This study provides proof of concept for cell-based heparin without heparin-induced thrombocytopenia side effects.
Abstract
Heparan sulfate (HS) polysaccharides are master regulators of diverse biological processes via sulfated motifs that can recruit specific proteins. 3-O-sulfation of HS/heparin is crucial for anticoagulant activity, but despite emerging evidence for roles in many other functions, a lack of tools for deciphering structure-function relationships has hampered advances. Here, we describe an approach integrating synthesis of 3-O-sulfated standards, comprehensive HS disaccharide profiling, and cell engineering to address this deficiency. Its application revealed previously unseen differences in 3-O-sulfated profiles of clinical heparins and 3-O-sulfotransferase (HS3ST)–specific variations in cell surface HS profiles. The latter correlated with functional differences in anticoagulant activity and binding to platelet factor 4 (PF4), which underlies heparin-induced thrombocytopenia, a known side effect of heparin. Unexpectedly, cells expressing the HS3ST4 isoenzyme generated HS with potent anticoagulant activity but weak PF4 binding. The data provide new insights into 3-O-sulfate structure-function and demonstrate proof of concept for tailored cell-based synthesis of next-generation heparins.
INTRODUCTION
The heparan sulfate (HS) family of polysaccharides found throughout metazoan lifeforms comprises the most anionic polysaccharides in nature ranging from 20 to 200 monosaccharide units in length, and HS is ubiquitously expressed on cell surfaces and in the extracellular matrix of mammals. HS is essential for life in all mammalian species and modulates numerous biological activities involving growth and development, inflammation and immune system regulation, angiogenesis and metabolism, as well as disease pathologies of cancer, infection, and neurodegenerative disorders (1, 2). The degree and patterns of their sulfation represent huge diversity for informational cues to direct and tightly regulate biological functions. They achieve this through selective interactions with protein partners via divergent sulfated binding motifs that bind to cognate protein-binding sites. HS is produced by a complex biosynthetic machinery that initially creates a repeating disaccharide unit of uronic acid (UA) and N-acetylglucosamine (GlcNAc), where the UA is either iduronic acid (IdoA) or glucuronic acid (GlcA). The glucosamines can be modified with an N-sulfate (NS) or remain as an N-acetyl (NAc) moiety [via action of N-deacetylase/N-sulfotransferases 1 to 4 (NDST1 to NDST4)]. UAs can subsequently be modified with an O-sulfate at the carbon-2 position by 2-O-sulfotransferase (HS2ST1). Further O-sulfates can be added to glucosamine residues at the carbon-6 position [via 6-O-sulfotransferases 1 to 3 (HS6ST1 to HS6ST3)] and more rarely at the carbon-3 position [via 3-O-sulfotransferases 1 to 6 (HS3ST1 to HS3ST6)]. Divergent patterns of sulfation created by the orchestration of these enzyme families are the key hallmarks of functionally specific protein-binding sites in HS. Deciphering the details of these sulfation patterns, including the apparently less common 3-O-sulfate modification, remains a major hurdle.
Heparin, a member of the HS family, is a widely used anticoagulant and is the world’s most sold biopharmaceutical by weight, yet it remains a poorly characterized heterogeneous animal-sourced product (3, 4). Most unfractionated heparin (UFH) is purified from porcine intestinal mucosa, with low–molecular weight heparins (LMWHs) being fractionated from UFH (5, 6). In addition, the supply and quality of heparins are causes for concern due to infection outbreaks in animal stocks, such as the ongoing swine flu in China, and the contamination of crude heparin with oversulfated glycosaminoglycans (GAGs) in 2007 that resulted in many deaths (7, 8).
The mechanism of heparin’s anticoagulant activity involves predominantly heparin binding and activation of antithrombin III (ATIII), which is then able to complex and inactivate thrombin, factor Xa (FXa), and other proteases (9). Heparin binds to ATIII through a specific pentasaccharide sequence, GlcNS6S-GlcA-GlcNS3S6S-IdoA2S-GlcNS6S, whereas the interaction of ATIII and thrombin requires heparin chains of at least 18 monosaccharide units in length (10). In contrast, FXa activity via ATIII activation requires only the pentasaccharide sequence, and a synthetic heparin mimetic (fondaparinux) has been created on the basis of this structure (11). Removal of the 3-O-sulfate group on the 3-O-sulfated glucosamine (GlcNS3S6S) within the pentasaccharide sequence was shown to result in limited ATIII activity (12), demonstrating the essential requirement for 3-O-sulfation for potent anticoagulant activity.
Major complications of heparin and LMWH in clinical use include both bleeding and thrombosis. The structural heterogeneity of heparins provides the avidity to complex with large numbers of proteins including plasma proteins, which can lead to adverse consequences of unpredictable anticoagulation and also life-threatening heparin-induced thrombocytopenia (HIT) (13). HIT can be nonimmune or immune-mediated, both resulting in decreased platelet counts. Platelets produce a protein called platelet factor 4 (PF4; also called CXCL4), which is capable of forming large heparin-PF4 complexes; in immune HIT, antibodies to these complexes are induced and platelets are activated, resulting in the formation of blood clots and low platelet levels (13, 14). Heparin has the highest incidence of HIT at around 5% of patients, whereas LMWH has an incidence of around 1% (15). Heparin/LMWH binding to PF4 has previously been demonstrated to require N-sulfation of the glucosamine (GlcNS) and 2-O-sulfation of the UA (UA2S) (16). This raises the possibility of better targeting of heparin therapeutics with reduced side effects by tailored generation of optimized structures.
Exploiting the evident biomedical potential of heparin and HS requires systematic structure-function relationship studies, which in turn requires detailed structural elucidation. Current methods for disaccharide analysis of HS, most commonly high-performance liquid chromatography (HPLC)–based methods, do not permit complete identification of disaccharide units containing 3-O-sulfation of the glucosamine, primarily due to lack of standards. Although a few isolated 3-O-sulfated disaccharides have been studied (17, 18), many are not available for disaccharide analyses. Digestion of heparins commonly results in digestion-resistant 3-O-sulfated tetrasaccharide structures (19), adding to the difficulties of retrieving the 3-O-sulfated disaccharide standards. The 3-O-sulfation step is catalyzed by seven isoenzymes, the expression of which is spatiotemporally regulated in a broad range of tissues (20–22). HS3ST1/5 are considered to be the main isoenzymes involved in 3-O-sulfation of the anticoagulant drug heparin (produced in mast cells and from which UFH is derived) (23, 24), and a number of isozymes have been implicated in herpes simplex virus entry to cells (24, 25). The HS3STs remain incompletely characterized, despite the fact that the 3-O-sulfate modification is increasingly being linked to much broader biological functions, including tissue formation, brain symmetry, and neuronal growth (2, 20, 22). Silencing of HS3ST expression is observed in cancers (26, 27), and 3-O-sulfation enhances tau phosphorylation and plaque formation in Alzheimer’s disease (28). Despite increasing insights into biological functions, the structural features of HS that drive these biological activities, and crucially the role of 3-O-sulfate groups, remain largely unknown (29) due to the limitations of analytical methods. This represents a major gap in knowledge, which must be closed in order to exploit HS/heparin in new therapeutic applications.
The numerous roles that GAGs play in protein interactions have led to initial efforts to systematically engineer GAG biosynthetic pathways using a combinatorial genetic approach for generating large libraries of individual cells that display different repertoires of HS, chondroitin sulfate (CS), and dermatan sulfate structures (30, 31). The genetic engineering approach has enabled the dissection of protein-binding features, as well as the expression of a large library of homogeneous GAGs with differences in sulfation patterns that can be expressed with and without a proteoglycan core. These can be used for studies on extracellular matrices, mechanical properties, hydrogels, technology platforms, and biotherapeutics. Chemical and chemoenzymatic synthesis of oligosaccharide structures linked to array platforms has also been initiated to elucidate specific binding motifs (17, 32), platforms for GAG sequencing methods (33–38), and development of technology to synthesize fondaparinux in fewer steps (39). These approaches have the potential to contribute to dissecting the binding specificities of HS for a range of HS-binding proteins (30–34), but 3-O-sulfation has not been tackled in a systematic manner to date.
Here, we demonstrate an integrated strategy for dissecting the roles of 3-O-sulfation in HS and heparin by combining an optimized HPLC method that enables comprehensive disaccharide analysis of HS and heparin/LMWH with genetic engineering in cells lacking expression of HS3STs (Fig. 1, A to D). The comprehensive HPLC method was validated with a complete set of chemoenzymatically synthesized 3-O-sulfated disaccharide standards (Fig. 1, A and B) and applied to demonstrate unique 3-O-sulfate profiles of clinical heparin/LMWHs. We further exploited the strategy to explore the functional properties of the HS3ST isoenzyme family by individual targeted knockin (KI) of all seven human HS3STs in Chinese hamster ovary (CHO) cells (Fig. 1D). The resulting HS species displayed divergent 3-O-sulfation and disaccharide compositions, and also modified functional properties (Fig. 1C). We demonstrate that CHO cells expressing HS3ST4 generated HS chains with potent anticoagulant activity similar to some LMWHs, yet very weak ability to bind PF4, indicating potential for reduced HIT. These results provide proof of concept for cell-based biosynthesis of an anticoagulant HS with improved therapeutic properties (Fig. 1E). Furthermore, the integrated strategy provides a new basis for dissecting the distinct sulfated motifs that drive the many biological functions of HS.
Fig. 1. Strategy for dissection of 3-O-sulfation structure-function and engineering of a cell-based HS anticoagulant.
