10S structure explains myosin-2 regulation.
Abstract
We solved the near-atomic resolution structure of smooth muscle myosin-2 in the autoinhibited state (10S) using single-particle cryo–electron microscopy. The 3.4-Å structure reveals the precise molecular architecture of 10S and the structural basis for myosin-2 regulation. We reveal the position of the phosphorylation sites that control myosin autoinhibition and activation by phosphorylation of the regulatory light chain. Further, we present a previously unidentified conformational state in myosin-2 that traps ADP and Pi produced by the hydrolysis of ATP in the active site. This noncanonical state represents a branch of the myosin enzyme cycle and explains the autoinhibition of the enzyme function of 10S along with its reduced affinity for actin. Together, our structure defines the molecular mechanisms that drive 10S formation, stabilization, and relief by phosphorylation of the regulatory light chain.
INTRODUCTION
Myosin-2 converts the energy derived from the hydrolysis of adenosine 5′-triphosphate (ATP) into force and directed movement on actin filaments. As the major contractile protein in eukaryotic cells, myosin-2 powers a repertoire of vital biological processes including the contraction of muscle, cell division, and cell adhesion (1, 2). Myosin-2 is composed of two heavy chains, two essential light chains (ELCs), and two regulatory light chains (RLCs) (fig. S1A) (3). The heavy chains fold into a globular head domain and an α-helical rod domain (3). The head domain is composed of the enzymatic motor domain and the light chain–binding neck domain (4). The head couples biochemical transitions during the adenosine triphosphatase (ATPase) cycle to structural transitions that generate force and translocation of actin filaments (4, 5). The rods of two heavy chains form a coiled-coil tail that can self-associate to form filaments (fig. S1B) (6).
The ATPase activity of myosin-2 is precisely regulated to meet the changing physiological demands of cells. The activity of vertebrate smooth and nonmuscle myosin-2 is determined by phosphorylation of the RLC on S19 that controls the interaction of the heads with actin and the assembly into filaments (fig. S1B) (7–9). In the phosphorylated active state, myosin-2 forms filaments and hydrolyzes ATP to power the movement of actin. In the dephosphorylated inactive state, myosin-2 adopts a compact conformation that is kinetically inert (>1000-fold reduced ATPase activity) (10, 11), compromised to interact with actin [Kd (dissociation constant) > 100 μM] (12) and to self-assemble into filaments (fig. S1B) (9, 13, 14). These properties are attributed to the complex folding of the myosin-2 molecule. In the inactive state, the myosin heads are docked against each other in an asymmetric configuration (15–17). This minimal autoinhibitory conformation is also known as the “interacting heads motif” (IHM) (18, 19) in relaxed muscle and is essential to myosin-2 function (20). It represents an ancient and universal mechanism to inhibit myosin-2 activity (20, 21). The tail can further fold back on itself and interact with the IHM in the dephosphorylated forms of smooth and nonmuscle myosin-2 (15, 17, 21, 22). This conformation was first described 40 years ago and is known as 10S in reference to its sedimentation coefficient (fig. S1B) (14, 23).
The current understanding of 10S architecture derives from early negative stain studies, cryo–electron microscopy (cryo-EM) studies of myosin fragments on lipid monolayers and, more recently, higher-resolution structures of intact molecules from single-particle cryo-EM (16, 22, 24, 25). Although the recent pseudo-atomic models of 10S have improved the resolution from ~20 to ~4.3 Å (17, 18, 24, 25), the resolution has been insufficient to describe key features of the molecule such as side chain interactions and the underlying molecular mechanisms that explain myosin-2 autoinhibition and the subsequent activation by phosphorylation. Several critical questions therefore remain. What are the conformations of the head domains? How are the ATP hydrolysis products trapped in the active sites? How does the unphosphorylated RLC stabilize and the phosphorylated RLC destabilize 10S? We addressed these questions by solving the near-atomic resolution structure of 10S at a global resolution of 3.4 Å, the highest for any 10S structure so far. This allowed us to build a complete model of the IHM including the bound adenosine 5′-diphosphate (ADP) and inorganic phosphate (Pi) produced by the hydrolysis of ATP. Further, we built de novo models of previously ambiguous regions including the hinge points and the register of the tail segments, the N and C termini of the RLC, loop-2, loop-4, and the CM-loop, as well as the pliant and the hook regions that contribute to the asymmetric binding of the light chains. We reveal a previously unidentified conformation of the myosin head domain, hereafter referred to as the “off state,” that traps the ATP hydrolysis products in the active site and prevents actin activation of the myosin ATPase. Together, our structure defines the molecular mechanisms that drive 10S formation, stabilization, and relief via phosphorylation dependent activation.
RESULTS
Molecular architecture of 10S
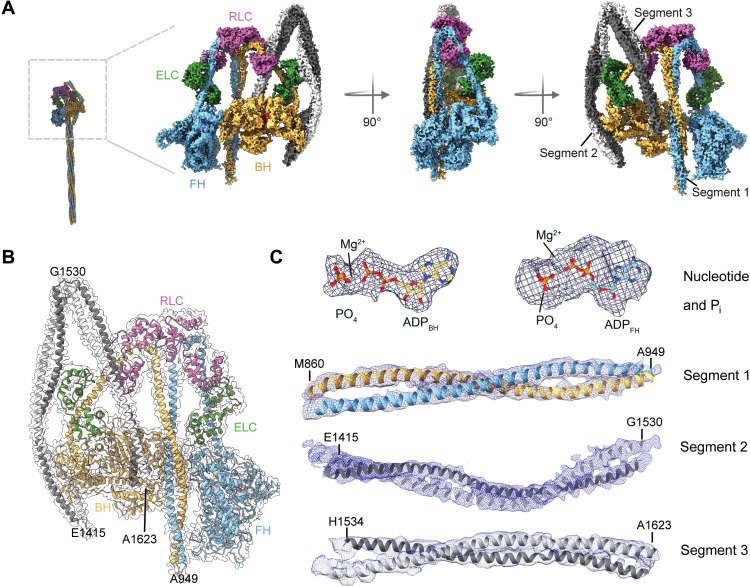
We solved the structure of chicken smooth muscle myosin-2 in the 10S state to a 3.4-Å resolution (Fig. 1, A and B; figs. S2 and S3; table S1; and movies S1 and S2). Our cryo-EM density map was of sufficient resolution to place the tail segments with confidence. We could resolve actin binding elements and surface loops in the head domains, both light chains, Pi, and the nucleotide in the myosin heads in the 10S structure (Fig. 1 and fig. S4). The distal, flexible parts of 10S such as the portions of the tail that are not interacting with the heads are not well resolved in our structure. We show that 10S has a flat, compact shape that is formed by asymmetric intra and intersubunit interactions between the two myosin heavy chains and bound light chains (Fig. 1A and movies S1 and S2) in agreement with structural and biochemical data (16, 17, 24–27). The heads dock against each other and are hereafter referred to as the blocked head (BH) and the free head (FH), as defined previously (16). Both heads assume a primed conformation with the lever arms in the “up” position. Each head contains clear densities for the trapped ADP and Pi in the active sites (Fig. 1C). The light chains stabilize and contribute to the asymmetry of 10S. The myosin tails are folded twice at two hinge points and assume a three-segment hairpin-like structure (Fig. 1, A and B) (14, 22). Segment 1 emanating from the BH and FH forms a coiled coil that runs asymmetrically between both heads (Figs. 1, A and B, and 2C). Segment 2 locks the N terminus and converter of the BH and further interacts with the ELCBH (Figs. 1, A and B, and 2, D and E). Segment 3 interacts with the RLCBH and folds back on the BH (Fig. 1, A and B).
