Abstract
The hexameric HOPS (homotypic fusion and protein sorting) complex is a conserved tethering complex at the lysosome-like vacuole, where it mediates tethering and promotes all fusion events involving this organelle. The Vps39 subunit of this complex also engages in a membrane contact site between the vacuole and the mitochondria, called vCLAMP. Additionally, four subunits of HOPS are also part of the endosomal CORVET tethering complex. Here, we analyzed the partition of HOPS and CORVET subunits between the different complexes by tracing their localization and function. We find that Vps39 has a specific role in vCLAMP formation beyond tethering, and that vCLAMPs and HOPS compete for the same pool of Vps39. In agreement, we find that the CORVET subunit Vps3 can take the position of Vps39 in HOPS. This endogenous pool of a Vps3-hybrid complex is affected by Vps3 or Vps39 levels, suggesting that HOPS and CORVET assembly is dynamic. Our data shed light on how individual subunits of tethering complexes such as Vps39 can participate in other functions, while maintaining the remaining subcomplex available for its function in tethering and fusion.
INTRODUCTION
The yeast vacuole is an acidic hydrolytic organelle that recycles macromolecules for reuse of cellular building blocks, and thus is equivalent to the lysosome of higher eukaryotes. Additionally, it plays a major role in the regulation of cellular pH, and is a storage place for amino acids and ions, including calcium and other metals, phosphate and polyphosphate (Thumm, 2000). The vacuole receives material through vesicular transport from the endosomal pathway, from the autophagic pathway, and from the biosynthetic pathway directly from the Golgi complex. The vesicle fusion machinery for all these routes includes Ypt7, the Rab7-like GTPase of the vacuole, and the HOPS tethering complex.
HOPS is a heterohexameric tethering complex that consists of a core of the four Class C subunits Vps11, Vps16, Vps33, and Vps18 and the Vps39 and Vps41 subunits at either end. Vps39 and Vps41 bind to GTP-bound Ypt7 and thus allow the HOPS complex to bridge Ypt7-positive membranes (see Figure 2A in Results and Discussion). The four core subunits are shared with another hexameric tethering complex, called CORVET, which acts in earlier steps of the endocytic pathway. This complex has two additional subunits, Vps3 and Vps8, which interact with the early endosomal Rab5-like GTPase Vps21 (Ungermann and Kümmel, 2019). Vps3 and Vps39 both interact with Vps11 in the same C-terminal region (see Figure 2A in Results and Discussion; Ostrowicz et al., 2010).
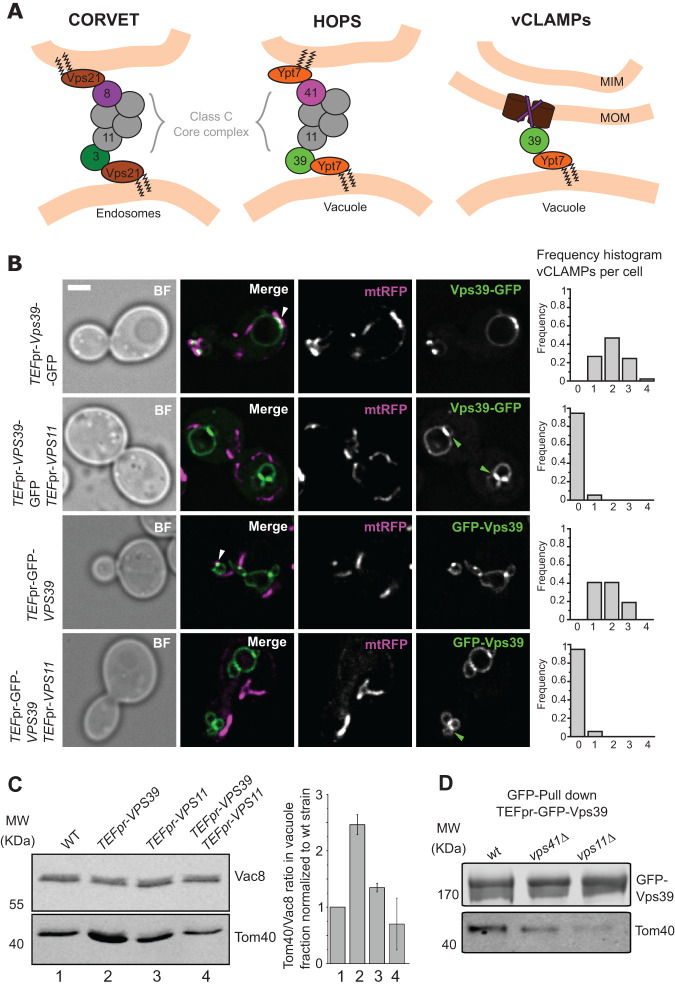
FIGURE 2:
HOPS and vCLAMPs compete for Vps39. (A) Diagram depicting the proteins that form the endolysosomal tethering complexes and the Vps39–vCLAMP tether. The Vps39 protein is present in both HOPS and vCLAMP, and the Class C-core complex, formed by Vps33, Vps16, Vps18, and Vps11, is part of both HOPS and CORVET. (B) Analysis of the effect of Vps11 overexpression on extended vCLAMP formation. Overexpressed Vps39 tagged with GFP at either the N- or C-terminus was imaged relative to mitochondrial targeted RFP (mtRFP). Where indicated, Vps11 was overexpressed under the control of TEF1 promoter. White arrowheads indicate overextended vCLAMPs, and the frequency of observation of these structures is represented in the frequency histogram to the right of the images. Green arrowheads indicate accumulations of Vps39 between two vacuoles. The scale bar represents 2 μm. (C) Examination of mitochondrial copurification in vacuole preparations. Vacuoles were purified from the indicated strains. Western blotting was used to evaluate the copurification of mitochondria by analyzing the levels of Tom40 (mitochondria marker) and Vac8 (vacuolar marker) in the purified vacuoles. The plot to the right displays the ratio Tom40/Vac8 normalized to WT, as mean ± SD of three independent experiments. (D) Coimmunoprecipitation of Vps39 and Tom40 is diminished in strains with defective HOPS. Vps39 was N-terminal GFP-tagged and overexpressed under control of TEF1pr in strains containing deletions in VPS41 of VPS11. Vps39 was immunoprecipitated with GFP-TRAP Sepharose beads, and eluates were analyzed by SDS–PAGE and Western blotting for levels of Vps39 and copurified Tom40.
