Abstract
Introduction:
Dogs with naturally occurring diabetes mellitus represent a potential model for human type 1 diabetes, yet significant knowledge voids exist in terms of the pathogenic mechanisms underlying the canine disorder. Untargeted metabolomic studies from a limited number of diabetic dogs identified similarities to humans with the disease.
Objective:
To expand and validate earlier metabolomic studies, identify metabolites that differ consistently between diabetic and healthy dogs, and address whether certain metabolites might serve as disease biomarkers.
Methods:
Untargeted metabolomic analysis via liquid chromatography-mass spectrometry was performed on serum from diabetic (n=15) and control (n=15) dogs. Results were combined with those of our previously published studies using identical methods (12 diabetic and 12 control dogs) to identify metabolites consistently different between the groups in all 54 dogs. Thirty-two candidate biomarkers were quantified using targeted metabolomics. Biomarker concentrations were compared between the groups using multiple linear regression (corrected P<0.0051 considered significant).
Results:
Untargeted metabolomics identified multiple persistent differences in serum metabolites in diabetic dogs compared with previous studies. Targeted metabolomics showed increases in gamma amino butyric acid (GABA), valine, leucine, isoleucine, citramalate, and 2-hydroxyisobutyric acid in diabetic versus control dogs while indoxyl sulfate, N-acetyl-L-aspartic acid, kynurenine, anthranilic acid, tyrosine, glutamine, and tauroursodeoxycholic acid were decreased.
Conclusion:
Several of these findings parallel metabolomic studies in both human diabetes and other animal models of this disease. Given recent studies on the role of GABA and branched chain amino acids in human diabetes, the increase in serum concentrations in canine diabetes warrants further study of these metabolites as potential biomarkers, and to identify similarity in mechanisms underlying this disease in humans and dogs.
Keywords: animal model, biomarkers, canine, diabetes, dog(s), metabolites, metabolomics
1. Introduction
Metabolomics is an emerging tool for novel biomarker discovery and investigation of disease pathology (Zhang et al., 2015). Novel biomarkers with the potential to improve early disease detection have been discovered in a variety of human diseases. Similarly, in veterinary medicine, metabolomics is a newly expanding field, and investigations into differences among dog breeds, response to nutritional changes, and evaluation for novel biomarkers in a variety of canine diseases have recently been reported (Carlos et al., 2020).
Metabolomic perturbations found in human type 1 diabetes (T1D) patients with poor glycemic control have included increases in branched chain amino acids (BCAAs), other amino acids (e.g., lysine, proline, serine, methionine), and anti-inflammatory short chain fatty acids (SCFAs), while metabolites involved in glycolysis were decreased (Dutta et al., 2016). Additionally, changes in multiple metabolites have been identified in young children that later progress to T1D, such as decreases in amino acids (including tryptophan, glutamic acid, and aspartic acid) (Lamichhane et al., 2019) and lipids (including sphingomyelins, triacylglycerols, and phosphatidylcholines) (Lamichhane et al., 2018). In human type 2 diabetes, a variety of metabolites have been associated with existing disease or disease prediction, most notably elevated BCAAs, elevated tyrosine and phenylalanine, and decreased glycine and glutamine (Guasch-Ferré et al., 2016).
Canine diabetes is similar to human T1D in certain ways, including a lifelong requirement for insulin (in most cases) along with marked β-cell loss at the time of diagnosis (Nelson and Reusch, 2014). However, many components of diabetes pathogenesis and other potential similarities to the human disease in dogs remain unknown, which may be due in part to the lack of ability to detect and study dogs at an early stage in disease (O'Kell et al., 2017b). Hence, our overall goal was to identify novel biomarkers that differ between diabetic and healthy dogs that may be candidates for future longitudinal studies of disease prediction in dogs. Our previous untargeted metabolomics studies in both fasted and unfasted cohorts identified altered profiles in sera from diabetic dogs compared to healthy control dogs, with some similarities to those found in the human disease (O'Kell et al., 2017a, O'Kell et al., 2019). Herein, we report an untargeted metabolomics effort to first, validate our preliminary findings (O'Kell et al., 2017a, O'Kell et al., 2019) in an independent cohort and, thereby, identify metabolites that differ between diabetic and healthy control dogs. Candidate metabolite biomarkers were then selected and quantitated by targeted metabolomics.
2. Methods
2.1. Study Enrollment
The study was approved by the University of Florida (UF) Institutional Animal Care and Use Committee (IACUC) and Veterinary Hospital Research Review Committee, and owners provided written informed consent prior to enrollment. Dogs were recruited from the client-owned population of the UF Small Animal Hospital. A diagnosis of diabetes was made prior to enrollment and was based on clinical signs (polyuria, polydipsia) concurrent with hyperglycemia and glucosuria. Diabetic dogs (n=15) were included if they were >1 year of age, had a body weight ≥5 kg, and, if female, were spayed prior to the diagnosis of diabetes to exclude dogs with insulin resistance diabetes caused by progesterone and growth hormone exposure that have pathogenic differences to the more typical form of adult onset diabetes in dogs(Gilor et al., 2016). Control dogs (n=15) were included if the history and physical exam excluded active disease and if they were >1 year of age, had a body weight ≥5 kg, received no medications other than routine parasite preventatives, and, if female, were spayed. Control dogs were breed and/or size matched to diabetic dogs whenever possible, and an equal number of dogs that were fasted (n=9) and non-fasted (n=6) were included in each group.
2.2. Sample Collection
Blood samples were collected routinely from the jugular vein using a needle and syringe into a red top tube containing clot activator (BD Serum Blood Collection Tubes). Serum was separated after 30 minutes and frozen immediately at −80°C for 3-24 months prior to the untargeted metabolomic analysis described in 2.3. The time of blood collection in relation to the most recent meal was recorded.
2.3. Metabolite Extraction and Untargeted Metabolomics Analysis
Metabolite extraction and analysis was performed as previously described (O'Kell et al., 2017a, O'Kell et al., 2019) using a Thermo Q-Exactive Orbitrap mass spectrometer with Dionex UHPLC and autosampler (ThermoScientific San Jose, CA). Metabolite identification was performed as previously described and represented level 1 identification (O'Kell et al., 2017a, O'Kell et al., 2019).
2.4. Statistical Analysis for Untargeted Metabolomics
Mann-Whitney U Tests were used to compare median age and body weight between the diabetes and control groups, and a Fisher’s Exact Test was used to compare sex distribution. For biomarker identification from untargeted metabolomics data, statistical analysis was performed using Metaboanalyst. Values not present in 80% of the data were removed, and missing values were imputed using k-nearest neighbor. Data was interquartile range filtered, sum normalized, log2 transformed, and autoscaled. T-tests (adjusted for multiple comparisons using false discovery rate) were used to compare metabolites between diabetic and control groups. The metabolite distributions were normally distributed following transformation based on evaluation of all metabolite expression. P < 0.05 was considered significant.
2.5. Targeted Metabolomics Biomarker Selection
Results from untargeted metabolomics analysis for the present study (n=30) and the combined prior untargeted metabolomics studies (n=24) (O'Kell et al., 2017a, O'Kell et al., 2019) were evaluated to identify candidate metabolite biomarkers. For both present and prior studies, metabolites that were significantly different (p<0.05) between diabetic and control groups based on a t-test (as described in section 2.4) were identified. Using Metaboanalyst (http://www.metaboanalyst.ca), classic univariate receiver operating characteristic (ROC) curves were generated (Xia et al., 2013), and metabolites with an area under the ROC curve (AUC) >0.8 were also identified. Additionally, biomarker meta-analysis of data from both studies was performed using a P-value combining method (Stouffer’s method) in Metaboanalyst. Metabolites that were identified as consistently associated with diabetes from the t-tests, ROC curve/AUC analysis, and biomarker meta-analysis were selected as candidate biomarkers to move forward to targeted metabolomics (Figure 1). Additional metabolites in the same metabolic pathway(s) as candidate metabolites were also included if assay methods were readily available.
