Loss of ATF4 intrinsically leads to functional aging-like attrition of adult hematopoietic stem cells via targeting HIF1α.
Abstract
Aging of hematopoietic stem cells (HSCs) directly contributes to dysfunction of hematopoietic and immune systems due to aging-associated alterations in HSC features. How the function of adult HSCs is regulated during aging so that relevant pathologic abnormalities may occur, however, remains incompletely understood. Here, we report that ATF4 deficiency provokes severe HSC defects with multifaceted aging-like phenotype via cell-autonomous mechanisms. ATF4 deletion caused expansion of phenotypical HSCs with functional attrition, characterized by defective repopulating and self-renewal capacities and myeloid bias. Moreover, the ATF4−/− HSC defects were associated with elevated mitochondrial ROS production by targeting HIF1α. In addition, loss of ATF4 significantly delayed leukemogenesis in the MLL-AF9–induced leukemia model. Mechanistically, ATF4 deficiency impaired HSC function with aging-like phenotype and alleviated leukemogenesis by regulating HIF1α and p16Ink4a. Together, our findings suggest a possibility of developing new strategies for the prevention and management of HSC aging and related pathological conditions.
INTRODUCTION
Aging is a complex and inevitable biological process characterized by a progressive decline in overall physiological functions, leading to decreased capacity to resist impairment and the increased susceptibility to pathogenesis of a variety of diseases such as neurodegenerative diseases, cardiovascular diseases, and cancers, and risk of death (1–4). It is well recognized that whole organismal aging can often be attributed to the aging process of adult stem cells (1, 4–10). In such a context, aging of hematopoietic stem cells (HSCs) has been found to be directly related to dysfunction of hematopoietic and immune systems due to aging-associated alterations in HSC features. Accumulating evidence demonstrates that both intrinsic or extrinsic factors thus far known to be involved in a wide variety of physiological or pathological events, such as oxidative stress, DNA damage, telomere shortening, and cell polarity, can contribute to HSC aging, linked to decreased HSC self-renewal and regeneration, impaired immune function, and increased cancer incidence (5, 8, 11, 12). Despite the above-described findings, however, driving mechanisms for HSC aging are yet to be fully understood, and identifying the regulatory genes key to the biological processes of HSC aging would facilitate understanding and intervening the mechanism underlying adult HSC aging.
The activating transcription factor 4 (ATF4) is a basic region leucine zipper transcription factor and plays pivotal roles in physiological responses to stresses including hypoxia, endoplasmic reticulum stress, amino acid deprivation, oxidation, and mitochondrial stress (13–16). Previous evidence revealed that ATF4 regulates fetal liver hematopoiesis (17, 18), bone formation (19), energy homeostasis (20), and cancer (16). ATF4 was recently reported to contribute to functional expansion of HSCs during embryonic development, as demonstrated in a mouse fetal liver model, in a cell-extrinsic manner through up-regulating cytokines such as Angptl3 secreted from stromal cells (18), and ATF4 up-regulation following amino acid deprivation could promote HSC survival through integrated stress response (ISR) (21). Of particular note is that previous reports have found that ATF4 modulates yeast lifespan extension and is up-regulated in the liver of aged mice (22–24), thus implicating a possible involvement of ATF4 in the regulation of adult aging. Nevertheless, in the context of general aging and adult HSC development, whether ATF4 is fundamentally involved in modulating adult HSC maintenance and functions remains unknown. To address this question, an appropriate mammal model with which contribution of ATF4 to the cellular phenotypes and functions of developing and adult HSCs can be assessed was needed.
Here, using constitutive and conditional ATF4-deficient mice, we found that ATF4 deletion led to functional defects of adult HSCs in association with aging-like phenotype through down-regulating hypoxia-inducible factor 1α (HIF1α) and consequently up-regulating mitochondrial reactive oxygen species (ROS). Our finding uncovers a previously unrecognized and direct role of ATF4 in regulating adult HSC aging and provides a foundation for further studying the significance of ATF4 in the pathogenesis of related conditions.
RESULTS
ATF4 is essential for the homeostasis of adult hematopoietic stem and progenitor cells
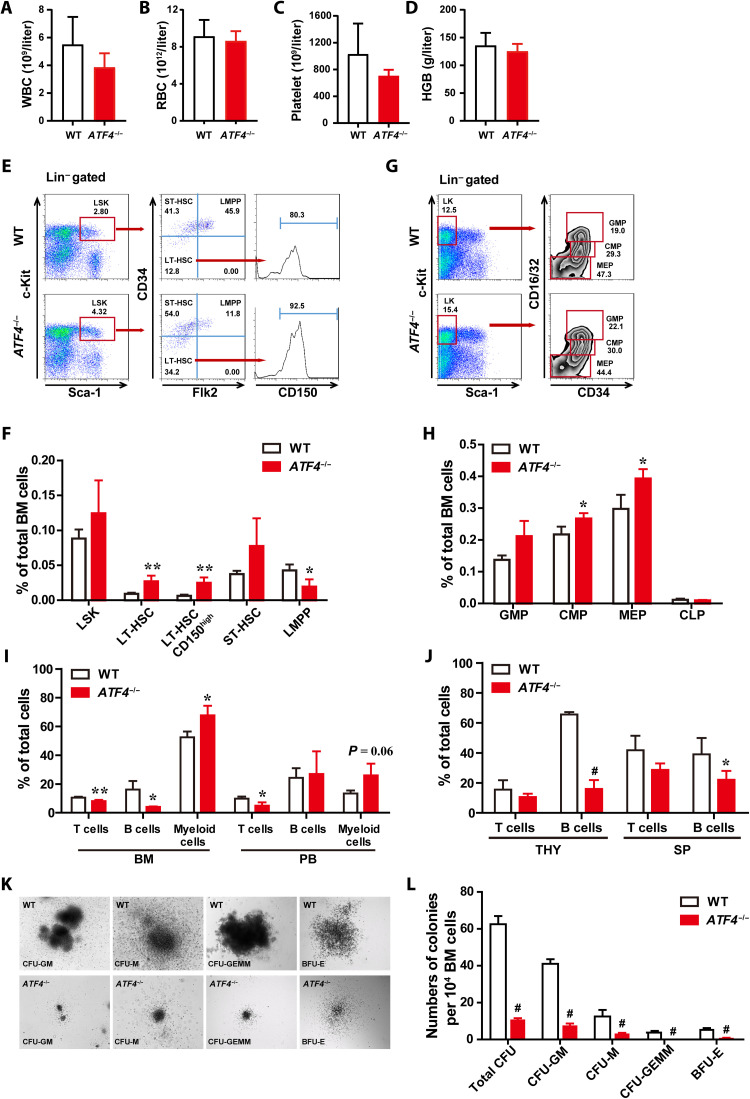
We first investigated the role of ATF4 in adult hematopoiesis under steady state using constitutive knockout mice (ATF4−/−). Compared to wild-type (WT) mice, ATF4−/− mice showed comparable blood counts and frequencies of different types of blood cells in peripheral blood (PB) (Fig. 1, A to D, and fig. S1, A to H). ATF4−/− mice displayed a significant increase in the percentages and numbers of LT-HSCs and CD150high–LT-HSCs (Long-term hematopoietic stem cells), which are characterized to myeloid-biased HSCs (25), and a marked decrease of lymphoid-primed multipotent progenitor (MPP) (LMPP) fraction in the bone marrow (BM; Fig. 1, E and F, and fig. S1I). Moreover, we found that the percentage of common myeloid progenitors (CMPs) and megakaryocyte-erythroid progenitors (MEPs) was elevated in ATF4−/− BM; however, there was no difference in total numbers of these populations (Fig. 1, G and H, and fig. S1J). Further analysis revealed that ATF4 deletion skewed toward myeloid lineage accompanied with decreased lymphoid lineages in ATF4−/− BM cells (Fig. 1I), and ATF4−/− mice exhibited significant decreases in lymphoid cells in the PB, thymus, and spleen (Fig. 1, I and J). To assess the differentiation potential of ATF4-deficient hematopoietic progenitors, we performed in vitro colony-forming unit (CFU) cell assays. As shown in Fig. 1 (K and L), the number and size of colonies derived from ATF4−/− BM MNCs were significantly reduced. In addition, similar results were obtained from ATF4−/− spleen cells (fig. S1, K to M). Collectively, our findings demonstrate that ATF4 plays an essential role in the homeostasis of adult hematopoiesis, and that its deficiency resulted in phenotypic expansion of HSCs, defective colony-forming ability, and myeloid differentiation skewing.
Fig. 1. ATF4 is essential for the homeostasis of adult hematopoietic stem and progenitor cells (HSPCs).
(A to D) PB cell counts of WT mice and ATF4−/− mice (4 months) (n = 6 to 7 mice per group). HGB, hemoglobin; RBC, red blood cells; WBC, white blood cells. (E and F) Representative FACS plots (E) of LSK cells, LT-HSCs, ST-HSCs, LMPPs, and CD150high-HSCs and histograms (F) showing the frequencies of the indicated cells in total BM cells of WT mice and ATF4−/− mice (2 to 4 months) (n = 4 mice per group). (G and H) Representative FACS plots (G) of GMPs, CMPs, and MEPs and histograms (H) showing the frequencies of the GMPs, CMPs, MEPs, and CLPs in total BM cells of WT mice and ATF4−/− mice (2 to 4 months) (n = 3 mice per group). (I and J) The frequencies of T, B, and myeloid cells in BM, PB, thymus (THY), and spleen (SP) from WT and ATF4−/− mice (4 months) (n = 3 to 5 mice per group). (K and L) Representative images (K) and the colony numbers (L) of total CFU, CFU-M, CFU-GM, CFU-GEMM, and BFU-E from BM cells of WT and ATF4−/− mice (n = 4 to 5). CFU-macrophages (CFU-M); CFU–granulocytes and macrophages (CFU-GM); CFU-granulocytes, erythrocytes, monocytes, and megakaryocytes (CFU-GEMM); and burst-forming unit–erythroid (BFU-E). *P < 0.05, **P < 0.01, and #P < 0.001 (t test).
