Abstract
CTF4 and CTF18 are required for high-fidelity chromosome segregation. Both exhibit genetic and physical ties to replication fork constituents. We find that absence of either CTF4 or CTF18 causes sister chromatid cohesion failure and leads to a preanaphase accumulation of cells that depends on the spindle assembly checkpoint. The physical and genetic interactions between CTF4, CTF18, and core components of replication fork complexes observed in this study and others suggest that both gene products act in association with the replication fork to facilitate sister chromatid cohesion. We find that Ctf18p, an RFC1-like protein, directly interacts with Rfc2p, Rfc3p, Rfc4p, and Rfc5p. However, Ctf18p is not a component of biochemically purified proliferating cell nuclear antigen loading RF-C, suggesting the presence of a discrete complex containing Ctf18p, Rfc2p, Rfc3p, Rfc4p, and Rfc5p. Recent identification and characterization of the budding yeast polymerase κ, encoded by TRF4, strongly supports a hypothesis that the DNA replication machinery is required for proper sister chromatid cohesion. Analogous to the polymerase switching role of the bacterial and human RF-C complexes, we propose that budding yeast RF-CCTF18 may be involved in a polymerase switch event that facilities sister chromatid cohesion. The requirement for CTF4 and CTF18 in robust cohesion identifies novel roles for replication accessory proteins in this process.
The establishment of sister chromatid cohesion during S phase is a critical step in the series of events leading to high-fidelity cell division. By holding sisters together, cohesion proteins enable kinetochores to face opposite poles of the mitotic spindle, facilitating capture by microtubules from opposite poles (99). The sister chromatid association is sufficient to resist the separating force of the mitotic spindle until each kinetochore has been captured, at which time sister chromatid associations are released at the initiation of anaphase (reviewed in references 50, 72, 77, and 88). Because cohesion tightly binds sisters together from their synthesis to their separation, it must be properly established and maintained in a flexible environment supporting chromatin alterations that permit transcription, replication, repair, and condensation of the genome.
Cohesion between sister chromatids is carried out by at least four classes of proteins. The core particle, cohesin, is composed of at least four subunits encoded in budding yeast by the SMC1, SMC3, MCD1 (SCC1), and SCC3 (IRR1) genes (33, 68). Fully assembled cohesin binds chromatin in vitro and in vivo (9, 68, 97, 101). Orthologs of cohesins have been identified in Xenopus laevis, Drosophila melanogaster, Schizosaccharomyces pombe, Arabidopsis thaliana, Mus musculus, and Homo sapiens (6, 18, 58, 59, 82, 94, 100, 109, 110). Interestingly, although Mcd1p is required for both cohesion and chromosome condensation in budding yeast, these processes are carried out by distinct protein complexes in the Xenopus experimental system (33, 40, 41, 58). In addition, Pds5p, which is also required for the maintenance of sister chromatid cohesion, genetically and physically interacts with the cohesin complex (36, 78, 94). Thus, interactions between the cohesin complex and Pds5p are required to mediate sister chromatid cohesion. A highly conserved mechanism governs sister chromatid separation at anaphase initiation, mediated by the action of a CDC20-associated form of the anaphase-promoting complex (APCCDC20), which provides a ubiquitin-conjugating activity directing the degradation of the anaphase inhibitor protein Pds1p (reviewed in reference 70). Upon release from a Pds1p-Esp1p complex, active Esp1p promotes the proteolysis of Mcd1p (94, 104, 107). This event is associated with loss of cohesion between sister chromatids and with poleward movement of the chromosomes (reviewed in references 73 and 116).
Scc2p and Scc4p, members of the second class of proteins, direct the binding of cohesin proteins to chromatin (16, 101). SCC2 and SCC4 associate with each other in coimmunoprecipitation experiments but are not core components of the cohesin particle (16). Scc2p may mediate cohesin complex interaction with chromatin via associations with Mcd1p and Scc3p. While the localizations of both Scc2 and Scc4 proteins on chromatin spreads are similar to one another, the two proteins seem to occupy different chromosomal loci from Mcd1p (16, 101). Furthermore, both Scc2p and Scc4p associate with chromatin in a nuclease- and salt-resistant manner, suggesting that they are tightly bound in a higher-order chromatin structure. Scc2p and Scc4p are required for establishment of cohesion early in the cell cycle but are not required for maintenance of cohesion in metaphase arrested cells (16). This function appears to be conserved, since a fission yeast homologue of Scc2p (Mis4p) is also required in S phase (26). SCC2 homologues have also been identified as the Coprinus RAD9 (83) and Drosophila Nipped-B (81) genes.
A third class of molecules, defined by CTF7(ECO1) function, is required to render the cohesin complex competent to mediate siser chromatid cohesion during S phase. Budding yeast are inviable in the absence of CTF7, and conditional alleles lead to precocious sister separation (87, 101). Although Ctf7p associates with chromatin, it does not stably associate with the core cohesin particle, nor is it requred for cohesin association with chromatin (87, 101). Execution point studies indicate a requirement for CTF7 in S phase (87, 101). Interestingly, the chromosome instability and temperature-sensitive lethality of ctf7 alleles are suppressed by high-copy-number expression of POL30, encoding budding yeast proliferating-cell nuclear antigen (PCNA) (87). One hypothesis is that Ctf7p is required at the replication fork to activate interactions between cohesins on sister chromatids for a functional “glue” to be formed. Budding yeast Ctf7p is homologous to a C-terminal domain of the fission yeast Eso1 protein, whose N-terminal segment is homologous to the budding yeast DNA repair polymerase RAD30 (96). The Eso1+ gene functions similarly during S phase and is required for sister chromatid cohesion, pointing to conservation of this activity as well as its association with replication of DNA.
Recent studies have identified proteins more directly involved in DNA replication as members of the fourth class of cohesion proteins. This category includes PCNA by genetic interaction with CTF7 as described above (87) and a new DNA polymerase family, designated polymerase κ, exemplified by budding yeast gene TRF4 (108). PCNA forms a homotrimeric ring structure (clamp) which encircles DNA and supports processive DNA replication by associated DNA polymerases δ and ɛ (reviewed in references 39 and 47). A “clamp-loader,” replication factor C (RF-C), is required to facilitate association of PCNA with DNA (reviewed in reference 71). RF-C is composed of five essential subunits that have a common core region of homology that may facilitate interactions with PCNA (1, 42, 65), as well as interact with DNA (103). RF-C may also mediate a switch from polymerase α-directed replication initiation to processive replication by polymerases δ and ɛ through competitive interactions with PCNA and the budding yeast single-stranded binding protein replication protein A (RPA) (reviewed in reference 19).
TRF4 and its paralog TRF5 are both members of the β-polymerase superfamily as defined by protein alignment (3). While neither the TRF4 nor TRF5 gene is essential, in combination they exhibit synthetic lethality. Recent work has provided direct evidence that Trf4p encodes a novel polymerase and that a trf4 trf5 double mutant exhibits highly inefficient S-phase DNA replication (108). Like other budding yeast genes that function in cohesion, TRF4 is required for both chromosome condensation (14) and sister chromatid cohesion (108). The chromosomal defects present in a trf4 mutant cell influence the maintenance of sister chromatid cohesion under mitotic arrest conditions, probably through an uncharacterized mechanism that operates during DNA replication (108).
In this work, we present an analysis of two genes, CTF4 and CTF18, that exhibit genetic and physical interactions with components of the replication fork and that are required for sister chromatid cohesion. Analysis of these genes provides independent evidence that cohesion-related functions are indeed carried out by proteins associated with the DNA synthesis machinery. Previous genetic analyses have indicated that neither CTF4 nor CTF18 is essential; however, absence of either gene increases chromosome instability and mitotic recombination rates and induces a strong preanaphase delay (51, 53, 69). CTF4 was first identified in concurrent studies as CTF4 (CHL15) (52) and POB1 (69) and encodes a 104-kDa polypeptide with motifs suggestive of three zinc finger structures, as well as a helix-loop-helix region (35, 51). Ctf4p exhibits high-affinity binding to DNA polymerase α in vitro (69), and ctf4 mutants show genetic interactions with conditional alleles of DNA polymerase α (encoded by POL1[CDC17]) and other genes intimately associated with DNA synthesis (24, 112). A ctf4Δ mutation also causes synthetic lethality with a null allele of CTF18, the other gene investigated in this study (24). CTF18 (CHL12) was independently identified in two screens for chromosome loss mutants (52, 91). CTF18 encodes a predicted 84-kDa polypeptide with homology to all five subunits of the RF-C complex from yeast and other organisms, with the most significant similarity to the large subunit Rfc1p (17, 53).
Here we find that both CTF4 and CTF18 are required for sister chromatid cohesion. This, rather than the presence of damaged DNA, is likely to be the major mechanism underlying chromosome loss in both ctf4 and ctf18 null mutants. The preanaphase delay exhibited by each of these mutants is due to a spindle assembly checkpoint arrest. Because we and others have observed that CTF4 and CTF18 exhibit genetic interactions with genes that function in DNA replication, we suggest that CTF4 and CTF18 act in association with the replication fork complex(es) to facilitate the establishment of robust sister chromatid cohesion. We propose a model in which Ctf18p, an Rfc1p paralog, may act within a complex similar to the previously characterized RF-C, consistent with physical and genetic evidence presented in this report.
The replication fork plays numerous roles in the chromosome cycle, including duplication of genomic DNA, detection and repair of lesions (85, 93), and regulation of transcriptional states (20, 89). The properties of CTF4 and CTF18 highlight a new function of the replication fork machinery that is essential for high-fidelity segregation of the genome. The characterization of these proteins, which interact with residents of the replication fork and whose loss of function compromises cohesion, presents new opportunities for investigation of this novel role of the replication machinery.
MATERIALS AND METHODS
Strains and media.