Fig.1.
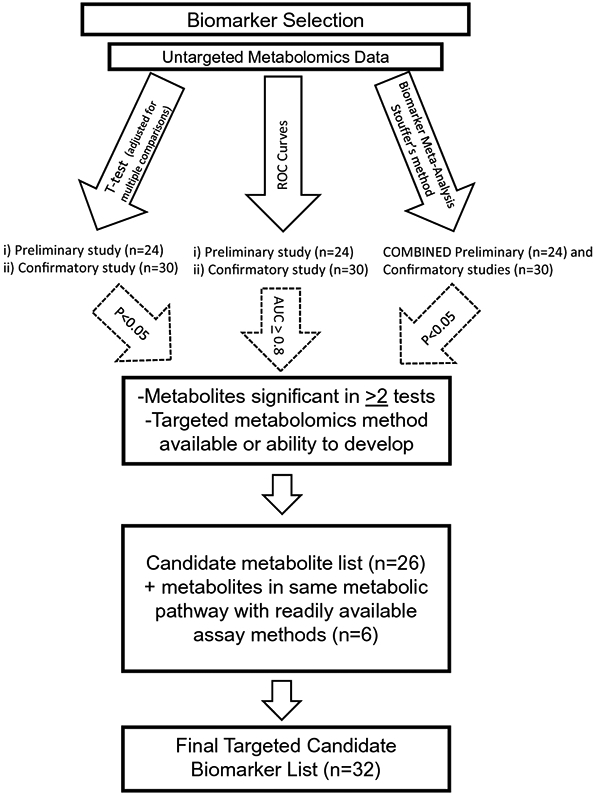
Flow chart for biomarker selection for targeted metabolomics analysis
2.6. Samples for Targeted Metabolomics
Fifty-four canine serum samples were used for targeted metabolomics, including those from the current study described above (n=30), as well as banked samples from all dogs enrolled in our previously reported untargeted metabolomics studies (n=24) (O'Kell et al., 2017a, O'Kell et al., 2019) (Figure 2). All serum samples were stored at −80°C. Samples from the previously published studies and current study were stored for 33-54 months and 11-51 months, respectively, prior to analysis.
Fig. 2.
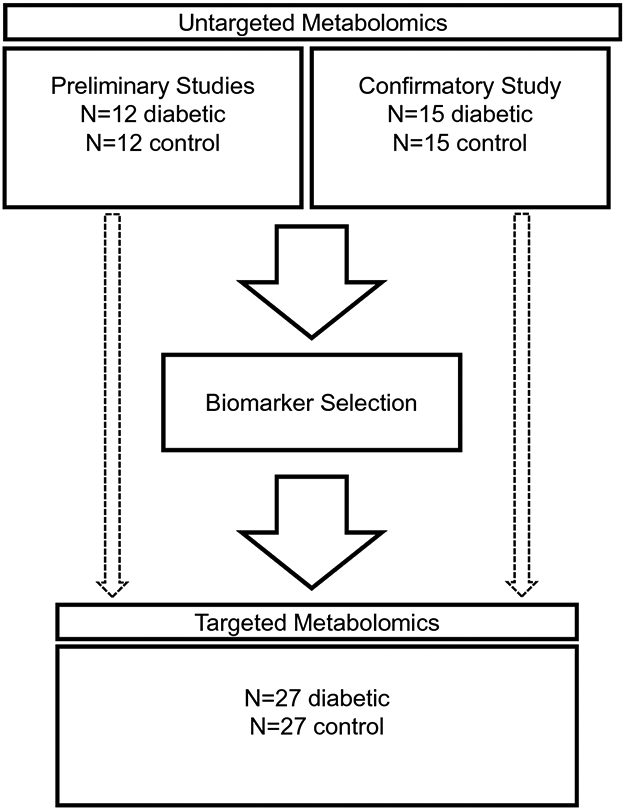
Study organizational flow chart
2.7. Targeted Metabolomics Metabolite Extraction and Analysis
Targeted metabolomics analysis was performed for 32 metabolites using liquid chromatography-high resolution mass spectrometry (LC-HRMS). Chromatographic separation of 27 metabolites was performed by reversed-phase (RP), and 5 by hydrophilic interaction liquid chromatography (HILIC). For each metabolite, 25 μL sample was spiked with 50 ng internal standard prior to extraction. Sample extraction was done by protein precipitation using 200 μL of 8:1:1 acetonitrile/methanol/acetone. Further protein precipitation was allowed at 4°C for 30 minutes. The samples were centrifuged at 20,000 x g at 4°C for 10 minutes. Supernatants were dried down under nitrogen gas at 30°C. Samples for RP-LC-HRMS were reconstituted with 25 μL of 0.1% formic acid in water. Samples for HILIC-LC-HRMS were reconstituted with 25 μL 90:10 acetonitrile/10mM ammonium acetate.
For both RP and HILIC, 9-point calibration curves (ranging from 0.5 to 16000 ng/mL) were prepared from a serially diluted standards mixture in appropriate solvent supplemented with 50 ng of corresponding internal standards (Online Resource Tables 1 and 6). A negative control and four positive controls were also included in the analysis at 0, 5, 500, 2000, and 8000 ng/mL spiked with 50 ng internal standard. All calibrators and controls were prepared at 25 μL in triplicate. Linearity was established for each compound following FDA Bioanalytical Method Validation. The lower limit of quantitation (LLOQ) was determined at the lowest concentration of analyte that was quantifiable with precision and accuracy of <20%.
Both RP and HILIC quantitation were performed on a Thermo Q-Exactive Orbitrap MS with Dionex UHPLC and autosampler. All samples, calibrators, and controls were analyzed in positive and negative heated electrospray ionization with a mass resolution of 35,000 at m/z 200 as separate injections. RP separation was achieved on an ACE 18-pfp 100 x 2.1 mm, 2 μm column with mobile phase A as 0.1% formic acid in water and mobile phase B as acetonitrile. The flow rate was 350 μL/min with a column temperature of 25°C. Injection volume was 2 μL for positive and 4 μL for negative polarity. HILIC separation was achieved on a Waters Acquity UPLC BEH Amide 150 x 2.1 mm, 1.7 μm column with mobile phase A as 20 mM ammonium formate pH 4 and mobile phase B as 0.1% acetic acid in acetonitrile. The flow rate was 300 μL/min with a column temperature of 25°C. Injection volume was 2 μL for positive and 4 μL for negative polarity. Peak area integration of analyte and corresponding internal standard was performed using Xcalibur Quan Browser (version 3.0.63). Calibration curves were constructed using the peak area ratio of analyte to internal standard against nominal concentrations.
2.8. Statistical Analysis for Targeted Metabolomics
For targeted metabolomics analysis, metabolite concentration values below the lower limit of quantification (LLOQ) were set at the LLOQ for statistical analysis. Scatter plots were used to evaluate for outliers. Outliers were checked for analytical issues (by assessing laboratory records) or patient factors (e.g., concurrent disease, lipemia, sample hemolysis), but neither analytic nor patient confounders were consistently present in outliers. Two extreme outliers that were above the upper limit of quantification even after 10x dilution were removed from the results of a single metabolite (2-hydroxyisobutyric acid). Metabolite concentrations were log transformed, and multiple linear regression analysis of metabolite concentration was performed for each metabolite to analyze the effect of group (diabetes or control) on metabolite concentration; age (continuous) was accounted for by including it in the model. A Holm-Sidak correction was performed to adjust for multiple comparisons, and a P-value of <0.0051 was considered significant for diabetes. We report the confidence intervals as estimated percentage increase or decrease in the diabetic group based on the original untransformed scale. The assumptions for linear regression were met by evaluating for independence between age and group using the variance inflation factor, by assessing scatter plots for linearity of age versus the log of the metabolites, by evaluating autocorrelation of residuals with the Durbin Watson test, and with the D’Agostino Pearson test for normality of residuals. The assumptions for linear regression were met or had minor deviations for which linear regression is generally robust to. Analysis was performed using GraphPad Prism 8.0 and 9.1 and Microsoft Office Excel 2019. Additional analysis for ROC curve generation was performed using Metaboanalyst to calculate the AUC.