Deletion of ATF4 impairs the function of adult HSCs
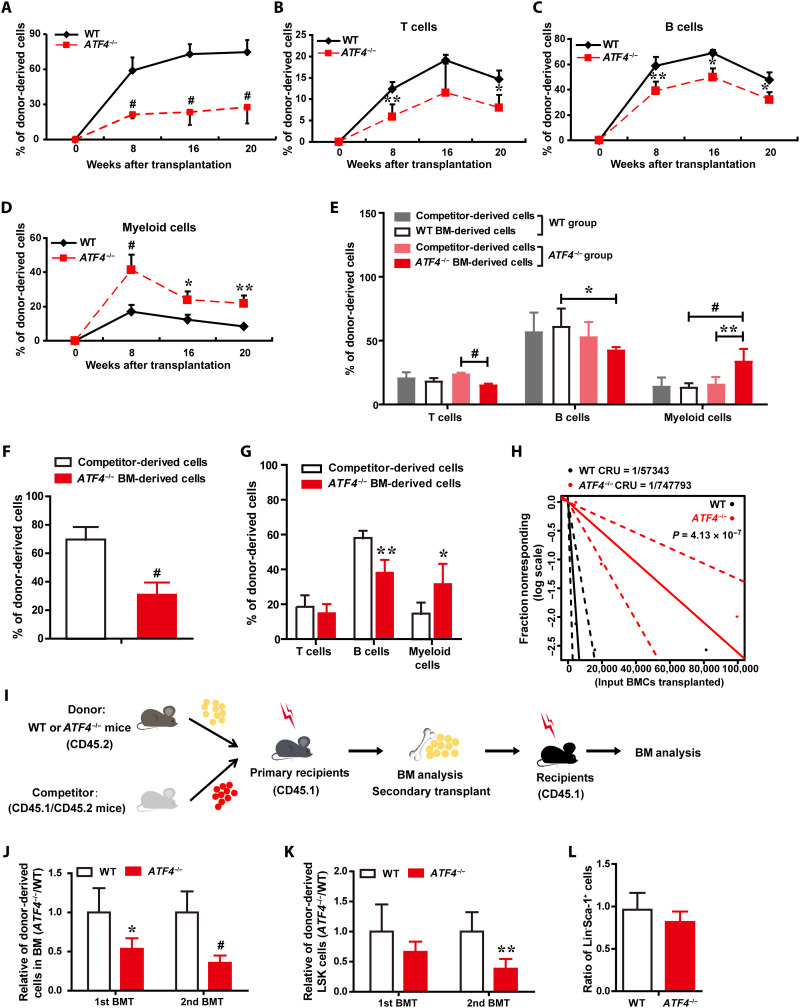
We next assessed whether ATF4 affects the function of adult HSCs. First, we performed competitive repopulation assay by transplanting an equal number of WT or ATF4−/− BM cells with competitors to evaluate the in vivo reconstitution capacity of ATF4−/− HSCs. Analysis of these chimeric mice was performed by flow cytometry over 8 to 20 weeks after the BM transplantation. Although we observed phenotypic expansion of ATF4−/− HSCs under steady state (Fig. 1), upon transplantation, the proportion of ATF4−/− donor BM cell–derived cells in PB was significantly lower in the reconstituted mice compared to WT control at the indicated time points (Fig. 2A), suggesting that ATF4 deficiency impairs the reconstitution capacity of HSCs, even though the ratio of phenotypic HSCs is increased.
Fig. 2. Deletion of ATF4 impairs the function of HSCs.
(A to D) The contribution of WT (CD45.2+) and ATF4−/− (CD45.2+) donor-derived cells (A) and percentages of WT and ATF4−/− donor-derived T (B), B (C), and myeloid cells (D) in the PB of recipient mice after competitive BMT (n = 3 to 5 mice per group). (E) Frequencies of donor-derived T, B, and myeloid cells in the PB of recipients at 3 months after BMT (n = 3 to 9 mice per group). (F and G) Donor BM cells from ATF4−/− mice (CD45.2+) mixed with competitor BM cells (CD45.1+/CD45.2+) at a 5:1 ratio were injected into lethally irradiated recipient mice (CD45.1+). (F) The contribution of WT and ATF4−/− donor-derived cells in recipient PB at 16 weeks after competitive BMT (n = 4 mice per group). (G) Bar graphs display percentages of donor-derived T, B, and myeloid cells of the recipient mice at 16 weeks after BMT (n = 4 mice per group). (H) HSC frequencies of WT or ATF4−/− mice were quantified with a limiting dilution BMT assay (n = 7 to 8 mice per cell dose). (I) Schematic illustration of serial and competitive BMT. (J and K) Relative of donor-derived BM cells (J) and donor-derived LSK cells (K) in serial BMT recipient mice (n = 3 to 6 mice per group). (L) The homing ability of Lin−Sca-1+ primitive hematopoietic cells obtained from ATF4−/− or WT mice (n = 5 mice per group). *P < 0.05, **P < 0.01, and #P < 0.001 (t test).
Next, we assessed the differentiation potential of ATF4−/− HSCs by examining the chimeric ratios of different lineage reconstitution in the PB at different time points after transplantation. Consistent with our observation at steady state (Fig. 1), we found that the frequencies of ATF4−/− donor-derived myeloid cells were significantly higher, whereas lymphocytes (T and B cells) were lower in the recipient PB at 8, 16, and 20 weeks after BM transplantation (BMT), compared to WT control group (Fig. 2, B to D), suggesting that ATF4 deletion induced hematopoietic differentiation skewing to myeloid lineage. To rule out the potential technical bias, we also analyzed the frequency of different lineages from the competitor donor mice. As expected, no significant difference was found in the competitor-derived lineage cells among different recipient groups; in addition, the percentages of WT (CD45.2) and competitor donor-derived total cells (CD45.1+/CD45.2+), including myeloid cells (Mac1+, Gr-1+), T cells (CD3e+), and B cells (B220+), in PB were also similar (Fig. 2E). Thus, these data indicate that ATF4−/− HSCs exhibited markedly enhanced myeloid differentiation but decreased lymphoid reconstitution.
Furthermore, we transplanted ATF4−/− BM cells along with WT competitor into recipients at a ratio of 5:1 (donor:competitor). Our results showed a significantly lower percentage of ATF4−/− donor-derived cells than that of WT competitors (Fig. 2F). Similarly, a higher percentage of ATF4−/−-derived myeloid cells, but a lower percentage of B lymphocytes, was also detected (Fig. 2G). These results revealed a severe, cell-autonomous repopulating defect of ATF4−/− HSCs. To directly enumerate the frequency of HSCs, we performed a limiting dilution assay. We found that the frequency of functional HSCs in ATF4−/− mice was 13-fold less than that of WT mice (Fig. 2H and fig. S2, A and B), strongly suggesting that ATF4 deficiency greatly decreased the functional HSCs.
To examine the effects of ATF4 on the self-renewal activity and long-term function of HSCs, we performed a serial BM transplantation experiment (Fig. 2I). The percentages of ATF4−/− donor-derived BM cells and LSK cells were lower in the primary transplants and progressively declined in the secondary transplants (Fig. 2, J and K), suggesting that ATF4 is important for the self-renewal capability of HSCs. Together with the data that ATF4 deletion did not affect HSC homing (Fig. 2L and fig. S2C), our results demonstrate that loss of ATF4 impairs the function of adult HSCs.
ATF4 deficiency provokes HSC impairment via cell-intrinsic mechanisms
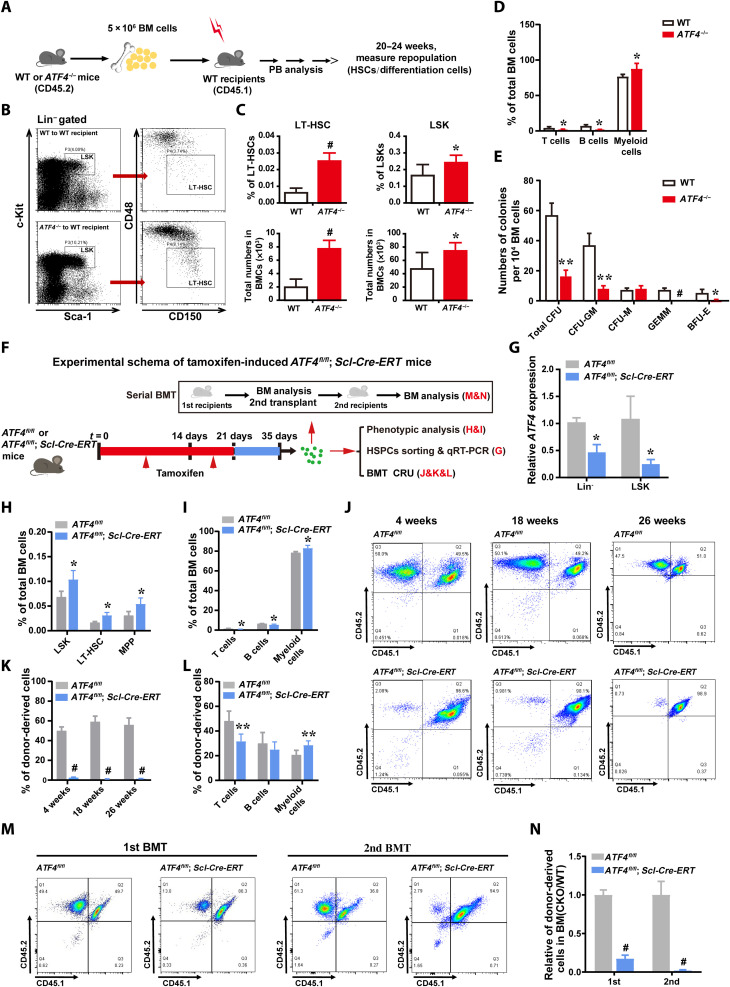
To determine whether the above-described deficiency of ATF4-deleted HSCs was due to cell-intrinsic or cell-extrinsic mechanisms, uncompetitive BMT was performed by transplanting ATF4−/− BM cells into irradiated WT mice or transplanting WT BM cells into irradiated ATF4−/− mice (Fig. 3A and fig. S3E). Consistent with our observation presented in Fig. 1, under the noncompetitive BMT conditions, expansion of HSCs (Fig. 3, B and C), skewed myeloid differentiation (Fig. 3D and fig. S3, A to D), and reduced colony-forming capacity (Fig. 3E) were detected in ATF4−/− donor-derived cells. However, in the reciprocal experiment of transplanting WT donor BM cells into WT or ATF4−/− recipients, we did not find significant differences in the percentages of WT hematopoietic stem and progenitor cells (HSPCs) and lineage cells between two groups (fig. S3, F to J), suggesting that ATF4−/− adult BM microenvironment does not affect hematopoiesis. These data support the notion that the defects of ATF4−/− HSCs were attributed to cell-intrinsic mechanisms.
Fig. 3. ATF4 deficiency provokes HSCs impairment via cell-intrinsic mechanisms.
(A) Schematic diagram of uncompetitive BMT. (B and C) Representative FACS plot (B) and the frequencies and total numbers (C) of LT-HSCs (LSK CD150+CD48−) and LSKs in BM from irradiated WT recipient mice transplanted with BM cells from ATF4−/− or WT mice, respectively (n = 4 mice per group). (D) The percentages of the donor-derived T, B, and myeloid cells in BM of WT recipients (n = 2 to 4 mice per group). (E) The quantification of the colony numbers of total CFU, CFU-M, CFU-GM, CFU-GEMM, and BFU-E from ATF4−/− or WT donor-derived BM cells in WT recipients (n = 3). (F) Experimental schema of tamoxifen-induced ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT mice. (G) ATF4 expression levels in Lin− cells and LSK cells after tamoxifen administration, as measured by qRT-PCR. (H and I) Frequencies of LT-HSCs (LSK CD150+CD48−), MPP (LSK CD150−CD48+), LSK cells (H), and T, B, and myeloid cells (I) in total BM cells of ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT mice (2 to 4 months) after tamoxifen administration (n = 3 mice per group). (J and K) Representative FACS plots (J) and the contribution of ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT donor-derived cells (K) in the recipient’s PB, which was analyzed at 4, 18, and 26 weeks after competitive BMT (n = 5 to 7 mice per group). (L) Frequencies of donor-derived T, B, and myeloid cells in the PB of primary recipients at 1 month after BMT (n = 4 to 8 mice per group). (M and N) Representative FACS plots (M) and the contribution of ATF4fl/fl (CD45.2+) and ATF4fl/fl; Scl-Cre-ERT (CD45.2+) donor-derived cells in BM (N) (n = 4 mice per group). The BMCs from first and second recipients were analyzed at 8 weeks after serial BMT, respectively. *P < 0.05, **P < 0.01, and #P < 0.001 (t test).
