Abstract
Staphylococcus aureus has to cope with oxidative and electrophile stress during host-pathogen interactions. The TetR-family repressor GbaA was shown to sense electrophiles, such as N-ethylmaleimide (NEM) via monothiol mechanisms of the two conserved Cys55 or Cys104 residues in vitro. In this study, we further investigated the regulation and function of the GbaA repressor and its Cys residues in S. aureus COL. The GbaA-controlled gbaAB-SACOL2595-97 and SACOL2592-nmrA-2590 operons were shown to respond only weakly 3-10-fold to oxidants, electrophiles or antibiotics in S. aureus COL, but are 57-734-fold derepressed in the gbaA deletion mutant, indicating that the physiological inducer is still unknown. Moreover, the gbaA mutant remained responsive to disulfide and electrophile stress, pointing to additional redox control mechanisms of both operons. Thiol-stress induction of the GbaA regulon was strongly diminished in both single Cys mutants, supporting that both Cys residues are required for redox-sensing in vivo. While GbaA and the single Cys mutants are reversible oxidized under diamide and allicin stress, these thiol switches did not affect the DNA binding activity. The repressor activity of GbaA could be only partially inhibited with NEM in vitro. Survival assays revealed that the gbaA mutant confers resistance under diamide, allicin, NEM and methylglyoxal stress, which was mediated by the SACOL2592-90 operon encoding for a putative glyoxalase and oxidoreductase. Altogether, our results support that the GbaA repressor functions in the defense against oxidative and electrophile stress in S. aureus. GbaA represents a 2-Cys-type redox sensor, which requires another redox-sensing regulator and an unknown thiol-reactive ligand for full derepression of the GbaA regulon genes.
Keywords: Staphylococcus aureus, GbaA, Thiol switches, Allicin, Diamide, Electrophiles
Graphical abstract
Highlights
-
•
The TetR repressor GbaA is not fully inactivated by oxidants and electrophiles in S. aureus.
-
•
The GbaA regulon is induced by thiol stress in the gbaA mutant, indicating additional mechanisms of redox regulation.
-
•
Cys55 and Cys104 are both involved in redox-sensing of GbaA in S. aureus.
-
•
The GbaA regulon functions in the defense against diamide, allicin, NEM and methylglyoxal.
1. Introduction
Staphylococcus aureus is an opportunistic human pathogen, which colonizes the skin and the nose of one quarter of the human population [1], but can also cause life-threatening infections especially in immunocompromised persons, ranging from skin and soft tissue infections to systemic septic shock syndrome, chronic osteomyelitis, endocarditis and pneumonia [[2], [3], [4]]. Moreover, S. aureus rapidly acquires new antibiotic resistant elements resulting in an increased prevalence of multi-resistant S. aureus isolates with limited treatment options [5,6].
During infections and antibiotics treatments, S. aureus has to cope with reactive oxygen, chlorine and electrophile species (ROS, RCS, RES) [7]. Activated macrophages and neutrophils produce ROS, such as superoxide anion and H2O2 as well as the strong microbicidal agent hypochlorous acid (HOCl) in large quantities to kill invading pathogens in the acidic phagosome [[8], [9], [10]]. ROS and HOCl lead to oxidation of amino acids, unsaturated fatty acids, carbohydrates and nucleotides, resulting in RES with electron-deficient centers as secondary reactive metabolites, such as quinones, epoxides and the highly toxic dicarbonyl compounds glyoxal and methylglyoxal (MG) [[11], [12], [13]]. Enhanced levels of reactive aldehydes and MG are produced during infections in inflamed tissues, in the blood and in activated neutrophils, causing alkylation of lysine, arginine and cysteine residues in proteins [14,15]. Moreover, the host-derived electrophilic metabolite itaconate reprograms the host metabolism to stimulate macrophage immune responses and to promote biofilm formation in bacterial pathogens [16,17]. Thus, host-pathogen interactions generate various reactive species, which require the expression of efficient protection, detoxification and repair systems in S. aureus for successful infection, spread and survival in the human body.
These defense mechanisms are often controlled by redox-sensing regulators, such as SarZ, MgrA, HypR and QsrR, which utilize conserved Cys residues to sense and respond to ROS, RCS, RES or antibiotics via post-translational thiol-modifications, leading to activation of their specific regulons in S. aureus [7,[18], [19], [20], [21], [22], [23], [24]]. Redox-sensing transcription factors sense ROS, RCS and RES via one-Cys-type and two-Cys-type mechanisms, depending on the number of Cys residues [18,19]. The Bacillus subtilis MarR-type OhrR repressor is the prototype of the one-Cys-type redox sensor, which is inactivated by organic hydroperoxides and HOCl via S-bacillithiolation of the single Cys residue in both subunits [18,25,26]. In contrast, OhrR of Xanthomonas campestris, YodB of B. subtilis and HypR of S. aureus harbor more than one Cys residue and are regulated by intersubunit disulfide formation between the N-terminal redox-sensing Cys and the C-terminal Cys of opposing subunits according to the two-Cys-type model [18,21,[27], [28], [29]]. Thus, the number of Cys residues determines the regulatory mechanism of thiol-based redox regulators owing to different thiol-modifications.
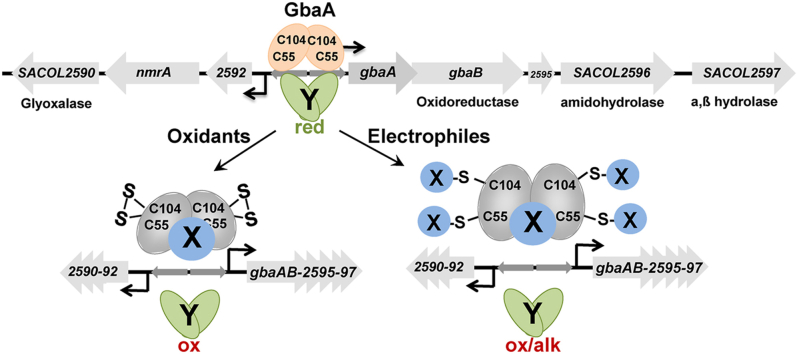
In S. aureus, the TetR-family GbaA regulator was characterized as a negative regulator of glucose-induced biofilm formation, since a gbaA mutation enhanced the production of poly-N-acetylglucosamine (PNAG), required for polysaccharide intracellular adhesin (PIA)-dependent biofilm formation in a super-biofilm-elaborating S. aureus isolate TF2758 [30,31]. GbaA was shown to repress transcription of two divergent operons, including the upstream SACOL2592-nmrA-90 and downstream gbaAB-SACOL2595-97 operons [30,31]. The upstream operon encodes a putative glyoxalase, the NAD+-dependent epimerase/dehydratase NmrA and the DUF2316 hypothetical protein. The downstream operon encodes the GbaA repressor, a short chain dehydrogenase/oxidoreductase GbaB, an amidohydrolase and an α,β-fold hydrolase [[30], [31], [32]]. GbaA binds to a 9-6-9 bp inverted repeat sequence ATAAACGGA-N6-TCCGTTTGT in the upstream promoter regions of both divergent operons [30,31]. However, the physiological inducer for GbaA inactivation and the functions of the GbaA regulon genes are unknown in S. aureus.
In transcriptomic studies, the GbaA regulon was weakly upregulated under various disulfide and electrophile stress conditions, such as AGXX®, allicin, HOCl, methylhydroquinone (MHQ) and lapachol stress [[33], [34], [35], [36]]. GbaA harbors two Cys55 and Cys104 residues, which are highly conserved across TetR/AcrR homologs of other Gram-positive bacteria (Fig. S1), and located close to the DNA binding and regulatory domains of the GbaA dimer as modelled based on the template of Escherichia coli AcrR (Fig. S2). Thus, we investigated the role of GbaA and its Cys residues for redox sensing under various thiol-stress conditions in vitro and in vivo. While our work was in progress, GbaA was described as monothiol electrophile sensor that senses N-ethylmaleimide (NEM) and oxidants via one of the two Cys residues in vitro, since DNA binding activity was only impaired in the single Cys55 and Cys104 mutants, but not in the two-Cys wild type protein [32]. Oxidation of GbaA by diamide, bacillithiol disulfide (BSSB) or S-nitroso glutathione (GSNO) led to formation of the intramolecular C55-C104 disulfide, which did not change the structure and DNA binding activity. Only the monothiol GbaA variants could be inactivated by S-bacillithiolation with BSSB or S-alkylation with NEM in vitro [32]. Therefore, GbaA was suggested to function as monothiol electrophile sensor under oxidative and electrophile stress. However, the physiological role of the GbaA regulon under electrophile stress, such as NEM and MG remained unclear in the previous study [32].
Here, we have further studied the function and regulation of GbaA under oxidative and electrophile stress in S. aureus COL. The GbaA regulon was only weakly induced under various disulfide, electrophile and antibiotics stress conditions in S. aureus. Moreover, the full derepression of the GbaA regulon depends on inactivation of a second thiol-redox regulator. While both Cys residues are required for redox sensing of GbaA in vivo, diverse thiol switches are not sufficient for inactivation of GbaA and the single Cys proteins in vitro. However, phenotype analyses revealed that the GbaA regulon conferred resistance under diamide, allicin, NEM and MG stress, indicating that the GbaA regulon functions in the defense under disulfide and electrophile stress.
