Dear Editor,
Mycobacterium tuberculosis (Mtb) is the leading cause of acquired immunodeficiency syndrome‐related death, with approximately 208 000 tuberculosis (TB) deaths occurring annually since 2019. 1 Moreover, human immunodeficiency virus (HIV) co‐infection is a major risk factor for progression to active TB disease. 1 The two pathogens act synergistically to accelerate the deterioration of the host immune system, posing a serious threat to public health. Therefore, questions have been raised regarding the phenotype of and changes in immunological function in individuals with HIV‐Mtb coinfection.
HIV infection dysregulates the Mtb‐specific T‐lymphocytic immune response. 2 Moreover, elevated CD4+CD25+FoxP3+ with highly expressed programmed cell death 1 (PD1) has been reported to affect T‐cell functions in the pathogenesis of TB. 3 However, the peripheral whole blood lymphocyte compartment and the possible pathologies of HIV‐Mtb coinfection have not been completely identified. Because of the large phenotypic diversity among lymphocyte populations, characterization of alterations across all immune cell subsets is difficult. Nevertheless, high‐dimensional, flow and mass cytometry, also known as CyTOF, could overcome the shortages and has been used to characterize immune populations in the field of infectious diseases and to predict biomarkers. 4 In this context, we aimed to establish the immune cell landscape of peripheral blood mononuclear cells (PBMCs) in patients with HIV infection alone, Mtb infection alone, HIV‐Mtb coinfection and healthy controls (HCs) (Figure 1A).
FIGURE 1.
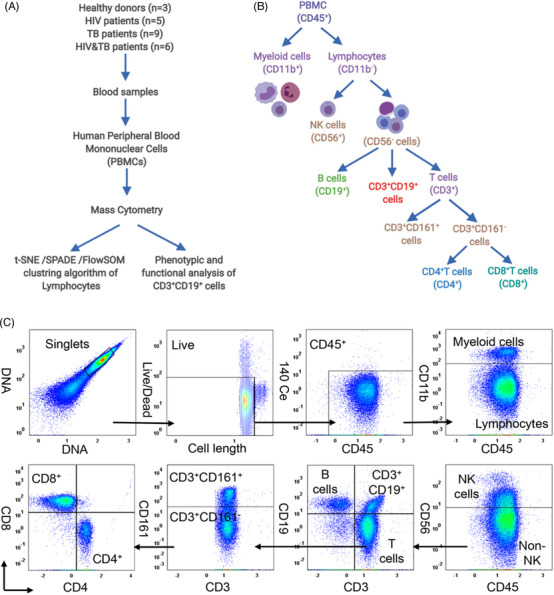
Schematic diagram of the experimental design and the gating strategy of time‐of‐flight cytometry (CyTOF). (A) Human peripheral blood specimens were collected from patients with human immunodeficiency virus (HIV) infection (n = 5), Mycobacterium tuberculosis (Mtb) infection (n = 9), HIV‐Mtb coinfection (n = 6) and healthy controls (HCs) (n = 3) at the Fifth People's Hospital of Wuxi and Shanghai Public Health Clinical Centre in accordance with the Declaration of Helsinki and with patients consent. All patients were aged between 20 and 60 years old. Cells were harvested and measured with the Helios system. Data were normalised in the software CyTOF using EQ Four Element Calibration Beads and analysed on Cytobank (https://www.cytobank.org/). Peripheral blood mononuclear cells (PBMCs) labelled with a metal‐tagged antibodies were divided into different immune populations. (B) A gating strategy was used to differentiate living intact single cells from debris, dead cells and cell aggregates. The immune cell population was gated with respective immune markers
Based on the combination of surface markers, PBMCs were manually gated as myeloid cells (CD45+CD11b+) and lymphocytes (CD45+CD11b–CD3+). Lymphocytes were further divided into nine major populations, including natural killer cells (CD56+), T cells (CD3+), B cells (CD19+), CD3+CD19+ cells, CD161+ cells, CD4+T cells, CD4+CD8+T cells, CD8+T cells and other cells (double‐negative lymphocytes) (Figure 1B,C). All samples were then subjected to viSNE and FlowSOM analysis, 5 , 6 and a new lymphocyte subset, CD3+CD19+ cells, was characterized with high CD45, CD3 and CD19 expression; slight CD11b and CD56 expression; and barely CD4 and CD8 expression (Figure 2A). To our knowledge, CyTOF has not previously been used to identify CD3+CD19+ subsets in a complex disease scenario, including HIV infection, Mtb infection and HIV‐Mtb coinfection. The CD3+CD19+ cell counts were observed markedly reduced in patients with infection compared to those in HCs (Figure 2B,C).
FIGURE 2.