To further investigate the role of ATF4 in HSCs, we constructed ATF4fl/fl mice and crossed them with an inducible Scl-Cre-ERT strain (fig. S3, K to M), which allowed us to specifically delete ATF4 in HSPCs after treatment with tamoxifen (26). Using ATF4fl/fl; Scl-Cre-ERT mice, serial phenotypic and functional assays were carried out (Fig. 3F), and in line with our previous data generated from the constitutive ATF4−/− mice, HSC-specific deletion of ATF4 led to the aging-like phenotypes, including expansion of phenotypic HSCs and progenitors (Fig. 3, G and H) and skewed myeloid differentiation (Fig. 3I). Similarly, competitive repopulation assay also displayed defective repopulating (Fig. 3, J and K) and myeloid bias (Fig. 3L) and self-renewal capacities of HSCs (Fig. 3, M and N), further suggesting that ATF4 deficiency provokes HSC impairment via cell-autonomous mechanisms. Thus, our findings from both constitutive and conditional ATF4−/− models showed HSC dysfunction with multifaceted HSC aging-like phenotype.
ATF4 deficiency leads to defective HSCs with aging-like phenotype
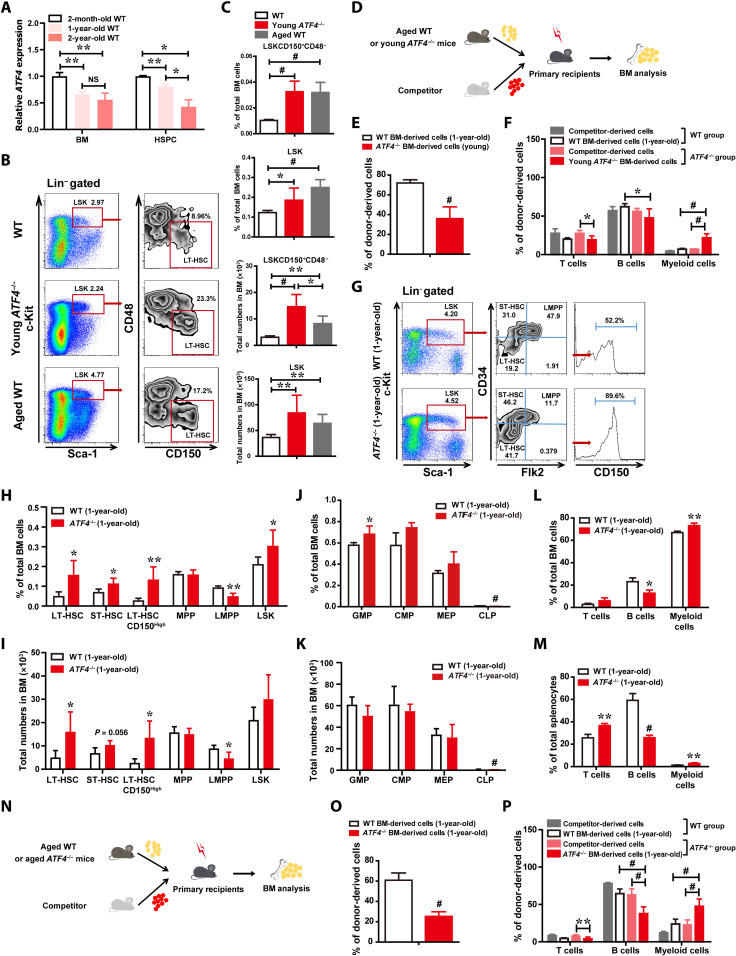
In young ATF4-deficient mice (4 months old), we found premature hair graying (fig. S4A). This observation, in combination with our aforementioned data showing the aging-like phenotype of ATF4−/− HSCs, prompted us to further investigate the role of ATF4 in regulating HSC aging. ATF4 expression levels in LSK cells progressively decreased with age, and there is a nearly 50 to 60% reduction in the aged BM and LSK cells as compared with the young group (Fig. 4A; 2 to 3 months old versus 24 months old), suggesting a pivotal role of ATF4 in modulating the aging process of HSCs.
Fig. 4. ATF4 deficiency leads to defective HSC with aging-like phenotype.
(A) ATF4 expression levels in BM cells and LSK cells. (B and C) Representative FACS plots (B) and the frequency and total numbers (C) of LT-HSCs (LSKCD150+CD48−) and LSK cells in BM from WT (4 months), ATF4−/−(4 months), and aged (24 months) mice (n = 3 mice per group). (D) Schematic illustration of competitive BMT (aged WT mice versus young ATF4−/− mice). (E and F) The contribution of WT (1-year-old) and ATF4−/− (3-month-old) donor BM-derived cells (E) and donor BM-derived T, B, and myeloid cells (F) in recipient PB at 8 weeks after competitive BMT (n = 5 mice per group). (G to I) Representative FACS plots (G) and histograms (H and I) showing the frequencies and total numbers of indicated populations in total BM cells from WT mice (1-year-old) and ATF4−/− mice (1-year-old) (n = 4 to 6 mice per group). (J and K) Frequencies and total numbers of GMPs, CMPs, MEPs, and CLPs in BM cells from WT mice (1-year-old) and ATF4−/− mice (1-year-old) (n = 3 to 4 mice per group). (L and M) Frequencies of T, B, and myeloid cells in BM (L) and spleen (M) from WT mice (1-year-old) and ATF4−/− mice (1-year-old) (n = 3 to 4 mice per group). (N to P) Schematic illustration of competitive BMT (aged WT mice versus aged ATF4−/− mice) (N). The contribution of WT and ATF4−/− donor BM-derived cells (O) and donor BM-derived T, B, and myeloid cells (P) in PB of recipients at 8 weeks after BMT (n = 7 to 8 mice per group). *P < 0.05, **P < 0.01, and #P < 0.001 (t test).
To determine whether ATF4 regulates physiological homeostasis during aging, we analyzed the frequency and number of phenotypic HSPCs in BM cells derived from ATF4−/− mice (4 months old), aged WT mice (24 months old), and WT mice (4 months old), respectively. Notably, both the BM obtained from ATF4−/− mice and from aged WT mice showed significant expansion of LSKs and LT-HSCs (Fig. 4, B and C). Moreover, when the repopulating and differentiating abilities of young ATF4−/− HSCs were compared to those of aged WT using a competitive repopulation unit assay with donor cells (1 × 106) and equal number of competitor cells (Fig. 4D), we found that the percentage of young ATF4−/− donor-derived PB cells was markedly lower in the transplants as compared to the aged group (Fig. 4E), while young ATF4−/− HSCs showed notably increased myeloid but decreased lymphoid reconstitution when compared with the aged WT HSCs (Fig. 4F). In addition, we also tested whether the aged ATF4−/− mice (1-year-old) also contained an expanded phenotypical HSC population and have shown myeloid bias. As shown in Fig. 4 (G to I), fluorescence-activated cell sorting (FACS) analysis illustrated that the frequencies and total numbers of LT-HSCs and CD150high-LT-HSCs were significantly increased, whereas the percentage and total numbers of LMPPs in the BM of aged ATF4−/− mice (1-year-old) were markedly decreased, as compared with those in the aged WT mice. Analysis of HSC markers exhibited that the phenotypic HSCs were four- to fivefold greater in aged ATF4−/− mice when compared with their WT counterparts. Consistently, we found that aged ATF4−/− mice also displayed an expansion of phenotypic HSCs in the BM during aging, similar to our observation in the young ATF4-deficient mice (2 to 4 months). Furthermore, there was a marked decrease in the frequency and numbers of CLPs and a modest increase in the frequency of granulocyte-macrophage progenitors (GMP) in aged ATF4−/− mice (Fig. 4, J and K, and fig. S4B), and a significant decrease in lymphoid cells and an increase in myeloid cells in aged ATF4−/− BM and spleen were found (Fig. 4, L and M), suggesting that ATF4 deficiency in aged mice also perturbs myeloid and lymphoid differentiation.
We next compared the repopulating and differentiating abilities of aged ATF4−/− HSCs to aged WT control (Fig. 4N). Consistent with the results from the young mice groups, the percentage of donor-derived PB cells from aged ATF4−/− mice was significantly lower and prone to myeloid differentiation when compared to those of the aged WT donor cells (Fig. 4, O and P). Furthermore, directly comparing young with aged ATF4−/− groups revealed that the effect of ATF4 deletion on the aging-like phenotype was age dependent, since there appeared decreased repopulating abilities and more myeloid bias in aged ATF4−/− mice as compared with those in young ATF4−/−mice (fig. S4, C and D), further suggesting that the deletion of ATF4 leads to HSC prematuration, and that the aged ATF4−/− mice displayed a progressive aging-like phenotype during aging. Together, our data demonstrate that ATF4 deletion in mice leads to aging characteristics of HSCs, including increased phenotypic HSCs, reduced repopulating capacities, and skewed myeloid lineage, supporting the hypothesis that ATF4 plays a role in regulating the aging of HSCs.
Mitochondrial ROS contributes to ATF4 deficiency–induced HSC defects
As ROS are believed to be important mediators of aging and numerous degenerative diseases, including those involving HSC dysfunction and senescence (12, 27), we asked whether ROS plays a role in the development of the ATF4-deficient HSC phenotype. Intracellular concentration of ROS was measured by 2′-7′-dichlorofluorescein diacetate (DCF-DA) staining. We found that compared to WT LT-HSCs, ATF4−/− LT-HSCs produced markedly higher levels of ROS (Fig. 5A). Moreover, MitoTracker Orange CM-H2TMRos analysis showed higher oxidative stress in ATF4−/− LT-HSCs than WT LT-HSCs (Fig. 5B), suggesting that mitochondria is the main source for the increased ROS in ATF4−/− HSCs. Consistent with a previous finding that high levels of ROS induce quiescence loss of HSCs (11, 28, 29), the cell cycle assay revealed that ATF4−/− LT-HSCs were less quiescent (Fig. 5C).
