Abstract
The family of Bro1 proteins coordinates the activity of the Endosomal Sorting Complexes Required for Transport (ESCRTs) to mediate a number of membrane remodeling events. These events culminate in membrane scission catalyzed by ESCRT-III, whose polymerization and disassembly is controlled by the AAA-ATPase, Vps4. Bro1-family members Alix and HD-PTP as well as yeast Bro1 have central “V” domains that noncovalently bind Ub and connect ubiquitinated proteins to ESCRT-driven functions such as the incorporation of ubiquitinated membrane proteins into intralumenal vesicles of multivesicular bodies. Recently, it was discovered that the V domain of yeast Bro1 binds the MIT domain of Vps4 to stimulate its ATPase activity. Here we determine the structural basis for how the V domain of human HD-PTP binds ubiquitin. The HD-PTP V domain also binds the MIT domain of Vps4, and ubiquitin binding to the HD-PTP V domain enhances its ability to stimulate Vps4 ATPase activity. Additionally, we found that V domains of both HD-PTP and Bro1 bind CHMP5 and Vps60, respectively, providing another potential molecular mechanism to alter Vps4 activity. These data support a model whereby contacts between ubiquitin, ESCRT-III, and Vps4 by V domains of the Bro1 family may coordinate late events in ESCRT-driven membrane remodeling events.
INTRODUCTION
The Endosomal Sorting Complexes Required for Transport (ESCRTs), particularly ESCRT-III, and the Vps4 AAA-ATPase mediate a variety of membrane remodeling processes typically characterized by bending and scission of membranes away from the cytosol (Vietri et al., 2020). Members of the Bro1 family of proteins work upstream of this process to correctly recruit and initiate polymerization of ESCRT-III and convey target proteins into forming the membrane structure. For instance, yeast Bro1 serves as a ubiquitin (Ub) receptor, helping sort ubiquitinated membrane proteins into intralumenal vesicles (ILVs) that bud into the endosome for later delivery and degradation in lysosomes/vacuoles (Pashkova et al., 2013; Tang et al., 2016). Bro1 accomplishes this task by binding ESCRT-III subunits, Ub, and clathrin, allowing it to operate in parallel with the ESCRT-0,I,II proteins that have similar protein:protein connectivity. The mammalian Alix protein plays a role in viral budding by connecting viral gag proteins to the ESCRT-III membrane severing apparatus (Votteler and Sundquist, 2013). Moreover, Alix mediates a number of other ESCRT-driven processes including exosome production (Baietti et al., 2012), abscission during cytokinesis, membrane repair, and multivesicular body (MVB) sorting of select cargoes (Carlton et al., 2008; Dores et al., 2012; Skowyra et al., 2018; Larios et al., 2020). HD-PTP, Brox, and rhophillin-1 and -2 are additional members of the Bro1 family of proteins in humans (Toyooka et al., 2000; Ichioka et al., 2008).
Both Alix and HD-PTP have a role in sorting proteins into MVBs (Bissig and Gruenberg, 2014; Tabernero and Woodman, 2018). However, Alix plays a more prominent role in the generation of tetraspanin-enriched exosomes, whereas HD-PTP plays a role in sorting ubiquitinated membrane proteins into ILVs for eventual lysosomal degradation (Matsuo et al., 2004; Schmidt et al., 2004; Cabezas et al., 2005; Bowers et al., 2006; Doyotte et al., 2008; Kharitidi et al., 2015; Parkinson et al., 2015; Sun et al., 2015; Wenzel et al., 2018; Larios et al., 2020). These functions are important for both development and tumor suppression (Gingras et al., 2009, 2017). The function of HD-PTP closely parallels that of Bro1 in yeast. Loss of Bro1 blocks ESCRT-III–dependent formation of endosomal ILVs and causes a “class E” phenotype in which ubiquitinated proteins accumulate on the limiting membrane of aberrant endosomal compartments (Odorizzi et al., 2003). Overall, the process of forming ILVs and sorting cargo into them involves the recognition of Ub-cargo by a cohort of Ub-binding ESCRT components such as ESCRT-0, ESCRT-I, and ESCRT-II, which cluster Ub-cargo in clathrin-enriched microdomains and initiate membrane deformation (Williams and Urbe, 2007; Vietri et al., 2020). These components then nucleate the formation of a polymeric ESCRT-III, producing filaments that drive invagination and scission of the ILVs themselves. Proper editing and disassembly of ESCRT-III is catalyzed by the AAA-ATPase Vps4, which uses its N-terminal MIT domain to interact with substrate ESCRT-III proteins containing a C-terminal MIT-interacting motif (McCullough et al., 2018). Bro1 appears to act in part as a receptor for Ub-cargo that functions in parallel with ESCRT-0 (Hrs/Stam). This role is clearly revealed when ESCRT-0 and ESCRT-I are compromised for their ability to bind Ub or clathrin or if the connection between ESCRT-I and ESCRT-II is broken (Pashkova et al., 2013; Tang et al., 2016). How well Bro1 and HD-PTP represent distinct cargo-sorting pathways that are separate from ESCRT-0,I,II or whether they function within a network of obligatory and redundant connections to ESCRT-0 and ESCRT-I is not yet clear.
Bro1 and HD-PTP share a common architecture that helps them achieve their common function in sorting Ub-cargo into ILVs. Both components accomplish this function by binding Ub and clathrin and connecting—either directly or indirectly—with ESCRT-III. Their N-terminal BODs (Bro1 homology domains), which form a crescent structure, bind the ESCRT-III component Snf7 and CHMP4, respectively (Kim et al., 2005; Gahloth et al., 2017b). The V domains, composed of two 3-helical coiled-coil arms, bind Ub (Pashkova et al., 2013). Both proteins also associate with deubiquitinating enzymes, Doa4 and UBPY (albeit via different modes), that can deubiquitinate cargo late in the sorting reaction (Ali et al., 2013; Richter et al., 2013). HD-PTP has additional interactions that may help it perform its endosomal functions. These include interactions with the ESCRT-0 component STAM2 via its BOD, the ESCRT-I component UBAP1 via its V domain, and TSG101 via its proline-rich region domain C-terminal to the V domain (Ichioka et al., 2008; Stefani et al., 2011; Kimura et al., 2014; Gahloth et al., 2016; Lee et al., 2016). HD-PTP also has a low-activity protein tyrosine phosphatase (PTP) domain with a noncanonical active site (Zhang et al., 2017). However, this feature is not required to support MVB sorting of Ub-cargo (Doyotte et al., 2008).
The ability of Bro1 and HD-PTP to both bind Ub-cargo and interface with ESCRT-III provides a rationale for how these proteins could coordinate cargo sorting with the final stages in ILV formation. Such a regulatory device would be important because it could ensure that ILVs are loaded specifically with Ub-cargo. Previous studies have shown that the presence of Ub-cargo is required for ESCRTs to assemble efficiently onto endosomal membranes, providing a way to ensure that ILVs are made in response to cargo (MacDonald et al., 2012). Recently, studies in yeast have discovered that uncoupling the ability of Bro1 to bridge its full cohort of protein interactions leads to formation of ILVs that lack Ub-cargo (Tseng et al., 2021). Moreover, the V domain alone can stimulate the activity of Vps4 in vitro through contact with the Vps4 MIT domain. This stimulation of Vps4 can be further enhanced when the Bro1 V domain (VBro1) binds Ub (Tseng et al., 2021). Bro1 mutants that fail to stimulate Vps4 are defective in MVB sorting, suggesting that this mode of regulation is critical for correctly orchestrating cargo sorting with the latter stages of ILV formation. To better understand the emerging roles of HD-PTP, we investigated the structural basis of how its V domain binds Ub. Our studies reveal two tandem Ub-binding sites on the N-terminal 3-helical bundle of the HD-PTP V domain. Modeling experiments based on nuclear magnetic resonance (NMR) paramagnetic relaxation enhancement (PRE) experiments as well as mutational studies suggest that this tandem array accommodates K63-linked Ub, which operates as a strong signal for MVB sorting (Piper and Lehner, 2011). We show that like the VBro1, the V domains of Rim20 (VRim20) and HD-PTP (VHDPTP) bind the MIT domain of Vps4. In addition, the Ub-binding domains of VHDPTP mediate Ub-enhanced stimulation of Vps4 ATPase activity in vitro, emphasizing the conserved nature of this regulatory mode. Finally, our analysis revealed that the VHDPTP and VBro1 bind CHMP5 and Vps60, respectively, which may constitute a specific ESCRT-III regulatory mechanism dedicated to the HD-PTP/Bro1 cargo-sorting pathway that operates in parallel with the canonical ESCRT-0,I,II MVB-sorting pathway.
RESULTS
PRE experiments locate Ub binding on the N-terminal arm of the HD-PTP V domain
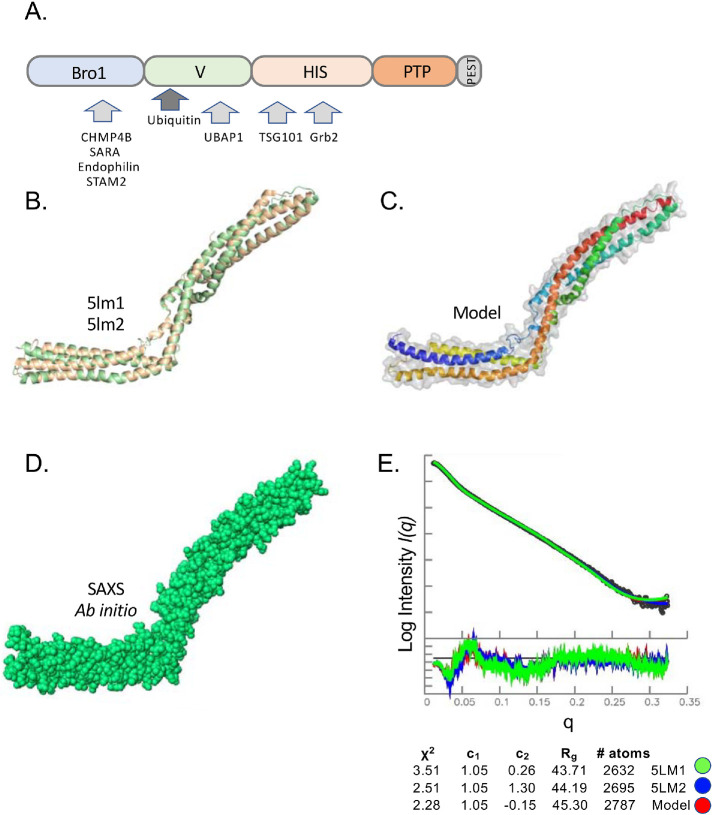
We sought to determine the basis for how the VHDPTP binds Ub. The V domain is a coiled-coil region between the N-terminal Bro1 domain and the histidine/proline-rich domain and phosphatase domains (Figure 1A). Recently, the crystal structure of the HD-PTP domain was solved, revealing a two-armed structure, each composed of a 3-helical bundle (Figure 1B). This structure is similar to the crystal structures of Bro1 and Alix, except that the HD-PTP domain adopts an extended conformation lacking the closed, “V”-shaped axis seen in Alix (Gahloth et al., 2016, 2017b). SAXS experiments have confirmed that HD-PTP adopts this extended shape in solution (Gahloth et al., 2017a). To guide our experiments in determining where Ub binds onto the HD-PTP domain, we produced a recombinant V domain (residues 361–711). We performed SAXS analysis to confirm its behavior in solution and to verify that our data matched the characteristics of previous studies (Figure 1, D and E). The ab initio modeling from our SAXS data produced an envelope that closely matched the known HD-PTP crystal structure. Chi-squared values determined with Crysol comparing our data with crystal structures of HD-PTP showed good agreement, especially for PDB:5lm2. We also built a model of HD-PTP that filled in missing loops from the crystal structure (Figure 1C) and, when compared with the experimental SAXS data, yielded a slight improvement to the Chi-squared values (Figure 1E). This model was then used throughout our studies.