(A) Chemical synthesis is used to generate 3-O-sulfated tetrasaccharides, which are subsequently digested to the 3-O-sulfated disaccharide standards required for complete analysis of HS and heparin/LMWHs. (B) A comprehensive HPLC method is developed for the analysis of HS and heparin/LMWH disaccharide composition including 3-O-sulfation. (C) To generate a cell-based anticoagulant, functional assays for anticoagulant activity are used for comparing HS from genetically engineered cells with clinical heparin/LMWHs. A major side effect of heparin is HIT due to PF4 binding to heparin chains; therefore, PF4 binding of HS/heparin/LMWHs is measured to identify low-binding variants that would not generate this side effect. (D) CHO cells produce CS/dermatan sulfate (DS) GAGs as well as HS, which necessitates laborious and problematic purification for isolation of HS chains. To avoid this, CRISPR-Cas9 can be used to KO genes to ablate CS/DS biosynthesis. Targeted stable KI of genes can be achieved using ZFNs, producing CHO cell lines individually expressing the seven HS3STs, resulting in a cell-based HS3ST library. (E) Heparin/LMWHs are widely used anticoagulant drugs, yet are complex, heterogeneous, and poorly characterized animal products derived predominantly from porcine intestinal mucosa. Cell-based production could offer reduced complexity, and genetic engineering of the heparin/HS biosynthetic pathway in cells can generate a more homogeneous population of HS/heparin chains with desired properties.
RESULTS
Production of 3-O-sulfated disaccharide standards
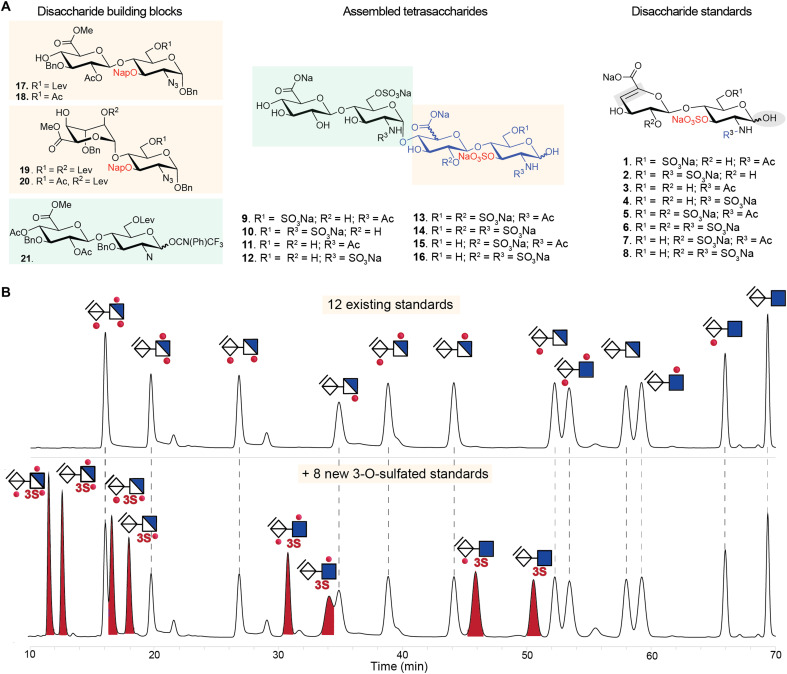
A widely used approach for evaluating structural composition of heparin/HS is profiling of disaccharide building blocks, typically by digestion with a mixture of heparin lyases followed by HPLC-based separation methods with ultraviolet, fluorescence, or mass spectrometry (MS) detection. Yet, no method to date is inclusive of all 3-O-sulfated disaccharides due to the lack of suitable standards. We set out to resolve this longstanding issue by producing 3-O-sulfated tetrasaccharides having different patterns of N-, 2-O-, and 6-O-sulfations (Fig. 2A, fig. S1, table S1, and supplementary experimental methods), which, after exposure to heparinases, were expected to provide the required disaccharide standards 1 to 8 having a 3-O-sulfate moiety. The strategy entailed the chemical synthesis of tetrasaccharides that are modified by protecting groups (9 to 16) that, at a late stage of synthesis, can selectively be removed to reveal alcohols that can then be sulfated. This strategy overcomes the incompatibility of chemical oligosaccharides assembly in the presence of sulfates. Thus, disaccharide donor 21 and disaccharide acceptors 17 to 20 were coupled to give four tetrasaccharides that are modified by different patterns of levulinoyl esters and a napthylmethyl ether. These protecting groups could selectively be removed by treatment with hydrazine acetate and 2,3-dichloro-5,6-dicyano-1,4-benzoquinone, respectively. The resulting alcohols were sulfated by sulfur trioxide-triethylamine complexes (SO3.NEt3). Next, the esters were saponified and the azido moieties were reduced to amines, which were either N-sulfated or N-acetylated and then subjected to hydrogenation to provide the targeted tetrasaccharides 9 to 16.
Fig. 2. Synthetic route to access 3-O-sulfated HS disaccharide standards underpinning a comprehensive HPLC-based disaccharide composition method for heparin/HS.
(A) A modular synthetic approach from common disaccharide building blocks was used to generate tetramers, followed by a sequence of O- and N-sulfation and global deprotection to yield sulfated tetrasaccharides, which, upon lyase digestion, afforded eight 3-O-sulfated authentic disaccharide standards (1 to 8 on the right of the figure). Supplementary experimental methods provide complete details of synthesis and analysis. (B) Upper chromatogram shows C18 HPLC separation of the 12 commonly used commercially available disaccharide standards, while lower chromatogram shows separation of an expanded panel of disaccharide standards including the eight 3-O-sulfated disaccharides synthesized as above in (A). AMAC-labeled disaccharides (20 pmol) were separated in each run using a gradient elution program of 0 to 18 min (18.5% buffer B), 18 to 30 min (18.5 to 19% B), 30 to 57 min (19 to 27% B), and 57 to 70 min (27 to 51% B) at 0.2 ml/min at 30°C and fluorescence detection at 525 nm. Dotted lines indicate elution times for disaccharide standards. Peaks at ~22, ~29, and ~32 min are derived from the ΔUA-GlcNS6S (D0S6), ΔUA2S-GlcNS (D2S0), and ΔUA2S-GlcNAc3S6S (D2A9) disaccharides, respectively.
These tetrasaccharides were subjected to heparinase digestion to provide the eight 3-O-sulfated disaccharides (1 to 8) required for compositional analysis of heparin/HS chains. A challenge, however, was that 3-O-sulfation can cause resistance to heparinase digestion, resulting in undigested tetrasaccharide products from HS/heparin substrates (19). To address this issue, the chemically synthesized tetrasaccharide GlcA-GlcNS6S–GlcA-GlcNS6S3S (G0S6-G0S9) (10, Fig. 2A), a known resistant structure, was used to develop a protocol that permitted heparin lyase conversion into the corresponding disaccharide products (fig. S2). Because heparinases are known to be unstable enzymes prone to loss of activity after freeze-thaw cycles (40), freshly resuspended lyophilized heparinases were used. Enzymatic digestion was carried out under optimized buffer conditions for heparinase activity with prevention of peeling reaction (41) and the stepwise addition of heparinase I (2 hours), heparinase III (2 hours), and heparinase II (2 hours), and complete digestion of tetrasaccharide 10 was accomplished. This protocol was used to digest all eight synthesized tetrasaccharides (fig. S3 and tables S1 and S2), generating four N-acetylated disaccharides [1—(ΔUA-GlcNAc3S6S (D0A9); 3—ΔUA-GlcNAc3S (D0A3); 5—ΔUA2S-GlcNAc3S6S (D2A9); 7—ΔUA2S-GlcNAc3S (D2A3)] and four N-sulfated disaccharides [2—ΔUA-GlcNS3S6S (D0S9); 4—ΔUA-GlcNS3S (D0S3); 6—ΔUA2S-GlcNS3S6S (D2S9); 8—ΔUA2S-GlcNS3S (D2S3)] (Fig. 2A). Note that digestion by heparinases (lyase enzymes) introduces a double bond between carbon atoms 4 and 5 on the GlcA/IdoA residues, resulting in loss of distinction between these ΔUA isomers, as is the case with standard heparinase digestion of heparin/HS for compositional analysis. To improve sensitivity of detection and facilitate isolation of the disaccharide standards, the tetrasaccharides and the digestion products were derivatized with fluorescent 2-aminoacridone (AMAC) via reductive amination. This was followed by purification by reversed-phase HPLC using C18, resulting in eight 3-O-sulfated disaccharide standards, the structures of which were verified by MS and MS/MS (figs. S4 and S5).
HPLC method for comprehensive HS disaccharide analysis
Next, we developed an HPLC protocol that can separate the 12 conventional and the 8 new 3-O-sulfated disaccharide standards expected to cover the complete range of disaccharides found in heparin/HS (Fig. 2B and table S3). C18 chromatography using 150 mM ammonium acetate with 46 mM dibutylamine (pH 5.6) as buffer A and 120 mM ammonium acetate with 36.8 mM dibutylamine and 20% acetonitrile as buffer B provided baseline separation of most standards. Partial overlaps of four disaccharide pairs were noted [ΔUA2S-GlcNS6S (D2S6)/ΔUA2S-GlcNS3S (D2S3), ΔUA-GlcNAc3S6S (D0A9)/ΔUA-GlcNS (D0S0), ΔUA2S-GlcNH2 (D2H0)/ΔUA2S-GlcNAc6S (D2A6), and ΔUA-GlcNH2 (D0H0)/ΔUA-GlcNAc6S (D0A6)], but quantitation could still be achieved.