Apart from its pivotal function in HOPS, Vps39 has an additional role in the formation of a membrane contact site between vacuoles and mitochondria, termed vCLAMP for vacuolar and mitochondrial patch (Elbaz-Alon et al., 2014; Hönscher et al., 2014). Membrane contact sites (MCSs) are structures that mediate communication between organelles through close proximity of their membranes that does not result in fusion. Proteins or protein complexes that interact with both membranes, called tethers, form this apposition (Eisenberg-Bord et al., 2016). Different metabolites are exchanged at these structures, providing a platform for organelle biogenesis, signaling and communication (Prinz et al., 2019). Recent screens have revealed that most organelles contact one another through membrane contact sites (Kakimoto et al., 2018; Shai et al., 2018). For many MCSs the machinery involved in their formation is not or is only partially described, and mechanistic insights into their function or regulation remain scarce (Scorrano et al., 2019).
This also applies to vCLAMPs. It is known that this MCS is under metabolic regulation, as it is present when cells grow with glucose as a carbon source but disassembles under respiratory conditions (Hönscher et al., 2014). vCLAMPs are established by the interaction of the protein Vps39 with vacuolar-localized Ypt7 and with the Tom40 subunit of the translocase of the outer membrane on mitochondria (Elbaz-Alon et al., 2014; Hönscher et al., 2014; González Montoro et al., 2018). Additionally, the Vps13 protein has been identified at the mitochondria- vacuole interface (Lang et al., 2015), and several other subcellular localizations (Park and Neiman, 2012; Park et al., 2016; De et al., 2017; Bean et al., 2018), yet does not seem to be part of the same structure formed by Vps39 (González Montoro et al., 2018). We previously identified a mutant in Vps39 that is impaired in vCLAMP formation yet functional in HOPS. This showed that the two functions of Vps39 are genetically separable. Cells carrying this vCLAMP-impaired Vps39 mutant showed reduced survival to starvation, and reduced growth in particular stress conditions (González Montoro et al., 2018). Additionally, vCLAMPs likely generate a specialized region within mitochondria, as the areas involved in vCLAMP formation are devoid of cristae (Hönscher et al., 2014) and concentrate assemblies of particular complexes in the inner mitochondrial membrane (Tirrell et al., 2020).
In this work, we analyze the cross-talk among vCLAMP and endolysosomal tethering complexes, which is mediated by shared subunits. Our data reveal that in vivo, the subunits are not always part of the thus far considered standard tethering complexes, indicative of additional functions in the cell. Furthermore, the interaction of Vps39 and Vps3 with the hexameric tethering complexes is part of a dynamic equilibrium, and this is linked to vCLAMP formation. We thus provide a first systematic analysis of the participation of shared subunits in their different functions to uncover their cellular regulation.
RESULTS AND DISCUSSION
The HOPS subunit Vps39 functions in vCLAMPs beyond tethering
The protein Vps39 is a component of both vCLAMP and the hexameric HOPS tethering complex, which mediates tethering of membranes during processes of organelle or vesicle fusion. We thus initially analyzed whether, as a parallel to its role within HOPS, the role of Vps39 at vCLAMPs is just the tethering of membranes. To address this, we used a strain carrying the Vps3912xM mutant, which we previously showed to be functional as a subunit of the HOPS complex but unable to mediate vCLAMP formation (González Montoro et al., 2018). In this strain, we generated artificial tethers between the vacuole and the mitochondria. This approach was the basis for the identification of the ERMES complex as a contact site tether between ER and mitochondria (Kornmann et al., 2009), and artificial tethers were also able to rescue the lack of tethering of mitochondria to the cell cortex mediated by Num1 (Klecker et al., 2013; Lackner et al., 2013).
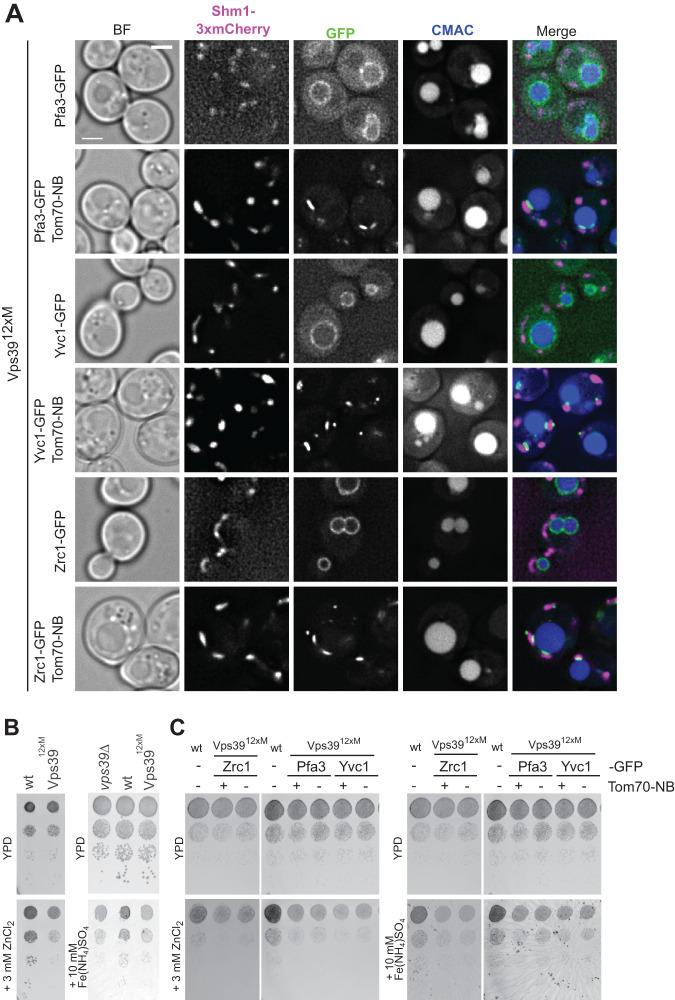
To generate the artificial tethers, we tagged different vacuolar membrane proteins with GFP and the Tom70 mitochondrial outer membrane protein with a nanobody that binds to GFP. As vacuolar membrane proteins we used the palmitoyltransferase Pfa3 (Smotrys et al., 2005), the calcium channel Yvc1 (Palmer et al., 2001), and the zinc channel Zrc1 (MacDiarmid et al., 2000). Adding a nanobody to Tom70 changed the distribution of the vacuolar proteins, which then accumulated in proximity to the mitochondria (Figure 1A). This shows that this strategy indeed allowed the generation of an artificial membrane contact site between the two organelles.
