ABSTRACT
Carbapenemase-producing Escherichia coli sequence type (ST) 648 strains were isolated from two international visitors without previous medical exposure from Southeast Asian countries in a hospital in Japan. One isolate, FUJ80154, carried blaNDM-5 in a complex class 1 integron on an IncFIB/FII plasmid; the other isolate, FUJ80155, carried two copies of blaOXA-48 on the chromosome flanked by IS1R on both sides. The core-genome based-phylogenetic analysis with publicly available genome data of E. coli ST648 carrying blaNDM-5 or blaOXA-48-like demonstrated high genetic similarity between FUJ80154 and NDM-5-prooducing E. coli ST648 strains isolated in South and Southeast Asian countries. On the other hand, no closely related isolates of FUJ80155 were identified. In the absence of prior hospitalization overseas, neither patient had qualified for routine screening of multidrug-resistant organisms, and the isolates were incidentally identified in cultures ordered at the discretion of the treating physician.
IMPORTANCE Although patients with history of international hospitalization are often subject to screening for multidrug-resistant organisms, it is unclear whether patients who reside in countries where carbapenemase-producing Enterobacterales (CPE) is endemic but have no history of local hospitalization contribute to the transmission of CPE. In this study, NDM-5-producing and OXA-48-producing Escherichia coli sequence type (ST) 648, a recently recognized high-risk, multidrug-resistant clone, were detected from two overseas visitors without previous medical exposure. The findings of this study suggest that active surveillance culture on admission to hospital may be considered for travelers from countries with endemicity of carbapenem-resistant organisms even without history of local hospitalization and underscore the need to monitor cross-border transmission of high-risk clones, such as carbapenemase-producing E. coli ST648.
KEYWORDS: Escherichia coli, NDM-5, OXA-48, ST648, carbapenemase
OBSERVATION
The spread of carbapenemase-producing Enterobacterales (CPE) is recognized as a global public health threat since the 2000s (1). CPE may be introduced into a country with low endemicity by patients with a history of hospitalization in a country where they are endemic. It has been reported that the frequency of isolation of CPE in a country may increase sharply when high-risk CPE clones are introduced from abroad and spread across health care facilities in a short period of time (2, 3). Japan is characterized by low rates of CPE, and most of these isolates produce imipenemase (IMP)-type carbapenemases (4, 5). On the other hand, CPE producing Klebsiella pneumoniae carbapenemase (KPC), NDM, or OXA-48-like enzymes are extremely rare in Japan even in recent years, and most are identified in patients with a history of overseas hospitalization (6, 7). Therefore, many major hospitals in Japan screen patients with history of overseas hospitalization for multidrug-resistant organisms (MDROs), including CPE (7).
In this study, we investigated the epidemiological characteristics of patients from whom CPE producing non-IMP enzymes (non-IMP CPEs) was isolated in 2019 in a 1,435-bed Japanese university hospital. This study was approved by the Ethics Committee of Fujita Health University School of Medicine (approval number: HM19-170). Bacterial identification and antimicrobial susceptibility testing in the hospital were performed using Vitek MS and Vitek 2 (bioMérieux, Marcy l'Etoile, France), respectively. Modified carbapenem inhibition method (mCIM) was performed on carbapenem-non-susceptible Enterobacterales according to the CLSI M100 guidelines, for the confirmation of carbapenemase production (8). Types of carbapenemases produced by CPE isolates were analyzed with NG-Test CARBA 5 (NG Biotech, Guipry, France).
Only two non-IMP CPE isolates were detected during the study period (Table S1 in the supplemental material). One isolate (FUJ80154) was identified as NDM-producing E. coli and the other isolate (FUJ80155) was identified as OXA-48-like-producing E. coli. Both patients were long-term residents of Southeast Asian countries but had no underlying medical conditions and no history of local hospitalization. The patients were not among the prespecified population for active surveillance culture at the hospital, but cultures were submitted at the discretion of the treating physicians. While FUJ80154 was resistant to all β-lactams, FUJ80155 was susceptible to carbapenems reflecting the relatively lower hydrolytic activity of OXA-48-like enzymes against carbapenems compared with that of NDM enzymes.
