Abstract
Gadd45γ, a family member of the growth arrest and DNA damage-inducible gene family 45 (Gadd45), is strongly induced by interleukin-2 (IL-2) in peripheral T cells. While in most tissues all Gadd45 family members are expressed, Gadd45γ is the only member that is induced by IL-2. Here we show that the IL-2-induced expression of Gadd45γ is dependent on a signaling pathway mediated by the tyrosine kinase Jak3 and the transcription factors Stat5a and Stat5b (signal transducer and activator of transcription). Previous studies with ectopically overexpressed Gadd45γ in various cell lines implicated its function in negative growth control. To analyze the physiological role of Gadd45γ we used homologous recombination to generate mice lacking Gadd45γ. Gadd45γ-deficient mice develop normally, are indistinguishable from their littermates, and are fertile. Furthermore, hematopoiesis in mice lacking Gadd45γ is not impaired and Gadd45γ-deficient T lymphocytes show normal responses to IL-2. These data demonstrate that Gadd45γ is not essential for normal mouse development and hematopoiesis, possibly due to functional redundancy among the Gadd45 family members. Gadd45γ is also dispensable for IL-2-induced T-cell proliferation.
Elucidating the molecular mechanisms of interleukin-2 (IL-2)-driven T-cell proliferation is critical to understanding the regulation of the immune system. The signaling pathways emerging from the IL-2 receptor require Jak3 and Jak1, two members of the JANUS family of tyrosine kinases (Jak), which bind to and phosphorylate the IL-2 receptor as well as substrates recruited to the receptor complex. Among the substrates are Stat5a and Stat5b (signal transducer and activator of transcription), both of which are phosphorylated and translocated to the nucleus where they activate gene transcription (reviewed in references 8 and 9). Mice lacking either Jak3 or both Stat5a and Stat5b (Stat5a/b) have revealed their essential function for IL-2-induced T-cell proliferation (13, 15). In an attempt to delineate the IL-2 signaling pathway downstream of Stat5a/b in T cells, we screened a subtracted cDNA library for Stat5 target genes. Applying representational difference analysis (RDA) and subsequent Northern blot analysis of activated wild-type versus Stat5a/b double knockout splenocytes, we were able to identify IL-2-inducible, Stat5-dependent genes. Among the clones obtained was the murine gene of Gadd45γ, which shows 97% identity at the amino acid level to its human counterpart (20). This gene had been previously identified by several approaches, including as an activator of MEKK4/MTK1 (17), an IL-2-induced immediate-early gene (cytokine response gene 6 [CR6]) (3), an oncostatin M-inducible gene (OIG37) (14), a gene induced by depletion of neurotrophic factor in PC12 neuronal cells (11), and a gene coding for a Gadd-related protein of 17 kDa (GRP17) (16).
In addition to Gadd45γ, the Gadd45 family (growth arrest and DNA damage-inducible gene family) consists of Gadd45α and Gadd45β, whose murine isoform is also designated MyD118 (1, 5). Gadd45γ shows high sequence similarity to Gadd45α and Gadd45β at the amino acid level (68 and 70%, respectively). All Gadd45 family members are small, acidic proteins that are expressed to different degrees in all tissues (14). Overexpression of any of the family members in various cell lines leads to association with proliferating cell nuclear antigen and p21, a cyclin-dependent kinase inhibitor, and results in growth suppression (4, 14). These findings led to the hypothesis that Gadd45 members play a critical role in negative growth control.
Individual Gadd45 family members are characterized by specific inducible patterns of expression (reviewed in reference 12). Gadd45α is induced in a p53-dependent manner by γ irradiation (10). Gadd45β is upregulated by transforming growth factor β, which induces growth arrest and apoptosis in the myeloid cell line M1 (1). Moreover, both Gadd45α and Gadd45β are induced by IL-6, a differentiation-inducing cytokine, and by DNA-damaging agents such as methyl methanesulfonate (MMS) (19). In contrast to the inducible expression pattern of Gadd45α and Gadd45β associated with growth arrest, differentiation, and apoptosis, Gadd45γ has been shown to be induced in the proliferative response of T cells to IL-2 (3, 20). This finding led to the hypothesis that IL-2 also mediates negative growth control elements such as Gadd45γ to ensure proper cell cycling and genomic stability (20). On the other hand, it could be hypothesized that Gadd45γ supports growth promotion when expressed at physiological levels. In order to elucidate the physiological function of Gadd45γ, we generated Gadd45γ-deficient mice by using targeted gene disruption. Here, we show that Gadd45γ is dispensable for normal mouse development, hematopoiesis, and in particular, IL-2-driven T-cell proliferation.
MATERIALS AND METHODS
Construction of the Gadd45γ targeting vector and generation of mutant mice.
A Gadd45γ genomic clone was isolated from a 129SV/J mouse genomic library (Genome Systems Inc.). An 8-kb genomic EcoRI fragment containing all four exons coding for Gadd45γ was used to construct a targeting vector. A 2-kb BstEI fragment encompassing most of the coding region was replaced with a neomycin-resistance gene driven by the thymidine kinase promoter. This strategy led only the first 13 amino acids of intact Gadd45γ to be translated, which, however, will not give rise to a stable product. A cDNA coding for the diphtheria toxin A driven by the thymidine kinase promoter (2) was then cloned 3′ of the genomic sequence to facilitate negative selection.
