Somatic PIGA gene mutations initiate pathogenesis of paroxysmal nocturnal hemoglobinuria (PNH) [1, 2] and exemplify evolutional somatic adaptability of hematopoietic stem cells (HSCs). The resultant glycosylphosphatidylinositol (GPI) anchor-deficient phenotype appears to offer a growth and/or survival advantage over other unaffected HSCs [3, 4] in the context of immune-mediated bone marrow failure [5] explaining the unique association of PNH with aplastic anemia (AA). Recently, additional mutations in genes other than PIGA were identified in PNH clones, offering an explanation for intrinsic expansion drive of PNH clones irrespective of or after the initial immune escape occurred [6]. Multiple PIGA mutations have been discovered at very low frequencies in healthy individuals [7], as well as in isolated reports of occasional PNH patients [8, 9] and thus observation did not lead to the appreciation that the phenomenon of an oligoclonal disease origin may be ubiquitous and potentially essential for the pathogenesis of PNH. PIGA mutant cells fail to expand in engineered animal models [10], suggesting that additional permissive conditions and factors are required for PNH clone evolution. Using next generation sequencing (NGS) of the PIGA gene and cell sorting by flow cytometry, our goal was to determine if we could improve PIGA mutant clone detection and gain insight into PNH clonal dynamics through longitudinal monitoring. According to theoretical stipulations ~20% patients may harbor >1 PIGA mutant hematopoietic stem cell (HSC) [11]. The recently described acquisition of multiple lesions at HLA loci in AA represents an analogous scenario of immune evasion and somatic adaptability of HSCs [12, 13].
We set out to experimentally challenge this hypothesis, and determine if, in addition to the classic two-hit scenario characterized by a founder PIGA mutation followed by a subclonal driver hit, there are more complex, potentially mosaic, clonal dynamics; whereby multiple PIGA clones compete for dominance. We also asked whether stratifying AA/PNH and classic PNH patients would provide additional insight as to the process of clonal selection.
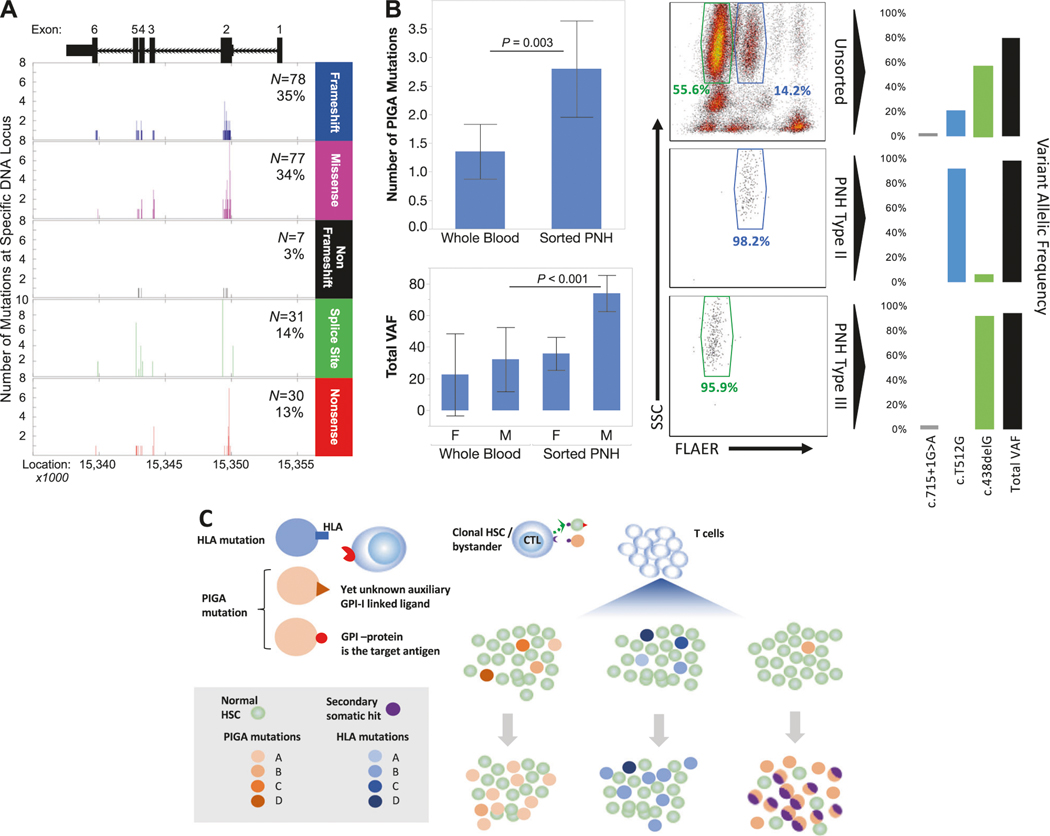
Patients with PNH and AA were diagnosed according to current clinical guidelines and following informed consent enrolled into this study, as detailed in supplemental data. Using flow cytometry to identify PNH clones, 133 patients with PNH (N = 33), AA with small PNH clones >0.5% of WBCs (N = 33), and AA/PNH (N = 67) were studied (Table S1). Mean WBC PNH clone sizes were 77%, 0.22%, and 27%, respectively. DNA was extracted and subjected to multi-amplicon deep sequencing of PIGA using primers covering all exons of the PIGA gene (Table S2). Using deep NGS analysis, 223 mutations in the PIGA gene were detected (Fig. 1a). Frameshift (35%, N = 78) and missense mutations (34%, N = 77) were most common, followed by splice site (14%, N = 31), nonsense (13%, N = 30), and nonframeshift insertion/deletion (3%, N = 7) mutations.