From the targeted metabolomics log transformed data, Linear Discriminant analysis machine learning was used to identify a biomarker panel for discriminating between diabetic and healthy control groups via forward feature selection incorporating repeated cross-validation to measure uncertainty. Statistical analysis indicated multiple biomarkers to be associated with diabetes, and machine learning is used to complement these results evaluating the use of these biomarkers to predict disease in individuals, which implies potential utility in clinical applications. (Varga et al., 2020) To assess the selected panel, 2/3 of the dog samples (18 diabetic and 18 control) were randomly selected for inclusion in a test set for metabolite feature selection, while the remaining 1/3 of the dogs (9 control and 9 diabetic) were randomly selected to serve as a validation set to test the final model. Each metabolite of the training set was evaluated via a Linear Discriminant classifier with 5-fold cross-validation where accuracy was measured as AUC. For the machine learning we used a highly conservative selection criteria for our initial candidate biomarker set. The metabolites were then filtered to include those that were significant based on a non-parametric Wilcoxon Rank Sum test after a Bonferroni correction on the training set and also had an AUC significantly larger than 0.75.
This training set consisting of 8 metabolites was then evaluated via forward feature selection using a Linear Discriminant classifier again with 5-fold cross-validation where accuracy was measured as AUC. The cross-validation procedure was repeated 100 times to assess the variability in the model accuracy. After all features were processed for inclusion into the model, each metabolite was assessed via a one-sided t-test to evaluate if its addition to the model significantly increased model performance from the prior model. If one or more of the metabolite features were identified as significantly increasing the AUC, an ANOVA with Tukey’s post-hoc test was used to evaluate all features and identify which feature(s) were optimal. In the case of a tie, the process was split and both models were evaluated in the next step.
3. Results
3.1. Patient Characteristics
The diabetic group (n=15) consisted of five females (all spayed) and ten males (nine neutered) with a median age of 8.5 years (range, 5-13 years) and a median body weight of 10.8 kg (range, 5.4-57.7 kg) (Table 1). The median duration of diabetes was 10 months, with a range of 1 day to 2.5 years. All but two recently diagnosed diabetic dogs were receiving treatment with exogenous insulin at the time of sample collection. There were four mixed breed dogs, two Rottweilers, and one each of Pomeranian, Miniature Pinscher, Bichon Frise, Silky Terrier, Maltese, Cavalier King Charles Spaniel, Labrador Retriever, Miniature Australian Shepherd, and Miniature Schnauzer. The healthy control group (n=15) consisted of nine females (all spayed) and six males (all neutered), with a median age of 5.5 years (range, 3-10 years) and a median body weight of 7.5 kg (range, 5.1-45.9 kg). This group included six mixed breed dogs, two Rottweilers, and one each of Pomeranian, Miniature Pinscher, Silky Terrier, Cavalier King Charles Spaniel, Labrador Retriever, Miniature Australian Shepherd, and Miniature Schnauzer. The diabetic group was significantly older than the control group (P=0.0012), while body weight was comparable between the two groups (P=0.25).
Table 1.
Patient Characteristics – Confirmatory Study
| Group | Breed | Age (years) |
Weight (kg) |
Sexa | Fasted/Non- fasted |
Duration since diagnosis of diabetes (months) |
|---|---|---|---|---|---|---|
| Diabetic | Pomeranian | 7.75 | 9.6 | NM | Fasted | 0.5 |
| Miniature Pinscher | 8.75 | 5.9 | SF | Fasted | 30 | |
| Bichon Frise | 13 | 6.7 | NM | Fasted | 10 | |
| Mix | 8.5 | 32.8 | SF | Fasted | 0.03 | |
| Silky Terrier | 13 | 6.8 | NM | Fasted | 5 | |
| Mix | 6.5 | 10.8 | SF | Fasted | 1.5 | |
| Rottweiler | 8 | 41.6 | SF | Fasted | 10 | |
| Maltese | 11 | 5.4 | NM | Fasted | 8 | |
| Mix | 10.5 | 8.2 | NM | Fasted | 14 | |
| Cavalier King Charles Spaniel | 7.5 | 17.4 | NM | Non-fasted | 24 | |
| Labrador retriever | 12 | 43.9 | MI | Non-fasted | 7 | |
| Miniature Australian Shepherd | 8 | 19 | NM | Non-fasted | 12 | |
| Mix (Pug/beagle) | 11 | 16 | NM | Non-fasted | 16 | |
| Rottweiler | 5 | 57.7 | NM | Non-fasted | 16 | |
| Miniature Schnauzer | 7.5 | 6.9 | SF | Non-fasted | 6 | |
| Control | Pomeranian | 4 | 5.4 | SF | Fasted | N/A |
| Miniature Pinscher | 8 | 5.5 | SF | Fasted | N/A | |
| Bichon/Poodle | 8.5 | 5.3 | SF | Fasted | N/A | |
| Mix | 6.5 | 32.5 | SF | Fasted | N/A | |
| Silky Terrier | 10 | 5.1 | SF | Fasted | N/A | |
| Mix | 3 | 7.5 | SF | Fasted | N/A | |
| Rottweiler | 3 | 45.9 | NM | Fasted | N/A | |
| Mix | 3.5 | 5.4 | NM | Fasted | N/A | |
| Mix | 3.5 | 5.9 | SF | Fasted | N/A | |
| Cavalier King Charles Spaniel | 3.5 | 6 | SF | Non-fasted | N/A | |
| Labrador retriever | 4 | 32.2 | NM | Non-fasted | N/A | |
| Miniature Australian Shepherd | 7 | 17.7 | NM | Non-fasted | N/A | |
| Mix | 9 | 16.6 | NM | Non-fasted | N/A | |
| Rottweiler | 6.5 | 44.1 | NM | Non-fasted | N/A | |
| Miniature Schnauzer | 5.5 | 8.4 | SF | Non-fasted | N/A |
SF=spayed female; NM= neutered male; MI= male intac
3.2. Untargeted Metabolomics and Biomarker Selection
Untargeted metabolomics analysis identified 4,067 features detected from the positive ion mode and 5,647 from the negative. Of these features, 398 were known metabolites. One hundred and seven known metabolites (Online Resource Table 2) were significantly different (P<0.05) between the groups. Significantly different metabolites from the present study, as well as our previously reported preliminary studies (O'Kell et al., 2017a, O'Kell et al., 2019), were assessed for metabolites significantly different in both. ROC curve analysis of sensitivity and specificity for correctly classifying dogs as diabetic versus non-diabetic identified 85 and 67 metabolites with AUC ≥ 0.8 in the present and prior studies, respectively, indicating these metabolites to have excellent predictive capacity (Mandrekar, 2010) (Online Resource Table 3). Biomarker meta-analysis identified 90 candidate metabolite biomarkers (Online Resource Table 4). The results of each type of analysis were assessed for metabolites that were identified repeatedly (at least twice) among the different studies and methods of analysis, and from these, 26 metabolites were chosen for targeted metabolomics. Six additional metabolites in the same metabolic pathway(s) as candidate metabolites were also included if assay methods were readily available (Figure 2; Table 2).