E14 embryonic stem (ES) cells (129/Ola mouse strain) (kindly provided by Jan van Deursen) and RW4 ES cells (129/SvJ mouse strain) (Genome Systems Inc.) were electroporated and selected for heterozygous targeted clones as described previously (18). To verify correct targeting by Southern blotting, a 5′ external XbaI-EcoRI fragment was used to probe XbaI-digested genomic DNA resulting in a 6-kb wild-type and a 9-kb mutant fragment. Correct recombination at the 3′ region was confirmed by PCR. Out of five targeted ES cell clones which were injected into C57BL/6 blastocysts, two clones were successfully integrated into the germ line. All experiments were performed with both mouse lines independently and gave the same results.
Genotypes of mice were routinely determined with tail DNA, by using PCR. A mutant allele-specific primer (5′-CCACACTGCTCGACATTGGGTGG), a wild type-specific primer (5′-GGACCCTGCCTTGGAGAAGCTC), and a common primer (5′-GGTCGGTTCTTCCCGGCCCTGG) were mixed in each reaction mixture, and PCR products of 150 and 350 bp in length appeared from the targeted and the wild-type alleles, respectively.
Northern blot analysis.
Whole-cell RNA was prepared using the RNAzol B reagent (Tel-Test) according to the directions of the manufacturer. For Northern blotting, 20 μg of RNA per lane was run on a 1% formaldehyde agarose gel and blotted onto nitrocellulose membranes. The cDNAs of Gadd45γ, Gadd45α, and Gadd45β, all cloned from mouse tissues, as well as the cDNAs from glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and gamma interferon (IFNγ) were used as probes, which were labeled by using a random priming kit (readyprime; Amersham).
JNK kinase assay.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and cell debris was removed by centrifugation. Supernatants were incubated with JNK1 antisera (sc-474; Santa Cruz) for 2 h at 4°C. The immune complexes were precipitated with protein A agarose (Roche) and washed twice with RIPA buffer and twice with JNK kinase buffer (10 mM MgCl2, 50 mM Tris-HCl [pH 7.4], 5 mM benzamidine, 0.5 mM dithiothreitol, 1 mM sodium vanadate). The kinase reaction was performed in kinase buffer supplemented with 5 μCi of [γ-32P]ATP, 0.1 mM ATP, and glutathione S-transferase (GST)–c-Jun(1–135) as a substrate for 20 min at 30°C. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto supported nitrocellulose membranes. After autoradiography, the membranes were probed with JNK1 antisera (sc-474; Santa Cruz) and subsequently with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G. To visualize the proteins, a standard enhanced chemiluminescence reaction was employed (ECL; Amersham).
Bone marrow colony assays.
Bone marrow cells were prepared from tibia and femur in α-MEM medium (Life Technologies) containing 2% fetal calf serum (StemCell Technologies) mixed with cytokines and MethoCult M3230 (StemCell Technologies) giving a final concentration of 0.9% methylcellulose. For the assays the following cytokines were used: recombinant human erythropoietin (Epo) (Amgen), recombinant murine IL-3, IL-6, stem cell factor (SCF), and granulocyte-macrophage colony-stimulating factor (GM-CSF) (R&D Systems), as well as recombinant human thrombopoietin (TPO) (Genzyme). Cultures were plated in 35-mm culture dishes in duplicates and cultured at 37°C. The assay conditions were described previously (18).
Growth curve and proliferation response assays.
A single-cell suspension, prepared from spleen and lymph nodes, was stimulated with 2 μg of anti-CD3 (2C11; PharMingen)/ml and 500 U of recombinant human IL-2 (Chiron)/ml in RPMI 1640 media (Life Technologies) supplemented with 10% fetal calf serum, l-glutamine, penicillin, streptomycin, sodium pyruvate, essential and nonessential amino acids (all from Life Technologies), and β-mercaptoethanol (Sigma). To determine the growth curve of T cells, cells were kept in IL-2-containing media and were split when they reached a cell density above 106/ml. Viable cell counts were determined by trypan blue staining every 48 h. Cell cycle arrest of the growing T-cell culture was achieved by IL-2 withdrawal for 20 h and confirmed by propidium iodide DNA staining.
To assess the proliferative response to IL-2, 2 × 105 growth-arrested T cells were plated into 1 well of a 96-well round-bottomed plate in 200 μl of T-cell medium (see above) with indicated amounts of IL-2. After 8 h of stimulation, 1 μCi of [3H]thymidine (Amersham Life Sciences) was added to the culture for another 16 h. Cells were harvested and the incorporated [3H]thymidine was measured as counts per minute in a beta counter.
Staining of cells with Annexin V.
