Abstract
Nuclear movement is a fundamental process of eukaryotic cell biology. Skeletal muscle presents an intriguing model to study nuclear movement because its development requires the precise positioning of multiple nuclei within a single cytoplasm. Furthermore, there is a high correlation between aberrant nuclear positioning and poor muscle function. Although many genes that regulate nuclear movement have been identified, the mechanisms by which these genes act are not known. Using Drosophila melanogaster muscle development as a model system and a combination of live-embryo microscopy and laser ablation of nuclei, we have found that clustered nuclei encompass at least two phenotypes that are caused by distinct mechanisms. Specifically, Ensconsin is necessary for productive force production to drive any movement of nuclei, whereas Bocksbeutel and Klarsicht are necessary to form distinct populations of nuclei that move to different cellular locations. Mechanistically, Ensconsin regulates the number of growing microtubules that are used to move nuclei, whereas Bocksbeutel and Klarsicht regulate interactions between nuclei.
INTRODUCTION
Since the identification of the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex (Crisp et al., 2006; Starr and Fridolfsson, 2010; Tapley and Starr, 2013), the question of how nuclei move has been a pressing question in biology. The process of moving this heavy organelle is conserved throughout evolution in all cell types (Mosley-Bishop et al., 1999; Starr et al., 2001; Starr and Han, 2002; Tran et al., 2001; Lee et al., 2002; Del Bene et al., 2008; Zhang et al., 2009; Yu et al., 2011), thus magnifying the importance of understanding the underlying mechanism. Although many mechanisms have been described for mononucleated cells (Gundersen and Worman, 2013), how nuclei are moved in a syncytium has remained a mystery. Many genes that regulate nuclear position in syncytial skeletal muscle cells have been identified (Roman and Gomes, 2018), but how these genes contribute to nuclear movement and whether these genes regulate nuclear positioning through a single mechanism is not known.
In most contexts, nuclear movement is dependent on the microtubule cytoskeleton and its associated proteins, which generate the force to move nuclei, and the LINC complex, which transmits force between the cytoskeleton and the nucleus. This is indeed true during the development of the syncytial abdominal musculature of Drosophila melanogaster embryos and larvae. Several microtubule-associated proteins including Ensconsin/MAP7 (Metzger et al., 2012), Bsg25D/Ninein (Rosen et al., 2019), and the motors Kinesin and Cytoplasmic dynein (Folker et al., 2012, 2014) have been suggested to contribute to nuclear movement by regulating Kinesin activity (Metzger et al., 2012), microtubule stability (Rosen et al., 2019), and the application of force both directly on (Folker et al., 2014) and at a distance from (Folker et al., 2012) the nucleus. Similar experiments have shown that the LINC complex genes klarsicht (Elhanany-Tamir et al., 2012; Collins, Mandigo, et al., 2017), Msp300 (Elhanany-Tamir et al., 2012), and klaroid (Tan et al., 2018), along with the emerin homologues bocksbeutel and Otefin (Collins, Mandigo, et al., 2017; Mandigo et al., 2019), are also critical for nuclear positioning during muscle development. Despite identifying many of the factors that are critical for nuclear position, we know little about the mechanisms by which they support nuclear movement during muscle development.
The limited mechanistic understanding is in part driven by the complexity that many nuclei in a single cytoplasm create. Although many studies investigating myonuclear movement have been done in cell culture (Cadot et al., 2012; Wilson and Holzbaur, 2012), such in vitro systems lack the complex signaling cascades that provide directionality cues to nuclei as they translocate, highlighting the importance of studying nuclear movement in an organismal context (Folker et al., 2014). Consequently, most in vivo work has relied on describing nuclei as mispositioned with little, if any, distinction between phenotypes (Elhanany-Tamir et al., 2012; Folker et al., 2012; Metzger et al., 2012; Collins, Mandigo, et al., 2017). To better understand the mechanisms by which each gene regulates nuclear movement, it is critical to establish methods that can characterize nuclear phenotypes in vivo and distinguish between those that appear similar by a basic phenotypic scoring system. Here we describe a new analytical approach centered on live-embryo time-lapse microscopy and careful characterization of nuclear position combined with laser ablation of nuclei to provide the first direct evidence that some factors necessary for nuclear movement are required to apply force to nuclei, whereas other factors are necessary for the utilization of that force to reach a specific position rather than to move.
RESULTS
Disruptions of bocksbeutel and klarsicht have distinct effects on myonuclear positioning compared with that of ensconsin in the Drosophila embryo
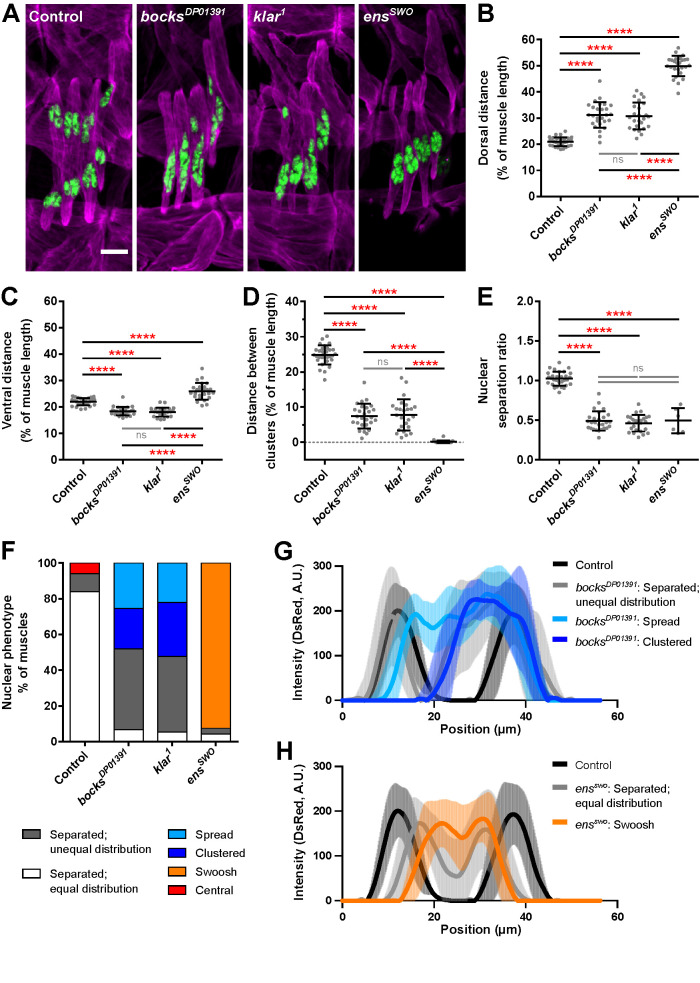
As a first approach, we have investigated the contributions of bocksbeutel (Drosophila emerin), klarsicht (Drosophila KASH-protein), and ensconsin (Drosophila MAP7). Each gene was zygotically removed in Drosophila embryos with the respective bocksDP01391 null (Collins, Mandigo, et al., 2017), klar1 null (Welte et al., 1998), or ensswo nonsense mutation (Metzger et al., 2012) alleles. Fixed images of stage 16 Drosophila embryos showed that in controls, nuclei were in two clusters positioned at either end of the lateral transverse (LT) muscle, whereas in ensswo, bocksDP01391, and klar1 embryos, most of the nuclei were in a single cluster (Figure 1A), as previously shown (Metzger et al., 2012; Collins, Mandigo, et al., 2017). Qualitatively, the nuclear positioning phenotypes in each of these genotypes were similar, and the phenotypes were previously described using very similar language. However, because the proteins encoded by klar and bocks are localized to the nucleus and the protein encoded by ens is localized to the cytoskeleton, we hypothesized that the phenotypes may in fact be distinct. To test this hypothesis, we quantified the precise position and morphology of the nuclei. These analyses mirror our previous characterization of nuclear position in klar and bocks mutants (Collins, Mandigo, et al., 2017), but similar detailed analysis of ens mutants that is necessary to compare these genotypes had not been completed. To quantitatively evaluate myonuclear position, the distance of each nuclear cluster from the dorsal and ventral muscle poles was measured. Because the LT muscles in all three mutants were significantly shorter (Supplemental Figure S1A; statistics summarized in Supplemental Table S1), we measured the raw distance between nuclei and the muscle end (Supplemental Figure S1, B and C) and the distance between nuclei and the muscle end as percent of muscle length (Figure 1, B and C). Compared with controls, nuclei in bocksDP01391 and klar1 embryos were positioned farther from the dorsal muscle pole (Figure 1B; Supplemental Figure S1B) yet closer to the ventral muscle pole (Figure 1C; Supplemental Figure S1C), as previously described (Collins, Mandigo, et al., 2017). However, nuclei in ensswo embryos were positioned in the cell center and significantly farther from both muscle poles when compared with controls or bocksDP01391 and klar1 embryos (Figure 1, B and C, and Supplemental Figure S1, B and C). Additionally, the distance between dorsal and ventral clusters was measured (Figure 1D and Supplemental Figure S1D). The distance between clusters was significantly decreased in bocksDP01391 and klar1 embryos because distinct clusters of nuclei formed in only a small fraction of muscles (Supplemental Figure S1, E and F). In controls, nuclei were evenly distributed between the two clusters. Although a percentage of bocksDP01391 and klar1 muscles (45% and 42%, respectively) had a distinct dorsal and ventral cluster, the distribution of nuclei between these two clusters was not even. Most nuclei were within the ventral region of the muscle, resulting in a much larger ventral cluster compared with control ventral clusters (Supplemental Figure S1H). In contrast, the dorsal cluster was much smaller compared with controls, consisting of just one or two nuclei (Supplemental Figure S1G). This uneven distribution of nuclei in bocksDP01391 and klar1 embryos significantly decreased the nuclear separation ratio. Similarly, in the 4% of ensswo embryos that had two distinct clusters of nuclei, the ventral cluster was larger than the dorsal cluster (Figure 1E). Although the total areas occupied by nuclei were similar between controls, bocksDP01391, and klar1, the area was significantly reduced in ensswo embryos (Supplemental Figure S1I). However, the number of nuclei was the same between controls and ensswo embryos, indicating that fusion is not affected, consistent with previous work (Supplemental Figure S2, A and B) (Metzger et al., 2012). Therefore, the reduced area in ensswo embryos is not attributed to a loss of nuclei. To determine whether nuclei in ensswo embryos are positioned deeper within the muscle, we measured the total volume of nuclei via three-dimensional imaging. The total volume occupied by nuclei was the same as in controls, indicating that the reduced nuclear area in ensswo embryos is due to nuclei occupying a greater depth of the muscle (Supplemental Figure S2, A and C, and Supplemental Movies S1 and S2).
