Significance
Proteins conduct numerous complex biological functions by use of tailored structural dynamics. The molecular details of how these emerged from ancestral peptides remains mysterious. How does nature utilize the same repertoire of folds to diversify function? To shed light on this, we analyzed bilobed proteins with a common structural core, which is spread throughout the tree of life and is involved in diverse biological functions such as transcription, enzymatic catalysis, membrane transport, and signaling. We show here that the structural dynamics of the structural core differentiate predominantly via terminal additions during a long-period evolution. This diversifies substrate specificity and, ultimately, biological function.
Keywords: biophysics, evolution, structural dynamics, protein structure, ligand binding
Abstract
Novel biophysical tools allow the structural dynamics of proteins and the regulation of such dynamics by binding partners to be explored in unprecedented detail. Although this has provided critical insights into protein function, the means by which structural dynamics direct protein evolution remain poorly understood. Here, we investigated how proteins with a bilobed structure, composed of two related domains from the periplasmic-binding protein–like II domain family, have undergone divergent evolution, leading to adaptation of their structural dynamics. We performed a structural analysis on ∼600 bilobed proteins with a common primordial structural core, which we complemented with biophysical studies to explore the structural dynamics of selected examples by single-molecule Förster resonance energy transfer and Hydrogen–Deuterium exchange mass spectrometry. We show that evolutionary modifications of the structural core, largely at its termini, enable distinct structural dynamics, allowing the diversification of these proteins into transcription factors, enzymes, and extracytoplasmic transport-related proteins. Structural embellishments of the core created interdomain interactions that stabilized structural states, reshaping the active site geometry, and ultimately altered substrate specificity. Our findings reveal an as-yet-unrecognized mechanism for the emergence of functional promiscuity during long periods of evolution and are applicable to a large number of domain architectures.
Proteins drive and maintain all fundamental cellular processes (1) by interactions with small molecules and/or other biopolymers. Important mechanistic information on proteins are accessible via structural analysis of their functional cycle (2). While classical approaches rely on the interpretation of static structure snapshots, the visualization of structural dynamics (i.e., to follow the interconversion of distinct structural states at high spatial and temporal resolution) (3–7) has been recognized as an essential complement. The folding funnel model (8), rooted in the free-energy landscape theory (9–11), has by now become a widely accepted way to describe the ensemble of such states (12–14).
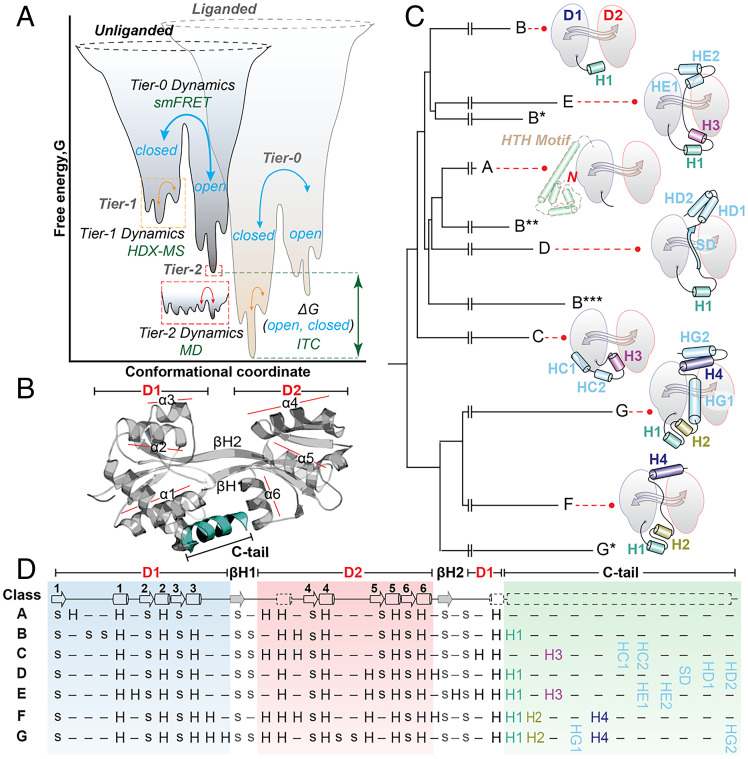
Distinct structural states can originate from local flexibility (i.e., bond vibrations, and side-chain rotations [Fig. 1A, Tier-2 dynamics]), changes in secondary structure (Fig. 1A, Tier-1 dynamics) or large-scale domain motions (Fig. 1A, Tier-0 dynamics). The free-energy landscape of a protein defines the lifetime of its structural states, ranging from nanoseconds (local flexibility) to seconds (large-scale motions). Transitions between the states are referred to as structural changes and are induced by interactions with ligands, posttranslational modifications (e.g., phosphorylation), or chemical events such as nucleotide hydrolysis. The coupling of the latter to structural changes enables proteins to perform a diverse range of functions.
Fig. 1.
Energetic funnel, structure, and evolution of the cherry-core. (A) One-dimensional cross-section of a hypothetical protein energy landscape adapted from Kern and coworkers (12) according to the Tier description and definitions introduced by Ansari and coworkers (15). A structural state is defined as the lowest point of a well on the energy surface. The populations of the Tier-0 states, “closed” and “open,” are defined as Boltzmann distributions, and their relative probabilities (pC, pO) are determined in this paper by smFRET, which follows large-scale domain motions. Tier-1 states describe local and fast structural fluctuations (e.g., changes in secondary structure elements like loop motions or loss of secondary structure). Tier-1 dynamics were probed in this study by HDX-MS. Rapid and localized Tier-2 dynamics (e.g., side-chain rotations) were not considered here but can be monitored via molecular dynamics (MD) simulations. Changes in the chemical environment (i.e., absence or presence of a ligand) modify the energy landscape via a bias for one of the two states. Typically, thermodynamic parameters such as the free energy difference of Tier-0 wells (ΔGOC) can be determined by ITC. (B) Structure of a representative bilobed protein, the substrate binding domain 2 (SBD2) of the ATP-binding cassette (ABC) amino acid transporter GlnPQ from Lactococcus lactis (PDB: 4KR5). (C) Summary of the structure-based phylogenetic tree with schematic representations of the different structural classes (class A through G) highlighting their termini. Complete sequence and structure-based phylogenetic trees are provided in SI Appendix, Fig. S1 and Datasets S1 and S2. We used the following notation: Secondary structure elements that are common between the different classes were assigned a number identifier (e.g., helix H4 in classes F and G), whereas the unique elements have a letter followed by a number identifier (e.g., HD1, first unique helix of class D proteins). An asterisk (*) marks a structural subclass (shown in detail in Dataset S3). Schematics are based on the crystal structures of the selected proteins. (D) Topology of bilobed proteins depicting the consensus of secondary structure elements: strands (s) or helices (H) belonging to the Domains (D1, D2), hinge-forming β-strands (βH1, βH2), and C-tails of all seven classes (A through G) are shown. Revised alignments of bilobed proteins are shown in Dataset S3. The secondary structure elements forming the consensus cherry-core structure are depicted on the Top row.
Tier-0 dynamics were observed and characterized in various settings (e.g., in motor proteins, in which they are used in propelling movement along filaments) (16), in the transport of molecules or biopolymers across biological membranes (17–21), or in the activity of proteins that perform mechanical work (22). Tier-1 dynamics drive the actions of various signaling proteins for transmission of signals (23–25). Structural and biochemical data indicate that enzymes also show varying degrees of structural dynamics (26), although it is not well understood what precise role this plays for catalytic activity. The current belief is that extensive structural dynamics in enzymes are not necessarily required for catalysis (27) but rather enact in regulation. For instance, many protein kinases exploit Tier-0 dynamics to generate active or inactive structural states (28). Tier-2 dynamics have been shown to be important for the evolution of enzymatic function (29). In addition to domain motions occurring within a structure, protein oligomerization and the possible quaternary dynamics can also be relevant for function (30). A well-characterized example is the allostery of hemoglobin that occurs on the transition between two distinct quaternary states (relaxed and tense) (31, 32). All this highlights that multi-Tier dynamics of proteins occur on various time- and length-scales (Fig. 1A) and are often the basis for function. It is, however, not yet well understood how structural dynamics are optimized during evolution to tailor protein function.
Analysis of protein sequences and structures has provided important insights into the evolution of protein function (33). A powerful approach is to assign the domain components of proteins to families and superfamilies on the basis of sequence alone (Pfam database for the protein family) (34) or in combination with structural information (class architecture topology homologous superfamily, CATH and structural classification of proteins, SCOP databases) (35, 36). The CATH and SCOP databases combined have identified ∼3,000 domain superfamilies comprising >50 million domains that account for ∼70% of the domains in completed genomes (37). More recently, the database ECOD (evolutionary classification of protein domains), groups domains by considering their evolutionary relationships (38). In ECOD, ∼760,000 domains have been assigned to 3,700 homologous groups (39). The most highly populated domain superfamilies/homologous groups are universal to all kingdoms of life (40, 41). The prevalence of proteins with multidomain architectures and the recurrent appearance of the same domain in nonhomologous proteins suggests that functional domains are reused when creating new proteins and functions (42, 43). Analyses of selected domain groups (44, 45), and more-recent large-scale investigations (46–48) have shown that domains in such groups generally share a common structural core (40 to 50% of the domain) that is highly conserved, even for relatives separated by billions of years. To achieve functional promiscuity of such a structural core, it has been proposed that larger structural embellishments (secondary structure elements or even entire domains) need to be added to the core for altered biochemical function (49). Such fusions occur frequently at the N or C termini (40), which has led to the belief that structural elements act as Lego bricks (47). Those are recombined in various ways for new functions to emerge during evolution (42, 49).
Much less is known on the role played by structural dynamics in the evolution of protein function (50–53). A recent model suggests that the native state of an evolved protein is the most abundant state of all possible structural states, which was selected for a specific function (54, 55). The “avant-garde” evolvability theory proposes the existence of a highly promiscuous primordial protein structure. It is assumed that for the emergence of the native state, the ability to evolve (evolvability) was traded for ligand-functional specificity (56, 57). These observations were experimentally verified (58) and agree with the proposal that changes in structural dynamics serve as a mechanism for the evolution of specialist enzymes from promiscuous generalists (59), though discordant examples are also observed in evolution (60). Furthermore, recent studies indicate that this evolvability theory can explain the short-period evolution (e.g., variation of enzyme local flexibility [variation of Tier-1/2 dynamics] to acquire new functions) particularly well (54–56, 61). Remarkably, this short-period evolution can be faithfully reproduced in vitro using directed evolutionary approaches based on consecutive rounds of single point mutations (54, 61). Ancestral protein reconstruction has also been useful in elucidating the role of structural dynamics in the emergence of specialized amino acid binding proteins from a promiscuous ancestor (62). However, it still remains unclear how, during longer periods of evolution, a primordial core structure evolves to modulate or diverge Tier-0 and/or quaternary dynamics, “generating” completely new functionalities.