FIGURE 1:
Artificial tethers cannot compensate for the lack of vCLAMP. (A) Generation of artificial vCLAMP tethers. Cells expressing different vacuolar membrane proteins C-terminally tagged with GFP were observed by fluorescence microscopy. Where indicated, the attachment of these proteins to mitochondria was induced by tagging Tom70 C-terminally with a nanobody that recognized GFP. Shm1–3xmCherry is used to visualize mitochondria and CMAC staining to label the vacuole lumen. The scale bar represents 2 μm. (B) vCLAMP impaired cells display a growth defect in the presence of zinc or iron. Wild type, vps39Δ, or vps3912xM strains were grown as serial dilutions on YPD, YPD containing 3 mM ZnCl2, and YPD containing 10 mM Fe(NH4)SO4. (C) Artificial tethers do not suppress the growth defects of vCLAMP-impaired strains. Strains containing the vCLAMP-impaired vps3912xM allele with or without the artificial tethers, as shown in A, were grown as serial dilutions on YPD, YPD containing 3 mM ZnCl2, and YPD containing 10 mM Fe(NH4)SO4.
To test if these artificial tethers compensate for the lack of Vps39-mediated vCLAMP, we took advantage of a growth deficiency of the vCLAMP-deficient Vps3912xM mutant on high concentrations of iron or zinc (Figure 1B). These ions are stored normally in the vacuole (MacDiarmid et al., 2000; Li et al., 2001). However, the artificial contacts were unable to suppress the growth phenotypes caused by the disruption of the Vps39-containing vCLAMP (Figure 1C). We conclude that Vps39-mediated vCLAMP does not just generate proximity between vacuoles and mitochondria, but requires Vps39 and possibly other specific proteins to function.
Even though this is a negative result, other MCSs can be replaced by such tethers (Kornmann et al., 2009; Lackner et al., 2012; Klecker et al., 2013). This suggests either that the dynamic nature of the Vps39-vCLAMP is important for its function or that the presence of these specific proteins is necessary to recruit further components or carry out another specific function beyond tethering. This prompted us to analyze the mechanisms involved in the dynamic recruitment of Vps39 to vCLAMPs.
Vps39 needs to disengage from HOPS to form vCLAMP
To understand the dynamics and regulation of Vps39 participation in its role as a component of both vCLAMP and HOPS (Figure 2A), we analyzed the effect of HOPS subunits on vCLAMP function in more detail. It was already shown that Vps39 can still interact with mitochondria in the absence of the other Ypt7-interacting HOPS subunit Vps41, and overexpression of Vps41 did not lead to overextended vCLAMPs (Hönscher et al., 2014). This suggested that Vps39 alone can establish vCLAMP, yet it was not clear if Vps39 needed to disassemble from the HOPS complex to engage in vCLAMP formation.
Overexpression of Vps39 results in the generation of overextended vCLAMPs between vacuoles and mitochondria, which can be observed by fluorescence microscopy (González Montoro et al., 2018; Figure 2B, white arrows). Vps39 directly binds Vps11 within HOPS (Ostrowicz et al., 2010; Plemel et al., 2011 Figure 2A). We therefore asked if cooverexpression of Vps11 would affect vCLAMP formation. While cells overexpressing Vps39–GFP or GFP-Vps39 display 1–3 extended vCLAMPs per cell, cooverexpression of Vps11 resulted in the disappearance of these accumulations (Figure 2B). In the strains where Vps11 was cooverexpressed, we often observed Vps39 accumulation between two vacuoles (Figure 2B, green arrows). This was confirmed by impaired copurification of mitochondria with vacuoles as a method of addressing the extent of the contact between these two organelles (Figure 2C). We conclude that Vps39 needs to disassemble from Vps11 within the HOPS complex to be able to establish vCLAMP, suggesting that the two functions compete for a pool of Vps39.
These results show that Vps39 needs to be free from its partners in HOPS to be able to engage in MCSs formation. We consider it likely that it is just the formation of the Vps39–Vps11 subcomplex that is able to bring Vps39 away from vCLAMPs, since the other subunits are not overexpressed. One important question is how this equilibrium is regulated in the cell. Interestingly, while the overexpression of Vps11 reduced the participation of overexpressed Vps39 in vCLAMP, it did not have the same effect on endogenous Vps39 (Figure 2C). We know that the levels of vCLAMP formation at endogenous levels are significant and can be reduced, because a Vps3912xM shows reduced levels (Gonzalez Montoro et al, 2018). This would indicate that the pool of Vps39 involved in vCLAMPs under endogenous conditions is protected by an as yet unknown mechanism from being incorporated into a Vps11–Vps39 subcomplex.
Furthermore, the assembly–disassembly dynamics of the HOPS complex could be influenced by other functions of the subunits. Vps39 has been reported to act as the guanine nucleotide exchange factor of the Gtr1 GTPase without participation of other HOPS subunits (Binda et al., 2009) and to mediate phosphatidylethanolamine transport to the mitochondria, independent of both HOPS and vCLAMPs (Iadarola et al., 2020). Likewise, the Vps41 alone is involved in the regulation of termination of vacuole inheritance (Wong et al., 2020), further suggesting that assembly–disassembly dynamics of the HOPS complex must exist in vivo.
We thus asked, if HOPS function is a prerequisite for vCLAMP engagement of Vps39 and therefore tested the interaction of Vps39 with Tom40 in HOPS deletion mutants, which can be tested by coimmunoprecipitation (Figure 2D). In both vps41∆ and vps11∆ cells, the copurification of Tom40 with Vps39 is diminished, although still detectable. This result indicates that HOPS function influences the formation of vCLAMPs. One possible explanation for this observation is that the formation of vCLAMPs requires a fully functional vacuole, which is not present in the absence of HOPS. Another possible explanation is that Vps39 is delivered to the vacuole as a part of HOPS and only then disassembles to engage in vCLAMP formation.