2. Material and methods
2.1. Bacterial strains and cultivations
Bacterial strains, plasmids and primers are listed in Tables S1 and S2. E. coli strains were cultivated in Luria broth (LB) for plasmid construction and protein expression. For stress experiments, S. aureus strains were cultivated in RPMI medium and treated with thiol-reactive compounds and antibiotics during the exponential growth at an optical density at 500 nm (OD500) of 0.5. Survival assays were performed by treatment of S. aureus cells with the thiol-reactive compounds at an OD500 of 0.5 and plating of 100 μl of serial dilutions onto LB agar plates, followed by counting of colony forming units (CFUs) after 24 h incubation. The statistics of the survival assays was calculated using Student's unpaired two-tailed t-test. Northern blot results were quantified with ImageJ and the statistics was calculated by the one-way ANOVA and Dunnet's multiple comparisons test using the Graph prism software.
2.2. Cloning, expression and purification of His-tagged GbaA, GbaAC55S and GbaAC104S proteins in E. coli
The gbaA (SACOL2593) gene was PCR amplified from chromosomal DNA of S. aureus COL using primers pET-gbaA-for-NheI and pET-gbaA-rev-BamHI (Table S2). The PCR product was digested with NheI and BamHI and cloned into plasmid pET11b, generating pET11b-gbaA. The plasmids pET11b-gbaAC55S and pET11b-gbaAC104S were constructed using PCR mutagenesis with primers including the cysteine-serine mutation as previously described [21,37]. For the gbaAC55S mutant, the primers pET-gbaA-for-NheI, gbaAC55S-rev, gbaAC55S-for and pET-gbaA-rev-BamHI were used in two first-round PCRs. For the gbaAC104S mutant, the primers pET-gbaA-for-NheI, gbaAC104S-rev, gbaAC104S-for and pET-gbaA-rev-BamHI were used for two first-round PCRs (Table S2). The PCR products of each Cys mutant were fused by overlap extension PCR with primers pET-gbaA-for-NheI and pET-gbaA-rev-BamHI to generate the gbaAC55S and gbaAC104S mutant alleles, which were cloned into pET11b as described above.
For expression and purification of His-tagged GbaA, GbaAC55S and GbaAC104S proteins, E. coli BL21(DE3) plysS with plasmids pET11b-gbaA, pET11b-gbaAC55S and pET11b-gbaAC104S was cultivated in 1.5 l LB medium until an OD600 of 0.8 followed by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h at 25 °C. Recombinant His-tagged proteins were purified using His Trap™ HP Ni-NTA columns and the ÄKTA purifier liquid chromatography system as described [21].
2.3. Construction of the S. aureus COL gbaA, gbaB and SACOL2590-92 mutants as well as the gbaA, gbaAC55S, gbaAC104S and gbaB complemented strains
The S. aureus gbaA, gbaB and SACOL2590-92 mutants were constructed using the temperature-sensitive E. coli-S. aureus shuttle vector pMAD as described [21,38]. Around 500 bp of the up- and downstream flanking regions of the corresponding genes were amplified using primers (Table S2) and fused by overlap extension PCR. The fusion products were digested with BglII and SalI and ligated into pMAD, which was cut with the same restriction enzymes. The methylated pMAD constructs from the intermediate strain S. aureus RN4220 were transferred into S. aureus COL by phage transduction. The clean deletion mutants of gbaA, gbaB and SACOL2590-92 were selected as described before [21,38].
For construction of the His-tagged S. aureus gbaA, gbaAC55S and gbaAC104S complemented strains as well as the untagged gbaB complementation, the coding sequences including the C-terminal His6-tag of gbaA, gbaAC55S and gbaAC104S were PCR amplified from plasmids pET11b-gbaA, pET11b-gbaAC55S and pET11b-gbaAC104S, whereas gbaB was amplified from S. aureus chromosomal DNA. The purified PCR products were ligated into plasmid pRB473 after digestion with BamHI and KpnI resulting in plasmids pRB473-gbaA, pRB473-gbaAC55S, pRB473-gbaAC104S and pRB473-gbaB (Table S2). The plasmids were introduced into the corresponding S. aureus gbaA and gbaB mutants via phage transduction as described [21,37].
2.4. RNA isolation and Northern blot analysis
To investigate transcriptional regulation of the GbaA-controlled up- and downstream operons, S. aureus COL strains were cultivated in RPMI and treated with various thiol-reactive compounds at an OD500 of 0.5 for 30 min as well as with different antibiotics for 60 min as previously described [21]. The applied concentrations of the compounds are indicated in the figure legends of the Northern blots.
Northern blot hybridizations were conducted with digoxigenin-labeled gbaB and SACOL2590-specific antisense RNA probes, which were synthesized by in vitro transcription with the T7 RNA polymerase and the primer pairs NB-gbaB-for and NB-gbaB-rev as well as NB-SACOL2590-for and NB-SACOL2590-rev, respectively (Table S2) as described [39,40].
2.5. Whole RNA-seq transcriptomics analysis and primary 5’ transcript mapping of the GbaA regulon genes
Whole RNA-seq transcriptomics was performed with RNA of S. aureus COL WT and the gbaA deletion mutant, which were harvested under control conditions at an OD500 of 0.5 as described [34]. Differential gene expression analysis of 3 biological replicates was performed using DESeq2 [41] with ReadXplorer v2.2 [42] as described [34]. Significant expression changes in the gbaA mutant versus WT cells were identified by an adjusted p-value cut-off of p ≤ 0.05 and a signal intensity ratio (M-value) cut-off of ≥0.6 or ≤ -0.6 (fold-change of ± 1.5) as in earlier studies [34].
The primary 5′ transcripts of the GbaA-controlled up- and downstream operons were mapped in untreated and allicin-treated cells using the 5′ end enriched RNA-seq dataset of untreated cells as reported earlier [33] and of cells exposed to 0.3 mM allicin stress. The cDNAs enriched for primary 5′-transcripts were prepared as described [43]. cDNAs were sequenced paired end on an Illumina MiSeq system (San Diego, CA, USA) using 75 bp read length. The R1 cDNA reads were mapped to the S. aureus COL genome with bowtie2 v2.2.7 [44] using the default settings for single-end read mapping and visualized with Read Explorer v.2.2 [42]. The whole transcriptome of the gbaA mutant versus WT under control conditions and the 5’ enriched RNA-seq raw data files of WT control and WT after allicin stress are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under the accession numbers E-MTAB-10887, E-MTAB-7385 and E-MTAB-10889, respectively.
2.6. Electrophoretic mobility shift assays (EMSAs) of GbaA and GbaA Cys mutant proteins
For DNA binding assays in vitro, EMSAs were performed with the DNA promoter probe containing the 150 bp upstream region of gbaA covering a region from −83 to +67 relative to the transcription start site (TSS). The DNA-binding reactions were performed with 15 ng of the promoter probe incubated with the purified His-tagged GbaA, GbaAC55S or GbaAC104S proteins for 45 min according to the EMSA protocol as described before [21].
2.7. Western blot analysis
S. aureus COL cells were collected before and after treatment with 2 and 5 mM diamide and 0.3 mM allicin for 30 min as described [21]. After harvesting, cells were washed and lysed in TE-buffer (pH 8.0) with 50 mM NEM using the ribolyzer. Protein lysates were separated using 15% non-reducing SDS-PAGE and subjected to Western blot analysis using His6 tag monoclonal antibodies (Sigma) as described previously [21,34,45].
2.8. MALDI-TOF mass spectrometry for identification of thiol-modifications of GbaA, GbaAC55S and GbaAC104S mutant proteins in vitro
The purified GbaA, GbaAC55S and GbaAC104S proteins were reduced with 10 mM DTT for 20 min, treated with 1 mM allicin or 1 mM diamide for 15 min, followed by alkylation of reduced thiols with 50 mM iodoacetamide (IAM) for 30 min in the dark. The post-translational thiol-modifications of GbaA and its Cys mutants were analyzed using non-reducing SDS–PAGE, in-gel tryptic digestion and mass spectrometry of the GbaA bands as described previously [21]. The peptides were measured using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) using an Ultraflex-II TOF/TOF instrument (Bruker Daltonics, Bremen, Germany) equipped with a 200 Hz solid-state Smart beam™ laser. The mass spectrometer was operated in the positive reflector mode. Mass spectra were acquired over an m/z range of 600–4,000. MS/MS spectra of selected peptides were acquired in the LIFT mode as described previously [21,46].