FIGURE 1:
Bocksbeutel, Klarsicht, and Ensconsin regulate myonuclear position in Drosophila embryos. (A) Immunofluorescence images of the LT muscles in one hemisegment from stage 16 (16 h after egg lay [AEL]) embryos for the indicated genotypes. Muscles in magenta, myonuclei in green. Scale bar, 10 µm. (B–D) Graphs indicating the distance between the dorsal end of the muscle and the nearest nucleus (B), the distance between the ventral end of the muscle and the nearest nucleus (C), and the distance between the dorsal and ventral clusters of nuclei (D). All distances were normalized to the muscle length. (E) Relative size of the dorsal cluster of nuclei compared with the ventral cluster of nuclei. It is important to note that in 21 out of the 27 ensswo embryos, there was only one cluster present. Thus, the nuclear separation ratio was calculated only for the six embryos that had two distinct clusters. Data points in B–E correspond to the average value within a single embryo. Error bars indicate the SD from ≥25 embryos for each genotype taken from at least three independent experiments. The one-way analysis of variance (ANOVA) with Tukey honest significant difference (HSD) post hoc test was used to assess the statistical significance of differences in measurements between all experimental groups. Not significant (ns) ≥ 0.05, ****P < 0.0001. (F) Frequency at which each nuclear positioning phenotype was observed in each of the indicated genotypes. (G, H) Averaged linescans of DsRed intensity for each nuclear phenotype observed in bocksDP01391 mutants (G) and ensswo mutants (H) compared with controls. Error bars indicate the SD from 10 LT muscles for each nuclear phenotype. Position correlates to the length of the muscle. The dorsal end position corresponds to 0 μm.
Movie S1.
Volumetric imaging of myonuclei in the lateral transverse muscle of a control Drosophila embryo. Movie of a three‐dimensional volumetric rendering of the dorsal and ventral nuclear clusters within a single LT muscle from a stage 16 (16 hours AEL) control embryo. Muscles in magenta, myonuclei in green. Scale bar, 5 μm. The LT muscle is rotated 360° along the x‐axis and 360° along the y‐axis.
Movie S2.
Volumetric imaging of myonuclei in the lateral transverse muscle of an ensswo mutant embryo. Movie of a three‐dimensional volumetric rendering of the nuclear cluster within a single LT muscle from a stage 16 (16 hours AEL) ensswo embryo. Muscles in magenta, myonuclei in green. Scale bar, 5 μm. The LT muscle is rotated 360° along the x‐axis and 360° along the y‐axis.
Based on these measurements, the most common phenotype observed in control embryos was nuclei that separated into two distinct groups of equal size (Figure 1F, “separated; equal distribution”). The most common phenotype in bocksDP01391 and klar1 embryos was a single cluster of nuclei near the ventral end of the muscle (50% and 52%, respectively) (Figure 1, F and G, “clustered” and “spread”) with a smaller percentage of muscles having a small cluster of dorsal nuclei and a larger cluster of ventral nuclei (Figure 1, F and G, “separated: unequal distribution”). Finally, the most common phenotype observed in ensswo embryos was a single cluster positioned near the center of the muscle (92%) (Figure 1, F and H, “swoosh”). In total, these data indicate that while Bocksbeutel, Klarsicht, and Ensconsin are all required for proper nuclear position as previously shown (Metzger et al., 2012; Collins, Mandigo, et al., 2017), measurements of the precise position of nuclear clusters and the distribution of nuclei between the clusters in each genotype revealed distinct phenotypes that were previously overlooked and distinguish the klar1 and bocksDP01391 phenotypes from the ensswo phenotype. These findings suggest that these genes have mechanistically distinct contributions to nuclear position.
Ensconsin is necessary for nuclear movement, whereas bocksbeutel and klarsicht are necessary to separate nuclei
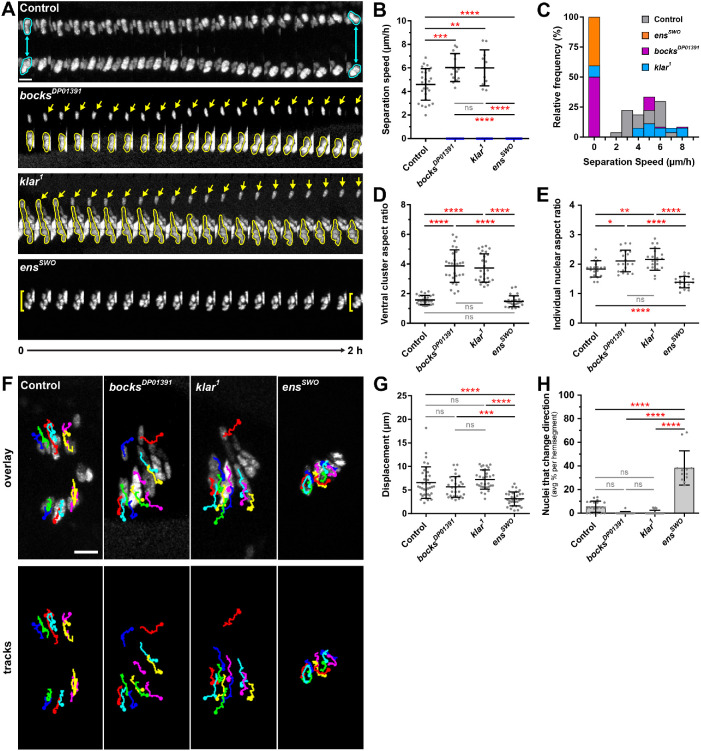
Because the nuclei were positioned differently in klar1 and bocksDP01391 mutants compared with ensswo mutants and the clusters themselves had a different shape, we hypothesized that the dynamics of nuclear movement would be different between these genotypes. To test this hypothesis, the position of nuclear clusters within the LT muscles was tracked over the course of 2 h. In control muscles, once all nuclei separated into two distinct clusters, these clusters migrated toward opposite muscle ends, steadily increasing the distance between themselves (Figure 2A and Supplemental Movie S3, left panel). However, 100% of all nuclei observed in ensswo muscles failed to separate over the time course (Figure 2A, yellow brackets, and Supplemental Movie S6, left panel), significantly reducing the separation speed to 0 μm/h (Figure 2, B and C). Similarly, nuclei that remained associated together in bocksDP01391 and klar1 muscles also failed to separate (Figure 2B, blue data points, and Supplemental Movies S4 and S5, left panels). However, the complete lack of separation was observed only in approximately 50% of muscles (Figure 2C). In the other 50% of muscles, a single nucleus separated and migrated toward the dorsal end of the muscle (Figure 2A, yellow arrows) at a rate slightly faster than in control nuclei (Figure 2B, gray data points). Furthermore, the morphology of the single clusters was different in bocksDP01391 and klar1 compared with ensswo. In bocksDP01391 and klar1 embryos, clustered nuclei were significantly elongated compared with controls. One simple explanation is that because nuclei fail to separate, all the nuclei within the muscle are now present within a single cluster, resulting in its elongated shape. However, in ensswo nuclear clusters as well as individual nuclei were spherical (Figure 2, D and E), despite having the same number of nuclei within single clusters as the klar1 and bocksDP01391 embryos (Supplemental Figure S2, A and B). Thus, these data further demonstrate that while depletion of bocks, klar, and ens results in superficially similar clustering of nuclei, the single clusters in bocksDP01391 and klar1 embryos are phenotypically distinct from those observed in ensswo. The elongated clusters in the klar1 and bocksDP01391 mutants suggested that nuclei in these mutants were under tension and may move, whereas the spherical morphology of single nuclei and resulting nuclear clusters in ensswo mutants suggested that they were not under tension and would therefore not move. Additionally, the stochastic escaper nuclei in klar1 and bocksDP01391 mutants moved rapidly to the proper cellular position, indicating that the machinery to move nuclei and the signaling mechanisms that dictate the direction of nuclear movement are intact. Conversely, the lack of escapers in ensswo mutants suggests a disruption of the machinery necessary to move nuclei.
