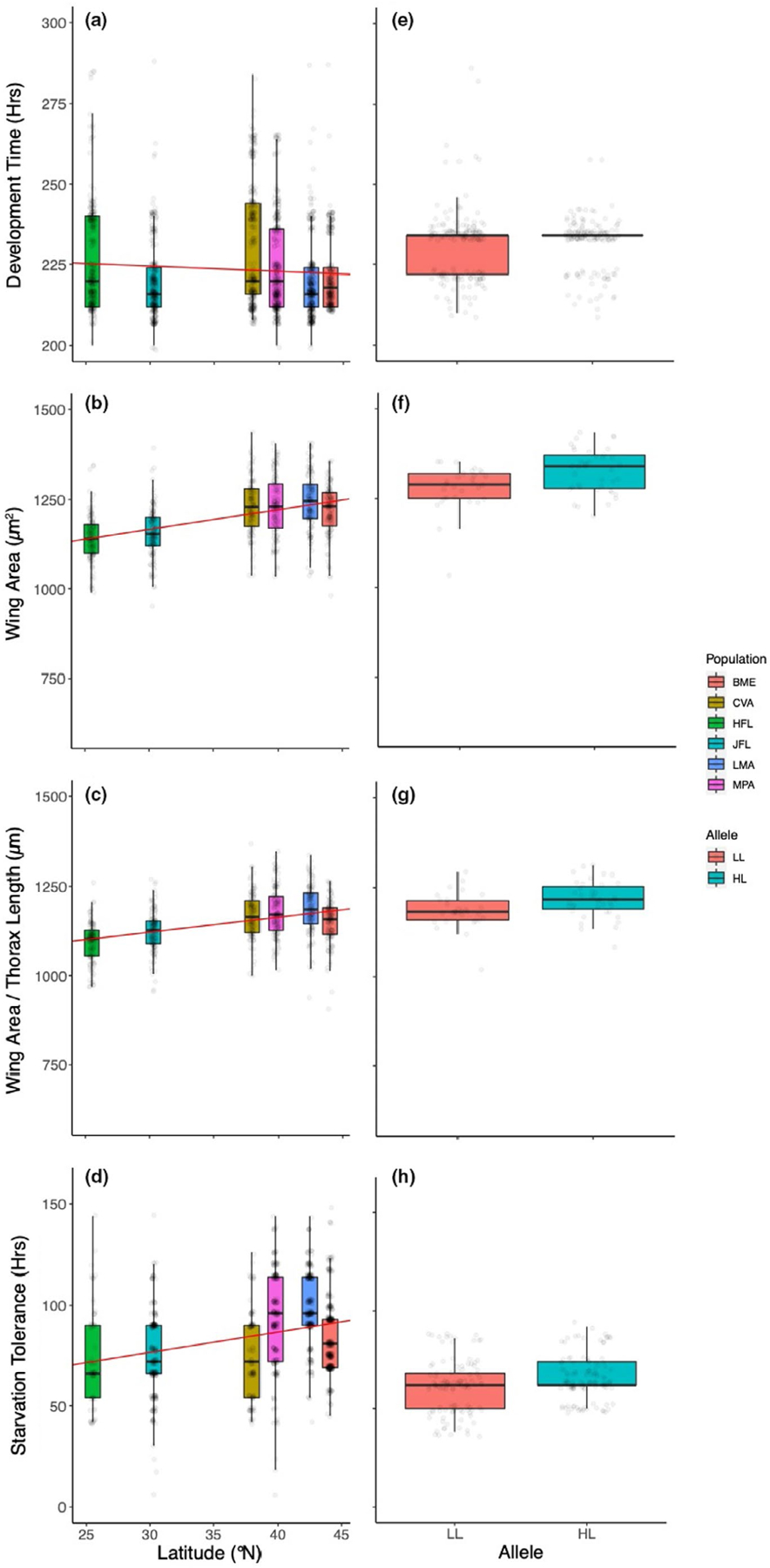
Phenotypic variation for females. Trait variation among natural populations collected across the latitudinal gradient in the eastern U.S. is plotted on the left (a–d) and traits exhibited by the homozygous high- and low-latitude foxo genotypes are depicted on the right (e–h). Box plots show the median (50th percentile; bold horizontal line in the box) and the 25th and 75th percentile (upper and lower horizontal edges of the box); the upper whisker represents the maximum value of the data that falls within 1.5 times the interquartile range over the 75th percentile; the lower whisker is the minimum value of the data that is within 1.5 times the interquartile range under the 25th percentile. In (a–d), regression lines (regression of means to latitude) are in red (Table S3) and indicate the extent of clinality. Development time did not vary predictably with latitude (a), and was also equivalent between foxo alleles (e). Wing area (c) and the ratio of wing area to thorax length (d) exhibit a positive latitudinal cline; these patterns of size variation in the natural populations were mirrored in both magnitude and direction by the observed differences in size parameters between the low and high-latitude foxo alleles (g, h). Starvation tolerance increased with increasing latitude (d); similarly, the high-latitude foxo allele was associated with increased starvation resistance (h) [Colour figure can be viewed at wileyonlinelibrary.com]
FIGURE 5.