Fig. 1. Cryo-EM structure of 10S.
(A) Cryo-EM reconstruction of 10S in the front, side, and back view. Individual subunits are segmented and color-coded. The two heavy chains (blue and yellow) and bound ELC (green) and RLC (pink) are shown. (B) Atomic model of 10S fitted in the final electron density map. Individual subunits are segmented and color-coded according to (A). (C) Representative cryo-EM densities of different regions of 10S. Densities of the nucleotide (3.4 Å) and Pi displayed at 12.5 σ contour level. Densities of segment 1 (4.5 Å), segment 2 (5 Å), and segment 3 (5.5 Å) displayed at 8.0 σ contour level.
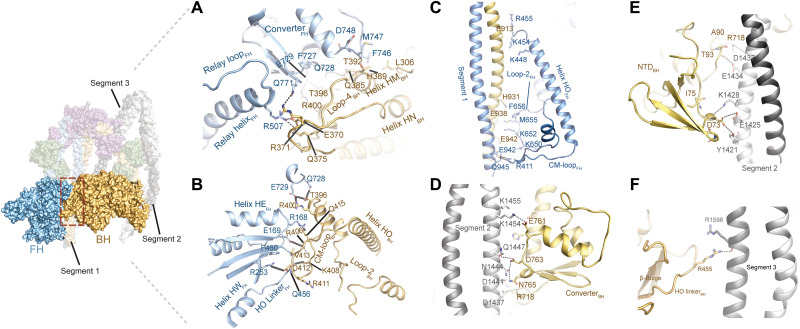
Fig. 2. Interactions of the myosin heads.
(A) Interactions of loop-4BH at the BH-FH interface. Potential interactions (distance > 4 Å) between R371BH and Q771FH and Q385BH and Q728FH are shown. (B) Interactions of CM-loopBH at the BH-FH interface. Potential interactions (distance > 4 Å) between D412BH and R253FH and R406BH and E169FH are shown. (C) Interaction between the FH and segment 1. (D) Interaction between the converterBH and segment 2. (E) Interaction between the NTDBH and segment 2. (F) Interaction between the HO linkerBH and segment 3. The views in (C) to (F) are rotated 180° relative to the overview figure to highlight key interactions at the respective interfaces.
Stabilization of the IHM
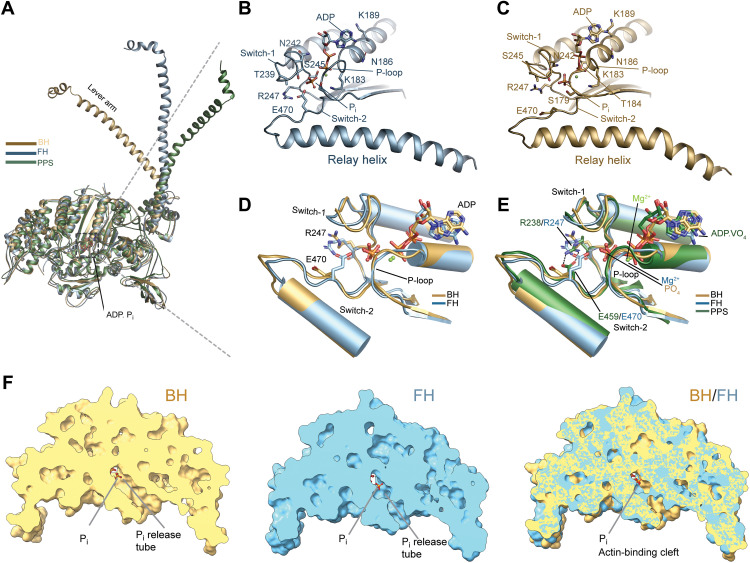
We show that the head-head interaction is stabilized by interactions between surface loops and allosteric elements of the motor domains. Notably, the structures of the two motor domains are not identical, and the cores deviate with an average root mean square deviation (RMSD) of 1.78 Å (fig. S5A). B-factor analysis (fig. S5E) further shows that the FH is more flexible than the BH. The increased flexibility is attributed to the lack of stabilizing interactions with segments 2 and 3 (Fig. 1, A and B). The lever arm angles of BH and FH vary by ~30° and are more acutely bent compared to myosins in the pre-powerstroke state (Fig. 3A, fig. S5C, table S2, and movie S3) (15, 17, 18). The outer clefts of both heads are more open (>20 Å) than has been seen in other myosin structures (fig. S5B and table S2) and contribute to the weak actin affinity of 10S (12).
Fig. 3. Active site and nucleotide interactions.
(A) Orientation of the lever arm in BH and FH in comparison to the pre-powerstroke state (PDB ID: 1QVI). (B) Active site and nucleotide interactions in the FH. Key residues are shown in stick representation. The distance between R247 and E470 is >6.1 Å. (C) Active site and nucleotide interactions in the BH. Key residues are shown in stick representation. (D) Comparison between the active sites in BH and FH. Key residues are shown in stick representation. (E) Comparison between the active sites in BH, FH, and the pre-powerstroke state (PDB ID: 2XEL). The salt bridge between switch-1 R247/R238 and switch-2 E470/E459 is only formed in the pre-powerstroke state and absent in BH and FH. Key residues are shown in stick representation. (F) Comparison of the phosphate release tube in the BH and FH.