Yeast transformation (29, 49) and genetic manipulations (34) were performed by published methods. rYPD is rich medium adjusted to pH 4 and supplemented with additional adenine hemisulfate (30 ng/ml), uracil (20 ng/ml), and l-tryptophan (30 ng/ml). Strain sets (Table 1) within each single experiment are composed of laboratory stocks related by transformation or genetic crosses maintaining background isogenicity. Complete oligonucleotide sequences will be provided on request.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YJH17.2 | MATaade2-1 can1-100 trp1-1 ura3-1 ctf18Δ::TRP1 his 3-11,15::GFP-Lac1-HIS3 leu2-3,112::lacO-LEU2 | This study |
| YJH18.3 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 ctf18Δ::LEU2 mad2Δ::HIS3 | This study |
| YCB624 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his2Δ200 leu2Δ1 rad9Δ::TRP1 | 10 |
| YJH35.1 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 his2Δ200 leu2Δ1 rad9Δ::TRP1 ctf18Δ::LEU2 | This study |
| YJH37 | MATaade2-1 can1-100 trp1-1 ura3-1 ctf4Δ::TRP1 his3-11,15::GFP-LacI-HIS3 leu2-3,112::lacO-LEU2 | This study |
| YJH38 | MATaura3-52 lys2-801 ade2-101 trp1 ars1ΔHIS3 leu2Δ1 CTF4::9Myc-klTRP1 | This study |
| YJH40.4 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 CTF18::9Myc-klTRP1 | This study |
| YJH41.1 | MATaura3-52 lys2-801 ade2-101 leu2Δ1 his3Δ200 mad2Δ::HIS3 ctf4-65 | This study |
| YJH48 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trpΔ63 leu2Δ1 ctf18Δ::LEU2 + pJH76.1 (GAL-CTF18-9myc) | This study |
| YJH62.6 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 RFC1::9Myc-klTRP1 | This study |
| YJH63.2 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 CTF18::9Myc-klTRP1 RFC2::6HA-KanMX | This study |
| YJH64.1 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 CTF18::9Myc-klTRP1 RFC4::6HA-KanMX | This study |
| YJH65.3 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 CTF18::9Myc-klTRP1 RFC5::6HA-KanMX | This study |
| YJH67.1 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 CTF18::9Myc-klTRP1 RFC3::6HA-KanMX | This study |
| YJH69.1 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 RFC2::6HA-KanMX | This study |
| YJH70.1 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 RFC3::6HA-KanMX | This study |
| YJH71.1 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 RFC4::6HA-KanMX | This study |
| YJH72.1 | MATaura3-52 lys2-801 ade2-101 trp1 ars1Δ HIS3 leu2Δ1 RFC5::6HA-KanMX | This study |
| AFS173 | MATaade2-1 can1-100 trp1-1 ura3-1 his3-11,15::GFP-LacI-HIS3 leu2-3,112::lacO-LEU2 | 92 |
| AFS387 | MATaade2-1 can1-100 trp1-1 ura3-1 mad2-1 his3-11,15::GFP-LacI-HIS3 leu2-3,112::lacO-LEU2 | 92 |
| s65 | MATaura3-52 lys2-801 ade2-101 leu2Δ1 his3Δ200 ctf4-65 | 91 |
| YE77 | MATa/aura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 ctf18Δ::LEU2/ctf18Δ::LEU2 | 53 |
| YE105 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trpΔ63 leu2Δ1 ctf18Δ::LEU2 | 53 |
| YRD501 | MATaleu2-3,112 ura3-52 trp1-289 cdc28-1 | Li laboratory |
| YRD510 | MATaleu2-3,112 ura3-52 trp1-289 cdc4-1 | Li laboratory |
| YRD543 | MATaleu2-3,112 ura3-52 trp1-289 his3Δ200 cdc7-4 | Li laboratory |
| YRD664 | MATaleu2-3,112 ura3-52 trp1-289 cdc34-2 | Li laboratory |
| YJL179 | MATaleu2-3,112 ura3-52 trp1-289 cdc46-1 | Li laboratory |
| YJL338 | MATaleu2-3,112 ura3-52 trp1-289 cdc2-1 | Li laboratory |
| YJL340 | MATaleu2-3,112 ura3-52 trp1-289 ade2 cdc6-1 | Li laboratory |
| YJL353 | MATaleu2-3,112 ura3-52 trp1-289 cdc17-1 | Li laboratory |
| YPH216 | MATaade2 ade3 his7 leu2 can1 sap1 cdc9-1 | Hartwell laboratory |
| YPH217 | MATaade2 ade3 his7 leu2 can1 sap1 cdc13-1 | Hartwell laboratory |
| YPH221 | MATahis7 leu2 ase2 ase3 sap3 gal1 ura1 cdc23-1 | Hartwell laboratory |
| YPH223 | MATahis7 leu2 ase2 ase3 sap3 gal1 cdc14-1 | Hartwell laboratory |
| YPH277 | MATaura3-52 lys2-801 ade2-101 trp1Δ63 + CFVII(RAD2.d.YPH277) URA3 SUP11 | 91 |
| YPH499 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 | 85a |
| YPH501 | MATa/aura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 his3Δ200/his3Δ200 trp1Δ63/trp1Δ63 leu2Δ1/leu2Δ1 | 85a |
| YPH1238 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ63 leu2Δ1 mad2Δ::HIS3 | Hieter laboratory |
| YDS489 | MATaura3-1 ade2-1 his3-11 trp1-1 rap1-5 | 61 |
All strains were grown to an optical density at 600 nm of 0.4 at 30°C unless otherwise noted. G1 arrest occurred after 2.5 h in 3 μM alpha mating pheromone; S-phase arrest was achieved by incubation for 2.5 h in 0.2 M hydroxyurea (HU); metaphase arrest was achieved by incubation for 4 h in 15 μg of nocadozole, per ml.
The CTF18 open reading frame (ORF) was disrupted by transforming with a SpeI-NotI fragment of pJH28::TRP. PCR primers flanking the inserted fragment were used to detect disruption of the CTF18 (OLFS278 plus OLFS279). The CTF4 ORF was disrupted using OLFS369 plus OLFS370 (10); verification PCR used OLFS371 plus OLFS372. A mad2Δ allele was transferred from YPH1238 using primers OLFS365 plus OLFS366; detection was performed using PCR primers OLFS367 plus OLFS368. Epitope tagging of CTF18, CTF4, RFC1, RFC2, RFC3, RFC4, and RFC5 was performed as described previously (49) using primer pairs OLFS359 plus OLFS360, OLFS373 plus OLFS374, OLFS491 plus OLFS492, OLFS510 plus OLFS511, OLFS506 plus OLFS507, OLFS508 plus OLFS509, and OLFS512 plus OLFS513, respectively; detection of integration used primer pairs OLFS344 plus OLFS345, OLFS371 plus OLFS372, OLFS493 plus OLFS494, OLFS514 plus OLFS515, OLFS437 plus OLFS438, OLFS439 plus OLFS440, and OLFS516 plus OLFS517.
Vector construction.
A SpeI-NotI fragment from BFS66, containing the CTF18 ORF and flank, was cloned into pBluescript II to give pJH28. pJH28::TRP was created by removal of the internal two-thirds of CTF18 ORF by digestion with NsiI and MluI, and its replacement by ligation of a PCR product from primers OLFS264 plus OLFS265, which amplified the TRP1 gene from pRS404 (10). pJH72.1 was constructed by cloning the entire CTF18 ORF into pCR2.1 (Invitrogen TOPO-TA) using primers OLFS274 plus OLFS362 and LTI Pfx Taq; the CTF18 ORF was then released using NcoI-EcoRI digestion and cloned into the same sites in pAS2-1 (Clontech), which created an in-frame fusion with the GAL4 DNA binding domain (pJH74.4). pJH73.2 was constructed from a PCR product (using LTI Pfx Taq, primers OLFS274 plus OLFS345, and YJH40.4 genomic DNA) digested with PvuI to liberate CTF18-9myc from the coamplified TRP1 marker. Taq polymerase modified the ends, and the product was TA cloned into pCR2.1. pJH76.1 was constructed by cloning the CTF18-9myc allele on an XhoI-HindIII fragment from pJH73.2 into the same sites in pRS316GU (74).
pJH78 was constructed using primers OLFS437 plus OLFS438 and genomic DNA from YJH40.4; the product was cloned into pCR2.1. pJH79 was constructed in similar fashion using primers OLFS439 plus OLFS440.
p414GEU1/12 was created by cloning a 2,236-bp SalI-EcoRI PCR fragment containing CTF18 into SalI- and EcoRI-cut p414GEU1 (54), placing the CTF18 ORF in frame with the vector ATG and double E1 epitope tag. The resulting plasmid conferred wild-type chromosome stability and growth at low temperature to a ctf18Δ strain in galactose-dependent fashion.
Flow cytometry.
A 1-ml volume of cells grown in rYPD was harvested and fixed in 500 μl of 0.2 M Tris (pH 7.5)–70% ethanol. After being washed in 1 ml of 0.2 M Tris (pH 7.5), samples were resuspended in 1 ml of 0.2 M Tris (pH 7.5) for >30 min, incubated in 100 μl of 0.2 M Tris (pH 7.5)–3 mg of RNase A per ml for 2.5 h at 37°C, washed with 1 ml of 0.2 M Tris (pH 7.5), and resuspended in 100 μl of 0.05% trypsin (Gibco 25300-054) at 37°C for 5 min. After a 1-ml wash in 0.2 M Tris (pH 7.5), samples were resuspended in 1 ml of 0.2 M Tris (pH 7.5)–9 μg of propidium iodide per ml.
Sister chromatid cohesion assay.
Log-phase cultures were resuspended in SC-HIS medium–40 mM 3-aminotriazole for 40 min to induce green fluorescent protein (GFP)-LacI expression, incubated in YPD–15 μg of nocodazole per ml for 3 h, fixed in 4% paraformaldehyde for 30 min, washed in 1 ml of SK (1 M sorbitol, 50 mM KPO4, pH 7.5), and resuspended in 50 μl of SK.
Chromosome spreads.
Chromosome spreads were prepared on slides essentially as described in references 48 and 68. Samples on cured slides were washed in phosphate-buffered saline (PBS) for 20 min, preincubated in PBS–1% bovine serum albumin (BSA) for 1 h, and incubated in polyclonal rabbit anti-myc antibody (sc-789; Santa Cruz Biotechnology) in PBS–1% BSA (at a 1:250 dilution for CTF18-9myc and a 1:500 dilution for CTF4-9myc) for 2 h. Samples were then washed three times with PBS, incubated for 2 h with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody (Sigma) for 2 h (1:1,000 in PBS–1% BSA), washed three times with PBS, and mounted in Fluorsave (Calbiochem)–2 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml.
Protein analysis.
Samples prepared as described previously (49) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide) (SDS-PAGE) and transferred to a nitrocellulose membrane. Polyclonal rabbit anti-myc antibody (sc-789) and a polyclonal goat anti-rabbit antibody–horseradish peroxidase (111-035-144; Jackson ImmunoResearch) were used for detection. Protein loading was determined by standardization to β-tubulin (polyclonal antibody R43; a gift of D. Koshland). Anti-Orc3p monoclonal antibody (SB3) was a gift of B. Stillman (57). Film scans were obtained with a Hewlett-Packard 4c/T scanner and analyzed using NIH Image 1.61.
Antisera against CTF18.
A C-terminal 18-kDa peptide of CTF18 expressed from pRSETA (InVitrogen) in bacterial strain BL21(DE3)/pLysE was affinity purified on a Ni2+-containing column (Qiagen), solubilized in 8 M urea, and used to immunize two New Zealand rabbits (Hazelton, Inc.). Antibodies from one (rabbit 10C) were purified against ctf18Δ protein extract bound to cyanogen bromide-activated Sepharose beads. The Western blot signal from purified bacterial peptide in serial dilution indicated that the 10C antibody was capable of detecting at least 2 ng of Ctf18p in the experiment in Fig. 6.
