Abstract
The Pharmacogene Variation Consortium (PharmVar) catalogues star (*) allele nomenclature for the polymorphic human CYP2B6 gene. Genetic variation within the CYP2B6 gene locus impacts the metabolism or bioactivation of clinically important drugs. Of particular importance are efficacy and safety concerns regarding: efavirenz, which is used for the treatment of HIV type-1 infection; methadone, a mainstay in the treatment of opioid use disorder and as an analgesic; ketamine, used as an antidepressant and analgesic; and bupropion, which is prescribed to treat depression and for smoking cessation. This GeneFocus provides a comprehensive overview and summary of CYP2B6 and describes how haplotype information catalogued by PharmVar is utilized by the Pharmacogenomics Knowledgebase (PharmGKB) and the Clinical Pharmacogenetics Implementation Consortium (CPIC).
CYP2B6 Brief History
The phenobarbital-induced microsomal drug metabolizing enzyme system in rat was recognized in the early 1960s (1) and facilitated cDNA cloning, as well as the identification of a similar CYP from a human liver cDNA library in 1985 and 1988 (2, 3). The gene, previously known as LM2, IIB1, or hIIB was later renamed CYP2B6 and was mapped to chromosome 19 (19q13.2) (4), located head-to-tail near the closely related non-functional pseudogene CYP2B7P (2, 3) within the large CYP2ABFGST gene cluster (5).
CYP2B6 is the only gene in the human CYP2B subfamily encoding functional enzyme. CYP2B6 is mainly expressed in the liver and, to substantially smaller extents, in extrahepatic tissues such as the alimentary tract, kidney, lung and brain according to the Genotype-Tissue Expression (GTEx) project (6). Until the end of 1990’s, CYP2B6 was thought to be an insignificant component of the hepatic P450 system as earlier studies reported very low or undetectable CYP2B6 protein expression in a significant proportion of human liver tissues, in part due to a lack of reliable antibodies (7). However, after specific antibodies were developed and protein was recombinantly expressed (8) microsomal CYP2B6 protein was detected in all livers. CYP2B6 accounted for ~2–5% of the total hepatic P450 content (9), but inter-individual variability ranged up to 100-fold (10).
The contribution of genetic polymorphisms to interindividual variability of CYP2B6 activity was recognized in the early 2000’s by reverse pharmacogenetics. A study using extensive sequencing and variation scanning in German human liver tissues led to the discovery of five nonsynonymous and four synonymous single nucleotide variants (SNVs) and these SNVs (alone or in combination) were assigned to six different CYP2B6 alleles, CYP2B6*2-*7 (11). One of these SNVs (c.516G>T, p.Q172H, rs3745274) was also reported in a Japanese population in the same year (12). The potential clinical impact of CYP2B6 genetic polymorphisms was first proposed in 2003 for c.785A>G (p.K262R, rs2279343) in a healthy volunteer study using bupropion as a substrate (13). Research on CYP2B6 genetic polymorphisms was accelerated after the discovery of CYP2B6 as the principal enzyme responsible for efavirenz clearance in 2003 (14). In 2004, investigators in the US and Japan showed a significant association of alleles harboring c.516G>T with efavirenz exposure and effect in HIV patients (15, 16). Later on, the c.516G>T containing CYP2B6*6 allele was demonstrated to be associated with a marked decrease in efavirenz metabolism (17).
In 2008, aberrant splicing was shown to be the mechanism by which c.516G>T (p.Q172H) confers reduced activity (18). Both c.516G>T and c.785A>G are part of the CYP2B6*6 allele but can also be found together on numerous other alleles, as well as occur individually in *4 (c.785A>G) and *9 (c.516G>T). Importantly, c.516G>T is relevant in compound heterozygous carriers with other SNVs causing decreased function or non-functional protein (19). In more recent studies, technical advancements allowed identification of several rare variants as additional sources of CYP2B6 interindividual variability (20–22). It is now established that the frequencies of the CYP2B6 genetic variants are highly variable among racial and ethnic populations, which may cause differences in treatment outcome of efavirenz-based HIV therapy and other CYP2B6 substrates.
Table 1 summarizes key resources referred to throughout this GeneFocus.
Table 1.
Online CYP2B6 Resources - Links to Sites and Online Resources Referenced Throughout the Review
| Sources and References | References |
|---|---|
| PharmVar | |
CYP2B6 important gene information
|
(88) |
| Standards | (96) |
| Allele Designation and Evidence Level Document | (92) |
| CYP2B6 Gene Expert Panel Roster | (93) |
| P450 Nomenclature Site – Archive | (94) |
| PharmGKB | |
| CYP2B6 gene page | (71) |
Gene-Specific Information Tables for CYP2B6
|
(77) |
| CYP2B6 Drug Label Annotations | (112) |
| CPIC | |
| Guidelines | (73) |
Gene/Drug Pairs
|
(74) |
| Other resources | |
| Genotype-Tissue Expression (GTEx) project | (6) |
| Drug Interactions Flockhart Table™ | (57) |
| FDA Pharmacogenomic Biomarkers in Drug Labeling | (113) |
| NCBI Reference Sequences Database | (114) |
| Locus Reference Genomic (LRG) Project | (95) |
Status of Nomenclature before PharmVar
CYP2B6 star (*) nomenclature was first described in the early 2000’s and catalogued by the Human Cytochrome P450 Allele Nomenclature Database (23). This nomenclature is focused on haplotypes to inform the combinatorial effect of all sequence variants on an allele on enzyme activity. This nomenclature system was widely embraced by the pharmacogenetics community as a global go-to resource for CYP variations, including CYP2B6. Thirty-seven CYP2B6 haplotypes, CYP2B6*2-*38, were submitted by investigators to this database and designated in sequential order they were received before it was transitioned to the Pharmacogene Variation (PharmVar) Consortium in 2017 (24). Although the original CYP nomenclature site only required submissions to provide sequence information for exons, investigators also deposited variants in the upstream gene region which were discovered searching for regulatory elements. Likewise, sequence variants were also identified in exon-flanking intronic regions. Although their coverage was inconsistent between submissions these variants were often included in haplotype definitions regardless of whether they were experimentally shown to impact CYP2B6 function. Although so-called ‘suballeles’ were also catalogued initially by the Human CYP allele nomenclature database, e.g., CYP2B6*1A-N or *4A-D, this practice eventually ceased.
The information provided by the CYP nomenclature site was utilized by researchers, as well as by knowledge resources and pharmacogenetics testing and implementation communities (25). PharmVar identified the need for a standardized allele submission system, well-defined allele definition criteria and a transparent review process to systematically catalog the increasing number of genetic variants and haplotypes being discovered. To provide the field with the most comprehensible data repository, PharmVar is cataloging CYP2B6 suballeles using a numeric-based system.
In this review, star allele sequence variants are provided relative to their positions in the CYP2B6 reference transcript (NM_000767.5); corresponding protein coordinates are also provided. For example, the CYP2B6*5 allele defining variant is referred to as c.1459C>T (p.R487C, rs3211371 [rs number provided when first mentioned]) and causes an arginine to cysteine change at amino acid position 487.
Clinical Relevance
Despite the identification by the late 1990s of over 70 compounds as substrates, CYP2B6 was thought to play a minor role in human drug metabolism due to the lack of reliable in vitro tools to study its regulation, comparative hepatic protein expression and function (26). However, the discoveries of selective molecular, biochemical, clinical and genomic tools in the past two decades demonstrated that CYP2B6 plays a much larger role in human drug metabolism than previously estimated. CYP2B6 was implicated in 4-hydroxylation of the prodrug cyclophosphamide in 1993, but this reaction was catalyzed by multiple enzymes and was not selective as a probe (27). The identification of S-mephenytoin N-demethylation by the end of the 1990’s (28) and the discovery of thioTEPA as the first selective inhibitor (29) catalyzed initial in vitro research on CYP2B6. In 2000, bupropion 4-hydroxylation was shown to be selectively catalyzed by CYP2B6 (30, 31), providing an opportunity to interrogate CYP2B6 activity in vitro and qualitatively in vivo.
The discovery of CYP2B6 as the principal P450 metabolism pathway of the antiretroviral drug efavirenz in 2003 (14) represented a major advance in the field as this initiated clinical pharmacogenomic studies. Efavirenz, then a cornerstone of HIV therapy, exhibits a narrow therapeutic range and high inter-patient variability in its exposure with attendant variability in both its beneficial and adverse effects (28). The initial link between CYP2B6 genetic polymorphisms and efavirenz exposure and adverse effects was reported in 2004 in HIV patients (15, 16). Since then, numerous studies conducted in healthy volunteers and HIV patients from across ethnogeographic groups have established that CYP2B6 genotype is associated with efavirenz exposure (32), CNS and psychiatric adverse effects (33), hepatic injury (34) and treatment discontinuation (35). A CPIC guideline of therapeutic recommendations for efavirenz prescribing based on CYP2B6 genotype was published in 2019 (36). It follows that efavirenz is not only a probe to measure CYP2B6 activity but also a prototype model drug for evaluating the clinical relevance of CYP2B6.
Currently, CYP2B6 is estimated to metabolize, fully or partially, approximately 8–13% of clinically important drugs and a long list of other xenobiotics of toxicological relevance (9, 37). Clinically important drugs for which CYP2B6 metabolic status is a major determinant of metabolism and response and/or toxicity include nevirapine (38), methadone (39), bupropion (40), artemisinin (41) and ketamine (42). The metabolism of certain environmental pollutants (43) and pesticides (21) is also highly dependent on CYP2B6 metabolic status.
Other Factors that can Influence CYP2B6 activity
The expression and function of the human CYP2B6 gene are highly variable among individuals, which can alter clinical outcomes in patients treated with CYP2B6 substrates. Besides CYP2B6 genetic variation, drug-drug interactions (DDIs) contribute to this variability. The expression of CYP2B6 is regulated primarily by the xenobiotic constitutive androstane receptor (CAR) and the pregnane X receptor (PXR) in the liver and is highly inducible by a variety of drugs including efavirenz and artemisinin (37). There appears to be a significant interplay between CYP2B6 variation and the extent of DDIs. For example, the promoter variant that causes increased function (-82T>C, CYP2B6*22, rs34223104) synergistically interplays with PXR-mediated induction in vitro (44). Some of the CYP2B6 inducers are substrates of this enzyme (e.g., cyclophosphamide, ifosfamide, efavirenz and artemisinin) and therefore accelerate their own metabolism affecting clearance or toxicity upon chronic dosing. The extent of efavirenz autoinduction is relevant in normal and intermediate metabolizers but not for poor metabolizers (45, 46). The magnitude of efavirenz-mediated DDIs, caused by enzymes other than CYP2B6, are also influenced by CYP2B6 genetic variation. For example, efavirenz inhibits CYP1A2, an interaction that is more pronounced in individuals with decreased or no CYP2B6 activity (47). Also, efavirenz-based HIV therapy reduces the exposure to lumefantrine and levonorgestrel (48, 49) via CYP3A4 induction probably increasing the risk of CYP2B6 poor/intermediate metabolizers of suboptimal lumefantrine and levonorgestrel exposure that may lead to treatment failure. This underscores the importance of characterizing the pharmacogenomics of the perpetrator drug (e.g. efavirenz) in assessing DDI risks.
In addition to induction, direct potent enzymatic inhibitors/inactivators of CYP2B6 have been identified in vitro, but only a small number of drugs have been tested and found to be in vivo inhibitors (e.g., voriconazole (50), ticlopidine and clopidogrel (51, 52)). Some herbal medicines may also impact CYP2B6 activity suggesting that herbal DDIs could contribute to adverse events or lack of efficacy. For example, Baicalin induces CYP2B6 activity while extracts from plants such as Hyptis suaveolens, Myrothamnus flabellifolius, Launaea taraxacifolia, Boerhavia diffusa and Newbouldia laevis, were shown to inhibit CYP2B6 activity in in vitro recombinant systems (53, 54). Moreover, recent evidence suggests that inflammation significantly reduces CYP2B6 expression (55) and activity as shown in type 2 diabetes patients (56). For more information on CYP2B6 inhibitors see the Drug Interactions Flockhart Table (57).