A network of electrostatic interactions and hydrogen bonds is formed between loop-4BH, relay helixFH, and converterFH (Fig. 2A). The HO linkerFH, helices HEFH, and HWFH block the CM-loopBH and establish an interlock between both heads (Fig. 2B). The CM-loopFH and loop-2FH interact with segment 1 (Fig. 2C). Rather than projecting toward the segments, loop-2BH is tucked inward to facilitate the positioning of segments 1 and 3 between both heads (Fig. 2C and fig. S5D). In addition, the converter of the FH is stacked against the base of the BH motor domain connected to helices HMBH and HNBH (Fig. 2A) and prevents the swing of its lever arm. Interactions between the converterBH (Fig. 2D) and the N-terminal domainBH (Fig. 2E) with segment 2 contribute to the asymmetry of 10S. In addition, the HO linkerBH interacts with segment 3 (Fig. 2F). This extensive network of intermolecular interactions not only establishes the interface between both heads but also contributes to compromised actin binding and hence the lack of actin-activation of the ATPase activity observed in biochemical studies (10, 12, 14, 28). Specifically, major elements of the actin-binding region of the BH, defined as loop-2, loop-3, loop-4, CM-loop, and helix-loop-helix (HLH), are either sequestered or sterically blocked by extensive interactions with the FH (Fig. 2, A to C, and fig. S6). Loop-2BH shields HLH and prevents the cleft closure required to establish a strong actin binding interface (fig. S5D). The actin binding interface of the FH is partially blocked by sequestration of the CM-loopFH and interactions of loop-2FH with segment 1 (Fig. 1A and fig. S5D). Loop-4FH and HLHFH are available for actin binding (figs. S6, D and F, and S7) and therefore contribute to the residual actin binding properties of 10S (10, 12). Together, the head-head interaction accounts for the compromised actin binding properties and ATPase activity of 10S.
A previously unknown conformation of the active site explains trapping of the ATP hydrolysis products
Our cryo-EM reconstruction of 10S shows that both heads retain the hydrolysis products, ADP and Pi, in agreement with biochemical data (Fig. 3, B to E, and movie S4) (10, 11). To the best of our knowledge, this is the first time that ADP and Pi produced by the hydrolysis of ATP have been resolved in any myosin structure without ambiguity. The ADPBH and ADPFH adopt distinct conformations (RMSD 2.15 Å) and are held in place through interaction with switch-1 and switch-2 and the P-loop. Switch-1 and switch-2 are in the closed conformation in both heads, reminiscent of the pre-powerstroke conformation (table S2). Notably, the repositioning of R247 in each head prevents the formation of the characteristic salt bridge between switch-1 R247 and switch-2 E470 (Fig. 3, B to E, and fig. S8A). The unique arrangement of structural elements in the active site exposes the blocked phosphate release tube at the back door in the FH (movie S5) (29, 30). Pi,FH is located at the entrance to the phosphate tube and has moved ~1.6 Å relative to Pi,BH (Fig. 3F). Sentinel residues S245 and S179 in switch-1FH and the P-loopFH together with the ADPFH form a cage for Pi,FH (Fig. 3B and fig. S8C) that prevents the movement of Pi into the phosphate tube and its subsequent release. The trapped Pi itself acts as a door stop and prevents the closure of the cleft that would lead to actin binding and product release in agreement with biochemical and in silico studies (11, 31, 32). The ADP.Pi state in the FH is further stabilized by interactions between the P-loop and ADP (Fig. 3B).
The phosphate tube is open in the BH (Fig. 3F and movie S5). Pi,BH is accommodated by interactions with the P-loopBH and switch-1BH (Fig. 3, C to E, and fig. S8, A to C). Interactions between the converter and the N terminus with segment 2 (17, 18) further inhibit active site rearrangements and prevent Pi release through the back door (Fig. 2, D and E) (31, 33). The front door route is blocked by the ADP and repulsive interactions with the α- and β-phosphates (Fig. 3, B to E). The relay helix that connects switch-2 with the converter is in the kinked conformation in both heads (fig. S8C), reminiscent of the pre-powerstroke state.
The observed rearrangement of the nucleotide switches together with the different positions of ADP and Pi in the active site, cleft, and lever positions contrast with structural states of isolated myosin motor domains obtained with x-ray crystallography and cryo-EM (table S2). This challenges the paradigm that the heads in 10S are in a bona fide pre-powerstroke state (25). The conformation of the primed heads in the off state instead suggests a branched kinetic scheme of the basic actomyosin ATPase cycle that is described by discrete structurally uncoupled states (table S2). The off state efficiently traps the nucleotide and prevents the actin-activation of the ATPase activity. Further, it prevents both, the reversal of the hydrolysis from the ADP.Pi to the ATP state and the transition into the ADP state, thereby contributing to the stability of 10S.
Both myosin light chains contribute to the asymmetry and stabilization of 10S
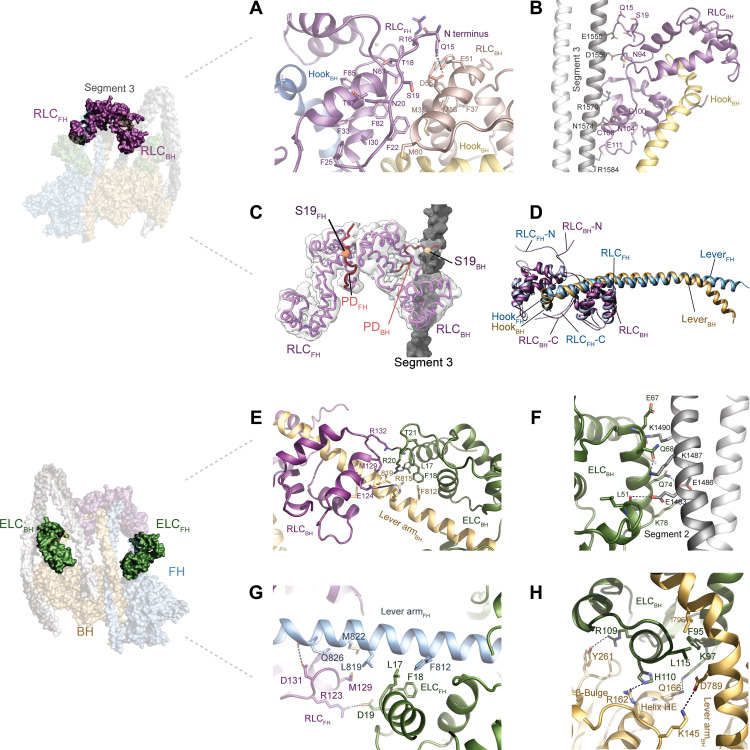
The RLC plays critical roles in the formation and stabilization of 10S (10, 34, 35). Further, it is the major regulator of the 10S/filament transition as phosphorylation of the RLC on S19 releases the tail from the IHM. This relieves the 10S state, promotes filament formation, and activates myosin motor activity (fig. S1B) (10, 12, 13, 36). At present, there are no high-resolution structures of the RLC that include the 24 N-terminal residues, also referred to as the phosphorylation domain (PD), to explain the details of the activation mechanism of 10S. We resolved amino acid 15 to the end of the RLCFH and amino acid 14 to the end of the RLCBH that include the S19 phosphorylation site (Fig. 4, A to C). The very N terminus of neither PD is visible in our structure, likely owing to its flexibility and the lack of stabilizing interactions. This observation is in line with previous biochemical studies that suggest dispensable roles for residues 1 to 14 in 10S formation (36–38). A network of hydrophobic interactions stabilizes the RLCBH-RLCFH (Fig. 4A) interface and electrostatic interactions stabilize the RLCBH:segment 3 interdomain interface of 10S (Fig. 4B). Specifically, the ~1215-Å2 solvent-accessible surface area at the RLCFH:RLCBH interface is stabilized by the PDFH including the phosphorylatable S19 (Fig. 4A). Key electrostatic interactions include Q15RLC,FH:D55RLC,BH and Q15RLC,FH:E51RLC,BH. Further, N20RLC,FH interacts with T87RLC,FH to lock in the position of the PD (Fig. 4A). The structural data agrees with biochemical studies that highlight the essential roles of RLC residues 15 to 20 and the dispensable roles for residues 1 to 14 in 10S formation (36, 38). S19FH is buried in the interface between both RLCs and is inaccessible for kinases (Fig. 4, A and C). The N terminus of the RLCBH docks against segment 3 (Fig. 4B). The ~1700-Å2 interface is largely stabilized by ionic interactions among D1559segment 3, N1574segment 3, and R1584segment 3 with N94RLC,BH, N104RLC,BH, and E111RLC,BH (Fig. 4B). S19RLC,BH is surface-exposed and not involved in interactions with segment 3, suggesting that it is amenable for phosphorylation by kinases including myosin light chain kinase (8). The presence of an accessible and inaccessible S19 phosphorylation site on the RLCBH and RLCFH (Fig. 4C and movie S6), respectively, agrees with biochemical data (10).