3. Results
3.1. The GbaA regulon is weakly induced by ROS, RES and antibiotics in S. aureus COL
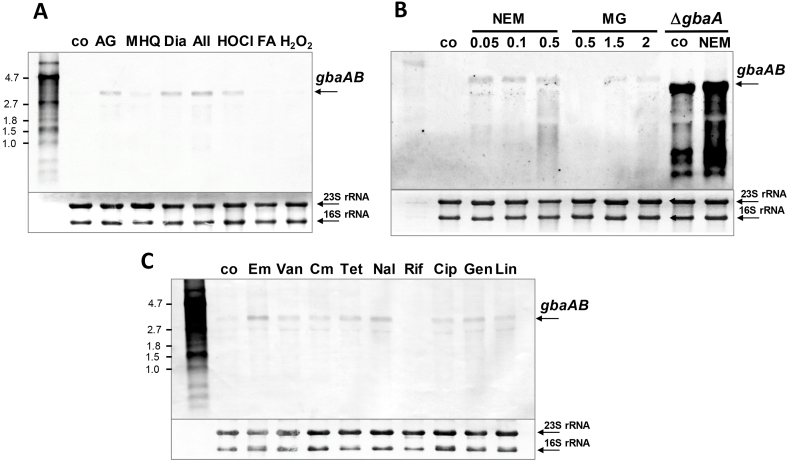
The TetR family GbaA repressor was previously characterized as monothiol electrophile sensor, which possibly senses NEM and MG via one of its two conserved Cys residues in S. aureus USA300 [32]. GbaA controls two divergent operons, the downstream gbaAB-SACOL2595-97 operon and the upstream SACOL2592-nmrA-2590 operon, which were previously shown to respond weakly to thiol-reactive compounds and antibiotics as revealed by transcriptome analyses [21,32,34,35,47]. We used Northern blot analyses to investigate transcription of the gbaAB-SACOL2595-97 operon after exposure to 5 μg/ml AGXX®, 50 μM MHQ, 2 mM diamide, 0.3 mM allicin, 1 mM HOCl, 0.75 mM formaldehyde (FA) and 10 mM H2O2, which were sub-lethal in growth and survival assays (Fig. 1A; Figs. S3A and S4A). Transcription of the gbaAB operon was further analyzed in S. aureus after treatment with sub-lethal and lethal doses of 0.05–0.5 mM NEM and 0.5–2 mM MG (Fig. 1B; Figs. S3B, S4B) as well as the antibiotics erythromycin, vancomycin, chloramphenicol, tetracycline, nalidixic acid, rifampicin, ciprofloxacin, gentamycin and linezolid (Fig. 1C, Figs. S3C and S4D). The Northern blot results revealed that transcription of the large 3.21 kb gbaAB-SACOL2595-97 operon was only weakly 3-10-fold upregulated by these strong oxidants, electrophiles and antibiotics in S. aureus COL, which is in the range of stress-induced ratios of previous RNA-seq transcriptome datasets [21,32,34,35,47] (Fig. 1A–C, Figs. S4A,B,D; Table S3). No significant up-regulation of the gbaAB operon was quantified under MHQ, H2O2, formaldehyde, chloramphenicol, rifampicin and ciprofloxacin exposure in S. aureus COL (Fig. 1A,C; Figs. S4A and D). However, much stronger >50-100-fold derepression of transcription of both GbaA-controlled operons was quantified in the gbaA mutant versus the WT under control conditions using Northern blots (Figs. 1B, Fig. 2C,D, Fig. S4C, Figs. S5A and B). In agreement with these data, the gbaAB-SACOL2595-97 and SACOL2592-nmrA-2590 operons were 57–194-fold and 401–734-fold upregulated in the RNA-seq transcriptome of the gbaA mutant compared to the WT control (Tables S3–S4) [30]. Together, the Northern blot and RNA-seq results clearly indicate that the tested oxidants, electrophiles and antibiotics cause only a very weak derepression of the GbaA regulon genes, while full derepression in the gbaA mutant leads to up-regulation of transcription in the range of 57-734-fold versus WT control. Thus, the tested compounds are clearly not the physiological inducers for complete inactivation of the GbaA repressor as already pointed out in earlier studies [32]. Thus, the identification of the GbaA inducer remains an open question.
Fig. 1.
Transcription of the gbaAB-SACOL2595-97 operon is weakly up-regulated by oxidative, electrophile and antibiotic treatments in S. aureus COL. Northern blot analysis was carried out using RNA isolated from S. aureus COL WT before (co) and 30 min after exposure to different thiol-reactive compounds (A, B) or 60 min after antibiotic treatments (C). For stress experiments, cells were treated with 5 μg/ml AGXX® (AG), 50 μM methylhydroquinone (MHQ), 2 mM diamide (Dia), 300 μM allicin (All), 1 mM HOCl, 0.75 mM formaldehyde (FA) and 10 mM H2O2 (A) or to 0.05–0.5 mM N-ethylmaleimide (NEM), 0.5–2 mM methylglyoxal (MG) for 30 min (B). For comparison of the weak transcriptional induction of the gbaAB operon after 0.5 mM NEM and 2 mM MG stress in the WT, the ΔgbaA mutant was analyzed under control and 0.5 mM NEM stress showing full derepression of the gbaAB operon in the control (B). For antibiotics experiments, S. aureus WT was exposed to 0.25 μg/ml erythromycin (Em), 0.5 μg/ml vancomycin (Van), 4 μg/ml chloramphenicol (Cm), 5 μg/ml tetracycline (Tet), 128 μg/ml nalidixic acid (Nal), 0.1 μg/ml rifampicin (Rif), 32 μg/ml ciprofloxacin (Cip), 2 μg/ml gentamicin (Gen) and 2 μg/ml linezolid (Lin) (C). The arrows point toward the size of the gbaAB-SACOL2595-97 specific operon transcript. The methylene blue stain is the RNA loading control indicating the 16S and 23S rRNAs. Band intensities of the gbaAB operon transcripts were quantified using ImageJ and the data shown in Figs. S4A–D.
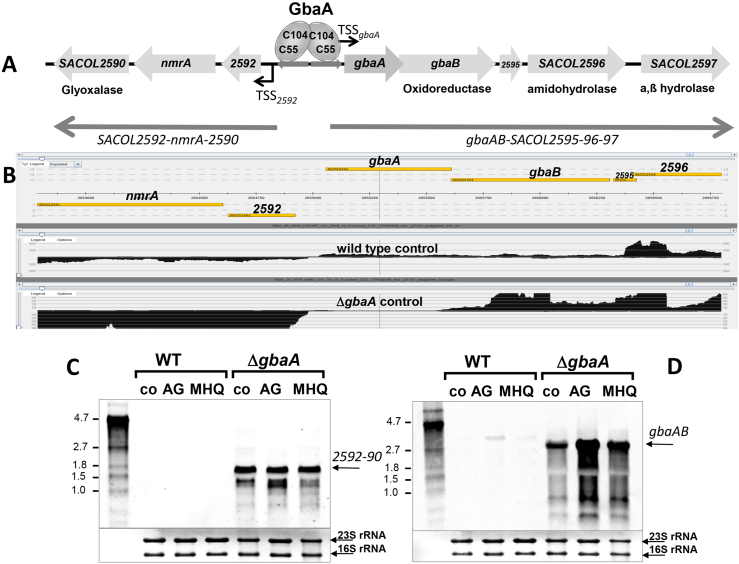
Fig. 2.
Deletion of gbaA results in derepression of transcription of the downstream gbaAB-SACOL2595-97 and upstream SACOL2592-90 operons. (A, B) Transcriptional organization of the divergent gbaAB-SACOL2595-97 and SACOL2592-90 operons in S. aureus. The upstream SACOL2592-nmrA-2590 operon encodes for a putative glyoxalase and NAD+-dependent epimerase/dehydratase (NmrA). The downstream gbaAB operon encodes for the GbaA repressor, a putative short chain oxidoreductase, an amidohydrolase and an a,ß hydrolase. (B) Both operons are negatively regulated by GbaA as displayed by the RNA-seq data of S. aureus COL WT and the gbaA mutant under control conditions using Read-Explorer. (C, D) Transcription of the SACOL2592-90 (C) and gbaAB-SACOL2595-97 operons (D) was analyzed in S. aureus COL WT and gbaA mutant strains before (co) and 30 min after treatment with 5 μg/ml AGXX® (AG) and 50 μM MHQ using Northern blots. Both operons remained inducible by AGXX® and MHQ stress in the gbaA mutant. The arrows point toward the transcript sizes of the gbaAB and SACOL2592-90 operons. The methylene blue bands denote the 16S and 23S rRNAs as RNA loading controls below the Northern blots. Band intensities of the gbaAB and SACOL2592-90 operon transcripts were quantified using ImageJ and the data are shown in Figs. S5A and B. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In addition, transcription of the gbaAB-SACOL2595-97 operon was still significantly 3-6-fold up-regulated in the gbaA mutant under oxidative and electrophile stress, including diamide, AGXX®, NEM and MG (Figs. 1B, Fig. 2C,D; Fig. 4A and B; Figs. S6A and B). This points to the presence of another redox-sensing regulator involved in the transcriptional control of the GbaA regulon genes, which senses and responds to thiol-stress conditions.
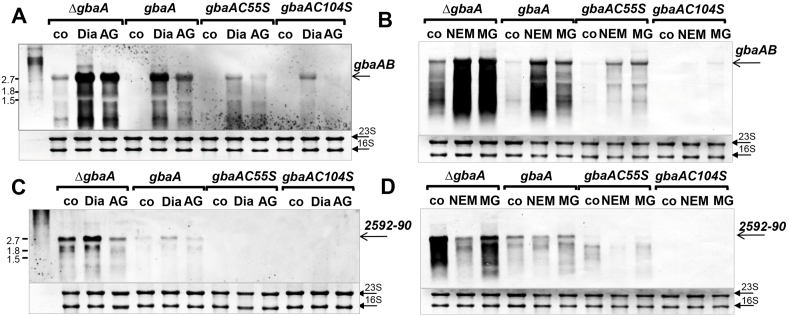
Fig. 4.
Northern blot analysis of transcription of the gbaAB-SACOL2595-97 and SACOL2592-90 operons under diamide, AGXX®, NEM and MG stress in the S. aureus COL gbaA mutant and the gbaA, gbaAC55S and gbaAC104S complemented strains. Transcription of the gbaAB-SACOL2595-97 (A,B) and SACOL2592-90 operons (C,D) was analyzed in the S. aureus gbaA deletion mutant and in the gbaA, gbaAC55S, gbaAC104S complemented strains before (co) and 30 min after treatment with 2 mM diamide (Dia), 5 μg/ml AGXX® (AG), 0.3 mM NEM and 2 mM MG using Northern blots. The arrows point toward the transcript sizes of the gbaAB-SACOL2595-97 or SACOL2592-90 operons. The methylene blue bands denote the 16S and 23S rRNAs as RNA loading controls below the Northern blots. Band intensities of the gbaAB and SACOL2592-90 operon transcripts were quantified using ImageJ and the data are shown in Figs. S6A–D.