Here, we test this aspect of the evolvability theory for proteins separated by long evolutionary periods during which Tier-0 or quaternary dynamics were introduced to an existing protein core structure. For this, we identified proteins with a bilobed domain structure with a high degree of plasticity and neutrality (29, 55), having 1) a conserved core structure, 2) large sequence diversity and related functional divergence, and 3) occurrence in all kingdoms of life. The selected structural core is composed of two Rossmann-like domains, which are believed to be among the most ancient architectures (63) and that are connected by a single β-sheet (Fig. 1B). We analyzed ∼600 proteins with this structural core that contain two related domains from the periplasmic-binding protein (PBP)–like II domain homologs (ECOD: X-, H- and T-groups PBP-like II). We show here that different members of these homologous proteins diverged with respect to domains or secondary structure elements, predominantly at their termini (Fig. 1 C and D). Using a combination of structural analysis and biophysical investigations, we demonstrate that such structural embellishments confer multi-Tier structural dynamics that diversify the function of the core structure to yield transcription factors, enzymes, or extracytoplasmic transport-related proteins. To understand both the mechanistic distinction of these proteins and the evolutionary trajectories, we used single-molecule Förster resonance energy transfer (smFRET) (64) and Hydrogen–Deuterium Exchange Mass Spectrometry (HDX-MS) (65). smFRET allows to monitor Tier-0 dynamics with a temporal resolution down to microseconds and subnanometer spatial resolution at the single-molecule level, even for highly heterogeneous structural ensembles (12, 66–68). HDX-MS complements smFRET, as it can probe Tier-1 dynamics throughout the structure that occur within the structural states identified by smFRET (Fig. 1A) (69). Using this combination of techniques, we show how the C-terminal extensions modulate Tier-0 dynamics in the structural core in a manner specific to their three-dimensional orientation. This arrangement also dictates specific geometrical criteria, crucial for establishing specific ligand interactions, all of which we detail in this work. On the other hand, we reveal the means by which N-terminal domain additions enable oligomerization to provide distinct quaternary dynamics in LysR-type transcriptional regulators (LTTRs). The remarkable modularity of these proteins permits us to confirm and expand the evolvability theory for the primordial core structure during a long-period evolution, which is largely facilitated, seemingly, by genetic recombination events.
Results
Structure, Classification, and Evolution of the Selected Bilobed Proteins.
In this study, our focus was on proteins composed of two globular lobes (bilobed), each of three layers (α/β/α), articulated around a central β-sheet hinge (Fig. 1B). This focus meant that not all multidomain architectures harboring the PBP-like II domains were included. To investigate the structural dynamics of the selected proteins, we constructed phylogenetic trees based on both sequence and structural information (Fig. 1C). This analysis indicated that the proteins evolved from a common ancestor that diversified into seven distinct structural classes A to G (Fig. 1 C and D), members of which are found throughout all kingdoms of life, in which some are even present in viruses (SI Appendix, Fig. S1A and Datasets S1 and S2). They have a consensus structure (Fig. 1B, Fig. 1 D, Top, and Dataset S3), which we have dubbed the cherry-core (hereafter CC, and proteins harboring this core, cherry-core proteins, CCPs) because of the bilobed structures’ resemblance to a cherry (SI Appendix, Fig. S2A), that contains two PBP-like II domains (70). According to ECOD database, the two domains of CCPs belong to the same X-, H-, and T-groups (PBP-like II; Dataset S3), supporting their common ancestry (38). In the consensus structure (Fig. 1 C and D), domains D1 and D2 adopt a face-to-face mirror-fashioned geometry with the active site, which is typically a ligand-binding site, located at their interface (SI Appendix, Fig. S2B).
Most of the selected proteins have distinct segments linked N-terminally to the CC (Fig. 1 C and D). Class A proteins have the addition of a LysR winged helix-turn-helix (HTH)–type DNA-binding domain (ECOD: X-, H-group: HTH, T-group: Winged; Fig. 1C). This element has been shown to be responsible for oligomerization and binding to promoter DNA (71–73). Most proteins of classes B through D, and F and G contain N-terminal localization signals for export via the general secretion (Sec) or the twin-arginine translocation (Tat) pathway (74–77). The presence of these signal peptides was obtained from UniProtKB (78) and verified manually by inspecting all 600 protein sequences with PRED-TAT, prediction of twin-arginine and secretory signal peptides (79). Class E proteins are predominantly cytosolic and lack an N-terminal signal peptide.
In addition, classes B through G have distinct C-terminal structural embellishments, hereafter termed C-tails (Fig. 1D). For example, Helical-tail 1 (H1) is common to all classes except C, and importantly, it has a similar placement in the three-dimensional architecture of the proteins. Other C-tails are unique to a specific class, such as the Helical-tails G (HG1 and 2), present only in class G proteins.
Function, Ligand Specificity, and Structural States of the Selected Bilobed Proteins.
The ligand specificity and function of the CCPs, as documented in UniProtKB (78), were found to correlate with the assigned structural class (SI Appendix, Fig. S1A). Class A proteins are bacterial transcription factors of the LTTR family. Class E proteins are predominantly eukaryotic single-turnover enzymes. The majority of the remainder (class B through D, F, and G) are found in prokaryotes and associate with the translocator domains of ABC transporters or with the membrane-embedded domains of chemoreceptors, in which they mediate unidirectional solute transport and signal transduction, respectively.
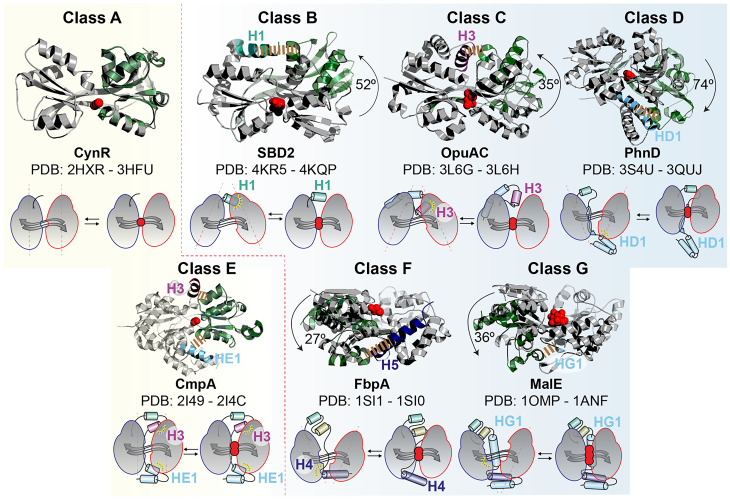
To investigate what role the distinct C-tails of these proteins might have played in structural dynamics and the evolution of new functions, we examined CCPs for which high-resolution structures of unliganded (apo) and liganded (holo) states were available (Fig. 2 and SI Appendix, Table S1). Interestingly, class A and E proteins, in most cases, exhibit nearly identical apo and holo structures. They also display the widest variety of substrates with little chemical structure similarity (SI Appendix, Fig. S3). For most members of the other classes, D1 and D2 of the CC undergo a rigid body rotation of varying degrees (Fig. 2 and SI Appendix, Table S1). For the solute-binding proteins, this mode of substrate binding has been termed the “Venus-Fly Trap mechanism” (80). These proteins recognize ligands with a specific pharmacophore (81): amino acids, ethanolamines, phosphonates, iron–phosphate complexes, and carbohydrates are recognized by classes B, C, D, F, and G, respectively (SI Appendix, Fig. S3). Another striking difference is that proteins in classes B through G are monomeric, whereas those in class A are oligomeric (see Fig. 4C).
Fig. 2.
Structures of CCPs highlighting their C-tails. Structures (Top) and schematics (Bottom) of the identified structural classes (A through G). The apo and holo structures of the indicated CCPs are superimposed to highlight the role of the C-tail. The domain to which the two structures were superimposed is represented in gray (D1 in A through E and D2 in F and G), whereas the one that is displaced relative to the C-tail in the open state is represented in green. Arrows and schematics indicate domain displacement (SI Appendix, Table S1). Interaction contact maps are presented in SI Appendix, Table S2. The most prominent contacts between the indicated secondary structure elements of the C-tail and D1 (F, G) or D2 (B through E) that stabilize the open state are indicated by a yellow dashed line. The PDB codes and protein names are indicated.
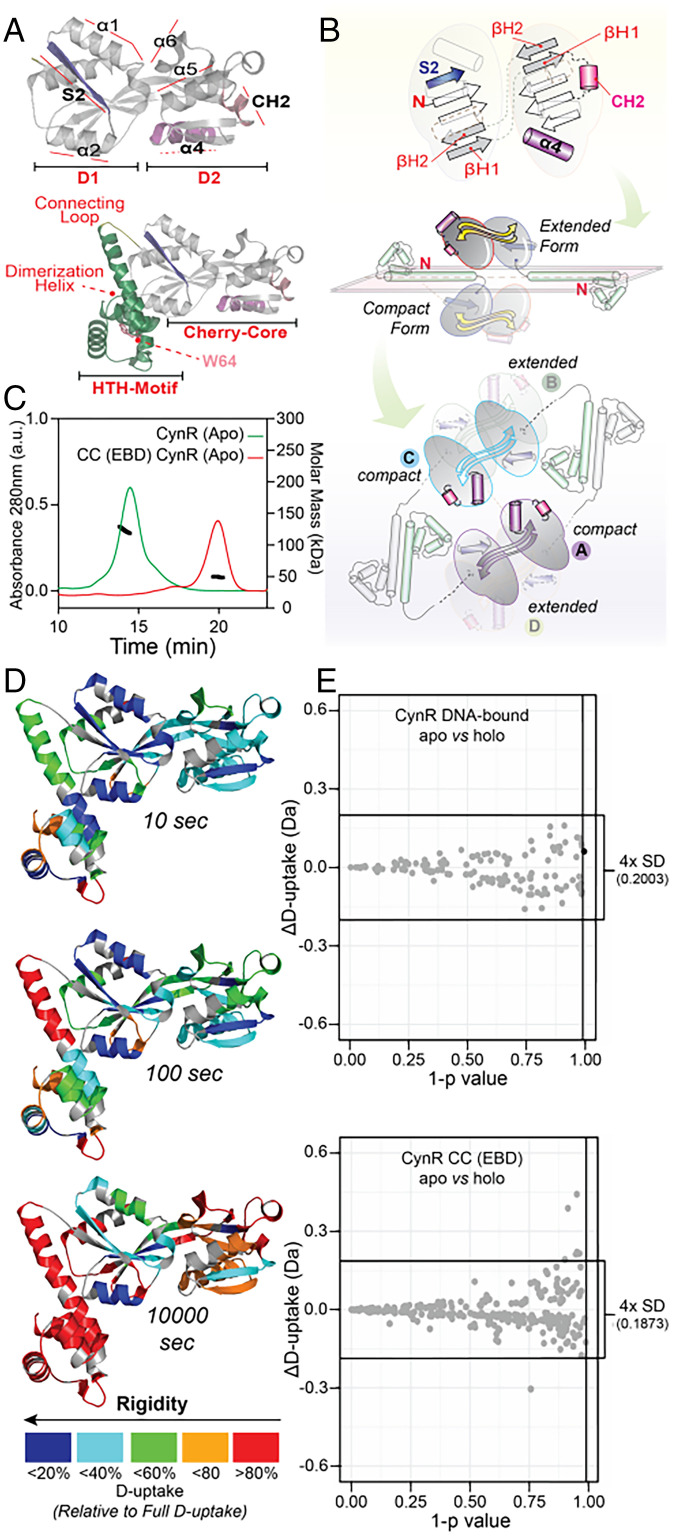
Fig. 4.