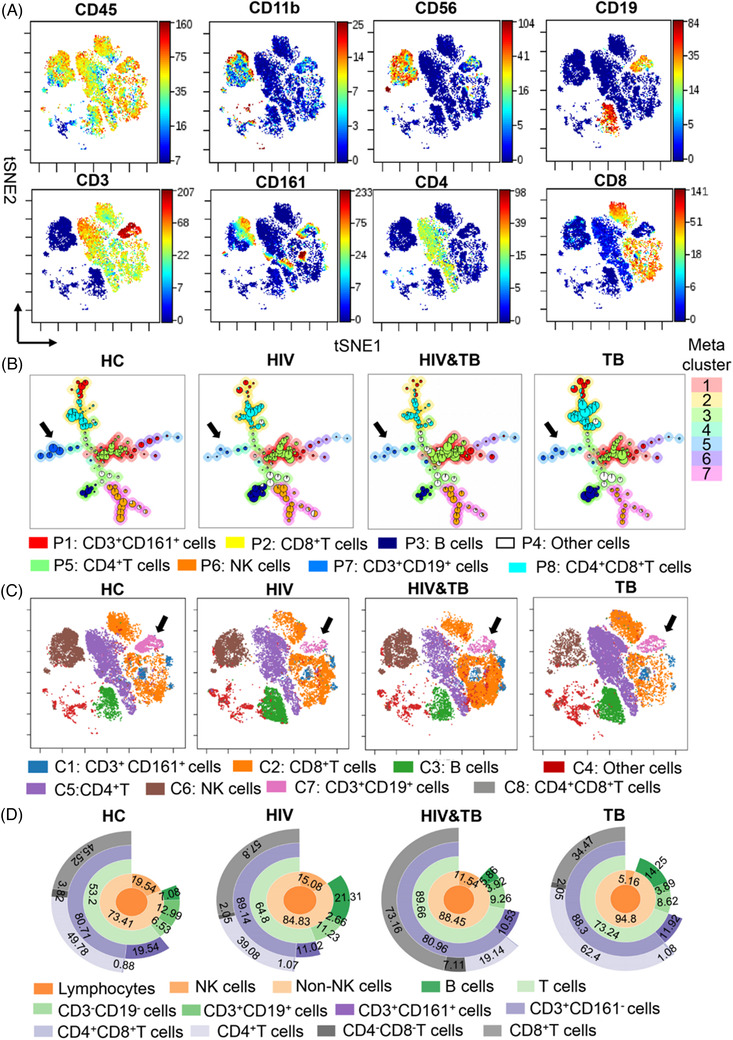
Validation of CD3+CD19+ cells and functional changes among patients with human immunodeficiency virus (HIV), Mycobacterium tuberculosis (Mtb) and HIV‐Mtb coinfection groups compared to healthy controls (HCs). (A) Eight surface markers (CD3, CD4, CD8, CD11b, CD19, CD45, CD56 and CD161) were used to construct a Visne map. (B and C) A FlowSOM and a Visne map of gated CD45+CD11b–CD56–CD3+ live single cells were used to depict the immune landscape. The black arrows in panels (B) and (C) indicate CD3+CD19+ subsets. (D) An abundance of lymphocyte cell subsets was generated using the sunburst method
The abundance of the 12 lymphocyte compartments is shown using a sunburst chart 7 in Figure 2D. The frequency of CD3+CD19+ cells was .20‐, .30‐ and .30‐fold lower in the lymphocytes of individuals with HIV alone, HIV‐Mtb coinfection and Mtb alone, respectively, than in HCs, but was 1.47‐fold higher in HIV‐Mtb co‐infection group than that in the HIV group and 1.01‐fold higher than that in the TB group (Figure 3A). Eight major lymphocyte populations were characterized by using spanning‐tree progression analysis of density‐normalized events (SPADE), 7 and a heatmap was shown to compare the level of expression of 41 immune functional markers in the lymphocyte populations of the three infected groups and HCs (Figure 3B,C). These markers were divided into four groups: (1) cluster differentiation markers that define distinct immune populations; (2) co‐stimulation markers including chemokine receptors, adhesion and activation molecules; (3) inhibitory markers; and (4) transcription factors. Expression of 20 immune functional markers by the novel CD3+CD19+ subset differed from that of cells expressing only CD3 or CD19 in the HCs (Figure 3B).
FIGURE 3.
