To the Editor,
The myelodysplastic syndromes (MDS) are a heterogeneous group of hematologic conditions characterized by dysplastic changes and ineffective hematopoiesis [1]. Almost all patients with MDS harbor at least one or more somatic molecular alterations [2]. Several of these mutations have an important impact on MDS pathophysiology, disease progression, and overall outcomes [2–4]. BCOR (BCL6 Corepressor) and BCORL1 (BCL6 Corepressor like 1) are two homologous genes located on chromosome X that have been described in MDS and other myeloid malignancies [5]. The BCOR gene is mutated (BCORMUT) in 4–6% of MDS patients and BCORL1 (BCORL1MUT) in <1% [5]. The clinical impact of these mutations on MDS prognosis has been controversial, with some studies showing a negative effect of BCOR on overall survival (OS), while others demonstrating a neutral effect on survival [2,5,6].
In this study, we examined the prognostic impact of these mutations on OS and acute myeloid leukemia transformation (AML), and further determined whether mutation characteristics (location, type, passenger, versus driver) can influence the outcome, explaining previous discordant mutation effects.
DNA from bone marrow and/or peripheral blood samples from 621 patients with MDS and related myeloid malignancies (including MDS/MPN (myeloproliferative neoplasms), and secondary acute myeloid leukemia (AML) were sequenced for the presence of BCOR and BCORL1 mutations, as well as 58 other genes that have been described as commonly mutated in MDS and other myeloid malignancies (Supplementary Table S1). Sample processing, DNA sequencing and mutational analyses are summarized in Supplementary data. Cases were classified according to the 2008 World Health Organization (WHO) schema [7]. The Revised International Prognostic Scoring System (IPSS-R) cytogenetic risk groups and prognostic scores were calculated as described previously [8]. OS was measured from the time of diagnosis to time of death or last follow-up and leukemia-free survival from the time of diagnosis to AML transformation or last follow-up. Variant allele frequencies (VAFs) adjusted by zygosity were used to define architecture of driver clones. Two-group comparisons were performed by Mann–Whitney test for continuous variables and Chi-squared test was performed for categorical variables. Survival curves were constructed using Kaplan–Meier method and compared using log-rank test. Two sided p<.05 was considered statistically significant. All statistical analyses were performed using R statistical programing language.
Of 621 included MDS patients, 29 (5%) had BCOR mutations and 13 (2%) had BCORL1 mutations (Supplementary Table S2). Patients with BCOR mutations were younger (median age 63 versus 68 years, p=.04), and had lower platelet counts at diagnosis (63 versus 93–109/L, p=.01) when compared to BCORWT patients, Supplementary Table S2. Cytogenetic risk categories per IPSS-R for BCORMUT were similar to BCORWT: very good 3 versus 2%, good 65 versus 62%, intermediate 10 versus 17%, poor 13 versus 7% and very poor 6 versus 9%, respectively (p=.62), (Supplementary Table S2). Risk categories per IPSS-R were also similar for BCORMUT compared to BCORWT: very low 10 versus 16%, low 31 versus 40%, intermediate 24 versus 18%, high 17 versus 14%, and very high 13 versus 10%, respectively (p=.69) (Supplementary Table S2).