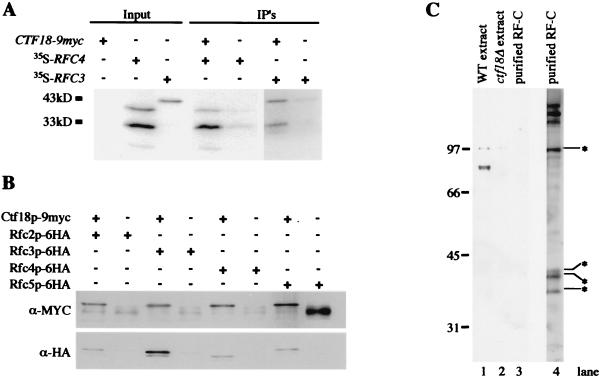
FIG. 6.
Ctf18p interacts with a subset of RF-C components but is not a component of purified RF-C. (a) Unlabeled Ctf18p-9myc was used to pull down [35S]methionine-labeled Rfc3p or Rfc4p. The slowest-migrating bands are approximately the size expected for full-length products; the faster-migrating bands are consistent with the positions of in-frame translational start sites. Similar results were obtained from four independent trials. (B) Immunoprecipitation experiments were performed in yeast whole-cell extracts from strains containing 6HA epitope-tagged alleles of Rfc2p, Rfc3p, Rfc4p, or Rfc5p in the presence and absence of a CTF18-9myc allele. Duplicate SDS-PAGE gels loaded as shown were transferred and probed with anti-myc and anti-HA antibodies. The slowest-migrating species detected by the anti-myc antibody corresponds to Ctf18p-9myc. Species detected by the α-HA antibody are consistent with the expected migrations for RF-C proteins as indicated. Note that excess immunoprecipitate was loaded in the rightmost lane, accounting for the increased intensity of the background cross-reactive band. (C) A 100-ng portion of purified RF-C complex was probed for the presence of Ctf18p using affinity-purified antibody 10C. The antibody detected a strong band at 85 kDa in protein from wild-type cells (WT extract, from YPH499) that was absent from ctf18Δ cells (ctf18Δ extract, from YE105). Lanes 1 to 3 show the Western blot, and lane 4 is silver stained SDS-PAGE. Starred species are those identified by Fien and Stillman (23) and Cullmann et al. (17).
Yeast two-hybrid.
pJH74 (BD-CTF18) does not autoactivate reporters in strain AH109 (Clontech). AH109 containing pJH74 was transformed with a Saccharomyces cerevisiae cDNA library (gift of S. Elledge) cloned into pACT2-1. A total of 180,000 transformants yielded 162 His+-positive strains, of which 65 were also Ade+. PCR analysis and plasmid isolation indicated the presence of four distinct genes. Two of these, RFC3 and RFC4, also conferred β-galactosidase activity by plate assay (Clontech).
In vitro immunoprecipitations.
Portions (1 μg) of circular pJH73.2 (CTF18-9myc), pJH78 (RFC3), or pJH79 (RFC4) were used as templates in a 50-μl linked transcription-translation system (Promega). A 24-μl volume of each product was used per immunoprecipitation in a final volume adjusted to 150 μl with 1× PBS–1% Triton X-100, and the mixtures were incubated for 2 h at 4°C. A 1-μl volume of anti-myc (sc-789) was added for a further 2-h incubation, followed by immune complex precipitation for 1 h using 10 μl of protein A-agarose beads (sc-2001; Santa Cruz). Following three washes (each with 1 ml of PBS–1% Triton X-100), the beads were resuspended in 20 μl of HU buffer (49) and boiled for 5 min. The entire supernatant was analyzed by SDS-PAGE (10% polyacrylamide).
Whole-cell extract immunoprecipitations.
A total of 5 × 108 logarithmically growing cells were washed twice with water and resuspended in 1.5 ml of ice-chilled buffer B60 (50 mM HEPES-NaOH [pH 7.3], 0.1% Triton X-100, 20 mM β-glycerophosphate, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin per ml, and 2 μg of pepstatin per ml 2× Complete [Roche Biochemicals], 60 mM potassium acetate), and 1.5 g of ice-chilled glass beads (400 to 600 μm in diametet was added. The tubes were vortexed eight times for 30 s with 30-s intervals on ice. After 10 min on ice, the lysate was decanted into ice-chilled 15-ml Corex tubes and centrifuged for 20 min at 18,000 × g at 4°C.
A 500-μl volume of clarified lysate was incubated with 25 μl of prewashed protein A-agarose beads at 4°C for 1 h. The beads were pelleted, and 450 μl of the lysate was transferred to a tube containing 7.5 μl of anti-myc antibody (Santa Cruz) and incubated at 4°C for 2 h. Then 25 μl of prewashed protein A-agarose-conjugated beads was added, and the mixture was incubated for 1 h at 4°C. The beads were then washed successively seven times: four times with B60 adjusted to 100 mM potassium acetate and once each with B60 adjusted to 210, 240, or 270 mM potassium acetate. The beads were boiled in HU buffer for 5 min and briefly pelleted at 13,000 rpm in an Eppendorf centrifuge before the supernatant was loaded for electrophoresis.
Identification of candidate CTF18 orthologues.
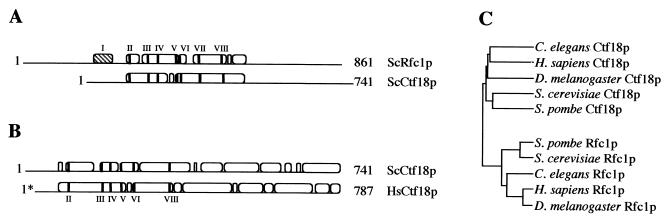
The protein sequence of budding yeast CTF18 was used as query for a TBLASTN search of the GenBank nonredundant protein database, yielding a significant match to human genomic clone HS321D2. GeneSCAN analysis of the genomic clone predicted three genes, including candidate HsCTF18, as depicted in Fig. 8. Verification between amino acids 356 and 1220 of the predicted protein is provided by sequence from the cDNA clone pJH3 (data not shown). The GenBank accession numbers for Rfc1p homologs and Ctf18p homologs in other species identified through PSI-BLAST are as follows: C. elegans Ctf18p, T23478; C. elegans Rfc1p, T20230; D. melanogaster Ctf18p, AAF51072.1; D. melanogaster Rfc1p, AAB58311.1; S. pombe Ctf18p, CAB62096.1; S. pombe Rfc1p, CAA18875; H. sapiens Rfc1p, NP_002904.1; S. cerevisiae Ctf18p, NP_013795.1; S. cerevisiae Rfc1p, NP_014860.1.
FIG. 8.
Candidate CTF18 orthologues and conserved RF-C boxes. (A and B) Modified output of National Center for Biotechnology Information pairwise BLAST (BLOSUM62) of representative sequences to illustrate the distribution of similarity. Roman numerals indicate RF-C homology boxes, the hatched box represents the ligase homology domain, and black bars mark the relative positions of RF-C boxes. (A) ScCtf18p versus ScRfc1p (e-value = 1e−15). (B) ScCtf18p versus HsCtf18p (e-value = 3e−35). (C) Proteins from C. elegans, D. melanogaster, S. pombe, and H. sapiens were identified among the top hits from a PSI-BLAST search using S. cerevisiae Ctf18p as the query. Clustal X v1.8 (44) was used to make a multiple alignment of proteins from C. elegans, D. melanogaster, S. pombe, and H. sapiens. Using the multiple alignment, a bootstrapped neighbor-joining tree was produced. The analysis shows that Ctf18p homologs cluster away from Rfc1p homologs.
RESULTS
The G2/M delay of ctf4 and ctf18 mutants is dependent on the spindle assembly checkpoint.
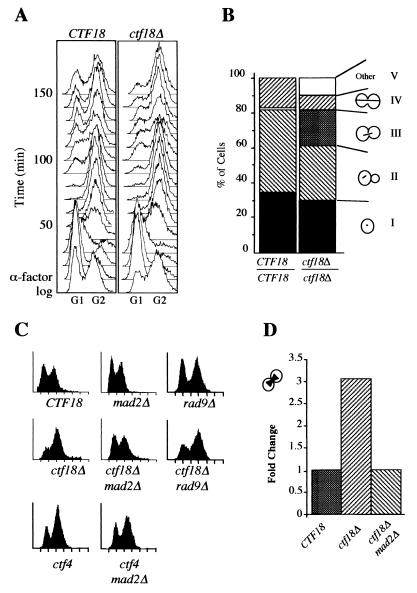
Previous work has shown that cells lacking CTF18 accumulate a large-budded morphology (53). In agreement, we found that analysis of log-phase cultures of ctf18Δ cells by flow cytometry revealed a substantial accumulation of cells with G2 DNA content. Time course analysis in synchronous cultures was performed to address the nature of the delay. After synchronization in G1, cells were released into rich medium and samples were taken every 10 min for flow cytometry (Fig. 1A). In comparison with the wild-type control, the progression of ctf18 cells was not detectably different for the first 80 min. New G1 cells appeared in wild-type and ctf18 strains at 90 and 100 min, respectively, indicating that a second cycle occurred with a ∼10-min delay in the mutant. Moreover, a large proportion of the ctf18 population remained in the G2 peak until the end of the experiment (150 min).
FIG. 1.
Absence of CTF4 or CTF18 leads to a MAD2-dependent preanaphase delay. (A) Log-phase cultures grown in rYPD were arrested in α-factor, released into rYPD, and processed for flow cytometry. The strains were ctf18Δ (YE105) and CTF18 (YPH499). (B) Log-phase cultures of ctf18Δ/ctf18Δ (YE77) or wild-type (YPH501) cells were fixed in formaldehyde and stained to visualize the DNA (DAPI) and microtubules (β-tubulin indirect immunofluorescence). Class III was defined as cells with a single DNA mass that crossed the neck and a spindle that did not extend beyond the center of either mother or bud. (C) Early-log-phase cells were processed for flow cytometry. Strains were CTF18 (YPH499), mad2Δ (YPH1238), rad9Δ (YCB624), ctf18Δ (YE105), ctf18Δ mad2Δ (YJH18.3), ctf18Δ rad9Δ (YJH35.1), ctf4 (s65), and ctf4 mad2Δ (YJH41.1). Similar results were obtained in five independent experiments; representative results are shown. (D) Formaldehyde-fixed log phase cells were stained with DAPI. Large-budded cells with a single nucleus located within the bud neck were scored (6% in wild type, n = 100). Strains were CTF18 (YPH499), ctf18Δ (YE105), and ctf18Δ mad2Δ (YJH18.3).