Age-related and sex differences
In human liver banks, CYP2B6 mRNA levels are low during fetal development (58), with CYP2B6 expression increasing after the neonatal period (birth to 30 days postnatal) (59). CYP2B6 activity appears as early as the first day of life, increases through infancy, and by 1 year of age, CYP2B6 levels and activity approach those of adults (58). Higher efavirenz clearance (~47%) was noted in younger children (<5 years old) than in older children (>5 years of age) (60) suggesting age-dependency of CYP2B6 activity. CYP2B6 activity is increased in pregnancy which may (61, 62) or may not (63) affect drug levels. Estradiol induces the expression of CYP2B6 mRNA and increases bupropion 4-hydroxylation in hepatocytes suggesting increased activity during the third trimester (64). There is an estrogen-response element in the regulatory region of CYP2B6, and a SNV (rs16974799) in intron 1 may contribute to variation in metabolism of CYP2B6 substrates (65). In vitro, females have similar (17, 59) or higher hepatic CYP2B6 levels (i.e., mRNA 3.9-fold; protein 1.7-fold and activity 1.6-fold) compared to males (66, 67). Among adults, compared to males, females have significantly faster in vivo propofol metabolism (68) and methadone metabolism (68). In contrast, findings are inconsistent for bupropion (40, 69) and efavirenz pharmacokinetics (30). Taken together, there is evidence supporting that estrogen can up-regulate CYP2B6, as well as the effect of sex and pregnancy on substrate drug concentrations, although this effect is not consistent between substrates.
CYP2B6 and the PharmGKB
The PharmGKB collects, curates, and disseminates knowledge about the impact of human genetic variation on drug response (70). The PharmGKB CYP2B6-dedicated webpage allows structured access to gene-specific PGx knowledge (71). Information is presented in sections including prescribing information, drug label annotations, clinical annotations, variant annotations, and curated pathways. Prescribing information encompasses annotations of clinical guidelines from sources such as the CPIC and the Royal Dutch Association for the Advancement of Pharmacy—Pharmacogenetics Working Group (DPWG) and “Rx study annotations” that provide genotype-based drug dosing or prescribing information reported in individual journal articles. One CPIC, one Dutch Pharmacogenetics Working Group and six Rx study annotations are available for CYP2B6 with CYP2B6-drug pairs.
CYP2B6 and the Clinical Pharmacogenetics Implementation Consortium (CPIC)
The CPIC develops freely available, evidence-based, clinical practice guidelines for drugs affected by genetic variations (72). To date, CPIC has published 25 guidelines (of which 11 have been updated), with genotype-based dosing recommendations for 19 genes and 57 drugs across various therapeutic areas. All guidelines and supplemental materials are freely available on the CPIC website (73). Each guideline has multiple components with genotype/phenotype-specific therapeutic recommendations at its core, access to the reviewed evidence, and implementation resources to support the translation of the guideline into electronic health records (EHRs), as well as an example of clinical decision support (CDS) text. The CPIC Guideline for CYP2B6 and efavirenz-containing antiretroviral therapy was published in 2019 (36). CYP2B6-drug pairs of interest for future guidelines are also listed on the CPIC website and include methadone and nevirapine (74).
The PharmVar site displays CPIC clinical allele function designations (increased, normal, decreased, no or uncertain function). Unlike other genes that have been extensively studied and several guidelines developed (e.g., CYP2C19 and CYP2D6), there is only one CPIC guideline for a CYP2B6 gene-drug pair. This guideline summarizes published evidence and provides therapeutic recommendations for efavirenz prescribing based on CYP2B6 genotypes (36). The development of this CPIC guideline includes the process for determining allele functionality based on published literature and input from guideline authors. A table describing allele function has been published as a Supplement to the CPIC guideline and can be found on PharmGKB [96]. Both in vivo (when available) and in vitro data were used to designate allele function (note that these function designations were made before CPIC developed a protocol for allele clinical function assignment). If functional information for an allele was not available, the SNV that is known to drive function was used by the author group to assign allele function. For instance, c.516G>T has been shown experimentally to reduce CYP2B6 expression via splicing (18). Thus, all haplotypes with c.516G>T were designated by CPIC as decreased clinical function (CYP2B6*6, *7, *9, *19, *20, *26, *34 and *36) or no function owing to the presence of other variants (CYP2B6*13, *37 and *38). There is variability in the detectable activity between and within alleles assigned decreased and no function, but any combination of these alleles results in the patient having a poor metabolizer (PM) phenotype. In contrast to other genes such as CYP2C19 or CYP2D6, CYP2B6 ‘no function’ alleles are rare. It is emphasized that CPIC assigns allele function based on clinical actionability, not solely on molecular or biochemical function and that alleles with no clinical function may have some residual activity. The filter option on the PharmVar display allows the user to sort alleles by CPIC functional status.
Genotype to Phenotype Translation
An individual has two CYP2B6 haplotypes, one on each chromosome, which constitutes their diplotype. For example, a CYP2B6*6/*18 diplotype assignment indicates that one chromosome (or allele) has SNVs defining the CYP2B6*6 haplotype and the second chromosome (or allele) has SNVs defining the CYP2B6*18 haplotype. The term “genotype” can refer either to the sum of all detected SNVs or to a person’s diplotype. In certain cases, assigning haplotypes to each chromosome may be difficult. If a person is heterozygous for e.g. c.516G>T, c.785A>G, and c.1459C>T, they can be assigned as CYP2B6*1/*7 (all variants in cis) or as CYP2B6*5/*6 (c.1459C>T being in trans with c.516G>T and c.785A>G). In order to distinguish haplotypes such as these, SNV linkage may need to be experimentally determined (75).
For phenotype classification, individuals are categorized into the five CPIC-recommended phenotype categories: poor (PM), intermediate (IM), and normal (NM) (formerly EM, extensive metabolizer), rapid (RM), and ultrarapid (UM) metabolizers (36). There is limited evidence that CYP2B6*4 and *22 alleles are associated with increased function. The phenotype categories of CYP2B6 RM (one normal function and one increased function allele, e.g. CYP2B6*1/*22) and CYP2B6 UM (two increased function alleles, e.g., *22/*22 and *4/*22) were created to allow for the possibility that these may be clinically relevant for some CYP2B6 substrates. Any combination of alleles classified by CPIC as no function and/or decreased function alleles are categorized as poor metabolizers. CYP2B6*6 is classified as ‘decreased function’ by CPIC, where a patient with a CYP2B6*1/*6 genotype is categorized as an IM, and a CYP2B6*6/*6 genotype as a PM. The CYP2B6*6/*6 genotype was reported to retain, on average, about 34% of activity compared to NM individuals with a reference CYP2B6*1/*1 genotype (36). In contrast, CYP2B6*28 is categorized by CPIC as a ‘no function’ allele due to c.1132C>T (p.R378X, rs34097093) that introduces a stop codon in exon 7 leading to a truncated protein that lacks the heme binding site (76). Thus, PM patients with a CYP2B6*28/*28 genotype have no enzyme activity. Patients genotyped as CYP2B6*6/*28 are also categorized as PM and are predicted to have activity between that observed for *6/*6 and *28/*28. Unlike CYP2D6 and CYP2C9 genes, which utilize the Activity Score system to facilitate genotype to phenotype translation, the process for CYP2B6 is based on the combination of increased, normal, decreased and no function alleles as provided in the Diplotype-Phenotype-Table (77) which provides all possible CYP2B6 genotypes (diplotypes) and their respective phenotypes.
Need for standardized genetic variation definitions and reporting of functional/clinical impact
To support research and clinical implementation, it is imperative to understand CYP2B6 variation, as well as allele and genotype function. While the function of the underlying genetic variant is known for several alleles, the function of others remains unknown or uncertain. In vitro functional analysis may produce inconsistent results which may be due, at least in part, to differences among test systems or substrates used (78, 79). We refer to the CYP2B6 functionality table for a detailed summary (77). Another challenge for function prediction is substrate dependent effects. For example, the CYP2B6*6 allele is associated with decreased efavirenz metabolism, but appears to confer enhanced cyclophosphamide metabolism (10, 80). Thus, except for true no function alleles, functional data should be extrapolated with caution. While CYP2B6 genetic variability for major ethnic groups are described (36), there is still a knowledge gap for many minority populations. Emerging evidence also suggests that co-medications may not affect all CYP2B6 variants equally and the extent of DDIs may be CYP2B6 substrate and genotype-dependent (see “Other Factors that can Influence CYP2B6 activity” for details).
For many genes encoding drug metabolizing enzymes including CYP2B6, haplotype information is critical to precisely predict a person’s metabolic capacity towards the large number of drugs metabolized by CYP2B6. As exemplified above for CYP2B6*6 and *9, it is sometimes difficult to unambiguously determine a person’s genotype (e.g. *1/*6 vs *4/*9 in which case the more common *1/*6 genotype is assigned by default). It is important to realize that these two SNVs also occur in CYP2B6*13, *37 and *38 which are no function alleles. Thus, if only c.516G>T/c.785A>G is tested these alleles may potentially be misclassified as CYP2B6*6 in rare cases. To complicate matters further, function altering SNVs such as c.516G>T/c.785A>G may also be linked with different combinations of SNVs in the upstream and 3’UTR regions which may contribute to variability among CYP2B6*6 alleles. Once these differences are better understood, alleles now catalogued as CYP2B6*6 suballeles may need to receive their own star designation to reflect these functional differences. This may also be true for other alleles. Thus, nomenclature is a dynamic process and will further evolve over time.
The need for standardized testing and reporting aligns with recent reports emphasizing that clinically actionable PGx information must be accurately represented in EHRs by using a harmonized system for genotype and phenotype information (81) (and references therein). Although many laboratories utilize star nomenclature as recommended by PharmVar and used by CPIC, there are interlaboratory differences in testing approaches and reporting (genotyping data may be described as chromosomal or genomic positions on a reference sequence, as an amino acid change, rsID, and/or using star allele nomenclature) (82). While test recommendations have been published for CYP2C9 and CYP2C19 (83, 84), no such guidance exists for CYP2B6. We believe that the utilization of star allele nomenclature as provided by PharmVar (25) will minimize “mis-interpretation” of a genotype result and its clinical implication(s).
The two end-user groups benefiting the most from standardized allele designations are clinicians and patients. Standardized terms and language will help clinicians to convey and explain results and patients to understand the meaning of test results. Consistent nomenclature is necessary for the integration of PGx into EHRs, the establishment of clinical decision support algorithms, and the design of clinical support tools and consistent test interpretation across healthcare systems. Finally, harmonized nomenclature also benefits basic and medical research.
The CYP2B6 Gene Locus
CYP2B6 and its closely related pseudogene CYP2B7 are located within a large CYP2 gene cluster that also includes the CYP2A and CYP2F genes (Figure 1A). The CYP2B6 gene is 27 kb long and comprises 9 exons encoding 491 amino acids. The gene harbors particularly large introns of which the 12 kb long intron 2 is most notable. The nearby presence of the highly homologous CYP2B7 gene requires careful attention to avoid inaccurate variant calls via off-target genotyping amplification or sequencing. CYP2B6 and CYP2B7 exon sequences differ only by 60 nucleotides (96% homology) (Figure 1) and many SNVs observed for CYP2B6 correspond to the reference nucleotide found in CYP2B7 including CYP2B6 c.983T>C (p.I328T; rs28399499).
Figure 1. Overview of the gene locus and allelic variation.