Compositional HPLC analysis of clinical heparins and LMWHs
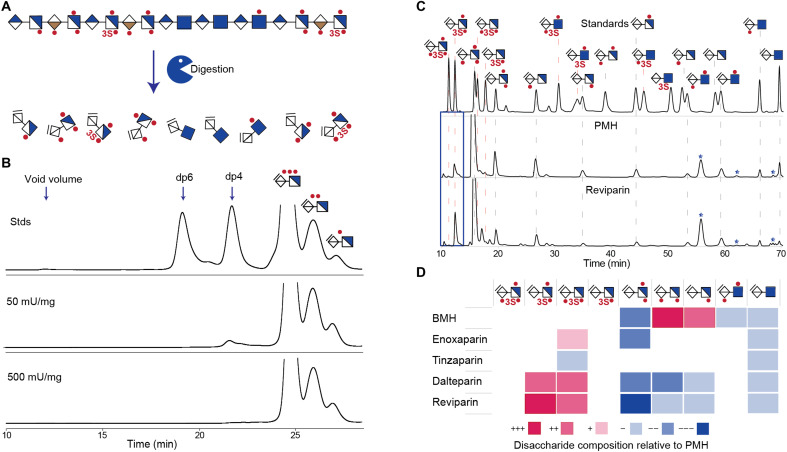
To verify that the optimized heparinase digestion protocol allows complete depolymerization of full-length 3-O-sulfate containing heparin as observed for the chemically synthesized tetrasaccharides (figs. S2 and S3), a dose-dependent heparinase digestion of porcine mucosal heparin (PMH) was evaluated using size exclusion chromatography (SEC) and C18 ion-pairing chromatography (Fig. 3, A and B, and figs. S6 and S7). Complete digestion without trace of resistant tetrasaccharides was achievable at 500 mU of each heparinase per milligram of PMH.
Fig. 3. Comprehensive disaccharide analysis of clinical heparins and LMWHs.
(A) Selective clinical UFHs and LMWHs were digested sequentially using heparinases I, III, and II, resulting in disaccharide products. (B) SEC of heparinase-digested products of PMH using heparinases at PMH concentrations of 50 and 500 mU/mg (elution was monitored at 232 nm). Chromatograms for the complete set of heparinase concentrations used for the experiment can be found in figs. S6 and S7. (C) Fluorescence C18 HPLC disaccharide analysis of PMH and reviparin (an LMWH). Disaccharide products of sequential heparinase digestion were labeled with AMAC before analysis and were compared with 20 pmol standards. Dotted lines indicate elution times for authentic disaccharide standards (red dotted lines, 3-O-sulfated disaccharides; truncated lines, disaccharides not found in the samples). The ΔUA2S-GlcNS3S (D2S3) and ΔUA-GlcNS3S (D0S3) disaccharides partially coelute with the highly abundant ΔUA2S-GlcNS6S (D2S6), reducing certainty of quantification for these disaccharides. Stars in the chromatograms indicate peaks that stem from the heparinases. Chromatograms for all analyzed heparins/LMWHs are provided in fig. S8. (D) Heat map of compositional differences in 3-O-sulfated disaccharide products from digested heparin/LMWHs compared to PMH. The heat map was constructed by quantifying the molar amounts of each disaccharide in a sample as percent of the total disaccharides and comparing this to the amounts found in the sample from PMH. The color coding illustrates increase/decrease observed in percentile ranges as indicated: +++, >5% increase; ++, 2.5 to 5% increase; +, 1 to 2.5% increase; −, 1 to 2.5% decrease; −−, 2.5 to 5% decrease; −−−, >5% decrease.
The optimized digestion protocol and HPLC method were used to analyze clinical-grade heparins (UFHs/LMWHs) (table S4). This is important because of their known heterogeneity, relatively incomplete characterization, and the pressing need for better quality control. Notably, distinct disaccharide profiles for each were observed (Fig. 3, C and D, and fig. S8). The UFHs [PMH and bovine mucosal heparin (BMH)] contain four 3-O-sulfated structures [ΔUA2S-GlcNS3S6S (D2S9)/ΔUA-GlcNS3S6S (D0S9)/ΔUA2S-GlcNS3S (D2S3)/ΔUA-GlcNS3S (D0S3)]. The main disaccharide for both was ΔUA2S-GlcNS6S (D2S6), while BMH was particularly rich in ΔUA2S-GlcNS (D2S0). We tested four PMH-derived LMWHs including enoxaparin (fractionated after benzoylation and alkaline hydrolysis), tinzaparin (produced by enzymatic digestion), and dalteparin and reviparin (produced by partial nitrous acid depolymerization) (42). Enoxaparin and tinzaparin had disaccharide profiles similar to PMH, with the main difference being tinzaparin having lower amounts of ΔUA2S-GlcNS3S (D2S3) (Fig. 3, C and D, and fig. S8). Dalteparin and reviparin both demonstrated larger peaks for the ΔUA-GlcNS3S6S (D0S9) disaccharide. BMH was found to contain slightly more ΔUA2S-GlcNS3S6S (D2S9) than the other heparins, indicating that this structure may not be as prevalent in porcine-derived heparins. None of the 3-O-sulfated N-acetylated structures were observed.
Next, we analyzed the anticoagulant activity of the clinical heparins by the commonly used anti-FXa assay, in which the ability to induce a conformational change in ATIII is measured via inhibition of cleavage of a FXa substrate (fig. S9A). As expected, the UFHs demonstrated a higher level of anticoagulant activity than the LMWHs. Next, we tested the propensity of the panel of heparins to bind the HIT-inducing agent PF4 (fig. S9B). LMWHs have reduced HIT side effects compared to UFHs due to their shorter chain lengths (43). All UFHs and LMWHs demonstrated PF4 binding, with UFHs demonstrating the strongest binding. An exception was the LMWH tinzaparin, which showed binding similar to BMH.
HS3ST-dependent alterations in HS profiles
It is very noteworthy that the apparently rare but functionally important step of the addition of 3-O-sulfate group to a HS chain is catalyzed by seven isoenzymes. HS3ST2/4/3A/3B/6 share a high sequence similarity in humans with ~70% sequence overlap, where HS3ST3A and HS3ST3B share identical catalytic domains, and HS3ST1 and HS3ST5 are less similar to the other HS3STs (fig. S10A) (20–22, 24, 44). As a further step toward systematic unraveling of the importance of 3-O-sulfation for the structure and bioactivity of HS, we used a genetic engineering approach to individually KI all seven human HS3STs into CHO cells (Fig. 4A and tables S5 to S11). CHO cells were selected as they are devoid of background HS3ST expression (45, 46). Furthermore, CHO cells express both HS and CS, and to avoid the presence of CS as a contaminating GAG, we used a genetically engineered cell line with knockout (KO) of CSGalNAcT1/CSGalNAcT2/Chsy1 (designated as CHO KO CS), where KI of the seven human HS3STs was performed by site-directed zinc finger nuclease (ZFN) gene KI (Fig. 4A). Expression of the HS3ST enzymes was confirmed by immunocytochemistry and sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) Western blot analysis (figs. S10, B and C, and S11). HS3ST1 lacks an apparent transmembrane domain and was predominantly detected as a secreted protein in the culture medium (figs. S10 and S11). This may question the role of this isoenzyme in intracellular sulfation of HS. It is presently unclear what the biological roles of secreted sulfotransferases and glycosyltransferases are given that these enzymes require high concentrations of donor substrates provided in the Golgi. Expression of secreted glycosyltransferases has, however, demonstrated variable efficiency in intracellular glycosylation (47, 48). It should be noted that replacing the signal peptide in HS3ST1 with a Golgi targeting transmembrane domain results in increased FXa activity of cellular HS (44), indicating that HS3ST1 at least primarily functions intracellularly. HS3ST2/6 was observed only in the cell lysate, and HS3ST5/4/3A/3B was observed in both lysate and culture medium, potentially indicating extracellular activities of these enzymes. The presence of sulfotransferases in the medium may be due to proteolytic cleavage of the transmembrane domains, which has previously been shown to result in secretion of HS3STs, NDSTs, and HS6STs (49, 50).
Fig. 4. Comprehensive analysis of HS from CHO cells expressing the human HS3ST family.
(A) Genetic engineering strategy for dissecting effects of specific HS3ST isoenzyme expression in CHO cells. (B) HS extracted from CHO KO CS and HS3ST4 KI cells was digested, and compositional disaccharide analysis was performed (a complete set of chromatograms can be found in fig. S12). Stars indicate heparinase enzyme peaks, and dotted lines indicate elution times for disaccharide standards. (C) Heat map analysis of the disaccharide products from the HS3ST-expressing cells. The heat map was constructed by quantifying the molar amounts of each disaccharide in a sample as percent of the total disaccharides and comparing this to the amounts found in the sample from the parental cell line. The color coding illustrates increase/decrease observed in percentile ranges as indicated: +++, >10% increase; ++, 5 to 10% increase; +, 1 to 5% increase; −, 1 to 5% decrease; −−, 5 to 10% decrease; −−−, >10% decrease. (D) FXa assay determining the anticoagulant activity of HS from the genetically engineered cells and select clinical heparin/LMWHs. The y axis shows absorbance at 405 nm, which is indicative of the amount of substrate cleaved by FXa for different concentrations of heparins/HS. All experiments were performed with duplicates and repeated a minimum of three times. (E) Biolayer interferometry assay for determining PF4 binding. The y axis shows the PF4-binding signal (nanometer) at two doses (125 and 500 nM). All experiments were performed a minimum of two times.