3.2. Mapping of strong SigmaA-dependent promoters upstream of the divergent gbaAB-SACOL2595-97 and SACOL2592-nmrA-2590 operons
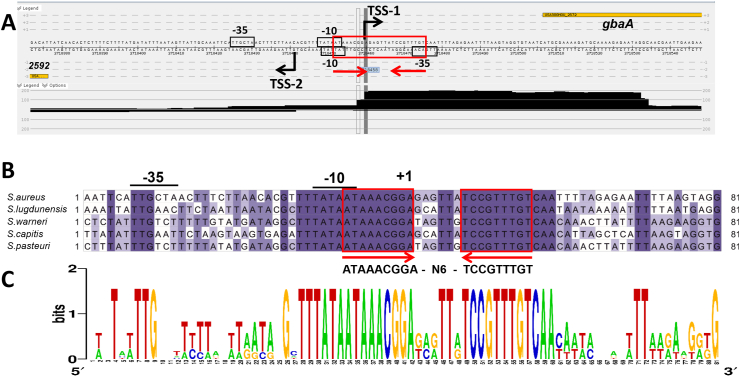
To study the transcriptional regulation by GbaA, we mapped the promoters of the upstream SACOL2592-nmrA-2590 and downstream gbaAB-SACOL2595-97 operons using RNA-seq of 5’ primary transcripts under allicin stress in S. aureus COL (Fig. 3A). The gbaA-specific transcription start site TSS-1 was identified as an adenine, which is located 45 bp upstream of the ATG start codon. TSS-1 is preceded by a strong SigmaA-dependent promoter with the consensus sequence TATAAT-N17-TTGCAT (Fig. 3A and B). The SACOL2592specific TSS-2 was mapped at a guanine located 65 bp upstream of the ATG start codon. TSS-2 is also preceded by a strong SigmaA-dependent promoter, which contains the consensus sequence TATTAT-N18-TTGACA. Thus, both −10 promoter regions overlap at the opposite strands upstream regions of the divergent gbaA and SACOL2592 genes (Fig. 3A). The GbaA repressor was previously shown to bind to the conserved 9-6-9 bp inverted repeat sequence ATAAACGGA-N6-TCCGTTTGT [31], which overlapped with the TSS-1 and the −10 region upstream of gbaA and with the −10 and −35 promoter elements upstream of SACOL2592 (Fig. 3A–C). Thus, transcription of both operons from the overlapping SigmaA-dependent promoters is strongly repressed by GbaA. The GbaA operator and the perfect −10 promoter elements upstream of gbaA and SACOL2592 are highly conserved across other staphylococci (Fig. 3B and C), indicating that both operons are highly transcribed under the specific inducing conditions.
Fig. 3.
Mapping of the 5′ ends of the gbaAB-SACOL2595-97 and SACOL2592-90 operons and the 9-6-9 bp inverted repeat as GbaA operator in S. aureus (A). 5′ RNA-seq was used to map TSS-1 and TSS-2 upstream of the divergent gbaAB and SACOL2592-90 operons, respectively, which is displayed with Read-Explorer. (B) The promoter sequence of the gbaAB operon and the 9-6-9 bp inverted repeat are highly conserved across different Staphylococcus species. The promoter regions were aligned using Clustal Omega and presented in Jalview. Intensity of the blue color gradient is based on 50% nucleotide sequence identity. (C) The conservation of the gbaA −10 promoter region and the GbaA operator is further displayed with WebLogo. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Cys55 and Cys104 are both important for redox sensing of GbaA in response to oxidants and electrophiles in vivo
TetR/AcrR family repressors are composed of N-terminal helix-turn-helix (HTH) DNA-binding domains (α1-α3 helices) and C-terminal regulatory domains (α4ab-α9) in each subunit of the dimer [[48], [49], [50]]. The C-terminal domain is involved in dimerization and senses specific inducers or ligands, leading to inactivation of the TetR/AcrR repressor activity [[48], [49], [50]]. GbaA shares the two conserved Cys55 and Cys104 residues with GbaA homologs across staphylococci and TetR/AcrR homologs in other Gram-positive bacteria (Fig. S1) [32]. The structural model of GbaA, which is based on the template of E. coli AcrR, suggests that Cys55 is located in the α4a domain close to the HTH motif, while Cys104 is in helix α6 of the C-terminal regulatory domain (Fig. S2) [[48], [49], [50]]. The distance of Cys55 and Cys104 in each subunit was calculated as ∼8.6 Å in this model, indicating that intramolecular disulfide formation will be possible as revealed previously [32].
To examine the function of the two Cys residues for DNA binding activity and redox-sensing of GbaA, the gbaA mutant was complemented with plasmid-encoded His-tagged gbaA, gbaAC55S and gbaAC104S alleles, expressed under a xylose-inducible promoter. Similar expression of GbaA and Cys mutant proteins in the complemented strains was verified by Western blot analyses using anti-His6 tag monoclonal antibodies (see section 3.5). Northern blots were used to study the transcriptional response of GbaA and the Cys mutants under oxidative and electrophile stress (Fig. 4A–D; Figs. S6A–D). Complementation of the gbaA mutant with gbaA and its Cys mutant alleles restored the repression of the gbaAB-SACOL2595-97 and SACOL2592-nmrA-2590 operons under control conditions, indicating that the Cys mutations do not affect the DNA binding activity of GbaA (Fig. 4A–D; Figs. S6A–D). In addition, the gbaAB-SACOL2595-97 operon was significantly 20-50-fold induced in the gbaA complemented strain, but non-significantly changed in the gbaAC55S and gbaAC104S mutants under AGXX®, diamide and NEM stress, indicating that both Cys residues are involved in redox sensing under disulfide and electrophile stress in vivo (Fig. 4A and B; Figs. S6A and B). In contrast to the gbaAB operon, the SACOL2592-nmrA-2590 operon was not significantly up-regulated in the gbaA complemented strain and no transcript visible in the gbaAC55S and gbaAC104S mutants (Fig. 4C and D; Figs. S6C and D). The non-significant thiol-stress induction of the SACOL2592-nmrA-2590 operon in the gbaA complemented strain is in agreement to the WT results (Fig. 2C, Fig. S5A). Together, these transcriptional results on the Cys55 and Cys104 mutants support that both Cys residues function in redox sensing of the GbaA repressor in vivo.
3.4. DNA binding activity of GbaA and the Cys mutants is not impaired under oxidative and MG stress in vitro
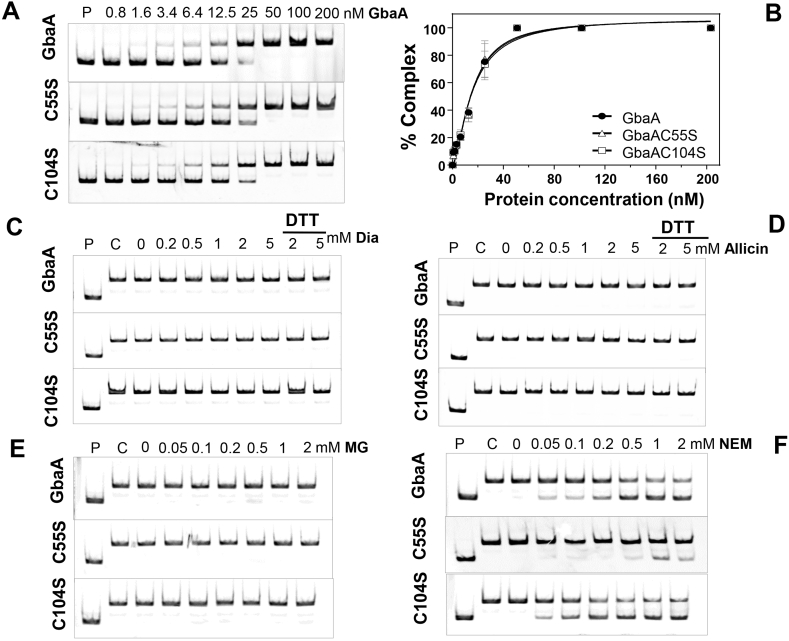
Next, gel-shift assays were used to study the effect of thiol-reactive compounds on DNA binding activity of purified GbaA and GbaA Cys mutant proteins to the gbaA promoter probe, which covered the −83 to +67 upstream region relative to TSS-1 (Fig. 3A and B). GbaA was shown to bind to the GbaA operator with a dissociation constant (KD) of 15.24 nM (Fig. 5A and B). Both GbaAC55S and GbaAC104S mutant proteins showed similar KD values, indicating that the Cys mutations do not affect the DNA binding activity of GbaA in vitro (Fig. 5A and B).
Fig. 5.
The DNA binding activity of GbaA and the Cys mutant proteins is not inhibited under disulfide stress (diamide, allicin) and MG, but partially affected by NEM in vitro. (A) EMSAs were used to analyze the DNA binding activity of increasing concentrations of GbaA, GbaAC55S and GbaAC104S proteins to the 150 bp gbaA promoter probe. (B) The percentage of the GbaA-DNA complex formation was determined according to the band intensities of five biological replicates of the EMSAs in A) and quantified using Image J 1.48v. Dissociation constants (KD) were calculated as 15.24 nM, 15.24 nM and 15.79 nM for GbaA, GbaAC55S and GbaAC104S mutant proteins, respectively using the Graph prism software version 6.01. (C-F) The DNA binding activity of GbaA, GbaAC55S and GbaAC104S proteins was not affected by diamide, allicin and MG (C–E), but partially inhibited with NEM (F).