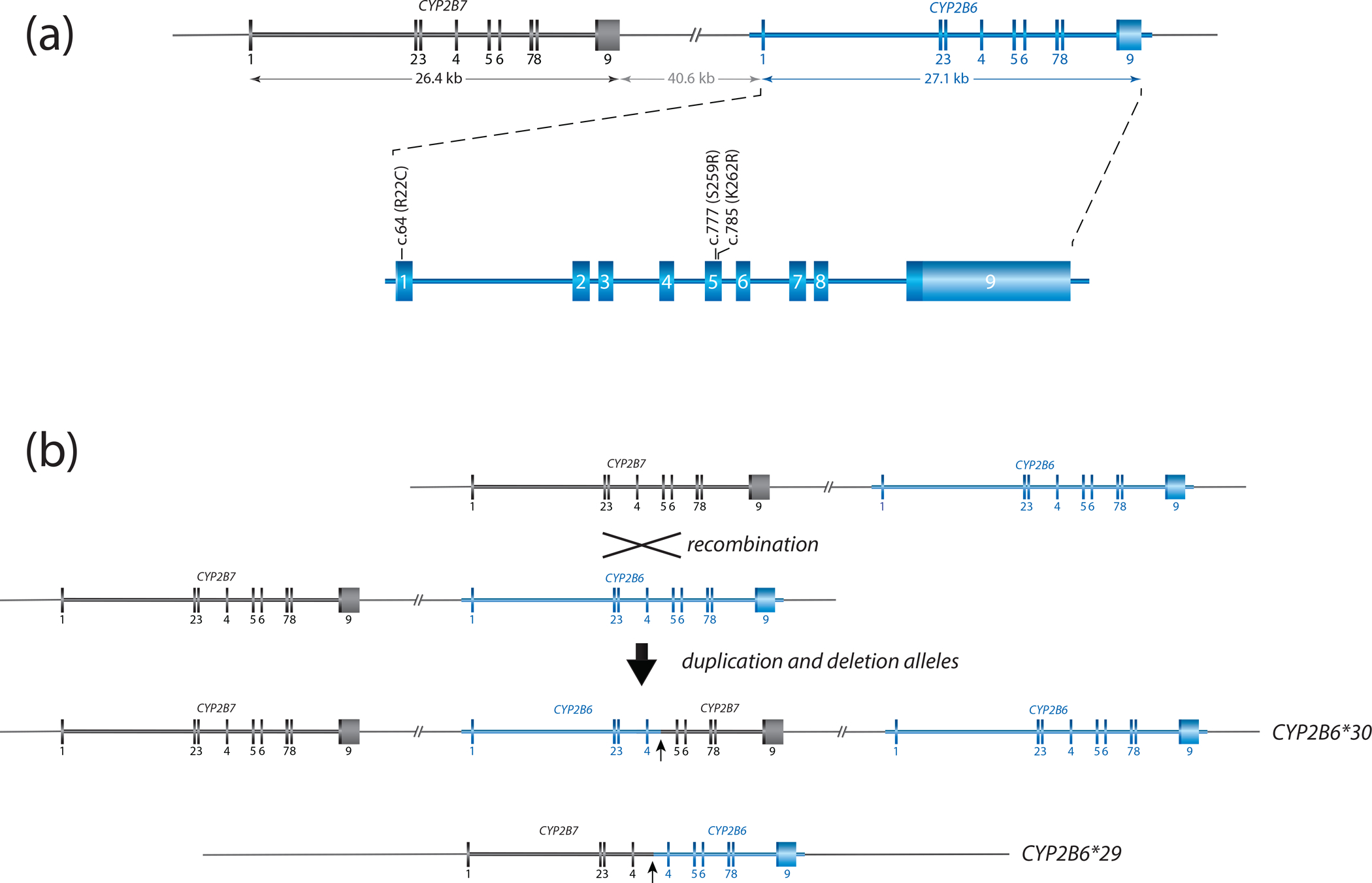
Panel A provides a graphical overview of the CYP2B gene locus containing CYP2B6 and CYP2B7. Both are encoded on the forward strand. CYP2B6 is composed of nine exons, covering 27.1 kb. The nonfunctional CYP2B7 pseudogene spans 26.4 kb. The two genes are interspersed by about 41 kb. Exonic sequences differ by only a few bases: exon 1 (9 base differences), exon 2 (9), exon 3 (6), exon 4 (13), exon 5 (2), exon 6 (4), exon 7 (10), exon 8 (3) and exon 9 (5). The enlarged region of CYP2B6 shows the location of selected sequence variants. c.64C>T (p.R22C) and c.777C>A (p.S259R) are SNVs defining CYP2B6*2 and *3 while c.785A>G (p.K262R) is part of multiple haplotypes including *4 and *6.
Panel B illustrates a reciprocal recombination event resulting in a partial deletion (CYP2B6*29) and a duplication (CYP2B6*30) event. CYP2B6*29 and *30 harbor 23 and 15 amino acid changes, respectively (see Structural Variation document for details). The recombination is facilitated by a 529 bp long intron 4 region with high homology between CYP2B6 and CYP2B7. Both recombination products are characterized by the presence of gene copies consisting of CYP2B7 and CYP2B6 as shown.
SNVs in the CYP2B6 locus have been identified in both coding and noncoding 5’ and 3’ regions. Since CYP2B6 is highly inducible and subject to transcriptional regulation (85), SNVs in the promoter region can impact expression levels and thereby alter function (66, 86). One example is the increased activity CYP2B6*22 allele which has four SNVs in the upstream region, of which one resides within the putative TATA box domain and impacts gene expression (86). Genetic variation in the 3’UTR has been shown to alter function by interfering with microRNA regulation (87). Consequently, the PharmVar gene experts recommended that CYP2B6 allele definitions should include −2330 bp of upstream region and 1500 bp of the 3’UTR (relative to the translation start and stop codons, respectively). This allows PharmVar to catalogue more detailed haplotype information on the suballele level. Should novel information arise regarding the impact of SNVs in the upstream or 3’UTR regions, alleles will be reclassified based on PharmVar standards.
The CYP2B6 gene is also characterized by structural variation. Unequal crossover recombination events within intron 4 of CYP2B6 and CYP2B7 have resulted in various hybrid forms of these two genes. PharmVar lists these deletion (CYP2B6*29) and duplication (CYP2B6*30) events as structural variations (Figure 1B). Additional information regarding CYP2B6 structural variants can be found in the “Structural Variation” document on the PharmVar CYP2B6 gene page (88).
CYP2B6 allele, genotype, and phenotype frequencies across populations
Substantial differences in allele frequencies occur across ancestrally diverse groups. The CYP2B6 frequency table available at PharmGKB (77) summarizes population-based allele frequencies reported in the literature. Studies were considered for inclusion if (i) the ethnicity of the population was clearly indicated, (ii) either allele or genotype frequencies were reported, (iii) the methodology by which the genes were genotyped was indicated, (iv) the sample population consisted of at least 50 individuals with a few exceptions, and (v) the study represented an original publication. The ethnicities/locations reported in the articles are mapped into seven geographically defined groups (American, Central/South Asian, East Asian, European, Near Eastern, Oceanian, and Sub-Saharan African) and two admixed groups (African American/Afro-Caribbean and Latino) using the biogeographical grouping system developed by PharmGKB (89). The CYP2B6 frequency table is periodically updated and contains multiple tabs summarizing “allele frequencies by biogeographical group”, “diplotype frequencies by biogeographical group”, “phenotype frequency”, and “references”; the latter describes allele frequencies for each publication included in the listing, which also allows the user to customize allele frequencies as needed. There are a number of limitations regarding the accuracy of allele frequencies including: (i) they are based on published allele frequency data (limited for some populations); (ii) most studies test for a limited number of allelic variants, and multiple SNVs reside in more than one star allele for CYP2B6 (e.g., c.516G>T is found in combination with other variants in 11 alleles: *6, *7, *13, *19, *20, *26, *34, *36, *37, *38, as well as the *29 hybrid); unless all the defining SNVs are tested, an allele may be mis-assigned; likewise, if no SNVs are found, CYP2B6*1 is assigned, which inflates the frequency of this allele; (iii) inadequate testing for CNVs; and (iv) errors translating SNV results into star alleles. Consequently, certain alleles may be over or underreported, or not detected at all. Therefore, all calculations based on allele frequencies are estimates and should be used with caution.
Observed and calculated frequencies of individual CYP2B6 SNVs, as well as star alleles frequencies markedly differ between ethnicities (10, 37, 90). The decreased function alleles CYP2B6*6 and *18, for example, are more prominent in the Sub-Saharan African, African American/Afro-Caribbean and Oceanian groups compared with other groups. In contrast, the CYP2B6*4 increased function allele is more common in the East Asian, Central/South Asian, Latino and Near Eastern groups and absent or very low in frequency in populations with African ancestry (91). Furthermore, populations including South Africans, African Americans, and others with diverse founding populations and admixed populations may show different allele frequency patterns. The frequency of CYP2B6*9 is inconclusive due to the challenge of distinguishing CYP2B6*1/*6 from *4/*9.
Genotype frequencies are the result of allele frequencies in each population and were calculated using the equation describing the Hardy Weinberg Equilibrium. Phenotype frequencies across populations are provided in the “Calculated phenotype frequency” tab in the PharmGKB/CPIC CYP2B6 Frequency Table (77). Oceanians have the highest frequency of poor metabolizers and Central/South and East Asians and Latinos the highest frequency of rapid metabolizers. We stress, however, that all phenotype group frequencies, including those shown in the PharmGKB/CPIC table, have to be viewed with caution due to the limitations regarding the accuracy of allele frequencies as well as the method used to translate genotype into phenotype and inconsistencies in the classification of “population”, “ethnicity” or “race.”
PharmVar Nomenclature and CYP2B6 allele designation
PharmVar stores and displays allelic data consistently across genes, relying on public standards and data sources wherever possible. The standardized nomenclature follows criteria developed by experts. The “Allele Designation and Evidence Level Criteria” document (92) describes the nomenclature system and provides examples. For example, a new star number is only issued if a haplotype contains a sequence variant that: i) results in an amino acid change, e.g., CYP2B6*2 which harbors an arginine to cysteine change (c.64C>T, p.R22C, rs8192709) or CYP2B6*28 which harbors a nonsense mutation changing an arginine to a premature stop codon (c.1132C>T, p.R378X); ii) abolishes a splice site (no examples for CYP2B6); or iii) changes expression levels causing decreased or increased function, which is exemplified by the CYP2B6*22 defining variant (c.-82C>T) which increases expression levels (86). Importantly, new haplotypes that contain previously characterized variants that eliminate function are catalogued under the original star allele number as a suballele. For example, any allele having a novel variant and c.1132C>T (p.R378X), will be designated as a CYP2B6*28 suballele and considered having no function, regardless of the functional status of the novel variant.
The PharmVar CYP2B6 gene expert panel
A panel of international experts was recruited from the PharmVar membership representing CYP2B6 research, clinical testing, and implementation. The panel also includes PharmGKB/CPIC representation to ensure that the nomenclature is consistent with CPIC guidelines and to facilitate dissemination to a greater audience through PharmGKB as well as other databases. Experts were tasked to assist with the transition of the gene into the PharmVar database which included review of existing haplotype definitions and the refinement of the regions to include into definitions. Moving forward, the panel is tasked with the review and designation of novel haplotypes submitted to PharmVar, as well as lending their expertise to resolve any issues that may arise regarding CYP2B6 nomenclature. The composition of the panel can be found on the PharmVar website (93).
The PharmVar CYP2B6 gene page
The content of the original nomenclature site for CYP2B6 was converted into its current format in September 2019 and remains accessible through the ‘Archive’ link (94).
Sequence variations are mapped to the genomic and transcript RefSeqs NG_007929.1 and NM_000767.5, respectively, and the GRCh37 (NC_000019.9) and GRCh38 (NC_000019.10) genome builds. A Locus Reference Genomic (LRG) record was requested by PharmVar from the LRG Project, a NCBI (RefSeq) and EMBL-EBI (Ensembl/GENCODE) initiative (95). LRGs are universally accepted reference standards that are created specifically for clinical reporting by manual curation. LRGs are stable entities that never change or are versioned after their release. The LRG for CYP2B6 (LRG_1267, released 8-24-2018) corresponds to the current NG_007929.1 reference sequence and is now used by PharmVar as the ‘gold-standard’ reference.