FIGURE 2:
Bocksbeutel, Klarsicht, and Ensconsin are necessary for the proper separation of myonuclei in Drosophila embryos. (A) Montages from time-lapse acquisitions showing the separation of the dorsal cluster from the ventral cluster of nuclei within a single LT muscle of a stage 15 (15 h AEL) embryo for the indicated genotypes. Nuclei outlined in cyan indicate the proper separation of nuclei into two distinct clusters (control). Yellow arrows indicate an escaper nucleus that separates from the ventral group in either bocksDP01391 or klar1 mutant embryos. Yellow brackets indicate nuclei that fail to separate and remain associated as a single cluster (ensswo). Scale bar, 10 µm. (B) Separation speed of nuclear clusters. Data points correspond to the speed measured from a single LT muscle. Gray data points indicate the speed at which the dorsal and ventral clusters of nuclei separate from one another, whereas blue data points indicate nuclei that failed to separate (speed = 0 µm/h). Error bars indicate the SD from ≥25 LT muscles for each genotype taken from independent experiments. (C) Relative distribution of nuclear separation speeds. (D) Aspect ratio of the ventral nuclear cluster measured at 0 h. Data points correspond to the ventral nuclear cluster within a single LT muscle. Error bars indicate the SD from ≥25 LT muscles for each genotype taken from independent experiments. (E) The aspect ratio of individual nuclei for the indicated genotypes. Data points correspond to a single nucleus. Error bars indicate the SD from 20 myonuclei for each genotype taken from independent experiments. (F) Tracks following the movement of individual nuclei within four LT muscles over the course of 2 h, superimposed over the first frame (t = 0 h). Scale bar, 10 µm. (G) Displacement of individual nuclei. Data points correspond to the displacement of a single nucleus. Error bars indicate the SD from 36 nuclei for each genotype taken from three independent experiments. (H) The number of nuclei that change direction as a percentage for the indicated genotypes. Data points correspond to the average percentage of nuclei that changed direction per hemisegment. Error bars indicate the SD from ≥10 hemisegments for each genotype taken from at least three different embryos. For B, D, E, G, and H, the one-way ANOVA with Tukey HSD post hoc test was used to assess the statistical significance of differences in measurements between all experimental groups. Not significant (ns) ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To test this, the trajectory of individual myonuclei within each cluster was tracked over the 2 h time course (Figure 2F). The total displacement of nuclei in bocksDP01391 and klar1 embryos was similar to that in controls, even in ventral clusters where more nuclei were present (Figure 2G and Supplemental Movies S3–S5, right panels). Although the displacement was similar to that in controls, aside from the occasional escaper, all of the nuclei within the cluster moved ventrally, suggesting that the interactions between nuclei within a cluster are restricting the movement toward the dorsal end of the muscle. The ventral movement of nuclei also suggests that the signaling cues that inform nuclei where to move are still present. This conclusion is further supported by the observation that in the event a nucleus does escape from the ventral cluster, it migrates to the proper dorsal position. Conversely, the displacement of nuclei in ensswo embryos was significantly decreased compared with that in controls, as nuclei rotated within the cluster, randomly changing directions, but did not translocate (Figure 2, G and H, and Supplemental Movie S6, right panel). Together these data suggest that in ensswo mutants, the ability of the cell to exert force on nuclei to resolve the single cluster into two distinct clusters is compromised. However, the movement of the nuclei in klar1 and bocksDP01391 suggests that force production is normal and that instead nuclei are being actively maintained in a single cluster.
Movie S3.
Nuclear migration in the lateral transverse muscle of a control Drosophila embryo. Time‐lapse acquisition showing the migration of myonuclei within four lateral transverse (LT) muscles of a control embryo. Tracks correspond to the movement of individual nuclei within each cluster over the course of two hours. Time‐lapse starts at stage 15 (15 hours AEL, t = 0 min), when nuclei have already separated into two distinct clusters. Each LT muscle has one dorsal cluster and one ventral cluster that migrate directionally to opposite ends of the muscle. At stage 16 (16 hours AEL), the dorsal and ventral clusters have reached their respective muscle pole, maximizing the distance between them. Scale bar, 10 μm.
Movie S4.
Altered nuclear migration in the lateral transverse muscle of a bocksDP01391 mutant embryo. Time‐lapse acquisition showing the migration of myonuclei within four lateral transverse (LT) muscles of a bocksDP01391 mutant embryo. Tracks correspond to the movement of individual nuclei over the course of two hours. Time‐lapse starts at stage 15 (15 hours AEL, t = 0 min), where a majority of nuclei failed to separate and remain clustered together in the ventral end of the muscle. Only two escaper nuclei separate from the ventral cluster and migrate directionally toward the dorsal muscle pole. Scale bar, 10 μm.
Movie S5.
Altered nuclear migration in the lateral transverse muscle of a klar1 mutant embryo. Time‐lapse acquisition showing the migration of myonuclei within four lateral transverse (LT) muscles of a klar1 mutant embryo. Tracks correspond to the movement of individual nuclei over the course of two hours. Time‐lapse starts at stage 15 (15 hours AEL, t = 0 min), where a majority of nuclei failed to separate and remain clustered together in the ventral end of the muscle. Only one escaper nucleus separates from the ventral cluster and migrates directionally toward the dorsal muscle pole. Scale bar, 10 μm.
Movie S6.
Altered nuclear migration in the lateral transverse muscle of an ensswo mutant embryo. Time‐lapse acquisition showing the migration of myonuclei within four lateral transverse (LT) muscles of an ensswo mutant embryo. Tracks correspond to the movement of individual nuclei over the course of two hours. Time‐lapse starts at stage 15 (15 hours AEL, t = 0 min). In each LT muscle, none of the nuclei separate and remain within a single cluster. Scale bar, 10 μm.
Laser ablation of myonuclei demonstrates that the application of force onto nuclei is ensconsin dependent
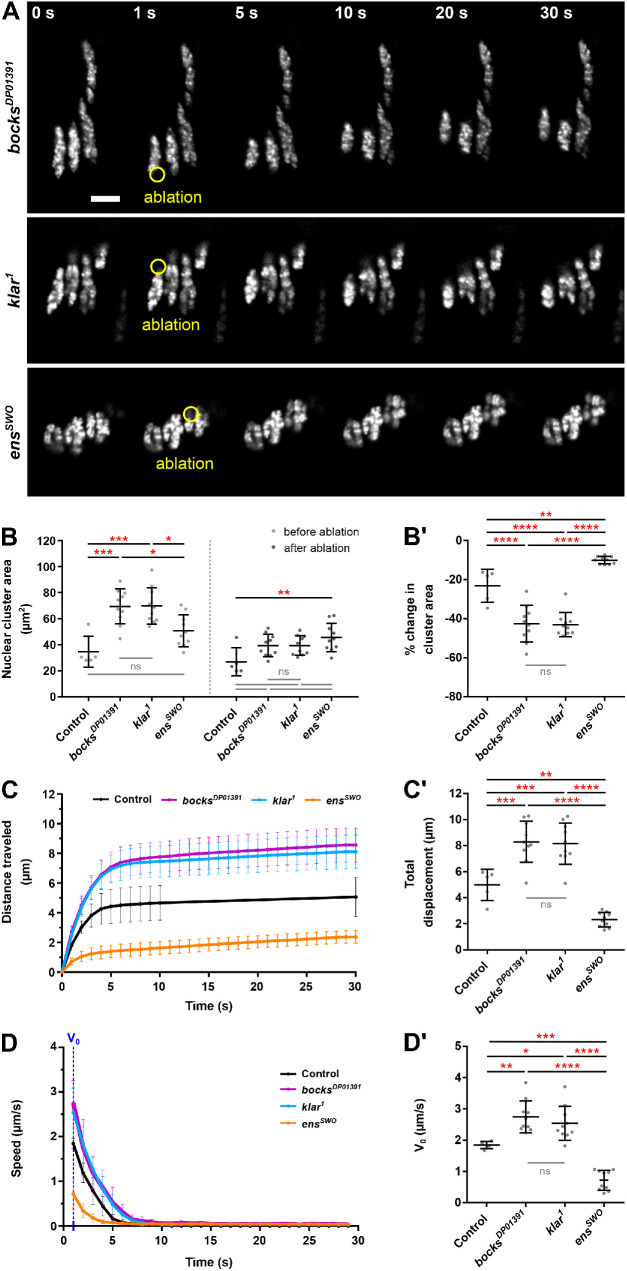
The fact that the nuclei were elongated in bocksDP01391 and klar1 mutants compared with controls and ensswo mutants suggested that they may be under tension. To test this hypothesis, we used 2-photon laser ablation to remove individual nuclei and measure the response of the neighboring nuclei within the syncytium (Supplemental Figure S3A). This approach has been used extensively to characterize the genes that are necessary for the production and maintenance of tension at cell–cell junctions (Rauzi and Lenne, 2015; Kong et al., 2019). In those studies, faster recoil of the cell membrane or its associated cytoskeletal elements was evidence of increased tension, whereas slower recoil was evidence of reduced tension. Because the nucleus serves as the microtubule-organizing center in muscle cells, we predicted that the ablation of a nucleus would disrupt its associated microtubule network and any interactions with neighboring nuclei. This would create an imbalance in the force between the dorsal and ventral ends, and therefore the speed of movement would be proportional to the degree of tension that the entire cluster of nuclei was experiencing. When a nucleus was ablated in controls (Supplemental Figure S3D, 1 s, yellow circle, and Supplemental Movie S7), the remaining nuclei within the cluster moved away from the ablation site, toward the center of the muscle fiber (Supplemental Figure S3D, 2–5 s). Nuclei in the opposite cluster within the same muscle also moved toward the muscle center, suggesting that the two clusters do interact. Additionally, the nuclei in the neighboring muscle do move after ablation. However, the movement is reduced compared with that of the nuclei in the muscle that has undergone ablation, suggesting that this movement is a consequence of the removal of the physical barrier dependent on the nuclei from the experimental muscle moving. Most importantly, ablation did not affect the health of the muscle or the animal. Imaging of the transmitted light demonstrated that there was no gross damage to the embryo. Furthermore, 3 h after ablation, nuclei returned to their proper position adjacent to the muscle end (Supplemental Figure S3E). Similar movements of nuclei toward the muscle center have been seen to occur due to muscle contractions, but in these cases when muscles detached from the tendon, the muscles formed a spheroid from which neither the muscle morphology nor the nuclear position recovered (Auld et al., 2018b). Thus, the return of nuclei to the end of the muscle is consistent with the nuclei moving and not movement of the muscle ends due to a loss of the myotendinous junction. Finally, ablation did not affect viability as embryos were able to developmentally progress to stage 17, initiate muscle contraction and hatching (Supplemental Figure S3E), and crawl out of the field of view.