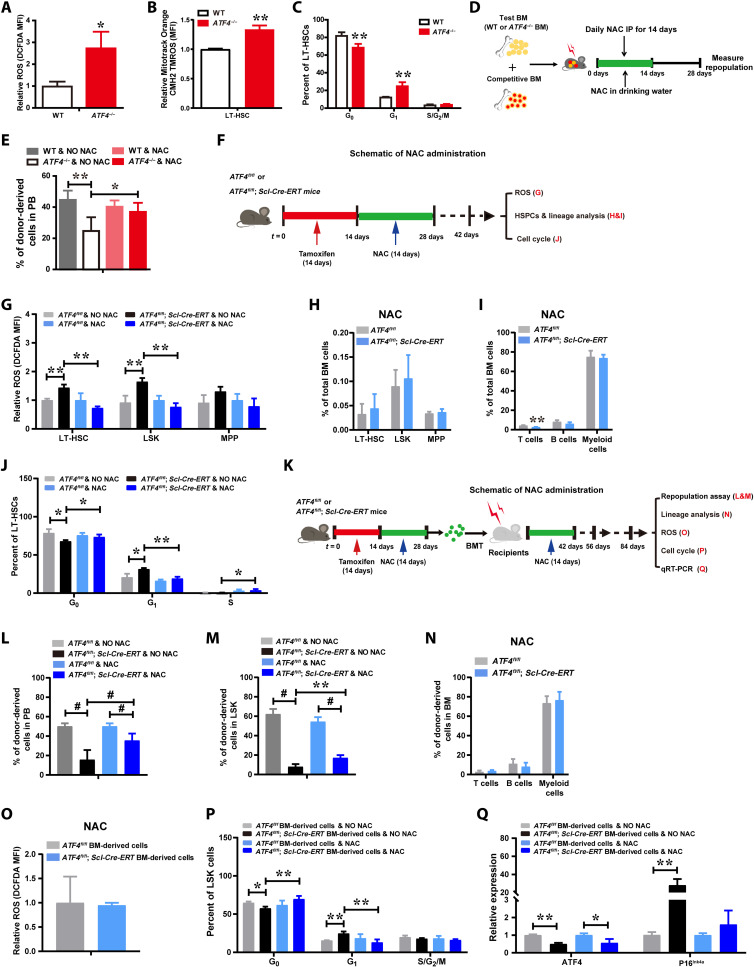
Fig. 5. Mitochondrial ROS contributes to ATF4 deficiency–induced HSC defects.
(A) ATF4-deficient LT-HSCs display increased ROS levels (n = 3 mice per group). (B) Increased redox-sensitive MitoTracker fluorescence in ATF4-deficient LT-HSCs (n = 2 to 3 mice per group). MFI, mean fluorescence intensity. (C) Cell cycle profiling of LT-HSCs from WT mice and ATF4−/− mice (n = 3 to 4 mice per group). (D) Schema of BMT and NAC administration (WT and ATF4−/− constitutive mutant mice). IP, immunoprecipitation. (E) Repopulation analysis in PB after NAC treatments (n = 4 to 6 mice per group). (F) Schematic of NAC administration for tamoxifen-induced ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT mice. (G) ROS levels in LT-HSCs, MPP, and LSK cells (n = 2 to 4 mice per group). (H and I) The frequency of LT-HSCs, MPP, and LSK cells (H) (n = 3 to 4 mice per group), and T, B, and myeloid cells (I) (n = 2 to 4 mice per group) in the BM. (J) Cell cycle profiling of LT-HSCs (n = 2 to 4 mice per group). (K) Schematic of NAC administration and BMT for tamoxifen-induced ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT mice. (L and M) Repopulation analysis of PB (L) and LSK in BM cells (M) from recipient mice 8 weeks after transplantation (n = 4 to 8 mice per group). (N) The percentages of donor BM-derived T, B, and myeloid cells in the BM (n = 3 to 4 mice per group). (O) ROS levels in LSK cells in donor-derived BMCs (n = 4 mice per group). (P) Cell cycle profiling of LSK cells in donor-derived BMCs (n = 3 to 4 mice per group). (Q) mRNA expression of ATF4 and p16Ink4a in ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT donor-derived BMCs. *P < 0.05, **P < 0.01 and #P < 0.001 (t test).
To evaluate whether increased ROS was causally involved in the functional attrition of HSCs caused by ATF4 deletion, we tested the effect of antioxidant on the presentation of ATF4−/− HSC phenotype in vivo. Tamoxifen-induced ATF4 Conditional Knockout (cKO) mice were administered daily with antioxidant N-acetyl-l-cysteine (NAC) or control saline solution for 2 weeks, and subsequently, BMCs were harvested to analyze for the levels of ROS produced, HSPCs and lineage frequencies, and cell cycle progression (Fig. 5F). Of note, NAC treatment reduced ROS levels in ATF4−/− LT-HSCs and LSKs (Fig. 5G), and reduction of ROS rescued the ATF4−/− HSC phenotype, including restoration of the frequencies of LT-HSCs and progenitors, and lineage cells in ATF4 cKO mice (Fig. 5, H and I, and fig. S5, A and B). NAC treatment also normalized cell cycle progression in ATF4−/− LT-HSCs (Fig. 5J). Moreover, to further evaluate the effect of ROS on ATF4−/− HSC engraftment defect, we performed competitive repopulation assay. Following transplantation, the reconstituted mice were further treated with or without NAC (Fig. 5K). Our results showed that ROS scavenging by NAC administration alleviated the engraftment defects of ATF4 cKO donors (Fig. 5, L and M). The same results were also observed using the constitutive ATF4−/− mice (Fig. 5, D and E). Furthermore, the percentages of lineage cells in ATF4 cKO donor BM-derived cells were similar to those in the control cells (Fig. 5N). In addition, elevated ROS and more active cell cycling in the ATF4 cKO donor cells (LSKs) were restored to the control level upon NAC treatments (Fig. 5O). Furthermore, NAC treatment restored the cell cycling status of the ATF4−/− HSPC donor cells (Fig. 5P). Notably, the expression of p16Ink4a transcript in sorted ATF4 cKO donor cells was markedly up-regulated, and clearance of ROS restored the expression of p16Ink4a transcript in ATF4 cKO donors to the control level (Fig. 5Q), which was consistent with a previous finding that p16Ink4a is a redox-sensitive cell cycle regulator. Together, our data support the notion that defects of ATF4-deficient HSCs can be attributed at least in part to the increased ROS.
ATF4 deficiency–induced HIF1α down-regulation is largely responsible for the defects of HSCs
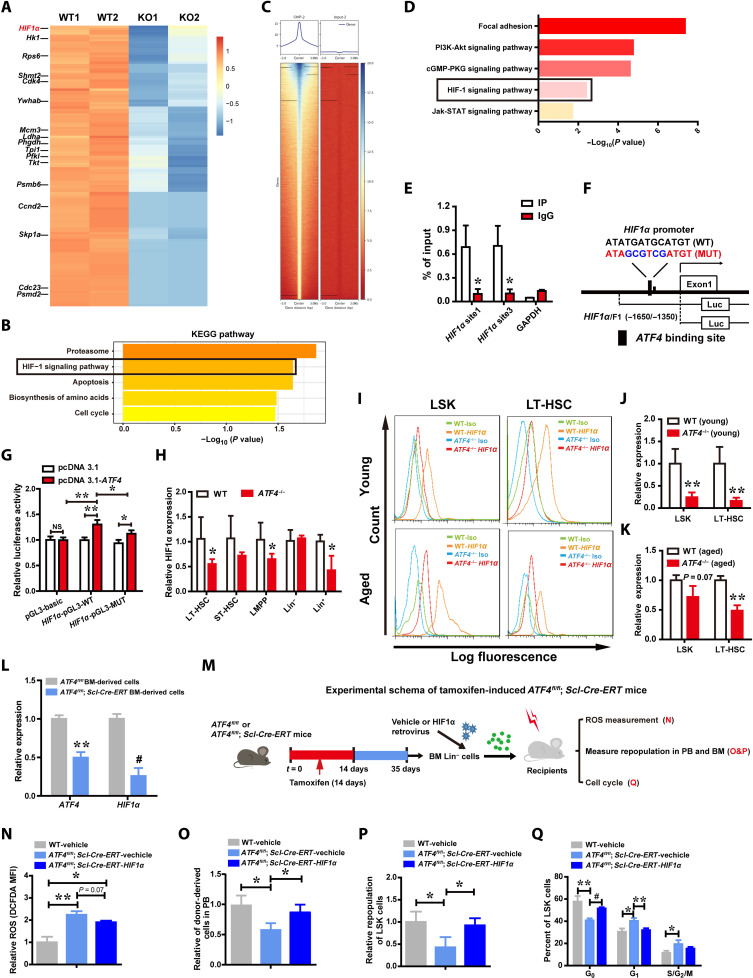
We next sought to explore the molecular mechanism underlying induction of the HSC defects with an aging-like phenotype by ATF4 deficiency. To comprehensively understand the regulatory role of ATF4 in HSCs, we sorted LT-HSCs from ATF4 cKO mice after inducing deletion of ATF4 and conducted an RNA sequencing (RNA-seq) assay to compare the gene expression profiles of ATF4−/− HSCs with WT HSCs. A total of 530 genes were found to be differentially expressed. KEGG enrichment analysis revealed that several terms (i.e., proteasome signaling pathway, HIF1 signaling pathway, and apoptosis signaling pathway) were significantly altered in ATF4−/− LT-HSCs (Fig. 6, A and B, and fig. S6A). To understand how ATF4 affects the mRNA expression of these altered genes, we performed ATF4 chromatin immunoprecipitation (ChIP) coupled to high-throughput sequencing (ChIP-seq) using mouse BM cells (Fig. 6C). Kyoto encyclopedia of genes and genomes (KEGG) analysis indicated that those genes whose promoter regions were bound by ATF4 in murine BM cells were highly enriched in the PI3K (phosphatidylinositol 3-kinase)–AKT pathway, HIF1 signaling pathway, and JAK (Janus tyrosine Kinase)–STAT (Signal Transducer and Activator of Transcription) pathway (Fig. 6D). As low HIF1α level has been previously shown to lead to oxidative stress and ROS accumulation (11, 28, 29), which is similar to the phenotype of ATF4−/− HSCs identified in our current study (Fig. 5), we further examined whether HIF1a can be a transcriptional target of ATF4. We found that ATF4 consensus-binding motifs were present in the promoter of the HIF1a gene (fig. S6B). ChIP analysis showed that ATF4 directly bound to the HIF1a promoter (Fig. 6E). Furthermore, we performed luciferase reporter assay and found that ectopic expression of ATF4 caused an increase in HIF1a promoter activity, and mutation of the ATF binding site abrogated this induction (Fig. 6, F and G). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay exhibited that ATF4 deletion reduced mRNA expression of HIF1a in LT-HSCs, LMPP, and Lin+ cells (Fig. 6H). Consistently, FACS analysis showed that ATF4 deletion led to decreased expression of HIF1α in both young and aged ATF4−/− mice (Fig. 6, I to K), indicating that HIF1α is a direct downstream target of ATF4 in HSCs.
Fig. 6. ATF4 deficiency–induced HIF1α down-regulation is largely responsible for the defects of HSCs.
(A and B) and Heatmap (A) of RNA-seq analysis of down-regulated genes in WT and ATF4−/− LT-HSCs (mouse) and selected KEGG pathway analysis (B). (C) Representative ChIP-seq data showing ATF4 binding peaks at promoter sequences of genes in mouse BM cells. (D) Selected KEGG pathway analysis for ATF4 target genes in mouse BM cells. (E) ChIP-qPCR assay was performed using anti-ATF4 antibody to identify ATF4 binding sites on the HIF1α promoter in BM cells (n = 3). (F) Schematic representation of the human HIF1a promoter. The proximal promoter region and its mutation construct were cloned into the pGL3 reporter plasmid. (G) Luciferase reporter activity in 293T cells upon ATF4 activation (n = 3). (H) mRNA expression of HIF1a in LT-HSCs, ST-HSCs, LMPP, Lin+, and Lin− cells from WT mice and ATF4−/− mice (2 to 4 months) was measured with qRT-PCR. (I) A representative FACS analysis of the expression of intracellular HIF1α in LSK cells and LT-HSCs stained with HIF1α antibody. (J and K) FACS analysis of intracellular HIF1α expression in LT-HSCs and LSK cells in BM from young ATF4−/− mice (2 to 4 months) (J) and ATF4−/−(1-year-old) mice (K) (n = 3 to 5 mice per group). (L) mRNA expression of ATF4 and HIF1a in ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT donor-derived BMCs. (M) Schematic design of rescuing experiment on HIF1a overexpression in Lin− cells from tamoxifen-induced ATF4fl/fl and ATF4fl/fl; Scl-Cre-ERT mice, respectively. (N to Q) ROS (N) (n = 2 to 3 mice per group), repopulation (O and P) (n = 3 to 4 mice per group), and cell cycle (Q) (n = 3 mice per group) analyses in donor cells transduced with HIF1α expression recombinant or control vectors from recipient mice. *P < 0.05, **P < 0.01, and #P < 0.001 (t test).
