PURPOSE
MET exon 14 skipping alterations (METex14) comprise a diverse set of actionable oncogene drivers in non–small-cell lung cancer (NSCLC). Recent studies have established the efficacy of tyrosine kinase inhibitors for this patient population. The landscape of co-occurring genetic alterations in METex14 NSCLC and their potential impact to therapeutic sensitivities has not yet been fully described.
MATERIALS AND METHODS
METex14 NSCLC cases were collected from three cohorts: the VISION trial, and data sets from Guardant360 and GenePlus. Clinicopathologic characteristics and METex14 mutation sites were analyzed and compared across data sets. Co-occurring genetic alterations and the clonality relationships to METex14 were evaluated.
RESULTS
Of 40,824 NSCLCs, 692 METex14 cases (1.7%) were identified, including 332 in Guardant360, 188 in VISION, and 172 in GenePlus. The demographics and mutation type and/or sites were similar in the Asian versus Western cohorts. MET amplification, which were found to be associated with sensitivity to MET kinase inhibitors, co-occurs in 7.6%-13.8% of cases, whereas kinase domain secondary mutation of MET co-occurs in 5%-6%. When co-occurring with METex14, EGFR mutations were often identified as the dominant clone (78%, 7 of 9), whereas when co-occurring, METex14 (39%, 7 of 18) and KRAS (44%, 8 of 18) had similar rates of clonal dominance. PIK3CA and PTEN mutations were almost always subclones (89%, 16 of 18) to METex14. Moreover, RET-CCDC6 fusion and EGFR mutation were detected following crizotinib treatment in two patients, suggesting novel mechanisms of resistance.
CONCLUSION
METex14 mutations frequently co-occur with other potential driver oncogenes with differing patterns of clonal dominance observed among the drivers. This cellular context can provide insights into whether METex14 is acting as a primary oncogenic driver or resistance mechanism and help guide treatment choices.
BACKGROUND
MET exon 14 skipping alterations (METex14) have recently been established as an actionable oncogene driver in non–small cell lung cancer (NSCLC).1 Small molecule MET tyrosine kinase inhibitors (TKIs) have shown efficacy in patients with METex14 NSCLC, with objective response rate ranging from 25% to 68% and median progression-free survival at 7.6-13.8 months.2-7 From 2020 to 2021, capmatinib and tepotinib received US Food and Drug Administration approval for METex14 NSCLC, representing a significant milestone in MET TKI development.
CONTEXT
Key Objective
To evaluate the mutational profile and co-occurring genetic alteration landscape of non–small-cell lung cancer harboring MET exon 14 skipping (METex14) and the clonality relationship between the METex14 and co-occurring mutations inferred from the variant allele frequency and dissect the potential resistance mechanisms in cases with longitudinal biopsies.
Knowledge Generated
The demographics and mutation type and/or sites of METex14 were similar in the Asian versus Western cohorts. METex14 mutations frequently co-occur with other potential driver oncogenes with differing patterns of clonal dominance observed among the drivers, and usually nondominant subcloned when co-occurred with EGFR and ERBB2.
Relevance
METex14 can act as a primary oncogenic driver or resistance mechanism, suggesting that appropriate treatment choices can be potentially guided by co-occurring alterations.
Obtaining a full understanding of co-occurring alterations with METex14 could be crucial in providing novel insights to increase our understanding of treatment sensitivity and resistance in METex14 NSCLC, and thus, guide future therapeutic strategy development. Acquired MET kinase domain (KD) mutations in residues D1228 and Y1230 have been shown to cause MET TKI resistance.8 Recent studies also indicate that some co-occurring alterations are detected in TP53, RAS-MAPK, and PI3K pathways.9 Gene amplifications (eg, EGFR, MDM2, and CDK4) were also observed in 6%-35% of METex14 NSCLC.10 Some of these genomic alterations have been confirmed as mechanisms of MET TKI resistance, especially RAS-MAPK and PI3K pathways.9,11-14 Furthermore, the efficacy of immunotherapy for patients with METex14 NSCLC was low despite high programmed death-ligand 1 expression.15 However, comprehensive landscape description of co-occurring mutations with METex14 in NSCLC is still missing.
Here, we leveraged three cohorts of METex14 NSCLCs and aimed to evaluate the mutational profile and co-occurring genetic alteration landscape of METex14 NSCLC across countries. We evaluated clonaility relationship between the METex14 and co-occurring mutations inferred from the variant allele frequency (VAF) and dissected the potential resistance mechanisms in cases with longitudinal biopsies.
MATERIALS AND METHODS
Study Population and Platform
Three data sets were queried for METex14 NSCLC: Guardant360 (July 2019 to July 2020), GenePlus (both circulating tumor DNA [ctDNA] and tissue, February 2017 to April 2020), and VISION trial ctDNA cohort (NCT02864992) detected by Guardant360. The Data Supplement shows Guardant360 ctDNA 74 gene and VISION ctDNA 73 gene (without CDK12) panels. The Data Supplement also shows GenePlus 1,021 and 59 gene panels for ctDNA or tissue. The panels used in tissue or blood samples from GenePlus were summarized in the Data Supplement. There were 48 genes covered in all patients across the three data sets (Data Supplement).
METex14 Detection
For Guardant360 (also VISION), single-nucleotide variant or indel variant that overlaps any of the two splice regions of MET exon 14 (chromosome 7:116411902-116412043; human genome [hg19]) defined as eight bp into the intron or three bp into the exon was identified with the Guardant360 assay. Detection of indels larger than 50 bp is described in previous publication.16 For GenePlus, the regions defined as METex14 were the same to Guardant360. Additionally, variants that affect bases as far as 26 bp into the intron were also identified as METex14.
Actionable Mutation Determination
The actionability of each mutation was determined when it was considered as pathogenic by Catalogue Of Somatic Mutations In Cancer (COSMIC) Score.17
Estimation of Mutation Clonality
Variant clonality was determined by normalizing VAF to the maximum somatic VAF in a sample. Variants were classified as clonal if the normalized value was ≥ 0.5, subclonal for values < 0.5 but ≥ 0.05, and subclonal minor if < 0.05.
Statistical Analysis
Group comparisons were performed using a 2-tailed chi-square test, with significance threshold of P value < .05. Analyses were performed using GraphPad Prism 8.0.
RESULTS
Clinicopathologic Characteristics of ctDNA Detected METex14 NSCLC
A total of 692 patients with NSCLC with METex14 were identified from three independent data sets of a combined total of 40,824 patients with NSCLC with an overall incidence of 1.7%, including Guardant360 (332 of 20,987, 1.6%), GenePlus (172 of 14,657, 1.2%), and VISION trial (188 of 5,180, 3.6%). Patient demographics and tumor characteristics were summarized in the Data Supplement. In all three data sets, METex14 occurred with higher frequency in adenocarcinoma, with increasing age and equal sex distribution.
When the GenePlus ctDNA cohort (n = 37) was compared with the Western data sets (Guardant360 plus VISION, n = 520), there were no differences in age (median 70.5 v 73 years) and sex distribution (female 54% v 57%), and similar patient demographic characteristics were noted in both Asian and Western data sets. Prevalence of METex14 was higher in the VISION trial (3.6%) than the other two real-world cohorts (Guardant360 [1.6%] and GenePlus [1.2%]), likely because VISION trial excluded EGFR and ALK-positive patients at initial screening.