Table 2:
Candidate Biomarkers for Targeted Metabolomic Analysis
| 2-hydroxyisobytyric acid |
| 5-hydroxymethyl-2-furaldehyde |
| 6-deoxy-hexose |
| Anthranilate |
| Aspartate |
| Citramalate |
| D-galacturonic acid/D-glucuronic acid/D-glucuronolactone |
| Erythritol |
| (fucose) |
| GABA |
| Glucosamine |
| Glycine |
| Indoxyl sulfate |
| (Kynurenic acid) |
| Kynurenine |
| LL-2,6-Diaminoheptanedioate |
| L-Glutamine |
| L-Histidine |
| L-Isoleucine |
| L-Proline |
| L-Tyrosine |
| (Leucine) |
| Mannosamine |
| Methyl-β-D-galactoside |
| N-acetyl-L-aspartic acid |
| Pyridoxal |
| (Serotonin) |
| (Tryptophan) |
| TUDCA |
| Uridine |
| Valine |
| (Xanthenuric acid) |
Metabolites in brackets represent metabolites that were not chosen based on data, but were within the same metabolic pathway as another chosen metabolite with a readily available analytic method.
3.3. Targeted Metabolomics
For the targeted metabolomics, serum samples from all 54 dogs enrolled in the current study and our published untargeted metabolomics studies (O'Kell et al., 2017a, O'Kell et al., 2019) were analyzed. The diabetic group consisted of 12 female (all spayed) and 15 males (all but one were neutered) with a median body weight of 10.8 kg (range, 5.4-57.7) and median age of 9.75 years (range, 1.3-14.25). The median duration of diabetes for all 54 dogs was 8 months, with a range of 1 day to 3 years. The control group consisted of 14 females (all spayed) and 13 males (all neutered) with a median body weight of 8.4 kg (range, 5.1-49.5) and a median age of 5.5 years (range, 2-11.75). As noted in the untargeted analysis above, only age was significantly different between the groups with the diabetic group being older (P<0.0001).
Of the 32 metabolites selected, 27 could be quantified by RP-LC-HRMS with 17 of those quantitated in positive polarity while 10 were quantitated in negative polarity. Seven metabolites including tryptophan, aspartate, glutamine, valine, 2-hydroxybutyric acid, tyrosine, and indoxyl sulfate were quantified from 10x dilution of the samples as they were all above the upper LOQ in the initial analysis of undiluted samples. There was significant ion suppression observed for the internal standard for aspartate that prevented quantitation and, thus, we could not rely on results in the canine samples. Results for xanthenuric acid, D-galacturonic acid/D-glucuronic acid/D-glucuronolactone, methyl-β-D-galactoside, and uridine showed concentrations that were below the LLOQ or not detectable for many samples, therefore no statistical analysis was performed. Five metabolites including erythritol, 6-deoxyhexose, glucosamine, mannosamine, and fucose were analyzed by HILIC-LC-HRMS. Erythritol, 6-deoxyhexose, and fucose could not be quantified accurately due to poor peak shape. In addition to poor peak shape, co-elution of 6-deoxyglucose and fucose also contributed to the high signal variability. Glucosamine and mannosamine could not be chromatographically resolved and, thus, are described together.
Of the 23 quantified metabolites for which statistical analysis was performed, the following were significantly different between dogs with diabetes and controls following multiple linear regression for group and controlled for age (P-value and 95% confidence interval of estimated percent increase/decrease in diabetic dogs): GABA (P<0.0001; increased 34-68%), valine (P<0.0001; increased 26-53%), indoxyl sulfate (P<0.0001; decreased 184-969%), 2-hydroxyisobutyric acid (P<0.0001; increased 49-80%), N-acetyl-L-aspartic acid (P=0.0015; decreased 13-48%), kynurenine (P=0.0018; decreased 45-162%), tauroursodeoxycholic acid (TUDCA) (P=0.0001; decreased 92-503%), anthranilic acid (P=0.0008; decreased 32-172%), isoleucine (P=0.001; increased 21-57%), leucine (P=0.0017; increased 15-51%), glutamine (P=0.0031; decreased 8-41%), tyrosine (P=0.0032; decreased 11-63%), tryptophan (P=0.0035; decreased 13-80%), and citramalate (P=0.005; increased 17-63%) (Figure 3). Linear regression models are available in Online Resource Table 5. The data is presented as a heat map in Online Resource Figure 1. Biomarker analysis identified AUC ≥ 0.8 for valine (0.90), 2-hydroxyisobutyric acid (0.90), GABA (0.88), N-acetyl-L-aspartic acid (0.88), indoxyl sulfate (0.85), L-glutamine (0.80), and kynurenine (0.80), anthranilic acid (0.8), and citramalate (0.8). Data for the 23 metabolites is summarized in Table 3.
Fig 3.
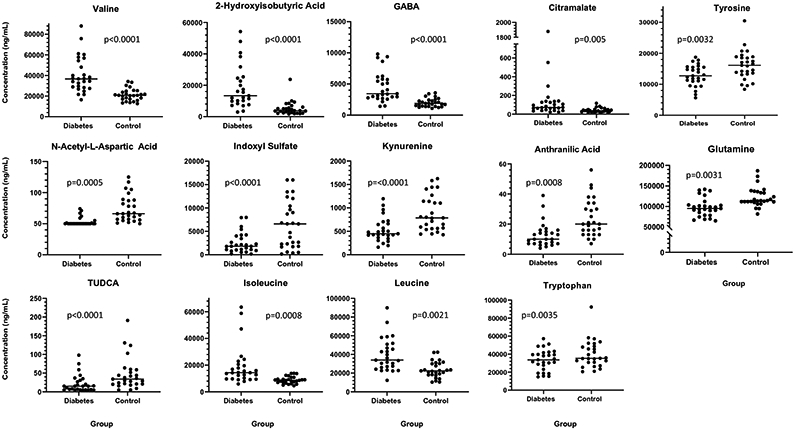
Scatter dot plots of metabolites significantly different between diabetes and control groups. Each circle represents an individual subject. The center line denotes the median. The lower limit of quantification for N-acetyl-L-aspartic acid and citramalate is 50 ng/mL and for TUDCA is 5 ng/mL. GABA=ɣ-aminobutyric acid; TUDCA = tauroursodeoxycholic acid
Table 3.