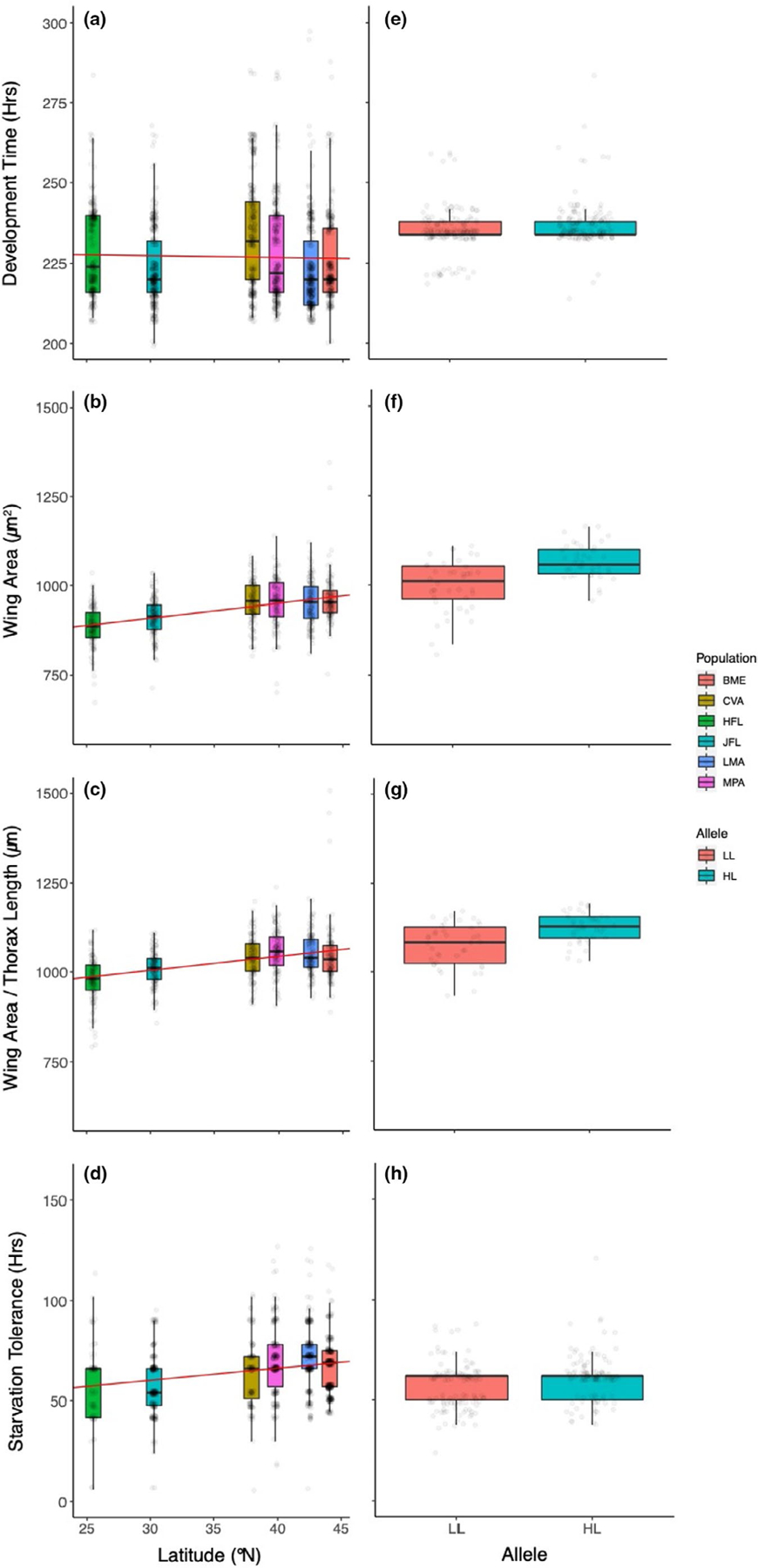
Patterns of phenotypic variation for males largely mirror those observed for females. Natural populations are depicted in (a–d) and are arranged by increasing latitude of origin; foxo genotypes are given in (e–h). Clines and concordant differences between foxo genotypes were observed for size-related traits (b,c and f,g) and starvation tolerance (d,h); development time was not clinal but was distinct between the assayed foxo alleles. Box plots show the median (50th percentile; bold horizontal line in the box) and the 25th and 75th percentile (upper and lower horizontal edges of the box); the upper whisker represents the maximum value of the data that falls within 1.5 times the interquartile range over the 75th percentile; the lower whisker is the minimum value of the data that is within 1.5 times the interquartile range under the 25th percentile. In (a–d), regression lines (regression of means to latitude) are in red (Table S3) and indicate the extent of clinality. [Colour figure can be viewed at wileyonlinelibrary.com]
3.2.2 |. Body size
In the six sampled natural populations that were assayed for trait variation, all measures of size (thorax length, wing area, and the ratio of wing area to thorax length) were highly distinct among populations (Table 1), and these patterns of differentiation exhibited a positive association with latitude (Table S3) for both females (Figure 4b,d) and males (Figure 5b,c). Sexes were analysed separately due to dimorphism and the potential for differential allometry, but exhibited qualitatively similar patterns of size variation among populations and as a function of latitude. As expected, these results are consistent with previous associations between latitude and body size in D. melanogaster (e.g., Coyne & Beecham, 1987; James et al., 1995; de Jong & Bochdanovits, 2003).