Mutation Characteristics of METex14 in NSCLC
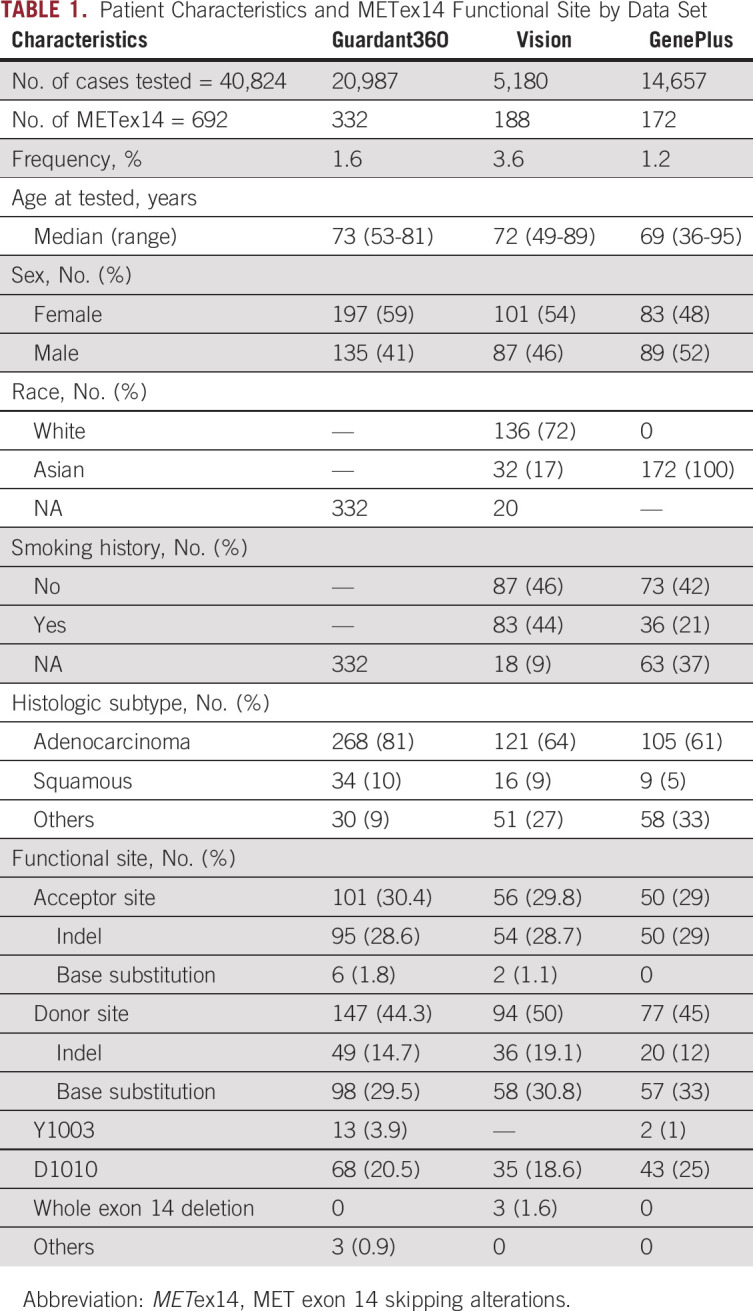
Next, we characterized the mutational landscape of METex14 NSCLC from the three data sets. The positions of MET mutations and the prevalence by functional alterations are shown in Figure 1 and Table 1. The functional sites of MET mutations were similar across the three data sets, allowing the differences among platforms and METex14 detection methods. In Guardant360, the prevalence by functional alteration sites were as follows: donor (44.3%), acceptor (30.4%), D1010 (20.5%), and Y1003 (3.9%). The most frequent mutation type was base substitution (55.7%), followed by indel (43.4%). Both the VISION and the GenePlus data sets revealed a remarkably similar pattern in terms of prevalence by functional alteration sites and most frequent mutation type (Fig 1, Table 1). Regarding acceptor versus donor sites and SNVs versus indels, there was no significant difference between the Asian and Western data sets.
FIG 1.
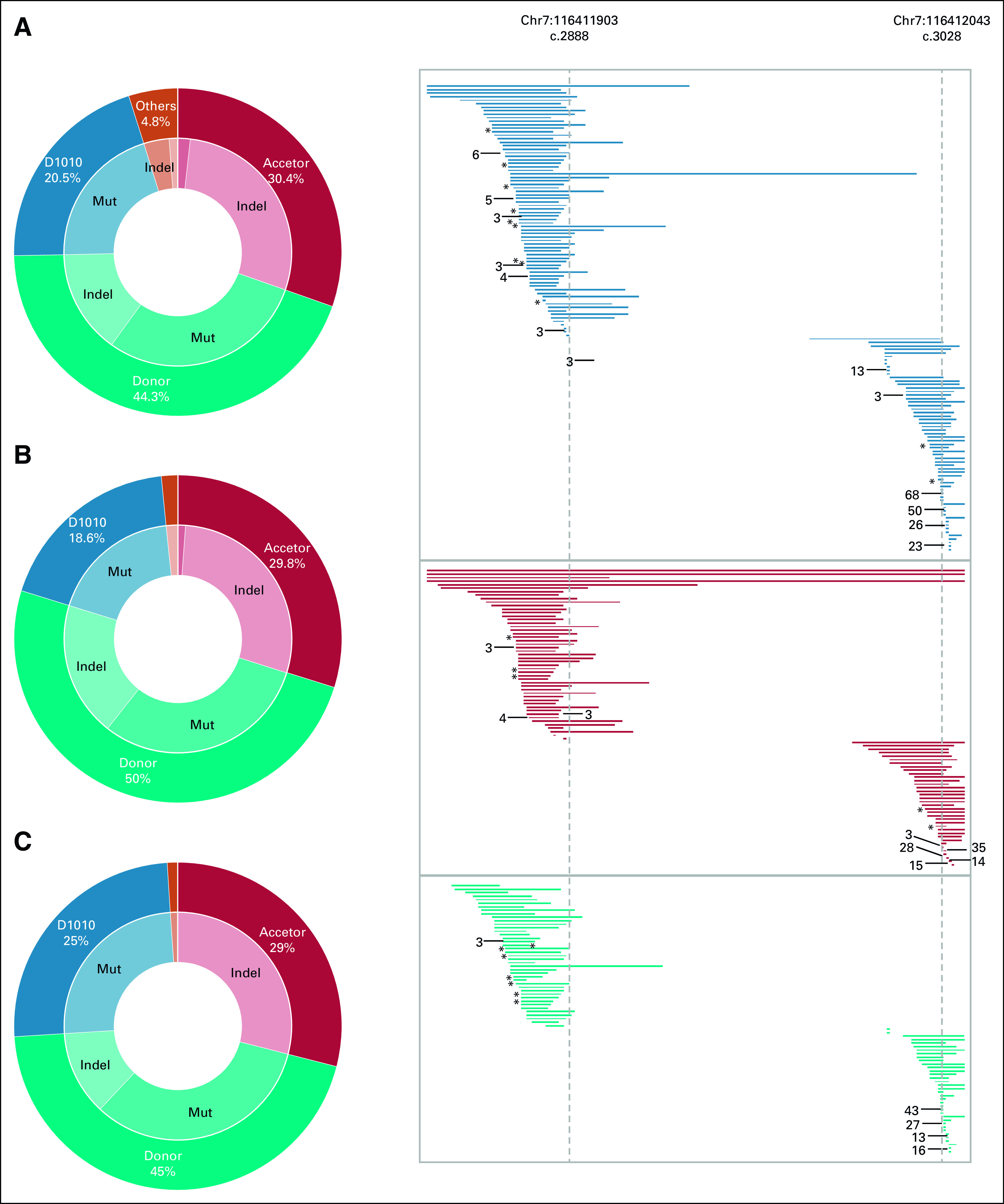
MET exon 14 skipping alterations mutation distribution in the three data sets: (A) Guardant360, (B) VISION, and (C) GenePlus. Genomic positions with alterations occurring in more than one case are indicated with an asterisk (*) for two and the number of cases is greater than two. Mut, mutation.