Our genetically engineered HS3ST-expressing CHO cells and the newly developed HS compositional analysis approach were combined to study the properties of the seven HS3STs by analyzing the HS produced in these cells at the disaccharide level (Fig. 4, B and C, and fig. S12). HPLC disaccharide analysis revealed distinct differences for most KI clones compared to the parent CHO cell. HS3ST1 primarily introduced ΔUA-GlcNS3S (D0S3), suggesting that ΔUA-GlcNS (D0S0) is the preferred substrate. In contrast, the related HS3ST5 (51, 52) mainly introduced ΔUA2S-GlcNS3S (D2S3) and ΔUA2S-GlcNS3S6S (D2S9). HS3ST2 displayed a preference for 2-O-sulfated epitopes as ΔUA2S-GlcNS3S (D2S3) was the main 3-O-sulfated disaccharide; however, note that HS3ST2 generated minimal ΔUA-GlcNS3S (D0S3) units. HS3ST4 displayed high levels of all four 3-O- and N-sulfated disaccharides, indicating that the enzyme is active on a wider range of substrates. As expected, HS3ST3A/3B showed similar disaccharide profiles, and in this case, ΔUA2S-GlcNS3S (D2S3) and ΔUA-GlcNS3S (D0S3) were the predominant 3-O-sulfated disaccharides. CHO cell HS from HS3ST6 showed a disaccharide profile very similar to the parental clone with minute levels of 3-O-sulfation detected. Although CHO cells are believed to lack expression of HS3STs, we unexpectedly noticed a peak corresponding to ΔUA-GlcNS3S6S (D0S9) in the digested HS from the parental CHO cell. To investigate this peak, we performed disaccharide profiling of a CHO cell with KO of all 6-O-sulfotransferases (CHO KO Hs6st1/2/3), which should thus be incapable of producing ΔUA-GlcNS3S6S (D0S9) (fig. S13). Because a peak corresponding to ΔUA-GlcNS3S6S (D0S9) was not observed for CHO KO Hs6st1/2/3, we suggest that, in CHO KO CS, the ΔUA-GlcNS3S6S (D0S9) disaccharide is not an artifact, and that CHO cells are capable of producing a minor amount of endogenous 3-O-sulfation through an undetermined mechanism. Together, the analysis of the HS3STs indicates that they all have preferences for N-sulfated substrates, and that HS3ST5/2 have a strong preference for 2-O-sulfated substrates, while HS3ST3A/3B/4 have more promiscuous substrate specificity for N/2-O/6-O-sulfated substrates.
Divergent bioactivities of HS from HS3ST-expressing cells
To relate different 3-O-sulfated HS structures derived by the action of the HS3ST family to resulting bioactivities, and to also exploit this approach for initial development of a cell-based heparin with low PF4 binding (Fig. 1E), we tested our HS3ST library of CHO cells for anticoagulant activity and PF4 binding. We first performed flow cytometry assays probing ATIII binding to the HS3ST-expressing CHO cells where HS3ST1/5/3A/3B demonstrated similar binding, while HS3ST4 demonstrated the highest binding, HS3ST2 demonstrated low ATIII binding, and HS3ST6 demonstrated no binding (fig. S14). Treatment with heparinases abolished ATIII binding to all cells. Next, we analyzed the anticoagulant activity of HS isolated from the engineered CHO cells and compared these to clinical heparin/LMWHs using anti-FXa assays (Fig. 4D and fig. S15A). HS from CHO KO CS demonstrated no measurable anticoagulant activity, while CHO cell HS produced by HS3ST1 showed anticoagulant activity in agreement with previous studies (44), further providing support for the prediction that HS3ST1 functions intracellularly in the HS biosynthesis despite being primarily secreted. CHO cell HS from HS3ST5/4/3A/3B demonstrated even higher levels of activity than HS3ST1, while CHO cell HS from HS3ST2 had comparatively low levels and HS3ST6 demonstrated no anticoagulant activity. To enable comparison of anticoagulant activity between the cellular-derived HS and heparins, we calculated half maximal inhibitory concentration (IC50) values from the FXa data, where lower IC5O values indicate more potent activity (fig. S16). Remarkably, the FXa activities of some of the CHO cell–derived HS were comparable to the activities found for some of the LMWHs. Thus, HS produced by CHO cells with KI of HS3ST4 exhibit almost 80% of the activity found for the LMWH reviparin. In clinical practice, the low IC50 of heparin is not necessary to achieve the anticoagulant effect, as LMWHs are efficient in the treatment of thrombosis and are clinically more commonly used than heparin. Comparing the results of ATIII binding and FXa assays with disaccharide analysis, it emerged that the inability of HS3ST2/6 to produce ΔUA-GlcNS3S (D0S3) may have had an impact on their ability to produce anticoagulant structures, which was an unexpected finding. HS from CHO cells with KI of N-sulfation (CHO KI NDST2) and 6-O-sulfation (CHO KI HS6ST1) (30) did not demonstrate anticoagulant activity, emphasizing the importance of 3-O-sulfation over N-/6-O-sulfation.
HIT is a potential life-threatening, immune-mediated adverse drug reaction to heparin due to the formation of PF4-heparin complexes. For the generation of cell-based HS as an alternative to animal heparins, we thus explored the PF4 binding of HS from the HS3ST1 to HS3ST6 KI cell lines (Fig. 4E and fig. S15B). CHO cell HS from HS3ST4-expressing cells demonstrated particularly low PF4 binding, despite exhibiting the highest anticoagulant activity, meaning that this isoenzyme is a potential candidate to use for bioengineering heparin with reduced potential to cause HIT. CHO cell HS from HS3ST5/3B also demonstrated comparatively low binding to PF4, while CHO cell HS from HS3ST1/2/3A/6 exhibited increased binding compared to CHO KO CS. HS from CHO cells with KI of HS6ST1 showed the strongest PF4 binding of all cellular HS, indicating that not only N-/2-O-sulfation but also 6-O-sulfation could be important for PF4-heparin complex formation. These data demonstrate the ability of the cell-based strategy to both identify and optimize HS bioactivities associated with distinct biosynthetic enzyme combinations.
DISCUSSION
The integrated strategies described here, incorporating an HPLC method for comprehensive disaccharide analysis, address a longstanding challenge in analysis of 3-O-sulfation in both HS and heparin. A comprehensive panel of 20 HS disaccharide standards, including eight 3-O-sulfated structures derived from synthetic compounds, made it possible to determine compositional differences in 3-O-sulfated disaccharides in both clinical heparins and HS derived from engineered CHO cells. The data provide insights into distinct action patterns of the seven human HS3ST isoenzymes. Our studies revealed that ΔUA-GlcNS3S (D0S3) is an important feature for anticoagulant activity, and 6-O-sulfation is important for PF4 binding. In addition, we identified that CHO cell HS from cells expressing HS3ST4 may be the optimal choice for cellular production of a heparin-like HS with high anticoagulant activity and low PF4 binding. This is a notable finding that could permit alleviation of the side effects of HIT often experienced with current animal-sourced heparins (Fig. 1E).
Although the critical role of 3-O-sulfation for anticoagulant activity of heparin was discovered in the 1980s, the importance of studying 3-O-sulfation has more recently emerged in the context of a much wider range of biological processes (2). To date, over 400 heparin/HS-binding proteins have been identified (53), yet only a handful of 3-O-sulfated structures have been identified or characterized (54). The HS3ST family has been most thoroughly studied at an organism level in zebrafish, where hs3st2 and hs3st4 were found to regulate brain symmetry and neuron growth, hs3st5 and hs3st6 play roles in cilia length and movement, and hs3st7 plays a role in cardiac contractile apparatus. In zebrafish, hs3st2 and hs3st3 generate the epitope for herpes simplex virus–1 (HSV-1) viral entry, and hs3st2 drives the epitope for tau plaques observed in Alzheimer’s disease (20). In mice, flies, rats, and worms, the importance of HS3STs in organ development and inflammatory processes is well documented (2). The strategy developed here now allows dissection of the structural features involved in 3-O-sulfated HS binding to proteins in these processes.
Heparin remains one of few pharmaceuticals still isolated from animal tissues without thorough structural characterization (4, 55). The new compositional analysis method described here, which includes all major 3-O-sulfate–containing disaccharides, will facilitate improved quality control of clinical heparins. It will make it possible to better understand their structure-activity profiles, as well as to tailor and develop new pharmaceutical applications of heparins. It was observed that LMWHs dalteparin and reviparin contain high levels of ΔUA-GlcNS3S6S (D0S9) and ΔUA2S-GlcNS3S (D2S3). In contrast, partial depolymerization and fractionation of PMH to generate enoxaparin and tinzaparin do not seem to affect disaccharide composition greatly, as disaccharide profiles are similar to those for parental PMH.