Approximately 5 × 105 cells were washed with phosphate-buffered saline, resuspended in 100 μl of binding buffer (10 mM HEPES, 0.9% NaCl, 2.5 mM CaCl2, 0.1% bovine serum albumin) containing 2 μl of Annexin V-Fluos (Annexin V bound to fluorescein isothiocyanate; Roche) and 1 μg of propidium iodide/ml, and incubated at room temperature for 15 min in the dark. The samples were analyzed on a Becton Dickinson FACScan flow cytometer (Becton Dickinson, San Jose, Calif.), collecting green fluorescence from bound Annexin V-fluorescein isothiocyanate molecules and red fluorescence from propidium iodide-DNA complexes in dead cells.
Irradiation and determination of the mitotic index.
Splenocytes were stimulated with anti-CD3 (2 μg/ml) and grown in IL-2-containing medium (200 U/ml) for 4 days. At this time the doubling time of the lymphocytes is approximately 16 h. Cells were then subjected to UV or γ irradiation. To determine the mitotic index, cells were incubated in 75 mM KCl for 15 min and fixed with methanol-acetic acid fixative. Cells were dropped on microscope slides and stained with Giemsa stain. Cells with condensed chromosomes were scored as mitotic cells.
RESULTS
IL-2-induced Gadd45γ transcription is Stat5 and Jak3 dependent.
In screening for IL-2-induced genes by RDA (6), we identified one clone which hybridized with two transcripts of 1.4 and 4 kb in IL-2-activated T cells (Fig. 1). Sequence analysis and a similarity search in GenBank identified this clone as the murine homolog of Gadd45γ, also designated CR6 (17, 20). The closest related genes to Gadd45γ are Gadd45α and Gadd45β, which, as illustrated in Fig. 1, are not regulated in T cells upon activation. Gadd45γ is also induced in primary splenic T cells activated by anti-CD3 and IL-2, but not in Stat5a/b-deficient T cells (Fig. 2, lanes 1 to 10), which have normal T-cell receptor signaling but fail to proliferate in response to IL-2 (13). Gadd45γ expression could be reconstituted in Stat5a/b-deficient T cells transduced by a retrovirus carrying the wild-type Stat5a gene (Fig. 2, lanes 11 to 13), demonstrating that the impaired Gadd45γ induction is due to the lack of Stat5. Since Stat5a/b activation by IL-2 is dependent on the tyrosine kinase Jak3, we compared Gadd45γ induction in wild-type versus Jak3-deficient splenocytes. As shown in Fig. 2 (lanes 14 to 23), Gadd45γ transcription is not elevated in Jak3-deficient splenocytes treated with anti-CD3 and IL-2. These data demonstrate that IL-2-induced Gadd45γ expression in peripheral T cells is dependent on signaling through Jak3 and Stat5a/b and that T cells uniquely express Gadd45γ among the Gadd45 family of genes.
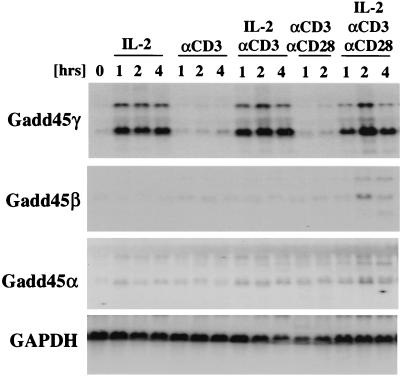
FIG. 1.
Gadd45γ is strongly induced in T cells via IL-2 but not TCR signaling. Splenocytes were incubated with anti-CD3 thereby activating T cells, which were then grown in IL-2 for 7 days. Cell cycle arrest in G1 phase of the pure T-cell culture was achieved by IL-2 withdrawal. Growth-arrested cells were then stimulated with IL-2, anti-CD3, and anti-CD28 as indicated in the figure. Northern blot analysis of starved (time point, 0) and stimulated T cells was performed using Gadd45γ-, Gadd45β-, and Gadd45α-specific probes. It is worth noticing that the blot was exposed for 6 days when probed with Gadd45β as well as Gadd45α but for only 4 h when probed with Gadd45γ. As a loading control the blot was probed with GAPDH.
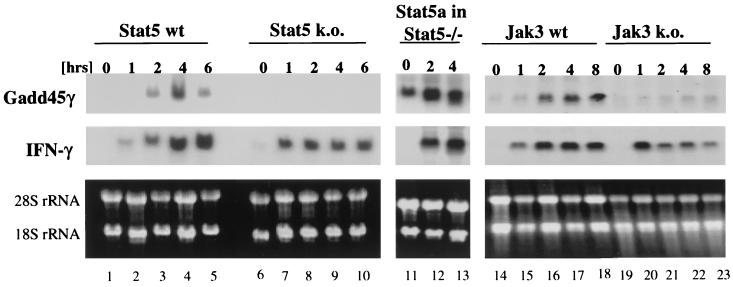
FIG. 2.