PharmVar is left-aligning, i.e. insertions and deletions of nucleotides in a repeat or homopolymer sequence are listed using the 3’ rule recommended by HGVS (see PharmVar ‘Standards’ for more details (96) or the CYP2D6 (97) and CYP2C19 (81) GeneFocus reviews for examples). Due to different alignment methods, coordinates for insertion/deletion polymorphisms may differ among databases. No such variants have, however, been described for CYP2B6 to date.
On the PharmVar CYP2B6 gene page, the user can easily cross-reference genomic and cDNA positions by choosing the respective reference sequence or genome build of interest. For LRG_1267 there is the option of two count modes, i.e. counting from the first nucleotide in the sequence or the ‘A’ of the ATG translation start codon being +1.
CYP2B6 haplotype evidence levels
PharmVar designates the “Haplotype Evidence Level” for each of the star alleles reported on the CYP2B6 gene page. Evidence levels are displayed as symbols indicating ‘definitive’ (Def), ‘moderate’ (Mod) or ‘limited’ (Lim) levels of support for a given haplotype. This three-tiered system represents a modified ClinVar classification system; more detailed information is provided in the ‘Allele Designation and Evidence Level’ document (92). This type of information (e.g., whether an allele was sequenced across the gene, how a haplotype was determined) was not always systematically captured prior to PharmVar. For existing haplotype definitions, a literature review was conducted in order to assign evidence levels. Most alleles are currently labeled as ‘Lim’ because their definitions are based on SNVs in the coding regions only and do not include the 2.3 kb of upstream or 1.5 kb of downstream sequence, or only parts thereof, which are now required for submission. The value of evidence levels is centered on providing users with as much information on haplotype reliability as possible and enabling users to quickly parse haplotypes based on robust, high evidence, versus other haplotypes with ‘Lim’ or ‘Mod’ evidence levels.
PharmVar solicits submissions for all alleles labeled ‘Lim’ and ‘Mod’ to ultimately raise their evidence levels to ‘Def’. Moreover, PharmVar also encourages additional submissions for alleles with single citations and shown as ‘Def’ to further corroborate a haplotype definition.
PharmVar IDs
PharmVar assigns an ID to each haplotype (PVID) which is a unique numeric identifier analogous to dbSNP rsIDs. Since star allele names are driven by functional grouping, an allele’s star number may not be permanent and be subject to change. Such changes may be necessary as new information becomes available, or during standardization efforts (see re-assignment of CYP2B6*16.001 to *18.002 described below). If an allele’s star designation is updated to a new star number, the PVID of the haplotype remains constant and does not change as exemplified by the revision of CYP2B6*16.001 to *18.002. However, if a star allele definition changes (e.g., through the addition or removal of variants, a new PVID will be assigned (examples are CYP2B6*1.007 and *6.002 to which SNVs were added). Original PVIDs and their haplotype definitions can be tracked in the database via the PVID Lookup function.
Curation efforts
Corrections, revisions, new alleles, and other updates:
Before the transition to PharmVar, coordinates were mapped to NG_000008.7, which has been removed from the National Center for Biotechnology Information (NCBI) database. Updates to the current CYP2B6 RefSeq NG_007929.1 (matching the LRG_1267) did not change position annotations of SNVs when counting from the translation start site.
During the process of transitioning CYP2B6 into the PharmVar database, comments and footnotes were removed, and some alleles were revised based on available information and to keep standardized annotations across genes (Table 3). References in support of allele definitions have been updated and those solely describing function removed (references for function are provided in the PharmGKB/CPIC CYP2B6 Allele Functionality table) (77). Changes and revisions made are detailed in the ‘Change Log’ document on the CYP2B6 gene page.
Table 3.
Summary of edits and changes made as alleles were transitioned into the PharmVar database in September 2019.
| Reason | Change | SNVs and Affected Alleles |
|---|---|---|
| Standardization | Intronic SNVs were removed | 12917A>T (*1D, *4C); 14593C>G (*1C); 15582C>T (*1C, *13B, *15A+B); 15837C>T (*28); 17897C>T (*18); 18273G>A (*11B, *12, *13A+B, *14, *15B, *17A+B, *18, *19, *20, *21, *27, *28, *38); 18627G>A (*18, *28); 21563C>T (*9, *13A+B, *19, *38). rs4803419 was significantly associated with increased plasma efavirenz exposure in GWAS studies; however, this SNV was not consistently genotyped and thus, it remains unknown which haplotypes carry this SNV |
| Standardization | Retired | After intronic SNVs were removed, the *1C, *13B and *15B haplotypes were identical to *1B (*1.002), *13A (*13.002) and *15A (*15.001) and thus were retired. |
| Other | Upstream SNVs were removed | -1456T>C and -750T>C were removed from *28 because it remained uncertain whether these SNVs are indeed on this haplotype (76) and personal communication with author) |
Several intronic SNVs have been removed from allele definitions due to the lack of evidence in the literature showing that they cause functional changes (Table 3). Consequently, rs4803419 was removed from CYP2B6*1C, *13B and *15B, making them identical to *1.002, *13.001 and *15.001, respectively, and resulting in their retirement. rs4803419 was described to associate with increased plasma efavirenz exposure (20); however, contradictory results have been reported (98, 99). Furthermore, the default assignment of rs4803419 to CYP2B6*1C rather than testing for other suballeles with this SNV (*13B and *15B), may have contributed to mixed findings. There are no data regarding a potential mechanism by which rs4803419 would impact function, and it remains speculative whether any of the observed associations are due to linkage with other, potentially distant SNV(s). Therefore, given the uncertain functional effect of this SNV, it was removed from allele definitions. In addition, removal of intronic SNV rs2279344 led to the retirement of CYP2B6*4C and *11B as these then matched *4.001 and *11.001, respectively. These haplotypes may be ‘unretired’ if new data become available.
The expert panel also recommended removal of two upstream SNVs c.-1456T>C (rs2054675) and c.-750T>C (rs4802101) from the CYP2B6*28 definition because it was uncertain whether they are present in this haplotype (76). CYP2B6*4B and *7B were also retired since they these suballeles did not differ by PharmVar standards.
Previously, CYP2B6*16 and CYP2B6*18 were categorized by CPIC as decreased function and no function alleles, respectively. CYP2B6*16 and *18 have now been consolidated under a single star designation due to recently published evidence showing no detectable protein expression and function in vitro (78, 100). The allele initially listed as CYP2B6*16.001 was revised to *18.002 with c.983T>C (p.I328T) being the core SNV (Table 2). Alleles identified in the future having c.983T>C (p.I328T) will therefore be listed as CYP2B6*18 suballeles, unless there is evidence that an additional variant rescues function.
Table 2.
Novel and confirmed suballeles and allele consolidation
| Core Allele Designation | Novel alleles/suballeles |
|---|---|
| *1 | *1.007, *1.013, *1.014, *1.015 |
| *2 | *2.002 |
| *6 | *6.002, *6.004 |
| *15 | *15.001 |
| *18 | *18.001 |
| Core Allele Designation | Consolidation |
| *16.001 | Consolidated under *18 as *18.002 |
In contrast, CYP2B6*8 and *13, both categorized by CPIC as no function alleles, were not consolidated. Although CYP2B6*8 and CYP2B6*13 do not appear to have function towards several substrates (79, 101), recombinant CYP2B6*13 protein retained appreciable activity towards artemether O-demethylation (~21% versus the *1 reference) (102). CYP2B6*13 activity for this and potentially other substrates, might be due to a combinatorial effect of the SNVs present on this haplotype. Until further evidence emerges, PharmVar will keep CYP2B6*8 and *13 as separate entities.
Core allele definitions
For many alleles there is a growing number of suballeles that share one or more ‘core’ defining sequence variant(s). Although suballele information can be valuable, e.g. design of test platforms (sequence or genotype-based) and interpretation of genotyping test results, there is no need to distinguish suballeles for phenotype prediction because all alleles under a star number are presumed to be functionally equivalent. Thus, even if a test is capable of distinguishing suballeles, from a functional standpoint, these can be simply reported using core allele definitions.
PharmVar and the PharmGKB have collaboratively developed criteria for core allele definitions. A core allele is defined only by sequence variations that cause an amino acid change or impact function by changing expression levels or interfere with splicing and are present in all suballeles within an allele group. With this rule-based system, suballeles are collapsed into a single ‘core’ definition representing all suballeles categorized under a star (*) number. For example, CYP2B6*6 suballeles share two SNVs that fulfill this rule, i.e. c.516G>T (p.Q172H) and c.785A>G (p.K262R). Thus, both SNVs constitute the CYP2B6*6 core allele definition (Figure 2). For CYP2B6*4, only c.785A>G (p.K262R) is shared among the two known subvariants and therefore is the only SNV of the *4 core allele definition (Figure 2).
Figure 2. Overview of core alleles and suballeles.
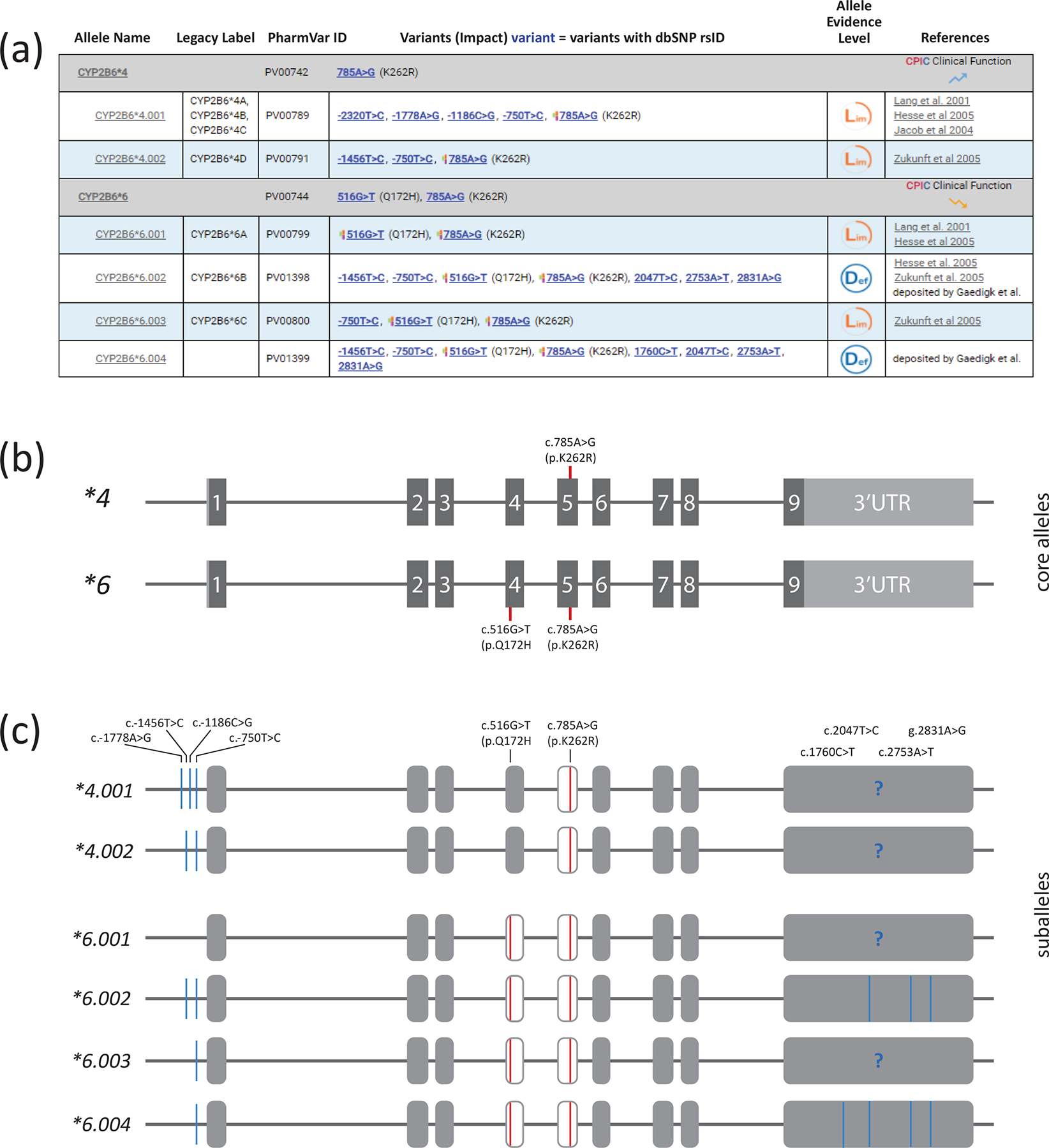
Panel A shows the CYP2B6*4 and *6 core allele definition (gray bar) with NM_000767.5 as the reference sequence. Core SNVs, PharmVar ID (PVID), and evidence level is shown for each allele. All currently defined suballeles are displayed underneath the core allele bar. Legacy allele designations are cross-referenced (e.g., *6.001 corresponds to *6A). Panel B is a graphical representation of the CYP2B6*4 and *6 core alleles and their core SNVs. c.785A>G (p.K262R) is present on both, while c.516G>T (p.Q172H) is present on *6. Note that each of these SNVs is also present on numerous other core alleles as shown in Figure 3. Core SNVs are shown by red lines; gray boxes represent the nine exons (scale is approximated). Panel C shows the CYP2B6*4 and *6 suballeles defined to date. As shown, suballeles may differ in the upstream and/or 3’UTR regions. “?” indicates that it remains unknown whether respective 3’UTR regions harbor SNVs. Therefore, these regions likely harbor additional SNVs. Core SNVs (causing an amino acid change) are shown in red, all other SNVs are highlighted in blue.