TABLE 1.
Patient Characteristics and METex14 Functional Site by Data Set
Co-occurring MET Amplification (METamp) With METex14 in NSCLC
The frequency of METamp co-occurred with METex14 was 8.4% (Guardant360), 13.8% (VISION), and 7.6% (GenePlus), respectively (Data Supplement). The mean VAF of METex14 in cases concomitant with METamp was significantly higher than those without METamp across three data sets (P < .001, Data Supplement). The distribution of gene copy number for Guardant360 and GenePlus is displayed in the Data Supplement. Most of the patients had an MET gene copy number between 2 and 4 (24 of 28 in Guardant360; 9 of 13 in GenePlus).
In VISION trial, four in five co-occurring METex14 and METamp cases (80%) had a partial response and one had target lesion tumor reduction, but progressive disease because of a new lesion. The response rate was numerically higher than in patients whose tumor did not have co-occurring METamp (4 of 5, 80% v 28 of 61, 46%), suggesting co-occurring METamp might be associated with responsiveness to targeted therapy,4 although the number in the METamp group was too small for a statistical comparison.
Secondary Mutations Within MET in METex14 NSCLC
Secondary mutations located in MET KDs, such as D1228 and Y1230, have been reported to be associated with resistance to MET TKI.8,11 In Guardant360, 46 (13.9%) had MET secondary mutations including 17 (5%) having at least one secondary mutation in the KDs, including H1094C/Y, D1228H/N, and Y1230C/H (Fig 2B). In GenePlus, secondary mutations were detected in 29 (16.9%) patients, including 11 (6%) in the KDs (Fig 2B). Most of the KD mutations were deemed pathogenic on the basis of COSMIC prediction score > 0.95. The VISION data set only included TKI-naive (part of trial eligibility criteria) patients and had no secondary mutations.
FIG 2.
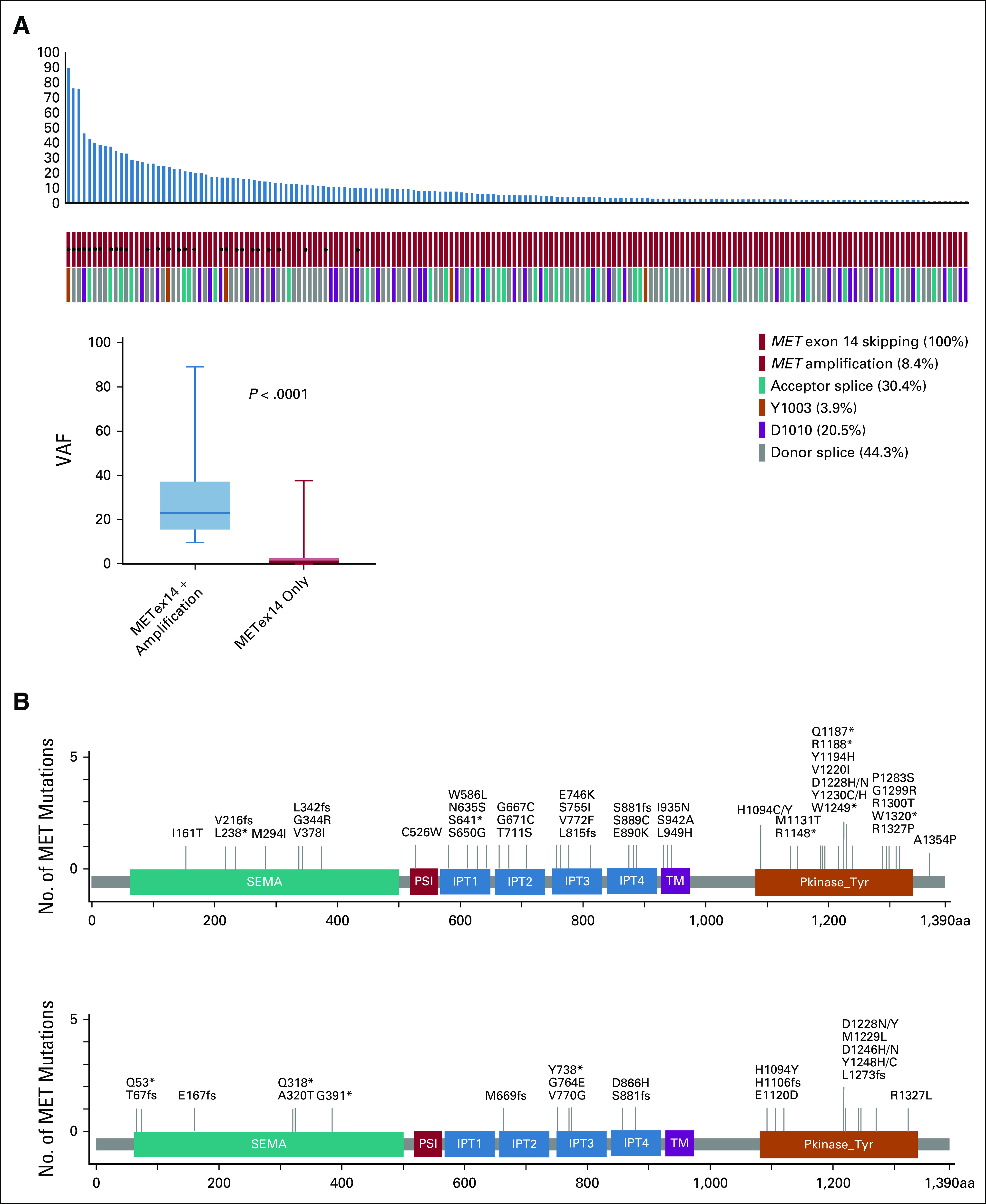
Co-occurring MET amplification and secondary mutations. (A) VAF of METex14 concurrent with MET amplification versus METex14 only in Guardant360. (B) Second site MET mutations in patients with METex14 non–small-cell lung cancer in Guardant360 and GenePlus data sets. IPT, immunoglobulin plexins transcription domains; METex14, MET exon 14 skipping alterations; PSI, plexins-semaphorin-integrin domain; TM, transmembrane domain; VAF, variant allele frequency.