Gadd45γ is induced in activated T cells via a Jak3-Stat5-mediated pathway. Splenocytes from Stat5a/b double knockout mice (Stat5 k.o., lanes 6 to 10) and their wild-type (wt) littermates (lanes 1 to 5) as well as from Jak3 knockout (lanes 19 to 23) and wild-type (lanes 14 to 18) mice were stimulated with anti-CD3 and IL-2 for the indicated times. In addition, Stat5a/b-deficient splenocytes were rescued with retroviral gene insertion of Stat5a while activated with anti-CD3 and then grown in IL-2 for 10 days. Reconstituted cells, which consisted of 90% retrovirus-harboring T cells, were then starved for 20 h and restimulated with IL-2 for the indicated times (lanes 11 to 13). Total RNA from each of these time points was loaded on an agarose gel and stained with ethidium bromide to show equal loading (lower panels) and thereafter blotted onto nitrocellulose membranes. Membranes were blotted with the cDNAs from Gadd45γ (upper panels) as well as from IFN-γ (middle panels), the latter of which is induced in primary T cells independent of Stat5 and Jak3 but is dependent on the T-cell receptor.
Generation of Gadd45γ-deficient mice.
To assess the physiological function of Gadd45γ we generated Gadd45γ-deficient mice. In the targeting construct most of the coding sequence was replaced by a neomycin resistance gene cassette (Fig. 3A) in order to generate a null mutation. Out of five homologous recombinant ES cell clones, two clones gave rise to germ line-transmitted mutant mouse strains as confirmed by Southern blot and PCR (Fig. 3B and C). Heterozygous mice of both lines were crossed to examine the phenotype caused by Gadd45γ deficiency. Intercrosses in both mouse lines gave birth to homozygous mutant Gadd45γ mice that were indistinguishable from their wild-type littermates and were fertile and viable for at least 1 year.
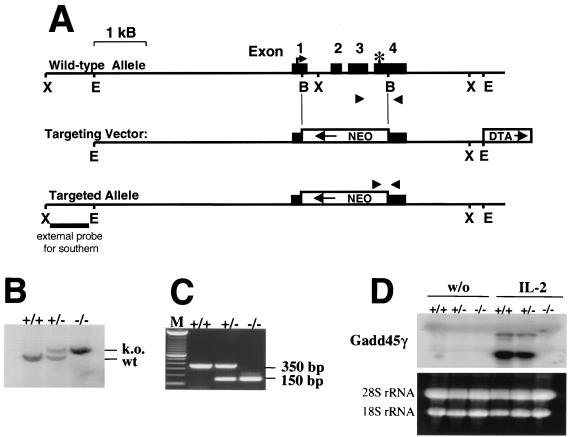
FIG. 3.
Targeted disruption of Gadd45γ in mice. (A) The murine Gadd45γ gene consists of 4 exons with the start codon in exon 1 (indicated by an arrow) and the stop codon in exon 4 (indicated by an asterisk). A targeting vector was constructed in which most of the coding region was replaced by a neomycin gene cassette. (B and C) Successful targeting was confirmed by Southern blot (B) using an external probe (indicated in panel A) and by PCR (C) with primers symbolized in panel A as triangles. (D) Northern blot analysis of IL-2-activated T cells with a Gadd45γ-specific probe from the coding region shows no hybridizing signal in knockout cells. As described in the Fig. 2 legend, growing T-cell cultures were starved and restimulated with or without IL-2 for 2 h and the RNA was harvested, subjected to electrophoresis, and blotted.
Northern blot analysis of IL-2-induced Gadd45γ-deficient T cells showed that neither transcript of Gadd45γ was detectable (Fig. 3D). Histological analysis of organs in which Gadd45γ is highly expressed, such as liver and kidney, revealed no abnormalities in Gadd45γ null mice (data not shown). The cellularity of hematopoietic tissues (bone marrow, thymus, spleen, and lymph nodes) and peripheral blood as well as the appearance of progenitor and mature T and B cells, NK cells, macrophages, granulocytes, and erythrocytes were comparable to wild-type littermates (data not shown). These data show that Gadd45γ is not essential for normal mouse development and hematopoiesis.
Cytokine response of Gadd45γ-deficient bone marrow cells.
Gadd45γ expression is induced by several cytokines, including IL-3, IL-6, and GM-CSF (20). Based on distinct expression patterns of the three Gadd45 family members, specific functions for each family member have been proposed. We, therefore, analyzed the contribution of Gadd45γ to the cytokine response of bone marrow cells. As shown in Table 1, Gadd45γ is dispensable for the ability of bone marrow cells to form colonies in response to IL-3, IL-6, SCF, Epo, TPO, and GM-CSF.
TABLE 1.
In vitro colony formation of bone marrow hematopoietic progenitor cells from Gadd45γ-deficient mice and wild-type littermates in response to various cytokinesa
| Colony type | Cytokine | No. of colonies formed from Gadd45γ genotype:b
|
|
|---|---|---|---|
| +/+ | −/− | ||
| CFC-Mix | IL-3, IL-6, SCF, Epo | 44 ± 5 | 43 ± 7 |
| CFU-Mix | IL-3 | 36 ± 11 | 40 ± 7 |
| BFU-E | IL-3, Epo | 19 ± 4 | 18 ± 5 |
| CFU-E | Epo | 329 ± 53 | 361 ± 56 |
| CFU-GM | GM-CSF | 50 ± 14 | 50 ± 12 |
| CFU-Meg | TPO | 30 ± 9 | 21 ± 9 |
Bone marrow cells from six animals of each genotype were prepared for colony assays.