Fig. 4. Light chain interactions.
(A) Interactions between the RLCBH (salmon) and the RLCFH (dark purple). The PD of the RLCFH forms extensive intra and intersubunit interactions. (B) RLCBH interacts with segment 3. (C) Position of the phosphorylatable residue S19 is shown for RLCBH (wheat spheres) and RLCFH (orange spheres). The resolved PDs of the RLCs are shown in red. S19FH is sandwiched between both RLCs; S19BH is in close proximity to segment 3 (gray). (D) Different binding modes of the RLCFH and the RLCFH to the respective lever arms. (E) Interactions between the ELCBH with the RLCBH and the lever armBH. (F) Interactions between the ELCBH and segment 2. (G) Interactions between the ELCFH and the RLCFH. (H) Interactions between the ELCBH, the BH, and the lever armBH.
Further, we resolved the C terminus of the RLC that plays important roles in stabilizing the 10S state (35). Most notably, the C termini of the RLCBH and RLCFH adopt a clamp-like conformation that stabilizes the interaction with the respective hooks of the lever arms at the head-tail junction (Fig. 4D). This conformation has not been observed in earlier structures of active and inactive myosins, suggesting that not only the PD but also the C terminus plays an active role in 10S formation and stabilization.
As with the RLC, we observed distinct binding modes of the ELCBH and the ELCFH to the respective myosin lever arms (Fig. 4 and fig. S9). The ELCBH forms intricate interactions with elements of the BH, segment 2, and the RLCBH (Fig. 4, E, F, and H) that contribute to the stability of the acutely bent lever arm (Fig. 3A and fig. S9). In contrast, the ELCFH only forms a single interaction with the RLCFH (D19ELC,BH:R123RLC,FH) (Fig. 4G). These differences in intersubunit interactions are imposed by the pliant region between the converter of the motor domain and the ELC that dictates the asymmetry of 10S (fig. S9).
Comparative analyses of the light chains in our structure show distinct conformations of the ELCs and RLCs bound to BH and FH that stabilize and contribute to the asymmetry of 10S (fig. S9). Notably, the conformations of the RLCs and ELCs differ from previous crystal structures of single-headed myosin-2s in the pre-powerstroke state by an RMSD of ~2.5 Å, suggesting that conformational changes in the light chains are required to assume and stabilize 10S (fig. S9).
DISCUSSION
Mechanism of 10S formation, stabilization, and phosphorylation activation
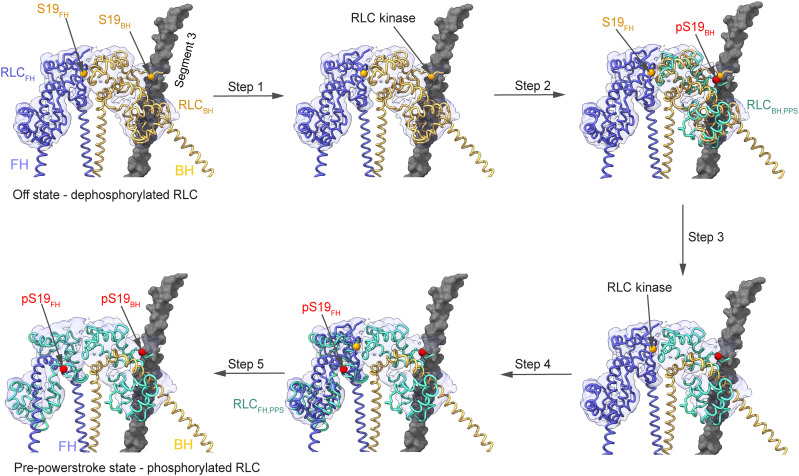
On the basis of our structure and previous biochemical data, we propose the following possible sequence of kinetic and structural events that result in the formation, stabilization, and activation of 10S by RLC phosphorylation (Fig. 5 and table S2).
Fig. 5. Model for 10S relief by RLC phosphorylation.
In the off state, phosphorylation of the RLCBH on S19 (pS19) causes structural transitions to the pre-powerstroke state (RLCBH,PPS, PDB ID: 1QVI; pS19 modeled) that make the RLCFH available for phosphorylation. Collectively, this is expected to result in 10S relief and the subsequent activation of myosin motor activity.
10S formation
Dephosphorylation of the RLC results in the formation of the IHM (7, 14, 16). This destabilizes the interactions of the rod that are necessary to form a filament. The formation of 10S occurs after dissociation of the RLC dephosphorylated myosin from a filament (39). The 10S state is formed by asymmetric head-head interactions in the IHM, unique conformations of both light chains, and the complex folding of the tail (Fig. 1, fig. S1, and movie S1). The 10S state is strongly disfavored in the presence of the phosphorylated RLC because a phosphate moiety on S19BH likely sterically clashes with segment 3 and a phosphate moiety on S19FH interferes with the formation and stabilization of the RLCBH-RLCFH interface (fig. S10). A distortion in the head domains that is induced by the binding of ATP and the hydrolysis of ATP to ADP and Pi in the unphosphorylated myosin transitions the motor in the off state rather than the pre-powerstroke state (Fig. 1, A and B). This kinetic step likely precedes folding of the heads into the IHM and subsequently the 10S conformation and prevents the release of the ATP hydrolysis products (Fig. 3, fig. S8, and movie S5) (40).
10S stabilization
In the off state, the myosin heads trap the ATP hydrolysis products ADP and Pi in their active sites (Fig. 3, fig. S8, and movie S5). The off state is best described by a structural uncoupling between the active site, the cleft, and the lever arm in BH and FH (Fig. 1, fig. S9, table S2, and movie S3). The off state is essential to 10S stability as it suppresses the release of the hydrolysis products ADP and Pi (11). The unique, wide open cleft and the sequestration of structural elements of the actin interface to stabilize the 10S conformation (figs. S5, B and D, and S6) result in weak actin binding (Kd > 100 μM) and a ~1000-fold reduced myosin ATPase activity (10, 28).