The delay indicated by flow cytometry may reflect late execution of any step after the completion of bulk DNA synthesis through daughter separation. The mitotic cycle was further characterized by analysis of asynchronous cultures using bud size and spindle morphology as indicators of cell cycle position. Comparison between ctf18Δ and wild-type controls revealed a dramatic increase in the number of cells with an intermediate-length spindle (Fig. 1B, class III), representing 20% of mutant cells versus 1% of wild-type cells. The abundant class III morphology indicates the presence of many cells with altered progression through metaphase or anaphase. We conclude that an early mitotic delay underlies the G2 accumulation observed by flow cytometry.
Early mitotic delay is often due to activation of either the DNA damage checkpoint or the spindle assembly checkpoint (reviewed in references 86 and 111). To investigate further, we constructed rad9Δ and mad2Δ mutants in a ctf18Δ background to remove the DNA damage and spindle assembly checkpoints, respectively. DNA content analysis of log-phase cultures grown at 30°C was used to monitor the cell cycle distributions in these double mutants (Fig. 1C). The results indicated that the preanaphase delay in the ctf18Δ mutant was dependent on the spindle assembly checkpoint and not on the DNA damage checkpoint. Similar analysis of ctf18Δ mec1Δ double mutants, in an sml1-1 mutant background which renders mec1Δ mutants viable (119), also indicated that the DNA damage checkpoint was not responsible for the ctf18Δ G2 peak accumulation (data not shown).
Morphological analysis also indicated a MAD2-dependent mitotic delay. A nuclear morphology in which the DAPI-stained chromosomal mass crossed the neck between mother and bud was present in higher levels in ctf18Δ cells than in wild-type cells (Fig. 1D). The frequency of this class of cells was reduced to wild-type levels in the ctf18Δ mad2Δ strain. Thus, in cultures grown at 30°C, the absence of ctf18 protein leads to activation of the spindle assembly checkpoint.
This conclusion suggested that a similar phenomenon could explain a non-RAD9-dependent delay observed in ctf4 mutant cells (69). Cells lacking CTF18 cannot survive in the absence of CTF4, consistent with the idea that these gene products separately contribute to the same essential function. To determine whether ctf4 mutant delay was dependent on the spindle assembly checkpoint, ctf4 mad2Δ double mutants were created and analyzed by flow cytometry. Again, the accumulation of ctf4 cells of G2 DNA content was dependent on MAD2 (Fig. 1C).
Sister chromatid cohesion failure in ctf18 and ctf4 mutants.
Most inducers of the spindle assembly checkpoint cause preanaphase delay or arrest associated with a short spindle morphology (reviewed in reference 86). However, an appreciable proportion of ctf18Δ cells contain partially elongated spindles and stretched DNA masses. Interestingly, a MAD2-dependent G2/M delay with an intermediate spindle morphology had recently been described for ctf7 mutants, which also exhibit a defect in sister chromatid cohesion. One hypothesis (87) is that precocious sister separation results in a loss of tension at the kinetochore-microtubule interface (triggering the spindle assembly checkpoint), allowing an increase in pole separation at metaphase arrest due to altered balance between opposing forces within the spindle. Based on the phenotypic similarity to ctf7, we were encouraged to assay ctf18 and ctf4 mutants for a sister chromatid cohesion defect.
In wild-type cells, the arms of duplicated sister chromatids remain tightly associated (32) until the dissolution of cohesins at the metaphase-anaphase transition. The CTF4 or CTF18 gene was deleted in a strain background containing a tandem array of lac operators integrated at LEU2 on chromosome III and expressing a GFP-LacI fusion (92). This configuration allows the assessment of sister chromatid cohesion on chromosome arms throughout the cell cycle: transition from one GFP signal spot to two indicates separation of sister chromatids. Detection of sister chromatid cohesion proficiency can be enhanced by inducing metaphase arrest using a microtubule-disrupting drug such as nocodazole (33, 68). Under these conditions, wild-type cells arrest with unseparated chromatids whereas mutants defective in cohesion exhibit separation.
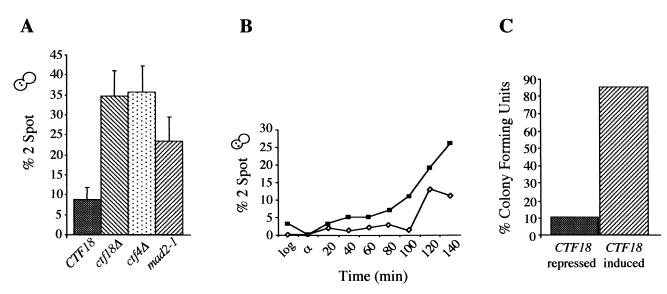
Log-phase ctf4Δ and ctf18Δ cultures were arrested in nocodazole for 3 h, fixed, and scored for the number of GFP spots per cell. Parallel analysis of wild-type and mad2-1 strains served as positive and negative controls for cohesion. Compared to wild-type cells, ctf4 and ctf18 mutants exhibited a high level of precociously separated sister chromatids (Fig. 2A). The number of cells with two GFP spots was comparable to that seen in mad2-1 cells, which cannot respond to the loss of spindle integrity and inappropriately proceed to anaphase (92). A time course experiment in which the number of GFP spots was observed through a synchronous cycle indicated that the number of cells containing separated sisters exhibited a steady accumulation over time (Fig. 2B). Wild-type and ctf18Δ strains were arrested with α-factor for 2.5 h, released into nocodazole-containing medium, and sampled every 10 min. The number of cell bodies containing two GFP signals was small for both wild-type and mutant strains early in the cycle, and increased as the cells traversed S phase and entered nocodazole arrest. The gradual, and early, appearance of cohesion failure suggests that mutant cells perform faulty cohesion establishment that results in slow decay of sister association.
FIG. 2.
CTF4 and CTF18 are required for sister chromatid cohesion. (A) Early-log-phase cultures were treated to induce GFP-LacI expression, incubated for 3 h in rYPD containing nocodazole, and fixed in paraformaldehyde. For each strain, 100 cells exhibiting GFP fluorescence were scored within each experiment. The histogram shows the mean and standard deviation for a minimum of three trials. Strains were CTF18 (AFS173), ctf18Δ (YJH17.2), ctf4Δ (YJH37), and mad2-1 (AFS387). Note that approximately 5% of ctf4Δ or ctf18Δ cells have >1 chromosome III as determined during G1 arrest. (B) Kinetics of sister chromatid separation in ctf18Δ mutants. Log-phase cultures were arrested in α-factor and released into rYPD-nocodazole. After α-factor release, both strains sychronously completed S phase at 60 min postrelease as determined by flow cytometry (data not shown). YJH17.2 data were normalized using the α-factor arrest time point, to remove the contribution of cells containing >1 copy of chromosome III. A total of 100 informative cells were counted at each time point. (C) A ctf18Δ strain containing GAL-CTF18 on a plasmid (YJH48) was grown in selective medium containing either galactose or glucose. Log-phase cells were then shifted to rYPD-nocadozole for 4 h and then plated onto SC-URA glucose medium. CFU were counted after 24 h. The experiment was performed twice with similar results (mean values are shown).
The sister chromatid cohesion defect predicts that many mutant cells should not be able to recover after exposure to nocodazole. ctf18Δ cells bearing a galactose-inducible copy of CTF18 were grown in repressing (glucose) or inducing (galactose) medium to early log phase and subjected to a 3-h nocodazole arrest. The cells were then spotted onto solid medium without nocodazole, and the ability to form microcolonies was scored the following day (Fig. 2C). Approximately 90% of ctf18Δ cells not expressing CTF18 failed to recover. This result is predicted by the data in Fig. 2A. If the GFP-marked chromosome separates with kinetics representative of other chromosomes, the appearance of ∼30% of cells with two GFP spots predicts that ∼5 of the 16 sister chromatid pairs in an average cell will have disassociated. Because dissociated sister chromatids are expected to segregate randomly, the mitotic division following drug removal is expected to result in very high inviability.
CTF18 is required for mitotic condensation of the rDNA array.
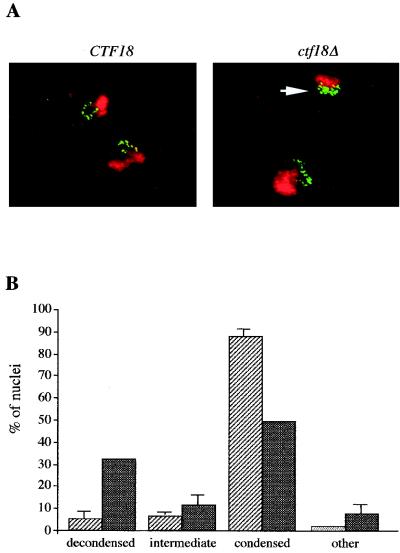
In budding yeast, the cohesion defect observed in mcd1, pds5, and trf4 mutants is accompanied by defective chromosome condensation (33, 36, 68, 108). To test whether CTF18 protein is required for chromosome condensation, wild-type and ctf18 cells were arrested in nocodazole-containing medium to obtain a uniformly staged culture at the point when condensation is complete. Formaldehyde-fixed cells were subjected to fluorescence in situ hybridization (32) with a digoxigenin-labeled DNA probe against the repetitive rDNA region. Bulk DNA was visualized with propidium iodide, and the rDNA probe was detected with antidigoxigenin and fluorescein isothiocyanate-conjugated secondary antibodies. Wild-type condensation of the rDNA region resulted in a distinct loop structure, as illustrated in the wild-type examples (Fig. 3A). However, 32% of ctf18Δ nuclei exhibited a decondensed rDNA staining pattern, indicating a condensation defect (Fig. 3). Thus, like the three other cohesion proteins for which mitotic chromosome condensation has been tested, Ctf18p is required for wild-type condensation of the rDNA as well as for sister chromatid cohesion.
FIG. 3.
Absence of CTF18 leads to a condensation defect. (A) Nocodazole-arrested CTF18 (YPH499) and ctf18Δ (YE105) cells were assayed by fluorescence in situ hybridization using an rDNA probe. rDNA is detected by immunofluorescence (green); chromosomal DNA is stained with propidium iodide (red). A nucleus with decondensed rDNA is indicated by the arrow. (B) A total of 100 nuclei were scored for rDNA condensation status in four categories. Data were collected twice (averages are shown).
Ctf4p and Ctf18p are stable proteins that associate with chromatin throughout the cell cycle.
To determine whether the steady-state levels or chromatin associations of Ctf4p or Ctf18p were regulated, epitope-tagged alleles of both genes were generated by integration of a 9myc epitope in frame after the last codon (Fig. 4A). Both epitope-tagged alleles conferred wild-type stability to a test chromosome, a sensitive indicator of protein function (data not shown).