The results of colony assays are shown as the mean with standard deviation of the colony number that arose from 105 bone marrow cells.
Ability of Gadd45γ-deficient T lymphocytes to proliferate.
Since Gadd45γ is strongly induced by IL-2 in peripheral T cells, we investigated the proliferative abilities of Gadd45γ-deficient T cells. For these studies, splenocytes were activated with anti-CD3 and IL-2 or IL-4 or without the addition of cytokine. Thereby, anti-CD3 stimulation activates all T cells, the addition of IL-4 favors the T helper 2 subset of CD4-positive T cells, and the presence of IL-2 favors CD8-positive T-cell subsets. The cell numbers of the growing cultures were monitored every 48 h. No differences were evident between wild-type and Gadd45γ-deficient T cells (data not shown). Irrespective of their genetic background, activated T cells proliferated in the presence of IL-2 for 14 to 18 days and exponential growth occurred between day 4 and day 9 (Fig. 4A). During this period the doubling times of the T-cell cultures from +/+, +/−, and −/− Gadd45γ mice from three independent experiments were 17.6 ± 1.4 h, 18.5 ± 2.7 h, and 18.0 ± 2.3 h, respectively. A slightly increased cell density of the Gadd45γ-deficient T cells was observed in the late phase of some experiments; however, wild-type as well as Gadd45γ-deficient cells stopped cycling after a comparable time in culture (Fig. 4A). Moreover, after 4 days the numbers of CD4- and CD8-positive T cells were similar in cultures of wild-type and Gadd45γ-deficient T cells (data not shown). To investigate the contribution of Gadd45γ to the ability of cultured T cells to arrest in the G1 phase after withdrawal of IL-2 and reenter the cell cycle after IL-2 restimulation, cultured T cells grown in the presence of IL-2 for 7 days were starved and restimulated with IL-2. Cell cycle analysis of growth-arrested and then IL-2-restimulated cells over a period of 6 to 25 h after stimulation was performed by propidium iodide staining of DNA. There was no significant difference between wild-type and knockout T cells with respect to growth arrest and IL-2-regulated cell cycle progression (data not shown). In addition, proliferation in response to increasing IL-2 concentrations was measured by thymidine incorporation. Fig. 4B illustrates that the proliferative response of cell cycle-arrested T cells from wild-type and Gadd45γ-deficient mice to increasing amounts of IL-2 was not statistically different. In order to exclude compensatory upregulation of Gadd45α and/or Gadd45β in Gadd45γ-deficient T cells, the expression of all three family members was analyzed by Northern blotting (Fig. 4C) and demonstrated the absence of upregulation of the other family members. Finally, as measured by Annexin V staining, the number of apoptotic cells did not differ between wild-type and Gadd45γ-deficient T cells (Fig. 4D).
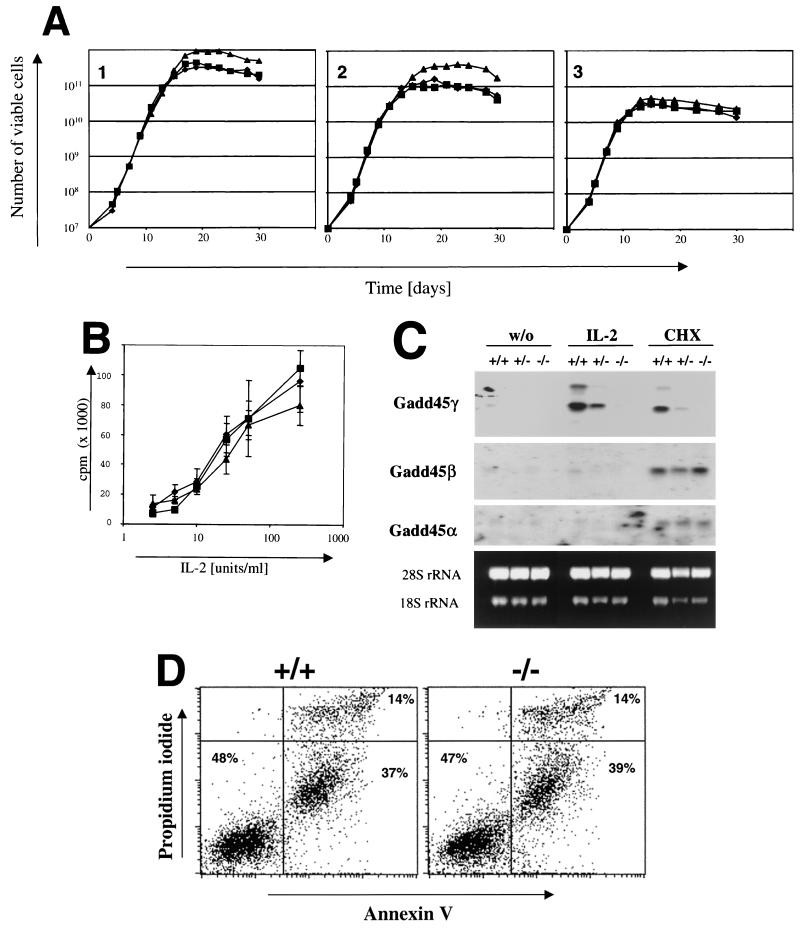
FIG. 4.