However, treatment of GbaA and the Cys mutants with diamide, allicin and the electrophile MG did not lead to dissociation of the proteins from the operator DNA in gel-shift assays in vitro (Fig. 5C–E). Thus, the oxidants and electrophiles do not cause major structural changes in the DNA binding domains of GbaA. This suggests that the second unknown regulator is responsive to thiol-stress conditions, while GbaA binds an unknown thiol-reactive compound as ligand. In contrast, exposure of GbaA and the GbaAC104S mutant protein to NEM resulted in partial relief from DNA binding, while the GbaAC55S mutant could be inactivated only weakly with NEM in vitro (Fig. 5F) [32]. These results confirm previous data [32], that GbaA responds only partially to NEM, but is not inactivated under disulfide and MG stress in vitro.
3.5. GbaA and the Cys mutants are oxidized to different thiol switches under diamide and allicin stress
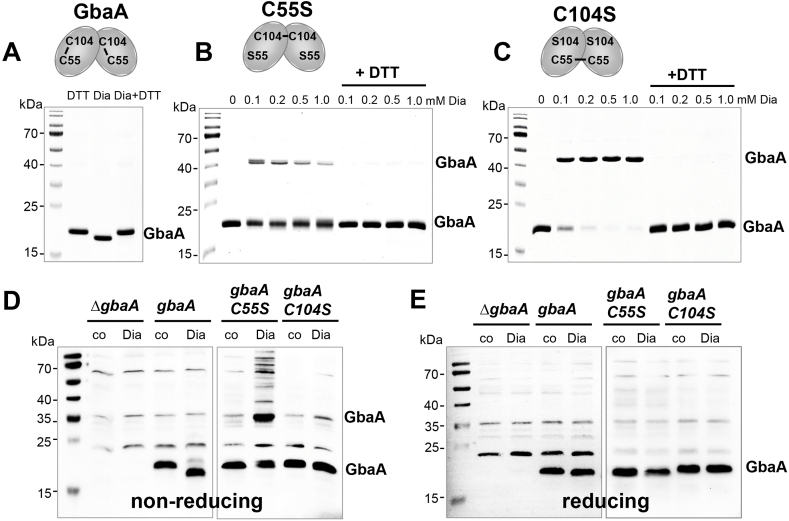
Non-reducing SDS-PAGE and MALDI-TOF-MS were used to monitor thiol-oxidation of GbaA and the Cys mutants after diamide and allicin stress in vitro (Fig. 6A–C, Fig. 7A–C, Fig. S7). The GbaA protein showed a slightly faster migration after diamide treatment compared to DTT-reduced GbaA, indicating the formation of an intramolecular disulfide between Cys55 and Cys104 in each subunit of the dimer (Fig. 6A). The intramolecular cross-link between Cys55 and Cys104 in the diamide-treated sample was confirmed by MALDI-TOF-MS, showing the corresponding mass peak of m/z= 2530.22 Da in the MS1 spectrum (Fig. S7C). In contrast, the diamide-treated GbaAC104S mutant was oxidized to the Cys55-Cys55′ disulfide-linked dimer, which migrates at the size of ∼40 kDa (Fig. 6C). In addition, a small fraction of the GbaAC55S mutant formed weakly intermolecular Cys104-Cys104’ disulfides, while the majority of the protein was not oxidized to the disulfide-linked dimer (Fig. 6B). These results demonstrate that GbaA responds to diamide by intramolecular disulfides, whereas the single Cys mutants are oxidized to intermolecular disulfides between both subunits. The weaker oxidation of the GbaAC55S mutant to intermolecular disulfides might indicate that the Cys104 residues in both subunits are less accessible for disulfide formation in vitro.
Fig. 6.
GbaA and the GbaA Cys mutants are oxidized to intra- and intermolecular disulfides by diamide in vitro and in vivo, respectively. (A–C) Purified GbaA was treated with 1 mM diamide (A), while the GbaAC55S (B) and GbaAC104S mutant proteins (C) were exposed to increasing concentrations of diamide for 15 min, followed by alkylation with 50 mM IAM for 30 min in the dark and separation by non-reducing SDS-PAGE. The non-reducing SDS-PAGE gels are stained with Coomassie Blue. To assess the reversibility, diamide-treated samples were reduced with 20 mM DTT for 15 min before alkylation and analysis by non-reducing SDS-PAGE. GbaA is oxidized to intramolecular C55-C104 disulfides by diamide as confirmed by MALDI-TOF MS (Fig. S7), while the C55S and C104S mutants form intermolecular disulfides as shown in the schematics above the gel images. (D, E) The S. aureus gbaA mutant and the gbaA complemented strain were treated with 5 mM diamide and the gbaAC55S and gbaAC104S complemented strains were exposed to 2 mM diamide for 30 min, alkylated with NEM and the protein extracts analyzed for thiol-oxidation of GbaA in vivo by non-reducing (D) and reducing (E) Western blot analysis with monoclonal anti-His6 tag antibodies. The protein loading controls are shown in Fig. S8.
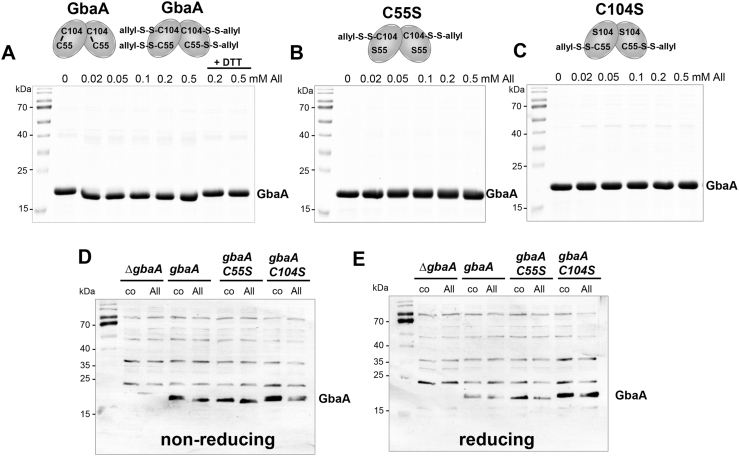
Fig. 7.
GbaA is oxidized to intramolecular disulfides and S-thioallylations by allicin in vitro. (A–C) Purified GbaA (A), GbaAC55S (B) and GbaAC104S mutant proteins (C) were treated with increasing concentrations of allicin for 15 min, followed by alkylation with 50 mM IAM for 30 min in the dark and separation by non-reducing SDS-PAGE. The non-reducing SDS-PAGE gels are stained with Coomassie Blue. For the analysis of reversibility, allicin-treated samples were reduced by 20 mM DTT for 15 min, alkylated and subjected to non-reducing SDS-PAGE. GbaA was oxidized to intramolecular disulfides and S-thioallylations. The GbaA Cys mutants are S-thioallylated under allicin stress as revealed by MALDI-TOF MS (Fig. S7) and shown in the schematics above the images. (D, E) The S. aureus gbaA mutant and gbaA, gbaAC55S and gbaAC104S complemented strains were treated with 0.3 mM allicin stress for 30 min, alkylated with NEM and the protein extracts were used to analyze thiol-oxidation of GbaA in vivo by non-reducing (D) and reducing (E) Western blot analysis with monoclonal anti-His6 tag antibodies. The protein loading controls are shown in Fig. S8.
To analyze thiol-oxidation of GbaA and its Cys mutants under diamide stress in vivo, cell extracts from the S. aureus gbaA mutant and the gbaA, gbaAC55S and gbaAC104 complemented strains were subjected to non-reducing anti-His-tag Western blot analysis (Fig. 6D). GbaA was oxidized to Cys55-Cys104 intramolecular disulfides by diamide as shown by the slower mobility band compared to reduced GbaA under control conditions. While the gbaAC55S mutant was strongly oxidized to Cys104-Cys104’ intermolecular disulfides, the gbaAC104S mutant did not form intermolecular disulfides (Fig. 6D). The Cys mutant results are in contrast to the in vitro disulfide data, but confirm that Cys104 is the more reactive and redox-sensing Cys in vivo. The reversibility of the intra- and intermolecular thiol switches in GbaA and GbaAC55S proteins was shown in the reducing Western blot analyses with DTT (Fig. 6E).
We further analyzed possible thiol-modifications of GbaA and the Cys mutants after allicin treatment (Fig. 7A–E). Allicin treatment of GbaA protein resulted in a slightly faster mobility, which might indicate intramolecular disulfide formation (Fig. 7A). The same slight mobility shift was also observed in the S. aureus gbaA strain in vivo (Fig. 7D). However, no intermolecular disulfide was detected in the monothiol GbaAC55S and GbaAC104S mutants after allicin exposure in vitro or in vivo (Fig. 7B–D). Since allicin causes S-thioallylation of protein thiols, the allicin-treated GbaA and the Cys mutant proteins were subjected to MALDI-TOF-MS (Fig. S7). Interestingly, allicin caused formation of the intramolecular C55-C104 disulfide peptide (m/z=2530.31) and S-thioallylations of the Cys55 and Cys104 peptides with mass shifts of 72 Da (m/z=1331.63 Da and m/z=1345.65 Da) (Fig. S7). In conclusion, our data support that GbaA is oxidized to intramolecular disulfides by diamide, while allicin causes S-thioallylation and intramolecular disulfides, which, however, does not affect the DNA binding activity of the GbaA repressor in vitro.