Whole genome sequencing of the non-IMP CPE isolates was performed with Illumina MiSeq (Illumina, Inc., San Diego, CA) and MinION (Oxford Nanopore Technologies, Oxford, UK). Hybrid de novo assembly using both MiSeq and MinION reads was conducted with Unicycler (version 0.4.9b). Genetic characterization of the genomes was performed using software available at the website of Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/) and ClermonTyping (9). Both isolates were E. coli of phylogroup F and sequence type (ST) 648, and carried multiple virulence genes, including those associated with uropathogenicity (chuA, fyuA, yfcV) and iron uptake (chuA, fyuA, irp2, iutA, sitA) (Table S1 in the supplemental material) (10, 11).
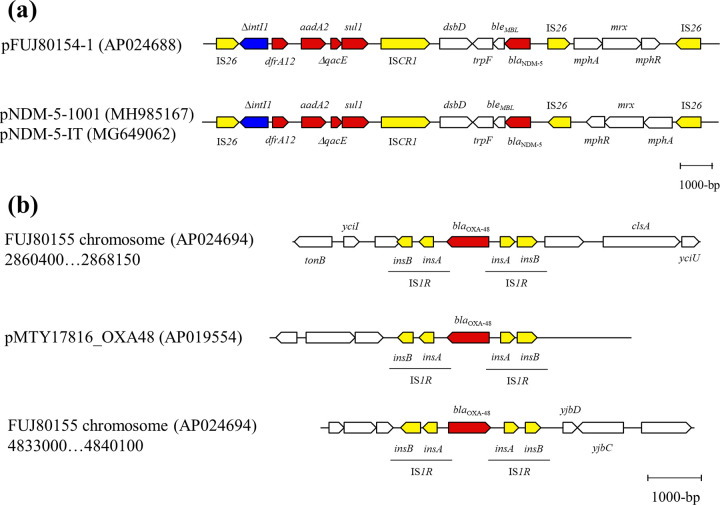
FUJ80154 possessed blaNDM-5 on an IncFIB/FII plasmid (pFUJ80154-1) and FUJ80155 carried blaOXA-48 on the chromosome. Additionally, both isolates possessed a number of resistance genes including blaCTX-M-15. blaNDM-5 was located on a complex class 1 integron and adjacent to the IS26-mediated composite transposon carrying mphA-mrx-mphR in FUJ80154 (Fig. 1a). A similar structure has been found on an IncFIA/FIB plasmid (pNDM-5-1001) of an E. coli ST410 clinical isolate from China (12) and on an IncFII plasmid (pNDM-5-IT) of an E. coli ST167 clinical isolate from Italy (13), although there is a difference in the direction of the mphA-mrx-mphR module. Two copies of blaOXA-48, both located between two copies of insA-insB (IS1R), were present on the chromosome of FUJ80155 (Fig. 1b). The 2,666-bp composite transposon with IS1R at both sides of blaOXA-48 was also found in a Col156-type plasmid (pMTY17816_OXA48) harbored by a Klebsiella pneumoniae isolate (TUM17186) detected at a Japanese hospital from a patient with history of recent hospitalization in Vietnam (6). In both isolates (FUJ80155 and TUM17186), no direct repeat sequences were found adjacent to the region, which suggested that these isolates acquired the 2,666-bp genetic region by homologous recombination.
FIG 1.
Genetic environment of carbapenemase genes. (a) Genetic environment of blaNDM-5 on pFUJ80154-1 plasmid and the similar genetic structure identified in previous studies and (b) genetic environment of two copies of blaOXA-48 on the chromosome of FUJ80155 and the identical genetic structure identified in a previous study. Block arrows indicate confirmed or putative open reading frames (ORFs) and their orientations. Arrow size is proportional to the predicted ORF length. The color codes are as follows: yellow, transposase genes; blue, integrase genes; red, antibiotic resistance genes; white, other.