Production of heparin in mammalian cells is considered a potential alternative to current animal sources, and advances have been made through overexpression and directed KI of enzymes functioning in the HS biosynthetic pathway (56). CHO cells have historically been chosen for genetic engineering (57, 58), and there is a need for custom-designed heparins with better safety profiles, such as reduced HIT. CHO cells stably expressing individual HS3STs had distinct differences in the composition of 3-O-sulfated HS disaccharides (Fig. 4, B and C). The analysis of the HS3STs indicates that they all have preferences for N-sulfated substrates and that HS3ST5/2 have a strong preference for 2-O-sulfated substrates, while HS3ST3A/3B/4 have more promiscuous substrate specificity for N/2-O/6-O-sulfated substrates. To explore the functional differences among the seven HS3STs, we tested the hypothesis that the HS3STs differentially regulate anticoagulant activity and PF4 binding (Fig. 4, D and E). This was clearly the case, and for example, CHO cell HS produced in HS3ST4 cells induced very strong anticoagulant activity and almost no PF4 binding, while CHO cell HS from HS3ST1 cells, the isoenzyme most commonly implicated in biosynthesis of anticoagulant heparin/HS (44, 57), induced relatively strong PF4 binding and only moderate anticoagulant activity. We found evidence that 6-O-sulfation can be important for PF4 binding because HS from CHO KI HS6ST1 induced particularly strong binding. This may be relevant for interpretation of the PF4-binding results, as heparins contain high levels of N-/6-/2-O-sulfated disaccharides. Our results highlight the need to further dissect the functions of the sulfotransferase isoenzyme families, their spatiotemporal expression, and their interplay in producing distinct binding motifs. Nevertheless, the findings here constitute an important step toward genetic engineering designs of CHO cells for production of heparin.
The study also increases the understanding of 3-O-sulfated motifs that lack anticoagulant activity, which may be desirable in the development of tailor-made GAG-based therapeutics where anticoagulant effects are unwanted. This may include GAG designs with anti-inflammatory, antitumor, antioxidant, and antiviral properties (59). Furthermore, the library of HS3ST KI cells can be applied beyond dissection of binding to ATIII and PF4 for determining binding specificities for known 3-O-sulfate–binding proteins such as neuropilin-1, cyclophilin B, stabilin, receptor for advanced glycation endproducts (RAGE), fibroblast growth factor–7 (FGF-7), FGF-9, FGF receptor–1 (FGFR-1), and bone morphogenetic protein 2 and 4 (BMP-2/4) (17, 60).
In summary, we describe an integrated strategy to analyze and determine biological properties of 3-O-sulfated HS structures. Proof of concept was demonstrated for the application of HS3ST-expressing cells for studying structure-function relationships and for production of heparins in mammalian host cells. This facilitated the development of improved heparins with potential for lower incidence of side effects. In combination with emerging HS sequencing methods, these approaches show promise for advancing discovery and characterization of the distinct motifs that direct the many biological interactions and functions of HS.
MATERIALS AND METHODS
Chemical synthesis of 3-O-sulfated tetrasaccharides and production of 3-O-sulfated disaccharide standards
General procedure for chemical glycosylation
Disaccharide acceptor (1.0 equiv), donor (1.2 equiv), and activated molecular sieves (4 Å) in dichloromethane (DCM) (0.2 M) were placed under an atmosphere of argon, and the resulting suspension was stirred at room temperature for 0.5 hour. The mixture was cooled (−20°C), trifilic acid (TfOH; 1.5 equiv) was added, and the resulting reaction mixture was stirred at −20°C for an additional 1 hour, after which it was quenched by the addition of pyridine. The mixture was filtered, the filtrate was concentrated under reduced pressure, and the residue was purified by silica gel column chromatography using a gradient of hexanes and EtOAc (from 4:1 to 1:9, v/v) to give a pure tetrasaccharide.
General procedure for O-sulfation
SO3.NEt3 complex (12.5 equiv per OH) was added to a solution of the hydroxyl-containing tetrasaccharide in N,N′-dimethylformamide (0.5 ml for 0.025 mmol), and the reaction mixure was stirred at 60°C for 16 hours. The reaction was quenched by the addition of a premixed solution of Et3N/MeOH [1:1 (v/v), 1.0 ml], and stirring was continued for another 0.5 hour. The mixture was concentrated in vacuo, and the resulting residue was purified by SEC over Sephadex LH-20 (GE healthcare) using MeOH/DCM (1:1, v/v) as eluent. The desired fractions were combined and concentrated in vacuo, and the residue was passed through a column of Dowex 50 × 8 Na+ resin (1.5 × 5 cm) using MeOH/H2O (9:1, v/v) as eluent.
General procedure for N-acetylation
Acetic anhydride (Ac2O; 10.0 equiv per amine) was added to a stirring solution of amino sugar in a mixture of MeOH (1.0 ml for 2.0 mg) and Et3N (20.0 equiv per amine) at 0°C. After 4 hours, another portion of Et3N and Ac2O was added. After stirring at 0°C for additional 4 hours, the reaction mixture was concentrated in vacuo, and the residue was passed through a column of Dowex 50 × 8 Na+ resin (0.6 × 5 cm) using H2O/AcCN (1:1, v/v) as eluent. Appropriate fractions were concentrated in vacuo and further purified by reversed-phase column chromatography (RP-18 silica gel column, H2O/AcCN, 9:1 to 1:1, v/v).
General procedure for N-sulfation
SO3.Py complex (5.0 equiv per amine) was added to a stirring solution of amino sugar in a mixture of MeOH (1.0 ml for 0.006 mmol), Et3N (0.3 ml), and aq. NaOH (0.1 M, 2.0 equiv) at 0°C. Additional portions of SO3.Py complex were added after 1, 2, 4, and 8 hours. After stirring at 0°C for additional 8 hours, the reaction mixture was concentrated in vacuo and the residue was passed through a column of Dowex 50 × 8 Na+ resin (0.6 × 5 cm) using H2O/AcCN (9:1, v/v) as eluent. Appropriate fractions were concentrated in vacuo and further purified by reversed-phase chromatography (RP-18 silica gel column, H2O/AcCN, 9.5:0.5 to 1:1, v/v).
General procedure for global deprotection
Pd(OH)2/C (10%, 1.5 times the weight of starting material) was added to a solution of tetrasaccharide in tBuOH/0.1 M aq. formic acid [1:1 (v/v), 1.0 ml for 1.0 mg]. The mixture was placed under an atmosphere of hydrogen, and after stirring for 16 hours, the mixture was filtered through a Polytetrafluoroethylene (PTFE) syringe filter and lyophilized. The residue was analyzed by nuclear magnetic resonance to confirm complete deprotection and identity of the tetrasaccharide. The compound was dissolved in sodium phosphate buffer (pH was adjusted to 3.5 with formic acid to avoid peeling reaction), aliquoted, and stored at −80°C.
Heparinase digestion
For enzymatic digestion of heparins, cellular HS, and synthetic tetrasaccharides with heparinases I, II, and III (IBEX Pharmaceuticals), a digestion buffer with a final concentration of 50 mM sodium acetate and 5 mM calcium acetate (pH 6.5) was used. Freshly resuspended lyophilized heparinase I was added first, heparinase III was added after 2 hours, and heparinase II was added 2 hours later, followed by incubation overnight at 37°C. A detailed description of reaction volumes, sample quantities, and enzyme doses used for all heparinase digestions is found in table S5.
Mass spectrometry
Disaccharide standards labeled with AMAC were subjected to offline MS analysis in an Orbitrap Fusion/Lumos MS (Thermo Fisher Scientific) and a Synapt G2-S modified with a drift tube Ion mobility spectrometry (IMS) cell. In the Orbitrap Fusion/Lumos MS (Thermo Fisher Scientific), an atmospheric pressure ionization heated electrospray ionization source (Thermo Fisher Scientific) was used, and the ion transfer tube was set at 275°C. Mass spectra were acquired in negative ion mode at a resolution of 120,000 [at mass/charge ratio (m/z) of 200] with a spray voltage of 3000 V. In the Synapt G2-S, the disaccharides were ionized using Pd/Pt-coated borosilicate capillaries fabricated in-house in negative mode at a capillary voltage of 800 V. MS/MS was performed on selected ions in the trap cell of the instrument at either 15- or 30-V collision voltage.
C18 HPLC disaccharide analysis of AMAC-labeled disaccharides
HS from CHO cells and pharmaceutical heparins were digested with heparinases I, II, and III, and disaccharide products were lyophilized. The disaccharides were then labeled with AMAC by resuspension in 10 μl of 0.1 M AMAC in 3:17 (v/v) acetic acid/dimethyl sulfoxide followed by incubation at room temperature for 15 min, before addition of 10 μl of 1 M NaCNBH3 and incubation at 45°C for 3 hours. The reactions were lyophilized, and excess AMAC was removed by two rounds of resuspension in 500 μl of acetone and pelleting by centrifugation at 20,000g for 20 min at 4°C. Samples were dissolved in 2% acetonitrile and analyzed on a Waters Acquity UPLC system equipped with a fluorescence detector with a BEH C18 column (2.1 × 150 mm, 1.7 μm; Waters) detecting the fluorescence signal at 525 nm. A standard mix of AMAC-labeled disaccharides (20 pmol of each) was analyzed immediately before samples. Commercially available disaccharide standards were purchased from Iduron and Sigma-Aldrich.
Size exclusion analysis of heparinase-digested PMH
For determining the enzyme concentration needed for complete digestion of heparin, PMH was digested with increasing concentrations of heparinases I, II, and III, and digested samples were purified using the Discovery BIO Wide Pore C5 HPLC Column (4.6 mm by 250 mm, 6-μm bead size; Sigma-Aldrich). Water was used as eluent A, and the flow-through was collected and lyophilized. The lyophilized samples were resuspended in 200 μl of 0.5 M ammonium bicarbonate and separated on a Superdex 30 (10 mm by 300 mm by 9 μm; GE Healthcare) SEC column using 0.5 M ammonium bicarbonate with monitoring at 232 nm. Standards of hexasaccharides (dp6), tetrasaccharides (dp4), and disaccharides (dp2) with one to three sulfate groups were used for calibration.