Targeted Metabolomics Results – Group Comparisons and Biomarker Analysis
| Diabetic Group | Control Group | |||||
|---|---|---|---|---|---|---|
| Metabolite | Median (ng/mL) |
Range (ng/mL) |
Median (ng/mL) |
Range (ng/mL) |
p-value± | AUC |
| Valine | 36672 | 16448-87972 | 20746 | 12648-34134 | <0.0001* | 0.90 |
| 2-hydroxyisobutyric acid | 13347 | 2953-54254 | 3856 | 1756-23773 | <0.0001* | 0.90 |
| GABA | 3441 | 1424-9801 | 1891 | 1089-3579 | <0.0001* | 0.88 |
| N-acetyl-L-aspartic acid | 50 | 50-74 | 66 | 50-125 | 0.0005* | 0.88 |
| Indoxyl sulfate | 1813 | 165-7975 | 6585 | 145-16000 | <0.0001* | 0.85 |
| Kynurenine | 448 | 160-1196 | 786 | 427-1625 | <0.0001* | 0.80 |
| Anthranilic acid | 10 | 4-39 | 20 | 7-56 | 0.0008* | 0.80 |
| Citramalate | 72 | 50-1877 | 50 | 50-116 | 0.005* | 0.80 |
| L-Glutamine | 95185 | 65313-141746 | 114128 | 81839-187058 | 0.0031* | 0.80 |
| Glucosamine/mannosamine | 28771 | 7879-71427 | 17489 | 6901-48921 | 0.012 | 0.76 |
| Tauroursodeoxycholic acid | 15 | 5-98 | 34 | 5-191 | <0.0001* | 0.75 |
| Tyrosine | 12730 | 5614-18737 | 16096 | 8369-30475 | 0.0032* | 0.72 |
| Isoleucine | 14296 | 5943-63445 | 8590 | 4603-13830 | 0.0008* | 0.71 |
| L-proline | 22402 | 9551-55733 | 32011 | 14221-52155 | 0.11 | 0.68 |
| Leucine | 33934 | 12285-89882 | 22438 | 10383-42380 | 0.0021* | 0.68 |
| 2,6-diaminoheptanedioate | 28 | 15-135 | 43 | 16-199 | 0.066 | 0.50 |
| 5-hydroxymethyl-2-furaldehyde | 7 | 5-16 | 6 | 5-14 | 0.45 | 0.62 |
| Glycine | 138 | 52-291 | 169 | 75-325 | 0.58 | 0.52 |
| Histidine | 556 | 326-1223 | 505 | 378-707 | 0.36 | 0.64 |
| Kynurenic acid | 671 | 500-5047 | 918 | 500-2812 | 0.16 | 0.57 |
| Pyridoxal | 51 | 50-393 | 73 | 53-151 | 0.74 | 0.61 |
| Serotonin | 1215 | 208-2222 | 1145 | 423-2058 | 0.62 | 0.5 |
| Tryptophan | 33636 | 14811-57219 | 35376 | 20522-92416 | 0.0035* | 0.62 |
p-value <0.0051 considered significant
AUC=area under the receiver operating characteristics curve
p-values were estimated from the regression model using log10 of the metabolites as the outcome
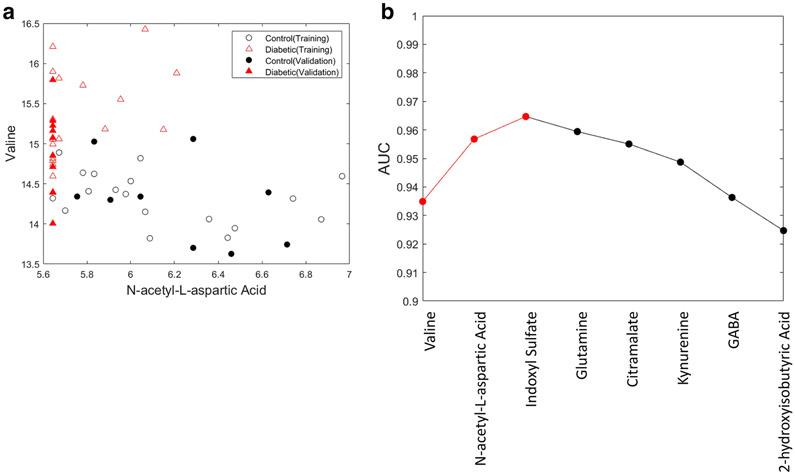
The machine learning training set included 8 metabolites: 1) valine, 2) N-acetyl-L-aspartic acid, 3) indoxyl sulfate, 4) L-glutamine, 5) citramalate, 6) kynurenine, 7) GABA, and 8) 2-hydroxyisobutyric acid (Table 4). Machine learning with forward feature selection identified the optimal Linear Discriminant classifier model as including three features (valine, n-acetyl-L-aspartic acid, and indoxyl sulfate) yielding a cross-validation AUC of 0.965 on the training data. The first iteration identified valine as the best single metabolite feature with an average AUC of 0.935. In the next iteration, either N-acetyl-L-aspartic acid or indoxyl sulfate, were the best second feature, AUC of 0.957 and 0.956, respectively. Figure 4A shows a scatter plot of the top two metabolites demonstrating excellent separation achieved by a clearly quantitative marker (valine) where the diabetic dogs have an elevated level of this metabolite, and a qualitative marker (N-acetyl-L-aspartic acid) where the diabetic dogs have a measured amount of this metabolite often below the LOQ. Following valine and N-acetyl-L-aspartic acid, the next best feature to add was indoxyl sulfate, which increased the average AUC to 0.965. At this point, the forward feature selection procedure indicated that adding more features to the model did not increase model performance based on a two-sample t-test comparing the 100 measured AUC values from the prior iteration to the current iteration. This is validated in Figure 4B where, for demonstration purposes, we continue to add metabolites to the model until all have been included. If all 8 measured metabolites are included in the model, the AUC drops to 0.924, which is less than utilizing only a single metabolite (valine). The final three-metabolite Linear Discriminant classifier model was generated from the full set of the training data and applied to the validation set of 18 dogs. This returned an AUC 0.975 on the validation set, which was not utilized to select or generate the model. This indicates both high performance of the three-feature metabolite model, but also that the model is not over-trained. Utilizing a standard probability of classification threshold of 0.5, these three markers yielded the correct classification of 77.8% with a false discovery of 0% on a validation set segregated from the training of the model.
Table 4.
Metabolites Included in Machine Learning Training Set
| Metabolite | Bonferroni p-value | Average AUC |
|---|---|---|
| Valine | 0.0001 | 0.935 |
| GABA | 0.0003 | 0.920 |
| 2-hydroxyisobutyric acid | 0.0005 | 0.896 |
| L-Glutamine | 0.0194 | 0.800 |
| N-acetyl-L-aspartic acid | 0.0229 | 0.765 |
| Citramalate | 0.0090 | 0.774 |
| Kynurenine | 0.0399 | 0.770 |
| Indoxyl sulfate | 0.0304 | 0.795 |
AUC=area under the receiver operating characteristics curve
Fig 4.
a. Visualization of the separation of control and diabetes groups utilizing two metabolite features valine and N-acetyl-L-aspartic acid, where open symbols indicate dogs that are part of the training data and the filled-in symbols are the dogs in the validation set
b. The change in area under the curve (AUC) as each metabolite feature identified via machine learning is added. Each point represents all features to the left on the x-axis included in the model. The optimal Linear Discriminant classifier model includes valine, n-acetyl-L-aspartic acid, and indoxyl sulfate
4. Discussion
Using an untargeted metabolomic approach, we found metabolic perturbations in diabetic compared to healthy control dogs, some of which were similar to our preliminary studies (O'Kell et al., 2017a, O'Kell et al., 2019). After selection of 32 candidate metabolite biomarkers based on these results, 23 were measurable reliably by targeted metabolomics analysis, which identified 14 metabolites with concentrations significantly different between diabetic and control groups: GABA, N-acetyl-L-aspartic acid, indoxyl sulfate, 2-hydroxyisobutyric acid, kynurenine, anthranilic acid, TUDCA, valine, isoleucine, leucine, glutamine, tyrosine, tryptophan, and citramalate. A machine learning approach also identified valine, indoxyl sulfate, and N-acetyl-L-aspartic acid as key metabolites in a Linear Discriminant Classifier model.
While GABA is well-known as an important inhibitory neurotransmitter in the mammalian nervous system, it is also present in other tissues including the pancreas (Tillakaratne et al., 1995). Within the pancreas, GABA predominates in the islets (Reetz et al., 1991, Menegaz et al., 2019). Previous studies have demonstrated that GABA acts in an autocrine manner in the islets to stimulate insulin secretion from β-cells (Braun et al., 2010, Bansal et al., 2011, Dong et al., 2006), and that this stimulation was suppressed at high glucose concentrations in vitro (Dong et al., 2006). Additionally, the presence of insulin decreased GABA induced insulin secretion, suggesting the presence of a negative feedback loop (Bansal et al., 2011). However, a more recent study contradicts these findings, showing that GABA is released in a pulsatile manner that is not dependent on glucose concentrations, and that endogenous GABA inhibits insulin secretion and regulates the periodicity of this secretion (Menegaz et al., 2019). This study also found that GABA is depleted in the β-cells of human patients with both type 1 and 2 diabetes compared to non-diabetic controls (Menegaz et al., 2019). The implications of the depletion of GABA from β-cells in diabetic patients is unclear at this time, but one proposed theory is that the loss of GABA increases insulin secretion at the expense of a loss of the periodicity of secretion at a time of high demand (Menegaz et al., 2019). Interestingly, one small study in humans found higher plasma GABA concentrations in patients with T1D compared with non-diabetic controls (Bhandage et al., 2018), similar to our findings in canine serum. The source of this increased circulating GABA in both canine and human diabetes patients is unknown, but possibilities may include the liver (Minuk, 1993) or endothelial cells (Sen et al., 2016). GABA has also been considered a promoter of β-cell regeneration, but these effects are controversial among studies in different species (Yi et al., 2020). Future studies to further investigate the role of GABA in diabetes in both humans and dogs are warranted.