FIGURE 1:
SAXS analysis of HD-PTP V domain. (A) Domain organization of HD-PTP including N-terminal Bro1 domain (residues 8–361), V domain (residues 366–704), histidine/PRR domain (residues 705–1128), phosphatase domain (residues 1192–1452), and C-terminal PEST domain (residues 1509–1636). Proteins known to interact with those domains are indicated. (B) Structure cartoon of the crystal structures of the HD-PTP V domain (PDB: 5LM1 [green], 5LM2 [wheat]). (C) Solution model for the HD-PTP V domain based on chimera of crystal structures and computational modeling missing loops colored from N- to C-termini using blue–red spectrum. (D) Ab initio envelope of the HD-PTP V domain in solution from SAXS data. (E) Guinier region of SAXS data of VHDPTP (4 mg/ml) as well as residuals to confirm little to no aggregation. At the bottom is the experimental and simulated pair distance distribution function P(r). Rg and Dmax determination from P(r) curves and χ2 values comparing experimental data with predicted P(r) curves of crystal structures and the solution model in C.
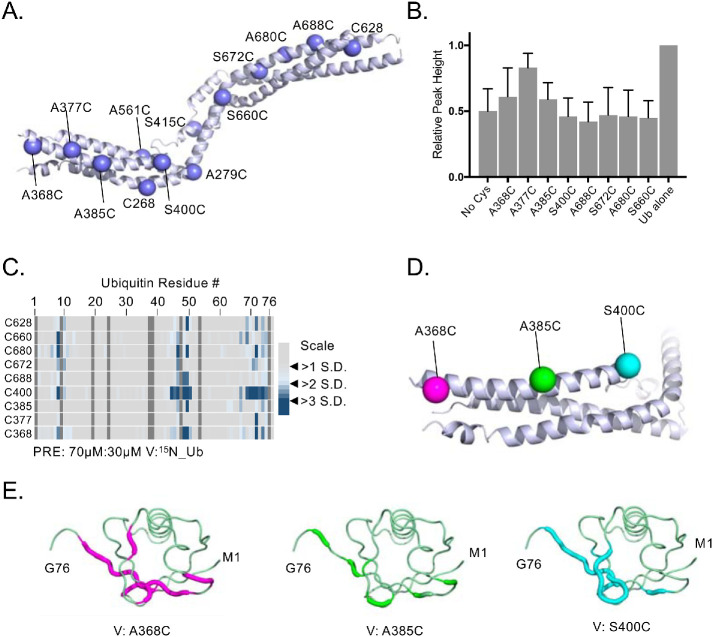
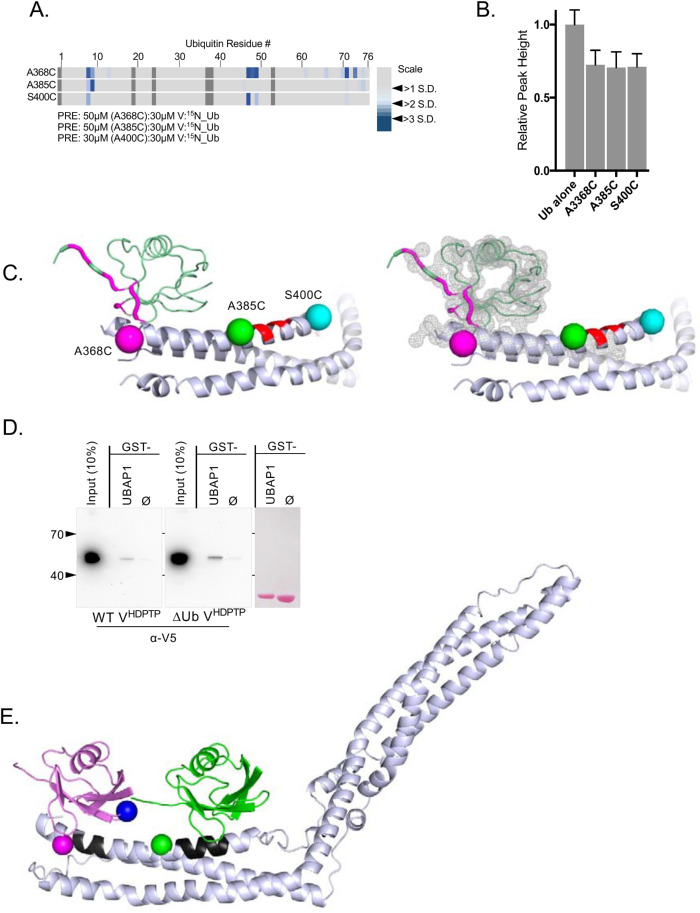
We next used recombinant V domains in a series of PRE experiments to find the region where Ub binds. This utilized a series of variant VHDPTP proteins, each containing a cysteine at distinct positions throughout the protein that could be covalently linked to a nitroxide spin label, MTSL (Figure 2A). This series of cysteine substitution mutants were made in the context of an VHDPTP in which the three native cysteine residues were changed to alanine. The PRE effect that induces peak broadening is observed in HSQC experiments when 15N-Ub is bound within ∼10–20 Å of the MTSL. These effects are highly localized as they diminish with distance to the 6th power. Titration experiments with unlabeled HD-PTP showed dramatic HSQC peak broadening for most of the amides of 15N-Ub without altering peak positions. This indicated that HD-PTP binds Ub in an intermediate to slow exchange, causing the peaks to broaden (Supplemental Figure 1). Therefore, we chose a concentration of HD-PTP that would partially bind the pool of 15N-Ub, leading to some peak broadening while still allowing additional peak broadening from PRE effects to be observed. Specific PRE effects were measured by difference in peak height in the absence and presence of ascorbate, which quenches the paramagnetic spin label. To ensure that differences in PRE effects were due to different positions of spin label rather than different levels of binding, we verified that the overall average peak broadening observed (e.g., the extent of binding) for each spin-labeled HD-PTP variant in the presence of ascorbate was comparable to the no-Cys HD-PTP (Figure 2B). All variants produced the comparable levels of Ub binding except the A377C mutant, which was defective in Ub-binding, possibly indicating that it was at or near the site of Ub interaction.
FIGURE 2:
Locating the binding regions of 15N-Ub on the HD-PTP V domain by PRE experiments. (A) Position of single-amino-acid replacement mutations comprising a set of single cysteine–containing VHDPTP variants. (B) Relative average HSQC peak heights (±SD) of all residues of 30 µM 15N-Ub HSQC spectra alone or in the presence of MTSL-labeled 70 µM HD-PTP variant proteins in the presence of 2 mM ascorbate. Binding of VHDPTP broadens 15N-Ub amide peaks, resulting in lower relative peak heights. (C) The indicated single cysteine–containing V domain variants were spin labeled with MTSL and incubated with 15N-Ub at a ratio of 70 µM:30 µM (V:Ub) in the absence and presence of reduction by ascorbate. PRE effects, measured by the ratio of peak intensities before and after reduction, are plotted by an increase in color intensity indicating those that were 2–4 SDs from the mean change. (D) Position of three cysteine substitutions on the N-terminal helix of HD-PTP that had the largest PRE effects on 15N-Ub labeled in magenta, green, and cyan, respectively. (E) Backbone amides of Ub undergoing largest PRE effects (>2 SD above the mean) when bound to HD-PTP spin labeled at residue 368, 385, or 400. These PRE effects were mapped onto Ub backbone (PDB:1UBQ) in magenta, green, and cyan, respectively.
Plotting the magnitude of PRE effects on detectable amide peaks as a function of amino acid position of Ub showed that large effects were derived from spin labels at residues 400 and 368 (Figure 2C). And plotting the largest PRE effects from the spin labels at position 368, 385, and 400 (Figure 2D) onto the structure of Ub (Figure 2E) indicated the same surface of Ub was near these positions. These PRE studies indicate that the major Ub-binding site(s) are located on the N-terminal arm but not on the C-terminal arm of the V domain. However, it was difficult to interpret the data as indicating a single Ub-binding site because PRE effects from a spin label placed at residue 385 were lower than those from spin labels placed on either side at residues 368 or 400. Because residues 368 and 400 are on opposite ends of the same arm, we hypothesized that there may be two Ub-binding sites in the VHDPTP domain clustered on its N-terminal proximal arm.
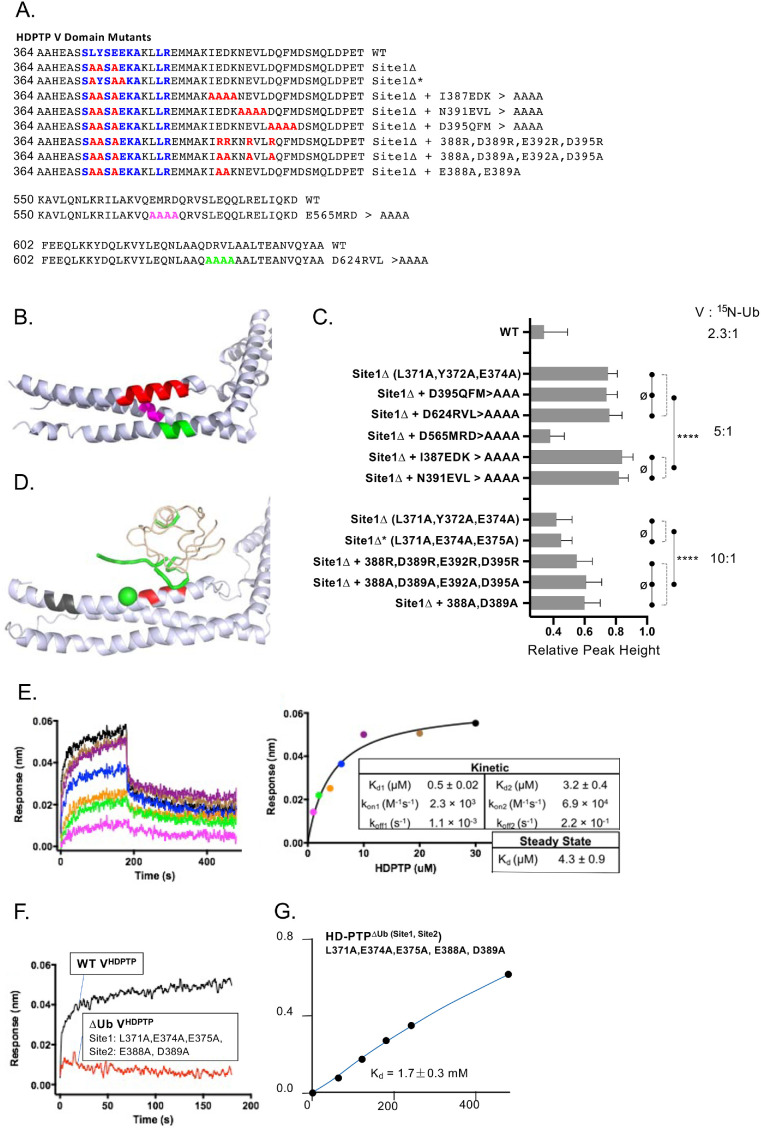
Ub binding at Site1
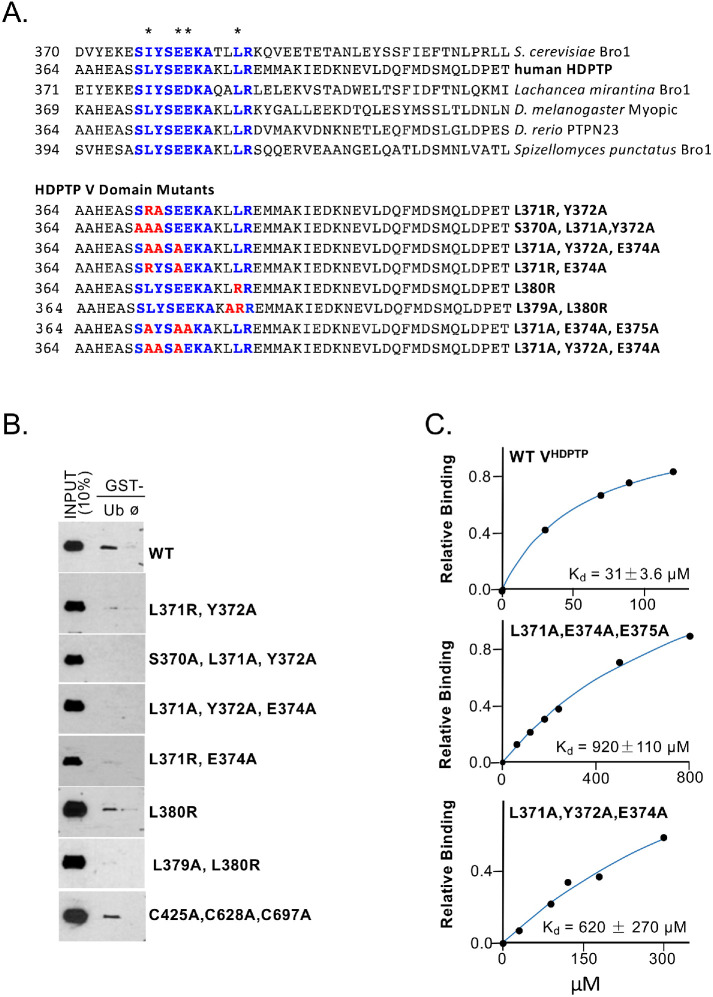
Aligning sequences and structures of VBro1 and VHDPTP revealed a conserved sequence motif at the interface were Ub binds Bro1VBro1 (Figure 3A). This binding site is close to residue A368, a position that gave large PRE effects, and residue A377, whose substitution to an MTSL-labeled cysteine compromised Ub binding (Figure 2, B and C). This region of the VHDPTP crystal structure also adopts the same conformation observed in the VBro1 crystal structure. Therefore, we tested a series of mutants within this region (Figure 3A, bottom) that altered conserved residues that mediate Ub binding in VBro1 including L371, E374, and E375 (Figure 3B). Pull-down experiments of recombinant V5 epitope–tagged V domains with GST-Ub beads showed that mutations in this conserved region dramatically diminished binding from that observed using wild-type (WT) VHDPTP or the VHDPTP lacking cysteine residues (C425A, C628A, C697A) that was used for PRE experiments. In addition, NMR titration experiments that measured overall peak broadening showed that the binding affinity (Kd) of VHDPTP for Ub was ∼31 µM, whereas mutants that ablate the conserved Ub-binding site (Site1) in HD-PTP had Kds of 0.6–1 mM (Figure 3C). Given that this site for Ub binding is conserved in sequence and structure with that of VBro1, we modeled the mode of binding of Ub to Site1 by overlaying the Ub-bound VBro1 structure onto VHDPTP (see below). This model agrees with the major PRE effects that we observed on the backbone of Ub when bound to VHDPTP spin label at position 368.