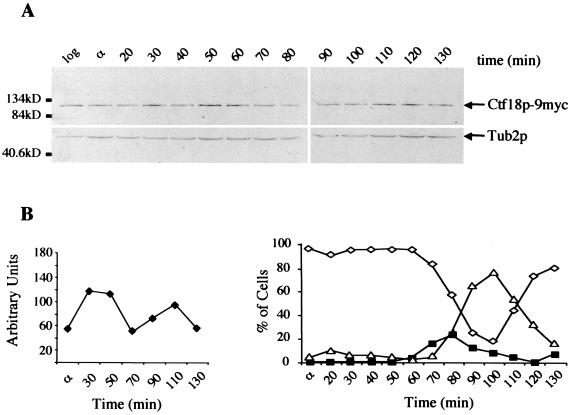
FIG. 4.
Ctf18p throughout the cell cycle. (A) α-factor arrest-release time course for CTF18-9myc cells (YJH40.4). CTF18-9myc was detected using anti-myc antibody and compared with β-tubulin (Tub 2p) on the same blot. (B) (Left) Fluctuation in Ctf18p-9myc accumulation was determined in normalized units by comparison of band intensity relative to β-tubulin. (Right) Separate aliquots from the same time course were prepared for flow cytometry and analyzed for DNA content (data not shown) as well as being scored for nuclear morphologies. These analyses indicated the execution of S phase by the majority of cells between 30 and 60 min and the initiation of M phase between 70 and 80 min.
Analysis of Ctf4p and Ctf18p levels indicate that they do not vary dramatically throughout the cell cycle. To assay the accumulation levels of Ctf18p-9myc and Ctf4p-9myc, cultures were grown to early log phase at 30°C, arrested for 2.5 h in α-factor, released into pheromone-free medium, and sampled every 10 min for 1.5 cell cycles. In a comparison with a β-tubulin standard, Ctf18p abundance varied over a three- to fivefold range, reaching a maximum during S phase and minimum during G2/M (Fig. 4B). These results are consistent with published mRNA levels (90), which show a shallow peak accumulation of CTF18 transcript near the G1/S boundary, and with the presence of two degenerate MluI cell cycle (MCB) boxes in the promoter region of CTF18 (53). Arrest release experiments did not reveal significant fluctuations in CTF4 protein levels in the cell cycle (data not shown).
Previous studies have shown that association between cohesion proteins and chromatin in budding yeast can be visualized by indirect immunofluorescence on spread chromosomes (48, 68). We detected both Ctf4p and Ctf18p association with chromatin in cells arrested in G1, S, or M phase (Fig. 5A and B) in this assay. Cells expressing epitope-tagged alleles were spheroplasted, lysed on glass slides in the presence of detergent, and washed to remove loosely adherent cellular contents. The presence and location of Ctf18p-9myc or Ctf4p9-myc proteins was compared with the location of DAPI-stained chromatin. Visible signals were observed in all samples of similar exposure, suggesting that some Ctf4p and Ctf18p is chromosome associated at each of these arrest points.
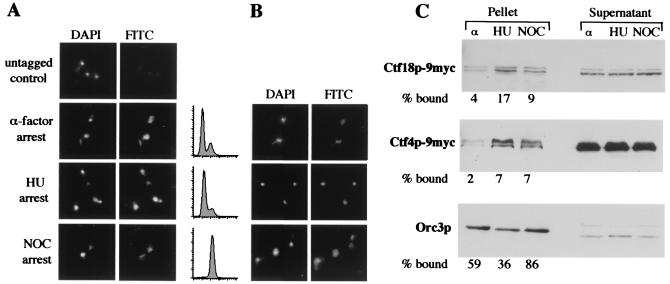
FIG. 5.
Ctf4p and Ctf18p are chromatin associated. Cells grown to early log phase in rich medium were arrested in α-factor (G1), HU (S), or nocadozole (M) for 2.5 h. (A) Ctf18p-9myc (in YJH40.4) was detected using indirect immunofluorescence on chromosome spreads. For comparison, an untagged control strain (YPH277) in log phase is shown in the top row. Drug-induced arrests were verified by flow cytometry as shown. (B) Ctf4p-9myc (YJH38) cells were similarly analyzed. (C) Chromatin pellet fractions from cells containing Ctf18p-9myc (YJH40.4), Ctf4p-9myc (YJH38), and Orc3p (YJH62.6) were generated from arrested cell populations as above and analyzed by Western blotting. Similar results were obtained for Ctf18p-9myc and Ctf4p-9myc in four independent experiments. Results for Orc3p are consistent with previous data from Liang and Stillman (57). In these experiments, a pellet-to-supernatant cell equivalent loading ratio of 4:1 was used.
In a quantitative analysis, the proportion of Ctf4p or Ctf18p that remained bound to chromatin in a simple fractionation protocol was analyzed (57). Whole-cell extracts obtained from spheroplast lysis were subjected to centrifugation through a sucrose cushion, which separates a pellet fraction (enriched for chromosome-associated proteins) from a supernatant fraction. For both Ctf4p and Ctf18p, there was a three- to fourfold increase in bound protein between G1-and S-phase arrests (Fig. 5C). In contrast, we and others (57) noted a different binding pattern for Orc3p, another known chromatin binding protein. Furthermore, we noted electrophoretic mobility variants for both Ctf4p and Ctf18p in this protocol, whose significance is not yet apparent. The increase in association with the pellet fraction at S-phase arrest (HU) and early M-phase arrest (nocodazole) is consistent with roles for Ctf4p and Ctf18p in DNA replication and/or cohesion.
CTF18 exhibits genetic interaction with replication mutants.
Genetic interaction between CTF18 and a subset of cell cycle genes involved in DNA metabolism as well as other aspects of the chromosome cycle was tested by using a synthetic dosage lethality phenotype. In synthetic dosage lethality, a process may be efficiently disrupted if one interacting factor is present at levels that interfere with complex formation in vivo and the function of another factor is limited by a hypomorphic mutation (25, 54, 55, 115). Since overexpression of CTF18 from a galactose-inducible promotor (in p414GEU1/12) did not cause a growth defect, it could be used in a synthetic dosage lethality screen.
A subset of cell cycle mutants were chosen that included genes involved in DNA metabolism as well as other aspects of the chromosome cycle. Each mutant was transformed under noninducing conditions with either a GAL-CTF18-expressing minichromosome (p414GEU1/12) or with empty vector (p414GEU1). After induction, growth of the transformants with or without overexpression of CTF18 was compared (Table 2). A temperature series ranging from 25 to 37°C was used to detect changes in maximum permissive growth temperature. A synthetic dosage lethal interaction was found with cdc2-1, cdc7-4, cdc17-1, and cdc46-1, each of which functions in an aspect of DNA synthesis. CDC2 and CDC17 encode catalytic subunits of polymerase δ and polymerase α, respectively (43). CDC7 encodes a protein-kinase whose activity controls replication initiation, probably through modification of targets in the prereplication complex (64). CDC46 encodes an essential protein of the prereplication complex, components of which are proposed to remain associated with the replicative fork (2). These genetic interactions extend the list of interactions already identified through traditional synthetic lethality (24, 87) and support the idea that Ctf18p interacts with replication fork proteins in vivo.
TABLE 2.
Synthetic dosage lethality with CTF18a
| Allele | Description | Phenotypeb |
|---|---|---|
| cdc2-1 | DNA polymerase δ large subunit | Lethal at 28°C |
| cdc7-4 | Protein kinase, controls initiation of DNA synthesis | Toxic at 30°C |
| cdc17-1 | DNA polymerase α large subunit | Toxic at 34°C |
| cdc46-1 | Acts at origins to initiate replication | Toxic at 32°C |
| rap1-5 | Repressor/activator protein, affects telomere structure | Lethal at 30°C |
| cdc9-1 | DNA ligase | NP |
| cdc13-1 | Binds telomeres | NP |
| cdc14-1 | PTPase required for mitotic exit | NP |
| cdc23-1 | Anaphase promoting complex subunit | NP |
| cdc28-1 | p34CDC2 kinase homolog | NP |
| cdc34-2 | Ubiquitin-conjugating enzyme, SCF subunit | NP |
| cdc4-1 | Ubiquitin-conjugating enzyme, SCF subunit | NP |
| cdc6-1 | Involved in DNA replication initiation | NP |
The growth of cells expressing GAL-CTF18 was compared with that of cells containing vector.
The lowest temperature with a GAL-CTF18-induced phenotype is given. NP, no synthetic phenotype; Lethal, no growth; Toxic, markedly slow growth. Descriptions are adapted from YPD (http://www.proteome.com) or SGD (http://genome-www.stanford.edu/Saccharomyces).
Ctf18p can interact with components of the RF-C complex.
To search for proteins with which Ctf18p interacts, we screened a yeast cDNA library using the two-hybrid system. The coding region of CTF18 was cloned in frame downstream of the GAL4 DNA binding domain (GAL4BD-CTF18). This fusion protein complemented the chromosome loss defect of a ctf18Δ mutant (data not shown). The screen identified two interacting genes whose fusion proteins were capable of conferring expression to three different GAL4-driven reporters: HIS3, ADE2, and the β-galactosidase gene (data not shown). Sequence analysis of the library plasmids indicated that they contained either RFC3 or RFC4, each present as full-length fusions.
To investigate further, Rfc3p, Rfc4p, and Ctf18p-9myc were produced in vitro using a coupled transcription-translation system. Rfc3p and Rfc4p were radiolabeled using [35S]methionine. Unlabeled Ctf18-9myc product was incubated with each, in the presence of a polyclonal anti-myc antibody. Immune complexes were captured on protein A-agarose beads, washed, and analyzed by SDS-PAGE. Labeled Rfc3p and Rfc4p were enriched specifically and reproducibly through coimmunoprecipitation with Ctf18p (Fig. 6A).
To directly address whether Ctf18p interacts with Rfc3p and Rfc4p in vivo as well as with other members of the RF-C complex, 6HA epitope-tagged alleles were generated of RFC2, RFC3, RFC4, and RFC5 in the presence and absence of the CTF18-9myc allele. In coimmunoprecipitation experiments, we found that Ctf18p-9myc precipitated with Rfc2p-6HA, Rfc3p-6HA, Rfc4p-6HA, and Rfc5p-6HA reproducibly (Fig. 6B). These results suggest the presence of an alternative RF-C complex, which we have termed RF-CCTF18 to distinguish it from the canonical, PCNA-loading RF-CRFC1. These results are consistent with the observed homology between CTF18 and RFC1 and the in vitro interactions observed between Ctf18p-9myc, Rfc3p, and Rfc4p. Interestingly, it has been demonstrated that RAD24, another RFC1 homolog, can also form a complex with the small RF-C subunits to facilitate alternative functions (31, 85).
Early purification methods for generating a biochemically defined RF-C complex (promoting PCNA association with DNA) involved a variable species that migrated near the predicted molecular weight of Ctf18p. Therefore we analyzed an active purified fraction graciously donated by the Stillman laboratory to ascertain whether Ctf18p might be present as a substoichiometric component. Affinity-purified polyclonal antibody (10C) that provided robust detection of 2 ng of Ctf18p by Western blot analysis (see Materials and Methods) did not detect a Ctf18p band in 100 ng of purified RF-C complex (Fig. 6C). Therefore we conclude that Ctf18p is not a detectable substoichiometric subunit of the biochemical activity characterized for processive DNA synthesis.
ctf18 mutant cells contain short telomeres.