We next applied this technique to clusters of nuclei that had failed to separate in klar1, bocksDP01391, and ensswo embryos to determine the relative tension that nuclei were under in each genotype (Figure 3A). When compared with controls, the area of the ventral clusters in bocksDP01391 (Supplemental Movie S8) and klar1 (Supplemental Movie S9) embryos before ablation was significantly larger (Figure 3B, before). After ablation, the remaining nuclei moved away from the ablation site and showed a 43% reduction in size in both genotypes (Figure 3, B and B′, after). The dramatic decrease in size suggests that the stretching of nuclei, in addition to the greater number of nuclei present, contributed to the difference in the size of the clusters. In contrast, nuclei in ensswo embryos (Supplemental Movie S10) moved only slightly after ablation (Figure 3A) and their size was reduced by only 10%, a value consistent with the removal of one nucleus of the eight that are present (Figure 3, B and B′). In addition, after ablation, clusters in bocksDP01391 and klar1 embryos traveled a greater distance compared with controls while clusters in ensswo embryos traveled a shorter distance (Figure 3, C and C′). Similarly, the clusters in bocksDP01391 and klar1 had a greater initial velocity compared with controls, whereas nuclei in ensswo embryos had a reduced initial velocity (Figure 3, D and D′). Together, the increased response of nuclei in bocksDP01391 and klar1 embryos demonstrate that nuclei in these genotypes are under more mechanical tension than nuclei in controls, while the decreased response of nuclei in ensswo embryos is consistent with them being under less mechanical tension compared with controls. This is consistent with the hypothesis that ensconsin is necessary for the application of force to nuclei but that klarsicht and bocksbeutel are necessary for the separation of nuclei and their directed movement in response to that force.
FIGURE 3:
Nuclei in bocksbeutel and klarsicht mutants are under more tension than nuclei in ensconsin mutants. (A) Montages from time-lapse acquisitions showing the ablation of a myonucleus within the LT muscles of a stage 16 (16 h AEL) embryo for the indicated genotypes. The first frame shows the nuclei before ablation (0 s). The next frame (1 s) shows the ablation of a single nucleus (yellow circle), followed by the subsequent response of the remaining nuclei after ablation (5–30 s). Scale bar, 10 µm. (B) Average area of nuclear clusters before and after ablation. (B′) The same data as in B represented as a percent change in cluster area. A negative change in area indicates that the size of the nuclear cluster decreased after the ablation. (C) Average displacement of nuclear clusters after ablation as a function of time. (C′) Average total displacement of nuclear clusters after ablation. (D) Average change in speed of nuclear clusters after ablation as a function of time. (D′) Average initial speed (V0) of nuclear clusters the first second after ablation. Data points in B–D′ correspond to an individual ablation event. Error bars indicate the SD from ≥5 ablation events performed in different embryos for each genotype. The one-way ANOVA with Tukey HSD post hoc test was used to assess the statistical significance of differences in measurements between all experimental genotypes to controls. Not significant (ns) ≥ 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Movie S7.
In vivo 2‐photon laser ablation of myonuclei in a control Drosophila embryo. Time‐lapse acquisition showing the ablation of a myonucleus within the lateral transverse (LT) muscles of a stage 16 (16 hours AEL) control embryo. The first frame shows the nuclei before ablation (0 s). The next frame (1 s) shows the ablation of a single nucleus (yellow circle), followed by the subsequent response of the remaining nuclei after ablation (2‐5 s). Myonuclei in green, transmitted light in gray. Scale bar, 10 μm.
Movie S8.
In vivo 2‐photon laser ablation of myonuclei in a bocksDP01391 mutant embryo. Time‐lapse acquisition showing the ablation of a myonucleus within the lateral transverse (LT) muscles of a stage 16 (16 hours AEL) bocksDP01391 mutant embryo. The first frame shows the nuclei before ablation (0 s). The next frame (1 s) shows the ablation of a single nucleus (yellow circle), followed by the subsequent response of the remaining nuclei after ablation (5‐30 s). Myonuclei in green, transmitted light in gray. Scale bar, 10 μm.
Movie S9.
In vivo 2‐photon laser ablation of myonuclei in a klar1 mutant embryo. Time‐lapse acquisition showing the ablation of a myonucleus within the lateral transverse (LT) muscles of a stage 16 (16 hours AEL) klar1 mutant embryo. The first frame shows the nuclei before ablation (0 s). The next frame (1 s) shows the ablation of a single nucleus (yellow circle), followed by the subsequent response of the remaining nuclei after ablation (5‐30 s). Myonuclei in green, transmitted light in gray. Scale bar, 10 μm.
Movie S10.
In vivo 2‐photon laser ablation of myonuclei in an ensswo mutant embryo. Time‐lapse acquisition showing the ablation of a myonucleus within the lateral transverse (LT) muscles of a stage 16 (16 hours AEL) ensswo mutant embryo. The first frame shows the nuclei before ablation (0 s). The next frame (1 s) shows the ablation of a single nucleus (yellow circle), followed by the subsequent response of the remaining nuclei after ablation (5‐30 s). Myonuclei in green, transmitted light in gray. Scale bar, 10 μm.
Bocksbeutel, klarsicht, and ensconsin are required for the organization of microtubules in Drosophila larval muscle
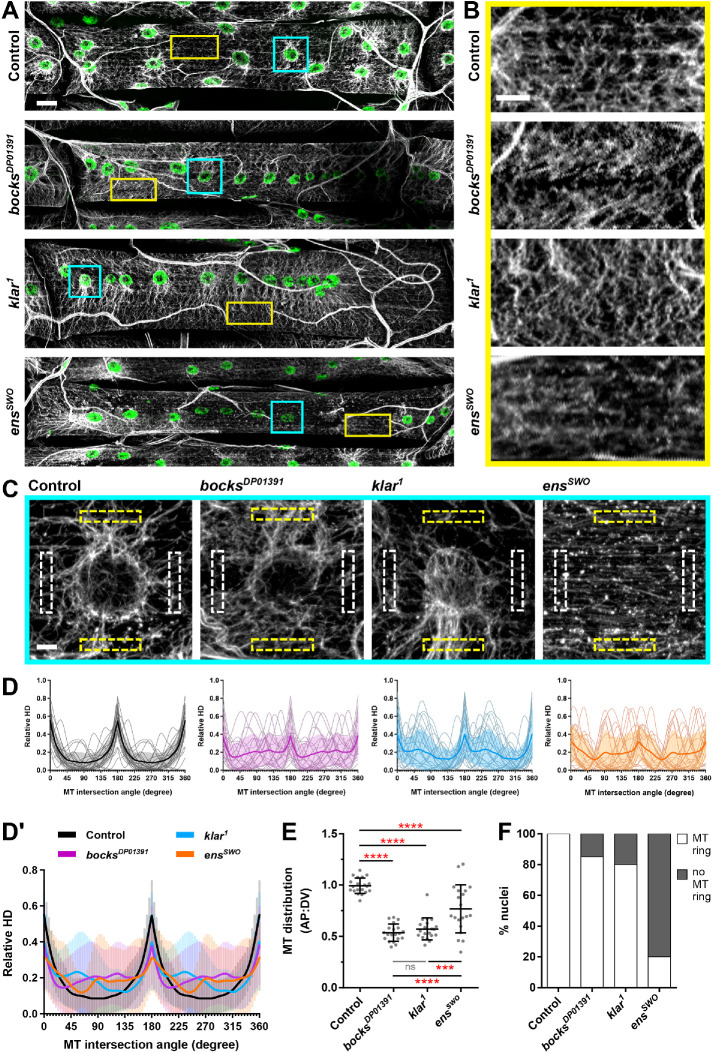
Because myonuclei are physically linked to the microtubule cytoskeleton (Tassin et al., 1985; Espigat-Georger et al., 2016), Ensconsin is a microtubule-binding protein (Bulinski and Bossler, 1994; Gallaud et al., 2014), and nuclear envelope proteins have been demonstrated to impact microtubule organization (Bugnard et al., 2005; Hale et al., 2008; Starr and Fridolfsson, 2010; Gimpel et al., 2017), we hypothesized that the differences in nuclear behaviors may be linked to variations in microtubule organization. To test this hypothesis, we used larvae in which the muscles are 100× larger and therefore provide greater resolution of microtubule organization. Additionally, we used the ventral longitudinal muscle 3 (VL3) of stage L3 larvae (Figure 4A), which are a large, flat, rectangular muscle group that is at the surface of a dissected larva. We focused on two distinct regions of microtubules that are uniquely organized. The first region pertained to areas of the muscle, distant from nuclei, where microtubules intersect to form a lattice (Figure 4, A, yellow box, and B) while the second region was adjacent to nuclei and consisted of microtubules that emanate from the nuclei (Figure 4, A, cyan box, and C). As previously reported (Elhanany-Tamir et al., 2012; Collins, Mandigo, et al., 2017), nuclei in bocksDP01391 and klar1 larvae were mispositioned in a single row along the anterior–posterior axis of the muscle compared with nuclei in controls, which were evenly distributed in two parallel lines. Analysis of the lattice network of microtubules (Figure 4B) was performed using the texture detection technique (TeDT), which detects the angles at which neighboring microtubules intersect (Liu and Ralston, 2014). In controls, the dominant intersection angles were parallel (0°, 180°, 360°) to the anterior–posterior axis of the muscle (Figure 4D, average in D′). Microtubules in bocksDP01391, klar1, and ensswo larval muscles were highly disorganized, with an overall reduction in the frequency of microtubules intersecting at every 180° (Figure 4D′), indicating a disruption of microtubule organization at positions far from nuclei in all three genotypes.