CynR tertiary and quaternary assemblies probed by MALS and HDX-MS. (A) Crystal structure of the CynR CC (PDB: 2HXR) with colored secondary structure elements discussed in the text that are critical for its quaternary dynamics (Top). A homology model of full-length CynR obtained from the SWISS-MODEL server with CbnR (PDB: 1IZ1) as a template (Bottom). The HTH domain is colored green, and the loop connecting it to the CC splitpea. W64 at the tip of the dimerization helix is indicated with an arrow. (B, Top) Schematic representation of the CC of CynR using the same color-coding as in A. (Middle) Schematic representation of the CynR homology model, with one of the protomers in the compact and the other in the extended configuration. The CC of the CynR protomers self-associate to form dimers or, for the full-length protein, tetramers. For clarity, two of the protomers have been omitted. (Bottom) The tetrameric assembly formed by the self-association of the dimerization helices and the CC. Of the two interacting protomers, one is present in the compact configuration, whereas the other is extended. The two protomers with the compact configuration are shown at the top of the plane, whereas those that are extended are at the bottom. (C) SEC-MALS analysis of full-length and CC of CynR (3 μM). Ultraviolet (UV) traces of the chromatograms were superimposed on the measured mass (black cycles). (D) Structural dynamics of full-length CynR in the absence of ligand and DNA by HDX-MS. Deuterium uptake values are reported for the incubation times in deuterated buffer and expressed relative to the fully deuterated control. These values are mapped onto the CynR homology model (as in A), using the indicated color gradient. Proline residues as well as the first residue of each peptide were excluded from mapping, as they do not contribute to the observed D-uptake. (E) Scatter plot visualization of the statistical analysis of D-uptake differences between the apo and holo states of full-length CynR (Upper) and the CC of CynR (Bottom). Three statistical criteria were used (SI Appendix, Material and Method), as described previously (98). Statistically significant differences would appear as black spheres (indicating that ΔD-uptake > 2×SD for a specific peptide), lying outside the 99% confidence threshold (1-P ≥ 0.99; indicated on y-axis) and outside the ± 4 × pooled average SD cut-off (indicated on x-axis; value given on the right).
smFRET to Monitor Large Domain Motions (Tier-0) in Bilobed Proteins.
To investigate structural dynamics of the CCPs, we first used smFRET (12, 66–68) to probe Tier-0 dynamics at near-physiological conditions in aqueous buffer at room temperature. One representative protein from each class was investigated: the effector binding domain (EBD) of CynR, representing the CC of full-length CynR (72) (class A), SBD2 (82) (class B), OpuAC (83) (class C), PhnD (84) (class D), CmpA (85) (class E), FbpA (86) (class F), and MalE (87) (class G). For these experiments, D1 and D2 were stochastically labeled with donor and acceptor fluorophores via cysteines that were substituted for nonconserved and surface-exposed residues, one in each domain (Fig. 3A). Fluorophore labeling was performed by maleimide–thiol conjugation (22, 88). Labeling positions were selected based on the crystal structures to show large changes in separation between the apo and holo states. smFRET was performed by confocal microscopy with alternating laser excitation (ALEX) (66).
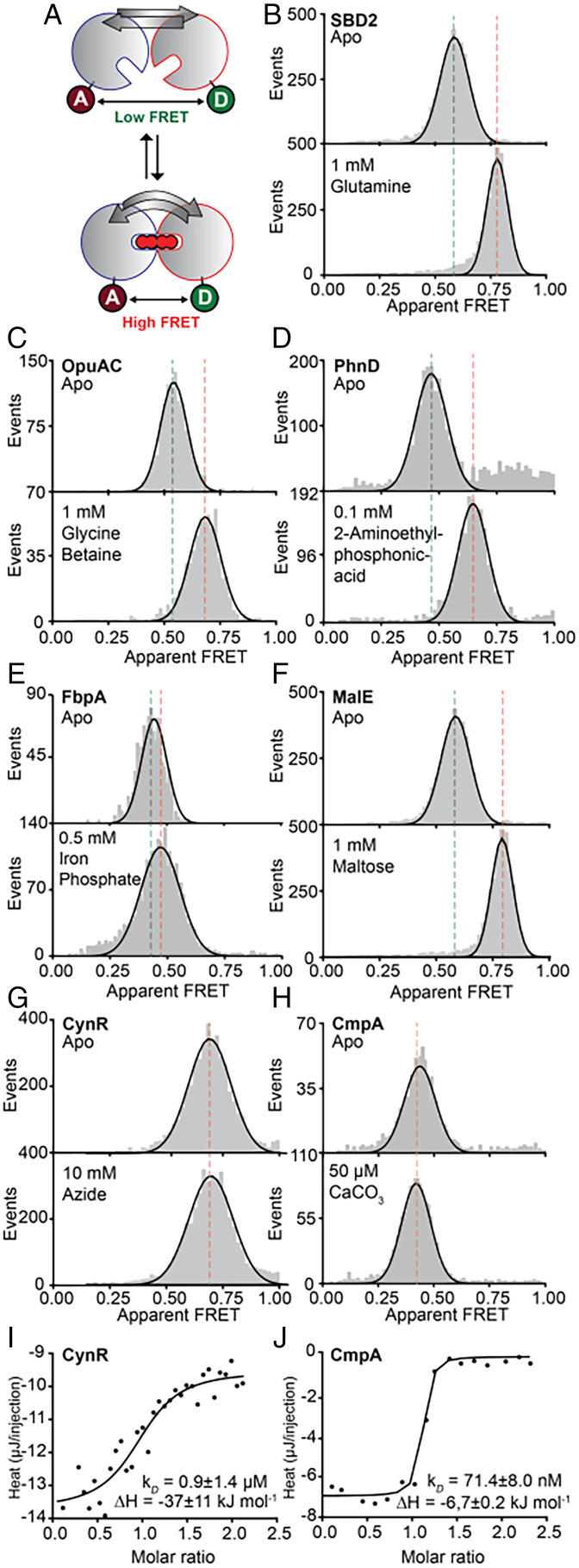
Fig. 3.
Monitoring structural changes and ligand binding in CCPs using smFRET and ITC. (A) Schematic of the experimental strategy to monitor structural states by FRET efficiency via stochastic labeling of D1 and D2 with donor and acceptor fluorophores. (B–H) Solution-based apparent FRET efficiency histograms in the absence (Top) or presence (Bottom) of saturating ligand concentrations for the indicated proteins. Gray bars are experimental data, and solid line is the fit. Centre position of Gaussian fits are given in SI Appendix, Table S5. (I and J) Binding isotherms of the calorimetric titration of azide (I) and calcium carbonate (J) to CynR (I) or CmpA (J), respectively, with the indicated thermodynamic parameters. For the apo condition of FbpA (E), the sample was treated extensively with citrate, as Iron (III) and the synergistic anion carbonate required for high-affinity binding to FbpA are removed efficiently by citrate treatment at low pH (92). According to high-resolution structural data (93), both Ca2+ and CO3− are present in the CmpA binding cleft. Indeed, we observed heat release upon titration of Ca2+ to a CO3− bound CmpA. Data points represent the heat of reaction per injection, and the line is the fit.
As predicted by our structural analysis (Fig. 2 and SI Appendix, Table S1) and in line with our previous observations (89), the FRET efficiency histograms and fitted distributions (Fig. 3 B–F) shifted toward higher FRET efficiency (E) values upon addition of saturating concentrations of ligand, for SBD2, OpuAC, PhnD, FbpA, and MalE. This indicates that in the apo state, the donor and acceptor dyes are further apart (Fig. 3 B–F, Upper, low FRET) compared to the holo state (Fig. 3 B–F, Lower, high FRET). Thus, our data suggest that ligand binding drives Tier-0 dynamics in these CCPs.
In contrast, the distributions of class A and E proteins [i.e., CynR (90) and CmpA (85), respectively] (Fig. 3 G and H), were virtually identical in the absence or presence of saturating ligand concentrations. Ligand binding was confirmed via isothermal titration calorimetry (ITC) showing binding affinities of both proteins in the micromolar range (Fig. 3 I and J). Thus, we conclude that the CC of class A and E proteins lack Tier-0 dynamics on the probed reaction coordinates for the selected FRET–distance pairs in contrast to the other structural classes.
Interestingly, this observation is in line with the known biological function. Class E proteins are predominantly single-turnover enzymes (SI Appendix, Fig. S1A), for which the rigidity of their active site is a prerequisite for catalysis (SI Appendix, Fig. S4A) (91). As previously suggested by us and others, Tier-0 dynamics in periplasmic-binding proteins (classes B, C, D, F, and G) are utilized in the regulation of transport in ABC importers (Discussion). A remaining question is, however, how ligand binding in class A proteins triggers transcriptional processes without major structural changes related to ligand binding.
HDX-MS to Examine Local Flexibility (Tier-1) of a Class A Protein, CynR.