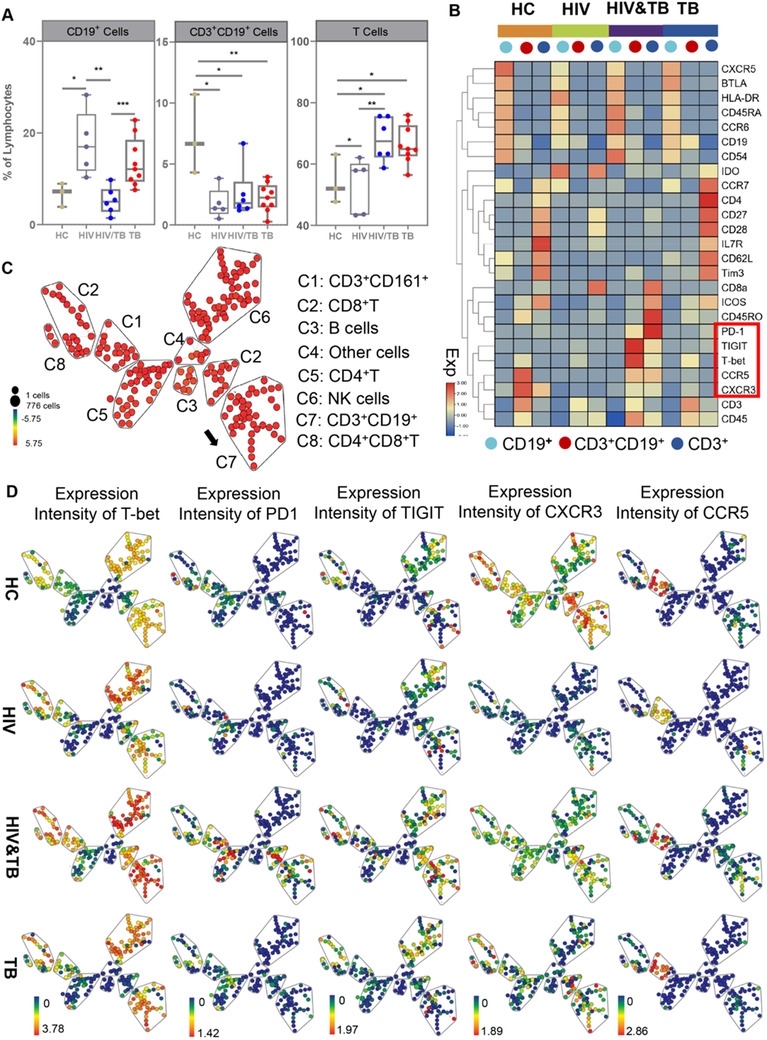
Characterization of the potential role of CD3+CD19+ cells and functional changes among the human immunodeficiency virus (HIV), Mycobacterium tuberculosis (Mtb) and HIV‐Mtb coinfection groups compared to healthy controls (HCs). (A) The frequency of CD19+ cells, CD3+CD19+ cells and CD3+ cells was further analysed using flow cytometry. Differences between each group were analysed using the Mann–Whitney U test. (B) A heat map representing median levels of marker expression for each population (CD3+ cells, CD3+CD19+ cells and CD19+ cells) was created using pheatmap. Differences in the levels of expression of 25 of 41 immune markers in samples from patients with HIV infection, Mtb infection, HIV‐Mtb coinfection and HCs were identified. (C) The SPADE algorithm was applied to gate CD45+CD11b–CD56–CD3+ from live and single cells to reveal the immune profile of eight major cell clusters for each cohort using the same surface markers in Figure 2A. (D) A SPADE plot was exhibited to show the median expression of T‐bet, PD1, TIGIT, CCR5 and CXCR3 in the eight lymphocyte subsets of HCs and patients with HIV infection, Mtb infection or HIV‐Mtb coinfection. Node size indicates the cell count, and colour gradient indicates the median value. Significant differences were indicated by *p < .05, **p < .01, ***p < .001
The cluster differentiation antigen CD45RA, the chemokine receptors CXCR5 and CCR6, the intercellular adhesion molecule CD54, the activation marker major histocompatibility complex, class II, DR (HLA‐DR) and the inhibitory molecule B‐ And T‐Lymphocyte attenuator (BTLA) were highly expressed in CD19+ cells. In addition, with the exception of HLA‐DR and indoleamine‐pyrrole 2,3‐dioxygenase (IDO), immune function markers including CXCR5, BTLA, CD45RA, CCR6 and CD54 were lower in the HIV group than in the other groups. Compared to the other groups, CCR6, CD45RA and CD54 were highly expressed on the CD19+ cells in the HIV‐Mtb co‐infection group. Compared to the HCs and the HIV‐Mtb coinfection group, HLA‐DR, CD45RA, CCR6 and CD54 were slightly expressed in the Mtb only group, but CXCR5 and BTLA were highly expressed in the Mtb group compared to the HIV‐Mtb coinfection group. The chemokine receptor CCR7, the cell adhesion molecule CD62L, the co‐simulation markers CD27, CD28, interleukin‐7 receptor (IL7R), inducible T‐cell costimulator (ICOS) and the co‐inhibitory molecules T cell immunoglobulin and mucin domain‐containing protein 3 (Tim3) were more highly expressed in CD3+ cells than the other two cell types. All of the markers except IL7R and ICOS were highly expressed in the Mtb group (Figure 3B).