10S relief
The RLCBH is phosphorylated by kinases including myosin light chain kinase (7, 41, 42) on S19BH (Figs. 1B and 5). The presence of the phosphate moiety on S19BH induces complex structural changes (movies S7 and S8). The RLCBH undergoes a pronounced conformational change into the pre-powerstroke conformation in agreement with biochemical studies (43). The PDBH likely disengages with segment 3, and the C terminus of the RLCBH is repositioned to adopt a pre-powerstroke conformation. This weakens the RLCBH-RLCFH interaction at the interface due to steric clashes and charge repulsions and leads to a major transition of the PDFH that exposes S19FH (Fig. 5, fig. S10, and movies S7 and S8). The increased accessible surface area of the PD of the RLCFH is expected to facilitate the interaction with RLC kinases. Phosphorylation of S19FH disrupts the RLCBH-RLCFH interface and triggers a conformational change of the RLCFH into the pre-powerstroke conformation (Fig. 5, fig. S10, and movies S7 and S8). Together, these transitions cause 10S relief and promote filament formation through the rod domain (28). This is necessary but not sufficient to activate myosin ATPase activity. Structural changes in the phosphorylated RLCs are likely transmitted via allosteric intersubunit communication through the ELC to the motor domain and trigger Pi release and hence the activation of myosin enzyme function.
Importance of the 10S structure for basic and translational science
The highly conserved IHM (20), the structural basis of the “super-relaxed state” (18, 44, 45), plays a key role in the regulation of striated muscle contraction (46) with special relevance to cardiac muscle contraction (45, 47). Many mutations in the ß-cardiac myosin-2 heavy chain gene associated with hypertrophic cardiomyopathy (HCM) cause hypercontractility of the heart by shifting the equilibrium between myosin-2 heads in the autoinhibited state to the active state (18, 48–51). HCM mutations cluster predominantly in the “myosin mesa,” a flat surface in the myosin head (49, 52), the converter (53, 54), and parts of segment 1 that contact the IHM (18, 48–50). These regions play key roles in the stabilization of 10S (Fig. 1) and are amenable for drug-discovery efforts, as seen in the development of the myosin inhibitor mavacamten that stabilizes the autoinhibited state of cardiac myosin-2 in vitro (50, 55) and in vivo (55, 56) and reverses the destabilizing effect of HCM mutations on the super-relaxed state (50, 57). The improved resolution of our 10S structure will facilitate future basic and translational research efforts to describe how mutations destabilize or dysregulate the IHM and lead to HCM and to exploit previously unidentified inter- and intramolecular interfaces for the development of mechanism-based therapeutics for HCM.
MATERIALS AND METHODS
Protein purification
Frozen chicken gizzards (100 g) (Pel-Freez Biologicals) were thawed, and the fascia was removed. The tissue was homogenized in 500 ml of buffer A [10 mM Mops (pH 7.0), 50 mM NaCl, 0.1 mM EGTA, 1 mM MgCl2, 1 mM dithiothreitol (DTT), and 0.1 mM phenylmethylsulfonyl fluoride containing 0.5% Triton X-100 in a Waring blender for 30 s. The homogenate was sedimented [10,000 rpm in a F-14-6 rotor (Thermo Fisher Scientific) for 10 min at 4°C], and the pellet was resuspended in 400 ml of buffer A. This step was repeated three times. The final pellet was homogenized in 400 ml of buffer B [40 mM Mops (pH 7.0), 40 mM NaCl, 1 mM EDTA, 2 mM EGTA, 1 mM DTT, and 10 mM ATP] and stirred for 30 min at 4°C. The extract was sedimented (20,000 rpm in a T29-8 rotor (Thermo Fisher Scientific) for 20 min at 4°C]. The supernatant was supplemented with 5 M NaCl and 0.5 M MgSO4 to final concentrations of 600 and 20 mM, respectively. Saturated (NH4)2SO4 was slowly added to 42% of saturation, and the solution was sedimented (12,000 rpm in a F-14 rotor for 10 min at 4°C). The supernatant was removed, and the volume was measured. Saturated (NH4)2SO4 was slowly added to 60% of saturation, and the solution was centrifuged (12,000 rpm in a F-14 rotor for 20 min at 4°C). The pellet was dissolved in 25 ml of buffer C [10 mM Mops (pH 7.0), 25 mM NaCl, 0.1 mM EGTA, and 1 mM DTT) and dialyzed overnight against 4 liters of the same buffer. The dialysate was sedimented [20,000 rpm in a T29-8 rotor (Thermo Fisher Scientific) for 10 min at 4°C], and the pellet was resuspended in 20 ml of buffer D [10 mM Mops (pH 7.0), 600 mM NaCl, 0.1 mM EGTA, and 1 mM DTT]. The protein was further purified with size exclusion chromatography on a HiLoad 16/600 Superose 6 pg (Cytiva) in size exclusion chromatography buffer [20 mM Mops (pH 7.5), 500 mM KCl, 0.1 mM EGTA, 1 mM MgCl2, and 2 mM DTT].
Sample preparation and Cryo-EM data collection
The purified protein was cross-linked for cryo-EM studies to stabilize the 10S conformation. In brief, the protein was diluted to 1 mg/ml in low-salt buffer [10 mM Mops (pH 7.3), 150 mM NaCl, 1 mM EGTA, and 2 mM MgCl2] and incubated with 1 mM ATP for 30 min. The protein was cross-linked using 0.1% glutaraldehyde for 1 min, and the reaction was quenched with 0.1 M tris (pH 8.0). The homogeneity of the cross-linked samples was assessed in negative staining EM (fig. S2). For cryo-EM, C-flat 1.2/1.3 300-mesh Au grids were glow-discharged with a PELCO easiGlow (Ted Pella Inc.) for 45 s. A volume of 4 μl of the cross-linked protein sample was applied to the grids, incubated for 1 min, and blotted for 4 s at 95% humidity. The grids were plunge-frozen into liquid ethane using a Leica EM Grid Plunger 2 (GP2) (Leica Microsystems) and stored in liquid nitrogen until data collection.
To assess sample stability and particle distribution, the grids were screened on a Glacios (Thermo Fisher Scientific) equipped with Falcon 3EC direct electron detector at 200 kV. Optimal grids based on the ice thickness and sample distribution were transferred to a Titan Krios G3i (Thermo Fisher Scientific) operated at 300 kV. The Krios was equipped with a K3 direct electron detector, a Bioquantum energy filter, and a Cs image corrector. A total of 2678 movies with a magnification of ×81,000, corresponding to a pixel size of 0.4495 Å, were collected in super-resolution mode at a defocus range from −1.5 to −3.5 μm with a total electron dose of 65 e−/Å2 per movie. Data collection was performed using EPU software (Thermo Fisher Scientific). Data collection statistics are shown in table S1.