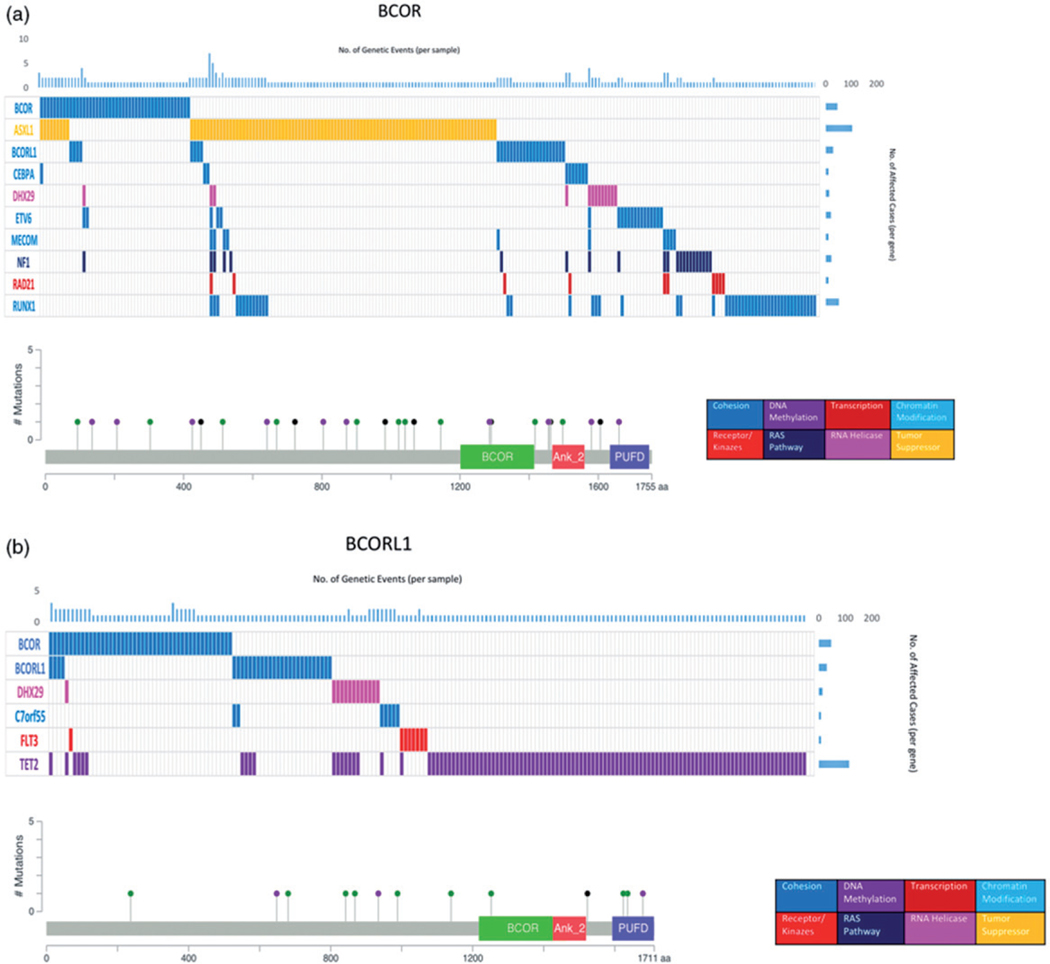
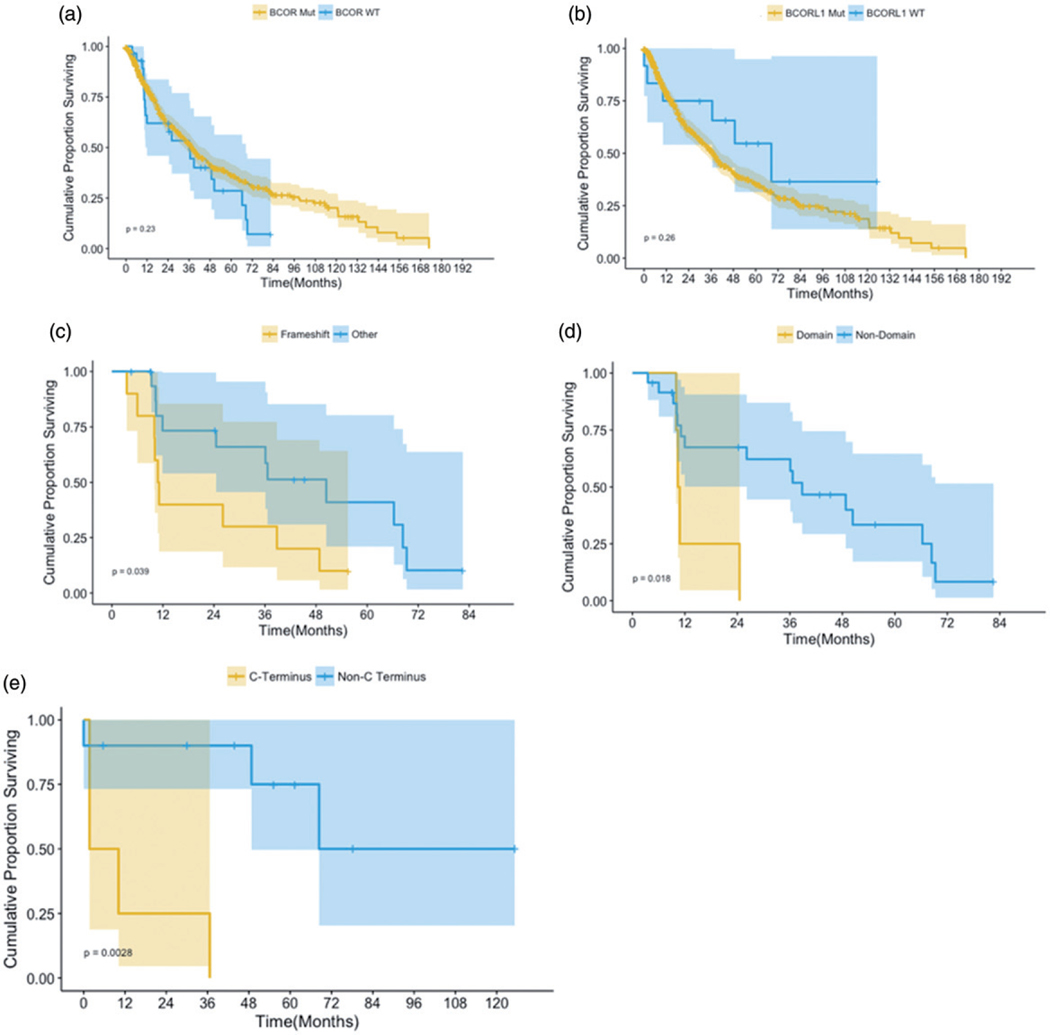
BCOR mutations were missense in nine patients (31%), frameshift insertion/deletion in 10 (34%), stopgain in 8 (28%), and nonsense in 2 (6%), Figure 1. BCOR mutations were commonly co-mutated with ASXL1 (p=.008), RUNX1 (p=.0001), NF1 (p=.002), ETV6 (p=.03), BCORL1 (p=.0001), MECOM (p=.021), RAD21 (p=.021), and CEBPA (p<.001), Figure 1. Clonal architecture analysis identified BCOR mutations as ancestral, subclonal, and mosaic in 41, 21, and 38% of cases, respectively. The median OS for BCORMUT patients was 24.5 months compared to 17.9 months for BCORWT (p=.23), Figure 2. While BCOR mutations did not impact OS even with adjustment for age and IPSS-R risk categories, mutation characteristics did: the median OS for patients with frameshift mutations was 10.9 months compared to 50.4 months for patients with other types of mutations (p=.03), Figure 2. The median OS for patients with mutations occurring in the binding domains was 10.6 months compared to 38.8 months for mutations outside of the binding domains (p=.01). There was no significant impact on OS based on the mutation order, whether they were ancestral, subclonal, or mosaic.
Figure 1.
Clustered mutation profiles and mutation maps for BCOR and BCORL1.
Figure 2.
Survival curves for BCOR and BCORL1.