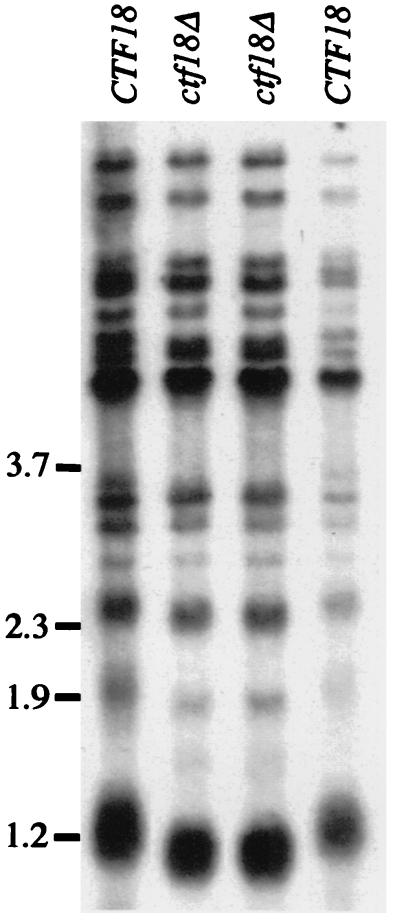
A screen for telomere length alteration in a collection of mutants exhibiting increased chromosome loss (91) identified a telomere length defect in all three alleles of ctf18 (V. Lundblad and F. Spencer, unpublished data). The screen assayed telomere length following cleavage of genomic DNA with XhoI, which cuts in both subtelomeric Y′ elements, to generate a broad 1.2-to 1.5-kb band, as well as in non-Y′-containing termini, to generate bands ranging in size from 2 to >6 kb. Both types of terminal restriction fragments exhibited a moderate reduction in size in the ctf18 null strain, indicating that a loss of CTF18 function influenced telomere length (Fig. 7).
FIG. 7.
CTF18 participates in telomere length control. Meiotic segregants from a single tetrad derived from a ctf18Δ/CTF18 heterozygote are shown. The broad band at 1.2 kb [detected by a radiolabeled poly(CA)n/(GT)n probe as in reference 84] contains termini from chromosomes with Y′ elements.
Alterations in telomere length maintenance can be a consequence of either reduced telomerase function or defects in other telomere-associated proteins that control chromosome end protection and/or replication (22). Telomerase null mutants die on extended outgrowth, accompanied by progressive telomere shortening. This phenotype is enhanced in rad52 mutants, which lack a compensatory recombinational pathway that maintains the telomere and hence cell viability (60). Clonal senesence was not observed in ctf18 or ctf18 rad52 mutants after 240 generations of outgrowth (data not shown). Therefore it is likely that the short telomere phenotype is not due to an absence of telomerase. Interestingly, the synthetic dosage lethality screen described above revealed a synthetic interaction with rap1-5 in the presence of excess Ctf18p (Table 2). RAP1 encodes a transcriptional repressor-activator DNA binding protein with effects on both gene expression and telomere length; cells bearing the rap1-5 mutation exhibit a decline in telomere length at semipermissive temperatures (61).
CTF18p homologues are detected in fission yeast and higher eukaryotes.
Although Ctf18p has homology to RF-C subunits, the similarity is concentrated within an approximately 250-amino-acid region containing RF-C homology boxes II through VIII and falls off sharply outside of this region. This is clearly seen in a pairwise-alignment diagram with Rfc1p from budding yeast, the most homologous RF-C subunit (Fig. 8A). Interestingly, Ctf18p is also homologous to predicted C. elegans, D. melanogaster, S. pombe, and H. sapiens proteins. While these predicted proteins have significant homology over six of the eight RF-C boxes, the similarity to Ctf18p extends outward from the central region, indicating that the predicted proteins are more closely related to Ctf18p than to Rfc1p. This is clearly seen in an alignment between S. cerevisiae Ctf18p and a predicted H. sapiens Ctf18p (Fig. 8B). This observation is supported by cluster analysis (Fig. 8C) of a multiple alignment containing the Ctf18p homologs and Rfc1 proteins from S. cerevisiae, C. elegans, D. melanogaster, S. pombe, and H. sapiens. The Rfc1 proteins have been experimentally defined (71, 98) with the exception of the C. elegans predicted Rfc1p. In cluster analysis, the Ctf18p homologs form a group apart from the Rfc1 proteins. Additional work is required to determine if the candidate Ctf18p homologs are involved in sister chromatid segregation during mitosis.
DISCUSSION
In this study, we have identified a role for CTF4 and CTF18 in sister chromatid cohesion. Consistent with a cohesion defect, ctf4 and ctf18 mutants induce the spindle assembly checkpoint. An increased fraction of cellular Ctf4p and Ctf18p appears to associate with chromatin during early S- and early M-phase arrests. ctf18Δ mutants exhibit a chromosome condensation defect, similar to other cohesion mutants tested to date (33, 36, 108). CTF18 is putative paralog of RFC1, and its encoded protein physically interacts with Rfc2p, Rfc3p, Rfc4p, and Rfc5p. However, it is not a component of the canonical PCNA clamp-loading RF-C complex. We and others have observed that CTF4 and CTF18 interact physically and genetically with many genes that function in DNA replication. These results directly address the hypothesis suggested by recent identification of a requirement for budding yeast DNA polymerase κ for the proper establishment of cohesion (108). The requirement for DNA polymerases and accessory factors in cohesion strongly supports the hypothesis that cohesion establishment is a job performed by proteins associated with replication fork complexes.
Checkpoints and cohesion.
The MAD2-dependent preanaphase delay observed in both ctf4 and ctf18 mutants is predicted after cohesion failure, because dissociation of a sister chromatid pair should relieve tension on the kinetochore-microtubule junction, thereby activating the spindle assembly checkpoint. As noted previously (87), cohesion failure is not repaired following activation of the spindle assembly checkpoint but, rather, appears to be augmented during mitotic arrest. In agreement, we have observed a marked reduction in the viability of ctf18 cells after a 3-h checkpoint-induced arrest. While the classic model of checkpoint function emphasizes the opportunity allowed for repair of an inducing lesion, in this case a checkpoint-induced delay favors removal of damaged cells from the dividing population.
Interestingly, Kouprina et al. (53) noted a RAD9-dependent arrest in a ctf18 null mutant at 11°C. In contrast, at the normal growth temperature (30°C) we detected no appreciable RAD9 or MEC1 dependence of the preanaphase delay in ctf18Δ cells; therefore we propose that the 11°C observation reveals an additional physiological consequence. Perhaps at low temperature ctf18Δ cells accumulate defects that are detected by the DNA damage checkpoint. Paradoxically, Kouprina et al. (53) found that ctf18Δ rad9Δ cells exhibit increased (rather than decreased) viability at the nonpermissive temperature. We propose an explanation based on the observed sister chromatid cohesion defect. A delay induced by the RAD9-dependent checkpoint will probably augment the frequency of cohesion failure in ctf18Δ cells, similar to a nocadozole-induced preanaphase delay (Fig. 2B). Thus, removal of the checkpoint may explain the enhanced cell survival.
If checkpoint arrest promotes cell death in cohesion-defective cells, why do viable cohesion mutants exhibit aneuploidy? One potential explanation is that adaptation of the spindle assembly checkpoint allows at least some cells with separated chromosomes to progress through mitosis. Cohesion-defective mutants are frequently isolated in chromosome loss screens. Within a collection of viable chromosome loss mutants encompassing approximately 60 genes (91), alleles of six genes required for cohesion (CTF4, CTF7, CTF18, SMC1, MCD1, and SCC2) have been identified to date. This observation suggests that cohesion defects may underlie a significant proportion of naturally occurring chromosome instability phenotypes.
CTF18 as an RFC1 paralog.
Similarity between Ctf18p and Rfc1p has been noted previously (Cullmann et al. 1995), and BLASTP alignment analyses using the entire predicted yeast proteome indicate that these two proteins exhibit the highest degree of homology to one another (e-value = 3e−15 [Fig. 8]). In a two-hybrid screen, we identified strong interactions of Ctf18p with both Rfc3p and Rfc4p. In vitro immunoprecipitation experiments confirmed the ability of Ctf18p to directly interact with in vitro-generated Rfc3p and Rfc4p. Furthermore, in vivo coimmunoprecipitation studies indicate that Ctf18p interacts with Rfc2p, Rfc3p, Rfc4, and Rfc5p. Independent studies have also identified a complex which contains Ctf18p, Rfc2p, Rfc3, Rfc4p, and Rfc5p protein (M. Mayer and P. Hieter, personal communication). Recent studies indicate that RF-C homology box IV, a highly conserved β-sheet found in both Rfc1p and Ctf18p, may be required to mediate interactions between Rfc1p and PCNA (1). In addition, the region between RF-C homology boxes IV and VIII has been implicated in RF-CRFC1 subunit interactions (5). However, Ctf18p is not detected in a purified, biochemically active RF-C fraction. Together, these observations support the concept of a novel CTF18-containing RF-C like complex (RF-CCTF18) distinct from PCNA-loading RF-CRFC1.
This is not the first reported instance of an alternative RF-C like protein complex. ScRAD24p exhibits limited homology to Rfc1p observed in alignments of multiple family members (62). Rad24p physically associates with Rfc2p, Rfc3p, Rfc4p, and Rfc5p (31, 85). Moreover, RAD24, RFC2, and RFC5 are required for DNA damage checkpoint activity (31, 75, 85), consistent with the formation of a functional RF-CRAD24 protein complex. Based on structural and genetic analyses, it has been proposed that the RF-CRAD24 complex may provide a clamp-loading activity to a PCNA-like trimer (80, 105).
Roles of CTF4 and CTF18 in association with DNA replication proteins.