Of importance, a sequence variant found in a core allele definition is not necessarily unique to that haplotype. As illustrated in Figure 3, c.785A>G (p.K262R) is part of 11 haplotypes including CYP2B6*4 and *6. Apart from CYP2B6*4, these haplotypes also have c.516G>T (p.Q172H), and there is one haplotype, CYP2B6*9, which has only c.516G>T, but not c.785A>G.
Figure 3. Comparison of alleles with common sequence variations: allele default assignment.
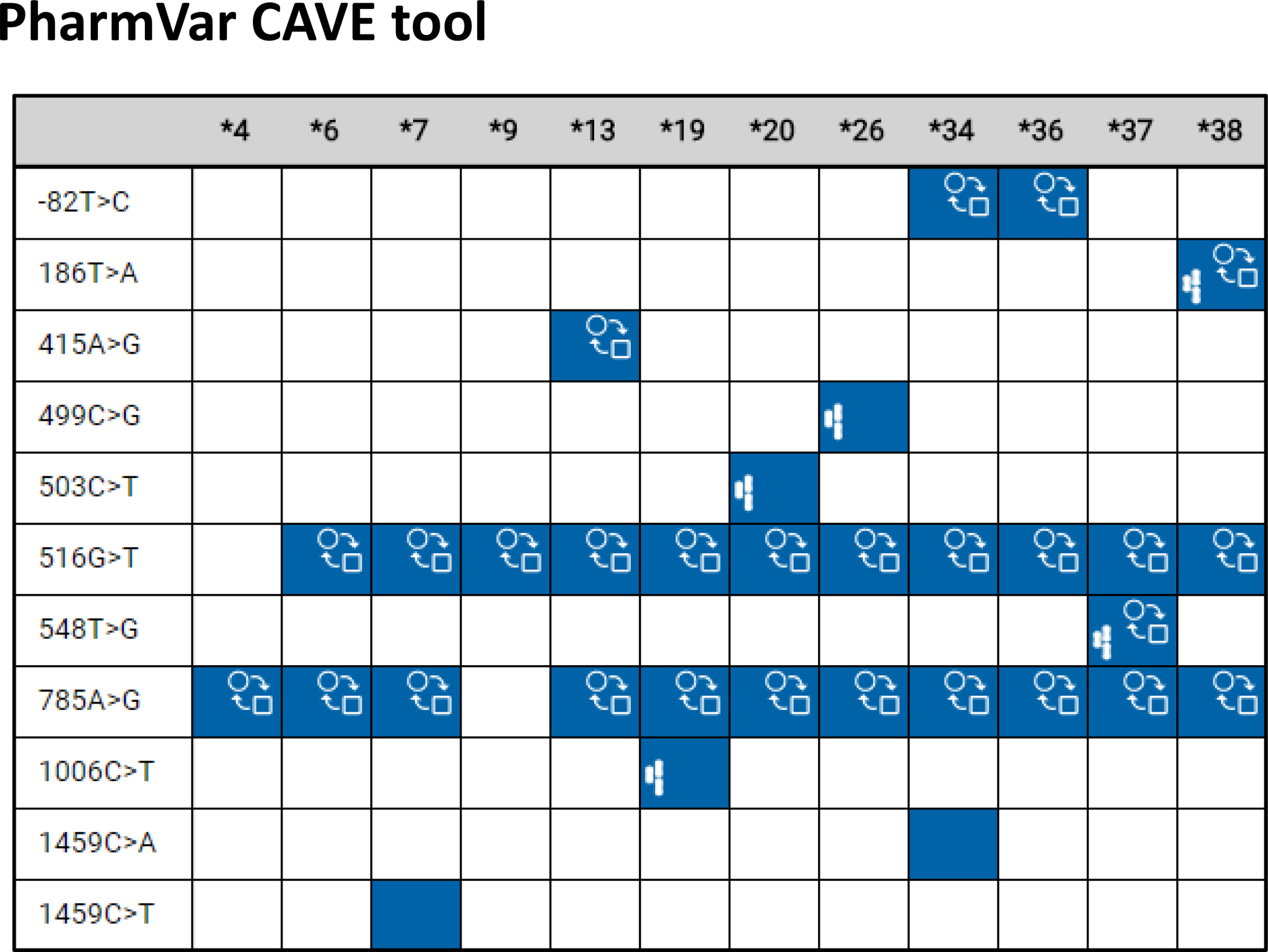
This comparison was generated with the comparative allele viewer (CAVE) tool and depicts all star alleles with c.516G>T (Q172H) and/or c.785A>G (K262R) except CYP2B6*29, a CYP2B7-2B6 hybrid that harbors 23 amino acid changes including Q152H. Blue boxes indicate the presence of a core SNV on all suballeles, while the gray box indicates that the core SNV is not present on all suballeles. The function ( ) symbol indicates that a core SNV alters function and the PharmVar (
) symbol indicates that a core SNV alters function and the PharmVar ( ) symbol highlights that a core SNV is unique to a star allele. Of note, since c.785A>G is increasing function of CYP2B6*4 it is thus annotated with the function symbol and consistently shown as such across alleles. The impact of this SNV on the function of an allele with an additional SNV(s) may be minor or unclear. SNV positions refer to transcript coordinates on the NM_000767.5 reference sequence.
) symbol highlights that a core SNV is unique to a star allele. Of note, since c.785A>G is increasing function of CYP2B6*4 it is thus annotated with the function symbol and consistently shown as such across alleles. The impact of this SNV on the function of an allele with an additional SNV(s) may be minor or unclear. SNV positions refer to transcript coordinates on the NM_000767.5 reference sequence.
One challenge with core allele definitions is that an allele definition may change over time as new information becomes available. For instance, based on the rules described above, the current core allele definition of CYP2B6*38 harbors c.186T>A (p.Y62X, rs281864907), c.516G>T (p.Q172H) and c.785A>G (p.K262R). According to allele designation criteria, all new alleles having c.186T>A are nonfunctional due to the ensuing p.Y62X stop codon and thus, will be assigned as CYP2B6*38 suballeles, regardless of the nature of the other SNV(s) present. Consequently, the CYP2B6*38 core allele definition may change, should a sequence containing c.186T>A, but not c.516G>T, and/or c.785A>G be identified.
The core alleles are the basis of the CYP2B6 allele definition table used in CPIC guidelines and by PharmGKB (see PharmGKB ‘Gene Specific Information tables’ [Table 1]). The CYP2B6 core allele definitions are also utilized for clinical annotations in PharmGKB.
The PharmVar Comparative Allele ViewEr
The Comparative Allele ViewEr (CAVE) tool was developed by PharmVar to easily compare core alleles (103). This tool can be accessed using the “Compare View” button on the CYP2B6 gene page. Figure 3 showcases this tool on CYP2B6 allelic variants having either c.516G>T, c.785A>G, or both. In this display mode it is easy to see which core SNVs are shared among haplotypes, whether they alter function and/or are unique to a haplotype, as well as to visualize ‘default’ allele and genotype assignments. As discussed above, these two variants are present in multiple CYP2B6 alleles, together or separately, and sometimes in combination with other variants. Therefore, if genetic testing does not assay all of the variants in these alleles, assumptions are made to assign the allele, usually based on frequency. If a particular genotype test only includes c.516G>T, an observed ‘T’ variant may be defaulted to CYP2B6*6 simply because it is the most commonly found allele with this variant even though c.516G>T on its own defines the CYP2B6*9 allele. If testing includes c.516G>T and c.785A>G, alleles positive for one of these SNVs will default to CYP2B6*9 and *4, respectively, but if both SNVs are present a CYP2B6*1/*6 genotype will likely be assigned because this genotype is statistically more likely than *4/*9, or any other combination that also contains these variants. It is important to keep in mind that defaulting to the most common allele and/or genotype can result in an incorrect phenotype assignment. This example demonstrates the complexity of CYP2B6 genotyping and allele assignments, as well as the utility of the CAVE tool.
Reporting genotype and translation into phenotype
PharmVar and PharmGKB have also collaboratively developed templates to facilitate more consistent and transparent reporting of genotype details and how genotype is translated into phenotype (this information can be provided as supplemental materials of a publication to facilitate access to important data for subsequent curation). The first template file (Table S1) collects information including methods or platforms used for genotyping and which SNVs were interrogated; the template also provides a standardized set-up for reporting genotype results for individual subjects, as well as allele frequencies. The second template file (Table S2) facilitates the reporting of how genotype is translated into phenotype as well as genotype frequencies. Although it is recommended by CPIC, as well as other groups to use their standardized translation method, not every investigator or laboratory adopts this method. Too often, papers state that ‘genotyping was performed as previously described’ or indicate that ‘CYP2B6 phenotype was correlated with the metabolism of a drug’ without specifying which SNVs or alleles were genotyped or how phenotype was assigned. The lack of such information can make it extremely difficult, if not impossible, for curators to compare findings or extract information for CPIC guideline development. Colleagues are therefore strongly encouraged to utilize the provided templates, or revised versions thereof, for publication of these types of information.
CYP2B6 reference materials
The GeT-RM is a combined effort among the Centers for Disease Control and Prevention–based Genetic Testing Reference Material Coordination Program, Coriell Institute for Medical Research, and members of the pharmacogenetic testing community. Established sets of well-characterized reference materials are needed for assay development, validation, quality control, and proficiency testing, especially for complex genes such as CYP2B6. To address this need, a set of 137 genomic DNA samples were characterized for 28 pharmacogenes, including CYP2B6 and “consensus” genotypes established (104). The methods employed for testing differed considerably regarding the number of alleles tested. Only four alleles, CYP2B6*2, *6, *7 and *18 were verified by more than two methods leaving many samples with ambiguous consensus assignments. The authors discuss that differences in platform design was one limitation, but also emphasize that many SNVs are found in many alleles. While the available data are valuable, there is clearly a need to characterize these materials more comprehensively. Reference materials can be acquired from the Coriell Institute (Camden, NJ, USA).
Inferring CYP2B6 diplotype from Next Generation Sequence data and public databases
Several bioinformatic tools (105–107) have been developed to allow for diplotype calling from next generation sequencing (NGS) data. These tools use probabilistic approaches based on the variants called to infer the most likely diplotype, which in turn are based on the catalog of known haplotypes, such as those defined by PharmVar. Most of these tools were originally optimized to call CYP2D6 diplotypes and, to date, only Stargazer (106) has been expanded to call diplotypes for the CYP2B6 gene. An important consideration is the requirement for high-quality NGS data and precise haplotype definitions to accurately call diplotypes. Due to the high sequence similarity, there is the potential for sequencing reads to misalign with the highly homologous adjacent CYP2B7 pseudogene. Misalignments can be reduced by using longer read lengths, careful sequencing design or improved NGS alignment tools.