3.2.3 |. Starvation resistance
As with body size, starvation tolerance was highly variable among isofemale lines within populations yet exhibited a robust association with geography for both sexes (Table 1, Figures 4d and 5d). The patterns of increasing tolerance with increasing latitude is consistent with other aspects of stress tolerance in North American populations (Schmidt & Paaby, 2008), but opposite to what has been observed on the Indian subcontinent (Karan et al., 1998). Starvation tolerance does not appear to vary predictably with latitude in other assayed geographic regions (Hoffmann et al., 2001; Robinson et al., 2000). All raw data are available at https://doi.org/10.5061/dryad.hhmgqnkgm.
3.3 |. foxo alleles may make a large contribution to body size clines
3.3.1 |. Development time
Development time varied significantly across sets of lines and replicates; in contrast to the other traits measured, development time was not distinct between the high- and low-latitude foxo alleles for females but was for males (Table 2, Figures 4e and 5e). Differences in average development time between foxo genotypes, measured as time to eclosion, were small in magnitude for each sex (less than one hour difference in mean development time for females, slightly more than two hours for males; Figures 4e and 5e). Overall these results suggest that there are no major functional differences associated with these naturally occurring foxo alleles with respect to development time. This is somewhat congruent with data on Australian clines, where the relationship between body size and development time is inconsistent across latitudes (e.g., James et al., 1995, 1997). Development time was the most variable of the traits studied here, both with respect to experimental replication and variation among natural as well as reconstituted outbred populations.
TABLE 2.
Analyses of variance (ANOVA) for the phenotypic effects of the low-latitude (LL) and high-latitude (HL) alleles at the foxo locus. The models show the effects of allelic state, set nested in allele, and of replicate cage nested in the combination of allele and set. See Materials and Methods for further details
| Females |
Males |
|||||
|---|---|---|---|---|---|---|
| F | df | p | F | df | p | |
| Developmental time | ||||||
| Allele | 0.67 | 1 | .41 | 8.73 | 1 | .0034 |
| Set (A) | 2.44 | 2 | .09 | 8.48 | 2 | .0003 |
| Cage (A,S) | 7.04 | 4 | <.0001 | 3.74 | 4 | .0055 |
| Thorax length | ||||||
| Allele | 2.85 | 1 | .096 | 9.97 | 1 | .0023 |
| Set (A) | 1.70 | 2 | .19 | 4.95 | 2 | .0097 |
| Cage (A,S) | 3.85 | 3 | <.014 | 3.99 | 4 | .0055 |
| Wing area | ||||||
| Allele | 8.72 | 1 | .0044 | 41.35 | 1 | <.0001 |
| Set (A) | 2.05 | 2 | .14 | 10.23 | 2 | .0001 |
| Cage (A,S) | 0.78 | 3 | .51 | 11.99 | 4 | <.0001 |
| Wing area/thorax length | ||||||
| Allele | 4.66 | 1 | .035 | 34.19 | 1 | <.0001 |
| Set (A) | 1.92 | 2 | .16 | 6.36 | 2 | .0029 |
| Cage (A,S) | 0.52 | 3 | .67 | 13.95 | 4 | <.0001 |
| Starvation tolerance | ||||||
| Allele | 8.38 | 1 | .0042 | 8.14 | 1 | .0047 |
| Set (A) | 9.41 | 2 | .0001 | 7.79 | 2 | .0005 |
| Cage (A,S) | 4.63 | 4 | .0013 | 2.36 | 4 | .055 |
3.3.2 |. Body size
Thorax length and wing area were both highly distinct between the high- and low-latitude foxo genotypes (Table 2). Significant heterogeneity was present between the replicate sets A and B, as expected based on distinct composition of founding inbred lines, as well as among experimental replicates that were cultured independently since cage initiation. Despite these two sources of cage effects, the differences between foxo genotypes were consistent with expectations based on geography: the genotypes homozygous for the high-latitude foxo allele were significantly larger than genotypes homozygous for the low-latitude allele (Figures 4f and 5f). The ratio of wing area to thorax length also demonstrated significant and predictable differences between the high and low-latitude foxo genotypes (Figures 4g and 5g). Furthermore, the observed differences between foxo genotypes are strikingly similar in effect size to the magnitude of trait differences observed between the populations sampled from the latitudinal extremes (Figure 4b,c vs. Figure 4f,g; Figure 5b,c vs. Figure 5f,g), suggesting that variation at the foxo locus makes a major contribution to body size clines.
3.3.3 |. Starvation resistance
Starvation resistance was also distinct between the foxo genotypes and varied predictably with geography (Table 2, Figures 4h and 5h). For both males and females, the genotype homozygous for the high-latitude foxo allele was associated with increased starvation tolerance, which may be associated with effects of the alleles on body size and/or lipid content (e.g., Chippindale et al., 1996). A significant amount of variance in starvation tolerance was also associated with cage effects for both sets of inbred lines and culture replicate (Table 2). The effect size associated with foxo genotype was approximately 10%, again similar to what was observed for associated differences in body size. However, unlike the patterns observed for size related traits, the differences between foxo genotypes appear to explain a small amount of the variance in this trait among natural populations across the sampled geographic range in the eastern U.S. (Figure 4d vs. Figure 4h; Figure 5d vs. Figure 5h). These distinct patterns also suggest that differences in starvation tolerance are not determined solely by differences in size.