Elevations of the BCAAs (valine, isoleucine, and leucine) in diabetic dogs are consistent with findings in the human disease. Indeed, elevated BCAAs are a marker of prediabetes, insulin resistance, and a predictor of the development of type 2 diabetes in humans (Gar et al., 2018). Interestingly, in dogs, serum BCAAs are higher in obese dogs than ideal weight control dogs; subsequently, serum BCAAS are lower compared with control in these same dogs after weight loss(Vendramini et al., 2021). Body condition and fat content was not evaluated in the present study and is an area for future evaluation given the association of obesity with insulin resistance in various species. In human patients with T1D, BCAAs are elevated in patients classified as having either good or poor diabetic control compared with non-diabetic control patients, and among all patients BCAAs are correlated with blood glucose and hemoglobin A1c (Dutta et al., 2016). Additionally, serum BCAAs are also increased prior to autoantibody seroconversion in children that later progress to T1D (Oresic et al., 2008). Hence, metabolomics studies in canine pre-diabetes are warranted to determine their utility as a disease predictive biomarker.
N-acetyl-aspartic acid is found in high concentrations in the brain and can also be found in adipose tissue and some cancers (Bogner-Strauss, 2017); however, N-acetyl-aspartic acid has received limited study in diabetes. In a mouse model of type 2 diabetes, increased activity of aspartoacylase, the enzyme that hydrolyzes N-acetyl-aspartic acid, was detected in the duodenum and brain compared with control mice (Surendran et al., 2006). In human subjects with T1D, decreased N-acetylaspartate/creatinine ratio in the brain measured via proton magnetic resonance spectroscopy was associated with peripheral neuropathy, proliferative retinopathy, and longer diabetes duration (Hansen et al., 2019). To the authors’ knowledge, systemic concentrations of this metabolite have not been studied in human patients or animal models of T1D. These prior reports, in concert with the findings in our canine population, suggest that N-acetyl-aspartic acid may be relevant as a biomarker of diabetes or disease complications.
Indoxyl sulfate is a uremic toxin and is elevated in humans (Holle et al., 2020) and dogs (Cheng et al., 2015) with kidney disease, and has been identified as a biomarker for atherosclerosis in human type 2 diabetes (Omori et al., 2020) and a predictor of cardiovascular events in patients with heart failure (Imazu et al., 2020). Interestingly, indoxyl sulfate was lower in diabetic dogs, which was also found in mice with type 2 diabetes (db/db mouse model) (Connor et al., 2010). Indoxyl sulfate is produced by metabolism of dietary tryptophan into indole by the gut microbiota, which is absorbed and converted to indoxyl sulfate by the liver (Leong and Sirich, 2016). Dietary or gut microbiome analysis were not performed in the present study, and it is therefore unknown if there were differences in dietary protein intake between the groups or if differences in gut microbiota affected indoxyl sulfate production.
Other significantly different metabolites have received some study in diabetes in other species. 2-hydroxyisobutyric acid is also increased in human patients with type 2 diabetes compared to control (Li et al., 2009, Chong et al., 2018), as well as patients with pre-diabetes (Chong et al., 2018). Kynurenine was found to be decreased in the pre-diabetic state of two different rat type 2 diabetes models (Yokoi et al., 2015). Additionally, kynurenine was shown to increase glucose induced insulin secretion in healthy rat islets (Liu et al., 2015). While it is unknown if dogs in a pre-diabetic state will have similar findings to the overtly diabetic dogs in our study, this decrease in kynurenine deserves further study as an early disease biomarker. On the contrary, in human type 1 diabetes, plasma anthranilic acid is higher than in control patients or those with type 2 diabetes, tryptophan is higher than in control patients, and there is no difference in kynurenine between type 1 diabetes patients and controls (Oxenkrug et al., 2015). These findings differ from the dogs in our study in which anthranilic acid was decreased the diabetic group. Tryptophan showed a statistically significant decrease in the diabetic group, however there was marked overlap between the groups and the clinical relevance of this finding is questionable. Various bile acids, including taurine conjugated bile acids, have been studied in the context of glucose metabolism and diabetes (Rajani and Jia, 2018). In humans with type 2 diabetes and in those with intermediate glucose tolerance, total taurine conjugated bile acids were elevated (Wewalka et al., 2014). Tauroursodeoxycholic acid was found to stimulate glucose induced insulin secretion in mouse islets in-vitro (Vettorazzi et al., 2016). The amino acid glutamine also stimulates glucose induced insulin secretion (Han et al., 2021). Our findings in diabetic dogs showing decreased TUDCA, kynurenine, and glutamine, all metabolites that stimulate glucose induced insulin secretion in rodents, are intriguing, though further study is needed to determine if these metabolites could be involved in a decrease or dysfunction in insulin secretion in this species.
A limitation of the present study is that many factors aside from disease state may affect serum metabolites. Metabolomic profiles have been shown to vary by dog breed (Lloyd et al., 2016, Lloyd et al., 2017) and size (Middleton et al., 2017). To address this, control dogs were breed matched to diabetic dogs whenever possible, and mixed breeds were size matched to other mixed breeds. Although differences in diet affect the metabolome, one study showed that breed was a stronger determinant of metabolic differences among individual dogs than environmental conditions such as diet (Lloyd et al., 2016). The post-prandial state may lead to additional changes in some metabolites. In one study of canine diets differing in calcium content, several metabolites showed significant changes within four hours post meal, although none were the key metabolites found in our study (Allaway et al., 2019). In the present study, we attempted to decrease this source of variability by matching fasted or fed status among dogs with similar characteristics, although exact timing after feeding was not standardized. Importantly, one of our goals is to identify robust biomarkers that can be measured in either the fasted or fed state, supporting our decision to include both fasted and fed dogs in our studies. An additional limitation of this study is that we cannot determine whether these differences in metabolites are a feature of the underlying disease etiology or a consequence of the loss of endogenous insulin and subsequent metabolic derangements, which may be particularly relevant for those dogs with newly diagnosed disease versus dogs with more established disease in this study. Future studies evaluating these metabolites in dogs in a pre-diabetic state are necessary to investigate such questions.
5. Conclusions
We believe this represents the first study to report a selection of candidate serum biomarkers in canine diabetes using untargeted metabolomics analysis followed by targeted metabolomics to obtain quantitative confirmation of results. Of the biomarkers found to be statistically significant, valine, GABA, N-acetyl-L-aspartic acid, and indoxyl sulfate were among those that distinguished diseased from healthy groups. GABA in particular is of interest given similar findings in the plasma of human subjects, as well as GABA’s depletion in β-cells of humans with both type 1 and type 2 diabetes (Menegaz et al., 2019, Bhandage et al., 2018). Further studies to investigate the role of GABA, BCAAs, and other key metabolites in canine diabetes and human T1D patients are warranted.
Supplementary Material
Acknowledgements
The authors sincerely thank Dr. Amanda Posgai and Dr. Sara Williams (University of Florida) for editorial assistance and Dr. Rhonda Bacher for her statistical expertise and advice.