Anti-FXa assay
Human plasma ATIII (1 μM) (Sigma-Aldrich) in 50 mM tris-HCl, 175 mM NaCl, and 7.5 mM EDTA (pH 8.4) and bovine FXa (1 μM) (Sigma-Aldrich) were both diluted 1:30 in 0.9% NaCl, and 8 mM FXa substrate (Sigma-Aldrich) was diluted 1:10 in 0.9% NaCl immediately before assay. ATIII (37.5 μl) was added to each well of a 96-well plate before adding heparin/HS samples at a range of concentrations diluted to 12.5 μl in 0.9% NaCl. Mixtures were incubated for 2 min at 37°C before addition of bovine FXa (37.5 μl) followed by 1-min incubation at 37°C. The FXa substrate (37.5 μl) was then added followed by incubation at 37°C for 10 min before 37.5 μl of acetic acid was used to stop the enzymatic reaction. Absorbance was read at 405 nm using a Synergy LX plate reader (BioTek), and IC50 values used for quantification of anticoagulant activity relative to PMH were determined with an online AAT Bioquest IC50 calculator.
Biolayer interferometry
GAGs were biotinylated at their reducing end as described previously (61). Briefly, biolayer interferometry was carried out using streptavidin biosensors (ForteBio) hydrated for 10 min before use in the assay buffer [10 mM Hepes, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween 20 (pH 7.4)]. Hydrated sensors were then submerged into wells of a black-walled 96-well plate containing 200 μl of biotinylated GAGs suspended at 2.5 μg/ml in assay buffer until saturated. Saturation was confirmed with an additional GAG immobilization step, where no further GAG was immobilized. Sensors were then cleaned by submersion in wells containing 200 μl of regeneration buffer [0.1 M glycine, 1 M NaCl, and 0.1% Tween (pH 9.5)] and equilibrated in assay buffer. GAG-coated sensors were submerged in wells containing 200 μl of PF4 resuspended in assay buffer for 180 s (association) and then transferred to wells containing assay buffer alone (dissociation), and data were recorded throughout. Background binding to non–GAG-coated sensors and signal produced by buffer alone was recorded and subtracted from the GAG-coated sensor signal. The maximum signal recorded during each cycle was then used as a measure of the degree of binding of PF4 to each GAG at different concentrations. Between cycles, bound PF4 was removed from GAG-coated sensors using regeneration buffer and sensors were then equilibrated in assay buffer. Data were acquired using an Octet Red96 system (ForteBio) at 5 Hz and analyzed using the Octet analysis program.
Genetic engineering of CHO cells
CHOZN GS−/− (Sigma-Aldrich) cells were maintained as suspension culture in T-flasks at 37°C and 5% CO2 using a 1:1 mix of EX-CELL CD CHO Fusion (Sigma-Aldrich) and BalanCD CHO Growth A (Irvine scientific), supplemented with 2 mM l-glutamine. For targeted KI at the CHO SafeHarbor locus, a modified ZFN ObLiGare method was used as previously described (58, 62). Full complementary DNAs of coding regions of the human HS3STs (Horizon Discovery and Harvard PlasmID Database) were used, a C-terminal S-tag or V5-tag was linked to HS3STs by polymerase chain reaction (PCR), and constructs were further cloned into the EPB69 donor plasmid. EPB69 contained inverted CHO SafeHarbor locus ZFN-binding sites flanking the cytomegalovirus promoter–ORF (open reading frame)–BGH (bovine growth hormone) polyA terminator and two tDNA insulator elements flanking the ZFN-binding sites, as previously described (58, 63). Transfection DNA mixes contained 4.5 μg of donor plasmid DNA and 1.5 μg of each of two ZFNs tagged with green fluorescent protein (GFP) and Crimson, respectively. For each transfection, 1.5 × 106 cells were electroporated using Amaxa Kit V and Amaxa Nucleofector 2B (Lonza) according to the manufacturer’s instructions. Forty-eight hours after transfection, 10 to 15% of cells with the highest labeling for both GFP and Crimson were enriched by fluorescence-activated cell sorting (FACS) on SH800 (Sony). One week later, the FACS-sorted cell pool was further single cell–sorted into round-bottom 96-well plates with Dulbecco’s Modified Eagle Medium F-12 Nutrient Mixture (Thermo Fisher Scientific) to obtain single clones. KI clones were screened by immunocytochemistry, and monoallelic-targeted KI clones were validated by PCR with primers specific for the junction area between the donor plasmid and the SafeHarbor locus. A primer set flanking the targeted KI locus was also used to characterize the allelic insertion status. A minimum of three clones were obtained for each HS3ST KI.
Immunocytochemistry
CHO cells were washed in phosphate-buffered saline (PBS), spotted onto Teflon printed diagnostic slides (Immuno-Cell International), air-dried, and permeabilized with ice-cold acetone for 5 min. Polyclonal antibodies to S-tag (GenScript) were used 1:200 in PBS with 0.1% bovine serum albumin (BSA) at 4°C overnight followed by fluorescein isothiocyanate (FITC)–conjugated rabbit anti-mouse immunoglobulin G (IgG) antibody (DAKO) 1:300 in 1× PBS with 0.1% BSA for 1 hour at room temperature. An FITC-conjugated monoclonal antibody to V5-tag (Thermo Fisher Scientific) was used 1:500 in PBS with 0.1% BSA at 4°C overnight. Slides were analyzed in an Axioskop 2 Plus (Zeiss) microscope, and images were obtained using an AxioCam MRc (Zeiss) camera.
SDS-PAGE Western blot
Cells (1 × 106) were seeded in a T25 flask in 6-ml medium and grown for 72 hours, before washing the cells three times in PBS and adding 700 μl of cold radioimmunoprecipitation assay (RIPA) buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.1% Na deoxycholate, and 1 mM EDTA] containing protease inhibitor cocktail (Sigma-Aldrich). Samples were thoroughly vortexed before incubation for 20 min on ice with vortexing every 5 min followed by ultrasonication (40% amplitude) for 3 × 5 s with 5-s pauses using the Fisherbrand Model 120 Sonic Dismembrator (Thermo Fisher Scientific). Samples were centrifuged at 20,000g at 4°C for 15 min, and the protein concentration of the supernatant was measured using a BCA protein assay kit (Thermo Fisher Scientific). Protein (10 μg) or the corresponding fraction of medium used for culturing cells to obtain 10 μg of protein was mixed with 10 mM DDT (Dichlorodiphenyltrichloroethane) and 1× loading buffer, heated to 90°C for 10 min, and separated on NuPAGE 4 to 12% Bis-Tris gels (Thermo Fisher Scientific). Proteins were transferred to nitrocellulose membranes at 320 mA for 60 min in MES buffer with 20% methanol. Membranes were blocked with 5% skimmed milk in TBS-T (tris-buffered saline tween-20) for 60 min before they were incubated with either horseradish peroxidase (HRP)–conjugated antibodies to V5-tag (Thermo Fisher Scientific) or S-tag in TBS-T with 5% skimmed milk at 4°C overnight. Membranes were washed 3 × 5 min in TBS-T followed by incubation of S-tag membranes with HRP-conjugated rabbit anti-mouse IgG antibody (DAKO) in 5% skimmed milk in TBS-T for 1 hour at room temperature and washing 3 × 5 min in TBS-T. Pierce ECL Plus Western Blotting Substrate (Thermo Fisher Scientific) was used according to the manufacturer’s instructions, and images were captured using ImageQuant Las 4000 (GE Healthcare).
Extraction and purification of GAGs from CHO cells
Cells were washed in PBS and diluted to 1 × 107 cells/ml in 50 mM tris-HCl (pH 7.4), 10 mM CaCl2, and 0.1% Triton X-100. Pronase (Roche) was added (1 mg/ml), and reactions were incubated overnight rotating tray in an incubator set at 37°C followed by heat inactivation. MgCl2 (5 mM) and deoxyribonuclease I (1 μg/ml; Sigma-Aldrich) were added, and samples were incubated at 37°C for 4 hours. Samples were treated with ribonuclease A (10 μg/ml; Sigma-Aldrich) and 5 mM EDTA at 37°C for 2 hours, followed by neuraminidase (0.5 mU/ml; Sigma-Aldrich) at 37°C overnight. For cells expressing CS, chABC (chondroitinase ABC) (20 mU/ml) was added and samples were incubated at 37°C for 4 hours. Samples were again incubated with pronase at 1 mg/ml for overnight digestion at 37°C. Samples were acidified to pH 4 to 5 with acetic acid, centrifuged at 20,000g for 20 min, filtered through 0.45-μm filters, and isolated on HiTrap DEAE FF columns (5 ml; GE Healthcare). Columns were equilibrated with 20 mM NaOAc and 0.5 M NaCl (pH 5.0), and samples were eluted with 1.25 M NaCl. GAGs were precipitated by addition of cold NaOAc-saturated 100% ethanol (3:1, v/v) and centrifuged at 20,000g for 20 min at 4°C, and the pellets were dried on SpeedVac. Samples were resuspended in deionized water, further purified using Discovery BIO Wide Pore C5-5 (Sigma-Aldrich), and desalted on 1-ml HiTrap desalting columns (GE Healthcare).