4 |. DISCUSSION
In D. melanogaster, there is abundant evidence for local adaptation. Natural populations exhibit rapid and predictable responses to environmental parameters that vary with season, both in terms of phenotypic (Behrman, Howick, et al., 2018; Behrman, Kawecki, et al., 2018; Behrman et al., 2015; Rajpurohit et al., ,2017, 2018; Schmidt & Conde, 2006) and allele frequency (Behrman, Kawecki, et al., 2018; Bergland et al., 2014; Cogni et al., 2014; Machado et al., 2021) change. Similarly, many fitness-associated traits have been shown to vary predictably with latitude, often in parallel across independent gradients (e.g., Oakeshott et al., 1982; Paaby et al., 2010; Yang & Edery, 2018). Latitudinal allele frequency clines at candidate loci (e.g., Cogni et al., 2017; Paaby et al., 2010; Schmidt et al., 2000; Sezgin et al., 2004) are now placed in a genomic context in which tens of thousands of SNPs are known to be clinal (e.g., Bergland et al., 2016; Fabian et al., 2012; Kapun et al., 2016; Kolackzowski et al., 2011). While allele frequency clines may be generated by demography (Bergland et al., 2016; Kao et al., 2015), at least some of the observed clines may be generated by spatially varying selection (Schmidt et al., 2008; Svetec et al., 2016). Two of the associated, major questions are: (i) how many, or what proportion of, allele frequency clines reflect spatially varying selection and thus local adaptation; and (ii) how are allele frequency and phenotypic clines integrated? Alleles exist in a genomic context, and complex traits are similarly correlated as well as affected by epistasis (Mackay, 2014); it is extremely unlikely that all allele and phenotypic clines are independent and reflect selection on single variants or traits. However, the effect size of individual adaptive polymorphisms and the architecture of local adaptation are, arguably, not well resolved. There is a paucity of detailed, mechanistic and comprehensive investigations as to the functional significance of segregating polymorphisms in natural populations. It is infeasible to assess the functional impact of all polymorphisms across the genome. However, multiple, intersecting methodologies (e.g., direct mapping, expression analyses, mutant analysis, patterns of variation in natural populations) can be used to identify a subset of variants that may be examined for functional significance. Ideally, investigation of a sufficient number of outliers could generate an empirical distribution of genic or allelic effect sizes for specific fitness-associated traits. Such investigations are essential in resolving the genetic architecture and dynamics of local adaptation.
The foxo locus is an example of a robust candidate suitable for such functional analysis and integration of genotype to phenotype to fitness. Genetic manipulations of the foxo gene have revealed pronounced effects on lifespan, multiple aspects of stress tolerance including starvation resistance, and growth phenotype (Giannakou et al., 2004; Hwangbo et al., 2004; Jünger et al., 2003; Kramer et al., 2003; Kramer et al., 2008; Puig et al., 2003; Slack et al., 2011). These traits vary with latitude in D. melanogaster (Coyne & Beecham, 1987; James et al., 1995; Schmidt et al., 2005); thus, the foxo gene is a logical candidate for determining variation for these traits in natural populations (de Jong & Bochdonavits, 2003). However, while it is clear that foxo laboratory mutants or transgenes have highly pleiotropic effects on life-history phenotypes, polymorphisms segregating in natural populations need not necessarily affect variance for traits related to gene function (e.g., Stern, 2011).
In previous work, we have performed independent assays of the natural foxo alleles studied here, with a focus on genotype by environment interactions (diet, temperature) and predictions regarding insulin signalling (Durmaz et al., 2019). Here, we extend this work by (i) demonstrating that these foxo alleles represent major clinal outlier loci probably subject, either directly or indirectly, to spatially varying selection; (ii) directly comparing the effects of the foxo alleles to new data on phenotypic clines and showing that they may make a significant contribution to these trait clines, particularly for body size; and (iii) by independently verifying the robustness of the allelic effects at foxo using different assay conditions in another laboratory.
Our present results demonstrate that allelic variation at foxo has large effects on body size and starvation tolerance. Durmaz et al. (2019) also show that these focal foxo variants are also associated with significant differences in viability, fat catabolism, and FOXO activity, as indicated by differences in transcript abundance of a FOXO target (InR). We show here that two measures of size, thorax length and wing area, as well as starvation tolerance, vary predictably with latitude in natural populations of D. melanogaster. Furthermore, the high-latitude foxo allele is associated with larger size and greater starvation resistance, whereas flies homozygous for the low-latitude foxo allele are smaller and less tolerant. Thus, the allelic effects parallel patterns of variation observed in natural populations, and are also consistent with predicted effects on phenotype due to genotypic differences in insulin/insulin-like signalling (Durmaz et al., 2019). This is distinct from the countergradient patterns that have been previously observed for allelic variants at the Drosophila insulin receptor (InR) (Paaby et al., 2014). The effect size of the foxo alleles is seemingly large, particularly for body size: the difference between high-and low-latitude foxo alleles is approximately the same as the observed size difference between flies sampled from Florida and Maine. This suggests that allelic variation at foxo underpins, at least in part, variance in body size in these populations. It remains to be determined whether foxo is a major-effect locus for size in other taxa.