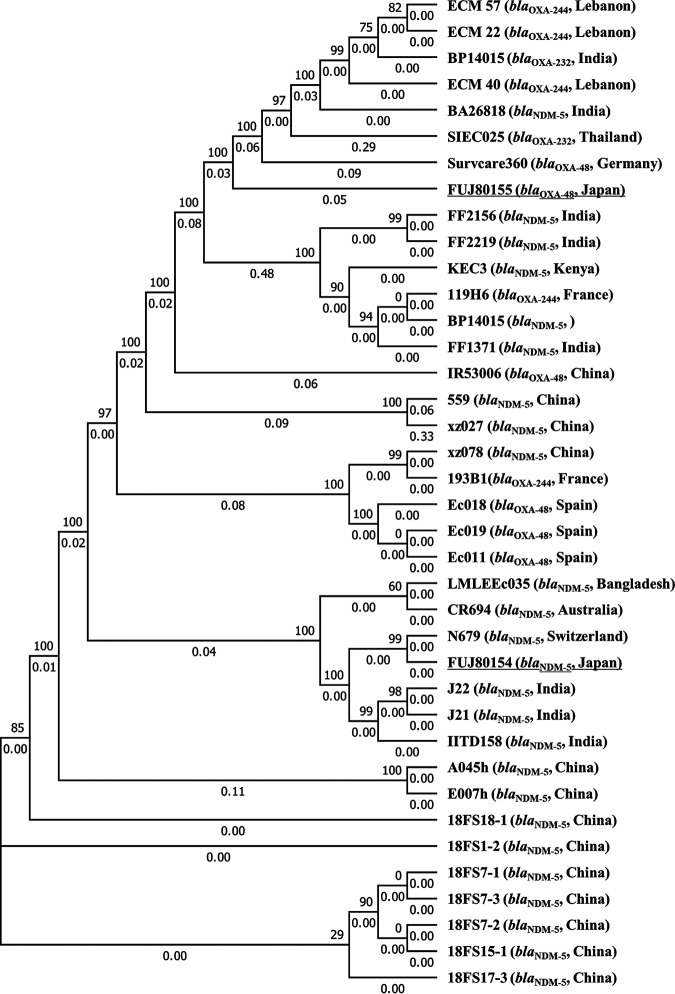
While ST648 has been recognized as a major global clone of extended-spectrum β-lactamase (ESBL)-producing E. coli (14, 15), ST648 isolates carrying carbapenemase genes have also been reported. The first NDM-5-producing strain ever reported was ST648 E. coli, which was detected from a patient transferred directly from India to the United Kingdom (16). In another study, one of 10 NDM-1-producing E. coli isolates from the United Kingdom and two of seven NDM-1-producing E. coli isolates from Pakistan were ST648 (17). All of these isolates also carried blaCTX-M. OXA-48-producing E. coli ST648 isolates have been documented in several countries, all of which also produced CTX-M-15 (18, 19). One of two patients with OXA-48-producing E. coli ST648 in Poland had history of hospitalization in Cambodia, and the other had history of numerous foreign travels (18). These reports suggest that CTX-M-15-producing ST648, which has selective advantage on its own, has acquired carbapenemase genes that are spreading in each region, and is also beginning to spread across borders. As of 11 October 2021, whole genome data for 24 ST648 isolates carrying blaNDM-5 and 12 ST648 isolates carrying blaOXA-48-like were available in GenBank (Table S2). The core-genome based-phylogenetic analysis of these genomes with those of the isolates analyzed in this study was performed using FUJ80155 as the reference genome as described previously (4). The phylogenetic tree was generated with FastTree 2.1.9 (https://bioweb.pasteur.fr/packages/pack@FastTree@2.1.9). Six isolates (LMLEEc035, CR694, N679, J22, J21, and IITD158) were closely related with FUJ80154 with 22–80 core genome SNPs (Fig. 2). Four of these isolates were from India and Bangladesh, and one of the remaining isolates from Australia was detected in a patient who had just returned from India (20). On the other hand, FUJ80155 had no closely related isolates with the SNPs to the closest isolate (ECM 22) being 2,147. This result supports the presumption that FUJ80155 independently acquired blaOXA-48 by homologous recombination.
FIG 2.
Phylogenetic tree of ST648 Escherichia coli isolates carrying blaNDM-5 or blaOXA-48-like. The carbapenemase genes carried by the isolates and the countries in which the isolates were detected are described in the parentheses after the isolate names. The isolates analyzed in this study are underlined.