FIGURE 3:
An N-terminal binding site for Ub. (A) Top, Amino acid alignment of the V domains of Bro1 and HD-PTP from the indicated species with conserved residues (blue) within the Ub-binding site of Saccharomyces cerevisiae Bro1. Residues in Bro1 that when mutated result in loss of Ub binding are indicated (*). Bottom, Alignment of residues within a set of human HD-PTP V domain mutants showing the conserved Ub-binding residues of Bro1 (blue) and amino acid substitutions within that region (red). (B) Pull-down experiments measuring binding of the indicated V5 epitope–tagged HD-PTP V domain mutant proteins to GST alone (ø) or GST-Ub preloaded onto glutathione beads. Bound fractions together with a 10% equivalent of input were immunoblotted with anti-V5 antibodies. (C) Binding curves of 15N-Ub to either WT or the mutant VHDPTP containing the indicated mutations in Ub-binding Site1.
Ub binding at Site2
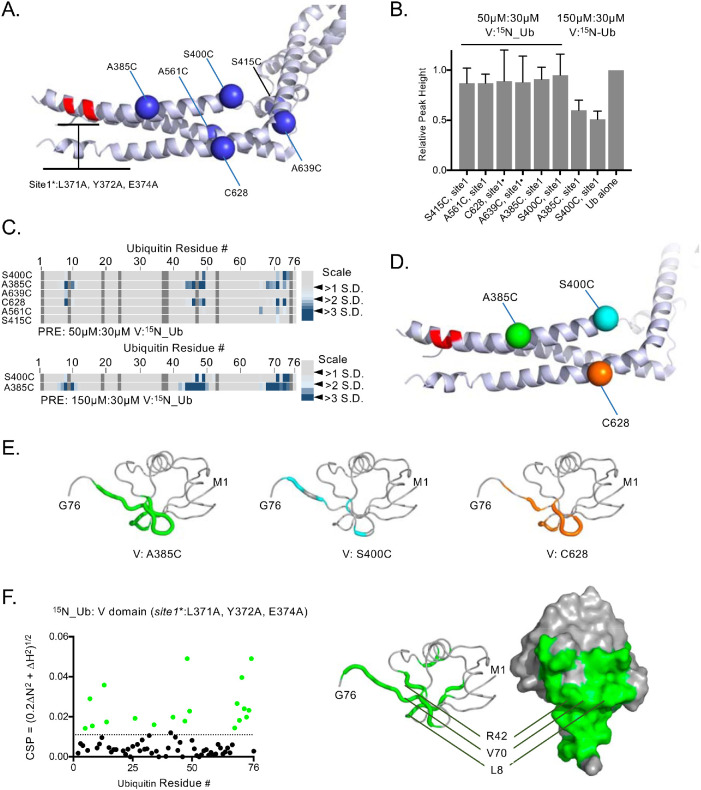
Eliminating the conserved Ub-binding Site1 from VHDPTP resulted in diminished binding of Ub but not complete loss, indicating the presence of at least one additional Ub-binding site. Moreover, the large PRE effects on 15N-Ub when bound to HD-PTP spin labeled at residue 400 indicated that this second binding site would be near that position, fairly close to Site1 (Figures 2 and 3). To find Site2, we performed additional PRE experiments with a set of spin-labeled VHDPTP proteins in which the binding activity of Site1 was ablated with a combination triple mutant: L371A, Y372A, E374A (Figure 4A). PRE effects when the V domains were present at a ratio of 1.6:1 with 15N-Ub were weak but clearly observed for spin-labeled residues at 385 and more weakly at 400 and 628, whereas the overall levels of binding assessed by average peak broadening were comparable (Figure 4, B and C). Increasing the VHDPTP: 15N-Ub ratio to 5:1 in these experiments greatly increased Ub binding and PRE effects (Figure 4, B and C). These effects were localized to the hydrophobic surface of Ub, which engages virtually all Ub-binding proteins (Piper et al., 2014) (Figure 4, D and E). Additional PRE experiments at a higher ratio of VHDPTP to Ub clearly showed stronger PRE effects from spin label at position 385 than at 400 (Figure 4C, bottom), even with slightly less binding overall to the S400C VHDPTP protein (Figure 4B). The weaker Ub binding by VHDPTP lacking Site1 allowed us to observe HSQC chemical shift perturbations within 15N-Ub as the complex was in intermediate-to-fast exchange. Plotting the largest chemical shift perturbations onto the surface of Ub indicated that Site2 contains residues L8, R42, I44, and V70 (Figure 4F). This confirmed the PRE experiments indicating that Ub uses its well-characterized hydrophobic surface to bind VHDPTP at Site2.
FIGURE 4:
Mapping a second Ub-binding site. (A) Position of single-amino-acid replacement mutations comprising a set of single cysteine–containing VHDPTP variants in the context of inactivated Ub-binding Site1. (B) Relative average HSQC peak heights (±SD) of all residues of 30 µM 15N-Ub HSQC spectra alone or in the presence of 50 or 150 µM HD-PTP variant proteins. (C) Spin-labeled single cysteine–containing V domain variants lacking Site1 Ub binding were incubated with 15N-Ub at the indicated ratios in the absence and presence of reduction by ascorbate. PRE effects, measured by change in ratio of peak intensity before and after reduction, are plotted by an increase in color intensity indicating those that were 2–4 SDs from the mean change. (D) Highlighted in green, cyan, and orange are positions giving maximal PRE on 15N-Ubin the absence of Site1. (E) Backbone amides of Ub undergoing largest PRE effects (>2 SD above the mean) when bound to HD-PTP spin labeled at residues indicated in D. (F) Left, HSQC data of 15N-Ub (30 mM) in the presence of (150 µM) the HD-PTP V domain lacking the Site1 Ub-binding site. The magnitudes of chemical shift perturbations ([0.2∆N2 + ∆H2]1/2) are plotted for each assignable backbone amide of Ub. Dashed line indicates level chemical shift perturbation 1 SD above the mean. Right, Residues undergoing significant chemical shift perturbations in F are plotted onto Ub structure (PDB: 1UBQ).
We investigated Ub-binding to Site2 using a set of mutations in VHDPTP that also contained mutations ablating Site1 binding (L371A, Y372A, E374A) (Figure 5). These mutations were distributed over each of the three helices within the coiled-coil that comprises the N-terminal arm (Figure 5, A and B). NMR experiments with 15N-Ub in the presence of a fivefold excess of VHDPTP mutants allowed us to assess the relative effects of binding by the degree of peak broadening, reflected by the decrease in relative peak height. A low level of binding was observed for VHDPTP carrying mutations in Site1 (L371A, Y372A, E374A) alone, and binding within this context was further diminished by combination mutations to I387A, E388A, D389A, and K390A as well as N391A, E392A, V393A, and L394A. In contrast, no significant decrease in binding was observed when the Site1∆ mutant (L371A, Y372A, E374A) was combined with mutations farther along the first alpha helix or within the other two helices of the first arm: (D395A, Q396A, F397A, M398A); (D624A, 625A, V626A, L627A); and (E565A, M366A, R367, D368A). Interestingly, the latter alanine substitution mutant resulted in far better binding of VHDPTP carrying the Site1∆ mutant, suggesting that the additional Ub-binding site(s) were sensitive to their immediate context that might allow for better rotation of helices to engage Ub with tighter binding. Together, these results prompted us to examine the area along the first helix that encompassed acidic residues E388, D389, E392, D395 using VHDPTP mutants at a ratio of 10:1 with 15N-Ub in HSQC experiments. Whereas Site1∆ mutants containing either the (L371A, Y372A, E374A) triple substitution or the (L371A, E374A, E375A) mutations had the same level of reduced Ub binding, additional alteration of all four acidic residues (E388, D389, E392, D395) or just E388 and D389 to alanine caused significantly diminished binding further. These data suggested that Site2 is centered on residues 387–392, and modeling Ub over this site rationalized the PRE data (Figure 4, C and E) that we obtained for 15N-Ub interacting with a Site1 mutant with a spin label attached at residue 385 (Figure 5D). To examine the impact when both Site1 and Site2 were altered, we performed binding experiments using biolayer interferometry. Sensors were coated with avidin and then bound with Ub tagged at the C-terminus with a biotinylated BirA site. Immobilized biotinylated Ub was then allowed to associate and dissociate with increasing concentrations of WT VHDPTP to yield both on- and off-rates as well as a Kd of 4.3 µM (Figure 5E). This value was similar to the Kd obtained by NMR (which was about sevenfold lower), and differences were likely due to the different environments and constraints of the two interacting proteins. Whereas the Kd of WT VHDPTP measuring by biolayer interferometry was somewhat tighter, no binding to biotinylated Ub could be observed by a mutant VHDPTP lacking its Ub-binding activity at both Site1 (L371A, E374A, E375A) and Site2 (E388A, D389A) (Figure 5F). We also measured Kd using titrations and monitoring changes in the HSQC spectra. Whereas Site1 mutation (L371A, E374A, E385A) alone gave a Kd of 0.9 mM, combining that with mutation of Site2 (E388A, D389A) diminished binding further to an estimated Kd of 1.7 mM (Figure 5G).