Flow cytometry assays
Cells at 50 to 80% confluence were detached with PBS containing 10 mM EDTA (Gibco) and washed in PBS containing 0.5% BSA. The cells were seeded into a 96-well plate at 105 cells per well. A portion of the cells were treated with a heparinase mix [heparinase II (2.5 mU/ml) and heparinase III (5 mU/ml); IBEX] for 30 min at 37°C in PBS containing 0.5% BSA. Staining with recombinant ATIII (Hyphen BioMed) (500 nM) was done for 1 hour at 4°C in PBS containing 0.5% BSA. Cells were then incubated with an anti-ATIII antibody (Serpin-C1, R&D Systems) for 30 min at 4°C in PBS containing 0.5% BSA. The bound complex was finally detected using anti-goat Alexa Fluor 488 (Thermo Fisher Scientific). Data analysis was performed using FlowJo software, and statistical analyses were done in Prism 8 (GraphPad).
Acknowledgments
We thank H. Birchenough and the Biomolecular analysis core facility at the University of Manchester for help in establishing the chemokine:GAG interaction methodology. We also thank E. Yates, M. Skidmore, and S. Guimond for supplying four commercially available heparins.
Funding: This work was funded by The Carlsberg Foundation CF20-0412 (R.L.M.); The Lundbeck Foundation, R223-2016-563 (H.C.); The Danish National Research Foundation DNRF107 (H.C.); The European Commission, GlycoImaging H2020-MSCA-ITN-721297 (H.C. and R.K.); NIH, P41GM103390 and HLBI R01HL151617 (G.-J.B.); The Sir Henry Dale Fellowship, Welcome Trust/Royal Society 218570/Z/19/Z (D.P.D.); The National Science Foundation, RAPID 2031989 (J.E.T.); NIH, HL131474 (J.D.E.); and The Deutsche Forschungsgemeinschaft, German Research Foundation SFB 1340 (K.P.).
Author contributions: Conceptualization: R.K., P.C., Z.Y., J.E.T., H.C., G.-J.B., and R.L.M. Methodology: R.K., P.C., A.J., Z.Y., S.Y.V., T.M.C., C.D.P., G.P.S., Y.-H.C., D.R.S., L.H., J.D.E., K.P., D.P.D., and R.L.M. Investigation: R.K., P.C., A.J., Z.Y., S.Y.V., T.M.C., C.D.P., G.P.S., Y.-H.C., D.R.S., L.H., J.D.E., K.P., D.P.D., and R.L.M. Funding acquisition: H.C., G.-J.B., and R.L.M. Project administration: H.C., G.-J.B., and R.L.M. Supervision: H.C., G.-J.B., and R.L.M. Writing (original draft): R.K., P.C., Z.Y., J.E.T., H.C., G.-J.B., and R.L.M. Writing (review and editing): R.K., P.C., A.J., Z.Y., S.Y.V., T.M.C., C.D.P., G.P.S., Y.-H.C., D.R.S., L.H., J.D.E., K.P., D.P.D., J.E.T., H.C., G.-J.B., and R.L.M.
Competing interests: Y.-H.C. is an employee of GlycoDisplay Aps, and Z.Y. and H.C. are cofounders of GlycoDisplay Aps and hold ownerships and financial interest in the company. The other authors declare no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All cell lines are available on request under a standard material transfer agreement with University of Copenhagen for academic research purposes.
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Tables S1 to S11
Supplementary experimental methods for chemical synthesis and analysis
References
REFERENCES AND NOTES
- 1.Soares da Costa D., Reis R. L., Pashkuleva I., Sulfation of glycosaminoglycans and its implications in human health and disorders. Annu. Rev. Biomed. Eng. 19, 1–26 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Xu D., Esko J. D., Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83, 129–157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glass C. A., Recombinant heparin-new opportunities. Front. Med. 5, 341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oduah E. I., Linhardt R. J., Sharfstein S. T., Heparin: Past, present, and future. Pharmaceuticals (Basel) 9, 38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spadarella G., di Minno A., Donati M. B., Mormile M., Ventre I., di Minno G., From unfractionated heparin to pentasaccharide: Paradigm of rigorous science growing in the understanding of the in vivo thrombin generation. Blood Rev. 39, 100613 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Fu L., Li G., Yang B., Onishi A., Li L., Sun P., Zhang F., Linhardt R. J., Structural characterization of pharmaceutical heparins prepared from different animal tissues. J. Pharm. Sci. 102, 1447–1457 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H., Zhang Z., Linhardt R. J., Lessons learned from the contamination of heparin. Nat. Prod. Rep. 26, 313–321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilanova E., Tovar A. M. F., Mourao P. A. S., Imminent risk of a global shortage of heparin caused by the African Swine Fever afflicting the Chinese pig herd. J. Thromb. Haemost. 17, 254–256 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Huntington J. A., Mechanisms of glycosaminoglycan activation of the serpins in hemostasis. J. Thromb. Haemost. 1, 1535–1549 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Kim Y. S., Linhardt R. J., Structural features of heparin and their effect on heparin cofactor II mediated inhibition of thrombin. Thromb. Res. 53, 55–71 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Petitou M., Duchaussoy P., Herbert J. M., Duc G., el Hajji M., Branellec J. F., Donat F., Necciari J., Cariou R., Bouthier J., Garrigou E., The synthetic pentasaccharide fondaparinux: First in the class of antithrombotic agents that selectively inhibit coagulation factor Xa. Semin. Thromb. Hemost. 28, 393–402 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Atha D. H., Lormeau J. C., Petitou M., Rosenberg R. D., Choay J., Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry 24, 6723–6729 (1985). [DOI] [PubMed] [Google Scholar]
- 13.Rauova L., Poncz M., McKenzie S. E., Reilly M. P., Arepally G., Weisel J. W., Nagaswami C., Cines D. B., Sachais B. S., Ultralarge complexes of PF4 and heparin are central to the pathogenesis of heparin-induced thrombocytopenia. Blood 105, 131–138 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Dyer D. P., Salanga C. L., Volkman B. F., Kawamura T., Handel T. M., The dependence of chemokine-glycosaminoglycan interactions on chemokine oligomerization. Glycobiology 26, 312–326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed I., Majeed A., Powell R., Heparin induced thrombocytopenia: Diagnosis and management update. Postgrad. Med. J. 83, 575–582 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stringer S. E., Gallagher J. T., Specific binding of the chemokine platelet factor 4 to heparan sulfate. J. Biol. Chem. 272, 20508–20514 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Chopra P., Joshi A., Wu J., Lu W., Yadavalli T., Wolfert M. A., Shukla D., Zaia J., Boons G. J., The 3-O-sulfation of heparan sulfate modulates protein binding and lyase degradation. Proc. Natl. Acad. Sci. U.S.A. 118, e2012935118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhurandhare V. M., Pagadala V., Ferreira A., Muynck L., Liu J., Synthesis of 3-O-sulfated disaccharide and tetrasaccharide standards for compositional analysis of heparan sulfate. Biochemistry 59, 3186–3192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamada S., Yoshida K., Sugiura M., Sugahara K., Khoo K. H., Morris H. R., Dell A., Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin III-binding site of porcine intestinal heparin. J. Biol. Chem. 268, 4780–4787 (1993). [PubMed] [Google Scholar]
- 20.Cadwallader A. B., Yost H. J., Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: I. The 3-O-sulfotransferase family. Dev. Dyn. 235, 3423–3431 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Lawrence R., Yabe T., Hajmohammadi S., Rhodes J., McNeely M., Liu J., Lamperti E. D., Toselli P. A., Lech M., Spear P. G., Rosenberg R. D., Shworak N. W., The principal neuronal gD-type 3-O-sulfotransferases and their products in central and peripheral nervous system tissues. Matrix Biol. 26, 442–455 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thacker B. E., Xu D., Lawrence R., Esko J. D., Heparan sulfate 3-O-sulfation: A rare modification in search of a function. Matrix Biol. 35, 60–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J., Shworak N. W., Fritze L. M., Edelberg J. M., Rosenberg R. D., Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J. Biol. Chem. 271, 27072–27082 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Xia G., Chen J., Tiwari V., Ju W., Li J. P., Malmström A., Shukla D., Liu J., Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J. Biol. Chem. 277, 37912–37919 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Moon A. F., Edavettal S. C., Krahn J. M., Munoz E. M., Negishi M., Linhardt R. J., Liu J., Pedersen L. C., Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1. J. Biol. Chem. 279, 45185–45193 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulberti S., Mao X., Bui C., Fournel-Gigleux S., The role of heparan sulfate maturation in cancer: A focus on the 3O-sulfation and the enigmatic 3O-sulfotransferases (HS3STs). Semin. Cancer Biol. 62, 68–85 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Hellec C., Delos M., Carpentier M., Denys A., Allain F., The heparan sulfate 3-O-sulfotransferases (HS3ST) 2, 3B and 4 enhance proliferation and survival in breast cancer MDA-MB-231 cells. PLOS ONE 13, e0194676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sepulveda-Diaz J. E., Alavi Naini S. M., Huynh M. B., Ouidja M. O., Yanicostas C., Chantepie S., Villares J., Lamari F., Jospin E., van Kuppevelt T. H., Mensah-Nyagan A. G., Raisman-Vozari R., Soussi-Yanicostas N., Papy-Garcia D., HS3ST2 expression is critical for the abnormal phosphorylation of tau in Alzheimer’s disease-related tau pathology. Brain 138, 1339–1354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjellen L., Lindahl U., Specificity of glycosaminoglycan-protein interactions. Curr. Opin. Struct. Biol. 50, 101–108 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Chen Y. H., Narimatsu Y., Clausen T. M., Gomes C., Karlsson R., Steentoft C., Spliid C. B., Gustavsson T., Salanti A., Persson A., Malmström A., Willén D., Ellervik U., Bennett E. P., Mao Y., Clausen H., Yang Z., The GAGOme: A cell-based library of displayed glycosaminoglycans. Nat. Methods 15, 881–888 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Qiu H., Shi S., Yue J., Xin M., Nairn A. V., Lin L., Liu X., Li G., Archer-Hartmann S. A., dela Rosa M., Galizzi M., Wang S., Zhang F., Azadi P., van Kuppevelt T. H., Cardoso W. V., Kimata K., Ai X., Moremen K. W., Esko J. D., Linhardt R. J., Wang L., A mutant-cell library for systematic analysis of heparan sulfate structure-function relationships. Nat. Methods 15, 889–899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zong C., Venot A., Li X., Lu W., Xiao W., Wilkes J. S. L., Salanga C. L., Handel T. M., Wang L., Wolfert M. A., Boons G. J., Heparan sulfate microarray reveals that heparan sulfate-protein binding exhibits different ligand requirements. J. Am. Chem. Soc. 139, 9534–9543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller R. L., Guimond S. E., Schwörer R., Zubkova O. V., Tyler P. C., Xu Y., Liu J., Chopra P., Boons G. J., Grabarics M., Manz C., Hofmann J., Karlsson N. G., Turnbull J. E., Struwe W. B., Pagel K., Shotgun ion mobility mass spectrometry sequencing of heparan sulfate saccharides. Nat. Commun. 