Funding
This research was supported by Zomedica Inc. and the National Institute of Diabetes and Digestive and Kidney Disease of the National Institutes of Health (AO – K08DK116735). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of interest/Competing interests Zomedica Inc. provided financial support for the study, but had no influence on study design, data analysis, or the decision to publish. No other potential conflicts of interest relevant to this article were reported.
Availability of data and material The datasets generated during and/or analyzed during the current study are available at the NIH Common Fund's National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org (Study ID ST001754, ST001742, and PR000396).
Compliance with Ethical Requirements All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Publisher's Disclaimer: This AM is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect postacceptance improvements, or any corrections. Use of this AM is subject to the publisher's embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. See here for Springer Nature's terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
References
- ALLAWAY D, GILHAM M, WAGNER-GOLBS A, MALDONADO SG, HAYDOCK R, COLYER A, STOCKMAN J & WATSON P 2019. Metabolomic profiling to identify effects of dietary calcium reveal the influence of the individual and postprandial dynamics on the canine plasma metabolome. J Nutr Sci, 8, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANSAL P, WANG S, LIU S, XIANG YY, LU WY & WANG Q 2011. GABA coordinates with insulin in regulating secretory function in pancreatic INS-1 beta-cells. PLoS One, 6, e26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHANDAGE AK, JIN Z, KOROL SV, SHEN Q, PEI Y, DENG Q, ESPES D, CARLSSON PO, KAMALI-MOGHADDAM M & BIRNIR B 2018. GABA Regulates Release of Inflammatory Cytokines From Peripheral Blood Mononuclear Cells and CD4(+) T Cells and Is Immunosuppressive in Type 1 Diabetes. EBioMedicine, 30, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOGNER-STRAUSS JG 2017. N-Acetylaspartate Metabolism Outside the Brain: Lipogenesis, Histone Acetylation, and Cancer. Front Endocrinol (Lausanne), 8, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAUN M, RAMRACHEYA R, BENGTSSON M, CLARK A, WALKER JN, JOHNSON PR & RORSMAN P 2010. Gamma-aminobutyric acid (GABA) is an autocrine excitatory transmitter in human pancreatic beta-cells. Diabetes, 59, 1694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARLOS G, DOS SANTOS FP & FROEHLICH PE 2020. Canine metabolomics advances. Metabolomics, 16, 16. [DOI] [PubMed] [Google Scholar]
- CHENG FP, HSIEH MJ, CHOU CC, HSU WL & LEE YJ 2015. Detection of indoxyl sulfate levels in dogs and cats suffering from naturally occurring kidney diseases. Vet J, 205, 399–403. [DOI] [PubMed] [Google Scholar]
- CHONG J, SOUFAN O, LI C, CARAUS I, LI S, BOURQUE G, WISHART DS & XIA J 2018. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res, 46, W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONNOR SC, HANSEN MK, CORNER A, SMITH RF & RYAN TE 2010. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Mol Biosyst, 6, 909–21. [DOI] [PubMed] [Google Scholar]
- DONG H, KUMAR M, ZHANG Y, GYULKHANDANYAN A, XIANG YY, YE B, PERRELLA J, HYDER A, ZHANG N, WHEELER M, LU WY & WANG Q 2006. Gamma-aminobutyric acid up- and downregulates insulin secretion from beta cells in concert with changes in glucose concentration. Diabetologia, 49, 697–705. [DOI] [PubMed] [Google Scholar]
- DUTTA T, KUDVA YC, PERSSON XM, SCHENCK LA, FORD GC, SINGH RJ, CARTER R & NAIR KS 2016. Impact of Long-Term Poor and Good Glycemic Control on Metabolomics Alterations in Type 1 Diabetic People. J Clin Endocrinol Metab, 101, 1023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAR C, ROTTENKOLBER M, PREHN C, ADAMSKI J, SEISSLER J & LECHNER A 2018. Serum and plasma amino acids as markers of prediabetes, insulin resistance, and incident diabetes. Crit Rev Clin Lab Sci, 55, 21–32. [DOI] [PubMed] [Google Scholar]
- GILOR C, NIESSEN SJ, FURROW E & DIBARTOLA SP 2016. What's in a Name? Classification of Diabetes Mellitus in Veterinary Medicine and Why It Matters. J Vet Intern Med, 30, 927–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUASCH-FERRÉ M, HRUBY A, TOLEDO E, CLISH CB, MARTÍNEZ-GONZÁLEZ MA, SALAS-SALVADÓ J & HU FB 2016. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care, 39, 833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAN G, TAKAHASHI H, MURAO N, GHENI G, YOKOI N, HAMAMOTO Y, ASAHARA SI, SEINO Y, KIDO Y & SEINO S 2021. Glutamate is an essential mediator in glutamine-amplified insulin secretion. J Diabetes Investig, 12, 920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANSEN TM, BROCK B, JUHL A, DREWES AM, VORUM H, ANDERSEN CU, JAKOBSEN PE, KARMISHOLT J, FROKJAER JB & BROCK C 2019. Brain spectroscopy reveals that N-acetylaspartate is associated to peripheral sensorimotor neuropathy in type 1 diabetes. J Diabetes Complications, 33, 323–328. [DOI] [PubMed] [Google Scholar]
- HOLLE J, KIRCHNER M, OKUN J, BAYAZIT AK, OBRYCKI L, CANPOLAT N, BULUT IK, AZUKAITIS K, DUZOVA A, RANCHIN B, SHROFF R, CANDAN C, OH J, KLAUS G, LUGANI F, GIMPEL C, BÜSCHER R, YILMAZ A, BASKIN E, ERDOGAN H, ZALOSZYC A, ÖZCELIK G, DROZDZ D, JANKAUSKIENE A, NOBILI F, MELK A, QUERFELD U & SCHAEFER F 2020. Serum indoxyl sulfate concentrations associate with progression of chronic kidney disease in children. PLoS One, 15, e0240446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMAZU M, FUKUDA H, KANZAKI H, AMAKI M, HASEGAWA T, TAKAHAMA H, HITSUMOTO T, TSUKAMOTO O, MORITA T, ITO S & KITAKAZE M 2020. Plasma indoxyl sulfate levels predict cardiovascular events in patients with mild chronic heart failure. Sci Rep, 10, 16528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMICHHANE S, AHONEN L, DYRLUND TS, KEMPPAINEN E, SILJANDER H, HYOTY H, ILONEN J, TOPPARI J, VEIJOLA R, HYOTYLAINEN T, KNIP M & ORESIC M 2018. Dynamics of Plasma Lipidome in Progression to Islet Autoimmunity and Type 1 Diabetes - Type 1 Diabetes Prediction and Prevention Study (DIPP). Sci Rep, 8, 10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMICHHANE S, KEMPPAINEN E, TROST K, SILJANDER H, HYOTY H, ILONEN J, TOPPARI J, VEIJOLA R, HYOTYLAINEN T, KNIP M & ORESIC M 2019. Circulating metabolites in progression to islet autoimmunity and type 1 diabetes. Diabetologia, 62, 2287–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEONG SC & SIRICH TL 2016. Indoxyl Sulfate-Review of Toxicity and Therapeutic Strategies. Toxins (Basel), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, XU Z, LU X, YANG X, YIN P, KONG H, YU Y & XU G 2009. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal Chim Acta, 633, 257–62. [DOI] [PubMed] [Google Scholar]