To directly analyze the impact of HIF1α on the repopulating ability of ATF4−/− HSCs, we overexpressed HIF1α in isolated ATF4−/− Lin− cells by transducing them with control Babe-puro retrovirus (Babe virus) or Babe-puro-HIF1α retrovirus (HIF1α virus) (29, 30) (fig. S6C). We found that the repopulation defect after BMT could be markedly restored by ectopic expression of HIF1α in ATF4−/− Lin− cells (fig. S6D). Moreover, HSC-specific deletion of ATF4 resulted in HIF1a down-regulation in sorted cKO donor cells (Fig. 6L). In addition, after the tamoxifen-induced ATF4 cKO Lin− cells were transduced with recombinant retrovirus carrying HIF1a, subsequent Competitive repopulating unit (CRU) assay (Fig. 6M) showed that overexpression of HIF1α reduced ROS production in ATF4−/− HSPCs (Fig. 6N) and that HIF1α overexpression markedly improved the repopulating ability and quiescence of HSCs (Fig. 6, O to Q), while the expression of p16Ink4a was restored to the control level after HIF1α rescue (fig. S6E). Together, our data indicate that HIF1a is a direct downstream target of ATF4 and may present a major mediator for the function of ATF4 in HSCs.
ATF4 is required for murine AML development
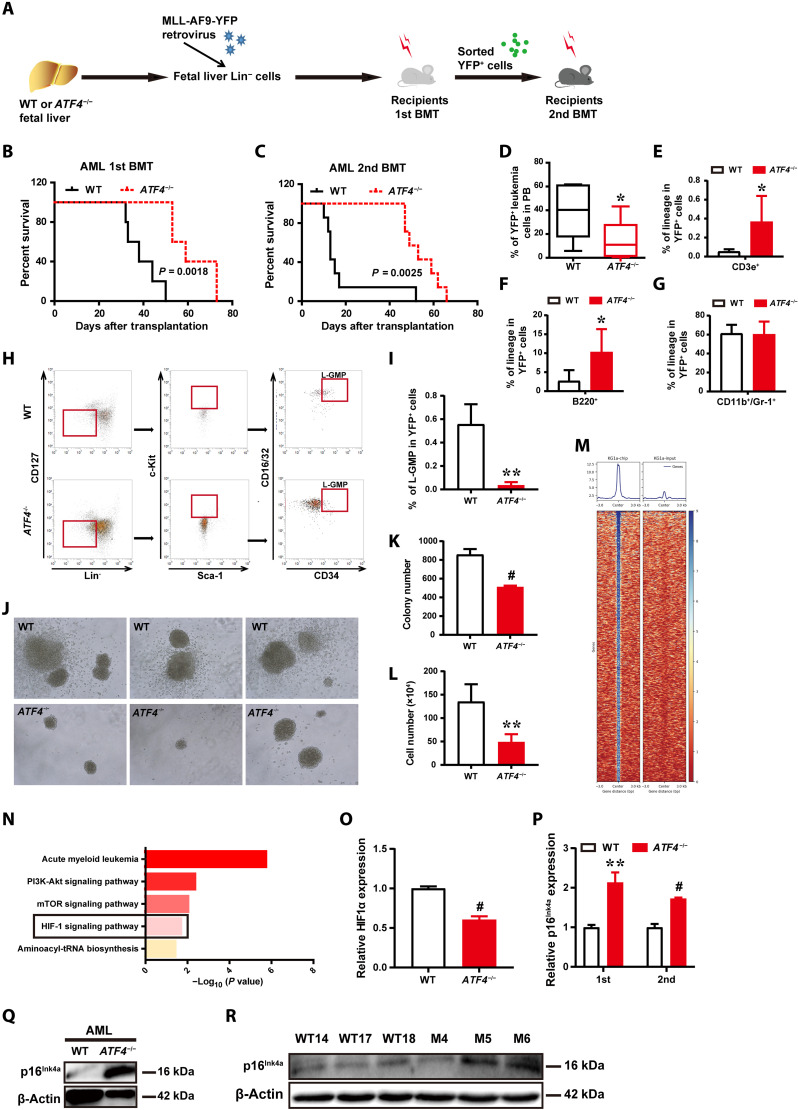
Although previous studies showed that ATF4 is overexpressed in various types of solid tumors and leukemia (16, 21, 31), the role of ATF4 in leukemogenesis remains elusive. Therefore, we further investigated the role of ATF4 in leukemia initiation and development using the MLL-AF9 fusion gene–induced acute myeloid leukemia (AML) model (32). Notably, MLL-AF9–transduced ATF4−/− donor cells displayed significantly extended overall survival after the first transplantation and second transplantation (Fig. 7, A to C). Furthermore, 6 weeks after transplantation, the frequency of YFP+ATF4−/− donor-derived leukemic cells was significantly lower than that from WT mice (15.79% versus 39.19%; Fig. 7D). These MLL-AF9–transduced ATF4−/− donor cells mainly expressed lymphoid markers of B220 and CD3e (Fig. 7, E to G).
Fig. 7. ATF4 deletion delays leukemogenesis in MLL-AF9–induced AML model.
(A) Schematic illustration of the MLL-AF9–induced murine AML model. (B and C) Survival data for recipient mice receiving WT or ATF4-null MLL-AF9+ fetal liver Lin− cells upon the first (B) and second (C) transplantation (n = 5 to 7, log-rank test). (D) Quantification of YFP+ leukemia cells in the PB of primary recipients transplanted with MLL-AF9–transduced WT or ATF4-null fetal liver Lin− cells (n = 9 to 10 mice per group). (E to G) Quantification of T (E), B (F), and myeloid cells (G) in the BM of primary leukemic recipients (n = 3 to 6 mice per group). (H) Representative plots of L-GMP (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32+) cells in the BM of leukemic recipients. (I) Quantification of the percentages of L-GMP cells in AML cells (n = 3 to 4 mice per group). (J) Representative images of colony formation of WT and ATF4−/− YFP+Mac-1+c-Kit+, LICs of the secondary recipients in the first plating. (K and L) Quantification of the colony numbers (K) and total cell numbers of colonies (L) were counted (n = 3 to 6). (M) Representative ChIP-seq data showing ATF4 binding peaks at promoter sequences of genes in KG1α cells. (N) Selected KEGG pathway analysis for ATF4 target genes in KG1α cells. (O) mRNA expression of HIF1a in MLL-AF9–induced control and ATF4-null AML cells. (P) mRNA expression of p16Ink4a in MLL-AF9–induced control and ATF4-null AML cells (first and second transplanted). (Q) Western blotting analysis of p16Ink4a expression in MLL-AF9–induced control and ATF4-null AML cells. (R) Western blotting analysis of the indicated proteins in MEFs from WT mice and ATF4−/− mice. *P < 0.05, **P < 0.01, and #P < 0.001 (t test).
Subsequently, we further examined whether ATF4 affected the frequency of leukemia-initiating cells (LICs). As shown in Fig. 7 (H and I), the frequency of ATF4−/− immunophenotypic L-GMP (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32+) cells, which were reported to be enriched for LICs (32), was approximately 14.8-fold lower than that of the WT counterparts. The clonogenic potential of ATF4-null LICs from the secondary recipients decreased drastically, as indicated by the remarkable reduction in colony sizes, colony numbers, and total cell numbers during primary plating (Fig. 7, J to L). Thus, these data suggest that ATF4 played an important role in leukemia development and that the ATF4-null LICs had severe functional defects.
As our aforementioned findings indicated that HIF1α was a direct downstream target gene regulated by ATF4, and it was also previously reported that HIF1α played a crucial role in maintaining LSC survival, we were prompted to test whether ATF4 deletion delays leukemogenesis through the HIF1α signaling pathway. To this end, ChIP-seq was performed in human AML KG1α cells (Fig. 7M and fig. S6, F and G). As expected, HIF1 signaling pathways were highly enriched, similarly to what was found in ChIP-seq analysis of mouse BMCs (Fig. 7N). Furthermore, we found that expression of HIF1α was markedly decreased in ATF4−/− AML cells (Fig. 7O). In addition, ATF4 deletion resulted in elevated mRNA expression of p16Ink4a in primary and secondary transplanted AML cells (Fig. 7P). Western blotting analysis demonstrated that ATF4-null AML cells displayed a significant increase in the levels of p16Ink4a expression (Fig. 7Q). Furthermore, we confirmed that p16Ink4a expression increased in ATF4−/− MEFs (Fig. 7R). Together, these data suggest that HIF1α and p16Ink4a appear to be associated with ATF4-regulated survival and proliferation of leukemia cells in AML development, and further investigation is warranted to determine their detailed roles in this context.
DISCUSSION
Balancing the processes of HSC self-renewal and differentiation is crucial to sustaining a lifelong production of blood cells (33–36). Progressive loss of the balance between HSC self-renewal capacity and multilineage differentiation, characterized by expanded HSC number, decreased HSC regenerative capacity, and myeloid bias, however, occurs during aging of HSCs (5, 8, 11, 12). Our current study reveals that ATF4 plays an essential role in adult HSC homeostasis via regulating the HIF1α pathway, and ATF4 loss leads to HSC defects associated with an aging-like phenotype.
In our study, under homeostatic conditions, we found that both young and aged ATF4−/− mice displayed an expansion of phenotypic LT-HSCs (CD34+Flk2+LSK and CD150+CD48+LSK). Previous studies showed that CD150high-LT-HSCs are myeloid-biased and robustly increase in the HSC population during aging, while CD150low-LT-HSCs are lymphoid-biased and decrease with age (25). Young and aged ATF4−/− mice displayed significantly increased CD150high–LT-HSCs (myeloid-biased HSCs, CD150highCD34+Flk2+LSK) and a marked decrease of LMPP (CD34+Flk2+LSK) in the BM. Furthermore, ATF4−/− HSCs exhibited markedly enhanced myeloid differentiation but decreased lymphoid reconstitution regardless of age. Together, our findings demonstrate that the loss of ATF4 resulted in aging characteristics of HSCs, including phenotypic expansion of HSCs and hematopoietic myeloid differentiation skewing, suggesting that ATF4 may play a pivotal role in the regulation of HSC aging.