Fig. 1.
Next generation sequencing (NGS) detection of PIGA gene mutations. The timepoint with the most PIGA mutations was selected from each patient, grouped by type, and graphed by DNA location. Frameshift and missense mutations were the most common, with exon 2 the most frequently affected in each mutation type (a). Significantly more PIGA mutations were found by NGS in DNA from the sorted PNH fraction compared to DNA from whole blood (b, top left, P=0.003, t-test, 95% CI) and the mean total VAF of all mutations detected in sorted samples compared to whole blood is significantly higher in males (b, bottom left, P<0.001, t-test, 95% CI). Flow results from a patient with a pronounced Type II PNH clone (b, center and right) with sort purity plots from the PNH Type II (b, center middle) and PNH Type III (b, center bottom) fractions. PIGA NGS performed on DNA isolated from unsorted whole blood (b, top right), Type II PNH cells (b, middle right), and Type III PNH cells (b, bottom right) demonstrates the nearly exclusive presence of a missense mutation (c. T521G) in the Type II fraction while a frameshift mutation (c.438delG) predominates in the Type III fraction. Hypothetical summary illustration showing that pre-existing PIGA mutations and HLA mutations or LOH allow for evasion of immune attack characteristic of AA and may lead to polyclonal AA/PNH with varying relative frequencies of distinct PIGA mutant clones that may or may not acquire additional non-PIGA mutations (c)
As detection rates correlate with PNH clone size in WBCs (P < 0.0001, chi-square, Figure S1), sorting PNH positive and negative cells by flow cytometry (Figure S2; PNH N = 12, AA/PNH N = 17; mean purity 97.3%, SD 4.2%, Table S3) significantly increased the number of mutations detected by NGS (Fig. 1b, top left). The number of mutations detected per patient in the flow sorted population ranged between 0 and 12 with a mean double that of whole blood (2.8 vs. 1.4, P = 0.003, t-test). This effect was statistically significant in males (Fig. 1b, bottom left, P < 0.001), as PIGA is located on chromosome X, and while a similar increase in mutation detection was observed in females, the difference was not statistically significant. Of the sorted subcohort, we detected PIGA mutations in 25 out of 29 patients (86%), with the undetected balance likely being PIGA deletions larger than 250 bp that are not detectable by NGS [14]. Thus, previous approaches to PNH may have underestimated the prevalence of multiple PIGA mutations, which we define as PIGA clonal mosaicism.