3.6. GbaA controls defense mechanisms against oxidative and electrophile stress
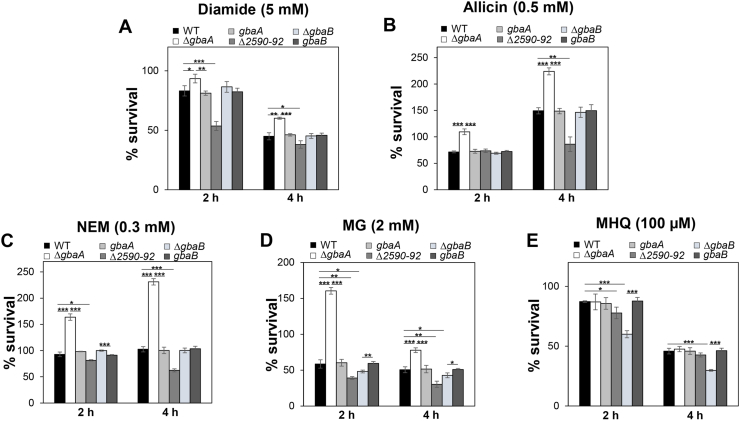
GbaA was shown to regulate two short chain dehydrogenases/oxidoreductases, GbaB and NmrA, and the putative glyoxalase (SACOL2590), which could be involved in the defense against oxidative and electrophile stress in S. aureus. The putative glyoxalase might be involved in MG detoxification in S. aureus. Since the GbaA regulon is induced by diamide, allicin, MG and NEM, we analyzed the survival phenotypes of the gbaA, gbaB and SACOL2592-nmrA-SACOL2590 deletion mutants under these thiol-stress conditions.
The survival assays revealed that the S. aureus COL gbaA mutant was significantly more resistant under diamide, allicin, MG and NEM stress as compared to the WT (Fig. 8A–E). While the % survival rate of the gbaA mutant was 1.5–2.7-fold increased with 0.5 mM allicin, 0.3 mM NEM and 2 mM MG compared to the WT, no significantly enhanced tolerance towards MHQ stress could be determined in the absence of gbaA (Fig. 8A–E). This enhanced survival of the gbaA mutant under diamide, allicin, NEM and MG could be reversed to the WT level in the gbaA complemented strain. In addition, both GbaA Cys mutants showed a significantly 1.6–3.4 -fold decreased survival after 4h of treatment with NEM and MG in relation to the gbaA complemented strain, while no difference in viability was measured with allicin (Fig. 9A–C). These results support the role of the Cys55 and Cys104 residues of the GbaA repressor in the control of electrophile resistance. To clarify the involvement of the GbaA-regulon genes in stress tolerance, the survival of the SACOL2590-92 and gbaB mutants was investigated (Fig. 8A–E). The SACOL2590-92 mutant was significantly impaired in viability after exposure to diamide, allicin, MG and NEM compared to the WT, whereas the gbaB mutant showed only a slight survival defect under MG and MHQ stress (Fig. 8A–E). However, we did not observed growth phenotypes of the gbaA and SACOL2590-92 mutants in response to these high concentrations of allicin, MG and NEM in comparison to the WT (Fig. S9). Overall, the survival results support that GbaA confers tolerance under disulfide and electrophile stress in S. aureus via control of the upstream SACOL2592-nmrA-SACOL2590 operon, which might be involved in allicin, diamide, MG and NEM detoxification. Future analyses will be directed to investigate the functions of these hypothetical proteins under oxidative and MG stress in S. aureus.
Fig. 8.
The GbaA regulon confers resistance under disulfide stress (diamide, allicin) and electrophiles (NEM, MG) in S. aureus. For survival assays, S. aureus COL WT, the gbaA, gbaB and SACOL2592-90 deletion mutants and gbaA, gbaB complemented strains were grown in RPMI medium until an OD500 of 0.5 and treated with 5 mM diamide (A), 0.5 mM allicin (B), 0.3 mM NEM (C), 2 mM MG (D) and 100 μM MHQ (E). CFUs were counted after plating 100 μl of serial dilutions onto LB agar plates after 2 and 4 h of stress exposure. The survival of treated cells was normalized to the untreated control, which was set to 100%. The results are from four biological replicates. Error bars indicate the standard deviation (SD) and the statistics was calculated using a Student's unpaired two-tailed t-test. Symbols are: *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001.
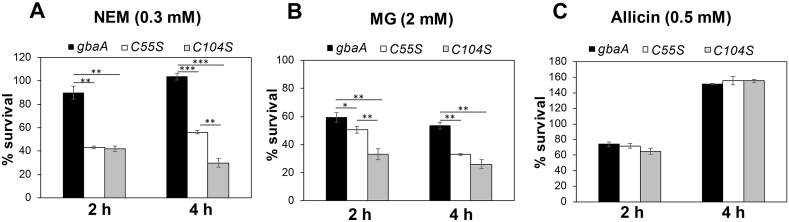
Fig. 9.
The Cys55 and Cys104 residues of GbaA are required for NEM and MG tolerance. Survival assays were performed for the S. aureus gbaA mutant complemented with gbaA, gbaAC55S and gbaAC104S alleles. Strains were grown in RPMI until an OD500 of 0.5 and treated with 0.3 mM NEM (A), 2 mM MG (B) and 0.5 mM allicin (C) to determine CFUs after 2 and 4 h of stress exposure. The percentage survival was normalized to the control. Error bars represent the standard deviation (SD) calculated from three biological replicates. The statistics was determined using a Student's unpaired two-tailed t-test. Symbols are: *p ≤ 0.05, **p ≤ 0.01 and ***p ≤ 0.001.
4. Discussion
The TetR family GbaA repressor controls the SACOL2592-nmrA-2590 and gbaAB-SACOL2595-97 operons [30,31]. Biochemical studies revealed that GbaA functions as monothiol redox sensor, which senses electrophiles via one of its two Cys residues in the single Cys mutants, while the intramolecular disulfide of GbaA did not play a regulatory role [32]. In this work, we investigated the regulation and function of GbaA and its Cys mutants in S. aureus COL under various thiol-stress conditions. Northern blot results revealed that the gbaAB-SACOL2595-97 operon is only weakly 3-10-fold induced under oxidative, electrophile and antibiotics stress in S. aureus COL (Fig. 1; Figs. S4A,B,D), which is far below the high level of 57-734-fold derepression as observed in the transcriptome of the gbaA deletion mutant and in the Northern blot analyses (Fig. 1B; Fig. 2C and D; Tables S3–S4). Moreover, induction of the SACOL2592-nmrA-2590 operon under these thiol-stress conditions was not visible in S. aureus COL using Northern blots, although this upstream operon is clearly regulated by GbaA (Fig. 2A–C).
Based on these findings, we conclude that GbaA does not sense directly any of these thiol-reactive compounds, including AGXX®, diamide, allicin, NEM and MG, which lead only to a weak inactivation of GbaA in vivo. The impact of the gbaA deletion and GbaA Cys mutants on transcriptional regulation of the gbaAB-SACOL2595-97 operon was studied under diamide, AGXX®, allicin, NEM and MG stress in vivo. Here, we made the surprising observation that the gbaA mutant still responds strongly to thiol-stress conditions, such as diamide, AGXX®, NEM and MG, indicating that regulation of the GbaA-controlled operons is more complex and involves another yet unknown (co)regulator. The transcriptional analyses suggest that inactivation of the GbaA repressor is the prerequisite for much faster inactivation of the secondary regulator under thiol-stress conditions, as requirement for full derepression of the gbaAB-SACOL2595-97 operon (Fig. 4A–D).
Using 5’ RNA-seq, TSS-1 and TSS-2 of the divergent transcripts were mapped at the opposing strands, respectively. Since both operons are transcribed from strong SigmaA-dependent promoters, which overlap at both strands in the −10 region, we hypothesize that the secondary regulator might represent another transcriptional repressor. One scenario could be that the primary regulator GbaA requires a specific ligand for inactivation as shown for other TetR-family regulators [49,51]. The secondary regulator likely senses thiol-stress conditions only, as shown by the full derepression of the gbaAB-SACOL2595-97 operon in the gbaA mutant under various disulfide and electrophile stress conditions. The physiological inducers of GbaA could be also combinations of electrophiles, antibiotics or oxidants.
GbaA belongs to the TetR/AcrR family of transcriptional regulators, consisting of a DNA binding HTH motif and a regulatory core domain, which is responsible for dimerization and interacts with different ligands, such as tetracycline and multiple antibiotics, disinfectants and other toxic compounds [48,49,51]. TetR/AcrR family regulators control various functions, including resistance to multiple antibiotics, catabolism of organic compounds, lipid metabolism, iron homeostasis, osmotic stress and virulence functions [48,49,51]. A main feature of the E. coli AcrR structure is the presence of a large cavity in the ligand binding pocket, which was shown to accommodate many different ligands, such as ethidium bromide, proflavin, and rhodamine 6G and ciprofloxacin to inhibit DNA binding activity [49,52]. Similarly, the multidrug efflux pump regulator QacR of S. aureus responds to many cationic lipophilic antiseptics and disinfectants, such as rhodamine 6G, crystal violet, palmatine, nitidine as well as antimicrobial plant alkaloids [[53], [54], [55]]. However, QacR and AcrR control multidrug resistance via their specific efflux pumps, which is not the case for GbaA. Thus, GbaA might be inactivated by specific thiol-reactive compounds or metabolites, which bind to the ligand pocket, leading to oxidation of GbaA and a second redox regulator to induce the upstream and downstream operons.