Despite the parallels we observed between the assayed foxo variants and the patterns in natural populations, we cannot conclude that it is these two focal SNPs (positions) that themselves cause the observed differences in size and starvation tolerance, or that they directly contribute to variance for these traits in natural populations. The linkage disequilibrium present in the founding inbred lines (DGRP) would decay to some extent by the eight generations of outcrossing but would remain pronounced; thus, without further characterization, these SNPs need to be interpreted as markers for functionally significant allelic variation segregating at this locus. Gene editing or similar techniques, in which the focal sites are manipulated in multiple common genetic backgrounds, would be essential in directly examining causality. Such investigations are the focus of future work.
5 |. CONCLUSION
Here, we have shown that both starvation tolerance and two estimates of body size exhibit pronounced latitudinal clines in the sampled natural populations, whereas development time exhibited no clear association with latitude. Similarly, we find that allelic variation at foxo has predictable effects on body size and starvation tolerance but minor and sex-specific effects on development time. The assayed alleles at foxo explain a small amount of the variance among natural populations for starvation tolerance, but may have a large impact on genetic variance for size. Our results suggest a distinct genetic architecture for correlated fitness traits, and that allelic variation at the foxo locus underlies, in part, patterns of local adaptation in natural populations of this genetic model.
Supplementary Material
ACKNOWLEDG EMENTS
We thank two anonymous reviewers for helpful comments on the manuscript and the members of the Flatt and Schmidt laboratories for their gracious assistance in all aspects of this work. This work was supported by the National Institutes of Health (R01GM100366 to PS), the U.S. National Science Foundation (DEB0921307 to PS), the Austrian Science Foundation (FWF P21498-B11 to TF), the Swiss National Science Foundation (grants PP00P3_133641, PP00P3_165836 and 31003A-182262 to TF), and a Mercator Fellowship from the German Research Foundation (DFG to TF), held as a EvoPAD Visiting Professor at the Institute for Evolution and Biodiversity, University of Munster.
Funding information
National Institute of General Medical Sciences, Grant/Award Number: NIGMSR01100366
Footnotes
DATA AVAILABILITY STATEMENT
The raw phenotypic data have been made available at Dryad at: https://doi.org/10.5061/dryad.hhmgqnkgm.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- Arnegard ME, McGee MD, Matthews B, Marchinko KB, Conte GL, Kabir S, Bedford N, Bergek S, Chan YF, Jones FC, Kingsley DM, Peichel CL, & Schluter D (2014). Genetics of ecological divergence during speciation. Nature, 511(7509), 307–311. 10.1038/nature13301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton KG (2002). Patterns of within-species body size variation of birds: Strong evidence for Bergmann's rule. Global Ecology and Biogeography, 11, 505–523. [Google Scholar]
- Azevedo RBR, James AC, McCabe J, & Partridge L (1998). Latitudinal variation of wing: Thorax size ratio and wing aspect ratio in Drosophila melanogaster. Evolution, 52, 1353–1362. 10.1111/j.1558-5646.1998.tb02017.x [DOI] [PubMed] [Google Scholar]
- Barton NH, Etheridge AM, & Veber A (2017). The infinitesimal model: Definition, derivation, and implications. Theoretical Population Biology, 118, 50–73. 10.1016/j.tpb.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Behrman EL, Howick VM, Kapun M, Staubach F, Bergland AO, Petrov DA, & Schmidt PS (2018). Rapid seasonal evolution in innate immunity of wild Drosophila melanogaster. Proceedings of the Royal Society of London B, 285, 20172599. 10.1098/rspb.2017.2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman EL, Kawecki TJ, & Schmidt P (2018). Natural variation in couch potato mediates rapid evolution of learning and reproduction in natural populations of Drosophila melanogaster. bioRxiv, 288696 10.1101/288696 [DOI] [Google Scholar]
- Behrman EL, Watson SS, O’Brien KR, Heschel MS, & Schmidt PS (2015). Seasonal variation in life history traits in two Drosophila species. Journal of Evolutionary Biology, 28(9), 1691–1704. 10.1111/jeb.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Behrman EL, O'Brien KR, Schmidt PS, & Petrov DA (2014). Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genetics, 10, e1004775. 10.1371/journal.pgen.1004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, González J, Schmidt P, & Petrov D (2016). Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Molecular Ecology, 25, 1157–1174. 10.1111/mec.13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanckenhorn WU (2000). The evolution of body size: What keeps organisms small? Quarterly Review of Biology, 75, 385–407. 10.1086/393620 [DOI] [PubMed] [Google Scholar]
- Blanckenhorn WU, & Demont M (2004). Bergmann and converse bergmann latitudinal clines in arthropods: Two ends of a continuum? Integrative and Comparative Biology, 44(6), 413–424. 10.1093/icb/44.6.413 [DOI] [PubMed] [Google Scholar]
- Bonnet T, Wandeler P, Camenisch G, & Postma E (2017). Bigger is fitter? Quantitative genetic decomposition of selection reveals an adaptive evolutionary decline of body mass in a wild rodent population. PLoS Biology, 15(1), e1002592. 10.1371/journal.pbio.1002592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, & Pritchard JK (2017). An expanded view of complex traits: From polygenic to omnigenic. Cell, 169(7), 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Marquet PA, & Taper ML (1993). Evolution of body size: Consequences of an energetic definition of fitness. American Naturalist, 142(4), 573–584. 10.1086/285558 [DOI] [PubMed] [Google Scholar]