T cells of Gadd45γ-deficient mice proliferate normally in response to IL-2. (A) Growth curves of +/+ (diamond), +/− (square), and −/− (triangle) T cells: splenocytes from the three genotypes were seeded at day 0 at a cell density of 5 × 105/ml in a volume of 4 ml and stimulated with anti-CD3 and IL-2. The cultures were kept in IL-2-containing media and were split when they reached a cell density above 106/ml. Viable cell counts were determined by trypan blue staining every 48 h. Three independently performed experiments (A1, A2, and A3) are shown in the figure. (B) Cultured T cells of +/+ (diamond), +/− (square), and −/− (triangle) mice which were grown in the presence of IL-2 for 7 days were starved for 20 h and then stimulated with increasing amounts of IL-2. The proliferation rate was determined via a [3H]thymidine incorporation assay and is given as counts per minute (cpm). The results are the averages of three experiments. (C) Northern blot analysis with starved (w/o) and IL-2-stimulated T cells of Gadd45γ mice revealed no compensatory upregulation of the family members Gadd45β and Gadd45α in Gadd45γ-deficient T cells. As a control, the same T-cell culture was incubated with 100 μg of cycloheximide (CHX)/ml for 2 h, which stabilizes the messages of Gadd45 family members. (D) Apoptotic T lymphocytes were determined by staining with Annexin V. Activated splenocytes were cultured in IL-2-containing media for 5 days, harvested and stained with fluorescein isothiocyanate-labeled Annexin V protein, which binds to the membranes of apoptotic cells. To distinguish between dead cells and cells undergoing apoptosis, cells were incubated in propidium iodide solution, which stains only dead cells. The cells were then analyzed on a Becton Dickinson FACScan in two-color mode using CellQuest software. From each genotype, six animals were analyzed; a representative set including the percentage of cells in each quadrant is shown in the figure.
JNK activity in Gadd45γ-deficient T lymphocytes.
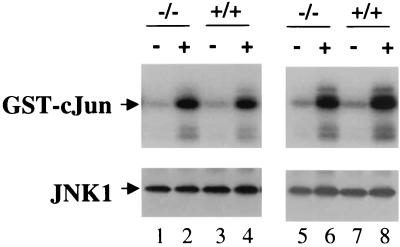
Gadd45 family members have been implicated in controlling the activity of the Jun N-terminal kinase, JNK, via binding to the ubiquitously expressed protein kinase MTK1/MEKK4 (17). The binding of Gadd45γ to MTK1/MEKK4 is thought to activate the kinase activity of MTK1, which in turn activates MEKK3 and MEKK6, upstream activators of JNK. We investigated the JNK activity in wild-type T cells cultured for 4 days in the presence of IL-2, at which time they expressed high levels of Gadd45γ. Comparison with Gadd45γ-deficient T cells cultured under the same conditions revealed no difference between the basal levels of JNK activities (Fig. 5). In addition, anisomycin-stimulated JNK activity is not altered in Gadd45γ-deficient T cells (Fig. 5), demonstrating that Gadd45γ is dispensable for JNK activity.
FIG. 5.
JNK activity of Gadd45γ-deficient T cells. Splenocytes of two Gadd45γ+/+ (lanes 3, 4, 7, and 8) and two Gadd45γ−/− (lanes 1, 2, 5, and 6) animals were stimulated with anti-CD3 and cultured for 4 days in IL-2-containing media. Half of the cells were stimulated with 100 μg of anisomycin/ml for 30 min (lanes 2, 4, 6, and 8), and the other half was left untreated (lanes 1, 3, 5, and 7). Cells were harvested and JNK1 was immunoprecipitated from cleared cell lysates. JNK1 immunocomplexes were then subjected to in vitro kinase assays using the N-terminal part of cJun as the substrate. Kinase reactions were stopped after 30 min and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting. The upper panels show the autoradiogram and lower panels the corresponding JNK1 Western blot demonstrating equal amounts of JNK1.
Checkpoint activation and genomic stability in Gadd45γ-deficient T lymphocytes.