Image processing and 3D reconstruction
All raw movies were aligned, drift-corrected, and dose-weighed using MotionCor2 software (58). Defocus and contrast transfer function (CTF) parameters were estimated using Gctf wrapper in cryoSPARC v2.15 (59). Particle picking routines in cryoSPARC v2.15 (59) were used to generate templates for particle picking. A small set of particles for reference-free two-dimensional (2D) classifications was selected and subsequently used as templates for four rounds of iterative 2D classifications to remove junk particles. Fifteen well-aligned 2D classes were used as templates for ab initio 3D reconstruction with 448,685 particles in cryoSPARC. A major class with 234,464 particles of three classes that showed clear 10S-like features was selected for homogenous 3D refinements and achieved a global resolution of 4.2 Å. A soft mask was applied to mask out the flexible segment regions distal to the IHM in Relion (60). The initial binary mask was extended three pixels in all directions and subsequently further extended with a raised-cosine soft edge of six pixels in width to improve the local resolution of the reconstructed map. Nonuniform refinement using this mask was performed after optimizing and refining the per-particle defocus parameters in cryoSPARC. A global resolution of 3.40 Å was achieved in cryoSPARC at a Fourier shell correlation (FSC) at 0.143, and the final FSC curve was corrected for the effect of a soft mask using high-resolution noise substitution. This 3D map was postprocessed with Locscale in CCP-EM (Collaborative Computational Project for Electron Cryo-Microscopy) (61). All reported resolutions are based on the gold-standard FSC = 0.143 criteria. Data collection statistics and image processing summary are shown in table S1 and fig. S3.
Model building, refinement, and validation
The postprocessed map was used to build a model of smooth muscle myosin-2 10S (sequence IDs: myosin heavy chain, PP10587; ELC, P02607; RLC, P02612) using templates of human nonmuscle myosin-2C [Protein Data Bank (PDB) entry 5I4E] and smooth muscle myosin-2 (PDB entry 1BR2) for the motor domains. The structure of chicken smooth muscle myosin-2 with bound light chains (PDB entry 3J04) was used as an initial template for building models for ELC and RLC using rigid body docking into the cryo-EM reconstructions of 10S with molecular dynamics flexible fitting function in Chimera (62). Iterative model building was performed using real-space refinement in Phenix (63) and Coot (64) to manually build the motor domains and the ELC. The C terminus of the RLC and part of the PD of both RLCs that includes T18 and S19 were manually built. ADP, magnesium, and phosphate molecules were placed in the cryo-EM densities using Coot (64). Elements of loop-2, CM-loop, and loop-4 in both motor domains were manually built in Coot (64) as they adopt distinct conformations compared to known myosin structures.
The structure of human ß-cardiac myosin S2 fragment (PDB entry 2FXM) was used as template to build segment 1 of 10S. α-Helical restraints in Coot (64) were used to refine the segment 1. To place the coiled-coil segments 2 and 3, residue G1530 was used as a marker. Sequence analysis revealed a single glycine (G1530) in the junction of segments 2 and 3. This residue was predicted to serve as a hinge point between both segments. The structures of human ß-cardiac myosin segments 2 and 3 (PDB entries 6PFP and 4XA4) were used as template to build segments 2 and 3 of smooth muscle myosin-2, respectively. In addition, the classic markers (N1532 from segment 3 and C108 from RLCBH) were selected for segment placing. Manual adjustments to the backbone and side chains for this model were performed in Coot (64) and real-space refinement was performed using Phenix (63). Models were further minimized and refined using Namdinator (65), which aids in automatic molecular dynamics flexible fitting of structural models into cryo-EM maps using group ADP and real-space refinements and validation tools in Phenix (63) together with the RosettaCommons software package (66). Although the distal parts of the segments of the model have local resolutions greater than 5 Å, these regions were placed with confidence using Coot (64) based on the known cross-linking markers (26) and G1530 as the sole hinge point at the junction of segments 2 and 3. Of note, at this resolution, the rotamer conformations of the side chains of the residues may not be faithfully determined. However, we used the high-resolution structures of segments 2 and 3 from human β-cardiac myosin as templates to build the best possible rotamers using Coot (64) and minimized the rotamers using Namdinator (65). All models were manually refined using postprocessed cryo-EM reconstruction and manually verified using Coot (64). The refined model was validated using MolProbity (67). The RMSD for BH and FH was calculated for residues 29 to 788. Proteins, Interfaces, Structures and Assemblies (PISA) analysis (68) was used to calculate the solvent-accessible surface areas. Of note, cryo-EM maps usually do not have isotropic resolution (69), as seen in fig. S2E for 10S. For this reason, only key side chains and their interactions are shown in regions of the molecules where the local resolution is <4 Å, if not stated otherwise, and the cryo-EM map is of sufficient quality to place side chains with high confidence. The nomenclature for secondary structure assignments and key structural elements in 10S is based on prior literature (70, 71). Figures were made with UCSF Chimera (62), PyMol (https://pymol.org), and BioRender (www.BioRender.com).
Acknowledgments
We thank Y. Narui and B. Deng [Center for Electron Microscopy and Analysis (CEMAS), The Ohio State University] for help with data collection. We thank the CEMAS Shared Resource at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for support, and use of the facility.
Funding: This study was supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant number R01GM143539 (to K.C.); the Ohio State University Comprehensive Cancer Center and the NIH under grant number P30 CA016058; and the Intramural Research Program of the National Heart, Lung, and Blood Institute of the NIH under grant number HL001786 (to J.R.S.).