Identification of an in vivo pool of the Vps41–Vps3–Class C complex
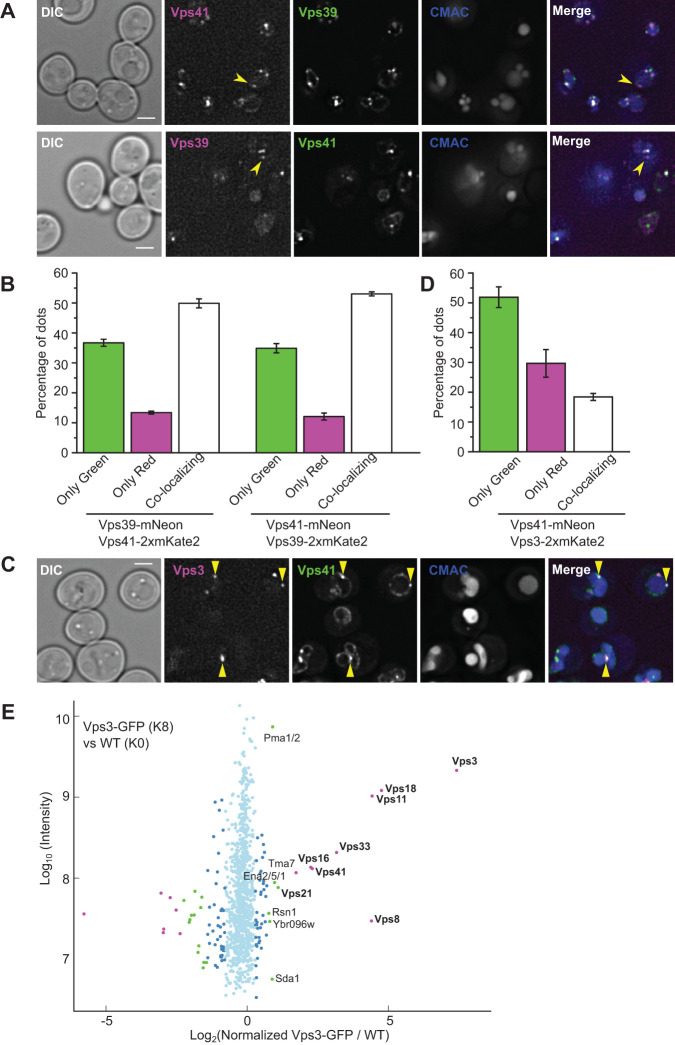
If an equilibrium of Vps39 between vCLAMPs and HOPS takes place, HOPS complexes in which Vps39 has disassembled should be present in cells. To assess if this is the case, we C-terminally tagged the HOPS-specific Vps39 and Vps41 subunits either with mNeonGreen (green) or with mKate2 (red) fluorophores to monitor both at endogenous levels. Both proteins are present at the vacuole and in dots proximal to the vacuole, yet they are hard to visualize due to their low expression (Figure 3A). We then analyzed them in both combinations for colocalization and observed that the majority of dots colocalized (Figure 3B). However, a significant portion did not. The portion depended on which protein carried the red fluorophore, even though we used a double red fluorescent protein to increase sensitivity, indicating that not all localizations of the protein are visible in the red channel. As we performed the experiment in both combinations, we considered the real number of not colocalizing structures as the ones seen in the red (less sensitive) channel, at around 10% (Figure 3B). We conclude that in vivo, a minor but significant portion of Vps41 is not in complex with Vps39.
FIGURE 3:
An intermediate ClassC–Vps3–Vps41 tethering complex exists in vivo. (A) Colocalization study of endogenously tagged Vps39 and Vps41. Representative fluorescence microscopy images of the localization Vps39 and Vps41 tagged C-terminally with either mNeonGreen or mKate2. The vacuolar lumen was stained with CMAC. The yellow arrowheads indicate structures positive for one subunit but not the other. The scale bar represents 2 μm. (B) Quantification of the experiment shown in A. The plots indicate the percentage of dots positive for one or the other marker or both, as mean ± SD from three independent experiments. (C) Assessment of Vps41 and Vps3 localization under endogenous level. Strains expressing C-terminally mNeonGreen-tagged Vps41 and mKate2-tagged Vps3 were observed by fluorescence microscopy. Yellow arrowhead indicates colocalizing dots. CMAC was used to stain the vacuole lumen. The scale bar represents 2 μm. (D) Quantification of the experiment described in C. The bar graph represents the percentage of dots positive for one or the other marker or both, as mean ± SD of three independent experiments. (E) SILAC-based GFP-Trap pull down of Vps3–GFP and mass spectrometry analysis. Light isotope-labeled control cells and heavy isotope-labeled cells expressing Vps3–GFP were used. The log10 of protein intensity is plotted against the log2 of the normalized heavy/light SILAC ratio. Significant outliers are colored in magenta (p < 10−11), green (p < 10−4), or dark blue (p < 0.05); other detected proteins are shown in light blue. All subunits of the CORVET tethering complex are significantly enriched, as well as the interacting Rab GTPase Vps21, and Vps41, previously described only as a subunit of HOPS.
We next asked the fate of the HOPS when Vps39 would dissociate. One possibility would be that Vps3, the other binding partner of Vps11 in the context of the CORVET complex, replaces Vps39 as both proteins bind to the same region of Vps11 (Ostrowicz et al., 2010, Figure 2A). We already know that such a heterohexameric complex containing both Vps3 and Vps41 can form if Vps3 is overexpressed, and is stable enough to be purified (Lürick et al., 2017; Ostrowicz et al., 2010; Peplowska et al., 2007). We thus asked if such a heterohexamer does exist in the cell. We first tagged Vps3 and Vps41 C-terminally with mNeonGreen and mKate2 fluorophores and analyzed their colocalization, and indeed observed that there is a significant population of structures positive for both subunits (Figure 3, C and D). Second, we used stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry to determine proteins copurifying with endogenously expressed Vps3–GFP. As expected, we observed a strong coenrichment of the other subunits of the CORVET complex. However, we also observed a clear enrichment of Vps41, indicating that these two proteins coexist in a complex in cells under normal conditions, albeit to a lower extent than the normal CORVET (Figure 3E).
Our results indicate the existence of an intermediate ClassC–Vps3–Vps41 complex in vivo. We knew from previous analyses that this complex is formed if Vps39 or Vps8 is absent (Peplowska et al., 2007) and can be obtained upon overexpression (Ostrowicz et al., 2010). An important, yet unresolved question is whether this is a functional tether or just a buffering mechanism for subunits. If such a tethering complex does mediate fusion events in vivo, this would mean that Ypt7- and Vps21-positive compartments fuse with each other, a mechanism that has been observed for Rab5 and Rab7 positive compartments in metazoan cells (Luzio et al., 2010). In yeast, Vps21-positive endosomes mature to Ypt7-positive late endosome by a mechanism of Rab transition that involves the recruitment of the Ypt7 GEF, the Mon1–Ccz1 complex, by Vps21 (Langemeyer et al., 2020). If, additionally, Vps21 and Ypt7 compartments can fuse with each other, this will provide a separate means of maturation, which could perhaps be followed by a specific subpopulation of endosomes. In addition to Vps21, Saccharomyces cerevisiae has three additional Rabs belonging to the Rab5 class, Ypt52, Ypt53, and Ypt10 (Singer-Krüger et al., 1994; Nickerson et al., 2012; Schmidt et al., 2017; Langemeyer et al., 2020); one possibility is that these different paralogs preferentially favor the different maturation mechanisms.