FIGURE 4:
Bocksbeutel, klarsicht, and ensconsin disrupt microtubule organization in Drosophila larval skeletal muscle. (A) Immunofluorescence images of ventral longitudinal muscle 3 from stage L3 larvae for the indicated genotypes. Microtubules (α-tubulin) in gray, myonuclei in green. Scale bar, 25 µm. (B) Magnified regions of the microtubule (MT) lattice taken from the images shown in A, as indicated by the yellow box. Scale bar, 10 µm. (C) Magnified regions of microtubules emanating from myonuclei taken from the images shown in A, as indicated by the cyan box. White dotted boxes indicate the locations of anterior and posterior fluorescence intensity measurements while yellow dotted boxes indicate the locations of dorsal and ventral fluorescence intensity measurements for microtubule polarity analysis. Scale bar, 5 µm. (D) TeDT analysis of MT lattice regions. Intersection angles are represented as directional histograms (HD) from 0° to 360°. Thin lines indicate TeDT analysis for individual MT lattice regions, while the thick color line indicates the average of 20 MT lattice regions for each genotype. (D′) Average TeDT analysis from 20 MT lattice regions as shown in D for bocksDP01391 (purple), klar1 (blue), and ensswo (orange) compared with controls (black). For D and D′, error bars indicate the SD from 20 MT lattice regions for each genotype. (E) Polarity of microtubules around myonuclei, represented as the microtubule distribution ratio for each nucleus. Data points correspond to the microtubule distribution ratio of a single nucleus. Error bars indicate the SD from 20 nuclei for each genotype from ≥10 VL3 muscles. The one-way ANOVA with Tukey HSD post hoc test was used to assess the statistical significance of differences in measurements between all experimental groups. Not significant (ns) ≥ 0.05, ***P < 0.001, ****P < 0.0001. (F) The frequency in which microtubule rings were observed around nuclei in each of the indicated genotypes. A total of 20 nuclei were analyzed for each genotype from ≥10 VL3 muscles.
To evaluate the organization of microtubules that extend off of nuclei, we determined the percentage of nuclei that have a dense ring of microtubules on the nuclear periphery (Figure 4F) and measured the proportion of microtubules extending on the dorsal–ventral axis of the muscle versus the anterior–posterior axis (Figure 4E). In controls, all nuclei had a ring of microtubules and the distribution ratio was close to 1.0, indicating that microtubules are uniformly emanating from nuclei. Although 85% of bocksDP01391 and 80% of klar1 nuclei had a ring of microtubules (Figure 4F), the distribution ratio was reduced to 0.535 and 0.572 in bocksDP01391 and klar1 larvae, respectively (Figure 4E), indicating that more microtubules are extending along the dorsal–ventral axis compared with the anterior–posterior axis. However, only 20% of nuclei in ensswo mutants had rings (Figure 4F). This was surprising because previous data suggested that ensconsin did not regulate microtubule organization in muscles (Metzger et al., 2012) and that ensconsin regulated only Kinesin activity in other cell types (Barlan et al., 2013). Furthermore, there was an increase in the polarization of microtubules on the dorsal–ventral axis compared with controls and an increased variation in that polarity compared with controls, bocksDP01391, and klar1 mutants (Figure 4E). Together, these data indicate that although Bocksbeutel, Klarsicht, and Ensconsin are necessary to maintain the link between myonuclei and microtubules, the disruption of bocks or klar results in the reorganization of microtubules around mispositioned nuclei, whereas the disruption of ens disrupts the association of microtubules with the nuclei and the organization of the microtubules around the low percentage of nuclei that do have associated microtubules.
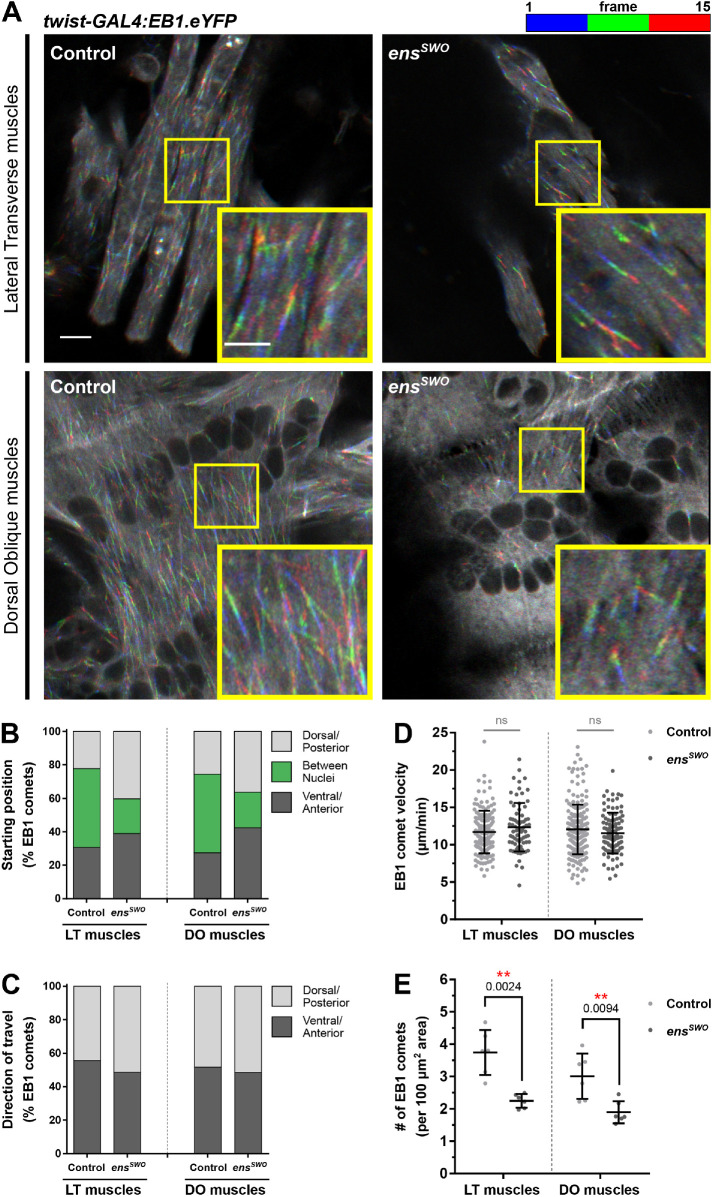
Our finding that microtubule organization is dependent on ensconsin differs from previous studies that suggested that the function of ensconsin was to activate Kinesin (Barlan et al., 2013). To determine whether the disruption of microtubule organization was evident during embryonic nuclear movement, we examined the behavior of EB1 during embryonic muscle development when nuclei are actively moving. EB1 comets were tracked for 1 min in the LT muscles (Supplemental Movies S11 and S12) and the dorsal oblique (DO) muscles (Supplemental Movies S13 and S14), a set of broad, flat muscles that are more amenable to fast, live-embryo imaging (Figure 5A). The location from which EB1 emerged, their direction of travel, and their speed were the same in controls and ensswo embryos in both muscle types (Figure 5, B–D). However, the number of EB1 comets was significantly decreased in both LT and DO muscles of ensswo embryos (Figure 5E), indicating that Ensconsin is critical to regulating the number of growing microtubules during Drosophila muscle development. Because most microtubules emanate from the nuclei in Drosophila larval muscles, the decrease in microtubule number (Figure 5E) is consistent with the decreased percentage of nuclei with microtubule rings (Figure 4F), further supporting a role for ensconsin in maintaining the general organization of microtubules within skeletal muscles.
FIGURE 5:
Depletion of ensconsin decreases the number of EB1 comets in Drosophila embryonic muscles. (A) Temporal overlays tracking EB1 comets for 15 s in the LT muscles and dorsal oblique (DO) muscles of stage 16 control and ensswo embryos. Scale bar, 5 μm. Inset in yellow box: Magnified regions of the temporal overlays tracking EB1 comets for 15 s. Scale bar, 3 μm. (B) Frequency of EB1 comets observed in controls and ensswo muscles starting in the dorsal/posterior muscle pole region, the ventral/anterior muscle pole region, or the region between nuclei. (C) Frequency of EB1 comets observed in controls and ensswo muscles traveling toward either the dorsal/posterior muscle pole or the ventral/anterior muscle pole. (D) Average velocity of EB1 comets in controls and ensswo muscles. Data points correspond to the velocity measured from a single EB1 comet. Error bars indicate the SD from EB1 comets measured from six different embryos for each muscle group taken from independent experiments. (E) Average number of EB1 comets counted in controls and ensswo muscles, normalized to the muscle area. Data points correspond to the total number of EB1 comets counted from a single embryo. Error bars indicate the SD from six different embryos for each muscle group taken from independent experiments. For D and E, Student’s t test with Welch’s correction was used to assess the statistical significance of differences in measurements between Ensconsin-depleted embryos and controls for each muscle group.
Movie S11.
In vivo imaging of EB1 comet dynamics in the lateral transverse muscles of a control Drosophila embryo. Time‐lapse acquisition of the lateral transverse muscles in a stage 16 (16 hours AEL) control embryo expressing EB1.eYFP. Time course, 60 s. Scale bar, 5 μm.
Movie S12.
In vivo imaging of EB1 comet dynamics in the lateral transverse muscles of an ensswo mutant embryo. Time‐lapse acquisition of the lateral transverse muscles in a stage 16 (16 hours AEL) ensswo mutant embryo expressing EB1.eYFP. Time course, 60 s. Scale bar, 5 μm.
Movie S13.
In vivo imaging of EB1 comet dynamics in the dorsal oblique muscles of a control Drosophila embryo. Time‐lapse acquisition of the dorsal oblique muscles in a stage 16 (16 hours AEL) control embryo expressing EB1.eYFP. Time course, 60 s. Scale bar, 5 μm.