- Chakraborty M, & Fry JD (2016). Evidence that environmental heterogeneity maintains a detoxifying enzyme polymorphism in Drosophila melanogaster. Current Biology, 26, 219–223. 10.1016/j.cub.2015.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Bachman GC, Connaty AD, McClelland GB, & Storz JF (2012). Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proceedings of the National Academy of Sciences USA, 109, 8635–8640. 10.1073/pnas.1120523109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Chu TJF, & Rose MR (1996). Complex tradeoffs and the evolution of starvation resistance in Drosophila melanogaster. Evolution, 50, 753–766. 10.1111/j.1558-5646.1996.tb03885.x [DOI] [PubMed] [Google Scholar]
- Clemente F, & Vogl C (2012). Unconstrained evolution in short introns? – An analysis of genome-wide polymorphism and divergence data from Drosophila. Journal of Evolutionary Biology, 25, 1975–1990. 10.1111/j.1420-9101.2012.02580.x [DOI] [PubMed] [Google Scholar]
- Cogni R, Kuczynski C, Koury S, Lavington E, Behrman EL, O'Brien KR, Schmidt PS, & Eanes WF (2014). The intensity of selection acting on the couch potato gene–spatial-temporal variation in a diapause cline. Evolution, 68(2), 538–548. 10.1111/evo.12291 [DOI] [PubMed] [Google Scholar]
- Cogni R, Kuczynski K, Koury S, Lavington E, Behrman EL, O’Brien KR, Schmidt PS, & Eanes WF (2017). On the long-term stability of clines in some metabolic Genes in Drosophila melanogaster. Scientific Reports, 7, 42766. 10.1038/srep42766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, Carré C, Noselli S, & Léopold P (2005). Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science, 310(5748), 667–670. 10.1126/science.1119432 [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G Jr, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, & Kingsley DM (2005). Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science, 307(5717), 1928–1933. 10.1126/science.1107239 [DOI] [PubMed] [Google Scholar]
- Comeault AA, Flaxman SM, Riesch R, Curran E, Soria-Carrasco V, Gompert Z, Farkas TE, Muschick M, Parchman TL, Schwander T, Slate J, & Nosil P (2015). Selection on a genetic polymorphism counteracts ecological speciation in a stick insect. Current Biology, 25(15), 1975–1981. 10.1016/j.cub.2015.05.058 [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB, Cardeno C, & Langley CH (2012). Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics, 192, 131–137. 10.1534/genetics.112.141622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, & Beecham E (1987). Heritability of two morphological characters within and among natural populations of Drosophila melanogaster. Genetics, 117, 727–737. 10.1093/genetics/117.4.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong G, & Bochdanovits Z (2003). Latitudinal clines in Drosophila melanogaster: Body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. Journal of Genetics, 82, 207–223. 10.1007/BF02715819 [DOI] [PubMed] [Google Scholar]
- Durmaz E, Rajpurohit S, Betancourt N, Fabian DK, Kapun M, Schmidt P, & Flatt T (2019). A clinal polymorphism in the insulin signaling transcription factor foxo contributes to life history adaptation in Drosophila. Evolution, 73, 1774–1792. 10.1111/evo.13759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlotterer C, & Flatt T (2012). Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Molecular Ecology, 21, 4748–4769. 10.1111/j.1365-294X.2012.05731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Lack JB, Mathur V, Schlötterer C, Schmidt PS, Pool JE, & Flatt T (2015). Spatially varying selection shapes life history clines among populations of Drosophila melanogaster from sub-Saharan Africa. Journal of Evolutionary Biology, 28, 826–840. 10.1111/jeb.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, & Antebi A (2008). C. elegans dauer formation and the molecular basis of plasticity. Genes & Development, 22, 2149–2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T (2020). Life-History Evolution and the Genetics of Fitness Components in Drosophila melanogaster. Genetics, 214(1), 3–48. 10.1534/genetics.119.300160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Juenger MA, Hafen E, Leevers SJ, & Partridge L (2004). Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science, 305, 361. 10.1126/science.1098219 [DOI] [PubMed] [Google Scholar]
- Gilchrist AS, Azevedo RBR, Partridge L, & O’Higgins P (2000). Adaptation and constraint in the evolution of Drosophila melanogaster wing shape. Evolution & Development, 2, 114–124. 10.1046/j.1525-142x.2000.00041.x [DOI] [PubMed] [Google Scholar]
- Hof AEV, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, & Saccheri IJ (2016). The industrial melanism mutation in British peppered moths is a transposable element. Nature, 534, 102–105. 10.1038/nature17951 [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Hallas R, Sinclair C, & Mitrovski P (2001). Levels of variation in stress resistance in Drosophila among strains, local populations and geographic regions: Patterns for desiccation, starvation, cold resistance, and associated traits. Evolution, 55, 1621–1630. 10.1111/j.0014-3820.2001.tb00681.x [DOI] [PubMed] [Google Scholar]
- Huey RB, Gilchrist GW, Carson ML, Berrigan D, & Serra L (2000). Rapid evolution of a geographic cline in size in an introduced fly. Science, 287, 308–309. 10.1126/science.287.5451.308 [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gersham B, Tu M-P, Palmer M, & Tatar M (2004). Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature, 429, 562–566. 10.1038/nature02549 [DOI] [PubMed] [Google Scholar]