The DNA-binding domain of class A proteins typically comprises a ∼58-aa HTH motif. This is followed by a ∼20-aa-long helix that provides a dimerization interface and a connecting loop that links the DNA-binding domain to the CC (Fig. 4A). The CC acts as the tetramerization interface within the full-length CCP or the dimerization interface within the EBD (Fig. 4B and SI Appendix, Fig. S4 B–E). Indeed, by using size exclusion chromatography with multi angle light scattering (SEC-MALS) we observed that (full-length) CynR is tetrameric, whereas its CC is dimeric (Fig. 4C).
To determine whether ligand binding to CynR resulted in structural changes that were not detectable along our selected reaction coordinate or that were too small or fast for smFRET (Fig. 3G), we performed HDX-MS. In contrast to smFRET, which reports on a single distance along a single reaction coordinate, HDX-MS probes structural dynamics at near–residue level resolution, providing global insights into Tier-1 dynamics. HDX-MS detects the exchange of hydrogens with deuterium at solvent-accessible and non–hydrogen-bonded backbone amides (94, 95). Hydrogens involved in stabilizing the secondary, tertiary, or quaternary structure of a protein via hydrogen bonds are exchanged more slowly through structural transitions that disrupt these bonds. Deuterium incorporation into the protein can then be determined, following proteolysis, by MS. The mass difference between hydrogen (1H) and deuterium (2H) results in a mass shift between nondeuterated and deuterated peptides that is a measure of the number of exchanged hydrogens (65, 96, 97).
For such investigations, the CC of CynR or full-length CynR were isotopically labeled (pD, 7.4; 25 °C) for different time periods (10 to 105 sec) either in free or DNA-bound states, and the peptic fragments were identified by MS (Fig. 4D and Dataset S4). For each peptide, the fraction of deuterium uptake relative to the maximum determined deuterium incorporation was calculated (Dataset S4). The data reveal a rigid character of the CC of CynR, whereas the dimerization helix and the rest of the DNA-binding domain turned out to be more flexible (Fig. 4D). To identify the regions in which pronounced structural changes were induced by azide binding, we performed comparative HDX-MS. For this, we determined the difference in deuterium uptake (ΔD) for each peptide between different conditions (e.g., CynR apo versus holo states). Observed differences would indicate a decrease (protection) or an increase (deprotection) of deuterium uptake upon azide binding (Dataset S4). No statistically significant change in the deuterium uptake was observed (Fig. 4E). From this, we can conclude that no detectable change in structural dynamics is induced by the ligand in the CC of CynR in its free or DNA-bound form (Fig. 4E and Dataset S4). This includes Tier-0 dynamics (detected by smFRET and HDX-MS) and Tier-1 or quaternary dynamics (both detected by HDX-MS).
An Asymmetric C-tail Drives Large-Scale Tertiary (Tier-0) Structural Changes.
From our results on the selected CCPs, only those with an asymmetric C-tail display Tier-0 dynamics (Figs. 2 and 3). To investigate how the C-tails introduced Tier-0 dynamics to the CC, we compared crystal structures of the apo and holo states of the CCPs to identify interactions between their C-tails and the CC. Fig. 2 summarizes the results of these comparisons. Interestingly, we found that the holo structures are similar for all CCPs, but the apo states are class specific. Contact mapping of the interactions between the CC and C-tail using the protein interaction calculator web-server (99) showed that the number and characteristics of the interactions depends on the structural class (Fig. 2 and SI Appendix, Table S2). Strikingly, these interactions stabilize predominantly the open state of the CC, the only exception being for classes A (C-tail-less) and E. In the former cases, stabilization is associated with an asymmetrical placement of the C-tail with respect to D1 and D2. In contrast, the C-tail of class E (predominantly H3 and HE1; Fig. 2) is placed symmetrically around D1 and D2 and thus cannot provide the required structural asymmetry needed to create a stable open state. The interactions of the C-tail to stabilize the open state involve the consensus CC-helices of D1 and D2 (Fig. 2 and SI Appendix, Table S2). In classes B, C, and D, such interactions involve D2, whereas D1 is contacted in classes F and G. These asymmetrical interactions create active sites with distinct geometries (Fig. 2 and SI Appendix, Fig. S2).
In alternative phylogenetic trees, based on the protein sequence using either D1/D2-domains or the C-tail, the clustering remains similar (SI Appendix, Fig. S1 B–D and Datasets S1 and S2), indicating that D1/D2 (CC domains) and the C-tail of a specific class coevolved to be part of the same polypeptide. This is in line with the role of the C-tail to interact with one specific domain, D1 or D2, to stabilize the open structural state.
Experimental Verification of the Role of C-tail Interactions in Stabilizing the Open State.
To confirm the role of the C-tail interactions with D1 or D2 for stabilization of the open state, we manipulated the relevant ones (Fig. 2 and SI Appendix, Table S2) in SBD2, SBD1, and MalE (Fig. 5 and SI Appendix, Figs. S5 and S6). The impact was tested via assessing structural states and ligand-binding affinities and monitored in smFRET experiments. Test cases were selected from class B and G, as these CCP classes are only remotely related by the first major clade in the evolutionary trees (Fig. 1C), and their open state is stabilized by distinct helices within the C-tails (Fig. 2), contacting either D2 (class B) or D1 (class G).
Fig. 5.
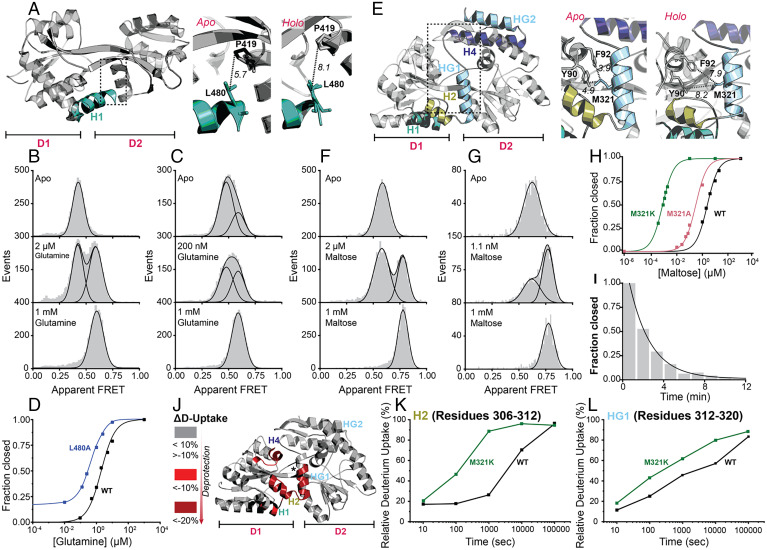
Experimental verification of the role of C-tail interactions in stabilizing the open conformation of SBD2 and MalE by smFRET and HDX-MS. (A) Dotted rectangles (Left) on the SBD2 (in A, PDB: 4KR5) structure highlights the critical contact region between the C-tail and the CC that stabilize the open state. Zoom in of rectangle regions (Middle and Right) depicting interactions between the C-tail helix H1 and D2 in the indicated apo (open) or holo (closed) states. Distances (Å) between L480 and P419 are shown as black dotted lines. (B and C) Solution-based apparent FRET efficiency histograms of SBD2 (B) and SBD2 (L480A) (C) at different conditions as indicated. (D) Fraction of the closed state (high-FRET state) of SBD2 and the indicated derivative as a function of glutamine concentration. (E) As in panel (A) for the MalE (PDB: 1OMP) structure with the indicated secondary structure elements and critical contacts. (F and G) Solution-based apparent FRET efficiency histograms of MalE (F) and MalE (M321K) (G) at different conditions as indicated. (H) Fraction of the closed state (high-FRET state) of MalE and its derivatives as a function of maltose concentration. (I) Maltose release from MalE (M321K) over time determined by solution-based smFRET (reference detailed values in SI Appendix, Fig. S6). Data points (panels D and H) and gray bars (panels B, C, F, G, and I) are the experimental data, and the solid line is the fit. (J) Map of regions in MalE structure that show statistically significant increase in deuterium uptake caused by M321K (numerical values and complete statistical analysis presented in Dataset S5). n = 3. (K and L) Deuterium uptake for the indicated MalE C-tail helices. SDs (SD) are shown in the deuterium uptake plots.
In SBD2, a hydrophobic interaction between L480 in the C-tail and P419 in D2 was weakened by substitution of L480 with alanine (L480A; Fig. 5A and SI Appendix, Table S2). The mutation resulted in the appearance of a subpopulation of molecules (∼20%) that were in the closed state in the absence of glutamine (Fig. 5 C versus B). To rule out the possibility that this might have been due to an artifact introduced by the choice of fluorophores, a second pair was tested, which showed a comparable result (SI Appendix, Fig. S5 A and B). We also examined whether residual endogenous ligand might account for this subpopulation by performing smFRET measurements on diluted samples, but these experiments displayed subpopulations of a similar size (SI Appendix, Fig. S5C). A closed-unliganded conformation was also observed for SBD2 previously but with a much lower abundance (∼1%). Detection of this small subpopulation required the use of confocal scanning microscopy (89), as populations <5% cannot be detected reliably with ALEX spectroscopy. We also observed small differences in the mean E values for apo and holo states of SBD2 (L480A) as compared to SBD2, suggesting that the structural landscape had been altered (SI Appendix, Fig. S5B). Destabilizing the open state is in line with the ∼10-fold increase in glutamine-binding affinity of SBD2 (L480A) as compared to SBD2 (Kd of 209 ± 64 nM and 1,990 ± 130 nM, respectively; Fig. 5D).
Determination of the dissociation constant () by smFRET measurements reports on the stability of the open state via Kd, where , is the Boltzmann constant, is the absolute temperature, and is the conformational (free) energy difference, where and are the (free) energies of the closed and open structural states without ligand bound, respectively (100). Thus, destabilizing the open state will decrease Kd and vice versa. From the percentages of open and closed in the absence of ligand, we obtain and for SBD2 (89) and SBD2 (L480A) (Fig. 5 B–D and SI Appendix, Fig. S5 A–C), respectively. Based on these results, the Kd value would be expected to decrease by ∼16 folds due to the L480A mutation, in close agreement with the estimated ∼10-fold difference. Similar trials on SBD1 did not show this trend (SI Appendix, Fig. S5 D–F), as the mutation had no impact on the structural dynamics of SBD1 (I249A). The absence of a subpopulation of molecules in the closed state at apoprotein conditions (alike SBD2) is based on the inability to evaluate all stabilizing open state interactions (SI Appendix, Table S2, compare SBD1 versus SBD2) so as to abolish the relevant ones. This is likely due to the fact that H1 residues of SBD1 (SI Appendix, Fig. S5G) participating in interprotomer contacts arose from crystallographic conditions (SI Appendix, Fig. S5H).