In silico mapping simulation of reference CYP2B6 reads, as previously reported for CYP2D6 (105), showed that single-end sequencing reads below 100 nucleotides in length led to several potential misalignments (Figure 4A). Potential read alignment errors were also possible using paired-end reads with up to read lengths of 150 nucleotides and insert sizes less than 200 bp, with exons 5 and 9 being the main culprits (Figure 4B). For reference, sequencing reads in the 1000 Genomes project are composed of single end reads reported to range between 70–160 nucleotides in length. Misalignment is increasingly problematic when a CYP2B6 polymorphism can reflect the CYP2B7 reference sequence, leading to a greater likelihood of sequencing read misalignment.
Figure 4. In-silico modelling highlighting regions with potential misalignments from simulated reference CYP2B6 short-read sequences.
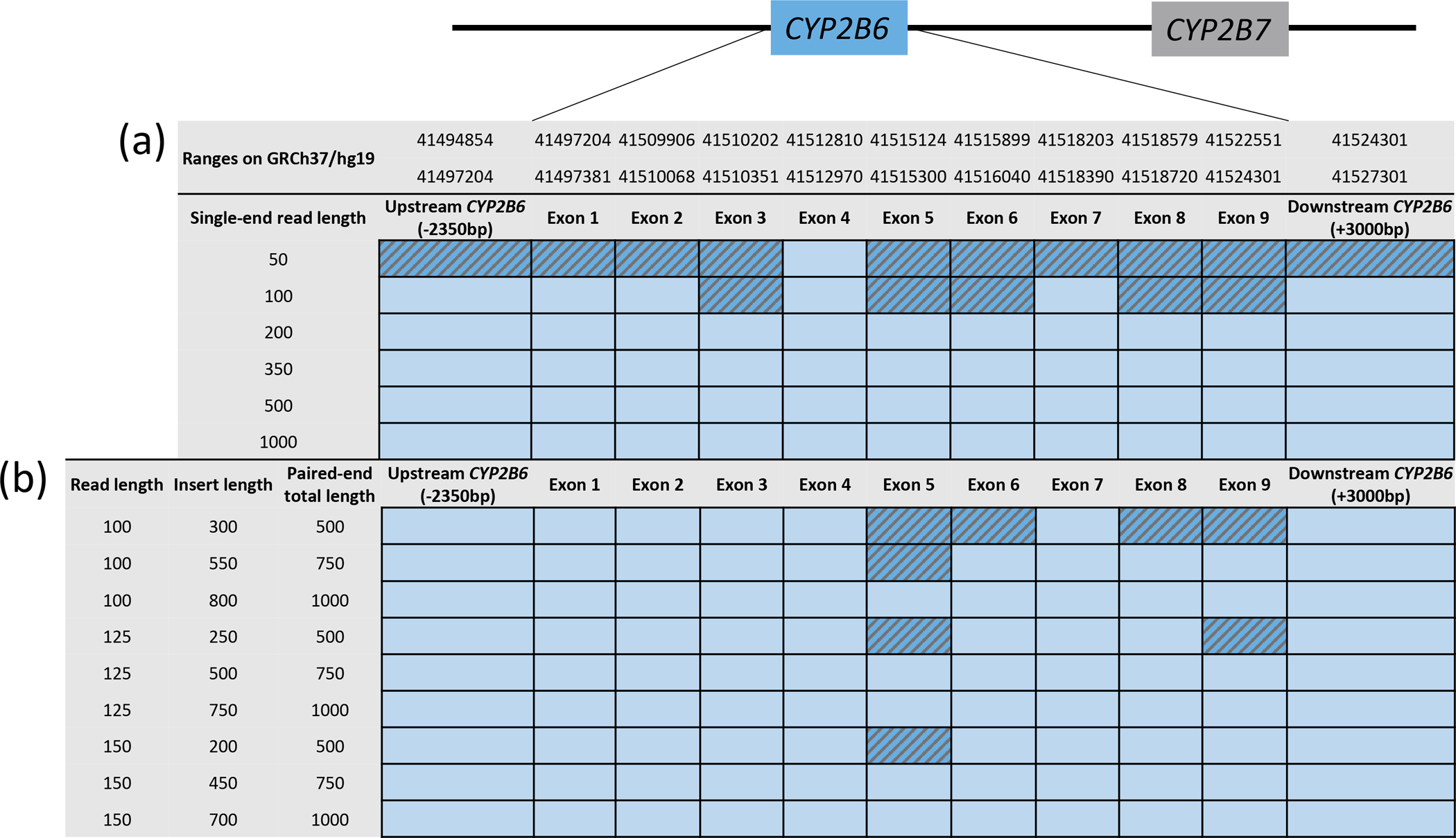
Panel A and B display simulated single-end reads ranging from 50 to 1,000 bases in length and paired-end reads from 100 to 150 bases and insert sizes from 300 to 800 bases, respectively. Reads that uniquely mapped to CYP2B6 with the BWA-mem aligner are shown in light blue and exons to which reads could tentatively also align elsewhere are hatched in darker blue.
Given these challenges, haplotype calls using 1000 Genomes Project data (and other public data resources such as ExAC, gnomAD, or ALFA) should be interpreted with caution. One example is c.785A>G (p.K262R), the functionally relevant missense variant in exon 5 that is part of the CYP2B6*4, *6, *7, *13, *16, *19, *20, *26, *34, *36, *37 and *38 alleles. The minor allele frequency (MAF) for c.785A>G varies widely between public sequencing databases reflecting the difficulty in accurately calling this variant from NGS. In European populations the MAF ranges from ‘not identified’ (1000 Genomes) to 3.7% (ExAC), 10.7% (gnomAD) and 22.5% (ALFA); this variability is also observed in African populations from ‘not identified’ (1000 Genomes), 6.5% (ExAC), 24.1% (gnomAD), 22.9% (ALFA). The true allele frequencies as determined through reliable PCR methods suggests that the MAF for this SNP is between ~25% and ~30% in Europeans (108) and Africans (91, 109), respectively. The improvement in capturing this variant in more recent databases (e.g. ALFA) is likely a reflection of the improvements in WGS accuracy using longer sequencing reads. Therefore, the linkage between c.785A>G and other variants cannot be estimated using conventional tools that leverage 1000 Genomes data, such as LDLink (110), despite its high true MAF. Furthermore, caution should be taken when interpreting WGS and WES data due to possible read alignment issues, with careful attention to exon 5 and 9 reads.
There is emerging data that WGS and NGS-based targeted gene panel sequencing produce sufficiently high-quality data and read alignments to support CYP2B6 the definition of new haplotypes. This is supported by samples including those shown in Figure 5, which have been subjected to two modes of NGS sequencing and produced consistent results among the methods.
Figure 5. Experimental approaches for phasing SNVs to establish haplotype.
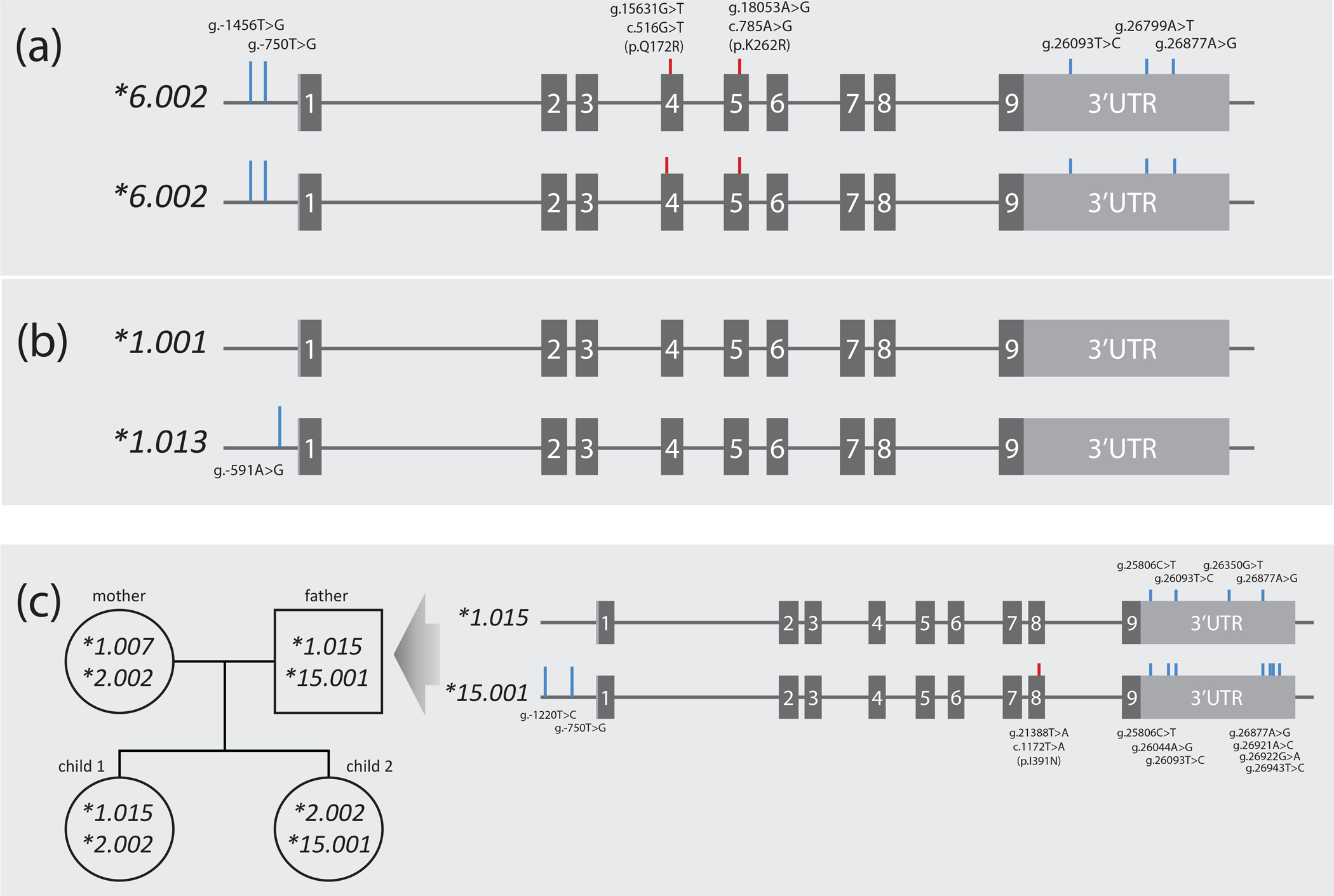
Panel A represents samples NA12003 and NA19176 both of which were homozygous for eight SNVs. Two are CYP2B6*6 core SNVs (c.516G>T (pQ172R) and c.785A>G (p.K262R); red lines) while the remaining SNVs sub-classify this haplotype as *6.002 (blue lines). Homozygosity across all SNVs unequivocally informs haplotype. Panel B depicts NA12156 which is heterozygous for a single SNV and thus unequivocally informs the haplotype of this novel suballele which was designated *1.013. Panel C exemplifies how haplotype can be inferred by inheritance (the example shown was part of a PharmVar submission). Haplotypes were determined using WGS data for a family with two offspring. Inferred haplotypes were corroborated with 10X Genomics Long-Read sequence technology for all family members. The CYP2B6*1.015 and *15.001 alleles of the father are detailed as indicated by the arrow. CYP2B6*1.007 found in the mother was found homozygous in NA18544, NA17227 and NA18952 (not shown) further supporting the use of these technologies to inform CYP2B6 haplotype.