Co-ocurring Genetic Alterations in Bypass Pathways in METex14 NSCLC
Next, we evaluated other co-occurring genentic alterations with METex14 in NSCLC. For a fair comparison, we focused only on the 48 cancer genes tested in all patients across the three data sets. First, we compared whether any comutations were enriched in METex14 NSCLC compared with NSCLC without METex14. In the Guardant360 and GenePlus cohorts, no gene alterations were enriched for co-occurring with METex14. EGFR, KRAS, and TP53 were significantly enriched in the non-METex14 tumors in both cohorts (Figs 3A and 3B), consistent with the notion that METex14 is a de novo oncogenic driver.
FIG 3.
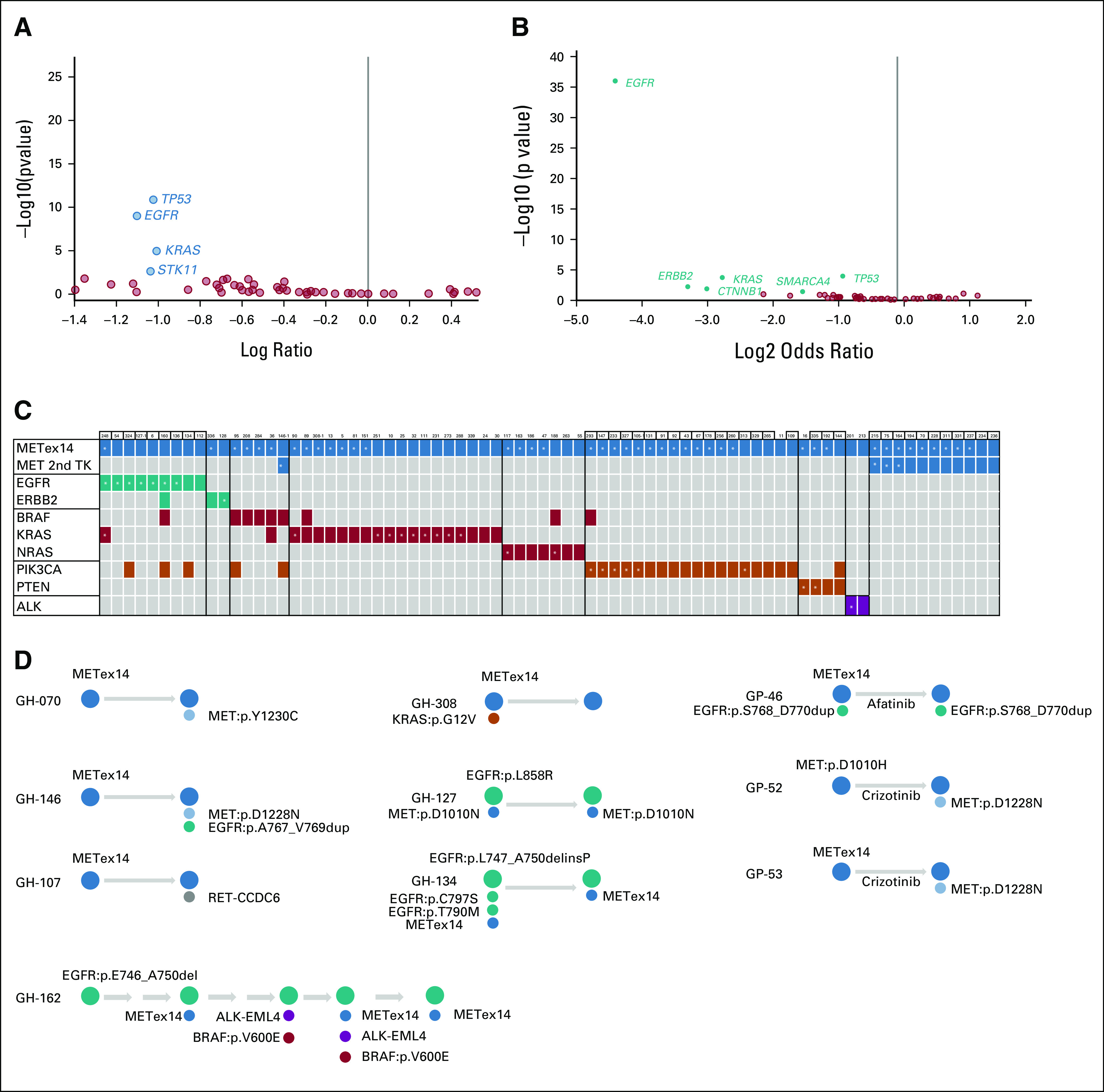
Co-occuring mutations with METex14 and longitudinal ctDNA cases. (A and B) Volcano plots showing the difference of co-occurring alterations between METex14 NSCLC and non-METex14 NSCLC in (A) Guardant360 and (B) GenePlus data sets. (C) Cases with co-occurring alterations with METex14 were shown, and the gene with dominant clone was labeled by star. (D) Genomic alterations of longitudinal cases at different time for patients with METex14 NSCLC in Guardant360 (GH) and GenePlus (GP) data sets. METex14, MET exon 14 skipping alterations; NSCLC, non–small-cell lung cancer; TK, tyrosine kinase domain.
We then focused on comutations in key bypass pathways, including RAS-MAPK, EGFR and ERBB2, PI3K and AKT, and cell cycle (CDK4/6) pathways (Data Supplement). In Guardant360, 136 (41%) had at least one co-occurring alteration in those pathways, including EGFR (24 of 332), ERBB2 (13 of 332); KRAS (20 of 332), BRAF (12 of 332), NRAS (7 of 332), NF1 (3 of 332), and PIK3CA (33 of 332), PTEN (7 of 332), and AKT1 (1 of 332) (Data Supplement). In the VISION trial, EGFR driver mutations were specifically excluded. Fifty-one (27%) patients had at least one co-occurring alteration; ALK and RET fusions, each occured once (Data Supplement). In GenePlus data set, 49 of 172 (28%) METex14 cases had at least one co-ocurring alteration (Data Supplement), with similar distribution to Guardant360 data set.
Clonality Relationship Between METex14 and Other Driver Oncogenes in NSCLC
Oncogene variant clonality can be deduced from VAF to infer dominant versus nondominant clonal relationship. We annotated comutations using COSMIC score to identify the potential activating mutations and found that 76 unique cases (23%) had at least one activating comutation in Guardant360 data set (Fig 3C). We then evaluated the relative clonality inferred by VAF for METex14 and the second activating mutations to dissect potential resistance mechanisms. In the EGFR cases, most cases (7 of 9, 78%) had EGFR as the dominant clone, as was the one case with ERBB2 mutation (A775_G776insYVMA, VAF 31.4%) being dominant compared with 0.56% in METex14 (GH#128, Fig 3C and Data Supplement). ALK fusions also had high VAF in both of two co-occurring cases (GH#201 and GH#213, Fig 3C). For PIK3CA and PTEN mutations, the clonality of METex14 was higher in 16 of 18 (89%) cases. Interestingly, for KRAS and METex14 co-occurring cases, 44% (8 of 18) had KRAS as the dominant clone, and 39% of cases were METex14-dominant. KRAS G12C and BRAF V600E were both detected in one single patient (GH#89, Fig 3C), and METex14 was the dominant mutation.