Dynamic formation of the Vps41–Vps3–Class C intermediate complex
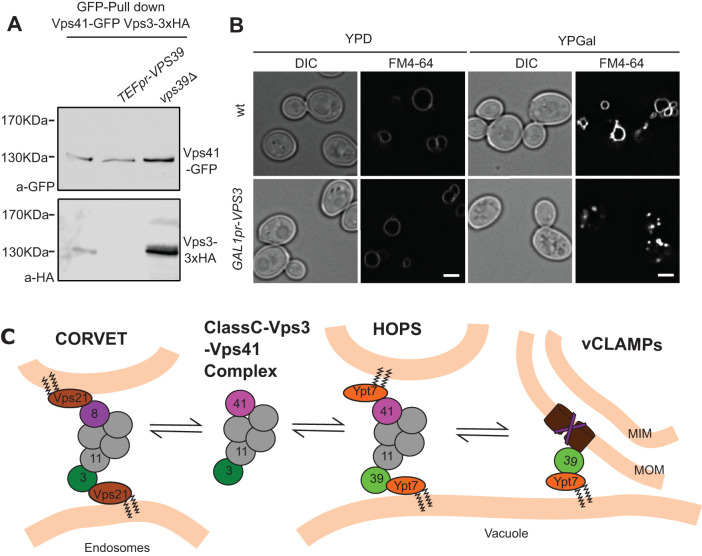
If, indeed, the disassembly of Vps39 from the HOPS complex to engage in vCLAMP results in the establishment of the Class C–Vps3–Vps41 complex, then the levels of Vps39 in the cell should influence the level of assembly of this complex. To test this, we analyzed the copurification between Vps41 and Vps3 upon overexpression or deletion of Vps39. Indeed, we saw that increasing the levels of Vps39 results in diminished copurification and deletion of Vps39 results in very increased copurification (Figure 4A). We then asked if the shift of this equilibrium also occurs in the opposite direction, and thus overexpressed Vps3, which should impair HOPS function, if it is able to sequester the Class C core complex from Vps39. Indeed, we observe that very high overexpression of Vps3 from the GAL1 promoter resulted in vacuolar fragmentation, indicating impaired HOPS function (Figure 4B), in agreement with our earlier findings (Peplowska et al., 2007). We conclude that the association of CORVET and HOPS subunits in a heterohexameric Class C–Vps3–Vps41 complex can explain Vps39 engagement in vCLAMP (Figure 4C). This working model is based on our deletion and overexpression analyses and requires further validation. Future studies need to address how Vps39 can partition between both functions, as a subunit of HOPS and vCLAMPs.
FIGURE 4:
The formation of different tethering complexes is determined by an equilibrium affected by the protein levels of the subunits. (A) Coimmunoprecipitation of Vps41 and Vps3 at different levels of Vps39. C-terminal GFP-tagged Vps41 was immunoprecipitated with a GFP-nanobody Sepharose affinity matrix from strains containing endogenous levels of Vps39, overexpressed Vps39 from the TEF1 promoter, or a deletion of VPS39. Eluates were analyzed by SDS–PAGE and Western blotting for levels of Vps41 and copurified Vps3–3xHA. (B) Vacuole morphology upon overexpression of Vps3. The morphology of vacuoles in cells overexpressing Vps3 from the strong GAL1 promoter was analyzed by FM4-64 staining of the vacuole membrane. The scale bar represents 2 μm. (C) Working model depicting the existence in vivo of an intermediate Class C–Vps3–Vps41 complex. Availability of the subunits shared with HOPS affect the levels of this complex, which could act as a buffering mechanism to allow the participation of Vps39 in the vCLAMP membrane contact site. For details see text.
In addition to Vps39, several other proteins that form contact site tethers have additional functions in the cell, which they undertake as part of other protein complexes. One examples of this is Mdm10, which forms part of the sorting and assembly machinery (SAM) complex of the mitochondrial outer membrane (Ellenrieder et al., 2016; Meisinger et al., 2004) and of the ERMES complex, a stable tether between the endoplasmic reticulum and the mitochondria (Kornmann et al., 2009). Another example is the protein Snd3, which is a component of the SND machinery, responsible for targeting proteins to the endoplasmic reticulum (Aviram et al., 2016), and was recently shown to be part of the contact site formed between the vacuole and the nuclear endoplasmic reticulum (Tosal-Castano et al., 2021). In both cases, as for Vps39, the proteins engage in these moonlighting functions in MCSs, independent of the other subunits of the complex of which they form part in their other functions. In the case of the vCLAMP, the Tom40 component is also a moonlighting protein, as it is the pore-forming subunit of the translocase of the outer membrane of mitochondria (TOM; Künkele et al., 1998; Becker et al., 2005; Shiota et al., 2015; Bausewein et al., 2017). Whether Tom40 engages in vCLAMPs as part of the TOM complex or whether full or partial disassembly is necessary is still unknown. For Mdm10 and Vps39, both functions are genetically separable by mutations (Ellenrieder et al., 2016; González Montoro et al., 2018).
The partition of moonlighting proteins between the different complexes may regulate contact formation or mediate cross-talk with the other processes in which the proteins are involved. MCS are increasingly recognized as regulated by different metabolic pathways. For example, the nuclear–vacuolar junction changes its size in response to glucose availability (Hariri et al., 2017; Tosal-Castano et al., 2021), and the vacuole–peroxisome MCSs expand under conditions of peroxisome proliferation (Wu et al., 2019). Whether these moonlighting proteins regulate cross-regulation of membrane contact sites with other cellular processes such as vesicular transport (Vps39) or protein biogenesis (Tom40, Snd3, Mdm10) is an appealing question that needs to be addressed in future research.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains, plasmids, molecular biology
Strains used in this study are listed in Supplemental Table S1. Saccharomyces cerevisiae strains were genetically manipulated by homologous recombination of PCR-amplified cassettes (Janke et al., 2004) using the methods described in the same study.
Plasmid pYM25 2xGAmKate2 was created by inserting the mKate2 sequence, amplified by PCR from a plasmid provided by Roland Wedlich-Söldner (University of Münster, Germany), into the SalI/BssHII sites of plasmid pYM25 (Janke et al., 2004). A second mKate2 sequence was inserted into the BssHII site. The resulting plasmid was confirmed by sequencing and was used to build the 2xGAmKate2 containing strains from this work. The plasmid expressing mitochondrial targeted RFP (pYX142–Mt–RFP) was kindly provided by Benedikt Westermann (University of Bayreuth, Germany) (Böckler and Westermann, 2014).