11, 1481 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller R. L., Dykstra A. B., Wei W., Holsclaw C., Turnbull J. E., Leary J. A., Enrichment of two isomeric heparin oligosaccharides exhibiting different affinities toward monocyte chemoattractant protein-1. Anal. Chem. 88, 11551–11558 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Lettow M., Grabarics M., Greis K., Mucha E., Thomas D. A., Chopra P., Boons G. J., Karlsson R., Turnbull J. E., Meijer G., Miller R. L., von Helden G., Pagel K., Cryogenic infrared spectroscopy reveals structural modularity in the vibrational fingerprints of heparan sulfate diastereomers. Anal. Chem. 92, 10228–10232 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Leary J. A., Miller R. L., Wei W., Schwörer R., Zubkova O. V., Tyler P. C., Turnbull J. E., Composition, sequencing and ion mobility mass spectrometry of heparan sulfate-like octasaccharide isomers differing in glucuronic and iduronic acid content. Eur. J. Mass Spectrom. 21, 245–254 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Wu J., Wei J., Chopra P., Boons G. J., Lin C., Zaia J., Sequencing heparan sulfate using HILIC LC-NETD-MS/MS. Anal. Chem. 91, 11738–11746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Q., Chopra P., Boons G. J., Sharp J. S., Improved de novo sequencing of heparin/heparan sulfate oligosaccharides by propionylation of sites of sulfation. Carbohydr. Res. 465, 16–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y., Masuko S., Takieddin M., Xu H., Liu R., Jing J., Mousa S. A., Linhardt R. J., Liu J., Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science 334, 498–501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohse D. L., Linhardt R. J., Purification and characterization of heparin lyases from Flavobacterium heparinum. J. Biol. Chem. 267, 24347–24355 (1992). [PubMed] [Google Scholar]
- 41.Huang Y., Mao Y., Zong C., Lin C., Boons G. J., Zaia J., Discovery of a heparan sulfate 3-O-sulfation specific peeling reaction. Anal. Chem. 87, 592–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baytas S. N., Linhardt R. J., Advances in the preparation and synthesis of heparin and related products. Drug Discov. Today 25, 2095–2109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merli G. J., Groce J. B., Pharmacological and clinical differences between low-molecular-weight heparins: Implications for prescribing practice and therapeutic interchange. P T 35, 95–105 (2010). [PMC free article] [PubMed] [Google Scholar]
- 44.Datta P., Li G., Yang B., Zhao X., Baik J. Y., Gemmill T. R., Sharfstein S. T., Linhardt R. J., Bioengineered Chinese hamster ovary cells with Golgi-targeted 3-O-sulfotransferase-1 biosynthesize heparan sulfate with an antithrombin-binding site. J. Biol. Chem. 288, 37308–37318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L., Lawrence R., Frazier B. A., Esko J. D., CHO glycosylation mutants: Proteoglycans. Methods Enzymol. 416, 205–221 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Xu X., Nagarajan H., Lewis N. E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., Andersen M. R., Neff N., Passarelli B., Koh W., Fan H. C., Wang J., Gui Y., Lee K. H., Betenbaugh M. J., Quake S. R., Famili I., Palsson B. O., Wang J., The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29, 735–741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho S. K., Cummings R. D., A soluble form of α1,3-Galactosyltransferase functions within cells to galactosylate glycoproteins. J. Biol. Chem. 272, 13622–13628 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Zhu G., Allende M. L., Jaskiewicz E., Qian R., Darling D. S., Worth C. A., Colley K. J., Young W. W., Two soluble glycosyltransferases glycosylate less efficiently in vivo than their membrane bound counterparts. Glycobiology 8, 831–840 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Kuhn P. H., Voss M., Haug-Kröper M., Schröder B., Schepers U., Bräse S., Haass C., Lichtenthaler S. F., Fluhrer R., Secretome analysis identifies novel signal Peptide peptidase-like 3 (Sppl3) substrates and reveals a role of Sppl3 in multiple Golgi glycosylation pathways. Mol. Cell. Proteomics 14, 1584–1598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habuchi H., Habuchi O., Kimata K., Purification and characterization of heparan sulfate 6-sulfotransferase from the culture medium of Chinese hamster ovary cells. J. Biol. Chem. 270, 4172–4179 (1995). [DOI] [PubMed] [Google Scholar]
- 51.Chen J., Liu J., Characterization of the structure of antithrombin-binding heparan sulfate generated by heparan sulfate 3-O-sulfotransferase 5. Biochim. Biophys. Acta 1725, 190–200 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Mochizuki H., Yoshida K., Gotoh M., Sugioka S., Kikuchi N., Kwon Y. D., Tawada A., Maeyama K., Inaba N., Hiruma T., Kimata K., Narimatsu H., Characterization of a heparan sulfate 3-O-sulfotransferase-5, an enzyme synthesizing a tetrasulfated disaccharide. J. Biol. Chem. 278, 26780–26787 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Ori A., Wilkinson M. C., Fernig D. G., A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 286, 19892–19904 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meissen J. K., Sweeney M. D., Girardi M., Lawrence R., Esko J. D., Leary J. A., Differentiation of 3-O-sulfated heparin disaccharide isomers: Identification of structural aspects of the heparin CCL2 binding motif. J. Am. Soc. Mass Spectrom. 20, 652–657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaidyanathan D., Williams A., Dordick J. S., Koffas M. A. G., Linhardt R. J., Engineered heparins as new anticoagulant drugs. Bioeng. Transl. Med. 2, 17–30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lord M. S., Cheng B., Tang F., Lyons J. G., Rnjak-Kovacina J., Whitelock J. M., Bioengineered human heparin with anticoagulant activity. Metab. Eng. 38, 105–114 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Baik J. Y., Gasimli L., Yang B., Datta P., Zhang F., Glass C. A., Esko J. D., Linhardt R. J., Sharfstein S. T., Metabolic engineering of Chinese hamster ovary cells: Towards a bioengineered heparin. Metab. Eng. 14, 81–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z., Wang S., Halim A., Schulz M. A., Frodin M., Rahman S. H., Vester-Christensen M. B., Behrens C., Kristensen C., Vakhrushev S. Y., Bennett E. P., Wandall H. H., Clausen H., Engineered CHO cells for production of diverse, homogeneous glycoproteins. Nat. Biotechnol. 33, 842–844 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Qiu M., Huang S., Luo C., Wu Z., Liang B., Huang H., Ci Z., Zhang D., Han L., Lin J., Pharmacological and clinical application of heparin progress: An essential drug for modern medicine. Biomed. Pharmacother. 139, 111561 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Thacker B. E., Seamen E., Lawrence R., Parker M. W., Xu Y., Liu J., Vander Kooi C. W., Esko J. D., Expanding the 3-O-sulfate proteome—Enhanced binding of neuropilin-1 to 3-O-sulfated heparan sulfate modulates its activity. ACS Chem. Biol. 11, 971–980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thakar D., Migliorini E., Coche-Guerente L., Sadir R., Lortat-Jacob H., Boturyn D., Renaudet O., Labbe P., Richter R. P., A quartz crystal microbalance method to study the terminal functionalization of glycosaminoglycans. Chem. Commun. (Camb.) 50, 15148–15151 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Maresca M., Lin V. G., Guo N., Yang Y., Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23, 539–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hintze J., Ye Z., Narimatsu Y., Madsen T. D., Joshi H. J., Goth C. K., Linstedt A., Bachert C., Mandel U., Bennett E. P., Vakhrushev S. Y., Schjoldager K. T., Probing the contribution of individual polypeptide GalNAc-transferase isoforms to the O-glycoproteome by inducible expression in isogenic cell lines. J. Biol. Chem. 293, 19064–19077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Domon B., Costello C. E., A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5, 397–409 (1988). [Google Scholar]
- 65.Arungundram S., al-Mafraji K., Asong J., Leach F. E. III, Amster I. J., Venot A., Turnbull J. E., Boons G. J., Modular synthesis of heparan sulfate oligosaccharides for structure-activity relationship studies. J. Am. Chem. Soc. 131, 17394–17405 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Tables S1 to S11
Supplementary experimental methods for chemical synthesis and analysis
References