Eighteen out of 188 (9.6%) patients in the VISION cohort had co-occurring alterations and the dominant clonality of METex14 was observed in 2 of 5 (40%) patients with co-occurring in EGFR and ERBB2, 0 of 1 in ALK (EML4-ALK fusion), 3 of 4 (75%) in K/NRAS, and 7 of 8 (87.5%) in PIK3CA and PTEN (Data Supplement). In GenePlus, similar pattern was seen (Data Supplement). In summary, when co-occurring with EGFR and ERBB2 mutations, METex14 alterations were more frequently observed as a subclone, whereas in the KRAS and BRAF co-occurring cases, KRAS and METex14 had similar frequency of being clonal.
We next explored the potential impact of clonality of METex14 and coalterations on response to MET inhibitors, leveraging the VISION cohort. In the 62 patients with outcome data, 52 had tumors with clonal METex14 and 10 had subclonal. The response rate for clonal group was 46.1% and subclonal group was 50% (Data Supplement), suggesting similar responses to tepotinib regardless of clonality; however, this analysis was underpowered because of the low number in the subclonal cohort.
Of the 18 patients with concomitant alterations, seven had clinical outcome information with tepotinib (Data Supplement). One case with both METex14 and ERBB2 (L796_V797del) had a reduction in tumor size. The METex14 was not the clonal mutation for this patient. No response was seen in the remaining six patients with point mutations in PI3KCA (n = 2), K/NRAS (n = 2), and PTEN (n = 2) at baseline, regardless of the clonality of METex14.
Longitudinal ctDNA Analysis for METex14 NSCLC
In Guardant360, 23 cases had more than one blood sample collected at different time points. Two cases had acquired secondary mutations in the MET KDs (Fig 3D), Y1230C (GH#070) and D1228N (GH#146). In GH#146 (Data Supplement) with METex14 (VAF 9.9%) and an acquired MET D1228N (VAF 1.5%), an EGFR exon 20 insertion (A767_V769dup, VAF 3.9%) and EGFR amplification were also observed, all following therapy with crizotinib, suggesting that alterations in EGFR can mediate polyclonal resistance to MET TKI therapy. Conversely, EGFR driver alterations (defined with high VAF) were detected in 3 cases (GH#127, #134, and #162, Fig 3D), suggesting METex14 could be a resistance mechanism to EGFR TKI, consistent with previous publications.18,19 In GH#162, nine different ctDNA samples was taken over three years, and METex14 was acquired at the fifth ctDNA. EML4-ALK and BRAF V600E were also observed over the course of treatment, suggesting a high heterogeneity of resistance mechanisms. In GH#107, RET-CCDC6 fusion was acquired, following crizotinib and chemoimmunotherapy, with VAF of 1.53% relative to METex14 VAF 16.5%. The emergence of RET-CCDC6 was also confirmed in tumor tissue by FISH assay, appearing only after crizotinib, indicating RET fusion as an acquired potential resistant mechanism to MET TKI.
In the GenePlus data set, six patients had samples collected at different time points. Five were collected before and after crizotinib treatment, and one was taken before and after afatinib. MET D1228N was identified in two post-crizotinib cases (GP#52 and #53, Fig 3D and Data Supplement). In GP# 46, EGFR exon 20 insertion (S768_D770dup, dominant clone), METex14, and EGFR amplification were present before treatment. Following afatinib treatment, EGFR exon 20 and METex14 remained detectable in ctDNA, with loss of EGFR amplification. Taken together, we confirm secondary MET mutations as a consistent resistance mechanism to MET TKI therapy, and provide evidence of novel resistance mechanisms, such as acquired RET fusion or EGFR mutation.
Tissue and Liquid Biopsy Concordance for METex14 NSCLC
Ten cases from GenePlus cohort had both tissue and ctDNA profiling at the same time from the same patients (Data Supplement). METex14 were identified both in tissue and ctDNA at identical functional sites for all, demonstrating perfect concordance. Among them, four had identical comutation results. The remaining six had different co-ocurring genomic alterations in TP53 (ctDNA only), MDM2 amplification (ctDNA only), PTEN mutation (tissue only), and four genomic mutations (EGFR, NF1, TP53, and RB1, tissue only, Data Supplement).
DISCUSSION
In this retrospective multicohort study, we identified 692 cases of METex14 NSCLC, including 557 ctDNA cases, which is, to our knowledge, the largest ctDNA METex14 NSCLC cohort reported to date. Our cohort confirms that patients with METex14 NSCLC are older with equal sex distribution,20-22 with a relatively high incidence in nonadenocarcinoma pathology. We separately confirmed these clinicopathologic features in both the Asian cohort and the predominantly Western data sets. The incidence of METex14 in GenePlus data set was 1.17% in 14,657 cases, similar to prior reports of 0.9% in 1,296 Chinese lung cancer cases,23,24 also similar to the Guardant360 real-world data set with an incidence of 1.6%. Furthermore, the types of METex14 alterations and the location were similar (Table 1), supporting that there are no significant differences between Asian and Western patients with METex14 NSCLC.
Co-occuring genetic alterations with an oncogene driver can associate with clinical response or resistance. We found METamp as the most frequent co-occuring alterations in METex14 NSCLC, at approximately 8%. A number of recent studies have reported on the outcomes of patients with METex14 and METamp NSCLC to MET TKI, including four of the five patients achieving partial response in the VISION study4 and 75%-80% partial response in cohort 4 and 5b in GEOMETRY mono-1 study.3 With the small sample sizes acknowledged, these data indicate that METex14 concurrent with METamp may have higher sensitivity to MET TKI. In our analysis, we found significantly higher VAF for METex14 when METamp is detected. Although it is likely that the high VAF of METex14 with METamp was related to increased copy number, as previously demonstrated in EGFR-mutant NSCLC,25 METex14 and METamp tumors represent a subgroup that are deeply addicted to aberrant MET signaling for tumorigenesis, and thus, a subgroup where the benefit of MET TKI is likely to be most pronounced. By contrast, secondary mutations in the MET KD are resistant mechanisms to MET TKI. In our study, a number of KD mutations, including D1228N in both Guadarnt360 and GenePlus, were detected, at a rate of 5%-6%, representing potential resistance mechanisms.
Using relative VAF to infer clonality, we analyzed other functional oncogenic alterations co-occurring with METex14. In the Guardant360 data set, most of the EGFR (9) and ERBB2 (1) mutations had higher clonality than METex14, indicating that EGFR and ERBB2 mutations were the dominant oncogene drivers, and suggesting that METex14 alterations were the potential drivers of resistance. This is consistent with the established notion that METex14 and METamp are resistance mechanisms to EGFR TKI for EGFR-mutant NSCLC.18,19,26
KRAS-activating mutations were found in only three cases in GenePlus cohort; however, 18 cases were identified in the Guardant data set,10,12,24 consistent with prior reports showing RAS alterations are more common in Western lung cancer populations.27 KRAS mutations and KRAS and BRAF amplification constitute a cause of resistance to MET TKI on the basis of previous clinical and preclinical studies.9,12,28 We found that KRAS mutation and METex14 demonstrate similar tendency to be the dominant clone when co-occurring, as opposed to EGFR and ERBB2, which are usually the dominant driver. Now that both KRAS G12C29 and METex14 have available targeted therapy options, it is of great importance to recognize the dominant clone to prioritize treatment: ultimately, it is likely that dual inhibition of KRAS and MET pathways may be needed for these cases.