FIGURE 5:
Mutagenesis of a second Ub-binding site. (A) Amino acid alignment of the WT HD-PTP V domain and various mutants (red) in the N-terminal helix or other helices (pink, green). Residues in blue highlight the region of homology with the Bro1 Ub-binding site. (B) Position of residues altered in the various mutant HD-PTP mutants shown in A (red: 387–390; pink: 565–568; green: 624–627). (C) Relative average peak heights (±SD) of 30 µM 15N-Ub HSQC spectra alone or in the presence of the purified V domain at the indicated ratio. Better binding results in lower peak height. Peak broadening was measured for all visible peaks in the 15N-Ub spectra to calculate the SD. One-way analysis of variance tests of groups that showed p > 0.1 between samples are designated (ø) vs. all samples within groups that each showed a difference from samples of the other groups with p < 0.0001 (****). (D) Position of Ub modeled on Ub-binding Site2 (red) near residue 385 (green) that when spin labeled produced the PRE effects mapped onto Ub in green. Residues in black indicate Site1. (E) Ub-biotin was loaded onto avidin-coated sensors and dipped into solutions containing the WT HD-PTP V domain. Octet Red96 sensorgrams are shown together with the calculated KD, Ka, and Kd of binding. (F) Octet Red96 sensorgrams for WT or ∆Ub mutant (Site1∆: L371, E374, EE75A; Site2∆: E388A, D389A) V domains. (G) Binding curves of the mutant V domain with mutations in Site1 (L371A, E374A, E374A) and Site2 (E388A, D389A). Binding was measured by the decrease in HSQC signal from 15N-Ub (30 µM) in the presence of increasing amounts of V domain.
Model for Ub binding
NMR experiments with a Site1 mutant allowed us to use chemical shift perturbations detected in HSQC spectra to map the residues of Ub that line the interaction site with VHDPTP (Figure 4F). This was because loss of Site1 changes the association of the V domain to one that is weaker and in fast exchange. Mutation of only Site2 resulted in an VHDPTP that still bound Ub in intermediate/slow exchange, preventing peak shifts from being observed. This indicates that Site1 is the higher-affinity binding site, which is consistent with the large decrease in binding affinity observed with Site1 mutated, whereas a lower fold change was observed when Site2 was mutated.
Identification of Site2 allowed us to refine our PRE analysis. Our initial set of single-cysteine substitution mutants showed PRE effects when spin labels were placed at 368, 385, and 400 (Figure 2C), which is likely from binding Ub at two distinct sites along that helix. Therefore we spin labeled those positions but in a Site2 mutant with the expectation that the PRE effects would be biased to just those positions near Site1 (Figure 6). To ensure the same level of overall binding to the spin-labeled variants, we monitored average peak broadening of 15N-Ub in HSQC spectra. To obtain comparable binding, spin-label variants at positions 368 and 385 were bound to 15N-Ub at a ratio of 1.6:1, whereas the variant labeled at S400 was bound to Ub at a ratio of 1:1 (Figure 6B). These experiments showed that major PRE effects of Site2 mutants were obtained with spin label at residue 368 (Figure 6A), consistent with the structural model if Ub bound at Site1 derived from homology modeling of the Bro1-Ub complex (Figure 6C). This allowed us to build a synthetic model where Ub occupies both Site1 and Site2 (Figure 6E and Supplemental Data). Here, both Ubs lie in tandem across the N-terminal helix in the coiled-coil arm. The orientation of Ub in the model accommodates di-Ub linked via a K63 linkage. Previous experiments have shown that VHDPTP binds avidly to K63-linked polyubiquitin, although unlike some proteins specific for K63-linked Ub polymers, the HD-PTP V domain also binds strongly to monoubiquitin (Pashkova et al., 2013).
FIGURE 6:
PRE mapping of Site1 Ub binding. (A) Paramagnetic relaxation effects on backbone amides of 15N-Ub using HD-PTP V domains containing single spin labels at the indicated positions (residues 368, 385, or 400) also containing mutations disrupting the Site2 Ub-binding site (E388A, D389A). The ratio of the V domain to 15N-Ub is shown. The largest paramagnetic enhancement effects, measured by the change in the ratio of peak intensity before and after reduction, are plotted by color indicating those that were 2–3 SDs from the mean change. (B) Relative average peak heights (±SD) of 30 µM 15N-Ub HSQC spectra alone or in the presence of 50 or 30 µM HD-PTP MTSL-labeled variant proteins in the presence of 2 mM ascorbate as described for A. (C) Positioning of Ub over Site1. Shown are positions of MTSL labeling used in A. The largest PRE effects on 15N Ub when bound to the V domain spin labeled at residue 368 were mapped onto the structure of Ub in magenta. Right, The fit of the HD-PTP:Ub model into the electron density map for the VBro1:Ub crystal structure (4JIO). (D) Pull-down experiments with WT or ∆Ub mutant HD-PTP V (Site1∆: L371A, Y372A, E374A; Site2∆: E388A, D389A, E392A) to assess binding to GST alone or GST fused to residues SNIKSLSFPKLDSDDSNQKT of UBAP1 that mediate binding to the distal C-terminal arm of the HD-PTP V domain. (E) Model of Ub bound to Site1 (black) and Site2 (black). Positions for PRE experiments at residue 368 (magenta) and 385 (green) of VHDPTP or position of K63 of Ub at Site1 (blue) are shown.
Loss of Site1/Site2 Ub binding does not affect interaction with partner proteins
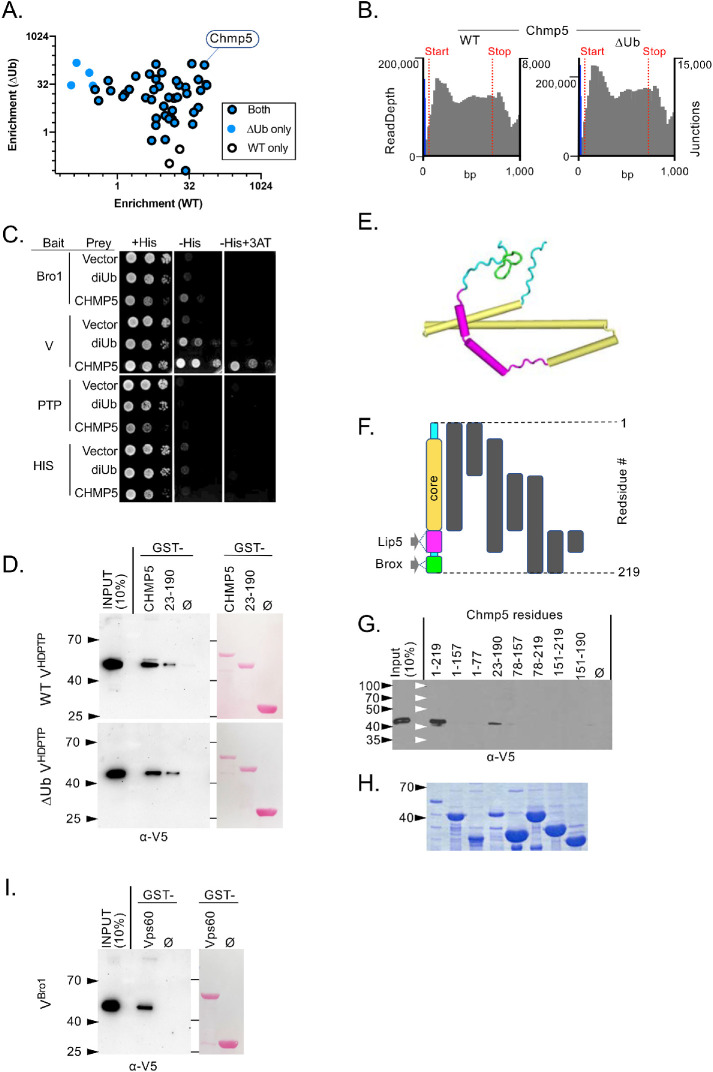
Determining the function of Ub binding by VHDPTP relies on testing mutants that are specifically defective in Ub binding but still retain interaction with other partners and maintain overall structural integrity. Size exclusion chromatography and 1H NMR experiments of WT and ∆Ub (Site1,2) VHDPTP proteins indicated that both were well folded and without aggregation. UBAP1, another Ub-binding protein, binds the distal C-terminal 3-helical bundle arm of VHDPTP at a site conserved in Alix that in turn binds LYPxL motifs found in viral gag and other proteins (Gahloth et al., 2016). As expected, both the WT and ∆Ub mutant V domains bound the LYPxL-containing interacting region of UBAP1 (Figure 6D). To explore whether other potential binding partners might be hampered by the ∆Ub mutations, we performed a comprehensive yeast 2-hybrid screen to compare interactomes of WT and ∆Ub VHDPTP domains using a batch method that we previously developed called DEEPN (dynamic enrichment for evaluation of protein networks). This approach uses deep sequencing of a population of “prey” plasmids to determine which are specifically enriched upon selection for a positive yeast 2-hybrid interaction. Because the prey library is verifiably delivered to cells that contain different “bait” proteins, one can compare the enrichment of genes upon selection between baits to determine whether there are differential interactions. For our analysis, we used a mouse cDNA library derived from multiple organs as previous described (Pashkova et al., 2016). Sequencing across library plasmid inserts showed that all the yeast populations with different prey plasmids contained comparable distributions of “preys” (Supplemental Figure 2). Positive yeast 2-hybrid interactions were then selected for by growing populations in the absence of histidine. We filtered for only genes that were enriched on either WT or ∆Ub V domains but not vector alone, met a probability threshold for enrichment by statistical modeling, and were in the proper reading frame and orientation with respect the Gal4-activation domain onto which their coding region was fused (Figure 7A). Very few interacting gene fragments showed differential interactions on WT VHDPTP versus ∆Ub VHDPTP, showing that at least some yeast 2-hybrid interactions could distinguish subtle differences on the surface of the VHDPTP domains due to the ∆Ub mutations (Figure 7A; Supplemental Figure 2). Yet, while these differential interactions were evident, their biological relevance has yet to be determined, and these interactions may suggest only a biochemical difference rather than a functional one. The majority of interacting proteins, however, interacted with both WT and ∆Ub VHDPTP proteins, supporting the idea that they had very similar conformations and surfaces.