In this study, CPE producing non-IMP carbapenemases, which are rare in Japan, was detected in two patients with a history of living abroad but no history of local hospitalization prior to the travel. Meanwhile, no CPE isolates were detected from routine screening of patients with history of inpatient care in foreign countries within 1 month of admission during the same period (data not shown). Although short-term travel has been reported to be a risk for acquisition of ESBL-producing Enterobacterales, acquisition of CPE was extremely rare in an analysis of the same travelers (21, 22). However, a recent French study demonstrated that international travel without history of hospitalization is a risk factor for detection of extensively drug-resistant organisms, such as CPE through inpatient screening (23). In recent years, community transmission of carbapenem-resistant Gram-negative organisms has been reported in several South and Southeast Asian countries, and it is assumed that the risk of acquiring CPE among travelers and residents without medical exposure is higher than before in these areas (24, 25). The reasons for the rarity of non-IPM carbapenemases in Japan are not clear, but the relative geographical isolation and low volume of cross-border traffic, along with early detection and isolation of patients carrying CPE of foreign origin may have contributed. As community transmission of CPE increases in some countries, international visitors without previous medical exposure who are currently not subjected to active screening may serve as a source of CPE that is uncommon locally.
In conclusion, E. coli ST648 isolates carrying carbapenemase genes were detected in two patients with a history of living abroad but with no history of local hospitalization. Although the cost of active screening and preemptive isolation must be balanced against the damage caused by the spread of MDROs, screening for MDROs on admission to hospital may be considered for international travelers and residents without history of hospitalization.
Accession numbers.
Nucleotide sequences of the chromosome and plasmids of non-IMP CPE isolates have been deposited in the NCBI database under accession number AP024687-AP024693 (FUJ80154) and AP024694-AP024701 (FUJ80155).
ACKNOWLEDGMENT
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material is available online only.
Contributor Information
Sohei Harada, Email: idharada@gmail.com.
Jeanette W.P. TEO, National University Hospital
REFERENCES
- 1.Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF. 2018. Carbapenemase-producing organisms: A global scourge. Clin Infect Dis 66:1290–1297. doi: 10.1093/cid/cix893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, Pantosti A, Pagani L, Luzzaro F, Rossolini GM, AMCLI-CRE Survey Participants. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill 18:20489. [PubMed] [Google Scholar]
- 3.Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y, Israeli KPC Kpn Study Group. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother 53:818–820. doi: 10.1128/AAC.00987-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki K, Harada S, Yahara K, Ishii Y, Motooka D, Nakamura S, Akeda Y, Iida T, Tomono K, Iwata S, Moriya K, Tateda K. 2018. Molecular characterization of IMP-1-producing Enterobacter cloacae complex isolates in Tokyo. Antimicrob Agents Chemother 62:e02091-17. doi: 10.1128/AAC.02091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto N, Asada R, Kawahara R, Hagiya H, Akeda Y, Shanmugakani RK, Yoshida H, Yukawa S, Yamamoto K, Takayama Y, Ohnishi H, Taniguchi T, Matsuoka T, Matsunami K, Nishi I, Kase T, Hamada S, Tomono K. 2017. Prevalence of, and risk factors for, carriage of carbapenem-resistant Enterobacteriaceae among hospitalized patients in Japan. J Hosp Infect 97:212–217. doi: 10.1016/j.jhin.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Honda NH, Aoki K, Kamisasanuki T, Matsuda N, To M, Matsushima H, Ishii Y, Haruki K. 2019. Isolation of three distinct carbapenemase-producing Gram-negative bacteria from a Vietnamese medical tourist. J Infect Chemother 25:811–815. doi: 10.1016/j.jiac.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Hayakawa K, Mezaki K, Sugiki Y, Nagamatsu M, Miyoshi-Akiyama T, Kirikae T, Kutsuna S, Takeshita N, Yamamoto K, Katanami Y, Ohmagari N. 2016. High rate of multidrug-resistant organism colonization among patients hospitalized overseas highlights the need for preemptive infection control. Am J Infect Control 44:e257–e259. doi: 10.1016/j.ajic.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing. CLSI document M100-S28. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. 