Through longitudinal ctDNA analysis, we identified, to our knowledge, the first reported case of acquired RET-CCDC6 fusion co-occurring with METex14 (GH #107; Fig 3C). Acquired RET fusion has been reported in osimertinib-resistant EGFR-mutant NSCLC, where acquired resistance was overcome by EGFR plus RET inhibition.30 Similar to ALK fusion concurrent with METex14, either combination therapy or a multikinase inhibitor would merit investigation as a therapeutic option.
Our study analyzed data sets from various sequencing platforms used in real-world and a clinical trial, which brings strengths as well as weaknesses. The addition of GenePlus cohort allowed a general comparison between Asian and Western populations, but the conclusion is limited by the heterogeneity of laboratory assays and sample source. Furthermore, the comutation and clonality analysis were only comparable across cohorts in the mutually tested 48 genes in all panels. Although they covered key oncogenic pathways in cancer, other genes of potential interest, such as SMAD4 and EZH2, cannot be compared because they were only tested in Guardant360 and GenePlus 1,021 platforms. One important inherited limitation of real-world study is the incomplete data. In our study, for example, we did not have clinical outcome data to different therapies in the Guardant360 and GenePlus cohorts. As such, future studies, both experimental and clinical, are warranted to validate these provocative genomics findings and their clinical implications.
In conclusion, we describe a large cohort of METex14 NSCLC, mostly identified through ctDNA detection. We demonstrate that co-occurring METamp is associated with high METex14 VAF and potential targeted therapy benefit, whereas MET KD secondary mutations are associated with targeted therapy resistance. When METex14 co-occurs with EGFR and ERBB2 mutations, METex14 most commonly serves as a nondominant subclone and is a potential mediator of EGFR TKI resistance. Finally, we reveal emerging novel resistance mechanisms to MET TKI, such as RET fusion, which warrant future translational and therapeutic studies to overcome resistance.
Xiuning Le
Consulting or Advisory Role: AstraZeneca, Lilly, EMD Serono, Spectrum Pharmaceuticals, Daiichi Sankyo/Lilly
Research Funding: Lilly (Inst), Boehringer Ingelheim (Inst)
Chuck Hensel
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Travel, Accommodations, Expenses: Guardant Health
Christine A. Ciunci
Honoraria: Imedex
Research Funding: Celgene (Inst), Merck (Inst), Bristol Myers Squibb (Inst), MacroGenics (Inst)
Stephen V. Liu
Consulting or Advisory Role: Genentech, Pfizer, Lilly, Bristol Myers Squibb, AstraZeneca, Takeda, Regeneron, G1 Therapeutics, Guardant Health, Janssen Oncology, MSD Oncology, Jazz Pharmaceuticals, Blueprint Medicines, Inivata, PharmaMar, Daiichi Sankyo/UCB Japan, BeiGene, Amgen, Turning Point Therapeutics, Elevation Oncology
Research Funding: Genentech/Roche (Inst), Pfizer (Inst), Bayer, Merck (Inst), AstraZeneca (Inst), Blueprint Medicines (Inst), Lilly (Inst), Rain Therapeutics (Inst), Alkermes (Inst), Bristol Myers Squibb (Inst), Turning Point Therapeutics (Inst), RAPT Therapeutics (Inst), Merus (Inst), Elevation Oncology (Inst), Erasca Inc (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Roche/Genentech, MSD Oncology
Marcelo V. Negrao
Consulting or Advisory Role: Mirati Therapeutics
Research Funding: Mirati Therapeutics, AstraZeneca, Novartis, Pfizer, ZIOPHARM Oncology, Checkmate Pharmaceuticals
Jennifer Yen
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Xuefeng Xia
Employment: Geneplus
Leadership: Geneplus
Juergen Scheuenpflug
Employment: Merck KGaA
Leadership: Merck KGaA
Stock and Other Ownership Interests: Merck KGaA
Christopher Stroh
Employment: Merck KGaA
Stock and Other Ownership Interests: Merck KGaA
Patents, Royalties, Other Intellectual Property: Predictive biomarker for anti-EGFR therapy
Dilafruz Juraeva
Employment: Merck
Stock and Other Ownership Interests: Merck KGaA
Anne Tsao
Consulting or Advisory Role: Novartis, Boehringer Ingelheim, Genentech/Roche, Lilly, Bristol Myers Squibb, Epizyme, AstraZeneca/MedImmune, ARIAD, EMD Serono, Takeda, HERON
Research Funding: Merck, Genentech/Roche, Seattle Genetics, Millennium, Bristol Myers Squibb, Boehringer Ingelheim, Polaris, EMD Serono, Takeda
Patents, Royalties, Other Intellectual Property: UptoDate
David Hong
Stock and Other Ownership Interests: OncoResponse, Telperian
Consulting or Advisory Role: Bayer, Guidepoint Global, GLG, Alphasights, Axiom Biotechnologies, Medscape, Numab, Pfizer, Seattle Genetics, Takeda, Trieza Therapeutics, WebMD, Infinity Pharmaceuticals, Amgen, Adaptimmune, Boxer Capital, EcoR1 Capital, Tavistock Life Sciences, Baxter, COG, Genentech, GroupH, Janssen, Acuta, HCW Precision, Prime Oncology, ST Cube, Alkermes, Aumbiosciences, Antheneum, Barclays, Bridgebio, CDR-Life, Cor2Ed, Gilead Sciences, Immunogen, Liberium, Oncologia Brasil, Pharma Intelligence, POET Congress, Turning Point Therapeutics, ZIOPHARM Oncology
Research Funding: Genentech (Inst), Amgen (Inst), Daiichi Sankyo (Inst), Adaptimmune (Inst), AbbVie (Inst), Bayer (Inst), Infinity Pharmaceuticals (Inst), Kite, a Gilead Company (Inst), MedImmune (Inst), National Cancer Institute (Inst), Fate Therapeutics (Inst), Pfizer (Inst), Novartis (Inst), Numab (Inst), Turning Point Therapeutics (Inst), Verastem (Inst), Kyowa (Inst), Loxo (Inst), Merck (Inst), Eisai (Inst), Genmab (Inst), Mirati Therapeutics (Inst), Mologen (Inst), Takeda (Inst), AstraZeneca (Inst), Navire (Inst), VM Pharma (Inst), Erasca Inc (Inst), Bristol Myers Squibb (Inst), Adlai Nortye (Inst), Seagen (Inst), Deciphera (Inst), Pyramid Biosciences (Inst), Lilly (Inst)
Travel, Accommodations, Expenses: Genmab, Society for Immunotherapy of Cancer, Bayer Schering Pharma, ASCO, AACR, Telperian
Victoria Raymond
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health, Trovagene
Paul Paik
Honoraria: Takeda, Calithera Biosciences, Boehringer Ingelheim, EMD Serono, AstraZeneca, Xencor, Bicara Therapeutics, GlaxoSmithKline
Consulting or Advisory Role: EMD Serono, Takeda, Calithera Biosciences, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Xencor, Bicara Therapeutics
Research Funding: EMD Serono, Boehringer Ingelheim, Bicara Therapeutics
Jianjun Zhang
Honoraria: Roche, Sino-USA Biomedical Platform, Geneplus, Origimed, Innovent Biologics, CancerNet, Zhejiang Cancer Hospital
Consulting or Advisory Role: AstraZeneca, Geneplus, Capital Medical University, AstraZeneca, Johnson & Johnson/Janssen, Novartis
Research Funding: Merck, Novartis
Travel, Accommodations, Expenses: Innovent Biologics, Zhejiang Cancer Hospital
John V. Heymach
Stock and Other Ownership Interests: Cardinal Spine, Bio-Tree
Consulting or Advisory Role: AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Lilly, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Catalyst Biotech, Foundation Medicine, Novartis, Mirati Therapeutics, BrightPath Biotheraputics, Janssen, Nexus Health Systems, Pneuma Respiratory, Kairos Ventures, Roche, Leads Biolabs
Research Funding: AstraZeneca (Inst), Spectrum Pharmaceuticals, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Licensing agreement between Spectrum and MD Anderson (including myself) regarding intellectual property for treatment of EGFR and HER2 exon 20 mutations
No other potential conflicts of interest were reported.