Fetal and adult HSCs have been distinctly characterized by their proliferation/renewal rates, lineage trends, and gene expression profiles (37). During embryogenesis, several anatomical sites, such as the yolk sac, the aorta-gonad mesonephric region, the placenta, and the fetal liver, provide a unique environment for hematopoiesis. Fetal liver HSCs rapidly divide and consequently lead to massive expansion to meet the demand of ontogenic development (38). After birth, HSCs colonize BM, and the adult HSCs are dormant, rarely divide, and function throughout the lifetime to maintain the homeostasis of the blood system. There is a remarkable switch in the molecular program and the functional behavior of HSCs during the transition from fetal liver HSCs to adult HSCs. Previous studies indicated that during fetal liver hematopoiesis, ATF4 maintains HSCs throughout ontogeny by mainly acting as an extrinsic factor to maintain an HSC-supportive niche via up-regulating cytokines such as Angptl3 in fetal liver stromal cells (18). Notably, our data first revealed that ATF4 expression progressively declines with age in adult BM and the LSK population. Moreover, our findings made in constitutive and conditional ATF4 (ATF4fl/fl; Scl-Cre-ERT mice) KO mice showed multifaceted aging-like properties of adult HSCs. Consistently, ATF4−/− HSC displayed a progressive aging-like phenotype during aging. Furthermore, our current data clearly demonstrate that ATF4 is critical in maintaining adult HSC aging in a cell-intrinsic manner, rather than a cell-extrinsic factor, suggesting that at embryonic and adult stages, ATF4 could play different roles in regulating HSC behavior through distinct molecular mechanisms, and that thereby modulation of ATF4 might contribute to adult HSC homeostasis and aging.
In light of the fact that ROS has been found to be an important driver of aging and that low ROS level contributes to HSC quiescence and long-term self-renewal capability (11, 12, 28, 29, 39, 40), we attempted to examine whether ROS production is associated with ATF4 depletion in our current study. Notably, aged HSCs are more vulnerable to extra physiologic oxygen shock/stress (EPHOSS)–linked ROS production, and inhibition of EPHOSS by cyclosporine A could increase recovery of transplantable and aged HSCs (39, 40). We found that ATF4−/− LT-HSCs produced markedly higher levels of ROS. Of note, high levels of ROS are known to induce lost quiescence of LT-HSCs (11, 28, 29). ATF4 deletion leads to enhanced cell cycling and a loss of quiescence in LT-HSCs, thereby inducing HSC hyper-proliferation, which might also explain the increased cell cycle entry in ATF4−/− HSCs as a compensatory effect. Nevertheless, such changes were unable to compensate for the functional defects of HSCs, which is similar to the age-associated increased number of functionally impaired HSCs. Cell cycle changes have been found in HSC aging (5, 8, 11). Aged HSCs have been shown to display delayed cell cycle kinetics, despite being recruited into an active cell cycle to a higher degree compared to young HSCs (41).
Our current findings support a notion that ATF4 loss in mice might trigger the development of HSC aging phenotype by causing mitochondrial ROS overproduction. Furthermore, our study also identifies HIF1a as a direct downstream target of ATF4. Of note, HIF1α has been shown to be essential for HSC function and their metabolic pathway (29), while HIF1α negatively modulates mitochondrial ROS production (28, 29). Our results showed that the repopulating defect of ATF4−/− HSCs and ROS could be restored by enforcing HIF1α expression, suggesting a cascade along which ATF4 loss triggers the development of HSC aging phenotype by causing mitochondrial ROS overproduction through targeting HIF1a. Previous studies showed that ROS could induce cellular senescence by up-regulating p16Ink4a and p19Arf, and that p16Ink4a was critical for the regulation of stem cell aging (42, 43). For instance, p16Ink4a was elevated in aged HSCs and contributes to the decline of HSCs (42, 44). Our present data suggest that ATF4 might regulate p16Ink4a through an indirect manner, ATF4 deficiency triggers mitochondrial ROS production by targeting HIF1α expression, and ROS might up-regulate p16Ink4a. We found that ATF4 deficiency has a stronger effect on HIF1α expression in young mice than in aged mice. Of note, ATF4 was reported to be a direct target of transcriptional repression of HIF1α in fetal cardiomyocytes (45). During the process of aging, there could be a negative feedback loop between ATF4 and HIF1α. Furthermore, we speculate that there might be a potential regulatory circuit among ATF4, HIF1α, p16Ink4a, and ROS, while the circuit could also be affected by the process of aging; further investigation in this context is needed.
HIF1α was not only required for HSC functions but also essential for cancer stem cells of mouse lymphoma and LSCs (46, 47). Deletion of HIF1a impaired the propagation of LSCs with elevated expression of p16Ink4a and p19Arf. Notably, our ChIP-seq data support the idea that the HIF1α pathway might play a role in the defects of ATF4−/− HSCs and in delaying leukemogenesis. Our ChIP and luciferase reporter assays suggest that HIF1a is a potential de novo and direct target of ATF4. In addition, we observed decreased HIF1a level and elevated p16Ink4a expression in ATF4-null YFP+ leukemia cells, suggesting that both of them may be pivotal in alleviating leukemia initiation and development. Therefore, ATF4 deletion might alleviate leukemia initiation and development associated with an involvement of HIF1a and p16Ink4a. Of note, Dick’s group developed the ATF4 reporter system to measure ISR activation and found that high ISR activity occurred in human HSC/MPPs and AML/LSCs; higher ATF4 protein levels maintain human LSC stemness, promoting LSC engraftment (21, 31). Thus, it appears that, under physiological conditions, ATF4 plays a critical role to maintain HSC homeostasis and stemness, whereas under complex oncogenic stress, ATF4 can be up-regulated to maintain LSC stemness, leading to a promoting effect on leukemogenesis, in which case inhibition of ATF4 delays AML development. Of note, other previous studies also have demonstrated a fragile balance between tumor suppression and stem cell aging. For instances, defect of several genes (Sirt1, Sirt3, and Wip1) leads to prematuration of HSCs, and inhibition of these genes could prevent tumorigenesis (48–53). Together, our current study suggests that ATF4 plays distinct and yet related roles in normal hematopoiesis and leukemogenesis.
Our findings suggest that up-regulating ATF4 may represent a strategy to alleviate aging-associated degeneration of HSC, and hence, investigation aimed at identifying effective approaches to increase ATF4 in HSPCs is of interest and underway in the laboratory. On the other hand, as our current study found that ATF4 might promote leukemogenesis, it would be of interest to further test whether ATF4 can be used as a therapeutic target for antileukemic treatment. New technologies of targeting a transcription factor are under active development, including proteolysis targeting chimaeras, use of cysteine reactive inhibitors, and targeting intrinsically disordered regions of transcription factors, in addition to other conventional methods (54).
In summary, our present study first identifies and highlights a previously unidentified and direct role of ATF4 in regulating adult HSC function and aging, with expanded implications in our understanding of the molecular mechanisms of HSC aging and age-related degeneration. Further investigation is warranted to explore strategies efficacious for achieving such an ATF4-dependent optimization of adult HSC functions.
MATERIALS AND METHODS
Animals
ATF4 heterozygous mice (ATF4+/− mice) (Stock# 013072) were originally obtained from the Jackson Laboratory. ATF4+/− mice were backcrossed onto a C57BL/6 background (CD45.2) for at least five generations (17). ATF4+/− mice were phenotypically normal and intercrossed to obtain ATF4−/− mice. Offspring of these mice were genotyped using PCR-based assays of genomic DNA samples (17). Littermates (WT) were used as controls in all experiments. ATF4 cKO mice were constructed via the CRISPR-Cas9 system by Nanjing Biomedical Research Institute of Nanjing University. To generate tamoxifen-induced, HSPC-specific deletion of ATF4, ATF4fl/fl; Scl-Cre-ERT mice were injected intraperitoneally with tamoxifen (20 mg/kg, T5648-1G; Sigma-Aldrich) daily for 14 to 21 days. The age-matched tamoxifen-induced ATF4fl/fl mice were used as controls. The genotyping of ATF4fl/fl mice and Scl-Cre-ERT mice was performed using the designed primers (table S3). WT C57BL/6 mice were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd and Model Animal Research Center, Nanjing University. Scl-Cre-ERT mice and CD45.1 B6.SJL mice were provided by J. Zheng (Shanghai Jiao Tong University, Shanghai, China) and Z. Ju (Institute of Aging and Regenerative Medicine, Jinan University, Guangzhou, China), respectively. C57BL/6-CD45.1 mice were crossed with CD45.2 mice to obtain CD45.1/CD45.2 mice. C57BL/6-CD45.1 congenic or C57BL/6-CD45.1/CD45.2 F1 mice were used for competitive repopulation assays. The animal experiment protocols were approved by the Ethics Committees for Animal Experimentation of Sun Yat-sen University and Southern Medical University. All procedures conformed to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication, 8th ed., 2011).
Blood cell counts
PB from the postorbital vein was collected and analyzed using a blood cell counter (Sysmex and Mindray).
Flow cytometry
Phenotypic analysis of lineage cells, hematopoietic progenitor, and stem cells was performed according to previous reports. LT-HSCs (Lin−Sca-1+c-Kit+CD34−Flk2−/Lin−Sca-1+c-Kit+CD150+CD48−), ST-HSCs (Lin−Sca-1+c-Kit+CD34+Flk2−), LSK cells (Lin−Sca-1+c-Kit+), MPPs (Lin−Sca-1+c-Kit+CD34+), LMPPs (Lin−Sca-1+c-Kit+CD34+Flk2+), GMPs (Lin−IL-7Rα−Sca-1−c-Kit+CD16/32hiCD34hi), CMPs (Lin−IL-7Rα−Sca-1−c-Kit+ CD16/32MedCD34hi), MEPs (Lin−IL-7Rα−Sca-1−c-Kit+CD16/32−CD34−), and CLPs (Lin−IL-7Rα+Sca-1Medc-KitMed) were analyzed with LSRII/Fortessa (BD Biosciences), CytoFLEX (Beckman Coulter), and Attune (Thermo Fisher Scientific) and sorted with FACSAria II and Influx cell sorter (BD Biosciences).
For analysis of chimerism in recipients, PB and BM were stained with CD45.1-PE/Alexa Fluor 700 and CD45.2–fluorescein isothiocyanate (FITC). To detect T cells (CD3e), B cells (B220), and myeloid cells (Mac-1/CD11b and Gr-1), samples were stained with CD3e-APC, B220-APC, Mac-1/CD11b-APC/PE, and Gr-1-APC. In all cases, FACS analysis of the above-listed lineages was also performed to confirm multilineage reconstitution as previously described. For HSC cell cycle analysis, BM cells were stained for HSC markers Sca-1-PB, c-Kit–PE-Cy7, CD34-FITC, and Flk2-PE, and lineage-APC-Cy7, and subsequently staining of Ki67 was done using anti-Ki67 antibody (B56) and 7-Aminoactinomycin D (7-AAD)/4′,6-diamidino-2-phenylindole, and fixation/permeabilization solutions (BD Pharmingen). All antibodies, unless stated, were otherwise purchased from BioLegend, eBioscence, and R&D systems (table S1). Analysis of flow cytometric data was performed using the FlowJo and Cytexpert software. Phenotypic analysis of lineage cells, hematopoietic progenitor, and stem cells was performed according to standard protocols, using the antibodies listed in table S1.