Patients with BCORL1MUT had lower WBC counts (median 2.7 versus 4–109/L; p=.02) and lower platelet counts at diagnosis (72 versus 91–109/L; p=.02) compared to BCORL1WT patients. BCORL1MUT were more likely to be classified in the intermediate risk group per IPSS-R (46 versus 18%; p=.01) and less likely to be classified in the low risk group (7 vs. 40%; p=.01) compared to BCORL1WT. BCORL1 mutations were missense in seven patients (53%), frameshift insertion/deletion in 3 (23%), stopgain in 1 (7%), and nonsense in 2 (15%), Figure 1. BCORL1MUT is commonly co-mutated with TET2 (p=.001), BCOR (p=.0001), DHX29 (p=.001), C7orf55 (p=.02), and FLT3 (p=.03), Figure 1. Clonal architecture analysis identified BCORL1 mutations as ancestral, subclonal, and mosaic in 61, 23, and 15% of patients, respectively. The median OS for BCORL1MUT patients was longer compared to BCORL1WT, but this was not significantly different (43.8 versus 19.9, respectively; p=.16) even after adjustment for age and IPSS-R risk categories. Subset survival analysis on BCORL1 mutation characteristics such as mutation type (frameshift versus others), and subclonal versus founder versus mosaic did not show any survival impact, with the exception of patients with mutations in the C-terminus, who had lower OS (5.9 versus 101.7 months) compared to patients with mutations outside the C-terminus (p=.003), Figure 2.
In this study, we have shown that BCOR and BCORL1 mutations occur with low frequency in MDS and can be ancestral or subclonal. Taken as a whole, the impact of BCOR mutations on OS was neutral, which may represent a form of random misclassification bias, as mutation characteristics such as location and type did have a significant impact on OS. The lack of difference in median OS for patients with BCORL1MUT may be due to the low number of patients in our patient cohort. In another study of 354 MDS patients, BCOR and BCORL1 were identified in 4.2 and 0.8% of the patients, respectively (5). In univariate analysis, BCOR mutations were associated with a poor prognosis in MDS even after adjustment for clinical variables such as age and IPSS scoring system, while the impact of BCORL1 was not estimated due to a low number of patients. This study included a small number of patients with BCOR mutations and did not take into account the mutation characteristics. The impact of BCOR mutations on OS has been neutral in multiple larger studies [3,4,6].
Our analysis highlights the importance of evaluating mutation characteristics such as location, ancestral/subclonal, and type of mutation when evaluating the prognostic impact of any mutation. For example, TP53 mutations have always been shown to have a negative impact on OS in MDS and AML patients, but several studies have shown that patients with lower VAF or mutations outside of the important functional domains of the mutations have different OS compared patients with higher VAF or with mutations in the important domain of the mutation [9,10].
In conclusion, BCOR mutation characteristics may have significant impact on outcomes in MDS patients and should be taken into account when evaluating the prognostic impact of mutational data on outcomes.
Supplementary Material
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article online at https://doi.org/10.1080/10428194.2018.1543885.
References
- [1].Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med. 2009;361:1872–1885. [DOI] [PubMed] [Google Scholar]
- [2].Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nazha A, Al-Issa K, Hamilton BK, et al. Adding molecular data to prognostic models can improve predictive power in treated patients with myelodysplastic syndromes. Leukemia. 2017;31:2848–2850. [DOI] [PubMed] [Google Scholar]
- [4].Nazha A, Narkhede M, Radivoyevitch T, et al. Incorporation of molecular data into the Revised International Prognostic Scoring System in treated patients with myelodysplastic syndromes. Leukemia. 2016;30:2214–2220. [DOI] [PubMed] [Google Scholar]
- [5].Damm F, Chesnais V, Nagata Y, et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122:3169–3177. [DOI] [PubMed] [Google Scholar]
- [6].Bejar R, Papaemmanuil E, Haferlach T, et al. Somatic mutations in MDS patients are associated with clinical features and predict prognosis independent of the IPSS-R: analysis of combined datasets from the International Working Group for Prognosis in MDS-Molecular Committee. Blood. 2015;126:907. [Google Scholar]
- [7].Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. [DOI] [PubMed] [Google Scholar]
- [8].Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sallman DA, Komrokji R, Vaupel C, et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia. 2016;30: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Al-Issa K, Sekeres MA, Nielsen A, et al. TP53 mutations and outcome in patients with myelodysplastic syndromes (MDS). Blood. 2016;128:4336. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.