- James AC, Azevedo RBR, & Partridge L (1995). Cellular basis and developmental timing in a size cline of Drosophila melanogaster. Genetics, 140, 659–666. 10.1093/genetics/140.2.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AC, Azevedo RBR, & Partridge L (1997). Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics, 146, 881–890. 10.1093/genetics/146.3.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJR, Jiggins FM, Jensen JD, Melo-Ferreira J, & Good JM (2018). Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science, 360(6395), 1355–1358. 10.1126/science.aar5273 [DOI] [PubMed] [Google Scholar]
- Jünger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, & Hafen E (2003). The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. Journal of Biology, 2, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Zubair A, Salomon MP, Nuzhdin SV, & Campo D (2015). Population genomic analysis uncovers African and European admixture in Drosophila melanogaster populations from the south-eastern United States and Caribbean Islands. Molecular Ecology, 24, 1499–1509. 10.1111/mec.13137 [DOI] [PubMed] [Google Scholar]
- Kapun M, Fabian DK, Goudet J, & Flatt T (2016). Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Molecular Biology and Evolution, 33, 1317–1336. 10.1093/molbev/msw016 [DOI] [PubMed] [Google Scholar]
- Karan D, Dahiya N, Munjal AK, Gibert P, Moreteau B, Parkash R, & David JR (1998). Desiccation and starvation tolerance of adult Drosophila: Opposite latitudinal clines in natural populations of three different species. Evolution, 52, 825–831. 10.1111/j.1558-5646.1998.tb03706.x [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, & Begun DJ (2011). Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics, 187, 245–260. 10.1534/genetics.110.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Davidge JT, Lockyer JM, & Staveley BE (2003). Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Developmental Biology, 3, 5. 10.1186/2F1471-213X-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Slade JD, & Staveley BE (2008). foxo is required for resistance to amino acid starvation in Drosophila. Genome, 51, 668–672. 10.1139/G08-047 [DOI] [PubMed] [Google Scholar]
- Lack JB, Monette MJ, Johanning EJ, Sprengelmeyer QD, & Pool JE (2016). Decanalization of wing development accompanied the evolution of large wings in high altitude Drosophila. Proceedings of the National Academy of Sciences USA, 113, 1014–1019. 10.1073/pnas.1515964113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Han F, Berglund J, Wang C, Almen MS, Webster MT, Grant BR, Grant PR, & Andersson L (2016). A beak size locus in Darwins finches facilitated character displacement during a drought. Science, 352(6284), 470–474. 10.1126/science.aad8786 [DOI] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, & Fang S (2012). Genomic variation in natural populations of Drosophila melanogaster. Genetics, 192, 533–598. 10.1534/genetics.112.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, & Stam LF (1988). Quantitative analysis of RNA produced by Slow and Fast alleles of Drosophila melanogaster. Proceedings of the National Academy of Sciences USA, 85, 5161–5165. 10.1534/genetics.112.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, & Kenyon C (2003). Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell, 115, 489–502. 10.1016/S0092-8674(03)00889-4 [DOI] [PubMed] [Google Scholar]
- Machado HE, Bergland AO, Taylor R, Tilk S, Behrman E, Dyer K, Fabian DK, Flatt T, González J, Karasov TL, Kozeretska I, Lazzaro BP, Merritt T, Pool J, O'Brien K, Rajpurohit S, Roy P, Schaeffer S, Serga S, … Petrov D (2021). Broad geographic sampling reveals predictable and pervasive seasonal adaptation in Drosophila. bioRxiv 10.1101/337543v4. Preprint, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC (2014). Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nature Reviews Genetics, 15(1), 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han YI, Magwire MM, Cridland JM, Richardson MF, Anholt RRH, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, … Gibbs RA (2012). The Drosophila melanogaster Genetic Reference Panel. Nature, 482, 173–178. 10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Mallarino R, & Hoekstra HE (2011). The developmental role of agouti in color pattern evolution. Science, 331(6020), 1062–1065. 10.1126/science.1200684 [DOI] [PubMed] [Google Scholar]
- Mattila J, Bremer A, Ahonen L, Kostiainen R, & Puig O (2009). Drosophila FoxO regulates organism size and stress resistance through an Adenylate cyclase. Molecular and Cellular Biology, 29, 5357–5365. 10.1128/MCB.00302-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown AD, Klápště J, Guy RD, Geraldes A, Porth I, Hannemann J, Friedmann M, Muchero W, Tuskan GA, Ehlting J, Cronk QC, El-Kassaby YA, Mansfield SD, & Douglas CJ (2014). Genome-wide association implicates numerous genes underlying ecological trait variation in natural populations of Populus trichocarpa. New Phytologist, 203(2), 535–553. 10.1111/nph.12815 [DOI] [PubMed] [Google Scholar]
- Oakeshott JG, Gibson JB, Anderson PR, Knibb WR, Anderson DG, & Chambers GK (1982). Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosophila melanogaster on different continents. Evolution, 36(1), 86–96. 10.1111/j.1558-5646.1982.tb05013.x [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, & Schmidt PS (2014). A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution, 68, 3395–3409. 10.1111/evo.12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, & Schmidt PS (2010). Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Molecular Ecology, 19, 760–774. 10.1111/j.1365-294X.2009.04508.x [DOI] [PubMed] [Google Scholar]