Fluorescence microscopy
Yeast cells were grown to exponential phase in yeast extract peptone dextrose (YPD) or synthetic complete medium supplemented with all amino acids (SDC). To stain the vacuolar membrane of the cells, 30 µM FM4-64 (Molecular Probes, Eugene, OR) was added and incubated for 30 min. The cells were washed twice with fresh medium and incubated in medium without dye for 1 h (Vida and Emr, 1995). For luminal staining of vacuoles, cells were incubated with 0.1 mM 7-amino-4-chloromethylcoumarin [CMAC] (Invitrogen) for 10 min, followed by washing with SDC medium.
Cells were imaged live in SDC media in a DeltaVision Elite imaging system based on an Olympus IX-71 inverted microscope equipped with a 100× N.A. 1.49 objective, a sCMOS camera (PCO, Kelheim, Germany), an InsightSSI illumination system, 40,6-diamidino-2-phenylindole, GFP, mCherry, and Cy5 filters, and SOftWoRx software (Applied Precision, Issaquah, WA). z-stacks images with 0.2-µm spacing were deconvolved (SOftWoRx). All images were processed and quantified with ImageJ software (NIH, Bethesda, MD). One representative plane of the z-stacks from deconvolved image is shown in each figure.
Vacuole isolation
Vacuoles were isolated as described by Haas (1995). Equivalent amounts of vacuolar proteins were resolved by SDS–PAGE and analyzed by Western blotting. The membrane was decorated against the rabbit polyclonal antibodies Vac8 and Tom40 (kindly provided by Klaus Pfanner, Freiburg, Germany) to detect vacuole and mitochondria fraction, respectively. DyLight 800 Goat anti Rabbit IgG (Thermo Scientific) was used as secondary antibody. The membrane was scanned by an Odyssey CLx infrared imaging system (LI-COR Biosciences).
Purification and coupling of GFP-binding protein
GFP-binding protein (GBP) was expressed and purified according to Rothbauer et al. (2008) with minor modifications. Competent Escherichia coli BL21 DE3 Rosetta cells (Novagen) expressing pET20–GFPnb (kindly provided by Michael Knop, Heidelberg, Germany) were grown in LB media supplemented with ampicillin and chloramphenicol at 30°C to reach OD600 = 0.8–1. The expression was induced by addition of 1 mM isopropyl-β-D-thiogalactopyranoside [IPTG] (Thermo Fisher Scientific) for 20 h at 23°C. Bacterial cells were harvested by centrifugation at 4000 × g for 10 min. Pellets were resuspended in 40 ml/l culture of binding buffer (1 × PBS, pH 8, 0.5 mM NaCl, 20 mM imidazole) containing 1 mM PMSF and 10 µg/ml lysozyme and incubated for 30 min at 4°C. The cell suspension was lysed using a M-110-L Microfluidizer processor (Microfluidics; 5 × 30 s pulse, 50% power, tip KE76) on ice. Soluble proteins were collected after centrifugation at 14,000 × g for 30 min and loaded onto a Protino Ni-NTA column (Machery-Nagel, Fisher Scientific), equilibrated with 10 column volumes of binding buffer without lysozyme. The His-tagged GBP proteins were eluted from the column by a gradient from 200 to 300 mM imidazole. The column was finally washed with 500 mM imidazole. Elution samples were pooled and concentrated and applied to a PD-10 desalting column (GE Healthcare Life Sciences) equilibrated in 1× PBS containing 0.5 mM EDTA. Proteins were finally loaded onto a Superdex 75 column (GE Healthcare Life Sciences) in the same buffer. Fractions containing protein were pooled and dialyzed via a PD-10 column using CB buffer (0,2M sodium bicarbonate [NaHCO3], 0.5 M NaCl, pH 8.3).
To bind protein to the beads, all centrifugation steps were at 300 × g for 1 min at 4°C. NHS–activated Sepharose 4 Fast Flow beads (GE Healthcare Life Sciences) were equilibrated with cold 1 mM HCl (15 volumes for 5 ml of beads). Purified GBP protein (5 mg) was loaded onto the beads in CB Buffer 50 ml total volume and incubated for 3 h at room temperature in a turning wheel. The supernatant was removed after centrifugation, and nonreacted groups were blocked with 50 ml of 0.1 M Tris/HCl, pH 8, for 2 h. Beads were washed three times with Tris buffer (0.1M Tris/HCl, pH 8) and acetate buffer (0.1 M sodium acetate, 0.5 M NaCl, pH 4) in alternation. Beads were then washed three times with PBS, resuspended in PBS containing 0.02% sodium azide, and stored at 4°C.
Immunoprecipitation of GFP-tagged proteins for Western blot analysis
Yeast cells expressing Vps41–GFP were grown in YPD to OD600 = 0.7–1, and 350 OD600 equivalent units of cells were collected by centrifugation at 3000 × g for 5 min. Pellets were washed with pull-down buffer (20 mM HEPES/NaOH, pH 7.4, 150 mM potassium acetate, 5% glycerol (vol/vol), 25 mM CHAPS, 1 mM phenylmethylsulfonyl fluoride [PMSF, Roth], and 1×FY protease inhibitor mix [Serva]) and lysed in the same buffer in a FastPrep homogenizer (MP Biomedicals). Lysates were centrifuged at 20000 × g for 20 min at 4°C. The cleared lysates were pooled and incubated with 40 µl of prewashed self-made anti-GFP Nanobody Sepharose affinity matrix beads for 1 h at 4°C. Beads were washed six times with pull-down buffer and boiled in SDS sample buffer for 5 min at 95°C. Proteins were resolved by SDS–PAGE and transferred to nitrocellulose membrane for Western Blotting. GFP-tagged proteins were detected by a mouse monoclonal anti-GFP antibody (Roche, product number 11814460001). Mouse monoclonal anti-HA antibody (Covance, MMS-101R-500) was used to detect copurified HA-tagged proteins. DyLight 800 goat anti mouse IgG (Invitrogen) was used as secondary antibody and analysis was done on an Odyssey CLx infrared imaging system (LI-COR Biosciences).