SUPPORT
This work was supported by the generous philanthropic contributions to The University of Texas MD Anderson Lung Moon Shot Program and the MD Anderson Cancer Center Support Grant P30 CA016672.
X.L. and L.H. contributed equally to this work. J.Z. and J.V.H. shared the senior authorship.
AUTHOR CONTRIBUTIONS
Conception and design: Xiuning Le, Lingzhi Hong, Chuck Hensel, Rongrong Chen, Christopher Stroh, Anne Tsao, Paul Paik, Jianjun Zhang, John Heymach
Provision of study materials or patients: Christine A. Ciunci, Xuefeng Xia, David Hong, Victoria Raymond, Paul Paik
Collection and assembly of data: Xiuning Le, Lingzhi Hong, Chuck Hensel, Rongrong Chen, Haley Kemp, Niamh Coleman, Stephen V. Liu, Christopher Stroh, David Hong, Victoria Raymond, Paul Paik, Jianjun Zhang, John Heymach
Data analysis and interpretation: Xiuning Le, Lingzhi Hong, Chuck Hensel, Rongrong Chen, Niamh Coleman, Christine A. Ciunci, Stephen V. Liu, Marcelo V. Negrao, Jennifer Yen, Xuefeng Xia, Juergen Scheuenpflug, Christopher Stroh, Dilafruz Juraeva, Anne Tsao, David Hong, Victoria Raymond, Paul Paik, Jianjun Zhang, John Heymach
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Xiuning Le
Consulting or Advisory Role: AstraZeneca, Lilly, EMD Serono, Spectrum Pharmaceuticals, Daiichi Sankyo/Lilly
Research Funding: Lilly (Inst), Boehringer Ingelheim (Inst)
Chuck Hensel
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Travel, Accommodations, Expenses: Guardant Health
Christine A. Ciunci
Honoraria: Imedex
Research Funding: Celgene (Inst), Merck (Inst), Bristol Myers Squibb (Inst), MacroGenics (Inst)
Stephen V. Liu
Consulting or Advisory Role: Genentech, Pfizer, Lilly, Bristol Myers Squibb, AstraZeneca, Takeda, Regeneron, G1 Therapeutics, Guardant Health, Janssen Oncology, MSD Oncology, Jazz Pharmaceuticals, Blueprint Medicines, Inivata, PharmaMar, Daiichi Sankyo/UCB Japan, BeiGene, Amgen, Turning Point Therapeutics, Elevation Oncology
Research Funding: Genentech/Roche (Inst), Pfizer (Inst), Bayer, Merck (Inst), AstraZeneca (Inst), Blueprint Medicines (Inst), Lilly (Inst), Rain Therapeutics (Inst), Alkermes (Inst), Bristol Myers Squibb (Inst), Turning Point Therapeutics (Inst), RAPT Therapeutics (Inst), Merus (Inst), Elevation Oncology (Inst), Erasca Inc (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Roche/Genentech, MSD Oncology
Marcelo V. Negrao
Consulting or Advisory Role: Mirati Therapeutics
Research Funding: Mirati Therapeutics, AstraZeneca, Novartis, Pfizer, ZIOPHARM Oncology, Checkmate Pharmaceuticals
Jennifer Yen
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Xuefeng Xia
Employment: Geneplus
Leadership: Geneplus
Juergen Scheuenpflug
Employment: Merck KGaA
Leadership: Merck KGaA
Stock and Other Ownership Interests: Merck KGaA
Christopher Stroh
Employment: Merck KGaA
Stock and Other Ownership Interests: Merck KGaA
Patents, Royalties, Other Intellectual Property: Predictive biomarker for anti-EGFR therapy
Dilafruz Juraeva
Employment: Merck
Stock and Other Ownership Interests: Merck KGaA
Anne Tsao
Consulting or Advisory Role: Novartis, Boehringer Ingelheim, Genentech/Roche, Lilly, Bristol Myers Squibb, Epizyme, AstraZeneca/MedImmune, ARIAD, EMD Serono, Takeda, HERON
Research Funding: Merck, Genentech/Roche, Seattle Genetics, Millennium, Bristol Myers Squibb, Boehringer Ingelheim, Polaris, EMD Serono, Takeda
Patents, Royalties, Other Intellectual Property: UptoDate
David Hong
Stock and Other Ownership Interests: OncoResponse, Telperian
Consulting or Advisory Role: Bayer, Guidepoint Global, GLG, Alphasights, Axiom Biotechnologies, Medscape, Numab, Pfizer, Seattle Genetics, Takeda, Trieza Therapeutics, WebMD, Infinity Pharmaceuticals, Amgen, Adaptimmune, Boxer Capital, EcoR1 Capital, Tavistock Life Sciences, Baxter, COG, Genentech, GroupH, Janssen, Acuta, HCW Precision, Prime Oncology, ST Cube, Alkermes, Aumbiosciences, Antheneum, Barclays, Bridgebio, CDR-Life, Cor2Ed, Gilead Sciences, Immunogen, Liberium, Oncologia Brasil, Pharma Intelligence, POET Congress, Turning Point Therapeutics, ZIOPHARM Oncology
Research Funding: Genentech (Inst), Amgen (Inst), Daiichi Sankyo (Inst), Adaptimmune (Inst), AbbVie (Inst), Bayer (Inst), Infinity Pharmaceuticals (Inst), Kite, a Gilead Company (Inst), MedImmune (Inst), National Cancer Institute (Inst), Fate Therapeutics (Inst), Pfizer (Inst), Novartis (Inst), Numab (Inst), Turning Point Therapeutics (Inst), Verastem (Inst), Kyowa (Inst), Loxo (Inst), Merck (Inst), Eisai (Inst), Genmab (Inst), Mirati Therapeutics (Inst), Mologen (Inst), Takeda (Inst), AstraZeneca (Inst), Navire (Inst), VM Pharma (Inst), Erasca Inc (Inst), Bristol Myers Squibb (Inst), Adlai Nortye (Inst), Seagen (Inst), Deciphera (Inst), Pyramid Biosciences (Inst), Lilly (Inst)
Travel, Accommodations, Expenses: Genmab, Society for Immunotherapy of Cancer, Bayer Schering Pharma, ASCO, AACR, Telperian
Victoria Raymond
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health, Trovagene
Paul Paik
Honoraria: Takeda, Calithera Biosciences, Boehringer Ingelheim, EMD Serono, AstraZeneca, Xencor, Bicara Therapeutics, GlaxoSmithKline
Consulting or Advisory Role: EMD Serono, Takeda, Calithera Biosciences, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Xencor, Bicara Therapeutics
Research Funding: EMD Serono, Boehringer Ingelheim, Bicara Therapeutics
Jianjun Zhang
Honoraria: Roche, Sino-USA Biomedical Platform, Geneplus, Origimed, Innovent Biologics, CancerNet, Zhejiang Cancer Hospital
Consulting or Advisory Role: AstraZeneca, Geneplus, Capital Medical University, AstraZeneca, Johnson & Johnson/Janssen, Novartis
Research Funding: Merck, Novartis
Travel, Accommodations, Expenses: Innovent Biologics, Zhejiang Cancer Hospital
John V. Heymach
Stock and Other Ownership Interests: Cardinal Spine, Bio-Tree
Consulting or Advisory Role: AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Lilly, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Catalyst Biotech, Foundation Medicine, Novartis, Mirati Therapeutics, BrightPath Biotheraputics, Janssen, Nexus Health Systems, Pneuma Respiratory, Kairos Ventures, Roche, Leads Biolabs
Research Funding: AstraZeneca (Inst), Spectrum Pharmaceuticals, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Licensing agreement between Spectrum and MD Anderson (including myself) regarding intellectual property for treatment of EGFR and HER2 exon 20 mutations
No other potential conflicts of interest were reported.