- Parsch J, Novozhilov S, Saminadin-Peter SS, Wong KM, & Andolfatto P (2010). On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Molecular Biology and Evolution, 27, 1226–1234. 10.1093/molbev/msq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Barrie B, Fowler K, & French V (1994). Evolution and development of body size and cell size in Drosophila melanogaster in response to temperature. Evolution, 48, 1269–1276. 10.1111/j.1558-5646.1994.tb05311.x [DOI] [PubMed] [Google Scholar]
- Partridge L, & Coyne JA (1997). Bergmann's rule in ectotherms: is it adaptive? Evolution, 51, 632–635. 10.1111/j.1558-5646.1997.tb02454.x [DOI] [PubMed] [Google Scholar]
- Promislow DE, Smith EA, & Pearse L (1998). Adult fitness consequences of sexual selection in Drosophila melanogaster. Proceedings of the National Academy of Sciences, 95(18), 10687–10692. 10.1073/pnas.95.18.10687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, & Tjian R (2003). Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes and Development, 17, 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2009). R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. R-project.org. [Google Scholar]
- Rajpurohit S, Gefen E, Bergland AO, Petrov DA, Gibbs AG, & Schmidt PS (2018). Spatiotemporal dynamics and genome-wide association analysis of desiccation tolerance in Drosophila melanogaster. Molecular Ecology, 27(17), 3525–3540. 10.1111/mec.14814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit S, Hanus R, Vrkoslav V, Behrman EL, Bergland AO, Petrov D, & Schmidt PS (2017). Adaptive dynamics of cuticular hydrocarbons in Drosophila. Journal of Evolutionary Biology, 30(1), 66–80. 10.1111/jeb.12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve MW, Fowler K, & Partridge L (2001). Increased body size confers greater fitness at low experimental temperature in male Drosophila melanogaster. Journal of Evolutionary Biology, 13, 836–844. 10.1046/j.1420-9101.2000.00216.x [DOI] [Google Scholar]
- Robinson SJW, Zwaan B, & Partridge L (2000). Starvation resistance and adult body composition in a latitudinal cline of Drosophila melanogaster. Evolution, 54, 1819–1824 [DOI] [PubMed] [Google Scholar]
- Rockman MV (2012). The QTN program and the alleles that matter for evolution: all thaťs gold does not glitter. Evolution, 66, 1–17. 10.1111/j.1558-5646.2011.01486.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff DA (2007). A centennial celebration for quantitative genetics. Evolution, 61(5), 1017–1032. 10.1111/j.1558-5646.2007.00100.x [DOI] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, & Merilä J (2013). Ecological genomics of local adaptation. Nature Reviews Genetics, 14(11), 807–820. 10.1038/nrg3522 [DOI] [PubMed] [Google Scholar]
- Schmidt PS, & Conde DR (2006). Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution, 60, 1602–1611. 10.1111/j.0014-3820.2006.tb00505.x [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Duvernell DD, & Eanes WF (2000). Adaptive evolution of a candidate gene for aging in Drosophila. Proceedings of the National Academy of Sciences USA, 97, 10861–10865. 10.1073/pnas.190338897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, & Eanes WF (2005). Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution, 59, 1721–1732. 10.1111/j.0014-3820.2005.tb01821.x [DOI] [PubMed] [Google Scholar]
- Schmidt PS, & Paaby AB (2008). Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution, 62, 1204–1215. 10.1111/j.1558-5646.2008.00351.x [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Zhu C-T, Das J, Batavia M, Yang L, & Eanes WF (2008). An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proceedings of the National Academy of Sciences USA, 105, 16207–16211. 10.1073/pnas.0805485105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Duvernell DD, Matzkin LM, Duan Y, Zhu C-T, Verrelli BC, & Eanes WF (2004). Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics, 168(2), 923–931. 10.1534/genetics.104.027649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C, Giannakou ME, Foley A, Goss M, & Partridge L (2011). dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging Cell, 10, 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL (2011). Evolution, development, and the predictable genome Roberts & Co. Publishers. [Google Scholar]
- Stillwell RC(2010).Arelatitudinalclinesinbodysizeadaptive? Oikos,119(9), 1387–1390. 10.1111/j.1600-0706.2010.18670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillwell RC, Morse GE, & Fox CW (2007). Geographic variation in body size and sexual size dimorphism of a seed-feeding beetle. American Naturalist, 170(3), 358–369. 10.1086/520118 [DOI] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, … Ostrander EA (2007). A Single IGF1 Allele is a major determinant of small size in dogs. Science, 316(5821), 112–115. 10.1126/science.1137045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetec N, Cridland JM, Zhao L, & Begun DJ (2016). The adaptive significance of natural genetic variation in the DNA damage response of Drosophila melanogaster. PLoS Genetics, 12(3), e1005869. 10.1371/journal.pgen.1005869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, & Cockerham CC (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38, 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Wellenreuther M, & Hansson B (2016). Detecting polygenic evolution: problems, pitfalls, and promises. Trends in Genetics, 32(3), 155–164, 10.1016/j.tig.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Yang Y, & Edery I (2018). Parallel clinal variation in the mid-day siesta of Drosophila melanogaster implicates continent-specific targets of natural selection. PloS Genetics, 14(9), e1007612. 10.1371/journal.pgen.1007612 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