For MalE, we constructed the derivatives MalE (M321A) (101) and MalE (M321K), with weakened interactions between the C-tail HG1 and D1, by disrupting the hydrophobic interaction of M321 with Y90 and F92 and the aromatic sulfur interactions with Y90 (Fig. 5E and SI Appendix, Table S2). For MalE (M321A), the stabilizing interactions of M321 are partially abolished. This resulted in an eightfold increase of maltose affinity (Kd from 2,400 ± 400 nM in MalE to 300 ± 50 nM in MalE [M321A] [Fig. 5H and SI Appendix, Fig. S6]). An even-stronger effect was observed for MalE (M321K) with an affinity enhanced by 3,000-fold (Kd of 0.81 ± 0.15 nM; Fig. 5 G versus F and Fig. 5H). Shilton and coworkers have also proposed the hydrophobic interactions of M321 as being important structural determinants of the open state (101), affecting the affinity of MalE for maltose, in agreement with our results.
We next compared the lifetime of the closed, maltose-bound conformation of MalE (M321K) with that of MalE. Addition of 10 nM maltose allowed MalE (M321K) to occupy the closed state exclusively (SI Appendix, Fig. S6H). A total 20 µM unlabeled MalE (M321K) protein was subsequently added to scavenge maltose, which is stochastically released from the labeled protein. In a time-course experiment, the decrease in the population of closed, maltose-bound MalE (M321K) was then followed as a function of time (102) (Fig. 5I and SI Appendix, Fig. S6H). From these experiments, we established that the lifetime of the closed, maltose-bound conformation of MalE (M321K) was 122 ± 12 s. This value was 2,500-fold higher than the value determined for MalE (0.048 ± 0.010 s; SI Appendix, Fig. S6E). This result is consistent with the observed increase affinity of the derivatives for maltose.
The C-tail Local Dynamics Impact the Structural Transitions of the cherry-core.
In the above-described MalE derivatives, the structural basis for the destabilization of the open state could not be addressed mechanistically. Our smFRET experiments report on Tier-0 dynamics that affect the proteins’ tertiary structure (Fig. 1A). Considering that we observed differences in the Tier-0 dynamics, as a consequence of C-tail-D1/D2 destabilization, only for SBD2, we performed comparative HDX-MS (Dataset S5) to monitor changes in the secondary structure of MalE in comparison to MalE (M321K). For this, we determined the difference in deuterium uptake (ΔD) for each peptide between MalE and MalE (M321K) in their apo states (Fig. 5J). Remarkably, comparative HDX-MS indicates that the differences between MalE and MalE (M321K) are localized almost exclusively at the C-tail and specifically in regions interacting with D1 (Dataset S5 and Fig. 5J). The D-uptake of the C-tail helices H2/HG1 that are critical for stabilizing the open state (Fig. 5E and SI Appendix, Table S2) denotes that their rigidity was significantly reduced in the MalE derivative (Fig. 5 K and L). As might be expected, reduced rigidity occurred also in a region within the CC containing Y90 and F92. Notably, the same regions become allosterically destabilized in MalE upon maltose binding (Dataset S5). These results support the idea that the mutation leads to a weaker interaction between the C-tail and the CC resulting in a destabilization of the open state, because these C-tail elements and the region containing contact residues were found to be more flexible and solvent exposed.
Taken together, we postulate that ligands attenuate the stability of the open state in the CCPs, adopting multiple allosteric models for signal propagation (103): either via Tier-0 (SBD2 [Fig. 5 C and D]) or Tier1/2 (MalE [Fig. 5 G, H, K, and L]) dynamics.
Discussion
Common (structural) origin represents the hallmark of Darwinian evolution. Homology or descent from a common ancestor is often deduced from similarities in protein sequences or better from structures, as the latter are more conserved during evolution (104). However, similar structures can originate from divergent, convergent or parallel evolution (105). The most common “tricks” nature uses to vary a protein domain are the following: β-strand invasion/withdrawal, insertions/deletions/substitutions of secondary structure elements, domain flip/swaps, and circular permutations (106, 107). The currently established evolvability theory relies on investigations involving a fixed-length polypeptide chain by observing the effects of sequence variations, accomplished primarily by directed evolutionary approaches or by investigating closely related functional homologs. The functional promiscuity originating from structural variability is altered by the few amino acid modifications that can yield alterations of local structural fluctuations. For this reason, this theory can well explain protein evolution during short time periods.
In this study, we focused on the analysis of structures that have diverged over longer evolutionary periods. We analyzed a group of ∼600 proteins that share a core structure with the same topology of secondary structure elements giving rise to identical three-dimensional structures. The structure was predominantly varied by terminal embellishments and exhibits detectable sequence identity, used for constructing the sequence-based phylogenetic trees (SI Appendix, Fig. S1). The identified proteins likely emerged from divergent long-term evolution from a common ancestor, which spreads throughout the tree of life. This common ancestor is seemingly represented by the consensus core structure (CC; SI Appendix, Fig. S2) and encountered within the type-II class of PBPs (70, 108). As proposed previously (70), the CC derived possibly from a gene duplication of a PBP-like II domain (ECOD; Dataset S3) connected by a β-sheet (Fig. 6A).
Fig. 6.
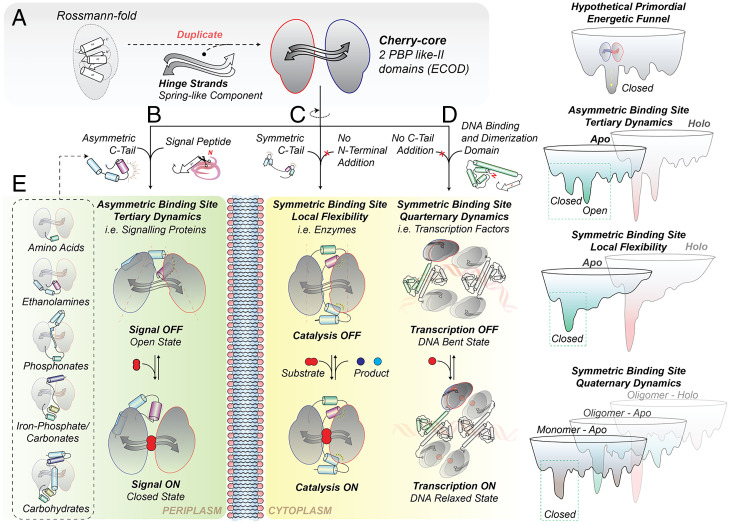
Model for the evolution of the cherry-core and hypothetical energetic funnels. (A) A gene duplication of a PBP-like II domain gave rise to the CC, being composed of a unique closed structural state, represented by a single well in the energetic funnel. The function of the CC was uniquely binding. Extant proteins acquired different extensions that altered their localization and dynamics and by that their function and specificity. (B) An N-terminal signal peptide and an asymmetrical C-terminal tail generated extracytoplasmic proteins evolving an additional open state. The different flavors of open states conferred distinct substrate specificities. The two states of CCPs, predominantly, signal substrate transport by their association to the membrane-embedded translocator domains of (ABC) transporters. (C) A symmetrical C-tail rigidified the closed state and yielded primarily single-turnover enzymes. (D) The N-terminal domain addition of a flexible dimerization helix and a DNA-binding motif (HTH-type) conferred distinct oligomeric assemblies with different quaternary dynamics, yielding transcription factors. (E) The different flavors of open states, conferred distinct substrate specificities. For details, refer to SI Appendix, Fig. S8.
CCPs with Asymmetric C-tails and N-Terminal Signal Peptides.
When the CC is fused N-terminally to a signal peptide and C-terminally to an asymmetric C-tail, the CC acts as an extracytoplasmic monomeric protein that associates with the translocator domains of ABC transporters or with chemoreceptors (SI Appendix, Fig. S1 and Fig. 6B). The two structural states are apo-open versus holo-closed and originate from Tier-0 dynamics, which are critical determinants of their biological function: the membrane-embedded partners can discriminate between open versus closed states to activate or inactivate a biological process (88, 109–112), such as solute transport. These structural transitions rely on the “generation” of an open state of the CC, accomplished by the C-tail, through its asymmetric interactions with either of the domains of the CC (D1 or D2; Fig. 2). Evidently, the open state is stabilized by such enthalpic contributions and destabilized by mutations or ligand binding that increase the flexibility of the interacting regions (Fig. 5J) (the C-tail elements with either D2 [classes B, C, and D] or D1 [classes F and G] [Fig. 2]). Seemingly, entropic contributions (protein conformational entropy) bias the structural equilibrium toward the closed state. Given the fact that the holo is the closed state driven by ligand binding (89), we anticipate that the interactions of the ligand with the CC cleft allosterically induce order-to-disorder transitions to alter the structural equilibrium. Depending on the placement of the asymmetric C-tails in the three-dimensional space, we identified five different “flavors” of open states, establishing distinct geometries of active sites; all resembling a triangle distinctly oriented in space (Figs. 2 and 6E). We verified that each active site geometry can recognize a specific chemical structure (SI Appendix, Fig. S3), in full agreement with ligand binding triggering closing.
In contrast to the C-tail, the presence of the N-terminal signal peptide does not affect the Tier-0 states of MalE (MalE versus proMalE; SI Appendix, Table S5). However, to render protein trafficking to extracytoplasmic locations possible, the presence of the signal peptide is known to delay MalE folding (113, 114). By that, preproteins are allowed to be secreted by the protein translocase (115).