REFERENCES
- 1.Awad MM, Oxnard GR, Jackman DM, et al. : MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-met overexpression. J Clin Oncol 34:721-730, 2016 [DOI] [PubMed] [Google Scholar]
- 2.Drilon A, Clark JW, Weiss J, et al. : Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med 26:47-51, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf J, Seto T, Han JY, et al. : Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med 383:944-957, 2020 [DOI] [PubMed] [Google Scholar]
- 4.Paik PK, Felip E, Veillon R, et al. : Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N Engl J Med 383:931-943, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S, Fang J, Li X, et al. : Phase II study of savolitinib in patients (pts) with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations (METex14+). J Clin Oncol 38, 2020. (suppl; abstr 9519) [Google Scholar]
- 6.Awad MM, Leonardi GC, Kravets S, et al. : Impact of MET inhibitors on survival among patients with non-small cell lung cancer harboring MET exon 14 mutations: A retrospective analysis. Lung Cancer 133:96-102, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Offin M, Luo J, Guo R, et al. : CNS metastases in patients with MET exon 14-altered lung cancers and outcomes with crizotinib. JCO Precis Oncol 4, 2020. doi:10.1200/PO.20.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujino T, Kobayashi Y, Suda K, et al. : Sensitivity and resistance of MET exon 14 mutations in lung cancer to eight MET tyrosine kinase inhibitors in vitro. J Thorac Oncol 14:1753-1765, 2019 [DOI] [PubMed] [Google Scholar]
- 9.Rotow JK, Gui P, Wu W, et al. : Co-occurring alterations in the RAS-MAPK pathway limit response to MET inhibitor treatment in MET exon 14 skipping mutation-positive lung cancer. Clin Cancer Res 26:439-449, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrock AB, Frampton GM, Suh J, et al. : Characterization of 298 patients with lung cancer harboring MET exon 14 skipping alterations. J Thorac Oncol 11:1493-1502, 2016 [DOI] [PubMed] [Google Scholar]
- 11.Recondo G, Bahcall M, Spurr LF, et al. : Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET exon 14-mutant NSCLC. Clin Cancer Res 26:2615-2625, 2020 [DOI] [PubMed] [Google Scholar]
- 12.Suzawa K, Offin M, Lu D, et al. : Activation of KRAS mediates resistance to targeted therapy in MET exon 14-mutant non-small cell lung cancer. Clin Cancer Res 25:1248-1260, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamme P, Fernandes M, Copin MC, et al. : Alterations in the PI3K pathway drive resistance to MET inhibitors in NSCLC harboring MET exon 14 skipping mutations. J Thorac Oncol 15:741-751, 2020 [DOI] [PubMed] [Google Scholar]
- 14.Jorge SE, Schulman S, Freed JA, et al. : Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with MET amplification or MET exon 14 skipping mutation. Lung Cancer 90:369-374, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabari JK, Leonardi GC, Shu CA, et al. : PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann Oncol 29:2085-2091, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odegaard JI, Vincent JJ, Mortimer S, et al. : Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res 24:3539-3549, 2018 [DOI] [PubMed] [Google Scholar]
- 17.Tate JG, Bamford S, Jubb HC, et al. : COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 47 (D1): D941-D947, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzawa K, Offin M, Schoenfeld AJ, et al. : Acquired MET exon 14 alteration drives secondary resistance to epidermal growth factor receptor tyrosine kinase inhibitor in EGFR-mutated lung cancer. JCO Precis Oncol 3, 2019. doi:10.1200/PO.19.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoenfeld AJ, Chan JM, Kubota D, et al. : Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res 26:2654-2663, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YL, Yuan JQ, Wang KF, et al. : The prevalence of EGFR mutation in patients with non-small cell lung cancer: A systematic review and meta-analysis. Oncotarget 7:78985-78993, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergethon K, Shaw AT, Ou SH, et al. : ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 30:863-870, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw AT, Yeap BY, Mino-Kenudson M, et al. : Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27:4247-4253, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu SY, Gou LY, Li AN, et al. : The unique characteristics of MET exon 14 mutation in Chinese patients with NSCLC. J Thorac Oncol 11:1503-1510, 2016 [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Zhou Z, Lin L, et al. : Characterization of MET exon 14 alteration and association with clinical outcomes of crizotinib in Chinese lung cancers. Lung Cancer 148:113-121, 2020 [DOI] [PubMed] [Google Scholar]
- 25.Lam VK, Zhang J, Wu CC, et al. : Genotype-specific differences in circulating tumor DNA levels in advanced NSCLC. J Thorac Oncol 16:601-609, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le X, Puri S, Negrao MV, et al. : Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res 24:6195-6203, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrot-Zhang J, Chambwe N, Damrauer JS, et al. : Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell 37:639-654.e6, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahcall M, Awad MM, Sholl LM, et al. : Amplification of wild-type KRAS imparts resistance to crizotinib in MET exon 14 mutant non-small cell lung cancer. Clin Cancer Res 24:5963-5976, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong DS, Fakih MG, Strickler JH, et al. : KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med 383:1207-1217, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piotrowska Z, Isozaki H, Lennerz JK, et al. : Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 8:1529-1539, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]