FIGURE 7:
CHMP5 binding to WT and ∆Ub HD-PTP V domain. (A) Batch yeast 2-hybrid (DEEPN) screen using the WT and ∆Ub HD-PTP domain against mouse cDNA whole body library. Diploid yeast containing the same cDNA library were placed under selection for positive yeast 2-hybrid interactions. Plotted are genes in the correct translational frame that were enriched upon selection for interaction with the HD-PTP V domain but not enriched for interaction with vector alone. Most genes were enriched on both the WT (open black circles) and ∆Ub HD-PTP V domain (blue dots), Site1∆: L371A, Y372A, E374A; Site2∆: E388A, D389A, E392A), some passed statistical probability for only WT or the ∆Ub V domain (open circles or solid circle, respectively). (B) Fusion point of Chmp5 fragment to Gal4-activation domain in the prey plasmid (blue line, right Y-axis) and the read-depth of sequences for Chmp5 (gray lines, left Y-axis) describing the fragment of Chmp5 enriched when selected for Y2H interaction on the WT or mutant HD-PTP V domain. (C) Binary yeast 2-hybrid assay with diploid cells containing the indicated “bait” fragments of HD-PTP fused to the Gal4-DNA–binding domain and the indicated “prey” fragments fused to the Gal4-activation domain wherein a positive Y2H interaction drives production of the His3 and His+ phenotype. Cells were serially diluted and plated onto media with histidine or without histidine also containing 1 mM 3-aminotriazole (3AT). (D) Pull-down experiments measuring binding of the indicated V5 epitope–tagged HD-PTP V domain proteins (WT or ∆Ub [Site1∆: L371A, E374A, E375A; Site2∆: E392A, D395A]) to GST alone (ø) or GST-Chmp5 preloaded onto glutathione beads. Bound fractions together with a 10% equivalent of input were immunoblotted with α-V5 antibodies. (E) Homology model of CHMP5 using PDB: 2GD5 as the template with unstructured regions (cyan) and binding regions for Brox (green) and Lip5 (magenta) and a core alpha-helical domain (orange). (F) Schematic of Chmp5 fragments used in binding experiments in G. (G) Pull-down experiments measuring binding of the indicated V5 epitope–tagged HD-PTP V domain to equal levels of GST alone (ø) or GST fused to the indicated fragments of Chmp5 described in F preloaded onto glutathione beads. Bound fractions together with a 10% equivalent of input were immunoblotted with α-V5 antibodies. (H) Coomassie gel of purified GST-Chmp5 fusion proteins. (I) Pull-down experiments showing binding of V5 epitope–tagged VBro1 on GST alone (ø) or GST-Vps60 preloaded onto glutathione beads. Bound fractions together with a 10% equivalent of input were immunoblotted with α-V5 antibodies.
Chmp5 binds the HD-PTP V domain
One of the genes that enriched on both WT and ∆Ub VHDPTP was CHMP5, an ESCRT-III subunit whose yeast analogue is Vps60 (Figure 7A). The CHMP5 sequences enriched upon selection showed that the entire CHMP5-coding region was encapsulated in the interacting “prey” plasmid with a fusion point just upstream of the ATG start codon. To verify the yeast 2-hybrid interaction, we built a new CHMP5 “prey” plasmid in which the entire ORF was fused in frame to the Gal4-activiation domain (Gal4AD). This and the Gal4AD vector alone or fused to two tandem copies of Ub were tested for interaction with the Bro1 domain, V domain, PTP domain, and His/PRR domain. CHMP5 showed a strong interaction with VHDPTP, and growth was markedly stronger than that supported by the interaction of the VHDPTP with di-Ub. A far weaker interaction was observed between CHMP5 and the Bro1 domain of HD-PTP, possibly indicating indirect association via endogenous ESCRT-III subunits or direct binding through a motif similar in HD-PTP and Brox that allows the Bro1 domain of Brox to bind Chmp5 (Lee et al., 2016). Glutathione S-transferase pull-down experiments using recombinant V5 epitope–tagged WT and ∆Ub HD-PTP V domains showed that both bound GST-Chmp5 (Figure 7D), consistent with yeast 2-hybrid DEEPN analysis where the same fragment CHMP5 was enriched upon selection for interaction across both WT and ∆Ub VHDPTP domains (Figure 7B).
We next mapped what regions of Chmp5 mediated the interaction with VHDPTP. Modeling of the CHMP5 primary sequence using the Phyre2 server (Kelley et al., 2015) on the known closed conformation of CHMP3 (PDB:2GD5) predicted a helical “core” region from residues 23–157 following an unstructured N-terminal region (Figure 7, E and F). This is followed by flexible regions that bind the double MIT-domain region of Lip5 and a beta turn motif at the very distal end of Chmp5 that binds the Bro1 domain of Brox. GST pull-down experiments on fragments of Chmp5 indicated that both the “core” alpha-helical region and the Lip5-binding region combined were sufficient for interaction, while each of these regions was necessary for interaction. GST pull-down experiments further showed that yeast VBro1 bound the Chmp5 orthologue Vps60 (Figure 7I). Dynamic light scattering of the GST fusion proteins containing the alpha-helical core domain showed polydispersity, indicating some aggregation or polymerization. This did not account for differences in binding VHDPTP because full-length CHMP5 and a version lacking the C-terminal Brox domain that bound VHDPTP and the further truncated form lacking the Lip5-binding region that did not bind VHDPTP had similar levels of polydispersity (18.6%, 57.1%, and 36.6%, respectively). Together, these results identify a new interaction between the VHDPTP and CHMP6 that is conserved across phyla.
The Ub-binding sites of HD-PTP V domain are important for Vps4 stimulation
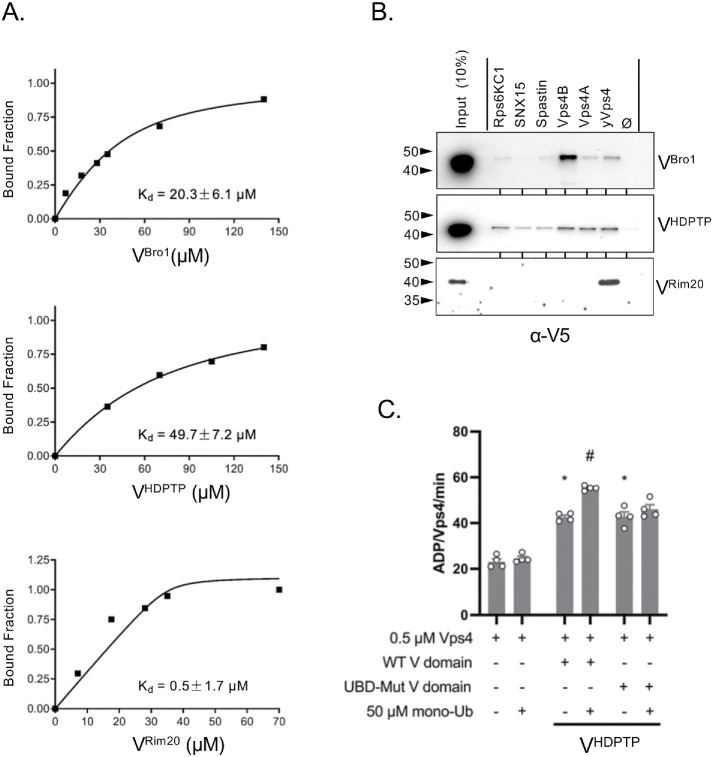
Previous experiments show that VBro1 stimulates the specific activity of the Vps4 AAA ATPase, which remodels and depolymerizes ESCRT-III subunit assemblies on membranes (Tseng et al., 2021). This is mediated by VBro1 binding the N-terminal MIT domain of Vps4. In addition, stimulation of Vps4 activity by VBro1 is further enhanced by the addition of Ub, which mediated its effects through the known Ub-interaction surface on the N-terminal helix of the VBro1. Those studies also showed that VHDPTP stimulated Vps4 activity, indicating a conserved regulatory feature (Tseng et al., 2021). NMR experiments confirmed binding of VHDPTP and VBro1 to the Vps4 MIT domain (Figure 8A; Supplemental Figure 3). Here 30 µM 15N-labeled Vps4 MIT domain was titrated, with increasing concentrations of VBro1 and VHDPTP yielding a binding curve that determined the Kds of 20 and 50 µM, respectively. We also found binding of the MIT domain by the VRim20, which associates with ESCRT components to mediate protease cleavage of the transcription factor RIM101 (Figure 8A). We next surveyed the ability of these V domains to bind to MITs from other proteins. Recombinant GST proteins fused to the MIT domains of VPS4A, VPS4B, Spastin, SNX15, and Rps6KC1 were used in pull-down assays using the V domains of Bro1, Rim20, and HD-PTP (Figure 8B). Both VBro1 and VHDPTP bound yeast Vps4 as well as human Vps4A and Vps4B MIT domains, whereas VRim20 showed strong but sole binding to yeast Vps4.
FIGURE 8:
Occupancy of HD-PTP Ub-binding sites stimulates Vps4 ATPase activity. (A) Binding curves of the indicated V domains from Bro1, HD-PTP, and RIM20 with yeast Vps4 MIT domain. Binding was measured by the decrease in HSQC signal from 15N-MIT (35 µM) in the presence of increasing amounts of the V domain. (B) Pull-down experiments showing binding of V5 epitope–tagged VBro1, VHDPTP, and VRim20 V on GST alone (ø) or GST fused to MIT domains from the indicated proteins. Bound fractions together with a 10% equivalent of input were immunoblotted with α-V5 antibodies. (C) Vps4 (0.5 µM) ATPase specific activity in the presence of 1 µM WT or ∆Ub (L371A, E374A, E375A, E388A, D389A) VHDPTP in the absence of presence of 50 µM mono-Ub. Error bars indicate SEM, (*) indicates p < 0.005 statistical significance compared with Vps4 alone, (#) indicates p < 0.005 statistical significance compared with HD-PTP V in the absence of Ub.
To evaluate the contribution of the Ub-binding sites in VHDPTP to modify the regulation of Vps4 by the V domain, we performed in vitro experiments that monitored the stimulation of Vps4 ATPase activity by VHDPTP in the presence and absence of Ub (Figure 8C). At a constant concentration of Vps4 MIT domain (500 nM), the addition of VHDPTP (4 µM) showed an approximately twofold increase in ATPase activity, which was enhanced with the addition of Ub (50 µM). The ∆Ub mutant of VHDPTP lacking both Ub-binding Site1 and Site2 also caused an approximately twofold stimulation of Vps4 ATPase activity. However, addition of Ub had no further effect, indicating that Ub stimulation was mediated through the Ub-binding sites that we identified and providing a biochemical role for the Ub-binding activity of VHDPTP.