The evolvability theory of Tawfik and coworkers (29, 57) explains the functional promiscuity encountered within the different structural classes. We selected two closely related proteins in our phylogenetic tree in class B (Fig. 1C) to illustrate this (Dataset S2, marked red asterisk). SBD1 (Protein Data Bank [PDB]: 4LA9) mediates the unidirectional transport of two substrates (glutamine and asparagine), whereas SBD2 (PDB: 4KR5) transports one of them (glutamine), though it captures it with higher affinity (88). SBD1 can be transformed to bind glutamine with higher affinity like SBD2 by mutating three amino acids (82). Clearly, functional promiscuity is traded seemingly with ligand specificity (62). However, only the modularity notion introduced in this study can explain a greater ligand-functional promiscuity. Class B proteins recognizing and mediating amino acid transport (SI Appendix, Fig. S1) would gain tertiary structural variability by acquiring four more helical elements at their C termini (H2, HG1, H4, and HG2; Fig. 1 C and D) (Fig. 6). This would potentially allow to switch them to class G to bind and mediate transport of carbohydrates (SI Appendix, Fig. S1A). According to another evolutionary trajectory, class B proteins, by acquiring helices H3, HE1, and HE2, could divert to class E and switch from transport-related proteins to enzymes (Fig. 6) by restricting their tertiary structural variability to a unique closed state. An extreme case of functional divergence has been experimentally verified in a recent study that restored the evolutionary history of the enzyme cyclohexadienyl dehydratase (116), a class B member according to our classification. The reconstructed ancestor of this enzyme was a highly promiscuous, transport-related protein possessing the open-unliganded and the closed-liganded structural states and was able to bind four different cationic amino acids. On the other side, cyclohexadienyl dehydratase binds a single substrate and forms a unique closed state for its catalytic function, since it is believed that structural sampling represents a constrain for catalytic activities (117, 118). Our study can now explain the means by which the addition of modular elements to a conserved core structure diversify its function (i.e., by modulating its structural landscape).
CCPs with Symmetric C-tails.
When the CC is combined C-terminally to a symmetric C-tail during evolution (Fig. 6C), the CC either operates as an enzyme or is associated with ABC transporters (SI Appendix, Fig. S1). Those proteins are present in a unique (apo and holo) closed state (Fig. 2), as asymmetrical interactions are an essential prerequisite for open state formation. Interestingly, these ABC transporter–associated proteins are both interacting with the actual translocator from the extracytoplasmic side (like the classes B, C, D, F, and G; described in the previous paragraph and in such a case also possess an N-terminal signal peptide) but are also tethered covalently to the translocator ATPase motor domains (93). As the switching (open to closed) behavior—to activate the transport cycle—is missing (88, 89, 109), the noncanonical arrangement of these ABC transporters could only trigger transport activation using a yet-uncharacterized mechanism. On the other hand, the lack of Tier-0 dynamics observed in the single-turnover enzymes is conforming to their enzymatic mechanism demanding an extremely rigid active site (SI Appendix, Fig. S4A) (91). Evidently, the rigidity required for the chemistry is so dramatic that the flexibility of a noncovalently (to the polypeptide chain) bound histidine to the cleft would render the reaction unproductive.
CCPs with N-Terminal Domains.
Lastly, the CC bearing an N-terminal domain harboring the HTH-type DNA-binding domain but no C-tail (Fig. 6D) yields transcription factors of the LTTR family. Apparently, in such a case, Tier-0 dynamics of the CC are not required for function (Fig. 3G). The rigidity of the CC (Fig. 4D) is evidently required for the stability of the quaternary assemblies, as those have large cavities and holes (SI Appendix, Fig. S7) (119–124).
Structural Basis of the Transcriptional Activation by the LysR-Type Regulators.
Our data and analysis gave insights into the steps taken during evolution to shape the CC into functional ligand receptor–associated proteins (classes B, C, D, F, and G) but also one-turn enzymes (class E). Yet, how the ligand-driven signal propagates within the LTTR family, such as CynR, remains largely elusive.
The CC of CynR constitutes only one part of its structure (i.e., the sensory EBD) of a full-length transcription factor of the LTTR family with an additional winged HTH-type DNA-binding domain (Fig. 4 A and B). Although only little direct experimental evidence is available, it has been proposed that changes in the tetrameric assembly of LysR-type transcription factors can be induced by ligand binding (72, 125). These quaternary dynamics were suggested to activate transcription by a transition from a bent DNA (transcription OFF) to an unbent state (transcription ON) (72, 125). Our smFRET and HDX results (Figs. 3G and 4E), however, did not reveal any detectable changes of the CC upon azide binding. This suggests that ligand-driven structural changes within the CC protomers are minor and might thus be very hard to detect.
To gain insights into potential quaternary dynamics, we analyzed the available high-resolution structures of the oligomeric assemblies belonging to the LysR transcription factors (119–124) to identify the evolutionary relevant ones by Evolutionary Protein–Protein Interface Classifier (EPIC) (126, 127). Subsequently, we modeled the CynR sequence after these structures in the presence or absence of DNA (SI Appendix, Fig. S7). The structural basis of the distinct tetrameric assemblies is the positioning of the dimerization helix with respect to the CC, dictated by the connecting loop (Fig. 4 A and B). In the extreme case that an additional helix (RD1-CH5; Dataset S3) is linked to and displaces the connecting loop, an octameric assembly is obtained (SI Appendix, Fig. S7H). To shed light on this, we monitored intrinsic Trp fluorescence during thermal melting (SI Appendix, Table S3). From the two tryptophans of CynR, one present in the dimerization helix (64 aa) and the other on D1 (274 aa) of the CC (Fig. 4 A and B), only the former one contributes to a fluorescent signal (SI Appendix, Table S3). Under DNA-free apo conditions, CynR displays a Tm(app) ∼55 °C that is significantly destabilized (∼6 °C) after binding to DNA. Addition of azide at free or DNA-bound CynR causes a ligand-characteristic signature throughout the Trp-temperature spectrums, giving rise to a secondary Tm(app) at ∼27 °C in both cases (SI Appendix, Table S3).
To understand the signal propagation originating from ligand binding, we inspected the available structural information (SI Appendix, Fig. S7). In agreement with the experimental findings (Figs. 3G and 4E), our structural analysis indicated that D1 and D2 motions are not required (SI Appendix, Table S4) for the oligomeric assemblies of the same transcription factor (OxyR) to differ dramatically in order to trigger structural changes on DNA (SI Appendix, Fig. S7 E–G). What varies between those assemblies is the order–disorder of specific secondary structure elements within D2 of the CC (S4 till α5; Dataset S3). We anticipate that such changes in the flexibility of secondary structure elements are induced by ligand binding, like in the case of MalE (Fig. 5 J–L).
We conclude that signal propagation in CCPs comprising large D1/D2 rearrangements (Fig. 2; classes B, C, D, F, and G) driven by the C-tail/D1 and D2 interactions involves bending/unbending of the “spring-like” hinge (108). On the other side, propagation in class A is initiated by small/localized rearrangements of secondary structure element within the CC somehow transmitted to the N-terminal HTH domain leading to global quaternary structural changes (SI Appendix, Figs. S7 and S8). Additional analysis regarding the molecular mechanisms of LTTR-transcriptional regulators will be the subject of future studies.
The Energy Landscape of the CCPs over Time.
Fig. 6 summarizes our proposed evolutionary path taken by the CC proteins over long time periods. The terminal modules impact and finetune the multi-Tier structural dynamics as can be described by the folding funnel model (8). The CCPs with asymmetric C-tails displaying Tier-0 dynamics (i.e., open and closed states) have the two characteristic wells in the funnel separated by a large energetic barrier (Fig. 6B and SI Appendix, Fig. S8C). For that reason, such proteins are found predominantly in the lowest–energetic level open state (apo energetic funnel; SI Appendix, Fig. S8C), with infrequent transitions to the closed one (Fig. 5B). Only in the SBD1/2 proteins (class B), ∼1% occurrence of a closed state has been experimentally observed (89). By destabilizing the open state of SBD2, we obtained a ∼20% occurrence of a closed state (Fig. 5C). As in the holo state energetic funnel, the lowest energy state is the closed one (SI Appendix, Fig. S8C); addition of the ligand shifts the equilibrium toward the closed state. Since the dissociation constant (KD) derives from the difference between the lowest energetic levels of the apo and holo funnels (SI Appendix, Fig. S8), destabilization of the C-tail (Fig. 5 D and H) that directs the open state of the apo funnel at a higher energetic level leads to an increased affinity.
Alternatively, CCPs with a symmetric C-tail (Fig. 6C) have a single main energetic well, defining the unique closed structural state both in the apo or holo funnels (SI Appendix, Fig. S8D).
The CCPs with N-terminal domain additions (Fig. 6D) have multiple energetic funnels corresponding to oligomerization states (SI Appendix, Fig. S8E). The energetic funnel of such CCPs is having multiple minima corresponding to the many different arrangements of the flexible N-terminal domain with respect to the CC (SI Appendix, Fig. S8E). Self-association (oligomer formation) and/or binding to their partners/ligands deepens specific wells that are required for function.
We anticipate the energetic funnel of the primordial consensus CC to be extremely rugged, with small energetic barriers between the wells, with a single well somewhat deeper (corresponding to the closed state), thus allowing sampling of multiple structural states (SI Appendix, Fig. S8A). Such a structural variability would lead to increased substrate promiscuity, however, exploiting extremely weak interactions (i.e., low binding affinities; SI Appendix, Fig. S8B).
Clearly, the structural promiscuity achieved by the modularity introduced in this study, expands the protein evolvability theory and establishes the notion that in order to comprehend protein evolution, it is essential to decode the energetic funnel of structural homologs. Structural elements or even domains added alike Lego bricks to a structural core, being the Lego Board, trigger distinct evolutionary trajectories.
Materials and Methods
Detailed materials and methods are included in SI Appendix, Material and Method. All proteins were expressed in Escherichia coli cells (BL21 DE3 or BL21 pLysS DE3) and grew in Luria Bertani or Terrific Broth media. Purification was based on affinity [Ni-NTA (Qiagen)], anion exchange [Q Sepharose (GE Healthcare)], and/or size-exclusion [Hi Load 26/60 Superdex 200 (GE Healthcare)] chromatography. Samples used for analysis (ITC, smFRET, HDX-MS, etc.) were single monodisperse peaks. Phylogenetic, structural analysis, alignments, and protein visualization were accomplished by widely accepted procedures via freely available software or servers: Structure similarity search-PDB webserver (128, 129), PDBeFold (130), Dali server (131), multidimensional QR factorization of multiple sequence and structure alignments (132)/visual molecular dynamics 1.9.2 software package (133), Protein blast (134), DynDom domain motion server (135, 136), Protein Interaction calculator webserver (99), EPIC server (126, 127), ECOD database (38), ConSurf-DB server (137, 138), SWISS-MODEL server (139), and PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC). smFRET experiments were performed with a custom-made confocal ALEX microscope that has been previously described in the literature (22, 89) and HDX-MS by a nanoACQUITY Ultra Performance Liquid Chromatography System with HDX technology (Waters, United Kingdom). Data and statistical analyses were performed by established procedures described in detail in the literature.