The presence of multiple mutant PNH clones in a single patient has been reported in the literature [8, 9], and has been inferred from the presence of both Type II and III GPI-deficient clones. In a representative patient from our cohort (Fig. 1b, center and right), Type II and III PNH cells are clearly derived from 2 codominant yet distinct clones; one characterized by a missense mutation (c.T512G, the Type II PNH clone), and a second with a frameshift deletion (c.438delG, the Type III PNH clone), and a 3rd splice site mutation in a minor clone within the Type III fraction. These results reinforce the notion that PNH is not a monoclonal disease, using current methods that allow for an ex vivo “snapshot” of a patient at a precise moment in time, thus supporting previous research using older techniques dependent on extensive in vitro T cell cultures [8, 9].
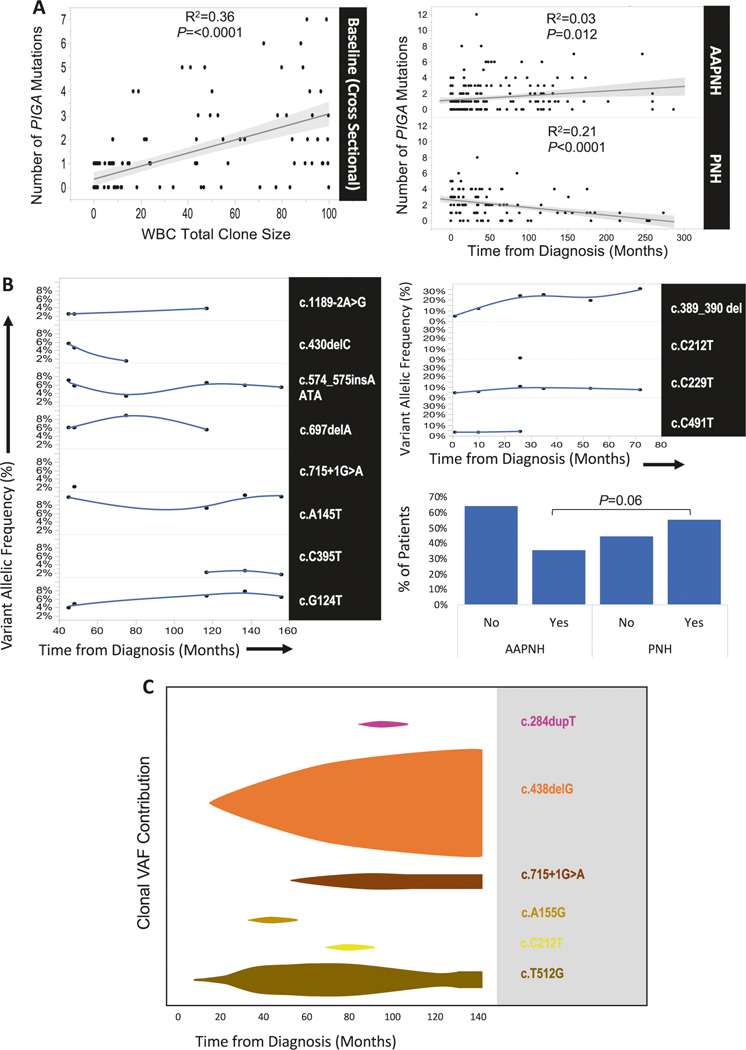
Although there are many hypotheses as to the pathogenesis of PNH, the association of PNH with AA strongly suggests that PNH emerges within immune-mediated bone marrow failure [5, 15]. Multiple PIGA mutations suggests that in AA/PNH, where immune selection pressure is putatively most pronounced, a higher number of PIGA mutant clones should be detectable. Cross sectional analysis at the initial sampling of the entire cohort demonstrates that there is an association by linear regression between WBC clone size by flow cytometry and the number of mutations detected by PIGA NGS in unsorted whole blood (Fig. 2a, left, linear regression R2 = 0.36, P < 0.0001). Regression analysis incorporating longitudinal sequencing results (AA/PNH, N = 29, observance range 3–127 months, median 41 months; PNH, N = 18, range 2–131 months, median 62 months, Table S1) suggests that, as time progresses, the number of mutations detected in AA/PNH increases or plateaus (Fig. 2a, right top, P = 0.012), although correlation is not strong (R2 = 0.03). In contrast, the trend is reversed in classic PNH, with the number of mutations detected decreasing over time (Fig. 2a, bottom, P < 0.0001), and a stronger inverse correlation between the time since diagnosis and the number of mutations (R2 = 0.21). Although this analysis warrants validation in a larger prospective study often precluded due to the rarity of PNH, our data suggest a general trend toward the eventual dominance of a single PIGA mutant clone in classic PNH. It is possible that such a mechanism also operates for AA with multiple HLA class-I mutant clones [12, 13].