SILAC labelling, GFP-Trap pull down, and mass spectrometry of Vps3–GFP
Wild type untagged control and mGFP-tagged Vps3 lysine auxotroph strains were grown in synthetic medium (SD) supplemented with 30 mg/l L-lysine or 30 mg/l 13C6, 15N2 L-lysine (Cambridge Isotope Laboratories, USA), respectively, for SILAC labelling. Cultures were grown to OD600 0.7 and 350 OD600 equivalent units were harvested. Cells were lysed in a FastPrep homogenizer (MP Biomedicals) in 750 μl of GFP-Trap buffer (20 mM HEPES/NaOH, pH 7.4,150 mM KOAc, 5% glycerol,1% β-octylglucoside, 1 mM PMSF, 1×FY). Lysates were centrifuged at 20,000 × g for 20 min at 4°C. Roti-Quant (Roth) was used to determine the protein concentration of cleared lysate. Equal protein amounts were incubated with 12.5 µl of GFP-Trap beads (Chromotek) for 30 min at 4°C. Beads were washed six times with GFP-Trap buffer. Beads from the heavy and light conditions were pooled in the last wash. Beads were processed for proteomics analysis with the iST proteomics kit (Preomics) according to the manufacturer’s instructions, but using LysC [Lysyl endopeptidase] (Wako).
Reversed-phase chromatography was performed on a Thermo Ultimate 3000 RSLCnano system connected to a Q Exactive-Plus mass spectrometer (Thermo) through a nanoelectrospray ion source. Peptides were separated on 50-cm PepMapC18 easy spray columns (Thermo) with an inner diameter of 75 mm. The column temperature was kept at 40°C. Peptides were eluted from the columns with a linear gradient of acetonitrile from 10 to 35% in 0.1% formic acid for 118 min at a constant flow rate of 300 nl/min. Eluted peptides from the column were directly electrosprayed into the mass spectrometer. Mass spectra were acquired on the Q Exactive-Plus in a data-dependent mode to automatically switch between full scan MS and up to 10 data-dependent MS/MS scans. The maximum injection time for full scans was 50 ms, with a target value of 3,000,000 at a resolution of 70,000 at m/z = 200. The 10 most intense multiply charged ions (z = 2) from the survey scan were selected with an isolation width of 1.6 Th and fragment with higher energy collision dissociation (Olsen et al., 2007) with normalized collision energies of 27. Target values for MS/MS were set at 100,000 with a maximum injection time of 80 ms at a resolution of 17,500 at m/z = 200. To avoid repetitive sequencing, the dynamic exclusion of sequenced peptides was set at 30 s. Saccharomyces cerevisiae Proteome (reference proteomes—EMBL—EBI) and MaxQuant software (version 1.6.17.0, www. maxquant.org/; Cox and Mann, 2008; Cox et al., 2011) were used to analyze the resulting MS and MS/MS spectra. Only proteins detected with at least one unique peptide and two peptides and for which a SILAC ratio can be quantified are shown in the graph. The complete list of identified proteins is shown is Supplemental Table S2.
Growth tests on solid media
For the growth test, cells were grown in YPD to logarithmic phase and diluted at OD600 = 0.25. Serial dilutions (1:10) were spotted onto plates made with different media. Plates were incubated for 1–4 d and imaged.
Supplementary Material
Acknowledgments
We thank Angela Perz for expert technical assistance. Fluorescence microscopy was carried out at the iBiOs facility, Osnabrück University, Germany. This work was supported by the DFG (UN111/11-1 to C.U.) and the SFB 944 (Project P24, to A.G.M.). A.G.M. is a recipient of an Advanced EMBO long-term fellowship (aALTF 609-2018). We are grateful to Roland Wedlich-Söldner, Benedikt Westermann, and Michael Knop for providing plasmids.
Abbreviations used:
- CORVET
Class C core vacuole/endosome tethering
- ERMES
ER-mitochondria encounter structure
- GEF
guanine nucleotide exchange factor
- HOPS
homotypic fusion and protein sorting
- MCSs
membrane contact sites
- MS
mass spectrometry
- SAM
sorting and assembly machinery
- SILAC
stable isotope labeling of amino acids in cell culture
- CTAGE5
cutaneous T-cell lymphoma-associated antigen 5
- D2GFP
destabilized green fluorescent protein
- DAPI
4’,6-diamidino-2-phenylindole
- SND
SRP-independent targeting
- TOM
translocase of the outer mitochondrial membrane
- vCLAMP
vacuolar and mitochondria patch.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-05-0227) on October 20, 2021.
REFERENCES
- Aviram N, Ast T, Costa EA, Arakel EC, Chuartzman SG, Jan CH, Haßdenteufel S, Dudek J, Jung M, Schorr S, et al. (2016). The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 540, 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausewein T, Mills DJ, Langer JD, Nitschke B, Nussberger S, Kühlbrandt W (2017). Cryo-EM structure of the TOM core complex from Neurospora crassa. Cell 170, 693–700.e7. [DOI] [PubMed] [Google Scholar]
- Bean BDM, Dziurdzik SK, Kolehmainen KL, Fowler CMS, Kwong WK, Grad LI, Davey M, Schluter C, Conibear E (2018). Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J Cell Biol 217, 3593–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Bannwarth M, Meisinger C, Hill K, Model K, Krimmer T, Casadio R, Truscott KN, Schulz GE, Pfanner N, et al. (2005). Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J Mol Biol 353, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Binda M, Péli-Gulli M, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 35, 563–573. [DOI] [PubMed] [Google Scholar]
- Böckler S, Westermann B (2014). Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell 28, 450–458. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- De M, Oleskie AN, Ayyash M, Dutta S, Mancour L, Abazeed ME, Brace EJ, Skiniotis G, Fuller RS (2017). The Vps13p–Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J Cell Biol 216, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Shai N, Schuldiner M, Bohnert M (2016). A tether is a tether is a tether: tethering at membrane contact sites. Dev Cell 39, 395–409. [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M (2014). A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell 30, 95–102. [DOI] [PubMed] [Google Scholar]
- Ellenrieder L, Opaliński Ł, Becker L, Krüger V, Mirus O, Straub SP, Ebell K, Flinner N, Stiller SB, Guiard B, et al. (2016). Separating mitochondrial protein assembly and endoplasmic reticulum tethering by selective coupling of Mdm10. Nat Commun 7, 13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Montoro A, Auffarth K, Hönscher C, Bohnert M, Becker T, Warscheid B, Reggiori F, van der Laan M, Fröhlich F, Ungermann C (2018). Vps39 interacts with Tom40 to establish one of two functionally distinct vacuole–mitochondria contact sites. Dev Cell 45, 621-636.e7. [DOI] [PubMed] [Google Scholar]
- Haas A (1995). A quantitative assay to measure homotypic vacuole fusion in vitro. Methods Cell Sci 17, 283–294. [Google Scholar]
- Hariri H, Rogers S, Ugrankar R, Liu YL, Feathers JR, Henne WM (2017). Lipid droplet biogenesis is spatially coordinated at ER–vacuole contacts under nutritional stress. EMBO Rep 19, 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, Ungermann C, et al. (2014). Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell 30, 86–94. [DOI] [PubMed] [Google Scholar]