Supplementary Material
Acknowledgments
This work was financed by an Netherlands Organization for Scientific Research (NWO Veni grant 722.012.012 to G.G.), an ERC Starting Grant (European Research Council StG No. 638536, SM-IMPORT to T.C.), Deutsche Forschungsgemeinschaft within GRK2062 (project C03 to T.C.), SFB863 (project A10 to M.Z. and A13 to T.C.), Research Foundation Flanders (FWO CARBS #G0C6814N to G.G. and A.E.). G.G. also acknowledges a fellowship from the European Molecular Biology Organization (EMBO long-term fellowship ALF 47-2012 to G.G.) and financial support by the Zernike Institute for Advanced Materials and the Rega foundation. Y.A.M. was supported by the Indonesia Endowment Fund for Education (Lembaga Pengelola Dana Pendidikan, Republik Indonesia, LPDP RI PhD scholarship). G.G. and Y.A.M. acknowledge financial support from the IMBB-FORTH (start-up grant to G.G.). N.Z. acknowledges an Alexander von Humboldt postdoctoral fellowship. N.E. acknowledges a fellowship from the Marie Sklodowska Curie Action (MSCA SoE FWO (195872). R.X. was supported by the Chinese Scholarship Council grant. T.C. was supported by the German Academic Exchange Service, Center of Nanoscience Munich, LMU excellent and the Center for Integrated Protein Science Munich. We thank Eitan Lerner for useful comments and critical reading of the manuscript, Monique Wiertsema for help with smFRET experiments, and Florence Husada for support with ITC experiments.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026165118/-/DCSupplemental.
Data Availability
Study data are included in the article, supporting information or SI Appendix and Datasets.
References
- 1.Cooper G. M., Hausman R. E., The Cell: A Molecular Approach (Sinauer Associates, Sunderland, MA, Washington, D.C., ed. 6, 2013). [Google Scholar]
- 2.Campbell I. D., Timeline: The march of structural biology. Nat. Rev. Mol. Cell Biol. 3, 377–381 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C., Dynamics of ligand binding to myoglobin. Biochemistry 14, 5355–5373 (1975). [DOI] [PubMed] [Google Scholar]
- 4.Frauenfelder H., McMahon B. H., Fenimore P. W., Myoglobin: The hydrogen atom of biology and a paradigm of complexity. Proc. Natl. Acad. Sci. U.S.A. 100, 8615–8617 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frauenfelder H., Sligar S. G., Wolynes P. G., The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Nienhaus K., Deng P., Kriegl J. M., Nienhaus G. U., Structural dynamics of myoglobin: Effect of internal cavities on ligand migration and binding. Biochemistry 42, 9647–9658 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Okazaki K., Koga N., Takada S., Onuchic J. N., Wolynes P. G., Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 103, 11844–11849 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leopold P. E., Montal M., Onuchic J. N., Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc. Natl. Acad. Sci. U.S.A. 89, 8721–8725 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G., Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 21, 167–195 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Liu S. Q., et al. , “Protein folding, binding and energy landscape: A synthesis” in Protein Engineering, Kaumaya P. P., Ed. (InTech, Rijeka, 2012), pp. 207–252. [Google Scholar]
- 11.Wales D. J., Energy Landscapes (Cambridge University Press, Cambridge, UK, 2003). [Google Scholar]
- 12.Henzler-Wildman K., Kern D., Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Schuler B., Hofmann H., Single-molecule spectroscopy of protein folding dynamics—Expanding scope and timescales. Curr. Opin. Struct. Biol. 23, 36–47 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Schuler B., Soranno A., Hofmann H., Nettels D., Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu. Rev. Biophys. 45, 207–231 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Ansari A., et al. , Protein states and proteinquakes. Proc. Natl. Acad. Sci. U.S.A. 82, 5000–5004 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vale R. D., The molecular motor toolbox for intracellular transport. Cell 112, 467–480 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Cabezon E., Lanza V. F., Arechaga I., Membrane-associated nanomotors for macromolecular transport. Curr. Opin. Biotechnol. 23, 537–544 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Husada F., et al. , Conformational dynamics of the ABC transporter McjD seen by single-molecule FRET. EMBO J. 37, e100056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Noinaj N., Buchanan S. K., Structural insights into the transport of small molecules across membranes. Curr. Opin. Struct. Biol. 27, 8–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang M., et al. , Single-molecule probing of the conformational homogeneity of the ABC transporter BtuCD. Nat. Chem. Biol. 14, 715–722 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Gouridis G., et al. , ABCE1 controls ribosome recycling by an asymmetric dynamic conformational equilibrium. Cell Rep. 28, 723–734.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox A. D., Der C. J., Ras history: The saga continues. Small GTPases 1, 2–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smock R. G., Gierasch L. M., Sending signals dynamically. Science 324, 198–203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzeng S. R., Kalodimos C. G., Dynamic activation of an allosteric regulatory protein. Nature 462, 368–372 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Kamerlin S. C., Warshel A., At the dawn of the 21st century: Is dynamics the missing link for understanding enzyme catalysis? Proteins 78, 1339–1375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loveridge E. J., Behiry E. M., Guo J., Allemann R. K., Evidence that a ‘dynamic knockout’ in Escherichia coli dihydrofolate reductase does not affect the chemical step of catalysis. Nat. Chem. 4, 292–297 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Huse M., Kuriyan J., The conformational plasticity of protein kinases. Cell 109, 275–282 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Petrović D., Risso V. A., Kamerlin S. C. L., Sanchez-Ruiz J. M., Conformational dynamics and enzyme evolution. J. R. Soc. Interface 15, 20180330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh J. A., Teichmann S. A., Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 84, 551–575 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Monod J., Wyman J., Changeux J. P., On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12, 88–118 (1965). [DOI] [PubMed] [Google Scholar]
- 32.Perutz M. F., Stereochemistry of cooperative effects in haemoglobin. Nature 228, 726–739 (1970). [DOI] [PubMed] [Google Scholar]
- 33.Reid A. J., Yeats C., Orengo C. A., Methods of remote homology detection can be combined to increase coverage by 10% in the midnight zone. Bioinformatics 23, 2353–2360 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Finn R. D., et al. , Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreeva A., et al. , Data growth and its impact on the SCOP database: New developments. Nucleic Acids Res. 36, D419–D425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sillitoe I., et al. , CATH: Comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 43, D376–D381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lees J. G., Dawson N. L., Sillitoe I., Orengo C. A., Functional innovation from changes in protein domains and their combinations. Curr. Opin. Struct. Biol. 38, 44–52 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Cheng H., et al. , ECOD: An evolutionary classification of protein domains. PLOS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaeffer R. D., Liao Y., Cheng H., Grishin N. V., ECOD: New developments in the evolutionary classification of domains. Nucleic Acids Res. 45, D296–D302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buljan M., Bateman A., The evolution of protein domain families. Biochem. Soc. Trans. 37, 751–755 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Chothia C., Gough J., Vogel C., Teichmann S. A., Evolution of the protein repertoire. Science 300, 1701–1703 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Das S., Smith T. F., Identifying nature’s protein Lego set. Adv. Protein Chem. 54, 159–183 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Söding J., Lupas A. N., More than the sum of their parts: On the evolution of proteins from peptides. BioEssays 25, 837–846 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Dessailly B. H., Redfern O. C., Cuff A. L., Orengo C. A., Detailed analysis of function divergence in a large and diverse domain superfamily: Toward a refined protocol of function classification. Structure 18, 1522–1535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galperin M. Y., Koonin E. V., Divergence and convergence in enzyme evolution. J. Biol. Chem. 287, 21–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuff A., et al. , The CATH hierarchy revisited-structural divergence in domain superfamilies and the continuity of fold space. Structure 17, 1051–1062 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferruz N., et al. , Identification and analysis of natural building blocks for evolution-guided fragment-based protein design. J. Mol. Biol. 432, 3898–3914 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves G. A., Dallman T. J., Redfern O. C., Akpor A., Orengo C. A., Structural diversity of domain superfamilies in the CATH database. J. Mol. Biol. 360, 725–741 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Das S., Dawson N. L., Orengo C. A., Diversity in protein domain superfamilies. Curr. Opin. Genet. Dev. 35, 40–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahar I., Chennubhotla C., Tobi D., Intrinsic dynamics of enzymes in the unbound state and relation to allosteric regulation. Curr. Opin. Struct. Biol. 17, 633–640 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Sharma N., General I. J., Schreiber G., Bahar I., Dynamic modulation of binding affinity as a mechanism for regulating interferon signaling. J. Mol. Biol. 429, 2571–2589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcos E., Crehuet R., Bahar I., Changes in dynamics upon oligomerization regulate substrate binding and allostery in amino acid kinase family members. PLoS Comput. Biol. 7, e1002201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S., Li H., Krieger J. M., Bahar I., Shared signature dynamics tempered by local fluctuations enables fold adaptability and specificity. Mol. Biol. Evol. 36, 2053–2068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otten R., et al. , Rescue of conformational dynamics in enzyme catalysis by directed evolution. Nat. Commun. 9, 1314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tokuriki N., Tawfik D. S., Protein dynamism and evolvability. Science 324, 203–207 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Aharoni A., et al. , The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37, 73–76 (2005). [DOI] [PubMed] [Google Scholar]
- 57.James L. C., Tawfik D. S., Conformational diversity and protein evolution – A 60-year-old hypothesis revisited. Trends Biochem. Sci. 28, 361–368 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Wouters M. A., Liu K., Riek P., Husain A., A despecialization step underlying evolution of a family of serine proteases. Mol. Cell 12, 343–354 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Zou T., Risso V. A., Gavira J. A., Sanchez-Ruiz J. M., Ozkan S. B., Evolution of conformational dynamics determines the conversion of a promiscuous generalist into a specialist enzyme. Mol. Biol. Evol. 32, 132–143 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Wheeler L. C., Lim S. A., Marqusee S., Harms M. J., The thermostability and specificity of ancient proteins. Curr. Opin. Struct. Biol. 38, 37–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harel M., et al. , Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 11, 412–419 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Clifton B. E., Jackson C. J., Ancestral protein reconstruction yields insights into adaptive evolution of binding specificity in solute-binding proteins. Cell Chem. Biol. 23, 236–245 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Hanukoglu I., Proteopedia: Rossmann fold: A beta-alpha-beta fold at dinucleotide binding sites. Biochem. Mol. Biol. Educ. 43, 206–209 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Ha T., et al. , Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc. Natl. Acad. Sci. U.S.A. 93, 6264–6268 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konermann L., Pan J., Liu Y. H., Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Hohlbein J., Craggs T. D., Cordes T., Alternating-laser excitation: Single-molecule FRET and beyond. Chem. Soc. Rev. 43, 1156–1171 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Kapanidis A. N., et al. , Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. U.S.A. 101, 8936–8941 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lerner E., et al. , Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 359, eaan1133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng J., Strutzenberg T., Pascal B. D., Griffin P. R., Protein dynamics and conformational changes explored by hydrogen/deuterium exchange mass spectrometry. Curr. Opin. Struct. Biol. 58, 305–313 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Fukami-Kobayashi K., Tateno Y., Nishikawa K., Domain dislocation: A change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C., A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. U.S.A. 85, 6602–6606 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maddocks S. E., Oyston P. C. F., Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology (Reading) 154, 3609–3623 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Schell M. A., Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47, 597–626 (1993). [DOI] [PubMed] [Google Scholar]