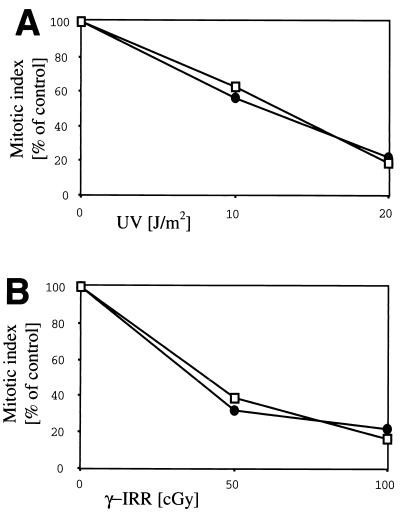
Since the Gadd45 family members have been implicated in genomic stability, we initially examined T cells that had been expanded for 14 days in culture for karyotypic abnormalities. Examination of 60 cells failed to identify any abnormal karyotypes (data not shown). Gadd45α-deficient lymphocytes have been shown to have defects in the cell cycle checkpoint activation after UV irradiation (7). Since Gadd45γ is uniquely induced by IL-2 in proliferating T cells, we wished to examine the potential contribution of this induction to G2M arrest of UV or γ-irradiated cells. For these studies, T lymphocytes from wild-type and Gadd45γ-deficient mice were expanded in vitro in the presence of IL-2 to allow the accumulation of Gadd45γ in the wild-type cells. As illustrated (Fig. 6), there were no differences between wild-type and Gadd45γ-deficient T cells in the induction of G2M arrest in response to increasing doses of either UV, as a DNA damaging agent, or γ irradiation, as a DNA double-strand break-inducing agent. Therefore, IL-2-induced expression of Gadd45γ in T cells is not essential for genomic stability or cell cycle arrest under the conditions examined.
FIG. 6.
Checkpoint activation of Gadd45γ-deficient T cells. Mitotic indices of proliferating T lymphocytes of Gadd45γ-deficient (open squares) and wild-type (closed circles) mice were determined after 1 h of UV radiation (A) or γ irradiation (γ-IRR) (B).
DISCUSSION
In this report we demonstrate that IL-2 stimulation of T cells leads to the induction of Gadd45γ in a Jak3- and Stat5-dependent manner. However, in contrast to Stat5, loss of Gadd45γ does not affect T-cell activation and proliferation. There are three possible explanations for the lack of any consequences. First, although Gadd45α and Gadd45β were not induced in IL-2-stimulated T cells, we cannot exclude compensation by another, unidentified member of the Gadd45 family with function overlapping that of Gadd45γ. However, this is unlikely since exhaustive searchs of murine or human expressed sequences, as well as sequencing the clones identified in our RDA screen, have failed to identify another family member. The second possibility is that critical functions mediated by Gadd45γ are redundantly provided by another, unrelated gene product. The third possibility is that the Stat5a/b-regulated expression of Gadd45γ and the functional consequences are not critical for T-cell proliferation under the conditions examined.
A number of genes have been identified that are induced with the activation of peripheral T cells, and many of these are not induced in Stat5a/b-deficient lymphocytes. For example, cyclin D2 and D3 are not expressed in Stat5-deficient T cells (13). In order to test the possibility that Gadd45γ is required for their expression, we examined the expression of D-type cyclin mRNAs and proteins in activated Gadd45γ-deficient T cells. Both cyclin D2 and D3 were upregulated normally in Gadd45γ null splenocytes when stimulated with anti-CD3 and IL-2 (data not shown).
The dependence on Stat5a/b for Gadd45γ expression in T cells and the reported requirement for Stat3 for the induction of Gadd45γ in myeloid cells would suggest that Stat binding sites exist in the promoter and mediate the induction (14). However, we have not found any putative Stat binding sites in the promoter sequence spanning 1 kb upstream of the putative TATA box nor in intronic sequences. It is possible that Stat transcription factors act on enhancer elements further upstream of the gene or indirectly. The lack of any consequence of deleting Gadd45γ might suggest that, irrespective of the mechanism, the induction of Gadd45γ may occur fortuitously because the gene shares genomic elements that are required for Stat5a/b regulation of a gene that is critical for T-cell function.
Since the identification of Gadd45γ, considerable effort has been invested in elucidating its functions including overexpression studies of Gadd45γ in diverse cell lines. One function of Gadd45γ, deduced from such studies, is the activation of MEKK4 in response to MMS-induced cell death of ML-1 cells and the subsequent activation of JNK and p38 MAPKs (17). This prompted the authors to speculate the pathway Gadd45γ → MEKK4 → p38/JNK, leading to apoptosis (17). However, other groups have not detected significant induction of Gadd45γ after MMS treatment in M1 cells (20) or in NIH-3T3 cells (14). In addition, we found no evidence that MMS induces Gadd45γ expression in T cells. Irrespective of that, neither the basal nor anisomycin-stimulated JNK activity is altered in Gadd45γ-deficient T cells (Fig. 5), demonstrating that Gadd45γ is dispensable for JNK activity.
The consequences of the overexpression of Gadd45γ have been variably reported to either induce apoptosis in HeLa cells (17) or not induce apoptosis but rather to reduce the growth rate of HeLa cells or NIH-3T3 and BaF3 cells (4, 14). In this regard, the percentage of apoptotic cells in cultures of proliferating Gadd45γ-null T cells is neither reduced nor enhanced in comparison to wild-type T cells (Fig. 4D). Nor is there any indication of a difference in the proliferative capacity, either increased or decreased, of Gadd45γ-deficient T cells. Therefore it is likely that the overexpression studies have not provided completely accurate information regarding the functions of Gadd45γ in cell growth and apoptosis regulation.