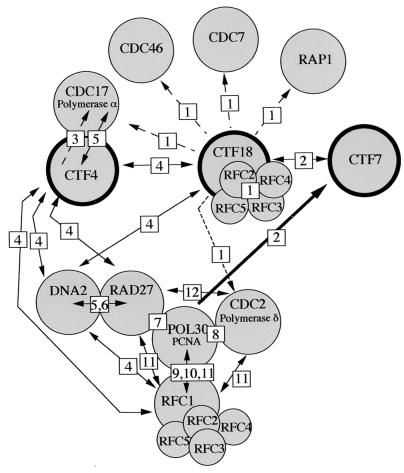
The synthetic dosage lethality results we report here, as well as the physical association of CTF18p with RFC3p and RFC4p, add to the growing list of genetic and physical interactions involving CTF4, CTF18, and components of the replication complex (Fig. 9). These interactions strongly suggest that the molecular functions of CTF4 and CTF18 are closely related to DNA synthesis. They also support a model in which sister chromatid cohesion defects may be among the consequences of mutations in other proteins that function at DNA replication forks. Although previous studies have suggested that Ctf4p and Ctf18p each play roles associated with DNA metabolism, these roles have not been defined. Intriguingly, CTF4 was isolated as a DNA polymerase α (Cdc17p) binding protein, and genetically interacts with with CDC17, RAD27, DNA2, and RFC1 (24). ctf4 mutants exhibited general properties consistent with DNA synthesis defects, including an elevated rate of mitotic recombination and an accumulation of preanaphase cells in log phase (51, 69). However, previous work indicated that the ctf4-induced preanaphase delay was not RAD9 or MEC1 dependent (69), and therefore it was unlikely that compromised DNA replication caused the observed preanaphase delay. The presence of a cohesion defect offers an explanation for the preanaphase delay and suggests a new direction for elucidation of the molecular role of Ctf4p.
FIG. 9.
Physical and genetic interactions among CTF4, CTF18, CTF7, and DNA synthesis proteins. The bold circles denote replication-associated proteins affecting sister chromatid cohesion. Physical associations are indicated by symbol overlap, synthetic lethality is indicated by solid lines with double arrows, and synthetic dosage lethality is indicated by dashed lines with single arrows. The interactions are described in this work [1] and references 87 [2], 112 [3], 24 [4], 69 [5], 11 [6], 56 [7], 4 [8], 28 [9], 67 [10], 114 [11], and 27 [12]. The physical interactions among RF-C subunits are described in references 17, 23, 75, and 104.
Similarly for CTF18, genetic interactions and mutant phenotypes have also supported an uncharacterized role in DNA metabolism. As noted by Kouprina et al. (53), CTF18 exhibited similarity to RF-C subunits and an elevated rate of mitotic recombination. Formosa and Nittis (24) observed traditional synthetic lethality between CTF18 and both DNA2 and CTF4. The synthetic dosage lethality reported here, with temperature-sensitive alleles of the replication genes CDC2, CDC17, CDC46, and CDC7, provide additional indication of a molecular function that influences the DNA synthesis machinery. Physical association with Rfc3p and Rfc4p in vivo and in vitro further supports this concept. However, the new observations of a spindle assembly checkpoint-dependent delay and a sister chromatid cohesion defect provide impetus to the search for a molecular role for Ctf18p in this aspect of chromosome metabolism.
CTF18 and telomere structure.
The short telomere length displayed by ctf18 null mutants, as well as the previously isolated ctf18 alleles, demonstrates that CTF18 is required for normal telomere structure. Since a ctf18 null mutant does not exhibit a telomere replication defect comparable to that of a telomerase null mutant, it is unlikely that this is due to an absence of telomerase. However, proteins important for full telomere replication may play structural rather than enzymatic roles. For example, the Mre11-Xrs2-Rad50 complex, which has been proposed to play a structural role in DNA repair of sister chromatids, may facilitate telomere replication by presenting chromosome ends to telomerase for replication (76). Ctf18p may also make a structural contribution to telomere length maintenance. Specifically, we suggest that loss of cohesion at chromosome ends in a ctf18 mutant decreases the efficiency of complete telomere replication. This hypothesis is attractive in light of the proposal that telomerase functions as a dimer at chromosome termini (79).
Cohesion and replication.
A temporal linkage between DNA synthesis and sister chromatid cohesion is suggested by the tight association between sister chromatids observed from the time of their generation until anaphase (32). Furthermore, the specificity of sister association must be derived from something other than sequence similarity because the efficiency of pairing between homologous chromosomes in a diploid does not indicate a comparable intimate physical relationship (12, 13). The establishment of sister chromatid cohesion as a part of the chromosomal duplication process provides an attractive model that satisfies the specificity requirement, as well as the time of appearance, of the association between sisters.
Indirect evidence for a relationship between DNA synthesis and sister chromatid cohesion is found in several recent studies, indicating that known cohesion proteins possess S-phase roles. S. cerevisiae Mcd1p, Ctf7p, Scc2p, and Scc4p must all be functional during S phase (16, 87, 101, 102), as must fission yeast Mis4p, an Scc2p homologue (26). In addition, S. pombe Eso1p includes extensive homology to both Ctf7p and polymerase η (96) in distinct domains. It has been recently proposed that this type of homology configuration, suggestive of an evolutionary gene fusion or splitting event, may indicate that where the two polypeptides are encoded separately, they nonetheless function together in the cell (21, 63). This view suggests that Ctf7p in S. cerevisiae acts together with a DNA polymerase activity. Finally, the novel polymerase κ protein family in budding yeast not only has a demonstrated requirement in replication of the genome but also plays a poorly understood role in establishing and/or maintaining robust cohesion between sisters (108). The independent observation that both CTF4 and CTF18 possess functions important for sister chromatid cohesion strongly supports the concept that replication forks directly mediate molecular events important for sister chromatid cohesion.
The proposed roles for CTF4 and CTF18 proteins in coupling S-phase DNA replication and cohesion do not preclude potential activities that are also important for cohesion maintenance. Several observations are consistent with roles for each of these proteins in cohesion establishment, such as genetic and physical interaction with replication proteins, the early defect in sister association observed for ctf18Δ cells, and the augmented chromatin association of both Ctf4p and Ctf18p during early S phase suggested by cell fractionation. However, a detailed analysis of rapid-response conditional alleles is required to evaluate functional execution points for these genes. Whether replication-associated proteins will contribute solely to the establishment of robust cohesion or will be important for later steps in the chromosome cycle that support the maintenance of cohesion until anaphase remains to be seen. We note that neither CTF4 nor CTF18 is an essential gene, although cohesion and replication are essential functions. The synthetic lethality observed for double null cells is consistent with each making a complementary but nonoverlapping molecular contribution to the essential function they share. Whether this represents spatial, temporal, or biochemical differentiation awaits further study.
The identification of polymerase κ, a new class of polymerase with a distinct function, is consistent with the previously observed division of labor among other characterized polymerases in budding yeast (reviewed in references 19 and 43). It has been proposed that cohesion may be established by polymerase κ through a mechanism whereby this specialized replication fork variant is required for replication through particular chromosomal regions (108; reviewed in reference 95). In support of this idea, the distribution of cohesion on chromatin is observed to be nonrandom (9, 30, 37, 66, 97), with evident areas of enrichment. Interestingly, recent studies have led to an attractive model for polymerase switching necessary for the transition from the primase function of polymerase α to the highly processive extension functions of polymerases δ and ɛ (106, 117, 118). Under this model, sequential competitions for common binding sites on single-stranded binding protein and the PCNA sliding clamp support the notion that the polymerase switch occurs in two steps: displacement of polymerase α by RF-CRFC1 followed by displacement of RF-CRFC1 by polymerase δ. It is tempting to speculate that RF-CCTF18 may promote a switch from the polymerase δ or ɛ complex to polymerase κ at sites of cohesion, in a role analogous to that of RF-CRFC1. This switch might incorporate chromatin association or activation of cohesion accessory factors such as CTF7 (87, 101) or the SCC2-SCC4 complex (16). We note that CTF18 is not an essential gene, although cohesion and replication are essential functions. Perhaps in a ctf18 null mutant, Rfc1p partially substitutes for Ctf18p, similar to the proposed functional substitutions between Trf4p and Trf5p (15, 108).
Taking a broad view, it is not yet clear whether a cohesion role is specific to polymerase κ or whether the regulation of sister chromatid association is a general job of replication forks. In favor of the general scenario, the tight association between Ctf4p and polymerase α large subunit (69) is consistent with a cohesion role for polymerase α-primase-mediated replication. Interestingly, bovine SMC1 and SMC3 candidate orthologues encode proteins characterized within the recombination protein complex RC-1, which also contains DNA polymerase ɛ (45, 46). Moreover, S. cerevisiae Mcd1p is required for wild-type radiation resistance (38), as is its candidate orthologue Rad21 in S. pombe (7, 8). The involvement of cohesion subunit proteins in DNA recombination and in repair suggests that these processes may incorporate cohesion remodeling activities in vivo. Perhaps the absence of a juxtaposed sister chromatid caused by compromised cohesion may lead to increased utilization of a homolog in recombinational DNA damage repair. This hypothesis may explain increased rates of mitotic recombination in cohesion-deficient mutants. Although not well understood, the association between cohesion and repair-recombination pathway activities strongly suggests a role for sister chromatid cohesion regulation in many aspects of DNA metabolism.
The requirement for CTF4 and CTF18 in cohesion identifies a role for these replication accessory proteins in high-fidelity chromosome segregation. The proposed intimate coupling between DNA synthesis and the establishment of sister chromatid cohesion by one or more DNA polymerase complexes provides a mechanism for the specificity of this tight and essential chromatin association. However, it is not yet clear how many distinct steps or molecular complexes operate in cohesion, which must incorporate sufficient flexibility to support both local and global dynamic chromatin restructuring events, including transcriptional responses, DNA repair, DNA replication, and mitotic chromosome condensation. It is to be anticipated that the protein complexes operating within mechanisms governing the establishment, maintenance, and release of sister chromatid cohesion are just coming to light.
ACKNOWLEDGMENTS
J.H. and E.K. contributed equally to this work.
We thank P. Hieter and M. Mayer for valuable discussion and for sharing unpublished results. We also thank V. Larionov, J. Li, L. Hartwell, A. Straight, A. Murray, S. Gasser, D. Koshland, B. Lavoie, S. Gould, M. Basrai, S. Devine, R. Krishnan, and B. Stillman for sharing reagents and thoughts. We are additionally grateful to R. Skibbens and C. Grieder for comments on the manuscript.
This work was supported by grants from the American Cancer Society and the NIH to F.S.