A Bro1 pathway utilizes Vps60
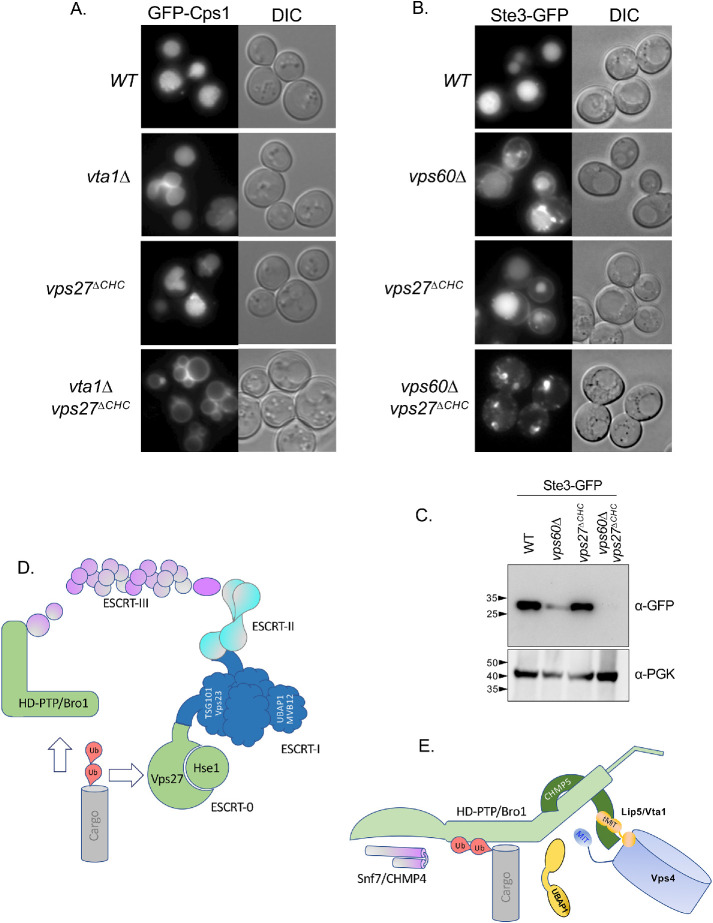
The conserved association between HD-PTP and CHMP5 and their yeast counterparts Bro1 and Vps10/Mos10 was further investigated in genetic experiments. Although Vps60/CHMP5 is structurally similar to other ESCRT-III subunits, it is not regarded as part of the core ESCRT-III but rather serves as an auxiliary ESCRT-III protein that may initiate a distinct functional step in the process of ILV biogenesis (McCullough et al., 2018). Vps60 interacts with Vta1, a regulatory protein of the Vps4 AAA-ATPase (Shiflett et al., 2004; Azmi et al., 2006). However, depending on the genetic background, their loss has a more moderate phenotype in comparison with the loss of any of the “core” components of ESCRT-0, I, II, or III (Babst et al., 2002; Shiflett et al., 2004; Azmi et al., 2006, 2008; Rue et al., 2008; Nickerson et al., 2010). The association of Vps60 with Bro1 suggested that this complex may serve as a dedicated parallel pathway for conveying Ub-cargo into an ESCRT-II–dependent ILV sorting pathway. Previous experiments suggested that ubiquitinated proteins might be processed by two distinct parallel pathways (Pashkova et al., 2013; Parkinson et al., 2015; Tang et al., 2016). One of these pathways is driven by ESCRT-0, which binds Ub-cargo and conveys it to polymerizing ESCRT-III via interactions via ESCRT-I and ESCRT-II. Another pathway uses Bro1 to bind Ub-cargo that in turn conveys it to ESCRT-III via interactions between the N-terminal Bro1 domain of Bro1 with Snf7 (Figure 9). These parallel pathways can be revealed when selective functions and connectivity of the ESCRTs are compromised (Pashkova et al., 2013; Tang et al., 2016), thereby shunting cargo so that it is more reliant on proper function of the alternate pathway (Figure 9C). For example, loss of the Ub-binding activity within Bro1 has little effect on MVB sorting; however, additional disruption of the association of ESCRT-0 with endosomal clathrin has a strong synthetic phenotype that demonstrated the role of Ub binding by Bro1 (Pashkova et al., 2013). We reasoned that if the major function of Vps60 is in a dedicated complex with Bro1, shifting cargo sorting into the Bro1 pathway would lead to an increased need for Vps60. As expected, we found that mutating the C-terminal clathrin-binding site of the ESCRT-0 component Vps27 (vps27∆CHC) had no effect on sorting of Ste3-GFP to the vacuole and that loss of Vps60 had a moderate defect for sorting Ste3-GFP into the vacuole (Figure 9B). However, major sorting defects were observed in vps60∆ vps27∆CHC double mutants. This synthetic defect was also appreciated when immunoblotting whole cell extracts for those transformants with anti-GFP antibodies to reveal the extent to which cells were able to cleave GFP from Ste3-GFP once it is delivered to the vacuole lumen (Figure 9B). These data support the idea that Vps60 functions along the Bro1-mediated MVB-sorting pathway, which is a specialized task in concert with the common function of “core” ESCRT-III subunits whose loss fully incapacitates all MVB sorting (Figure 9C). Because Vps60 also interacts with Vta1 (Figure 9E), we also assessed whether there were similar synthetic defects in MVB sorting when vta1∆ mutants were combined with mutations predicted to shunt Ub-cargo into the Bro1-dependent pathway. Here we followed GFP-Cps1, which sorts largely normally in vta1∆ mutants in the parental strain used here (Azmi et al., 2006). Yet, combining vta1∆ mutants with vps27∆CHC mutants resulted in clear defects in the delivery of GFP-Cps1 to the vacuole interior, whereas the single vps27∆CHC mutants were no different from wild-type cells (Figure 9A).
FIGURE 9:
Synthetic effects of CHMP5 and ESCRT-0. (A) Fluorescence and paired differential interference contrast (DIC) images demonstrating the extent of GFP-Cps1 sorting to the vacuole lumen in yeast strains carrying the indicated mutations. (B) Fluorescence and paired DIC images demonstrating the extent of Ste3-GFP sorting to the vacuole lumen in yeast strains carrying the indicated mutations. (C) Immunoblot of whole cell lysates from strains in B expressing Ste3-GFP using α-GFP antibodies to reveal the Pep4-dependent cleaved GFP fragment of ∼28 kDa or α-PGK as a load control. (D) Model showing different cargo sorting pathways. One uses ESCRT-0, which binds ubiquitinated cargo and associates with ESCRT-I, which in turn associates with ESCRT-II, which instigates ESCRT-III polymer nucleation through binding of the ESCRT-III component, Vps20. Alternatively, cargo can be routed through Bro1 or HD-PTP, which nucleate ESCRT-III by directly binding Snf7. (E) Cartoon of HD-PTP/Bro1 interactions depicting hypothesized connection between the V domain, Ub, and CHMP5, which in turn can associate with the tandem MIT domains (tMIT) of Vta1. The V domains can also bind the MIT domain of Vps4, shown unbound but in close proximity to the V domain in this cartoon. VHDPTP also binds UBAP1, depicted as unbound but near the V domain as well. Because Ub can cause hyperstimulation of Vps4 via the V domain through MIT binding, simultaneous occupancy of the V domain by binding Ub and MIT is likely. However, which sets of these interactions are complementary or competitive is not explicitly proposed in this model.
DISCUSSION
HD-PTP plays a pivotal role in sorting ubiquitinated proteins into endosomal ILVs that are targeted for degradation in lysosomes. Similar structure, interactions, and function make HD-PTP the closest human homologue of Bro1, the founding member of the Bro1 family of proteins. All of the central coiled-coil “V” domains among members of the Bro1 family noncovalently bind Ub, positioning them to associate with ubiquitinated targets to effect a variety of functions. While repeated attempts to cocrystalize Ub with VHDPTP were unsuccessful, we were able to use mutagenesis and PRE NMR approaches to define two Ub-interacting regions within VHDPTP, with the strongest site occupying the same position and similar sequence composition as the Ub-binding site in Bro1. Combined loss of Site1 and Site2 resulted in a V domain with exceptionally weak Ub-binding, with a Kd of ∼1.7 mM, supporting the concept that the main Ub-binding sites are encompassed in the two sites defined here. Modeling the tandem orientation of these binding sites with Ub (Figure 6D) implied that both might be occupied simultaneously in an orientation whereby the Ub moieties could be linked through K63. This possibility is rationalized by the fact that K63-linked polyubiquitin chains are used to modify cargo sorted into MVBs. Importantly, while HD-PTP may better accommodate a K63-linked di-Ub chain, it does not specifically require K63-linked Ubs to bind Ub because it binds mono-Ub with high good affinity (Figures 3C and 5F). This is in contrast to K63-specific interactions that are exemplified by proteins such as AMSH or TAB2, which use two very low affinity Ub-binding sites that must simultaneously bind two Ub moieties properly spaced by the K63 linage (Piper et al., 2014). The ability to accommodate K63 chains might serve other purposes such as protection from deubiquitination or inducing a conformational change in VHDPTP used for downstream functions.
Bro1 and HD-PTP have been proposed as an alternate Ub-sorting receptor that works in parallel with ESCRT-0 (Pashkova et al., 2013; Parkinson et al., 2015; Tang et al., 2016; Tabernero and Woodman, 2018). Preliminary experiments replacing endogenous HD-PTP with a version lacking both Ub-binding sites did not reveal major defects in EGFR sorting, a phenotype similar to that of yeast cells carrying a Bro1 mutant lacking its Ub-binding site. In yeast, the importance of the Ub-binding by Bro1 site was best revealed when alternative routes of Ub-cargo though the canonical ESCRT-0>I>II route was altered. Such parallel pathways have been proposed in mammalian cells as well, potentially explaining why loss of ESCRT-II in some circumstances does not alter MVB sorting of particular cargoes (Bowers et al., 2006; Parkinson et al., 2015). Thus, if HD-PTP also works as a parallel Ub-sorting receptor with ESCRT-0, specific loss of its Ub-binding capacity might apply only to a subset of MVB cargo or be fully revealed experimentally only when the canonical ESCRT-0,I,II pathway is partially compromised. Still, besides the Ub-binding sites defined here, HD-PTP might still associate with Ub via its binding to UBAP1, a Ub-binding ESCRT-I subunit that can also occupy a binding site on the distal 3-helical bundle arm of HD-PTP (Stefani et al., 2011; Gahloth et al., 2016). Mutating the UBAP1-binding site severely compromises the MVB-sorting function HD-PTP, suggesting that the additional Ub-recognition capacity or association with ESCRT-I, which are both afforded by associating with UBAP1, is important. How these interactions are integrated with those of CHMP5 and the MIT domain remain future goals of study and will help define the respective functions of these interactions.
Ub-binding also modulates the regulation of Vps4 activity by VHDPTP. Experiments in vitro have shown that VBro1 can stimulate Vps4 ATPase activity though association with its MIT domain that in turn likely promotes Vps4 oligomerization. This stimulation is increased when the V domain binds Ub. Similarly, HD-PTP binds the MIT domains of yeast and mammalian Vps4 isoforms (Figure 8, A and B) and has been shown to also stimulate Vps4 activity in vitro (Tseng et al., 2021). Our structural studies that defined Ub-binding sites on the VHDPTP allow us to show that Ub binding via HD-PTP enhances its ability stimulate Vps4 ATPase activity. This supports the idea that we have identified the major binding sites for Ub and highlights a conserved feature in the function of the V domains in general. Previous work defined mutations in VBro1 that still bound the MIT domain of Vps4 but lacked the ability to stimulate Vps4 activity by Ub, and these alleles in vivo had major defects in cargo sorting (Tseng et al., 2021). Thus, the phenotypes associated with loss of Ub binding by Bro1 could be due to either the inability to serve as a receptor to bind and sort Ub-cargo or the inability to have that Ub-cargo—or some other ubiquitinated component (Watanabe and Hatakeyama, 2017; Crespo-Yanez et al., 2018)—control Vps4 activity.