Author contributions: Conceptualization: K.C. Investigation: S.M.H., A.S.A., N.B., J.R.S., and K.C. Formal analysis: S.M.H. and K.C. Supervision: K.C. Validation: K.C. Visualization: S.M.H. and K.C. Writing—original draft: S.M.H. and K.C. Writing—review and editing: S.M.H., A.S.A., N.B., J.R.S., and K.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Atomic coordinates generated in this study have been deposited in the PDB under accession number 7MF3. The cryo-EM map generated in this study has been deposited in the Electron Microscopy Data Bank under accession number MDB-23810.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S2
Legends for movies S1 to S8
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S8
REFERENCES AND NOTES
- 1.Heissler S. M., Sellers J. R., Kinetic adaptations of myosins for their diverse cellular functions. Traffic 17, 839–859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sellers J. R., Regulation of cytoplasmic and smooth muscle myosin. Curr. Opin. Cell Biol. 3, 98–104 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Sellers J. R., Myosins: A diverse superfamily. Biochim. Biophys. Acta 1496, 3–22 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Rayment I., Holden H., Whittaker M., Yohn C., Lorenz M., Holmes K., Milligan R., Structure of the actin-myosin complex and its implications for muscle contraction. Science 261, 58–65 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Geeves M. A., Review: The ATPase mechanism of myosin and actomyosin. Biopolymers 105, 483–491 (2016). [DOI] [PubMed] [Google Scholar]
- 6.McLachlan A. D., Karn J., Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature 299, 226–231 (1982). [DOI] [PubMed] [Google Scholar]
- 7.Craig R., Smith R., Kendrick-Jones J., Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature 302, 436–439 (1983). [DOI] [PubMed] [Google Scholar]
- 8.Scholey J. M., Taylor K. A., Kendrick-Jones J., Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature 287, 233–235 (1980). [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H., Onishi H., Takahashi K., Watanabe S., Structure and function of chicken gizzard myosin. J. Biochem. 84, 1529–1542 (1978). [DOI] [PubMed] [Google Scholar]
- 10.Sellers J. R., Mechanism of the phosphorylation-dependent regulation of smooth muscle heavy meromyosin. J. Biol. Chem. 260, 15815–15819 (1985). [PubMed] [Google Scholar]
- 11.Cross R. A., Jackson A. P., Citi S., Kendrick-Jones J., Bagshaw C. R., Active site trapping of nucleotide by smooth and non-muscle myosins. J. Mol. Biol. 203, 173–181 (1988). [DOI] [PubMed] [Google Scholar]
- 12.Sellers J. R., Eisenberg E., Adelstein R. S., The binding of smooth muscle heavy meromyosin to actin in the presence of ATP. Effect of phosphorylation. J. Biol. Chem. 257, 13880–13883 (1982). [PubMed] [Google Scholar]
- 13.Kendrick-Jones J., Cande W. Z., Tooth P. J., Smith R. C., Scholey J. M., Studies on the effect of phosphorylation of the 20,000 Mr light chain of vertebrate smooth muscle myosin. J. Mol. Biol. 165, 139–162 (1983). [DOI] [PubMed] [Google Scholar]
- 14.Trybus K. M., Huiatt T. W., Lowey S., A bent monomeric conformation of myosin from smooth muscle. Proc. Natl. Acad. Sci. U.S.A. 79, 6151–6155 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Wendt T., Taylor D., Taylor K., Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J. Mol. Biol. 329, 963–972 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Wendt T., Taylor D., Messier T., Trybus K. M., Taylor K. A., Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J. Cell Biol. 147, 1385–1390 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendt T., Taylor D., Trybus K. M., Taylor K., Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. U.S.A. 98, 4361–4366 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alamo L., Wriggers W., Pinto A., Bártoli F., Salazar L., Zhao F. Q., Craig R., Padrón R., Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J. Mol. Biol. 384, 780–797 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodhead J. L., Zhao F.-Q., Craig R., Egelman E. H., Alamo L., Padrón R., Atomic model of a myosin filament in the relaxed state. Nature 436, 1195–1199 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Lee K. H., Sulbarán G., Yang S., Mun J. Y., Alamo L., Pinto A., Sato O., Ikebe M., Liu X., Korn E. D., Sarsoza F., Bernstein S. I., Padrón R., Craig R., Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc. Natl. Acad. Sci. U.S.A. 115, E1991–E2000 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung H. S., Burgess S. A., Billington N., Colegrave M., Patel H., Chalovich J. M., Chantler P. D., Knight P. J., Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 6022–6026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S. A., Yu S., Walker M. L., Hawkins R. J., Chalovich J. M., Knight P. J., Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol. 372, 1165–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Onishi H., Wakabayashi T., Electron microscopic studies of myosin molecules from chicken gizzard muscle I: The formation of the intramolecular loop in the myosin tail. J. Biochem. 92, 871–879 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Scarff C. A., Carrington G., Casas-Mao D., Chalovich J. M., Knight P. J., Ranson N. A., Peckham M., Structure of the shutdown state of myosin-2. Nature 588, 515–520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., Tiwari P., Lee K. H., Sato O., Ikebe M., Padrón R., Craig R., Cryo-EM structure of the inhibited (10S) form of myosin II. Nature 588, 521–525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olney J. J., Sellers J. R., Cremo C. R., Structure and function of the 10 S conformation of smooth muscle myosin. J. Biol. Chem. 271, 20375–20384 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Onishi H., Maita T., Matsuda G., Fujiwara K., Interaction between the heavy and the regulatory light chains in smooth muscle myosin subfragment 1. Biochemistry 31, 1201–1210 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Cross R. A., Citi S., Kendrick-Jones J., How phosphorylation controls the self-assembly of vertebrate smooth and non-muscle myosins. Biochem. Soc. Trans. 16, 501–503 (1988). [DOI] [PubMed] [Google Scholar]
- 29.Cecchini M., Alexeev Y., Karplus M., Pi release from myosin: A simulation analysis of possible pathways. Structure 18, 458–470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson J. D., Pate E., Rayment I., Yount R. G., Molecular dynamics analysis of structural factors influencing back door Pi release in myosin. Biophys. J. 86, 3794–3803 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yount R. G., Lawson D., Rayment I., Is myosin a "back door" enzyme? Biophys. J. 68, 44S-47S; discussion 47S–49S (1995). [PMC free article] [PubMed] [Google Scholar]
- 32.Llinas P., Isabet T., Song L., Ropars V., Zong B., Benisty H., Sirigu S., Morris C., Kikuti C., Safer D., Sweeney H. L., Houdusse A., How actin initiates the motor activity of myosin. Dev. Cell 33, 401–412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robert-Paganin J., Pylypenko O., Kikuti C., Sweeney H. L., Houdusse A., Force generation by myosin motors: A structural perspective. Chem. Rev. 120, 5–35 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Trybus K. M., Lowey S., The regulatory light chain is required for folding of smooth muscle myosin. J. Biol. Chem. 263, 16485–16492 (1988). [PubMed] [Google Scholar]
- 35.Trybus K. M., Chatman T. A., Chimeric regulatory light chains as probes of smooth muscle myosin function. J. Biol. Chem. 268, 4412–4419 (1993). [PubMed] [Google Scholar]