REFERENCES
- 1.Amin N S, Tuffo K M, Holm C. Dominant mutations in three different subunits of replication factor C suppress replication defects in yeast PCNA. Genetics. 1999;153:1617–1628. doi: 10.1093/genetics/153.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Koonin E V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayyagari R, Impellizzeri K J, Yoder B L, Gary S L, Burgers P M. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckwith W, McAlear M A. Allele-specific interactions between the yeast RFC1 and RFC5 genes suggest a basis for RFC subunit-subunit interactions. Mol Gen Genet. 2000;264:378–391. doi: 10.1007/s004380000339. [DOI] [PubMed] [Google Scholar]
- 6.Bhatt A M, Lister C, Page T, Fransz P, Findlay K, Jones G H, Dickinson H G, Dean C. The DIF1 gene of Arabidopsisis required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesion gene family. Plant J. 1999;19:463–472. doi: 10.1046/j.1365-313x.1999.00548.x. [DOI] [PubMed] [Google Scholar]
- 7.Birkenbihl R P, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombeinvolved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenbihl R P, Subramani S. The rad21 gene product of Schizosaccharomyces pombeis a nuclear, cell cycle-regulated phosphoprotein. J Biol Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- 9.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 10.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiaeS288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Budd M E, Campbell J L. A yeast replicative DNA helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgess S M, Kleckner N. Collisions between yeast chromosomal loci in vivo are governed by three layers of organization. Genes Dev. 1999;13:1871–1883. doi: 10.1101/gad.13.14.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess S M, Kleckner N, Weiner B M. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castano I B, Brzoska P M, Sadoff B U, Chen H, Christman M F. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- 15.Castano I B, Heath-Pagliuso S, Sadoff B U, Fitzhugh D J, Christman M F. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 1996;24:2404–2410. doi: 10.1093/nar/24.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 17.Cullmann G, Fien F, Kobayashi R, Stillman B. Characterization of the fivereplication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwiche N, Freeman L A, Strunnikov A. Characterization of the components of the putative mammalian sister chromatid cohesion complex. Gene. 1999;233:39–47. doi: 10.1016/s0378-1119(99)00160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey M J, O'Donnell M. Mechanisms of DNA replication. Curr Opin Chem Biol. 2000;4:581–586. doi: 10.1016/s1367-5931(00)00134-4. [DOI] [PubMed] [Google Scholar]
- 20.Ehrenhofer-Murray A E, Kamakaka R T, Rine J. A role for the replication proteins PCNA, RF-C, polymerase epsilon and cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics. 1999;153:1171–1182. doi: 10.1093/genetics/153.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright A J, Iliopoulos I, Kyrpides N C, Ouzounis C A. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 22.Evans S K, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J Cell Sci. 2000;113:3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 23.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Formosa T, Nittis T. Dna2 mutants reveal interactions with Dna polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics. 1999;151:1459–1470. doi: 10.1093/genetics/151.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman D B, Hollingsworth N M, Byers B. Insertional mutations in the yeast HOP1 gene: evidence for multimeric assembly in meiosis. Genetics. 1994;136:449–464. doi: 10.1093/genetics/136.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gary R, Park M S, Nolan J P, Cornelius H L, Kozyreva O G, Tran H T, Lobachev K S, Resnick M A, Gordenin D A. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol Cell Biol. 1999;19:5373–1582. doi: 10.1128/mcb.19.8.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerik K J, Gary S L, Burgers P M. Overproduction and affinity purification of Saccharomycesreplication factor C. J Biol Chem. 1997;272:1256–1262. doi: 10.1074/jbc.272.2.1256. [DOI] [PubMed] [Google Scholar]
- 29.Gietz, R. D., and R. A. Woods. Genetic transformation of yeast. BioTechniques, in press. [DOI] [PubMed]
- 30.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 31.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 1999;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 32.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 35.Harris S D, Hamer J E. sepB: an Aspergillus nidulansgene involved in chromosome segregation and the initiation of cytokinesis. EMBO J. 1995;14:5244–5257. doi: 10.1002/j.1460-2075.1995.tb00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He X, Asthana S, Sorger P K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 38.Heo S-J, Tatebayashi K, Kato J, Ikeda H. The RHC21 gene of budding yeast, a homologue of the fission yeast rad21+ gene, is essential for chromosome segregation. Mol Gen Genet. 1998;257:149–156. doi: 10.1007/s004380050634. [DOI] [PubMed] [Google Scholar]
- 39.Hingorani M M, O'Donnell M. Sliding clamps: a (tail)ored fit. Curr Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 40.Hirano T, Mitchison T J. A heterodimeric coiled-coli protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 41.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the DrosophilaBarren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 42.Howell E A, McAlear M A, Rose D, Holm C. CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol Cell Biol. 1994;14:255–267. doi: 10.1128/mcb.14.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubscher U, Nasheuer H, Syvaoja J. Eukaryotic DNA polymerases, a growing family. Trends Biochem Sci. 2000;25:143–147. doi: 10.1016/s0968-0004(99)01523-6. [DOI] [PubMed] [Google Scholar]
- 44.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 45.Jessberger R, Riwar B, Baechtold H, Akhmedov A T. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996a;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- 46.Jessberger R, Chui G, Linn S, Kemper B. Analysis of the mammalian recombination protein complex RC-1. Mutat Res. 1996;350:217–227. doi: 10.1016/0027-5107(95)00106-9. [DOI] [PubMed] [Google Scholar]
- 47.Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 48.Klein F, Laroche T, Cardenas M E, Hofmann J F, Schweizer D, Gasser S M. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 50.Koshland D E, Guacci V. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr Opin Cell Biol. 2000;12:297–301. doi: 10.1016/s0955-0674(00)00092-2. [DOI] [PubMed] [Google Scholar]
- 51.Kouprina N, Kroll E, Bannikov V, Bliskovsky V, Gizatullin R, Kirillov A, Shestopalov B, Zakharyev V, Hieter P, Spencer F, Larionov V. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kouprina N, Tsouladze A, Koryabin M, Hieter P, Spencer F, Larionov V. Identification and genetic mapping of CHL genes controlling mitotic chromosome transmission in yeast. Yeast. 1993;9:11–19. doi: 10.1002/yea.320090103. [DOI] [PubMed] [Google Scholar]
- 53.Kouprina N, Kroll E, Kirillov A, Bannikov V, Zakharyev V, Larionov V. CHL12, a gene essential for the fidelity of chromosome transmission in the yeast Saccharomyces cerevisiae. Genetics. 1994;138:1067–1079. doi: 10.1093/genetics/138.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kroll E S, Hyland K M, Hieter P, Li J J. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J J, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 56.Li X, Li J, Harrington J, Lieber M R, Burgers P M. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 57.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Losada A, Hirano M, Hirano T. Identification of XenopusSMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopusand human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 61.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 62.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 63.Marcotte E M, Pellegrini M, Ng H, Rice D W, Yeates T O, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;280:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 64.Masai H, Sato N, Takeda T, Arai K. CDC7 kinase complex as a molecular switch for DNA replication. Front Biosci. 1999;4:834–840. doi: 10.2741/masai. [DOI] [PubMed] [Google Scholar]
- 65.McAlear M A, Howell E A, Espenshade K K, Holm C. Proliferating cellnuclear antigen (pol30) mutations suppress cdc44 mutations and identify potential regions of interaction between the two encoded proteins. Mol Cell Biol. 1994;14:4390–4397. doi: 10.1128/mcb.14.7.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Megee P C, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- 67.Merrill B J, Holm C. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 69.Miles J, Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol. 1992;12:5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morgan D O. Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- 71.Mossi R, Hubscher U. Clamping down on clamps and clamp loaders—the eukaryotic replication factor C. Eur J Biochem. 1998;254:209–216. [PubMed] [Google Scholar]
- 72.Nasmyth K. Separating sister chromatids. Trends Biochem Sci. 1999;24:98–104. doi: 10.1016/s0968-0004(99)01358-4. [DOI] [PubMed] [Google Scholar]
- 73.Nasmyth K, Peters J M, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- 74.Nigro J M, Sikorski R, Reed S I, Vogelstein B. Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1357–1365. doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Noskov V N, Araki H, Sugino A. The RFC2 gene, encoding the third-largest subunit of the replication factor-C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4914–4923. doi: 10.1128/mcb.18.8.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 77.Orr-Weaver T L. The ties that bind: localization of the sister chromatid cohesin complex on yeast chromosomes. Cell. 1999;99:1–4. doi: 10.1016/s0092-8674(00)80055-0. [DOI] [PubMed] [Google Scholar]
- 78.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 79.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rauen M, Burtelow M A, Default V M, Karnitz L M. The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRAD9. J Biol Chem. 2000;275:29767–29771. doi: 10.1074/jbc.M005782200. [DOI] [PubMed] [Google Scholar]
- 81.Rollins R A, Morcillo P, Dorsett D. Nipped-B, a Drosophilahomologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schmiesing J A, Ball A R, Gregson H C, Alderton J M, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci USA. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seitz L C, Tang K, Cummings W J, Zolan M E. The rad9 gene of Coprinus cinereusencodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics. 1996;142:1105–1117. doi: 10.1093/genetics/142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shampay J, Blackburn E H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between RAD24 and RFC5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85a.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skibbens R V, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 87.Skibbens R V, Corson L B, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitoticchromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Skibbens R V. Holding your own: establishing sister chromatid cohesion. Genome Res. 2000;10:1664–1671. doi: 10.1101/gr.153600. [DOI] [PubMed] [Google Scholar]
- 89.Smith J S, Caputo E, Boeke J D. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors Mol. Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiaeby microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spencer F, Gerring S L, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Straight A F, Belmont A S, Robinett C C, Murray A W. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 93.Sugimoto K, Seiko A, Shimomura T, Matsumoto K. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sumara I, Vorlaufer E, Gieffers C, Peters B H, Peters J-M. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–761. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takahashi K, Yanagida M. Cell cycle. Replication meets cohesion. Science. 2000;289:735–736. doi: 10.1126/science.289.5480.735. [DOI] [PubMed] [Google Scholar]
- 96.Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tanaka T, Cosma M P, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 98.Tanaka T, Tanaka K, Murakami H, Okayama H. Fission yeast cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol Cell Biol. 1999;19:1038–1048. doi: 10.1128/mcb.19.2.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 100.Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey S E, Uhlmann F, Nasmyth K, Yanagida M. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 103.Uhlmann F, Cai J, Gibbs E, O'Donnell M, Hurwitz J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- 104.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 105.Venclovas C, Thelen M P. Structure-based predictions of Rad1, Rad9, Hus1, and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 107.Waizenegger I C, Hauf S, Meinke A, Peters J M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 108.Wang Z, Castano I B, De Las Penas A, Adams C, Christman M F. Pol kappa: A DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- 109.Warren W D, Lin E, Nheu T V, Hime G R, McKay M J. Drad21, a Drosophilarad21 homologue expressed in S-phase cells. Gene. 2000;250:77–84. doi: 10.1016/s0378-1119(00)00166-9. [DOI] [PubMed] [Google Scholar]
- 110.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 111.Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 112.Wittmeyer J, Formosa T. The SaccharomycesDNA polymerase alpha catalytic subunit interacts with CDC68/SPT16, andwith POB3, a protein similar to an HMG-1-like protein. Mol Cell Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiaeform an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- 114.Xie Y, Counter C, Alani E. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics. 1999;151:499–509. doi: 10.1093/genetics/151.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yan H, Gibson S, Tye B K. Mcm2 and Mcm3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991;5:944–957. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- 116.Yanagida M. Cell cycle mechanisms of sister chromatid separation; roles of Cut1/separin and Cut2/securin. Genes Cells. 2000;5:1–8. doi: 10.1046/j.1365-2443.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- 117.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 118.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao X, Muller E G, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]