Consistent with the decreased frequency of CD3+CD20+ cells in patients with HIV, 8 CD3+CD19+ cells were also less abundant in the Mtb and HIV‐Mtb co‐infection groups than in the HCs. Moreover, a marked variation in the frequency of T and B cells was noted in the three infected groups, while a slight variation in the frequency of CD3+CD19+ cells was noted in the three infected groups. Notably, CCR5 and CXCR3 were highly expressed in CD3+CD19+ cells from HCs but were minimally expressed in the HIV group. 9 CCR5 and CXCR3 expression was also lower in the HIV‐Mtb co‐infection group than in the HCs but was much higher than that in the other two groups (Figure 3D). The inhibitory molecules PD1 and T cell immunoreceptor with Ig and ITIM domains (TIGIT) were highly expressed in the CD3+CD19+ cells of the HIV‐Mtb coinfection group, indicating a phenotype involving greater suppression of the adaptive immune response. Notably, the transcription factor Tbx21 (T‐bet) was highly expressed as a result of stimulation by Mtb antigens, especially in the HIV‐Mtb coinfection group. 10
CONCLUSION
To our knowledge, this is the first study to use mass cytometry to reveal the adaptive immune landscape in samples from patients with HIV infection, Mtb infection and HIV‐Mtb coinfection and to discover a novel immune subset (CD3+CD19+ cells). The comprehensive analysis revealed the complex landscape of host immune response in response to pathogens including HIV, Mtb and HIV‐Mtb co‐infection. Refined analysis indicated that CD3+CD19+ cells in the HIV‐Mtb co‐infection group apparently had deceased expression of CCR5 and CXCR3 and increased expression of the inhibitory receptors PD1 and TIGIT. These findings may help to improve understanding of the immunopathogenesis of TB and HIV, and especially in individuals with HIV‐Mtb coinfection. Detailed study of the CD3+CD19+ subset and exploration of the underlying mechanisms for its involvement in HIV and Mtb infection and HIV‐Mtb coinfection may help to develop precise targeted treatments.
CONFLICT OF INTEREST
The authors declare no competing interests.
ACKNOWLEDGEMENTS
This work was supported by the grants from the Sanming Project of Medicine in Shenzhen (grant number: SZSM201512029); the National Science Foundation of China (grant number: 82072281),the Program to Produce Academic Personnel of the Shanghai 3‐year Action Plan to Create a Public Health System (2020–2022)–Health Education and Health Communication (grant number: GWV‐10.1‐XK02) and the Top Talent Support Program for young and middle‐aged people of Wuxi Health Committee (BJ2020091). The authors wish to thank PLTTECH in Zhejiang Province for technical support and kind assistance with flow cytometry and CyTOF analysis.
Qian Li and Jun Wang contributed equally to this work.
REFERENCES
- 1. World Health Organization . Global tuberculosis report 2020. Accessed August 1, 2021. http://www.who.int/publications/i/item/9789240013131.
- 2. Amelio P, Portevin D, Hella J, et al. HIV infection functionally impairs Mycobacterium tuberculosis‐specific CD4 and CD8 T‐cell responses. J Virol. 2019;93(5):e01728‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angerami MT, Suarez GV, Vecchione MB, et al. Expansion of CD25‐negative forkhead box P3‐positive T cells during HIV and Mycobacterium tuberculosis infection. Front Immunol. 2017;8:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165(4):780‐791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amir Ea, Davis K, Tadmor M, et al. viSNE enables visualization of high dimensional single‐cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gassen SV, Callebaut B, Helden MJ, et al. FlowSOM: using self‐organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87(7):636‐45. [DOI] [PubMed] [Google Scholar]
- 7. Spada F, Fuoco C, Pirrò S, et al. Characterization by mass cytometry of different methods for the preparation of muscle mononuclear cells. N Biotechnol. 2016;33(5 Pt A):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Förster F, Singla A, Arora SK, et al. CD20+T cell numbers are decreased in untreated HIV‐1 patients and recover after HAART. Immunol Lett. 2012;146(1‐2):74‐8. [DOI] [PubMed] [Google Scholar]
- 9. Mackay CR. CXCR3⁺CCR5⁺T cells and autoimmune diseases: guilty as charged? J Clin Invest. 2014;124(9):3682‐3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Austin JW, Buckner CM, Kardava L, et al. Overexpression of T‐bet in HIV infection is associated with accumulation of B cells outside germinal centers and poor affinity maturation. Sci Transl Med. 2019;11(520):eaax0904. [DOI] [PMC free article] [PubMed] [Google Scholar]