Movie S14.
In vivo imaging of EB1 comet dynamics in the dorsal oblique muscles of an ensswo mutant embryo. Time‐lapse acquisition of the dorsal oblique muscles in a stage 16 (16 hours AEL) ensswo mutant embryo expressing EB1.eYFP. Time course, 60 s. Scale bar, 5 μm.
DISCUSSION
All together, these data demonstrate that seemingly similar nuclear positioning phenotypes can be quite different and highlight the need to quantitatively examine nuclear positioning defects. Specifically, the nuclear positioning phenotypes in ensswo, bocksDP01391, and klar1 were all previously described in similarly descriptive terms, but we have demonstrated that these phenotypes are quantitatively distinct. Careful analysis of the precise position, shape, and movement of nuclei clearly indicates that there are distinct defects (Supplemental Figure S4). We found that loss of Ensconsin contributes to the reduced application of force to nuclei as they remain spherical and display no productive movement. Mechanistically, we correlate this behavior with a reduction in the number of growing microtubules and suggest that the number of growing microtubules is insufficient to generate the force necessary to move several large nuclei. These data also demonstrate a novel role for the LINC complex in facilitating the separation of nuclei from their neighbors. Surprisingly, the nuclei are under tension in the absence of the KASH-domain protein Klarsicht or the Emerin homologue Bocksbeutel. Consequently, nuclei moved a similar total distance to those nuclei in control embryos. However, nuclei could not separate and remained attached and therefore were all moved toward the ventral end of the muscle. Interestingly, in bocksDP01391 and klar1 mutants, nuclei did rarely separate from the single cluster and move as individuals to the dorsal end of the muscle. This observation is consistent with the phenotype being based in aberrant associations between nuclei and not a disruption of directional cues. The molecular nature of these interactions is still to be determined. However, models suggest that microtubule growth dynamics are sufficient to separate nuclei from their neighbors (Manhart et al., 2020). Therefore, the change in the organization of microtubules around the nuclei that we observe later in development may be present during nuclear movement and critically contribute to the separation of nuclei from their neighbors. Alternatively, defined roles for the LINC complex in the recruitment of Kinesin (Starr and Fridolfsson, 2010; Wilson and Holzbaur, 2012) may suggest that motor-dependent directional movement of nuclei is critical for the separation of nuclei independent of nuclear translocation. Finally, we used laser ablation of individual nuclei to demonstrate that nuclei in bocksDP01391 and klar1 mutants are under increased tension compared with controls, whereas those in ensswo mutants are under decreased tension compared with controls, to confirm that sustained force is applied to nuclei in bocksDP01391 and klar1 mutants but not in ensswo mutants. More broadly, these data present the first direct evidence that regulation of interactions between nuclei is a critical determinant of nuclear movement and that nucleus–nucleus interactions are LINC complex dependent. Thus, these data raise the possibility that aligned nuclei in the center of a developing or regenerating muscle are physically linked and that this linkage is critical for nuclear functions.
The molecular mechanisms by which Klarsicht and Bocksbeutel regulate separation of nuclei from their neighbors and the molecular mechanisms by which Ensconsin regulates the number of growing microtubules necessitate further investigation. However, we hypothesize that Ensconsin may contribute, either directly or indirectly, to microtubule nucleation and anchoring at the nuclear envelope. Recent work found that Bsg25D, the Drosophila homologue of Ninein, interacts with Ensconsin and that Bsg25D contributed to Ensconsin-dependent nuclear positioning (Rosen et al., 2019). Together with our data showing a reduction in the number of growing microtubules, we hypothesize that perhaps Bsg25D recruits Ensconsin to participate in microtubule nucleation. Alternatively, both Bsg25D and Ensconsin may anchor microtubules to the nuclear envelope. Release of microtubule minus ends from the nuclear envelope may potentiate microtubule instability and the reduction in growing microtubules. Indeed, Ninein does contribute to both nucleation and anchoring of microtubules to the centrosome (Delgehyr et al., 2005), and the loss of either function is consistent with the data here and previously published (Rosen et al., 2019).
The molecular mechanism by which Bocksbeutel and Klarsicht regulate nuclear position is harder to predict. The simplest explanation might be that they are required to recruit microtubule motors as has been seen in other systems (Starr et al., 2001; Cadot et al., 2012; Wilson and Holzbaur, 2012). However, the phenotype seen here is distinct from the phenotypes observed in animals null for either Cytoplasmic dynein or Kinesin (Folker et al., 2014). Alternatively, work in Caenorhabditis elegans found that loss of nucleus anchoring resulted in a similar clustering of nuclei (Starr et al., 2001). But the data we present are from developmental stages that require active movement of nuclei rather than anchoring. When combined with our finding that the clusters of nuclei still move in these genotypes, the simplest explanation is that these factors are required for nuclei to separate from one another. Because it is the loss of Bocksbeutel or Klarsicht that results in the phenotype, these data suggest that either the recruitment of a separation factor or LINC complex–dependent cytoskeletal organization is critical for the separation of nuclei. We speculate that this is based on variations in microtubule organization, consistent with our finding that microtubules are asymmetrically organized around nuclei in animals with mutations in either gene. Furthermore, it is likely that the microtubules that emanate from adjacent nuclei can interact with each other and with other nuclei. Thus, the ablation of individual nuclei will ablate the associated microtubule network. Thus, if the molecular glue is the microtubules either directly or indirectly, the data would be similar.
Altogether these data demonstrate that seemingly similar phenotypes are mechanically distinct and provide an approach along with some of the tools necessary to push beyond this basic understanding toward a molecular comprehension of how the movement of many nuclei is coordinated within a single cytoplasm.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Drosophila genetics
All stocks were grown under standard conditions at 25°C. Stocks used were apRed (Richardson et al., 2007), bocksDP01391 (Bloomington Drosophila Stock Center; 21846), klar1 (Bloomington Drosophila Stock Center; 3256), ensswo (Metzger et al., 2012), and UAS-EB1.eYFP (Rogers et al., 2008). Mutants were balanced and identified using TM6b, DGY. The UAS-EB1.eYFP construct was specifically expressed in the mesoderm using the twist-GAL4, apRed driver. Flies carrying apRed express a nuclear localization signal fused to the fluorescent protein DsRed downstream of the apterous mesodermal enhancer. This results in the specific labeling of the myonuclei within the LT muscles of the Drosophila embryo (Richardson et al., 2007). Thus, only nuclei within the LT muscles are labeled using this reporter. The twist-GAL4, apRed Drosophila line was made by recombining the apRed promoter and the specific GAL4 driver, with both elements on the second chromosome.
Immunohistochemistry
Embryos were collected at 25°C and washed in 50% bleach to remove the outer chorion membrane, washed with water, and then fixed in 50% Formalin (Sigma; Product #HT501128) diluted in 1:1 heptane for 20 min. Embryos were then devitellinized by vortexing in a 1:1 methanol:heptane solution. Primary antibodies for embryo staining were used at the following final dilutions: rabbit anti-DsRed (1:400; Clontech; 632496), rat antitropomyosin (1:200; Abcam; ab50567), mouse anti-GFP (1:50; Developmental Studies Hybridoma Bank; GFP-G1). The conjugated fluorescent secondary antibodies used were Alexa Fluor 555 donkey anti-rabbit (1:200), Alexa Fluor 488 donkey anti-rat (1:200), and Alexa Fluor 647 donkey anti-mouse (1:200) (all Life Technologies). Larvae at stage L3 were dissected as previously described (Collins, Mandigo, et al., 2017; Auld et al., 2018a). In brief, larvae were dissected in ice-cold PIPES dissection buffer containing 100 mM PIPES (Sigma-Aldrich; P6757), 115 mM d-sucrose (Fisher Scientific; BP220-1), 5 mM trehalose (Acros Organics; 182550250), 10 mM sodium bicarbonate (Fisher Scientific; BP328-500), 75 mM potassium chloride (Fisher Scientific; P333-500), 4 mM magnesium chloride (Sigma-Aldrich; M1028), and 1 mM EGTA (Fisher Scientific; 28-071-G) and then fixed with 10% Formalin (Sigma-Aldrich; HT501128). For larval staining, mouse anti–α-tubulin (1:200; Sigma-Aldrich; T6199) was used. Acti-stain 555 phalloidin (1:400; Cytoskeleton PHDH1-A) and Hoechst 33342 (1 μg/ml) were added with the fluorescent secondary antibody Alexa Fluor 488 donkey anti-mouse (1:200; Life Technologies). Both embryos and larvae were mounted in ProLong Gold (Life Technologies; P36930).
Analysis of myonuclear position in Drosophila embryos
Embryos at stage 16 were selected to be imaged based on overall embryo shape, the intensity of the apRed and tropomyosin signals, gut morphology, and the morphology of the trachea as previously described (Folker et al., 2012; Collins, Mandigo, et al., 2017; Auld et al., 2018a). Confocal z-stacks of fixed embryos were acquired on a Zeiss LSM 700 using a Plan-APOCHROMAT 40×, 1.4 NA oil objective with a 1.0× optical zoom (see Supplemental Table S2 for image acquisition settings). Images were processed as maximum-intensity projections and oriented such that the top is dorsal, the bottom is ventral, the left is anterior, and the right is posterior. Measurements were made using the Segmented Line tool in Fiji software (Schindelin et al., 2012). Muscle length measurements were taken starting from the dorsal tip and following through the center of each LT muscle, down to the ventral tip. Dorsal and ventral end distances were taken from each LT muscle by measuring the distance between the closest group of nuclei to the dorsal or ventral muscle pole, respectively. Internuclear distances were taken by measuring the shortest distance in between the dorsal and ventral clusters of nuclei within each LT muscle. Internuclear distances were also plotted according to relative frequency. All three measurements are reported as distances normalized to the muscle length (Figure 1) and as raw values (Supplemental Figure S1). All four LT muscles were measured in four hemisegments from each embryo. Statistical analysis was performed with Prism 4.0 (GraphPad).