Methods for CYP2B6 allele characterization
To date, Sanger sequencing has been the principal approach for identifying and characterizing novel CYP2B6 alleles (91, 109). Haplotypes can unequivocally be determined regardless of whether Sanger or short read-based NGS methods were utilized if a sample is homozygous for all SNVs (Figure 5A) or is heterozygous for only one SNV (Figure 5B). If a sample is heterozygous for more than one SNV, additional analyses will be necessary to establish whether SNVs are in cis or in trans using techniques such as allele-specific PCR or digital droplet PCR (111). Haplotypes may also be computationally inferred from sequencing data, but only receive a ‘Lim’ or “Mod’ evidence level depending on the number of subjects used for phasing, the frequency of the allele and/or whether uncertainty of SNVs being in cis or trans affects core SNVs. We refer to the recently updated criteria for computationally inferred haplotypes (see the allele definition criteria and evidence level document for more details and examples).
These challenges may be overcome by inferring haplotypes using inheritance (pedigree analysis/trio studies). Figure 5C demonstrates this approach in a pedigree with two offspring in which four different haplotypes were identified. Haplotype can also be determined by sequencing long-range PCR products with single molecule real-time (SMRT) sequencing, Nanopore sequencing, or 10X Genomics Linked-Read technology, or using whole genome data generated with these technologies. These methods have been detailed in the CYP2D6 and CYP2C19 GeneFocus reviews (81, 97) and successfully employed to resolve haplotypes for these genes. The 10X Genomics Linked-Read technology has been utilized to corroborate the haplotypes in the pedigree shown in Figure 5C.
Conclusions
This CYP2B6 GeneFocus provides essential information for the understanding of this highly polymorphic gene to facilitate basic and medical research, as well as to facilitate the implementation of this functionally complex pharmacogene into clinical practice. This GeneFocus complements the information provided by the CPIC guideline on efavirenz and by other pharmacogenetic resources. We are highlighting PharmVar efforts and challenges of systematically cataloging CYP2B6 allelic variation, genotype analysis and allele calling, as well as collaborative efforts with PharmGKB to make the information useful and easily accessible to the entire pharmacogenetics community.
Supplementary Material
Acknowledgement
We thank Marelize Swart, PhD, for serving on the gene expert panel and the preparation of the Read Me and Structural Variation documents and Roger Gaedigk, PhD, for assisting with artwork.
Funding
This work was funded by the National Institutes of Health for the Pharmacogene Variation Consortium (R24 GM123930; PI, A.G.), PharmGKB (U24 HG010615; PI, T.E.K.) and a Public Health Services Grant (R01GM121707, Z.D.). We also acknowledge funding from the Canada Research Chairs program (CRC in Pharmacogenomics) (R.F.T), CIHR grant PJY-159710 (R.F.T) and the Centre for Addiction and Mental Health (R.F.T), the Australian Centre for HIV and Hepatitis Virology Research (ACH2) (A.A.S.), the Swedish Research Council (grants 2016-01153, 2016-01154 and 2019-01837 (V.M.L), the EU/ EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (grant 875510) (V.M.L.), the European Union’s Horizon 2020 Research and Innovation Program U-PGx (grant 668353) (V.M.L.), the Strategic Research Programmes in Diabetes and Stem Cells and Regenerative Medicine (V.M.L.), the Robert Bosch Stiftung, Stuttgart, Germany (K.K.) and the National Research Foundation of South Africa and the Medical Research Council of South Africa (C.D.).
Conflicts of Interest:
V.M.L is a co-founder and shareholder of HepaPredict AB; consultancy work for Enginzyme AB. R.F.T. has consulted for Quinn Emanuel and Ethismos Research Inc and has received an annual honorarium for being an associate editor for Clinical Pharmacology and Therapeutics. All other authors declared no competing interests for this work.
References
- (1).Remmer H, Schoene B & Fleischmann RA Induction of the unspecific microsomal hydroxylase in the human liver. Drug Metab Dispos 1, 224–30 (1973). [PubMed] [Google Scholar]
- (2).Miles JS et al. A novel human cytochrome P450 gene (P450IIB): chromosomal localization and evidence for alternative splicing. Nucleic Acids Res 16, 5783–95 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Santisteban I, Povey S, Shephard EA & Phillips IR The major phenobarbital-inducible cytochrome P-450 gene subfamily (P450IIB) mapped to the long arm of human chromosome 19. Ann Hum Genet 52, 129–35 (1988). [DOI] [PubMed] [Google Scholar]
- (4).Nelson DR et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42 (1996). [DOI] [PubMed] [Google Scholar]
- (5).Hoffman SM, Fernandez-Salguero P, Gonzalez FJ & Mohrenweiser HW Organization and evolution of the cytochrome P450 CYP2A-2B-2F subfamily gene cluster on human chromosome 19. J Mol Evol 41, 894–900 (1995). [DOI] [PubMed] [Google Scholar]
- (6).Genotype-Tissue Expression (GTEx) Project <https://gtexportal.org/home/>.
- (7).Shimada T, Yamazaki H, Mimura M, Inui Y & Guengerich FP Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270, 414–23 (1994). [PubMed] [Google Scholar]
- (8).Gonzalez FJ, Crespi CL, Czerwinski M & Gelboin HV Analysis of human cytochrome P450 catalytic activities and expression. Tohoku J Exp Med 168, 67–72 (1992). [DOI] [PubMed] [Google Scholar]
- (9).Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH & Schwab M Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics 8, 743–59 (2007). [DOI] [PubMed] [Google Scholar]
- (10).Zanger UM & Klein K Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet 4, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lang T et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11, 399–415 (2001). [DOI] [PubMed] [Google Scholar]
- (12).Ariyoshi N, Miyazaki M, Toide K, Sawamura Y & Kamataki T A single nucleotide polymorphism of CYP2b6 found in Japanese enhances catalytic activity by autoactivation. Biochem Biophys Res Commun 281, 1256–60 (2001). [DOI] [PubMed] [Google Scholar]
- (13).Kirchheiner J et al. Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics 13, 619–26 (2003). [DOI] [PubMed] [Google Scholar]
- (14).Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA & Desta Z The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306, 287–300 (2003). [DOI] [PubMed] [Google Scholar]
- (15).Haas DW et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 18, 2391–400 (2004). [PubMed] [Google Scholar]
- (16).Tsuchiya K et al. Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun 319, 1322–6 (2004). [DOI] [PubMed] [Google Scholar]
- (17).Desta Z et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8, 547–58 (2007). [DOI] [PubMed] [Google Scholar]
- (18).Hofmann MH et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther 325, 284–92 (2008). [DOI] [PubMed] [Google Scholar]
- (19).Telenti A & Zanger UM Pharmacogenetics of anti-HIV drugs. Annu Rev Pharmacol Toxicol 48, 227–56 (2008). [DOI] [PubMed] [Google Scholar]
- (20).Holzinger ER et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genomics 22, 858–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lind L et al. Genetic and methylation variation in the CYP2B6 gene is related to circulating p,p’-dde levels in a population-based sample. Environ Int 98, 212–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Klein K et al. A New Panel-Based Next-Generation Sequencing Method for ADME Genes Reveals Novel Associations of Common and Rare Variants With Expression in a Human Liver Cohort. Front Genet 10, 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ingelman-Sundberg M, Daly AK, Oscarson M & Nebert DW Human cytochrome P450 (CYP) genes: recommendations for the nomenclature of alleles. Pharmacogenetics 10, 91–3 (2000). [DOI] [PubMed] [Google Scholar]
- (24).Gaedigk A et al. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharm Ther 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gaedigk A, Whirl-Carrillo M, Pratt VM, Miller NA & Klein TE PharmVar and the Landscape of Pharmacogenetic Resources. Clin Pharmacol Ther 107, 43–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ekins S, VandenBranden M, Ring BJ & Wrighton SA Examination of purported probes of human CYP2B6. Pharmacogenetics 7, 165–79 (1997). [DOI] [PubMed] [Google Scholar]
- (27).Chang TK, Weber GF, Crespi CL & Waxman DJ Differential activation of cyclophosphamide and ifosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res 53, 5629–37 (1993). [PubMed] [Google Scholar]
- (28).Heyn H, White RB & Stevens JC Catalytic role of cytochrome P4502B6 in the N-demethylation of S-mephenytoin. Drug Metab Dispos 24, 948–54 (1996). [PubMed] [Google Scholar]
- (29).Rae JM, Soukhova NV, Flockhart DA & Desta Z Triethylenethiophosphoramide is a specific inhibitor of cytochrome P450 2B6: implications for cyclophosphamide metabolism. Drug Metab Dispos 30, 525–30 (2002). [DOI] [PubMed] [Google Scholar]
- (30).Faucette SR et al. Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos 28, 1222–30 (2000). [PubMed] [Google Scholar]
- (31).Hesse LM et al. CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos 28, 1176–83 (2000). [PubMed] [Google Scholar]
- (32).Ayuso P, Neary M, Chiong J & Owen A Meta-analysis of the effect of CYP2B6, CYP2A6, UGT2B7 and CAR polymorphisms on efavirenz plasma concentrations. J Antimicrob Chemother 74, 3281–90 (2019). [DOI] [PubMed] [Google Scholar]
- (33).Cheng L et al. Meta-analysis of the associations of CYP2B6–516G>T polymorphisms with efavirenz-induced central nervous system side effects and virological outcome in HIV-infected adults. Pharmacogenomics J 20, 246–59 (2020). [DOI] [PubMed] [Google Scholar]
- (34).Yimer G et al. High plasma efavirenz level and CYP2B6*6 are associated with efavirenz-based HAART-induced liver injury in the treatment of naive HIV patients from Ethiopia: a prospective cohort study. Pharmacogenomics J 12, 499–506 (2012). [DOI] [PubMed] [Google Scholar]
- (35).Wyen C et al. Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J Antimicrob Chemother 66, 2092–8 (2011). [DOI] [PubMed] [Google Scholar]
- (36).Desta Z et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin Pharmacol Ther 106, 726–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang H & Tompkins LM CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9, 598–610 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Rotger M et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics 15, 1–5 (2005). [DOI] [PubMed] [Google Scholar]
- (39).Kharasch ED, Regina KJ, Blood J & Friedel C Methadone Pharmacogenetics: CYP2B6 Polymorphisms Determine Plasma Concentrations, Clearance, and Metabolism. Anesthesiology 123, 1142–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Benowitz NL, Zhu AZ, Tyndale RF, Dempsey D & Jacob P 3rd. Influence of CYP2B6 genetic variants on plasma and urine concentrations of bupropion and metabolites at steady state. Pharmacogenet Genomics 23, 135–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Asimus S & Ashton M Artemisinin--a possible CYP2B6 probe substrate? Biopharm Drug Dispos 30, 265–75 (2009). [DOI] [PubMed] [Google Scholar]
- (42).Li Y et al. CYP2B6*6 allele and age substantially reduce steady-state ketamine clearance in chronic pain patients: impact on adverse effects. Br J Clin Pharmacol 80, 276–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ng E et al. Genome-wide association study of plasma levels of polychlorinated biphenyls disclose an association with the CYP2B6 gene in a population-based sample. Environ Res 140, 95–101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Li H, Ferguson SS & Wang H Synergistically enhanced CYP2B6 inducibility between a polymorphic mutation in CYP2B6 promoter and pregnane X receptor activation. Mol Pharmacol 78, 704–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ngaimisi E et al. Effect of rifampicin and CYP2B6 genotype on long-term efavirenz autoinduction and plasma exposure in HIV patients with or without tuberculosis. Clin Pharmacol Ther 90, 406–13 (2011). [DOI] [PubMed] [Google Scholar]
- (46).Chung JY et al. Effects of pregnane X receptor (NR1I2) and CYP2B6 genetic polymorphisms on the induction of bupropion hydroxylation by rifampin. Drug Metab Dispos 39, 92–7 (2011). [DOI] [PubMed] [Google Scholar]
- (47).Metzger IF, Dave N, Kreutz Y, Lu JBL, Galinsky RE & Desta Z CYP2B6 Genotype-Dependent Inhibition of CYP1A2 and Induction of CYP2A6 by the Antiretroviral Drug Efavirenz in Healthy Volunteers. Clin Transl Sci 12, 657–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Maganda BA, Minzi OM, Ngaimisi E, Kamuhabwa AA & Aklillu E CYP2B6*6 genotype and high efavirenz plasma concentration but not nevirapine are associated with low lumefantrine plasma exposure and poor treatment response in HIV-malaria-coinfected patients. Pharmacogenomics J 16, 88–95 (2016). [DOI] [PubMed] [Google Scholar]
- (49).Neary M et al. The Effect of Gene Variants on Levonorgestrel Pharmacokinetics When Combined With Antiretroviral Therapy Containing Efavirenz or Nevirapine. Clin Pharmacol Ther 102, 529–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Desta Z et al. Inhibition of Cytochrome P450 2B6 Activity by Voriconazole Profiled Using Efavirenz Disposition in Healthy Volunteers. Antimicrob Agents Chemother 60, 6813–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Richter T et al. Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Ther 308, 189–97 (2004). [DOI] [PubMed] [Google Scholar]
- (52).Turpeinen M, Tolonen A, Uusitalo J, Jalonen J, Pelkonen O & Laine K Effect of clopidogrel and ticlopidine on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Clin Pharmacol Ther 77, 553–9 (2005). [DOI] [PubMed] [Google Scholar]
- (53).Fan L et al. Induction of cytochrome P450 2B6 activity by the herbal medicine baicalin as measured by bupropion hydroxylation. Eur J Clin Pharmacol 65, 403–9 (2009). [DOI] [PubMed] [Google Scholar]
- (54).Thomford NE et al. Inhibition of CYP2B6 by Medicinal Plant Extracts: Implication for Use of Efavirenz and Nevirapine-Based Highly Active Anti-Retroviral Therapy (HAART) in Resource-Limited Settings. Molecules 21, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Klein M, Thomas M, Hofmann U, Seehofer D, Damm G & Zanger UM A systematic comparison of the impact of inflammatory signaling on absorption, distribution, metabolism, and excretion gene expression and activity in primary human hepatocytes and HepaRG cells. Drug Metab Dispos 43, 273–83 (2015). [DOI] [PubMed] [Google Scholar]
- (56).Gravel S, Chiasson JL, Turgeon J, Grangeon A & Michaud V Modulation of CYP450 Activities in Patients With Type 2 Diabetes. Clin Pharmacol Ther 106, 1280–9 (2019). [DOI] [PubMed] [Google Scholar]
- (57).Drug Interactions Flockhart Table™ <https://drug-interactions.medicine.iu.edu/Main-Table.aspx >.