Transcriptional analyses further revealed that both GbaA single Cys mutants showed non-significant induction of the gbaAB-SACOL2595-97 operon under diamide, AGXX® and NEM stress as compared to the gbaA complemented strain (Fig. 4A and B; Figs. S6A and B). In addition, the Cys55 and Cys104 mutants showed decreased resistance under NEM and MG stress (Fig. 9), indicating that both Cys55 and Cys104 are required for redox sensing in vivo, supporting previous findings [32]. In previous biochemical studies, Cys104 was shown to be more reactive towards electrophiles compared to Cys55 [32], which is in line with the abolished transcription of the gbaAB-SACOL2595-97 operon and the increased susceptibility of the Cys104 mutant after NEM and MG stress compared to the Cys55 mutant (Fig. 4B; Fig. S6B; Fig. 9). While diamide leads to formation of the Cys55–Cys104 intramolecular disulfide in GbaA in vivo and in vitro, allicin caused a mix of intramolecular disulfides and S-thioallylation of both Cys residues (Fig. S7). However, the different thiol switches are not sufficient for GbaA inactivation in vitro, which probably explains the weak transcription of the gbaAB-SACOL2595-97 operon under thiol stress conditions in S. aureus COL WT. Thus, our results demonstrate that thiol switches occur in GbaA and the single Cys mutants in vivo, but they do not alter the structure and abolish the DNA binding activity completely, which is in agreement with previous results [32]. However, NEM caused partial inhibition of the DNA binding activity of GbaA and the GbaAC104S mutant in vitro, while the GbaAC55S mutant was less responsive to NEM (Fig. 5F) [32]. Our in vitro results suggest that in the GbaAC104S mutant, Cys55 is more accessible for C55–C55′ intersubunit disulfide formation by diamide or NEM alkylation, while the C104–C104’ disulfide or C104 alkylation are not favored in the GbaAC55S mutant in vitro. We further were unable to detect any effect of MG on the DNA binding activity of GbaA or the GbaA Cys mutants in vitro, perhaps since MG might cross-link amino-acid side chains with cytosine bases [56]. Altogether, our data show that GbaA functions as two-Cys-type redox sensor, which senses disulfide and electrophile stress via both Cys residues in vivo. However, an unknown thiol-reactive ligand and an additional redox-sensing (co)regulator are required for full derepression of the GbaA regulon, which are subjects of our future studies.
The phenotype analyses of GbaA regulon mutants support that strong oxidants or electrophiles could serve as physiological ligands and are perhaps detoxified by GbaA-controlled genes. The gbaA mutant was resistant to diamide, allicin, NEM and MG, while the deletion of the upstream SACOL2592-nmrA-2590 operon enhanced the susceptibility of S. aureus towards oxidants and electrophiles (Fig. 8). However, deletion of gbaB did not confer sensitivity to diamide, allicin and NEM, while the gbaB mutant was more sensitive to quinones and MG. The sensitivity of the SACOL2592-nmrA-2590 deletion mutant under MG stress is intriguing, since SACOL2590 encodes a glyoxalase enzyme, which could be involved in detoxification of MG. MG is a toxic α, β-unsaturated dicarbonyl compound, which is generated as a byproduct of glycolysis [[57], [58], [59]]. In B. subtilis, MG detoxification involves a bacillithiol-dependent glyoxalase pathway, consisting of the glyoxalase-I (GlxA) generating S-lactoyl bacillithiol, which is hydrolyzed by the glyoxalase-II (GlxB) to lactate [59,60]. The glyoxalase encoded by SACOL2590 belongs to the vicinal oxygen chelate (VOC) family of enzymes, which includes glyoxalase-I enzymes involved in the first step of MG detoxification.
In addition, NmrA and GbaB are both annotated as NAD(P)+-dependent oxidoreductases/short chain dehydrogenases (SDR). SDR enzymes were shown to catalyze oxidation-reduction reactions of various compounds, such as aldehydes, steroids, alcohols, sugars, xenobiotics and aromatic compounds using NAD(P)+ or NADP(H) cofactors [58,60]. Increasing intracellular NADH concentrations were previously determined in the gbaA mutant, suggesting that NmrA or GbaB might catalyze the oxidation of an electrophilic metabolite leading to NADH production [30]. However, since NmrA is lacking the essential tyrosine in the YxxxK active-site motif, it was suggested to function rather as regulator of the NAD(P)+/NADP(H) redox balance [61]. Future analyses will reveal the roles of the glyoxalase and SDR/oxidoreductases in detoxification of MG, allicin, diamide and unknown thiol-reactive metabolites to maintain the cellular redox homeostasis.
Similarly, the redox-sensitive TetR-family regulator NemR of E. coli was shown to sense oxidants and electrophiles, such as HOCl, NEM and MG [58,62,63]. NemR contains 6 cysteine residues and was inactivated by intermolecular disulfides, resulting in induction of the NEM reductase NemA and the glyoxalase I (GloA) to confer resistance under HOCl and MG stress in E. coli [63]. While there are functional links to MG detoxification between NemR of E. coli and GbaA of S. aureus, the regulation of GbaA is far more complex, since the physiological inducer and the additional redox-sensitive (co)regulator are unknown and both are required for full derepression of the GbaA regulon genes. Future studies will be directed to investigate combinations of thiol-reactive antimicrobials as inducers and to utilize gbaA promoter mutations to shed light on the genetic basis for full derepression.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgements
This work was supported by an European Research Council (ERC) Consolidator grant (GA 615585) MYCOTHIOLOME and grants from the Deutsche Forschungsgemeinschaft (AN746/4-1 and AN746/4-2) within the SPP1710 on “Thiol-based Redox switches”, by the SFB973 project C08 and TR84 project B06 to H.A. For mass spectrometry (C.W.) we would like to acknowledge the assistance of the Core Facility BioSupraMol supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.freeradbiomed.2021.10.024.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Foster T.J. The Staphylococcus aureus "superbug. J. Clin. Invest. 2004;114(12):1693–1696. doi: 10.1172/JCI23825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Boucher H.W., Corey G.R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 2008;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 4.Archer G.L. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 1998;26(5):1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 5.Chambers H.F., Deleo F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7(9):629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vestergaard M., Frees D., Ingmer H. Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 2019;7(2) doi: 10.1128/microbiolspec.gpp3-0057-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linzner N., Loi V.V., Fritsch V.N., Antelmann H. Thiol-based redox switches in the major pathogen Staphylococcus aureus. Biol. Chem. 2021;402(3):333–361. doi: 10.1515/hsz-2020-0272. [DOI] [PubMed] [Google Scholar]
- 8.Ulfig A., Leichert L.I. Cell Mol Life Sci; 2020. The Effects of Neutrophil-Generated Hypochlorous Acid and Other Hypohalous Acids on Host and Pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winterbourn C.C., Kettle A.J. Redox reactions and microbial killing in the neutrophil phagosome. Antioxidants Redox Signal. 2013;18(6):642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 10.Winterbourn C.C., Kettle A.J., Hampton M.B. Reactive oxygen species and neutrophil function. Annu. Rev. Biochem. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 11.Marnett L.J., Riggins J.N., West J.D. Endogenous generation of reactive oxidants and electrophiles and their reactions with DNA and protein. J. Clin. Invest. 2003;111(5):583–593. doi: 10.1172/JCI18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs A.T., Marnett L.J. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc. Chem. Res. 2010;43(5):673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmastro-Greenwood M., Freeman B.A., Wendell S.G. Redox-dependent anti-inflammatory signaling actions of unsaturated fatty acids. Annu. Rev. Physiol. 2014;76:79–105. doi: 10.1146/annurev-physiol-021113-170341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vetter S.W. Glycated serum albumin and AGE receptors. Adv. Clin. Chem. 2015;72:205–275. doi: 10.1016/bs.acc.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Hazen S.L., d'Avignon A., Anderson M.M., Hsu F.F., Heinecke J.W. Human neutrophils employ the myeloperoxidase-hydrogen peroxide-chloride system to oxidize alpha-amino acids to a family of reactive aldehydes. Mechanistic studies identifying labile intermediates along the reaction pathway. J. Biol. Chem. 1998;273(9):4997–5005. doi: 10.1074/jbc.273.9.4997. [DOI] [PubMed] [Google Scholar]