Analysis of myonuclear cluster area in Drosophila embryos
The areas of nuclear clusters were measured in fixed stage 16 embryos as previously described (Collins, Mandigo, et al., 2017). In brief, the area of each cluster of nuclei near either the dorsal or ventral muscle pole was measured in Fiji (Schindelin et al., 2012). The total area of nuclear clusters in each LT muscle was calculated by adding the dorsal and ventral areas. The nuclear separation ratio was calculated by dividing the area of the dorsal cluster by the area of the ventral cluster. Nuclear clusters from all four LT muscles were measured in four hemisegments from each embryo. Statistical analysis was performed with Prism 4.0 (GraphPad).
For qualitative nuclear phenotype analysis, embryos were scored on how nuclei were positioned within the first three LT muscles of each hemisegment. LT 4 was excluded for this analysis due to its variable muscle morphology. Nuclear phenotypes were categorized as either “separated; equal distribution” (nuclei properly segregated into two distinct, even clusters with a nuclear separation ratio ≥0.85 and ≤1.15), “separated; unequal distribution” (nuclei that segregated into two disproportionate clusters with a nuclear separation ratio <0.85 or >1.15), “central” (a nucleus that is not associated with either the dorsal or ventral group located in the middle of the myofiber), “clustered” (nuclei remained in a single cluster toward the ventral end of the myofiber), “spread” (nuclei are distributed through the myofiber with no distinct dorsal or ventral clusters), or “swoosh” (nuclei remained in a single cluster within the middle of the myofiber). Linescans of DsRed intensity were performed on 10 LT muscles for each nuclear phenotype and averaged to determine the typical distribution of nuclei in bocksDP01391 and ensswo genotypes compared with controls.
Volumetric imaging and analysis of nuclear clusters
Fixed stage 16 embryos were imaged on a Zeiss LSM 880 with Airyscan (superresolution acquisition, 2× Nyquist sampling) using a Plan-APOCHROMAT 40×, 1.3 NA oil objective at a 1.0× optical zoom and 0.15 µm step size interval through the entire depth of the muscle (Supplemental Table S2). Postprocessing of Airyscan images was completed in ZEN Blue 2016 software. The quantitative volumetric analysis was performed in Imaris version 9.2.1 (Bitplane AG). Images were first processed as maximum-intensity projections of confocal z-stacks and oriented such that the top is dorsal, the bottom is ventral, the left is anterior, and the right is posterior. A volumetric rendering of each nuclear cluster was created using the Surface Visualization tool of the DsRed channel. Volume measurements were automatically computed from the Surface renderings by Imaris. Statistical analysis was performed with Prism 4.0 (GraphPad).
Live-embryo imaging and analysis
Embryos for live-imaging were prepared as previously described (Collins, Mandigo, et al., 2017; Auld et al., 2018a). In brief, embryos were collected at 25°C, washed in 50% bleach to remove the outer membrane, washed with water, and mounted with halocarbon oil (Sigma; Product #H8898). For time-lapse imaging of nuclear movement, stage 15 embryos were selected for imaging based on gut morphology, the position of nuclei, and the intensity of the apRed signal as previously described (Folker et al., 2012; Collins, Mandigo, et al., 2017; Auld et al., 2018a) with the following modifications. Time-lapse images were acquired on a Zeiss LSM 700 using a Plan-APOCHROMAT 40×, 1.4 NA oil objective with a 1.0× optical zoom at an acquisition rate of 1 min/stack for 2 h (Supplemental Table S2). Movies were processed in Fiji (Schindelin et al., 2012) as maximum-intensity projections of confocal z-stacks and corrected for drift using the Correct 3D drift plug-in. To calculate the separation speed of nuclei, the Line tool was used to measure the distance between dorsal and ventral nuclear clusters at time 0 h and again at time 2 h. Separation speeds were also plotted according to relative frequency.
The aspect ratio of individual nuclei was measured as previously described (Folker et al., 2014; Auld et al., 2018b). In brief, the length of each nucleus that could be confidently distinguished from its neighbors was measured along the dorsal–ventral axis and divided by the length of each nucleus along the anterior–posterior axis by using the Line tool in Fiji. A total of 20 nuclei were examined per genotype from at least three independent experiments. The aspect ratio for each nucleus was measured at a single time point when the edges of the individual nucleus could be clearly seen as distinct from other nuclei within a cluster. The aspect ratio of ventral clusters was measured at time 0 h using the Shape Descriptors plug-in, which calculates the aspect ratio of an ellipse by dividing the major axis of the ellipse by its minor axis. An aspect ratio value closer to 1 indicates a more spherical cluster. Tracks following the movement of individual nuclei within clusters were generated using the Manual Tracking plug-in.
The displacement of each nucleus was calculated as the difference between the final and initial positions. The directionality of individual nuclei was measured as previously described (Folker et al., 2014). In brief, single nuclei were examined and each nucleus was counted once throughout the time course. A nucleus was determined to have changed direction if it persistently moved a distance of at least one nuclear radius in the direction opposite to its previous direction of translocation. Nuclear directionality was reported as a percentage of nuclei that changed direction. All statistical analysis was performed with Prism 4.0 (GraphPad).
To assess for potential fusion defects, the nuclei in the LT muscles were counted from live stage 17 embryos when nuclei have separated and maximized their distance from their neighbors. Nuclei within the LT muscles were identified by expression of DsRed. The nuclei were counted from all four LT muscles within a single hemisegment, with a total of four hemisegments analyzed for each embryo.
2-Photon ablation of myonuclei
Embryos were collected at 25°C, washed in 50% bleach to remove the outer membrane, washed with water, and mounted with halocarbon oil (Sigma; Product #H8898). Stage 16 embryos were selected for ablation based on gut morphology, the position of nuclei, and the intensity of the apRed signal as previously described (Folker et al., 2012; Collins, Mandigo, et al., 2017; Auld et al., 2018a). Time-lapse images of embryos before, during, and after ablation were acquired on a Zeiss LSM 710 NLO using a Plan-APOCHROMAT 40×, 1.1 NA water objective with a 1.0× optical zoom at an acquisition rate of 1 s/frame for 30 s. Ablation was performed using the Coherent Chameleon Ultra II femtosecond pulsed-IR laser at 860 nm with 15%–17% laser power (Supplemental Table S2). As shown in Supplemental Figure S3, a nucleus was selected for ablation by drawing a region of interest (ROI) in ZEN Black 2012 software. For each ablation time lapse, the first frame (time = 0 s) was taken before the ablation event. The next frame (time = 1 s) shows the ablation of the targeted nucleus, followed by the subsequent response of the remaining nuclei present. Because no muscle marker is present, transmitted light images were also collected during the time lapse to ensure that ablation did not destroy the surrounding tissue. An ablation was considered successful by the loss of the DsRed signal accompanied by the movement of nuclei. Nuclei that were simply photobleached were characterized by just the loss of DsRed fluorescence without any subsequent response from the embryo (Supplemental Figure S3B). A failed ablation attempt that resulted in boiling of the embryo was identified by a hole burned through the membrane (Supplemental Figure S3C, arrowhead), as seen through the transmitted light channel.
Movies were processed in Fiji (Schindelin et al., 2012) as single confocal slices and oriented such that the top is dorsal, the bottom is ventral, the left is anterior, and the right is posterior. The area of clusters in which a nucleus was ablated was measured before and after the ablation event. The areas of nuclear clusters before and after ablation were plotted as percentage change. The displacement and velocity of nuclear clusters were measured using the centroid measurement, which calculates the center point of a cluster based on the average x and y coordinates of all pixels in the cluster. The total displacement of each cluster was calculated as the cumulative distance traveled over the 30 s after ablation. The initial velocity was defined as the speed a cluster traveled the first second after ablation. Statistical analysis was performed with Prism 4.0 (GraphPad).
Analysis of microtubule organization in Drosophila larvae
Confocal z-stacks of dissected stage L3 larvae were acquired on a Zeiss LSM 700 using a Plan-APOCHROMAT 40×, 1.4 NA oil objective lens at a 0.5× optical zoom for whole muscle images and at a 2.0× optical zoom for regions around myonuclei (Supplemental Table S2). Images were processed as maximum-intensity projections and oriented such that the top is dorsal, the bottom is ventral, the left is anterior, and the right is posterior. Microtubule organization was assessed in two distinct ROIs within the ventral longitudinal muscle 3 (VL3). The first region consists of microtubules that intersect at regions between nuclei to form a lattice. For these regions, the TeDT was used (Liu and Ralston, 2014). TeDT is a robust tool that can assess the orientation of the microtubule network by detecting the dominant angles at which microtubules intersect one another. For TeDT analysis, 200 × 100 square pixel regions of the microtubule lattice that excluded nuclei were cropped from whole muscle images. TeDT analysis on cropped regions was performed in MATLAB (MathWorks), which presented the resulting intersection angles detected as directional histograms (HD) from 0° to 360°.