- (58).Pearce RE et al. Developmental Expression of CYP2B6: A Comprehensive Analysis of mRNA Expression, Protein Content and Bupropion Hydroxylase Activity and the Impact of Genetic Variation. Drug Metab Dispos 44, 948–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Croom EL, Stevens JC, Hines RN, Wallace AD & Hodgson E Human hepatic CYP2B6 developmental expression: the impact of age and genotype. Biochem Pharmacol 78, 184–90 (2009). [DOI] [PubMed] [Google Scholar]
- (60).Saitoh A et al. Efavirenz pharmacokinetics in HIV-1-infected children are associated with CYP2B6-G516T polymorphism. J Acquir Immune Defic Syndr 45, 280–5 (2007). [DOI] [PubMed] [Google Scholar]
- (61).Pond SM, Kreek MJ, Tong TG, Raghunath J & Benowitz NL Altered methadone pharmacokinetics in methadone-maintained pregnant women. J Pharmacol Exp Ther 233, 1–6 (1985). [PubMed] [Google Scholar]
- (62).Olagunju A et al. Pharmacogenetics of pregnancy-induced changes in efavirenz pharmacokinetics. Clin Pharmacol Ther 97, 298–306 (2015). [DOI] [PubMed] [Google Scholar]
- (63).Fokina VM et al. Pharmacokinetics of Bupropion and Its Pharmacologically Active Metabolites in Pregnancy. Drug Metab Dispos 44, 1832–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Dickmann LJ & Isoherranen N Quantitative prediction of CYP2B6 induction by estradiol during pregnancy: potential explanation for increased methadone clearance during pregnancy. Drug Metab Dispos 41, 270–4 (2013). [DOI] [PubMed] [Google Scholar]
- (65).Chiang YC et al. Reduced dosing and liability in methadone maintenance treatment by targeting oestrogen signal for morphine addiction. J Cell Mol Med 21, 3552–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Lamba V et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther 307, 906–22 (2003). [DOI] [PubMed] [Google Scholar]
- (67).Al Koudsi N & Tyndale RF Hepatic CYP2B6 is altered by genetic, physiologic, and environmental factors but plays little role in nicotine metabolism. Xenobiotica 40, 381–92 (2010). [DOI] [PubMed] [Google Scholar]
- (68).Choong E et al. Sex difference in formation of propofol metabolites: a replication study. Basic Clin Pharmacol Toxicol 113, 126–31 (2013). [DOI] [PubMed] [Google Scholar]
- (69).Ilic K et al. The influence of sex, ethnicity, and CYP2B6 genotype on bupropion metabolism as an index of hepatic CYP2B6 activity in humans. Drug Metab Dispos 41, 575–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Whirl-Carrillo M et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).PharmGKB CYP2B6 gene page <https://www.pharmgkb.org/gene/PA123>.
- (72).Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM & Caudle KE The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin Pharmacol Ther 107, 171–5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).CPIC Guidelines <https://cpicpgx.org/guidelines/>.
- (74).CPIC Gene/drug pairs <https://cpicpgx.org/genes-drugs/>.
- (75).Futatsugawa Y, Kubota T, Ishiguro A, Suzuki H, Ishikawa H & Iga T PCR-based haplotype determination to distinguish CYP2B6*1/*7 and *5/*6. Clin Chem 50, 1472–3 (2004). [DOI] [PubMed] [Google Scholar]
- (76).Rotger M et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther 81, 557–66 (2007). [DOI] [PubMed] [Google Scholar]
- (77).PharmGKB Gene-specific Information Tables for CYP2B6 <https://www.pharmgkb.org/page/cyp2b6RefMaterials>.
- (78).Watanabe T et al. Functional characterization of 40 CYP2B6 allelic variants by assessing efavirenz 8-hydroxylation. Biochem Pharmacol 156, 420–30 (2018). [DOI] [PubMed] [Google Scholar]
- (79).Watanabe T et al. Functional characterization of 26 CYP2B6 allelic variants (CYP2B6.2-CYP2B6.28, except CYP2B6.22). Pharmacogenet Genomics 20, 459–62 (2010). [DOI] [PubMed] [Google Scholar]
- (80).Ariyoshi N et al. Q172H replacement overcomes effects on the metabolism of cyclophosphamide and efavirenz caused by CYP2B6 variant with Arg262. Drug Metab Dispos 39, 2045–8 (2011). [DOI] [PubMed] [Google Scholar]
- (81).Botton MR et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther Epub June 29 (2020). doi: 10.1002/cpt.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Kalman LV et al. Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther 99, 172–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Pratt VM et al. Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J Mol Diagn 20, 269–76 (2018). [DOI] [PubMed] [Google Scholar]
- (84).Pratt VM et al. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn 21, 746–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Sueyoshi T & Negishi M Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol 41, 123–43 (2001). [DOI] [PubMed] [Google Scholar]
- (86).Zukunft J et al. A natural CYP2B6 TATA box polymorphism (-82T--> C) leading to enhanced transcription and relocation of the transcriptional start site. Mol Pharmacol 67, 1772–82 (2005). [DOI] [PubMed] [Google Scholar]
- (87).Burgess KS et al. Variants in the CYP2B6 3’UTR Alter In Vitro and In Vivo CYP2B6 Activity: Potential Role of MicroRNAs. Clin Pharmacol Ther 104, 130–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).PharmVar CYP2B6 gene page <https://www.pharmvar.org/gene/CYP2B6>.
- (89).Huddart R et al. Standardized Biogeographic Grouping System for Annotating Populations in Pharmacogenetic Research. Clin Pharmacol Ther 105, 1256–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Colic A, Alessandrini M & Pepper MS Pharmacogenetics of CYP2B6, CYP2A6 and UGT2B7 in HIV treatment in African populations: focus on efavirenz and nevirapine. Drug Metab Rev 47, 111–23 (2015). [DOI] [PubMed] [Google Scholar]
- (91).Klein K et al. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genomics 15, 861–73 (2005). [DOI] [PubMed] [Google Scholar]
- (92).PharmVar Allele Designation and Evidence Level document <www.pharmvar.org/documents/Allele_Designation_and_Evidence_Level_Criteria_V2.1.pdf >.
- (93).PharmVar CYP2B6 Gene Expert Panel <https://www.pharmvar.org/expert-panels>.
- (94).PharmVar Legacy P450 Nomenclature site content <https://www.pharmvar.org/htdocs/archive/index_original.htm>; previoulsy at cypalleles.ki/se.
- (95).Locus Reference Genomic (LRG) Records <https://www.lrg-sequence.org/>.
- (96).PharmVar Standards <https://www.pharmvar.org/genes>.
- (97).Nofziger C et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther 107, 154–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Evans J, Swart M, Soko N, Wonkam A, Huzair F & Dandara C A Global Health Diagnostic for Personalized Medicine in Resource-Constrained World Settings: A Simple PCR-RFLP Method for Genotyping CYP2B6 g.15582C>T and Science and Policy Relevance for Optimal Use of Antiretroviral Drug Efavirenz. OMICS 19, 332–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Dickinson L et al. Pharmacokinetic and Pharmacodynamic Comparison of Once-Daily Efavirenz (400 mg vs. 600 mg) in Treatment-Naive HIV-Infected Patients: Results of the ENCORE1 Study. Clin Pharmacol Ther 98, 406–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Wang PF, Neiner A & Kharasch ED Stereoselective Bupropion Hydroxylation by Cytochrome P450 CYP2B6 and Cytochrome P450 Oxidoreductase Genetic Variants. Drug Metab Dispos 48, 438–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Lang T et al. Multiple novel nonsynonymous CYP2B6 gene polymorphisms in Caucasians: demonstration of phenotypic null alleles. J Pharmacol Exp Ther 311, 34–43 (2004). [DOI] [PubMed] [Google Scholar]
- (102).Honda M et al. Functional characterization of CYP2B6 allelic variants in demethylation of antimalarial artemether. Drug Metab Dispos 39, 1860–5 (2011). [DOI] [PubMed] [Google Scholar]
- (103).Gaedigk A et al. The Evolution of PharmVar. Clin Pharmacol Ther 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Pratt VM et al. Characterization of 137 Genomic DNA Reference Materials for 28 Pharmacogenetic Genes: A GeT-RM Collaborative Project. J Mol Diagn 18, 109–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Twist GP et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genom Med 1, 15007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Lee SB et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med 21, 361–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Numanagic I et al. Allelic decomposition and exact genotyping of highly polymorphic and structurally variant genes. Nat Commun 9, 828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Lee AM et al. CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry 62, 635–41 (2007). [DOI] [PubMed] [Google Scholar]
- (109).Rohrich CR et al. CYP2B6*6 and CYP2B6*18 Predict Long-Term Efavirenz Exposure Measured in Hair Samples in HIV-Positive South African Women. AIDS Res Hum Retroviruses 32, 529–38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Machiela MJ & Chanock SJ LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Boone EC et al. Long-Distance Phasing of a Tentative “Enhancer” Single-Nucleotide Polymorphism With CYP2D6 Star Allele Definitions. Front Pharmacol 11, 486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).PharmGKB CYP2B6 Drug Label Annotations <https://www.pharmgkb.org/gene/PA123/labelAnnotation>.
- (113).FDA Pharmacogenomic Biomarkers in Drug Labeling <https://www.fda.gov/drugs/science-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling >.
- (114).NCBI. Reference Sequences database <https://www.ncbi.nlm.nih.gov/refseq/>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