2018. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom 4:e000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malberg Tetzschner AM, Johnson JR, Johnston BD, Lund O, Scheutz F. 2020. In silico genotyping of escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58:e01269-20. doi: 10.1128/JCM.01269-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaufler K, Semmler T, Wieler LH, Trott DJ, Pitout J, Peirano G, Bonnedahl J, Dolejska M, Literak I, Fuchs S, Ahmed N, Grobbel M, Torres C, McNally A, Pickard D, Ewers C, Croucher NJ, Corander J, Guenther S. 2019. Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob Agents Chemother 63:e00243-19. doi: 10.1128/AAC.00243-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou H, Jia X, Liu H, Li S, Wu X, Huang S. 2020. Emergence of NDM-5-Producing Escherichia coli in a Teaching Hospital in Chongqing, China: IncF-Type Plasmids May Contribute to the Prevalence of blaNDM-5. Front Microbiol 11:334. doi: 10.3389/fmicb.2020.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giufrè M, Errico G, Accogli M, Monaco M, Villa L, Distasi MA, Del Gaudio T, Pantosti A, Carattoli A, Cerquetti M. 2018. Emergence of NDM-5-producing Escherichia coli sequence type 167 clone in Italy. Int J Antimicrob Agents 52:76–81. doi: 10.1016/j.ijantimicag.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother 69:1224–1230. doi: 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 15.Peirano G, van der Bij AK, Gregson DB, Pitout JD. 2012. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 50:294–299. doi: 10.1128/JCM.06025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 18.Izdebski R, Baraniak A, Zabicka D, Machulska M, Urbanowicz P, Fiett J, Literacka E, Bojarska K, Kozinska A, Zieniuk B, Hryniewicz W, Gniadkowski M, OXA-48-PL Study Group. 2018. Enterobacteriaceae producing OXA-48-like carbapenemases in Poland, 2013-January 2017. J Antimicrob Chemother 73:620–625. doi: 10.1093/jac/dkx457. [DOI] [PubMed] [Google Scholar]
- 19.Solgi H, Giske CG, Badmasti F, Aghamohammad S, Havaei SA, Sabeti S, Mostafavizadeh K, Shahcheraghi F. 2017. Emergence of carbapenem resistant Escherichia coli isolates producing blaNDM and blaOXA-48-like carried on IncA/C and IncL/M plasmids at two Iranian university hospitals. Infect Genet Evol 55:318–323. doi: 10.1016/j.meegid.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Wailan AM, Paterson DL, Caffery M, Sowden D, Sidjabat HE. 2015. Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia. Genome Announc 3:e00194-15. doi: 10.1128/genomeA.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arcilla MS, van Hattem JM, Haverkate MR, Bootsma MCJ, van Genderen PJJ, Goorhuis A, Grobusch MP, Lashof AMO, Molhoek N, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J. 2017. Import and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis 17:78–85. doi: 10.1016/S1473-3099(16)30319-X. [DOI] [PubMed] [Google Scholar]
- 22.van Hattem JM, Arcilla MS, Bootsma MC, van Genderen PJ, Goorhuis A, Grobusch MP, Molhoek N, Oude Lashof AM, Schultsz C, Stobberingh EE, Verbrugh HA, de Jong MD, Melles DC, Penders J. 2016. Prolonged carriage and potential onward transmission of carbapenemase-producing Enterobacteriaceae in Dutch travelers. Future Microbiol 11:857–864. doi: 10.2217/fmb.16.18. [DOI] [PubMed] [Google Scholar]
- 23.Macaux L, Ndoye O, Cordel H, Pomares TB, Seytre D, Bouchaud O, Cohen Y, Zahar JR, Carbonnelle E. 2018. Extensively-drug-resistant bacteria carriers among overseas travellers: One-third had not been hospitalized previously. Int J Antimicrob Agents 52:385–389. doi: 10.1016/j.ijantimicag.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Mave V, Chandanwale A, Kagal A, Khadse S, Kadam D, Bharadwaj R, Dohe V, Robinson ML, Kinikar A, Joshi S, Raichur P, McIntire K, Kanade S, Sachs J, Valvi C, Balasubramanian U, Kulkarni V, Milstone AM, Marbaniang I, Zenilman J, Gupta A. 2017. High burden of antimicrobial resistance and mortality among adults and children with community-onset bacterial infections in India. J Infect Dis 215:1312–1320. doi: 10.1093/infdis/jix114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugawara Y, Hagiya H, Akeda Y, Takeuchi D, Sakamoto N, Matsumoto Y, Motooka D, Nishi I, Tomono K, Hamada S. 2021. Community spread and acquisition of clinically relevant Escherichia coli harbouring blaNDM among healthy Japanese residents of Yangon, Myanmar. J Antimicrob Chemother 76:1448–1454. doi: 10.1093/jac/dkab070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01827-21_Supp_1_seq3.xlsx, XLSX file, 0.01 MB (12KB, xlsx)
Supplemental material. Download SPECTRUM01827-21_Supp_2_seq4.xlsx, XLSX file, 0.01 MB (11.8KB, xlsx)