- LIU JJ, RAYNAL S, BAILBE D, GAUSSERES B, CARBONNE C, AUTIER V, MOVASSAT J, KERGOAT M & PORTHA B 2015. Expression of the kynurenine pathway enzymes in the pancreatic islet cells. Activation by cytokines and glucolipotoxicity. Biochim Biophys Acta, 1852, 980–91. [DOI] [PubMed] [Google Scholar]
- LLOYD AJ, BECKMANN M, TAILLIART K, BROWN WY, DRAPER J & ALLAWAY D 2016. Characterisation of the main drivers of intra- and inter- breed variability in the plasma metabolome of dogs. Metabolomics, 12, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLOYD AJ, BECKMANN M, WILSON T, TAILLIART K, ALLAWAY D & DRAPER J 2017. Ultra high performance liquid chromatography-high resolution mass spectrometry plasma lipidomics can distinguish between canine breeds despite uncontrolled environmental variability and non-standardized diets. Metabolomics, 13, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANDREKAR JN 2010. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol, 5, 1315–6. [DOI] [PubMed] [Google Scholar]
- MENEGAZ D, HAGAN DW, JOANA A, CIANCIARUSO C, RODRIGUEZ-DIAZ R, MOLINA J, DOLAN RM, BECKER MW, SCHWALIE PC, NANO R, LEBRETON F, KANG C, SAH R, GAISANO HY, BERGGREN P-O, BAEKKESKOV S, CAICEDO A & PHELPS EA 2019. Mechanism and effects of pulsatile GABA secretion from cytosolic pools in the human beta cell. Nature Metabolism, 1, 1110–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIDDLETON RP, LACROIX S, SCOTT-BOYER MP, DORDEVIC N, KENNEDY AD, SLUSKY AR, CARAYOL J, PETZINGER-GERMAIN C, BELOSHAPKA A & KAPUT J 2017. Metabolic Differences between Dogs of Different Body Sizes. J Nutr Metab, 2017, 4535710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MINUK GY 1993. Gamma-aminobutyric acid and the liver. Dig Dis, 11, 45–54. [DOI] [PubMed] [Google Scholar]
- NELSON RW & REUSCH CE 2014. Animal models of disease: classification and etiology of diabetes in dogs and cats. J Endocrinol, 222, T1–9. [DOI] [PubMed] [Google Scholar]
- O'KELL AL, GARRETT TJ, WASSERFALL C & ATKINSON MA 2017a. Untargeted metabolomic analysis in naturally occurring canine diabetes mellitus identifies similarities to human Type 1 Diabetes. Sci Rep, 7, 9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'KELL AL, GARRETT TJ, WASSERFALL C & ATKINSON MA 2019. Untargeted metabolomic analysis in non-fasted diabetic dogs by UHPLC-HRMS. Metabolomics, 15, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'KELL AL, WASSERFALL C, CATCHPOLE B, DAVISON LJ, HESS RS, JUSHNER J & ATKINSON MA 2017b. Comparative pathogenesis of autoimmune diabetes in humans, NOD mice, and canines: has a valuable animal model of type 1 diabetes been overlooked? Diabetes, 66, 1443–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMORI K, KATAKAMI N, ARAKAWA S, YAMAMOTO Y, NINOMIYA H, TAKAHARA M, MATSUOKA TA, TSUGAWA H, FURUNO M, BAMBA T, FUKUSAKI E & SHIMOMURA I 2020. Identification of Plasma Inositol and Indoxyl Sulfate as Novel Biomarker Candidates for Atherosclerosis in Patients with Type 2 Diabetes. -Findings from Metabolome Analysis Using GC/MS. J Atheroscler Thromb, 27, 1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORESIC M, SIMELL S, SYSI-AHO M, NÄNTÖ-SALONEN K, SEPPÄNEN-LAAKSO T, PARIKKA V, KATAJAMAA M, HEKKALA A, MATTILA I, KESKINEN P, YETUKURI L, REINIKAINEN A, LÄHDE J, SUORTTI T, HAKALAX J, SIMELL T, HYÖTY H, VEIJOLA R, ILONEN J, LAHESMAA R, KNIP M & SIMELL O 2008. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med, 205, 2975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OXENKRUG G, VAN DER HART M & SUMMERGRAD P 2015. Elevated anthranilic acid plasma concentrations in type 1 but not type 2 diabetes mellitus. Integr Mol Med, 2, 365–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAJANI C & JIA W 2018. Bile acids and their effects on diabetes. Front Med, 12, 608–623. [DOI] [PubMed] [Google Scholar]
- REETZ A, SOLIMENA M, MATTEOLI M, FOLLI F, TAKEI K & DE CAMILLI P 1991. GABA and pancreatic beta-cells: colocalization of glutamic acid decarboxylase (GAD) and GABA with synaptic-like microvesicles suggests their role in GABA storage and secretion. Embo j, 10, 1275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEN S, ROY S, BANDYOPADHYAY G, SCOTT B, XIAO D, RAMADOSS S, MAHATA SK & CHAUDHURI G 2016. gamma-Aminobutyric Acid Is Synthesized and Released by the Endothelium: Potential Implications. Circ Res, 119, 621–34. [DOI] [PubMed] [Google Scholar]
- SURENDRAN S, MATALON R & TYRING SK 2006. Upregulation of aspartoacylase activity in the duodenum of obesity induced diabetes mouse: implications on diabetic neuropathy. Biochem Biophys Res Commun, 345, 973–5. [DOI] [PubMed] [Google Scholar]
- TILLAKARATNE NJ, MEDINA-KAUWE L & GIBSON KM 1995. gamma-Aminobutyric acid (GABA) metabolism in mammalian neural and nonneural tissues. Comp Biochem Physiol A Physiol, 112, 247–63. [DOI] [PubMed] [Google Scholar]
- VARGA TV, NISS K, ESTAMPADOR AC, COLLIN CB & MOSELEY PL 2020. Association is not prediction: A landscape of confused reporting in diabetes - A systematic review. Diabetes Res Clin Pract, 170, 108497. [DOI] [PubMed] [Google Scholar]
- VENDRAMINI THA, MACEDO HT, ZAFALON RVA, MACEGOZA MV, PEDRINELLI V, RISOLIA LW, OCAMPOS FMM, JEREMIAS JT, PONTIERI CFF, FERRIOLLI E, COLNAGO LA & BRUNETTO MA 2021. Serum metabolomics analysis reveals that weight loss in obese dogs results in a similar metabolic profile to dogs in ideal body condition. Metabolomics, 17, 27. [DOI] [PubMed] [Google Scholar]
- VETTORAZZI JF, RIBEIRO RA, BORCK PC, BRANCO RC, SORIANO S, MERINO B, BOSCHERO AC, NADAL A, QUESADA I & CARNEIRO EM 2016. The bile acid TUDCA increases glucose-induced insulin secretion via the cAMP/PKA pathway in pancreatic beta cells. Metabolism, 65, 54–63. [DOI] [PubMed] [Google Scholar]
- WEWALKA M, PATTI ME, BARBATO C, HOUTEN SM & GOLDFINE AB 2014. Fasting serum taurine-conjugated bile acids are elevated in type 2 diabetes and do not change with intensification of insulin. J Clin Endocrinol Metab, 99, 1442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIA J, BROADHURST DI, WILSON M & WISHART DS 2013. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics, 9, 280–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YI Z, WASEEM GHANI M, GHANI H, JIANG W, WASEEM BIRMANI M, YE L, BIN L, CUN LG, LILONG A & MEI X 2020. Gimmicks of gamma-aminobutyric acid (GABA) in pancreatic beta-cell regeneration through transdifferentiation of pancreatic alpha- to beta-cells. Cell Biol Int, 44, 926–936. [DOI] [PubMed] [Google Scholar]
- YOKOI N, BEPPU M, YOSHIDA E, HOSHIKAWA R, HIDAKA S, MATSUBARA T, SHINOHARA M, IRINO Y, HATANO N & SEINO S 2015. Identification of putative biomarkers for prediabetes by metabolome analysis of rat models of type 2 diabetes. Metabolomics, 11, 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG A, SUN H, YAN G, WANG P & WANG X 2015. Metabolomics for Biomarker Discovery: Moving to the Clinic. Biomed Res Int, 2015, 354671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.