The second ROI has microtubules emanating directly from the myonuclei. The polarity of these microtubules was analyzed as previously described (Collins, Mandigo, et al., 2017). The fluorescence intensity was measured from a 10 μm × 2 μm region positioned 15 μm anteriorly and 15 μm posteriorly from the center of the nucleus, using the Plot Profile tool in Fiji (Schindelin et al., 2012). Similarly, the fluorescence intensity was also measured from a 2 μm × 10 μm region positioned 15 μm dorsally and 15 μm ventrally from the center of the nucleus. Average fluorescence intensities were calculated for the anterior/posterior (AP) positions as well as the dorsal/ventral (DV) positions. A ratio between the average AP and DV fluorescence intensities was used to determine the microtubule distribution ratio. A value of 1 indicates a uniform distribution of microtubules around the nucleus. Values >1 indicate that there are more microtubules distributed within the AP regions relative to the nucleus, while values <1 indicate that there are more microtubules distributed within the DV regions relative to the nucleus. The organization of microtubules emanating from nuclei was also qualitatively assessed based on the presence of a dense microtubule ring around the nuclear periphery. Images of nuclei were blindly scored for the presence or absence of a microtubule ring. A nucleus was considered to have a microtubule ring based on the contiguous presence of α-tubulin intensity around the perimeter of the nucleus. Statistical analysis was performed with Prism 4.0 (GraphPad).
Analysis of microtubule dynamics in Drosophila embryos
Embryos for live imaging of EB1 comets were collected and prepared similarly. Stage 16 embryos were selected for imaging based on gut morphology, the position of nuclei, and the intensity of the apRed signal as previously described (Collins, Mandigo, et al., 2017; Auld et al., 2018a; Folker et al., 2012). Time-lapse images of EB1-eYFP were acquired on a Zeiss LSM 880 with Airyscan Fast mode (super resolution acquisition, 2× Nyquist sampling) using a Plan-APOCHROMAT 40×, 1.3 NA oil objective at a 4.0× optical zoom at an acquisition rate of 1 s/frame for 60 s (Supplemental Table S2). Postprocessing of Airyscan Fast images was done in ZEN Blue 2016 software. EB1 comets were imaged within the LT muscles as well as the dorsal oblique (DO) muscles, which are a flatter muscle group, ideal for imaging quick dynamics. Movies were processed as single confocal slices in Fiji (Schindelin et al., 2012). Time-lapse images taken in the LT muscles were oriented such that the top is dorsal, the bottom is ventral, the left is anterior, and the right is posterior. Time-lapse images taken in the DO muscles were oriented such that the top is posterior, the bottom is anterior, the left is dorsal, and the right is ventral. Trajectories of EB1 comets were made from time-lapse images using the Temporal-Color Code plugin, which sums up the first 15 consecutive frames (1 s each), and then overlays the resulting image to a blue-green-red color sequence, with each color representing a total of 5 s. All quantifications of EB1 dynamics was performed on temporal overlays by hand. Only comets that were visible for the full 15 s were used in this analysis. The starting position of each comet was categorized within the LT muscles as either starting within the dorsal pole region, ventral pole region, or between nuclei. Similarly, the starting position of each comet was categorized within the DO muscles as either starting within the anterior pole region, posterior pole region, or between nuclei. The direction of EB1 comets was also determined as either traveling dorsally/posteriorly or ventrally/anteriorly and whether the comets move toward or away from the nearest myotendinous junction. The length of EB1 trajectories over the 15 s timeframe was measured to calculate EB1 comet velocity over the 1 min time lapse. The EB1 comets were counted, and the number was normalized to the muscle area. Statistical analysis was performed with Prism 4.0 (GraphPad).
Data and material availability
All Drosophila stocks are available upon request. All data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary Material
Acknowledgments
We thank Wendy C. Salmon and the W. M. Keck Microscopy Facility at the Whitehead Institute for infrastructure and support with regard to all 2-photon laser ablation experiments. Additionally, we thank Bret Judson and the Boston College Imaging Core for infrastructure and support with regard to superresolution imaging using the Zeiss LSM 880 Airyscan microscope. Drosophila stocks obtained from the Bloomington Drosophila Stock Center (NIHP400D018537) were used in this study. This work was supported by an American Heart Association (AHA) Scientist Development Grant (15SDG22460004) to E.S.F. and institutional funds provided by Boston College.
Abbreviations used:
- apRed
apterous red
- bocks
bocksbeutel
- DGY
deformed, GMR:YFP
- DO
dorsal oblique
- ens
ensconsin
- eYFP
enhanced yellow fluorescent protein
- KASH
Klarsicht, ANC-1, Syne homology
- klar
klarsicht
- L3
3rd instar larval stage
- LINC
linker of nucleoskeleton and cytoskeleton
- LSM
laser scanning microscope
- LT
lateral transverse
- MT
microtubule
- NA
numerical aperture
- SD
standard deviation
- TeDT
texture detection technique
- VL3
ventral longitudinal muscle 3.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-06-0324) on September 15, 2021.
REFERENCES
Boldface names denote co–first authors.
- Auld AL, Collins MA, Mandigo TR, Folker ES (2018a). High-resolution imaging methods to analyze LINC complex function during Drosophila muscle development. Methods Mol Biol 1840, 181–203. [DOI] [PubMed] [Google Scholar]
- Auld AL, Roberts SA, Murphy CB, Camuglia JM, Folker ES (2018b). Aplip1, the Drosophila homolog of JIP1, regulates myonuclear positioning and muscle stability. J Cell Sci 131, jcs.205807. [DOI] [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI (2013). The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol 23, 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnard E, Zaal KJM, Ralston E (2005). Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton 60, 1–13. [DOI] [PubMed] [Google Scholar]
- Bulinski JC, Bossler A (1994). Purification and characterization of ensconsin, a novel microtubule stabilizing protein. J Cell Sci 107, 2839–2849. [DOI] [PubMed] [Google Scholar]
- Cadot B, Gache V, Vasyutina E, Falcone S, Birchmeier C, Gomes ER (2012). Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep 13, 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Mandigo TR, Camuglia JM, Vazquez GA, Anderson AJ, Hudson CH, Hanron JL, Folker ES (2017). Emery–Dreifuss muscular dystrophy–linked genes and centronuclear myopathy–linked genes regulate myonuclear movement by distinct mechanisms. Mol Biol Cell 28, 2303–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H (2008). Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell 134, 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M (2005). Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 118, 1565–1575. [DOI] [PubMed] [Google Scholar]
- Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T (2012). Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol 198, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espigat-Georger A, Dyachuk V, Chemin C, Emorine L, Merdes A (2016). Nuclear alignment in myotubes requires centrosome proteins recruited by nesprin-1. J Cell Sci 129, 4227–4237. [DOI] [PubMed] [Google Scholar]
- Folker ES, Schulman VK, Baylies MK (2012). Muscle length and myonuclear position are independently regulated by distinct dynein pathways. Development 139, 3827–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Schulman VK, Baylies MK (2014). Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development 141, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud E, Caous R, Pascal A, Bazile F, Gagné J-P, Huet S, Poirier GG, Chrétien D, Richard-Parpaillon L, Giet R (2014). Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells. J Cell Biol 204, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel P, Lee YL, Sobota RM, Calvi A, Koullourou V, Patel R, Mamchaoui K, Nédélec F, Shackleton S, Schmoranzer J, et al. (2017). Nesprin-1α-dependent microtubule nucleation from the nuclear envelope via Akap450 Is necessary for nuclear positioning in muscle cells. Curr Biol 27, 2999–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ (2013). Nuclear positioning. Cell 3, 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D (2008). Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J 95, 5462–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Loison O, Chavadimane Shivakumar P, Chan EHY, Saadaoui M, Collinet C, Lenne PF, Clément R (2019). Experimental validation of force inference in epithelia from cell to tissue scale. Sci Rep 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Starr DA, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y (2002). Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell 13, 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ralston E (2014). A new directionality tool for assessing microtubule pattern alterations. Cytoskeleton 71, 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandigo TR, Turcich BD, Anderson AJ, Hussey MR, Folker ES (2019). Drosophila emerins control LINC complex localization and transcription to regulate myonuclear position. J Cell Sci 132, jcs.235580. [DOI] [PubMed] [Google Scholar]
- Manhart A, Azevedo M, Baylies M, Mogilner A (2020). Reverse-engineering forces responsible for dynamic clustering and spreading of multiple nuclei in developing muscle cells. Mol Biol Cell 31, 1802–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK (2012). MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley-Bishop KL, Li Q, Patterson K, Fischer JA (1999). Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol 9, 1211–1220. [DOI] [PubMed] [Google Scholar]
- Rauzi M, Lenne P-F (2015). Probing cell mechanics with subcellular laser dissection of actomyosin networks in the early developing Drosophila embryo. In: Tissue Morphogenesis: Methods and Protocols, ed. Nelson C. M., New York: Springer, 209–218. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK (2007). SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134, 4357–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Peifer M, Rogers SL (2008). A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell 19, 3163–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W, Gomes ER (2018). Nuclear positioning in skeletal muscle. Semin Cell Dev Biol 82, 51–56. [DOI] [PubMed] [Google Scholar]
- Rosen JN, Azevedo M, Soffar DB, Boyko VP, Brendel MB, Schulman VK, Baylies MK (2019). The Drosophila Ninein homologue Bsg25D cooperates with Ensconsin in myonuclear positioning. J Cell Biol 218, 524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26, 421–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M (2002). Role of ANC-1 in tethering nuclei to the actin. Science 298, 406–409. [DOI] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M (2001). Unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development 128, 5039–5050. [DOI] [PubMed] [Google Scholar]
- Tan KL, Haelterman NA, Kwartler CS, Regalado ES, Lee PT, Nagarkar-Jaiswal S, Guo DC, Duraine L, Wangler MF, Bamshad MJ, et al. (2018). Ari-1 regulates myonuclear organization together with Parkin and is associated with aortic aneurysms. Dev Cell 45, 226–244.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley EC, Starr DA (2013). Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol 25, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Maro B, Bornens M (1985). Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol 100, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoué S, Chang F (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol 153, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF (1998). Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell 92, 547–557. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Holzbaur ELF (2012). Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci 125, 4158–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M (2011). KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet 20, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M (2009). SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.