Taken together, our results show that the Stat5a/b-regulated gene, Gadd45γ, is dispensable for T-cell function and, in general, demonstrate that the identification of a target gene does not necessarily mean that the target gene mediates an essential function. More generally, Gadd45γ is not required for hematopoiesis or for normal development. The existence of two additional family members raises the possibility that they function redundantly to Gadd45γ in some critical cell lineages. As with many gene families, it will be important to generate strains of mice that lack two or all of the family members to explore the possibility of redundancy.
ACKNOWLEDGMENTS
We thank John Raucci and Christie Nagy for injection of ES cells into blastocytes as well as Neena Carpino and Richard Ashmun for FACS analysis and Linda Snyder and Kristen Rothammer for technical support. We also thank John Cleveland, Gery Zambetti, Veronika Sexl, Nick Carpino, Christopher Duntsch, Demin Wang, and Jean-Chris Marine for helpful discussion.
This work was supported by the Cancer Center CORE grant CA21765, by grants RO1 DK42932 and PO1 HL53749, and by the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Abdollahi A, Lord K A, Hoffman-Liebermann B, Liebermann D A. Sequence and expression of a cDNA encoding MyD118: a novel myeloid differentiation primary response gene induced by multiple cytokines. Oncogene. 1991;6:165–167. [PubMed] [Google Scholar]
- 2.Adachi M, Suematsu S, Kondo T, Ogasawara J, Tanaka T, Yoshida N, Nagata S. Targeted mutation in the Fas gene causes hyperplasia in peripheral lymphoid organs and liver. Nat Genet. 1995;11:294–300. doi: 10.1038/ng1195-294. [DOI] [PubMed] [Google Scholar]
- 3.Beadling C, Johnson K W, Smith K A. Isolation of interleukin 2-induced immediate-early genes. Proc Natl Acad Sci USA. 1993;90:2719–2723. doi: 10.1073/pnas.90.7.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan W, Richter G, Cereseto A, Beadling C, Smith K A. Cytokine response gene 6 induces p21 and regulates both cell growth and arrest. Oncogene. 1999;18:6573–6582. doi: 10.1038/sj.onc.1203054. [DOI] [PubMed] [Google Scholar]
- 5.Fornace A J, Jr, Alamo I, Jr, Hollander M C. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frazer J K, Pascual V, Capra J D. RDA of lymphocyte subsets. J Immunol Methods. 1997;207:1–12. doi: 10.1016/s0022-1759(97)00091-4. [DOI] [PubMed] [Google Scholar]
- 7.Hollander M C, Sheikh M S, Bulavin D V, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro T A, Kim K E, Tolosa E, Ashwell J D, Rosenberg M P, Zhan Q, Fernandez-Salguero P M, Morgan W F, Deng C X, Fornace A J., Jr Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 8.Ihle J N. Cytokine receptor signalling. Nature. 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 9.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 10.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 11.Kojima S, Mayumi-Matsuda K, Suzuki H, Sakata T. Molecular cloning of rat GADD45gamma, gene induction and its role during neuronal cell death. FEBS Lett. 1999;446:313–317. doi: 10.1016/s0014-5793(99)00234-3. [DOI] [PubMed] [Google Scholar]
- 12.Liebermann D A, Hoffman B, Steinman R A. Molecular controls of growth arrest and apoptosis: p53-dependent and independent pathways. Oncogene. 1995;11:199–210. [PubMed] [Google Scholar]
- 13.Moriggl R, Topham D J, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster M Y, Bunting K D, Grosveld G C, Ihle J N. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama K, Hara T, Hibi M, Hirano T, Miyajima A. A novel oncostatin M-inducible gene OIG37 forms a gene family with MyD118 and GADD45 and negatively regulates cell growth. J Biol Chem. 1999;274:24766–24772. doi: 10.1074/jbc.274.35.24766. [DOI] [PubMed] [Google Scholar]
- 15.Nosaka T, van Deursen J M, Tripp R A, Thierfelder W E, Witthuhn B A, McMickle A P, Doherty P C, Grosveld G C, Ihle J N. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki M, Watanabe T K, Fujiwara T, Nakamura Y, Takahashi E, Tanigami A. Molecular cloning, expression, and mapping of a novel human cDNA, GRP17, highly homologous to human gadd45 and murine MyD118. J Hum Genet. 1999;44:300–303. doi: 10.1007/s100380050164. [DOI] [PubMed] [Google Scholar]
- 17.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 18.Teglund S, McKay C, Schuetz E, van Deursen J M, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle J N. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhan Q, Lord K A, Alamo I, Jr, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Fornace A J., Jr The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Bae I, Krishnaraju K, Azam N, Fan W, Smith K, Hoffman B, Liebermann D A. CR6: a third member in the MyD118 and Gadd45 gene family which functions in negative growth control. Oncogene. 1999;18:4899–4907. doi: 10.1038/sj.onc.1202885. [DOI] [PubMed] [Google Scholar]