Seven structurally similar proteins comprise the ESCRT-III apparatus that mediates MVB biogenesis through crafting the final membrane remodeling events that lead to scission of ILVs. The functional steps they undergo are nucleation, polymerization, and an additional remodeling/elaboration step that may alter flat spherical arrays into assemblies that can promote membrane deformation and scission (Remec Pavlin and Hurley, 2020). These remodeling steps coincide with the arrival of Vps4, which may edit the composition of the polymer as well as disassemble it entirely (Adell et al., 2017; Schoneberg et al., 2018; Pfitzner et al., 2020). Components from the ESCRT-III polymer as well as ESCRT-III components have been divided into those that form the “core” complex that is critical for MVB biogenesis and accessory/auxiliary subunits that may modify the function of the main ESCRT-III polymer (Babst et al., 2002). These auxiliary subunits include Did2/CHMP1, Ist1/IST1, and Vps60/CHMP5, the latter of which we find associates with the VBro1 and VHDPTP, respectively. Only moderate defects in MVB formation are observed with the loss of any of these auxiliary components in yeast and mammalian cells and all regulate the activity of Vps4 (Shiflett et al., 2004; Shim et al., 2006; Azmi et al., 2008; Dimaano et al., 2008; Rue et al., 2008; Nickerson et al., 2010; Tan et al., 2015; Vild et al., 2015; Crespo-Yanez et al., 2018). A key role for some of these subunits has emerged from observations that Vps4 can edit the core ESCRT-III polymer with Did2/CHMP1, which is then stabilized by the addition of Ist1 to cause the shape change required for the final stages of scission (McCullough et al., 2018; Nguyen et al., 2020; Pfitzner et al., 2020). A similar molecular role for CHMP5 may exist given previous genetic analysis of double mutants that shows synthetic defects between the Did2-Ist1 (CHMP1-IST1) complex and the Vps60-Vta1 (CHMP5-Lip5) complex, suggesting that they work in parallel on a common process (Dimaano et al., 2008; Rue et al., 2008; Nickerson et al., 2010; Brune et al., 2019). Our genetic studies reveal a critical role for Vps60 as well as Vta1 when cargo sorting is diminished through the canonical ESCRT-0,I,II pathway and shunted more through the Bro1 pathway (Figure 9). Our findings that physically connect Vps60 with Bro1 and CHMP5 with HD-PTP (Figure 7) support a satisfying hypothesis that the ESCRT-III auxiliary components Did2∼Ist1 and CHMP5∼Vta1 divide their function along the parallel Ub-cargo sorting pathways headed by ESCRT-0 and Bro1/HD-PTP, respectively. Stimulatory and inhibitory effects on Vps4 ATPase activity have been observed in vitro through Vps60/CHMP5∼Vta1/Lip5 interactions (Skalicky et al., 2012; Yang et al., 2012). In addition, the V domains from HD-PTP and Bro1 also mediate Ub-enhanced Vps4 stimulation as shown in this study (Figure 8) and previous studies (Tseng et al., 2021). These interactions provide a number of potential molecular mechanisms that could specify targets and timing of Vps4 activity to not only ensure proper ILV scission but also that cargo is retained in these ILVs and can undergo a proper degree of deubiquitination and allow HD-PTP/Bro1 to escape incorporation into ILVs. How these molecular interactions of the V domains, including the association with UBAP1, are orchestrated should provide insight into what and how these events unfold in MVB biogenesis.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Plasmids and strains
Plasmids, strains, and antibodies utilized in this study are described in Supplemental Table 1. DNA of human HDPTP V-domain protein encoding residues 361–711 was created by gene synthesis (Genscript), amplified by PCR, and cloned into the pET151/D-TOPO vector (ThermoFisher), which encodes 6xHis tag at the N-terminus of VHDPTP or into the pTEF-GBD vector, which encodes the Gal4 DNA-binding domain at the N-terminus of VHDPTP as a bait for the yeast 2-hybrid DEEPN screen. CHMP5-encoding fragments were amplified from the mouse cDNA yeast 2-hybrid library obtained from Clontech (Mountain View, CA). Vps60-endocoding fragments were amplified from yeast genomic DNA. Amino acid mutations were generated by quick-change PCR-based mutagenesis (Xia et al., 2015).
Protein expression and purification
Recombinant proteins were expressed in Escherichia coli BL21(DE3) with 0.5 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) induction at 20°C for 18 h. Cells producing HDPTP V-domain proteins with the N-terminal 6xHis tag were lysed by French press or OneShot cell disruptor (Constant Systems) in the buffer containing 287 mM NaCl, 2.7 mM KCl, 12 mM NaPO4 (pH 7.4), and protease inhibitors without EDTA (Roche). Bacterial cell lysates were clarified by centrifugation and applied on TALON Co2+ affinity resin (Clontech, Mountain View, CA). Proteins were eluted in the lysis buffer containing 150 mM imidazole (pH 7.7) (Talon-elution buffer) and dialyzed against 40 mM NaPO4, 50 mM NaCl (pH 6.95) buffer for NMR or phosphate-buffered saline (PBS) (pH 7.4) for GST pull down. For Octet binding, V domains were purified by size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare Biosciences, Pittsburgh, PA) equilibrated with PBS, 10 mM dithiothreitol buffer (pH 7.4). GST-fusion proteins were purified from bacterial lysate using GSH affinity resin (GE Healthcare) in PBS buffer containing protease inhibitors and 1 mM EDTA and then eluted with 25 mM glutathione in PBS (pH 7.4) followed by dialysis against PBS. Ub (15N-labeled) was produced in cells grown in M9 media supplemented with 15N components. For biotinylated Ub-avitagged protein expression, biotin (Sigma) was added in the Luria Broth media (100 mg/l) upon addition of 1 mM IPTG (Li and Sousa, 2012). 15N-Ub and Ub-biotin proteins were purified from cell lysates by precipitation with 3.5% perchloric acid and CMC cation exchange chromatography followed by dialysis into 40 mM NaPO4, 50 mM NaCl (pH 6.95) buffer for NMR or PBS (pH 7.4) for biolayer interferometry binding experiments.
Yeast 2-hybrid studies
Protein interaction screening using the DEEPN method with WT and the ∆Ub mutant VHDPTP was performed with a pan-tissue mouse cDNA library as previously described (Pashkova et al., 2016) using modified low-copy “bait” plasmids and a dedicated software package described previously to evaluate gene enrichments as well as to computationally reconstruct plasmids encoding Y2H-interacting proteins (Krishnamani et al., 2018; Peterson et al., 2018). Briefly, yeast MAT A (strain Y3581) expressing Gal4-BD HD-PTP fusion proteins from p6649 (WT) or p6650 (Ub mutant) was grown in SD-Trp liquid media. Y187 yeast expressing mouse PAN+brain library from the pGAD vector were grown in SD-Leu liquid media. After mating on YPDA media, diploids were collected and grown in SD-Leu-Trp media at 30°C until saturation. Cells were pelleted to extract genomic DNA, PCR amplified across cDNA library inserts, and prepared for 150 base pair paired-end Illumina sequencing. Binary Y2H interactions were performed in diploid yeast as described previously (Pashkova et al., 2006) using plasmids reconstructed in low-copy “bait” and “prey” plasmids as previously described (Peterson et al., 2018).
Protein-binding assays
For GST pull-down experiments, 0.15 mg GST or Ub-GST was immobilized on 50 µl GSH resin and incubated with cell lysates expressing recombinant V5-tagged HDPTP V-domain proteins as described previously (Pashkova et al., 2013). Monoclonal anti-V5 antibodies were used for Western blotting (Life Technology, Carlsbad, CA).
NMR analysis
For NMR binding experiments, 15N/1H HSQC spectra of 15N-Ub at 30 µM in the presence or absence of VHDPTP were collected at 25°C on a Bruker Avance II 800 or 500 MHz spectrometer. Measurements of chemical shift perturbations and PRE effects were previously described (Pashkova et al., 2013). For PRE experiments, single cysteine mutations were introduced in the HDPTP V-domain. The mutant proteins were labeled with spin label MTSL overnight at 4°C in Talon elution buffer and then dialyzed against NMR buffer (50 mM NaPO4, pH 7.0). For each protein sample of mutant HDPTP V mixed with 15N-Ub, two HSQC spectra were collected. In one sample the spin label was in oxidized form, and in the second sample MTSL was reduced by addition of 2 mM ascorbic acid. Data were analyzed with SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) and NMR View (One Moon Scientific, Westfield, NJ). The PRE effects were calculated as 1 – (peak height in oxidized conditions/peak height in reduced conditions) and displayed as increments of SD of the mean PRE for all peaks.
Biolayer interferometry
Steptavidin Biosensors (Pall ForteBio) were used with an Octet RED96 Biolayer Interferometry (BLI) System (Pall ForteBio, Menlo Park, CA). Experiments were performed at 37°C in black 96-well plates (Greiner Bio-One, Monroe, NC) with shaking at 1000 rpm. Serial dilutions of VHDPTP as the analyte were used to ensure that the assay was responsive in the µM range for Kd. Data were analyzed using Octet Analysis software. Curves were fitted to obtain the Kd value using a simple single-site binding equation as binding strength was too low to ascertain stoichiometry accurately.
ATPase assays
Measurement of Vps4 ATPase activity was performed in ATPase buffer (20 mM HEPES, 100 mM KOAc, 5 mM MgOAc, pH 7.5) as previously described (Babst et al., 1998; Azmi et al., 2006, 2008; Norgan et al., 2013; Davies et al., 2014; Tan et al., 2015). All reactions were incubated at 30°C for 30 min before initiation by the addition of ATP (4 mM final concentration). Images were captured using a Typhoon FLA 7000 (GE Healthcare, United Kingdom). Vps4 titration data represent ATPase activities from a minimum of three independent experiments, with each experiment performed in duplicate. Data represent ATPase activities from a minimum of three independent experiments, with each experiment performed in replicate within the experiment. Data were graphed, and the statistical significance was assessed by T tests using Prism 5 (GraphPad, San Diego, CA). The Vps4 concentration used was 0.5 µM because this concentration of Vps4 exhibits submaximal specific activity. ATPase assays in the presence of Ub were performed with the minimal concentration (50 µM) yielding maximal enhancement.
SAXS and molecular modeling
SAXS data were collected at the 12-ID-B beamline at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, using solutions of VHDPTP ranging from 2 to 8 mg/ml. Data were processed with PRIMUS. The radius of gyration (Rg) estimated from Guinier plots was 40.4 + 0.5 Å for VHDPTP. The maximum diameter (Dmax) determined with GNOM was 148 Å. The ab initio models were generated with GASBOR (Svergun et al., 2001). FoXS, along with its Minimal Ensemble Search algorithm, was used to calculate the weighted fit of SAXS data to PDB:5LM1, 5LM2, and a synthetic model of VHDPTP (Schneidman-Duhovny et al., 2010). The synthetic model of VHDPTP was computed using YASARA to model in missing loops. Where possible, ordered regions in either 5LM1 or 5LM2 were used to make a complete chain. Modeling of Ub occupying Site1 of VHDPTP was done by superimposing the N-terminal helix of VHDPTP onto the crystal structure of the N-terminal helix of VBro1 in a complex with Ub. Modeling Site2 was accomplished with HADDOCK (Dominguez et al., 2003) using the VHDPTP model bound to di-Ub bound at Site1. The di-Ub was a translational fusion with three flexible residues between G76 and M1 to allow for multiple conformations including those that can assume K63-linked di-Ub bound to K63-specific Ub-binding proteins. Chemical shift perturbations for residues on the Ub surface when bound to VHDPTP lacking Site1 as well as large PRE effects from spin labels at position 385 were used as constraints. The model for CHMP5 was recreated from the alphafold2 server (https://alphafold.ebi.ac.uk/entry/Q9NZZ3).
Supplementary Material
Acknowledgments
We acknowledge Tabitha A. Peterson for her help with the DEEPN assay and Brian A. Davies for experiments and communications regarding the function of the HD-PTP V domain mutants in cells. We acknowledge the contributions of the Iowa Institute for Human Genetics (IIHG) genomic sequencing core and the Carver College of Medicine NMR and protein crystallography cores. This research was supported, in whole or in part, by National Institutes of Health Grants R01 GM116826 (to D.J.K.) and R01 GM58202 (to R.C.P.).
Abbreviations used:
- AAA
ATPases associated with diverse cellular activities
- Alix
ALG-2-interacting protein X
- AMSH
associated molecule with the SH3 domain of STAM
- BOD
Bro1 domain
- Bro1
BCK1-like resistance to osmotic shock
- CHC
clathrin heavy chain
- CHMP1
charged multivesicular body protein 1
- CHMP4
charged multivesicular body protein 4
- CHMP5
charged multivesicular body protein 5
- DEEPN
dynamic enrichment for evaluation of protein networks
- ESCRT
endosomal sorting complex required for transport
- Gal4AD
Gal4-activiation domain
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HD-PTP
His domain-containing protein tyrosine phosphatase
- HSQC
heteronuclear single quantum coherence
- IPTG
Isopropyl β- d-1-thiogalactopyranoside
- IST1
increased sodium tolerance 1
- MIT
Microtubule Interacting and Trafficking
- MTSL
(S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate)
- NMR
nuclear magnetic resonance
- PRE
paramagnetic relaxation enhancement
- SAXS
small angle x-ray scattering
- STAM2
signal transducing adaptor molecule 2
- TAB2
TGF-beta activated kinase 1 (MAP3K7) Binding Protein 2
- TSG101
tumor susceptibility 101
- Ub-cargo
ubiquitinated cargo
- Ub
ubiquitin
- UBAP1
ubiquitin associated protein 1
- UBPY
USP8
- Vps4
vacuolar protein sorting 4
- Vps60
vacuolar protein sorting 60
- Y2H
yeast 2-hybrid
- YPD
yeast extract–peptone–dextrose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-04-0219) on September 29, 2021.