Fig. 2.
Longitudinal analysis and clonal dynamics of PIGA mutations. Cross sectional analysis at the earliest available sample by linear regression demonstrates an association between the number of PIGA mutations detected in whole blood and the WBC clone size by flow cytometry (a, left). Despite this association, linear regression of the number of mutations against the time from diagnosis differs in AA/PNH patients (a, top right) when compared to patients with a classic PNH diagnosis (a, bottom right). Representative AA/PNH patient followed longitudinally with complex clonal dynamics more common in AA/PNH (b, left) in contrast to the emergence of a dominant clone in classic PNH (b, right top), a trend more prevalent in PNH than AA/PNH (b, bottom right, 55% (10/18) vs. 38% (10/28), respectively, P=0.06). Schematic fish plot derived from data illustrating the emergence of a dominant clone from six distinct PIGA mutations in classic PNH (c)
Analysis of representative examples with long-term longitudinal monitoring illustrates a contrast between AA/PNH (Fig. 2b, left) and classic PNH (Fig. 2b, top right) (additional patients depicted in Figure S3). Mutations can fluctuate in the contribution to the overall PNH clone size over time with some mutations increasing while others decrease in AA/PNH (Fig. 2b, left). When we examined the entire longitudinal cohort, in PNH patients a dominant clone (defined as >2×the remaining VAF) was present at the baseline measurement or emerged over time (10 out of 18, 55%), while in AA/PNH only 10 out of 28 (36%) exhibited this trend (Fig. 2b, bottom right, P = 0.06, t-test), suggesting that AA/PNH patients tend to display more complex clonal dynamics whereby multiple clones contract or expand. While there are generally exceptions to a rule, it may be that active immune pressure in AA/PNH may allow for observation of complex clonal dynamics that are not present in classic PNH, and it remains to be understood if cases of AA/PNH and classic PNH exist in a continuum.
Overall, our data suggest that PNH begins in an oligoclonal state in which multiple PIGA mutant clones compete to emerge. Thus PIGA mutation and the resultant GPI-deficiency exemplifies another way of immune escape in AA similar to that proposed for somatic HLA mutations or LOH (Fig. 1c), while in classic PNH the evolution may follow a different path. Surviving PIGA mutant clones may gain intrinsic additional growth advantages due to secondary mutations in other genes [6], knowledge of which may determine the best treatment modality. In other cases, ongoing immune attack may be the dominant pathology requiring treatment. Whether HLA mutant clones also experience intrinsic amplification by additional somatic events remains unknown. It would be interesting to investigate whether similar immune selection pressure is exerted during immune checkpoint inhibitor therapy.
PIGA mutation detection by NGS is more sensitive if sequencing analysis of PIGA mutant clones is performed on sorted rather than unsorted cell populations. This approach was used here to detect multiple mutations with the same phenotype, as well as those with coexistent Type II and Type III phenotypes, a result consistent with studies in healthy individuals and PNH patients demonstrating the pre-existence of multiple PIGA mutations [7–9]. In contrast to otherwise reasonable speculation whereby new clones evolve to replace exhausted, older hematopoietic clones, we illustrate that it is more likely that there is a preexisting PIGA mutated pool initially, and that over time, a single dominant clone emerges in classic PNH.