BMT assay
All transplant experiments were performed by retro-orbitally injecting freshly isolated BM cells into lethally irradiated [8 gray (Gy)], 8- to 10-week-old recipient mice. For the BMT assay, BM cells from donor mice were injected into lethally irradiated recipient mice with or without an equal number of competitor cells. For the second transplantations, the same number of sorted test and competitor donor cells from the first recipients was serially transplanted into the next recipient mice.
For the limiting dilution competitive repopulation assay, three cell doses (5 × 104, 2 × 105, and 1 × 106) of test donor BM cells isolated from WT and ATF4−/− mice were mixed with a constant dose (2 × 105) of WT competitor BM cells (CD45.1/CD45.2) and subsequently transplanted into lethally irradiated recipient mice. PB in each recipient was analyzed for the engraftment levels of donor cells at the indicated time points 4 months after transplantation, using anti-CD45.1 and anti-CD45.2 antibodies. Anti-CD3e, anti-B220, anti–Mac-1/CD11b, and anti–Gr-1 antibodies were used for lineage analysis. For each individual animal, positive engraftment was defined as 1% or more of CD45.2 cells containing T, B, and myeloid cells. The number of mice negative for reconstitution in each cell dose was measured, and HSC frequency was estimated using Poisson statistics (L-Calc software; STEMCELL Technologies). Animals that died during the course were excluded and not counted in the limiting dilution analysis.
Homing assay for Lin−Sca-1+ primitive hematopoietic cells
Homing assay was performed according to the protocol of a standard method described previously (55).
CFU assay
Fresh 2 × 104 BM cells or 1 × 105 splenocytes were seeded into M3434 methylcellulose (STEMCELL Technologies) for 12 days, respectively. The total number of colonies was counted, and the types of colonies were quantified via microscopy. The sorted YFP+Mac-1+c-Kit+ LICs of secondary AML recipients were seeded in M3534 methylcellulose (STEMCELL Technologies) for 7 to 10 days. The total number of colonies was counted, and the types of colonies were quantified via microscopy.
Retroviral infection, transplantation, and flow cytometric analysis
MLL-AF9–expressing retroviruses were produced in 293T cells with an MSCV-MLL-AF9-IRES-YFP encoding plasmid (32). Lin− fetal liver cells were isolated from WT and ATF4−/− mice, respectively, and then infected with MLL-AF9 retroviruses. Infected cells (1 × 105) were transplanted into lethally irradiated (8 Gy) C57BL/6 mice by retro-orbital injection. Indicated YFP+ BM cells (3 × 103) from primarily transplanted mice were further infused into recipient mice for secondary transplantation. For analysis of lineages, PB cells were stained with Mac-1/CD11b-PE, Gr-1-APC, CD3e-APC, and B220-PE. For analysis of L-GMPs, BM cells were stained with lineage–APC-Cy7, Sca-1-PB, c-Kit–PE-Cy7, CD34-FITC, IL-7Rα–APC, and CD16/32-PE (BioLegend).
For the rescue experiments, the vector of pBabe-puro-HA-HIF1α-WT (Addgene plasmid #19365) and its control vector pBabe-puro (Addgene plasmid #1764) were purchased from Addgene (Addgene). To produce recombinant retrovirus, plasmid DNA was transfected into 293T cells. WT and ATF4−/− Lin− fetal liver cells (fig. S6C) or WT and ATF4−/− adult BM cells (Fig. 6M) were infected with retroviruses, respectively. Infected cells and helper cells were transplanted into lethally irradiated recipients. PB in each recipient was analyzed for the engraftment level, and BM cells were analyzed for cell cycle analysis, engraftment, and ROS level of donor cells at the indicated time points after transplantation.
Measurement of intracellular ROS with CMH2TMRos
MitoTracker Orange CM-H2TMRos (Thermo Fisher Scientific) was used to in vivo determine the oxidative activity of mitochondria according to a standard method described.
Intracellular ROS assay
To measure intracellular ROS levels, HSCs from WT mice and ATF4−/− mice were incubated with 5 nM DCF-DA CM-H2–DCF-DA (Thermo Fisher Scientific) at 37°C for 25 min, followed by fluorescence analysis by flow cytometry.
BMT and NAC administration in vivo
After 14 days of tamoxifen injection, control and ATF4 cKO mice were administered with the antioxidant NAC (500 mg kg−1 day−1 in drinking water; Sigma-Aldrich) and were provided with drinking water daily for 2 weeks; NAC (100 mg/kg) was also administered by intraperitoneal injection daily for 2 weeks, and subsequently, BMCs were harvested and isolated for analysis of ROS levels, HSPCs and lineage frequency, and the status of cell cycle (Fig. 5F). Meanwhile, we performed competitive BMT, and the reconstituted mice were treated with or without NAC (Fig. 5K). Repopulation, lineage analysis, ROS levels, and cell cycle were then examined at the indicated time point after BMT.
RNA isolation and purification and qRT-PCR
HSPCs were directly sorted into reaction buffer. AML GFP+ cells were sorted into Trizol. We performed RT and real-time quantitative PCR with SYBR Green PCR Master Mix using designed primers. All reactions were carried out at least in triplicate. In each experiment, cell populations were sorted from three individual mice. Data were analyzed using BioMark Real-Time PCR Analysis Software v.2.0 (Fluidigm). The results from each experiment were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase. The primers used for the amplification of the indicated genes are listed in table S2.
RNA-seq and bioinformatics analysis
To compare the gene expression profiles between ATF4−/− and WT HSCs, total RNA was extracted from sorted HSC populations from the WT and ATF4−/− BM cells, respectively. The procedures of enrichment and cell staining for isolating HSCs (Lin−c-Kit+Sca-1+CD48−CD150+) from mouse BM were performed as previously described (56). Library for sequencing was prepared following a modified Smart-seq 2 protocol. Briefly, RNA was reverse-transcribed using Supercript II. First-strand complementary DNA (cDNA) was synthesized and amplified before constructing a cDNA library. The Ampure XP beads were used to purify cDNA. The following sequences were processed and analyzed by GENEWIZ. Raw data of RNA-seq were deposited in the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive (SRA), with accession number GSE#132682.
Assay of intracellular HIF1α expression
For intracellular expression of HIF1α in HSCs, BM cells were stained for HSC markers, subsequently staining with PE-conjugated mouse anti-HIF1α (R&D Systems) compared with PE-conjugated mouse immunoglobulin G (IgG).
ChIP analysis and ChIP-seq assay
Human AML KG1α cells (1 × 108) or mouse BM cells (2 × 107) were used for each ChIP assay. The ChIP procedure was performed according to a previously described protocol (57, 58), using 10 μg of anti-ATF4 antibody (Abcam) (table S4). Enriched DNA fragments were subjected to the preparation of standard libraries for ChIP-seq according to the manufacturer’s instructions, and the resulting ChIP-seq libraries were sequenced on an Illumina Hi-seq X-Ten (Annoroad Gene Technology Co. Ltd). ChIP-seq data were mapped to mouse genome (GRCm38) by using Bowtie2, and ChIP peaks were called using model-based analysis of ChIP-seq 14 (MACS2), with the input sample as the control. Enrichment heatmaps that surround ChIP peaks were produced by using deeptools, and signal plotting of individual genes was generated using the Integrated Genome Viewer (Novel Bioinformatics Co. Ltd for the support of bioinformatics analysis). The raw data from ChIP-seq can be downloaded from the NCBI’s SRA using accession number GSE132681.
ChIP assay was performed to identify ATF4 binding sites on the HIF1a promoter in mouse BM cells. The DNA isolated from input chromatin fragments and from the precipitated chromatin fragments by anti-ATF4 antibody (Abcam) or control IgG was subjected to real-time PCR (ChIP-qPCR) using primers flanking the consensus ATF4 binding sites on the HIF1a promoter (BersinBio Bioscience Co. Ltd for the technique service).
Western blotting analysis
Western blotting was performed according to the protocol described previously (59). The primary antibodies used for Western blot were as follows: anti-CDKN2A/p16Ink4a (1:1000; Abcam) and anti–β-actin (1:5000; Proteintech). For the origin and description of all antibodies used in this study, see table S4.
Luciferase reporter assays
293T cells were cultured in 96-well plates and cotransfected with pcDNA3.1-ATF4 vector or pcDNA3.1 vector, together with luciferase reporter plasmid that contained the WT (HIF1α-pGL3-WT) or mutant (HIF1α-pGL3-MUT) promoter of HIF1a. Transcriptional activation of HIF1a by ATF4 was evaluated using a luciferase reporter system (Promega).
Statistical analysis
All quantitative data obtained from repeated experiments are expressed as means ± SD. The statistical significance of the differences between the groups was calculated using an unpaired Student’s t test, and the survival curves were analyzed using a log-rank (Mantel-Cox) test. *P < 0.05, **P < 0.01, and #P < 0.001. NS, not significant.
Acknowledgments
We thank the following individuals: J. Zheng (Shanghai Jiao Tong University) for sharing the MLL-AF9 plasmid; W. Cai (Laboratory Animal Centre, Zhongshan School of Medicine, Sun Yat-Sen University) for care of the mice in accordance with guidelines; T. Zhang for technical assistance of mouse irradiation; and X. Li and C. Rong (Zhongshan School of Medicine, Sun Yat-Sen University), J. Wang and X. Liu (Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou), and Hongfen Shen (Cancer Research Institute, Southern Medical University) for flow cytometric analysis and FACS sorting of cells. We thank the members of the Li laboratories for helpful comments and discussion.
Funding: This work was supported by the National Key Research and Development Program of China (2017YFA0106300), the National Natural Science Foundation of China (grant nos. 81600086 and 81770100, to Y.S.), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (grant no. 81621004), Projects of International Cooperation and Exchanges NSFC (grant no. 81820108025), and the Fundamental Research Funds for the Central Universities (grant no. 50000-31620026).
Ethics statement: This investigation was carried out according to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH Publication No. 85-23, revised 1996). Ethical approval was given by the Medical Ethics Committee of Animal Experimentation of Sun Yat-sen University, with the following reference number: 2016-099.
Author contributions: Y.S. conceived the ideas, designed all experiments, performed most and major experiments, analyzed data, and wrote paper. X.L.L. contributed to the statistical analyses of the data and some experiments. B.L.,Y.Z., and W.L. assisted in ChIP-seq and some experiments. S.Z. provided technical assistance and expertise in the ATF4 mouse colony recovery and maintenance. F.H., H.T., X.Z., X.L., and J.W. assisted in some experiments. J.C. assisted in some experiments, and discussed and edited the manuscript. M.L. conceived the ideas, designed and discussed experiments, supervised progress, and extensively edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw data from RNA-seq and ChIP-seq can be downloaded from the NCBI’s SRA using accession number GSE132683 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132683).