REFERENCES
- Adell MAY, Migliano SM, Upadhyayula S, Bykov YS, Sprenger S, Pakdel M, Vogel GF, Jih G, Skillern W, Behrouzi R, et al. (2017). Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 6, e31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali N, Zhang L, Taylor S, Mironov A, Urbé S, Woodman P (2013). Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol 23, 453–461. [DOI] [PubMed] [Google Scholar]
- Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ (2006). Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol 172, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ (2008). ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell 14, 50–61. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD (2002). Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17, 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. (2012). Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat Cell Biol 14, 677–685. [DOI] [PubMed] [Google Scholar]
- Bissig C, Gruenberg J (2014). ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24, 19–25. [DOI] [PubMed] [Google Scholar]
- Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, Luzio JP (2006). Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem 281, 5094–5105. [DOI] [PubMed] [Google Scholar]
- Brune T, Kunze-Schumacher H, Kolling R (2019). Interactions in the ESCRT-III network of the yeast Saccharomyces cerevisiae. Curr Genet 65, 607–619. [DOI] [PubMed] [Google Scholar]
- Cabezas A, Bache KG, Brech A, Stenmark H (2005). Alix regulates cortical actin and the spatial distribution of endosomes. J Cell Sci 118, 2625–2635. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Agromayor M, Martin-Serrano J (2008). Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA 105, 10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-Yanez X, Aguilar-Gurrieri C, Jacomin AC, Journet A, Mortier M, Taillebourg E, Soleilhac E, Weissenhorn W, Fauvarque MO (2018). CHMP1B is a target of USP8/UBPY regulated by ubiquitin during endocytosis. PLoS Genet 14, e1007456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BA, Norgan AP, Payne JA, Schulz ME, Nichols MD, Tan JA, Xu Z, Katzmann DJ (2014). Vps4 stimulatory element of the cofactor Vta1 contacts the ATPase Vps4 α7 and α9 to stimulate ATP hydrolysis. J Biol Chem 289, 28707–28718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M (2008). Ist1 regulates Vps4 localization and assembly. Mol Biol Cell 19, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM (2003). HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125, 1731–1737. [DOI] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J (2012). ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol 197, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P (2008). The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci USA 105, 6308–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahloth D, Heaven G, Jowitt TA, Mould AP, Bella J, Baldock C, Woodman P, Tabernero L (2017a). The open architecture of HD-PTP phosphatase provides new insights into the mechanism of regulation of ESCRT function. Sci Rep 7, 9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahloth D, Levy C, Heaven G, Stefani F, Wunderley L, Mould P, Cliff MJ, Bella J, Fielding AJ, Woodman P, Tabernero L (2016). Structural basis for selective interaction between the ESCRT regulator HD-PTP and UBAP1. Structure 24, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahloth D, Levy C, Walker L, Wunderley L, Mould AP, Taylor S, Woodman P, Tabernero L (2017b). Structural basis for specific interaction of TGFbeta signaling regulators SARA/endofin with HD-PTP. Structure 25, 1011–1024.e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras MC, Kazan JM, Pause A (2017). Role of ESCRT component HD-PTP/PTPN23 in cancer. Biochem Soc Trans 45, 845–854. [DOI] [PubMed] [Google Scholar]
- Gingras MC, Kharitidi D, Chenard V, Uetani N, Bouchard M, Tremblay ML, Pause A (2009). Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP). Int J Dev Biol 53, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Ichioka F, Kobayashi R, Katoh K, Shibata H, Maki M (2008). Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4). FEBS J 275, 682–692. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitidi D, Apaja PM, Manteghi S, Suzuki K, Malitskaya E, Roldan A, Gingras MC, Takagi J, Lukacs GL, Pause A (2015). Interplay of endosomal pH and ligand occupancy in integrin alpha5beta1 ubiquitination, endocytic sorting, and cell migration. Cell Rep 13, 599–609. [DOI] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH (2005). Structural basis for endosomal targeting by the Bro1 domain. Dev Cell 8, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Kawawaki J, Kakiyama Y, Shimoda A, Tanaka K (2014). The ESCRT-III adaptor protein Bro1 controls functions of regulator for free ubiquitin chains 1 (Rfu1) in ubiquitin homeostasis. J Biol Chem 289, 21760–21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamani V, Peterson TA, Piper RC, Stamnes MA (2018). Informatic analysis of sequence data from batch yeast 2-hybrid screens. J Vis Exp 2018, 57802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios J, Mercier V, Roux A, Gruenberg J (2020). ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol 219, jcb.201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Oh KJ, Lee D, Kim BY, Choi JS, Ku B, Kim SJ (2016). Structural study of the HD-PTP Bro1 domain in a complex with the core region of STAM2, a subunit of ESCRT-0. PLoS One 11, e0149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sousa R (2012). Novel system for in vivo biotinylation and its application to crab antimicrobial protein scygonadin. Biotechnol Lett 34, 1629–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC (2012). Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep 13, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534. [DOI] [PubMed] [Google Scholar]
- McCullough J, Frost A, Sundquist WI (2018). Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu Rev Cell Dev Biol 34, 85–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HC, Talledge N, McCullough J, Sharma A, Moss FR 3rd, Iwasa JH, Vershinin MD, Sundquist WI, Frost A (2020). Membrane constriction and thinning by sequential ESCRT-III polymerization. Nat Struct Mol Biol 27, 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, West M, Henry R, Odorizzi G (2010). Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell 21, 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgan AP, Davies BA, Azmi IF, Schroeder AS, Payne JA, Lynch GM, Xu Z, Katzmann DJ (2013). Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J Biol Chem 288, 26147–26156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD (2003). Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci 116, 1893–1903. [DOI] [PubMed] [Google Scholar]
- Parkinson MD, Piper SC, Bright NA, Evans JL, Boname JM, Bowers K, Lehner PJ, Luzio JP (2015). A non-canonical ESCRT pathway, including histidine domain phosphotyrosine phosphatase (HD-PTP), is used for down-regulation of virally ubiquitinated MHC class I. Biochem J 471, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Sunshine AB, Rich M, Dunham MJ, Yu L, Piper RC (2013). The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev Cell 25, 520–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Jin Y, Ramaswamy S, Weisman LS (2006). Structural basis for myosin V discrimination between distinct cargoes. EMBO J 25, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Peterson TA, Krishnamani V, Breheny P, Stamnes M, Piper RC (2016). DEEPN as an approach for batch processing of yeast 2-hybrid interactions. Cell Rep 17, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TA, Stamnes MA, Piper RC (2018). A yeast 2-hybrid screen in batch to compare protein interactions. J Vis Exp 2018, 57801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfitzner A-K, Mercier V, Jiang X, Moser von Filseck J, Baum B, Šarić A, Roux A (2020). An ESCRT-III polymerization sequence drives membrane deformation and fission. Cell 182, 1140–1155.e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Dikic I, Lukacs GL (2014). Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol 6, a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Lehner PJ (2011). Endosomal transport via ubiquitination. Trends Cell Biol 21, 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remec Pavlin M, Hurley JH (2020). The ESCRTs—converging on mechanism. J Cell Sci 133, jcs.240333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CM, West M, Odorizzi G (2013). Doa4 function in ILV budding is restricted through its interaction with the Vps20 subunit of ESCRT-III. J Cell Sci 126, 1881–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue SM, Mattei S, Saksena S, Emr SD (2008). Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell 19, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MHH, Hoeller D, Yu J, Furnari FB, Cavenee WK, Dikic I, Bogler O (2004). Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Mol Cell Biol 24, 8981–8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A (2010). FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38, W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoneberg J, Pavlin MR, Yan S, Righini M, Lee IH, Carlson LA, Bahrami AH, Goldman DH, Ren X, Hummer G, et al. (2018). ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 362, 1423–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J (2004). Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem 279, 10982–10990. [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Hayden MS, Lee KY, Trombetta ES, Pypaert M, Nara A, Yoshimori T, Wilm B, Erdjument-Bromage H, et al. (2006). CHMP5 is essential for late endosome function and down-regulation of receptor signaling during mouse embryogenesis. J Cell Biol 172, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, Uter NT, Bajorek M, Myszka DG, Sundquist WI (2012). Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport. J Biol Chem 287, 43910–43926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra ML, Schlesinger PH, Naismith TV, Hanson PI (2018). Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360, eaar5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani F, Zhang L, Taylor S, Donovan J, Rollinson S, Doyotte A, Brownhill K, Bennion J, Pickering-Brown S, Woodman P (2011). UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol 21, 1245–1250. [DOI] [PubMed] [Google Scholar]
- Sun S, Zhou X, Zhang W, Gallick GE, Kuang J (2015). Unravelling the pivotal role of Alix in MVB sorting and silencing of the activated EGFR. Biochem J 466, 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001). Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80, 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero L, Woodman P (2018). Dissecting the role of His domain protein tyrosine phosphatase/PTPN23 and ESCRTs in sorting activated epidermal growth factor receptor to the multivesicular body. Biochem Soc Trans 46, 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Davies BA, Payne JA, Benson LM, Katzmann DJ (2015). Conformational changes in the endosomal sorting complex required for the transport III subunit Ist1 lead to distinct modes of ATPase Vps4 regulation. J Biol Chem 290, 30053–30065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Buchkovich NJ, Henne WM, Banjade S, Kim YJ, Emr SD (2016). ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis. eLife 5, e15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, Nakamura A, Shimizu N, Shimizu K (2000). HD-PTP: a novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun 278, 671–678. [DOI] [PubMed] [Google Scholar]
- Tseng CC, Dean S, Davies BA, Azmi IF, Pashkova N, Payne JA, Staffenhagen J, West M, Piper RC, Odorizzi G, Katzmann DJ (2021). Bro1 stimulates Vps4 to promote intralumenal vesicle formation during multivesicular body biogenesis. J Cell Biol 220, e202102071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, Stenmark H (2020). The many functions of ESCRTs. Nat Rev Mol Cell Biol 21, 25–42. [DOI] [PubMed] [Google Scholar]
- Vild CJ, Li Y, Guo EZ, Liu Y, Xu Z (2015). A novel mechanism of regulating the ATPase VPS4 by its cofactor LIP5 and the endosomal sorting complex required for transport (ESCRT)-III protein CHMP5. J Biol Chem 290, 7291–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI (2013). Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Hatakeyama S (2017). Fine-tuning of thymocyte development by ubiquitination-mediated stability control of the ESCRT protein CHMP5. Cell Mol Immunol 14, 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel EM, Schultz SW, Schink KO, Pedersen NM, Nahse V, Carlson A, Brech A, Stenmark H, Raiborg C (2018). Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat Commun 9, 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL, Urbe S (2007). The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol 8, 355–368. [DOI] [PubMed] [Google Scholar]
- Xia Y, Chu W, Qi Q, Xun L (2015). New insights into the QuikChange process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res 43, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Vild C, Ju J, Zhang X, Liu J, Shen J, Zhao B, Lan W, Gong F, Liu M, et al. (2012). Structural basis of molecular recognition between ESCRT-III-like protein Vps60 and AAA-ATPase regulator Vta1 in the multivesicular body pathway. J Biol Chem 287, 43899–43908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Fan G, Hao Y, Hammell M, Wilkinson JE, Tonks NK (2017). Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes Dev 31, 1939–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.