Supplementary Material
Acknowledgements
This work was supported by grants from the NIH (RO1HL-082983, U54 RR019391, and K24 HL-077522, to J.P. Maciejewski); the Aplastic Anemia & MDS International Foundation; the Robert Duggan Charitable Fund; Scott Hamilton CARES.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41375-018-0138-5) contains supplementary material, which is available to authorized users.
References
- 1.Takeda J, Miyata T, Kawagoe K, Iida Y, Endo Y, Fujita T, et al. Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell. 1993;73:703–11. [DOI] [PubMed] [Google Scholar]
- 2.Bessler M, Mason PJ, Hillmen P, Miyata T, Yamada N, Takeda J, et al. Paroxysmal nocturnal haemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene. EMBO J. 1994;13:110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagakura S, Ishihara S, Dunn DE, Nishimura J, Kawaguchi T, Horikawa K, et al. Decreased susceptibility of leukemic cells with PIG-A mutation to natural killer cells in vitro. Blood. 2002;100:1031–7. [DOI] [PubMed] [Google Scholar]
- 4.Bessler M, Mason P, Hillmen P, Luzzatto L. Somatic mutations and cellular selection in paroxysmal nocturnal haemoglobinuria. Lancet. 1994;343:951–3. [DOI] [PubMed] [Google Scholar]
- 5.Young NS, Maciejewski JP, Sloand E, Chen G, Zeng W, Risitano A, et al. The relationship of aplastic anemia and PNH. Int J Hematol. 2002;76:168–72. [DOI] [PubMed] [Google Scholar]
- 6.Shen W, Clemente MJ, Hosono N, Yoshida K, Przychodzen B, Yoshizato T, et al. Deep sequencing reveals stepwise mutation acquisition in paroxysmal nocturnal hemoglobinuria. J Clin Invest. 2014;124:4529–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araten DJ, Nafa K, Pakdeesuwan K, Luzzatto L. Clonal populations of hematopoietic cells with paroxysmal nocturnal hemoglobinuria genotype and phenotype are present in normal individuals. Proc Natl Acad Sci USA. 1999;96:5209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Endo M, Ware RE, Vreeke TM, Singh SP, Howard TA, Tomita A, et al. Molecular basis of the heterogeneity of expression of glycosyl phosphatidylinositol anchored proteins in paroxysmal nocturnal hemoglobinuria. Blood. 1996;87:2546–57. [PubMed] [Google Scholar]
- 9.Nishimura J, Inoue N, Wada H, Ueda E, Pramoonjago P, Hirota T, et al. A patient with paroxysmal nocturnal hemoglobinuria bearing four independent PIG-A mutant clones. Blood. 1997;89:3470–6. [PubMed] [Google Scholar]
- 10.Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, et al. Glycosylphosphatidylinositol-anchor-deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood. 1996;87:3600–6. [PubMed] [Google Scholar]
- 11.Hill A, DeZern AE, Kinoshita T, Brodsky RA. Paroxysmal nocturnal haemoglobinuria. Nat Rev Dis Prim. 2017;3:17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaimoku Y, Takamatsu H, Hosomichi K, Ozawa T, Nakagawa N, Imi T, et al. Identification of an HLA class I allele closely involved in the autoantigen presentation in acquired aplastic anemia. Blood. 2017;129:2908–16. [DOI] [PubMed] [Google Scholar]
- 13.Babushok DV, Duke JL, Xie HM, Stanley N, Atienza J, Perdigones N, et al. Somatic HLA mutations expose the role of class I-mediated autoimmunity in aplastic anemia and its clonal complications. Blood Adv. 2017;1:1900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Keefe CL, Sugimori C, Afable M, Clemente M, Shain K, Araten DJ, et al. Deletions of Xp22.2 including PIG-A locus lead to paroxysmal nocturnal hemoglobinuria. Leukemia. 2011;25:379–82. [DOI] [PubMed] [Google Scholar]
- 15.Maciejewski JP, Rivera C, Kook H, Dunn D, Young NS. Relationship between bone marrow failure syndromes and the presence of glycophosphatidyl inositol-anchored protein-deficient clones. Br J Haematol. 2001;115:1015–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.