- 16.Riquelme S.A., Liimatta K., Wong Fok Lung T., Fields B., Ahn D., Chen D., Lozano C., Saenz Y., Uhlemann A.C., Kahl B.C., Britto C.J., DiMango E., Prince A. Pseudomonas aeruginosa utilizes host-derived itaconate to redirect its metabolism to promote biofilm formation. Cell Metabol. 2020;31(6):1091–1106 e6. doi: 10.1016/j.cmet.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomlinson K.L., Lung T.W.F., Dach F., Annavajhala M.K., Gabryszewski S.J., Groves R.A., Drikic M., Francoeur N.J., Sridhar S.H., Smith M.L., Khanal S., Britto C.J., Sebra R., Lewis I., Uhlemann A.C., Kahl B.C., Prince A.S., Riquelme S.A. Staphylococcus aureus induces an itaconate-dominated immunometabolic response that drives biofilm formation. Nat. Commun. 2021;12(1):1399. doi: 10.1038/s41467-021-21718-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antelmann H., Helmann J.D. Thiol-based redox switches and gene regulation. Antioxidants Redox Signal. 2011;14(6):1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillion M., Antelmann H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015;396(5):415–444. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji Q., Zhang L., Jones M.B., Sun F., Deng X., Liang H., Cho H., Brugarolas P., Gao Y.N., Peterson S.N., Lan L., Bae T., He C. Molecular mechanism of quinone signaling mediated through S-quinonization of a YodB family repressor QsrR. Proc. Natl. Acad. Sci. U. S. A. 2013;110(13):5010–5015. doi: 10.1073/pnas.1219446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loi V.V., Busche T., Tedin K., Bernhardt J., Wollenhaupt J., Huyen N.T.T., Weise C., Kalinowski J., Wahl M.C., Fulde M., Antelmann H. Redox-sensing under hypochlorite stress and infection conditions by the Rrf2-family repressor HypR in Staphylococcus aureus. Antioxidants Redox Signal. 2018;29(7):615–636. doi: 10.1089/ars.2017.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P.R., Nishida S., Poor C.B., Cheng A., Bae T., Kuechenmeister L., Dunman P.M., Missiakas D., He C. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 2009;71(1):198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poor C.B., Chen P.R., Duguid E., Rice P.A., He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 2009;284(35):23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen P.R., Bae T., Williams W.A., Duguid E.M., Rice P.A., Schneewind O., He C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2006;2(11):591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.W., Soonsanga S., Helmann J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U. S. A. 2007;104(21):8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi B.K., Gronau K., Mäder U., Hessling B., Becher D., Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics. 2011;10(11) doi: 10.1074/mcp.M111.009506. M111 009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mongkolsuk S., Helmann J.D. Regulation of inducible peroxide stress responses. Mol. Microbiol. 2002;45(1):9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 28.Chi B.K., Albrecht D., Gronau K., Becher D., Hecker M., Antelmann H. The redox-sensing regulator YodB senses quinones and diamide via a thiol-disulfide switch in Bacillus subtilis. Proteomics. 2010;10(17):3155–3164. doi: 10.1002/pmic.201000230. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.J., Lee I.G., Lee K.Y., Kim D.G., Eun H.J., Yoon H.J., Chae S., Song S.H., Kang S.O., Seo M.D., Kim H.S., Park S.J., Lee B.J. Two distinct mechanisms of transcriptional regulation by the redox sensor YodB. Proc. Natl. Acad. Sci. U. S. A. 2016;113(35):E5202–E5211. doi: 10.1073/pnas.1604427113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You Y., Xue T., Cao L., Zhao L., Sun H., Sun B. Staphylococcus aureus glucose-induced biofilm accessory proteins, GbaAB, influence biofilm formation in a PIA-dependent manner. Int J Med Microbiol. 2014;304(5–6):603–612. doi: 10.1016/j.ijmm.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Yu L., Hisatsune J., Hayashi I., Tatsukawa N., Sato'o Y., Mizumachi E., Kato F., Hirakawa H., Pier G.B., Sugai M. A novel repressor of the ica Locus discovered in clinically isolated super-biofilm-elaborating Staphylococcus aureus. mBio. 2017;8(1) doi: 10.1128/mBio.02282-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray A., Edmonds K.A., Palmer L.D., Skaar E.P., Giedroc D.P. Staphylococcus aureus glucose-induced biofilm accessory protein A (GbaA) is a monothiol-dependent electrophile sensor. Biochemistry. 2020;59(31):2882–2895. doi: 10.1021/acs.biochem.0c00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritsch V.N., Loi V.V., Busche T., Sommer A., Tedin K., Nürnberg D.J., Kalinowski J., Bernhardt J., Fulde M., Antelmann H. The MarR-type repressor MhqR confers quinone and antimicrobial resistance in Staphylococcus aureus. Antioxidants Redox Signal. 2019;31(16):1235–1252. doi: 10.1089/ars.2019.7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loi V.V., Busche T., Preuss T., Kalinowski J., Bernhardt J., Antelmann H. The AGXX antimicrobial coating causes a thiol-specific oxidative stress response and protein S-bacillithiolation in Staphylococcus aureus. Front. Microbiol. 2018;9:3037. doi: 10.3389/fmicb.2018.03037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loi V.V., Huyen N.T.T., Busche T., Tung Q.N., Gruhlke M.C.H., Kalinowski J., Bernhardt J., Slusarenko A.J., Antelmann H. Staphylococcus aureus responds to allicin by global S-thioallylation - role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress. Free Radic. Biol. Med. 2019;139:55–69. doi: 10.1016/j.freeradbiomed.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Linzner N., Fritsch V.N., Busche T., Tung Q.N., Van Loi V., Bernhardt J., Kalinowski J., Antelmann H. The plant-derived naphthoquinone lapachol causes an oxidative stress response in Staphylococcus aureus. Free Radic. Biol. Med. 2020;158:126–136. doi: 10.1016/j.freeradbiomed.2020.07.025. [DOI] [PubMed] [Google Scholar]
- 37.Loi V.V., Harms M., Müller M., Huyen N.T.T., Hamilton C.J., Hochgräfe F., Pane-Farre J., Antelmann H. Real-time imaging of the bacillithiol redox potential in the human pathogen Staphylococcus aureus using a genetically encoded bacilliredoxin-fused redox biosensor. Antioxidants Redox Signal. 2017;26(15):835–848. doi: 10.1089/ars.2016.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnaud M., Chastanet A., Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70(11):6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wetzstein M., Volker U., Dedio J., Lobau S., Zuber U., Schiesswohl M., Herget C., Hecker M., Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 1992;174(10):3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tam le T., Eymann C., Albrecht D., Sietmann R., Schauer F., Hecker M., Antelmann H. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ. Microbiol. 2006;8(8):1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 41.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilker R., Stadermann K.B., Schwengers O., Anisiforov E., Jaenicke S., Weisshaar B., Zimmermann T., Goesmann A. ReadXplorer 2-detailed read mapping analysis and visualization from one single source. Bioinformatics. 2016;32(24):3702–3708. doi: 10.1093/bioinformatics/btw541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeifer-Sancar K., Mentz A., Ruckert C., Kalinowski J. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genom. 2013;14:888. doi: 10.1186/1471-2164-14-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chi B.K., Roberts A.A., Huyen T.T., Basell K., Becher D., Albrecht D., Hamilton C.J., Antelmann H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxidants Redox Signal. 2013;18(11):1273–1295. doi: 10.1089/ars.2012.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suckau D., Resemann A., Schuerenberg M., Hufnagel P., Franzen J., Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003;376(7):952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 47.Mäder U., Nicolas P., Depke M., Pane-Farre J., Debarbouille M., van der Kooi-Pol M.M., Guerin C., Derozier S., Hiron A., Jarmer H., Leduc A., Michalik S., Reilman E., Schaffer M., Schmidt F., Bessieres P., Noirot P., Hecker M., Msadek T., Völker U., van Dijl J.M. Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet. 2016;12(4) doi: 10.1371/journal.pgen.1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos J.L., Martinez-Bueno M., Molina-Henares A.J., Teran W., Watanabe K., Zhang X., Gallegos M.T., Brennan R., Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69(2):326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng W., Li C., Xie J. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell. Signal. 2013;25(7):1608–1613. doi: 10.1016/j.cellsig.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Li M., Gu R., Su C.C., Routh M.D., Harris K.C., Jewell E.S., McDermott G., Yu E.W. Crystal structure of the transcriptional regulator AcrR from Escherichia coli. J. Mol. Biol. 2007;374(3):591–603. doi: 10.1016/j.jmb.2007.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuthbertson L., Nodwell J.R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 2013;77(3):440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su C.C., Rutherford D.J., Yu E.W. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem. Biophys. Res. Commun. 2007;361(1):85–90. doi: 10.1016/j.bbrc.2007.06.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeuchi K., Imai M., Shimada I. Conformational equilibrium defines the variable induction of the multidrug-binding transcriptional repressor QacR. Proc. Natl. Acad. Sci. U. S. A. 2019;116(40):19963–19972. doi: 10.1073/pnas.1906129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grkovic S., Brown M.H., Roberts N.J., Paulsen I.T., Skurray R.A. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 1998;273(29):18665–18673. doi: 10.1074/jbc.273.29.18665. [DOI] [PubMed] [Google Scholar]
- 55.Grkovic S., Hardie K.M., Brown M.H., Skurray R.A. Interactions of the QacR multidrug-binding protein with structurally diverse ligands: implications for the evolution of the binding pocket. Biochemistry. 2003;42(51):15226–15236. doi: 10.1021/bi035447+. [DOI] [PubMed] [Google Scholar]
- 56.Kalapos M.P. The tandem of free radicals and methylglyoxal. Chem. Biol. Interact. 2008;171(3):251–271. doi: 10.1016/j.cbi.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 57.Ferguson G.P., Totemeyer S., MacLean M.J., Booth I.R. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 1998;170(4):209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 58.Gray M.J., Wholey W.Y., Parker B.W., Kim M., Jakob U. NemR is a bleach-sensing transcription factor. J. Biol. Chem. 2013;288(19):13789–13798. doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandrangsu P., Loi V.V., Antelmann H., Helmann J.D. The role of bacillithiol in Gram-positive firmicutes. Antioxidants Redox Signal. 2018;28(6):445–462. doi: 10.1089/ars.2017.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chandrangsu P., Dusi R., Hamilton C.J., Helmann J.D. Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol. Microbiol. 2014;91(4):706–715. doi: 10.1111/mmi.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stammers D.K., Ren J., Leslie K., Nichols C.E., Lamb H.K., Cocklin S., Dodds A., Hawkins A.R. The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 2001;20(23):6619–6626. doi: 10.1093/emboj/20.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gray M.J., Li Y., Leichert L.I., Xu Z., Jakob U. Does the transcription factor NemR use a regulatory sulfenamide bond to sense bleach? Antioxidants Redox Signal. 2015;23(9):747–754. doi: 10.1089/ars.2015.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee C., Shin J., Park C. Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coli K-12. Mol. Microbiol. 2013;88(2):395–412. doi: 10.1111/mmi.12192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.