- 36.Ikebe M., Morita J., Identification of the sequence of the regulatory light chain required for the phosphorylation-dependent regulation of actomyosin. J. Biol. Chem. 266, 21339–21342 (1991). [PubMed] [Google Scholar]
- 37.Ikebe M., Hartshorne D. J., Conformation-dependent proteolysis of smooth-muscle myosin. J. Biol. Chem. 259, 11639–11642 (1984). [PubMed] [Google Scholar]
- 38.Ikebe M., Ikebe R., Kamisoyama H., Reardon S., Schwonek J. P., Sanders C. R. II, Matsuura M., Function of the NH2-terminal domain of the regulatory light chain on the regulation of smooth muscle myosin. J. Biol. Chem. 269, 28173–28180 (1994). [PubMed] [Google Scholar]
- 39.Rosenfeld S. S., Xing J., Rener B., Lebowitz J., Kar S., Cheung H. C., Structural and kinetic studies of the 10 S<==>6 S transition in smooth muscle myosin. J. Biol. Chem. 269, 30187–30194 (1994). [PubMed] [Google Scholar]
- 40.Cross R. A., Cross K. E., Sobieszek A., ATP-linked monomer-polymer equilibrium of smooth muscle myosin: The free folded monomer traps ADP.Pi. EMBO J. 5, 2637–2641 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellers J. R., Pato M. D., Adelstein R. S., Reversible phosphorylation of smooth muscle myosin, heavy meromyosin, and platelet myosin. J. Biol. Chem. 256, 13137–13142 (1981). [PubMed] [Google Scholar]
- 42.Trybus K. M., Filamentous smooth muscle myosin is regulated by phosphorylation. J. Cell Biol. 109, 2887–2894 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kast D., Espinoza-Fonseca L. M., Yi C., Thomas D. D., Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proc. Natl. Acad. Sci. U.S.A. 107, 8207–8212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Khayat H. A., Kensler R. W., Squire J. M., Marston S. B., Morris E. P., Atomic model of the human cardiac muscle myosin filament. Proc. Natl. Acad. Sci. U.S.A. 110, 318–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hooijman P., Stewart M. A., Cooke R., A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 100, 1969–1976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phung L. A., Foster A. D., Miller M. S., Lowe D. A., Thomas D. D., Super-relaxed state of myosin in human skeletal muscle is fiber-type dependent. Am. J. Phys. Cell Physiol. 319, C1158–C1162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robert-Paganin J., Auguin D., Houdusse A., Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 9, 4019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nag S., Trivedi D. V., Sarkar S. S., Adhikari A. S., Sunitha M. S., Sutton S., Ruppel K. M., Spudich J. A., The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24, 525–533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trivedi D. V., Adhikari A. S., Sarkar S. S., Ruppel K. M., Spudich J. A., Hypertrophic cardiomyopathy and the myosin mesa: Viewing an old disease in a new light. Biophys. Rev. 10, 27–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson R. L., Trivedi D. V., Sarkar S. S., Henze M., Ma W., Gong H., Rogers C. S., Gorham J. M., Wong F. L., Morck M. M., Seidman J. G., Ruppel K. M., Irving T. C., Cooke R., Green E. M., Spudich J. A., Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. U.S.A. 115, E8143–E8152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adhikari A. S., Trivedi D. V., Sarkar S. S., Song D., Kooiker K. B., Bernstein D., Spudich J. A., Ruppel K. M., β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 10, 2685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spudich J. A., The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem. Soc. Trans. 43, 64–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Giustiniani D., Arad M., Ortíz-Genga M., Barriales-Villa R., Fernández X., Rodríguez-García I., Mazzanti A., Veira E., Maneiro E., Rebolo P., Lesende I., Cazón L., Freimark D., Gimeno-Blanes J. R., Seidman C., Seidman J., Kenna W. M., Monserrat L., Phenotype and prognostic correlations of the converter region mutations affecting the β myosin heavy chain. Heart 101, 1047–1053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawana M., Sarkar S. S., Sutton S., Ruppel K. M., Spudich J. A., Biophysical properties of human β-cardiac myosin with converter mutations that cause hypertrophic cardiomyopathy. Sci. Adv. 3, e1601959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green E. M., Wakimoto H., Anderson R. L., Evanchik M. J., Gorham J. M., Harrison B. C., Henze M., Kawas R., Oslob J. D., Rodriguez H. M., Song Y., Wan W., Leinwand L. A., Spudich J. A., McDowell R. S., Seidman J. G., Seidman C. E., A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science 351, 617–621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stern J. A., Markova S., Ueda Y., Kim J. B., Pascoe P. J., Evanchik M. J., Green E. M., Harris S. P., A small molecule inhibitor of sarcomere contractility acutely relieves left ventricular outflow tract obstruction in feline hypertrophic cardiomyopathy. PLOS ONE 11, e0168407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohde J. A., Thomas D. D., Muretta J. M., Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. Proc. Natl. Acad. Sci. U.S.A. 114, E1796–E1804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burnley T., Palmer C. M., Winn M., Recent developments in the CCP-EM software suite. Acta Crystallogr. D Struct. Biol. 73, 469–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kidmose R. T., Juhl J., Nissen P., Boesen T., Karlsen J. L., Pedersen B. P., Namdinator—Automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leman J. K., Weitzner B. D., Lewis S. M., Adolf-Bryfogle J., Alam N., Alford R. F., Aprahamian M., Baker D., Barlow K. A., Barth P., Basanta B., Bender B. J., Blacklock K., Bonet J., Boyken S. E., Bradley P., Bystroff C., Conway P., Cooper S., Correia B. E., Coventry B., Das R., de Jong R. M., DiMaio F., Dsilva L., Dunbrack R., Ford A. S., Frenz B., Fu D. Y., Geniesse C., Goldschmidt L., Gowthaman R., Gray J. J., Gront D., Guffy S., Horowitz S., Huang P.-S., Huber T., Jacobs T. M., Jeliazkov J. R., Johnson D. K., Kappel K., Karanicolas J., Khakzad H., Khar K. R., Khare S. D., Khatib F., Khramushin A., King I. C., Kleffner R., Koepnick B., Kortemme T., Kuenze G., Kuhlman B., Kuroda D., Labonte J. W., Lai J. K., Lapidoth G., Leaver-Fay A., Lindert S., Linsky T., London N., Lubin J. H., Lyskov S., Maguire J., Malmström L., Marcos E., Marcu O., Marze N. A., Meiler J., Moretti R., Mulligan V. K., Nerli S., Norn C., Ó’Conchúir S., Ollikainen N., Ovchinnikov S., Pacella M. S., Pan X., Park H., Pavlovicz R. E., Pethe M., Pierce B. G., Pilla K. B., Raveh B., Renfrew P. D., Burman S. S. R., Rubenstein A., Sauer M. F., Scheck A., Schief W., Schueler-Furman O., Sedan Y., Sevy A. M., Sgourakis N. G., Shi L., Siegel J. B., Silva D.-A., Smith S., Song Y., Stein A., Szegedy M., Teets F. D., Thyme S. B., Wang R. Y.-R., Watkins A., Zimmerman L., Bonneau R., Macromolecular modeling and design in Rosetta: Recent methods and frameworks. Nat. Methods 17, 665–680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Lyumkis D., Challenges and opportunities in cryo-EM single-particle analysis. J. Biol. Chem. 294, 5181–5197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cope M. J., Whisstock J., Rayment I., Kendrick-Jones J., Conservation within the myosin motor domain: Implications for structure and function. Structure 4, 969–987 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Geeves M. A., Fedorov R., Manstein D. J., Molecular mechanism of actomyosin-based motility. Cell. Mol. Life Sci. 62, 1462–1477 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S2
Legends for movies S1 to S8
Movies S1 to S8