Supplementary Materials
This PDF file includes:
Tables S1 to S4
Figs. S1 to S6
REFERENCES AND NOTES
- 1.Calcinotto A., Kohli J., Zagato E., Pellegrini L., Demaria M., Alimonti A., Cellular senescence: Aging, cancer, and injury. Physiol. Rev. 99, 1047–1078 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Segura A., Nehme J., Demaria M., Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Kubben N., Misteli T., Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat. Rev. Mol. Cell Biol. 18, 595–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan G., Lazare S. S., Aging of hematopoietic stem cells. Blood 131, 479–487 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Ermolaeva M., Neri F., Ori A., Rudolph K. L., Cellular and epigenetic drivers of stem cell ageing. Nat. Rev. Mol. Cell Biol. 19, 594–610 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Neves J., Sousa-Victor P., Jasper H., Rejuvenating strategies for stem cell-based therapies in aging. Cell Stem Cell 20, 161–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shlush L. I., Age-related clonal hematopoiesis. Blood 131, 496–504 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., Menzies K. J., Auwerx J., The role of mitochondria in stem cell fate and aging. Development 145, dev143420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Revuelta M., Matheu A., Autophagy in stem cell aging. Aging Cell 16, 912–915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akunuru S., Geiger H., Aging, clonality, and rejuvenation of hematopoietic stem cells. Trends Mol. Med. 22, 701–712 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantel C., Messina-Graham S., Moh A., Cooper S., Hangoc G., Fu X.-Y., Broxmeyer H. E., Mouse hematopoietic cell-targeted STAT3 deletion: Stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging-like phenotype. Blood 120, 2589–2599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams C. M., Ebert S. M., Dyle M. C., Role of ATF4 in skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 20, 164–168 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Ameri K., Harris A. L., Activating transcription factor 4. Int. J. Biochem. Cell Biol. 40, 14–21 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Kasai S., Yamazaki H., Tanji K., Engler M. J., Matsumiya T., Itoh K., Role of the ISR-ATF4 pathway and its cross talk with Nrf2 in mitochondrial quality control. J. Clin. Biochem. Nutr. 64, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wortel I. M. N., van der Meer L. T., Kilberg M. S., van Leeuwen F. N., Surviving stress: Modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol. Metab. 28, 794–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuoka H. C., Townes T. M., Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99, 736–745 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y., Zhou J., Liu D., Dong F., Cheng H., Wang W., Pang Y., Wang Y., Mu X., Ni Y., Li Z., Xu H., Hao S., Wang X., Ma S., Wang Q. F., Xiao G., Yuan W., Liu B., Cheng T., ATF4 plays a pivotal role in the development of functional hematopoietic stem cells in mouse fetal liver. Blood 126, 2383–2391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G., Cao H., Wu H., Song J., Pan X., Sun Q., Ling S., Li Y., Zhu M., Zhang P., Peng S., Xie X., Tang T., Hong A., Bian Z., Bai Y., Lu A., Li Y., He F., Zhang G., Li Y., miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 19, 93–100 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Deng J., Yuan F., Guo Y., Xiao Y., Niu Y., Deng Y., Han X., Guan Y., Chen S., Guo F., Deletion of ATF4 in AgRP neurons promotes fat loss mainly via increasing energy expenditure. Diabetes 66, 640–650 (2017). [DOI] [PubMed] [Google Scholar]
- 21.van Galen P., Mbong N., Kreso A., Schoof E. M., Wagenblast E., Ng S. W. K., Krivdova G., Jin L., Nakauchi H., Dick J. E., Integrated stress response activity marks stem cells in normal hematopoiesis and leukemia. Cell Rep. 25, 1109–1117.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Li W., Li X., Miller R. A., ATF4 activity: A common feature shared by many kinds of slow-aging mice. Aging Cell 13, 1012–1018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W., Miller R. A., Elevated ATF4 function in fibroblasts and liver of slow-aging mutant mice. J. Gerontol. A Biol. Sci. Med. Sci. 70, 263–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., Fox L. A., Dang N., Johnston E. D., Oakes J. A., Tchao B. N., Pak D. N., Fields S., Kennedy B. K., Kaeberlein M., Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133, 292–302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beerman I., Bhattacharya D., Zandi S., Sigvardsson M., Weissman I. L., Bryder D., Rossi D. J., Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. U.S.A. 107, 5465–5470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Göthert J. R., Gustin S. E., Hall M. A., Green A. R., Göttgens B., Izon D. J., Begley C. G., In vivo fate-tracing studies using the Scl stem cell enhancer: Embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood 105, 2724–2732 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Ergen A. V., Goodell M. A., Mechanisms of hematopoietic stem cell aging. Exp. Gerontol. 45, 286–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suda T., Takubo K., Semenza G. L., Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Takubo K., Goda N., Yamada W., Iriuchishima H., Ikeda E., Kubota Y., Shima H., Johnson R. S., Hirao A., Suematsu M., Suda T., Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391–402 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Yan Q., Bartz S., Mao M., Li L., Kaelin W. G. Jr., The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol. Cell. Biol. 27, 2092–2102 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Galen P., Kreso A., Mbong N., Kent D. G., Fitzmaurice T., Chambers J. E., Xie S., Laurenti E., Hermans K., Eppert K., Marciniak S. J., Goodall J. C., Green A. R., Wouters B. G., Wienholds E., Dick J. E., The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature 510, 268–272 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Krivtsov A. V., Twomey D., Feng Z., Stubbs M. C., Wang Y., Faber J., Levine J. E., Wang J., Hahn W. C., Gilliland D. G., Golub T. R., Armstrong S. A., Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Crane G. M., Jeffery E., Morrison S. J., Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17, 573–590 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Dzierzak E., Bigas A., Blood development: Hematopoietic stem cell dependence and independence. Cell Stem Cell 22, 639–651 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Haas S., Trumpp A., Milsom M. D., Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell 22, 627–638 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Jacobsen S. E. W., Nerlov C., Haematopoiesis in the era of advanced single-cell technologies. Nat. Cell Biol. 21, 2–8 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Kong W., Yang W., Patel R. M., Casey E. B., Okeyo-Owuor T., White J. M., Porter S. N., Morris S. A., Magee J. A., Single-cell analysis of neonatal HSC ontogeny reveals gradual and uncoordinated transcriptional reprogramming that begins before birth. Cell Stem Cell 27, 732–747.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi K., Kondo M., Developmental switch of mouse hematopoietic stem cells from fetal to adult type occurs in bone marrow after birth. Proc. Natl. Acad. Sci. U.S.A. 103, 17852–17857 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantel C. R., O’Leary H. A., Chitteti B. R., Huang X., Cooper S., Hangoc G., Brustovetsky N., Srour E. F., Lee M. R., Messina-Graham S., Haas D. M., Falah N., Kapur R., Pelus L. M., Bardeesy N., Fitamant J., Ivan M., Kim K.-S., Broxmeyer H. E., Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell 161, 1553–1565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capitano M. L., Mohamad S. F., Cooper S., Guo B., Huang X., Gunawan A. M., Sampson C., Ropa J., Srour E. F., Orschell C. M., Broxmeyer H. E., Mitigating oxygen stress enhances aged mouse hematopoietic stem cell numbers and function. J. Clin. Invest. 131, e140177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flach J., Bakker S. T., Mohrin M., Conroy P. C., Pietras E. M., Reynaud D., Alvarez S., Diolaiti M. E., Ugarte F., Forsberg E. C., Le Beau M. M., Stohr B. A., Mendez J., Morrison C. G., Passegue E., Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512, 198–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janzen V., Forkert R., Fleming H. E., Saito Y., Waring M. T., Dombkowski D. M., Cheng T., DePinho R. A., Sharpless N. E., Scadden D. T., Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443, 421–426 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ju Z., Choudhury A. R., Rudolph K. L., A dual role of p21 in stem cell aging. Ann. N. Y. Acad. Sci. 1100, 333–344 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Attema J. L., Pronk C. J. H., Norddahl G. L., Nygren J. M., Bryder D., Hematopoietic stem cell ageing is uncoupled from p16INK4A-mediated senescence. Oncogene 28, 2238–2243 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Guimaraes-Camboa N., Stowe J., Aneas I., Sakabe N., Cattaneo P., Henderson L., Kilberg M. S., Johnson R. S., Chen J., McCulloch A. D., Nobrega M. A., Evans S. M., Zambon A. C., HIF1α represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev. Cell 33, 507–521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Liu Y., Malek S. N., Zheng P., Liu Y., Targeting HIF1α eliminates cancer stem cells in hematological malignancies. Cell Stem Cell 8, 399–411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Li H., Xi H. S., Li S., HIF1α is required for survival maintenance of chronic myeloid leukemia stem cells. Blood 119, 2595–2607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Osdal T., Ho Y., Chun S., McDonald T., Agarwal P., Lin A., Chu S., Qi J., Li L., Hsieh Y. T., Dos Santos C., Yuan H., Ha T. Q., Popa M., Hovland R., Bruserud O., Gjertsen B. T., Kuo Y. H., Chen W., Lain S., McCormack E., Bhatia R., SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell 15, 431–446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rimmele P., Bigarella C. L., Liang R., Izac B., Dieguez-Gonzalez R., Barbet G., Donovan M., Brugnara C., Blander J. M., Sinclair D. A., Ghaffari S., Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Reports 3, 44–59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown K., Xie S., Qiu X., Mohrin M., Shin J., Liu Y., Zhang D., Scadden D. T., Chen D., SIRT3 reverses aging-associated degeneration. Cell Rep. 3, 319–327 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J., Liu B., Yu D., Zuo Y., Cai R., Yang J., Cheng J., SIRT3 deacetylase activity confers chemoresistance in AML via regulation of mitochondrial oxidative phosphorylation. Br. J. Haematol. 187, 49–64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z., Yi W., Morita Y., Wang H., Cong Y., Liu J. P., Xiao Z., Rudolph K. L., Cheng T., Ju Z., Wip1 deficiency impairs haematopoietic stem cell function via p53 and mTORC1 pathways. Nat. Commun. 6, 6808 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Fontana M. C., Nanni J., di Rorà A. G. L., Petracci E., Padella A., Ghetti M., Ferrari A., Marconi G., Soverini S., Iacobucci I., Papayannidis C., Curti A., Audisio E., Giannini M. B., Rondoni M., Lanza F., Cavo M., Martinelli G., Simonetti G., Pharmacological inhibition of WIP1 sensitizes acute myeloid leukemia cells to the MDM2 inhibitor Nutlin-3a. Biomedicine 9, 388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bushweller J. H., Targeting transcription factors in cancer—From undruggable to reality. Nat. Rev. Cancer 19, 611–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Y., Shao L., Bai H., Wang Z. Z., Wu W.-S., Slug deficiency enhances self-renewal of hematopoietic stem cells during hematopoietic regeneration. Blood 115, 1709–1717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H., Kozono D. E., O’Connor K. W., Vidal-Cardenas S., Rousseau A., Hamilton A., Moreau L., Gaudiano E. F., Greenberger J., Bagby G., Soulier J., Grompe M., Parmar K., D’Andrea A. D., TGF-β inhibition rescues hematopoietic stem cell defects and bone marrow failure in fanconi anemia. Cell Stem Cell 18, 668–681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee T. I., Johnstone S. E., Young R. A., Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ying Z., Tian H., Li Y., Lian R., Li W., Wu S., Zhang H. Z., Wu J., Liu L., Song J., Guan H., Cai J., Zhu X., Li J., Li M., CCT6A suppresses SMAD2 and promotes prometastatic TGF-β signaling. J. Clin. Invest. 127, 1725–1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai J., Li R., Xu X., Zhang L., Lian R., Fang L., Huang Y., Feng X., Liu X., Li X., Zhu X., Zhang H., Wu J., Zeng M., Song E., He Y., Yin Y., Li J., Li M., CK1α suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat. Cell Biol. 20, 465–478 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4
Figs. S1 to S6