- Iadarola DM, Basu W, Trivedi PP, Fu G, Nan B (2020). Vps39 is required for ethanolamine-stimulated elevation in mitochondrial phosphatidylethanolamine. BBA - Mol Cell Biol Lipids 1865, 158655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Kakimoto Y, Tashiro S, Kojima R, Morozumi Y, Endo T, Tamura Y (2018). Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci Rep 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecker T, Scholz D, Förtsch J, Westermann B (2013). The yeast cell cortical protein Numl integrates mitochondrial dynamics into cellular architecture. J Cell Sci 126, 2924–2930. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P (2009). An ER–mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkele K, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W (1998). The preprotein translocation channel of the outer membrane of mitochondria. Cell 93, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Lackner LL, Ping H, Graef M, Murley A, Nunnari J (2012). Endoplasmic reticulum-associated mitochondria–cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci 110, E458–E467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Ping H, Graef M, Murley A, Nunnari J (2013). Endoplasmic reticulum-associated mitochondria–cortex tether functions in the distribution and inheritance of mitochondria. Proc Natl Acad Sci USA 110, E458–E467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AB, Peter ATJ, Walter P, Kornmann B (2015). ER–mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol 210, 883–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer L, Borchers AC, Herrmann E, Füllbrunn N, Han Y, Perz A, Auffarth K, Kümmel D, Ungermann C (2020). A conserved and regulated mechanism drives endosomal Rab transition. Elife 9, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen OS, Ward DMV, Kaplan J (2001). CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem 276, 29515–29519. [DOI] [PubMed] [Google Scholar]
- Lürick A, Gao J, Kuhlee A, Yavavli E, Langemeyer L, Perz A, Raunser S, Ungermann C (2017). Multivalent Rab interactions determine tether-mediated membrane fusion. Mol Biol Cell 28, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Gray SR, Bright NA (2010). Endosome–lysosome fusion. Biochem Soc Trans 38, 1413–1416. [DOI] [PubMed] [Google Scholar]
- MacDiarmid CW, Gaither LA, Eide D, Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D, et al. (2000). Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J 19, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Rissler M, Chacinska A, Sanjuán Szklarz LK, Milenkovic D, Kozjak V, Schönfisch B, Lohaus C, Meyer HE, Yaffe MP, et al. (2004). The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell 7, 61–71. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, Russell MRG, Lo SY, Chapin HC, Milnes JM, Merz AJ (2012). Termination of isoform-selective Vps21/Rab5 signaling at endolysosomal organelles by Msb3/Gyp3. Traffic 13, 1411–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M (2007). Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods 4, 709–712. [DOI] [PubMed] [Google Scholar]
- Ostrowicz CW, Bröcker C, Ahnert F, Nordmann M, Lachmann J, Peplowska K, Perz A, Auffarth K, Engelbrecht-Vandré S, Ungermann C (2010). Defined subunit arrangement and Rab interactions are required for functionality of the HOPS tethering complex. Traffic 11, 1334–1346. [DOI] [PubMed] [Google Scholar]
- Palmer CP, Zhou X, Lin J, Loukin SH, Kung C, Saimi Y (2001). A TRP homolog in Saccharomyces cerevisiae forms an intracellular Ca 2 % -permeable channel in the yeast vacuolar membrane. Proc Natl Acad Sci USA 98, 7801–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-S, Neiman AM (2012). VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J Cell Sci 125, 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-S, Thorsness MK, Policastro R, McGoldrick LL, Hollingsworth NM, Thorsness PE, Neiman AM (2016). Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol Biol Cell 27, 2435–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C (2007). The CORVET tethering complex interacts with the Yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell 12, 739–750. [DOI] [PubMed] [Google Scholar]
- Plemel RL, Lobingier BT, Brett CL, Angers CG, Nickerson DP, Paulsel A, Sprague D, Merz AJ, Barr FA (2011). Subunit organization and Rab interactions of Vps–C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell 22, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Toulmay A, Balla T (2019). The functional universe of membrane contact sites. Nat Rev Mol Cell Biol 21, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7, 282–289. [DOI] [PubMed] [Google Scholar]
- Schmidt O, Weyer Y, Fink MJ, Müller M, Weys S, Bindreither M, Teis D. (2017). Regulation of Rab5 isoforms by transcriptional and post-transcriptional mechanisms in yeast. FEBS Lett 591, 2803–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnóczky G, Kornmann B, Lackner LL, Levine TP, Pellegrini L, Reinisch K, et al. (2019). Coming together to define membrane contact sites. Nat Commun 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai N, Yifrach E, van Roermund CWT, Cohen N, Bibi C, Ijlst L, Cavellini L, Meurisse J, Schuster R, Zada L, et al. (2018). Systematic mapping of contact sites reveals tethers and a function for the peroxisome–mitochondria contact. Nat Commun 9, 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen H-HH, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M, et al. (2015). Molecular architecture of the active mitochondrial protein gate. Science 349, 1544–1548. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger B, Stenmark H, Düsterhöft A, Philippsen P, Yoo JS, Gallwitz D, Zerial M (1994). Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol 125, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotrys JE, Schoenfish MJ, Stutz MA, Linder ME (2005). The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J Cell Biol 170, 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M (2000). Structure and function of the yeast vacuole and its role in autophagy. Microsc Res Tech 51, 563–572. [DOI] [PubMed] [Google Scholar]
- Tirrell PS, Nguyen KN, Luby-phelps K, Friedman JR (2020). MICOS subcomplexes assemble independently on the mitochondrial inner membrane in proximity to ER contact sites. J Cell Biol 219, e202003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosal-Castano S, Peselj C, Kohler V, Habernig L, Berglund LL, Ebrahimi M, Vögtle FN, Höög J, Andréasson C, Büttner S (2021). Snd3 controls nucleus–vacuole junctions in response to glucose signaling. Cell Rep. 34, 108637. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Kümmel D (2019). Structure of membrane tethers and their role in fusion. Traffic 20, 479–490. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Hepowit NL, Port SA, Yau RG, Peng Y, Azad N, Habib A, Harpaz N, Schuldiner M, Hughson FM, et al. (2020). Cargo release from myosin V requires the convergence of parallel pathways that phosphorylate and ubiquitylate the cargo adaptor. Curr Biol 30, 4399–4412.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, de Boer R, Krikken AM, Aks˛it A, Yuan W, van der Klei IJ (2019). Peroxisome development in yeast is associated with the formation of Pex3-dependent peroxisome–vacuole contact sites. Biochim Biophys Acta Mol Cell Res 1866, 349–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.