- 74.Albiniak A. M., Baglieri J., Robinson C., Targeting of lumenal proteins across the thylakoid membrane. J. Exp. Bot. 63, 1689–1698 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Frain K. M., Gangl D., Jones A., Zedler J. A., Robinson C., Protein translocation and thylakoid biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857, 266–273 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Natale P., Brüser T., Driessen A. J., Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—Distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Orfanoudaki G., Economou A., Proteome-wide subcellular topologies of E. coli polypeptides database (STEPdb). Mol. Cell. Proteomics 13, 3674–3687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.UniProt Consortium, UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bagos P. G., Nikolaou E. P., Liakopoulos T. D., Tsirigos K. D., Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26, 2811–2817 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Mao B., Pear M. R., McCammon J. A., Quiocho F. A., Hinge-bending in L-arabinose-binding protein. The “Venus’s-flytrap” model. J. Biol. Chem. 257, 1131–1133 (1982). [PubMed] [Google Scholar]
- 81.Wermuth C. G., Ganellin C. R., Lindberg P., Mitscher L. A., Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998). Pure Appl. Chem. 70, 1129–1143 (1998). [Google Scholar]
- 82.Fulyani F., et al. , Functional diversity of tandem substrate-binding domains in ABC transporters from pathogenic bacteria. Structure 21, 1879–1888 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Wolters J. C., et al. , Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA. PLoS One 5, e10361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alicea I., et al. , Structure of the Escherichia coli phosphonate binding protein PhnD and rationally optimized phosphonate biosensors. J. Mol. Biol. 414, 356–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maeda S., Price G. D., Badger M. R., Enomoto C., Omata T., Bicarbonate binding activity of the CmpA protein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in active transport of bicarbonate. J. Biol. Chem. 275, 20551–20555 (2000). [DOI] [PubMed] [Google Scholar]
- 86.Shouldice S. R., et al. , Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens. J. Bacteriol. 186, 3903–3910 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shilton B. H., The dynamics of the MBP-MalFGK(2) interaction: A prototype for binding protein dependent ABC-transporter systems. Biochim. Biophys. Acta 1778, 1772–1780 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Gouridis G., et al. , Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat. Struct. Mol. Biol. 22, 57–64 (2015). [DOI] [PubMed] [Google Scholar]
- 89.de Boer M., et al. , Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. eLife 8, e44652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lamblin A. F., Fuchs J. A., Functional analysis of the Escherichia coli K-12 cyn operon transcriptional regulation. J. Bacteriol. 176, 6613–6622 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lai R. Y., et al. , Thiamin pyrimidine biosynthesis in Candida albicans : A remarkable reaction between histidine and pyridoxal phosphate. J. Am. Chem. Soc. 134, 9157–9159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo M., et al. , Synergistic anion and metal binding to the ferric ion-binding protein from Neisseria gonorrhoeae. J. Biol. Chem. 278, 2490–2502 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Koropatkin N. M., Koppenaal D. W., Pakrasi H. B., Smith T. J., The structure of a cyanobacterial bicarbonate transport protein, CmpA. J. Biol. Chem. 282, 2606–2614 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Ramisetty S. R., Washburn M. P., Unraveling the dynamics of protein interactions with quantitative mass spectrometry. Crit. Rev. Biochem. Mol. Biol. 46, 216–228 (2011). [DOI] [PubMed] [Google Scholar]
- 95.Wales T. E., Engen J. R., Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006). [DOI] [PubMed] [Google Scholar]
- 96.Engen J. R., Smith D. L., Investigating the higher order structure of proteins. Hydrogen exchange, proteolytic fragmentation, and mass spectrometry. Methods Mol. Biol. 146, 95–112 (2000). [DOI] [PubMed] [Google Scholar]
- 97.Englander S. W., Kallenbach N. R., Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 16, 521–655 (1983). [DOI] [PubMed] [Google Scholar]
- 98.Tsirigotaki A., Papanastasiou M., Trelle M. B., Jørgensen T. J., Economou A., Analysis of translocation-competent secretory proteins by HDX-MS. Methods Enzymol. 586, 57–83 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Tina K. G., Bhadra R., Srinivasan N., PIC: Protein interactions calculator. Nucleic Acids Res. 35, W473–W476 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Boer M., Gouridis G., Muthahari Y. A., Cordes T., Single-molecule observation of ligand binding and conformational changes in FeuA. Biophys. J. 117, 1642–1654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Telmer P. G., Shilton B. H., Insights into the conformational equilibria of maltose-binding protein by analysis of high affinity mutants. J. Biol. Chem. 278, 34555–34567 (2003). [DOI] [PubMed] [Google Scholar]
- 102.de Boer M., Maximum likelihood analysis of non-equilibrium solution-based single-molecule FRET data. arXiv [Preprint] (2020). https://arxiv.org/abs/2008.12278. Accessed 27 August 2020.
- 103.Motlagh H. N., Wrabl J. O., Li J., Hilser V. J., The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Illergård K., Ardell D. H., Elofsson A., Structure is three to ten times more conserved than sequence--a study of structural response in protein cores. Proteins 77, 499–508 (2009). [DOI] [PubMed] [Google Scholar]
- 105.Elias M., Tawfik D. S., Divergence and convergence in enzyme evolution: Parallel evolution of paraoxonases from quorum-quenching lactonases. J. Biol. Chem. 287, 11–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grishin N. V., Fold change in evolution of protein structures. J. Struct. Biol. 134, 167–185 (2001). [DOI] [PubMed] [Google Scholar]
- 107.Peisajovich S. G., Rockah L., Tawfik D. S., Evolution of new protein topologies through multistep gene rearrangements. Nat. Genet. 38, 168–174 (2006). [DOI] [PubMed] [Google Scholar]
- 108.Scheepers G. H., Lycklama A Nijeholt J. A., Poolman B., An updated structural classification of substrate-binding proteins. FEBS Lett. 590, 4393–4401 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Cui J., Davidson A. L., ABC solute importers in bacteria. Essays Biochem. 50, 85–99 (2011). [DOI] [PubMed] [Google Scholar]
- 110.Falke J. J., Hazelbauer G. L., Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26, 257–265 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Higgins C. F., ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 (1992). [DOI] [PubMed] [Google Scholar]
- 112.Zhang Y., et al. , Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. U.S.A. 96, 939–944 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beena K., Udgaonkar J. B., Varadarajan R., Effect of signal peptide on the stability and folding kinetics of maltose binding protein. Biochemistry 43, 3608–3619 (2004). [DOI] [PubMed] [Google Scholar]
- 114.Randall L. L., Hardy S. J., Correlation of competence for export with lack of tertiary structure of the mature species: A study in vivo of maltose-binding protein in E. coli. Cell 46, 921–928 (1986). [DOI] [PubMed] [Google Scholar]
- 115.Tsirigotaki A., De Geyter J., Šoštaric N., Economou A., Karamanou S., Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 15, 21–36 (2017). [DOI] [PubMed] [Google Scholar]
- 116.Clifton B. E., et al. , Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nat. Chem. Biol. 14, 542–547 (2018). [DOI] [PubMed] [Google Scholar]
- 117.Bar-Even A., Milo R., Noor E., Tawfik D. S., The moderately efficient enzyme: Futile encounters and enzyme floppiness. Biochemistry 54, 4969–4977 (2015). [DOI] [PubMed] [Google Scholar]
- 118.Campbell E., et al. , The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 12, 944–950 (2016). [DOI] [PubMed] [Google Scholar]
- 119.Jiang Y. L., et al. , Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR. Proc. Natl. Acad. Sci. U.S.A. 115, 403–408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jo I., et al. , Structural details of the OxyR peroxide-sensing mechanism. Proc. Natl. Acad. Sci. U.S.A. 112, 6443–6448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Monferrer D., et al. , Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol. Microbiol. 75, 1199–1214 (2010). [DOI] [PubMed] [Google Scholar]
- 122.Muraoka S., et al. , Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: Unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328, 555–566 (2003). [DOI] [PubMed] [Google Scholar]
- 123.Pedre B., et al. , Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing. Proc. Natl. Acad. Sci. U.S.A. 115, E11623–E11632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sainsbury S., et al. , The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res. 37, 4545–4558 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim Y., et al. , Crystal structure of the ligand-binding domain of a LysR-type transcriptional regulator: Transcriptional activation via a rotary switch. Mol. Microbiol. 110, 550–561 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baskaran K., Duarte J. M., Biyani N., Bliven S., Capitani G., A PDB-wide, evolution-based assessment of protein-protein interfaces. BMC Struct. Biol. 14, 22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bliven S., Lafita A., Parker A., Capitani G., Duarte J. M., Automated evaluation of quaternary structures from protein crystals. PLOS Comput. Biol. 14, e1006104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Prlic A., et al. , Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics 26, 2983–2985 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ye Y., Godzik A., Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19 (suppl. 2), ii246–ii255 (2003). [DOI] [PubMed] [Google Scholar]
- 130.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 131.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roberts E., Eargle J., Wright D., Luthey-Schulten Z., MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics 7, 382 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, (1996). [DOI] [PubMed] [Google Scholar]
- 134.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 135.Poornam G. P., Matsumoto A., Ishida H., Hayward S., A method for the analysis of domain movements in large biomolecular complexes. Proteins 76, 201–212 (2009). [DOI] [PubMed] [Google Scholar]
- 136.Taylor D., Cawley G., Hayward S., Quantitative method for the assignment of hinge and shear mechanism in protein domain movements. Bioinformatics 30, 3189–3196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ben Chorin A., et al. , ConSurf-DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 29, 258–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goldenberg O., Erez E., Nimrod G., Ben-Tal N., The ConSurf-DB: Pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 37, D323–D327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arnold K., Bordoli L., Kopp J., Schwede T., The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Study data are included in the article, supporting information or SI Appendix and Datasets.